- 1Department of Clinical Laboratory, Dalian Women and Children’s Medical Group, Dalian, China
- 2Centre for Reproductive and Genetic Medicine, Dalian Women and Children’s Medical Group, Dalian, China
- 3Department of Clinical Laboratory, Central Hospital of Dalian University of Technology, Dalian Municipal Central Hospital, Dalian, China
- 4Maternity Ward, Dalian Women and Children’s Medical Group, Dalian, China
- 5Child Health Care Clinic, Dalian Women and Children’s Medical Group, Dalian, China
Background: The interaction between the human breast milk microbiota and human milk oligosaccharides (HMOs) plays a crucial role in the healthy growth and development of infants. We aimed to clarify the link between the breast milk microbiota and HMOs at two stages of lactation.
Methods: The microbiota and HMOs of 20 colostrum samples (C group, 1–5 days postpartum) and 20 mature milk samples (S group, 42 days postpartum) collected from postpartum mothers were analyzed using 16S rRNA gene high-throughput sequencing and high-performance liquid chromatography–tandem mass spectrometry.
Result: The total average HMO content was significantly higher in the C group than in the S group (6.76 ± 1.40 g/L vs. 10.27 ± 2.00 g/L, p < 0.05). Among the HMOs, the average values of 2′-fucosyllactose (2′-FL, 1.64 ± 1.54 g/L vs. 3.03 ± 1.79 g/L), 3′-sialyllactose (3′-SL, 0.10 ± 0.02 g/L vs. 0.21 ± 0.06), 6′-SL (0.22 ± 0.09 g/L vs. 0.33 ± 0.11 g/L), and lacto-N-triaose 2 (LNT2, 0.03 ± 0.01 g/L vs. 0.16 ± 0.08 g/L) were significantly lower in the S group than in the C group (p < 0.05), while that of 3′-FL was significantly higher in the S group than in the C group (1.35 ± 1.00 g/L vs. 0.41 ± 0.43 g/L, p < 0.05). The diversity and structure of the microbiota in the S and C groups were also significantly different (p < 0.05). Comparative analysis of the microbial communities revealed that Proteobacteria and Firmicutes were the most abundant phyla, in both groups, with the keystone species (Serratia, Streptococcus and Staphylococcus) of breast milk closely interacting with HMOs, including 3′-SL, 6′-SL, and LNT2. In PICRUSt2 functional prediction analysis, the S group exhibited significant reduction in the expression of genes involved in several infectious disease pathways.
Discussion: Our findings support the recognition of human milk as a synbiotic comprising beneficial bacteria and prebiotic HMOs.
1 Introduction
Breast milk is the most natural and complete food for infant growth (1). In addition to being rich in protein, fat, carbohydrates, vitamins, and water, breast milk also contains immunoglobulins, human milk oligosaccharides (HMOs), cytokines, and hormones. As a group of polysaccharide substances that are unique to humans, with diverse functions and structures, HMOs are characteristically difficult to digest (1). The content of HMOs accounts for about 20% of the total carbohydrate content in breast milk (2). While there are individual changes in HMO over the entire period of breastfeeding (up to 24 months postpartum), the absolute concentration and relative abundance of most of the examined HMOs decreased during lactation. In addition to the previously documented differences in HMO concentrations between secretors and non-secretors, many of the changes in HMO that occur during lactation differ between secretors and non-secretors (3). After ingestion of HMOs, approximately 0.05% of the HMOs enter the bloodstream with the vast majority of HMOs entering the large intestine (4, 5). Owing to their unique carbohydrate structure, they are capable of playing a role in the infant’s resistance to low gastric pH, as well as pancreatic and brush border enzymes (6). Hence, the majority of HMOs are eliminated from the body through feces, with a portion of the HMOs being metabolized—mainly by the infant gut microbiota—to produce various beneficial effects (7). HMOs serve as metabolic substrates for the growth of probiotics such as Bifidobacterium in the infant’s gut (8, 9), thereby reducing intestinal pH and inhibiting pathogen growth. Important products of the metabolism of HMOs, including acetic acid and butyric acid, can lower the pH value in the intestine and inhibit the growth of pathogens. In addition to affecting bifidobacteria, other bacteria that have also been studied, including Lactobacillus acidophilus and Eubacterium hallii, exhibit physiological effects that are all related to HMOs (10). In addition to having nutritional effects on the gut microbiota of infants, there is increasing evidence that HMOs can interact with pathogens to reduce the occurrence of infections (2). The structure of HMOs, which are similar to the surface glycoconjugates on which microorganisms attach to human intestinal cells, can act as bait receptors for binding to pathogens, reducing the adhesion of pathogenic microorganisms to the intestinal wall (11).
Importantly, breast milk also has a microbial ecosystem. Breast milk has traditionally been considered to be a sterile liquid (12). However, using culture medium screening methods, researchers have confirmed the presence of Staphylococcus and Streptococcus in breast milk collected under sterile conditions. Studies using microbial identification techniques such as metagenomic sequencing and 16S rRNA gene amplification sequencing have shown that breast milk also contains various other bacteria, including Lactobacillus and Bifidobacterium (13). A systematic review of the bacterial bank in breast milk showed the presence of approximately 820 types of bacteria, mainly belonging to the Gram-positive Firmicutes and Gram-negative Proteobacteria phyla (14). Current studies have shown that the breast milk microbiota is a decisive factor in the development of the gut microbiota and plays a pioneering role in the healthy development of infants (15). From the initial contact of the infant’s gut with the microbiota transferred from breast milk, the microorganisms begin to colonize and grow in the intestine. This is a gradual process that plays a crucial role in the maturation of the intestinal lymphoid tissue and the growth and development of intestinal epithelial cells for infants (16). The microbiota in breast milk mainly reduces the risk of infant infection and disease through competition and inhibition among the microbiota (8). Importantly, maternal diet, age, and different periods of lactation affect the nutrients, bioactive compounds, immune factors, and microbiota contained in breast milk, which further affect the structural changes in the gut microbiota for infants (17, 18).
Human milk oligosaccharides and the breast milk microbiota play important roles in the establishment and development of the infant gut microbiota. The relationship between the changes in HMO concentration that occur during lactation and the microbiota of breast milk is not well-studied. The purpose of this study were to: (1) investigate the differences in the microbial community and the content and types of HMOs in breast milk between colostrum samples and mature milk samples at 42 days postpartum; and (2) analyze correlations between HMOs and the breast milk microbiota. Our findings provide specific experimental evidence for the importance of breastfeeding, which is of great significance for the development of infant formula foods that are closer to breast milk.
2 Experimental methods
2.1 Participants
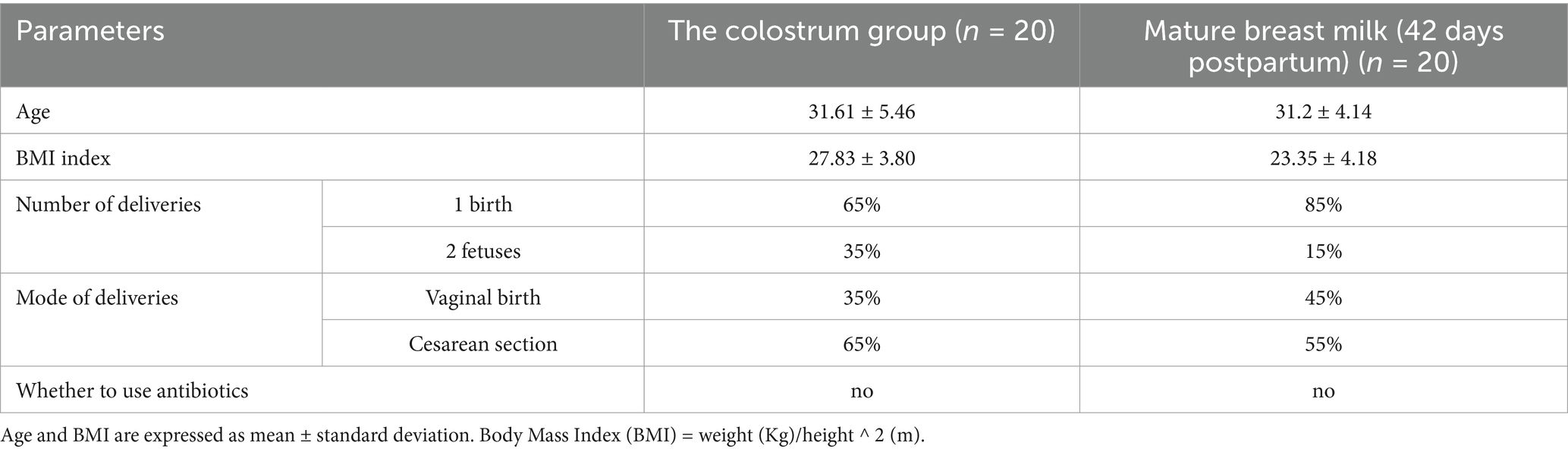
In this study, 40 healthy postpartum mothers were recruited at Dalian Women and Children’s Medical Center (Group) for the collection of breast milk samples. Twenty human colostrum samples (C group) were donated within 1–5 days postpartum by 20 mothers, and 20 mature milk samples (S group) were donated by the remaining 20 mothers at 42 days postpartum (19). We also collected maternal demographic and clinical data including age, height, weight, body mass index, number of deliveries, and delivery method (Supplementary Table S1).
Inclusion criteria were as follows: (1) no underlying metabolic disorders; (2) no gastrointestinal diseases, immunological disorders, infectious diseases, or organic diseases. (3) Received no antibiotics within 1 month before breast milk sampling. The study was conducted following the principles of the Declaration of Helsinki, and reviewed by the Medical Ethics Committee of Dalian Women and Children’s Medical Center (approval number: 2022017). Written informed consent was obtained from the mother before the participation in the study.
2.2 Breast milk samples collection
Before collecting breast milk samples, the mothers were required to clean their hands with soap and use disinfectant wipes or 75% alcohol to sanitize the nipple and areola surfaces. They were also instructed to discard the first drop of breast milk to prevent contamination. Breast milk samples (more than 5 mL) were collected, placed in a pre-prepared sterile container, sealed, and stored in a −80°C freezer until analysis (20).
