- Department of Biology, University of Vermont, Burlington, VT, United States
Across species, taste provides important chemical information about potential food sources and the surrounding environment. As details about the chemicals and receptors responsible for gustation are discovered, a complex view of the taste system is emerging with significant contributions from research using the fruit fly, Drosophila melanogaster, as a model organism. In this brief review, we summarize recent advances in Drosophila gustation and their relevance to taste research more broadly. Our goal is to highlight the molecular mechanisms underlying the first step of gustatory circuits: ligand-receptor interactions in primary taste cells. After an introduction to the Drosophila taste system and how it encodes the canonical taste modalities sweet, bitter, and salty, we describe recent insights into the complex nature of carboxylic acid and amino acid detection in the context of sour and umami taste, respectively. Our analysis extends to non-canonical taste modalities including metals, fatty acids, and bacterial components, and highlights unexpected receptors and signaling pathways that have recently been identified in Drosophila taste cells. Comparing the intricate molecular and cellular underpinnings of how ligands are detected in vivo in fruit flies reveals both specific and promiscuous receptor selectivity for taste encoding. Throughout this review, we compare and contextualize these Drosophila findings with mammalian research to not only emphasize the conservation of these chemosensory systems, but to demonstrate the power of this model organism in elucidating the neurobiology of taste and feeding.
Introduction
The chemical sense of taste allows animals to evaluate their food options to encourage the consumption of beneficial nutrients and avoidance of potential toxins. Since gustation links the environment to nutrition and fitness, it is not surprising that this sense is well-conserved across a wide range of animals, from humans to fruit flies (1). The concept that certain chemicals elicit distinct taste perceptions can be traced back to the earliest philosophers, but a clear understanding of the molecular and cellular basis of taste only started to emerge in the early 2000s. Over the last two decades, there has been extensive research into identifying the receptors responsible for the “five basic tastes”: sweet, bitter, salty, sour, and umami (2). Many details of these canonical taste modalities are well-established in both mammalian and non-mammalian model organisms, including the fruit fly, Drosophila melanogaster (1, 3–5). Drosophila is a powerful model organism in neurobiology research that has continued to advance our understanding of gustation due to the ability to record taste cell activity in vivo from a single neuron or a complete set of specific taste cells (6–8). Readily available genetic tools also allow for investigation into the role of taste receptors in cellular physiology and chemosensory behaviors (9–11). This review introduces the Drosophila taste system and describes recent insights into novel tastant-receptor interactions for both canonical and non-canonical taste modalities with comparisons to mammalian gustation.
The fruit fly taste system
In both mammals and Drosophila, primary chemosensory cells initiate taste sensation by evaluating a food source’s chemical properties. The mammalian gustatory system uses taste receptor cells (TRCs), modified epithelial cells found in taste buds throughout the oral cavity. TRCs detect chemicals and relay this information to afferent gustatory nerves (1), but the Drosophila peripheral nervous system directly detects tastants via gustatory receptor neurons (GRNs) (12). GRNs are distributed throughout the fly body, but the highest concentration of taste cells involved in feeding is located in the labellum, the Drosophila tongue homolog (5). Labellar GRNs express taste receptors that allow for the rapid identification of chemicals, promoting selectivity for compounds that represent specific taste modalities (13, 14), akin to lingual taste cells in mammals.
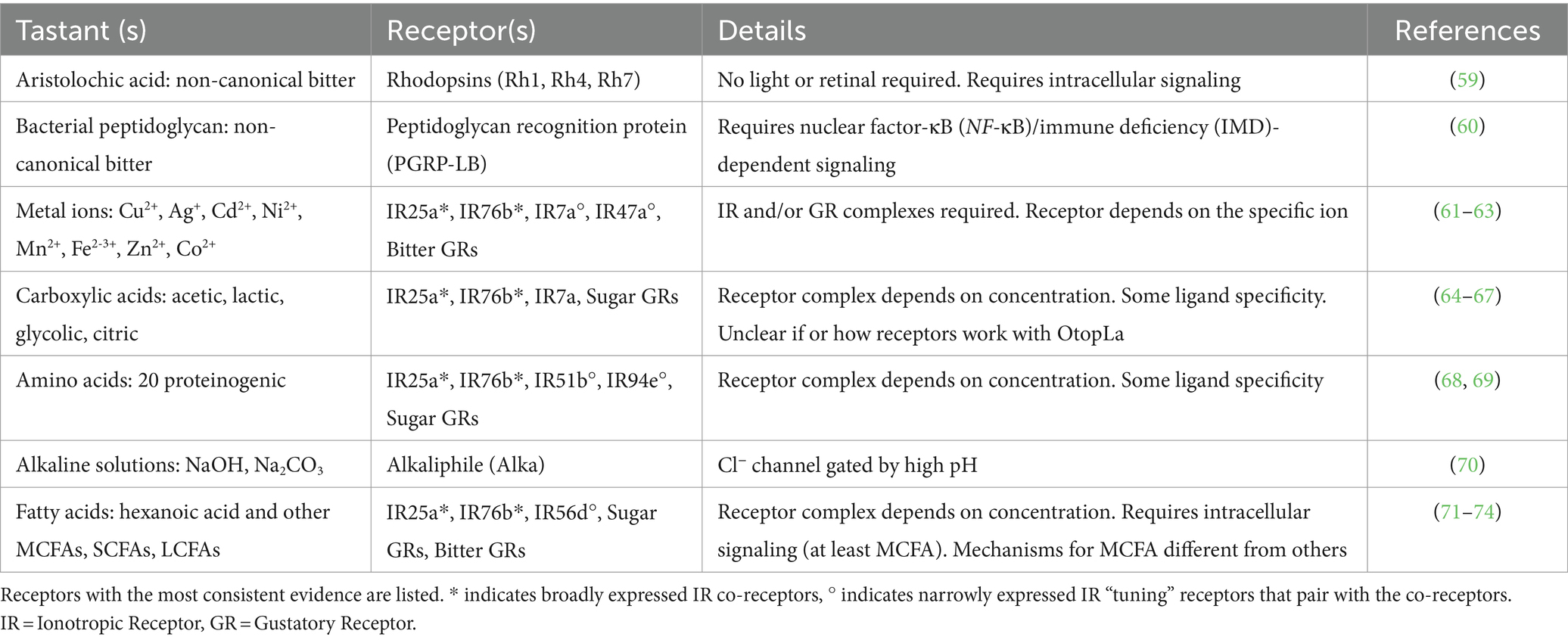
GRNs in the fruit fly labellum have been categorized into five groups based on their distinct receptor profiles and taste modality responsiveness: “sweet,” “bitter,” “water,” “salty,” and “IR94e” (15). These five GRN classes can be consistently mapped on a fly’s labellum across ~62 gustatory sensilla that are classified by size, each containing two or four GRNs (10, 16, 17) (Figure 1A). GRN axons project to the sub-esophageal zone in the brain (17, 18), where arborizations of both GRNs and motor neurons generate local circuits for taste-induced behavioral responses (19). The Drosophila whole-brain connectome (20–23) allows neural circuits to be traced from tastant-receptor activation through behavioral output to enhance our understanding of how taste information is encoded and modulated (24–28).
