- 1Division of Geriatric Medicine, Department of Medicine, University of Alberta, Edmonton, AB, Canada
- 2Kaye Edmonton Clinic, Alberta Health Services, Edmonton, AB, Canada
- 3Department of Psychiatry, University of Alberta, Edmonton, AB, Canada
All microorganisms like bacteria, viruses and fungi that reside within a host environment are considered a microbiome. The number of bacteria almost equal that of human cells, however, the genome of these bacteria may be almost 100 times larger than the human genome. Every aspect of the physiology and health can be influenced by the microbiome living in various parts of our body. Any imbalance in the microbiome composition or function is seen as dysbiosis. Different types of dysbiosis are seen and the corresponding symptoms depend on the site of microbial imbalance. The contribution of the intestinal and extra-intestinal microbiota to influence systemic activities is through interplay between different axes. Whole body dysbiosis is a complex process involving gut microbiome and non-gut related microbiome. It is still at the stage of infancy and has not yet been fully understood. Dysbiosis can be influenced by genetic factors, lifestyle habits, diet including ultra-processed foods and food additives, as well as medications. Dysbiosis has been associated with many systemic diseases and cannot be diagnosed through standard blood tests or investigations. Microbiota derived metabolites can be analyzed and can be useful in the management of dysbiosis. Whole body dysbiosis can be addressed by altering lifestyle factors, proper diet and microbial modulation. The effect of these interventions in humans depends on the beneficial microbiome alteration mostly based on animal studies with evolving evidence from human studies. There is tremendous potential for the human microbiome in the diagnosis, treatment, and prognosis of diseases, as well as, for the monitoring of health and disease in humans. Whole body system-based approach to the diagnosis of dysbiosis is better than a pure taxonomic approach. Whole body dysbiosis could be a new therapeutic target in the management of various health conditions.
1 Introduction
Microbial cells involve different microorganisms like bacteria, fungi, protozoa, and viruses, which maintain balance in the microbial environment (1). The human body contains both human and microbial cells and as such, “Human beings are now considered as hybrid organisms” (2). Microbial communities inhabiting our body are known as the human microbiota. These microbiotas are seen in the skin, oral cavity, conjunctiva, respiratory tract, genitourinary (GU) and gastrointestinal (GI) tracts. The microbiota in different body surfaces has the ability to repel pathogens, a property known as colonization resistance (3). The microbiome is composed of the microbiota, its genes, and its products, which includes microbial structural products as well as microbial metabolites. It is the second genome of our body. Microbiome can be considered as an acquired invisible organ to the naked eye and present throughout the body. The human gut microbiome is made up of two or more microbiota that is organized to carry out a particular metabolic function and groups/ colonies of microbiome (within the body) with related function similar to an organ system. The connectivity of the microbiome is by integrating different microbiota such as eukaryote, prokaryote, archaea, and viruses and also includes host- microbiome interaction. Full metagenomic DNA sequencing is the basis of microbiome-based diseases (2). Understanding the entire view of the microbiome is not just learning about the colonies of microbiota, but also looking at the metabolic potential which can affect the microbial functioning including the host- microbiome interaction (4). This virtual organ has been a neglected organ till recently. Not only in Modern Medicine, but also in the traditional medicine like Chinese Medicine, Indian Medicine, like Ayurveda, are looking at the relationship of gut microbiota with host health and diseases. The holistic approach in traditional medicine is also now viewed in modern medicine with interplay of various organs with the spirit in the body. Most of these traditional systems give importance mainly to diet. By understanding the role of microbiome, we can appreciate the above-mentioned holistic concept in clinical practice with different medicinal systems (5, 6).
Non-pathogenic bacteria in the body have an effect on health. When there is microbial imbalance or compositional change, dysbiosis can result. Understanding the non-pathogenic microorganisms, microbial genes, and microbiota-derived metabolites will provide a more complete understanding of the microbiome. Different host factors affect the microbial environment (7). Microbiota composition varies with individual genotype, diet, and environment. Diet is the most important contributor of microbial flora. Microbiota plays a crucial role for energy extraction from nutrients through unique enzyme and biochemical pathways (8–10). The composition of the microbiome is host specific and changes throughout an individual’s lifetime (11). With environmental conditions especially with urbanizations, humans are exposed to different environmental exposures including pollution. Air pollutants like carbon monoxide (CO), nitrogen dioxide (NO2) which comes from vehicle exhaust and industrial wastes can play a role (12).
When dysbiosis occurs in the body, the pathogenic bacteria override the beneficial ones potentially causing diseases (13, 14). Whole body dysbiosis is a term to describe the changes in the quantity, variety, and/or location of microorganisms in the human body. This could include both intestinal tract dysbiosis and extraintestinal dysbiosis which have been linked to many human diseases. Malfunction of the microbiota virtual organ can affect even distant organs. The difficulty in explaining dysbiosis is due to the fact that there is no clear definition of a healthy gut microbiota with huge interindividual variation existing in the normal healthy population (15). In eubiosis, there is a preponderance of beneficial bacteria (Phyla Firmicutes and Bacteroidetes) over pathogenic bacteria (Phylum Proteobacteria) (16). Whereas in dysbiosis, changes in different components can be seen such as: (1) loss of beneficial bacteria, (2) overgrowth of potentially pathogenic bacteria, and (3) loss of overall bacterial diversity which can all occur simultaneously (15, 17).
2 Classification of whole body dysbiosis
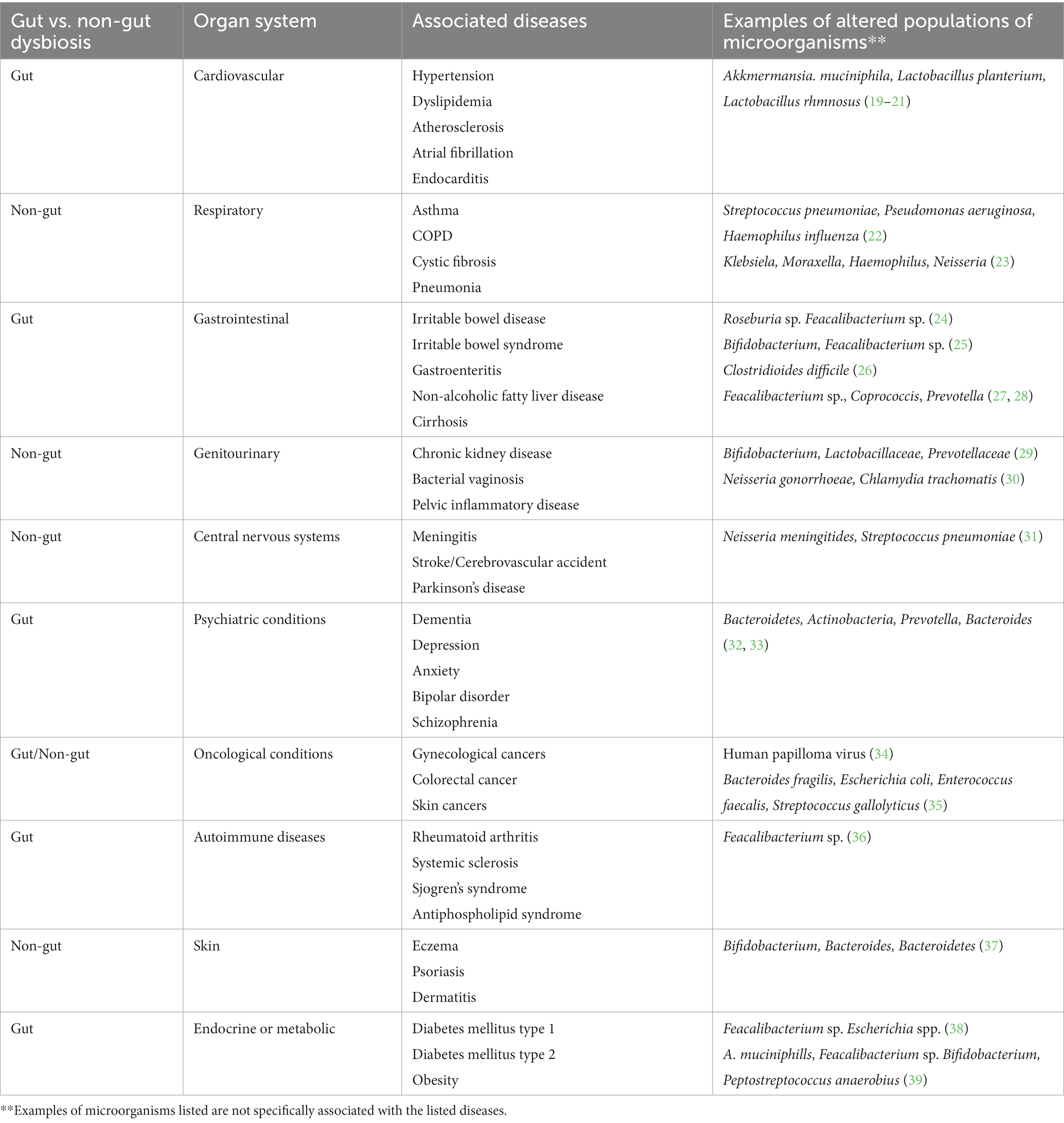
The microbiome in the gut, skin, lung and genitourinary tract is quite distinct and plays a role in health and disease (16, 18) (Table 1). The whole body dysbiosis can be classified into 1. Gut microbial dysbiosis including oral dysbiosis and 2. Non-gut microbial dysbiosis.
2.1 Gut microbial dysbiosis including oral dysbiosis
Huge colonies of microbiota reside within the gastrointestinal tract and produce metabolites which can enter into the blood circulation and affect extraintestinal organs (40–42) Dysbiosis of the oral microbiome is commonly seen with gingivitis, periodontitis, dental caries and oral candidiasis and is associated with systemic diseases (40).
In general, commensal microbiota is very important in maintaining health (43). The role that commensals and pathobionts play in their interaction with the microbial dysbiosis and host is so critical to shifts from health to disease in the oral cavity (44).
Imbalance of oral microbiome is related to disease states. The studies done with saliva showed decreased levels of pyruvate and N-acetylglucosamine in chronic periodontitis (45, 46).
Also, with aging, oral microbiome transformation occurs and lead to systemic diseases. After the age of 60 years, genus Lactobacillus can increase in the oral microbiome and is suspected to contribute to neurodegenerative disorders (47). A study by Jo et al. identified a distinctive connection between the oral and gut microbiota through lactobacilli, which might lead to functional alterations of the Parkinson Disease (PD)-associated microbiome (48).
2.1.1 Changes occurring with dysbiosis in the gut
2.1.1.1 When the normal gut microbiota becomes pathogenic with loss of beneficial bacteria
Normal gut microbiota may act like opportunistic pathogens, when host resistance fails by a gut infection or when the immune resistance becomes deficient. Gut bacteria are less abundant in the stomach and upper intestine and become more populated in the lower GI tract. Both gastric acid and bile have antibacterial properties and prevent pathological bacterial colonization in the upper GI tract. In addition, mechanical factors like peristalsis and the presence of antibacterial substances like bacteriocins and fatty acids also prevent pathological adherence. Antibiotics can inhibit or kill the normal microbiome, leading to pathological overgrowth resulting in dysbiosis (49). With a decrease in peristalsis and lower oxidation–reduction potential, higher numbers of gut bacteria are seen in the ileum and colon. The majority of colonic bacteria are obligate anaerobes (50), however, there are many facultative anaerobes, such as the Enterobacteria, that can contribute to significant negative metabolomic changes.
2.1.1.2 Overgrowth of potentially pathogenic bacteria/loss of overall bacterial diversity representing the microbial signature of dysbiosis
There are more than 1,000 different species of microbiota in the gastrointestinal tract. Most of them are good and essential for optimum health such as Bifidobacterium and Lactobacillus. The gut microbiome is predominantly composed of Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria. When there is an imbalance of either of these phyla of bacteria, dysbiosis can result. The typical signature of dysbiosis is the expansion of Proteobacteria (51). However, even certain Firmicutes, such as Ruminococcus gnavus and Bacteroides fragilis from the Bacteroidetes phyla have been found to play a role in bacterial dysbiosis, which can be seen if Inflammatory Bowel Disease (IBD) and Crohn’s disease (52). Bacteria that display pathogenic properties are referred to as pathobionts, and individual may be naturally colonized with these types of bacteria. Some examples of pathobionts include Clostridioides difficile (formally Clostridium difficile), Enterococcus faecalis and Campylobacter are considered as harmful and pathogenic. The above organisms have a relatively small infective doses, C. difficile at less than 10 spores, E. faecalis at 10 colony-forming units (CFU), and Campylobacter at 500–800 CFUs, that can lead to a disruption of the normal gut microbiome. With dysbiosis, two variations can occur with human microorganisms. 1. An abundance of good bacteria can raise the pH and lead to uncomfortable symptoms of gas, bloating and/or diarrhea. 2. With an abundance of bad bacteria, good bacteria can get diminished with loss of entire species that were present which leads to a reduction in the variety of organisms present (microbial diversity). This abundance of bad bacteria can cause widespread health concerns with depressed immune function, as well as an increase in inflammatory responses. In critical illness, “severe reduction in “health-promoting” commensal intestinal bacteria (such as Firmicutes or Bacteroidetes) and an increase in potentially pathogenic bacteria (e.g., Proteobacteria like Salmonella, Vibrio)” can occur (51). Gut bacteria dynamics vary based on location and the surface within the gastrointestinal tract. Penetration of pathogenic bacteria like Shigella, Salmonella, and Campylobacter throughout the gut surface needs a large bacterial exposure to cause illness (53).
2.2 Non-gut microbial dysbiosis
Outside of the gastrointestinal tracts, there have been other body systems that have been associated with microbial dysbiosis:
2.2.1 Lung dysbiosis including ear, nose, throat tract dysbiosis
The lung microbiota commonly seen in healthy human lungs are Firmicutes, Proteobacteria, Bacteroidetes and Actinobacteria (54). With dysbiosis, altered microbial patterns are seen in the lungs (55). Evidence of microbial dysbiosis is seen with both ears, nose, throat (ENT) and lung conditions (56). Lung dysbiosis occurs with asthma, chronic obstructive pulmonary disease (COPD), idiopathic pulmonary fibrosis as well as lung cancers through the gut-lung axis (57–63).
Cigarette smoking is a known risk factor for COPD and lung cancer, and studies have shown that it can affect the microbiota both in the gut and lungs (64, 65).
A prospective cohort study by Szmidt et al. of 35,339 Swedish women found long-term (10 years) of high fiber intake (from cereal and fruit but not vegetable sources) to be linked with a 30% lower risk of COPD (66). A meta-analysis study showed daily consumption of 10 grams of dietary fiber, cereal fiber and fruit fiber reduced the risk of developing COPD but that effect is not seen with vegetable fiber (67).
The growing evidence points out that gut microbiota can influence the lung microbiome systemically through the gut-lung axis. This opens the potential for intranasal aerosol microbiota therapy in lung dysbiosis subjects (68).
The lung microbiome has been showed to play a role in the progression of lung cancer or its risk for recurrence (69).
2.2.2 Skin dysbiosis including conjunctival and eye dysbiosis
Skin is the largest organ and the skin microbiome contributes to immunity. Microbial composition varies with dry, moist, or oily areas of the skin (70). Recent evidence supports the association between ocular diseases and gut microbiota through gut-eye axis (71). Skin dysbiosis occurs through the skin-gut axis (72), which can be affected by diet (71). Western diet (high fat, higher amount of sugar and salt and processed food ingredients) has been related with psoriasis and atopic dermatitis. Dermatitis herpetiformis associated with celiac disease has shown improvement when changed to a gluten- free diet (73). Ultraviolet B exposure which increases the serum Vitamin D levels by altering the alpha and beta diversity of the gut microbiome (73).
In addition to probiotics (74), prebiotics can also help with skin conditions. An extract from Probiotic Lactobacillus planatarum helps in the management of acne lesions by improving skin barrier function (75). A combination of a probiotic and prebiotic like Bifidobacterium and Glucooligosaccharides (GOS) reduce the transepidermal water loss of skin and prevent erythema (76). GOS have been used in many skin conditions like atopic dermatitis, eczema and photo aging diseases (77). Metabolites produced by probiotics (also known as postbiotics), like sodium butyrate, are used to treat psoriasis which is a proliferative skin disease (78). Other postbiotics, like short chain fatty acids (SCFA), produce anti-inflammatory activities in various skin disorders (76). Through the gut-skin axis, gut dysbiosis is associated with skin conditions, such as atopic dermatitis, psoriasis, acne and rosacea (79–81). Microbials metabolites can affect the immune system via the gut-skin-axis (73). Imbalance of the microbiome can increase the chance of skin infections and diseases, whereas restoring balance with a healthy microbiome may help in the recovery of skin diseases including wound healing (82, 83). Skin cancers can happen because of the microbial toxins causing cellular damage (84).
Microbiota with distinct characteristics is seen in the gut and skin, and this specific microbial composition is affected by a range of other individual attributes, such as age, ethnicity, genetics, climate and skincare (85, 86). Aging, diabetes and skin diseases, can cause microbial dysbiosis and increase infection risk (87).
2.2.3 Genitourinary dysbiosis
Lactobacillus is a common microbiota seen in the healthy vagina (88). With menopause there is reduced estrogen which can increase the vaginal pH, altering the vaginal microbiome and lead to reduced levels of this genus of bacteria. Lactobacillus prevents proliferation of pathogenic microorganisms/vaginal dysbiosis. Postmenopausal changes in the gut microbiome are associated with increased short-chain fatty acids and hydrogen sulfide levels and may play a role in the gut -vagina- bladder- axis. Lactobacillus function to protect the vaginal mucosa against the colonization and proliferation of pathogenic microorganisms. Urinary microbiome dysbiosis is associated with interstitial cystitis, urinary tract infection (UTI), bladder pain syndrome and different types of urinary incontinence (29, 88–94).
Microbiome in the urinary and the genital tract may arise from the gut or vaginal microbiota in females as well as from the environment (95).
3 Concept of whole body dysbiosis
Various extraintestinal organs play a role in the physiological function of the gut microbiome. Gut and non-gut dysbiosis communicate through different axes in a bidirectional manner. This highlights the concept of the gut–organ axis (Figure 1) (11, 96). When there is a disruption in the gut microbiome, there can be a reduction in SCFA producing bacteria along with an increase in toxin producing ones. The intestinal barrier can also be weakened, contributing to bacterial translocation that can influence systemic inflammation. This has been associated with various health conditions, such as diabetes, cardiovascular disease, and neurocognitive disorders (Figure 1). Immune signaling and metabolic reactions contribute to these pathways. This interrelationship can lead to various diseases. Interplay between gut and non-gut dysbiosis, and as such, the different axes is shown in Figure 1. This has opened a new concept, and we coin a new terminology called “whole body dysbiosis.” This concept should help to better understand the pathogenic links between different organs and different medical conditions. (A) The Gut-Brain axis may be affected by dysbiosis leading to altered levels of neurotransmitters and bacterial metabolites. Bacterial translocation can influence neuroinflammation which may play a role in neurocognitive and psychiatric conditions. (B) Gut-Heart Axis may be impacted by altered bacterial population that may generate increased levels of trimethylamine (TMA) that is oxidized in the gut to trimethylamine-N-oxide (TMAO) that can increase the risk of cardiovascular disease. (C) Gut-Skin Axis can be influenced by increases immune response from bacterial translocation which results in increased sebum leading to skin disorders, such as acne. (D) Gut-Kidney Axis can be influenced by dysbiosis by increased toxin forming bacteria leading to uremic toxins damaging the kidneys. The production of TMAO can also contribute to renal insufficiency. (E) Gut-Bone Axis can be impacted by increased immune response and signaling that can affect bone resorption. (F) Gut-Genitourinary Axis is influenced by changes in the gut microbiome can lead to increased SCFA and hydrogen sulfide levels. Postmenopausal reduction in estrogen leads to increased vaginal pH causes a decrease in Lactobacillus. This in turn can contribute to urinary tract infections (UTI), interstitial cystitis, and different type of urinary incontinence. (G) Gut-Liver Axis can be influenced by bacteria metabolites through the hepatoenteric circulation which can activate hepatic stellate and Kupffer cells leading to cytokine and chemokine production resulting in liver damage, insulin resistance, and metabolic disorders. (H) Gut-Adipose Axis can be affected through increased lipopolysaccharide exposure and causing metabolic endotoxemia. (I) Gut-Lung Axis is altered due to increased inflammation and immune signaling leading to conditions such as asthma.

