
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 30 September 2021
Sec. Molecular and Cellular Oncology
Volume 11 - 2021 | https://doi.org/10.3389/fonc.2021.742149
 Gaofeng Li1†
Gaofeng Li1† Jun Gong2†
Jun Gong2† Shulong Cao1†
Shulong Cao1† Zhaoyang Wu1
Zhaoyang Wu1 Dong Cheng3
Dong Cheng3 Jia Zhu4
Jia Zhu4 Xuqun Huang5
Xuqun Huang5 Jingyi Tang1
Jingyi Tang1 Yuning Yuan1
Yuning Yuan1 Wenqi Cai1
Wenqi Cai1 Haiyuan Zhang1*
Haiyuan Zhang1*Ovarian cancer, a common malignant tumor, is one of the primary causes of cancer-related deaths in women. Systemic chemotherapy with platinum-based compounds or taxanes is the first-line treatment for ovarian cancer. However, resistance to these chemotherapeutic drugs worsens the prognosis. The underlying mechanism of chemotherapeutic resistance in ovarian cancer remains unclear. Non-coding RNAs, including long non-coding RNAs, microRNAs, and circular RNAs, have been implicated in the development of drug resistance. Abnormally expressed non-coding RNAs can promote ovarian cancer resistance by inducing apoptosis inhibition, protective autophagy, abnormal tumor cell proliferation, epithelial-mesenchymal transition, abnormal glycolysis, drug efflux, and cancer cell stemness. This review summarizes the role of non-coding RNAs in the development of chemotherapeutic resistance in ovarian cancer, including their mechanisms, targets, and potential signaling pathways. This will facilitate the development of novel chemotherapeutic agents that can target these non-coding RNAs and improve ovarian cancer treatment.
Ovarian cancer is a relatively common malignant tumor in women that easily metastasizes in its advanced stages (1–3). Systemic chemotherapy remains the mainstay treatment of advanced ovarian cancer (4, 5). However, the emergence of drug resistance has limited its clinical application (6).
Ovarian cancer cells have developed multiple resistance mechanisms, including impairment in cellular copper transporters, intracellular detoxification, DNA damage repair (DDR), and non-coding RNA (ncRNA)-mediated drug resistance. Copper transporter 1 (CTR1), ATP7A, and ATP7B participate in the uptake or efflux of platinum (7, 8). Interestingly, when their expression is dysregulated, drug concentration within ovarian cancer cells decrease, resulting in drug resistance. Glutathione S-transferase π (GST-π) is an intracellular detoxification enzyme that promotes the conjugation of glutathione (GSH) with chemotherapeutic drugs, and such conjugated drugs are easily excreted and their toxic effects are eliminated; overall, this process also results in drug resistance in ovarian cancer (9–11). The cellular DDR system detects and repairs damaged DNA to maintain a stable genome, and this inhibits cisplatin-mediated DNA damage (12, 13). These abovementioned three mechanisms of drug resistance in ovarian cancer have been studied in depth. However, the mechanism by which ncRNA causes drug resistance is unclear.
NcRNAs account for a majority of cellular RNAs and do not encode any functional proteins (14). After transcription, they mainly perform their biological functions at the RNA level (15). Unexpectedly, some ncRNAs promote drug resistance by inducing multiple cell phenotypes. Among the ncRNAs, long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs) are thought to be mainly responsible for causing drug resistance in ovarian cancer. With a length of more than 200 nucleotides, lncRNAs regulate gene expression at the transcriptional, post-transcriptional, and epigenetic levels (16). They induce tumor cell stemness, apoptosis inhibition, abnormal tumor cell proliferation, drug efflux, protective autophagy, and epithelial-mesenchymal transition (EMT) to facilitate drug resistance in ovarian cancer. MiRNAs can be 20–25 nucleotides long (17). They recognize and bind mRNA by complementary base pairing, leading to mRNA degradation or translational inhibition (18). The resultant abnormally expressed mRNA can promote drug resistance by inducing apoptosis inhibition, abnormal glycolysis, drug efflux, and EMT in ovarian cancer. CircRNAs have a covalently closed loop structure and are relatively stable (19). They mainly act as competing endogenous RNAs (ceRNAs) to reverse the inhibitory effect of miRNAs on mRNA expression (20). Therefore, circRNAs regulate the expression of resistance-related proteins by regulating their mRNA expression, which promotes drug resistance in ovarian cancer.
This review summarizes the functional mechanisms and signaling pathways of lncRNAs, miRNAs, and circRNAs in ovarian cancer.
At the transcriptional level, lncRNAs bind transcription factors to promote or inhibit the transcription of target genes (Figure 1). LncRNA HOTAIR recruits and binds the transcription factor SNAIL, which prevents it from binding to the hepatocyte nuclear factor 4 alpha (HNF4α) promoter, reducing its expression (21).
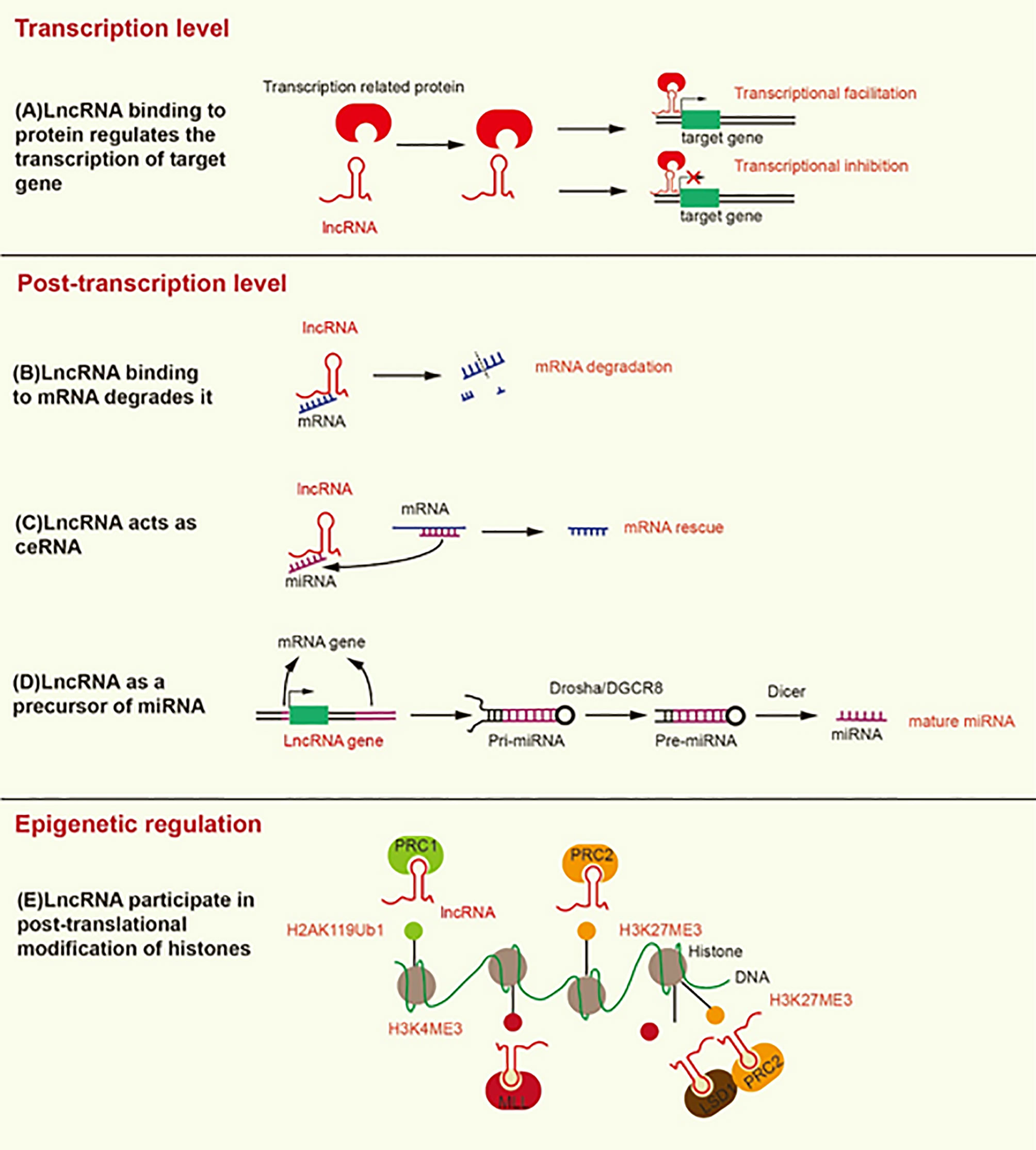
Figure 1 Mechanisms underlying the modification induced by lncRNA at the transcriptional, post-transcriptional, and epigenetic levels. (A) LncRNA binding to transcription-related proteins mediates transcriptional activation or transcriptional inhibition. (B) LncRNA promotes the degradation of mRNA by recruiting STAU1 to the SBS of dsRNA. (C) LncRNA, as a ceRNA, separates mRNA from its corresponding miRNA target. (D) LncRNA as a precursor of miRNA, develops into mature miRNA under the action of Drosha/DGCR8 and Dicer. (E) LncRNA contributes to the formation of H2AK119Ub1 and H3K27ME3, which inhibit gene transcription. In addition, lncRNA contributes to the formation of H3K4ME3, which promotes gene transcription.
