
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CLINICAL TRIAL article
Front. Nutr., 23 January 2024
Sec. Nutrition and Microbes
Volume 11 - 2024 | https://doi.org/10.3389/fnut.2024.1307734
This article is part of the Research TopicProbiotics and Nutrient Bioavailability: Enhancing Gut Health to Combat MalnutritionView all 4 articles
The current randomized, double-blind, crossover clinical trial was conducted to evaluate changes in the amino acid absorption and gut microbiota on consumption of pea protein supplemented with an enzymes-probiotics blend (Pepzyme Pro). A total of 15 healthy subjects were instructed to take test (pea protein + Pepzyme Pro) or placebo (pea protein + maltodextrin) for 15 days with a 30-day washout period. Blood samples were analyzed for plasma-free amino acids, insulin, and C-reactive protein (CRP). Additionally, nitrogen levels in urine and feces, along with the composition of gut microbiota, were evaluated. On day 15, the test arm showed a tendency to increase the rate of absorption and total absorption (AUC) of amino acids compared with the placebo arm, though the increase was statistically insignificant. In addition, 15-day test supplementation showed a tendency to reduce Tmax of all the amino acids (statistically insignificant except alanine, p = 0.021 and glycine, p = 0.023) in comparison with the placebo supplementation. There were no changes in urine and fecal nitrogen levels as well as serum CRP levels in the test and placebo arm. The increase in serum insulin level after 4 h was statistically significant in both arms, whereas the insulin level of the placebo and test arm at 4 h was not statistically different. Supplementation showed changes with respect to Archaea and few uncharacterized species but did not show statistically significant variations in microbiome profile at the higher taxonomic levels. A study with large sample size and detailed gut microbiome analysis is warranted to confirm the results statistically as well as to characterize altered species. However, the current study could provide an inkling of a positive alteration in protein digestibility, amino acid absorption, and gut microbiome with regular consumption of protein and enzymes-probiotics blend.
Clinical Trial Registration:clinicaltrials.gov/; identifier [CTRI/2021/10/037072].
The increasing clinical evidence highlights the potential health benefits and significance of proteins. It has not only increased awareness but also heightened the popularity of protein supplements among athletes and recreationally active adults. Currently, numerous protein products are marketed as sports nutrients, muscle recovery supplements, medical and dietary formulations, weight management products, geriatric supplements, and maternal/infant dietary supplements (1–3). The sources of these dietary and therapeutic proteins could be from either animal or plant origins. Proteins from animal origin are considered complete proteins and are in high demand, but they come with some ethical and regulatory concerns. On the other hand, proteins from plant sources are abundantly available as good-quality proteins and overcome the ethical and regulatory issues that are associated with animal-sourced proteins (4, 5).
Pea protein is generally considered a high-quality protein as it satisfies FAO/WHO/UNU recommendations in terms of the availability of amino acids in a balanced ratio and the presence of all essential amino acids (EAA) except methionine (6). The digestible indispensable amino acid score (DIAAS) for the pea protein is nearly one, which makes it suitable to fulfill the amino acid requirements of the body (7). Furthermore, it has comparatively lower allergenic responses, negligible health controversies, and natural availability of biologically active peptides over the other plant proteins (7–9). Regular consumption of pea protein could offer a series of benefits including muscle growth, weight management, diabetes management, and healthy heart functions (10).
The nutritive benefits of the proteins mainly depend on the composition and bioavailability of the amino acids (11). Based on this fact, protein with high digestibility and bioavailability is always a preferred choice among athletes and recreationally active adults. Consumption of protein hydrolysate is another simplest and most widely used strategy to get the benefits of improved digestibility and bioavailability of the protein. On the other hand, these approaches come with the challenges such as limited sources of protein with high digestibility and possibilities of the presence of endogenous protease inhibitory peptides in the hydrolysate (12). Combining a protein of choice with a blend of enzymes and probiotics can be an alternate approach to enhance the protein digestibility and amino acid bioavailability. Earlier studies have shown that the co-ingestion of an exogenous enzyme blend along with plant protein increases protein digestibility (13) and reduces the difference between animal and plant protein in terms of nutritive benefits (14).
Previously, several clinical studies have demonstrated a positive impact of enzyme supplementation on protein digestion and absorption (15–17). Exogenous digestive enzymes can degrade dietary proteins simultaneously or sequentially with endogenous proteases to improve digestion and increase the availability of amino acids/smaller peptides for absorption (13). Furthermore, the effect of probiotics on the protein digestibility is of current interest. Jager et al. (18) have shown that the oral supplementation of probiotics along with the protein improved the postprandial changes in blood amino acids and can be a suitable approach to overcome compositional shortcomings of the plant protein. The positive influence of probiotics on the digestion of proteins is mainly associated with their ability to ameliorate the gut microbiota (19). This resulted in an improvement in protein digestion and its utilization, as well as a reduction in the harmful protein fermentation and associated health risks (20). Furthermore, Garcia et al. (21) hydrolyzed pea protein with proteases followed by fermentation with lactic acid bacteria and found an increase in the degree of hydrolysis and a reduction in the immunogenicity of pea protein.
Pepzyme Pro was used as a model blend of enzymes and probiotics in the current interventional study. The study was initiated with the hypothesis that the addition of blend of enzymes and probiotics in pea protein would increase the protein digestibility and bioavailability. Additionally, it would possibly contribute in positive alteration in the gut microbiota. Based on the stated hypothesis, this study aimed to evaluate the effect of supplementation of an enzymes-probiotics blend with pea protein on the digestion of proteins and the bioavailability of amino acids. Furthermore, the impact of supplementation on the gut microbiota was also under the scope of this study. Additionally, the safety assessment was done by measuring fecal and urine nitrogen content, as well as insulin and C-reactive protein (CRP) level.
