- 1Department of Pediatrics, University of Minnesota, Minneapolis, MN, United States
- 2Department of Psychology, University of Minnesota, Minneapolis, MN, United States
- 3Division of Epidemiology and Community Health, School of Public Health, University of Minnesota, Minneapolis, MN, United States
- 4Harold Hamm Diabetes Center, University of Oklahoma Health Sciences Center, Oklahoma City, OK, United States
- 5Department of Computer Science and Engineering, University of Minnesota, Minneapolis, MN, United States
- 6Division of Maternal-Fetal Medicine, University of Minnesota, Minneapolis, MN, United States
- 7Pregnancy and Child Health Research Center, HealthPartners Institute, Bloomington, MN, United States
Introduction: Early life gut microbiomes are important for brain and immune system development in animal models. Probiotic use has been proposed as a strategy to promote health via modulation of microbiomes. In this observational study, we explore if early life exposure to probiotics via the mother during pregnancy and lactation, is associated with decreased inflammation in breastmilk, maternal and infant microbiome variation, and altered infant neurodevelopmental features.
Methods: Exclusively breastfeeding mother-infant dyads were recruited as part of the “Mothers and Infants Linked for Healthy Growth (MILk) Study.” Probiotic comparison groups were defined by exposure to maternal probiotics (NO/YES) and by timing of probiotic exposure (prenatal, postnatal, total). C-reactive protein (CRP) and IL-6 levels were determined in breastmilk by immunoassays, and microbiomes were characterized from 1-month milk and from 1- and 6-month infant feces by 16S rDNA sequencing. Infant brain function was profiled via electroencephalogram (EEG); we assessed recognition memory using event-related potential (ERP) responses to familiar and novel auditory (1 month) and visual (6 months) stimuli. Statistical comparisons of study outcomes between probiotic groups were performed using permutational analysis of variance (PERMANOVA) (microbiome) and linear models (all other study outcomes), including relevant covariables as indicated.
Results: We observed associations between probiotic exposure and lower breastmilk CRP and IL-6 levels, and infant gut microbiome variation at 1- and 6-months of age (including higher abundances of Bifidobacteria and Lactobacillus). In addition, maternal probiotic exposure was associated with differences in infant ERP features at 6-months of age. Specifically, infants who were exposed to postnatal maternal probiotics (between the 1- and 6-month study visits) via breastfeeding/breastmilk, had larger differential responses between familiar and novel visual stimuli with respect to the late slow wave component of the EEG, which may indicate greater memory updating potential. The milk of mothers of this subgroup of infants had lower IL-6 levels and infants had different 6-month fecal microbiomes as compared to those in the “NO” maternal probiotics group.
Discussion: These results support continued research into “Microbiota-Gut-Brain” connections during early life and the role of pre- and postnatal probiotics in mothers to promote healthy microbiome-associated outcomes in infants.
1 Introduction
The early life gut microbiome co-develops in concert with aspects of host physiology such as mucosal barrier function, metabolism, immunity and brain function. The significance of this co-development is highlighted by studies in animal models that have shown that disruptions in early-life microbiomes are mechanistically linked to abnormal development of these physiological systems (1–4). In humans, infant gut microbiome composition is affected by maternal microbiomes (via transfer of microbes), birth mode, infant diet and environmental exposures (5–8). Based on the importance of microbiomes in health, there has been increasing research interest in designing strategies to promote healthy microbiomes during infancy, a sensitive period for brain development. The proposed goal of early microbiome modulation is, ideally, to improve both short and long-term health outcomes, particularly in infants that are at risk for microbiome disruption. Consistent with the Developmental Origins of Health and Disease (DOHaD) hypothesis, modulation strategies that can be targeted to the birth parent at very early times in development (e.g., prior to or during pregnancy) offer the possibility for greatest offspring health benefit (9–12).
One therapeutic strategy that has been proposed to promote healthy early life microbiomes is probiotic administration to women during pregnancy and lactation. Probiotic supplementation during pregnancy has been demonstrated to be safe for both mother and infant (13, 14), with several studies showing that probiotic use during pregnancy and lactation modulates breastmilk and/or infant gut microbiota (15–17). In pregnant women, prenatal probiotics are associated with improved metabolism and pregnancy outcomes including reduced incidences of gestational diabetes, preeclampsia, vaginal infections, and preterm labor (18–21). Prenatal probiotics have also been associated with health benefits in offspring, specifically with respect to gut health, the development of immunity, and reduced atopic disease (22–26). An association has been shown between a mother’s intestinal microbiota and their serum inflammation biomarkers, both of which may contribute to newborn early life immune and metabolic programming, independent from probiotic exposure, highlighting a potential broader role for maternal microbes and inflammation to affect offspring health (27). Connections between probiotic use during pregnancy/lactation and host and offspring inflammatory markers is an emerging theme of current research (28–30).
Research in animal models supports a role for gut microbes in modulating brain function (i.e., the Microbiome-Gut-Brain Axis) across the lifespan (31–33). Germ-free mice, as well as mice with perturbed gut microbiomes due to antibiotic exposure, exhibit deficits in social functioning and memory (34, 35) and altered anxiety and motor responses (1). Importantly, developing (young) brains have been shown to be far more susceptible to the effects of microbiome modulation than mature brains (1), reinforcing the important concept that there is an early-life sensitive period for brain developmental programming by the microbiome. The hippocampus, a brain structure critical to recognition memory and cognitive function, is the target of many of these gut microbial-mediated effects (1, 35, 36). For example, adult germ-free mice exhibit defects in memory (35), altered hippocampus levels of the neurotransmitter serotonin (36), and altered expression of hippocampus gene transcripts (1). Dietary supplementation with specific bacteria (e.g., Lactobacillus and Bifidobacteria) has been shown to enhance memory in adult rats (37). These exciting mechanistic connections among gut microbes, probiotics, and early life brain function in animal models have not yet been translated to human infants.
In human infants, functional maturation of the hippocampus and other medial temporal lobe structures responsible for recognition memory processing is evaluated by recording and analyzing the brain’s electrophysiologic response to familiar vs. novel stimuli using electroencephalogram (EEG), or event-related potential (ERP) responses (38, 39). The ERP waveform is the portion of the EEG that reflects cognitive processing of a stimulus. It is embedded in the raw EEG data and is extracted by averaging the EEG across multiple presentations of the stimulus (38). Studies of infants at risk for neurodevelopmental impairment have demonstrated that recognition memory can be evaluated in very young (newborn) infants and, importantly, that early recognition memory is associated with later recognition and behavioral memory performance (40, 41) and cognitive function (42). Previous research has also shown the predictive value of early ERP for later language development, reading ability, and risk for autism (43–46).
In this pilot observational study, we explore associations between maternal probiotic exposure (and its timing) with infant brain (recognition memory) function as measured by ERPs, specifically the electrophysiological responses recorded at the scalp in response to familiar and novel auditory and visual stimuli. We also compare inflammatory markers in breastmilk as well as milk and infant fecal microbiome features with respect to maternal probiotic exposure to gain insight into potential mechanistic links among inflammation, microbiomes, and brain function.
2 Materials and methods
2.1 Subject enrollment and inclusion criteria
Mother-infant dyads were enrolled as part of the Mothers and Infants Linked for Healthy Growth (MILk) study, a multi-site study in Oklahoma City, Oklahoma and the Minneapolis/St. Paul metropolitan area in Minnesota as previously described (47). The participants in the present study included a subset of MILk Study dyads who consented to additional microbiome assessments of their milk and their infants’ feces. Briefly, dyads were healthy, with mothers meeting the following inclusion criteria: 21–45 years of age, non-smoking, non-alcohol drinking, non-diabetic, English speaking and understanding, and delivered singleton infants at term gestation (37 0/7–42 1/7 weeks) who were appropriately grown for gestational age (between the 10th and 90th percentile on WHO growth charts). Infants were exclusively breastfed (EBF) through 6 months of age, with EBF defined as being fed only human milk and < 24 oz. (720 mL) of formula or other liquids since birth or their last study visit. None of the women reported symptoms of mastitis or breast infection at the time of breastmilk collection.
2.2 Clinical and demographic variable definitions and collection methods
Clinical and demographic data were obtained via the electronic health record from prenatal care and the birth hospitalization as well as from electronic questionnaires administered to mothers at the study visits at 1 and 6 months of infant age and are listed in Supplementary Table 1. Study questionnaires did not ask about parent gender identity and readers of this paper should be encouraged to read/use the gender-associated terms in this paper according to those with which they most identify, per the guidance of the Academy of Breastfeeding Medicine. Maternal pre-pregnancy body mass index (BMI) was defined as healthy (18.5–25), overweight (25–30), or obese (> 30). Normal versus excess gestational weight gain (GWG) was defined as developed by the American College of Obstetrics and Gynecology (American College of Obstetrics and Gynecology, 2013). The healthy eating index (HEI) is a measure of diet quality as developed by the USDA (48–50). Maternal and infant antibiotic exposure as yes/no variables were defined as follows. Maternal prenatal antibiotic exposure was considered “yes” if occurring during pregnancy and up until (not including) the birth hospitalization based on participant report. Maternal and infant postnatal antibiotics were considered “yes” at 1 month of infant age if occurring during the birthing process (including antibiotics administered due to C-section and/or Group B Streptococcus (GBS) positive maternal status) and up to and including the 1-month sample collection time. Maternal and infant postnatal antibiotic exposures were considered “yes” at 6-months of infant age if occurring after the 1-month and up to the 6-month sample collection time.