At the post-transcriptional level, the Alu element of lncRNA binds the Alu element of the 3’-untranslated region (3’UTR) in the target mRNA to form staufen1-binding site (SBS) by incomplete base-pairing. SBS binds Staufen1 (STAU1) to degrade the target mRNA (22). STAU1, as an RNA-binding protein, binds to SBS, leading to the degradation of mRNA in mammals (23). Therefore, lncRNA can directly interact with specific mRNA to degrade mRNA (Figure 1). On the contrary, as a type of ceRNA, lncRNA can sponge miRNA to release mRNA and restore the function of mRNA (Figure 1) (24). For example, LINC01118 sponges miR-134 to rescue the mRNA of ABCC1 (25). LncRNA also functions as a precursor of miRNA (Figure 1). Most miRNAs are derived from protein-coding genes in the human genome, but some miRNAs are derived from ncRNA-coding genes (26). Specifically, the genes encoding lncRNA are processed to form primary miRNA (pri-miRNA), and then pri-miRNA is further processed in the nucleus by Drosha/DGCR8 into a double-hairpin precursor miRNA (pre-miRNA) (27). Finally, the pre-miRNA is transported to the cytoplasm and cut into mature miRNA by Dicer (28). For instance, the exon of lncRNA H19 contains miR-675, and H19 acts as a precursor of miR-675 to regulate its expression (29). LncRNA can also change the cellular localization of proteins. For instance, lncRNA MALAT1 can bind YAP to inhibit its translocation from the nucleus to the cytoplasm (30).
LncRNA can mediate histone modification, resulting in epigenetic regulation (Figure 1). Both lncRNA ANRIL and lncRNA H19 can interact with enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) and suppressor of zeste 12 (SUZ12) (31, 32). EZH2 and SUZ12 are subunits of polycomb repressive complex 2 (PRC2), and PRC2 inhibits the transcription of target genes by trimethylation at lysine 27 of histone H3 (H3K27Me3) (33). The interaction of ANRIL and H19 with EZH2 and SUZ12 can promote H3K27Me3 to inhibit the transcription of target genes (34, 35). In addition, lncRNA HOTTIP interacts with WD repeat domain 5 (WDR5) to promote trimethylation at lysine 4 of histone H3 (H3K4Me3), thereby promoting the transcription of target genes (36). WDR5 is a core component of mixed lineage leukemia (MLL), which catalyzes the formation of H3K4Me3 (36). LncRNA HOTAIR can interact with EZH2 and lysine specific demethylase 1 (LSD1), which is a histone demethylase that prevents the formation of H3K4Me3 to inhibit gene transcription. HOTAIR coordinates the interaction between EZH2 and LSD1 to detach the methyl groups from H3K4 to transform transcriptional activation into transcriptional inhibition of the target gene (37). Likewise, lncRNA ultraconserved element 338 (uc.338) binds BMI1, a subunit of PRC1, to monoubiquitinate histone H2A on lysine 119 (H2AK119ub1), which inhibits the transcription of target genes (38). We summarize the mechanisms and signaling pathways of lncRNAs that lead to platinum or taxane chemotherapeutic resistance in ovarian cancer (Table 1).
ABC transporters are transmembrane proteins that can expel drugs from within cells through a process called drug efflux. LncRNAs promote the expression of some ABC transporters by sponging miRNA, which promotes drug efflux and induces drug resistance in ovarian cancer (Figure 2) (25, 40, 44). In ovarian cancer, both lncRNA MALAT1 and LINC01118 are upregulated, and they both promote drug efflux by increasing the expression of ABCC1 to induce drug resistance (25, 40). The difference is that MALAT1 can interact with the Notch1 protein to activate the Notch1 signaling pathway and promote the expression of ABCC1 to induce cisplatin resistance, while LINC01118 upregulates the expression of ABCC1 by sponging miR-134 to promote paclitaxel resistance. Moreover, the silencing of MALAT1 reduced tumor growth when ovarian tumor xenograft model mice were treated with cisplatin. Although lncRNA UCA1 promotes cisplatin resistance by inhibiting apoptosis (45), UCA1 can also promote paclitaxel resistance by inducing drug efflux through sponging of miR-129 to rescue ABCB1 expression (44). ABCB1 facilitates the elimination of chemotherapeutic drugs from cancer cells (46). UCA1 is highly upregulated in paclitaxel-resistant ovarian cancer cells. Drug efflux caused by ABC transporters is a very important drug resistance pathway. Given the lack of related studies, further studies are needed to understand how lncRNA makes ovarian cancer cells resistant to chemotherapeutic drugs through its action on ABC transporters. It will allow us to identify novel strategies to overcome the drug resistance of ovarian cancer.
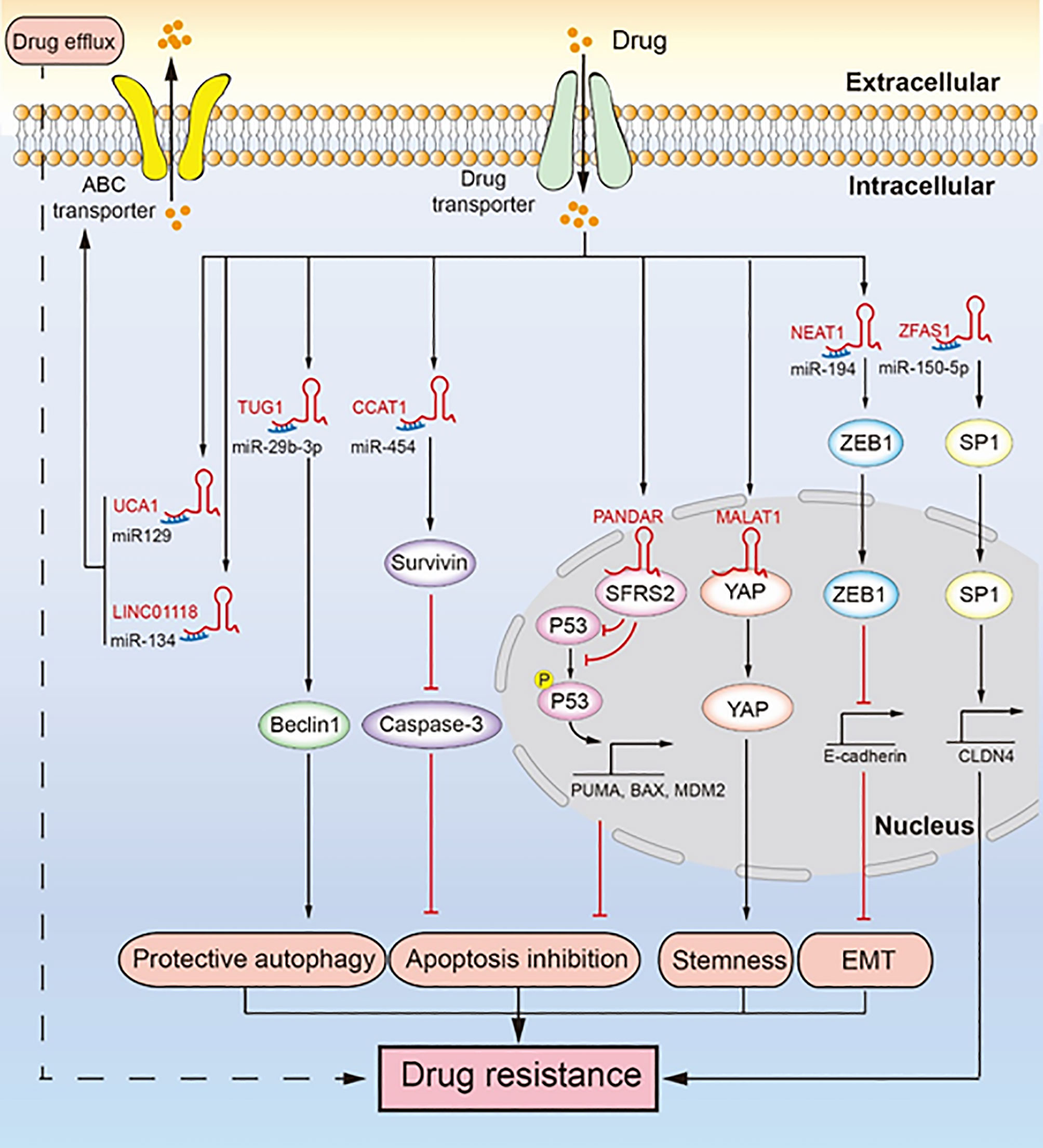
Figure 2 LncRNAs regulate chemotherapeutic resistance through diverse signaling pathways. Multiple lncRNAs promote drug resistance by inducing drug efflux, protective autophagy, apoptosis inhibition, tumor stemness, and epithelial-mesenchymal transition (EMT). LINC01118 and UCA1 regulate ABC transporters promoting drug resistance. TUG1 regulates Beclin1 leading to drug resistance induced by protective autophagy. CCAT1 upregulates survivin to inhibit the expression of caspase-3, which inhibits apoptosis and leads to drug resistance. PANDAR reduces the expression and phosphorylation of P53, inhibiting apoptosis and promoting drug resistance. MALAT1 binds YAP and inhibits its translocation from the nucleus to the cytoplasm, which facilitates cancer cell stemness leading to drug resistance. NEAT1 upregulates the expression of ZEB1, which promotes EMT leading to drug resistance. ZFAS1 promotes the transcription of CLDN4 to upregulate SP1, which leads to drug resistance.
EMT involves transformation of epithelial cells, through loss of their polarity, into mesenchymal cells, which gives them the ability of invasion, migration, and anti-apoptosis (47). EMT induced by lncRNA promotes the malignancy of tumors and makes them resistant to chemotherapeutics (Figure 2) (42). For example, lncRNA NEAT1 is upregulated in paclitaxel-resistant ovarian cancer cells, which sponges miR-194 to restore the high expression of ZEB1. ZEB1 is essential for EMT because it inhibits the transcription of E-cadherin (48). In addition, NEAT1 knockdown significantly inhibited tumor growth in an ovarian tumor xenograft mouse model. EMT plays an important role in cancer metastasis and drug resistance, and the role of lncRNA-mediated EMT in the drug resistance of ovarian cancer needs further research.