This prospective, interventional, double-blinded, randomized, placebo-controlled, and crossover study was conducted in conformity with ICH-GCP (E6 R2) guidelines, the Helsinki Declaration, and the local regulatory requirements (Indian GCP, Indian Council of Medical Research, and New Drugs and Clinical Trials Rules-2019). There were no further changes or amendments made after the approval of the protocol. The trial included 15 healthy adult subjects with an age range of 18–35 years, normal body weight (body mass index (BMI) of 19–24.99 kg/m2), and the willingness to provide written informed consent and comply with study instructions for its duration. Subjects with a known history of (i) smoking or tobacco consumption, (ii) clinically significant physiological, neurological, or psychiatric disease, (iii) organ transplantation or surgery in the past 6 months, (iv) known hypersensitivity or idiosyncratic reaction, intolerance to any ingredients in the formulation or any related product, as well as severe hypersensitivity reactions (like angioedema) to any drugs or food products, and/or (v) difficulty in donating blood were excluded from the study. Participants who met the necessary inclusion criteria were further encouraged not to change their current physical activity levels and to refrain from exercise for 24 h before starting the clinical trial.
The enzymes-probiotics blend (Pepzyme Pro) was obtained as a gift sample from Specialty Enzymes and Probiotics, Chino, USA, and was used as an IP. It is a commercial formulation of proteolytic enzymes (acid proteases from Aspergillus niger) and probiotics (Bacillus coagulans LBSC DSM (17654), Bacillus clausii 088AE (MCC 0538), Bacillus subtilis PLSSC (ATCCSD 7280), Lactobacillus acidophilus 033AE, and Lactobacillus plantarum 022AE (MCC 0537). A sachet of pea protein (30 g) with the enzymes-probiotics blend (1% of the protein) and maltodextrin (1% of the protein) was used as a test supplement and a placebo supplement, respectively. Maltodextrin was used as a placebo to nullify the effect of excipient (maltodextrin) present in the Pepzyme Pro. The labeling and packaging of the test and placebo were the same, except for the coded batch numbers mentioned on the sachet. The participants, investigators, and entire study team were blinded for the treatment allocation until the end of the study.
The study schedule is provided in Table 1. The study comprised of initial screening, baseline testing, and two 15-day supplementation periods separated by a 30-day washout phase. Block randomization was done on online randomization tool1 using a pseudorandom number generator. All the subjects were instructed to open a sachet of supplement (test or placebo) daily in the morning and mix it in lukewarm water (300–500 mL). They were further instructed to drink supplement mixture daily in the morning on empty stomach. Occurrence of adverse events and concomitant medication were recorded throughout the trial period. Site visit was planned on days 1 and 15 of each supplementation period. Upon visit, each subject was instructed to consume their respective protein supplementation, and blood samples were withdrawn at 0, 0.5, 1, 2, 3, and 4 h for the analysis of plasma-free amino acids. On day 15 of both the supplementation period, the blood samples were withdrawn at 0 h and 4 h for measuring serum levels of insulin and CRP. Urine (24 h collection) and fecal samples were collected on days 1 and 15 on each supplementation period for the analysis of nitrogen content. In addition, collected fecal samples were also used for the gut microbiota analysis.
The concentrations of amino acids, insulin, and CRP in plasma were determined at Thyrocare Mumbai, India, using liquid chromatography tandem mass spectrometry method, electrochemiluminescence immunoassay, and immunoturbidimetry, respectively. Urine and fecal nitrogen content were analyzed by using Dumas combustion method. Leucine Rich Bio Pvt. Ltd., India, analyzed the gut microbiota of all the test samples using the shotgun microbiome sequencing method.
Stool samples were collected using Invitek Molecular Stool Collection Module [Cat. No. 1038111300, Berlin, Invitek Molecular GmbH]. All participants were instructed appropriately to use the kit for the sample collection. The samples once collected were shipped under room temperature to the processing unit for DNA extraction.
DNA from stool samples was extracted using QIAamp® Fast DNA Stool Mini (Cat No./ID: 51604, QIAGEN) following the manufacturer’s “Fast DNA Stool Mini Handbook” for fast purification of genomic DNA. Eluted DNA was collected in 1.5 mL DNA Lo-Bind microcentrifuge tubes, and the quantity and quality of DNA were assessed by Qubit 2.0 DNA HS Assay (Thermo Fisher, Massachusetts, USA) and NanoDrop® (Roche, USA) to meet the sequencing requirements.
Whole metagenome sequencing of all the samples was performed using long-read sequencing technology. Briefly, the DNA library was prepared with the Ligation sequencing kit (SQK-LSK109) (Oxford Nanopore Technologies (ONT), Oxford, UK), then loaded onto a R9.4.1 MinION flow cell (FLO-MIN106), and sequenced on the ONT MinION Mk1B device (MIN-101B). Basecalling and demultiplexing of sequence reads were performed with Guppy v4.2.2 and with assistance from MinKNOW GUI v4.1.22. Raw sequencing reads were stored in FastQ format for further computational analysis. The upstream analysis involved measurement of quality checks and quality improvement, including but not limited to host [human] sequence removal. This was followed by the alignment of quality-processed reads to a reference database of microbial genomes. The raw and % normalized abundances of all the microorganisms identified within these samples, were quantified, and later used for downstream analysis involving various statistical measures. Data filtering and data normalization steps were performed for the removal of low-quality or uninformative features from raw abundance data to improve downstream statistical analysis. Taxonomic composition of communities across samples and comparing groups were analyzed for direct quantitative comparison of abundances. Both the alpha and beta diversity analyses were performed using the phyloseq package (22, 23), and the results were plotted as box and whisker plots for alpha diversity and PCoA plot for beta diversity.