2.3 Development of probiotic groups and timing categories
Data regarding maternal probiotic exposure was collected via questionnaires administered during pregnancy and lactation (at 1-month and 6-month study visits) and included: use of probiotics (NO/YES), timing of probiotic use (before birth, between birth and 1-month study visit, and between 1- and 6-month study visits), and name of probiotic supplement (if known). Questionnaires did not specifically ask about yogurt consumption in the diet. In preliminary analyses, mothers and infants with less than 1 month of maternal probiotic exposure (5% of the cohort) had the same outcomes as those with NO maternal probiotic exposure (data not shown), and thus were included in the NO group. For all fecal and breastmilk analyses, probiotic groups (NO/YES) were further categorized by timing of exposure: prenatal, postnatal, and total (prenatal + postnatal) (Figure 1). For 6-month sample analyses, an additional exposure group was defined as those exposed to maternal probiotics after 1 month of age, to explore the association of more recent probiotic exposure on 6-month outcomes.
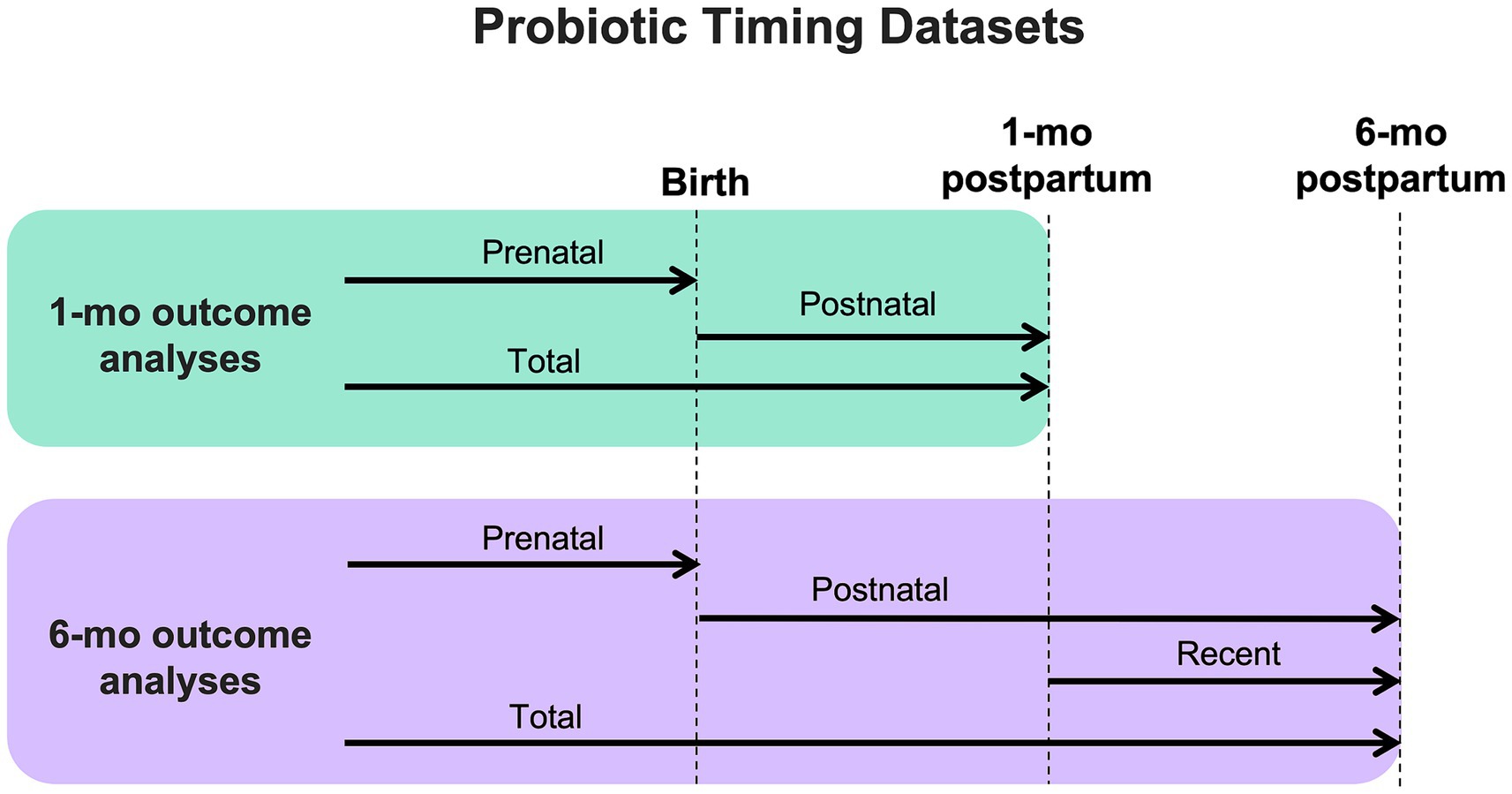
Figure 1. Diagram showing how outcome datasets were organized for analysis based on timing of maternal probiotic exposure.
Due to the number of different probiotic supplements that the women in this cohort were exposed to (28 total), with only a small number of participants reporting use of any one of them, we did not assess differences in outcomes by specific probiotic supplement. A breakdown of known components in the probiotic supplements reported by subjects is detailed in Supplementary Table 2.1.
2.4 Sample collection and storage
Breastmilk and fecal sample collection and storage were performed as previously described (47). Briefly, breastmilk samples were obtained at a study visit when infants were 1 month old using a hospital-grade electric breast pump (Medela Symphony; Medela, Inc., Zug, Switzerland) and stored at −80°C within 20 min of collection. Fecal samples were collected at 1 and 6 months of infant age. Samples were either collected at a study visit, when feces were produced on site, or via a home collection kit. Parents and study staff were instructed to collect a pea-sized amount of freshly formed feces from diapers and place in a vial containing ethanol. Samples were transported or mailed to the study laboratory and then stored at −80°C upon arrival.
2.5 Inflammatory marker determinations
C-reactive protein (CRP) and interleukin-6 (IL-6) were quantified in breastmilk as described previously (51). Briefly, breastmilk was thawed, 1 mL of milk was centrifuged, and the fat layer was removed. Skimmed milk was then used with commercially available immunoassay kits to detect and quantify CRP and IL-6 (Abcam, Cambridge, MA; and R&D Systems, Minneapolis, MN; kits, respectively).
2.6 DNA isolation and microbial sequencing
DNA isolation from breastmilk and feces was performed as previously described (47). Briefly, DNA was extracted from 1 mL of skimmed breastmilk with the PowerSoil Pro kit (QIAGEN, Germantown, MD) and was stored in 100 μL of the kit elution solution at −80°C. DNA was extracted from thawed fecal samples using the PowerSoil kit (QIAGEN, Germantown, MD) and stored in 100 μL of kit elution solution at −80°C as previously described (47).
16S bacterial rDNA (V4 region) amplicons, generated by PCR, were sequenced by the University of Minnesota Genomics Center (UMGC, Minneapolis, MN) as previously described (52). Sequencing was carried out on an Illumina MiSeq system (Illumina, San Diego, CA) using V2 2×250 bp paired end chemistry. Raw sequences that passed quality control procedures were processed with the Shi7 (53) learning program, were aligned to reference databases using BURST (54, 55) generated from the 16S RefSeq collection compiled by the National Center for Biotechnology Information (NCBI), and taxonomy (genus level or higher) and reference tables were imported into RStudio.
2.7 Assessment of recognition memory function by event-related potentials (ERPs)
ERP measures of infant recognition memory performance were obtained at 1 and 6 months of age during follow up visits as previously described (56, 57). Briefly, a Hydrocel Geodesic sensor Net (Electrical Geodesics Incorporated (EGI), Eugene, OR) with 64 electrodes was placed on the infant’s scalp. For this study, we focused on ERP feature extractions from 4 scalp regions (left frontal, left central, right frontal, right central) that included 3 adjacent lead sensors each. These lead sensors were chosen based on beneficial characteristics for analysis: increased activity with least background noise for recognition memory task assessment, and with electrical activity at these leads expected to reflect synchronous activities of adjacent populations of neurons. At 1 month of age, infants were presented with a 750 msec recording of their mother’s voice or a stranger’s voice pronouncing the word “baby” alternating with equal but random probability. Voice recordings were digitized and edited using Creative Wave Studio; recordings were presented at 75.0 dB sound pressure. A total of 100 trials (50 of each voice) were presented. At 6 months of age, infants were presented randomly, one at a time for 500 msec each, with pictures of their mother’s face and stranger’s face, for a total of 100 trials (50 of each face). Pictures are standardized with respect to race, hair color, face and total picture size, neutral facial expression, and background color. Testing occurred in an electrically shielded room, and lights in the room were dimmed during stimulus presentation. EEG data were collected and recorded on-line using NetAmps Amplifiers (EGI, Inc., Eugene, OR) and NetStation software and referenced to a single vertex electrode (gain = 10,000x; filter = 0.1–100 Hz bandpass; sampling rate = 250 Hz).
Using NetStation analysis software (EGI), EEG data were filtered offline using a 30-Hz low-pass filter. Filtered EEG data from 1-month-old infants were divided into 2,100 msec segments (100 msec pre-event baseline, 750 msec stimulus presentation, and 1,250 msec post-stimulus onset, and baseline corrected to the average pre-stimulus voltage. Filtered EEG data from 6-month-old infants were divided into 1,600 msec segments) 100 msec baseline, 500 msec stimulus presentation, 1,000 msec post-stimulus onset. Data were visually inspected and excluded if they contained electrocardiographic or motion artifact. Consistent with previous research (58), trials were rejected if they contained more than nine artifact-contaminated channels, and these individual rejected channels on remaining trials were replaced using spherical spline interpolation. Subjects that contributed a minimum of 15 acceptable trials per condition were included in analyses; individual subject averages were calculated for each condition (mother/stranger) and re-referenced to the average reference.