Cancer cell stemness contributes to self-renewal and differentiation of cancer cells, which help tumor cells to regenerate and resist the toxicity of chemotherapeutic drugs (49). LncRNA can promote cancer cell stemness of ovarian cancer to promote drug resistance (Figure 2) (30). Specifically, MALAT1 is upregulated in cisplatin-resistant cells as well as in non-adherent spheres; it interacts with the YAP protein to inhibit its translocation to the cytoplasm from the nucleus, which leads to enhanced cancer cell stemness. While cancer cell stemness is a predominant mechanism contributing to drug resistance in ovarian cancer cells, not many studies have been conducted on cancer cell stemness induced by lncRNA. Therefore, there is further scope for research.
Abnormal cell proliferation in the presence of chemotherapeutic drugs indicates that the treatment is ineffective or that the cells have become resistant. LncRNA HOTAIR reduces the sensitivity of ovarian cancer to cisplatin by inducing abnormal tumor cell proliferation (50, 51). Mechanically, HOTAIR activates the Wnt/β-catenin signaling pathway to enhance the expression of cyclinD1 and CDK4. CyclinD1 is a marker of cell proliferation (50). It activates CDK4-expressing cells to progress from the G1 phase into the S phase, which accelerates cell cycle progression (52). In an ovarian tumor xenograft mouse model, HOTAIR downregulation inhibited tumor growth and cyclinD1 expression, and this inhibition effect was more remarkable when cisplatin was administered. The role of lncRNA HOTAIR in chemotherapeutic resistance caused by the abnormal proliferation of ovarian cancer cells and other tumor cells is understudied, necessitating further studies.
Protective autophagy is another mechanism contributing to chemotherapeutic resistance (53). Specifically, cancer cells undergo protective autophagy to obtain nutrients and promote their survival by degrading metabolic waste, damaged proteins, and damaged organelles, thereby increasing their resistance against chemotherapeutic drugs. LncRNA TUG1 mediates paclitaxel resistance by inducing protective autophagy in ovarian cancer (Figure 2) (43). Specifically, TUG1 is upregulated in cisplatin-resistant cells and sponges miR-29b-3p to indirectly upregulate the expression of Beclin1, which increases autophagosome formation in ovarian cancer (43). In an ovarian tumor xenograft mouse model, TUG1 promoted tumor growth by resisting the effect of paclitaxel, and downregulation of TUG1 decreased the tumor size and weight. Drug resistance due to protective autophagy in tumors is a hot topic of current research, and the role of lncRNA in this mechanism has gained attention. Although autophagy has long been known to contribute to drug resistance in cancer cells, the ability of lncRNA to induce protective autophagy and resulting in ovarian cancer drug resistance seems to be a new research direction.
Apoptosis is the self-destructive mechanism of cells, and apoptosis inhibition rescues cancer cells and induces chemotherapeutic resistance. LncRNAs regulate apoptosis-related proteins to induce apoptosis inhibition, thereby inducing cisplatin resistance in ovarian cancer (Figure 2) (39, 41, 45). Mechanistically, lncRNA CCAT1 is upregulated in cisplatin-resistant ovarian cancer cells, which enhances the expression of survivin as lncRNA CCAT1 sponges miR-454 (39). Survivin, an inhibitor of apoptosis, can bind and inhibit caspase-9, caspase-3, and caspase-7, which hinder apoptosis and cause cisplatin resistance (54). In the nucleus, lncRNA PANDAR binding the SFRS2 protein downregulates the expression of P53 and its phosphorylation at serine 15 (Ser-15), which inhibits the transcription of P53-mediated pro-apoptotic genes, including MDM2, BAX, and PUMA (41). In addition, PANDAR is upregulated by cisplatin. Likewise, lncRNA UCA1 induces cisplatin resistance by indirectly promoting the expression of SPRK1 and BCL-2 and inhibiting the expression of BAX, caspase-3, and caspase-9 (45). LncRNA NEAT1 regulates miR-770-5p/PARP1 signaling to induce cisplatin resistance in ovarian cancer (55). NEAT1 is overexpressed in cisplatin-resistant ovarian cancer cells and sponges miR-770-5P to upregulate the expression of poly adenosine diphosphate-ribose polymerase 1 (PARP1), which leads to chemotherapeutic resistance in cancer (56). Recent studies have shown that lncRNA SNHG22 is highly expressed in ovarian cancer tissues and promotes cisplatin resistance (57). SNHG22 sponges miR-2467 to enhance Gal-1 expression, and Gal-1 activates the H-Ras/Raf/ERK pathway to induce apoptosis inhibition and drug resistance (58). Likewise, lncRNA EPEI upregulation indirectly downregulates P53 expression, leading to carboplatin resistance in ovarian endometrioid adenocarcinoma (59). Inactivation of P53, a tumor suppressor gene, can promote carcinogenesis and inhibit cell apoptosis (60). LncRNA NCK adaptor protein 1 (NCK1)-AS1 not only sponges miR-137 to upregulate NCK1 expression but also directly interacts with c-CBI to inhibit its degradation caused by ubiquitination (61). This inhibits the apoptosis of ovarian cancer cells and makes them resistant to cisplatin (61, 62). In an ovarian tumor xenograft mouse model, the knockdown of CCAT1 reduced tumor weight, whereas PANDAR overexpression increased tumor volume (39, 41). In addition, P53 and PUMA were downregulated in a PANDAR-overexpressing xenograft mouse model. In ovarian cancer apoptosis inhibition is a well-known drug resistance mechanism. Here, lncRNA does not directly regulate the classic apoptosis pathway to induce drug resistance caused by apoptosis inhibition. Therefore, in-depth study of lncRNA may provide new insights into the classic apoptosis pathway.
LncRNAs induce chemotherapeutic resistance of ovarian cancer by acting on special DNA regulatory proteins (57, 63–67). LncRNA HOTAIR can facilitate the expression of Homeobox A7 (HOXA7) and sponge miR-138-5p to rescue EZH2 and sirtuin 1 (SIRT1) expression in ovarian cancer, both of which contribute to cisplatin resistance (63, 68). EZH2 is involved in histone methylation, while SIRT1 mediates histone deacetylation (69, 70). In addition, HOTAIR influences the DNA damage response to promote cisplatin resistance in ovarian cancer (64). ANRIL overexpression confers makes ovarian cancer cells resistant to cisplatin through the let-7a/high-mobility group protein A2 (HMGA2) axis (65). HMGA2 can also influence the proliferation and differentiation of cells by upregulating PRC2 (71–73). LncRNA ZFAS1 is overexpressed in epithelial ovarian cancer cells and directly targets miR-150-5p to enhance the expression of specificity protein 1 (SP1), which makes ovarian cancer cells resist to cisplatin and paclitaxel (66). Another study showed that SP1, as a transcription factor, facilitates the transcription of claudin-4 (CLDN4), which causes low DNA methylation and high histone H3 acetylation in the CLDN4 promoter region (74, 75). In an ovarian tumor xenograft mouse model, downregulation of HOTAIR and ANRIL could slow down tumor growth (63, 65). In addition, HOTAIR downregulation decreased the protein levels of HOXA7 in vivo. Understanding the role of DNA regulation in organisms is still a difficult problem, and research on lncRNA-mediated DNA regulatory proteins to promote drug resistance remains at a relatively superficial level, which requires more researches to reveal it.
MiRNA is a type of endogenous ncRNA with regulatory functions (76). It is processed by nucleases from longer primary transcripts and is 20-25 nucleotides long. MiRNA mainly forms a silencing complex, which binds to the 3’UTR region of the target mRNA through complementary base pairing and controls the stability and translation of the mRNA (18). The specific mechanism of miRNA-induced drug resistance in ovarian cancer is described below. Here, we summarize the mechanisms and signaling pathways of miRNAs that lead to platinum or taxane chemotherapeutic resistance in ovarian cancer (Table 2).
Multiple miRNAs have been found to regulate the expression of ABC transporters to mediate drug efflux, leading to chemotherapeutic resistance (84, 91, 95, 96). Specifically, miR-130a, miR-1307, and miR-27a promote drug resistance in ovarian cancer by increasing the expression of P-glycoprotein (P-gp) (91, 95, 96). P-gp, a drug transporter encoded by multi-drug resistance-1 (MDR1), is also called ABCB1. It promotes drug resistance through its drug efflux function. MiR-130a is overexpressed in cisplatin-resistant ovarian cancer cells, and it indirectly enhances the expression of P-gp. MiR-1307 and miR-27a are highly expressed in paclitaxel-resistant ovarian cancer cells. Mechanically, miR-1307 relieves the transcriptional repression of ETV4 by directly downregulating CIC expression, and ETV4 upregulates the transcription of MDR1 by binding to the MDR1 promoter region (97, 98). MiR-27a targets homeodomain-interacting protein kinase-2 (HIPK2), which reduces the transcriptional repression of MDR1 caused by HIPK2 (99). Downregulation of miR-411, mediated by low levels of SLC27A2, enhances the expression of ABCG2, which promotes drug efflux to induce cisplatin resistance in ovarian cancer (84). The common members of the ABC transporter family are ABCC1, ABCB1, and ABCG2. ABCC1 and ABCG2 are less frequently reported in studies related to miRNA and ABC transporters. Therefore, more studies on the miRNA-induced ABCB1 or ABCG2 expression, which leads to drug resistance in ovarian cancer, will improve our understanding of the drug resistance mechanism of ABC transporters.
EMT has been widely implicated in the malignant behavior of tumors. It promotes tumor invasion and migration. Li et al. (100) found that miR-181a is overexpressed in paclitaxel-resistant ovarian cancer cells, and it induces paclitaxel resistance by facilitating EMT through the upregulation of N-cadherin and downregulation of E-cadherin. N-cadherin is a positive regulator, and E-cadherin is a negative regulator of EMT (101). The malignant phenotype induced by miRNA-induced EMT may be the key to ovarian cancer drug resistance, which requires further research.
