- 1Department of Food Science and Technology, University of Nebraska-Lincoln, Lincoln, NE, United States
- 2Nebraska Food for Health Center, University of Nebraska, Lincoln, NE, United States
- 3Complex Biosystems, University of Nebraska-Lincoln, Lincoln, Nebraska, United States
- 4Medical Sciences, Reckitt/Mead Johnson Nutrition Institute, Evansville, IN, United States
- 5Department of Pediatrics, Indiana University School of Medicine, Indianapolis, IN, United States
- 6Department of Internal Medicine, University of Nebraska Medical Center, Omaha, NE, United States
- 7Frederick F. Paustian Inflammatory Bowel Disease Center, University of Nebraska Medical Center, Omaha, NE, United States
- 8Fred and Pamela Buffett Cancer Center, University of Nebraska Medical Center, Omaha, NE, United States
Introduction: Few studies have evaluated the effects of milk fat globule membrane (MFGM) on microbiota and immune markers in early infant nutrition.
Methods: In this double-blind randomized study, infants (7–18 days of age) received either bovine milk-based infant formula (Control) or similar formula with an added source (5 g/L) of bovine MFGM (INV-MFGM) for 60 days. A reference group received mother’s own human milk over the same period (HM). Oral and stool samples were collected (Baseline and Day 60) to evaluate microbiota, immune markers, and metabolites.
Results: At Day 60, stool bacterial diversity and richness were higher in formula groups vs HM, as were Bifidobacterium bifidum and B. catenulatum abundance. Compared to HM, stool pH was higher in Control, while acetate, propionate, isovalerate, and total short- and branched-chain fatty acids were higher in INV-MFGM. Butyrate and lactate increased for INV-MFGM from baseline to Day 60. No group differences in oral microbiota or immune markers (α- and β-defensin, calprotectin, or sIgA) were detected, although sIgA increased over time in all study groups. Added bovine MFGM in infant formula modulated stool microbiota and short- and branched-chain fatty acids compared to human milk; changes were modest relative to control formula.
Discussion: Overall, distinct patterns of stool metabolites and microbiota development were observed based on early nutrition.
Clinical trial registration: ClinicalTrials.gov, identifier NCT04059666.
Introduction
The maturation of the infant gastrointestinal (GI) tract, including establishment of the GI microbiota and development of the immune system, has long-term implications on health (1). Several factors affect infant GI microbiota composition, including birth mode, environment, antibiotics, and diet (2, 3), with diet one of the most significant factors (4–6). Because of the importance of diet on infant health, the World Health Organization recommends exclusive feeding of human milk (breastfeeding) over the first six months of life to reduce risk of adverse short and long-term health outcomes for infants (7). However, a significant number of infants worldwide are fed infant formula necessitating improvements to infant formulas to more closely replicate the functions of human milk (8). Human milk components considered to have relevant functionality for incorporation and improvement of infant formula include human milk oligosaccharides, lactoferrin, and long chain poly-unsaturated fatty acids.
Evidence of the importance of milk fat globule membrane (MFGM) has increased over the past decade. MFGM is a secretory structure produced by mammary epithelial cells composed of a phospholipid trilayer that contains proteins, glycoproteins, glycolipids, and triacylglycerols (9–11). The lipid composition of human and bovine MFGM (bMFGM) is highly similar (12–15). Previous studies demonstrated MUC-1 and lactadherin, functional bMFGM components, enhance immune responses and reduce microbial adhesion within the GI tract (15, 16). In animal models, feeding formulas with added bMFGM led to significant differences in fecal microbiota (17, 18).
Addition of bMFGM could help bring the composition and functionality of infant formula closer to human milk. In infant studies, addition of bMFGM to infant formula was associated with healthy growth throughout the first year of life (19–21) and reduced susceptibility to infection (22, 23). Multiple studies demonstrated beneficial effects on neurodevelopment in infants (24–27) and young children (28). Prospective studies examining effects of bMFGM in infants showed moderate modulation of the gut microbiota (29–31) and metabolome (29–33), including increased abundance of Bifidobacterium species (29). bMFGM also reduced adverse health events (34) and generated serum cytokine profiles more similar to breastfed infants (35). One study found that infant formula with added bMFGM moderately affected the infant oral microbiota and reduced the presence of the common inner ear pathogen Moraxella catarrhalis (23). The current study was designed to investigate effects of bMFGM on development of stool and oral microbiotas and stool immune biomarkers in infants receiving a routine bovine milk-based infant formula compared to a similar formula with added bMFGM. A reference group of infants exclusively receiving mother’s-own milk was also registered.
Materials and methods
Study design and participants
Healthy 7- to 18-day old infants were recruited at five clinical sites in the United States (Altamonte Springs, FL; Owensboro, Ky; Birmingham, AL; Kingsport and Memphis, TN; ClinicalTrials.gov: NCT04059666)1 in a multicenter, double-blind, randomized, controlled, prospective trial. Participants were enrolled between March 2019 and October 2021. The study was suspended March 19, 2020 due to the COVID-19 pandemic and was re-initiated in October 2020 with an amended protocol to allow minimal in-person contact of parents and participants with the study site. The CONSORT 2010 checklist of information to include when reporting a randomized trial is included as Supplementary Figure 1. Full inclusion and exclusion criteria are described in Supplementary Table 2. COVID-19 was not assessed in participants. Parent(s) or the participant’s legal guardian(s) provided written informed consent prior to enrollment. The research protocol (protocol #3390-1) and informed consent forms observing the Declaration of Helsinki were approved by Advarra (Columbia, MD, United States; IRB #Pro00027443). Briefly, eligible infants exclusively receiving mother’s-own human milk since Day 1 of life (i.e., breastfed) with the intent to continue through the duration of the study were registered in a human milk (HM) reference group. Eligible infants whose mothers had chosen to initiate infant formula feeding and were exclusively receiving marketed infant formula for at least 24 h prior to randomization were assigned to one of two study formulas (Mead Johnson Nutrition, Evansville, IN): Control, a routine cow’s milk-based infant formula (similar to previously marketed Enfamil®) or investigational formula (INV-MFGM), which was similar in composition and had an added source of bMFGM (5 g/L; Lacprodan MFGM-10, Arla Foods Ingredients P/S, Denmark). Both formulas had a prebiotic blend of polydextrose and galactooligosaccharides (Table 1).
The study sponsor created a computer-generated randomization schedule, provided in sealed, consecutively numbered envelopes. Each study formula was designated by its own unique code (only known by the sponsor) and assigned by opening the next sequential envelope. Formulas were provided directly to parents for the study period. Neither the product labels nor the sealed envelopes permitted unblinding by the study site. Additionally, the monitoring personnel were blinded to product identification. Blinding could only be broken in the event of a medical emergency that required knowledge of the study formula for managing the participant’s health. However, it was not necessary to break the study code prematurely in the current study. To maintain balance in enrollment of each of the study arms, it was suggested to randomize at least two infants receiving formula for each registration of an infant receiving human milk. Participants were assigned to exclusive study feeding over a 60-day period. Study visits occurred at 4 to 16 days of age (Baseline) and Day 60 (60–64 days following the start of study feeding). Researchers were blinded to the identity of the samples (identified by unique codes) until initial statistical analyses of data was completed.
Study outcomes
The primary outcome was changes in stool microbiota. Two participant stool samples were collected by parents/caregivers at Baseline and Day 60 and returned to the study site. Samples could be obtained from more than one diaper over a 24-h period to meet the minimum amount needed for analysis: ∼0.5 g for microbial community composition (OMNIgene®∙GUT Collection Kit, DNA Genotek, Ottawa, Canada) and ∼5 g in a tube without stabilizer to measure pH, SCFA, and immune biomarkers. The OMNIgene®∙GUT Collection Kit was selected for collection of stool microbial DNA samples as previous studies had demonstrated sample stability at room temperature (36). After collection, samples were stored at 4°C until transported the study site, at which point they were stored at −20°C. Samples were shipped on dry ice to the University of Nebraska-Lincoln and subsequently stored at −80°C until further processed.
Secondary outcomes included oral microbiota; stool color, consistency, pH and SCFA; 24-h formula intake recall; and medically-confirmed adverse events. Buccal swabs were collected at Baseline and Day 60 by study site personnel using a specialized swab and tube (OMNIgene∙ORAL Collection Kit, DNA Genotek). Participants had not eaten within 30 min prior to collection. All samples were stored at a minimum of −20°C at the study site, shipped on dry ice to the University of Nebraska-Lincoln, and subsequently stored at −80°C until further processed.
Body weight was recorded at Baseline. Parents completed a 24-h recall of study formula intake (fluid oz/day) by phone at Day 30 (± 5 days) and at the Day 60 study visit. A 48-h recall of stool consistency (responses scaled from 1 to 5: hard, formed, mushy, unformed or seedy, watery) was collected at Baseline, Day 30, and Day 60. Adverse events (categorized as: Body as a Whole; Eyes, Ears, Nose, and Throat; Gastrointestinal; Metabolic and Nutrition; Musculoskeletal; Respiratory; and Skin) were recorded throughout the study.
Community sequencing and analysis
DNA from stool and oral samples was extracted using a previously-described bead-beating phenol-chloroform method (37). 16S rRNA gene sequencing of the V4 variable region was performed as described previously (38). Initial sequence analysis was conducted with DADA2 (39) in R (ver. 4.2.1). Forward and reverse reads were truncated to 240 and 210 bp, respectively. Sequences were de-replicated into unique amplicon sequence variants (ASV) and a list of representative sequences with 1,484 features was generated. Taxonomy was successfully assigned to 1,359 features using the SILVA database (40) (release 1.38.1) with a classifier based on 99% sequence identity, filtering out Archaeal, Chloroplast, or Mitochondrial sequences. The raw 16S rRNA sequences were deposited in the NCBI database under BioProject ID PRJNA1005334.2
pH and S/BCFA
Fecal samples were diluted (1:10, deionized water) and homogenized prior to pH measurement as described (37). Short chain fatty acids (SCFA; acetate, butyrate, propionate) and branched-chain fatty acids (BCFA; isovalerate and isobutyrate) were measured using gas chromatography as described and are reported per wet weight of stool (37). To measure lactic acid, stool samples were homogenized, diluted 1:10 in 10 mM sulfuric acid and filtered through 0.45 μm filters. Samples (10 μL) were injected into a HPLC (Aligent 1260 Infinity, Waldbronn, Germany) containing Aminex HPX-87H column (300 x 7.8 mm, Bio-Rad) equipped with a diode array detector. The column was held at 50°C with a 0.6 mL/min flow rate; 10 mM sulfuric acid was the mobile phase. Lactic acid concentrations were calculated relative to a standard curve.
Immune biomarkers
Enzyme-linked immunosorbent assay (ELISA) kits were used to determine stool secretory immunoglobulin A (sIgA), β-defensin, and calprotectin (Immunodiagnostik AG, Bensheim, Germany) and α-defensin (Hycult Biotech, Wayne, PA USA). Assays were performed according to manufacturers’ protocols on two biological replicates in technical duplicate.