2.3 High-performance liquid chromatography (HPLC)-tandem mass spectrometry (MS/MS) of HMOs in breast milk
Breast milk samples were blended to mix and brought to room temperature. A 1-mL aliquot of each sample was transferred to a clean, 2-mL centrifuge tube and spun at 4°C under 12,000 rpm for 10 min. After removing the upper oil layer, 1 mL of anhydrous ethanol was added, and the mixture was allowed to stand for 4 min to precipitate protein. After centrifugation at 12,000 rpm for 5 min, the 200-μL supernatant was loaded onto an HPLC-MS/MS machine (Quantum Hi-Tech [Guangdong] Biological Co., Ltd.) for quantitative analysis of each component, alongside the following 16 HMO standards (20): 2′-fucosyllactose (2′-FL), 3′-fucosyllactose (3′-FL), lacto-N-tetraose (LNT), lacto-N-neotetraose (LNnT), 3′-sialyllactose (3′-SL), 6′-sialyllactose (6′-SL), lactose-N-difucohexose I (LNDFH I), lactose-N-difucotetraose (LDFT), lactoyl-N-fucopentaose I (LNFP I), disialylated lactose-N-tetrasaccharide (DSLNT), sialyl-lactose-N-tetrasaccharide (LSTc), bisfucosyllactose-N-hexasaccharide (DFLNH), lactose-N-difucohexose II (LNDFH II), and lactoyl-N-fucopentaose II (LNFP II). HMOs are mainly divided into three structural types: neutral fucosylated HMOs, neutral non-fucosylated HMOs, and acidic sialylated HMOs. Neutral fucosylated HMOs mainly include 2′-FL and 3′-FL; neutral non-fucosylated HMOs include LNT and LNnT; and acidic sialylated HMOs include 3′-SL and 6′-SL (21, 22).
2.4 Nucleic acid extraction and Illumina sequencing of human breast milk microbiota DNA
Total DNA was isolated from breast milk samples (1.5–2.0 mL) using the E.Z.N.A.® Stool DNA Kit (Epicentre), in accordance with the manufacturer’s instructions. The V3–V4 hypervariable variable regions of genomic DNA samples were amplified by polymerase chain reaction (PCR) using primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). PCR products of breast milk microbiota composition were determined by V3–V4 variable regions of the 16S rRNA gene high-throughput sequencing by Shanghai Maggi Biomedical Technology Co., Ltd., using an Illumina MiSeq PE300 following Illumina protocols. The 16S rRNA gene sequences were defined as one operational taxonomic unit (OTU) based on 97% similarity. The abundance-based coverage estimator (Ace) index, observed species (Sobs) index, and Shannon diversity index were used to reflect the alpha diversity of samples. A hierarchical clustering tree at the OTU level, principal component analysis (PCoA), and non-metric multidimensional scaling analysis (NMDS) were used to analyze beta diversity. Linear discriminant analysis (LDA) was used to screen for dominant microbial communities (23). Random Forest model analysis was performed on the Maggi Platform under default settings through the utilization of an out-of-bag error within the R environment (version 3.3.1). The data were standardized at the genus level by the Z-score method, and the area under the curve (AUC) was verified under the condition that the number of with 500 decision trees (24). Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was applied to predict functional profiles of the gut microbiota resulting from reference-based OTU picking against the Greengenes database. The predicted genes were then summarized using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway categorization. The difference-rich OTUs were employed to calculate Pearson’s correlation coefficients, which were used to conduct network analysis. Gephi was used for topology analysis and visualization purposes (23).
2.5 Statistical analysis
The experimental data and statistical charts in this study were analyzed and processed using SPSS 22 software (IBM, United States) and Graph Pad Prism Version 8 (Graph Pad Software Inc., United States). Statistical analysis of KEGG pathway data was performed with STAMP v2.1.3 using Welsh’s t-test (p < 0.05). The experimental data were expressed as mean ± standard deviation (SD), with a p-value <0.05 indicating statistical significance; *p < 0.05, **p < 0.01 and ***p < 0.001 vs. the M group.
3 Results
3.1 Basic clinical information
Analysis of the basic clinical data of the study participants showed no significant differences in age or body mass index (BMI) between the C and S groups (Table 1 and Supplementary Table S1).
3.2 Analysis of HMOs in breast milk
As shown in Figure 1A, there were differences in the concentrations of individual HMOs in breast milk between the C and S groups. In both study groups, the top five most abundant individual HMOs identified were all found to belong to the fucosylated group: 2′-FL (30%), LNFPI (18%), LSTc (13%), LNT (6%), and LDFT (6%) in the C group; and 2′-FL (24%), 3′-FL (20%), LNFP I (10%), LNT (9%), and LNFP II (8%) in the M group. The average total HMO content in breast milk was significantly lower in the S group than in the C group (6.76 ± 1.40 g/L vs. 10.27 ± 2.00 g/L, p < 0.05; Figure 1B). Among the neutral fucosylated HMOs, the average concentration of 2′-FL was significantly lower in the S group than in the C group (1.64 ± 1.54 g/L vs. 3.03 ± 1.79 g/L, p < 0.05), whereas the average concentration of 3′-FL was significantly higher in the S group than in the C group (1.35 ± 1.00 g/L vs. 0.41 ± 0.43 g/L, p < 0.05; Figure 1B). Among the neutral non-fucosylated HMOs, the average concentration of LNT2 was significantly lower in the S group than in the C group (0.03 ± 0.01 g/L vs. 0.16 ± 0.08 g/L, p < 0.05), but there was no significant differences in those of LNT (0.59 ± 0.27 g/L vs. 0.59 ± 0.39 g/L) and LNnT (0.34 ± 0.14 g/L vs. 0.40 ± 0.12 g/L) between the two groups (p > 0.05). The average concentrations of the acidic sialylated HMOs 3′-SL (0.10 ± 0.02 g/L vs. 0.21 ± 0.06 g/L) and 6′-SL (0.22 ± 0.09 g/L vs. 0.33 ± 0.11 g/L) were significantly lower in the S group than in the C group (p < 0.05). The average concentrations of LNDFH I (0.25 ± 0.25 g/L vs. 0.47 ± 0.26 g/L), LDFT (0.19 ± 0.21 g/L vs. 0.65 ± 0.71 g/L), LNFP I (0.70 ± 0.67 g/L vs. 1.90 ± 0.96 g/L), DSLNT (0.19 ± 0.07 g/L vs. 0.36 ± 0.14 g/L), and LSTc (0.23 ± 0.18 g/L vs. 1.37 ± 0.56 g/L) were significantly lower in the S group than in the C group (p < 0.05). The average concentration of DFLNH was significantly higher in the S group than in the C group (0.28 ± 0.30 g/L vs. 0.05 ± 0.11 g/L, p < 0.05). There were no significant differences in the average concentrations of LNDFH II (0.12 ± 0.15 g/L vs. 0.08 ± 0.10 g/L) and LNFP II (0.54 ± 0.52 g/L vs. 0.28 ± 0.37 g/L) between the C and S groups, respectively (p > 0.05).
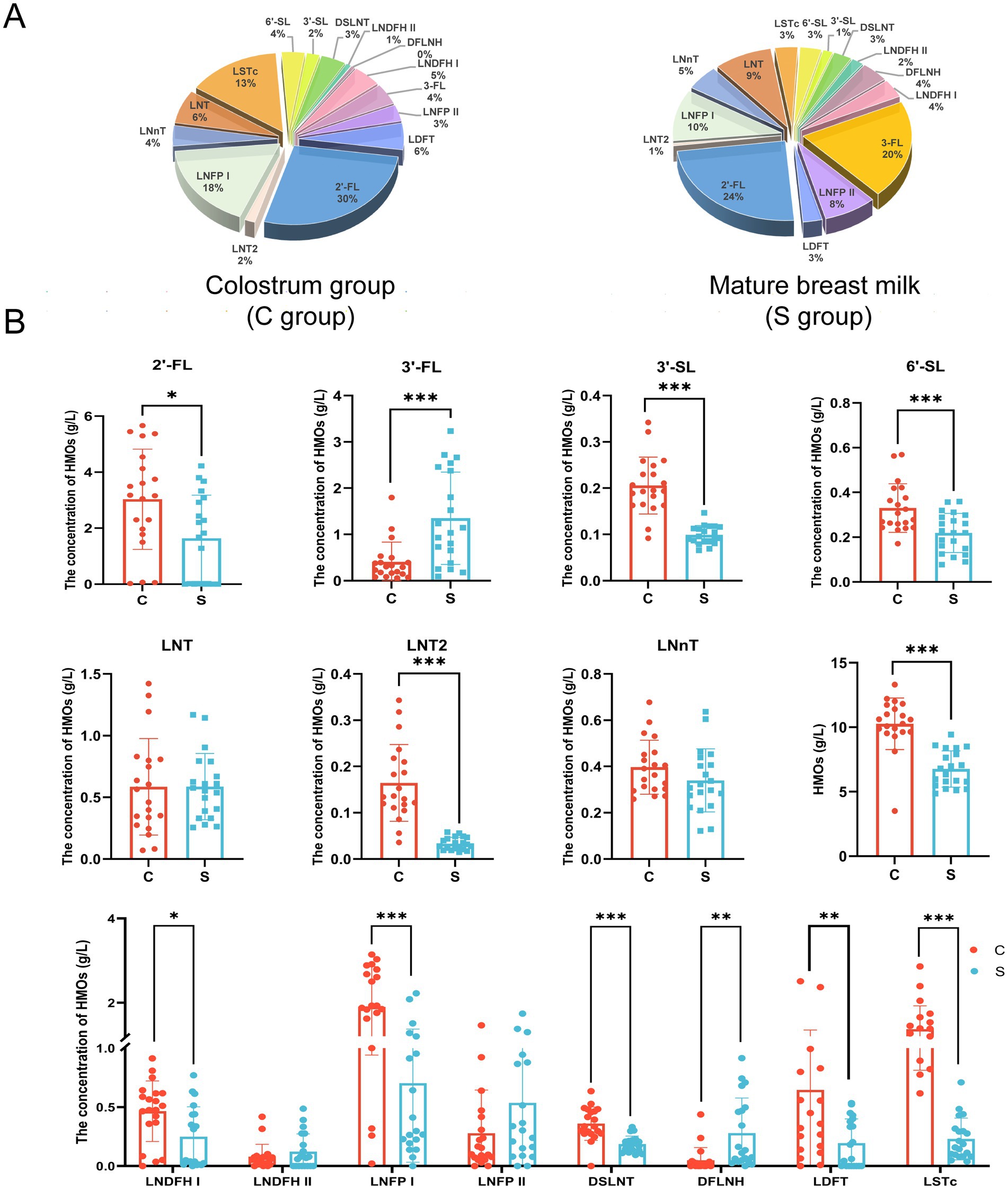
Figure 1. The proportion (A) and composition (B) of oligosaccharides in breast milk. C: the colostrum group (n = 20); S: the mature milk (42 days postpartum) group (n = 20). *p < 0.05, **p < 0.01, ***p < 0.001 compared with the C group.