Figure 1. Overview of the gut-organ axis. Disruptions in the gut microbiome can lead to a decrease number of short-chain fatty acid producing bacteria and increased toxin producing bacteria. Along with this, there may be disruptions in the intestinal barriers leading to bacterial translocation that may influence systemic inflammation. (A) The gut-brain axis, (B) gut-heart axis, (C) gut-skin axis, (D) gut-kidney axis, (E) gut-bone, (F) gut-genitourinary axis, (G) gut-liver axis, (H) gut-adipose, and (I) gut-lung axis.
4 Metabolic consequences of dysbiosis
Short chain fatty acids are the metabolic end products of bacterial fermentation, which may have an effect on host health. Short chain fatty acids like propionate, acetate, and butyrate affect carbohydrate fermentation and play a role in the regulation of intestinal motility, as well as, anti-inflammatory function with prevention of leaky gut barrier. Indole degradation of the amino acid tryptophan increases epithelial-cell tight-junction resistance and reduces inflammatory markers. The gut microbiome plays a role in different vitamin synthesis like Vitamin K2, B12, biotin, folate which are co-factors for various metabolic pathways. Ceramide induces degradation of sphingomyelin via alkaline sphingomyelinase and in the prevention of tumorigenesis. Ceramide also plays a role in the regulation of gut-liver axis (97).
5 Dysbiosis and different diseases
Dysbiosis has been associated with a growing list of diseases (Table 1) with complex pathologies. Dysbiosis occurs commonly in GI and non-GI diseases. Human microbiota is linked to different diseases including noncommunicable diseases and autoimmune diseases (33, 98–100). The microbiota may also play a role in cancer through immune modulation and activation of signaling pathways for cell proliferation (101–103). Under conditions of dysbiosis, there can be a reduction of protective bacteria with a switch to more abundant pathogenic and cancer-promoting bacteria, which can include Streptococcus bovis, Sulfidogenic bacteria, Fusobacterium nucleatum, Bacteroides fragilis, Clostridium septicum, Escherichia coli, Helicobacter pylori, Enterococcus faecalis, Human papilloma virus, John Cunnigham virus, and Epstein Barr virus. This can include the promotion of particular functions such as angiogenesis, loss or apoptosis, and cell proliferation (104). There have been other studies that have shown certain microorganism can potentially contribute to colorectal cancer via the production of toxic metabolites, interactions with the immune system, and the release of genotoxic virulence factors (105).
6 Risk factors for dysbiosis
Whole body dysbiosis could be a risk factor for many diseases. As shown in Figure 2, the first human microbiome is inherited at birth and is highly stable, whereas the acquired microbiome after birth depends on environmental factors (98). In some cases, studies have linked dysbiosis to being born via C-section and being formula fed among newborns (106).
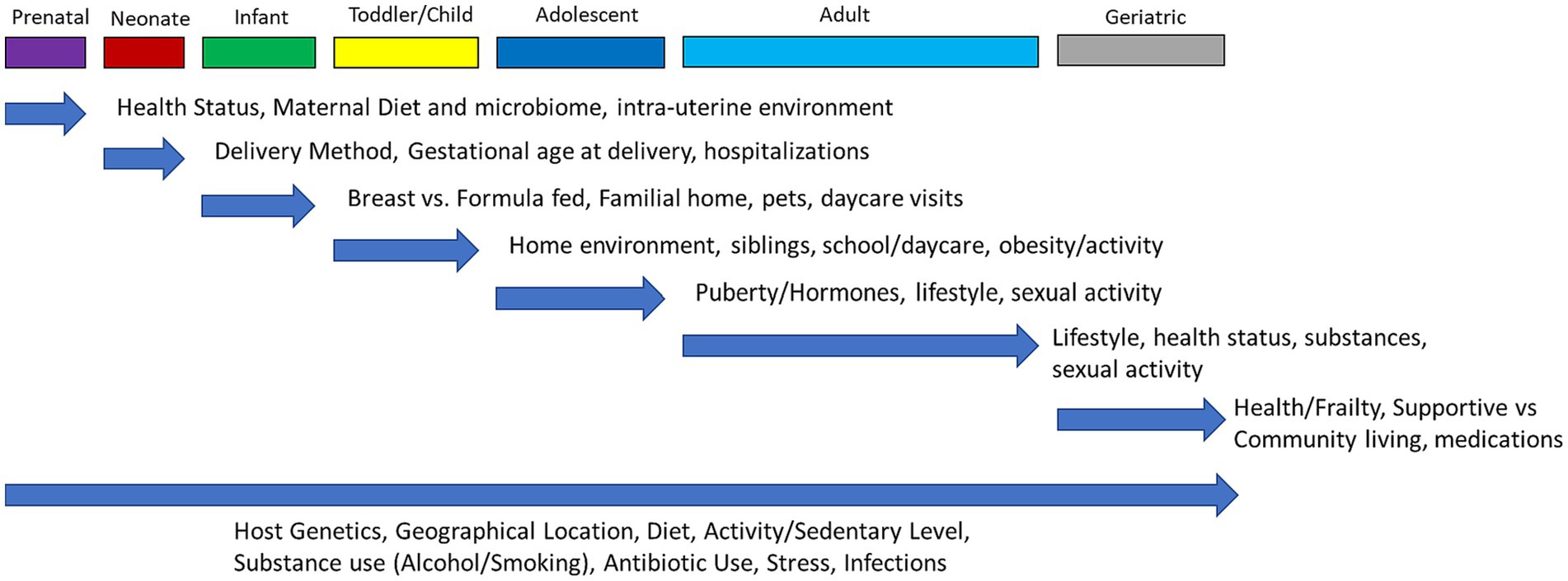
Figure 2. Dynamic changes of microbiome over the life span of humans. Throughout each stage of life (top row) there are factors that can have an influence and impact the human microbiome. Each blue arrow highlights what specific factors are most likely to alter the microbiome at that stage of life, with the effects potentially remaining throughout the rest of life. The bottom arrow indicates influences on the microbiome that are non-modifiable (host genetics) or that can occur throughout all stages of life.
6.1 Modifiable risk factors
Before birth, the fetus is considered to be sterile. Starting from birth, different modifiable risk factors like type of birth, breast feeding, antibiotic use, life style factors (dental hygiene, alcohol, smoking), environmental factors (air pollution) and also unprotected sex can influence the microbiome composition and diversity.
There are many factors that can lead to the state of dysbiosis, including the excessive or wrong use of antibiotics, excessive alcohol consumption, increased intake of sugar or protein, frequent use of antacids, exposure to pesticides on unwashed fruits and vegetables, and chronic stress with weakening of the immune system (48) Also, poor dental hygiene, unprotected sex, and anxiety can lead to dysbiosis. With sexual intercourse, there is emerging evidence that there is a transmission and exchange of the microbiome found in vaginal and seminal fluids (107).
The composition of our microbiota is influenced by host genotype, environment, lifestyle and diet (108). There is a dysbiosis risk stratification scale called INDIS survey which helps stratification of intestinal dysbiosis in adult patients (109).
7 Medications and dysbiosis
Most commonly, it is expected that when treating patients with antibiotics, there will be an impact on the gut microbiome. However, there have been studies that found other types of medications that can have antimicrobial effects (110). Since it is an exhaustive list, we have focused on selected medications to explain the dysbiosis effects of drugs. We have highlighted only some of the typically prescribed medications in older adults, such as antibiotics, proton pump inhibitors, metformin, psychotropics, statins, and opioids, were discussed with its affect on gut microbiome.
7.1 Antibiotics
In general, antibiotic treatment reduces the diversity of gut microbiota species, which leads to metabolic shifts, increases gut colonization, which can lead to bacterial antibiotic resistance (111–113) In humans antibiotic use is associated with Antibiotic-Associated Diarrhea, C. difficile-associated Diarrhea, Helicobacter Pylori Infection in the short term. Where as in the long term, antibiotic use can contribute to the development of obesity, asthma, allergy, and IBD (114, 115).
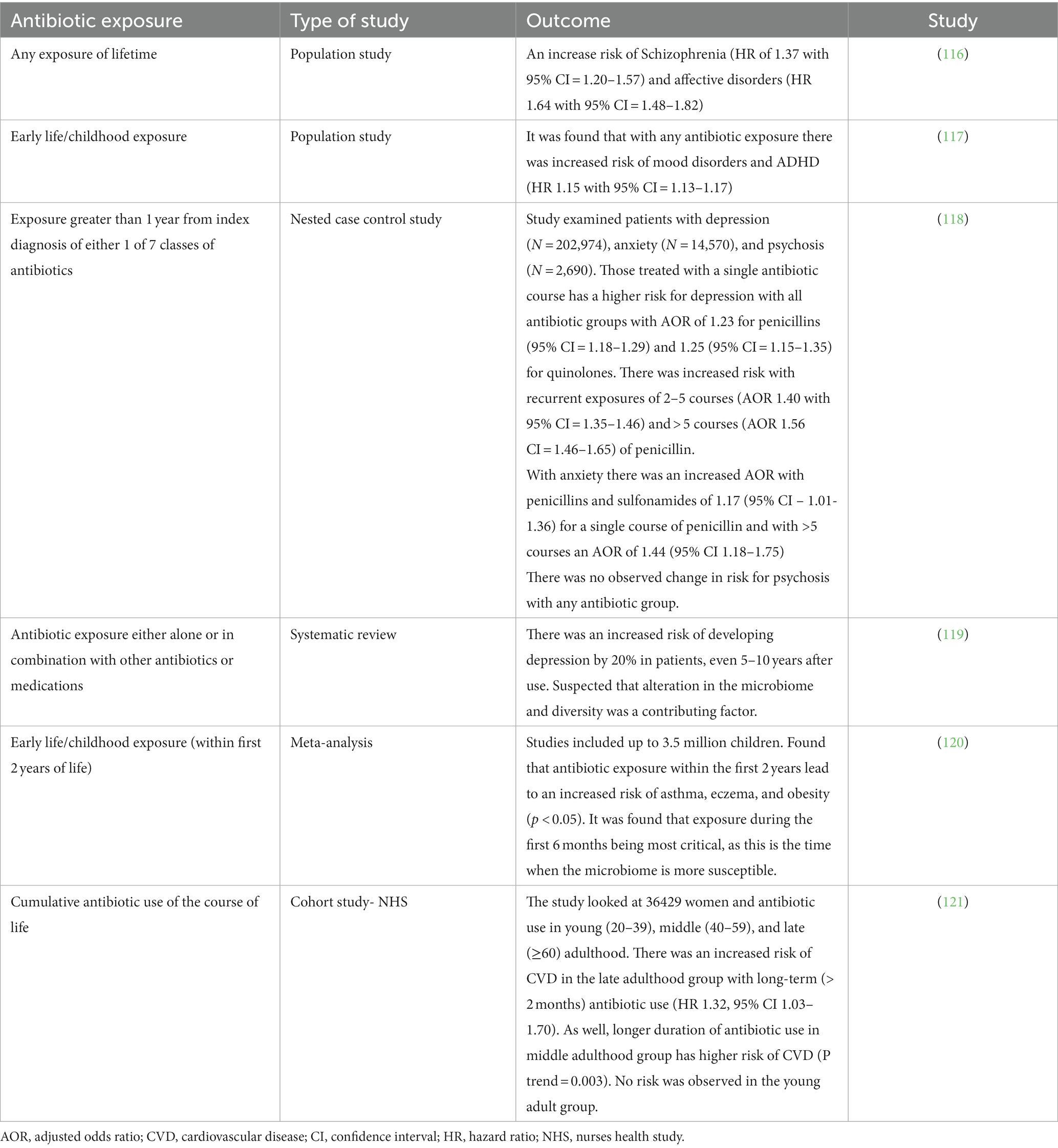
There have been various human studies that examined the impact of antibiotics on the gut microbiome (Table 2). Even a single exposure of antibiotic use, even in childhood, can have a lasting effect on gut microbiome, more so with broad-spectrum antibiotics. It is hypothesized that the use of antibiotic regimens, both single and multiple use, may influence mental health conditions, such as depression and Alzheimer’s dementia, by changing the population of the gut microbiome (32, 112).
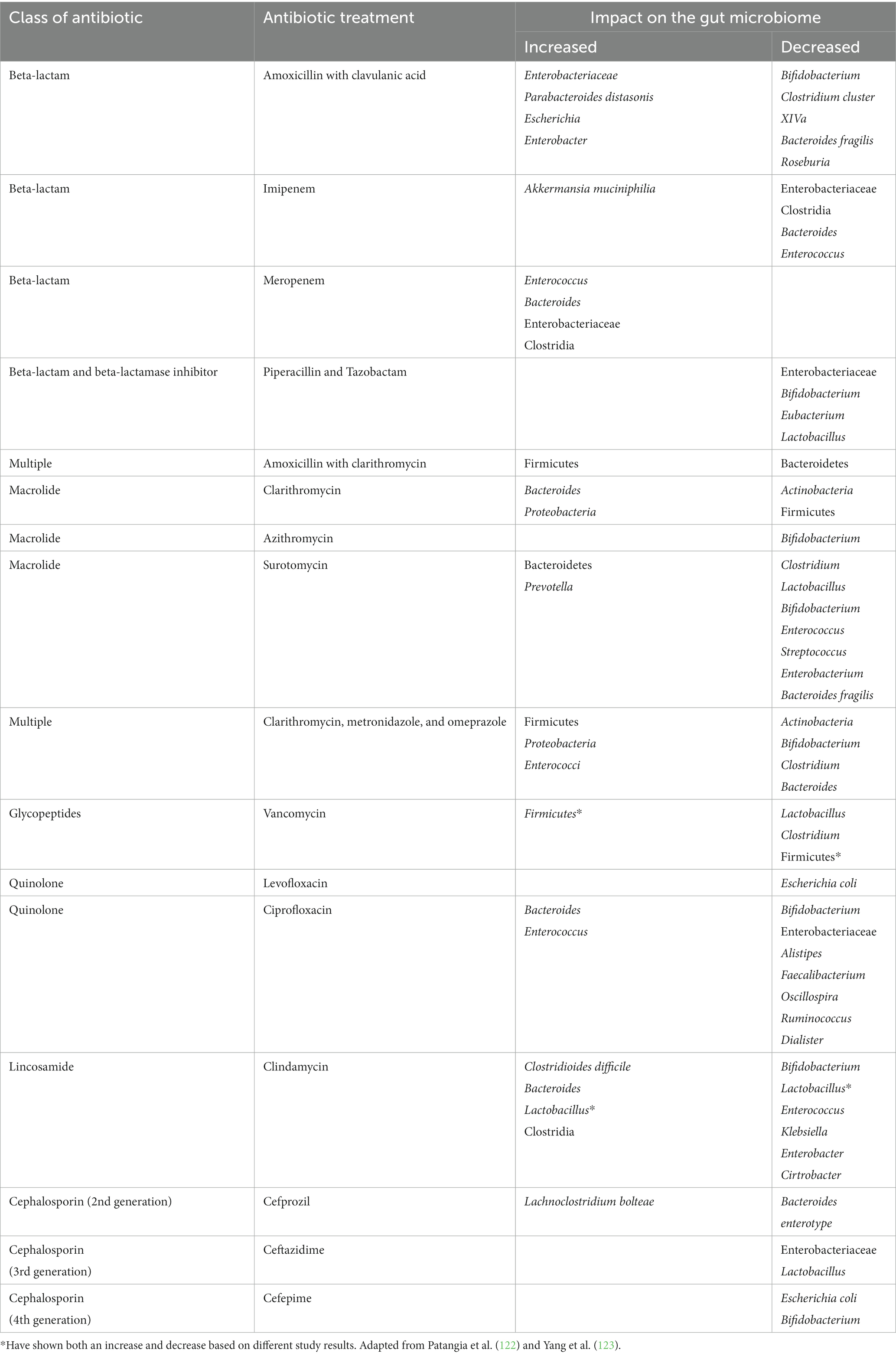
Different classes of antibiotics have been examined for their potential impact on the gut microbiome. Beta-lactam and glycopeptide, such as amoxicillin and ceftriaxone, have been found to cause dysbiosis in the gut (113–115) As well, changes in the community composition in the gut has been found to be caused by DNA replication inhibitors or DNA-damaging antibiotic, including quinoline and nitrofurantoin as examples (113–115). Alteration in mucus secretion, ion transport, and inflammatory response has been found related to the non-antimicrobial effects of macrolides. Other transcription and protein synthesis inhibitors have also been found to cause the distress of the gut microbiome network (113–115). With respect to the vaginal and urinary microbiome, there has been found a decrease in the overall diversity with increased abundance of Lactobacillus iners when exposed to nitroimidazole antibiotics, such as metronidazole and azithromycin (113–115). With different classes of antibiotics, changes in the gut microbiome can be seen, however, specific microbial changes are not consistently seen across studies (Table 3).
When the gut microbiome is exposed to antibiotics, the changes can persist from weeks to years. Broad spectrum antibiotics more commonly cause dysbiosis. It was found that treatment with ciprofloxacin, clindamycin, and clarithromycin with metronidazole left changes to the gut microbiome lasting 1, 2, and 4 years, respectively (113–115). It may be that the long-term consequences from antibiotic exposure may play a role in the development of obesity, allergies, and even asthma (113–115). At present, due to the heterogenicity of the study designs, there remains limitations on determining the effects of antibiotics on dysbiosis. Future antibiotic studies should control for medical comorbidities, age, and diet to get a better understanding of the impact of just antibiotics alone.
7.2 Proton pump inhibitors
One of the common medications used to treat gastroesophageal reflux and peptic ulcer disease are proton pump inhibitors (PPIs). However, there is evidence that these medications can contribute to dysbiosis, primarily through Clostridioides difficile infections, with higher rates found in hospitalized patients (124, 125). Studies showed that the use of PPIs can lead to a decrease in the alpha-diversity in those prescribed the medication compared to those not using them (125, 126). There have been multiple studies that found individuals using PPIs who had a significant increase in various bacterial genera, including, Enterococcus, Streptococcus, Staphylococcus, and Rothia; as well as, the species such as Lactobacillus salivarius and a potentially pathogenic species of Escherichia coli (127). Another study by Bruno et al. also found that PPIs can lead to dysbiosis throughout various segments of the GI tract with increased Enterobacteriaceae, Enterococcaceae, and Lactobacillaceae while there was a decrease in Ruminococcaceae and Bifidobacteriaceae in the colon (128). It is suspected that individuals who are on long-term PPI use can be at risk for enteric infection through dysbiosis which can lead to irritable bowel syndrome (IBS) development (129).
7.3 Metformin
It is a common medication, used in the treatment of diabetes. It has been found that patients treated with this medication, when compared to non-users, did not have a significant difference seen in the alpha-diversity, but rather some difference in beta-diversity (130). Another study also found that metformin can lead to an alteration in the gut microbiome, causing an increase in various bacteria, such as Akkermansia muciniphila, Escherichia spp., and Lactobacillus, while other bacteria, like Intestinibacter were found to have decreased levels (131). This study also highlighted that metformin can promote SCFA production, which has been found to help support the intestinal barrier and regulate the secretion of gut peptides (131). A study by Wu et al. found that metformin can exhibit a positive influence on the gut microbiome and when fecal samples of metformin-treated individuals were transferred to germ-free mice, the mice had improved glucose tolerance (132).
7.3.1 Antidepressants
Chronic exposure of antidepressants in preclinical studies have shown a decrease in richness of gut bacteria compared to controls (133). This study, by Lukić et al., included mice treated for 21 days with either fluoxetine (10 mg/kg), escitalopram (10 mg/kg), venlafaxine (10 mg/kg), duloxetine (10 mg/kg), or desipramine (20 mg/kg) and found that all these antidepressants, except desipramine, lead to a reduced richness of the gut microbiome (133). The authors also looked at the genus Ruminococcus and duloxetine and found that mice treated with the antidepressant along with a supplementation of the R. flavefacians, showed an attenuation of the antidepressant effects (133). When looking at the gene expression, R. flavefaciens, was found to decrease the expression of synaptic signaling and neurodegenerative genes, similar to that of patients with depression (133). A study by Cussotto et al. found that mice given fluoxetine had an inhibited growth of Succinivibrio and Prevotella (134). In humans, it has been found that treatment with antidepressants can affect the composition of the gut microbiome (135). Among the selective serotonin reuptake inhibitors, the antidepressant sertraline has been found to have the most potent antimicrobial activity and even a synergistic effect with antibiotics (136).