Statistical analyses
The sample size was chosen to investigate stool microbiota composition as the primary variable in infants receiving one of two study formulas or mother’s-own breast milk for a 60-day feeding period. Specifically, group differences in infant stool microbiota between human milk and formula fed infants had been previously demonstrated in studies with three groups of 20 participants (37). Group differences in children’s stool microbiota were also observed between participants that differed in the presence or absence of diabetes-associated autoantibodies with two groups of 18 participants (41). To have a total of 20 infants per group with protocol-compliant samples at Baseline and Day 60, the planned sample size for the study was originally 111 participants (37 per group; allowing for a potential 45% drop-out rate). However, enrollment was interrupted and subsequently complicated by the COVID-19 pandemic. As a result, the study concluded prematurely and the final sample size was reduced. Nevertheless, sample sizes were sufficiently powered to observe differences in stool microbiota at Day 60 between HM and formula feeding groups. Differences in subject age at baseline was analyzed by Kruskal-Wallis test with Dunn’s correction for multiple comparisons. Formula intake was analyzed by ANOVA. Stool consistency was analyzed using the Cochran-Mantel-Haenszel row means score test. Incidence of adverse events was analyzed using Fisher’s exact test. Unadjusted pairwise comparisons were performed if the overall test was statistically significant. These tests were conducted at α = 0.05 using SAS version 9.4 (Cary, NC), except for differences in subject age at baseline, which was performed with Graphpad Prism 10.2 (Boston, MA).
Statistical analysis of 16S rRNA gene sequencing was performed in R (ver. 4.1.0 and 4.3.1). Shannon and Observed richness indices were computed at the ASV level. Pairwise comparisons of Baseline and Day 60 variation in each group, and between group variation at Day 60 were performed through Kruskal-Wallis testing. Holm FDR correction was integrated, and significance was determined at α = 0.05. No significant differences in Observed ASV richness were observed between study groups at Baseline. Data was plotted with GraphPad Prism (ver 9.5). Bray-Curtis, Binary Jaccard, and Unweighted UniFrac dissimilarities were calculated on relative abundance data and visualized with Principal Coordinates Analysis plots (PCoA), with significance of differences determined by pairwise permutational multivariate analysis of variance (PERMANOVA) with Bonferroni FDR correction.
Prior to assessing taxonomic differences, features were filtered to remove those detected in a single sample or in the lowest 10% of sequences, for a total of 270 features. Taxa abundance was compared by Wilcoxon sign rank testing with Benjamini-Hochberg FDR correction on log2-transformed data; heat maps were generated using Metacoder (42) for genera with > 10 reads and significant differences detected. Significantly different ASVs were also identified by comparing Baseline to Day 60 and between groups at Day 60 using DESeq2 (43). To account for high positive fold-changes present in low abundance features, ASVs with significant differences but a mean abundance < 100 reads ( < 0.25% relative abundance) were removed. Significant differences in Bifidobacterium species abundance were determined by ANOVA with Brown and Forsythe correction for unequal variance on log2-transformed data floored at minimum relative non-zero abundance using GraphPad Prism (ver 9.5).
Changes in pH, S/BCFA, and immune biomarkers were analyzed using SAS ver. 3.8; all tests were conducted with α = 0.05. Least square means (LSM) of fixed effects (pH, S/BCFA, and immune biomarkers) for each group between Baseline and Day 60 with a repeated measure ANOVA were compared using a Toeplitz covariance structure. Bonferroni adjustment was used for multiple-comparison correction. For this model, ‘groups’ and ‘visits’ were the fixed effects while ‘visit’ was the repeated measure, and subject within group was the assigned random error. To assess group differences, analysis of covariance (ANCOVA) was performed, with baseline measurements assigned as the covariate. Group differences between the LSM of Day 60 measurements were compared. Double square root transformations were applied to all pH, S/BCFA, and immune biomarkers to adjust for normality. An analysis of probability of a zero observation was performed on measurements of isobutyrate and isovalerate, resulting in a modified log transformation used to adjust for normality after the removal of zeros from the dataset. Data was plotted with GraphPad Prism (ver 9.5).
Results
Participant characteristics
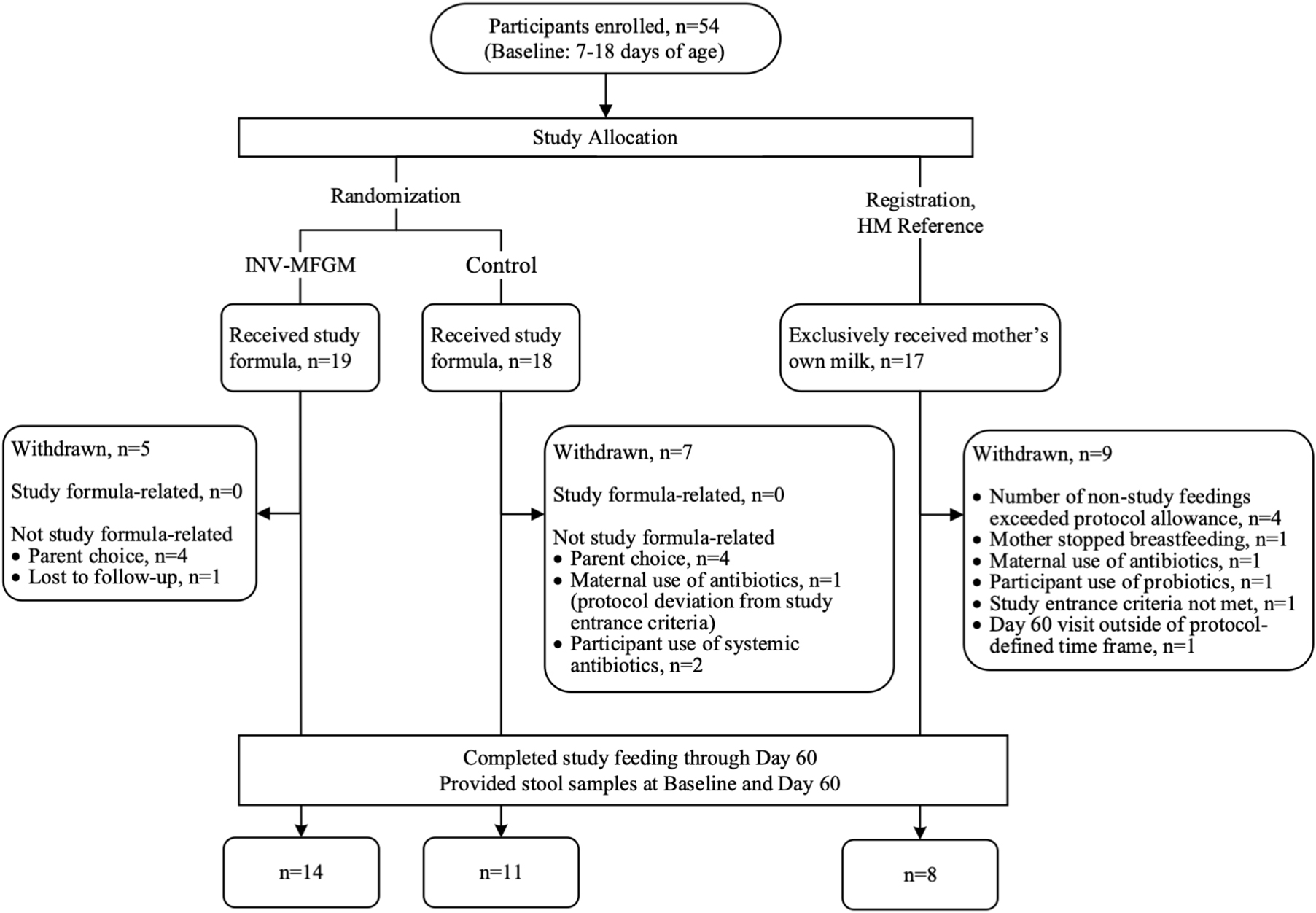
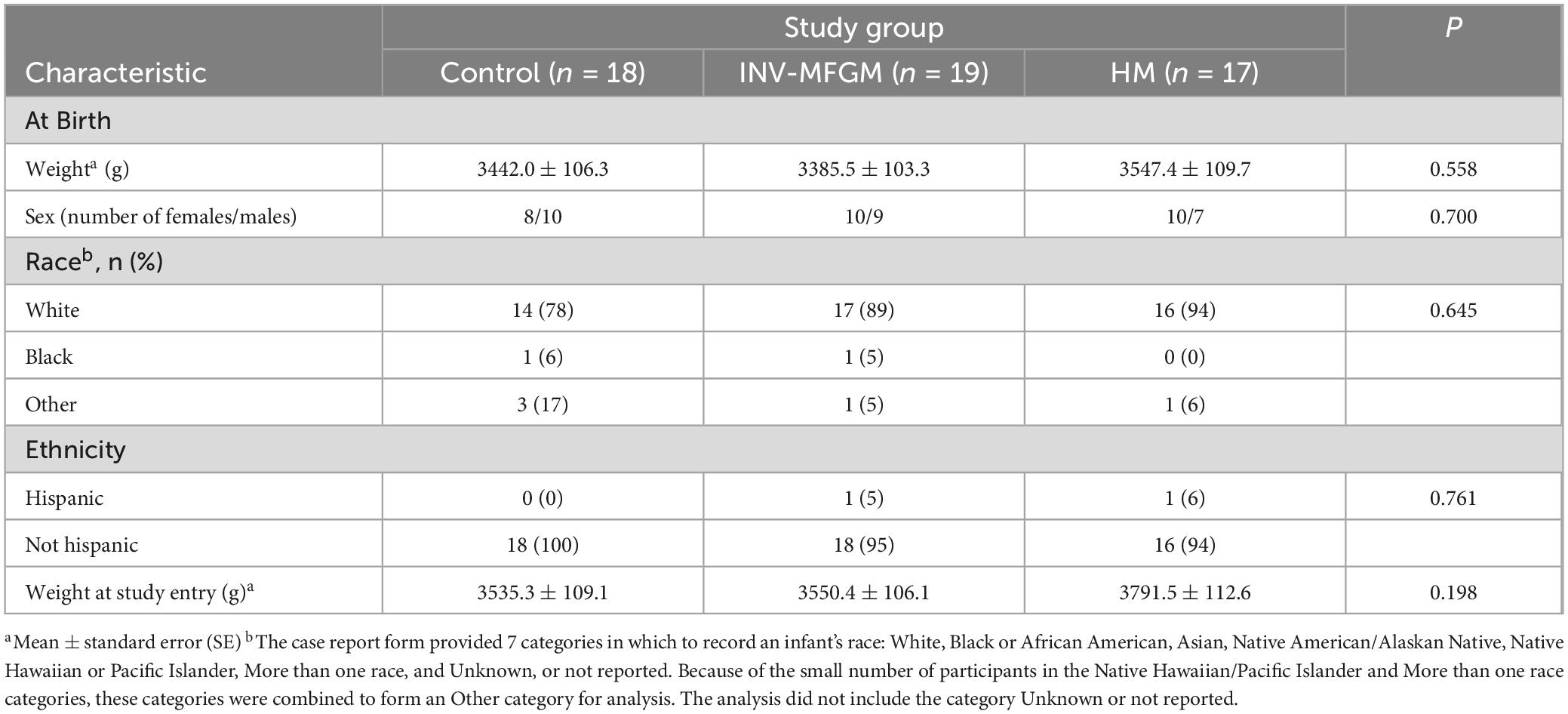
A total of 54 participants were enrolled in the study (Figure 1); 33 participants completed the study (Control: n = 11; INV-MFGM: n = 14; HM: n = 8). Infants were 37 to 42 weeks gestational age at birth and all were delivered vaginally. Mothers of eligible infants had chosen to initiate formula feeding prior to study enrollment, and infants in formula feeding groups had exclusively received infant formula for at least 24 h prior to randomization. Supplementary Table 2 lists full inclusion and exclusion criteria. No differences in weight (mean ± SE) at birth or study entry, sex, race, or ethnicity were detected (Table 2). No significant differences between study groups were observed in days of age when baseline samples were collected (Supplementary Table 3). No group differences were detected in parent-reported study formula intake (mean fluid oz/day) at Days 30 and 60. At Day 30, control and INV-MFGM had an intake of 27.6 ± 1.5 oz and 26.3 ± 1.3 oz, respectively (p = 0.544). Similarly, at Day 60, control and INV-MFGM had an intake of 29.8 ± 1.7 oz and 29.2 ± 1.5 oz, respectively (p = 0.792). For stool consistency, no group differences were detected at Baseline (Supplementary Table 4). At Day 30 significant differences between Control and HM groups were detected; primary differences were more infants with stool categorized as “mushy” in the Control and stool categorized as “unformed or seedy” in the HM group. At Day 60, significant differences in distribution included more infants in Control and INV-MFGM groups with “mushy” stool vs more infants in the HM group with “unformed or seedy” or “watery” stool. No group differences were detected in the number of participants with at least one adverse event reported (Control: n = 8, 44%; INV-MFGM: n = 13, 68%; HM: n = 6, 35%; p = 0.131). Incidences of adverse events (categorized as: Body as a Whole; Eyes, Ears, Nose, and Throat; Gastrointestinal; Metabolic and Nutrition; Musculoskeletal; Respiratory; and Skin) were low with no significant differences by group. No serious adverse events were reported. No significant differences in study discontinuation were detected and no study discontinuations were related to study formula.