Differential abundance (DA) analysis was performed with five different DA tools, viz., univariate analysis (24), metagenomeSeq (25–27), EdgeR (v3.12) (28), DeSeq2 (29), and LEfSe (linear discriminant analysis effect size) (30). Once the DA analysis was performed using individual tools, we identified those microbial species that were called significantly differentially abundant in “consensus” by all five DA tools, ensuring the robustness of the DA characterization.
Primary endpoints were set to assess the efficacy of enzymes-probiotics blend in the digestion and absorption of pea protein. It includes the estimation of bioavailability of plasma amino acids within 4 h of consumption of protein in placebo arm and test arm, and pairwise comparison between both the arms. Secondary endpoints were set to evaluate the changes in nitrogen level in urine and feces, changes in insulin and CRP levels in serum, and changes in fecal gut microbiome. Additionally, an assessment of adverse events was done to evaluate the safety of the enzymes-probiotics blend after oral supplementation.
Data were examined using SAS software, version 9.1, by keeping a 5% significance level (confidence interval 95%) and maintaining a minimum statistical power of 80%. Primary and secondary endpoints were analyzed separately. The concentration vs. time curve (AUC) for each amino acid was calculated at all available time points using the linear trapezoidal rule. The difference between test arm and placebo arm was statistically analyzed using Student’s t-test. All the data were represented as mean ± standard error (SE). p ≤ 0.05 denotes statistical significance unless specified.
This randomized, double-blinded, crossover clinical study was initiated on 07 November 2021 and completed on 08 January 2022 (Figure 1) on 15 healthy subjects with average age of 21.40 ± 3.20 years. The other demographic details of the subjects, such as height, weight, and BMI, are presented in Table 2. All the patients completed the study.
The effect of supplementation of enzymes-probiotics blend along with the pea protein for 15 days on the rate of amino acid absorption was measured for 4 h post-consumption on day 1 and day 15. On day 1, the rate of amino acid absorption in test and placebo arm was similar (Figure 2A). However, on day 15, variation was observed in the rate of amino acid absorption in the test and placebo arm (Figure 2B). After test supplementation, the rate of absorption of arginine, glutamine, isoleucine, leucine, lysine, methionine, serine, threonine, tryptophan, and tyrosine was positively altered by 1.25, 1.68, 1.25, 1.27, 1.26, 1.39, 1.35, 1.57, 1.44, and 1.41 folds, respectively (statistically insignificant). The analysis of essential amino acids (EAA) and branched chain amino acids (BCAA) illustrated no difference in the test and placebo arm on day 1. However, the appreciable difference though not statistically significant was observed in the increase of EAA and BCAA on day 15 within 1 h post-consumption (Figure 3). On day 15, test arm showed tendency to increase the plasma concentration of histidine (4.19%), isoleucine (25.09%), leucine (27.09%), lysine (26.37%), methionine (40.92%), phenylalanine (7.12%), threonine (57.21%), tryptophan (43.87%), and valine (5.36%), 1 h post-consumption of protein, over the placebo arm. In addition, overall absorption rate of EAA and BCAA on day 15 was in increasing trend than that of on day 1 in both placebo and test arm, though statistically insignificant.
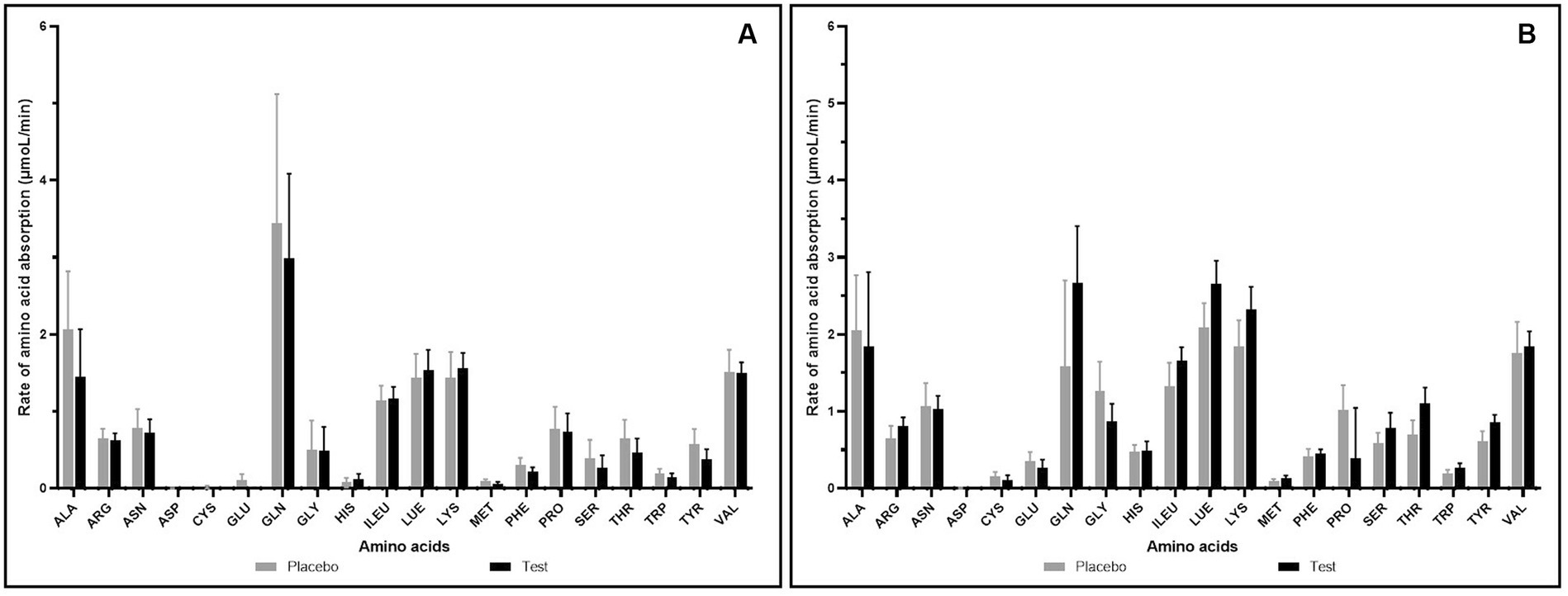
Figure 2. Rate of amino acid absorption (μmoL/min) after consumption of pea protein (placebo) Vs pea protein + enzymes-probiotics blend (test) on day 1 (A) and day 15 (B). Values represented as mean ± standard error. Data were statistically analyzed using the Student’s t-test, and p ≤ 0.05 was considered statistically significant.