7.3.2 Antipsychotics
There are research studies looking at the influence of antipsychotics on the gut microbiome in both animals and humans. It was found that germ-free mice treated with olanzapine did not exhibit the same weight gain as their colonized counterparts. When these mice were colonized, the weight gain was then seen, suggesting that the gut microbiome may be involved and play a role with the side effect of olanzapine (137). In a human study involving patients with schizophrenia, when treated with risperidone for 24 weeks, there was a change in the gut composition that included increased Bifidobacterium and Escherichia coli, with decreased Clostridium coccoides and Lactobacillus (138). It was also found that female patients treated with atypical antipsychotics had decreased species diversity with Lachnospiraceae, Akkermansia, and Sutterella, compared to those treated with non-atypical antipsychotics; interestingly male patients did not show a significant diversity difference (139). It appears that patients treated with antipsychotic showed an altered ratio of Firmicutes: Bacteroidetes, resembling that seen in obese patients, which may provide evidence to the associated weight gain seen with these medications (140).
7.4 Non-steroidal anti-inflammatory
There have been various studies looking at the impact of NSAIDs medications on gut bacteria through dysbiotic changes. Specific NSAIDs, such as celecoxib and ibuprofen lead to an increase in certain bacterial families such as Enterococcaceae, Enterobacteriaceae, Erysipelotrichaceae, Acidaminococcaceae, and Desulfovibrionaceae (141). In elderly patients who are prescribed NSAIDs a depletion in Lactobacillus and Collinsella aerofaciens and an enrichment in Roseburia is seen, compared to non-users (142).
7.5 Opioids
Opioids have been commonly prescribed to treat moderate to severe pain. Studies have found that they may play a role in bacterial translocation through disruption of the gut barrier (143). When examining hospital patients, opioid use was associated with increased alpha-diversity, particularly with Parabacteroides, Propionimicrobium, Alistipes, Sutterella, Clostridium, Bifidobacterium, unclassified Lachnospiraceae, and Pyramidobacter; with a negative association with Polyomavirus, Pseudomonas, unclassified Ruminococcaceae, Candida, and Megamonas (144). It is hypothesized that since opioids tend to delay GI transit time, this may be more conducive to bacterial growth in the colon and allow for the increased diversity seen in certain microbial populations.
7.6 Statins
Statins are medications that are commonly and routinely used to help treat dyslipidemia that often include some GI side effects. There has been evidence that the use of statins can contribute to changes in the beta-diversity of the gut microbiome (145). In a study looking at idiopathic Parkinson patients, the use of statins leads to an increased relative abundance of Burkholderiaceae, Propionibacteriaceae, Enterococcaceae, Actinomycetaceae, and Enterobacteriaceae (146). As well, viruses were found to be increased in participants that were treated with a statin (146). Variation in statin response has been attributed to the effect of microbiota (147). However, when controlling for statin exposure, no significant difference was observed between the participants and controls, which was felt to be due to a small sample size. A study looking at human subjects found that individuals treated with rosuvastatin for 4–8 weeks had a significantly altered gut microbiome (148). In particular, the phyla Firmicutes and Fusobacteria showed a negative correlation to the lowering of the low-density lipoprotein cholesterol (LDL-C) level while Cyanobacteria and Lentisphaerae were positively associated with the lower LDL-C level (148).
In conclusion, the end results of these medication induced microbiome alterations provide a significant impact on dysbiosis and contributes to many diseases.
8 Clinical features of dysbiosis
Most patients with dysbiosis present with gastrointestinal symptoms like halitosis or bad breath, frequent flatus, bloating, food intolerances, food sensitivity, abdominal cramping, diarrhea and/ or mucus in the stool. They can have other symptoms like vaginal or rectal itching, skin conditions, fatigue, mood symptoms like depression or anxiety, and problems with memory. These symptoms depend on the system impacted by dysbiosis (149–151).
9 Investigations for dysbiosis
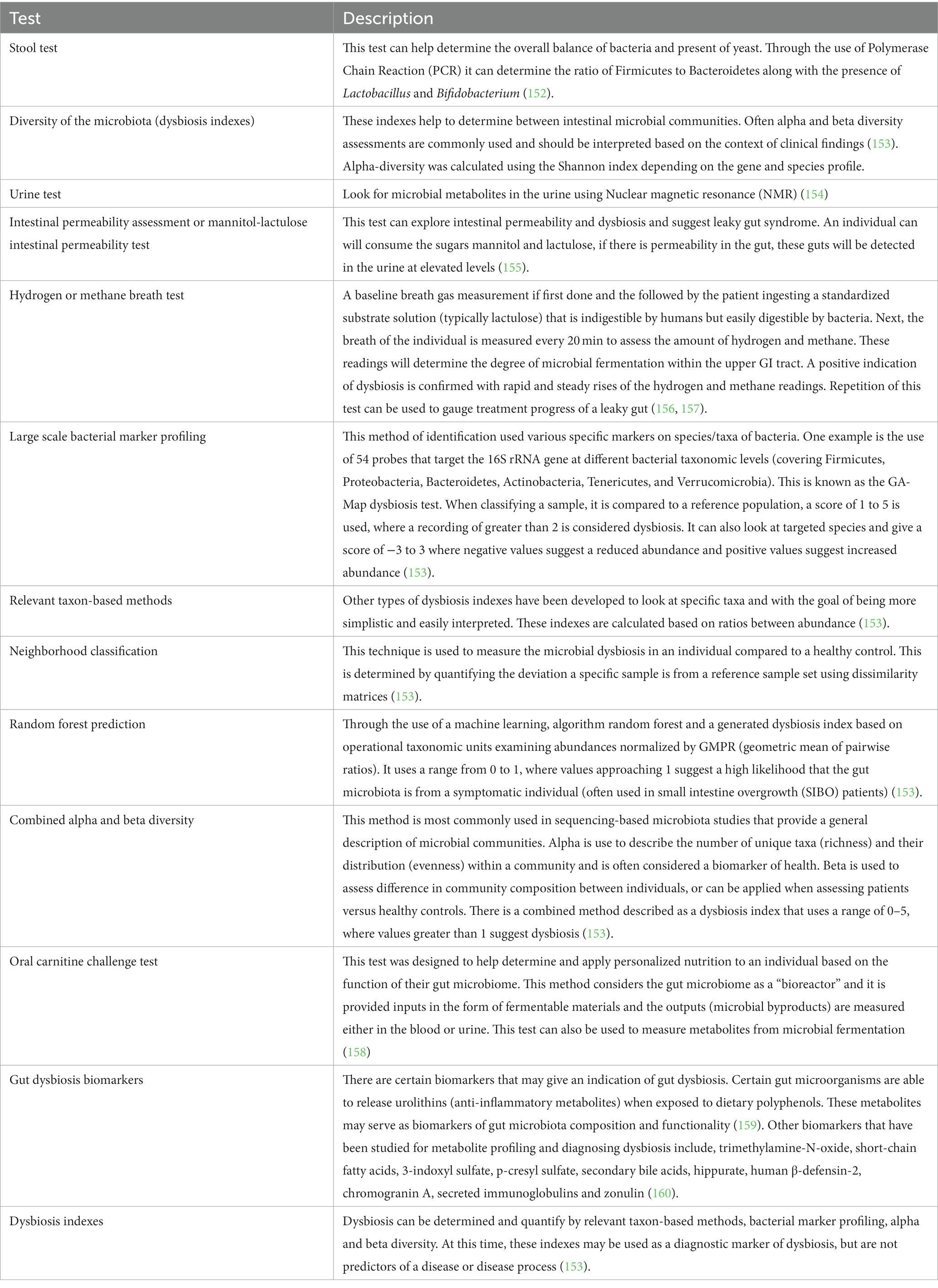
Dysbiosis cannot be diagnosed through standard blood tests or through scopes (endoscopy or colonoscopy), but many tests (Table 4) may aid with diagnosing dysbiosis, which is not commonly done in clinical practice at this point. Generally, it is known as CDSA (Comprehensive Digestive Stool Analysis).
9.1 Comprehensive digestive stool analysis
CDSA include analysis of different microbiota such a lactobacilli, bifidobacteria, E. coli, Proteus, Pseudomonas, Salmonella, Shigella, Vibrio, yeast and microbiome analysis including sequencing technologies, dysbiosis indexes, metagenomics, metatranscriptomics as well as assessment of microbial metabolites like Short Chain Fatty Acids. It also includes Hydrogen Breadth test, which detects the presence of gases produced by bacteria and excessive gases indicate imbalance of bacteria (161, 162).
Stool or fecal specimens can be used to look at gut microbiota and microbiome because of relative ease of access of the sample (152).
Sequencing technologies are usually based on samples collected from inner—colonic (mucosal biopsy/capture microdissection, luminal brushing, intestinal fluid lavage), which gives a better view of the colon’s microbial diversity. 16S ribosomal RNA (rRNA) amplification and whole-genome shotgun sequencing (WGS), are the two typical sequencing technologies used to diagnose gut microbiota diversity (152).
9.1.1 Dysbiosis indexes
Microbiome analysis using metrics of markers of dysbiosis included alpha-diversity and beta-diversity as well as distributions of predominant phyla. The three alpha-diversity indices (Shannon index, Simpson’s Index, Chao-1 Index) and beta- diversity metrics like Bray-Curtis distance will be done. Alpha diversity, which indicates the relative abundance of microbial species in a biological sample, where as beta diversity and gamma diversity measures species diversity over time (165) Dysbiosis indexes have to be interpreted in the context of the clinical findings (166). Dysbiosis is measured by using dysbiosis indexes. To quantify dysbiosis, large-scale bacterial marker profiling, relevant taxon-based methods, neighborhood classification, random forest prediction, and combined alpha and beta diversity indexes are used (166). Studies using these indexes showed among chronic respiratory conditions, cystic fibrosis is the one which had a link between alpha diversity and lung function (163). Another study showed the alpha diversity of gut microbiota could be a promising predictor for Alzheimer’s Dementia (AD), Schizophrenia, and Multiple Sclerosis (MS), but not for all neurological diseases (164) (Table 4).
9.1.1.1 Metagenomics
Metagenomics is the study of the genomes in a microbial community and constitutes the first step to study the microbiome (165). Metatranscriptomics helps to identify the genes that are expressed. The sequencing of hypervariable regions and shotgun sequencing are technologies that enable the taxonomic classification of microorganisms from the DNA present in microbial communities. However, they are not capable of measuring what is actively expressed. Conversely, we advocate that metatranscriptomics is a “new” technology that makes the identification of the mRNAs of a microbial community possible, quantifying gene expression levels and active biological pathways. Furthermore, it can be also used to characterize symbiotic interactions between the host and its microbiome (166).
9.2 Mannitol-lactulose intestinal permeability test
Dysbiosis results in increased inflammation, elevated levels of zonulin, destruction of intestinal tight junctions, and intestinal permeability, which allow lipopolysaccharides (LPS) to leak into systemic circulation. LPS is a powerful endotoxin that causes chronic inflammation throughout the body. Chronic inflammation is associated with chronic diseases and the acceleration of biological aging (151).
Urinary excretion of lactulose and mannitol after oral intake is a good test for evaluating intestinal permeability and altered ratio indicates leaky gut syndrome (155) (Table 4).
9.3 Hydrogen or methane breath test
This common test is used to assess for small intestinal dysbiosis and also to assess the effectiveness of leaky gut treatment (167).
9.4 Identification of gut microbial metabolites: (metabolomics)
After taxonomic identification and genomic insights of microbiota and microbiome, we will focus on the functional capabilities and metabolomic characterizations using the technique of metabolomics. In simple terms it is functional readout of microbial activity (168, 169).
After that taxonomic identification, untargeted metabolomics profiling, and targeted metabolomics focusing on short chained fatty acids (SCFAs) analysis and others were done. Correlations between SCFAs and gut microbiota were also examined. Microbiome derived metabolites, such as lipopolysaccharides, SCFAs, secondary bile acids, or tryptophan-related metabolites play a role in the pathology of dysbiosis and can be measured from CSF (Cerebrospinal Fluid), plasma, urine, feces with NMR (Nuclear Medicine Resonance) spectroscopy analysis to measure quantitative metabolomics (170, 171) (Table 4).
Gut microbiota can function like an endocrine organ with bioactive metabolites like SCFA, trimethylamine N-oxide (TMAO), tryptophan metabolites (TRP) which can circulate in the human blood and be delivered to different target tissues. Trimethylamine N-oxide, p-cresyl sulfate and indoxyl sulfate have pro-inflammatory effects and may contribute to chronic inflammatory diseases. Tryptophan and its metabolites, indole acetic acid and indole-3-propionic acid, have been reported to enhance sensitivity of chemotherapy against cancer. To treat certain chronic diseases, a strategy using gut microbiota derived metabolites may be helpful.
9.5 Selected targeted metabolomics-measurement of SCFA
Three major SCFAs are acetic acid, propionic acid, butyric acid, and two less abundant SCFA are valeric acid and caproic acid. They are produced in the large intestine through the anaerobic fermentation of indigestible carbohydrates (172, 173). These microbial by-products can be measured using gas chromatography (156) and more specifically, gas chromatography–mass spectrometry can analyze SCFA in stools.
9.6 Trimethylamine N oxide
Carnitine and choline are commonly found in red meat and eggs, which were once thought to be semi-essential nutrients for the human body. However, these nutrients can be utilized by microorganisms in the gut to produce trimethylamine (TMA) as a byproduct. The TMA absorbed from the gut is then oxidized into TMAO in the liver and has proven to be a strong risk factor for cardiovascular disease (CVD) (174). Biomarker TMAO plays a role in cardiovascular disease, renal disease, type II diabetes and colorectal cancer (174).
Resveratrol may reduce the level of plasma TMAO and help in treating atherosclerosis in an animal study by acting like a prebiotic (175). Oral carnitine challenge tests are used to measure metabolites after gut microbial fermentation and to help identify TMAO-producer phenotype (158). Other gut metabolite biomarkers could be relevant to prodromal disease. Urolithins are anti-inflammatory metabolites produced from some dietary polyphenols by specific gut microbial ecologies (urolithin metabotypes) and have been proposed as biomarkers of gut microbiota composition and functionality (159). Thus, trimethylamine-N-oxide, short-chain fatty acids, 3-indoxyl sulfate, p-cresyl sulfate, secondary bile acids, hippurate, human β-defensin-2, chromogranin A, secreted immunoglobulins, and zonulin may serve as biomarkers for metabolite profiling with diagnostic suitability for dysbiosis and diseases (176).
9.7 Tryptophan metabolites
Tryptophan (TRP), the essential amino acid obtained from diet, is mainly metabolized through the kynurenine (KYN) pathway and it plays a role in different metabolic disorders. The gut microbiome can convert tryptophan into indole, and its derivatives, which can contribute to GI function, inflammation, antioxidation, and immune system regulation. Disorders in tryptophan metabolism can impact various diseases such as irritable bowel syndrome, colitis, depression, Alzheimer dementia, schizophrenia, and Parkinson disease. There is growing research about tryptophan metabolism disruption in neoplastic diseases, such as colorectal, liver, lung, and breast cancer (177). High-performance liquid chromatography-mass spectrometry, and gas chromatography–mass spectrometry can be used to measure tryptophan metabolites (178).
In conclusion, metagenomics and metatranscriptomics data are generated using sequencing data, whereas metabolomics data is analyzed using liquid and gas chromatography techniques, mass spectrometry (MS) and nuclear magnetic resonance (NMR) techniques. Integrating all metagenomics, metatranscriptomics, and metabolomics—would provide a complete picture from genes to phenotype (179).
From the authors point of view, doing CDSA and identification of gut microbial metabolites as the starting workup for dysbiosis and the next step is to use tests better than the taxonomic indicators to define microbiomes in health and disease.
9.8 Microbiome health index
Microbiome Health Index (MHI) was developed by Blount et al. to diagnose post-antibiotic dysbiosis. It is a promising biomarker of post-antibiotic dysbiosis and subsequent restoration of microbiota (180).
10 Genetics of microbial dysbiosis (non-modifiable risk factor)
There are a variety of factors that can contribute to alterations and differences in the gut microbiome seen with individuals. A study by Zoetendal et al. compared adult monozygotic twins to their unrelated marital partners and found that there were greater similarities between the gut microbiome among the monozygotic twins; this was hypothesized due to the influence of their genotype on the microbial diversity (181). Another interpretation of this was that the microbial similarities were due to the twins having a shared mother (181). Another study found that marital partners had different microbial communities colonized in their ear canal, however within families there were common dominant bacterial species (182). At this time, there is emerging evidence that there may be an interplay between host genetics and the gut microbiome, however the mechanisms are not completely understood.
In a genome-wide association study of 7,738 patients (from the Dutch Microbiome Project), the authors examined 207 taxa and 205 pathways and found a significant signal (p < 1.89 × 10−10) near the Lactase (LCT) and ABO genes that were associated with multiple microbial taxa and pathways (183). In particular, there were able to narrow down an association with Bifidobacterium adolescentis at the LCT loci and Bifidobacterium bifidum, and Collinsella aerofaciens at the ABO loci. Animal studies in pigs have found that a deletion at the ABO locus, that inactivates the ABO acetylglucosaminyltransferase (enzymes in glycoprotein biosynthesis), led to a change in the porcine microbiome composition (184). The study by Lopera-Maya et al. also found 22 other loci that may have an association with microbial taxa and pathways and be correlated with trait heritability, however a larger sample size is needed to further explore the role of host genetics on the gut microbiome (183).
Using metagenomic sequencing a genome-wide analysis using 1,514 subjects was done and found 9 loci with microbial taxonomies and 33 loci with microbial pathways and gene ontology terms (p < 5 × 10–8) (185). It was found that LCT single nucleotide polymorphisms (SNP) with the Bifidobacterium genus (p < 3.45 × 10–8) may in fact be a gene-diet interaction that can influence the abundance of Bifidobacterium (185). Other investigations looked at SNP-based heritability and used microbiome genome wide association to determine host genetic variants related with the gut microbiome. The group of Xu et al. found that Saccharibacteria could lead to a decreased serum creatinine concentration and potentially increase the estimated glomerular filtration rate through the interplay between host genetics and the gut microbiome (186).
11 Management of dysbiosis including risk factor modification
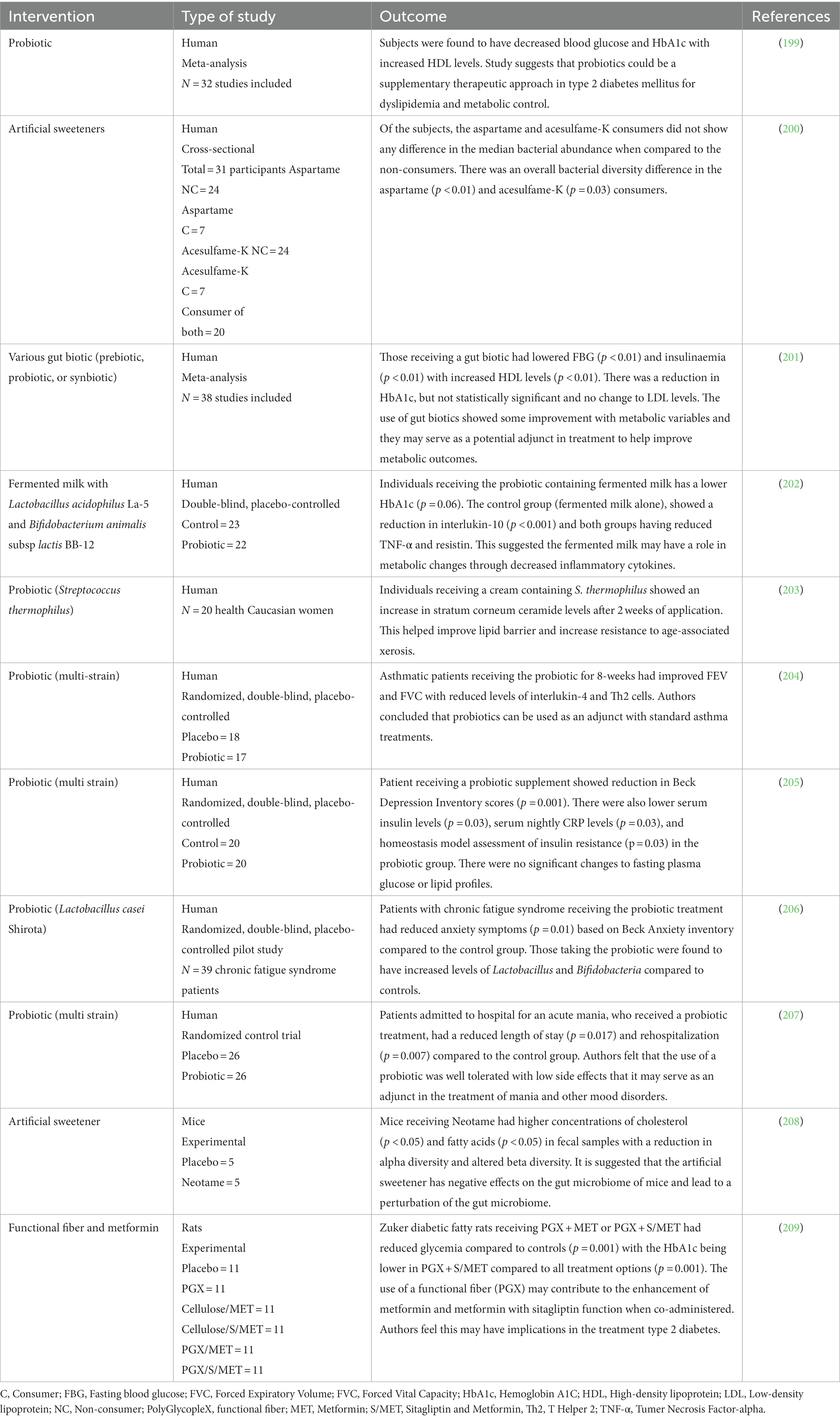
Various treatments can be used in managing dysbiosis and diet is an important step to improve dysbiosis (Tables 5, 6). Addressing the risk factors for dysbiosis, like avoiding medications that cause dysbiosis, stress management, avoiding ultra processed foods and alcohol, can help in the management.