Effects of infant feeding duration and type on microbial community structure
Overall changes in stool and oral microbiota within and between study groups were assessed (Figure 2). No differences in stool richness (Observed amplicon sequence variants (ASVs) or diversity (Shannon Diversity) were detected at Baseline. Richness and diversity for the HM group remained stable across the feeding period, whereas richness increased significantly from Baseline to Day 60 in both formula groups and Shannon diversity increased significantly in the INV-MFGM group. Diversity and Richness were significantly higher for both INV-MFGM and Control compared to the HM group at Day 60. For oral microbiota, no group differences in diversity and richness were detected at Baseline or Day 60 or from Baseline to Day 60.
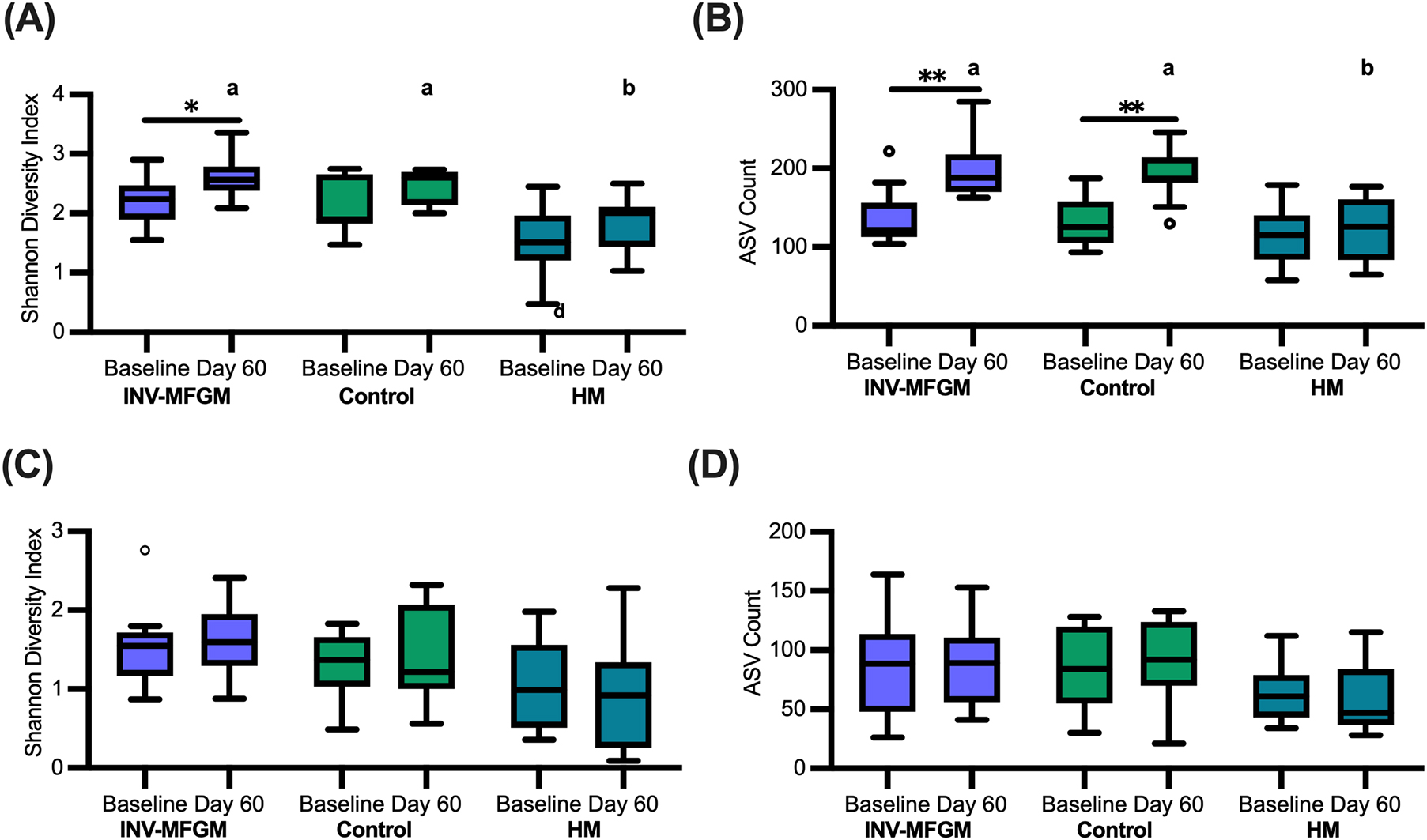
Figure 2. Diversity and richness of microbiotas. For stool (A,B) and oral microbiotas (C,D), pairwise comparisons of (A,C) Shannon diversity and (B,D) ASV richness were performed by study time point (Baseline and Day 60) and feeding group. Boxplots show the median, first and third quartiles with the whiskers extending to 1.5 x IQR (Interquartile Range). ° Used to indicate points outside of whiskers. Unique letters above Day 60 boxplots indicate significant difference between study groups. *p < 0.05; **p < 0.01.
To assess overall differences in community composition, Bray-Curtis, Jaccard, and UniFrac dissimilarities were calculated and visualized using PCoA (Supplementary Figure 1). For stool, there were no significant differences between study groups based on phylogenetic diversity (UniFrac). For comparisons based upon ASV composition (Jaccard), both formula groups were significantly different from the HM reference group, although the magnitude of differences was low (INV-MFGM: R2 = 0.089, p = 0.003; Control: R2 = 0.120, p = 0.003) and there were no significant differences between formula groups. A small amount of variation was observed between INV-MFGM and HM reference groups (R2 = 0.083, p = 0.030) based upon ASV abundance (Bray-Curtis), but no other differences between study groups were significant. For oral microbiota, there were no significant differences in phylogenetic diversity or composition between study groups; small differences in overall ASV abundance were observed between INV-MFGM and HM groups (R2 = 0.134, p = 0.039). Spearman correlation performed on amplicon sequence variants (ASVs, equivalent to unique species or strains) with > 0.25% abundance identified 22 ASVs conserved across oral and stool (Supplementary Figure 2).
Composition of infant stool and oral microbiotas
Relative abundance of stool (Supplementary Figure 3) and oral (Supplementary Figure 4) genera at Baseline and Day 60 varied across individuals. Abundance of Bifidobacterium in stool microbiota varied widely by individual and study time point. Oral microbiotas were dominated by two main genera, Streptococcus, present in all samples at Baseline and Day 60, and Veillonella, which was more common at Day 60.
Heat trees were used to visualize differences in stool microbiota taxa for each study group between Baseline and Day 60 (Figures 3A–C) and between study groups at Day 60 (Figures 3D–F). In both formula groups, Enterobacteriaceae and Streptococcus spp. decreased from Baseline to Day 60, while Dorea, Ruminococcus gnavus, Intestinabcter, Granulicatella, Akkermansia and Actinomyces paecansis increased. In the HM group, Rothia mucilaginosa and B. breve increased from Baseline to Day 60, whereas Gemella decreased. At Day 60, formula groups had higher Ruminococcus, Flavonifractor, Granulicatella, Gemella, Veillonella, Bifidobacterium, Akkermansia, and Actinomyces and lower Enterobacteriaceae, Haemophilus, and Streptococcus anginosus compared to HM. Higher Intestinibacter and Bacteroides and lower Staphylococcus, Xanthomonadaceae and Burkholderiales were detected for the INV-MFGM vs HM group, whereas higher Erysipelotrichaceae, Enterococcus, Streptococcus salivarus, Blautia, and Dorea and lower Bacteroides stercoris were detected for the Control vs HM group. Few differences were detected between formula study groups. Compared to the HM group, different ASVs that classify as Bifidobacterium at the species-level were enriched in each study formula group: B. bifidum in the INV-MFGM group and B. catenulatum in the Control group. Few group differences for oral communities at Day 60 were detected (Supplementary Figure 5).
