- Department of Pharmacology, School of Pharmacy, Southwest Medical University, Luzhou, Sichuan, China
High sugar, high-fat diets and unhealthy lifestyles have led to an epidemic of obesity and obesity-related metabolic diseases, seriously placing a huge burden on socio-economic development. A deeper understanding and elucidation of the specific molecular biological mechanisms underlying the onset and development of obesity has become a key to the treatment of metabolic diseases. Recent studies have shown that the changes of bile acid composition are closely linked to the development of metabolic diseases. Bile acids can not only emulsify lipids in the intestine and promote lipid absorption, but also act as signaling molecules that play an indispensable role in regulating bile acid homeostasis, energy expenditure, glucose and lipid metabolism, immunity. Disorders of bile acid metabolism are therefore important risk factors for metabolic diseases. The farnesol X receptor, a member of the nuclear receptor family, is abundantly expressed in liver and intestinal tissues. Bile acids act as endogenous ligands for the farnesol X receptor, and erroneous FXR signaling triggered by bile acid dysregulation contributes to metabolic diseases, including obesity, non-alcoholic fatty liver disease and diabetes. Activation of FXR signaling can reduce lipogenesis and inhibit gluconeogenesis to alleviate metabolic diseases. It has been found that intestinal FXR can regulate hepatic FXR in an organ-wide manner. The crosstalk between intestinal FXR and hepatic FXR provides a new idea for the treatment of metabolic diseases. This review focuses on the relationship between bile acids and metabolic diseases and the current research progress to provide a theoretical basis for further research and clinical applications.
1 Introduction
Obesity is a chronic condition characterized by excessive fat accumulation in the body, resulting from a combination of genetic, environmental, and endocrine factors and presenting as an excess of body weight. With changes in economic conditions and improvements in living standards, it has emerged as a significant societal issue. The 2023 report from the World Obesity Federation predicts that by 2035, half of the global population will be overweight or obese (1). The prevalence of metabolic disorders associated with obesity, such as type 2 diabetes mellitus (T2DM), non-alcoholic fatty liver disease (NAFLD), hyperlipidemia (HLD), and hypertension (HTN), has experienced a significant increase. This has emerged as a pressing public health issue and places a substantial strain on healthcare systems globally (2, 3). There is accumulating evidence linking obesity and its related metabolic disorders to an perturbation in bile acid homeostasis, encompassingT2DM (4), NAFLD (5), HTN (6), and other conditions characterized by altered bile acid profiles. Bile acid, the primary constituents of bile, represent a broad category of acids synthesized by the liver through cholesterol metabolism (7–9). It plays a pivotal role in lipid metabolism, functioning as intestinal cleansers by emulsifying lipids to facilitate their digestion and absorption (10–13). Inadequate secretion of bile acids can impact lipid absorption and metabolism. Recent research has revealed that bile acids act as signaling molecules on their receptors to regulate glucose and lipid metabolism, inflammatory response, energy metabolism, and other pathways essential for maintaining body homeostasis (14–17). FXR, a well-studied receptor belonging to the nuclear receptor (NR) superfamily., serves as a transcription factor activated by BAs and tightly regulates the synthesis of BAs and their enterohepatic circulation. Disruption of bile acid metabolism can lead to aberrant FXR signaling, which is a significant predisposing factor in the pathogenesis of metabolic diseases, such as hyperlipidemia. Current investigations are focused on targeting BAs-FXR for the treatment of metabolic diseases by regulating bile acid levels to balance and restore the FXR signaling mechanism. It is important to note that FXR exhibits tissue specificity in regulating metabolic diseases, and its physiological functions being complex or even contradictory in different tissues. This article will first discuss the biological properties of bile acids and their relationship with metabolic diseases. Subsequently, we will focus on the latest research on the bile acid receptor FXR as a therapeutic target.
2 Biological characterization of bile acids
2.1 Bile acids synthesis
Bile acids are classified into primary and secondary bile acids based on their source (18–20). They can also be categorized as either conjugated or free, depending on their structural composition. Primary bile acids are synthesized in the liver from cholesterol through both the classical (neutral) and alternative (acidic) pathways. The classical pathway is initiated by cholesterol 7 alpha-hydroxylase (CYP7A1) and catalyzed by sterol 12 alpha-hydroxylase (CYP8B1) to produce bile acids (CA) and to a lesser extent, deoxycholic acid (CDCA) by sterol 27-hydroxylase (CYP27A1) (21). Production of chenodeoxycholic acid (CDCA) by the alternative pathway is catalyzed by CYP27A1 and oxysterol 7αhydroxylase (CYP7B1) (22–24) (Figure 1). It is noteworthy that bile acid species are closely related to species. In humans, CDCA remains unchanged, whereas in mice it is converted to α-murine cholic acid through the action of sterol 6β-hydroxylase (25, 26). Alpha-muricholic acid (α-MCA) forms beta-muricholic acid by isomerization of 7α-OH to 7β-OH (27, 28). Primary bile acids produced in hepatocytes have a C-24 carboxyl group that is bound to either taurine or glycine (29), resulting in the main type of bile acid in the liver being bound bile acid. Bile salt efflux pump (BSEP, coded by ABCB11) transports and stores bound bile acids in the gallbladder (30, 31). The intake of food regulates the synthesis of bile acids (32). Following meals, there is an increase in bile acid synthesis, leading to the production of 12-OH bile acids (bile acids with a hydroxyl structure at the C12 position, CA), which further facilitate the absorption of fat and cholesterol from intestinal foods for metabolic needs (33). Prolonged consumption of high-fat diets significantly elevates levels of 12-OH bile acids, contributing to the development of obesity phenotype (34, 35). Conversely, individuals who are resistant to obesity show increased concentrations of non-12-OH-bile acids, specifically CDCA (bile acids without a hydroxyl structure at the C12 position) (36). These studies suggest that activation of the bile acid replacement pathway may offer potential improvements for metabolic disorders.
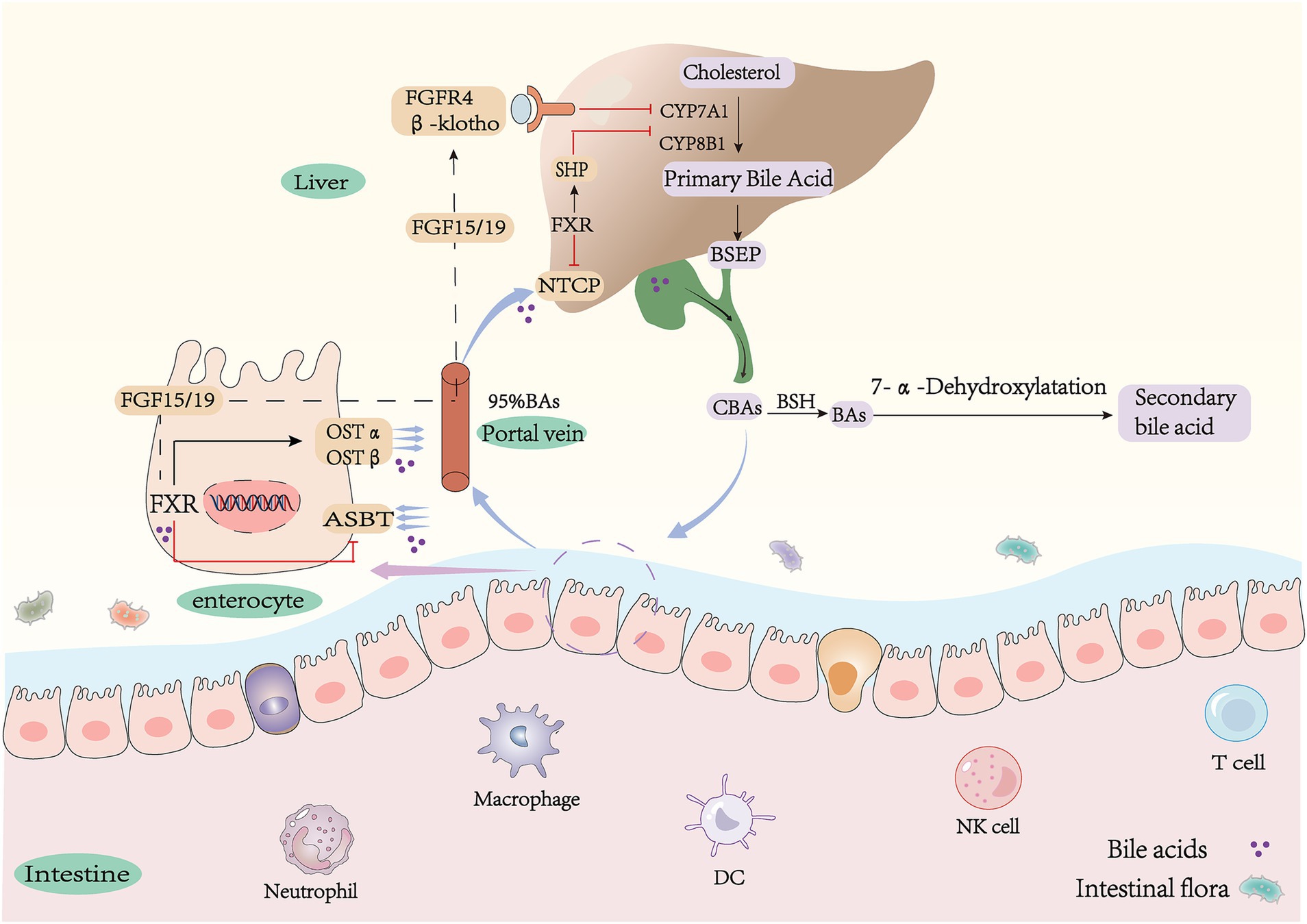
Figure 1. Synthesis and metabolism of bile acids. CA, cholic acid; CDCA, chenodeoxycholic acid; G(T)CA, glyco(tauro)- cholic acid; G(T)CDCA, glyco(tauro)- chenodeoxycholic acid; α(β)MCA, α(β)-muricholic acid; T(α/β)-MCA, tauro-α/β-muricholic acid; DCA, deoxychoic acid; LCA, lithocholic-acid; ω-MCA, ω-muricholic-acid; UDCA, ursodeoxycholic; HDCA, hyodeoxycholic acid; MDCA, murideoxy acid; CYP7A1, cholesterol 7α-hydroxylase; CYP8B1, sterol 12α-hydroxylase; CYP27A1, sterol 27-hydroxylase; CYP7B1, oxysterol 7α-hydroxylase; FGF15/19, fibroblast growth factor15/19; FXR, farnesoid X receptor; SHP, small heterodimer partner; NTCP, Na + −taurocholate co-transporting polypeptide; ASBT, apical sodium-dependent bile acid transporter; OSTα/β, organic solute transporter α/β.