Components of the electrical response reflecting hippocampus-based memory recognition and updating were the focus of this analysis. At 1 month of age, components of interest included P2 amplitude (adaptive mean within 200-500 msec window), an early sensory component modulated by memory, and the slow wave (SW, measured as mean amplitude of 900–2000 msec window). Following a familiar stimulus, the typical response is a return of the waveform to baseline after the P2 peak, reflecting that no further processing is needed. A negative slow wave after the P2 peak is seen in response to “new” stimuli and demonstrates novelty detection. Further, the slow wave difference score (familiar minus novel mean area amplitude from 900 to 2000 ms post stimulus) indexes discrimination between familiar and novel stimuli (38, 56, 59). Components of interest at 6 months of age included the negative component deflection (NC, adaptive mean within 350–700 msec window) which represents attentional response mediated by memory, and the SW (mean amplitude of 900–1,500 msec window). Like the 1-month paradigm, the SW reflects cognitive processing of the stimulus. At 6 months of age, we anticipate that infants would have fully encoded their mothers’ face, giving rise to a return to baseline after the NC while the stranger’s face would be partially encoded into memory (due to presentation in 50% of trials), evoking a positive slow wave, indexing memory updating (60). We also calculated a slow wave difference score (familiar minus novel SW mean amplitude from 900 to 1,500 msec) to indicate discrimination between the two stimuli.
2.8 Statistical analyses and data visualization
Welch’s t-tests were used to compare inflammatory marker levels between probiotic groups, with effect size (Cohen’s d) and 95% confidence intervals reported. RStudio (1.1.463) was used for microbiome feature extraction and to analyze microbiome data. Data analysis and visualization libraries used included: vegan, ape, nlme, ggplot2, ggbeeswarm, tidyverse, textshape, reshape2, randomcoloR, limma, ggsignif, Rtsne, igraph, gtools, and BiodiversityR. Euclidean dissimilarity beta-diversity measurements, using centered log ratio (CLR) normalized taxonomic counts (Aitchison’s distance), were compared using permutational analysis of variance (PERMANOVA) tests (Ashbury, et al.). Lactobacillus and Bifidobacteria abundances were compared between probiotic groups using a linear model with p-values, beta-coefficients, and 95% confidence intervals determined. Infant ERP features were analyzed only in the cohort of infants that provided fecal microbiome data at the corresponding time point. ERP features were compared in probiotic groups using linear models with p-values, beta-coefficients, and 95% confidence intervals reported. Clinical and demographic covariables were compared between probiotic (NO/YES) groups, for each probiotic timing category and by specific outcome, using Welch’s t-test (for comparison of HEI, a continuous variable) and chi-square test (for all other comparisons, categorical values) (Supplementary File 2; Supplementary Tables 2, 3). For all outcome analyses, covariables that differed between comparison groups were included in linear statistical models as indicated in the Results section. For all analyses, p-values <0.05 were considered statistically significant.
3 Results
3.1 Cohort description and comparison of clinical and demographic variables in probiotic groups and timing categories
The majority of women in the study cohort delivered infants vaginally (~83%). Pre-pregnancy BMI in the normal weight range was observed in ~38% of women, and the mean HEI was ~66, which is above the average reported for women by the U.S. National Center for Health Statistics (2017–2018). Similar numbers of women had optimal versus excess gestational weight gain (~48% vs. ~52%, respectively). Infant sex was balanced (~48% males vs. ~52% females) in the total cohort. White race was reported by mothers for ~80% of women and infants in the study cohort. With respect to antibiotic exposure, ~23% of women reported exposure to prenatal antibiotics. Postnatal antibiotic exposure between birth and the 1-month study visit was reported by ~48% of mothers, primarily related to antibiotics given at the time of birth for Cesarean section or positive screening for Group B Streptococcus. Exposure to antibiotics between the 1- and 6-month study visit was reported by ~14% of women. For infant postnatal antibiotic exposure, ~2% of infants received postnatal antibiotics prior to the 1-month study visit, and 15% prior to the 6-month study visits. Infant (as opposed to maternal) probiotic exposure was reported for ~2% of infants at 1 month and ~ 4% of infants at 6 months of age.
Maternal probiotic exposure (NO/YES) groups described 79 and 21% of women-infant dyads, respectively, in this study. Statistical comparisons of demographic and clinical variables were performed between the two probiotic groups (NO vs. YES), for each analysis type and time point and are shown in Supplementary Table 2.2 (1-month timepoint) and Supplementary Table 2.3 (6-month timepoint). This was done because not every subject provided data for all outcomes analyzed. Overall, the vast majority of covariates did not differ between probiotic groups. The few covariates that did differ were included in adjusted statistical models to understand their contribution to any significant differences in outcomes between probiotic groups, as detailed in the Results sections below.
3.2 Maternal probiotic exposure corresponds to lower levels of inflammatory markers in breastmilk
Elevated levels of bioactive markers of inflammation have been implicated in mechanisms of how microbes contribute to negative health outcomes. Thus, we measured CRP and IL-6 levels in breastmilk, collected at 1 month postpartum, to understand how probiotic exposure is associated with milk inflammatory markers. Probiotic exposure (total) was associated with lower breastmilk C-reactive protein and IL-6 levels (Figure 2; Table 1) as compared to NO probiotic exposure. To explore how timing of probiotic exposure may be associated with inflammatory marker levels, subgroup analyses were performed for pre- and postnatal timing categories. Both pre- and postnatal maternal probiotic use were significantly associated with lower milk CRP concentrations. Differences in milk IL-6 levels were not observed for probiotic timing sub-categories (Table 1).
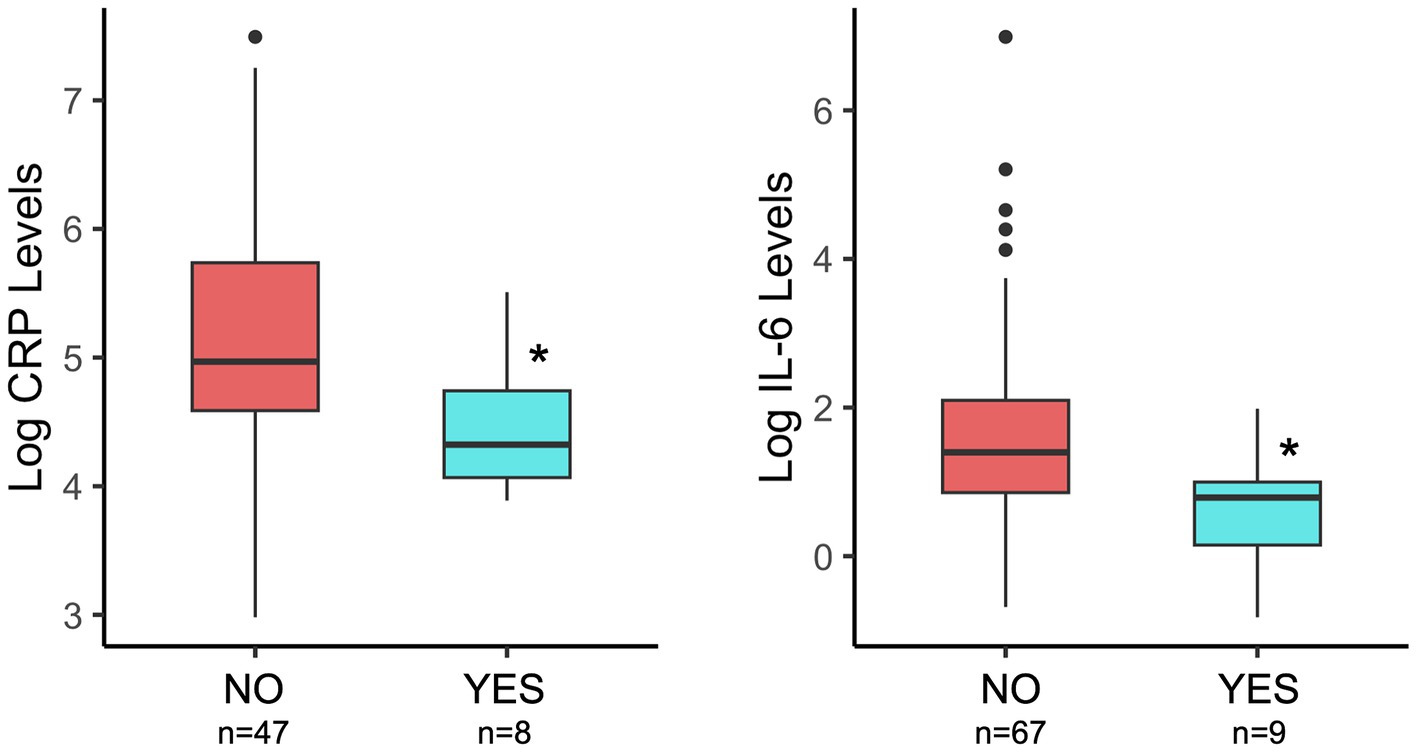
Figure 2. Comparison of breastmilk CRP and IL-6 levels by maternal probiotic group (NO/YES) and considering the total time of potential exposure (pre- plus post-natal). Details of statistical analyses are included in the Table 1. *p = 0.02 (CRP) and *p = 0.01 (IL-6).