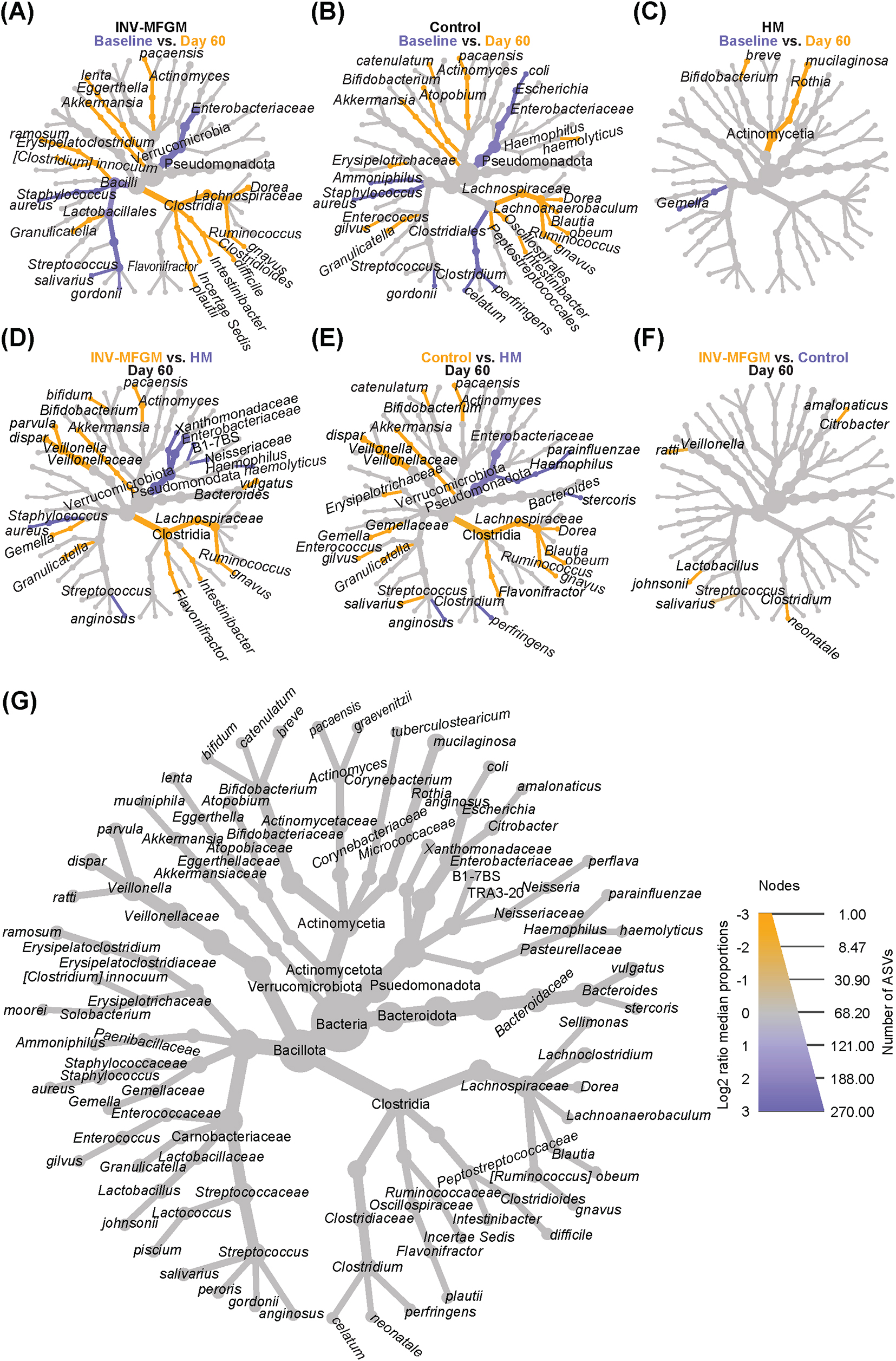
Figure 3. Differential heat trees comparing infant stool taxa. Difference in relative abundance by study time point (Baseline vs. Day 60) shown by feeding group: (A) INV-MFGM, (B) Control, and (C) HM. Differences in relative abundance between study groups at Day 60 shown by comparisons of: (D) INV-MFGM vs HM, (E) Control vs HM, and (F) INV-MFGM vs Control. (G) A reference taxonomic tree representing all taxa with differential abundance and key indicating the correlations between node size and total ASVs and color intensity with relative abundance.
DESeq2 analysis was used to compare differences in ASV abundance in stool between Baseline and Day 60 for each study group (Figure 4A) and between study groups at Day 60 (Figure 4B). At Day 60, higher Veillonella, Lachnoclostridium, Flavonifractor, Blautia, Bifidobacterium, Ruminococcus, and Clostridium innocuum and lower Streptococcus, Klebsiella, and Clostridium sensu stricto were detected in formula groups vs HM. Compared to the Control, the INV-MFGM group had higher Hungatella, Bacteroides, and Akkermansia ASVs, and lower Streptococcus. B. catenulatum and B. bifidum were higher in both study formula groups vs HM. Wilcoxon rank sum tests were performed to investigate differences in abundance of the Bifidobacterium genus and species between study groups at Day 60 (Figure 5). No differences in abundance of the Bidobacterium genus and the most abundant Bifidobacterium species, B. breve, were detected between study groups. Significantly higher B. catenulatum and B. bifidum were detected in formula groups compared to HM.
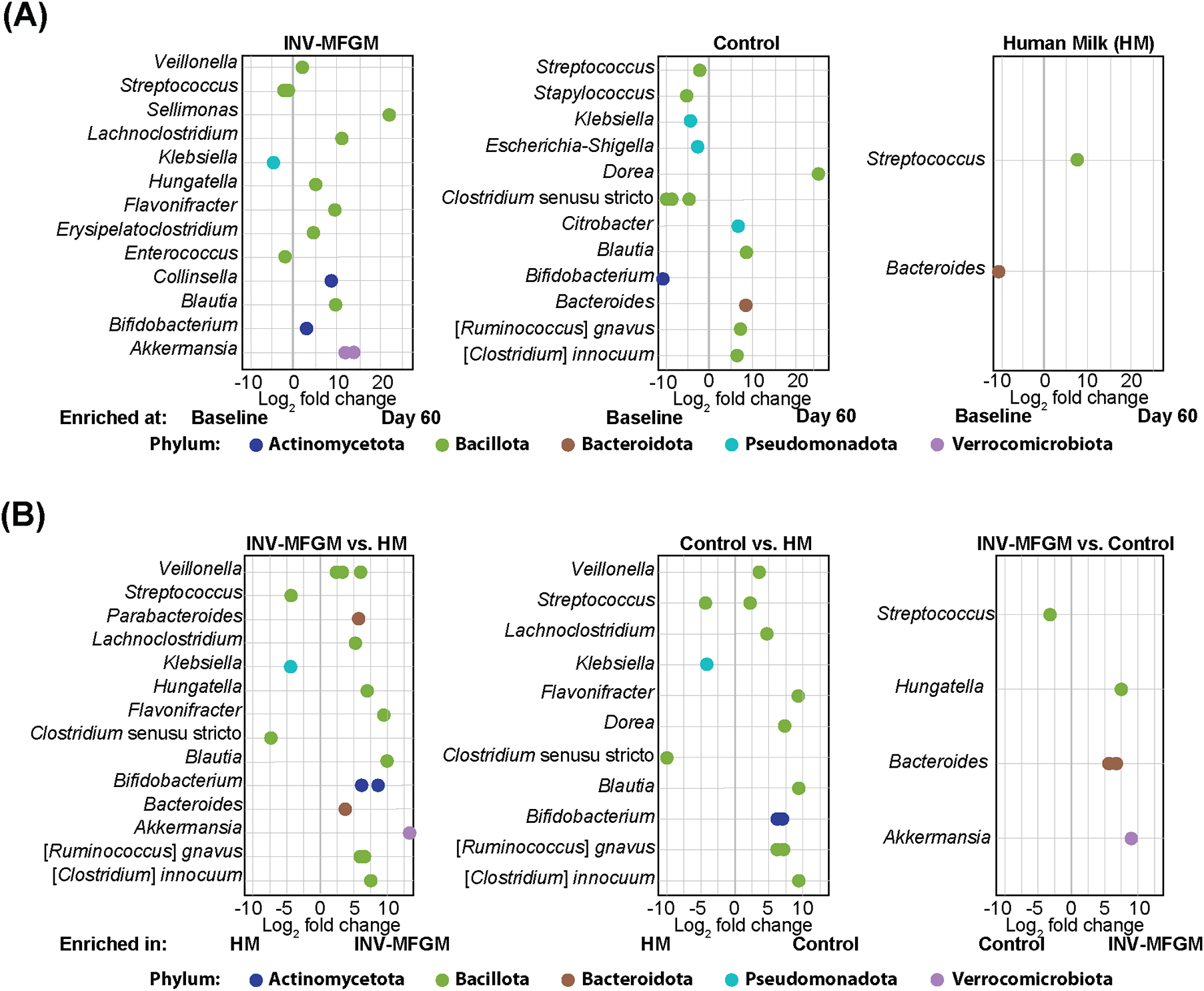
Figure 4. Analysis of differentially abundant infant stool ASVs. ASVs with differential abundance between (A) Baseline and Day 60 and (B) study groups at Day 60.
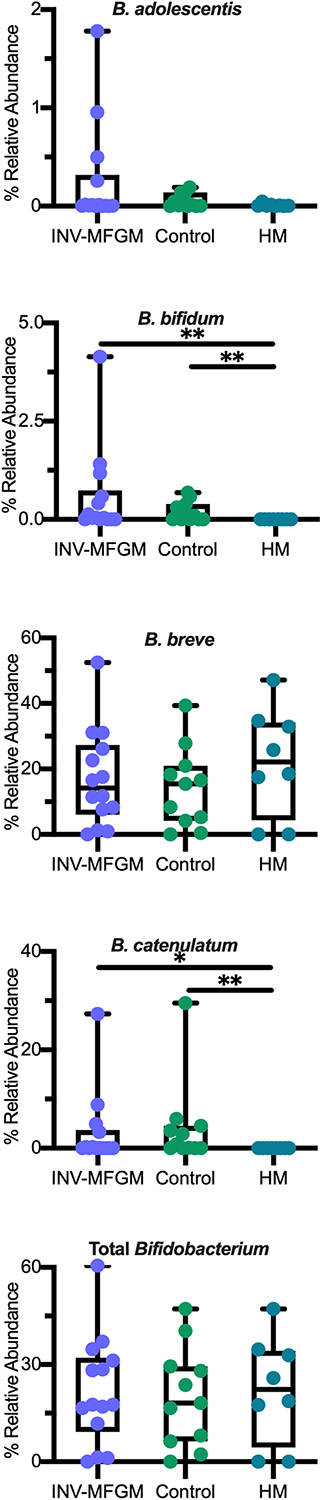
Figure 5. Differences in relative abundance of individual Bifidobacterium species and total Bifidobacterium between study groups at Day 60. Boxplots show the median, first and third quartiles with the whiskers extending through minimum and maximum. *p < 0.05; **p < 0.01.