Figure 1. Neural encoding of novel tastant-receptor interactions in Drosophila. (A) Fruit flies will explore their chemosensory environment by extending their proboscis and interacting with tastants via sensillum on the labellum. (A inset) Three types of sensilla (S = short, I = intermediate, L = long) each contain two or four gustatory receptor neurons (GRNs) from five different cell types (color depicts each class, grouped by modality and receptor expression). Mechanosensory neurons are depicted in black. (B) Each GRN has a diverse array of chemosensory receptors (colored to match the GRN that houses it). Recently investigated tastants are depicted (gray boxes) with their specific receptors (colored arrows). * indicates “tuning” IRs that generally work with the ionotropic co-receptors. (*) indicates receptors that work alongside ionotropic co-receptors for only some of the taste modalities depicted. Tastant-receptor pairings that require intracellular pathways are depicted (gray arrows). Created with Biorender.com.
Gustatory processing commonly starts with two main classes of taste receptors in Drosophila: gustatory receptors (GRs) and ionotropic receptors (IRs). GRs consist of seven transmembrane domains, an intracellular N-terminus, and an extracellular C-terminus (29–32). Earlier studies that disrupted G-protein subunits in GR-expressing cells found a reduction in taste responses (33, 34), but recent research elucidating the structure of two sugar GRs has determined that they form tetrameric ligand-gated cation channels with peripheral ligand binding sites and a single central pore (35). The other class of Drosophila taste receptors, IRs, share structural similarities with synaptic, glutamate-gated ion channels: 3 transmembrane domains and a 2-lobed extracellular binding domain (36–38). IRs form heteromeric receptor complexes comprised of both co-receptors and “tuning” receptors that function as ligand-gated ion channels (39). In contrast to IRs and GRs, mammalian T1Rs and T2Rs are G-protein coupled receptors (GPCRs) (40–42), yet the repertoire of tastants acting via these receptors and their impact on behavior is remarkably conserved (1).
Sweet, bitter, and salty taste
Direct activation of Drosophila “sweet” GRNs leads to appetitive feeding behaviors whereas “bitter” GRN activation produces avoidance (15, 19, 43), consistent with mammalian studies on these canonical tastes (1, 44, 45). Drosophila sugar receptors formed from 9 GR genes (sugar GRs) (46–51) detect mono- and disaccharides along with artificial sweeteners and molecules perceived as sweet to humans (52–54). The remaining 30+ GR genes form receptors in “bitter” GRNs (bitter GRs), detecting a range of bitter compounds (e.g., caffeine, lobeline, denatonium, and quinine) (55–58). Recent work in Drosophila has identified two non-canonical bitter signaling pathways for the detection of specific ligands, using rhodopsins and a peptidoglycan recognition protein (PGRP), that open new avenues for taste transduction (Table 1; Figure 1B). Rhodopsin GPCRs are typically light-sensitive with an opsin protein and retinal chromophore, but three rhodopsins (Rh1, Rh4, and Rh7) were found to function as taste receptors that do not require light or retinal (59). These rhodopsins detect aristolochic acid and activate “bitter” GRNs at particularly low concentrations through a phospholipase C (PLC) signaling cascade that involves TRPA1 (59). Mouse taste buds express some opsin RNA (75), suggesting these channels may have a conserved role in chemosensation. The other non-canonical pathway involves PGRPs, pattern recognition receptors traditionally involved in the immune response to pathogens. TRPA1 and canonical bitter GRs (Gr33a, Gr66a) were previously implicated in the detection of bacterial components (76, 77), but the newly described PGRP (PGRP-LB) expressed in the labellum specifically detects bacterial peptidoglycans. Unexpectedly, this receptor uses nuclear factor-κB (NF-κB)/immune deficiency (IMD)-dependent signaling to activate “bitter” GRNs (60). An interest in the role of oral taste receptors in microbial detection has emerged in mammalian research (78, 79), and this recent work in fruit flies highlights an unexpected role for NF-κB/IMD signaling in taste cells that impacts feeding choices (60).
Recent advances in salt taste have revealed a complex taste transduction system that allows for concentration-dependent salt feeding in both mammals and fruit flies (80). A set of “high salt” or “salty” GRNs in the Drosophila labellum are specifically activated by high concentrations of various salt ions (15, 81, 82). Salt also activates other GRNs (“sweet,” “bitter,” and “IR94e”) while inhibiting “water” GRNs, producing a combinatorial code that can lead to flexible behaviors (15). Salt taste research highlights the role of IRs that use the broadly expressed co-receptors, IR25a and IR76b, plus a narrowly expressed “tuning” IR to form functional receptors that detect specific salt ions (15, 82–84). Canonical salt taste centers around NaCl and occasionally other mono- or divalent ions (80), but recent research has shifted focus to identify the taste mechanisms for other ions.
Metal taste
Metals, including divalent and trivalent salt ions, have complex taste profiles (85–87) that have garnered increasing attention due to their accumulation in soil, crops, and foods from human activities (88, 89). Recent studies established that the human bitter taste receptor TAS2R7 acts as a metal cation receptor for detecting zinc and copper (90), yet this can only be demonstrated in vitro. Fruit flies avoid consuming metals and in vivo quantifications of neuronal activity reveal that metal ions activate taste cells through multiple receptors (Table 1; Figure 1B).
In Drosophila, some metal ions require only bitter GRs (Cu2+, Ag+) or IRs (Mn2+, Ni2+, Cd2+) for detection, while others require both (Zn2+, Co2+). Interestingly, cellular responses to iron involve both receptor types or solely IRs depending if it is in the Fe2+ or Fe3+ form, respectively (61). Cadmium sensitivity requires an IR complex in two types of GRNS: co-receptors (IR25a, IR76b) plus IR7a in “bitter” GRNs and the same co-receptors plus IR47a in “salty” GRNs (63). A recent brief report found an additional “tuning” receptor, IR56b, to be necessary for zinc avoidance (62), however, this receptor complex detects NaCl in “sweet” GRNs for attraction (83), so this is an unexpected result. Overall, this research in flies provides clear evidence that a range of individual metal ions have specific taste detection mechanisms. As metal contamination continues to rise, understanding gustatory pathways for metal ligands will become increasingly important across animals with relevance to environmental health and food safety.
Sour taste
pH is an important indicator of food quality and sour taste describes the gustatory detection of acids. Recently, the Otop1 proton channel was identified as the “sour receptor” in mammals (91, 92), and a homolog, OtopLa, is expressed in fruit fly GRNs in the labellum (93, 94). While the discovery of Otop channels was an important breakthrough for sour taste, different acids have distinct taste qualities even at the same pH, suggesting there is more to sour taste than pH alone (95, 96).