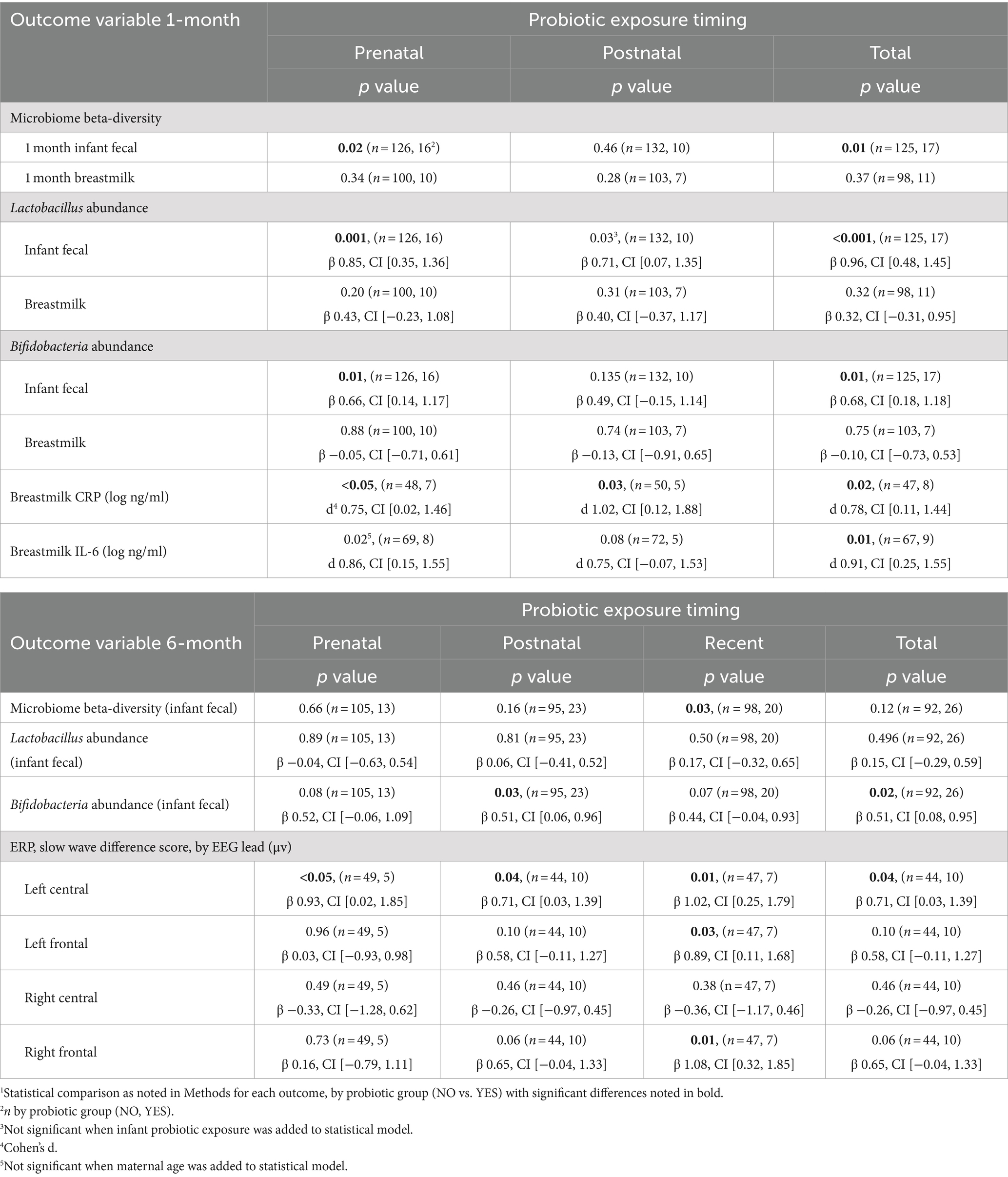
Table 1. Associations between maternal probiotic exposure at different timepoints (prenatal, postnatal, recent, and total) and infant fecal microbiome features, breastmilk inflammatory protein levels, and infant neurocognitive features at 1 and 6 months of age1
3.3 Maternal probiotic exposure is associated with variation in infant fecal, but not breastmilk, microbiome composition
Infant fecal microbiome composition (beta-diversity) at 1 month of age significantly differed between probiotic exposure (NO/YES) groups (Table 1) when considering total time (prenatal and postnatal) of exposure. In subgroup analyses, prenatal, and not postnatal, exposure was significantly associated with infant fecal microbiome variation (Table 1). Maternal age was higher in the probiotic “YES” groups for the prenatal and total timing categories at 1-month Supplementary Table 2.2. When maternal age was included in the statistical models, we observed that probiotic groups remained different with respect to microbiome compositions, indicating that maternal age is likely not contributing to these associations. We also compared the fecal abundances of “beneficial” bacterial taxa, Lactobacillus and Bifidobacteria, between probiotic groups. Of note, Lactobacillus was a component in the majority of probiotic supplements that were self-reported by the women in this cohort (~94%, Supplementary Table 2.1). At the 1-month timepoint, the probiotic (YES) group of infants had significantly higher abundances of fecal Lactobacillus and Bifidobacteria in the prenatal and total timing categories (Figure 3; Table 1). These differences remained significant when maternal age, the only covariable that differed for these probiotic groups and timing categories, was included in the statistical models. In contrast to the results with infant feces, 1-month breastmilk microbiomes did not differ with respect to composition or differential abundances of taxa, based on probiotic exposure, for any probiotic timing category.
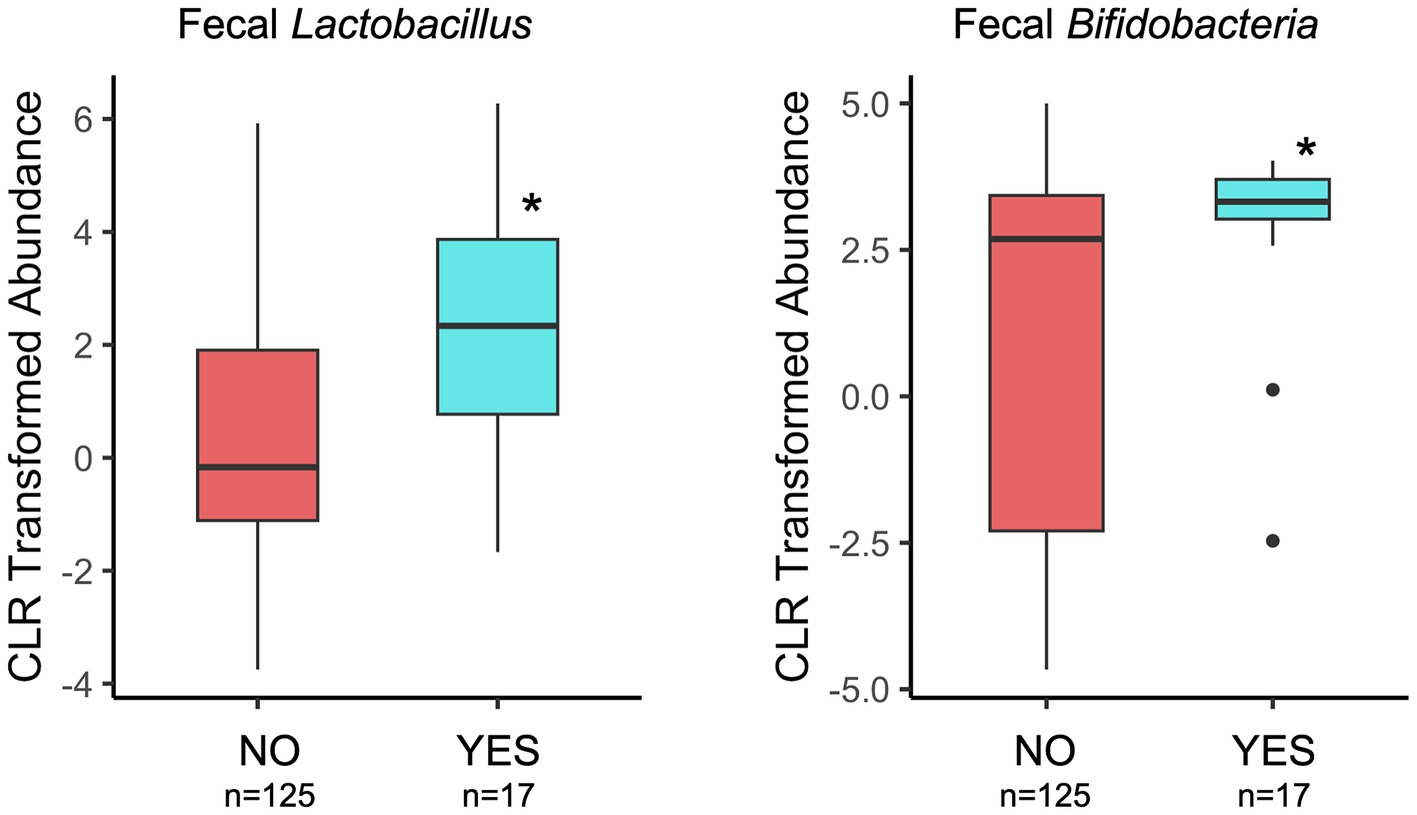
Figure 3. Comparison of Lactobacillus and Bifidobacteria relative abundances in 1-month infant feces by maternal probiotic group and considering the total time of potential exposure (pre- and post-natal). Details of statistical analyses are included in the Table 1. *p < 0.001 (genus Lactobacillus) and *p = 0.01 (genus Bifidobacteria).