For oral microbiotas, Staphylococcus decreased across study groups from Baseline to Day 60 (Supplementary Figure 6A). Study formula groups also demonstrated increased Granulicatella and Alloprevotella levels and decreased Streptococcus from Baseline to Day 60. At Day 60 (Supplementary Figure 6B) both formula groups had increased Prevotella, Porphyromonas, Granulicatella and Lachnoanaerobaculum compared to HM; Alloprevotella was higher for INV-MFGM compared to HM; and Rothia and Veillonella were higher for the Control group compared to HM. There were no significant differences between formula groups at Day 60.
Stool pH and S/BCFA
Stool S/BCFA are shown in Figure 6. No significant group differences in S/BCFA were detected at Baseline, with the exception of significantly lower propionate in the HM vs INV-MFGM group. Butyrate, total BCFA, and lactate significantly increased in the INV-MFGM group from Baseline to Day 60. Acetate, propionate and total SCFA were significantly higher in the INV-MFGM vs HM group at Day 60. Total BCFA and isovalerate were significantly higher for INV-MFGM vs Control and HM groups at Day 60 and isobutyrate was significantly different among groups (INV-MFGM > control > HM). No correlations between stool genera and metabolites were detected. No group differences in stool pH were detected at Baseline or from Baseline to Day 60 (Figure 7). However, stool pH was significantly higher for the control vs HM group at Day 60.
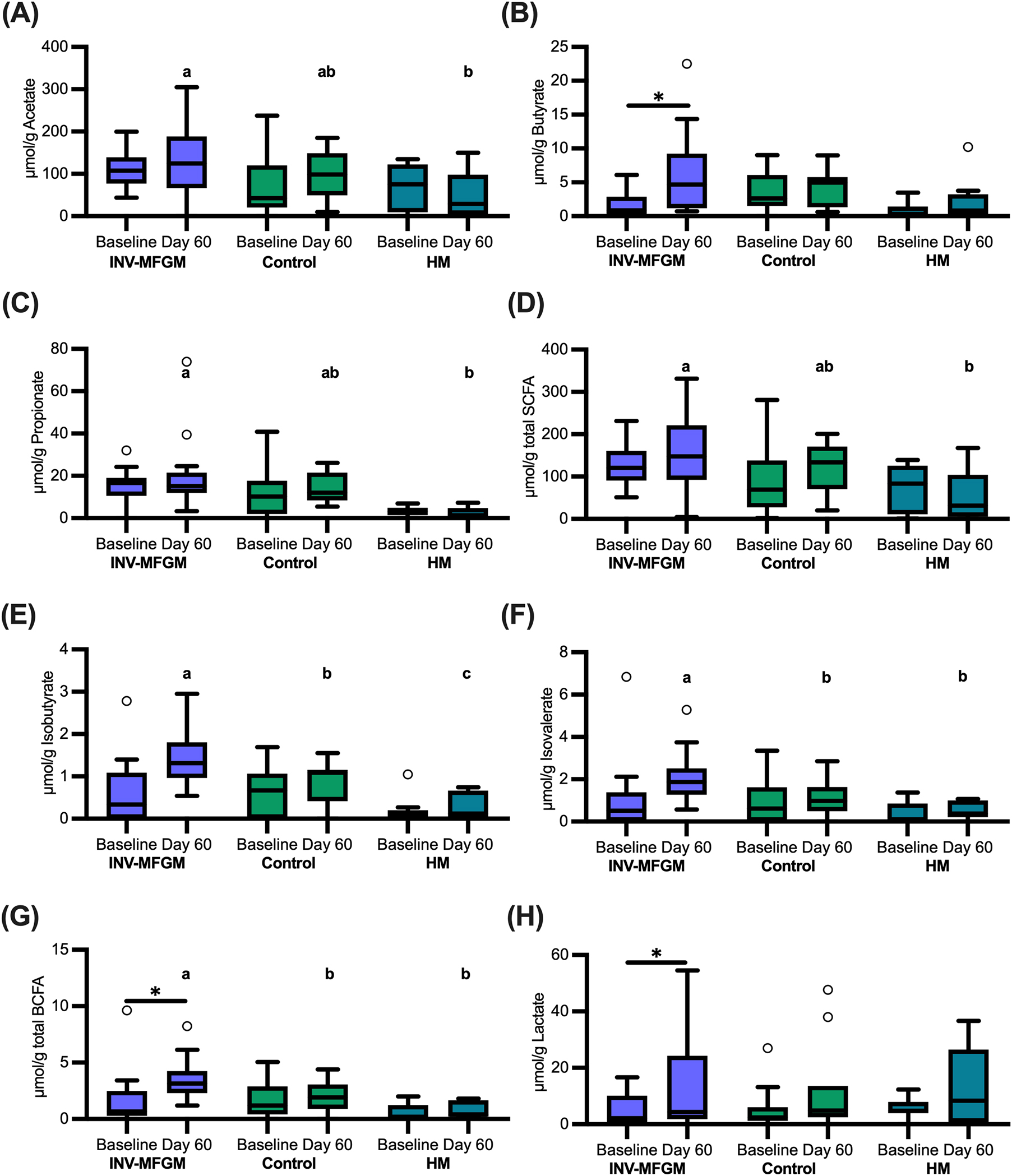
Figure 6. Stool S/BCFA and lactate concentrations. (A) Acetate, (B) butyrate, (C) propionate, (D) total SCFA, (E) isobutyrate, (F) isovalerate, (G) total BCFA, and (H) lactate were measured at Baseline and Day 60 and were reported as μmol/g of wet weight stool. Boxplots show the median, first and third quartiles with the whiskers extending to 1.5 x IQR (Interquartile Range). ° Used to indicate points outside of whiskers. Unique letters above Day 60 boxplots indicate study groups with significantly different values. *p < 0.05.
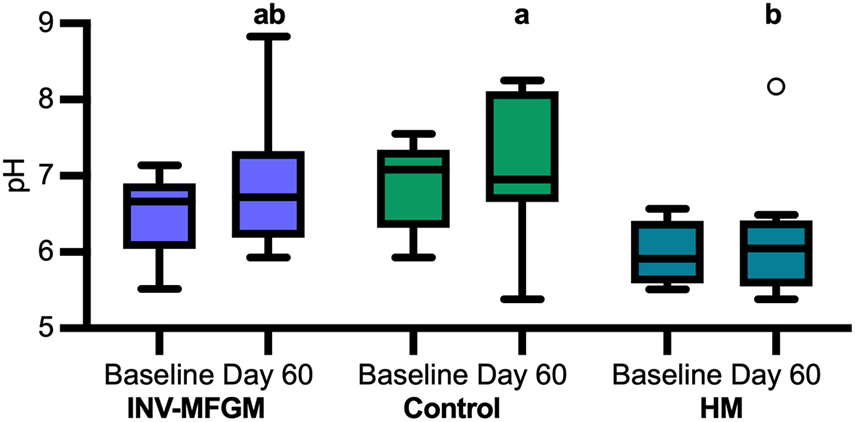
Figure 7. pH of infant stool. pH was measured at Baseline and Day 60. Boxplots show the median, first and third quartiles with the whiskers extending to 1.5 x IQR (Interquartile Range). ° Used to indicate points outside of whiskers. Unique letters above Day 60 boxplots indicate study groups with significantly different values.
Immune biomarkers
Immune biomarkers are shown in Figure 8. No group differences were detected for α-defensin, β-defensin, and calprotectin at Baseline or Day 60 or between Baseline and Day 60. No significant group differences were detected at Baseline or Day 60 for sIgA; however, sIgA concentrations increased from Baseline to Day 60 for Control and HM.
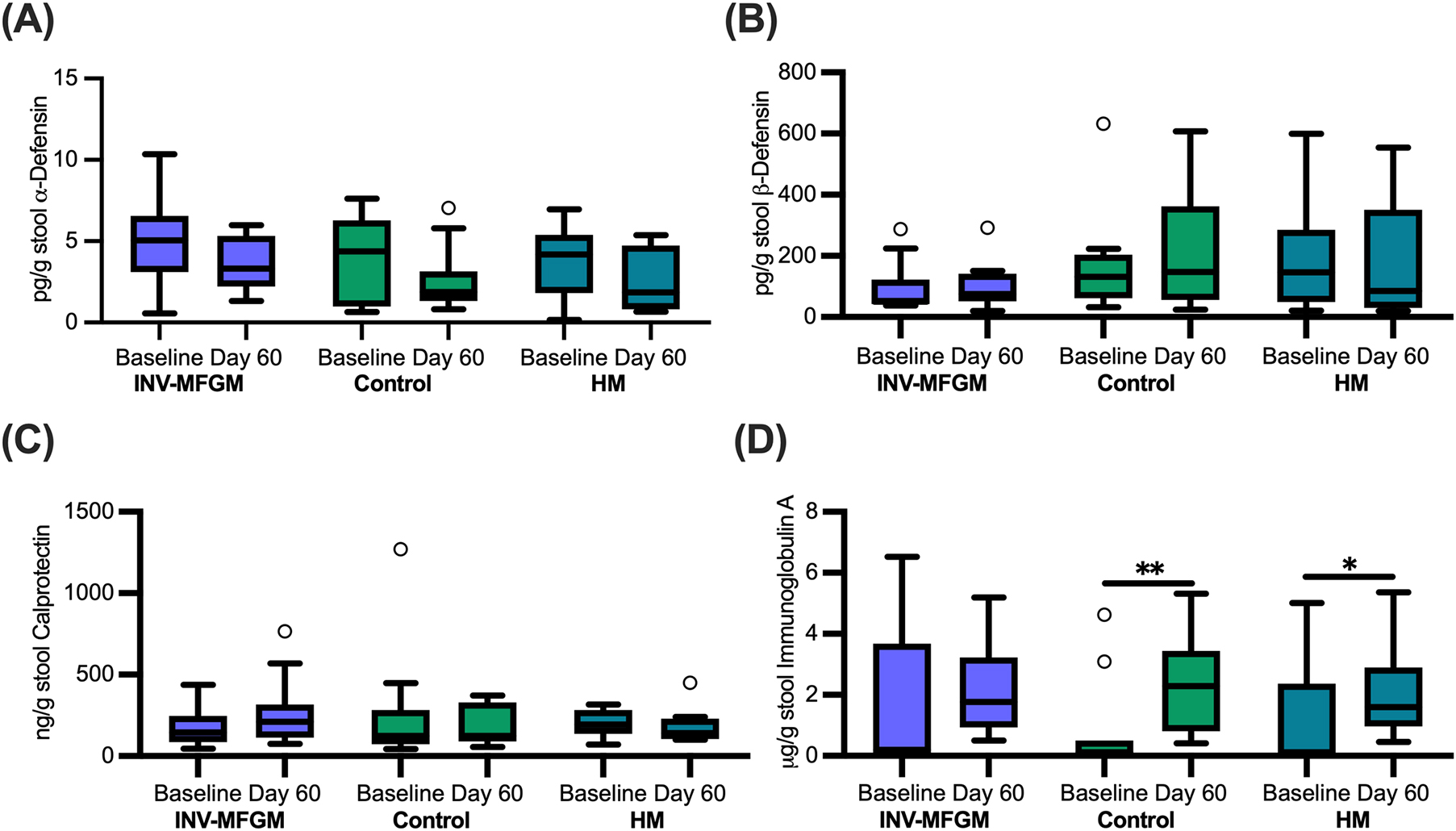
Figure 8. Infant stool immune biomarkers. Concentrations of (A) α-defensin, (B) β-defensin, (C) calprotectin, and (D) secretory immunoglobulin A measured at Baseline and Day 60. Boxplots show the median, first and third quartiles with the whiskers extending to 1.5 x IQR (Interquartile Range). ° Used to indicate points outside of whiskers. No significant differences were observed between groups at Day 60. *p = 0.03; **p = 0.01.