2.2 Enterohepatic circulation of bile acids
Primary conjugated bile acids in the liver are transported and secreted into the gallbladder for storage by BSEP and multidrug resistance-associated protein 2 (MRP2). The stored bile acids flow into the intestine with bile. The majority of conjugated bile acids are reabsorbed into enterocytes through apical membrane sodium-dependent bile acid transporters (ASBT) in the terminal ileum. They then enter the portal circulation via basolateral heteromeric organic solute transporters α/β (OSTα/β) in the ileal epithelium (37–41). Only a small amount of bile acids are converted to more hydrophobic secondary bile acids through uncoupling and dehydroxylation, which is influenced by the gut microbiota (42, 43). Reabsorbed BAs are absorbed into hepatocytes via the sodium taurocholate cotransporter (NTCP) across the sinusoidal space surface (44). They are then converted to conjugated bile acids by hepatocytes and flow into the intestine with bile, which constitutes the enterohepatic circulation of bile acids (24, 45) (Figure 1). Bile acids facilitate the connection between the intestine and the liver through this process, which plays a significant role in maintaining the body’s balance of bile acid pool (46).
2.3 Gut flora mediates bile acid metabolism
The gut, as the largest digestive organ, harbors a diverse community of microbes, including bacteria, viruses, archaea, and fungi (47–49). The intestinal microbiota is composed of four major phyla: Bacteroidetes, Proteobacteria, Firmicutes, and Actinobacteria. Among these phyla, Bacteroidetes and Firmicutes are the most predominant and collectively account for approximately 90% of the total microbial population (50). This complex ecosystem plays a crucial role in food digestion and nutrient absorption while also producing various small molecule metabolites such as secondary bile acids, short-chain fatty acids, and indoles. These metabolites interact with the host to modulate immunity, energy metabolism, and other physiological pathways (51). Bile acids enter the lower part of the intestine and undergo further metabolism by the gut microbiota to form secondary bile acids (52), These modifications include deconjugation, dehydroxylation, oxidation, or epimerization (10, 53). Bile salt hydrolase (BSH) deconjugation plays a pivotal role in the metabolism of bile acids (54–56). Conjugated bile acids in the ileum and colon are hydrolyzed into unconjugated forms by BSH (57, 58), leading to their conversion into free bile acids. BSH is predominantly expressed in Lactobacillus, Clostridium, Bifidobacterium, and Bacteroidetes (59–62). Uncoupled free bile acids are converted to secondary bile acids through 7α-dehydroxylation by microorganisms (63, 64). The bacterial 7α dehydroxylase responsible for this process mainly comes from Clostridium XI and Clostridium XIVa and is catalyzed by the bacterial Bai operon (65–67). Secondary bile acids varied between species. In humans, CA and CDCA were converted to deoxycholic acid (DCA) and lithocholic acid (LCA) by removing the C7 hydroxyl group (57). In mice, α/β-MCA was converted to the common product murine deoxycholic acid (MDCA). β-MCA was isomerized by C6 to form ω-MCA, which was then dehydroxylated by 7α to form porcine deoxycholic acid (HDCA) (Figure 1). It is important to note that in mice, ursodeoxycholic acid (UDCA) is a primary bile acid because its Cyp2c70 oxidizes CDCA directly in the liver to produce UDCA. However, in humans, UDCA is a secondary bile acid derived from CDCA by redox in response to bacterial flora and is a differential isomer of CDCA (68, 69). The study shows that the bile acid metabolites in CYP2C9 humanized mice are similar to those in Cyp2c−/− mice, supporting this view (70). The composition of bile acids in different segments of the intestine is highly influenced by the presence of intestinal flora. Primary bile acids are predominantly found in the upper part of the intestine, whereas secondary bile acids are predominant in the lower part of the intestine (71). Depletion of intestinal flora results in a reduction of primary unconjugated bile acids and secondary bile acids. Consequently, intestinal flora increases the diversity of bile acids (72).
3 Bile acid-FXR axis regulates body metabolism
3.1 FXR regulates bile acid homeostasis
Bile acid receptors are divided into two major classes: nuclear receptors, including farnesoid X receptor (FXR), vitamin D receptor (VDR), pregnane X receptor (PXR) and constitutive androstane receptor (CAR); and membrane receptors, mainly including Takeda G protein-coupled receptor 5(TGR5) and sphingosine 1-phosphate receptor 2 (S1PR2). Among these, the farnesol X receptor (FXR) is one of the most studied. The gene encoding for FXRα is expressed in various tissues, such as the liver, intestine, kidney, and adrenal gland (73, 74). The highest expression is observed in the liver and intestine (10, 75). In humans, there are two genes, FXRα and FXRβ, with FXRα encoding four isoforms (FXRα1, FXRα2, FXRα3, and FXRα4). The expression of each isoform in different tissues was variable. FXRα1/2 was predominantly expressed in the liver (76), while FXRα3/4 isoforms were mainly expressed in the intestine (77). FXRβ is a pseudogene in humans (78). FXR has structures common to nuclear receptors, a ligand-independent transcriptional activation domain (AF1), a core DNA-binding domain (DBD), a hinge region, a C-terminal ligand-binding domain (LBD), and a ligand-dependent activation function domain (AF2) (79). Bile acids act as endogenous ligands for FXR, with varying abilities to activate the receptor (80, 81). CDCA was the most potent FXR agonist (EC50 10 μM), whereas DCA (EC50 50uM) and LCA (EC50 50uM) were much less able to activate FXR than CDCA (79). Due to the agonistic effect of bile acids on FXR, researchers have developed steroidal FXR agonists, which are mostly derivatives of bile acids. Among them, obeticholic acid (OCA) is the ethylated product of CDCA at the six-position, exhibiting a higher potency in binding to FXR and being 100 times more active than CDCA (79), and INT-767 is a semi-synthetic bile acid derivative that selectively agonises FXR (82). Currently, FXR antagonists are relatively backward, and most of them are natural products. For example, Tα-MCA and Tβ-MCA have been shown to be natural antagonists of FXR (22, 83, 84). Additionally, UDCA has been shown to inhibit FXR activity (85). The regulation of bile acid synthesis and distribution by FXR plays a crucial role in the body (86). Activation of ileal FXR leads to increased expression and secretion of enterocyte fibroblast growth factor 15 (FGF15, human homolog FGF19), which reaches the liver through the blood circulation and binds to fibroblast growth factor FGF receptor 4 (FGFR4) on the hepatocyte cell membrane to inhibit the expression of CYP7A1, and the activation of the hepatic FXR-dimeric chaperone (SHP) signaling pathway also reduces the expression of the enzyme which reduces hepatic bile acid synthesis and attenuates the toxic effects of bile acid overaccumulation on cells. FXR regulates both bile acid synthesis and transport. When there is an excess of bile acids in the liver, hepatic FXR is activated, leading to upregulation of BSEP to induce bile acid efflux and downregulation of NTCP expression to reduce reabsorption, thereby alleviating hepatic cholestasis (87). The negative feedback regulation of ASBT by FXR results in decreased intestinal and hepatic bile acid reabsorption, ultimately leading to increased excretion (88). Metabolic disease models have reported significant changes in bile acid profiles, which can be effectively alleviated by activating the FXR receptor (89). FXR plays a crucial role in regulating bile acid synthesis and enterohepatic circulation to maintain balanced distribution throughout the body and prevent potential health hazards.
3.2 FXR and glycolipid homeostasis
Bile acids and bile acid receptors play a crucial role in regulating glucolipid metabolism. The hepatic FXR is a significant therapeutic target for addressing NAFLD. Nonalcoholic fatty liver disease (NAFLD) is a liver metabolic disorder characterized by the excessive deposition of fatty liver cells due to factors such as alcohol and high-fat diet (12, 90, 91). Steatosis, which precedes the onset of NAFLD, resulting from increased hepatic uptake of fatty acids and lipid synthesis. During steatosis, low expression of FXR upregulates the expression of cholesterol regulatory element-binding protein-1C (SREBP-1C) through an SHP-dependent mechanism (92, 93), which in turn upregulates the expression of stearoyl coenzyme A desaturase 1 (SCD1), fatty acid synthase (FAS), and acetyl-coenzyme A carboxylase (ACC), ultimately leading to increased hepatic lipid synthesis (94, 95). Peroxisome proliferator-activated receptor α (PPAR-α), a key transcriptional regulator of lipid metabolism that promotes β-oxidation of fatty acids (96), is mainly expressed in the liver and its activity is also regulated by FXR. Carnitine palmitoyltransferase 1 (CPT1) acts as a rate-limiting enzyme for fatty acid β-oxidation, and low expression of FXR reduces PPARα and CPT1 expression on the one hand to decrease fatty acid oxidation, which reduces fat consumption resulting in hepatic lipid accumulation (97, 98), and on the other hand, promotes the expression of the scavenger receptor (CD36) to increase fatty acid uptake and directly regulates ApoC-II to decrease triglyceride hydrolysis thereby accelerating NAFLD (99, 100). Therefore, regulation of gene expression related to hepatic lipid metabolism could be a therapeutic target for metabolic diseases. It was found that FXR expression was reduced in the Male Wistar rats diabetes model. Phosphoenolpyruvate carboxykinase (PEPCK), glucose-6-phosphatase (G6Pase), and fructose-1,6-bisphosphatase (FBP1) are three crucial enzymes in the gluconeogenic pathway (101). They are all negatively regulated by FXR (Figure 2). Reduced hepatic FXR expression leads to an upregulation of gluconeogenesis, ultimately resulting in elevated blood glucose levels. Treatment with CDCA has been shown to reverse this phenomenon (102). Hepatic FXR−/− mice have been observed to exhibit dysregulated glucose homeostasis due to insulin resistance (103). Furthermore, activation of the enterohepatic FXR-Fgf15 axis inhibits hepatic gluconeogenesis and promotes glycogen synthesis (104). Recent studies have demonstrated that inhibition of intestinal FXR paradoxically increases glucagon-like peptide-1 (GLP-1) production and regulates glucose homeostasis (105, 106). GLP-1 is produced by intestinal cells as a hormone that stimulates insulin secretion and inhibits glucagon secretion. The inhibition of FXR-promoted GLP-1 production in enteroendocrine L cells is associated with this effect. It is noteworthy that activating FXR in pancreatic β-cells can enhance insulin secretion. Acarbose treatment has been shown to effectively reduce β-cell proliferation in diabetic mice, leading to a significant increase in insulin content and a decrease in insulin resistance. However, these effects were not observed in FXR-KO mice (107). The contradictory effects of FXR in intestinal L cells and pancreatic β-cells suggest that the FXR signaling pathway is tissue-specific, and different tissues may have opposing roles.