At 6 months of infant age, probiotic (NO/YES) groups also had different fecal microbiome compositions, but only for the subgroup that had postnatal maternal probiotic exposure after 1 month of age (Table 1). This may indicate that probiotic exposure closer to the time of sample collection is more important to fecal microbiome variation. The association of 6-month microbiome variation with more recent exposure of infants to maternal postnatal probiotics remained significant when the statistical model was adjusted for infant probiotics, the only covariable that differed between probiotic groups for this exposure time. With respect to taxa abundances, maternal postnatal and total probiotic exposure were associated with higher abundance of fecal Bifidobacteria in 6-month-olds. There were no differences in Lactobacillus abundances for any timing category (Table 1).
3.4 Recent postnatal maternal probiotic exposure was associated with variation in ERP responses in 6-month-old infants
To compare infant recognition memory function in probiotic groups, ERP components of interest were extracted from infant EEGs that were obtained in response to auditory (1 month) or visual (6 month) stimuli as described in the Methods. We observed larger slow wave difference scores in 3 of 4 scalp EEG regions, for 6-month-old infants exposed to more recent maternal probiotics (after 1 month of age) (Figure 4; Table 1), potentially indicating a greater stimulus discrimination. This subgroup of infants with larger difference scores and more recent exposure to maternal probiotics also had different 6-month fecal microbiome compositions (PERMANOVA, p = 0.02, n = 39 NO probiotic and n = 7 YES probiotic) and lower breastmilk IL-6 levels (Welch’s t-test, p = 0.004, Cohen’s d 1.14, CI [0.35, 1.90], n = 39 NO probiotic and n = 7 YES probiotic). In this subgroup, Lactobacillus and Bifidobacteria abundances did not differ in 6-month feces between probiotic NO/YES groups. We were unable to test for associations with other outcomes (milk CRP and 1-month fecal microbiome features) in this subgroup due to a lack of adequate subject numbers in the probiotics YES group to enable a statistical comparison. We also observed ERP differences (in late slow wave difference scores) at 6 months of age for the prenatal probiotic timing category, but only in the left central scalp lead (Table 1). For 1 month ERP testing, we did not find any significant associations between probiotic exposure (any timing group) and differences in ERP features in any scalp electrode region.
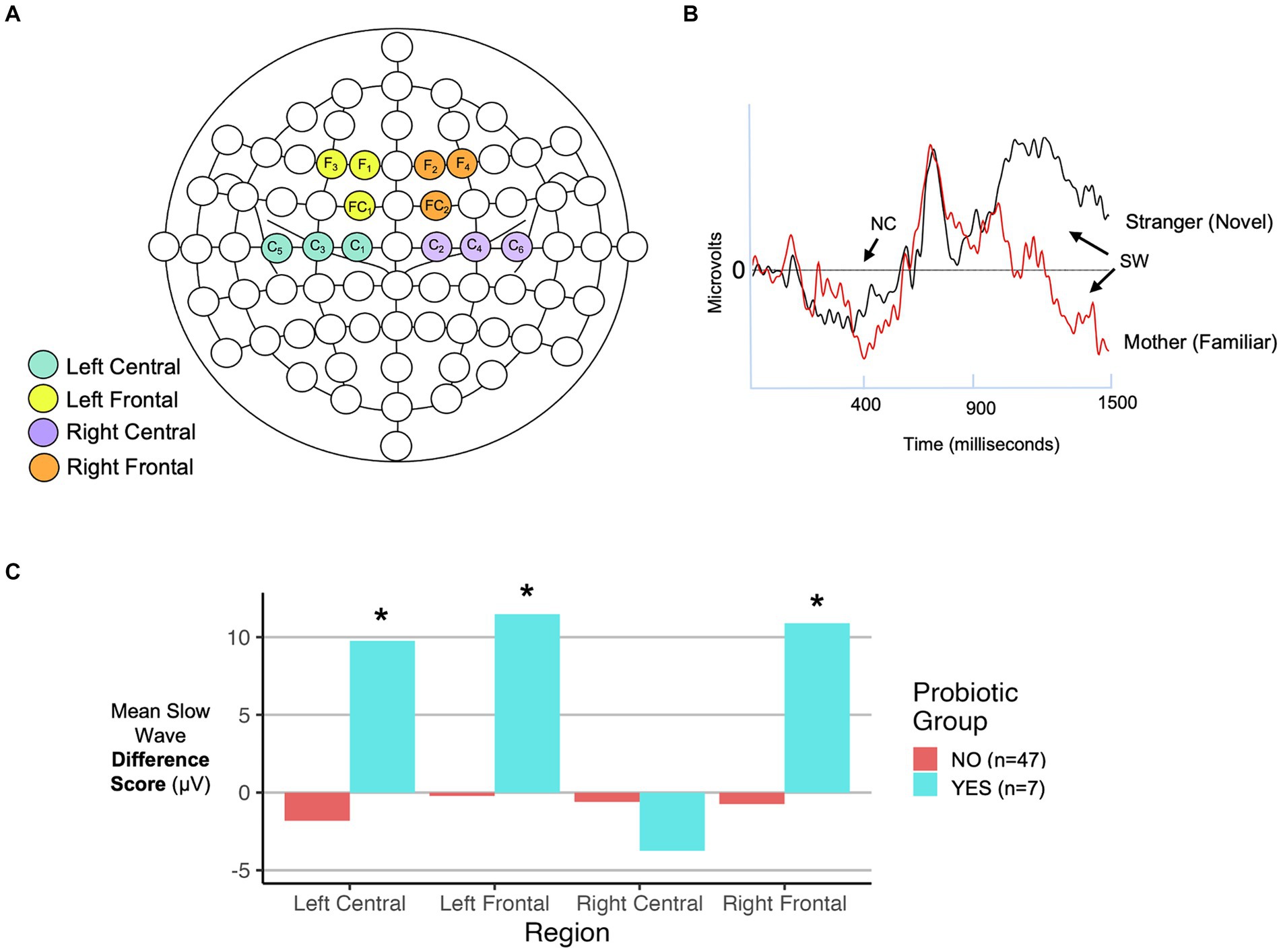
Figure 4. Infant ERP analysis features. (A) EEG net map with electrodes highlighted that correspond to scalp regions analyzed in this study. (B) Representative ERP recording with locations of the negative component (NC) peak at approximately 400 ms (350–700 ms time range analyzed) and slow wave (SW) from 900-1500 ms indicated with arrows. Two responses are shown: one to a familiar (own mother-based) and one to a novel (stranger-based) stimulus. (C) Comparison of SW difference scores (familiar - novel) by probiotic exposure group in 6-month-old infants for each of the four scalp regions indicated in panel (A). Results are shown for infants exposed to recent postnatal maternal probiotics. Details of statistical analyses are included in the Table 1. *p < 0.05.
4 Discussion
Consistent with the Developmental Origins of Health and Disease Hypothesis, nutritional factors are one type of environmental exposure during early life that have the potential to affect long-term health outcomes. Establishment of gut microbial communities during infancy is largely shaped by diet, and probiotics are thought to promote healthy microbiomes that confer health benefits. The overarching objective of our research is to explore how microbes, a type of nutritional outcome/factor, are linked to the development of brain function during early human life. In this pilot observational study, we aimed to characterize how maternal dietary supplementation with probiotics is associated with several features that have been proposed to be important for the function of the Microbiome-Gut-Brain Axis (1, 31, 33) namely inflammation (via assay of breastmilk inflammation-related proteins), the gut microbiome of infants, and infant brain function. To enhance the potential, for hypothesis generation for future studies, we maximized comparison group sizes by initially analyzing for associations between maternal probiotic exposure and each outcome individually. We found associations between maternal probiotic use and (1) decreased breastmilk CRP and IL-6 levels, (2) differences in infant fecal microbiomes at 1- and 6- months of age, and (3) differences in ERP responses for a subgroup of infants with more recent exposure to maternal probiotics. In an exploratory subgroup analysis of these infants with larger ERP difference scores at 6 months of age and a complete data set for all outcomes, maternal probiotic exposure was also associated with decreased IL-6 levels as well as variation in 6-month fecal microbiome composition. These findings suggest that probiotic usage in pregnant and lactating mothers may alter the Gut-Microbiome-Brain Axis in human infants.
Probiotic supplementation during pregnancy and lactation has been associated with a reduction in maternal inflammation-associated proteins. Pregnant women who consumed probiotic yogurt had decreased levels of CRP in their blood (27, 28), and decreased levels of breastmilk IL-6 were reported in women using probiotics during late pregnancy and lactation (61). We also found decreased CRP and IL-6 levels in the breastmilk of mothers exposed to probiotics, consistent with these prior reports. The mechanism by which maternal probiotics influence breastmilk inflammation-associated proteins has not been clearly defined. One potential hypothesis is that probiotic organisms, or altered microbiomes resulting from probiotic organisms, modulate inflammatory responses in the maternal gut which are then transferred from the gut to the systemic circulation and then to the breastmilk. Alternatively, probiotic-altered maternal gut microbiomes could be transferred to the breast, as has been proposed in the enteromammary circulation mechanism (62), to affect inflammation responses directly at that site. Although we did not find modulation of breastmilk microbiomes to be associated with probiotic exposure, as has been reported by other studies (63), this does not eliminate the possibility that changes to milk microbes and/or their functions (not able to be detected by our methods) are playing a role in the regulation of milk cytokine levels by maternal probiotics.