11.1 Food and food products
11.1.1 Dietary interventions-diet/food modifications
Various diets have been examined in relation to their impact on the human microbiome.
11.1.1.1 Fermented foods
Fermented foods are unique products that have many potential benefits that range from food safety to human health. Increased shelf life and stability of foods is a long-standing safety benefit of the fermentation process (210). Various methods to obtain fermented foods include spontaneous fermentation, specific starter culture use, and back slopping (utilization of previously fermented foods to start fermentation in a new batch) (192, 211). Fermented foods have the capacity to contain probiotic cultures that could directly confer potential human health benefits. It is important to consider various factors, including the number of live cultures present at the time of food consumption, as well as the specific strains present within the food (192). Other elements, including food matrix, packaging, food formulation and others can have an impact on the potential of these foods to benefit human health. To be considered a fermented probiotic food, various thresholds for consideration need to be met as noted in the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement from 2021 (192). Some potential benefits of these foods can include: displacement of pathogenic bacteria within the gut through microbiome compositional change, alterations to the digestibility/tolerability of foods (examples include reduced concentrations of phytates, lactose and fermentable sugars), metabolite benefits directly related to immune function (211).
11.1.1.2 Plant-based fiber rich foods
A reduction in opportunistic bacteria and inflammatory bacteria were seen, along with an increase in good gut bacteria and their metabolites with a plant-based dietary approach (193). Subjects in an interventional pilot study consumed red beet root juice over 14 days showed changes in gut microbiome with statistically significant increases in Akkermansia muciniphila and decreases in Bacteroides fragilis potentially conferring metabolic benefits and possible reduction in the risk of diabetes and obesity. Statistically significant increases in some SCFA were also observed in this pilot study with isobutyric and butyric acid that may support those metabolic benefits (194). There are various studies examining orange juice and possible benefits to the gut. One study with functional orange juice showed growth of emerging probiotics such as Bacteroides xylanisolvens and decrease in other strains, such as Clostridia sp. Therefore, this prebiotic orange juice may enhance gut microbiota composition and be a potential functional food (212). In another human study examining the intake of blood orange juice, significant changes were seen regarding SCFA production (particularly propanoic acid and isobutyric acid) and improved cardiometabolic risk factors (213). An animal study compared two orange juices with 100% fruit juice (high sucrose and flavonoids) and fruit beverage (higher glucose and fructose) being offered to rats. Of note, the rats offered the 100% orange fruit juice showed improved microbial diversity with altered Firmicutes/Bacteroidetes (F/B) ratio (decrease) and insulin resistance improvement while the fruit beverage group showed no diversity change with an increased F/B ratio (214). Whole fruit in themselves can have considerable impacts on the microbiome with implications to GI transit time and constipation. The exact constituents responsible and the most ideal fruit type remains to be determined (215). In a systematic review and meta-analysis on different fruits, Huo et al. found kiwi fruit had a predominant effect on microbial culture amounts as well as improvements in functional constipation (216). Other studies showed that a vegan diet rich in fiber will increase SCFA and inhibit pathogenic bacterial colonization (217, 218).
11.1.1.3 Mediterranean diet
The Mediterranean Diet has been examined more broadly in relation to health and the microbiome (195, 196). This diet is generally described as having a greater focus on minimally processed fruits and vegetables with the inclusion of pulses, nuts, seeds, and fish in relative abundance. Meat is included, although there is a reduction in frequency of this with particular limitation to processed meat, and foods rich in saturated fatty acids. Polyunsaturated fatty acids (PUFAs), monounsaturated fatty acids (MUFAs) with special focus on olive oil, phenolic compounds, omega 3 fatty acids, fiber and low glycemic index foods tend to be consumed in higher amounts as compared to a “Western Diet.” The implications of the above include a reduction in the risk of cardiovascular disease, diabetes, metabolic complications, cancer, inflammatory conditions among other health concerns (219–221). The diet itself has also been associated with improvement in microbiome composition and diversity which can lead to lower risk of gut dysbiosis. From the Mediterranean Diet, the specific constituents that lend themselves to health benefits include: a variety of minimally or unprocessed whole grains/cereals, legumes, a variety of produce with vegetables and salads, dried fruit, nuts/seeds, honey, and olive oil. Low to moderate consumption of poultry, eggs, fish, wine, unprocessed or minimally processed cheese and yogurt also play a role while red and processed meats are consumed in very low frequency. There is evidence to support various microbiome impacts from this diet with increased microbial diversity, and increases in the abundance of Bacteroides, Prevotella, Lactobacillus, Faecalibacterium, Clostridium, and Oscillospira. In contrast a decrease in the abundance of Firmicutes is noted (196).
11.1.1.4.Western diet
A Western Diet generally is defined as a diet that has an abundance of processed foods leading to increased intake of salt, saturated fat (possibly trans fats), and added sugars. Along with this, there is generally a reduced intake of fiber rich foods, whole grains, and fish. The consequences of this leads to lower intakes of PUFAs, MUFAs, phenolic compounds, omega 3 fatty acids, fiber and low glycemic index foods. This dietary pattern has the potential to erode human health in many ways including the gut microbiome. With this dietary approach increased opportunistic bacteria and inflammatory markers are seen with gut dysbiosis (221).
In an animal study, comparing Mediterranean diet (MD) to Western diet (WD) there was an abundance of mammary gland Lactobacilli in monkeys who take MD with a resulting increase in bile acid metabolites and decrease in reactive oxygen metabolites (221). Another study in humans showed subjects who adhere to MD were found to have higher levels of SCFA (222).
11.1.1.5 Ketogenic diet
This diet has also been examined regarding its impact on human health as well as the microbiome itself. A Ketogenic diet typically has a considerable limitation in the amount of carbohydrates consumed with diets containing 20–50 g per day or less (5–10% energy intake). The purpose of this is to promote ketone body production (acetone, beta-hydroxybutyrate, acetoacetate) to be used as a fuel source as opposed to glucose impacting the microbiome and host metabolism (197). Regarding the microbiome, some animal and human studies have shown positive impacts (re-shaped gut microbiome and biological functions) and negative impacts (decreased variability in gut bacteria with increased pro-inflammatory strains) (223). It is possible that the modified gut microbiome may be critical to potential outcomes in relation to the ketogenic diet as seen in seizure management (224). Complimentary dietary modifications such as the inclusion of prebiotics, probiotics, fermented foods and others may minimize some potential drawbacks that the ketogenic diet may have on the microbiome as noted in this study (223). There is some potential promise for treatment or prevention of dementia with the ketogenic diet although human studies are few and in early stages (197). A review article by Dowis et al. points out that the ketogenic diet may have therapeutic benefits “helping with weight loss, improving lipid markers for cardiovascular health, healing a disrupted microbiome, improving epigenetic markers, reversing diabetes, or reducing the need for medication, and improving responses to cancer treatments.” But the article stressed the need for well-designed randomized controlled trials that should be done to confirm the therapeutic possibilities provided by this dietary intervention (225). It is important to highlight the relatively complicated nature of this diet in relation to more conventional dietary approaches in order to achieve ketosis where ketone bodies are promoted as an energy source. Some of the possible complications of this dietary approach can include nausea, vomiting, changes to satiety along with implications to bone mineral density, hepatic function, pancreatic function, blood glucose management, cardiovascular disease risk among other health concerns (226). These impacts do bear careful consideration prior to long term ketogenic diet implementation.
11.1.1.6 Gut biotics
11.1.1.6.1 Probiotics
Probiotics (such as Bifidobacterium and Lactobacillus) and prebiotics are known to improve gut health and restore bacterial gut balance to achieve eubiosis. There is some evidence that probiotics have been shown to alleviate functional gastrointestinal symptoms (FGID) which is commonly seen in dysbiosis (227).
While most probiotics show safety and recovery efficacy, the impacts in relation to disease improvements are statistically marginal (188). However, typical probiotics are not applied to specific diseases. Therefore, the selection and detailed description of new and disease-specific next-generation probiotics (NGP) are crucially necessary (188). NGP are individual bacterial strains through gene sequencing and bioinformatics tools. They are designed to better understand colonization, efficacy and safety of the probiotic bacteria (188, 189).
Nanoprobiotics and nanoprebiotics represent promising future strategies to target dysbiosis (228). Durazzo et al., showed in their meta-analysis that probiotics showed improvement with body weight in overweight individuals and improvements in various metabolic diseases including fatty liver and type 2 diabetes mellitus (229, 230).
Probiotics are shown in animal studies to help with wound healing (231). This might happen through the “brain-intestine-skin axis” by improving systemic immune response and affecting peripheral tissue response (232).
Since the strains introduced by probiotic intake may not colonize the gut permanently, probiotics may need to be taken periodically in order to sustain their benefits, but more research is needed in this angle. Various methods for probiotic foods to exert their actions exist as included in the ISAPP consensus statement (233, 234).
Probiotics may not be safe for all individuals. In immunocompromised or critically ill people, probiotics can increase opportunistic infections and so a risk benefit assessment should be done before recommending these products (235).
11.1.1.6.2 Prebiotics
To be considered as a prebiotic, a food must provide a benefit directly to microorganisms that can improve human health (190). There are many potential food products that can meet this definition including fruits, vegetables, pulses, tubers, whole grains, and sourdough bread. Some caution is needed in individuals with inflammatory bowel disease and other digestive concerns. It is of value to consider increasing these foods in incremental amounts to limit digestibility issues and to improve tolerability. Other factors such as activity and hydrational status will also have considerable impacts in this regard. Dietary fiber has been shown in randomized controlled trial (RCT) to promote the growth of SCFAs producing bacteria which may impact type 2 diabetes management (236).
11.1.1.6.3 Synbiotics
Synbiotic foods are an intentional combination between a prebiotic food source and probiotic microorganism (191). It is important to emphasize that both of these components are required to confer human health benefit. Two definitions of synbiotic foods have been considered. Complimentary synbiotics are foods that contain both a prebiotic and probiotic food component that work independently of each other to benefit human health. Another category to consider include synergistic synbiotic foods which also contain prebiotic fibers and probiotic microorganisms. The distinction here is that the prebiotic substrates must be selectively chosen to directly nourish the live bacterial cultures being included in the same food product with human health benefit as a result (191). The intentional prebiotic and probiotic combination can multiply potential health benefits to the host organism beyond impacts that could be reasonably expected from either component taken alone. The possibility of harnessing benefits that are greater than the sum of its parts poses a very intriguing possibility to human health improvements that can include microbiome modulation, and immune impacts to name a few (237).
11.1.1.7 Foods to avoid to improve dysbiosis
11.1.1.7.1 Processed foods
When the natural state of a food is changed for a specific reason, this can be considered a processed food. Some typical purposes of food processing include shelf stability, enhancements to food safety, improvements to food palatability/taste, and increase in nutritional value. To achieve these purposes foods may be pasteurized, canned, chemically altered, fermented, frozen, and dried, among other techniques.
Processed foods can be defined in various ways but are perhaps best defined via the NOVA classification system which divides food products into four groups based on the degree of food processing. NOVA Classifications: (1) Unprocessed or minimally processed foods, (2) Processed culinary ingredients, (3) Processed foods, (4) Ultra-processed foods (238). Some of the examples for (1) Unprocessed or minimally processed foods: Milk, Eggs, Carrots, Broccoli, Potatoes, Chicken, Oats, Rice, Dried Pulses, Unsalted nuts, (2) Processed culinary ingredients: olive oil, sugar, honey, salt, (3) Processed foods: canned tuna, canned pulses, salted/flavored nuts, tomato paste, homemade bread, wine, (4) Ultra-processed foods: chocolate, candies, potato chips, ice cream, pre-made pizza/burgers, carbonated soda beverages (238).
There is robust evidence to support the harms of ultra processed foods to human health with connections between ultra processed foods and dysbiosis which highlights the importance of identifying these foods within individual diets (239–242). The intake of ultra-processed foods can help promote a microbial environment that tends toward inflammation and oxidative change that increases the risk of gastrointestinal health concerns like inflammatory bowel disease, neurodegenerative diseases, and metabolic health consequences including obesity and beyond (239–242). The specific dietary components of ultra processed foods that can relate to human gut microbiome harm include higher intake amounts of sugar, fat, salt and food additives with reduction in dietary fiber, polyunsaturated fatty acids and phenolic compounds. Impacts to the microbiome seen from these constituents included an increase in the genus phyla Firmicutes with reductions in Bacteroidetes. Increases in Lactobacillus, Faecalibacterium of the Clostridium cluster IV are seen with ultra processed foods. Depletions in dietary fiber led to reductions in Bifidobacterium and some Clostridium subgroups (Roseburia and Eubacterium rectale) (241).
11.1.1.7.2 Food additives/preservatives
There is growing evidence to show that food additives and preservatives also likely play a role in disturbing the gut microbiome (243). Non-caloric sweeteners, emulsifiers, antimicrobial preservatives, food colorants and other additives can promote dysbiosis leading to many potential consequences which may include impairments to glucose metabolism, inflammation and/or increased chronic disease risk (244, 245). The impact of food additives to the microbiome can be vast with impacts to gut microbiota across various species including Firmicutes, Bacteroidetes, Barnesiella, Prevotella, Ruminococcaceae, and Bifidobacterium. Whether these constituents are decreased or increased does seem to vary widely based on the food additive being studied as noted by Song et al. (246) and Zhou et al. (247).
A human randomized control trial study was done showing that emulsifier use (Carboxymethylcellulose) impacted the microbiome with decreases in Faecalibacterium prausnitzii and Ruminococcus sp., and increases in Roseburia sp. and Lachnospiraceae (248).
In another human trial examining microbiome impacts of food additives to human fecal samples, sodium benzoate increased the amounts of Bifidobacterium while sodium sulphite decreased Bifidobacterium while increasing Escherichia coli and Shigella (249).
It is clear that these dietary components have a definitive impact on the gut microbiome with further human studies needed to delineate health consequences.
11.2 Lifestyle changes
Smoking, alcoholism, physical activity, stress and sleep deprivation contribute to dysbiosis. It has been found that cigarette smoking can lead to intestinal and microbial dysbiosis (64, 250). Other studies have found that smoking cessation improved intestinal dysbiosis (251, 252). A study by Leclercq et al. found that with chronic alcohol consumption there are changes in the gut microbiome and decreased intestinal barrier integrity which can lead to increased depression, anxiety, and craving through the microbiome-brain-gut axis (253, 254). Muthu et al. in their study showed subjects with chronic alcoholic consumption had lower percentage of Clostridia, Bacilli and Bacteroidetes whereas a higher percentage of Gammaproteobacteria (254). A meta-analysis showed alcohol can affect the microbiome derived metabolites like neurotransmitters which are associated with mood and behavioral disorders secondary to alcohol intake (256). Alcohol is shown to damage the microbiome but with abstinence, a reduction in gut dysbiosis can be seen (253).
Disruptions in sleep can have an impact on the gut microbiome (257), whereas improvements with sleep lead to positive changes in microbial diversity (258). Though more research is needed, a meta-analysis found that patients using a gut biotic reported better perceived sleep health (259). Lifestyle can also have an impact on gut microbiome health and diversity. Individuals with a more sedentary lifestyle were found to have less microbial diversity and more bacterial species associated with disease, such as Escherichia coli (260). In comparison, individuals that have a more active lifestyle had a richer bacterial diversity and reduced dysbiosis with more SCFA producing bacteria (260). When looking at the role of stress on the gut microbiome, psychological stress can lead to altered bacterial composition (261). In stressful events, the Hypothalamic Pituitary Adrenal (HPA) axis becomes temporarily active leading to the release of various hormones. With prolonged activation, this can lead to heightened inflammation that can impact gut barrier permeability and lead to dysbiosis (262).
11.3 Impact of food processing technology on dysbiosis
11.3.1 Microwave treatments
One of the major factors that can influence the gut microbiome is our diet. Along with this, emerging research is highlighting that it is not just the food items, but the ways in which we prepare and process our food that can impact the microbiome. In particular, the use of microwave technology has been linked to the utilization of dietary fibers by the gut microbiome.
Microwave treatments may provide a beneficial impact to the fermentability and health impact of dietary fibers leading to an improvement in SCFA production and impacts on bacterial changes (263, 264). Microwave impact on specific dietary fibers is noted with some improvement to fermentability although impacts to whole meals remain to be seen. A study using microwave treatment in combination with enzymatic processing showed an increased availability of dietary fiber. This processing promotes an increase in the F/B ratio. Overall, this processing technique increases the availability of insoluble fiber for fermentation (265).
11.4 Microbiome-based therapies
11.4.1 Fecal microbiota transplantation
Fecal microbiota transplantation (FMT) is the transfer of fecal bacteria from a healthy donor to a recipient. The purpose of this is to repopulate the recipient’s GI tract with beneficial bacteria. It is most notably used in patients with Clostridioides difficile infection, but the principle may be applied to microbial dysbiosis to help restore healthy bacteria. For the management of recurrent C. difficile, the use of FMT has been approved in the USA as the infection can occur 25–35% during index infections and up to 60% with recurrent cases (266). CDC (Centre for Disease Control) recommends microbiome sparing antibiotic Fidaxomicin as the first line therapy, which also helps to prevent recurrence. It also recommends microbiome therapeutics like Fecal Microbiota Transplantation (FMT) in recurrent C. difficile infection, which hope to reduce the dependence on antibiotics for recurrent infection (267). Safety concerns in different type of populations should be explored for FMT in future research studies. There have been various animal studies looking at the effects of mood and behavior through the use of FMT. One study took fecal samples from patients with depression and transferred them to germ-free mice. These mice began to exhibit more depressive-like behavior (268). When looking at some human studies, there are emerging case-reports of patients with a diagnosis of Alzheimer dementia showing improvement in memory and mood after receiving a FMT for a C. difficile infection (269, 270). A double blind RCT looked at patients with irritable bowel syndrome (IBS) who received a FMT and found a reduction of symptoms, such as fatigue, up to 3 months following treatment with reduction in the dysbiosis index (187). A recent meta-analysis looked at the efficacy of FMT in IBS and found the mode of delivery may have an impact on benefit, with colonoscopy and nasojejunal tube more impactful than oral capsules (271). At this time, further research is still needed about the role of FMT in the treatment of various diseases.
11.4.2 SCFA
SCFA derived from indigestible carbohydrates can participate in the metabolism of bile acid (BA) and lipopolysaccharide (LPS) (272). SCFA has been shown to suppress the proliferation and induce apoptosis of tumor cells (273, 274). SCFA can also be used in the treatment of auto immune disorders (275). Probiotics, prebiotics and synbiotics can modulate the growth and metabolic activity of the microbiota. Use of prebiotics and probiotics that modulate local and systemic SCFA concentrations appears to be a promising therapy in infections (276). Recent preliminary evidence points out that SCFA has the potential for treating type 2 DM (277). More research is needed in this area.
SCFA may have a role in the management and treatment of chronic kidney disease owing to reduction in inflammation and oxidative stress (278). Valerate or valeric acid is another short chain fatty acid produced in small amounts during the fermentation of dietary fiber. This short chain fatty acid is depleted from the gut following antibiotics and restored with fecal microbiota transplantation. In a pre-clinical study valerate decreased the incidence of C. difficile in a mouse model of infection (279). In another study examining valeric acid level, it was noted that more depleted valeric acid amounts were present in ultra high-risk groups prior to conversion to schizophrenia and in those already with the mental health disorder. This suggests that valeric acid may be involved in the conversion to schizophrenia (280).