3.3 Quality assessment of human breast milk microbiota DNA samples and 16S rRNA gene amplification and sequencing
Comparison of the results of 16S rRNA gene high-throughput sequencing analysis of the human breast milk microbiota in the C and S groups (Supplementary Table S2) showed stabilization of the refraction curve of the Sobs and Shannon indexes (Supplementary Figure S1), indicating sufficient depth of sequencing data for each group of samples. Similarly, the Good’s coverage index of the breast milk microbiota reached above 0.99 (Supplementary Table S2). The Ace, Sobs, and Chao indexes of the S group showed significantly higher alpha-diversity compared with those of the C group (p < 0.01; Figure 2A). There was no difference between the Shannon index and Simpson index values (Supplementary Table S3).
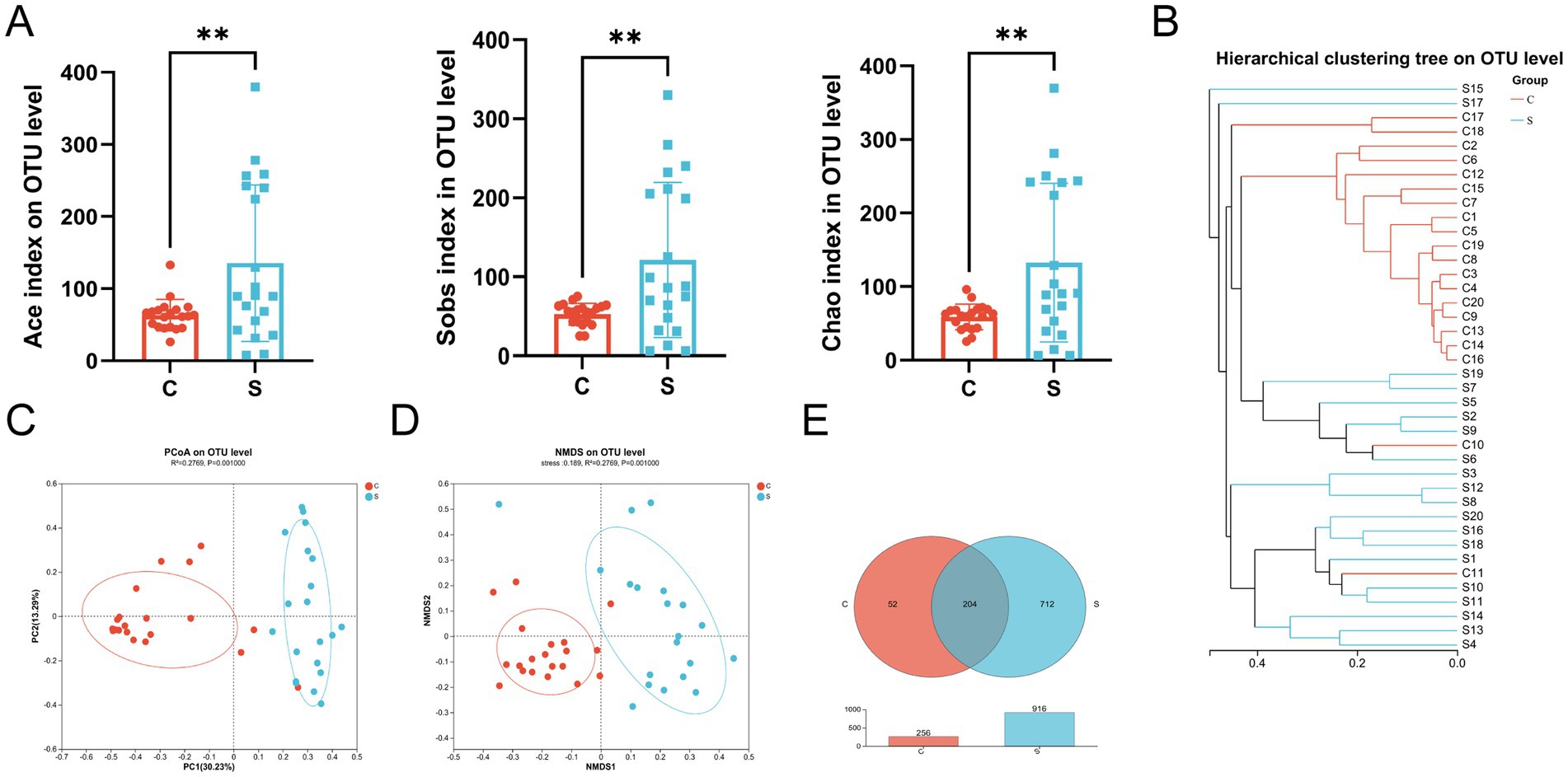
Figure 2. The microbial diversity and composition of the breast milk microbiota in the C and S group. (A) Alpha diversity (Ace and Sobs and Chao index) on the OTU level. (B) Hierarchical clustering tree on the OTU level. (C) Principal component analysis (PCoA) with cluster. (D) Non-metric multidimensional scaling analysis (NMDS) with cluster. (E) Venn diagram. C: the colostrum group (n = 20); S: the mature milk (42 days postpartum) group (n = 20). *p < 0.05, **p < 0.01, ***p < 0.001 compared with the C group.
The beta-diversity of the breast milk microbiota was demonstrated by the formation of two distinct clusters between the C and S groups, as illustrated by hierarchical clustering, PCoA (R2 = 0.2769, p = 0.001) and NMDS (Stress = 0.189, R2 = 0.2769, p = 0.001) analysis at the OTU level (Figures 2B–D). Meanwhile, there were 204 shared OTUs between the groups. The S group had 712 unique OTUs, which far exceeded the 53 OTUs in the C group, indicating that the number of OTUs in the S group increased with time (p < 0.01; Figure 2E).
3.4 Analysis of the composition of microbial communities in colostrum and mature milk
At the phylum level, the microbial community of the C group was mainly composed of Proteobacteria, Firmicutes, and Bacteroidota, while that of the S group was mainly composed of Proteobacteria, Firmicutes, and Actinobacteria (Supplementary Figure S2A). The relative abundance of Proteobacteria in the breast milk was significantly higher in the C group than in the S group (p < 0.05; Figures 3A,B). The relative abundance of Firmicutes and Actinobacteria in the breast milk is significantly higher in the S group than in the C group (p < 0.05; Figures 3A,B). At the Family level, the relative abundance of Alcaligenaceae, Staphylococcaceae, Bacillaceae, Streptococcaceae and Gemellaceae in the S group was significantly higher than that in the C group (p < 0.05, Figure 3C). The abundance of Yersiniaceae and Pseudomonas was significantly higher in the C group than in the S group (p < 0.05, Figure 3C). Combining Random Forest analysis and the Wilcoxon rank-sum test at the genus level, the abundance of Serratia, Acinetobacter, and Pseudomonas were significantly higher in the C group than in the S group (p < 0.05; Figure 3D and Supplementary Figures S2B,D). The abundances of the genera Achromobacter, Staphylococcus, Bacillus, Streptococcus, Escherichia-Shigella, and Gemella genera were significantly higher in the S group than that in the C group (p < 0.05).