Maternal probiotic supplementation was also associated with variation in the composition of infant gut microbiomes at both 1 and 6 months of age in our study cohort, consistent with previous reports. Of note, Lactobacillus and Bifidobacteria were more highly abundant in 1-month fecal microbiomes of infants exposed to maternal probiotics, and Bifidobacteria was more abundant in 6-month-old infants exposed to postnatal maternal probiotics. Lactobacillus and Bifidobacteria, components of many of the probiotic supplements used by the women in our cohort, are beneficial bacteria that have been associated with decreased gut inflammation and improved health outcomes in infants, including atopic dermatitis and food allergy (64), necrotizing enterocolitis (65) and infantile colic (66). Consistent with our results, a recent meta-analysis reported that probiotic supplementation of mothers during pregnancy and lactation is associated with increased abundances of the probiotic bacteria in their infants’ feces, with abundances peaking near the first month of life (67). Although it appears that probiotic organisms are not sustained long-term in the infant gut, it remains unclear if early, transient colonization could be sufficient to modulate long-term outcomes.
The hippocampus, a brain region involved in recognition learning and memory, has been shown in rodent studies to be a primary brain structure affected by gut microbes (1, 35, 36). In addition, a recent study (68) reported that species within the Bifidobacterium genus isolated from infant feces along with specific human milk oligosaccharides from their mothers’ milk, were associated with early cognitive development, highlighting the idea that specific microbes may play a role in neurodevelopment during critical periods (69–71). Based on these prior studies, we hypothesized that ERP measures of recognition memory may differ based on exposure to maternal probiotics in human infants. We observed that in 6-month-old infants who were exposed to more recent maternal probiotics, the difference in ERP response (slow wave mean area amplitude) to familiar and novel stimuli was significantly larger for the majority of scalp regions measured. The larger difference score in these infants may indicate a greater ability to discriminate between familiar and novel stimuli in the probiotic-exposed group, or, in other words, enhanced recognition memory function (56, 59). This altered recognition memory could be due to structural or functional changes in hippocampus-based brain circuits associated with the Microbiome-Gut-Brain axis. The exact processes underlying how gut microbes affect brain function are not yet completely understood, but proposed mechanisms in animals include modulation of expression of host and/or microbial genes and proteins involved in neurotransmission, synaptic plasticity [e.g., Brain Derived Neurotrophic Factor (BDNF)], metabolism (e.g., short-chain fatty acids), stress hormones, gut-brain neuronal signaling via the vagus nerve, and brain structure (1, 2, 36, 72).
This pilot study has limitations. First, because of the observational study design, our results could be affected by a lack of consistency in the type and duration of probiotic exposure as well as the relative low number of infants exposed to maternal probiotics as compared to those who were not. Follow-on controlled clinical trial studies are needed to validate and expand on the findings of our study. Second, the lower subject numbers from which we had a complete set of outcome data limited our ability to characterize complex interactions among all outcomes. Third, complete sample and outcome collection at more frequent time points and over a longer period of time (e.g., first 2 years of life) would have afforded the opportunity to better identify time-dependent outcome associations with probiotic exposure. Our study also has key strengths. First, we leveraged a unique cohort of exclusively breastfeeding mother-infant dyads; thus, infant results were not confounded by variation in their diet (addition of formula or solid foods). Second, we used ERPs to assess brain (recognition memory) function in infants. ERP can be performed in much younger (preverbal) infants than traditional cognitive testing; assessment closer to the time of birth has the potential to identify relationships between exposures (relevant specifically to the breastfeeding period) and brain function with minimum confounding or dilution of effects by subsequent exposures. ERP responses have been validated in small cohort sizes to be able to detect important differences between groups (38, 59, 73). Overall, we argue that this study adds to the growing literature regarding maternal probiotic use to optimize infant outcomes. These preliminary data are important in that they have provided additional hypotheses that can be used to design future studies aiming to develop early probiotic supplementation strategies to optimize infant neurodevelopmental outcomes.
Data availability statement
Sequence and code data sets presented in this study can be found in online repositories. Raw DNA sequence data sets for this study are available in BioProject PRJNA880162 at NCBI (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA880162/). Computer code for statistical models and experimental data tables are available at: https://github.com/CGaleLab/Probiotic_Microbiome_Infant_Neurodevelopment.
Ethics statement
The studies involving humans were approved by University of Minnesota Institutional Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
SG: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. TH: Conceptualization, Data curation, Methodology, Writing – review & editing. NM: Data curation, Formal analysis, Investigation, Writing – review & editing. JH: Data curation, Writing – review & editing. LH: Conceptualization, Writing – review & editing. MG: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. DF: Funding acquisition, Investigation, Resources, Writing – review & editing. DK: Conceptualization, Methodology, Supervision, Writing – review & editing. KJ: Resources, Supervision, Writing – review & editing. ES: Methodology, Project administration, Writing – review & editing. ED: Conceptualization, Data curation, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. CG: Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing. MS: Conceptualization, Funding acquisition, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. We acknowledge the support of the following: the U of MN Department of Pediatrics Progressive grant (MHS), NIH R01 HD080444 (MILk study) (DF and EWD), NIH R01 HD109830 (CAG and EWD), NIH R21 HD099473 (CAG, DK, MKG), the Masonic Children’s Hospital Research Fund for Neurodevelopment and Child Mental Health Research, (CAG, DK, EWD, MKG) and the University of MN Department of Pediatrics “R” Award (CAG, DK, MKG).
Acknowledgments
The authors would like to acknowledge and thank all the women and health care providers who contributed to the MILk study. We also acknowledge the valuable assistance of Lauren Asfaw and Stephanie Mackenthun in the University of Minnesota Division of Maternal- Fetal Medicine, Kristin Sandness and the resources of the Center for Neurobehavioral Development, Rebecca Hollister from the Center for Pediatric Obesity at the University of Minnesota, the Clinical and Translational Research Services support team at the Clinical and Translational Science Institute at the University of Minnesota (supported by grant number UL1TR002494 from the National Institutes of Health’s National Center for Advancing Translational Sciences), laboratory resources from the University of Oklahoma Health Sciences Center, and Elisabeth Seburg, and the data collection and information technology team at the HealthPartners Institute. The authors also acknowledge use of the following resources: Diet*Calc Analysis Program, Version 1.5.0. National Cancer Institute, Epidemiology and Genomics Research Program. October 2012. Diet History Questionnaire, Version 2.0. National Institutes of Health, Epidemiology and Genomics Research Program, National Cancer Institute. 2010, and the DHQ Nutrient Database. dhq2.database.092914.csv. National Cancer Institute, Epidemiology and Genomics Research Program. Study data were collected and managed using REDCap electronic data capture tools hosted at the University of Minnesota. REDCap (Research Electronic Data Capture) is a secure, web-based software platform designed to support data capture for research studies (Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)-A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381.; Harris PA, Taylor R, Minor BL, et al. The REDCap consortium: Building an international community of software platform partners. J Biomed Inform. 2019;95).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2024.1456111/full#supplementary-material
References
1. Heijtz, RD, Wang, S, Anuar, F, Qian, Y, Björkholm, B, Samuelsson, A, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci. (2011) 108:3047–52. doi: 10.1073/pnas.1010529108
2. Sudo, N, Chida, Y, Aiba, Y, Sonoda, J, Oyama, N, Yu, XN, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice: commensal microbiota and stress response. J Physiol. (2004) 558:263–75. doi: 10.1113/jphysiol.2004.063388