Discussion
The effects of MFGM on stool and oral microbiota and immune markers in early infant nutrition were investigated in healthy infants receiving routine cow’s milk-based infant formulas with or without added bovine MFGM. This study complements the growing foundation of clinical data supporting the safety, tolerability, and potential functional benefit of adding bMFGM to infant formula (19–23). Multiple studies have also demonstrated beneficial effects on behavior and neurodevelopment in infants (24–27) and in young children through 5.5 years of age (28). The current study extends the body of clinical data supporting MFGM to the earliest period of nutrition in infancy. B. bifidum and B. catenulatum species were higher in infants receiving study formulas compared to a reference group receiving mother’s-own milk. In addition, distinct patterns of stool S/BCFA were observed based on study feeding.
These results suggest one or more components of bMFGM could be metabolized by the digestive tract microbiota, leading to modulation of S/BCFA. An earlier study that examined infant gut maturation reported higher stool isobutyrate, isovalerate, and propionate in infants receiving bMFGM in formula compared to breastfed infants (30). In the current study, similar results were observed, as well as higher acetate and total S/BCFA in the INV-MFGM group compared to HM. Differences in isobutyrate were also detected in study formula groups and higher isovalerate and total BCFA were observed in the INV-MFGM vs Control and HM groups. While variations in stool consistency between HM and formula-fed groups may contribute to differences in total branched and short chain fatty acids measured in stool normalized to wet weights, the effect of stool consistency is likely to be modest based on previous correlations between stool consistency and water content (44). We previously reported feeding extensively hydrolyzed protein formula led to higher stool BCFA in infants as well as higher stool pH (37). In this study, stool pH values in infants receiving Control formula were 0.9 pH units higher compared to infants receiving mother’s own milk, while pH was not significantly different in infants fed formula with added bMFGM compared to infants fed mother’s own milk. In addition to production of BCFA, protein fermentation can lead to increased production of amines which can increase pH (45). Alternatively, differences in stool pH between Control and HM groups could be due to HMO fermentation in the HM group which lowers pH (46). Further studies would be needed to determine whether these differences in stool pH are significant and what mechanism(s) potentially contribute to these differences.
Richness and diversity in stool was significantly higher in infants receiving either study formula compared to HM at Day 60, similar to previous reports (37, 47, 48). High diversity in the GI microbiota across the majority of the lifespan is generally considered to reflect a healthier state (49). However, low diversity is considered to reflect a healthier state in infants, as low diversity is often due to enrichment of HMO-consuming Bifidobacterium taxa (50). As no differences in overall Bifidobacterium levels between groups were detected in our study, it is unclear how these differences in microbiota richness and diversity would affect function.
The primary stool taxa detected across all groups and timepoints belonged to phyla typically associated with the infant gut microbiota (51, 52). Consistent with previous studies (51, 53, 54), Bifidobacterium was abundant for many participants. However, Bifidobacterium appeared to be absent or below detection for several participants at Baseline and Day 60 in all study groups. The absence of Bifidobacterium in infants has been observed previously (55, 56) and may be due to several factors, including mode of delivery (57, 58), normal daily fluctuation of Bifidobacteria in stool, founder effects, and displacement by other taxa (59).
Infant-associated Bifidobacterium usually include B. breve, B. bifidum and B. longum (60, 61). While we observed B. breve and B. bifidum, we also observed B. adolescentis, B. catenulatum, and B. dentium. B. breve was the most abundant species detected in all groups. B. catenulatum and B. bifidum were higher in both formula groups compared to the HM group. Although B. catenulatum has been associated with adult microbiotas (62), some strains also appear in the gastrointestinal tract of infants (63), especially post-weaning (64). A subspecies of B. catenulatum, B. catenulatum subsp. kashiwanohense, is found to primarily colonize infants and encodes genes for metabolism of human milk oligosaccharides (HMO) (63). B. bifidum has been reported to colonize infants via maternal transfer and to have co-evolved with the human host (65). The ability of B. bifidum to efficiently degrade complex glycans, such as those found in human milk oligosaccharides (65), and as plant-associated carbohydrates (66), allow B. bifidum to enhance the growth of other Bifidobacterium species through cross-feeding. The absence of B. longum (particularly B. longum subsp. infantis) in the HM group is consistent with previous observations that this subspecies with unique abilities to consume HMOs is often absent in infants receiving human milk in resource-rich countries (55). However, accurate species-level identification using the V4 region is challenging which could have resulted in unclassified Bifidobacterium at the species level (67).
The inclusion of a prebiotic blend of polydextrose and galactooligosaccharides likely contributed to the enrichment of Bifidobacterium taxa in formula-fed infants as has been previously reported (68). A previous study that observed enrichment of Bifidboacterium taxa in response to supplementation of formula with bovine MFGM compared to control formula did not appear to contain prebiotics (29). In contrast, a second study where formula composition was more similar to that described here (supplemented with galactooligosacchardes at 2.3 g/100g) did not observe significant differences in Bifidobacterium between study groups fed formula with or without bovine MFGM (30). Neither study reported change in Bifidobacterium species; rather differences were reported for the Bifidobacterium genus.
Previous reports suggest lower stool α- and β- defensin concentrations in infants receiving human milk vs standard formula (69). In contrast, higher stool calprotectin and sIgA were reported in infants receiving HM compared to exclusive formula or mixed feeding (69, 70). Immune biomarkers concentrations in this current study were within the ranges reported in previous studies, but no group differences were detected at Baseline or Day 60. While increased sIgA concentrations from Baseline to Day 60 for Control and HM groups were detected, there were no group differences detected either at Baseline or Day 60. Lack of differences between study groups at Day 60 is consistent with a previous study assessing effects of consumption of MFGM-supplemented formulas which found no significant differences in fecal sIgA levels in infants that were consuming formula with and without MFGM supplementation at 4 months of age (19).
The few studies to consider effects of diet on the infant oral microbiota have suggested breastfeeding reduces bacterial diversity (71–73). However, no differences in diversity from Baseline to Day 60 or between study feeding groups at Day 60 were detected in the current study. Inability to detect differences in oral microbiota diversity between study feeding groups may be due to small sample sizes. Previous studies that demonstrated significant differences between breastfed and formula fed infants or between infants consuming standard formula and formula supplemented with MFGM have had much larger numbers of participants, although one study observed significant differences between breastfed and formula fed infants in a similarly small study. Consistent with this hypothesis, statistical significance of differences in Day 60 ASV were detected between HM and INV-MFGM (p = 0.138) and HM and Control p = 0.137). The primary bacterial taxa detected across all study time points and feeding groups belong to phyla typically associated with the infant oral microbiota (74). Consistent with previous studies (74–76), Streptococcaceae were dominant members of the oral microbiota. No species were identified as cariogenic Streptococcus mutans, which has been associated with negative impacts on oral and overall health (77). Although a total of 22 ASVs correlated between oral and fecal microbiotas, the oral-gut microbial axis remains an area of interest not extensively studied in infants (76).
Results from this study suggest that formulas with different lipid ingredients affect the microbiota differently. Compared to previous studies, this study limited the impact of covariables such as use of probiotics, prebiotics, or other immune-modulating ingredients in addition to bMFGM. Other key strengths of this study include enrollment of infants at a very early age, inclusion of a human milk reference, and strict entry criteria which excluded Caesarian section born infants or those who received antibiotics or probiotics peri-partum. These criteria reduced potential variability that could be introduced by these environmental factors. Limitations of the study included small sample size due to difficulty in recruitment following the outbreak of the worldwide COVID pandemic. While having the planned sample size would have been ideal, the data reported here is valuable in demonstrating the development of oral and stool microbiota populations in infants. Data on infant feeding history (such as breastfeeding or use of infant formula prior to randomization), materinal pre-gestational health, and perinatal antibiotic use may also have provided further insights into variation between study groups at baseline. Further studies with a larger sample size, that include both Caesarian-section and vaginal births to evaluate effects of real-world practices, and longitudinal studies that examine both microbiota and physiological outcomes may be needed to fully understand the potential systemic benefits that result from the incorporation of functional components in infant formulas.
Conclusion
Infants exclusively receiving a routine cow’s milk-based infant formula with or without added bMFGM or mother’s own milk from early infancy (7–18 days of age) had distinct patterns of gut and oral microbiota development over a 60-day feeding period. Bifidobacterium abundance was generally similar across study feeding groups, with B. breve the dominant species. B. bifidum and B. catenulatum were enriched in formula-fed groups. Additionally, Akkermansia species were higher in infants receiving bMFGM in formula compared to other groups consistent with a previous report on bMFGM supplementation (30). Although some health-associated taxa were identified in infants receiving bMFGM in formula, microbiota composition was generally most similar between formula groups compared to infants receiving mother’s own human milk. In addition, infants in the HM group had consistently lower stool pH and microbial metabolite concentrations. Infants receiving added bMFGM in formula had significantly higher butyrate and lactate at Day 60 compared to baseline and significantly higher BCFA compared to control formula at Day 60. Microbial metabolite profiles were also differentiated by study feeding group, however, further studies are needed to determine if outcomes were mediated through changes in the functional activity of the gut microbiota. This study complements the growing body of clinical data that supports the safety, tolerability, and potential functional benefit bMFGM to infant formula by extending foundational understanding to the earliest period of infant nutrition and microbiota development.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/1005334, PRJNA1005334.