Like humans, fruit flies show dose-dependent attraction or aversion to certain carboxylic acids (67). Weak organic acids, such as acetic acid, may have the ability to cross the membrane of taste cells to impact transduction by altering intracellular pH, but through unknown mechanisms (97, 98). In Drosophila, attractive concentrations of specific organic acids—acetic, lactic, glycolic, and citric—require taste receptors for the activation of “sweet” GRNs (64–66). At least one broadly expressed co-receptor, IR25a, is involved, along with sugar GRs for detecting organic acids (64–66) (Table 1; Figure 1B). Even at the same pH, these acids differentially activate Drosophila taste cells in vivo, indicating diverse receptor binding and/or abilities to cross cell membranes. Attempts to distinguish between the detection of pH and anion species show that IRs are largely involved with anion detection, whereas the sugar GRs are responsive to the change in pH (66). Ascorbic acid (Vitamin C), a distinct but related acidic compound, was also found to activate “sweet” GRNs through similar mechanisms (65). High concentrations of carboxylic acids are aversive (99), and IR7a in “bitter” GRNs is specifically required for acetic acid avoidance, without the need for IR co-receptors (67). While the cooperative role of OtopLa channels along with these receptors remains unclear, these findings underscore the dual activation of taste cells by acids through both receptors and proton influx.
Alkaline taste
Since pH influences food quality, the ability to detect both basic and acidic pH levels would be advantageous. Previous mammalian studies on basic pH sensation focused on somatosensation (100), but humans show alkaline sensitivity on the tip of the tongue (101) and a recent study in rats found that sodium carbonate (Na2CO3) solutions activate taste nerves significantly more than Na+ alone. However, alkaline taste has not been well described. A recent study in Drosophila established the existence of alkaline taste, and identified a novel receptor required for the detection of basic solutions (70) (Table 1; Figure 1B). Alkaliphile (Alka) is a Cl− channel gated by high pH that is necessary for alkaline taste (70). The Alka receptor is expressed in a subset of the “bitter” GRNs (~21%), but it is currently unclear what other cell types may express this receptor. Regardless, this study in flies establishes a novel tastant-receptor interaction for alkaline taste that may be relevant to mammals. Interestingly, the Otop1 proton channel for sour taste was recently found to be a candidate alkaline receptor in vitro (102), indicating a need for future comparative studies on basic and acidic pH detection mechanisms.
Umami (amino acid) taste
Protein feeding is coupled with the chemical detection of amino acids. Umami taste is a specific savory sensation, usually associated with monosodium L-glutamate (MSG), an amino acid often found in foods at higher concentrations (103–105). The mammalian GPCR complex consisting of T1R1 + T1R3 is referred to as the “umami receptor” (45) and has a high sensitivity to glutamate in humans (106). In most vertebrates, this receptor is broadly responsive to amino acid ligands and amino acids can also activate sugar taste receptors, bitter taste receptors, or act through metabotropic glutamate receptors in multiple cell types (107–114). This combinatorial coding likely occurs in response to individual amino acids in a dose-dependent manner. Through in vitro assays, mammalian bitter receptors display dose-dependent activation by amino acids, however, some inconclusive results are attributed to the possibility of endogenous amino acid receptors in the cell line used for these experiments (109). In vivo studies in Drosophila circumvent these concerns and allow for a deeper understanding of the combinatorial coding for amino acid taste (Table 1).
Fruit flies require and consume amino acids based on internal state, such as mating status or nutritional deficiency (115). The IR co-receptors (IR76b and IR25a) are necessary for detecting most of the proteinogenic amino acids at various concentrations (68, 69, 116), and the “tuning” receptors identified for amino acid sensation to date are IR51b and IR94e. IR51b is a bitter cell-specific receptor that detects high concentrations of arginine, valine, leucine, tryptophan, isoleucine, lysine, and proline (68). IR94e receptors are integral for the detection of glutamate in various forms, and this “tuning” receptor is expressed in a newly described set of taste cells that induce mild feeding aversion (27, 69). A thorough description of the combinatorial coding for low concentrations (25 mM) of arginine reveals that “sweet” GRNs are activated through both sugar GRs (Gr5a, Gr61a, and Gr64f) and IR co-receptors (68) (Figure 1B). The overlap between sugar-sensing and amino acid-sensing resembles a pattern found in mammals (117).
A feature of the mammalian “umami receptor” is enhancement by purine-5′-nucleotides (IMP and GMP) (111, 114, 118), but this feature is not known to occur in fruit flies. Additionally, while no metabotropic glutamate receptors have been identified in fruit fly amino acid taste, the IRs are ancestrally related to ionotropic glutamate receptors (36–38, 119), suggesting a conserved use of glutamate receptors in chemosensation (107, 113, 120). In Drosophila, another intriguing element is that an odorant binding protein (OBP19b) secreted from nearby cells can bind certain amino acids to impact their detection by taste cells (121), but it is unclear how conserved this mechanism may be. Despite some differences from the mammalian system, the Drosophila model offers a way to study dose-dependent encoding of individual or groups of amino acids to better understand this canonical yet complex taste modality.
Fatty acid taste
Fatty acids are highly energetic essential nutrients that are attractive to both mammals and Drosophila (71, 122–124). Initially, fat palatability was thought to be driven by texture and olfaction (125), but more recent research has highlighted the importance of gustation (126–128). In mice, CD36 is a fatty acid transporter expressed in taste buds that contributes to fatty acid preferences (129), and two GPCRs (GPR40 and GPR120) appear to function as lingual fat receptors (130). Although Drosophila homologs have not been discovered, GRNs in the labellum do detect fatty acids (71). Similar to carboxylic and amino acids, the cellular and behavioral responses to fatty acids in flies depend on concentration.
At low concentrations (~0.1%), hexanoic acid elicits appetitive responses in Drosophila, while at high concentrations (~1–2%), it prompts aversion (74). Hexanoic acid attraction is driven by “sweet” GRNs, requiring both IR56d and Gr64d (72–74, 131, 132). Aversion to hexanoic acid is controlled by “bitter” GRNs via three bitter receptors: Gr32a, Gr33a, and Gr66a (74) (Table 1). Recent work has also demonstrated that the fly gustatory system can distinguish between different classes of fatty acids based on chain length (73). While all classes of fatty acids require the IR co-receptors (IR25a and IR76b) for detection, medium-chain fatty acid (MCFA) taste requires “sweet” GRNs and the IR56d receptor, whereas short-chain (SCFA) and long-chain (LCFA) fatty acid taste does not (73). These findings indicate that IR56d is selective for MCFAs, while the co-receptors may function more broadly. However, a recent study questioned the involvement of IR25a and IR76b in the labellar response to the MCFA hexanoic acid (74). The molecular and cellular underpinnings of SCFA/LCFA detection and fatty acid discrimination remain unclear, but these complexities reflect a nuanced fatty acid taste encoding system that is sensitive to both concentration and subtle variations in molecular structure.