The benefits of anti-inflammatory impacts related to SCFA may even extend to the epithelium including treatment of various conditions such as psoriasis and acne (281). Mental health including epilepsy may even benefit from SCFA through various pathways including neurotransmitter impacts, the protection of the blood brain barrier, reduction of oxidative stress to neural tissue and downregulation of psychosocial stress (282) Studies have shown the role of SCFA in treating cancers, autoimmune diseases, infections, type 2 diabetes mellitus, chronic kidney disease, epilepsy and inflammatory skin diseases. Although SCFA impacts are quite encouraging across many health conditions, human studies in this area remains limited. More research and clinical trials are needed to reveal the therapeutic potential of SCFA.
11.4.3 Postbiotics
Postbiotics are soluble components of microbial cells or their derived metabolites that can provide therapeutic benefits (198). Species other than those belonging to the traditionally safe genus Bifidobacterium or the family Lactobacillaceae, which could not be administered live due to concerns about their safety, have been explored as potential postbiotics (283).
12 Dysbiosis and related costs
The impact of microbial dysbiosis can lead to increased health care costs related to both acute and chronic conditions. In particular, antibiotic-associated diarrhea can lead to increased morbidity and lengthier hospital admissions, requiring more healthcare resources (284). In the United Kingdom, the resulting intensive care unit stays and need for readmission was speculated to cost £13,272.53 per patient with antibiotic-associated diarrhea (284). As well, patients with a C. difficile associated diarrhea often require extended hospitalization and multiple medical treatments, including laboratory tests. In the United States, data from 2014 found that patients with a primary C. difficile associated diarrhea would incur $24,205 USD in health care costs while a recurrent C. difficile associated diarrhea patient would require US$10,580 (284).
As previously noted, dysbiosis can influence many chronic diseases with considerable implications among these illnesses. Chronic health conditions are associated with increasing resource costs to society with the CDC indicating that “90% of the nation’s $4.1 trillion in annual health care expenditures are for people with chronic and mental health conditions”(285). Dietary and lifestyle approaches possess a great deal of promise to combat chronic conditions that may be influenced in considerable ways by dysbiosis and microbiome imbalance. Making use of these relatively non-invasive strategies seems prudent to minimize both health risks and societal costs.
13 Benefits and limitations to dysbiosis diagnosis and management
There is currently no specific method or gold standard technique to diagnosis microbial dysbiosis in a patient. To date, the use of a stool sample analysis is the most common way to interpret the state of a patients gut microbiome and if a potential dysbiotic state exists. By continuing to develop more specific tools and methods, such as microbial metabolite detection, a better comprehension of changes in the gut microbiome can be gained. From this, there may be further understanding in how the gut microbiome may play a role in the physiology and pathology of certain human diseases. Further, gene-level and bioactive microbial protein analyses of microbiome-disease is better than taxonomic analysis.
There remains certain limitation in our knowledge around the gut microbiome, including that there is no one consistent model that serves as a means to capture the phenotypic diversity and complexity of the microbiome. As well, the concept of an “ideal microbiome” has not been established. Thus, the beneficial bacteria for one individual may not serve the same benefit for another (286). As well, clear guidelines or protocol on the treatment of a dysbiotic state, as well as ways to maintain a healthy gut microbiome has not been established. Even through various lifestyle and dietary interventions, there may be a need for a more personalized therapeutic approach for the treatment of gut dysbiosis (286).
In recent years many publications have highlighted the role of microbiota and dysbiosis in different diseases. Like any other diseases, genetic, epigenetic, lifestyle and environmental factors play a role in the medical condition of dysbiosis. Systemic screening of microbiota and measuring metabolites is now possible. In recent years, there are many targeted studies investigating gut microbiome alternations in different human diseases. Abnormal metabolites levels have been linked to certain diseases. For example, trimethylamine levels are associated with cardiovascular disorders, bile acids like deoxycholic acid and lithocholic acids with colorectal cancer and SCFA butyrate with cognitive disorder. Use of simple supplemental therapies like probiotics, prebiotics, synbiotics with regular treatment can potentiate the effect or reduce the toxicity of treatment for diseases. Obviously more interventional study research is needed in humans. Role of diet in shaping microbiota is also changing the view of strategies of improving systemic and whole-body health. With microbiome-based therapies, dysbiosis can also be treated by transplanting bacteria or bacterial-derived byproducts (SCFA, post biotics) to ameliorate the microbiome and restore health. For wide spread use of these therapies more research is needed. Microbiome testing is still in its infancy and has limited value for day-to-day practice at this point. A new form of microbiome therapeutics is the evolving phage therapy.
Overall, with new emerging microbiome studies with different medical conditions, analyzing the microbiome with conventional methods of diagnosis and using the different strategies for the management of dysbiosis along with traditional management may improve healthcare, especially where conventional approaches have failed (287). Broad adoption by medical communities will help with the advancement of ways to treat diseases using the microbiome-based approaches.
14 Conclusion
Human gut microbiome in multiple studies has been shown to play an important role in health. Dysbiosis can be considered as a medical condition. Whole body dysbiosis causes imbalance in the composition or function of gut and non-gut related microbiome and can have a broad clinical presentation as a medical disorder from metabolic syndromes to cancer. Definition of normal gut microbiota has not been clearly defined so far. It varies between individuals based on genetics, food preferences, lifestyle, geographic and environmental factors. Much of the understanding about dysbiosis comes from animal studies. Considerable evidence from both animal and human studies has accumulated showing a clear link between the microbiome imbalance in diseases. Understanding the communication and pathways involved in these interactions are essential to improve our knowledge of dysbiosis, and our ability to treat or prevent dysbiosis. There are different interactions between the gut and different organs that regulate function, called the gut–organ axis. Understanding the microbiota–gut–organ-axis has opened the door for better appreciation of different disease pathologies and offers opportunities to study microbial therapeutics through the regulation of the microbiome. Dysbiosis can be seen as an initiation, perpetuation or outcome of diseases and can be the target for treating these conditions. It can affect any body part that has its own special ecosystem of microorganisms throughout the whole individual. Dysbiosis may have different impacts on different hosts depending on the nature of the dysbiotic community and underlying genetic predispositions for disease.
Gut microbiota metabolites like SCFA play a role in dysbiosis. Tests used to diagnose dysbiosis are the urine test, hydrogen breath test, comprehensive digestive stool analysis, intestinal permeability test, microbiome diversity test, and measurement of SCFA levels. Many medically necessary treatments including medications impact the microbiome.
Diet and lifestyle changes should be considered as a therapeutic approaches to improve dysbiosis. The microbiome may vary daily, weekly and monthly depending on diet and life style factors. Metabolites derived from gut microbiota like SCFA can play a role in the development of diseases. Certain diets can influence different gut bacteria and metabolites. Treating dysbiosis with lifestyle changes and diet modification (including avoiding ultra processed foods) can alter the gut microbiome composition and function. Diet and lifestyle alterations appear to be the most obvious, non-invasive, and immediate way of altering the microbiome composition and function. There are ongoing research and clinical trials in the field of microbial therapeutics. Identifying and reversing dysbiosis can be life-changing for many people. In a dysbiotic condition, dietary and lifestyle modifications, treatment with gut antibiotics, and, with more severe cases, faecal transplantation, are the interventions used to correct this state. There are only a limited number of human research studies to show this relationship. Further human studies are needed in this area to more clearly elucidate pathways, mechanisms and benefits to human health. Overall evidence at this point shows dysbiosis as a probable new therapeutic target in the management of diseases.
Author contributions
KA: Conceptualization, Writing – original draft, Writing – review & editing. JM: Conceptualization, Writing – original draft, Writing – review & editing. TH: Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Litvak, Y, and Bäumler, AJ. The founder hypothesis: a basis for microbiota resistance, diversity in taxa carriage, and colonization resistance against pathogens. PLoS Pathog. (2019) 15:e1007563. doi: 10.1371/journal.ppat.1007563
2. Baquero, F, and Nombela, C. The microbiome as a human organ. Clin Microbiol Infect. (2012) 18:2–4. doi: 10.1111/j.1469-0691.2012.03916.x
3. Sekirov, I, and Finlay, BB. Human and microbe: united we stand. Nat Med. (2006) 12:736–7. doi: 10.1038/nm0706-736
4. Berg, G, Rybakova, D, Fischer, D, Cernava, T, Vergès, MC, Charles, T, et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome. (2020) 8:103. doi: 10.1186/s40168-020-00875-0
5. Li, J, Li, D, Chen, Y, Chen, W, Xu, J, and Gao, L. Gut microbiota and aging: traditional Chinese medicine and modern medicine. Clin Interv Aging. (2023) 18:963–86. doi: 10.2147/CIA.S414714
6. Wallace, RK. The microbiome in health and disease from the perspective of modern medicine and ayurveda. Medicina. (2020) 56:462. doi: 10.3390/medicina56090462
7. Integrative HMP (iHMP) Research Network Consortium. The integrative human microbiome project. Nature. (2019) 569:641–8. doi: 10.1038/s41586-019-1238-8
8. Montalto, M, D’Onofrio, F, Gallo, A, Cazzato, A, and Gasbarrini, G. Intestinal microbiota and its functions. Dig Liver Dis. (2009) Suppl. 3:30–4. doi: 10.1016/S1594-5804(09)60016-4
9. Ley, RE, Peterson, DA, and Gordon, JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. (2006) 124:837–48. doi: 10.1016/j.cell.2006.02.017
10. Schuijt, TJ, Lankelma, JM, Scicluna, BP, de Sousa e Melo, F, Roelofs, JJTH, de Boer, JD, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. (2016) 65:575–83. doi: 10.1136/gutjnl-2015-309728
11. Sekirov, I, Russell, SL, Antunes, LCM, and Finlay, BB. Gut microbiota in health and disease. Physiol Rev. (2010) 90:859–904. doi: 10.1152/physrev.00045.2009
12. Abjani, F, Madhavan, P, Chong, PP, Chinna, K, Rhodes, CA, and Lim, YAL. Urbanization and its associated factors affecting human gut microbiome. Where are we heading to? Ann Hum Biol. (2023) 50:137–47. doi: 10.1080/03014460.2023.2170464
13. Battson, ML, Lee, DM, Weir, TL, and Gentile, CL. The gut microbiota as a novel regulator of cardiovascular function and disease. J Nutr Biochem. (2018) 56:1–15. doi: 10.1016/j.jnutbio.2017.12.010
14. Hawrelak, JA, Cattley, T, and Myers, SP. Essential oils in the treatment of intestinal dysbiosis: a preliminary in vitro study. Altern Med Rev. (2009) 14:380–4.
15. Petersen, C, and Round, JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. (2014) 16:1024–33. doi: 10.1111/cmi.12308
16. Zhang, YJ, Li, S, Gan, RY, Zhou, T, Xu, DP, and Li, HB. Impacts of gut Bacteria on human health and diseases. Int J Mol Sci. (2015) 16:7493–519. doi: 10.3390/ijms16047493
17. Belizário, JE, and Faintuch, J. Microbiome and gut dysbiosis. Exp Suppl. (2018) 109:459–76. doi: 10.1007/978-3-319-74932-7_13
18. Carding, S, Verbeke, K, Vipond, DT, Corfe, BM, and Owen, LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. (2015) 26:26191. doi: 10.3402/mehd.v26.26191
19. Wang, Z, Klipfell, E, Bennett, BJ, Koeth, R, Levison, BS, Dugar, B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. (2011) 472:57–63. doi: 10.1038/nature09922
20. Fava, S. Glucagon-like peptide 1 and the cardiovascular system. Curr Diabetes Rev. (2014) 10:302–10. doi: 10.2174/1573399810666141030125830
21. Sabaté, J-M, Jouët, P, Harnois, F, Mechler, C, Msika, S, Grossin, M, et al. High prevalence of small intestinal bacterial overgrowth in patients with morbid obesity: a contributor to severe hepatic steatosis. Obes Surg. (2008) 18:371–7. doi: 10.1007/s11695-007-9398-2
22. Karakasidis, E, Kotsiou, OS, and Gourgoulianis, KI. Lung and gut Microbiome in COPD. J Pers Med. (2023) 13:804. doi: 10.3390/jpm13050804
23. Barcik, W, Boutin, RCT, Sokolowska, M, and Finlay, BB. The role of lung and gut microbiota in the pathology of asthma. Immunity. (2020) 52:241–55. doi: 10.1016/j.immuni.2020.01.007
24. Morgan, XC, Tickle, TL, Sokol, H, Gevers, D, Devaney, KL, Ward, DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. (2012) 13:R79. doi: 10.1186/gb-2012-13-9-r79
25. Dionne, J, Ford, AC, Yuan, Y, Chey, WD, Lacy, BE, and Saito, YA. A systematic review and meta-analysis evaluating the efficacy of a gluten-free diet and a low fodmaps diet in treating symptoms of irritable bowel syndrome. Am J Gastroenterol. (2018) 113:1290–300. doi: 10.1038/s41395-018-0195-4
26. Sehgal, K, and Khanna, S. Gut microbiome and Clostridioides difficile infection: a closer look at the microscopic interface. Ther Adv Gastroenterol. (2021) 14:175628482199473. doi: 10.1177/1756284821994736
27. Li, F, Ye, J, Shao, C, and Zhong, B. Compositional alterations of gut microbiota in nonalcoholic fatty liver disease patients: a systematic review and meta-analysis. Lipids Health Dis. (2021) 20:22. doi: 10.1186/s12944-021-01440-w
28. Brandl, K, and Schnabl, B. Intestinal microbiota and nonalcoholic steatohepatitis. Curr Opin Gastroenterol. (2017) 33:128–33. doi: 10.1097/MOG.0000000000000349
29. Neugent, ML, Hulyalkar, NV, Nguyen, VH, Zimmern, PE, and De Nisco, NJ. Advances in understanding the human urinary Microbiome and its potential role in urinary tract infection. MBio. (2020) 11:e00218–20. doi: 10.1128/mBio.00218-20
30. Brotman, RM. Vaginal microbiome and sexually transmitted infections: an epidemiologic perspective. J Clin Invest. (2011) 121:4610–7. doi: 10.1172/JCI57172
31. Janowski, A, and Newland, J. From the microbiome to the central nervous system, an update on the epidemiology and pathogenesis of bacterial meningitis in childhood. F1000Res. (2017) 6, 6:F1000 Faculty Rev-86. doi: 10.12688/f1000research.8533.1
32. Halverson, T, and Alagiakrishnan, K. Gut microbes in neurocognitive and mental health disorders. Ann Med. (2020) 52:423–43. doi: 10.1080/07853890.2020.1808239
33. Vijay, A, and Valdes, AM. Role of the gut microbiome in chronic diseases: a narrative review. Eur J Clin Nutr. (2022) 76:489–501. doi: 10.1038/s41430-021-00991-6
34. Sharifian, K, Shoja, Z, and Jalilvand, S. The interplay between human papillomavirus and vaginal microbiota in cervical cancer development. Virol J. (2023) 20:73. doi: 10.1186/s12985-023-02037-8
35. Rebersek, M. Gut microbiome and its role in colorectal cancer. BMC Cancer. (2021) 21:1325. doi: 10.1186/s12885-021-09054-2
36. Alpizar-Rodriguez, D, Lesker, TR, Gronow, A, Gilbert, B, Raemy, E, Lamacchia, C, et al. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann Rheum Dis. (2019) 78:590–3. doi: 10.1136/annrheumdis-2018-214514
37. Kim, JE, and Kim, HS. Microbiome of the skin and gut in atopic dermatitis (AD): understanding the pathophysiology and finding novel management strategies. J Clin Med. (2019) 8:444. doi: 10.3390/jcm8040444
38. Siljander, H, Honkanen, J, and Knip, M. Microbiome and type 1 diabetes. EBioMedicine. (2019) 46:512–21. doi: 10.1016/j.ebiom.2019.06.031
39. Remely, M, Hippe, B, Zanner, J, Aumueller, E, Brath, H, and Haslberger, AG. Gut microbiota of obese, type 2 diabetic individuals is enriched in Faecalibacterium prausnitzii, Akkermansia muciniphila and Peptostreptococcus anaerobius after weight loss. Endocr Metab Immune Disord Drug Targets. (2016) 16:99–106. doi: 10.2174/1871530316666160831093813
40. Georges, FM, Do, NT, and Seleem, D. Oral Dysbiosis and systemic diseases. Front Dental Med. (2022) 3:1933. doi: 10.3389/fdmed.2022.995423
41. Claesson, MJ, Cusack, S, O'Sullivan, O, Greene-Diniz, R, de Weerd, H, Flannery, E, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA. (2011) 108:4586–91. doi: 10.1073/pnas.1000097107
42. Bik, EM, Ugalde, JA, Cousins, J, Goddard, AD, Richman, J, and Apte, ZS. Microbial biotransformations in the human distal gut. Br J Pharmacol. (2018) 175:4404–14. doi: 10.1111/bph.14085
43. Lloyd- Price, J, Abu-Ali, G, and Huttenhower, C. The healthy human microbiome. Genome Med. (2016) 8:51. doi: 10.1186/s13073-016-0307-y
44. Cugini, C, Ramasubbu, N, Tsiagbe, VK, and Fine, DH. Dysbiosis from a microbial and host perspective relative to oral health and disease. Front Microbiol. (2021) 12:617485. doi: 10.3389/fmicb.2021.617485
45. Aimetti, M, Cacciatore, S, Graziano, A, and Tenori, L. Metabonomic analysis of saliva reveals generalized chronic periodontitis signature. Metabolomics. (2012) 8:465–74. doi: 10.1007/s11306-011-0331-2
46. Singh, MP, Saxena, M, Saimbi, CS, Siddiqui, MH, and Roy, R. Post-periodontal surgery propounds early repair salivary biomarkers by (1)H NMR based metabolomics. Metabolomics. (2019) 15:141. doi: 10.1007/s11306-019-1593-3
47. Kazarina, A, Kuzmicka, J, Bortkevica, S, Zayakin, P, Kimsis, J, Igumnova, V, et al. Oral microbiome variations related to ageing: possible implications beyond oral health. Arch Microbiol. (2023) 205:116. doi: 10.1007/s00203-023-03464-5
48. Jo, S, Kang, W, Hwang, YS, Lee, SH, Park, KW, Kim, MS, et al. Oral and gut dysbiosis leads to functional alterations in Parkinson’s disease. npj Parkinsons Dis. (2022) 8:87. doi: 10.1038/s41531-022-00351-6
49. Simon, GL, and Gorbach, SL. Normal alimentary tract microflora In: MJ Blaser, PD Smith, JI Ravdin, HB Greenberg, and RL Guerrant, editors. Infections of the gastrointestinal tract. New York, NY: Raven Press (1995). 53.