3. Cho, I, Yamanishi, S, Cox, L, Methé, BA, Zavadil, J, Li, K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. (2012) 488:621–6. doi: 10.1038/nature11400
4. Cox, LM, Yamanishi, S, Sohn, J, Alekseyenko, AV, Leung, JM, Cho, I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. (2014) 158:705–21. doi: 10.1016/j.cell.2014.05.052
5. Dominguez-Bello, MG, Costello, EK, Contreras, M, Magris, M, Hidalgo, G, Fierer, N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci. (2010) 107:11971–5. doi: 10.1073/pnas.1002601107
6. Fouhy, F, Guinane, CM, Hussey, S, Wall, R, Ryan, CA, Dempsey, EM, et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother. (2012) 56:5811–20. doi: 10.1128/AAC.00789-12
7. Mueller, NT, Bakacs, E, Combellick, J, Grigoryan, Z, and Dominguez-Bello, MG. The infant microbiome development: mom matters. Trends Mol Med. (2015) 21:109–17. doi: 10.1016/j.molmed.2014.12.002
8. Singh, SB, Madan, J, Coker, M, Hoen, A, Baker, ER, Karagas, MR, et al. Does birth mode modify associations of maternal pre-pregnancy BMI and gestational weight gain with the infant gut microbiome? Int J Obes. (2020) 44:23–32. doi: 10.1038/s41366-018-0273-0
9. Franzago, M, La Rovere, M, Guanciali, P, Vitacolonna, E, and Stuppia, L. Epgenetics and human reproduction: the primary prevention of the noncommunicable diseases. Epigenomics. (2019) 11:1441–60. doi: 10.2217/epi-2019-0163
10. Li, Y . Epigenetic mechanisms link maternal diets and gut microbiome to obesity in the offspring. Front Genet. (2018) 9:342. doi: 10.3389/fgene.2018.00342
11. Indrio, F, Martini, S, Francavilla, R, Corvaglia, L, Cristofori, F, Mastrolia, SA, et al. Epigenetic matters: the link between early nutrition, microbiome, and Long-term health development. Front Pediatr. (2017) 5:178. doi: 10.3389/fped.2017.00178
12. Vitacolonna, E, Masulli, M, Palmisano, L, Stuppia, L, and Franzago, M. Inositols, probiotics, and gestational diabetes: clinical and epigenetic aspects. Nutrients. (2022) 14:1543. doi: 10.3390/nu14081543
13. Jarde, A, Lewis-Mikhael, AM, Moayyedi, P, Stearns, JC, Collins, SM, Beyene, J, et al. Pregnancy outcomes in women taking probiotics or prebiotics: a systematic review and meta-analysis. BMC Pregnancy Childbirth. (2018) 18:14. doi: 10.1186/s12884-017-1629-5
14. Sheyholislami, H, and Connor, KL. Are probiotics and prebiotics safe for use during pregnancy and lactation? A systematic review and Meta-analysis. Nutrients. (2021) 13:2382. doi: 10.3390/nu13072382
15. Mastromarino, P, Capobianco, D, Miccheli, A, Praticò, G, Campagna, G, Laforgia, N, et al. Administration of a multistrain probiotic product (VSL#3) to women in the perinatal period differentially affects breast milk beneficial microbiota in relation to mode of delivery. Pharmacol Res. (2015) 95-96:63–70. doi: 10.1016/j.phrs.2015.03.013
16. Grzeskowiak, L, Gronlund, M-M, Beckmann, C, Salminen, S, von Berg, A, and Isolauri, E. The impact of perinatal probiotic intervention on gut microbiota: double blind placebo-controlled trials in Finland and Germany. Anaerobe. (2012) 18:7–13. doi: 10.1016/j.anaerobe.2011.09.006
17. Dotterud, CK, Avershina, E, Sekelja, M, Simpson, MR, Rudi, K, Storrø, O, et al. Does maternal perinatal probiotic supplementation Alter the intestinal microbiota of mother and child? J Pediatr Gastroenterol Nutr. (2015) 61:200–7. doi: 10.1097/MPG.0000000000000781
18. Luoto, R, Laitinen, K, Nermes, M, and Isolauri, E. Impact of maternal probiotic-supplemented dietary counselling on pregnancy outcome and prenatal and postnatal growth: a double-blind, placebo-controlled study. Br J Nutr. (2010) 103:1792–9. doi: 10.1017/S0007114509993898
19. Laitinen, K, Poussa, T, and Isolauri, Ethe Nutrition, Allergy, Mucosal immunology and intestinal microbiota group. Probiotics and dietary counselling contribute to glucose regulation during and after pregnancy: a randomised controlled trial. Br J Nutr. (2008) 101:1679–87. doi: 10.1017/S0007114508111461
20. Brantsaeter, AL, Myhre, R, Haugen, M, Myking, S, Sengpiel, V, Magnus, P, et al. Intake of probiotic food and risk of preeclampsia in Primiparous women: the Norwegian mother and child cohort study. Am J Epidemiol. (2011) 174:807–15. doi: 10.1093/aje/kwr168
21. Reid, G, and Bocking, A. The potential for probiotics to prevent bacterial vaginosis and preterm labor. Am J Obstet Gynecol. (2003) 189:1202–8. doi: 10.1067/S0002-9378(03)00495-2
22. Doege, K, Grajecki, D, Zyriax, BC, Detinkina, E, Zu Eulenburg, C, and Buhling, KJ. Impact of maternal supplementation with probiotics during pregnancy on atopic eczema in childhood – a meta-analysis. Br J Nutr. (2012) 107:1–6. doi: 10.1017/S0007114511003400
23. Bertelsen, RJ, Brantsæter, AL, Magnus, MC, Haugen, M, Myhre, R, Jacobsson, B, et al. Probiotic milk consumption in pregnancy and infancy and subsequent childhood allergic diseases. J Allergy Clin Immunol. (2014) 133:165–171.e8. doi: 10.1016/j.jaci.2013.07.032
24. Zuccotti, G, Meneghin, F, Aceti, A, Barone, G, Callegari, ML, Di Mauro, A, et al. Probiotics for prevention of atopic diseases in infants: systematic review and meta-analysis. Allergy. (2015) 70:1356–71. doi: 10.1111/all.12700
25. Wickens, K, Barthow, C, Mitchell, EA, Kang, J, Van Zyl, N, Purdie, G, et al. Effects of Lactobacillus rhamnosus HN001 in early life on the cumulative prevalence of allergic disease to 11 years. Pediatr Allergy Immunol. (2018) 29:808–14. doi: 10.1111/pai.12982
26. Baldassarre, M, Palladino, V, Amoruso, A, Pindinelli, S, Mastromarino, P, Fanelli, M, et al. Rationale of probiotic supplementation during pregnancy and neonatal period. Nutrients. (2018) 10:1693. doi: 10.3390/nu10111693
27. Kwok, KO, Fries, LR, Silva-Zolezzi, I, Thakkar, SK, Iroz, A, and Blanchard, C. Effects of probiotic intervention on markers of inflammation and health outcomes in women of reproductive age and their children. Front Nutr. (2022) 9:889040. doi: 10.3389/fnut.2022.889040
28. Asemi, Z, Jazayeri, S, Najafi, M, Samimi, M, Mofid, V, Shidfar, F, et al. Effect of daily consumption of probiotic yoghurt on lipid profiles in pregnant women: a randomized controlled clinical trial. Pak J Biol Sci. (2011) 14:476–82. doi: 10.3923/pjbs.2011.476.482
29. Meyer, AL, Elmadfa, I, Herbacek, I, and Micksche, M. Probiotic, as well as conventional yogurt, can enhance the stimulated production of proinflammatory cytokines. J Hum Nutr Diet. (2007) 20:590–8. doi: 10.1111/j.1365-277X.2007.00807.x
30. Lin, YP, Thibodeaux, CH, Peña, JA, Ferry, GD, and Versalovic, J. Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun. Inflamm Bowel Dis. (2008) 14:1068–83. doi: 10.1002/ibd.20448
31. Borre, YE, O’Keeffe, GW, Clarke, G, Stanton, C, Dinan, TG, and Cryan, JF. Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol Med. (2014) 20:509–18. doi: 10.1016/j.molmed.2014.05.002
32. Moloney, RD, Desbonnet, L, Clarke, G, Dinan, TG, and Cryan, JF. The microbiome: stress, health and disease. Mamm Genome. (2014) 25:49–74. doi: 10.1007/s00335-013-9488-5
33. Guida, F, Turco, F, Iannotta, M, De Gregorio, D, Palumbo, I, Sarnelli, G, et al. Antibiotic-induced microbiota perturbation causes gut endocannabinoidome changes, hippocampal neuroglial reorganization and depression in mice. Brain Behav Immun. (2018) 67:230–45. doi: 10.1016/j.bbi.2017.09.001
34. Desbonnet, L, Clarke, G, Shanahan, F, Dinan, TG, and Cryan, JF. Microbiota is essential for social development in the mouse. Mol Psychiatry. (2014) 19:146–8. doi: 10.1038/mp.2013.65
35. Gareau, MG, Wine, E, Rodrigues, DM, Cho, JH, Whary, MT, Philpott, DJ, et al. Bacterial infection causes stress-induced memory dysfunction in mice. Gut. (2011) 60:307–17. doi: 10.1136/gut.2009.202515
36. Clarke, G, Grenham, S, Scully, P, Fitzgerald, P, Moloney, RD, Shanahan, F, et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry. (2013) 18:666–73. doi: 10.1038/mp.2012.77
37. O’Hagan, C, Li, JV, Marchesi, JR, Plummer, S, Garaiova, I, and Good, MA. Long-term multi-species Lactobacillus and Bifidobacterium dietary supplement enhances memory and changes regional brain metabolites in middle-aged rats. Neurobiol Learn Mem. (2017) 144:36–47. doi: 10.1016/j.nlm.2017.05.015
38. deRegnier, RA, Nelson, CA, Thomas, KM, Wewerka, S, and Georgieff, MK. Neurophysiologic evaluation of auditory recognition memory in healthy newborn infants and infants of diabetic mothers. J Pediatr. (2000) 137:777–84. doi: 10.1067/mpd.2000.109149
39. de Haan, M, and Nelson, CA. Recognition of the Mother’s face by six-month-old infants: a neurobehavioral study. Child Dev. (1997) 68:187–210.
40. Nelson, CA, Wewerka, SS, Borscheid, AJ, deRegnier, RA, and Georgieff, MK. Electrophysiologic evidence of impaired cross-modal recognition memory in 8-month-old infants of diabetic mothers. J Pediatr. (2003) 142:575–82. doi: 10.1067/mpd.2003.210
41. Riggins, T, Miller, NC, Bauer, PJ, Georgieff, MK, and Nelson, CA. Consequences of low neonatal Iron status due to maternal diabetes mellitus on explicit memory performance in childhood. Dev Neuropsychol. (2009) 34:762–79. doi: 10.1080/87565640903265145
42. Rose, SA, Feldman, JF, and Wallace, IF. Infant information processing in relation to six-year cognitive outcomes. Child Dev. (1992) 63:1126–41.