MCFA taste also requires intracellular signaling, as flies with a mutant norpA, a Drosophila homolog for PLC, have disrupted MCFA detection (71) (Figure 1B). Whether or not PLC signaling is necessary for SCFA and LCFA sensation is unknown. Furthermore, one study showed that the sugar GR, Gr64e, is an essential component of MCFA signal transduction, unexpectedly serving as a downstream component in the PLC pathway within “sweet” GRNs (133). Notably, a recent investigation found that Gr64e mutation did not affect electrophysiological responses to the MCFA hexanoic acid (74). Despite this discrepancy, activation of a secondary receptor via PLC mimics the mammalian fatty acid signaling cascade. Mice lacking PLC or TRPM5, a downstream receptor in the PLC cascade, lose their taste preference for fatty acids (134). Collectively, these results imply that PLC-mediated intracellular mechanisms underpin fatty acid gustation in both Drosophila and mammals, despite mammalian research primarily focusing on LCFAs which remain attractive at higher concentrations (130). Drosophila fatty acid taste emphasizes the conserved nature of macronutrient taste encoding and may prove valuable for informing future fat perception research to uncover more about this non-canonical taste modality that has many health implications.
Discussion
Recent advances in gustation research using Drosophila melanogaster as a model organism have revealed several unexpected ligand-receptor interactions within the taste system that play crucial roles in chemosensation and behavior. The discovery of two novel receptor signaling types in bitter cells, through non-canonical rhodopsin and immune signaling, has revealed unexpected transducers for contact chemical cues. Moreover, the fly gustatory system contains a markedly complex set of receptors to detect specific metals, which may become increasingly relevant in this Anthropocene Epoch. The identification of receptors for carboxylic acid anions suggests a mechanism for sour taste that extends beyond proton detection, while a novel receptor for alkaline solutions highlights the role of gustation in discerning a broader pH spectrum. The ability to study intact taste cells in awake flies has provided key insights into the concentration-dependent nature of ligand detection across multiple receptors and cell types for carboxylic, amino, and fatty acids that imply combinatorial taste coding mechanisms to specific molecules. Future work can apply these insights to continue understanding the repertoire of tastant-receptor interactions behind basic, canonical tastes and emerging, non-canonical taste modalities.
Author contributions
CA: Writing – review & editing, Writing – original draft, Visualization, Conceptualization. JG: Writing – review & editing, Writing – original draft, Visualization, Conceptualization. KA: Writing – review & editing, Visualization. MS: Writing – review & editing, Writing – original draft, Visualization, Supervision, Funding acquisition, Conceptualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The University of Vermont new lab startup funds were used to support this publication.
Acknowledgments
BioRender was used to generate Figure 1.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Yarmolinsky, DA, Zuker, CS, and Ryba, NJ. Common sense about taste: from mammals to insects. Cell. (2009) 139:234–44. doi: 10.1016/j.cell.2009.10.001
2. Beauchamp, GK. Basic Taste: A Perceptual Concept. J Agric Food Chem. (2019) 67:13860–9. doi: 10.1021/acs.jafc.9b03542
3. Chandrashekar, J, Hoon, MA, Ryba, NJP, and Zuker, CS. The receptors and cells for mammalian taste. Nature. (2006) 444:288–94. doi: 10.1038/nature05401
4. Montell, C. A taste of the Drosophila gustatory receptors. Curr Opin Neurobiol. (2009) 19:345–53. doi: 10.1016/j.conb.2009.07.001
5. Scott, K. Gustatory processing in Drosophila melanogaster. Annu Rev Entomol. (2018) 63:15–30. doi: 10.1146/annurev-ento-020117-043331
6. Delventhal, R, Kiely, A, and Carlson, JR. Electrophysiological recording from Drosophila labellar taste sensilla. J Vis Exp. (2014) 2014:e51355. doi: 10.3791/51355
7. Dweck, HKM, and Carlson, JR. Diverse mechanisms of taste coding in Drosophila. Science. Advances. (2023) 9:eadj7032. doi: 10.1126/sciadv.adj7032
8. Marella, S, Mann, K, and Scott, K. Dopaminergic modulation of sucrose acceptance behavior in Drosophila. Neuron. (2012) 73:941–50. doi: 10.1016/j.neuron.2011.12.032
9. Chen, Y-CD, and Dahanukar, A. Recent advances in the genetic basis of taste detection in Drosophila. Cell Mol Life Sci. (2020) 77:1087–101. doi: 10.1007/s00018-019-03320-0
10. Freeman, EG, and Dahanukar, A. Molecular neurobiology of Drosophila taste. Curr Opin Neurobiol. (2015) 34:140–8. doi: 10.1016/j.conb.2015.06.001
11. Montell, C. Drosophila sensory receptors-a set of molecular Swiss Army knives. Genetics. (2021) 217:1–34. doi: 10.1093/genetics/iyaa011
12. Singh, RN. Neurobiology of the gustatory systems of Drosophila and some terrestrial insects. Microsc Res Tech. (1997) 39:547–63. doi: 10.1002/(SICI)1097-0029(19971215)39:6<547::AID-JEMT7>3.0.CO;2-A
13. Fujishiro, N, Kijima, H, and Morita, H. Impulse frequency and action potential amplitude in labellar chemosensory neurones of Drosophila melanogaster. J Insect Physiol. (1984) 30:317–25. doi: 10.1016/0022-1910(84)90133-1
14. Ishimoto, H, and Tanimura, T. Molecular neurophysiology of taste in Drosophila. Cell Mol Life Sci. (2004) 61:10–8. doi: 10.1007/s00018-003-3182-9
15. Jaeger, AH, Stanley, M, Weiss, ZF, Musso, PY, Chan, RC, Zhang, H, et al. A complex peripheral code for salt taste in Drosophila. eLife. (2018) 7:e37167. doi: 10.7554/eLife.37167
16. Falk, R, Bleiser-Avivi, N, and Atidia, J. Labellar taste organs of Drosophila melanogaster. J Morphol. (1976) 150:327–41. doi: 10.1002/jmor.1051500206