Glycolysis is beneficial for the malignant phenotype of tumors as it provides energy for the metabolism of ovarian cancer cells to induce cisplatin resistance (Figure 3) (77, 102). MiR-1180 induces the abnormal upregulation of glycolysis by activating the Wnt signaling pathway and its downstream components including Wnt5a, β-catenin, c-Myc, and CyclinD1 proteins that can enhance glycolysis. MiR-1180 targets SFRP1 to relieve its inhibitory effect on Wnt5a, which activates the Wnt/β-catenin signaling pathway to upregulate PDK1 expression. PDK1 is a key enzyme required for the glycolysis of tumor cells and is transcribed by the combination of lymphoid enhancer factor/T-cell factor (LEF/TCF) and β-catenin (103). This abnormal upregulation of glycolysis is a cause of cisplatin resistance in ovarian cancer. Drug resistance in ovarian cancer cells induced by miRNA-mediated abnormal glycolysis is not extensively studied; therefore, further research on this topic may reveal a new link between miRNA and drug resistance.
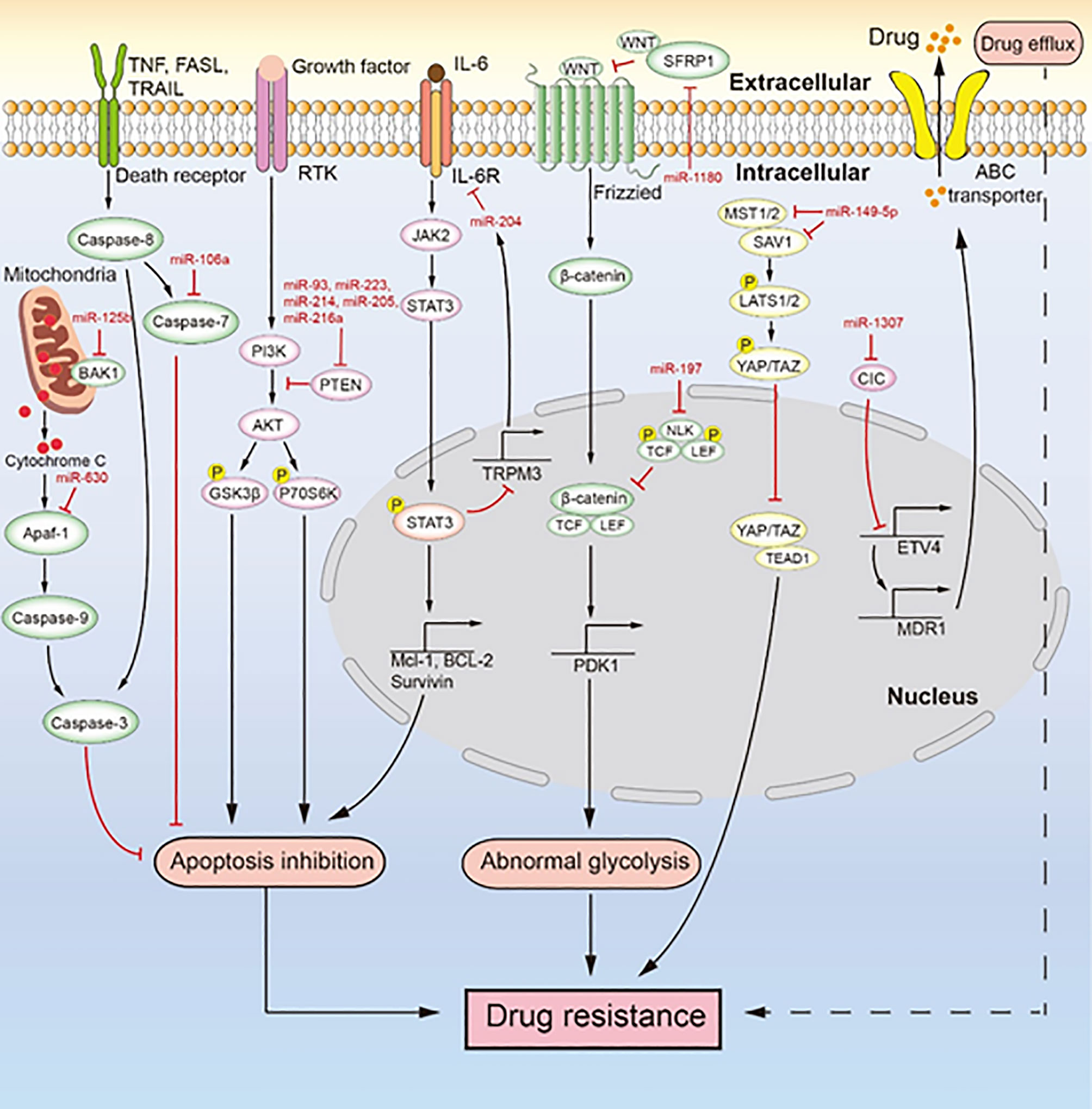
Figure 3 MiRNAs mediate chemotherapeutic resistance by multiple signaling pathways. Multiple miRNAs lead to drug resistance by inducing drug efflux, apoptosis inhibition, and abnormal glycolysis. MiR-106a, miR-630, and miR-125b inhibit apoptosis to induce drug resistance. MiR-93, miR-223, miR-214, miR-205, and miR-216a regulate the PI3K/AKT signaling pathway to inhibit apoptosis, which induces drug resistance. The IL-6/STAT3/miR-204 feedback loop inhibits apoptosis to promote drug resistance. MiR-1180 and miR-197 act via the Wnt signaling pathway to promote drug resistance. MiR-149-5p regulates the expression of MST1/2 and SAV1 to facilitate drug resistance. MiR-1307 upregulates the expression of ABCB1 to promote drug resistance.
As the main reason for chemotherapeutic resistance, apoptosis inhibition is induced by the abnormal expression of miRNAs to cause chemotherapeutic resistance in ovarian cancer (Figure 3). For instance, Xu et al. (79) reported that miR-149-5p is highly expressed in cisplatin-resistant ovarian cancer cells, and inactivation of the Hippo signaling pathway induces cisplatin resistance by directly inhibiting the expression of MST1 and SAV1. The downregulation of MST1 and SAV1 reduces the phosphorylation of YAP and TAZ through inhibition of the phosphorylation of LAST1/2. This enhances the nuclear levels of YAP and TAZ, and their upregulation inhibits the activity of caspase-3 and caspase-9, leading to apoptosis inhibition. MiR-106a is highly expressed in cisplatin-resistant and paclitaxel-resistant ovarian cancer cells (80, 93). It not only targets PDCD4 to downregulate the level of cleaved caspase-8 and cleaved caspase-3 in the death receptor pathway but also directly inhibits the expression of caspase-7 and BCL10. Therefore, miR-106a inhibits cell apoptosis by downregulating the expression of apoptosis-related proteins and makes ovarian cancer cells resistant to cisplatin and paclitaxel. MiR-214 targets PTEN to induce cisplatin resistance in ovarian cancer (82). The overexpression of miR-214 in ovarian cancer and the inhibition of PTEN expression reduce the activation of the AKT pathway, which promotes the phosphorylation of glycogen synthase kinase 3h (GSK3β) and p70 Ribosomal Protein S6 Kinase (p70S6K). Thus, miR-214 facilitates apoptosis inhibition to induce cisplatin resistance. Similarly, the upregulation of miR-93, miR-223, or miR-216a expression can promote the cisplatin resistance of ovarian cancer through activation of the PI3K/AKT pathway caused by the sponging of PTEN (78, 81, 88, 89, 104). In addition, miR-21, miR-130a, miR-205, and miR-93 can negatively regulate the expression of PTEN, which makes ovarian cancer cells resistant to cisplatin (83, 90, 96, 105). MiR-125b is highly expressed in cisplatin-resistant ovarian cancer cells and targets BAK1 to induce cisplatin resistance (87). As a pro-apoptotic protein, BAK1 enhances the mitochondrial permeability and promotes the release of cytochrome C, thereby promoting the occurrence of mitochondrial apoptosis (106). PRKCD is considered to induce apoptosis (107). MiR-204, as a key factor of the IL-6R/STAT3/miR-204 feedback loop, causes cisplatin resistance in epithelial ovarian cancer (86). Mechanistically, the binding of interleukin-6(IL-6) to IL-6R activates JAK2 to promote the level and nuclear translocation of p-STAT3, which enhances the transcription of anti-apoptotic proteins (MCL-1, BCL-2, and survivin). It also binds to the promoter region of TRPM3 to inhibit the transcription of miR-204, which the then enhances the level of its target protein IL-6R, further activating the transcription of anti-apoptotic proteins induced by p-STAT3 to facilitate cisplatin resistance. The high expression of miR-630 in paclitaxel-resistant ovarian cancer cells induces apoptosis inhibition by directly downregulating APAF-1 (92, 108). As an activator of mitochondrial apoptosis, APAF-1 induces apoptosis in ovarian cancer. In an ovarian tumor xenograft mouse model, the upregulation of miR-223 and miR-205 decreased tumor growth and the expression of PTEN (78, 83). Likewise, the high expression of miR-204 enhanced cisplatin resistance, and the low expression of miR-630 increased paclitaxel sensitivity in an ovarian tumor xenograft mouse model (86, 92). Both miR-204 and miR-630 promote the malignant phenotype of tumor cells. MiRNA inhibits the apoptosis of ovarian cancer cells mainly through inhibition of the death receptor pathway, inhibition of the mitochondrial apoptosis pathway, and activation of the PI3K/AKT pathway. These three signaling pathways are extensively studied, but the role of miRNA in these pathways seems like an unexplored area of research. Studies in this area will deepen our understanding of the drug resistance of ovarian cancer caused by apoptosis inhibition.