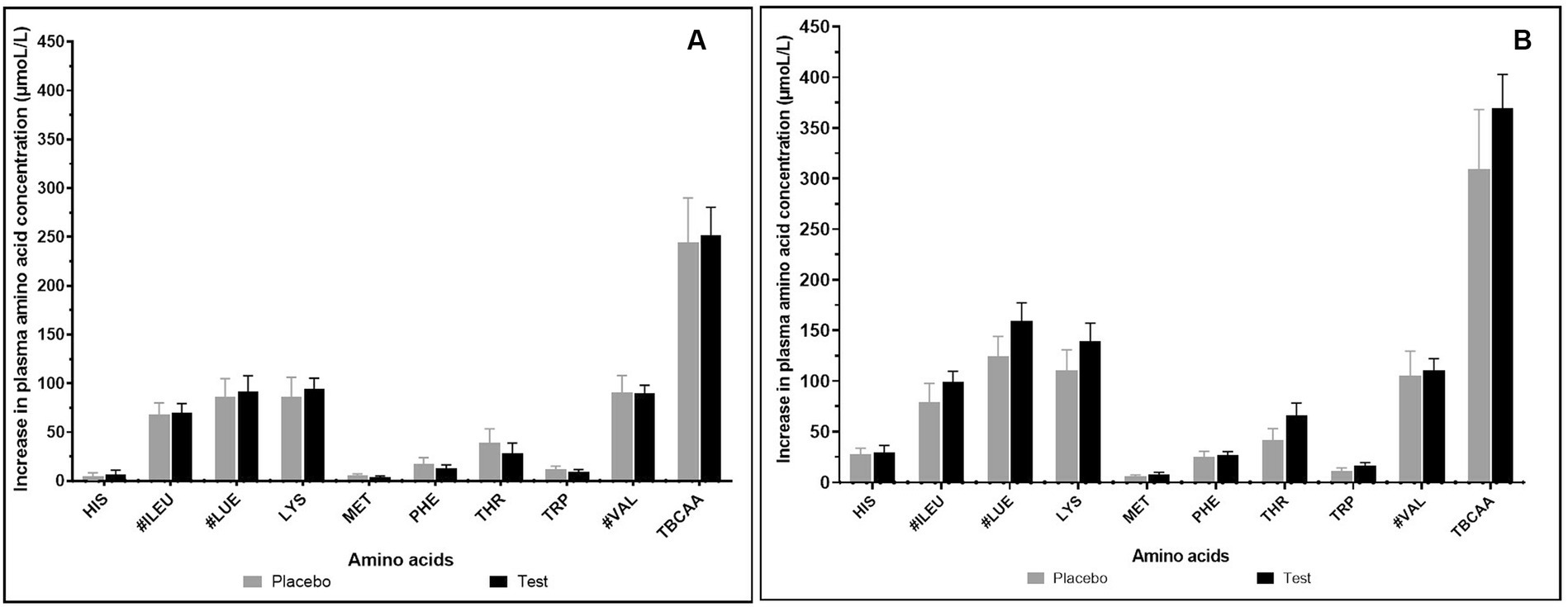
Figure 3. Increase in essential and branched chain amino acid concentration (μmol/L) after 1 h of consumption of pea protein (placebo) Vs pea protein + enzymes-probiotics blend (test) on day 1 (A) and day 15 (B).Values represented as mean ± standard error. TBCAA: Total branched chain amino acids. # represents individual branched chain amino acid. Data were statistically analyzed using the Student’s t-test, and p ≤ 0.05 was considered statistically significant.