17. Stocker, RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. (1994) 275:3–26. doi: 10.1007/BF00305372
18. Stocker, R, and Schorderet, M. Cobalt filling of sensory projections from internal and external mouthparts in Drosophila. Cell Tissue Res. (1981) 216:513–23. doi: 10.1007/BF00238648
19. Gordon, MD, and Scott, K. Motor control in a Drosophila taste circuit. Neuron. (2009) 61:373–84. doi: 10.1016/j.neuron.2008.12.033
20. Dorkenwald, S, Matsliah, A, Sterling, AR, Schlegel, P, Yu, SC, Mckellar, CE, et al. Neuronal wiring diagram of an adult brain. bioRxiv. (2023) 11:2023.06.27.546656. doi: 10.1101/2023.06.27.546656
21. Dorkenwald, S, Mckellar, CE, Macrina, T, Kemnitz, N, Lee, K, Lu, R, et al. FlyWire: online community for whole-brain connectomics. Nat Methods. (2022) 19:119–28. doi: 10.1038/s41592-021-01330-0
22. Scheffer, LK, Xu, CS, Januszewski, M, Lu, Z, Takemura, S-Y, Hayworth, KJ, et al. A connectome and analysis of the adult Drosophila central brain. eLife. (2020) 9:e57443. doi: 10.7554/eLife.57443
23. Zheng, Z, Lauritzen, JS, Perlman, E, Robinson, CG, Nichols, M, Milkie, D, et al. A complete Electron microscopy volume of the brain of adult Drosophila melanogaster. Cell. (2018) 174:730–743.e22. doi: 10.1016/j.cell.2018.06.019
24. Deere, JU, Sarkissian, AA, Yang, M, Uttley, HA, Martinez Santana, N, Nguyen, L, et al. Selective integration of diverse taste inputs within a single taste modality. eLife. (2023) 12:e84856. doi: 10.7554/eLife.84856
25. Jacobs, RV, Wang, CX, Lozada-Perdomo, FV, Nguyen, L, Deere, JU, Uttley, HA, et al. Overlap and divergence of neural circuits mediating distinct behavioral responses to sugar. bioRxiv. (2023). doi: 10.1101/2023.10.01.560401
26. Shiu, PK, Sterne, GR, Engert, S, Dickson, BJ, and Scott, K. Taste quality and hunger interactions in a feeding sensorimotor circuit. eLife. (2022) 11:e79887. doi: 10.7554/eLife.79887
27. Shiu, PK, Sterne, GR, Spiller, N, Franconville, R, Sandoval, A, Zhou, J, et al. A leaky integrate-and-fire computational model based on the connectome of the entire adult Drosophila brain reveals insights into sensorimotor processing. bioRxiv. (2023). doi: 10.1101/2023.05.02.539144
28. Sterne, GR, Otsuna, H, Dickson, BJ, and Scott, K. Classification and genetic targeting of cell types in the primary taste and premotor center of the adult Drosophila brain. eLife. (2021) 10:e71679. doi: 10.7554/eLife.71679
29. Clyne, PJ, Warr, CG, and Carlson, JR. Candidate taste receptors in Drosophila. Science. (2000) 287:1830–4. doi: 10.1126/science.287.5459.1830
30. Dunipace, L, Meister, S, Mcnealy, C, and Amrein, H. Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Curr Biol. (2001) 11:822–35. doi: 10.1016/S0960-9822(01)00258-5
31. Robertson, HM, Warr, CG, and Carlson, JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci. (2003) 100:14537–42. doi: 10.1073/pnas.2335847100
32. Scott, K, Brady, R, Cravchik, A, Morozov, P, Rzhetsky, A, Zuker, C, et al. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell. (2001) 104:661–73. doi: 10.1016/S0092-8674(01)00263-X
33. Bredendiek, N, Hütte, J, Steingräber, A, Hatt, H, Gisselmann, G, and Neuhaus, EM. Goα is involved in sugar perception in Drosophila. Chem Senses. (2010) 36:69–81. doi: 10.1093/chemse/bjq100
34. Ueno, K, Kohatsu, S, Clay, C, Forte, M, Isono, K, and Kidokoro, Y. Gsα is involved in sugar perception in Drosophila melanogaster. J Neurosci. (2006) 26:6143–52. doi: 10.1523/JNEUROSCI.0857-06.2006
35. Ma, D, Hu, M, Yang, X, Liu, Q, Ye, F, Cai, W, et al. Structural basis for sugar perception by Drosophila gustatory receptors. Science. (2024) 383:eadj2609. doi: 10.1126/science.adj2609
36. Benton, R, Vannice, KS, Gomez-Diaz, C, and Vosshall, LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. (2009) 136:149–62. doi: 10.1016/j.cell.2008.12.001
37. Croset, V, Rytz, R, Cummins, SF, Budd, A, Brawand, D, Kaessmann, H, et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. (2010) 6:e1001064. doi: 10.1371/journal.pgen.1001064
38. Rytz, R, Croset, V, and Benton, R. Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem Mol Biol. (2013) 43:888–97. doi: 10.1016/j.ibmb.2013.02.007
39. Abuin, L, Bargeton, B, Ulbrich, MH, Isacoff, EY, Kellenberger, S, and Benton, R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. (2011) 69:44–60. doi: 10.1016/j.neuron.2010.11.042
40. Andres-Barquin, PJ, and Conte, C. Molecular basis of bitter taste: the T2R family of G protein-coupled receptors. Cell Biochem Biophys. (2004) 41:099–112. doi: 10.1385/CBB:41:1:099
41. Sainz, E, Cavenagh, MM, Lopezjimenez, ND, Gutierrez, JC, Battey, JF, Northup, JK, et al. The G-protein coupling properties of the human sweet and amino acid taste receptors. Dev Neurobiol. (2007) 67:948–59. doi: 10.1002/dneu.20403
42. Wong, GT, Gannon, KS, and Margolskee, RF. Transduction of bitter and sweet taste by gustducin. Nature. (1996) 381:796–800. doi: 10.1038/381796a0
43. Marella, S, Fischler, W, Kong, P, Asgarian, S, Rueckert, E, and Scott, K. Imaging taste responses in the fly brain reveals a functional map of taste category and behavior. Neuron. (2006) 49:285–95. doi: 10.1016/j.neuron.2005.11.037
44. Mueller, KL, Hoon, MA, Erlenbach, I, Chandrashekar, J, Zuker, CS, and Ryba, NJ. The receptors and coding logic for bitter taste. Nature. (2005) 434:225–9. doi: 10.1038/nature03352
45. Zhao, GQ, Zhang, Y, Hoon, MA, Chandrashekar, J, Erlenbach, I, Ryba, NJ, et al. The receptors for mammalian sweet and umami taste. Cell. (2003) 115:255–66. doi: 10.1016/S0092-8674(03)00844-4
46. Dahanukar, A, Lei, Y-T, Kwon, JY, and Carlson, JR. Two gr genes underlie sugar reception in Drosophila. Neuron. (2007) 56:503–16. doi: 10.1016/j.neuron.2007.10.024
47. Fujii, S, Yavuz, A, Slone, J, Jagge, C, Song, X, and Amrein, H. Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Curr Biol. (2015) 25:621–7. doi: 10.1016/j.cub.2014.12.058