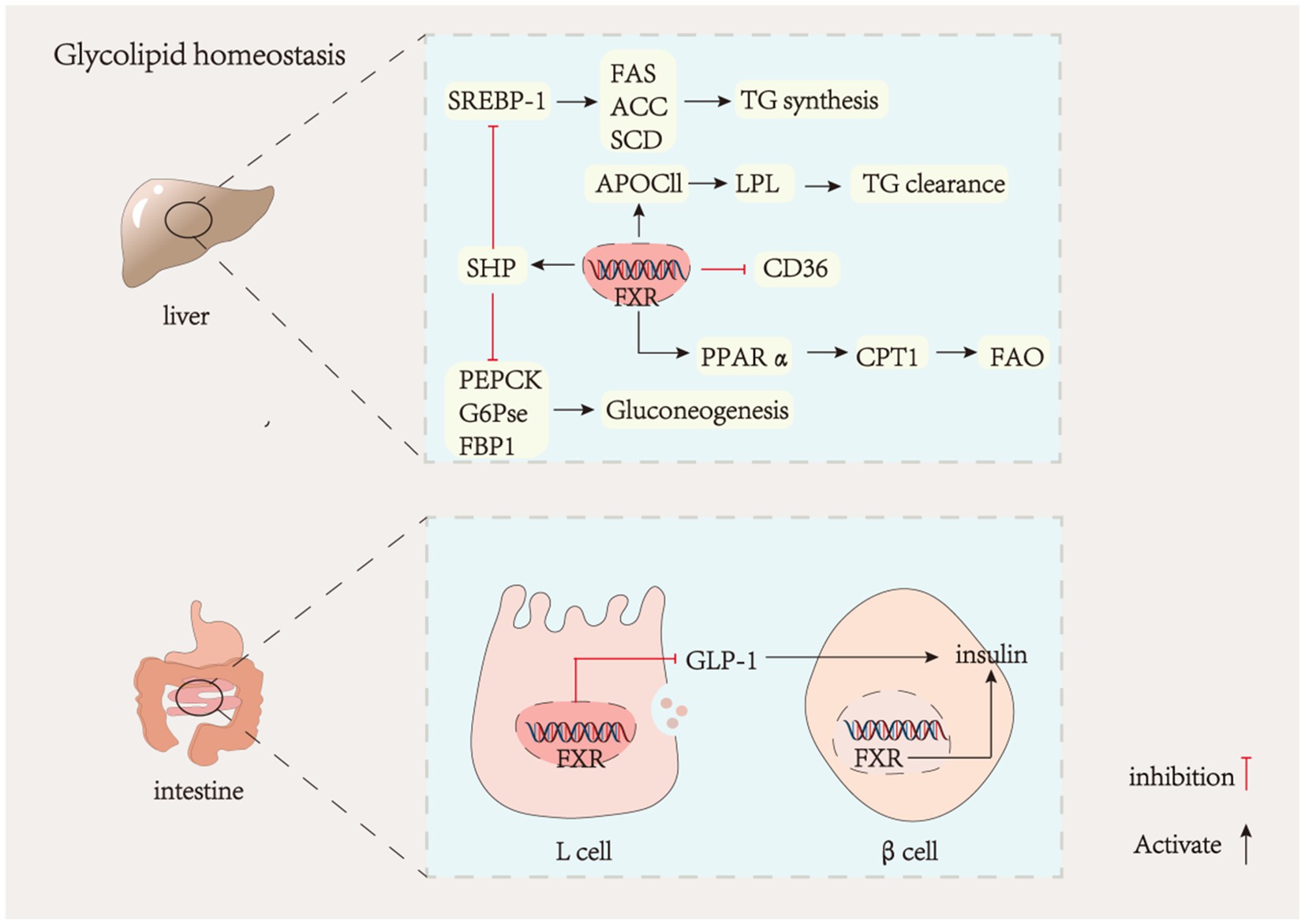
Figure 2. Liver FXR and glycolipid metabolism. GLP-1, glucagon-like peptide-1; SREBP-1, sterol regulatory element-binding protein-1; FAS, fatty acid synthase; ACC, acetyl-CoA carboxylase; SCD, stearoyl-CoA desaturase; APOCII, apolipoprotein C-II; CD36, platelet glycoprotein; PPAα, peroxisome proliferators-activated receptors; CPT1, carnitine acyl transferase I; FAO, fatty acid oxidation; PEPCK, phosphoenolpyruvate carboxy kinase; G6PCE, glucose-6-phosphatase; FBP1, phosphofructokinase.
3.3 FXR and immunity
Metabolic diseases are characterized by chronic inflammation. Research has demonstrated that excessive lipid accumulation in the body results in the release of numerous inflammatory factors, which hastens the onset of metabolic diseases (108–110). Obese individuals may experience a significant increase in secondary bile acids, specifically DCA. High concentrations of DCA can damage the microbiota membrane, leading to the release of lipopolysaccharide (LPS) and an increase in intestinal permeability. LPS can then cross the intestine and enter the bloodstream, activating Toll-like receptor (TLR) and causing endotoxemia, which results in the release of inflammatory factors (111, 112). Bile acids have a significant role in regulating inflammatory processes (113–115). The absence of FXR in mice causes inflammation in the body (116). FXR is believed to be associated with intestinal defenses against microbes and barrier function. The intestinal inflammation caused by Dextran Sulfate Sodium Salt (DSS) can be improved by the FXR agonist INT-747 (117). FXR is expressed in various immune cells, such as monocytes, macrophages, dendritic cells (DCs), and natural killer T cells (NKT) (118). FXR expression in immune cells has implications for their differentiation and cytokine production. Activation of FXR in macrophages inhibits NF-κB activation and reduces the release of inflammatory factors such as IL-1β and IL-6 (119). Meanwhile, it promotes the expression of IL-10, which contributes to the transformation of macrophages from M1-type to M2-type. FXR reduces the release of TNF-α in DC, inhibits its inflammatory signaling and promotes peripheral Treg cell production. In hepatic NKT cells, activation of FXR inhibits the release of the inflammatory factor IL-1β (63). Microbial-bile acids play a crucial role in the protective effect of intestinal flora against intestinal inflammation. Secondary bile acids are involved in immune cell differentiation and regulate the balance of Treg and Th17. Oral administration of bile acids inhibits the differentiation of TH17 cells, thereby alleviating DSS-induced colitis (120, 121). Activation of FXR inhibits inflammation by blocking the TLR4/NF-kB signaling pathway (122, 123). These findings suggest that FXR is crucial in maintaining normal intestinal barrier function and reducing hepatic inflammation (124, 125).
3.4 FXR and gut microbiota
The gut microbiota is widely recognized as a pivotal factor in regulating host health. Bile acids and gut microbiota have a mutually inhibitory and interdependent relationship. Bile acids exhibit an antibacterial effect, preventing the overgrowth of gut microbiota. High concentrations of hydrophobic bile acids exert direct antibacterial activity, primarily through cell membrane damage (126). Additionally, bile acids can also modulate the composition of intestinal flora by interacting with receptors. The activation of FXR by bile acids prevents bacterial overgrowth and ectopic (127). Reduced levels of bile acid may lead to intestinal bacterial overgrowth. Bile acid supplementation can protect the intestinal barrier by remodeling the composition of the intestinal flora (128). Conversely, the gut microbiota can alter the composition of the bile acid pool and rely on FXR signaling to regulate body metabolism (129). There is increasing evidence that gut flora and metabolites are significant factors in the development of metabolic diseases (130, 131). Recently, research on the interaction between gut microbiota and bile acid in host metabolism has gained popularity (132). A long-term Western diet can lead to an increased Firmicutes to Bacteroidetes ratio in gut microbiota, which can result in obesity (133, 134). Alterations in gut flora further affect bile acid metabolism and FXR signaling pathways, and the gut flora-BAs-FXR axis has been gradually elucidated, with levels of BSH-associated microbial enzymes negatively correlating with intestinal levels of bound bile acids. Pu-erh tea (135), diammonium glycyrrhizinate (136), and triterpene saponins (137) have been shown to improve metabolic disorders by altering the levels of bile salt hydrolase-containing flora. This results in an elevation of bound bile acids concentration, which counteracts the intestinal FXR pathway. Therefore, intestinal FXR is an important drug target for metabolic disease intervention and acts as a mediator of intestinal flora, which plays a crucial role in the body’s metabolism.
4 Metabolic diseases and the BA-FXR signaling pathway
4.1 Activation of bile acid receptor FXR improves metabolic disease mechanisms
There is a significant association between FXR and metabolic disease. FXR activation improves clinical markers, such as lowering cholesterol (TC) and triglyceride (TG) levels. The mechanism of nuclear receptor FXR in regulating hepatic glucose-lipid metabolism has been elucidated, and activation of FXR alleviates non-alcoholic fatty liver disease induced by high-fat diet in C57BL/6 male mice (138). This is consistent with the conclusion that GW4064 and CDCA (the most potent agonist of FXR) ameliorate bacitracin-induced ICR (CD-1) and obesity in C57BL/6 male mice (139). The mechanisms involved in this process include the inhibition of lipid synthesis via FXR-SHP-SREBP-1c and the promotion of fatty acid oxidation by FXR-PPAR-α. The regulation of both hepatic gluconeogenesis and intestinal FXR and hepatic FXR are also involved. The researchers from the University of California discovered that hepatic FXRs are primarily responsible for decreasing lipid synthesis, while intestinal FXRs decrease lipid absorption to safeguard against NAFLD and therefore have a lipid-lowering function. This demonstrates the collaborative action of hepatic and small intestinal FXR receptors in distinct mechanisms to reduce triglyceride accumulation in the liver (140, 141). Fexaramine, an FXR agonist targeting the gut, has been shown to induce the conversion of white adipose tissue to brown adipose tissue. This conversion is associated with an increase in the levels of LCA-producing bacteria, such as Acetobacter and Anaplasma spp. These bacteria activate the TGR5/GLP-1 signaling pathway (142, 143) (Figure 3). This leads to an improvement in obesity and insulin resistance. The activation of FGF15/19 by bile acids can trigger SHP, which in turn inhibits hepatic lipogenesis (144). Therefore, a compound known as OCA may serve as an effective treatment for NAFLD through its induction of 1-deoxy sphingolipid degradation and inhibition of bile acid synthesis (145).