The change in the absorption of free amino acids in test and placebo arm was analyzed by assessing the maximum plasma concentration (Cmax), the corresponding time (Tmax), and area under the curve (AUC). The AUC (concentration vs. time) was calculated for the plasma amino acids through the linear trapezoidal rule and using all available time points. Cmax was defined as the highest observed concentration of amino acid, and Tmax was the time when Cmax was reached. On day 1, there was no noticeable difference in the AUC and Cmax in the test and placebo arm, (Table 3A), whereas on day 15, test supplementation showed proclivity to increase the absorption of alanine, arginine, glutamine, glycine, isoleucine, leucine, lysine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, and valine as determined by the increase in the AUC and Cmax values (Table 3B). Furthermore, the Tmax of all the amino acids in test and placebo arm was studied on day 1 and day 15. The Tmax of tryptophan (4%) and tyrosine (11%) showed statistically insignificant decrease after consumption of test supplement on day 1 (Table 4A), whereas on day 15, test supplementation demonstrated the tendency to decrease Tmax of the all amino acids except histidine (Table 4B). In particular, alanine (40%, p = 0.021) and glycine (40%, p = 0.023) showed statistically significant decrease in Tmax after supplementation of the test product.
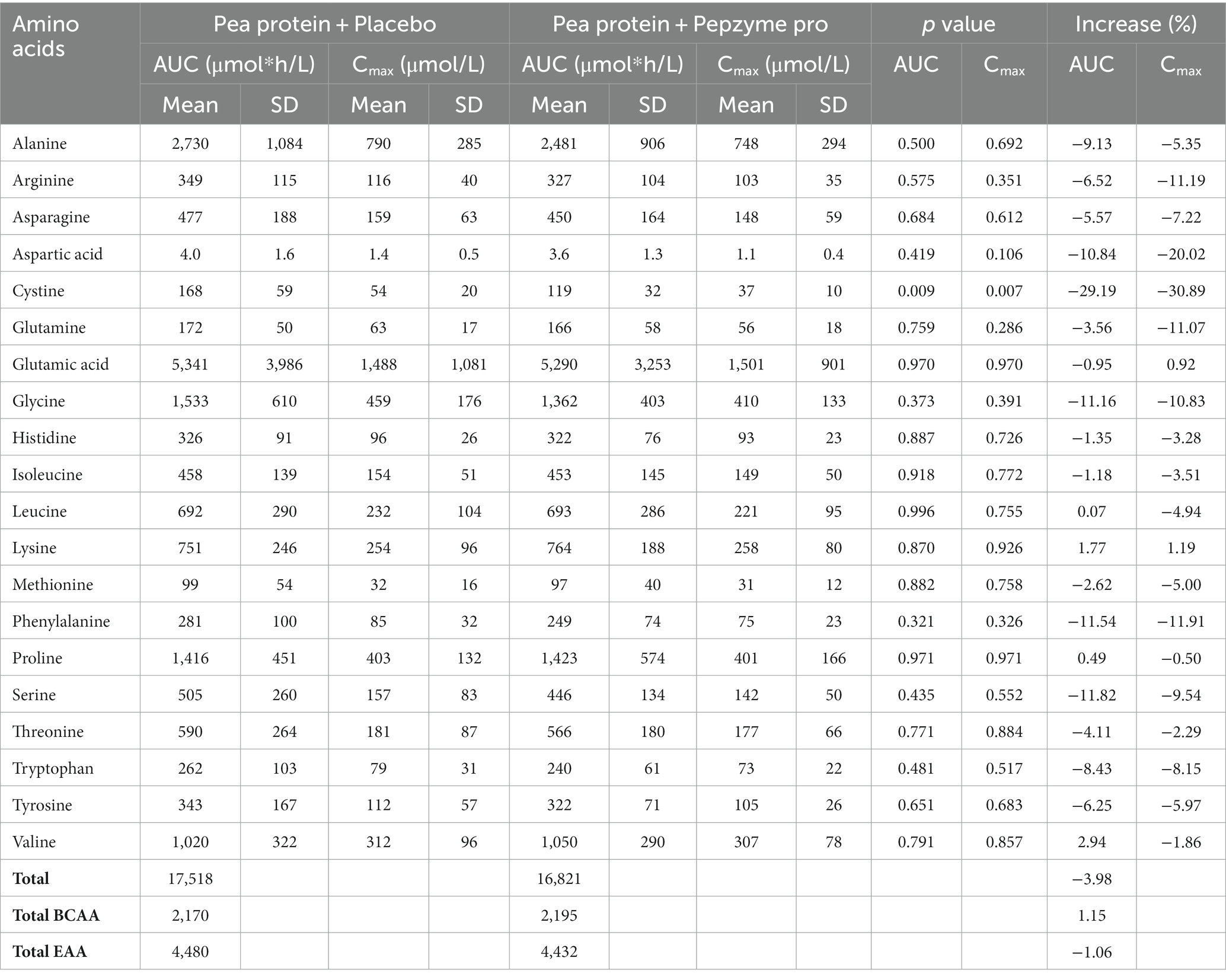
Table 3A. AUC and Cmax for plasma amino acid concentration of placebo (Pea protein) and test (Pea protein + enzymes-probiotics blend) on day 1.