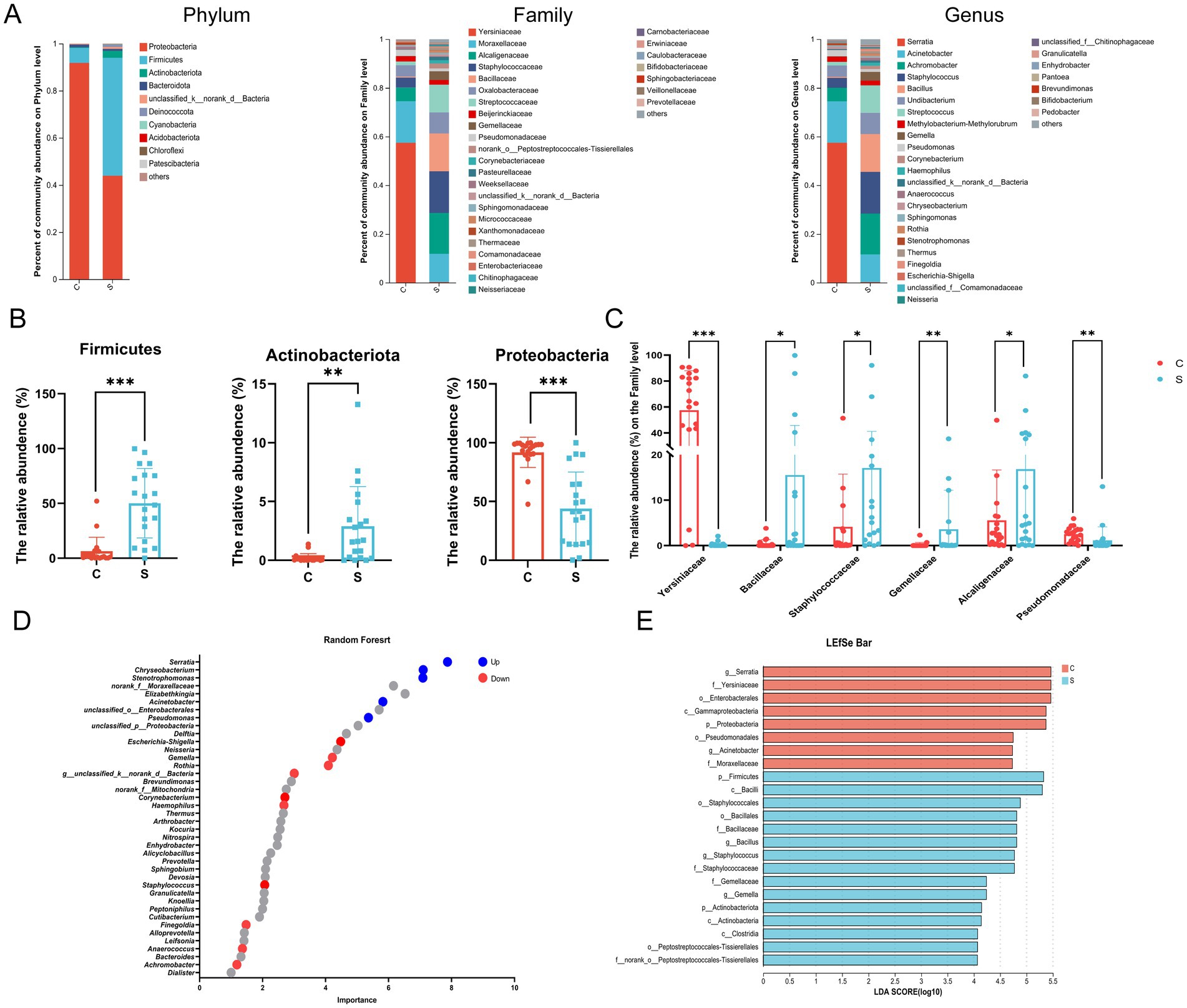
Figure 3. Analysis of the structure and communities of the breast milk microbiota in the N and M group. (A) Microbial distributions of different groups at the phylum, Family and Genus level. (B) The relative abundance of Firmicutes, Actinobacteria and Proteobacteria at the Phylum level. (C) The relative abundance of Alcaligenaceae, Bacillaceae, Gemellaceae, Pseudomonadaceae, Staphylococcaceae, and Yersiniaceae at the Family level. (D) Random forest at the genus level. Up (red) represented the S group raised; down (blue) represented the S group decreased (p < 0.05). (E) Linear discriminant analysis (LDA) distribution by analyzing Linear discriminate analysis effect size (LEfSe). The threshold of the LDA score was 4.0. C: the colostrum group (n = 20); S: the mature milk (42 days postpartum) group (n = 20). *p < 0.05, **p < 0.01, ***p < 0.001 compared with the C group.
Linear discriminant analysis effect size (LefSe) analysis was employed to identify biomarker bacterial genera among two groups using a LDA score = 4. The significant biomarker bacteria in the C group were identified as p_Proteobacteria, o_Enterobacteriales, o_Pseudomonas, f_Yersiniaceae, f_Moraxellaceae, g_Serratia and g_Acinetobacter. The relative abundance of p_Firmicutes, p_Actinobacteriota, f_Bacillaceae, f_Staphylococcaceae, f_Gemellaceae, f_Histostreptococcales, g_Bacillus, g_Staphylococcus, and g_Gemella were enriched in the S group (Figure 3E).
3.5 Different potential functions and identification of co-abundance networks of colostrum and mature milk at the OTU level
The impact of the stage of breast milk on various KEGG pathways was assessed via PICRUST2 analysis of the 16s rRNA amplicon sequencing data using PICRUST2. This revealed significant reductions in the expression of genes related to the following KEGG level 2 functional pathways in mature milk compared with those in colostrum: infectious disease (bacteria, parasitic), cancer (specific types, overview), cardiovascular disease, and nervous system (p < 0.05; Figure 4A). The analysis of level 3 KEGG pathways revealed a significant increase in biosynthesis of amino acids, aminoacyl-tRNA biosynthesis, pyruvate metabolism, Staphylococcus aureus infection, lysine biosynthesis, nucleotide excision repair, D-alanine metabolism and pantothenate and CoA biosynthesis in the S group compared with those in the C group (p < 0.05; Figure 4B). In contrast, several infection-related pathways were reduced in the S group, including biofilm formation-Escherichia coli, lipopolysaccharide biosynthesis, bacterial secretion system, biofilm formation-Vibrio cholera, pertussis, phosphonate and phosphinate metabolism, plant–pathogen interaction, Shigellosis, phenylpropanoid biosynthesis, biosynthesis of ansamycins, monobactam biosynthesis, alpha-linolenic acid metabolism, African trypanosomiasis, chagas disease (American trypanosomiasis), glutamatergic synapse, and GABAergic synapse (p < 0.05; Figure 4B).
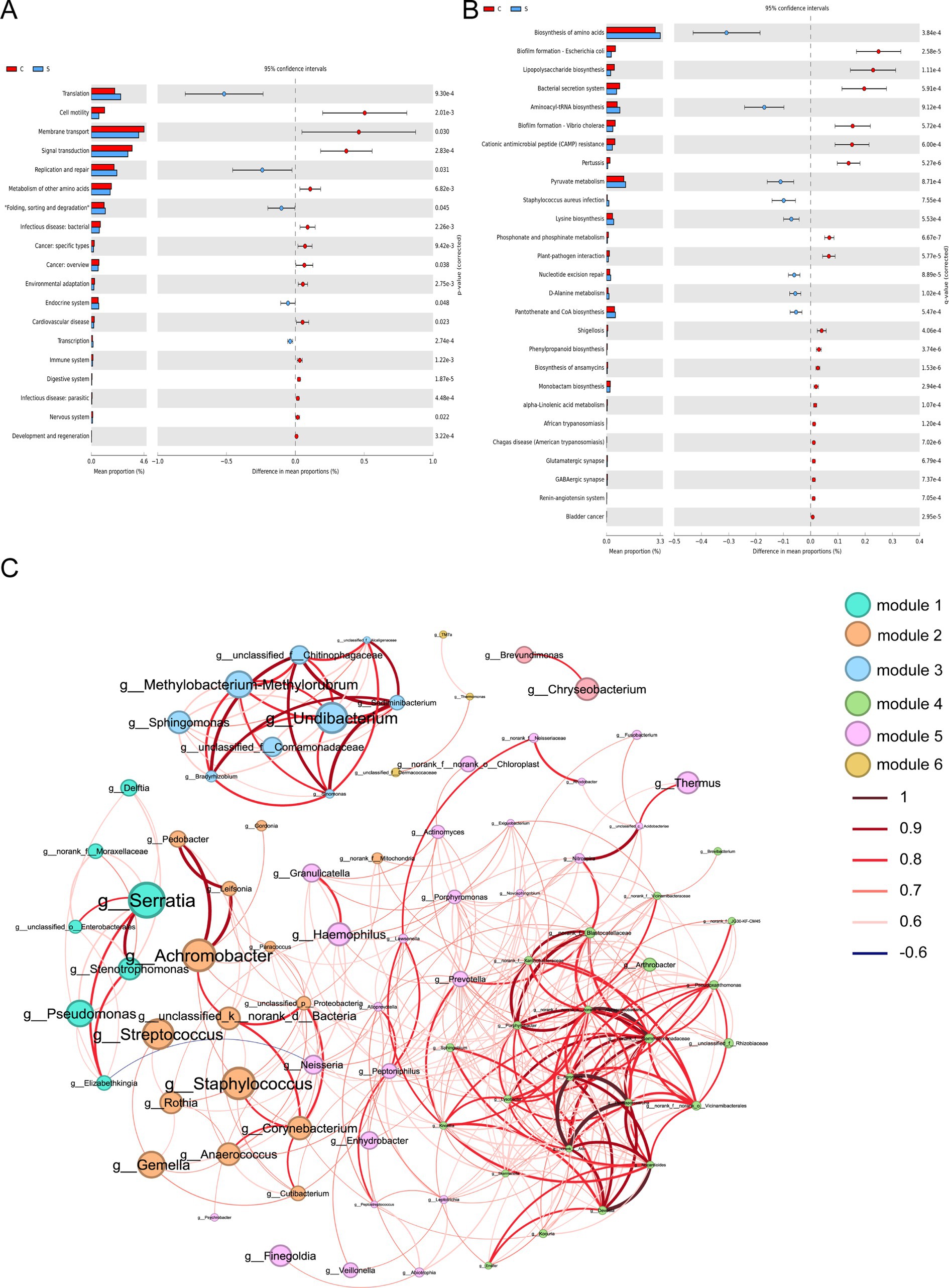
Figure 4. Functional prediction and co-abundance networks at the genus level between the C group and the S group. (A) STAMP analysis for the inferred metabolic pathway in level 2. (B) STAMP analysis for the inferred metabolic pathway in level 3. (C) The co-abundance networks. C: the colostrum group (n = 20); S: the mature milk (42 days postpartum) group (n = 20). *p < 0.05, **p < 0.01, ***p < 0.001 compared with the C group.
Functional prediction analysis fails to capture the complexity of bacterial interactions in microbial communities. Therefore, we sought to explore the inherent patterns of Co-occurrence or Co-exclusion of specific microbial communities driven by spatial and temporal changes and environmental processes at the genus level. This network analysis revealed 84 nodes and 377 edges. Among these, Serratia, Streptococcus and Staphylococcus as the keystone species exhibiting a close-positive association with other genera including Achromobacter, Leifsonia, Neisseria, and Delfia (Figure 4C and Supplementary Table S4).
3.6 Correlation analysis between HMOs and the breast milk microbiota
In the heatmap based on Spearman correlation analysis at the family level (Supplementary Figure S3), Lactobacillaceae was negatively correlated with LNFP II (p < 0.05). Staphylococcaceae and Gemellaceae were negatively correlated with 3′-SL, LNT2, LSTc, and total HMOs (p < 0.05). Yersiniaceae, Pseudomonadaceae, and Xanthomonadaceae were positively correlated with 2′-FL, DSLNI, 3′-FL, LNT2, LSTc, LNFP I and HMOs, and negatively correlated with LNFP II, DFLNH, and 3′-FL (p < 0.05). Alcaligenaceae and Bacillaceae was positively correlated with 6’-SL (p < 0.05) and DFLNH (p < 0.05), respectively.