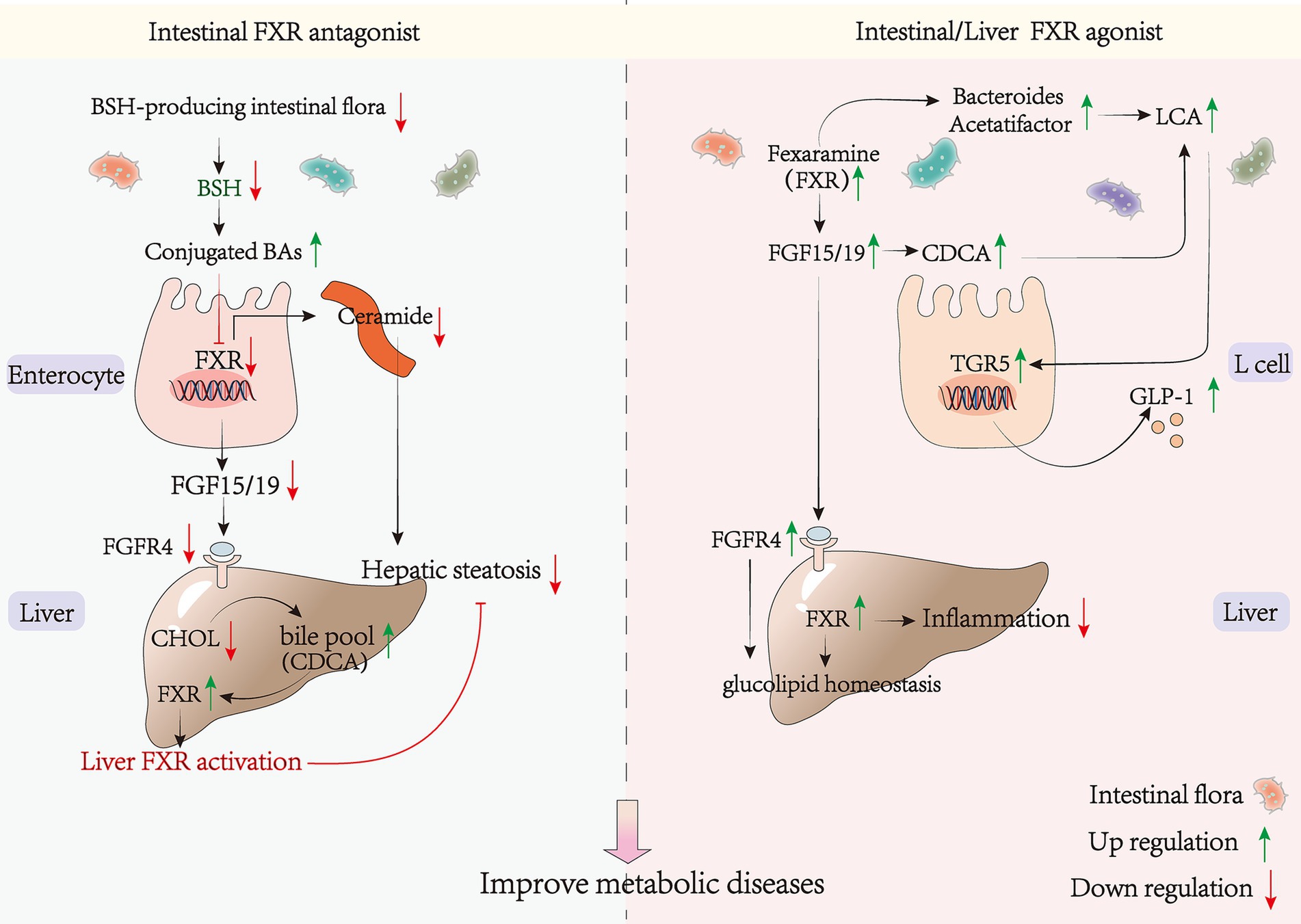
Figure 3. FXR agonists/antagonists and metabolic diseases. BSH, bile salt hydrolase; SMPD3, sphingomyelin phosphodiesterase 3; SPLC2, serine palmitoyltransferase; CERS6, ceramide synthase 6.
4.2 Inhibition of bile acid receptor FXR improves metabolic disease mechanisms
GW4064 is a synthetic non-steroidal FXR agonist. However, it has been found to treat mice on a high-fat diet while also accelerating the induction of obesity production and lipid accumulation (146). This suggests that the presence of other FXR signaling in the organism, with different regulatory mechanisms from the liver, may accelerate the development of metabolic diseases. Recent research has demonstrated that inhibition of intestinal FXR signaling can improve metabolic disorders by increasing hepatic bile acid pool, reducing cholesterol accumulation, and alleviating fatty liver (147). Inhibition of intestinal FXR can increase the hepatic bile acid pool, reduce cholesterol accumulation, and alleviate metabolic diseases (148–150). Furthermore, repression of intestinal FXR signaling through inhibition of bacterial bile salt hydrolase activity has shown promise in ameliorating metabolic disorders (151). Additionally, various interventions such as Pu-erh tea (135), chlorogenic acid (149), Lactobacillus plantarum LP104171 (150) and metformin (152) have been found to inhibit the intestinal FXR-FGF15 signaling pathway. This was achieved by decreasing the level of microorganisms containing BSH enzymes and increasing the level of bound bile acids such as T-α-MCA. T-β-MCA and GUDCA can increase bile acid biosynthesis, which in turn activates hepatic FXR and leads to a lipid-lowering effect (153). Therefore, promoting bile acid synthesis can reduce hypercholesterolemia (154) (Figure 3). Pu-erh tea inhibits the intestinal FXR signal and activates the liver replacement pathway to produce CDCA, which in turn activates hepatic FXR. This establishes a link between intestinal and hepatic FXR and elucidates the mechanism of the alternative pathway to ameliorate metabolic diseases. Ceramide, as a lipotoxic inducer of metabolic disorders, has been shown to increase ER stress through elevated serum levels (155–157). Sphingosine phosphodiesterase 3 (SMPD3) has been identified as a target gene of FXR (158). Inhibiting intestinal FXR can downregulate the expression of ceramide synthetase S6 (CERS6) and Serine Palmitoyltransferase Long Chain Base Subunit 2 (SPTLC2), leading to reduced ceramide synthesis. This decrease in ceramide levels may alleviate ER stress, upregulate citrate synthase (CS) expression, downregulate pyruvate carboxylase (PC)expression, and further inhibit gluconeogenesis, ultimately preventing the onset of diabetes (159) (Figure 3). Selective inhibition of the FXR signaling pathway in the gut, rather than the liver, or cross-organ enhancement of FXR signaling in the liver, can effectively avoid potential side effects and increase therapeutic efficacy. This also elucidates the potential for divergent therapeutic effects resulting from different routes of administration. For instance, the oral FXR agonist GW4064 has been shown to exacerbate weight gain and insulin resistance, while intraperitoneal injection yields opposite effects (146). This can be attributed to the pharmacokinetic understanding that oral administration may stimulate intestinal FXR through absorption, while intraperitoneal injection directly activates liver FXR via the circulatory system. Determining the role of FXR in the body’s metabolism is a complex task, especially in regards to whether activating intestinal FXR could improve metabolic diseases. Previous studies have produced conflicting results, indicating that the previously proposed mechanisms are insufficient to explain the diverse physiological functions of FXR in various tissues. It remains uncertain whether the adverse impacts of agonists are confined to specific tissues or if the overall effect of various mechanisms is influenced by governing factors. Further investigation is necessary to determine the specific molecular mechanisms involved.
5 Conclusion
Bile acids, derived from hepatic cholesterol metabolism, play a pivotal role in maintaining the body’s homeostasis. They serve not only as intestinal surfactants to facilitate the absorption of dietary lipids but also as signaling molecules that modulate pathways involved in insulin resistance, lipid metabolism, and inflammation. There is mounting evidence suggesting that individuals with diabetes, hyperlipidemia, and obesity display perturbed bile acid profiles and elevated levels of secondary bile acids. This leads to aberrant modulation of the intestinal FXR signaling pathway, resulting in lipid accumulation and insulin resistance, which exacerbates metabolic symptoms.
Currently, the treatment options for metabolic diseases are limited. Activation of liver FXR has shown potential in improving metabolic diseases by inhibiting lipogenesis and glycogenesis, making FXR agonists a promising new target for the treatment of conditions such as obesity and diabetes. The investigation of the FXR agonist OCA, along with nonsteroidal agonists, as potential therapeutic drugs for NAFLD/NASH is ongoing. However, it has been observed that the use of GW4064 agonist exacerbates metabolic symptoms, possibly because the agonist is not tissue-specific and activates FXR signaling at various sites throughout the body. Nonetheless, it is important to note that the intestinal FXR signaling pathway operates through different mechanisms. On one hand, activating intestinal FXR reduces lipid absorption and promotes hepatic gluconeogenesis across organs. On the other hand, inhibiting intestinal FXR increases GLP-I secretion from intestinal L cells, improving insulin sensitivity and increasing hepatic cholesterol metabolism. In recent years, the mechanism of the intestinal flora-BAs-FXR axis has been further elucidated. To improve metabolic diseases, the intestinal FXR pathway can be inhibited by decreasing the level of BSH enzyme-containing bacteria, which increases the binding of bile acids. This reveals the connection between intestinal flora, bile acid interactions, and body metabolism, constituting a well-established intestinal flora-BAs-FXR metabolism network. This presents a novel perspective for enhancing and managing. Nevertheless, the molecular mechanisms underlying the contradictory impact between this approach and the treatment of metabolic diseases with FXR agonists remain ambiguous, and the correlation between hepatic and intestinal FXR has not been firmly established. Given the diversity in bile acid pool composition in both mice and humans, further comprehensive investigation is imperative prior to translating findings from mouse experiments into clinical therapy. The above indicates that FXR may have opposing effects in various tissues and cells, highlighting the importance of considering tissue specificity when studying the paradoxical effects of FXR. It is anticipated that ligand drugs targeting FXR will prove efficacious in addressing metabolic diseases.