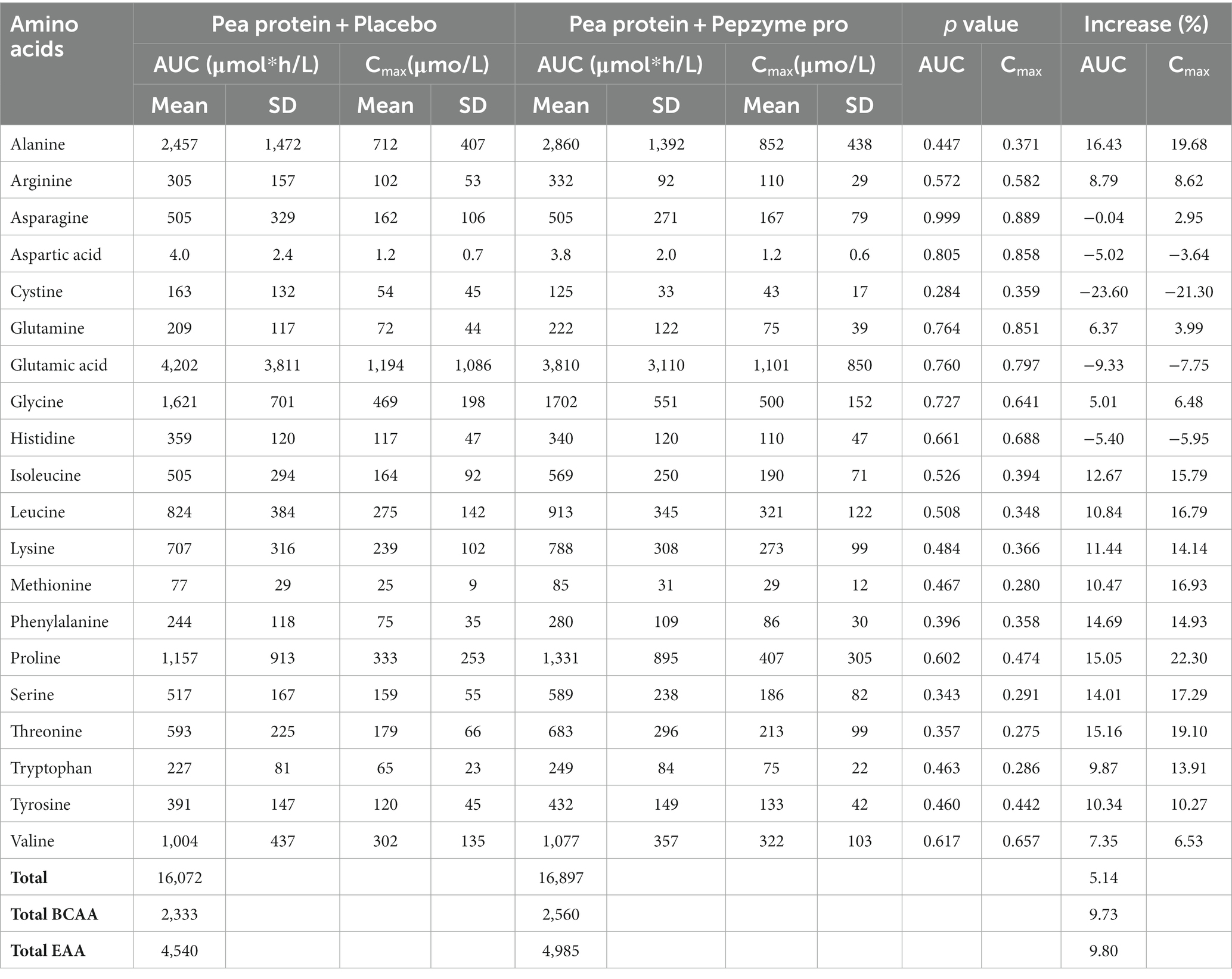
Table 3B. AUC and Cmax for plasma amino acid concentration of placebo (Pea protein) and test (Pea protein + enzymes-probiotics blend) on day 15.
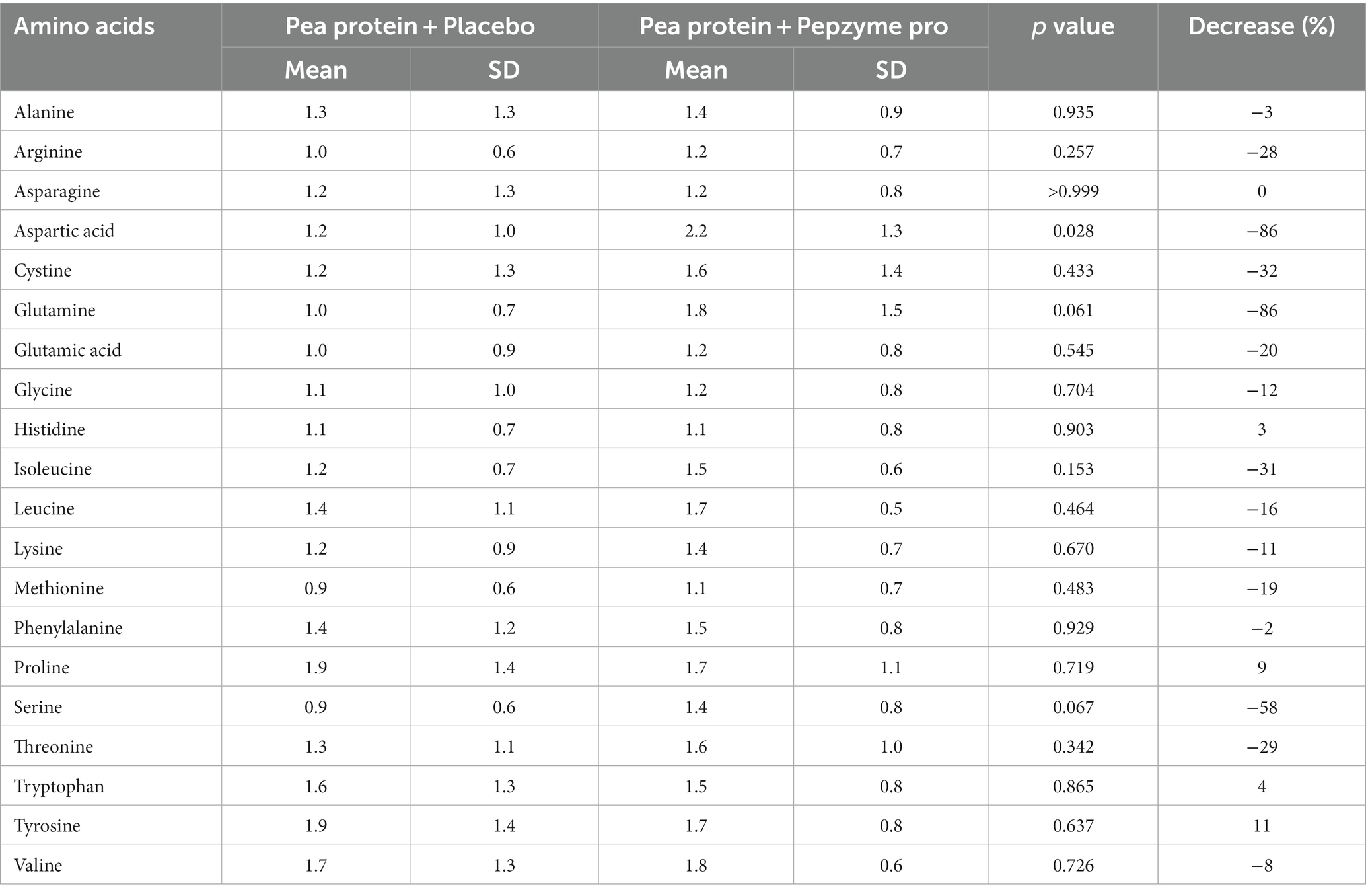
Table 4A. Tmax (h) for plasma amino acid concentration of placebo (pea protein) and test (Pea protein + enzymes-probiotics blend) on day 0.