43. Friedrich, M, and Friederici, AD. Early N400 development and later language acquisition. Psychophysiology. (2006) 43:1–12. doi: 10.1111/j.1469-8986.2006.00381.x
44. Molfese, DL, and Molfese, VJ. Discrimination of language skills at five years of age using event-related potentials recorded at birth. Dev Neuropsychol. (1997) 13:135–56. doi: 10.1080/87565649709540674
45. Van Zuijen, TL, Plakas, A, Maassen, BAM, Maurits, NM, and Van Der Leij, A. Infant ERPs separate children at risk of dyslexia who become good readers from those who become poor readers. Dev Sci. (2013) 16:554–63. doi: 10.1111/desc.12049
46. Seery, AM, Vogel-Farley, V, Tager-Flusberg, H, and Nelson, CA. Atypical lateralization of ERP response to native and non-native speech in infants at risk for autism spectrum disorder. Dev Cogn Neurosci. (2013) 5:10–24. doi: 10.1016/j.dcn.2012.11.007
47. Heisel, T, Johnson, AJ, Gonia, S, Dillon, A, Skalla, E, Haapala, J, et al. Bacterial, fungal, and interkingdom microbiome features of exclusively breastfeeding dyads are associated with infant age, antibiotic exposure, and birth mode. Front Microbiol. (2022) 13:1050574. doi: 10.3389/fmicb.2022.1050574
48. Tahir, M, Haapala, J, Foster, L, Duncan, K, Teague, A, Kharbanda, E, et al. Higher maternal diet quality during pregnancy and lactation is associated with lower infant weight-for-length, body fat percent, and fat mass in early postnatal life. Nutrients. (2019) 11:632. doi: 10.3390/nu11030632
49. Krebs-Smith, SM, Pannucci, TE, Subar, AF, Kirkpatrick, SI, Lerman, JL, Tooze, JA, et al. Update of the healthy eating index: HEI-2015. J Acad Nutr Diet. (2018) 118:1591–602. doi: 10.1016/j.jand.2018.05.021
50. Reedy, J, Lerman, JL, Krebs-Smith, SM, Kirkpatrick, SI, Pannucci, TE, Wilson, MM, et al. Evaluation of the healthy eating Index-2015. J Acad Nutr Diet. (2018) 118:1622–33. doi: 10.1016/j.jand.2018.05.019
51. Whitaker, KM, Marino, RC, Haapala, JL, Foster, L, Smith, KD, Teague, AM, et al. Associations of maternal weight status before, during, and after pregnancy with inflammatory markers in breast Milk. Obesity. (2017) 25:2092–9. doi: 10.1002/oby.22025
52. Gohl, DM, Vangay, P, Garbe, J, MacLean, A, Hauge, A, Becker, A, et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol. (2016) 34:942–9. doi: 10.1038/nbt.3601
53. Al-Ghalith, GA, Hillmann, B, Ang, K, Shields-Cutler, R, and Knights, D. SHI7 is a self-learning pipeline for multipurpose short-read DNA quality control. mSystems. (2018) 3:e00202–17. doi: 10.1128/mSystems.00202-17
54. Vangay, P, Hillmann, BM, and Knights, D. Microbiome learning repo (ML repo): a public repository of microbiome regression and classification tasks. GigaScience. (2019) 8:giz042. doi: 10.1093/gigascience/giz042
55. Al-Ghalith, G, and Knights, D. BURST enables mathematically optimal short-read alignment for big data. (2020). Available at: http://biorxiv.org/lookup/doi/10.1101/2020.09.08.287128
56. Pfister, KM, Zhang, L, Miller, NC, Hultgren, S, Boys, CJ, and Georgieff, MK. ERP evidence of preserved early memory function in term infants with neonatal encephalopathy following therapeutic hypothermia. Pediatr Res. (2016) 80:800–8. doi: 10.1038/pr.2016.169
57. Hickey, MK, Miller, NC, Haapala, J, Demerath, EW, Pfister, KM, Georgieff, MK, et al. Infants exposed to antibiotics after birth have altered recognition memory responses at one month of age. Pediatr Res. (2021) 89:1500–7. doi: 10.1038/s41390-020-01117-7
58. DeBoer, T, Scott, LS, and Nelson, CA. In: Methodological handbook for research using event-related potentials. The MIT Press; Cambridge, MA: (2005). p. 263–297.
59. deRegnier, R, Wewerka, S, Georgieff, MK, Mattia, F, and Nelson, CA. Influences of postconceptional age and postnatal experience on the development of auditory recognition memory in the newborn infant. Dev Psychobiol. (2002) 41:216–25. doi: 10.1002/dev.10070
60. de Haan, M, and Nelson, CA. Brain activity differentiates face and object processing in 6-month-old infants. Dev Psychobiol. 35:1113–21. doi: 10.1037//0012-1649.35.4.1113
61. Baldassarre, M, Di Mauro, A, Mastromarino, P, Fanelli, M, Martinelli, D, Urbano, F, et al. Administration of a Multi-Strain Probiotic Product to women in the perinatal period differentially affects the breast Milk cytokine profile and may have beneficial effects on neonatal gastrointestinal functional symptoms. A randomized clinical trial. Nutrients. (2016) 8:677. doi: 10.3390/nu8110677
62. Selvamani, S, Dailin, D, Gupta, V, Wahid, M, Keat, H, Natasya, K, et al. An insight into probiotics bio-route: translocation from the Mother’s gut to the mammary gland. Appl Sci. (2021) 11:7247. doi: 10.3390/app11167247
63. Alemu, B, Azeze, G, Wu, L, Lau, S, Wang, C, and Wang, Y. Effects of maternal probiotic supplementation on breast milk microbiome and infant gut microbiome and health: a systematic review and meta-analysis of randomized controlled trials. Am J Obstet Gynecol MFM. (2023) 5:101148. doi: 10.1016/j.ajogmf.2023.101148
64. Cukrowska, B, Ceregra, A, Maciorkowska, E, Surowska, B, Zegadło-Mylik, MA, Konopka, E, et al. The effectiveness of probiotic Lactobacillus rhamnosus and Lactobacillus casei strains in children with atopic dermatitis and Cow’s Milk protein allergy: a multicenter, randomized, double blind, placebo controlled study. Nutrients. (2021) 13:1169. doi: 10.3390/nu13041169
65. Morgan, RL, Preidis, GA, Kashyap, PC, Weizman, AV, Sadeghirad, B, Chang, Y, et al. Probiotics reduce mortality and morbidity in preterm, low-birth-weight infants: a systematic review and network Meta-analysis of randomized trials. Gastroenterology. (2020) 159:467–80. doi: 10.1053/j.gastro.2020.05.096
66. Savino, F, Montanari, P, Galliano, I, Daprà, V, and Bergallo, M. Lactobacillus rhamnosus GG (ATCC 53103) for the Management of Infantile Colic: a randomized controlled trial. Nutrients. (2020) 12:1693. doi: 10.3390/nu12061693
67. Zaidi, AZ, Moore, SE, and Okala, SG. Impact of maternal nutritional supplementation during pregnancy and lactation on the infant gut or breastmilk microbiota: a systematic review. Nutrients. (2021) 13:1137. doi: 10.3390/nu13041137
68. Cho, S, Samuel, TM, Li, T, Howell, BR, Baluyot, K, Hazlett, HC, et al. Interactions between Bifidobacterium and Bacteroides and human milk oligosaccharides and their associations with infant cognition. Front Nutr. (2023) 10:1216327. doi: 10.3389/fnut.2023.1216327
69. Carlson, AL, Xia, K, Azcarate-Peril, MA, Goldman, BD, Ahn, M, Styner, MA, et al. Infant gut microbiome associated with cognitive development. Biol Psychiatry. (2018) 83:148–59. doi: 10.1016/j.biopsych.2017.06.021
70. Gao, W, Salzwedel, AP, Carlson, AL, Xia, K, Azcarate-Peril, MA, Styner, MA, et al. Gut microbiome and brain functional connectivity in infants-a preliminary study focusing on the amygdala. Psychopharmacology. (2019) 236:1641–51. doi: 10.1007/s00213-018-5161-8
71. Tamana, SK, Tun, HM, Konya, T, Chari, RS, Field, CJ, Guttman, DS, et al. Bacteroides-dominant gut microbiome of late infancy is associated with enhanced neurodevelopment. Gut Microbes. (2021) 13:1–17. doi: 10.1080/19490976.2021.1930875
72. Luczynski, P, Whelan, SO, O’Sullivan, C, Clarke, G, Shanahan, F, Dinan, TG, et al. Adult microbiota-deficient mice have distinct dendritic morphological changes: differential effects in the amygdala and hippocampus. Gaspar P, editor. Eur J Neurosci. (2016) 44:2654–66. doi: 10.1111/ejn.13291
Keywords: probiotics, gut microbiome, Infant, Inflammation, neurodevelopment, event related potential, breastmilk
Citation: Gonia S, Heisel T, Miller N, Haapala J, Harnack L, Georgieff MK, Fields DA, Knights D, Jacobs K, Seburg E, Demerath EW, Gale CA and Swanson MH (2024) Maternal oral probiotic use is associated with decreased breastmilk inflammatory markers, infant fecal microbiome variation, and altered recognition memory responses in infants—a pilot observational study. Front. Nutr. 11:1456111. doi: 10.3389/fnut.2024.1456111
Edited by:
Longgang Jia, Tianjin University of Science and Technology, ChinaReviewed by:
Qiong Jia, Peking University Third Hospital, ChinaAlena Sidenkova, Ural State Medical University, Russia
Copyright © 2024 Gonia, Heisel, Miller, Haapala, Harnack, Georgieff, Fields, Knights, Jacobs, Seburg, Demerath, Gale and Swanson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marie H. Swanson, aGljazAyNDVAdW1uLmVkdQ==
†Present addresses: Timothy Heisel, University of Minnesota Genomics Center (UMGC), Minneapolis, MN, United StatesJacob Haapala, HealthPartners Institute, Minneapolis, MN, United States
 Sara Gonia1
Sara Gonia1 Timothy Heisel
Timothy Heisel Lisa Harnack
Lisa Harnack David A. Fields
David A. Fields Dan Knights
Dan Knights Ellen W. Demerath
Ellen W. Demerath Cheryl A. Gale
Cheryl A. Gale Marie H. Swanson
Marie H. Swanson