Author contributions
YL: Conceptualization, Writing – original draft. LW: Writing – review & editing. QY: Writing – review & editing. LL: Writing – review & editing. YX: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Project of the National Natural Science Foundation of China (Nos. 81873067 and 81102868), Luzhou Science and Technology Bureau (No. 2022-JYJ-124), and Southwest Medical University (No. 2020ZRQNB006).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Lobstein, T, Jackson-Leach, R, Powis, J, Brinsden, H, and Gray, M. World obesity atlas 2023. (2023). Available at: https://data.worldobesity.org/publications/?cat=19
2. Eley, VA, Thuzar, M, Navarro, S, Dodd, BR, and van Zundert, AA. Obesity, metabolic syndrome, and inflammation: An update for anaesthetists caring for patients with obesity. Anaesth Crit Care Pain Med. (2021) 40:100947. doi: 10.1016/j.accpm.2021.100947
3. Chew, NWS, Ng, CH, Tan, DJH, Kong, G, Lin, C, Chin, YH, et al. The global burden of metabolic disease: data from 2000 to 2019. Cell Metab. (2023) 35:414–428.e3. doi: 10.1016/j.cmet.2023.02.003
4. Tawulie, D, Jin, L, Shang, X, Li, Y, Sun, L, Xie, H, et al. Jiang-Tang-san-Huang pill alleviates type 2 diabetes mellitus through modulating the gut microbiota and bile acids metabolism. Phytomedicine. (2023) 113:154733. doi: 10.1016/j.phymed.2023.154733
5. Hong, T, Zou, J, He, Y, Zhang, H, Liu, H, Mai, H, et al. Bisphenol A induced hepatic steatosis by disturbing bile acid metabolism and FXR/TGR5 signaling pathways via remodeling the gut microbiota in CD-1 mice. Sci Total Environ. (2023) 889:164307. doi: 10.1016/j.scitotenv.2023.164307
6. Lei, S, He, S, Li, X, Zheng, B, Zhang, Y, and Zeng, H. Effect of lotus seed resistant starch on small intestinal flora and bile acids in hyperlipidemic rats. Food Chem. (2023) 404:134599. doi: 10.1016/j.foodchem.2022.134599
7. de Boer, JF, Bloks, VW, Verkade, E, Heiner-Fokkema, MR, and Kuipers, F. New insights in the multiple roles of bile acids and their signaling pathways in metabolic control. Curr Opin Lipidol. (2018) 29:194–202. doi: 10.1097/mol.0000000000000508
8. Lefort, C, and Cani, PD. The liver under the spotlight: bile acids and oxysterols as pivotal actors controlling metabolism. Cells. (2021) 10:400. doi: 10.3390/cells10020400
9. Portincasa, P, Di Ciaula, A, Garruti, G, Vacca, M, De Angelis, M, and Wang, DQ. Bile acids and GPBAR-1: dynamic interaction involving genes, environment and gut microbiome. Nutrients. (2020) 12:3709. doi: 10.3390/nu12123709
10. McGlone, ER, and Bloom, SR. Bile acids and the metabolic syndrome. Ann Clin Biochem. (2019) 56:326–37. doi: 10.1177/0004563218817798
11. Xue, R, Su, L, Lai, S, Wang, Y, Zhao, D, Fan, J, et al. Bile acid receptors and the gut-liver Axis in nonalcoholic fatty liver disease. Cells. (2021) 10:2806. doi: 10.3390/cells10112806
12. Jiao, TY, Ma, YD, Guo, XZ, Ye, YF, and Xie, C. Bile acid and receptors: biology and drug discovery for nonalcoholic fatty liver disease. Acta Pharmacol Sin. (2022) 43:1103–19. doi: 10.1038/s41401-022-00880-z
13. Simbrunner, B, Trauner, M, and Reiberger, T. Review article: therapeutic aspects of bile acid signalling in the gut-liver axis. Aliment Pharmacol Ther. (2021) 54:1243–62. doi: 10.1111/apt.16602
14. Chiang, JYL, and Ferrell, JM. Bile acid metabolism in liver pathobiology. Gene Expr. (2018) 18:71–87. doi: 10.3727/105221618x15156018385515
15. Chen, M, Guo, WL, Li, QY, Xu, JX, Cao, YJ, Liu, B, et al. The protective mechanism of Lactobacillus plantarum FZU3013 against non-alcoholic fatty liver associated with hyperlipidemia in mice fed a high-fat diet. Food Funct. (2020) 11:3316–31. doi: 10.1039/c9fo03003d
16. Krautkramer, KA, Fan, J, and Bäckhed, F. Gut microbial metabolites as multi-kingdom intermediates. Nat Rev Microbiol. (2021) 19:77–94. doi: 10.1038/s41579-020-0438-4
17. Ferrell, JM, and Chiang, JYL. Understanding bile acid signaling in diabetes: from pathophysiology to therapeutic targets. Diabetes Metab J. (2019) 43:257–72. doi: 10.4093/dmj.2019.0043
18. Wang, K, Liao, M, Zhou, N, Bao, L, Ma, K, Zheng, Z, et al. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. (2019) 26:222–235.e5. doi: 10.1016/j.celrep.2018.12.028
19. Singh, J, Metrani, R, Shivanagoudra, SR, Jayaprakasha, GK, and Patil, BS. Review on bile acids: effects of the gut microbiome, interactions with dietary Fiber, and alterations in the bioaccessibility of bioactive compounds. J Agric Food Chem. (2019) 67:9124–38. doi: 10.1021/acs.jafc.8b07306
20. Deng, X, Xiao, L, Luo, M, Xie, P, and Xiong, L. Intestinal crosstalk between bile acids and microbiota in irritable bowel syndrome. J Gastroenterol Hepatol. (2023) 38:1072–82. doi: 10.1111/jgh.16159
21. Li, D, Cui, Y, Wang, X, Liu, F, and Li, X. Apple polyphenol extract improves high-fat diet-induced hepatic steatosis by regulating bile acid synthesis and gut microbiota in C57BL/6 male mice. J Agric Food Chem. (2021) 69:6829–41. doi: 10.1021/acs.jafc.1c02532
22. Wahlström, A, Sayin, SI, Marschall, HU, and Bäckhed, F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. (2016) 24:41–50. doi: 10.1016/j.cmet.2016.05.005
23. Chiang, JYL, and Ferrell, JM. Bile acid receptors FXR and TGR5 signaling in fatty liver diseases and therapy. Am J Physiol Gastrointest Liver Physiol. (2020) 318:G554–73. doi: 10.1152/ajpgi.00223.2019
24. Li, R, Andreu-Sánchez, S, Kuipers, F, and Fu, J. Gut microbiome and bile acids in obesity-related diseases. Best Pract Res Clin Endocrinol Metab. (2021) 35:101493. doi: 10.1016/j.beem.2021.101493
25. Jia, W, Xie, G, and Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. (2018) 15:111–28. doi: 10.1038/nrgastro.2017.119
26. Honda, A, Miyazaki, T, Iwamoto, J, Hirayama, T, Morishita, Y, Monma, T, et al. Regulation of bile acid metabolism in mouse models with hydrophobic bile acid composition. J Lipid Res. (2020) 61:54–69. doi: 10.1194/jlr.RA119000395
27. de Boer, JF, Verkade, E, Mulder, NL, de Vries, HD, Huijkman, N, Koehorst, M, et al. A human-like bile acid pool induced by deletion of hepatic Cyp2c70 modulates effects of FXR activation in mice. J Lipid Res. (2020) 61:291–305. doi: 10.1194/jlr.RA119000243
28. Guo, GL, and Chiang, JYL. Is CYP2C70 the key to new mouse models to understand bile acids in humans? J Lipid Res. (2020) 61:269–71. doi: 10.1194/jlr.C120000621
29. Poland, JC, and Flynn, CR. Bile acids, their receptors, and the gut microbiota. Physiology (Bethesda). (2021) 36:235–45. doi: 10.1152/physiol.00028.2020
30. Chiang, JYL, and Ferrell, JM. Discovery of farnesoid X receptor and its role in bile acid metabolism. Mol Cell Endocrinol. (2022) 548:111618. doi: 10.1016/j.mce.2022.111618
31. Ahmad, TR, and Haeusler, RA. Bile acids in glucose metabolism and insulin signalling – mechanisms and research needs. Nat Rev Endocrinol. (2019) 15:701–12. doi: 10.1038/s41574-019-0266-7
32. Chiang, JYL, and Ferrell, JM. Bile acids as metabolic regulators and nutrient sensors. Annu Rev Nutr. (2019) 39:175–200. doi: 10.1146/annurev-nutr-082018-124344
33. Li, R, Palmiotti, A, de Vries, HD, Hovingh, MV, Koehorst, M, Mulder, NL, et al. Low production of 12α-hydroxylated bile acids prevents hepatic steatosis in Cyp2c70(−/−) mice by reducing fat absorption. J Lipid Res. (2021) 62:100134. doi: 10.1016/j.jlr.2021.100134
34. Hori, S, Abe, T, Lee, DG, Fukiya, S, Yokota, A, Aso, N, et al. Association between 12α-hydroxylated bile acids and hepatic steatosis in rats fed a high-fat diet. J Nutr Biochem. (2020) 83:108412. doi: 10.1016/j.jnutbio.2020.108412
35. Yoshitsugu, R, Kikuchi, K, Iwaya, H, Fujii, N, Hori, S, Lee, DG, et al. Alteration of bile acid metabolism by a high-fat diet is associated with plasma transaminase activities and glucose intolerance in rats. J Nutr Sci Vitaminol (Tokyo). (2019) 65:45–51. doi: 10.3177/jnsv.65.45
36. Wei, M, Huang, F, Zhao, L, Zhang, Y, Yang, W, Wang, S, et al. A dysregulated bile acid-gut microbiota axis contributes to obesity susceptibility. EBioMedicine. (2020) 55:102766. doi: 10.1016/j.ebiom.2020.102766
37. Ferrebee, CB, Li, J, Haywood, J, Pachura, K, Robinson, BS, Hinrichs, BH, et al. Organic solute transporter α-β protects Ileal enterocytes from bile acid-induced injury. Cell Mol Gastroenterol Hepatol. (2018) 5:499–522. doi: 10.1016/j.jcmgh.2018.01.006
38. van de Wiel, SMW, de Waart, DR, Oude Elferink, RPJ, and van de Graaf, SFJ. Intestinal Farnesoid X receptor activation by pharmacologic inhibition of the organic solute transporter α-β. cell Mol. Gastroenterol Hepatol. (2018) 5:223–37. doi: 10.1016/j.jcmgh.2017.11.011
39. Harnisch, LO, Mihaylov, D, Bein, T, Apfelbacher, C, Kiehntopf, M, Bauer, M, et al. Determination of individual bile acids in acute respiratory distress syndrome reveals a specific pattern of primary and secondary bile acids and a shift to the acidic pathway as an adaptive response to the critical condition. Clin Chem Lab Med. (2022) 60:891–900. doi: 10.1515/cclm-2021-1176
40. Tveter, KM, Mezhibovsky, E, Wu, Y, and Roopchand, DE. Bile acid metabolism and signaling: emerging pharmacological targets of dietary polyphenols. Pharmacol Ther. (2023) 248:108457. doi: 10.1016/j.pharmthera.2023.108457
41. Zhang, J, Lyu, A, and Wang, C. The molecular insights of bile acid homeostasis in host diseases. Life Sci. (2023) 330:121919. doi: 10.1016/j.lfs.2023.121919