At the genus level, unclassified_k_norank_d_Bacteria, Staphylococcus, Corynebacterium, Anaerococcus, Rothia, Gemella, Neisseria, Acinetobacter and Haemophilus were negatively correlated with 3′-SL, LNT2, and LSTc (p < 0.05; Figure 5). Staphylococcus was negatively correlated with HMOs (p < 0.05), while Finegoldia was negatively correlated with LNT2 and LSTc (p < 0.05). Serratia, Pseudomonas, and Stenotrophomonas were positively correlated with 3′-SL, LNT2, LSTc, DSLNT, and HMOs (p < 0.05), and were negatively correlated with DFLNH and 3′-FL (p < 0.05; Figure 5). Serratia and Pseudomonas were positively correlated with 6′-SL (p < 0.05; Figure 5). Bifidobacterium was negatively correlated with LNDFH II (p < 0.05; Figure 5).
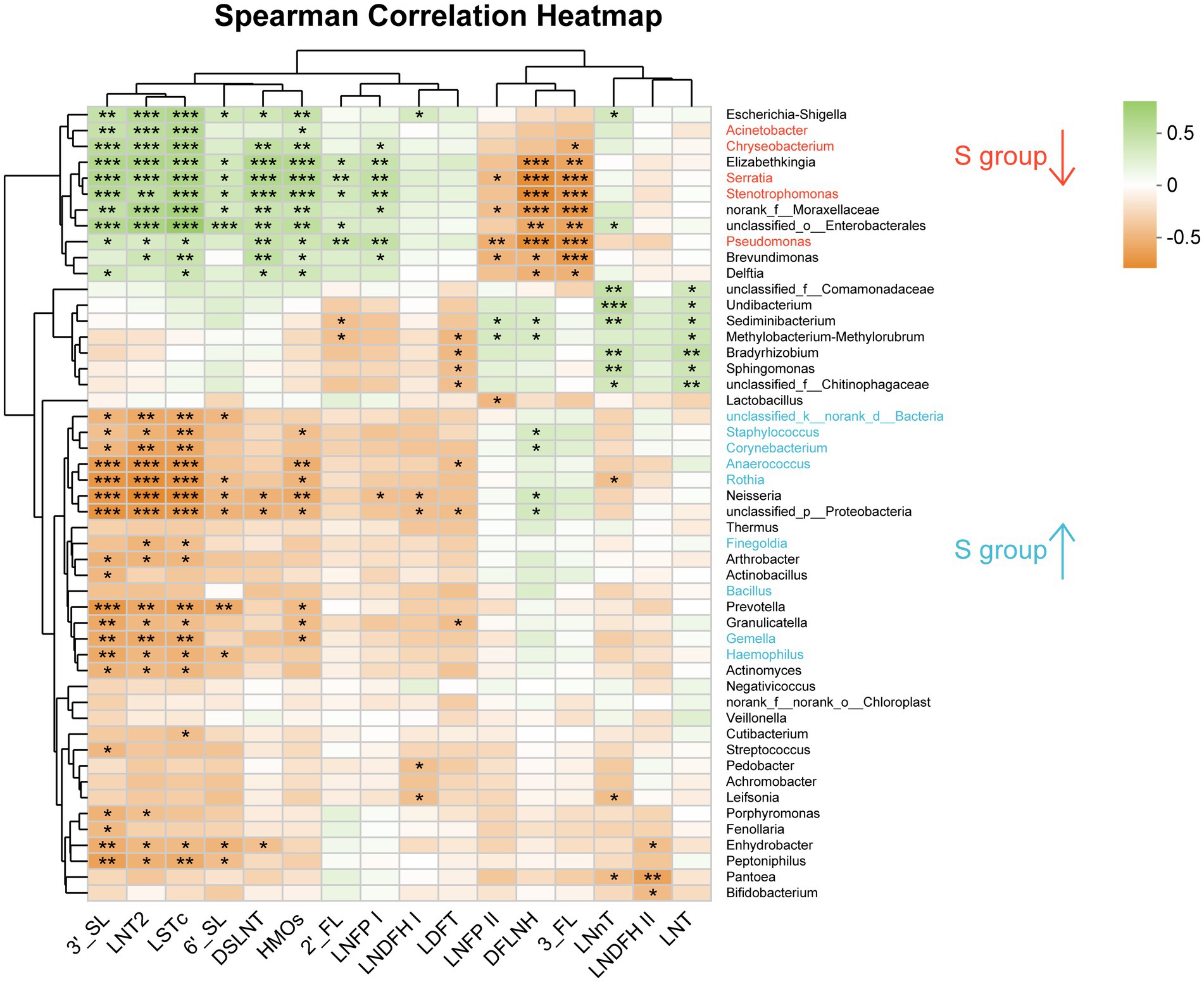
Figure 5. Analysis of the correlation between oligosaccharides and the breast milk microbiota in the C group and the S group at the Genus level. C: the colostrum group (n = 20); S: the mature milk (42 days postpartum) group (n = 20). *p < 0.05, **p < 0.01, ***p < 0.001.
4 Discussion
Breast milk is the cornerstone of healthy growth for infants, offering a wide range of benefits. HMOs, which are considered prebiotics (25, 26), are the third most abundant nutrient in breast milk, after lactose and lipids (27). Previous data have shown significant differences in the total HMO concentration in breast milk among individuals. Additionally, previous studies have shown that the concentration of HMOs is highest in colostrum (average 9–22 g/L) and slightly lower in transitional milk (average 8–19 g/L from post-birth days 8 to day 15). As lactation progresses, the concentration of HMOs in mature breast milk gradually decreases, from 6 to 15 g/L in milk collected within 1 month to 4–6 g/L in milk collected within 6 months post birth (2, 7). This is consistent with the results of this study, in which we found a significant difference in the average concentrations of HMOs between colostrum (9.7 g/L) and mature milk (6.8 g/L). However, a study conducted by Kunz et al. (28) found no difference in total HMO concentrations between colostrum, transition milk, and mature milk. This discrepancy may be attributable to differences in sample preparation, detection methods, and data analysis. The current detection methods for HMOs are not uniform. Elwakiel et al. (29) used the capillary laser-induced fluorescence method, Ma et al. (30) employed the HPLC-MRM-MS method, and Huang et al. (31) used the ultra-HPLC-fluorometric detection method.
The most abundant HMO is 2′-FL, accounting for approximately 20–40% of the total HMO content in colostrum (30). This is consistent with the results of our study, in which 2′-FL was found to account for 27% of the total HMO content. As lactation progresses, the concentration of HMOs in breast milk continuously changes with most HMOs decreasing over time. Six HMOs (2′-FL, LNnT, difucosyllactose, lacto-N-tetraose, 3′- and 6′-SL) have been observed to enrich butyrate-producing bacteria and Bifidobacteriaceae in vitro simulating microbial ecology and the gut epithelium (32). One exception is 3′-FL, which increases along the length of the intestine and stimulates the production of mucin and antimicrobial peptides in intestinal goblet cells, benefiting the immunity of the infant’s gut (33). Consistent with the results of this study, previous studies reported a negative correlation between 2′-FL and 3′-FL (2, 34), mainly attributable to the two HMOs containing similar molecular structures and sharing guanosine 5′-diphosphate-1-amylose (GDP) as a substrate (35). LNT2 can be obtained through the acid hydrolysis of LNnT, which can quickly increase the abundance of beneficial bacteria and reduce the abundance of pathogenic bacteria, affecting the healthy balance of the human colon microbiota (36). In a study by Kong et al. (36), 90.1% of LNT2 was utilized by the infant fecal microbiome in vitro, effectively increasing the production of acetic acid, succinic acid, lactic acid, and butyric acid, and significantly increasing the number of Bifidobacterium cells (37). Although the trend of its concentration change was barely mentioned by the authors, the study showed that LNT2 concentration increased with the prolongation of lactation time. Conversely, the concentration of DFLNH reportedly gradually decreases with lactation (38). This is inconsistent with the results of our study, in which the concentration of DFLNH was significantly higher in mature milk than in colostrum, and may be attributable to factors including race and lifestyle habits. Furthermore, we found that the concentrations of two acidic sialylated HMOs (3′-SL and 6′-SL) decreased with the duration of lactation. The 6′-SL and 2′-FL have previously been shown to protect against necrotizing enterocolitis (severity) by reducing inflammation through inhibition of the Toll-like receptor 4 activation pathway (39). Additionally, in the newborn piglet model, 3′-SL and 6′-SL administration were shown to up-regulate the level of glia-derived neurotrophic factor (GDNF) and the phosphorylation of CAMP response element binding protein (CREB) on the extracellular domain of neural cell adhesion molecule (NCAM) in the ileum, thereby controlling neuronal function to promote intestinal maturation (40, 41).
Recent studies have demonstrated that, while breast milk contributes to approximately 25% of an infant’s gut microbiota (41), its impact on short-and long-term infant health remains uncertain (42). Nonetheless, limited research has established a definitive correlation between the breast milk microbiota and HMOs. This prompted our interest in investigating the regulation of the human milk microbiota and its association with HMOs during lactation. Over time, the diversity and richness of the breast milk microbiota continue to decrease, mainly due to changes in the nutritional composition of breast milk (43, 44). The major bacterial phyla identified in this study included Firmicutes and Proteobacteria consistent with published results (45–47). The breast milk microbiota is the main source of early colonization in the gut of infants (48–50). Here, we found that Achromobacter, Staphylococcaceae, Bacillaceae, Streptococcaceae, Gemellaceae, Yersiniaceae, and Pseudomonas accounted for most of the microbiota in breast milk at the family level consistent with those at the genus level. During the course of lactation, we observed increases in the relative abundance of Achromobacter, Acinetobacter, Staphylococcus, Bacillus, and Streptococcus within the human milk microbiota. The gut microbiota of neonates was positively correlated with most of the microorganisms in the human milk microbiota, which included unclassified_Enterobacteriaceae, Escherichia-Shigella, Streptococcus, and Bifidobacterium, suggesting that human milk microbiota may promote neonatal intestinal colonization (51). In contrast, a previous study found the dominant genera, including Acinetobacter, Streptococcus, Enterobacteriaceae, Serratia, Chryseobacterium, Prevotella, and Corynebacterium, did not change significantly between the different lactation periods (52). Our finding showed that significant negative correlation between Acinetobacter and 3′-SL, LNT2, and LSTc. Studies have confirmed the antibacterial and anti-biofilm activities of HMOs against various bacteria in vivo, including S. agalactiae, Staphylococcus aureus, and Pseudomonas aeruginosa (53). We also found a negative correlation between HMOs and Staphylococcus, consistent with the findings of Moossavi et al. (54). In contrast, Williams et al. found a positive correlation between the relative abundance of S. aureus and HMO content of healthy breast milk samples (55).