48. Jiao, Y, Moon, SJ, and Montell, C. A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc Natl Acad Sci. (2007) 104:14110–5. doi: 10.1073/pnas.0702421104
49. Jiao, Y, Moon, SJ, Wang, X, Ren, Q, and Montell, C. Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila. Curr Biol. (2008) 18:1797–801. doi: 10.1016/j.cub.2008.10.009
50. Slone, J, Daniels, J, and Amrein, H. Sugar receptors in Drosophila. Curr Biol. (2007) 17:1809–16. doi: 10.1016/j.cub.2007.09.027
51. Yavuz, A, Jagge, C, Slone, J, and Amrein, H. A genetic tool kit for cellular and behavioral analyses of insect sugar receptors. Fly (Austin). (2014) 8:189–96. doi: 10.1080/19336934.2015.1050569
52. Dus, M, Min, S, Keene, AC, Lee, GY, and Suh, GS. Taste-independent detection of the caloric content of sugar in Drosophila. Proc Natl Acad Sci USA. (2011) 108:11644–9. doi: 10.1073/pnas.1017096108
53. Gordesky-Gold, B, Rivers, N, Ahmed, OM, and Breslin, PAS. Drosophila melanogaster prefers compounds perceived sweet by humans. Chem Senses. (2008) 33:301–9. doi: 10.1093/chemse/bjm088
54. Stafford, JW, Lynd, KM, Jung, AY, and Gordon, MD. Integration of taste and calorie sensing in Drosophila. J Neurosci. (2012) 32:14767–74. doi: 10.1523/JNEUROSCI.1887-12.2012
55. Lee, Y, Moon, SJ, Montell, C, and Snyder, SH. Multiple gustatory receptors required for the caffeine response in Drosophila. Proc Natl Acad Sci USA. (2009) 106:4495–500. doi: 10.1073/pnas.0811744106
56. Moon, SJ, Lee, Y, Jiao, Y, and Montell, C. A Drosophila gustatory receptor essential for aversive taste and inhibiting male-to-male courtship. Curr Biol. (2009) 19:1623–7. doi: 10.1016/j.cub.2009.07.061
57. Shim, J, Lee, Y, Jeong, YT, Kim, Y, Lee, MG, Montell, C, et al. The full repertoire of Drosophila gustatory receptors for detecting an aversive compound. Nat Commun. (2015) 6:8867. doi: 10.1038/ncomms9867
58. Weiss, LA, Dahanukar, A, Kwon, JY, Banerjee, D, and Carlson, JR. The molecular and cellular basis of bitter taste in Drosophila. Neuron. (2011) 69:258–72. doi: 10.1016/j.neuron.2011.01.001
59. Leung, NY, Thakur, DP, Gurav, AS, Kim, SH, Di Pizio, A, Niv, MY, et al. Functions of opsins in Drosophila taste. Curr Biol. (2020) 30:e1366:1367–1379.e6. doi: 10.1016/j.cub.2020.01.068
60. Masuzzo, A, Manière, G, Grosjean, Y, Kurz, L, and Royet, J. Bacteria-derived peptidoglycan triggers a non-canonical NF-κB dependent response in Drosophila gustatory neurons. J Neurosci. (2022) 42:JN-RM-2437-21–823. doi: 10.1523/JNEUROSCI.2437-21.2022
61. Xiao, S, Baik, LS, Shang, X, and Carlson, JR. Meeting a threat of the Anthropocene: taste avoidance of metal ions by Drosophila. Proc Natl Acad Sci. (2022) 119:e2204238119. doi: 10.1073/pnas.2204238119
62. Luo, R, Zhang, Y, Jia, Y, Zhang, Y, Li, Z, Zhao, J, et al. Molecular basis and homeostatic regulation of zinc taste. Protein Cell. (2022) 13:462–9. doi: 10.1007/s13238-021-00845-8
63. Li, X, Sun, Y, Gao, S, Li, Y, Liu, L, and Zhu, Y. Taste coding of heavy metal ion-induced avoidance in Drosophila. iScience. (2023) 26:106607. doi: 10.1016/j.isci.2023.106607
64. Devineni, AV, Sun, B, Zhukovskaya, A, and Axel, R. Acetic acid activates distinct taste pathways in Drosophila to elicit opposing, state-dependent feeding responses. eLife. (2019) 8:e47677. doi: 10.7554/eLife.47677
65. Shrestha, B, Aryal, B, and Lee, Y. The taste of vitamin C in Drosophila. EMBO Rep. (2023) 24:e56319. doi: 10.15252/embr.202256319
66. Stanley, M, Ghosh, B, Weiss, ZF, Christiaanse, J, and Gordon, MD. Mechanisms of lactic acid gustatory attraction in Drosophila. Curr Biol. (2021) 31:3525–3537.e6. doi: 10.1016/j.cub.2021.06.005
67. Rimal, S, Sang, J, Poudel, S, Thakur, D, Montell, C, and Lee, Y. Mechanism of acetic acid gustatory repulsion in Drosophila. Cell Rep. (2019) 26:1432–1442.e4. doi: 10.1016/j.celrep.2019.01.042
68. Aryal, B, Dhakal, S, Shrestha, B, and Lee, Y. Molecular and neuronal mechanisms for amino acid taste perception in the Drosophila labellum. Curr Biol. (2022) 32:1376–1386.e4. doi: 10.1016/j.cub.2022.01.060
69. Guillemin, J, Li, V, Davis, G, Audette, K, Li, J, Jelen, M, et al. Taste cells expressing Ionotropic Receptor 94e reciprocally impact feeding and egg laying in Drosophila. bioRxiv. (2024). doi: 10.1101/2024.01.23.576843
70. Mi, T, Mack, JO, Koolmees, W, Lyon, Q, Yochimowitz, L, Teng, ZQ, et al. Alkaline taste sensation through the alkaliphile chloride channel in Drosophila. Nat Metab. (2023) 5:466–80. doi: 10.1038/s42255-023-00765-3
71. Masek, P, and Keene, AC. Drosophila fatty acid taste signals through the PLC pathway in sugar-sensing neurons. PLoS Genet. (2013) 9:e1003710. doi: 10.1371/journal.pgen.1003710
72. Tauber, JM, Brown, EB, Li, Y, Yurgel, ME, Masek, P, and Keene, AC. A subset of sweet-sensing neurons identified by IR56d are necessary and sufficient for fatty acid taste. PLoS Genet. (2017) 13:e1007059. doi: 10.1371/journal.pgen.1007059
73. Brown, EB, Shah, KD, Palermo, J, Dey, M, Dahanukar, A, and Keene, AC. Ir56d-dependent fatty acid responses in Drosophila uncover taste discrimination between different classes of fatty acids. eLife. (2021) 10:e67878. doi: 10.7554/eLife.67878
74. Pradhan, RN, Shrestha, B, and Lee, Y. Molecular basis of Hexanoic acid taste in Drosophila melanogaster. Mol Cells. (2023) 46:451–60. doi: 10.14348/molcells.2023.0035
75. Sukumaran, SK, Lewandowski, BC, Qin, Y, Kotha, R, Bachmanov, AA, and Margolskee, RF. Whole transcriptome profiling of taste bud cells. Sci Rep. (2017) 7:7595. doi: 10.1038/s41598-017-07746-z
76. Charroux, B, Daian, F, and Royet, J. Drosophila aversive behavior toward Erwinia carotovora carotovora is mediated by bitter neurons and Leukokinin. iScience. (2020) 23:101152. doi: 10.1016/j.isci.2020.101152
77. Soldano, A, Alpizar, YA, Boonen, B, Franco, L, López-Requena, A, Liu, G, et al. Gustatory-mediated avoidance of bacterial lipopolysaccharides via TRPA1 activation in Drosophila. eLife. (2016) 5:e13133. doi: 10.7554/eLife.13133