50. Hao, WL, and Lee, YK. Microflora of the gastrointestinal tract: a review. Methods Mol Biol. (2004) 268:491–502. doi: 10.1385/1-59259-766-1:491
51. Human Microbiome Project Consortium Structure. Function and diversity of the healthy human microbiome. Nature. (2012) 486:207–14. doi: 10.1038/nature11234
52. Zafar, H, and Saier, MH. Gut Bacteroides species in health and disease. Gut Microbes. (2021) 13:1–20. doi: 10.1080/19490976.2020.1865706
53. Gorbach, SL. Microbiology of the gastrointestinal tract In: S Baron, editor. Medical Microbiology. 4th ed. Galveston, TX: University of Texas Medical Branch at Galveston (1996) Chapter 95. Available at: https://www.ncbi.nlm.nih.gov/books/NBK7670/
54. Yang, D, Xing, Y, Song, X, and Qian, V. The impact of lung microbiota dysbiosis on inflammation. Immunology. (2020) 159:156–66. doi: 10.1111/imm.13139
55. Marsland, BJ, and Gollwitzer, ES. Host- microorganism interactions in lung diseases. Nat Rev Immunol. (2014) 14:827–35. doi: 10.1038/nri3769
56. Rohde, R, and Friedland, DR. Clinical perspectives on nasopharyngeal morphology in humans. Anat Rec. (2022) 305:2065–74. doi: 10.1002/ar.24926
57. Matheau, E, Escribano- Vazquez, U, Descamps, D, Cherbuy, C, Langella, P, Riffault, S, et al. Paradigms of lung microbiota functions in health and disease, particularly in asthma. Front Physiol. (2018) 9:1168. doi: 10.3389/fphys.2018.01168
58. Huang, YJ, Sethi, S, Murphy, T, Nariya, S, Boushey, HA, and Lynch, SV. Airway microbiome dynamics in exacerbations of chronic obstructive pulmonary disease. J Clin Microbiol. (2014) 52:2813–23. doi: 10.1128/JCM.00035-14
59. O’Dwyer, DN, Ashley, SL, Gurczynski, SJ, Xia, M, Wilke, C, Falkowski, NR, et al. Lung microbiota contribute to pulmonary inflammation and disease progression in pulmonary fibrosis. Am J Respir Crit Care Med. (2019) 199:1127–38. doi: 10.1164/rccm.201809-1650OC
60. Han, MK, Zhou, Y, Murray, S, Tayob, N, Noth, I, Lama, VN, et al. Lung Microbiome and disease progrsession in idiopathic pulmonary fibrosis: an analysis of the COMET study. Lancet Respir Med. (2014) 2:548–56. doi: 10.1016/S2213-2600(14)70069-4
61. Mao, Q, Jiang, F, Yin, R, Wang, J, Xia, W, Dong, G, et al. Interplay between the lung microbiome and lung cancer. Cancer Lett. (2018) 415:40–8. doi: 10.1016/j.canlet.2017.11.036
62. Dang, AT, and Marsland, BJ. Microbes, metabolites, and the gut - lung axis. Mucosal Immunol. (2019) 12:843–50. doi: 10.1038/s41385-019-0160-6
63. Anand, S, and Diet Mande, SS. Microbiota and gut- lung connection. Front Microbiol. (2018) 9:2147. doi: 10.3389/fmicb.2018.02147
64. Lee, SH, Yun, Y, Kim, SJ, Lee, EJ, Kim, SJ, Lee, EJ, Chang, Y, Ryu, S, et al. Association between cigarette smoking status and composition of gut microbiota: population-based cross- sectional study. J Clin Med. (2018) 7:282. doi: 10.3390/jcm7090282
65. Antinozzi, M, Giffi, M, Sini, N, Gallè, F, Valeriani, F, De Vito, C, et al. Cigarette smoking and human gut microbiota in healthy adults: a systematic review. Biomedicines. (2022) 10:510. doi: 10.3390/biomedicines10020510
66. Szmidt, MK, Kaluza, J, Harris, HR, Linden, A, and Wolk, A. Long-term dietary fiber intake and risk of chronic obstructive pulmonary disease: a prospective cohort study of women. Eur J Nutr. (2020) 59:1869–79. doi: 10.1007/s00394-019-02038-w
67. Valisoltani, N, Ghoreishy, SM, Imani, H, Rajabi Harsini, A, Jowshan, M, Travica, N, et al. Fiber intake and risk of chronic obstructive pulmonary disease: a systematic review and dose response meta-analysis. Food Sci Nutr. (2023) 11:6775–88. doi: 10.1002/fsn3.3640
68. Budden, KF, Gellatly, SL, Wood, DL, Cooper, MA, Morrison, M, Hugenholtz, P, et al. Emerging pathogenic links between microbiota and the gut- lung axis. Nat Rev Microbiol. (2017) 15:55–63. doi: 10.1038/nrmicro.2016.142
69. Natalini, JG, Singh, S, and Segal, LN. The dynamic lung microbiome in health and disease. Nat Rev Microbiol. (2023) 21:222–35. doi: 10.1038/s41579-022-00821-x
70. Gallo, RL. Human skin is the largest epithelial surface for interaction with microbes. J Invest Dermatol. (2017) 137:1213–4. doi: 10.1016/j.jid.2016.11.045
71. Campagnoli, LIM, Varesi, A, Barbieri, A, Marchesi, N, and Pascale, A. Targeting the gut-eye axis: an emerging strategy to face ocular diseases. Int J Mol Sci. (2023) 24:13338. doi: 10.3390/ijms241713338
72. O’Neill, CA, Monteleone, G, McLaughlin, JT, and Paus, R. The gut-skin axis in health and disease: a paradigm with therapeutic implications. Bioessays. (2016) 38:1167–76. doi: 10.1002/bies.201600008
73. Mahmud, MR, Akter, S, Tamanna, SK, Mazumder, L, Esti, IZ, Banerjee, S, et al. Impact of gut microbiome on skin health: gut-skin axis observed through the lenses of therapeutics and skin diseases. Gut Microbes. (2022) 14:2096995. doi: 10.1080/19490976.2022.2096995
74. Yu, Y, Dunaway, S, Champer, J, Kim, J, and Alikhan, A. Changing our microbiome: probiotics in dermatology. Br J Dermatol. (2020) 182:39–46. doi: 10.1111/bjd.18088
75. Muizzuddin, N, Maher, W, Sullivan, M, Schnittger, S, and Mammone, T. Physiological effect of a probiotic on skin. J Cosmet Sci. (2012) 63:385–95.
76. Lolou, V, and Panayiotidis, MI. Functional role of probiotics and prebiotics on skin health and disease. Fermentation. (2019) 5:41. doi: 10.3390/fermentation5020041
77. Davani-Davari, D, Negahdaripour, M, Karimzadeh, I, Seifan, M, and Mohkam, M. Masoumi S prebiotics: definition, types, sources, mechanisms, and clinical applications. Food Secur. (2019) 8:92. doi: 10.3390/foods8030092
78. Krejner, A, Bruhs, A, Mrowietz, U, Wehkamp, U, Schwarz, T, and Schwarz, A. Decreased expression of G-protein-coupled receptors GPR43 and GPR109a in psoriatic skin can be restored by topical application of sodium butyrate. Arch Dermatol Res. (2018) 310:751–8. doi: 10.1007/s00403-018-1865-1
79. De Pessemier, B, Grine, L, Debaere, M, Maes, A, Paetzold, B, and Callewaert, C. Gut-skin Axis: current knowledge of the interrelationship between microbial Dysbiosis and skin conditions. Microorganisms. (2021) 9:353. doi: 10.3390/microorganisms9020353
80. Lee, SY, Lee, E, Park, YM, and Hong, SJ. Microbiome in the gut-skin axis in atopic dermatitis. Allergy Asthma Immunol Res. (2018) 10:354–62. doi: 10.4168/aair.2018.10.4.354
81. Lee, HJ, and Kim, M. Skin barrier function and the Microbiome. Int J Mol Sci. (2022) 23:13071. doi: 10.3390/ijms232113071
82. Aguwa, C, Enwereji, N, Santiago, S, Hine, A, Kels, GG, McGee, J, et al. Targeting dysbiosis in psoriasis, atopic dermatitis, and hidradenitis suppurativa: the gut-skin axis and microbiome-directed therapy. Clin Dermatol. (2023) 41:640–9. doi: 10.1016/j.clindermatol.2023.09.019
83. Wilkinson, HN, and Hardman, MJ. Cellular senescence in acute and chronic wound repair. Cold Spring Harb Perspect Biol. (2022) 14:a041221. doi: 10.1101/cshperspect.a041221
84. Woo, YR, Cho, SH, Lee, JD, and Kim, HS. The human microbiota and skin Cancer. Int J Mol Sci. (2022) 23:1813. doi: 10.3390/ijms23031813
85. Lu, HP, Lai, YC, Huang, SW, Chen, HC, Hsieh, CH, and Yu, HT. Spatial heterogeneity of gut microbiota reveals multiple bacterial communities with distinct characteristics. Sci Rep. (2014) 4:6185. doi: 10.1038/srep06185
86. Costello, EK, Lauber, CL, Hamady, M, Fierer, N, Gordon, JI, and Knight, R. Bacterial community variation in human body habitats across space and time. Science. (2009) 326:1694–7. doi: 10.1126/science.1177486
87. Smythe, P, and Wilkinson, HN. The skin Microbiome: current landscape and future opportunities. Int J Mol Sci. (2023) 24:3950. doi: 10.3390/ijms24043950
88. Petrova, MI, Lievens, E, Malik, S, Imholz, N, and Lebeer, S. Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Front Physiol. (2015) 6:81. doi: 10.3389/fphys.2015.00081
89. Park, MG, Cho, S, and Oh, MM. Menopausal changes in the Microbiome—a review focused on the genitourinary Microbiome. Diagnostics. (2023) 13:1193. doi: 10.3390/diagnostics13061193
90. Whiteside, SA, Razvi, H, Dave, S, Reid, G, and Burton, JP. The microbiome of the urinary tract—a role beyond infection. Nat Rev Urol. (2015) 12:81–90. doi: 10.1038/nrurol.2014.361
91. Lewis, DA, Brown, R, Williams, J, White, P, Jacobson, SK, Marchesi, JR, et al. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front Cell Infect Microbiol. (2013) 3:41. doi: 10.3389/fcimb.2013.00041
92. Aragón, IM, Herrera-Imbroda, B, Queipo-Ortuño, MI, Castillo, E, del Moral, JSG, Gómez-Millán, J, et al. The urinary tract microbiome in health and disease. Eur Urol Focus. (2018) 4:128–38. doi: 10.1016/j.euf.2016.11.001
93. Greenbaum, S, Greenbaum, G, Moran-Gilad, J, and Weintraub, AY. Ecological dynamics of the vaginal microbiome in relation to health and disease. Am J Obstet Gynecol. (2019) 220:324–35. doi: 10.1016/j.ajog.2018.11.1089
94. Leue, C, Kruimel, J, Vrijens, D, Masclee, A, van Os, J, and van Koeveringe, G. Functional urological disorders: a sensitized defence response in the bladder-gut-brain axis. Nat Rev Urol. (2017) 14:153–63. doi: 10.1038/nrurol.2016.227
95. Čeprnja, M, Hadžić, E, Oros, D, Melvan, E, Starcevic, A, and Zucko, J. Current viewpoint on female urogenital microbiome—the cause or the consequence? Microorganisms. (2023) 11:1207. doi: 10.3390/microorganisms11051207
96. Wozniak, H, Beckmann, TS, Fröhlich, L, Soccorsi, T, Le Terrier, C, de Watteville, A, et al. The central and biodynamic role of gut microbiota in critically ill patients. Crit Care. (2022) 26:250. doi: 10.1186/s13054-022-04127-5
97. Li, Y, Nicholson, RJ, and Summers, SA. Ceramide signaling in the gut. Mol Cell Endocrinol. (2022) 544:111554. doi: 10.1016/j.mce.2022.111554
98. Cani, PD. Gut microbiota—at the intersection of everything? Nat Rev Gastroenterol Hepatol. (2017) 14:321–2. doi: 10.1038/nrgastro.2017.54
99. Shen, Y, Xu, J, Li, Z, Huang, Y, Yuan, Y, Wang, J, et al. Analysis of gut microbiota diversity and auxiliary diagnosis as a biomarker in patients with schizophrenia: a cross-sectional study. Schizophr Res. (2018) 197:470–7. doi: 10.1016/j.schres.2018.01.002
100. De Luca, F, and Shoenfeld, Y. The microbiome in autoimmune diseases. Clin Exp Immunol. (2019) 195:74–85. doi: 10.1111/cei.13158
101. Vimal, J, Himal, I, and Kannan, S. Role of microbial dysbiosis in carcinogenesis and cancer therapies. Indian J Med Res. (2020) 152:553–61. doi: 10.4103/ijmr.IJMR_1026_18
102. Hanus, M, Parada-Venegas, D, Landskron, G, Wielandt, AM, Hurtado, C, Alvarez, K, et al. Immune system, microbiota, and microbial metabolites: the unresolved triad in colorectal Cancer microenvironment. Front Immunol. (2021) 12:612826. doi: 10.3389/fimmu.2021.612826
103. Raza, MH, Gul, K, Arshad, A, Riaz, N, Waheed, U, Rauf, A, et al. Microbiota in cancer development and treatment. J Cancer Res Clin Oncol. (2019) 145:49–63. doi: 10.1007/s00432-018-2816-0
104. Artemev, A, Naik, S, Pougno, A, Honnavar, P, and Shanbhag, NM. The association of microbiome dysbiosis with colorectal cancer. Cureus. (2022) 14:e22156. doi: 10.7759/cureus.22156
105. Dahmus, JD, Kotler, DL, Kastenberg, DM, and Kistler, CA. The gut microbiome and colorectal cancer: a review of bacterial pathogenesis. J Gastrointest Oncol. (2018) 9:769–77. doi: 10.21037/jgo.2018.04.07
106. Hawrelak, JA, and Myers, SP. The causes of intestinal dysbiosis. A review. Altern Med Rev. (2004) 9:180–97.
107. Ma, ZS. Microbiome transmission during sexual intercourse appears stochastic and supports the red queen hypothesis. Front Microbiol. (2022) 12:789983. doi: 10.3389/fmicb.2021.789983
108. Hooper, LV, and Gordon, JI. Commensal host-bacterial relationships in the gut. Science. (2001) 292:1115–8. doi: 10.1126/science.1058709
109. Okipney, A, Amorim de Souza, JR, Ligocki Campos, AC, Campos, LF, Anjo, PR, and Abreu, C. Risk stratification for intestinal dysbiosis in hospitalized adult patients according to the National Dysbiosis Survey (INDIS). BRASPEN J. (2020) 2:149–59. doi: 10.37111/braspenj.2020352008
110. Vich Vila, A, Collij, V, Sanna, S, Sinha, T, Imhann, F, Bourgonje, AR, et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun. (2020) 11:1–11. doi: 10.1038/s41467-019-14177-z
111. Lange, K, Buerger, M, Stallmach, A, and Bruns, T. Effects of antibiotics on gut microbiota. Dig Dis. (2016) 34:260–8. doi: 10.1159/000443360
112. Dahiya, D, and Nigam, PS. Antibiotic-therapy-induced gut Dysbiosis affecting gut microbiota-brain Axis and cognition: restoration by intake of probiotics and Synbiotics. Int J Mol Sci. (2023) 24:3074. doi: 10.3390/ijms24043074
113. Korpela, K, Salonen, A, Virta, LJ, Kekkonen, RA, Forslund, K, Bork, P, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun. (2016) 7:10410. doi: 10.1038/ncomms10410
114. Ramirez, J, Guarner, F, Bustos Fernandez, L, Maruy, A, Sdepanian, VL, and Cohen, H. Antibiotics as major disruptors of gut microbiota. Front Cell Infect Microbiol. (2020) 10:572912. doi: 10.3389/fcimb.2020.572912
115. Zimmermann, P, and Curtis, N. The effect of antibiotics on the composition of the intestinal microbiota - a systematic review. J Infect. (2019) 79:471–89. doi: 10.1016/j.jinf.2019.10.008
116. Kohler, O, Petersen, L, Mors, O, et al. Infections and exposure to anti-infective agents and the risk of severe mental disorders: a nationwide study. Acta Psychiatr Scand. (2017) 135:97–105. doi: 10.1111/acps.12671
117. Lavebratt, C, Yang, LL, Giacobini, M, Forsell, Y, Schalling, M, Partonen, T, et al. Early exposure to antibiotic drugs and risk for psychiatric disorders: a population-based study. Transl Psychiatry. (2019) 9:317. doi: 10.1038/s41398-019-0653-9
118. Lurie, I, Yang, Y-X, Haynes, K, Mamtani, R, and Boursi, B. Antibiotic exposure and the risk for depression, anxiety, or psychosis: a nested case-control study. J Clin Psychiatry. (2015) 76:1522–8. doi: 10.4088/JCP.15m09961
119. Pouranayatihosseinabad, M, Bezabih, Y, Hawrelak, J, Peterson, GM, Veal, F, and Mirkazemi, C. Antibiotic use and the development of depression: a systematic review. J Psychosom Res. (2023) 164:111113. doi: 10.1016/j.jpsychores.2022.111113
120. Bejaoui, S, and Poulsen, M. The impact of early life antibiotic use on atopic and metabolic disorders: meta-analyses of recent insights. Evol Med Public Health. (2020) 1:279–89. doi: 10.1093/emph/eoaa039
121. Heianza, Y, Zheng, Y, Ma, W, Rimm, EB, Albert, CM, Hu, FB, et al. Duration and life-stage of antibiotic use and risk of cardiovascular events in women. Eur Heart J. (2019) 40:3838–45. doi: 10.1093/eurheartj/ehz231
122. Patangia, DV, Anthony Ryan, C, Dempsey, E, Paul Ross, R, and Stanton, C. Impact of antibiotics on the human microbiome and consequences for host health. Microbiol Open. (2022) 11:e1260. doi: 10.1002/mbo3.1260
123. Yang, L, Bajinka, O, Jarju, PO, Tan, Y, Taal, AM, and Ozdemir, G. The varying effects of antibiotics on gut microbiota. AMB Express. (2021) 11:116. doi: 10.1186/s13568-021-01274-w
124. Kwok, CS, Arthur, AK, Anibueze, CI, Singh, S, Cavallazzi, R, and Loke, YK. Risk of Clostridium difficile infection with acid suppressing drugs and antibiotics: meta-analysis. Am J Gastroenterol. (2012) 107:1011–9. doi: 10.1038/ajg.2012.108
125. McDonald, EG, Milligan, J, Frenette, C, and Lee, TC. Continuous proton pump inhibitor therapy and the associated risk of recurrent Clostridium difficile infection. JAMA Intern Med. (2015) 175:784–91. doi: 10.1001/jamainternmed.2015.42
126. Jackson, MA, Goodrich, JK, Maxan, ME, Freedberg, DE, Abrams, JA, Poole, AC, et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut. (2016) 65:749–56. doi: 10.1136/gutjnl-2015-310861
127. Seto, CT, Jeraldo, P, Orenstein, R, Chia, N, and DiBaise, JK. Prolonged use of a proton pump inhibitor reduces microbial diversity: implications for Clostridium difficile susceptibility. Microbiome. (2014) 2:42. doi: 10.1186/2049-2618-2-42
128. Bruno, G, Zaccari, P, Rocco, G, Scalese, G, Panetta, C, Porowska, B, et al. Proton pump inhibitors and dysbiosis: current knowledge and aspects to be clarified. World J Gastroenterol. (2019) 25:2706–19. doi: 10.3748/wjg.v25.i22.2706
129. Xia, B, Yang, M, Nguyen, LH, He, Q, Zhen, J, Yu, Y, et al. Regular use of proton pump inhibitor and the risk of inflammatory bowel disease: pooled analysis of 3 prospective cohorts. Gastroenterology. (2021) 161:1842–1852.e10. doi: 10.1053/j.gastro.2021.08.005
130. de la Cuesta-Zuluaga, J, Mueller, NT, Corrales-Agudelo, V, Velásquez-Mejía, EP, Carmona, JA, Abad, JM, et al. Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care. (2017) 40:54–62. doi: 10.2337/dc16-1324
131. Rodriguez, J, Hiel, S, and Delzenne, NM. Metformin: old friend, new ways of action-implication of the gut microbiome? Curr Opin Clin Nutr Metab Care. (2018) 21:294–301. doi: 10.1097/MCO.0000000000000468
132. Wu, H, Esteve, E, Tremaroli, V, Khan, MT, Caesar, R, Mannerås-Holm, L, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. (2017) 23:850–8. doi: 10.1038/nm.4345
133. Lukić, I, Getselter, D, Ziv, O, Oron, O, Reuveni, E, Koren, O, et al. Antidepressants affect gut microbiota and Ruminococcus flavefaciens is able to abolish their effects on depressive-like behavior. Transl Psychiatry. (2019) 9:133. doi: 10.1038/s41398-019-0466-x
134. Cussotto, S, Strain, CR, Fouhy, F, Strain, RG, Peterson, VL, Clarke, G, et al. Differential effects of psychotropic drugs on microbiome composition and gastrointestinal function. Psychopharmacology. (2019) 236:1671–85. doi: 10.1007/s00213-018-5006-5
135. Donoso, F, Cryan, JF, Olavarría-Ramírez, L, Nolan, YM, and Clarke, G. Inflammation, lifestyle factors, and the Microbiome-gut-brain Axis: relevance to depression and antidepressant action. Clin Pharmacol Ther. (2023) 113:246–59. doi: 10.1002/cpt.2581