Serratia and Escherichia-Shigella have been shown to be abundant in human breast milk and infant feces (56). The gradual decrease in Serratia in break milk during lactation may reduce its pathogenic risk to infants, especially for premature infants, who face a higher risk of infection. However, a study of prodigiosin produced by Serratia sp. C6LB and isolated from milk yield inconsistent findings, exhibiting significant biological activities including antibacterial properties, anti-biofilm effects, and cytotoxicity (57). By combining the results from Random Forest, LefSe, and network analyses, we found that Serratia, Staphylococcus, and Streptococcus were the core genera of the breast milk microbiota, consistent with the results of Lackey et al. (17, 58). Furthermore, at the genus level, the top 50 bacteria, which included Serratia, Staphylococcus, and Streptococcus, were almost associated with 3′-SL, LNT2, LSTc, 6′-SL, and DSLNT. Additionally, KEGG analysis identified several infectious disease pathways with significantly decreased activity in mature milk, suggesting that the microbial community in breast milk may possess anti-infective properties, which could be closely related to the abundant HMOs in breast milk. HMOs and the human milk microbiota complement each other to impact the gut microbiota of infants, benefiting specific bacterial genera (55). The HMOs can directly mediate Staphylococcus, Streptococcus, Lactobacillus, and Enterococcus in the infant intestine to regulate maternal mammary epithelial cell responses and local immune responses (59). Neonatal breast milk HMOs such as 3′-SL and 6′-SL have the higher correlation with the gut microbiota compared with those of 5-month-old breast milk (60). We also observed a negative correlation between Lactobacillaceae or Bifidobacterium, which had a low relative abundance (about 2–3%), and the mother’s own HMOs (LNDFH II or LNFP II) in both colostrum and mature milk groups. It was previously shown that Lactobacillus levels were negatively correlated with LNFP II and LNDFH II, which is consistent with the results presented here (61). LNDFH II can be enzymatically produced from LNT by α1, 3/4-focusing transferase (FucT14) derived from Helicobacter pylori DMS 6709 (62, 63). A study demonstrated Bifidobacterium longum was negatively correlated with LNT by analyzing 170 breast milk HMOs samples and children’s fecal microbiota samples (64). Importantly, we cannot ignore the beneficial effects of these bacteria on the intestinal and immune health of infants. Both Lactobacillus and Bifidobacterium have the ability to metabolize and break down glycans such as HMOs, which is beneficial for the absorption of HMOs and thereby provides important nutritional support for infants (65). Conversely, HMOs in breast milk can selectively stimulate the proliferation of beneficial bacteria (Bifidobacterium and Lactobacillus), which is conducive to maintaining intestinal microecological balance and inhibiting the growth of harmful pathogens (66). Therefore, the microbial community together with the HMOs in breast milk constitute a complex, effective defense mechanism that provides the newborn with protection against a variety of potential infections.
The addition of prebiotics, such as 2′-FL, 3′-FL, 3′-SL, and 6′-SL, to infant formula has been approved in the USA (66–69), European Union, and other jurisdictions (70–73). Indeed, HMOs are currently the focus of much research (74–80). Several studies have investigated the safety and tolerability of infant formulas supplemented with HMOs, including those containing one (74, 76), two (77, 78), or five (79, 80) types of HMOs. This study has demonstrated a significant decrease in the total content of HMOs with increasing lactation time, highlighting the importance of colostrum-derived HMOs in breast feeding. We also described the roles of 3′-SL and 6′-SL, which are closely related to breast milk, as well as LNT2. Our findings also provide new evidence and serve as a reference for the addition of certain prebiotic HMO molecules to the development of the next generation of infant milk powders formulated to meet the needs of infants during the transition from breastfeeding to formula.
This study has certain limitations. First, we studied a small number of samples collected at two of the three lactation stages (colostrum and mature milk) from mothers living in only one region. While previous studies investigated the human milk microbiota or HMO content at certain time points during lactation, the focus of this study was the association of the human milk microbiota with the expression of the corresponding HMOs during these two periods. It should also be acknowledged that our analysis did not consider the potential effects of age, lifestyle, maternal diet, presence or absence of the cesarean section, or geographical location on the concentration of HMOs in breast milk (58, 81, 82). Future studies should examine these different parameters on the changes in the human milk microbiota and HMOs during lactation in larger and more diverse cohorts. Additional studies are also needed to understand the associations between the biological functions of specific HMOs and the human milk microbiota over time. Multi-omics analyses (e.g., metabolomics) in combination with culture techniques and multi-site studies, as well as information on infant health outcomes, could be useful to analyze the structure and function of the human milk microbiota in various ways. Further research is urgently needed to elucidate the complex links between the HMO–microbiome and health through microbes, metabolism, and immunity.
In summary, breast milk, which is recognized as a synbiotic, is a highly complex matrix containing a beneficial, dynamic bacterial community and biologically active molecules including HMOs. This study provides new evidence and a fresh perspective on the relationship between breast milk bacteria and HMOs. It is necessary to further explore the mechanisms that drive the establishment and maturation of the intestinal microbiota and the immune system in infants during different lactation periods. Our findings may be useful for the development of powdered infant formula that more accurately mimics the nutritional composition of breast milk.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The studies involving humans were approved by Medical Ethics Committee of Dalian Women and Children’s Medical Center. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
HG: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. WZ: Formal analysis, Methodology, Software, Writing – review & editing. JZ: Formal analysis, Methodology, Software, Writing – review & editing. ZW: Formal analysis, Methodology, Software, Writing – review & editing. HS: Supervision, Writing – review & editing. JS: Writing – review & editing. MS: Conceptualization, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the PhD career start-up foundation of Dalian Women and Children’s Medical Group, Dalian, China (grant number 2022-BSQD-0156).
Acknowledgments
This work would not have been possible without the participants and Dalian Women and Children’s Medical Center in Dalian.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2024.1512700/full#supplementary-material
References
1. Cheng, L, Akkerman, R, Kong, C, Walvoort, MTC, and de Vos, P. More than sugar in the milk: human milk oligosaccharides as essential bioactive molecules in breast milk and current insight in beneficial effects. Crit Rev Food Sci Nutr. (2021) 61:1184–200. doi: 10.1080/10408398.2020.1754756
2. Thurl, S, Munzert, M, Boehm, G, Matthews, C, and Stahl, B. Systematic review of the concentrations of oligosaccharides in human milk. Nutr Rev. (2017) 75:920–33. doi: 10.1093/nutrit/nux044
3. Plows, JF, Berger, PK, Jones, RB, Alderete, TL, Yonemitsu, C, Najera, JA, et al. Longitudinal changes in human milk oligosaccharides (HMOs) over the course of 24 months of lactation. J Nutr. (2021) 151:876–82. doi: 10.1093/jn/nxaa427
4. Engfer, MB, Stahl, B, Finke, B, Sawatzki, G, and Daniel, H. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am J Clin Nutr. (2000) 71:1589–96. doi: 10.1093/ajcn/71.6.1589
5. Marriage, BJ, Buck, RH, Goehring, KC, Oliver, JS, and Williams, JA. Infants fed a lower calorie formula with 2’FL show growth and 2’FL uptake like breast-fed infants. J Pediatr Gastroenterol Nutr. (2015) 61:649–58. doi: 10.1097/MPG.0000000000000889
6. Gabrielli, O, Zampini, L, Galeazzi, T, Padella, L, Santoro, L, Peila, C, et al. Preterm milk oligosaccharides during the first month of lactation. Pediatrics. (2011) 128:e1520–31. doi: 10.1542/peds.2011-1206
7. Bode, L. The functional biology of human milk oligosaccharides. Early Hum Dev. (2015) 91:619–22. doi: 10.1016/j.earlhumdev.2015.09.001
8. Lyons, KE, Ryan, CA, Dempsey, EM, Ross, RP, and Stanton, C. Breast milk, a source of beneficial microbes and associated benefits for infant health. Nutrients. (2020) 12:1039. doi: 10.3390/nu12041039