CircRNA is a new research hotspot in the regulation of drug resistance of ovarian cancer by ncRNAs, but research on this topic is still in its nascent stages (109–112). Luo et al. (109) found that circFoxp1 upregulates the level of CCAAT enhancer binding protein gamma (CEBPG) and formin-like 3 (FMNL3) to promote cisplatin resistance by sponging miR-22 or miR-150-3p. CEBPG and FMNL3, two oncogenes in ovarian cancer, are common targets of miR-22 and miR-150-3p, respectively. Similarly, circTNPO3, circCELSR1, and circNRIP1 are highly expressed in paclitaxel-resistant ovarian cancer cells (110–112). CircTNPO3 increases the expression of NIMA-related kinase 2 (NEK2) by directly targeting miR-1299, which then contributes to chemotherapeutic resistance in ovarian cancer (113). CircCELSR1 regulates the miR-1252/forkhead box R2 (FOXR2) axis to facilitate paclitaxel resistance in ovarian cancer. CircNRIP1 upregulates the expression of HOXC8 by sponging miR-211-5p, which makes ovarian cancer cells less sensitive to paclitaxel. HOXC8 enhances the expression of PCNA, CyclinD1, Bcl-2, MMP2, and MMP9, thereby inhibiting the expression of cleaved caspase-3. In an ovarian tumor xenograft mouse model, the knockdown of circTNPO3, circCELSR1, or circNRIP1 slowed down tumor growth (110–112). In addition, the knockdown of circTNPO3 or circNRIP1 increased paclitaxel sensitivity in vivo. In recent years, research on circRNA has been gaining momentum, but there is a lot of scope for further research. Therefore, future research could focus on the relationship between circRNA and ovarian cancer resistance, which will open avenues for targeting ncRNAs to overcome drug resistance in ovarian cancer.
Many types of ncRNAs exist, and each type has many different members. In this review, we have covered most members of lncRNA, miRNA, and circRNA families that have been found to be associated with drug resistance in ovarian cancer in recent years and their mechanisms leading to drug resistance. Studies on ncRNAs in ovarian cancer drug resistance have mainly focused on the well-known drug resistance pathways of ncRNA, such as apoptosis inhibition, drug efflux, and EMT. Through these studies, we can understand the role of ncRNAs in tumors, their function at the molecular level, and their response to chemotherapeutic drugs. Additional in-depth studies on these ncRNAs in terms of the signaling pathways that they are involved in, including identifying the specific upstream and downstream factors, will help us understand how ncRNAs induce ovarian cancer drug resistance and lead to poor prognosis. However, the main challenge is to screen and isolate the most prominent ncRNAs related to drug resistance from a large pool of ncRNAs.
Systemic chemotherapy is currently the main treatment strategy for ovarian cancer patients. Unfortunately, drug resistance remains an inevitable problem in the long-term chemotherapy of cancer. Interventions aimed at abnormally expressed ncRNAs have shown promise in reversing the drug resistance of ovarian cancer. Currently, several ways have been proposed to achieve ncRNA-targeted therapy including the upregulation ofncRNAs by utilizing mimics, the exogenous expression or downregulation of ncRNAs by using small interfering RNAs (siRNAs) or short hairpin RNAs (shRNAs), and inhibition of ncRNA function through antisense oligonucleotides. These methods will allow us to regulate the expression of ncRNAs restore the drug sensitivity of drug-resistant ovarian cancer cells. Therefore, the combination of ncRNA-targeted therapy and chemotherapy may be a promising method for the treatment of ovarian cancer in the future. However, it is a major challenge to accurately and effectively apply these ncRNA modulators to the human body. It is reported that incorporation of specific oligonucleotides into nanoparticles can improve their delivery efficiency, thereby achieving optimal therapeutic effect on tumors (114). In addition, GalNAc-siRNA conjugates have been shown to accurately act as siRNAs against mRNAs in cells; therefore, this technology can be explored to counter ovarian cancer drug resistance (115). However, prior to the clinical application of such technology, research on its safety and practicality is indispensable. More relevant clinical trials are required to understand the true clinical potential of the above-mentioned ncRNAs, so that we can develop novel and effective treatment strategies for ovarian cancer.
GL, JG, SC, ZW, and HZ: contributed to this article with the design. GL, JG, ZW, DC, JZ, XH, and JT: literature search. GL, JG, SC, YY, and WC: drafting. GL, JG, SC, ZW, DC, WC, and HZ: revision. GL, JG, ZW, JZ, XH, JT, WC, and HZ: editing. HZ: final approval. All authors contributed to the article and approved the submitted version.
Our work was supported by the Key Project of Joint Funds of Hubei Health and Family Planning Commission (Grant No. WJ2018H174), the Postgraduate Innovation Fund Project of Yangtze University Health Science Center (Grant No. 202006), and the Natural Science Foundation of Hubei Province (Grant No. 2017CFB703).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
All authors of this manuscript apologize to researchers whose works were not cited in this review article due to space limitations.
1. Scilimati A, Ferorelli S, Iaselli MC, Miciaccia M, Pati ML, Fortuna CG, et al. Targeting COX-1 by Mofezolac-Based Fluorescent Probes for Ovarian Cancer Detection. Eur J Med Chem (2019) 179:16–25. doi: 10.1016/j.ejmech.2019.06.039
2. Momenimovahed Z, Tiznobaik A, Taheri S, Salehiniya H. Ovarian Cancer in the World: Epidemiology and Risk Factors. Int J Womens Health (2019) 11:287–99. doi: 10.2147/IJWH.S197604
3. Ko SY, Naora H. HOXA9 Promotes Homotypic and Heterotypic Cell Interactions That Facilitate Ovarian Cancer Dissemination via Its Induction of P-Cadherin. Mol Cancer (2014) 13:170. doi: 10.1186/1476-4598-13-170
4. Fujiwara K, Hasegawa K, Nagao S. Landscape of Systemic Therapy for Ovarian Cancer in 2019: Primary Therapy. Cancer (2019) 125(Suppl 24):4582–6. doi: 10.1002/cncr.32475
5. Karam A, Ledermann JA, Kim JW, Sehouli J, Lu K, Gourley C, et al. Fifth Ovarian Cancer Consensus Conference of the Gynecologic Cancer InterGroup: First-Line Interventions. Ann Oncol (2017) 28(4):711–7. doi: 10.1093/annonc/mdx011
6. Christie EL, Bowtell DDL. Acquired Chemotherapy Resistance in Ovarian Cancer. Ann Oncol (2017) 28(suppl_8):viii13–5. doi: 10.1093/annonc/mdx446
7. Lv X, Song J, Xue K, Li Z, Li M, Zahid D, et al. Core Fucosylation of Copper Transporter 1 Plays a Crucial Role in Cisplatin-Resistance of Epithelial Ovarian Cancer by Regulating Drug Uptake. Mol Carcinog (2019) 58(5):794–807. doi: 10.1002/mc.22971
8. Lukanović D, Herzog M, Kobal B, Cerne K. The Contribution of Copper Efflux Transporters ATP7A and ATP7B to Chemoresistance and Personalized Medicine in Ovarian Cancer. BioMed Pharmacother (2020) 129:110401. doi: 10.1016/j.biopha.2020.110401
9. Chen S, Jiao JW, Sun KX, Zong ZH, Zhao Y. MicroRNA-133b Targets Glutathione S-Transferase π Expression to Increase Ovarian Cancer Cell Sensitivity to Chemotherapy Drugs. Drug Des Devel Ther (2015) 9:5225–35. doi: 10.2147/DDDT.S87526
10. Tong X, Zhao J, Zhang Y, Mu P, Wang X. Expression Levels of MRP1, GST-π, and GSK3β in Ovarian Cancer and the Relationship With Drug Resistance and Prognosis of Patients. Oncol Lett (2019) 18(1):22–8. doi: 10.3892/ol.2019.10315
11. Dong SC, Sha HH, Xu XY, Hu TM, Lou R, Li H, et al. Glutathione S-Transferase π: A Potential Role in Antitumor Therapy. Drug Des Devel Ther (2018) 12:3535–47. doi: 10.2147/DDDT.S169833
12. Yap TA, Plummer R, Azad NS, Helleday T. The DNA Damaging Revolution: PARP Inhibitors and Beyond. Am Soc Clin Oncol Educ Book (2019) 39:185–95. doi: 10.1200/EDBK_238473
13. Wu Z, Li S, Tang X, Wang Y, Guo W, Cao G, et al. Copy Number Amplification of DNA Damage Repair Pathways Potentiates Therapeutic Resistance in Cancer. Theranostics (2020) 10(9):3939–51. doi: 10.7150/thno.39341