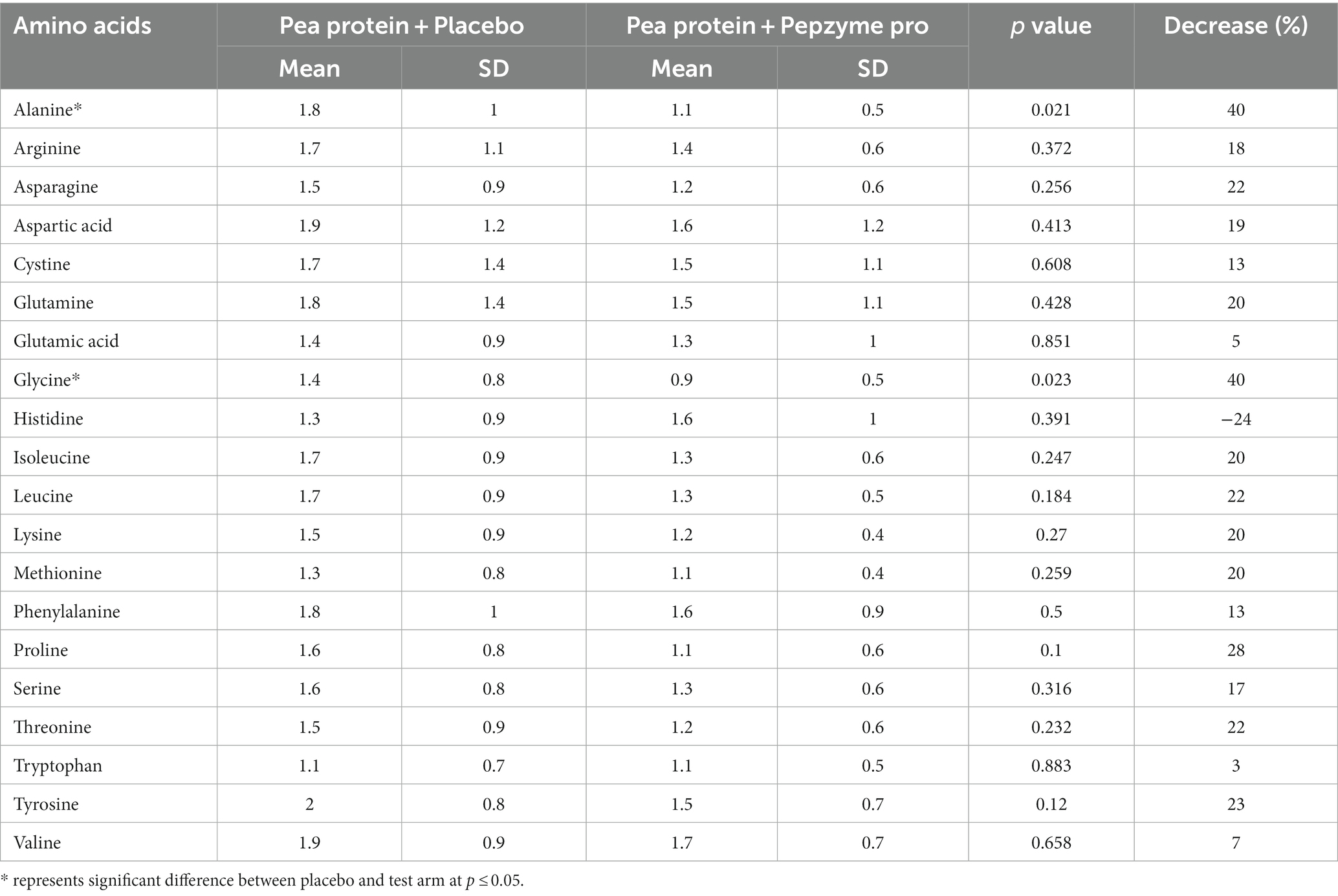
Table 4B. Tmax (h) for plasma amino acid concentration of placebo (Pea protein) and test (Pea protein + enzymes-probiotics blend) at day 15.