9. Wiciński, M, Sawicka, E, Gębalski, J, Kubiak, K, and Malinowski, B. Human milk oligosaccharides: health benefits, potential applications in infant formulas, and pharmacology. Nutrients. (2020) 12:266. doi: 10.3390/nu12010266
10. Kulinich, A, and Liu, L. Human milk oligosaccharides: the role in the fine-tuning of innate immune responses. Carbohydr Res. (2016) 432:62–70. doi: 10.1016/j.carres.2016.07.009
11. Plaza-Díaz, J, Fontana, L, and Gil, A. Human milk oligosaccharides and immune system development. Nutrients. (2018) 10:1038. doi: 10.3390/nu10081038
12. Ojo-Okunola, A, Nicol, M, and du Toit, E. Human breast milk bacteriome in health and disease. Nutrients. (2018) 10:1643. doi: 10.3390/nu10111643
13. Martín, R, Jiménez, E, Heilig, H, Fernández, L, Marín, ML, Zoetendal, EG, et al. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl Environ Microbiol. (2009) 75:965–9. doi: 10.1128/AEM.02063-08
14. Togo, A, Dufour, JC, Lagier, JC, Dubourg, G, Raoult, D, and Million, M. Repertoire of human breast and milk microbiota: a systematic review. Future Microbiol. (2019) 14:623–41. doi: 10.2217/fmb-2018-0317
15. Rautava, S. Early microbial contact, the breast milk microbiome and child health. J Dev Orig Health Dis. (2016) 7:5–14. doi: 10.1017/S2040174415001233
16. Gueimonde, M, Laitinen, K, Salminen, S, and Isolauri, E. Breast milk: a source of bifidobacteria for infant gut development and maturation? Neonatology. (2007) 92:64–6. doi: 10.1159/000100088
17. Wan, Y, Jiang, J, Lu, M, Tong, W, Zhou, R, Li, J, et al. Human milk microbiota development during lactation and its relation to maternal geographic location and gestational hypertensive status. Gut Microbes. (2020) 11:1438–49. doi: 10.1080/19490976.2020.1760711
18. Nunes, M, da Silva, CH, Bosa, VL, Bernardi, JR, ICR, W, Goldani, MZ, et al. Could a remarkable decrease in leptin and insulin levels from colostrum to mature milk contribute to early growth catch-up of SGA infants? BMC Pregnancy Childbirth. (2017) 17:410. doi: 10.1186/s12884-017-1593-0
19. Li, M, Chen, J, Shen, X, Abdlla, R, Liu, L, Yue, X, et al. Metabolomics-based comparative study of breast colostrum and mature breast milk. Food Chem. (2022) 384:132491. doi: 10.1016/j.foodchem.2022.132491
20. Huang, T, Zeng, Z, Liang, X, Tang, X, Luo, H, Wang, D, et al. Effect of breast milk with or without bacteria on infant gut microbiota. BMC Pregnancy Childbirth. (2022) 22:595. doi: 10.1186/s12884-022-04930-6
21. Matharu, D, Ponsero, AJ, Lengyel, M, Meszaros-Matwiejuk, A, Kolho, KL, de Vos, WM, et al. Human milk oligosaccharide composition is affected by season and parity and associates with infant gut microbiota in a birth mode dependent manner in a Finnish birth cohort. EBioMedicine. (2024) 104:105182. doi: 10.1016/j.ebiom.2024.105182
22. Urashima, T, Hirabayashi, J, Sato, S, and Kobata, A. Human milk oligosaccharides as essential tools for basic and application studies on galectins. Trends Glycosci Glycotechnol. (2018) 30:SJ11–24. doi: 10.4052/tigg.1734.1SJ
23. Huang, J, Zhou, H, Song, T, Wang, B, Ge, H, Zhang, D, et al. Fecal microbiota transplantation from sodium alginate-dosed mice and normal mice mitigates intestinal barrier injury and gut dysbiosis induced by antibiotics and cyclophosphamide. Food Funct. (2023) 14:5690–701. doi: 10.1039/d3fo01193c
24. Sun, WL, Hua, S, Li, XY, Shen, L, Wu, H, and Ji, HF. Microbially produced vitamin B12 contributes to the lipid-lowering effect of silymarin. Nat Commun. (2023) 14:477. doi: 10.1038/s41467-023-36079-x
25. Lemoine, A, Tounian, P, Adel-Patient, K, and Thomas, M. Pre-, pro-, syn-, and postbiotics in infant formulas: what are the immune benefits for infants? Nutrients. (2023) 15:1231. doi: 10.3390/nu15051231
26. Okburan, G, and Kızıler, S. Human milk oligosaccharides as prebiotics. Pediatr Neonatol. (2023) 64:231–8. doi: 10.1016/j.pedneo.2022.09.017
27. Selvamani, S, Kapoor, N, Ajmera, A, El Enshasy, HA, Dailin, DJ, Sukmawati, D, et al. Prebiotics in new-born and children’s health. Microorganisms. (2023) 11:2453. doi: 10.3390/microorganisms11102453
28. Kunz, C, Meyer, C, Collado, MC, Geiger, L, García-Mantrana, I, Bertua-Ríos, B, et al. Influence of gestational age, secretor, and Lewis blood group status on the oligosaccharide content of human milk. J Pediatr Gastroenterol Nutr. (2017) 64:789–98. doi: 10.1097/MPG.0000000000001402
29. Elwakiel, M, Hageman, JA, Wang, W, Szeto, IM, van Goudoever, JB, Hettinga, KA, et al. Human milk oligosaccharides in colostrum and mature milk of Chinese mothers: Lewis positive secretor subgroups. J Agric Food Chem. (2018) 66:7036–43. doi: 10.1021/acs.jafc.8b02021
30. Ma, L, McJarrow, P, Jan Mohamed, HJB, Liu, X, Welman, A, and Fong, BY. Lactational changes in the human milk oligosaccharide concentration in Chinese and Malaysian mothers’ milk. Int Dairy J. (2018) 87:1–10. doi: 10.1016/j.idairyj.2018.07.015
31. Huang, X, Zhu, B, Jiang, T, Yang, C, Qiao, W, Hou, J, et al. Improved simple sample pretreatment method for quantitation of major human milk oligosaccharides using ultrahigh pressure liquid chromatography with fluorescence detection. J Agric Food Chem. (2019) 67:12237–44. doi: 10.1021/acs.jafc.9b03445
32. Natividad, JM, Marsaux, B, Rodenas, CLG, Rytz, A, Vandevijver, G, Marzorati, M, et al. Human milk oligosaccharides and lactose differentially affect infant gut microbiota and intestinal barrier in vitro. Nutrients. (2022) 14:2546. doi: 10.3390/nu14122546
33. Cheng, L, Kong, C, Walvoort, MTC, Faas, MM, and de Vos, P. Human milk oligosaccharides differently modulate goblet cells under homeostatic, proinflammatory conditions and ER stress. Mol Nutr Food Res. (2020) 64:e1900976. doi: 10.1002/mnfr.201900976
34. Austin, S, De Castro, CA, Bénet, T, Hou, Y, Sun, H, Thakkar, SK, et al. Temporal change of the content of 10 oligosaccharides in the milk of Chinese urban mothers. Nutrients. (2016) 8:346. doi: 10.3390/nu8060346
35. Thum, C, Wall, CR, Weiss, GA, Wang, W, Szeto, IMY, and Day, L. Changes in HMO concentrations throughout lactation: influencing factors, health effects and opportunities. Nutrients. (2021) 13:2272. doi: 10.3390/nu13072272
36. Kong, C, Akkerman, R, Klostermann, CE, Hou, Y, Sun, H, Thakkar, SK, et al. Distinct fermentation of human milk oligosaccharides 3-FL and LNT2 and GOS/inulin by infant gut microbiota and impact on adhesion of Lactobacillus plantarum WCFS1 to gut epithelial cells. Food Funct. (2021) 12:12513–25. doi: 10.1039/d1fo02563e
37. Sodhi, CP, Wipf, P, Yamaguchi, Y, Fulton, WB, Kovler, M, Niño, DF, et al. The human milk oligosaccharides 2′-fucosyllactose and 6′-sialyllactose protect against the development of necrotizing enterocolitis by inhibiting toll-like receptor 4 signaling. Pediatr Res. (2021) 89:91–101. doi: 10.1038/s41390-020-0852-3
38. Ren, X, Yan, J, Bi, Y, Wang, Y, Jiang, S, Wang, J, et al. Human milk oligosaccharides are associated with lactation stage and Lewis phenotype in a Chinese population. Nutrients. (2023) 15:1408. doi: 10.3390/nu15061408
39. Yang, C, Zhang, P, Fang, W, Chen, Y, Zhang, N, Qiao, Z, et al. Molecular mechanisms underlying how sialyllactose intervention promotes intestinal maturity by upregulating GDNF through a CREB-dependent pathway in neonatal piglets. Mol Neurobiol. (2019) 56:7994–8007. doi: 10.1007/s12035-019-1628-9
40. Zhu, Y, Zhang, J, Zhang, W, and Mu, W. Recent progress on health effects and biosynthesis of two key sialylated human milk oligosaccharides, 3′-sialyllactose and 6′-sialyllactose. Biotechnol Adv. (2023) 62:108058. doi: 10.1016/j.biotechadv.2022.108058
41. Pannaraj, PS, Li, F, Cerini, C, Bender, JM, Yang, S, Rollie, A, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. (2017) 171:647–54. doi: 10.1001/jamapediatrics.2017.0378
42. Gómez-Gallego, C, Morales, JM, Monleón, D, du Toit, E, Kumar, H, Linderborg, KM, et al. Human breast milk NMR metabolomic profile across specific geographical locations and its association with the milk microbiota. Nutrients. (2018) 10:1355. doi: 10.3390/nu10101355
43. Lyons, KE, Shea, CAO, Grimaud, G, Ryan, CA, Dempsey, E, Kelly, AL, et al. The human milk microbiome aligns with lactation stage and not birth mode. Sci Rep. (2022) 12:5598. doi: 10.1038/s41598-022-09009-y
44. Murphy, K, Curley, D, O’Callaghan, TF, O'Shea, CA, Dempsey, EM, O'Toole, PW, et al. The composition of human milk and infant faecal microbiota over the first three months of life: a pilot study. Sci Rep. (2017) 7:40597. doi: 10.1038/srep40597
45. Kumar, H, du Toit, E, Kulkarni, A, Aakko, J, Linderborg, KM, Zhang, Y, et al. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front Microbiol. (2016) 7:1619. doi: 10.3389/fmicb.2016.01619
46. Cabrera-Rubio, R, Mira-Pascual, L, Mira, A, and Collado, MC. Impact of mode of delivery on the milk microbiota composition of healthy women. J Dev Orig Health Dis. (2016) 7:54–60. doi: 10.1017/S2040174415001397
47. Hunt, KM, Foster, JA, Forney, LJ, Schütte, UM, Beck, DL, Abdo, Z, et al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One. (2011) 6:e21313. doi: 10.1371/journal.pone.0021313
48. Biagi, E, Quercia, S, Aceti, A, Beghetti, I, Rampelli, S, Turroni, S, et al. The bacterial ecosystem of mother’s milk and infant’s mouth and gut. Front Microbiol. (2017) 8:1214. doi: 10.3389/fmicb.2017.01214