14. Slack FJ, Chinnaiyan AM. The Role of Non-Coding RNAs in Oncology. Cell (2019) 179(5):1033–55. doi: 10.1016/j.cell.2019.10.017
15. Amaral PP, Dinger ME, Mercer TR, Mattick JS. The Eukaryotic Genome as an RNA Machine. Science (2008) 319(5871):1787–9. doi: 10.1126/science.1155472
16. Statello L, Guo CJ, Chen LL, Huarte M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat Rev Mol Cell Biol (2020) 22(2):1–23. doi: 10.1038/s41580-020-00315-9
17. Ha M, Kim VN. Regulation of microRNA Biogenesis. Nat Rev Mol Cell Biol (2014) 15(8):509–24. doi: 10.1038/nrm3838
18. Kim D, Sung YM, Park J, Kim S, Kim J, Park J, et al. General Rules for Functional microRNA Targeting. Nat Genet (2016) 48(12):1517–26. doi: 10.1038/ng.3694
19. Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The Landscape of Circular RNA in Cancer. Cell (2019) 176(4):869–881.e13. doi: 10.1016/j.cell.2018.12.021
20. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The Biogenesis, Biology and Characterization of Circular RNAs. Nat Rev Genet (2019) 20(11):675–91. doi: 10.1038/s41576-019-0158-7
21. Jin L, Pan YL, Zhang J, Cao PG. LncRNA HOTAIR Recruits SNAIL to Inhibit the Transcription of HNF4α and Promote the Viability, Migration, Invasion and EMT of Colorectal Cancer. Transl Oncol (2021) 14(4):101036. doi: 10.1016/j.tranon.2021.101036
22. Gong C, Maquat LE. lncRNAs Transactivate STAU1-Mediated mRNA Decay by Duplexing With 3’ UTRs via Alu Elements. Nature (2011) 470(7333):284–8. doi: 10.1038/nature09701
23. Park E, Maquat LE. Staufen-Mediated mRNA Decay. Wiley Interdiscip Rev RNA (2013) 4(4):423–35. doi: 10.1002/wrna.1168
24. Zhao Y, Wang H, Wu C, Yan M, Wu H, Wang J, et al. Construction and Investigation of lncRNA-Associated ceRNA Regulatory Network in Papillary Thyroid Cancer. Oncol Rep (2018) 39(3):1197–206. doi: 10.3892/or.2018.6207
25. Shi C, Wang M. LINC01118 Modulates Paclitaxel Resistance of Epithelial Ovarian Cancer by Regulating miR-134/Abcc1. Med Sci Monit (2018) 24:8831–9. doi: 10.12659/MSM.910932
26. Godnic I, Zorc M, Jevsinek Skok D, Calin GA, Horvat S, Dovc P, et al. Genome-Wide and Species-Wide in Silico Screening for Intragenic MicroRNAs in Human, Mouse and Chicken. PloS One (2013) 8(6):e65165. doi: 10.1371/journal.pone.0065165
27. Li S, Le TN, Nguyen TD, Trinh TA, Nguyen TA. Bulges Control pri-miRNA Processing in a Position and Strand-Dependent Manner. RNA Biol (2020) 1–11. doi: 10.1080/15476286.2020.1868139
28. Sheng P, Fields C, Aadland K, Wei T, Kolaczkowski O, Gu T, et al. Dicer Cleaves 5’-Extended microRNA Precursors Originating From RNA Polymerase II Transcription Start Sites. Nucleic Acids Res (2018) 46(11):5737–52. doi: 10.1093/nar/gky306
29. Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, et al. The H19 lincRNA is a Developmental Reservoir of miR-675 That Suppresses Growth and Igf1r. Nat Cell Biol (2012) 14(7):659–65. doi: 10.1038/ncb2521
30. Wu X, Wang Y, Zhong W, Cheng H, Tian Z. The Long Non-Coding RNA MALAT1 Enhances Ovarian Cancer Cell Stemness by Inhibiting YAP Translocation From Nucleus to Cytoplasm. Med Sci Monit (2020) 26:e922012. doi: 10.12659/MSM.922012
31. Thomas AA, Feng B, Chakrabarti S. ANRIL Regulates Production of Extracellular Matrix Proteins and Vasoactive Factors in Diabetic Complications. Am J Physiol Endocrinol Metab (2018) 314(3):E191–e200. doi: 10.1152/ajpendo.00268.2017
32. Fan B, Pan W, Wang X, Wei M, He A, Zhao A, et al. Long Noncoding RNA Mediates Stroke-Induced Neurogenesis. Stem Cells (2020) 38(8):973–85. doi: 10.1002/stem.3189
33. Youmans DT, Gooding AR, Dowell RD, Cech TR. Competition Between PRC2.1 and 2.2 Subcomplexes Regulates PRC2 Chromatin Occupancy in Human Stem Cells. Mol Cell (2021) 81(3):488–501.e9. doi: 10.1016/j.molcel.2020.11.044
34. Batool A, Jin C, Liu YX. Role of EZH2 in Cell Lineage Determination and Relative Signaling Pathways. Front Biosci (Landmark Ed) (2019) 24:947–60. doi: 10.2741/4760
35. Xia R, Jin FY, Lu K, Wan L, Xie M, Xu TP, et al. SUZ12 Promotes Gastric Cancer Cell Proliferation and Metastasis by Regulating KLF2 and E-Cadherin. Tumour Biol (2015) 36(7):5341–51. doi: 10.1007/s13277-015-3195-7
36. Yang YW, Flynn RA, Chen Y, Qu K, Wan B, Wang KC, et al. Essential Role of lncRNA Binding for WDR5 Maintenance of Active Chromatin and Embryonic Stem Cell Pluripotency. Elife (2014) 3:e02046. doi: 10.7554/eLife.02046
37. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science (2010) 329(5992):689–93. doi: 10.1126/science.1192002
38. Bo C, Li N, Li X, Liang X, An Y. Long Noncoding RNA Uc.338 Promotes Cell Proliferation Through Association With BMI1 in Hepatocellular Carcinoma. Hum Cell (2016) 29(4):141–7. doi: 10.1007/s13577-016-0140-z
39. Wang DY, Li N, Cui YL. Long Non-Coding RNA CCAT1 Sponges miR-454 to Promote Chemoresistance of Ovarian Cancer Cells to Cisplatin by Regulation of Surviving. Cancer Res Treat (2020) 52(3):798–814. doi: 10.4143/crt.2019.498
40. Bai L, Wang A, Zhang Y, Xu X, Zhang X. Knockdown of MALAT1 Enhances Chemosensitivity of Ovarian Cancer Cells to Cisplatin Through Inhibiting the Notch1 Signaling Pathway. Exp Cell Res (2018) 366(2):161–71. doi: 10.1016/j.yexcr.2018.03.014
41. Wang H, Fang L, Jiang J, Kuang Y, Wang B, Shang X, et al. The Cisplatin-Induced lncRNA PANDAR Dictates the Chemoresistance of Ovarian Cancer via Regulating SFRS2-Mediated P53 Phosphorylation. Cell Death Dis (2018) 9(11):1103. doi: 10.1038/s41419-018-1148-y
42. An J, Lv W, Zhang Y. LncRNA NEAT1 Contributes to Paclitaxel Resistance of Ovarian Cancer Cells by Regulating ZEB1 Expression via miR-194. Onco Targets Ther (2017) 10:5377–90. doi: 10.2147/OTT.S147586
43. Gu L, Li Q, Liu H, Lu X, Zhu M. Long Noncoding RNA TUG1 Promotes Autophagy-Associated Paclitaxel Resistance by Sponging miR-29b-3p in Ovarian Cancer Cells. Onco Targets Ther (2020) 13:2007–19. doi: 10.2147/OTT.S240434
44. Wang J, Ye C, Liu J, Hu Y. UCA1 Confers Paclitaxel Resistance to Ovarian Cancer Through miR-129/ABCB1 Axis. Biochem Biophys Res Commun (2018) 501(4):1034–40. doi: 10.1016/j.bbrc.2018.05.104
45. Wang F, Zhou J, Xie X, Hu J, Chen L, Hu Q, et al. Involvement of SRPK1 in Cisplatin Resistance Related to Long Non-Coding RNA UCA1 in Human Ovarian Cancer Cells. Neoplasma (2015) 62(3):432–8. doi: 10.4149/neo_2015_051
46. Dastvan R, Mishra S, Peskova YB, Nakamoto RK, Mchaourab HS. Mechanism of Allosteric Modulation of P-Glycoprotein by Transport Substrates and Inhibitors. Science (2019) 364(6441):689–92. doi: 10.1126/science.aav9406
47. Dongre A, Weinberg RA. New Insights Into the Mechanisms of Epithelial-Mesenchymal Transition and Implications for Cancer. Nat Rev Mol Cell Biol (2019) 20(2):69–84. doi: 10.1038/s41580-018-0080-4
48. Miro C, Di Cicco E, Ambrosio R, Mancino G, Di Girolamo D, Cicatiello AG, et al. Thyroid Hormone Induces Progression and Invasiveness of Squamous Cell Carcinomas by Promoting a ZEB-1/E-Cadherin Switch. Nat Commun (2019) 10(1):5410. doi: 10.1038/s41467-019-13140-2
49. Najafi M, Farhood B, Mortezaee K. Cancer Stem Cells (CSCs) in Cancer Progression and Therapy. J Cell Physiol (2019) 234(6):8381–95. doi: 10.1002/jcp.27740
50. Li J, Yang S, Su N, Wang Y, Yu J, Qiu H, et al. Overexpression of Long non-Coding RNA HOTAIR Leads to Chemoresistance by Activating the Wnt/beta-Catenin Pathway in Human Ovarian Cancer. Tumour Biol (2016) 37(2):2057–65. doi: 10.1007/s13277-015-3998-6
51. Wang Y, Wang H, Song T, Zou Y, Jiang J, Fang L, et al. HOTAIR is a Potential Target for the Treatment of Cisplatinresistant Ovarian Cancer. Mol Med Rep (2015) 12(2):2211–6. doi: 10.3892/mmr.2015.3562
52. Karim R, Tse G, Putti T, Scolyer R, Lee S. The Significance of the Wnt Pathway in the Pathology of Human Cancers. Pathology (2004) 36(2):120–8. doi: 10.1080/00313020410001671957
53. Chang H, Zou Z. Targeting Autophagy to Overcome Drug Resistance: Further Developments. J Hematol Oncol (2020) 13(1):159. doi: 10.1186/s13045-020-01000-2