42. Ridlon, JM, Harris, SC, Bhowmik, S, Kang, DJ, and Hylemon, PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. (2016) 7:22–39. doi: 10.1080/19490976.2015.1127483
43. Joyce, SA, and Gahan, CG. Bile acid modifications at the microbe-host Interface: potential for nutraceutical and pharmaceutical interventions in host health. Annu Rev Food Sci Technol. (2016) 7:313–33. doi: 10.1146/annurev-food-041715-033159
44. Dawson, PA, Lan, T, and Rao, A. Bile acid transporters. J Lipid Res. (2009) 50:2340–57. doi: 10.1194/jlr.R900012-JLR200
45. Thomas, C, Pellicciari, R, Pruzanski, M, Auwerx, J, and Schoonjans, K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. (2008) 7:678–93. doi: 10.1038/nrd2619
46. Sivamaruthi, BS, Fern, LA, Rashidah Pg Hj Ismail, DSN, and Chaiyasut, C. The influence of probiotics on bile acids in diseases and aging. Biomed Pharmacother. (2020) 128:110310. doi: 10.1016/j.biopha.2020.110310
47. Di Ciaula, A, Bonfrate, L, Khalil, M, Garruti, G, and Portincasa, P. Contribution of the microbiome for better phenotyping of people living with obesity. Rev Endocr Metab Disord. (2023) 24:839–70. doi: 10.1007/s11154-023-09798-1
48. Qu, R, Zhang, Y, Ma, Y, Zhou, X, Sun, L, Jiang, C, et al. Role of the gut microbiota and its metabolites in tumorigenesis or development of colorectal Cancer. Adv Sci (Weinh). (2023) 10:e2205563. doi: 10.1002/advs.202205563
49. Barber, TM, Valsamakis, G, Mastorakos, G, Hanson, P, Kyrou, I, Randeva, HS, et al. Dietary influences on the microbiota-gut-brain Axis. Int J Mol Sci. (2021) 22:3502. doi: 10.3390/ijms22073502
50. de Vos, WM, Tilg, H, Van Hul, M, and Cani, PD. Gut microbiome and health: mechanistic insights. Gut. (2022) 71:1020–32. doi: 10.1136/gutjnl-2021-326789
51. Debnath, N, Kumar, R, Kumar, A, Mehta, PK, and Yadav, AK. Gut-microbiota derived bioactive metabolites and their functions in host physiology. Biotechnol Genet Eng Rev. (2021) 37:105–53. doi: 10.1080/02648725.2021.1989847
52. Frost, F, and Lerch, MM. Gut microbial pathways for bile acid metabolism. Hepatobiliary Surg Nutr. (2021) 10:379–81. doi: 10.21037/hbsn-21-11
53. Guzior, DV, and Quinn, RA. Review: microbial transformations of human bile acids. Microbiome. (2021) 9:140. doi: 10.1186/s40168-021-01101-1
54. Foley, MH, O'Flaherty, S, Barrangou, R, and Theriot, CM. Bile salt hydrolases: gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. (2019) 15:e1007581. doi: 10.1371/journal.ppat.1007581
55. Fogelson, KA, Dorrestein, PC, Zarrinpar, A, and Knight, R. The gut microbial bile acid modulation and its relevance to digestive health and diseases. Gastroenterology. (2023) 164:1069–85. doi: 10.1053/j.gastro.2023.02.022
56. Yang, Y, and Wu, C. Targeting gut microbial bile salt hydrolase (BSH) by diet supplements: new insights into dietary modulation of human health. Food Funct. (2022) 13:7409–22. doi: 10.1039/d2fo01252a
57. Sah, DK, Arjunan, A, Park, SY, and Jung, YD. Bile acids and microbes in metabolic disease. World J Gastroenterol. (2022) 28:6846–66. doi: 10.3748/wjg.v28.i48.6846
58. Adhikari, AA, Ramachandran, D, Chaudhari, SN, Powell, CE, Li, W, McCurry, MD, et al. A gut-restricted Lithocholic acid analog as an inhibitor of gut bacterial bile salt hydrolases. ACS Chem Biol. (2021) 16:1401–12. doi: 10.1021/acschembio.1c00192
59. Song, Z, Cai, Y, Lao, X, Wang, X, Lin, X, Cui, Y, et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome. (2019) 7:9. doi: 10.1186/s40168-019-0628-3
60. Winston, JA, and Theriot, CM. Diversification of host bile acids by members of the gut microbiota. Gut Microbes. (2020) 11:158–71. doi: 10.1080/19490976.2019.1674124
61. Foley, MH, O'Flaherty, S, Allen, G, Rivera, AJ, Stewart, AK, Barrangou, R, et al. Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization. Proc Natl Acad Sci USA. (2021) 118:e2017709118. doi: 10.1073/pnas.2017709118
62. Sun, L, Zhang, Y, Cai, J, Rimal, B, Rocha, ER, Coleman, JP, et al. Bile salt hydrolase in non-enterotoxigenic Bacteroides potentiates colorectal cancer. Nat Commun. (2023) 14:755. doi: 10.1038/s41467-023-36089-9
63. Jia, W, Li, Y, Cheung, KCP, and Zheng, X. Bile acid signaling in the regulation of whole body metabolic and immunological homeostasis. Sci China Life Sci. (2023) 67:865–78. doi: 10.1007/s11427-023-2353-0
64. Wang, D, Doestzada, M, Chen, L, Andreu-Sánchez, S, van den Munckhof, ICL, Augustijn, HE, et al. Characterization of gut microbial structural variations as determinants of human bile acid metabolism. Cell Host Microbe. (2021) 29:1802–1814.e5. doi: 10.1016/j.chom.2021.11.003
65. Funabashi, M, Grove, TL, Wang, M, Varma, Y, McFadden, ME, Brown, LC, et al. A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature. (2020) 582:566–70. doi: 10.1038/s41586-020-2396-4
66. Liu, TC, Kern, JT, Jain, U, Sonnek, NM, Xiong, S, Simpson, KF, et al. Western diet induces Paneth cell defects through microbiome alterations and farnesoid X receptor and type I interferon activation. Cell Host Microbe. (2021) 29:988–1001.e6. doi: 10.1016/j.chom.2021.04.004
67. Wang, L, Gong, Z, Zhang, X, Zhu, F, Liu, Y, Jin, C, et al. Gut microbial bile acid metabolite skews macrophage polarization and contributes to high-fat diet-induced colonic inflammation. Gut Microbes. (2020) 12:1819155–20. doi: 10.1080/19490976.2020.1819155
68. Xie, C, Huang, W, Young, RL, Jones, KL, Horowitz, M, Rayner, CK, et al. Role of bile acids in the regulation of food intake, and their dysregulation in metabolic disease. Nutrients. (2021) 13:1104. doi: 10.3390/nu13041104
69. Wahlström, A, Kovatcheva-Datchary, P, Ståhlman, M, Bäckhed, F, and Marschall, HU. Crosstalk between bile acids and gut microbiota and its impact on Farnesoid X receptor Signalling. Dig Dis. (2017) 35:246–50. doi: 10.1159/000450982
70. Takahashi, S, Fukami, T, Masuo, Y, Brocker, CN, Xie, C, Krausz, KW, et al. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J Lipid Res. (2016) 57:2130–7. doi: 10.1194/jlr.M071183
71. Lin, H, An, Y, Tang, H, and Wang, Y. Alterations of bile acids and gut microbiota in obesity induced by high fat diet in rat model. J Agric Food Chem. (2019) 67:3624–32. doi: 10.1021/acs.jafc.9b00249
72. Wang, Q, Lin, H, Shen, C, Zhang, M, Wang, X, Yuan, M, et al. Gut microbiota regulates postprandial GLP-1 response via ileal bile acid-TGR5 signaling. Gut Microbes. (2023) 15:2274124. doi: 10.1080/19490976.2023.2274124
73. Rao, AS, Wong, BS, Camilleri, M, Odunsi-Shiyanbade, ST, McKinzie, S, Ryks, M, et al. Chenodeoxycholate in females with irritable bowel syndrome-constipation: a pharmacodynamic and pharmacogenetic analysis. Gastroenterology. (2010) 139:1549–1558.e1. doi: 10.1053/j.gastro.2010.07.052
74. Fiorucci, S, Distrutti, E, Carino, A, Zampella, A, and Biagioli, M. Bile acids and their receptors in metabolic disorders. Prog Lipid Res. (2021) 82:101094. doi: 10.1016/j.plipres.2021.101094
75. Panzitt, K, and Wagner, M. FXR in liver physiology: multiple faces to regulate liver metabolism. Biochim Biophys Acta Mol basis Dis. (2021) 1867:166133. doi: 10.1016/j.bbadis.2021.166133
76. Ramos Pittol, JM, Milona, A, Morris, I, Willemsen, ECL, van der Veen, SW, Kalkhoven, E, et al. FXR isoforms control different metabolic functions in liver cells via binding to specific DNA motifs. Gastroenterology. (2020) 159:1853–1865.e10. doi: 10.1053/j.gastro.2020.07.036
77. Jiang, L, Zhang, H, Xiao, D, Wei, H, and Chen, Y. Farnesoid X receptor (FXR): structures and ligands. Comput Struct Biotechnol J. (2021) 19:2148–59. doi: 10.1016/j.csbj.2021.04.029
78. Tian, SY, Chen, SM, Pan, CX, and Li, Y. FXR: structures, biology, and drug development for NASH and fibrosis diseases. Acta Pharmacol Sin. (2022) 43:1120–32. doi: 10.1038/s41401-021-00849-4
79. Massafra, V, Pellicciari, R, Gioiello, A, and van Mil, SWC. Progress and challenges of selective Farnesoid X receptor modulation. Pharmacol Ther. (2018) 191:162–77. doi: 10.1016/j.pharmthera.2018.06.009
80. Greimel, T, Jahnel, J, Pohl, S, Strini, T, Tischitz, M, Meier-Allard, N, et al. Bile acid-induced tissue factor activity in hepatocytes correlates with activation of farnesoid X receptor. Lab Investig. (2021) 101:1394–402. doi: 10.1038/s41374-021-00628-z
81. Wang, Y, Ai, Z, Xing, X, Fan, Y, Zhang, Y, Nan, B, et al. The ameliorative effect of probiotics on diet-induced lipid metabolism disorders: A review. Crit Rev Food Sci Nutr. (2022) 64:3556–72. doi: 10.1080/10408398.2022.2132377
82. Anfuso, B, Tiribelli, C, Adorini, L, and Rosso, N. Obeticholic acid and INT-767 modulate collagen deposition in a NASH in vitro model. Sci Rep. (2020) 10:1699. doi: 10.1038/s41598-020-58562-x
83. Sayin, SI, Wahlström, A, Felin, J, Jäntti, S, Marschall, HU, Bamberg, K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. (2013) 17:225–35. doi: 10.1016/j.cmet.2013.01.003
84. Sun, L, Cai, J, and Gonzalez, FJ. The role of farnesoid X receptor in metabolic diseases, and gastrointestinal and liver cancer. Nat Rev Gastroenterol Hepatol. (2021) 18:335–47. doi: 10.1038/s41575-020-00404-2
85. Trabelsi, MS, Lestavel, S, Staels, B, and Collet, X. Intestinal bile acid receptors are key regulators of glucose homeostasis. Proc Nutr Soc. (2017) 76:192–202. doi: 10.1017/s0029665116002834