136. Bohnert, JA, Szymaniak-Vits, M, Schuster, S, and Kern, WV. Efflux inhibition by selective serotonin reuptake inhibitors in Escherichia coli. J Antimicrob Chemother. (2011) 66:2057–60. doi: 10.1093/jac/dkr258
137. Morgan, AP, Crowley, JJ, Nonneman, RJ, Quackenbush, CR, Miller, CN, Ryan, AK, et al. The antipsychotic olanzapine interacts with the gut microbiome to cause weight gain in mouse. PLoS One. (2014) 9:e115225. doi: 10.1371/journal.pone.0115225
138. Yuan, X, Zhang, P, Wang, Y, Liu, Y, Li, X, Kumar, BU, et al. Changes in metabolism and microbiota after 24-week risperidone treatment in drug naïve, normal weight patients with first episode schizophrenia. Schizophrenia Res. (2018) 201:299–306. doi: 10.1016/j.schres.2018.05.017
139. Flowers, SA, Evans, SJ, Ward, KM, McInnis, MG, and Ellingrod, VL. Interaction between atypical antipsychotics and the gut Microbiome in a bipolar disease cohort. Pharmacotherapy. (2017) 37:261–7. doi: 10.1002/phar.1890
140. Le Bastard, Q, Al-Ghalith, GA, Grégoire, M, Chapelet, G, Javaudin, F, Dailly, E, et al. Systematic review: human gut dysbiosis induced by non-antibiotic prescription medications. Aliment Pharmacol Ther. (2018) 47:332–45. doi: 10.1111/apt.14451
141. Rogers, MAM, and Aronoff, DM. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect. (2016) 22:178.e1–9. doi: 10.1016/j.cmi.2015.10.003
142. Mäkivuokko, H, Tiihonen, K, Tynkkynen, S, Paulin, L, and Rautonen, N. The effect of age and non-steroidal anti-inflammatory drugs on human intestinal microbiota composition. Br J Nutr. (2010) 103:227–34. doi: 10.1017/S0007114509991553
143. Meng, J, Yu, H, Ma, J, Wang, J, Banerjee, S, Charboneau, R, et al. Morphine induces bacterial translocation in mice by compromising intestinal barrier function in a TLR-dependent manner. PLoS One. (2013) 8:e54040. doi: 10.1371/journal.pone.0054040
144. Vincent, C, Miller, MA, Edens, TJ, Mehrotra, S, Dewar, K, and Manges, AR. Bloom and bust: intestinal microbiota dynamics in response to hospital exposures and Clostridium difficile colonization or infection. Microbiome. (2016) 4:12. doi: 10.1186/s40168-016-0156-3
145. Dias, AM, Cordeiro, G, Estevinho, MM, Veiga, R, Figueira, L, Reina-Couto, M, et al. The clinical pharmacology unit, São João hospital university Centre. Gut bacterial microbiome composition and statin intake-a systematic review. Pharmacol Res Perspect. (2020) 8:e00601. doi: 10.1002/prp2.601
146. Bedarf, JR, Hildebrand, F, Coelho, LP, Sunagawa, S, Bahram, M, Goeser, F, et al. Functional implications of microbial and viral gut metagenome changes in early-stage L-DOPA-naïve Parkinson's disease patients. Genome Med. (2017) 9:39. doi: 10.1186/s13073-017-0428-y
147. Wilmanski, T, Kornilov, SA, Diener, C, Conomos, MP, Lovejoy, JC, Sebastiani, P, et al. Heterogeneity in statin responses explained by variation in the human gut microbiome. Med. (2022) 3:388–405.e6. doi: 10.1016/j.medj.2022.04.007
148. Liu, Y, Song, X, Zhou, H, Zhou, X, Xia, Y, Dong, X, et al. Gut microbiome associates with lipid-lowering effect of rosuvastatin in vivo. Front Microbiol. (2018) 9:530. doi: 10.3389/fmicb.2018.00530
149. Saffouri, GB, Shields-Cutler, RR, Chen, J, Yang, Y, Lekatz, HR, Hale, VL, et al. Small intestinal microbial dysbiosis underlies symptoms associated with functional gastrointestinal disorders. Nat Commun. (2019) 10:2012. doi: 10.1038/s41467-019-09964-7
150. Shah, A, Talley, NJ, and Holtmann, G. Current and future approaches for diagnosing Small intestinal Dysbiosis in patients with symptoms of functional dyspepsia. Front Neurosci. (2022) 16:830356. doi: 10.3389/fnins.2022.830356
152. Tang, Q, Jin, G, Wang, G, Liu, T, Liu, X, Wang, B, et al. Current sampling methods for gut microbiota: a call for more precise devices. Front Cell Infect Microbiol. (2020) 10:151. doi: 10.3389/fcimb.2020.00151
153. Wei, S, Bahl, MI, Baunwall, SMD, Hvas, CL, and Licht, TR. Determining gut microbial dysbiosis: a review of applied indexes for assessment of intestinal microbiota imbalances. Appl Environ Microbiol. (2021) 87:e00395–21. doi: 10.1128/AEM.00395-21
154. Tynkkynen, T, Wang, Q, Ekholm, J, Anufrieva, O, Ohukainen, P, Vepsäläinen, J, et al. Proof of concept for quantitative urine NMR metabolomics pipeline for large-scale epidemiology and genetics. Int J Epidemiol. (2019) 48:978–93. doi: 10.1093/ije/dyy287
155. Musa, MA, Kabir, M, Hossain, MI, Ahmed, E, Siddique, A, Rashid, H, et al. Measurement of intestinal permeability using lactulose and mannitol with conventional five hours and shortened two hours urine collection by two different methods: HPAE-PAD and LC-MSMS. PLoS One. (2019) 14:e0220397. doi: 10.1371/journal.pone.0220397
156. Ribeiro, WR, Vinolo, M, Calixto, LR, and Ferreira, CM. Use of gas chromatography to quantify short chain fatty acids in the serum, colonic luminal content and feces of mice. Bio Protoc. (2018) 8:e3089. doi: 10.21769/BioProtoc.3089
157. Rana, SV, and Malik, A. Hydrogen breath tests in gastrointestinal diseases. Indian J Clin Biochem. (2014) 29:398–405. doi: 10.1007/s12291-014-0426-4
158. Wu, WK, Chen, CC, Liu, PY, Panyod, S, Liao, BY, Chen, PC, et al. Identification of TMAO-producer phenotype and host-diet-gut dysbiosis by carnitine challenge test in human and germ-free mice. Gut. (2019) 68:1439–49. doi: 10.1136/gutjnl-2018-317155
159. Romo-Vaquero, M, Fernández-Villalba, E, Gil-Martinez, AL, Cuenca-Bermejo, L, Espín, JC, Herrero, MT, et al. Urolithins: potential biomarkers of gut dysbiosis and disease stage in Parkinson's patients. Food Funct. (2022) 13:6306–16. doi: 10.1039/D2FO00552B
160. Rüb, AM, Tsakmaklis, A, Gräfe, SK, Simon, MC, Vehreschild, MJ, and Wuethrich, I. Biomarkers of human gut microbiota diversity and dysbiosis. Biomark Med. (2021) 15:137–48. doi: 10.2217/bmm-2020-0353
161. Blue Cross Blue Shield Association Evidence Positioning System®. 2.04.26- Fecal Analysis in the Diagnosis of Intestinal Dysbiosis, 01/2.
162. Jeffery, IB, Das, A, O'Herlihy, E, et al. Differences in fecal microbiomes and metabolomes of people with vs without irritable bowel syndrome and bile acid malabsorption. Gastroenterology. (2020) 158:1016–1028.e8. doi: 10.1053/j.gastro.2019.11.301
163. Fernandez, M, Alin, T, Métayer, C, Thiébaut, R, Enaud, R, and Delhaes, L. The respiratory microbiota alpha-diversity in chronic lung diseases: first systematic review and meta-analysis. Respir Res. (2022) 23:214. doi: 10.1186/s12931-022-02132-4
164. Li, Z, Zhou, J, Liang, H, Ye, L, Lan, L, Lu, F, et al. Differences in alpha diversity of gut microbiota in neurological diseases. Front Neurosci. (2022) 16:879318. doi: 10.3389/fnins.2022.879318
165. Qin, J, Li, R, Raes, J, Arumugam, M, Burgdorf, KS, Manichanh, C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. (2010) 464:59–65. doi: 10.1038/nature08821
166. Terrón-Camero, LC, Gordillo-González, F, Salas-Espejo, E, and Andrés-León, E. Comparison of metagenomics and Metatranscriptomics tools: a guide to making the right choice. Genes. (2022) 13:2280. doi: 10.3390/genes13122280
167. Tansel, A, and Levinthal, DJ. Understanding our tests: hydrogen-methane breath testing to diagnose Small intestinal bacterial overgrowth. Clin Transl Gastroenterol. (2023) 14:e00567. doi: 10.14309/ctg.0000000000000567
168. Adak, A, and Khan, MR. An insight into gut microbiota and its functionalities. Cell Mol Life Sci. (2019) 76:473–93. doi: 10.1007/s00018-018-2943-4
169. Zierer, J, Jackson, MA, Kastenmüller, G, Mangino, M, Long, T, Telenti, A, et al. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet. (2018) 50:790–5. doi: 10.1038/s41588-018-0135-7
170. Nicholson, JK, Holmes, E, Kinross, J, Burcelin, R, Gibson, G, Jia, W, et al. Host-gut microbiota metabolic interactions. Science. (2012) 336:1262–7. doi: 10.1126/science.1223813
171. Dettmer, K, Aronov, PA, and Hammock, BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. (2007) 26:51–78. doi: 10.1002/mas.20108
172. Miller, TL, and Wolin, MJ. Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl Environ Microbiol. (1996) 62:1589–92. doi: 10.1128/aem.62.5.1589-1592.1996
173. Blaak, EE, Canfora, EE, Theis, S, Frost, G, Groen, AK, Mithieux, G, et al. Short chain fatty acids in human gut and metabolic health. Benef Microbes. (2020) 11:411–55. doi: 10.3920/BM2020.0057
174. Farhangi, MA, and Vajdi, M. Novel findings of the association between gut microbiota-derived metabolite trimethylamine N-oxide and inflammation: results from a systematic review and dose-response meta-analysis. Crit Rev Food Sci Nutr. (2020) 60:2801–23. doi: 10.1080/10408398.2020.1770199
175. Chen, ML, Yi, L, Zhang, Y, Zhou, X, Ran, L, Yang, J, et al. Resveratrol attenuates trimethylamine-N-oxide (TMAO)-induced atherosclerosis by regulating TMAO synthesis and bile acid metabolism via remodeling of the gut microbiota. MBio. (2016) 7:e02210–5. doi: 10.1128/mBio.02210-15
176. Wu, WK, Hsu, CC, Sheen, LY, and Wu, MS. Measurement of gut microbial metabolites in cardiometabolic health and translational research. Rapid Commun Mass Spectrom. (2020) 34:e8537. doi: 10.1002/rcm.8537
177. Xue, C, Li, G, Zheng, Q, Gu, X, Shi, Q, Su, Y, et al. Tryptophan metabolism in health and disease. Cell Metab. (2023) 35:1304–26. doi: 10.1016/j.cmet.2023.06.004
178. Cseh, EK, Veres, G, Szentirmai, M, Nánási, N, Szatmári, I, Fülöp, F, et al. HPLC method for the assessment of tryptophan metabolism utilizing separate internal standard for each detector. Anal Biochem. (2019) 574:7–14. doi: 10.1016/j.ab.2019.03.005
179. Aguiar-Pulido, V, Huang, W, Suarez-Ulloa, V, Cickovski, T, Mathee, K, and Narasimhan, G. Metagenomics, Metatranscriptomics, and metabolomics approaches for Microbiome analysis. Evol Bioinformatics Online. (2016) 12:5–16. doi: 10.4137/EBO.S36436
180. Blount, K, Jones, C, Walsh, D, Gonzalez, C, and Shannon, WD. Development and validation of a novel Microbiome-based biomarker of post-antibiotic Dysbiosis and subsequent restoration. Front Microbiol. (2022) 12:781275. doi: 10.3389/fmicb.2021.781275
181. Zoetendal, EG, Akkermans, ADL, Akkermans-van Vliet, WM, de Visser, JAGM, and De Vos, WM. The host genotype affects the bacterial community in the human gastrointestinal tract. Microb Ecol Health Dis. (2001) 2001:129–34. doi: 10.1080/089106001750462669
182. Frank, DN, Spiegelman, GB, Davis, W, Wagner, E, Lyons, E, and Pace, NR. culture-independent molecular analysis of microbial constituents of the healthy human outer ear. J Clin Microbiol. (2003) 41:295–303. doi: 10.1128/JCM.41.1.295-303.2003
183. Lopera-Maya, EA, Kurilshikov, A, van der Graaf, A, Hu, S, Andreu-Sánchez, S, Chen, L, et al. Effect of host genetics on the gut microbiome in 7,738 participants of the Dutch microbiome project. Nat Genet. (2022) 54:143–51. doi: 10.1038/s41588-02100992-y
184. Yang, H, Wu, J, Huang, X, Zhou, Y, Zhang, Y, Liu, M, et al. ABO genotype alters the gut microbiota by regulating GalNAc levels in pigs. Nature. (2022) 606:358–67. doi: 10.1038/s41586-022-04769-z
185. Bonder, MJ, Kurilshikov, A, Tigchelaar, EF, Mujagic, Z, Imhann, F, Vila, AV, et al. The effect of host genetics on the gut microbiome. Nat Genet. (2016) 48:1407–12. doi: 10.1038/ng.3663
186. Xu, F, Fu, Y, Sun, T, Jiang, Z, Miao, Z, Shuai, M, et al. The interplay between host genetics and the gut microbiome reveals common and distinct microbiome features for complex human diseases. Microbiome. (2020) 8:145. doi: 10.1186/s40168-020-00923-9
187. El-Salhy, M, Hatlebakk, JG, Gilja, OH, et al. Efficacy of faecal microbiota transplantation for patients with irritable bowel syndrome in a randomised, double-blind, placebo-controlled study. Gut. (2020) 69:859–67. doi: 10.1136/gutjnl-2019-319630
188. Mandarino, FV, Sinagra, E, Raimondo, D, and Danese, S. The role of microbiota in upper and lower gastrointestinal functional disorders. Microorganisms. (2023) 11:980. doi: 10.3390/microorganisms11040980
189. Chang, CJ, Lin, TL, Tsai, YL, Wu, TR, Lai, WF, Lu, CC, et al. Next generation probiotics in disease amelioration. J Food Drug Anal. (2019) 27:615–22. doi: 10.1016/j.jfda.2018.12.011
190. Gibson, GR, Hutkins, R, Sanders, ME, Prescott, SL, Reimer, RA, Salminen, SJ, et al. Expert consensus document: the international scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. (2017) 14:491–502. doi: 10.1038/nrgastro.2017.75
191. Swanson, KS, Gibson, GR, Hutkins, R, Reimer, RA, Reid, G, Verbeke, K, et al. The international scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat Rev Gastroenterol Hepatol. (2020) 17:687–701. doi: 10.1038/s41575-020-0344-2
192. Marco, ML, Sanders, ME, Gänzle, M, Arrieta, MC, Cotter, PD, De Vuyst, L, et al. The international scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on fermented foods. Nat Rev Gastroenterol Hepatol. (2021) 18:196–208. doi: 10.1038/s41575-020-00390-5
193. Beam, A, Clinger, E, and Hao, L. Effect of Diet and dietary components on the composition of the gut microbiota. Nutrients. (2021) 13:2795. doi: 10.3390/nu13082795
194. Wang, Y, Do, T, Marshall, LJ, and Boesch, C. Effect of two-week red beetroot juice consumption on modulation of gut microbiota in healthy human volunteers - a pilot study. Food Chem. (2023) 406:134989. doi: 10.1016/j.foodchem.2022.134989
195. Nagpal, R, Shively, CA, Register, TC, Craft, S, and Yadav, H. Gut microbiome-Mediterranean diet interactions in improving host health. F1000Res. (2019) 8:699. doi: 10.12688/f1000research.18992.1
196. Mazza, E, Ferro, Y, Pujia, R, Mare, R, Maurotti, S, Montalcini, T, et al. Mediterranean Diet in healthy aging. J Nutr Health Aging. (2021) 25:1076–83. doi: 10.1007/s12603-021-1675-6
197. Davis, JJ, Fournakis, N, and Ellison, J. Ketogenic Diet for the treatment and prevention of dementia: a review. J Geriatr Psychiatry Neurol. (2021) 34:3–10. doi: 10.1177/0891988720901785
198. Homayouni Rad, A, Aghebati Maleki, L, Samadi Kafil, H, and Abbasi, A. Postbiotics: a novel strategy in food allergy treatment. Crit Rev Food Sci Nutr. (2021) 61:492–9. doi: 10.1080/10408398.2020.1738333
199. Kocsis, T, Molnár, B, Németh, D, Hegyi, P, Szakács, Z, Bálint, A, et al. Probiotics have beneficial metabolic effects in patients with type 2 diabetes mellitus: a meta-analysis of randomized clinical trials. Sci Rep. (2020) 10:11787. doi: 10.1038/s41598-020-68440-1
200. Frankenfeld, CL, Sikaroodi, M, Lamb, E, Shoemaker, S, and Gillevet, PM. High-intensity sweetener consumption and gut microbiome content and predicted gene function in a cross-sectional study of adults in the United States. Ann Epidemiol. (2015) 25:736–742.e4. doi: 10.1016/j.annepidem.2015.06.083
201. Bock, PM, Telo, GH, and Rea, R. The effect of probiotics, prebiotics or synbiotics on metabolic outcomes in individuals with diabetes: a systematic review and meta-analysis. Diabetologia. (2021) 64:26–41. doi: 10.1007/s00125-020-05295-1
202. Tonucci, LB, dos Santos, O, Maria, K, Licursi de Oliveira, L, Rocha Ribeiro, SM, and Duarte Martino, HS. Clinical application of probiotics in type 2 diabetes mellitus: a randomized, double-blind, placebo-controlled study. Clin Nutr. (2015) 36:85–92. doi: 10.1016/j.clnu.2015.11.011
203. Di Marzio, L, Cinque, B, Cupelli, F, De Simone, C, Cifone, MG, and Giuliani, M. Increase of skin-ceramide levels in aged subjects following a short-term topical application of bacterial sphingomyelinase from Streptococcus thermophilus. Int J Immunopathol Pharmacol. (2008) 21:137–43. doi: 10.1177/039463200802100115
204. Sadrifar, S, Abbasi-Dokht, T, Forouzandeh, S, Malek, F, Yousefi, B, Salek Farrokhi, A, et al. Immunomodulatory effects of probiotic supplementation in patients with asthma: a randomized, double-blind, placebo-controlled trial. Allergy Asthma Clin Immunol. (2023) 19:1. doi: 10.1186/s13223-022-00753-4
205. Akkasheh, G, Kashani-Poor, Z, Tajabadi-Ebrahimi, M, Jafari, P, Akbari, H, Taghizadeh, M, et al. Clinical and metabolic response to probiotic administration in patients with major depressive disorder: a randomized, double-blind, placebo-controlled trial. Nutrition. (2016) 32:315–20. doi: 10.1016/j.nut.2015.09.003
206. Rao, AV, Bested, AC, Beaulne, TM, Katzman, MA, Iorio, C, Berardi, JM, et al. A randomized, double-blind, placebo-controlled pilot study of a probiotic in emotional symptoms of chronic fatigue syndrome. Gut Pathog. (2009) 1:6. doi: 10.1186/1757-4749-1-6
207. Dickerson, F, Adamos, M, Katsafanas, E, Khushalani, S, Origoni, A, Savage, C, et al. Adjunctive probiotic microorganisms to prevent rehospitalization in patients with acute mania: a randomized controlled trial. Bipolar Disord. (2018) 20:614–21. doi: 10.1111/bdi.12652
208. Chi, L, Bian, X, Gao, B, Tu, P, Lai, Y, Ru, H, et al. Effects of the artificial sweetener neotame on the gut microbiome and fecal metabolites in mice. Molecules. (2018) 23:367. doi: 10.3390/molecules23020367
209. Reimer, RA, Grover, GJ, Koetzner, L, Gahler, RJ, Lyon, MR, and Wood, S. Combining sitagliptin/metformin with a functional fiber delays diabetes progression in zucker rats. J Endocrinol. (2014) 220:361–73. doi: 10.1530/JOE-13-0484
210. Leeuwendaal, NK, Stanton, C, O'Toole, PW, and Beresford, TP. Fermented foods, health and the gut Microbiome. Nutrients. (2022) 14:1527. doi: 10.3390/nu14071527
211. Dimidi, E, Cox, SR, Rossi, M, and Whelan, K. Fermented foods: definitions and characteristics, impact on the gut microbiota and effects on gastrointestinal health and disease. Nutrients. (2019) 11:1806. doi: 10.3390/nu11081806