54. Chen X, Duan N, Zhang C, Zhang W. Survivin and Tumorigenesis: Molecular Mechanisms and Therapeutic Strategies. J Cancer (2016) 7(3):314–23. doi: 10.7150/jca.13332
55. Zhu M, Yang L, Wang X. NEAT1 Knockdown Suppresses the Cisplatin Resistance in Ovarian Cancer by Regulating miR-770-5p/PARP1 Axis. Cancer Manag Res (2020) 12:7277–89. doi: 10.2147/CMAR.S257311
56. Appleman LJ, Beumer JH, Jiang Y, Lin Y, Ding F, Puhalla S, et al. Phase 1 Study of Veliparib (ABT-888), a Poly (ADP-Ribose) Polymerase Inhibitor, With Carboplatin and Paclitaxel in Advanced Solid Malignancies. Cancer Chemother Pharmacol (2019) 84(6):1289–301. doi: 10.1007/s00280-019-03960-w
57. Zhang PF, Wu J, Luo JH, Li KS, Wang F, Huang W, et al. SNHG22 Overexpression Indicates Poor Prognosis and Induces Chemotherapy Resistance via the miR-2467/Gal-1 Signaling Pathway in Epithelial Ovarian Carcinoma. Aging (Albany NY) (2019) 11(19):8204–16. doi: 10.18632/aging.102313
58. Zhang P, Zhang P, Shi B, Zhou M, Jiang H, Zhang H, et al. Galectin-1 Overexpression Promotes Progression and Chemoresistance to Cisplatin in Epithelial Ovarian Cancer. Cell Death Dis (2014) 5(1):e991. doi: 10.1038/cddis.2013.526
59. Zhao L, Yang J, Liu X, Fu X, Ding Y, Wang C. Overexpression of Long Noncoding RNA E2F-Mediated Cell Proliferation Enhancing Long Noncoding RNA Is Involved in the Development of Chemoresistance of Cancer Cells to Carboplatin in Ovarian Endometrioid Adenocarcinoma. Cancer Biother Radiopharm (2019) 34(9):566–71. doi: 10.1089/cbr.2019.2851
60. Parmakhtiar B, Burger RA, Kim JH, Fruehauf JP. HIF Inactivation of P53 in Ovarian Cancer Can Be Reversed by Topotecan, Restoring Cisplatin and Paclitaxel Sensitivity. Mol Cancer Res (2019) 17(8):1675–86. doi: 10.1158/1541-7786.MCR-18-1109
61. Chang H, Li B, Zhang X, Meng X. NCK1-AS1 Promotes NCK1 Expression to Facilitate Tumorigenesis and Chemo-Resistance in Ovarian Cancer. Biochem Biophys Res Commun (2020) 522(2):292–9. doi: 10.1016/j.bbrc.2019.11.014
62. Li B, Pi Z, Liu L, Zhang B, Huang X, Hu P, et al. FGF-2 Prevents Cancer Cells From ER Stress-Mediated Apoptosis via Enhancing Proteasome-Mediated Nck Degradation. Biochem J (2013) 452(1):139–45. doi: 10.1042/BJ20121671
63. Liu S, Lei H, Luo F, Li Y, Xie L. The Effect of lncRNA HOTAIR on Chemoresistance of Ovarian Cancer Through Regulation of HOXA7. Biol Chem (2018) 399(5):485–97. doi: 10.1515/hsz-2017-0274
64. Ozes AR, Miller DF, Ozes ON, Fang F, Liu Y, Matei D, et al. NF-kappaB-HOTAIR Axis Links DNA Damage Response, Chemoresistance and Cellular Senescence in Ovarian Cancer. Oncogene (2016) 35(41):5350–61. doi: 10.1038/onc.2016.75
65. Miao JT, Gao JH, Chen YQ, Chen H, Meng HY, Lou G. LncRNA ANRIL Affects the Sensitivity of Ovarian Cancer to Cisplatin via Regulation of Let-7a/HMGA2 Axis. Biosci Rep (2019) 39(7). doi: 10.1042/BSR20182101
66. Xia B, Hou Y, Chen H, Yang S, Liu T, Lin M. Long Non-Coding RNA ZFAS1 Interacts With miR-150-5p to Regulate Sp1 Expression and Ovarian Cancer Cell Malignancy. Oncotarget (2017) 8(12):19534–46. doi: 10.18632/oncotarget.14663
67. Li ZY, Wang XL, Dang Y, Zhu XZ, Zhang YH, Cai BX, et al. Long Non-Coding RNA UCA1 Promotes the Progression of Paclitaxel Resistance in Ovarian Cancer by Regulating the miR-654-5p/SIK2 Axis. Eur Rev Med Pharmacol Sci (2020) 24(2):591–603.
68. Zhang Y, Ai H, Fan X, Chen S, Wang Y, Liu L. Knockdown of Long Non-Coding RNA HOTAIR Reverses Cisplatin Resistance of Ovarian Cancer Cells Through Inhibiting miR-138-5p-Regulated EZH2 and SIRT1. Biol Res (2020) 53(1):18. doi: 10.1186/s40659-020-00286-3
69. Sun J, Cai X, Yung MM, Zhou W, Li J, Zhang Y, et al. miR-137 Mediates the Functional Link Between C-Myc and EZH2 That Regulates Cisplatin Resistance in Ovarian Cancer. Oncogene (2019) 38(4):564–80. doi: 10.1038/s41388-018-0459-x
70. Li D, Wu QJ, Bi FF, Chen SL, Zhou YM, Zhao Y, et al. Effect of the BRCA1-SIRT1-EGFR Axis on Cisplatin Sensitivity in Ovarian Cancer. Am J Transl Res (2016) 8(3):1601–8.
71. Xi YN, Xin XY, Ye HM. Effects of HMGA2 on Malignant Degree, Invasion, Metastasis, Proliferation and Cellular Morphology of Ovarian Cancer Cells. Asian Pac J Trop Med (2014) 7(4):289–92. doi: 10.1016/S1995-7645(14)60040-7
72. Sashida G, Wang C, Tomioka T, Oshima M, Aoyama K, Kanai A, et al. The Loss of Ezh2 Drives the Pathogenesis of Myelofibrosis and Sensitizes Tumor-Initiating Cells to Bromodomain Inhibition. J Exp Med (2016) 213(8):1459–77. doi: 10.1084/jem.20151121
73. Ueda K, Ikeda K, Ikezoe T, Harada-Shirado K, Ogawa K, Hashimoto Y, et al. Hmga2 Collaborates With JAK2V617F in the Development of Myeloproliferative Neoplasms. Blood Adv (2017) 1(15):1001–15. doi: 10.1182/bloodadvances.2017004457
74. Honda H, Pazin MJ, Ji H, Wernyj RP, Morin PJ. Crucial Roles of Sp1 and Epigenetic Modifications in the Regulation of the CLDN4 Promoter in Ovarian Cancer Cells. J Biol Chem (2006) 281(30):21433–44. doi: 10.1074/jbc.M603767200
75. Yoshida H, Sumi T, Zhi X, Yasui T, Honda K, Ishiko O. Claudin-4: A Potential Therapeutic Target in Chemotherapy-Resistant Ovarian Cancer. Anticancer Res (2011) 31(4):1271–7.
76. Kim K, Baek SC, Lee YY, Bastiaanssen C, Kim J, Kim H, et al. A Quantitative Map of Human Primary microRNA Processing Sites. Mol Cell (2021) 81(16):3422–3439.e11. doi: 10.1016/j.molcel.2021.07.002.
77. Gu ZW, He YF, Wang WJ, Tian Q, Di W. MiR-1180 From Bone Marrow-Derived Mesenchymal Stem Cells Induces Glycolysis and Chemoresistance in Ovarian Cancer Cells by Upregulating the Wnt Signaling Pathway. J Zhejiang Univ Sci B (2019) 20(3):219–37. doi: 10.1631/jzus.B1800190
78. Zhu X, Shen H, Yin X, Yang M, Wei H, Chen Q, et al. Macrophages Derived Exosomes Deliver miR-223 to Epithelial Ovarian Cancer Cells to Elicit a Chemoresistant Phenotype. J Exp Clin Cancer Res (2019) 38(1):81. doi: 10.1186/s13046-019-1095-1
79. Xu M, Xiao J, Chen M, Yuan L, Li J, Shen H, et al. Mir−149−5p Promotes Chemotherapeutic Resistance in Ovarian Cancer via the Inactivation of the Hippo Signaling Pathway. Int J Oncol (2018) 52(3):815–27. doi: 10.3892/ijo.2018.4252
80. Li H, Xu H, Shen H, Li H. microRNA-106a Modulates Cisplatin Sensitivity by Targeting PDCD4 in Human Ovarian Cancer Cells. Oncol Lett (2014) 7(1):183–8. doi: 10.3892/ol.2013.1644
81. Fu X, Tian J, Zhang L, Chen Y, Hao Q. Involvement of microRNA-93, a New Regulator of PTEN/Akt Signaling Pathway, in Regulation of Chemotherapeutic Drug Cisplatin Chemosensitivity in Ovarian Cancer Cells. FEBS Lett (2012) 586(9):1279–86. doi: 10.1016/j.febslet.2012.03.006
82. Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, et al. MicroRNA Expression Profiling in Human Ovarian Cancer: miR-214 Induces Cell Survival and Cisplatin Resistance by targeting PTEN. Cancer Res (2008) 68(2):425–33. doi: 10.1158/0008-5472.CAN-07-2488
83. Li J, Hu K, Gong G, Zhu D, Wang Y, Liu H, et al. Upregulation of MiR-205 Transcriptionally Suppresses SMAD4 and PTEN and Contributes to Human Ovarian Cancer Progression. Sci Rep (2017) 7:41330. doi: 10.1038/srep41330
84. Chen FD, Chen HH, Ke SC, Zheng LR, Zheng XY. SLC27A2 Regulates miR-411 to Affect Chemo-Resistance in Ovarian Cancer. Neoplasma (2018) 65(6):915–24. doi: 10.4149/neo_2018_180122N48
85. Su J, Ruan S, Dai S, Mi J, Chen W, Jiang S. NF1 Regulates Apoptosis in Ovarian Cancer Cells by Targeting MCL1 via miR-142-5p. Pharmacogenomics (2019) 20(3):155–65. doi: 10.2217/pgs-2018-0161
86. Zhu X, Shen H, Yin X, Long L, Chen X, Feng F, et al. IL-6r/STAT3/miR-204 Feedback Loop Contributes to Cisplatin Resistance of Epithelial Ovarian Cancer Cells. Oncotarget (2017) 8(24):39154–66. doi: 10.18632/oncotarget.16610
87. Kong F, Sun C, Wang Z, Han L, Weng D, Lu Y, et al. miR-125b Confers Resistance of Ovarian Cancer Cells to Cisplatin by Targeting Pro-Apoptotic Bcl-2 Antagonist Killer 1. J Huazhong Univ Sci Technolog Med Sci (2011) 31(4):543. doi: 10.1007/s11596-011-0487-z
88. Gong J, Xing C, Wang LY, Xie SS, Xiong WD. L-Tetrahydropalmatine Enhances the Sensitivity of Human Ovarian Cancer Cells to Cisplatin via microRNA-93/PTEN/Akt Cascade. J buon (2019) 24(2):701–8.