Ethics statement
The studies involving humans were approved by Advarra Institutional Review Board (Columbia, MD, United States; IRB #Pro00027443). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
CC: Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review and editing. CK: Data curation, Formal analysis, Methodology, Visualization, Writing – review and editing. CH: Conceptualization, Writing – review and editing. NM: Conceptualization, Writing – review and editing. JW: Conceptualization, Data curation, Writing – review and editing, Project administration. WZ: Conceptualization, Formal analysis, Writing – review and editing. SW: Conceptualization, Project administration, Writing – review and editing. RH: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Supervision, Writing – review and editing. JI: Conceptualization, Funding acquisition, Supervision, Writing – review and editing. JA: Data curation, Formal analysis, Methodology, Supervision, Visualization, Writing – review and editing.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. CK was partially supported by a Nebraska Food for Health Center Fellowship and received additional support for Writing, Editing, and Analysis under the Auspices of the U.S. Department of Energy from Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344. CC was partially supported by the Department of Food Science and Technology, University of Nebraska-Lincoln.
Acknowledgments
We thank the participants, their families, and the study staff and Devin Rose and Carmen Perez-Donado for assistance with HPLC. We also thank Maciej Chichlowski for advice on microbiome analysis. Sequence analysis was completed utilizing the Holland Computing Center, which receives support from the UNL Office of Research and Economic Development and the Nebraska Research Initiative.
Conflict of interest
RH is a founder, and JA has financial interest in Synbiotic Health. NM, JW, and WZ are employed by the study sponsor. CH and SW were previously employed by the study sponsor.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2024.1465174/full#supplementary-material
Footnotes
References
1. Stewart C, Ajami N, O’Brien J, Hutchinson D, Smith D, Wong M, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. (2018) 562:583–8. doi: 10.1038/s41586-018-0617-x
2. Martino C, Dilmore A, Burcham Z, Metcalf J, Jeste D, Knight R. Microbiota succession throughout life from the cradle to the grave. Nat Rev Microbiol. (2022) 20:707–20. doi: 10.1038/s41579-022-00768-z
3. Tamburini S, Shen N, Wu H, Clemente J. The microbiome in early life: Implications for health outcomes. Nat Med. (2016) 22:713–22. doi: 10.1038/nm.4142
4. Brink L, Mercer K, Piccolo B, Chintapalli S, Elolimy A, Bowlin A, et al. Neonatal diet alters fecal microbiota and metabolome profiles at different ages in infants fed breast milk or formula. Am J Clin Nutr. (2020) 111:1190–202. doi: 10.1093/ajcn/nqaa076
5. Ma J, Li Z, Zhang W, Zhang C, Zhang Y, Mei H, et al. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: A study of 91 term infants. Sci Rep. (2020) 10:15792. doi: 10.1038/s41598-020-72635-x
6. Penders J, Thijs C, Vink C, Stelma F, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. (2006) 118:511–21. doi: 10.1542/peds.2005-2824
7. World Health Organization. Global Strategy for Infant and Young Child Feeding. Geneva: World Health Organization (2003).
8. Victora C, Bahl R, Barros A, França G, Horton S, Krasevec J, et al. Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet. (2016) 387:475–90. doi: 10.1016/S0140-6736(15)01024-7
9. Jiang X, Zou X, Chao Z, Xu X. Preparation of human milk fat substitutes: A review. Life (Basel). (2022) 12:187. doi: 10.3390/life12020187
10. Lin T, Meletharayil G, Kapoor R, Abbaspourrad A. Bioactives in bovine milk: Chemistry, technology, and applications. Nutr Rev. (2021) 79:48–69. doi: 10.1093/nutrit/nuab099
11. Meng F, Uniacke-Lowe T, Ryan A, Kelly A. The composition and physico-chemical properties of human milk: A review. Trends Food Sci. (2021) 112:608–21. doi: 10.1016/j.tifs.2021.03.040
12. Delplanque B, Gibson R, Koletzko B, Lapillonne A, Strandvik B. Lipid quality in infant nutrition: Current knowledge and future opportunities. J Pediatr Gastroenterol Nutr. (2015) 61:8–17. doi: 10.1097/MPG.0000000000000818
13. Pan Y, Liu L, Tian S, Li X, Hussain M, Li C, et al. Comparative analysis of interfacial composition and structure of fat globules in human milk and infant formulas. Food Hydrocoll. (2022) 124:107290. doi: 10.1016/j.foodhyd.2021.107290
14. Wei W, Li D, Jiang C, Zhang X, Zhang X, Jin Q, et al. Phospholipid composition and fat globule structure II: Comparison of mammalian milk from five different species. Food Chem. (2022) 388:132939. doi: 10.1016/j.foodchem.2022.132939
15. Lönnerdal B. Infant formula and infant nutrition: Bioactive proteins of human milk and implications for composition of infant formulas. Am J Clin Nutr. (2014) 99:712S–7S. doi: 10.3945/ajcn.113.071993
16. Peterson J, Patton S, Hamosh M. Glycoproteins of the human milk fat globule in the protection of the breast-fed infant against infections. Biol Neonate. (1998) 74:143–62.
17. Bhinder G, Allaire J, Garcia C, Lau J, Chan J, Ryz N, et al. Milk fat globule membrane supplementation in formula modulates the neonatal gut microbiome and normalizes intestinal development. Sci Rep. (2017) 7:45274. doi: 10.1038/srep45274
18. Huërou-Luron I, Bouzerzour K, Ferret-Bernard S, Ménard O, Normand L, Perrier C, et al. A mixture of milk and vegetable lipids in infant formula changes gut digestion, mucosal immunity and microbiota composition in neonatal piglets. Eur J Nutr. (2018) 57:463–76. doi: 10.1007/s00394-016-1329-3
19. Billeaud C, Puccio G, Saliba E, Guillois B, Vaysse C, Pecquet S, et al. Safety and tolerance evaluation of milk fat globule membrane-enriched infant formulas: A randomized controlled multicenter non-inferiority trial in healthy term infants. Clin Med Insights Pediatr. (2014) 8:51–60. doi: 10.4137/cmped.s16962
20. Hedrick J, Yeiser M, Harris C, Wampler J, London H, Patterson A, et al. Infant formula with added bovine milk fat globule membrane and modified iron supports growth and normal iron status at one year of age: A randomized controlled trial. Nutrients. (2021) 13:4541. doi: 10.3390/nu13124541
21. Jaramillo-Ospina A, Toro-Campos R, Murguía-Peniche T, Wampler J, Wu S, Berseth C, et al. Added bovine milk fat globule membrane in formula: Growth, body composition, and safety through age 2: An RCT. Nutrition. (2022) 97:111599. doi: 10.1016/j.nut.2022.111599
22. Zavaleta N, Kvistgaard A, Graverholt G, Respicio G, Guija H, Valencia N, et al. Efficacy of an MFGM-enriched complementary food in diarrhea, anemia, and micronutrient status in infants. J Pediatr Gastroenterol Nutr. (2011) 53:561–8. doi: 10.1097/MPG.0b013e318225cdaf
23. Timby N, Domellöf M, Holgerson P, West C, Lönnerdal B, Hernell O, et al. Oral microbiota in infants fed a formula supplemented with bovine milk fat globule membranes -A randomized controlled trial. PLoS One. (2017) 12:e0169831. doi: 10.1371/journal.pone.0169831
24. Gurnida D, Rowan A, Idjradinata P, Muchtadi D, Sekarwana N. Association of complex lipids containing gangliosides with cognitive development of 6-month-old infants. Early Hum Dev. (2012) 88:595–601. doi: 10.1016/j.earlhumdev.2012.01.003
25. Timby N, Domellöf E, Hernell O, Lönnerdal B, Domellöf M. Neurodevelopment, nutrition, and growth until 12 mo of age in infants fed a low-energy, low-protein formula supplemented with bovine milk fat globule membranes: A randomized controlled trial. Am J Clin Nutr. (2014) 99:860–8. doi: 10.3945/ajcn.113.064295
26. Li F, Wu S, Berseth C, Harris C, Richards J, Wampler J, et al. Improved neurodevelopmental outcomes associated with bovine milk fat globule membrane and Lactoferrin in infant formula: A randomized, controlled trial. J Pediatr. (2019) 215:24–31.e8. doi: 10.1016/j.jpeds.2019.08.030
27. Xia Y, Jiang B, Zhou L, Ma J, Yang L, Wang F, et al. Neurodevelopmental outcomes of healthy Chinese term infants fed infant formula enriched in bovine milk fat globule membrane for 12 months - A randomized controlled trial. Asia Pac J Clin Nutr. (2021) 30:401–14. doi: 10.6133/apjcn.202109_30(3).0007
28. Colombo J, Harris C, Wampler J, Zhuang W, Shaddy D, Liu B, et al. Improved neurodevelopmental outcomes at 5.5 years of age in children who received bovine milk fat globule membrane and Lactoferrin in infant formula through 12 months: A randomized controlled Trial. J Pediatr. (2023) 261:113483. doi: 10.1016/j.jpeds.2023.113483
29. Zhao J, Yi W, Liu B, Dai Y, Jiang T, Chen S, et al. MFGM components promote gut Bifidobacterium growth in infant and in vitro. Eur J Nutr. (2022) 61:277–88. doi: 10.1007/s00394-021-02638-5
30. He X, Parenti M, Grip T, Lönnerdal B, Timby N, Domellöf M, et al. Fecal microbiome and metabolome of infants fed bovine MFGM supplemented formula or standard formula with breast-fed infants as reference: A randomized controlled trial. Sci Rep. (2019) 9:11589. doi: 10.1038/s41598-019-47953-4
31. Chichlowski M, Bokulich N, Harris C, Wampler J, Li F, Berseth C, et al. Effect of bovine milk fat globule membrane and Lactoferrin in infant formula on gut microbiome and metabolome at 4 months of age. Curr Dev Nutr. (2021) 5:nzab027. doi: 10.1093/cdn/nzab027
32. He X, Parenti M, Grip T, Domellöf M, Lönnerdal B, Hernell O, et al. Metabolic phenotype of breast-fed infants, and infants fed standard formula or bovine MFGM supplemented formula: A randomized controlled trial. Sci Rep. (2019) 9:339. doi: 10.1038/s41598-018-36292-5
33. Lee H, Slupsky C, Heckmann A, Christensen B, Peng Y, Li X, et al. Milk fat globule membrane as a modulator of infant metabolism and gut microbiota: A formula supplement narrowing the metabolic differences between breastfed and formula-fed infants. Mol Nutr Food Res. (2021) 65:e2000603. doi: 10.1002/mnfr.202000603
34. Li X, Peng Y, Li Z, Christensen B, Heckmann A, Stenlund H, et al. Feeding infants formula with probiotics or milk fat globule membrane: A double-blind, randomized controlled trial. Front Pediatr. (2019) 7:347. doi: 10.3389/fped.2019.00347