86. Liu, Y, Chen, K, Li, F, Gu, Z, Liu, Q, He, L, et al. Probiotic Lactobacillus rhamnosus GG prevents liver fibrosis through inhibiting hepatic bile acid synthesis and enhancing bile acid excretion in mice. Hepatology. (2020) 71:2050–66. doi: 10.1002/hep.30975
87. Song, Y, Sun, L, Ma, P, Xu, L, and Xiao, P. Dihydromyricetin prevents obesity via regulating bile acid metabolism associated with the farnesoid X receptor in Ob/Ob mice. Food Funct. (2022) 13:2491–503. doi: 10.1039/d1fo03971g
88. Liu, C, Cai, T, Cheng, Y, Bai, J, Li, M, Gu, B, et al. Postbiotics prepared using Lactobacillus reuteri ameliorates ethanol-induced liver injury by regulating the FXR/SHP/SREBP-1c Axis. Mol Nutr Food Res. (2024) 68:e2300927. doi: 10.1002/mnfr.202300927
89. Sheng, Y, Meng, G, Zhang, M, Chen, X, Chai, X, Yu, H, et al. Dan-shen Yin promotes bile acid metabolism and excretion to prevent atherosclerosis via activating FXR/BSEP signaling pathway. J Ethnopharmacol. (2024) 330:118209. doi: 10.1016/j.jep.2024.118209
90. Chalasani, N, Younossi, Z, Lavine, JE, Charlton, M, Cusi, K, Rinella, M, et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Clin Liver Dis (Hoboken). (2018) 11:81. doi: 10.1002/cld.722
91. Shah, RA, Alkhouri, N, and Kowdley, KV. Emerging drugs for the treatment of non-alcoholic steatohepatitis: a focused review of farnesoid X receptor agonists. Expert Opin Emerg Drugs. (2020) 25:251–60. doi: 10.1080/14728214.2020.1796968
92. Peng, A, Liu, S, Fang, L, Zhu, Z, Zhou, Y, Yue, S, et al. Inonotus obliquus and its bioactive compounds alleviate non-alcoholic fatty liver disease via regulating FXR/SHP/SREBP-1c axis. Eur J Pharmacol. (2022) 921:174841. doi: 10.1016/j.ejphar.2022.174841
93. Carino, A, Marchianò, S, Biagioli, M, Fiorucci, C, Zampella, A, Monti, MC, et al. Transcriptome analysis of dual FXR and GPBAR1 Agonism in rodent model of NASH reveals modulation of lipid droplets formation. Nutrients. (2019) 11:1132. doi: 10.3390/nu11051132
94. Kumari, A, Pal Pathak, D, and Asthana, S. Bile acids mediated potential functional interaction between FXR and FATP5 in the regulation of lipid metabolism. Int J Biol Sci. (2020) 16:2308–22. doi: 10.7150/ijbs.44774
95. Ferré, P, Phan, F, and Foufelle, F. SREBP-1c and lipogenesis in the liver: an update1. Biochem J. (2021) 478:3723–39. doi: 10.1042/bcj20210071
96. Duan, R, Guan, X, Huang, K, Zhang, Y, Li, S, Xia, J, et al. Flavonoids from whole-grain oat alleviated high-fat diet-induced hyperlipidemia via regulating bile acid metabolism and gut microbiota in mice. J Agric Food Chem. (2021) 69:7629–40. doi: 10.1021/acs.jafc.1c01813
97. Chen, H, Cheng, J, Zhou, S, Chen, D, Qin, W, Li, C, et al. Arabinoxylan combined with different glucans improve lipid metabolism disorder by regulating bile acid and gut microbiota in mice fed with high-fat diet. Int J Biol Macromol. (2021) 168:279–88. doi: 10.1016/j.ijbiomac.2020.12.036
98. Gu, M, Song, H, Li, Y, Jiang, Y, Zhang, Y, Tang, Z, et al. Extract of Schisandra chinensis fruit protects against metabolic dysfunction in high-fat diet induced obese mice via FXR activation. Phytother Res. (2020) 34:3063–77. doi: 10.1002/ptr.6743
99. Ghosh Laskar, M, Eriksson, M, Rudling, M, and Angelin, B. Treatment with the natural FXR agonist chenodeoxycholic acid reduces clearance of plasma LDL whilst decreasing circulating PCSK9, lipoprotein(a) and apolipoprotein C-III. J Intern Med. (2017) 281:575–85. doi: 10.1111/joim.12594
100. Wang, XX, Xie, C, Libby, AE, Ranjit, S, Levi, J, Myakala, K, et al. The role of FXR and TGR5 in reversing and preventing progression of Western diet-induced hepatic steatosis, inflammation, and fibrosis in mice. J Biol Chem. (2022) 298:102530. doi: 10.1016/j.jbc.2022.102530
101. Yamagata, K, Daitoku, H, Shimamoto, Y, Matsuzaki, H, Hirota, K, Ishida, J, et al. Bile acids regulate gluconeogenic gene expression via small heterodimer partner-mediated repression of hepatocyte nuclear factor 4 and Foxo1. J Biol Chem. (2004) 279:23158–65. doi: 10.1074/jbc.M314322200
102. Zhang, HM, Wang, X, Wu, ZH, Liu, HL, Chen, W, Zhang, ZZ, et al. Beneficial effect of farnesoid X receptor activation on metabolism in a diabetic rat model. Mol Med Rep. (2016) 13:2135–42. doi: 10.3892/mmr.2016.4761
103. Qiang, S, Tao, L, Zhou, J, Wang, Q, Wang, K, Lu, M, et al. Knockout of farnesoid X receptor aggravates process of diabetic cardiomyopathy. Diabetes Res Clin Pract. (2020) 161:108033. doi: 10.1016/j.diabres.2020.108033
104. Sun, R, Kong, B, Yang, N, Cao, B, Feng, D, Yu, X, et al. The hypoglycemic effect of Berberine and Berberrubine involves modulation of intestinal Farnesoid X receptor signaling pathway and inhibition of hepatic gluconeogenesis. Drug Metab Dispos. (2021) 49:276–86. doi: 10.1124/dmd.120.000215
105. Yan, Y, Niu, Z, Sun, C, Li, P, Shen, S, Liu, S, et al. Hepatic thyroid hormone signalling modulates glucose homeostasis through the regulation of GLP-1 production via bile acid-mediated FXR antagonism. Nat Commun. (2022) 13:6408. doi: 10.1038/s41467-022-34258-w
106. Tian, F, Chen, T, Xu, W, Fan, Y, Feng, X, Huang, Q, et al. Curcumin compensates GLP-1 deficiency via the microbiota-bile acids Axis and modulation in functional crosstalk between TGR5 and FXR in Ob/Ob mice. Mol Nutr Food Res. (2023) 67:e2300195. doi: 10.1002/mnfr.202300195
107. Qiu, Y, Shen, L, Fu, L, Yang, J, Cui, C, Li, T, et al. The glucose-lowering effects of α-glucosidase inhibitor require a bile acid signal in mice. Diabetologia. (2020) 63:1002–16. doi: 10.1007/s00125-020-05095-7
108. Pan, MX, Zheng, CY, Deng, YJ, Tang, KR, Nie, H, Xie, JQ, et al. Hepatic protective effects of Shenling Baizhu powder, a herbal compound, against inflammatory damage via TLR4/NLRP3 signalling pathway in rats with nonalcoholic fatty liver disease. J Integr Med. (2021) 19:428–38. doi: 10.1016/j.joim.2021.07.004
109. Biao, Y, Chen, J, Liu, C, Wang, R, Han, X, Li, L, et al. Protective effect of Danshen Zexie decoction against non-alcoholic fatty liver disease through inhibition of ROS/NLRP3/IL-1β pathway by Nrf2 signaling activation. Front Pharmacol. (2022) 13:877924. doi: 10.3389/fphar.2022.877924
110. Li, F, Li, Y, Li, Q, and Shi, X. Eriobotrya japonica leaf triterpenoid acids ameliorate metabolic syndrome in C57BL/6J mice fed with high-fat diet. Biomed Pharmacother. (2020) 132:110866. doi: 10.1016/j.biopha.2020.110866
111. Zhu, Y, Cai, PJ, Dai, HC, Xiao, YH, Jia, CL, and Sun, AD. Black chokeberry (Aronia melanocarpa L.) polyphenols attenuate obesity-induced colonic inflammation by regulating gut microbiota and the TLR4/NF-κB signaling pathway in high fat diet-fed rats. Food Funct. (2023) 14:10014–30. doi: 10.1039/d3fo02177g
112. Ding, YY, Lan, J, Fang, Y, Pan, Y, Gu, Z, Xue, J, et al. Dityrosine aggravates hepatic insulin resistance in obese mice by altering gut microbiota and the LPS/TLR4/NF-κB inflammatory pathway. Mol Nutr Food Res. (2023) 67:e2300373. doi: 10.1002/mnfr.202300373
113. Campbell, C, McKenney, PT, Konstantinovsky, D, Isaeva, OI, Schizas, M, Verter, J, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature. (2020) 581:475–9. doi: 10.1038/s41586-020-2193-0
114. Song, X, Sun, X, Oh, SF, Wu, M, Zhang, Y, Zheng, W, et al. Microbial bile acid metabolites modulate gut RORγ(+) regulatory T cell homeostasis. Nature. (2020) 577:410–5. doi: 10.1038/s41586-019-1865-0
115. Hang, S, Paik, D, Yao, L, Kim, E, Trinath, J, Lu, J, et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature. (2019) 576:143–8. doi: 10.1038/s41586-019-1785-z
116. Han, CY, Rho, HS, Kim, A, Kim, TH, Jang, K, Jun, DW, et al. FXR inhibits endoplasmic reticulum stress-induced NLRP3 Inflammasome in hepatocytes and ameliorates liver injury. Cell Rep. (2018) 24:2985–99. doi: 10.1016/j.celrep.2018.07.068
117. Gadaleta, RM, van Erpecum, KJ, Oldenburg, B, Willemsen, EC, Renooij, W, Murzilli, S, et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. (2011) 60:463–72. doi: 10.1136/gut.2010.212159
118. Campbell, C, Marchildon, F, Michaels, AJ, Takemoto, N, van der Veeken, J, Schizas, M, et al. FXR mediates T cell-intrinsic responses to reduced feeding during infection. Proc Natl Acad Sci USA. (2020) 117:33446–54. doi: 10.1073/pnas.2020619117
119. Fuchs, CD, and Trauner, M. Role of bile acids and their receptors in gastrointestinal and hepatic pathophysiology. Nat Rev Gastroenterol Hepatol. (2022) 19:432–50. doi: 10.1038/s41575-021-00566-7
120. Yan, Y, Lei, Y, Qu, Y, Fan, Z, Zhang, T, Xu, Y, et al. Bacteroides uniformis-induced perturbations in colonic microbiota and bile acid levels inhibit TH17 differentiation and ameliorate colitis developments. NPJ Biofilms Microbiomes. (2023) 9:56. doi: 10.1038/s41522-023-00420-5
121. Wang, J, Zhu, N, Su, X, Gao, Y, and Yang, R. Gut-microbiota-derived metabolites maintain gut and systemic immune homeostasis. Cells. (2023) 12:793. doi: 10.3390/cells12050793
122. Biagioli, M, and Carino, A. Signaling from intestine to the host: how bile acids regulate intestinal and liver immunity. Handb Exp Pharmacol. (2019) 256:95–108. doi: 10.1007/164_2019_225
123. Liu, S, Kang, W, Mao, X, Ge, L, Du, H, Li, J, et al. Melatonin mitigates aflatoxin B1-induced liver injury via modulation of gut microbiota/intestinal FXR/liver TLR4 signaling axis in mice. J Pineal Res. (2022) 73:e12812. doi: 10.1111/jpi.12812