89. Jin P, Liu Y, Wang R. STAT3 Regulated miR-216a Promotes Ovarian Cancer Proliferation and Cisplatin Resistance. Biosci Rep (2018) 38(4). doi: 10.1042/BSR20180547
90. Yu X, Chen Y, Tian R, Li J, Li H, Lv T. miRNA-21 Enhances Chemoresistance to Cisplatin in Epithelial Ovarian Cancer by Negatively Regulating PTEN. Oncol Lett (2017) 14(2):1807–10. doi: 10.3892/ol.2017.6324
91. Zhou Y, Wang M, Shuang T, Liu Y, Zhang Y, Shi C. MiR-1307 Influences the Chemotherapeutic Sensitivity in Ovarian Cancer Cells Through the Regulation of the CIC Transcriptional Repressor. Pathol Res Pract (2019) 215(10):152606. doi: 10.1016/j.prp.2019.152606
92. Eoh KJ, Lee SH, Kim HJ, Lee JY, Kim S, Kim SW, et al. MicroRNA-630 Inhibitor Sensitizes Chemoresistant Ovarian Cancer to Chemotherapy by Enhancing Apoptosis. Biochem Biophys Res Commun (2018) 497(2):513–20. doi: 10.1016/j.bbrc.2018.02.062
93. Huh JH, Kim TH, Kim K, Song JA, Jung YJ, Jeong JY, et al. Dysregulation of miR-106a and miR-591 Confers Paclitaxel Resistance to Ovarian Cancer. Br J Cancer (2013) 109(2):452–61. doi: 10.1038/bjc.2013.305
94. Sugio A, Iwasaki M, Habata S, Mariya T, Suzuki M, Osogami H, et al. BAG3 Upregulates Mcl-1 Through Downregulation of miR-29b to Induce Anticancer Drug Resistance in Ovarian Cancer. Gynecol Oncol (2014) 134(3):615–23. doi: 10.1016/j.ygyno.2014.06.024
95. Li Z, Hu S, Wang J, Cai J, Xiao L, Yu L, et al. MiR-27a Modulates MDR1/P-Glycoprotein Expression by Targeting HIPK2 in Human Ovarian Cancer Cells. Gynecol Oncol (2010) 119(1):125–30. doi: 10.1016/j.ygyno.2010.06.004
96. Li N, Yang L, Wang H, Yi T, Jia X, Chen C, et al. MiR-130a and MiR-374a Function as Novel Regulators of Cisplatin Resistance in Human Ovarian Cancer A2780 Cells. PloS One (2015) 10(6):e0128886. doi: 10.1371/journal.pone.0128886
97. LeBlanc VG, Firme M, Song J, Chan SY, Lee MH, Yip S, et al. Comparative Transcriptome Analysis of Isogenic Cell Line Models and Primary Cancers Links Capicua (CIC) Loss to Activation of the MAPK Signalling Cascade. J Pathol (2017) 242(2):206–20. doi: 10.1002/path.4894
98. Qi T, Qu Q, Li G, Wang J, Zhu H, Yang Z, et al. Function and Regulation of the PEA3 Subfamily of ETS Transcription Factors in Cancer. Am J Cancer Res (2020) 10(10):3083–105.
99. Nardinocchi L, Puca R, Sacchi A, D'Orazi G. Inhibition of HIF-1alpha Activity by Homeodomain-Interacting Protein Kinase-2 Correlates With Sensitization of Chemoresistant Cells to Undergo Apoptosis. Mol Cancer (2009) 8:1. doi: 10.1186/1476-4598-8-1
100. Li L, Xu QH, Dong YH, Li GX, Yang L, Wang LW, et al. MiR-181a Upregulation is Associated With Epithelial-to-Mesenchymal Transition (EMT) and Multidrug Resistance (MDR) of Ovarian Cancer Cells. Eur Rev Med Pharmacol Sci (2016) 20(10):2004–10.
101. Marchetti S, Bengalli R, Floris P, Colombo A, Mantecca P. Combustion-Derived Particles From Biomass Sources Differently Promote Epithelial-to-Mesenchymal Transition on A549 Cells. Arch Toxicol (2021). doi: 10.1007/s00204-021-02983-8
102. Icard P, Shulman S, Farhat D, Steyaert JM, Alifano M, Lincet H. How the Warburg Effect Supports Aggressiveness and Drug Resistance of Cancer Cells? Drug Resist Update (2018) 38:1–11. doi: 10.1016/j.drup.2018.03.001
103. Pate KT, Stringari C, Sprowl-Tanio S, Wang K, TeSlaa T, Hoverter NP, et al. Wnt Signaling Directs a Metabolic Program of Glycolysis and Angiogenesis in Colon Cancer. EMBO J (2014) 33(13):1454–73. doi: 10.15252/embj.201488598
104. Sen T, Sen N, Brait M, Begum S, Chatterjee A, Hoque MO, et al. DeltaNp63alpha Confers Tumor Cell Resistance to Cisplatin Through the AKT1 Transcriptional Regulation. Cancer Res (2011) 71(3):1167–76. doi: 10.1158/0008-5472.CAN-10-1481
105. Chen Q, Qin R, Fang Y, Li H. Berberine Sensitizes Human Ovarian Cancer Cells to Cisplatin Through miR-93/PTEN/Akt Signaling Pathway. Cell Physiol Biochem (2015) 36(3):956–65. doi: 10.1159/000430270
106. Todt F, Cakir Z, Reichenbach F, Emschermann F, Lauterwasser J, Kaiser A, et al. Differential Retrotranslocation of Mitochondrial Bax and Bak. EMBO J (2015) 34(1):67–80. doi: 10.15252/embj.201488806
107. Dashzeveg N, Yoshida K. Crosstalk Between Tumor Suppressors P53 and Pkcδ: Execution of the Intrinsic Apoptotic Pathways. Cancer Lett (2016) 377(2):158–63. doi: 10.1016/j.canlet.2016.04.032
108. Li Y, Zhou M, Hu Q, Bai XC, Huang W, Scheres SH, et al. Mechanistic Insights Into Caspase-9 Activation by the Structure of the Apoptosome Holoenzyme. Proc Natl Acad Sci USA (2017) 114(7):1542–7. doi: 10.1073/pnas.1620626114
109. Luo Y, Gui R. Circulating Exosomal Circfoxp1 Confers Cisplatin Resistance in Epithelial Ovarian Cancer Cells. J Gynecol Oncol (2020) 31(5):e75. doi: 10.3802/jgo.2020.31.e75
110. Xia B, Zhao Z, Wu Y, Wang Y, Zhao Y, Wang J. Circular RNA Circtnpo3 Regulates Paclitaxel Resistance of Ovarian Cancer Cells by miR-1299/NEK2 Signaling Pathway. Mol Ther Nucleic Acids (2020) 21:780–91. doi: 10.1016/j.omtn.2020.06.002
111. Zhang S, Cheng J, Quan C, Wen H, Feng Z, Hu Q, et al. Circcelsr1 (Hsa_Circ_0063809) Contributes to Paclitaxel Resistance of Ovarian Cancer Cells by Regulating FOXR2 Expression via miR-1252. Mol Ther Nucleic Acids (2020) 19:718–30. doi: 10.1016/j.omtn.2019.12.005
112. Li M, Cai J, Han X, Ren Y. Downregulation of Circnrip1 Suppresses the Paclitaxel Resistance of Ovarian Cancer via Regulating the miR-211-5p/HOXC8 Axis. Cancer Manag Res (2020) 12:9159–71. doi: 10.2147/CMAR.S268872
113. Liu X, Gao Y, Lu Y, Zhang J, Li L, Yin F. Upregulation of NEK2 Is Associated With Drug Resistance in Ovarian Cancer. Oncol Rep (2014) 31(2):745–54. doi: 10.3892/or.2013.2910
114. Yang T, Zhao P, Rong Z, Li B, Xue H, You J, et al. Anti-Tumor Efficiency of Lipid-Coated Cisplatin Nanoparticles Co-Loaded With MicroRNA-375. Theranostics (2016) 6(1):142–54. doi: 10.7150/thno.13130
Keywords: non-coding RNAs, ovarian cancer, chemotherapy, drug resistance (DR), cisplatin
Citation: Li G, Gong J, Cao S, Wu Z, Cheng D, Zhu J, Huang X, Tang J, Yuan Y, Cai W and Zhang H (2021) The Non-Coding RNAs Inducing Drug Resistance in Ovarian Cancer: A New Perspective for Understanding Drug Resistance. Front. Oncol. 11:742149. doi: 10.3389/fonc.2021.742149
Received: 15 July 2021; Accepted: 10 September 2021;
Published: 30 September 2021.
Edited by:
Nan-Shan Chang, National Cheng Kung University, TaiwanReviewed by:
Anamaria Brozovic, Rudjer Boskovic Institute, CroatiaCopyright © 2021 Li, Gong, Cao, Wu, Cheng, Zhu, Huang, Tang, Yuan, Cai and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiyuan Zhang, aHl6aGFuZ184OEAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.