78. Kouakou, YI, and Lee, RJ. Interkingdom detection of bacterial quorum-sensing molecules by mammalian taste receptors. Microorganisms. (2023) 11:1295. doi: 10.3390/microorganisms11051295
79. Qin, Y, Palayyan, SR, Zheng, X, Tian, S, Margolskee, RF, and Sukumaran, SK. Type II taste cells participate in mucosal immune surveillance. PLoS Biol. (2023) 21:e3001647. doi: 10.1371/journal.pbio.3001647
80. Taruno, A, and Gordon, MD. Molecular and cellular mechanisms of salt taste. Annu Rev Physiol. (2023) 85:25–45. doi: 10.1146/annurev-physiol-031522-075853
81. Lee, Y, Poudel, S, Kim, Y, Thakur, D, and Montell, C. Calcium taste avoidance in Drosophila. Neuron. (2018) 97:e64:67–74.e4. doi: 10.1016/j.neuron.2017.11.038
82. Mcdowell, SAT, Stanley, M, and Gordon, MD. A molecular mechanism for high salt taste in Drosophila. Curr Biol. (2022) 32:3070–3081.e5. doi: 10.1016/j.cub.2022.06.012
83. Dweck, HKM, Talross, GJS, Luo, Y, Ebrahim, SAM, and Carlson, JR. Ir56b is an atypical ionotropic receptor that underlies appetitive salt response in Drosophila. Curr Biol. (2022) 32:1776–1787.e4. doi: 10.1016/j.cub.2022.02.063
84. Zhang, YV, Ni, J, and Montell, C. The molecular basis for attractive salt-taste coding in Drosophila. Science. (2013) 340:1334–8. doi: 10.1126/science.1234133
85. Lawless, HT, Schlake, S, Smythe, J, Lim, J, Yang, H, Chapman, K, et al. Metallic taste and retronasal smell. Chem Senses. (2004) 29:25–33. doi: 10.1093/chemse/bjh003
86. Lawless, HT, Stevens, DA, Chapman, KW, and Kurtz, A. Metallic taste from electrical and chemical stimulation. Chem Senses. (2005) 30:185–94. doi: 10.1093/chemse/bji014
87. Tansaraviput, S, and Nolden, AA. Sucrose, NaCl, and citric acid suppress the metallic sensation of FeSO4. Chem Senses. (2024) 49:bjad052. doi: 10.1093/chemse/bjad052
88. Clemens, S, and Ma, JF. Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu Rev Plant Biol. (2016) 67:489–512. doi: 10.1146/annurev-arplant-043015-112301
89. Hou, D, O’connor, D, Igalavithana, AD, Alessi, DS, Luo, J, Tsang, DCW, et al. Metal contamination and bioremediation of agricultural soils for food safety and sustainability. Nat Rev Earth Environ. (2020) 1:366–81. doi: 10.1038/s43017-020-0061-y
90. Wang, Y, Zajac, AL, Lei, W, Christensen, CM, Margolskee, RF, Bouysset, C, et al. Metal ions activate the human taste receptor TAS2R7. Chem Senses. (2019) 44:339–47. doi: 10.1093/chemse/bjz024
91. Teng, B, Wilson, CE, Tu, Y-H, Joshi, NR, Kinnamon, SC, and Liman, ER. Cellular and neural responses to sour stimuli require the proton channel Otop1. Curr Biol. (2019) 29:e3645:3647–3656.e5. doi: 10.1016/j.cub.2019.08.077
92. Zhang, J, Jin, H, Zhang, W, Ding, C, O’keeffe, S, Ye, M, et al. Sour sensing from the tongue to the brain. Cell. (2019) 179:e315:392–402.e15. doi: 10.1016/j.cell.2019.08.031
93. Ganguly, A, Chandel, A, Turner, H, Wang, S, Liman, ER, and Montell, C. Requirement for an Otopetrin-like protein for acid taste in Drosophila. Proc Natl Acad Sci. (2021) 118:e2110641118. doi: 10.1073/pnas.2110641118
94. Mi, T, Mack, JO, Lee, CM, and Zhang, YV. Molecular and cellular basis of acid taste sensation in Drosophila. Nat Commun. (2021) 12:3730. doi: 10.1038/s41467-021-23490-5
95. Da Conceicao Neta, ER, Johanningsmeier, SD, and Mcfeeters, RF. The chemistry and physiology of sour taste—a review. J Food Sci. (2007) 72:R33–8. doi: 10.1111/j.1750-3841.2007.00282.x
96. Harvey, R. The relation between the total acidity, the concentration of the hydrogen ion, and the taste of acid solutions. J Am Chem Soc. (1920) 42:712–4. doi: 10.1021/ja01449a005
97. Liman, ER, Zhang, YV, and Montell, C. Peripheral coding of taste. Neuron. (2014) 81:984–1000. doi: 10.1016/j.neuron.2014.02.022
98. Lyall, V, Alam, RI, Phan, DQ, Ereso, GL, Phan, T-HT, Malik, SA, et al. Decrease in rat taste receptor cell intracellular pH is the proximate stimulus in sour taste transduction. Am J Phys Cell Phys. (2001) 281:C1005–13. doi: 10.1152/ajpcell.2001.281.3.C1005
99. Charlu, S, Wisotsky, Z, Medina, A, and Dahanukar, A. Acid sensing by sweet and bitter taste neurons in Drosophila melanogaster. Nat Commun. (2013) 4:2042. doi: 10.1038/ncomms3042
100. Bryant, BP. Mechanisms of somatosensory neuronal sensitivity to alkaline pH. Chem Senses. (2005) 30:i196–7. doi: 10.1093/chemse/bjh182
101. Kloehn, NW, and Brogden, W. The alkaline taste: a comparison of absolute thresholds for sodium hydroxide on the tip and mid-dorsal surfaces of the tongue. Am J Psychol. (1948) 61:90–3. doi: 10.2307/1417296
102. Tian, L, Zhang, H, Yang, S, Luo, A, Kamau, PM, Hu, J, et al. Vertebrate OTOP1 is also an alkali-activated channel. Nat Commun. (2023) 14:26. doi: 10.1038/s41467-022-35754-9
103. Dai, Z, Zheng, W, and Locasale, JW. Amino acid variability, tradeoffs and optimality in human diet. Nat Commun. (2022) 13:6683. doi: 10.1038/s41467-022-34486-0
105. Maga, JA, and Yamaguchi, S. Flavor potentiators. Crit Rev Food Sci Nutr. (1983) 18:231–312. doi: 10.1080/10408398309527364
106. Toda, Y, Hayakawa, T, Itoigawa, A, Kurihara, Y, Nakagita, T, Hayashi, M, et al. Evolution of the primate glutamate taste sensor from a nucleotide sensor. Curr Biol. (2021) 31:4641–4649.e5. doi: 10.1016/j.cub.2021.08.002
107. Chaudhari, N, Pereira, E, and Roper, SD. Taste receptors for umami: the case for multiple receptors. Am J Clin Nutr. (2009) 90:738S–42S. doi: 10.3945/ajcn.2009.27462H
108. Diepeveen, J, Moerdijk-Poortvliet, TC, and Van Der Leij, FR. Molecular insights into human taste perception and umami tastants: a review. J Food Sci. (2022) 87:1449–65. doi: 10.1111/1750-3841.16101
109. Kohl, S, Behrens, M, Dunkel, A, Hofmann, T, and Meyerhof, W. Amino acids and peptides activate at least five members of the human bitter taste receptor family. J Agric Food Chem. (2013) 61:53–60. doi: 10.1021/jf303146h