49. Milani, C, Duranti, S, Bottacini, F, Beghetti, I, Rampelli, S, Turroni, S, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. (2017) 81:e00036–17. doi: 10.1128/MMBR.00036-17
50. Dzidic, M, Collado, MC, Abrahamsson, T, Artacho, A, Stensson, M, Jenmalm, MC, et al. Oral microbiome development during childhood: an ecological succession influenced by postnatal factors and associated with tooth decay. ISME J. (2018) 12:2292–306. doi: 10.1038/s41396-018-0204-z
51. Ding, J, Ouyang, R, Zheng, S, Wang, Y, Huang, Y, Ma, X, et al. Effect of breastmilk microbiota and sialylated oligosaccharides on the colonization of infant gut microbial community and fecal metabolome. Meta. (2022) 12:1136. doi: 10.3390/metabo12111136
52. Liu, B, Zhao, J, Liu, Y, Qiao, W, Jiang, T, and Chen, L. Diversity and temporal dynamics of breast milk microbiome and its influencing factors in Chinese women during the first 6 months postpartum. Front Microbiol. (2022) 13:1016759. doi: 10.3389/fmicb.2022.1016759
53. Jarzynka, S, Spott, R, Tchatchiashvili, T, Ueberschaar, N, Martinet, MG, Strom, K, et al. Human milk oligosaccharides exhibit biofilm eradication activity against matured biofilms formed by different pathogen species. Front Microbiol. (2022) 12:794441. doi: 10.3389/fmicb.2021.794441
54. Moossavi, S, Atakora, F, Miliku, K, Sepehri, S, Robertson, B, Duan, QL, et al. Integrated analysis of human milk microbiota with oligosaccharides and fatty acids in the CHILD cohort. Front Nutr. (2019) 6:58. doi: 10.3389/fnut.2019.00058
55. Williams, JE, Price, WJ, Shafii, B, Yahvah, KM, Bode, L, McGuire, MA, et al. Relationships among microbial communities, maternal cells, oligosaccharides, and macronutrients in human milk. J Hum Lact. (2017) 33:540–51. doi: 10.1177/0890334417709433
56. Wang, K, Xia, X, Sun, L, Wang, H, Li, Q, Yang, Z, et al. Microbial diversity and correlation between breast milk and the infant gut. Food Secur. (2023) 12:1740. doi: 10.3390/foods12091740
57. Jardak, M, Atoissi, A, Msalbi, D, Atoui, D, Bouizgarne, B, Rigane, G, et al. Antibacterial, antibiofilm and cytotoxic properties of prodigiosin produced by a newly isolated Serratia sp. C6LB from a milk collection center. Microb Pathog. (2022) 164:105449. doi: 10.1016/j.micpath.2022.105449
58. McGuire, MK, Meehan, CL, McGuire, MA, Williams, JE, Foster, J, Sellen, DW, et al. What’s normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am J Clin Nutr. (2017) 105:1086–100. doi: 10.3945/ajcn.116.139980
59. Porro, M, Kundrotaite, E, Mellor, DD, and Munialo, CD. A narrative review of the functional components of human breast milk and their potential to modulate the gut microbiome, the consideration of maternal and child characteristics, and confounders of breastfeeding, and their impact on risk of obesity later in life. Nutr Rev. (2023) 81:597–609. doi: 10.1093/nutrit/nuac072
60. Laursen, MF, Pekmez, CT, Larsson, MW, Lind, MV, Yonemitsu, C, Larnkjær, A, et al. Maternal milk microbiota and oligosaccharides contribute to the infant gut microbiota assembly. ISME Commun. (2021) 1:21. doi: 10.1038/s43705-021-00021-3
61. Cabrera-Rubio, R, Kunz, C, Rudloff, S, García-Mantrana, I, Crehuá-Gaudiza, E, Martínez-Costa, C, et al. Association of maternal secretor status and human milk oligosaccharides with milk microbiota: an observational pilot study. J Pediatr Gastroenterol Nutr. (2019) 68:256–63. doi: 10.1097/MPG.0000000000002216
62. Baumgärtner, F, Jurzitza, L, Conrad, J, Beifuss, U, Sprenger, GA, and Albermann, C. Synthesis of fucosylated lacto-N-tetraose using whole-cell biotransformation. Bioorg Med Chem. (2015) 23:6799–806. doi: 10.1016/j.bmc.2015.10.005
63. Wang, L, Zhu, Y, Zhao, C, Zhao, M, Li, Z, Xu, W, et al. Engineering Escherichia coli for highly efficient biosynthesis of lacto-N-difucohexaose II through de novo GDP-l-fucose pathway. J Agric Food Chem. (2024) 72:10469–76. doi: 10.1021/acs.jafc.4c01264
64. Cho, S, Samuel, TM, Li, T, Howell, BR, Baluyot, K, Hazlett, HC, et al. Interactions between Bifidobacterium and Bacteroides and human milk oligosaccharides and their associations with infant cognition. Front Nutr. (2023) 10:1216327. doi: 10.3389/fnut.2023.1216327
65. Zúñiga, M, Monedero, V, and Yebra, MJ. Utilization of host-derived glycans by intestinal Lactobacillus and Bifidobacterium species. Front Microbiol. (2018) 9:1917. doi: 10.3389/fmicb.2018.01917
66. FDA. FDA. U.S., GRAS notice no. GRN 000650. 2-o-fucosvllactose Glycom A/S (2014). Available at: https://www.fda.gov/media/99125/download
67. FDA. FDA. U.S., GRAS notice no. GRN 000833, lacto-N-tetraose Glycom A/S (2019). Available at: https://www.fda.gov/media/133202/download
68. FDA. FDA. U.S., GRAS notice no. GRN 000881, 6’-sialyllactose Glycom A/S (2020). Available at: https://www.fda.gov/media/138495/download
69. FDA. FDA. U.S., GRAS notice no. GRN 000880, 3’-sialyllactose Glycom A/S (2020). Available at: https://www.fda.gov/media/138493/download
70. EFSA Panel on Nutrition, Novel Foods and Food Allergens (NDA)Turck, D, Castenmiller, J, de Henauw, S, Hirsch-Ernst, KI, Kearney, J, et al. Safety of lacto-N-tetraose (LNT) as a novel food pursuant to regulation (EU) 2015/2283. EFSA J. (2019) 17:e05907. doi: 10.2903/j.efsa.2019.5907
71. EFSA Panel on Nutrition, Novel Foods and Food Allergens (NDA)Turck, D, Castenmiller, J, de Henauw, S, Hirsch-Ernst, KI, Kearney, J, et al. Safety of 2′-fucosyllactose/difucosyllactose mixture as a novel food pursuant to regulation (EU) 2015/2283. EFSA J. (2019) 17:e05717. doi: 10.2903/j.efsa.2019.5717
72. EFSA Panel on Nutrition, Novel Foods and Food Allergens (NDA)Turck, D, Castenmiller, J, de Henauw, S, Hirsch-Ernst, KI, Kearney, J, et al. Safety of 3'-Sialyllactose (3'-SL) sodium salt as a novel food pursuant to regulation (EU) 2015/2283. EFSA J. (2020) 18:e06098. doi: 10.2903/j.efsa.2020.6098
73. EFSA Panel on Nutrition, Novel Foods and Food Allergens (NDA)Turck, D, Castenmiller, J, de Henauw, S, Hirsch-Ernst, KI, Kearney, J, et al. Safety of 6'-Sialyllactose (6'-SL) sodium salt as a novel food pursuant to regulation (EU) 2015/2283. EFSA J. (2020) 18:e06097. doi: 10.2903/j.efsa.2020.6097
74. Goehring, KC, Marriage, BJ, Oliver, JS, Wilder, JA, Barrett, EG, and Buck, RH. Similar to those who are breastfed, infants fed a formula containing 2'-Fucosyllactose have lower inflammatory cytokines in a randomized controlled trial. J Nutr. (2016) 146:2559–66. doi: 10.3945/jn.116.236919
75. Leung, TF, Ulfman, LH, Chong, MKC, Hon, KL, Khouw, I, Chan, PKS, et al. A randomized controlled trial of different young child formulas on upper respiratory and gastrointestinal tract infections in Chinese toddlers. Pediatr Aller Immunol. (2020) 31:745–54. doi: 10.1111/pai.13276
76. Ramirez-Farias, C, Baggs, GE, and Marriage, BJ. Growth, tolerance, and compliance of infants fed an extensively hydrolyzed infant formula with added 2'-Fl fucosyllactose (2'-Fl) human milk oligosaccharide. Nutrients. (2021) 13:186. doi: 10.3390/nu13010186
77. Gold, MS, Quinn, PJ, Campbell, DE, and Peake, J. Effects of an amino acid-based formula supplemented with two human milk oligosaccharides on growth, tolerability, safety, and gut microbiome in infants with Cow's milk protein allergy. Nutrients. (2022) 14:2297. doi: 10.3390/nu14112297
78. Román, E, Moreno Villares, JM, Domínguez Ortega, F, Carmona Martínez, A, Picó Sirvent, L, Santana Sandoval, L, et al. Real-world study in infants fed with an infant formula with two human milk oligosaccharides. Nutricion Hospitalaria. (2020) 37:698–706. doi: 10.20960/nh.03084
79. Lasekan, J, Choe, Y, Dvoretskiy, S, Devitt, A, Zhang, S, Mackey, A, et al. Growth and gastrointestinal tolerance in healthy term infants fed milk-based infant formula supplemented with five human milk oligosaccharides (Hmos): a randomized multicenter trial. Nutrients. (2022) 14:2625. doi: 10.3390/nu14132625
80. Parschat, K, Melsaether, C, Jäpelt, KR, and Jennewein, S. Clinical evaluation of 16-week supplementation with 5hmo-mix in healthy-term human infants to determine tolerability, safety, and effect on growth. Nutrients. (2021) 13:2871. doi: 10.3390/nu13082871
81. Davis, JC, Lewis, ZT, Krishnan, S, Bernstein, RM, Moore, SE, Prentice, AM, et al. Growth and morbidity of gambian infants are influenced by maternal milk oligosaccharides and infant gut microbiota. Sci Rep. (2017) 7:40466. doi: 10.1038/srep40466
Keywords: breast milk, human milk oligosaccharides, human milk microbiota, colostrum, mature milk
Citation: Ge H, Zhu W, Zhang J, Wang Z, Shi H, Sun J and Shi M (2024) Human milk microbiota and oligosaccharides in colostrum and mature milk: comparison and correlation. Front. Nutr. 11:1512700. doi: 10.3389/fnut.2024.1512700
Edited by:
Xinran Liu, Peking University People’s Hospital, ChinaReviewed by:
Boyuan Guan, Shenyang Agricultural University, ChinaJennifer Angeline Gaddy, Vanderbilt University Medical Center, United States
Copyright © 2024 Ge, Zhu, Zhang, Wang, Shi, Sun and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Shi, emhzaDY2NTVAc29odS5jb20=
 Hongda Ge
Hongda Ge Wenxiu Zhu2
Wenxiu Zhu2