124. Portincasa, P, Bonfrate, L, Khalil, M, Angelis, M, Calabrese, FM, D'Amato, M, et al. Intestinal barrier and permeability in health, obesity and NAFLD. Biomedicines. (2021) 10:83. doi: 10.3390/biomedicines10010083
125. Liu, X, Zhang, Y, Li, W, Zhang, B, Yin, J, Liuqi, S, et al. Fucoidan ameliorated dextran sulfate sodium-induced ulcerative colitis by modulating gut microbiota and bile acid metabolism. J Agric Food Chem. (2022) 70:14864–76. doi: 10.1021/acs.jafc.2c06417
126. Long, XQ, Liu, MZ, Liu, ZH, Xia, LZ, Lu, SP, Xu, XP, et al. Bile acids and their receptors: potential therapeutic targets in inflammatory bowel disease. World J Gastroenterol. (2023) 29:4252–70. doi: 10.3748/wjg.v29.i27.4252
127. Yang, M, Gu, Y, Li, L, Liu, T, Song, X, Sun, Y, et al. Bile acid-gut microbiota Axis in inflammatory bowel disease: from bench to bedside. Nutrients. (2021) 13:3143. doi: 10.3390/nu13093143
128. Xia, R, Zhang, Q, Xia, D, Hao, Q, Ding, Q, Ran, C, et al. The direct and gut microbiota-mediated effects of dietary bile acids on the improvement of gut barriers in largemouth bass (Micropterus salmoides). Anim Nutr. (2023) 14:32–42. doi: 10.1016/j.aninu.2023.03.008
129. Parséus, A, Sommer, N, Sommer, F, Caesar, R, Molinaro, A, Ståhlman, M, et al. Microbiota-induced obesity requires farnesoid X receptor. Gut. (2017) 66:429–37. doi: 10.1136/gutjnl-2015-310283
130. Wu, J, Wang, K, Wang, X, Pang, Y, and Jiang, C. The role of the gut microbiome and its metabolites in metabolic diseases. Protein Cell. (2021) 12:360–73. doi: 10.1007/s13238-020-00814-7
131. Barber, TM, Hanson, P, and Weickert, MO. Metabolic-associated fatty liver disease and the gut microbiota. Endocrinol Metab Clin N Am. (2023) 52:485–96. doi: 10.1016/j.ecl.2023.01.004
132. Zhang, C, Fang, R, Lu, X, Zhang, Y, Yang, M, Su, Y, et al. Lactobacillus reuteri J1 prevents obesity by altering the gut microbiota and regulating bile acid metabolism in obese mice. Food Funct. (2022) 13:6688–701. doi: 10.1039/d1fo04387k
133. Koliada, A, Syzenko, G, Moseiko, V, Budovska, L, Puchkov, K, Perederiy, V, et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. (2017) 17:120. doi: 10.1186/s12866-017-1027-1
134. Bell, DS. Changes seen in gut bacteria content and distribution with obesity: causation or association? Postgrad Med. (2015) 127:863–8. doi: 10.1080/00325481.2015.1098519
135. Huang, F, Zheng, X, Ma, X, Jiang, R, Zhou, W, Zhou, S, et al. Theabrownin from Pu-erh tea attenuates hypercholesterolemia via modulation of gut microbiota and bile acid metabolism. Nat Commun. (2019) 10:4971. doi: 10.1038/s41467-019-12896-x
136. Li, Y, Hou, H, Wang, X, Dai, X, Zhang, W, Tang, Q, et al. Diammonium Glycyrrhizinate ameliorates obesity through modulation of gut microbiota-conjugated BAs-FXR signaling. Front Pharmacol. (2021) 12:796590. doi: 10.3389/fphar.2021.796590
137. Xie, Z, Jiang, H, Liu, W, Zhang, X, Chen, D, Sun, S, et al. The triterpenoid sapogenin (2α-OH-Protopanoxadiol) ameliorates metabolic syndrome via the intestinal FXR/GLP-1 axis through gut microbiota remodelling. Cell Death Dis. (2020) 11:770. doi: 10.1038/s41419-020-02974-0
138. Shen, C, Pan, Z, Wu, S, Zheng, M, Zhong, C, Xin, X, et al. Emodin palliates high-fat diet-induced nonalcoholic fatty liver disease in mice via activating the farnesoid X receptor pathway. J Ethnopharmacol. (2021) 279:114340. doi: 10.1016/j.jep.2021.114340
139. Meng, Z, Yan, S, Sun, W, Yan, J, Teng, M, Jia, M, et al. Chlorothalonil induces obesity in mice by regulating host gut microbiota and bile acids metabolism via FXR pathways. J Hazard Mater. (2023) 452:131310. doi: 10.1016/j.jhazmat.2023.131310
140. Clifford, BL, Sedgeman, LR, Williams, KJ, Morand, P, Cheng, A, Jarrett, KE, et al. FXR activation protects against NAFLD via bile-acid-dependent reductions in lipid absorption. Cell Metab. (2021) 33:1671–1684.e4. doi: 10.1016/j.cmet.2021.06.012
141. Yang, C, Yang, L, Yang, Y, Wan, M, Xu, D, Pan, D, et al. Effects of flaxseed powder in improving non-alcoholic fatty liver by regulating gut microbiota-bile acids metabolic pathway through FXR/TGR5 mediating. Biomed Pharmacother. (2023) 163:114864. doi: 10.1016/j.biopha.2023.114864
142. Fang, S, Suh, JM, Reilly, SM, Yu, E, Osborn, O, Lackey, D, et al. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat Med. (2015) 21:159–65. doi: 10.1038/nm.3760
143. Pathak, P, Xie, C, Nichols, RG, Ferrell, JM, Boehme, S, Krausz, KW, et al. Intestine farnesoid X receptor agonist and the gut microbiota activate G-protein bile acid receptor-1 signaling to improve metabolism. Hepatology. (2018) 68:1574–88. doi: 10.1002/hep.29857
144. Kim, YC, Seok, S, Zhang, Y, Ma, J, Kong, B, Guo, G, et al. Intestinal FGF15/19 physiologically repress hepatic lipogenesis in the late fed-state by activating SHP and DNMT3A. Nat Commun. (2020) 11:5969. doi: 10.1038/s41467-020-19803-9
145. Gai, Z, Gui, T, Alecu, I, Lone, MA, Hornemann, T, Chen, Q, et al. Farnesoid X receptor activation induces the degradation of hepatotoxic 1-deoxysphingolipids in non-alcoholic fatty liver disease. Liver Int. (2020) 40:844–59. doi: 10.1111/liv.14340
146. Collins, SL, Stine, JG, Bisanz, JE, Okafor, CD, and Patterson, AD. Bile acids and the gut microbiota: metabolic interactions and impacts on disease. Nat Rev Microbiol. (2023) 21:236–47. doi: 10.1038/s41579-022-00805-x
147. Duan, Y, Zhang, F, Yuan, W, Wei, Y, Wei, M, Zhou, Y, et al. Hepatic cholesterol accumulation ascribed to the activation of ileum Fxr-Fgf15 pathway inhibiting hepatic Cyp7a1 in high-fat diet-induced obesity rats. Life Sci. (2019) 232:116638. doi: 10.1016/j.lfs.2019.116638
148. Wang, R, Fan, X, Lu, Y, Chen, D, Zhao, Y, and Qi, K. Dietary acetic acid suppress high-fat diet-induced obesity in mice by altering taurine conjugated bile acids metabolism. Curr Res Food Sci. (2022) 5:1976–84. doi: 10.1016/j.crfs.2022.10.021
149. Ye, X, Li, J, Gao, Z, Wang, D, Wang, H, and Wu, J. Chlorogenic acid inhibits lipid deposition by regulating the enterohepatic FXR-FGF15 pathway. Biomed Res Int. (2022) 2022:4919153–11. doi: 10.1155/2022/4919153
150. Wang, Y, Xing, X, Ma, Y, Fan, Y, Zhang, Y, Nan, B, et al. Prevention of high-fat-diet-induced dyslipidemia by Lactobacillus plantarum LP104 through mediating bile acid enterohepatic Axis circulation and intestinal Flora. J Agric Food Chem. (2023) 71:7334–47. doi: 10.1021/acs.jafc.2c09151
151. Zhong, XC, Liu, YM, Gao, XX, Krausz, KW, Niu, B, Gonzalez, FJ, et al. Caffeic acid phenethyl ester suppresses intestinal FXR signaling and ameliorates nonalcoholic fatty liver disease by inhibiting bacterial bile salt hydrolase activity. Acta Pharmacol Sin. (2023) 44:145–56. doi: 10.1038/s41401-022-00921-7
152. Sun, L, Xie, C, Wang, G, Wu, Y, Wu, Q, Wang, X, et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat Med. (2018) 24:1919–29. doi: 10.1038/s41591-018-0222-4
153. Liu, H, Sun, Y, Nie, C, Xie, X, Yuan, X, Ma, Q, et al. Highland barley β-glucan alleviated western diet-induced non-alcoholic fatty liver disease via increasing energy expenditure and regulating bile acid metabolism in mice. Food Funct. (2022) 13:11664–75. doi: 10.1039/d2fo01167k
154. Luo, Z, Li, M, Yang, J, Li, J, Zhang, Y, Liu, F, et al. Ferulic acid attenuates high-fat diet-induced hypercholesterolemia by activating classic bile acid synthesis pathway. Front Nutr. (2022) 9:976638. doi: 10.3389/fnut.2022.976638
155. Zhai, Y, Zhou, W, Yan, X, Qiao, Y, Guan, L, Zhang, Z, et al. Astragaloside IV ameliorates diet-induced hepatic steatosis in obese mice by inhibiting intestinal FXR via intestinal flora remodeling. Phytomedicine. (2022) 107:154444. doi: 10.1016/j.phymed.2022.154444
156. Jiang, J, Ma, Y, Liu, Y, Lu, D, Gao, X, Krausz, KW, et al. Glycine-β-muricholic acid antagonizes the intestinal farnesoid X receptor-ceramide axis and ameliorates NASH in mice. Hepatol Commun. (2022) 6:3363–78. doi: 10.1002/hep4.2099
157. Pathak, P, and Chiang, JYL. Sterol 12α-hydroxylase aggravates dyslipidemia by activating the ceramide/mTORC1/SREBP-1C pathway via FGF21 and FGF15. Gene Expr. (2019) 19:161–73. doi: 10.3727/105221619x15529371970455
158. Wu, Q, Sun, L, Hu, X, Wang, X, Xu, F, Chen, B, et al. Suppressing the intestinal farnesoid X receptor/sphingomyelin phosphodiesterase 3 axis decreases atherosclerosis. J Clin Invest. (2021) 131:e142865. doi: 10.1172/jci142865
159. Zhou, Q, Song, N, Wang, SQ, Wang, Y, Zhao, YK, and Zhu, XD. Effect of Gegen Qinlian decoction on hepatic gluconeogenesis in ZDF rats with type 2 diabetes mellitus based on the Farnesol X receptor/ceramide signaling pathway regulating mitochondrial metabolism and endoplasmic reticulum stress. Evid Based Complement Alternat Med. (2021) 2021:9922292–13. doi: 10.1155/2021/9922292
Keywords: bile acid, metabolome, metabolic disease, farnesoid X receptor, metabolize
Citation: Li Y, Wang L, Yi Q, Luo L and Xiong Y (2024) Regulation of bile acids and their receptor FXR in metabolic diseases. Front. Nutr. 11:1447878. doi: 10.3389/fnut.2024.1447878
Edited by:
John D. Imig, University of Arkansas for Medical Sciences, United StatesReviewed by:
Ya’Nan Zhang, Zhejiang A&F University, ChinaWei Guo, University of North Carolina at Greensboro, United States
Copyright © 2024 Li, Wang, Yi, Luo and Xiong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuxia Xiong, eHl4X2NlbGxAc3dtdS5lZHUuY24=
 Yao Li
Yao Li Lulu Wang
Lulu Wang Yuxia Xiong
Yuxia Xiong