110. Mcgrane, SJ, Gibbs, M, Hernangomez De Alvaro, C, Dunlop, N, Winnig, M, Klebansky, B, et al. Umami taste perception and preferences of the domestic cat (Felis catus), an obligate carnivore. Chem Senses. (2023) 48:bjad026. doi: 10.1093/chemse/bjad026
111. Nelson, G, Chandrashekar, J, Hoon, MA, Feng, L, Zhao, G, Ryba, NJ, et al. An amino-acid taste receptor. Nature. (2002) 416:199–202. doi: 10.1038/nature726
112. Pal Choudhuri, S, Delay, RJ, and Delay, ER. L-amino acids elicit diverse response patterns in taste sensory cells: a role for multiple receptors. PLoS One. (2015) 10:e0130088. doi: 10.1371/journal.pone.0130088
113. San Gabriel, A, Uneyama, H, Yoshie, S, and Torii, K. Cloning and characterization of a novel mGluR1 variant from vallate papillae that functions as a receptor for L-glutamate stimuli. Chem Senses. (2005) 30:i25–6. doi: 10.1093/chemse/bjh095
114. Zhang, J, Sun-Waterhouse, D, Su, G, and Zhao, M. New insight into umami receptor, umami/umami-enhancing peptides and their derivatives: a review. Trends Food Sci Technol. (2019) 88:429–38. doi: 10.1016/j.tifs.2019.04.008
115. Toshima, N, and Schleyer, M. Neuronal processing of amino acids in Drosophila: from taste sensing to behavioural regulation. Curr Opin Insect Sci. (2019) 36:39–44. doi: 10.1016/j.cois.2019.07.007
116. Steck, K, Walker, SJ, Itskov, PM, Baltazar, C, Moreira, J-M, and Ribeiro, C. Internal amino acid state modulates yeast taste neurons to support protein homeostasis in Drosophila. eLife. (2018) 7:e31625. doi: 10.7554/eLife.31625
117. Kalyanasundar, B, Blonde, GD, Spector, AC, and Travers, SP. Electrophysiological responses to sugars and amino acids in the nucleus of the solitary tract of type 1 taste receptor double-knockout mice. J Neurophysiol. (2020) 123:843–59. doi: 10.1152/jn.00584.2019
118. Zhang, F, Klebansky, B, Fine, RM, Xu, H, Pronin, A, Liu, H, et al. Molecular mechanism for the umami taste synergism. Proc Natl Acad Sci USA. (2008) 105:20930–4. doi: 10.1073/pnas.0810174106
119. Koh, TW, He, Z, Gorur-Shandilya, S, Menuz, K, Larter, NK, Stewart, S, et al. The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron. (2014) 83:850–65. doi: 10.1016/j.neuron.2014.07.012
120. Kurihara, K. Umami the fifth basic taste: history of studies on receptor mechanisms and role as a food flavor. Biomed Res Int. (2015) 2015:189402:1–10. doi: 10.1155/2015/189402
121. Rihani, K, Fraichard, S, Chauvel, I, Poirier, N, Delompré, T, Neiers, F, et al. A conserved odorant binding protein is required for essential amino acid detection in Drosophila. Commun Biol. (2019) 2:425. doi: 10.1038/s42003-019-0673-2
122. Bowen, D, Green, P, Vizenor, N, Vu, C, Kreuter, P, and Rolls, B. Effects of fat content on fat hedonics: cognition or taste? Physiol Behav. (2003) 78:247–53. doi: 10.1016/S0031-9384(02)00973-3
123. Tsuruta, M, Kawada, T, Fukuwatari, T, and Fushiki, T. The orosensory recognition of long-chain fatty acids in rats. Physiol Behav. (1999) 66:285–8. doi: 10.1016/S0031-9384(98)00299-6
124. Warwick, ZS, and Synowski, SJ. Effect of food deprivation and maintenance diet composition on fat preference and acceptance in rats. Physiol Behav. (1999) 68:235–9. doi: 10.1016/S0031-9384(99)00192-4
125. Rolls, ET, Critchley, HD, Browning, AS, Hernadi, I, and Lenard, L. Responses to the sensory properties of fat of neurons in the primate orbitofrontal cortex. J Neurosci. (1999) 19:1532–40. doi: 10.1523/JNEUROSCI.19-04-01532.1999
126. Pittman, D, Crawley, ME, Corbin, CH, and Smith, KR. Chorda tympani nerve transection impairs the gustatory detection of free fatty acids in male and female rats. Brain Res. (2007) 1151:74–83. doi: 10.1016/j.brainres.2007.03.027
127. Running, CA, Craig, BA, and Mattes, RD. Oleogustus: the unique taste of fat. Chem Senses. (2015) 40:507–16. doi: 10.1093/chemse/bjv036
128. Takeda, M, Sawano, S, Imaizumi, M, and Fushiki, T. Preference for corn oil in olfactory-blocked mice in the conditioned place preference test and the two-bottle choice test. Life Sci. (2001) 69:847–54. doi: 10.1016/S0024-3205(01)01180-8
129. Laugerette, F, Passilly-Degrace, P, Patris, B, Niot, I, Febbraio, M, Montmayeur, J-P, et al. CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J Clin Invest. (2005) 115:3177–84. doi: 10.1172/JCI25299
130. Cartoni, C, Yasumatsu, K, Ohkuri, T, Shigemura, N, Yoshida, R, Godinot, N, et al. Taste preference for fatty acids is mediated by GPR40 and GPR120. J Neurosci. (2010) 30:8376–82. doi: 10.1523/JNEUROSCI.0496-10.2010
131. Ahn, J-E, Chen, Y, and Amrein, H. Molecular basis of fatty acid taste in Drosophila. eLife. (2017) 6:e30115. doi: 10.7554/eLife.30115
132. Sánchez-Alcañiz, JA, Silbering, AF, Croset, V, Zappia, G, Sivasubramaniam, AK, Abuin, L, et al. An expression atlas of variant ionotropic glutamate receptors identifies a molecular basis of carbonation sensing. Nat Commun. (2018) 9:4252. doi: 10.1038/s41467-018-06453-1
133. Kim, H, Kim, H, Kwon, JY, Seo, JT, Shin, DM, and Moon, SJ. Drosophila Gr64e mediates fatty acid sensing via the phospholipase C pathway. PLoS Genet. (2018) 14:e1007229. doi: 10.1371/journal.pgen.1007229
Keywords: taste, gustation, chemosensation, Drosophila melanogaster, taste receptor, gustatory receptor, tastant
Citation: Arntsen C, Guillemin J, Audette K and Stanley M (2024) Tastant-receptor interactions: insights from the fruit fly. Front. Nutr. 11:1394697. doi: 10.3389/fnut.2024.1394697
Edited by:
Gabriella Morini, University of Gastronomic Sciences, ItalyReviewed by:
Wenli Tian, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2024 Arntsen, Guillemin, Audette and Stanley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Molly Stanley, bW9sbHkuc3RhbmxleUB1dm0uZWR1
†These authors share first authorship
 Christian Arntsen
Christian Arntsen Jacqueline Guillemin†
Jacqueline Guillemin† Molly Stanley
Molly Stanley