212. Leite, Ana Karoline Ferreira, Fonteles, Thatyane Vidal, Filho, Elenilson Godoy Alves, and Andrea, Francisca da Silva Oliveira, Sueli Rodrigues. Impact of orange juice containing potentially prebiotic ingredients on human gut microbiota composition and its metabolites, Food Chem (2023) 405:134706. doi: 10.1016/j.foodchem.2022.134706
213. Corrêa, TAF, Tobaruela, EC, Capetini, VC, Quintanilha, BJ, Cortez, RV, Taddei, CR, et al. Blood orange juice intake changes specific bacteria of gut microbiota associated with cardiometabolic biomarkers. Front Microbiol. (2023) 14:1199383. doi: 10.3389/fmicb.2023.1199383
214. Wang, K, Zhao, Y, Xu, L, Liao, X, and Xu, Z. Health outcomes of 100% orange juice and orange flavored beverage: a comparative analysis of gut microbiota and metabolomics in rats. Curr Res Food Sci. (2023) 6:100454. doi: 10.1016/j.crfs.2023.100454
215. Katsirma, Z, Dimidi, E, Rodriguez-Mateos, A, and Whelan, K. Fruits and their impact on the gut microbiota, gut motility and constipation. Food Funct. (2021) 12:8850–66. doi: 10.1039/d1fo01125a.34505614
216. Huo, J, Wu, L, Lv, J, Cao, H, and Gao, Q. Effect of fruit intake on functional constipation: a systematic review and meta-analysis of randomized and crossover studies. Front Nutr. (2022) 9:1018502. doi: 10.3389/fnut.2022.1018502
217. Makki, K, Deehan, EC, Walter, J, and Bäckhed, F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. (2018) 23:705–15. doi: 10.1016/j.chom.2018.05.012
218. Tomova, A, Bukovsky, I, Rembert, E, Yonas, W, Alwarith, J, Barnard, ND, et al. The effects of vegetarian and vegan diets on gut microbiota. Front Nutr. (2019) 6:47. doi: 10.3389/fnut.2019.00047
219. Rees, K, Takeda, A, Martin, N, Ellis, L, Wijesekara, D, Vepa, A, et al. Mediterranean-style diet for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst Rev. (2019) 2019:CD009825. doi: 10.1002/14651858.CD009825.pub3
220. Estruch, R, Ros, E, Salas-Salvadó, J, Covas, MI, Corella, D, Arós, F, et al. Primary prevention of cardiovascular disease with a Mediterranean Diet supplemented with extra-virgin olive oil or nuts. N Engl J Med. (2018) 378:e34. doi: 10.1056/NEJMoa1800389
221. Shively, CA, Register, TC, Appt, SE, Clarkson, TB, Uberseder, B, Clear, KYJ, et al. (2018). Consumption of mediterranean versus western diet leads to distinct mammary gland microbiome populations. Cell Rep. (2018) 25:47–56.e3. doi: 10.1016/j.celrep.2018.08.078
222. Garcia-Mantrana, I, Selma-Royo, M, Alcantara, C, and Collado, MC. Shifts on gut microbiota associated to Mediterranean Diet adherence and specific dietary intakes on general adult population. Front Microbiol. (2018) 9:890. doi: 10.3389/fmicb.2018.00890
223. Paoli, A, Mancin, L, Bianco, A, Thomas, E, Mota, JF, and Piccini, F. Ketogenic Diet and microbiota: friends or enemies? Genes. (2019) 10:534. doi: 10.3390/genes10070534
224. Olson, CA, Vuong, HE, Yano, JM, Liang, QY, Nusbaum, DJ, and Hsiao, EY. The gut microbiota mediates the anti-seizure effects of the ketogenic Diet. Cell. (2018) 173:1728–1741.e13. doi: 10.1016/j.cell.2018.04.027
225. Dowis, K, and Banga, S. The potential health benefits of the ketogenic Diet: a narrative review. Nutrients. (2021) 13:1654. doi: 10.3390/nu13051654
226. Włodarek, D. Role of ketogenic diets in neurodegenerative diseases (Alzheimer's disease and Parkinson's disease). Nutrients. (2019) 11:169. doi: 10.3390/nu11010169
227. Wang, Z, Roberts, AB, Buffa, JA, Levison, BS, Zhu, W, Org, E, et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. (2015) 163:1585–95. doi: 10.1016/j.cell.2015.11.055
228. Suez, J, Zmora, N, and Elinav, E. Probiotics in the next-generation sequencing era. Gut Microbes. (2020) 11:77–93. doi: 10.1080/19490976.2019.1586039
229. Durazzo, A, Nazhand, A, Lucarini, M, Atanasov, AG, Souto, EB, Novellino, E, et al. An updated overview on nanonutraceuticals: focus on nanoprebiotics and nanoprobiotics. Int J Mol Sci. (2020) 21:2285. doi: 10.3390/ijms21072285
230. Koutnikova, H, Genser, B, Monteiro-Sepulveda, M, Faurie, J-M, Rizkalla, S, Schrezenmeir, J, et al. Impact of bacterial probiotics on obesity, diabetes and nonalcoholic fatty liver disease related variables: a systematic review and metaanalysis of randomised controlled trials. BMJ Open. (2019) 9:e017995. doi: 10.1136/bmjopen-2017-017995
231. Tsiouris, CG, Kelesi, M, Vasilopoulos, G, Kalemikerakis, I, and Papageorgiou, EG. The efficacy of probiotics as pharmacological treatment of cutaneous wounds: meta-analysis of animal studies. Eur J Pharm Sci. (2017) 104:230–9. doi: 10.1016/j.ejps.2017.04.002
232. Campos, LF, Tagliari, E, Casagrande, TAC, Noronha, L, Campos, ACL, and Matias, JEF. Effects of probiotics supplementation on skin wound healing in diabetic rats. ABCD Arq Bras Cir Dig. (2020) 33:e1498. doi: 10.1590/0102-672020190001e1498
233. Zmora, N, Zilberman-Schapira, G, Suez, J, Mor, U, Dori-Bachash, M, Bashiardes, S, et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and Microbiome features. Cell. (2018) 174:1388–1405.e21. doi: 10.1016/j.cell.2018.08.041
234. Hill, C, Guarner, F, Reid, G, Gibson, GR, Merenstein, DJ, Pot, B, et al. Expert consensus document. The international scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. (2014) 11:506–14. doi: 10.1038/nrgastro.2014.66
235. Didari, T, Solki, S, Mozaffari, S, Nikfar, S, and Abdollahi, M. A systematic review of the safety of probiotics. Expert Opin Drug Saf. (2014) 13:227–39. doi: 10.1517/14740338.2014.872627
236. Zhao, L, Zhang, F, Ding, X, Wu, G, Lam, YY, Wang, X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. (2018) 359:1151–6. doi: 10.1126/science.aao5774
237. Gomez Quintero, DF, Kok, CR, and Hutkins, R. The future of synbiotics: rational formulation and design. Front Microbiol. (2022) 13:919725. doi: 10.3389/fmicb.2022.919725
238. Huybrechts, I, Rauber, F, Nicolas, G, Casagrande, C, Kliemann, N, Wedekind, R, et al. Characterization of the degree of food processing in the European prospective investigation into cancer and nutrition: application of the Nova classification and validation using selected biomarkers of food processing. Front Nutr. (2022) 9:1035580. doi: 10.3389/fnut.2023.1207555
239. Monteiro, CA, Cannon, G, Moubarac, JC, Levy, RB, Louzada, MLC, and Jaime, PC. The UN decade of nutrition, the NOVA food classification and the trouble with ultra-processing. Public Health Nutr. (2018) 21:5–17. doi: 10.1017/S1368980017000234
240. Atzeni, A, Martínez, MÁ, Babio, N, Konstanti, P, Tinahones, FJ, Vioque, J, et al. Association between ultra-processed food consumption and gut microbiota in senior subjects with overweight/obesity and metabolic syndrome. Front Nutr. (2022) 9:976547. doi: 10.3389/fnut.2022.976547
241. Martínez Leo, EE, and Segura Campos, MR. Effect of ultra-processed diet on gut microbiota and thus its role in neurodegenerative diseases. Nutrition. (2020) 71:110609. doi: 10.1016/j.nut.2019.110609
242. Srour, B, Kordahi, MC, Bonazzi, E, Deschasaux-Tanguy, M, Touvier, M, and Chassaing, B. Ultra-processed foods and human health: from epidemiological evidence to mechanistic insights. Lancet Gastroenterol Hepatol. (2022) 7:1128–40. doi: 10.1016/S2468-1253(22)00169-8
243. Hrncirova, L, Machova, V, Trckova, E, Krejsek, J, and Hrncir, T. Food preservatives induce Proteobacteria Dysbiosis in human-microbiota associated Nod2-deficient mice. Microorganisms. (2019) 7:383. doi: 10.3390/microorganisms7100383
244. Li, P, Li, M, Wu, T, Song, Y, Li, Y, Huang, X, et al. Systematic evaluation of antimicrobial food preservatives on glucose metabolism and gut microbiota in healthy mice. NPJ Sci Food. (2022) 6:42. doi: 10.1038/s41538-022-00158-y
245. Laudisi, F, Stolfi, C, and Monteleone, G. Impact of food additives on gut homeostasis. Nutrients. (2019) 11:2334. doi: 10.3390/nu11102334
246. Song, Z, Song, R, Liu, Y, Wu, Z, and Zhang, X. Effects of ultra-processed foods on the microbiota-gut-brain axis: the bread-and-butter issue. Food Res Int. (2023) 167:112730. doi: 10.1016/j.foodres.2023.112730
247. Zhou, X, Qiao, K, Wu, H, and Zhang, Y. The impact of food additives on the abundance and composition of gut microbiota. Molecules. (2023) 28:631. doi: 10.3390/molecules28020631
248. Chassaing, B, Compher, C, Bonhomme, B, Liu, Q, Tian, Y, Walters, W, et al. Randomized controlled-feeding study of dietary emulsifier Carboxymethylcellulose reveals detrimental impacts on the gut microbiota and metabolome. Gastroenterology. (2022) 162:743–56. doi: 10.1053/j.gastro.2021.11.006
249. Gerasimidis, K, Bryden, K, Chen, X, Papachristou, E, Verney, A, Roig, M, et al. The impact of food additives, artificial sweeteners and domestic hygiene products on the human gut microbiome and its fibre fermentation capacity. Eur J Nutr. (2020) 59:3213–3230. doi: 10.1007/s00394-019-02161-8
250. Savin, Z, Kivity, S, Yonath, H, and Yehuda, S. Smoking and the intestinal microbiome. Arch Microbiol. (2018) 200:677–84. doi: 10.1007/s00203-018-1506-2
251. Gui, X, Yang, Z, and Li, MD. Effect of cigarette smoke on gut microbiota: state of knowledge. Front Physiol. (2021) 12:673341. doi: 10.3389/fphys.2021.673341
252. Biedermann, L, Zeitz, J, Mwinyi, J, Sutter-Minder, E, Rehman, A, Ott, SJ, et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One. (2013) 8:e592605. doi: 10.1371/journal.pone.0059260
253. Ames, NJ, Barb, JJ, Schuebel, K, Mudra, S, Meeks, BK, Tuason, RTS, et al. Longitudinal gut microbiome changes in alcohol use disorder are influenced by abstinence and drinking quantity. Gut Microbes. (2020) 11:1608–31. doi: 10.1080/19490976.2020.1758010
254. Leclercq, S, Matamoros, S, Cani, PD, Neyrinck, AM, Jamar, F, Stärkel, P, et al. Intestinal permeability, gut-bacterial Dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci USA. (2014) 111:E4485–93. doi: 10.1073/pnas.1415174111
255. Mutlu, EA, Gillevet, PM, Rangwala, H, Sikaroodi, M, Naqvi, A, Engen, PA, et al. Colonic microbiome is altered in alcoholism am. Am J Physiol Gastrointest Liver Physiol. (2012) 302:G966–78. doi: 10.1152/ajpgi.00380.2011
256. Qamar, N, Castano, D, Patt, C, Chu, T, Cottrell, J, and Chang, SL. Meta-analysis of alcohol induced gut dysbiosis and the resulting behavioral impact. Behav Brain Res. (2019) 376:112196. doi: 10.1016/j.bbr.2019.112196
257. Smith, RP, Easson, C, Lyle, SM, Kapoor, R, Donnelly, CP, Davidson, EJ, et al. Gut microbiome diversity is associated with sleep physiology in humans. PLoS One. (2019) 14:e0222394. doi: 10.1371/journal.pone.0222394
258. Bowers, SJ, Summa, KC, Thompson, RS, González, A, Vargas, F, Olker, C, et al. A prebiotic Diet alters the fecal microbiome and improves sleep in response to sleep disruption in rats. Front Neurosci. (2022) 16:889211. doi: 10.3389/fnins.2022.889211
259. Irwin, C, McCartney, D, Desbrow, B, and Khalesi, S. Effects of probiotics and paraprobiotics on subjective and objective sleep metrics: a systematic review and meta-analysis. Eur J Clin Nutr. (2020) 74:1536–49. doi: 10.1038/s41430-020-0656-x
260. Clauss, M, Gérard, P, Mosca, A, and Leclerc, M. Interplay between exercise and gut microbiome in the context of human health and performance. Front Nutr. (2021) 8:637010. doi: 10.3389/fnut.2021.637010
261. Gubert, C, Kong, G, Renoir, T, and Hannan, AJ. Exercise, diet and stress as modulators of gut microbiota: implications for neurodegenerative diseases. Neurobiol Dis. (2020) 134:104621. doi: 10.1016/j.nbd.2019.104621
262. Madison, A, and Kiecolt-Glaser, JK. Stress, depression, diet, and the gut microbiota: human-bacteria interactions at the core of psychoneuroimmunology and nutrition. Curr Opin Behav Sci. (2019) 28:105–10. doi: 10.1016/j.cobeha.2019.01.011
263. Cantu-Jungles, TM, Zhang, X, Kazem, AE, Iacomini, M, Hamaker, BR, and Cordeiro, LMC. Microwave treatment enhances human gut microbiota fermentability of isolated insoluble dietary fibers. Food Res Int. (2021) 143:110293. doi: 10.1016/j.foodres.2021.110293
264. Luo, S, Hou, Y, Xie, L, Zhang, H, Liu, C, and Chen, T. Effects of microwave on the potential microbiota modulating effects of agro-industrial by-product fibers among different individuals. LWT. (2023) 178:114621. doi: 10.1016/j.lwt.2023.114621
265. Lamothe, LM, Cantu-Jungles, TM, Chen, T, Green, S, Naqib, A, Srichuwong, S, et al. Boosting the value of insoluble dietary fiber to increase gut fermentability through food processing. Food Funct. (2021) 12:10658–66. doi: 10.1039/D1FO02146J
266. Sorbara, MT, and Pamer, EG. Microbiome-based therapeutics. Nat Rev Microbiol. (2022) 20:365–80. doi: 10.1038/s41579-021-00667-9
267. Normington, C, Chilton, CH, and Buckley, AM. Clostridiodes difficile infections. New treatments and future perspectives. Curr Opin Gastroenterol. (2024) 40:7–13. doi: 10.1097/MOG.0000000000000989
268. Liu, S, Guo, R, Liu, F, Yuan, Q, Yu, Y, and Ren, F. Gut microbiota regulates depression-like behavior in rats through the neuroendocrine-immune-mitochondrial pathway. Neuropsychiatr Dis Treat. (2020) 16:859–69. doi: 10.2147/NDT.S243551
269. Hazan, S. Rapid improvement in Alzheimer's disease symptoms following fecal microbiota transplantation: a case report. J Int Med Res. (2020) 48:300060520925930. doi: 10.1177/0300060520925930
270. Park, SH, Lee, JH, Shin, J, Kim, JS, Cha, B, Lee, S, et al. Cognitive function improvement after fecal microbiota transplantation in Alzheimer's dementia patient: a case report. Curr Med Res Opin. (2021) 37:1739–44. doi: 10.1080/03007995.2021.1957807
271. Ianiro, G, Eusebi, LH, Black, CJ, Gasbarrini, A, Cammarota, G, and Ford, AC. Systematic review with meta-analysis: efficacy of faecal microbiota transplantation for the treatment of irritable bowel syndrome. Aliment Pharmacol Ther. (2019) 50:240–8. doi: 10.1111/apt.15330
272. Gomaa, EZ. Human gut microbiota/microbiome in health and diseases: a review. Antonie Leeuwenhoek. (2020) 113:2019–40. doi: 10.1007/s10482-020-01474-7
273. Yang, Q, Wang, B, Zheng, Q, Li, H, Meng, X, and Zhou, F. Zhang L a review of gut microbiota-derived metabolites in tumor progression and cancer therapy. Adv Sci. (2023) 10:2207366. doi: 10.1002/advs.202207366
274. Son, MY, and Cho, HS. Anticancer effects of gut microbiota-derived short-chain fatty acids in cancers. J Microbiol Biotechnol. (2023) 33:849–56. doi: 10.4014/jmb.2301.01031
275. Golpour, F, Abbasi-Alaei, M, Babaei, F, Mirzababaei, M, Parvardeh, S, Mohammadi, G, et al. Short chain fatty acids, a possible treatment option for autoimmune diseases. Biomed Pharmacother. (2023) 163:114763. doi: 10.1016/j.biopha.2023.114763
276. Machado, MG, Sencio, V, and Trottein, F. Short-chain fatty acids as a potential treatment for infections: a closer look at the lungs. Infect Immun. (2021) 89:e0018821. doi: 10.1128/IAI.00188-21
277. Tang, R, and Li, L. Modulation of short-chain fatty acids as potential therapy method for type 2 diabetes mellitus. Can J Infect Dis Med Microbiol. (2021) 2021:6632266. doi: 10.1155/2021/9756586
278. Magliocca, G, Mone, P, Di Iorio, BR, Heidland, A, and Marzocco, S. Short-chain fatty acids in chronic kidney disease: focus on inflammation and oxidative stress regulation. Int J Mol Sci. (2022) 23:5354. doi: 10.3390/ijms23105354
279. McDonald, JAK, Mullish, BH, Pechlivanis, A, Liu, Z, Brignardello, J, Kao, D, et al. Inhibiting growth of Clostridioides difficile by restoring Valerate, produced by the intestinal microbiota. Gastroenterology. (2018) 155:1495–1507.e15. doi: 10.1053/j.gastro.2018.07.014
280. Peng, H, Ouyang, L, Li, D, Li, Z, Yuan, L, Fan, L, et al. Short-chain fatty acids in patients with schizophrenia and ultra-high risk population. Front Psych. (2022) 13:977538. doi: 10.3389/fpsyt.2022.977538
281. Xiao, X, Hu, X, Yao, J, Cao, W, Zou, Z, Wang, L, et al. The role of short-chain fatty acids in inflammatory skin diseases. Front Microbiol. (2023) 13:1083432. doi: 10.3389/fmicb.2022.1083432
282. Kim, S, Park, S, Choi, TG, and Kim, SS. Role of short chain fatty acids in epilepsy and potential benefits of probiotics and prebiotics: targeting "health" of epileptic patients. Nutrients. (2022) 14:2982. doi: 10.3390/nu14142982
283. Vinderola, G, Sanders, ME, and Salminen, S. The concept of postbiotics. Food Secur. (2022) 11:1077. doi: 10.3390/foods11081077
284. Kesavelu, D, and Jog, P. Current understanding of antibiotic-associated dysbiosis and approaches for its management. Ther Adv Infect Dis. (2023) 10:20499361231154443. doi: 10.1177/20499361231154443
285. Health and Economic Costs of Chronic Diseases. Available at: https://www.cdc.gov/chronicdisease/about/costs/index.htm (Accessed December 26, 2023).
286. Matthewman, C, Narin, A, Huston, H, and Hopkins, CE. Systems to model the personalized aspects of microbiome health and gut dysbiosis. Mol Asp Med. (2023) 91:101115. doi: 10.1016/j.mam.2022.101115
Keywords: microbiota, microbiome, dysbiosis, gut biotics, virtual organ
Citation: Alagiakrishnan K, Morgadinho J and Halverson T (2024) Approach to the diagnosis and management of dysbiosis. Front. Nutr. 11:1330903. doi: 10.3389/fnut.2024.1330903
Edited by:
George Grant, University of Aberdeen, United KingdomReviewed by:
Kalyani Barve, SVKM’s Narsee Monjee Institute of Management Studies, IndiaLaura Abisai Pazos-Rojas, Escuela de Ingeniería y Ciencias, Tecnológico de Monterrey, Mexico
Copyright © 2024 Alagiakrishnan, Morgadinho and Halverson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kannayiram Alagiakrishnan, a2E5QHVhbGJlcnRhLmNh
†ORCID: Kannayiram Alagiakrishnan orcid.org/0000-0003-1444-2451
 Kannayiram Alagiakrishnan
Kannayiram Alagiakrishnan Joao Morgadinho2
Joao Morgadinho2 Tyler Halverson
Tyler Halverson