35. Li X, Peng Y, Li Z, Christensen B, Heckmann A, Lagerqvist C, et al. Serum cytokine patterns are modulated in infants fed formula with probiotics or milk fat globule membranes: A randomized controlled trial. PLoS One. (2021) 16:e0251293. doi: 10.1371/journal.pone.0251293
36. Wang Z, Zolnik C, Qiu Y, Usyk M, Wang T, Strickler H, et al. Comparison of fecal collection methods for microbiome and metabolomics studies. Front Cell Infect Microbiol. (2018) 8:301. doi: 10.3389/fcimb.2018.0030
37. Kok C, Brabec B, Chichlowski M, Harris C, Moore N, Wampler J, et al. Stool microbiome, pH and short/branched chain fatty acids in infants receiving extensively hydrolyzed formula, amino acid formula, or human milk through two months of age. BMC Microbiol. (2020) 20:337. doi: 10.1186/s12866-020-01991-5
38. Christensen C, Kok C, Auchtung J, Hutkins R. Prebiotics enhance persistence of fermented-food associated bacteria in in vitro cultivated fecal microbial communities. Front Microbiol. (2022) 13:908506. doi: 10.3389/fmicb.2022.908506
39. Callahan B, McMurdie P, Rosen M, Han A, Johnson A, Holmes S. DADA2: High resolution sample inference from Illumina amplicon data. Nat Methods. (2016) 13:581–3. doi: 10.1038/nmeth.3869
40. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. (2013) 41:D590–6. doi: 10.1093/nar/gks1219
41. Goffau M, Luopajärvi K, Knip M, Ilonen J, Ruohtula T, Härkönen T, et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes. (2013) 62:1238–44. doi: 10.2337/db12-0526
42. Foster Z, Sharpton T, Grünwald N. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput Biol. (2017) 13:e1005404. doi: 10.1371/journal.pcbi.1005404
43. Love M, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
44. Wenzl H, Fine K, Schiller L, Fordtran J. Determinants of decreased fecal consistency in patients with diarrhea. Gastroenterology. (1995) 108:1729–38. doi: 10.1016/0016-5085(95)90134-5
45. Diether N, Willing B. Microbial fermentation of dietary protein: An important factor in diet–microbe–host interaction. Microorganisms. (2019) 7:19. doi: 10.3390/microorganisms7010019
46. Estorninos E, Lawenko R, Palestroque E, Sprenger N, Benyacoub J, Kortman G, et al. Term infant formula supplemented with milk-derived oligosaccharides shifts the gut microbiota closer to that of human milk-fed infants and improves intestinal immune defense: A randomized controlled trial. Am J Clin Nutr. (2022) 115:142–53. doi: 10.1093/ajcn/nqab336
47. Lee S, Lim J, Kim B, Cho S, Kim N, Kim O, et al. Comparison of the gut microbiota profile in breast-fed and formula-fed Korean infants using pyrosequencing. Nutr Res Pract. (2015) 9:242–8. doi: 10.4162/nrp.2015.9.3.242
48. Yatsunenko T, Rey F, Manary M, Trehan I, Dominguez-Bello M, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. (2012) 486:222–7. doi: 10.1038/nature11053
49. Manor O, Dai C, Kornilov S, Smith B, Price N, Lovejoy J, et al. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat Commun. (2020) 11:5206. doi: 10.1038/s41467-020-18871-1
50. Duar R, Henrick B, Casaburi G, Frese S. Integrating the ecosystem services framework to define dysbiosis of the breastfed infant gut: The role of B. infantis and human milk oligosaccharides. Front Nutr. (2020) 7:33. doi: 10.3389/fnut.2020.00033
51. Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. (2017) 81:e36–17. doi: 10.1128/MMBR.00036-17
52. Palmer C, Bik E, Digiulio D, Relman D, Brown P. Development of the human infant intestinal microbiota. PLoS Biol. (2007) 5:177. doi: 10.1371/journal.pbio
53. Lewis Z, Mills D. Differential establishment of bifidobacteria in the breastfed infant gut. Nestle Nutr Inst Workshop Ser. (2017) 88:149–59. doi: 10.1159/000455399
54. Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. (2015) 17:852. doi: 10.1016/j.chom.2015.05.012
55. Frese S, Hutton A, Contreras L, Shaw C, Palumbo M, Casaburi G, et al. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere. (2017) 2:e501–17. doi: 10.1128/mSphere.00501-17
56. Nagpal R, Kurakawa T, Tsuji H, Takahashi T, Kawashima K, Nagata S, et al. Evolution of gut Bifidobacterium population in healthy Japanese infants over the first three years of life: A quantitative assessment. Sci Rep. (2017) 7:10097. doi: 10.1038/s41598-017-10711-5
57. Dominguez-Bello M, Costello E, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. (2010) 107:11971–5. doi: 10.1073/pnas.1002601107
58. Enav H, Bäckhed F, Ley R. The developing infant gut microbiome: A strain-level view. Cell Host Microbe. (2022) 30:627–38. doi: 10.1016/j.chom.2022.04.009
59. Tannock G, Lee P, Wong K, Lawley B. Why don’t all infants have Bifidobacteria in their stool? Front Microbiol. (2016) 7:834. doi: 10.3389/fmicb.2016.00834
60. Turroni F, Milani C, Duranti S, Ferrario C, Lugli G, Mancabelli L, et al. Bifidobacteria and the infant gut: An example of co-evolution and natural selection. Cell Mol Life Sci. (2018) 75:103–18. doi: 10.1007/s00018-017-2672-0
61. Mariat D, Firmesse O, Levenez F, Guimaraes VD, Sokol H, Doré J, et al. The firmicutes/bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. (2009) 9:123. doi: 10.1186/1471-2180-9-123
62. Ojima M, Yoshida K, Sakanaka M, Jiang L, Odamaki T, Katayama T. Ecological and molecular perspectives on responders and non-responders to probiotics and prebiotics. Curr Opin Biotechnol. (2022) 73:108–20. doi: 10.1016/j.copbio.2021.06.023
63. Liu J, Li W, Yao C, Yu J, Zhang H. Comparative genomic analysis revealed genetic divergence between Bifidobacterium catenulatum subspecies present in infant versus adult guts. BMC Microbiol. (2022) 22:158. doi: 10.1186/s12866-022-02573-3
64. Kato K, Odamaki T, Mitsuyama E, Sugahara H, Xiao J, Osawa R. Age-related changes in the composition of gut Bifidobacterium species. Curr Microbiol. (2017) 74:987–95. doi: 10.1007/s00284-017-1272-4
65. Duranti S, Lugli G, Milani C, James K, Mancabelli L, Turroni F, et al. Bifidobacterium bifidum and the infant gut microbiota: An intriguing case of microbe-host co-evolution. Environ Microbiol. (2019) 21:3683–95. doi: 10.1111/1462-2920.14705
66. Turroni F, Özcan E, Milani C, Mancabelli L, Viappiani A, van Sinderen D, et al. Glycan cross-feeding activities between bifidobacteria under in vitro conditions. Front Microbiol. (2015) 6:1030. doi: 10.3389/fmicb.2015.01030
67. Johnson J, Spakowicz D, Hong B, Petersen L, Demkowicz P, Chen L, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. (2019) 10:5029. doi: 10.1038/s41467-019-13036-1
68. Scalabrin D, Mitmesser S, Welling G, Harris C, Marunycz J, Walker D, et al. New prebiotic blend of polydextrose and galacto-oligosaccharides has a bifidogenic effect in young infants. J Pediatr Gastroenterol Nutr. (2012) 54:343–52. doi: 10.1097/MPG.0b013e318237ed95
69. Roggero P, Liotto N, Pozzi C, Braga D, Troisi J, Menis C, et al. Analysis of immune, microbiota and metabolome maturation in infants in a clinical trial of Lactobacillus paracasei CBA L74-fermented formula. Nat Commun. (2020) 11:2703. doi: 10.1038/s41467-020-16582-1
70. Asgarshirazi M, Shariat M, Nayeri F, Dalili H, Abdollahi A. Comparison of fecal calprotectin in exclusively breastfed and formula or mixed fed infants in the first six months of life. Acta Med Iran. (2017) 55:53–8.
71. Al-Shehri S, Sweeney E, Cowley D, Liley H, Ranasinghe P, Charles B, et al. Deep sequencing of the 16S ribosomal RNA of the neonatal oral microbiome: A comparison of breast-fed and formula-fed infants. Sci Rep. (2016) 6:38309. doi: 10.1038/srep38309
72. Holgerson P, Vestman N, Claesson R, Öhman C, Domellöf M, Tanner A, et al. Oral microbial profile discriminates breast-fed from formula-fed infants. J Pediatr Gastroenterol Nutr. (2013) 56:127–36. doi: 10.1097/MPG.0b013e31826f2bc6
73. Kageyama S, Furuta M, Takeshita T, Ma J, Asakawa M, Yamashita Y. High-level acquisition of maternal oral bacteria in formula-fed infant oral microbiota. mBio. (2022) 13:e0345221. doi: 10.1128/MBIO.03452-21
74. Kennedy B, Peura S, Hammar U, Vicenzi S, Hedman A, Almqvist C, et al. Oral microbiota development in early childhood. Sci Rep. (2019) 9:19025. doi: 10.1038/s41598-019-54702-0
75. Sulyanto R, Thompson Z, Beall C, Leys E, Griffen A. The predominant oral microbiota is acquired early in an organized pattern. Sci Rep. (2019) 9:10550. doi: 10.1038/s41598-019-46923-0
76. Butler C, Adams G, Blum J, Byrne S, Carpenter L, Gussy M, et al. Breastmilk influences development and composition of the oral microbiome. J Oral Microbiol. (2022) 14:2096287. doi: 10.1080/20002297.2022.2096287
Keywords: milk fat globule membrane, infant formula, human milk, microbiome, short chain fatty acids, clinical trial
Citation: Christensen C, Kok CR, Harris CL, Moore N, Wampler JL, Zhuang W, Wu SS, Hutkins R, Izard J and Auchtung JM (2024) Microbiota, metabolic profiles and immune biomarkers in infants receiving formula with added bovine milk fat globule membrane: a randomized, controlled trial. Front. Nutr. 11:1465174. doi: 10.3389/fnut.2024.1465174
Received: 15 July 2024; Accepted: 16 September 2024;
Published: 04 October 2024.
Edited by:
Carlos Gómez-Gallego, University of Eastern Finland, FinlandReviewed by:
Sharon Marie Donovan, University of Illinois Urbana-Champaign, United StatesGabriel Vinderola, National University of Littoral, Argentina
Copyright © 2024 Christensen, Kok, Harris, Moore, Wampler, Zhuang, Wu, Hutkins, Izard and Auchtung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jacques Izard, aml6YXJkQHVubWMuZWR1; Jennifer M. Auchtung, amF1Y2h0dW5nMkB1bmwuZWR1
†Present address: Car Reen Kok, Physical and Life Sciences Directorate, Lawrence Livermore National Laboratory, Livermore, CA, United States
‡These authors have contributed equally to this work and share first authorship
§These authors have contributed equally to this work and share senior authorship
 Chloe Christensen1,2‡
Chloe Christensen1,2‡ Jennifer L. Wampler
Jennifer L. Wampler Jacques Izard
Jacques Izard Jennifer M. Auchtung
Jennifer M. Auchtung