During this clinical trial, no adverse events were recorded in both the study arms, ensuring safety upon consumption of supplement in human subjects.
The effect of supplementation with pea proteins and enzymes-probiotics blend on fecal and urine nitrogen levels was evaluated in the test and placebo arms on days 1 and 15 of the study. The results indicated that supplementation did not alter the nitrogen levels in test compared with the placebo. Additionally, there was no statistically significant difference in the fecal nitrogen and urine nitrogen between the initial day (day 1) and the final day (day 15) of supplementation (Figure 4).
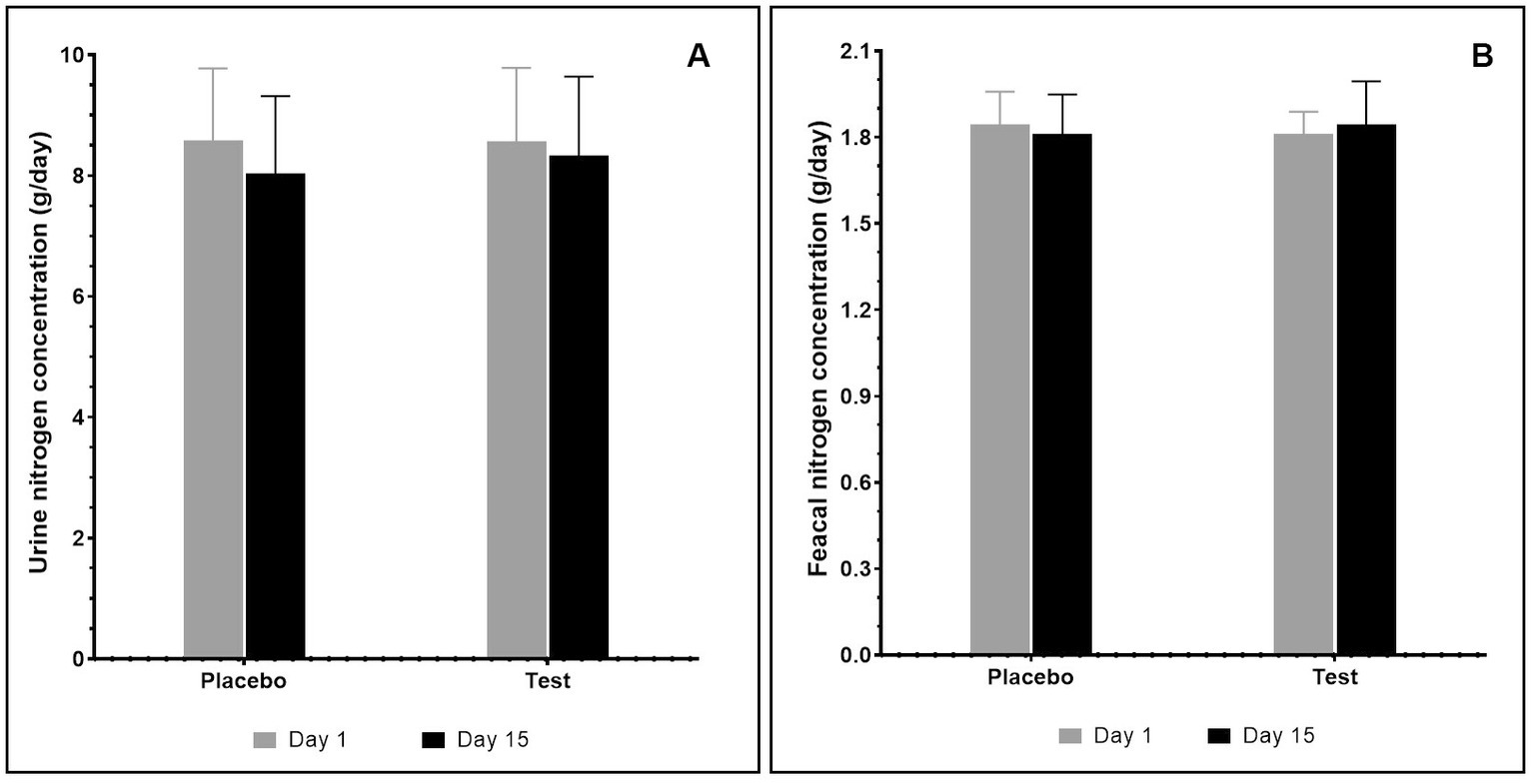
Figure 4. Total nitrogen content (g/day) after consumption of pea protein (placebo) and pea protein + enzymes-probiotics blend (test) present in urine (A) and fecal (B) samples. Values represented as mean ± standard error. Data were statistically analyzed using the Student’s t-test, and p ≤ 0.05 was considered statistically significant.
The changes in levels of serum insulin and CRP after the supplementation with pea proteins and enzymes-probiotics blend were quantified in the test and placebo arms on day 15 of the study (Figure 5). A considerable rise in the serum insulin level from 9.03 ± 2.43 μU/mL to 34.56 ± 9.98 μU/mL (p = 0.019) and 6.73 ± 1.11 μU/mL to 24.26 ± 6.48 μU/mL (p = 0.012) after 4 h of protein consumption was seen in the placebo and test arms, respectively. Consumption of protein has elevated the insulin level in both the placebo and test arms. The insulin level at 4 h in both arms was not statistically different. The CRP level did not show variation between the placebo and test arms as well as between the 0 h and 4 h samples, demonstrating the safety of the supplement.
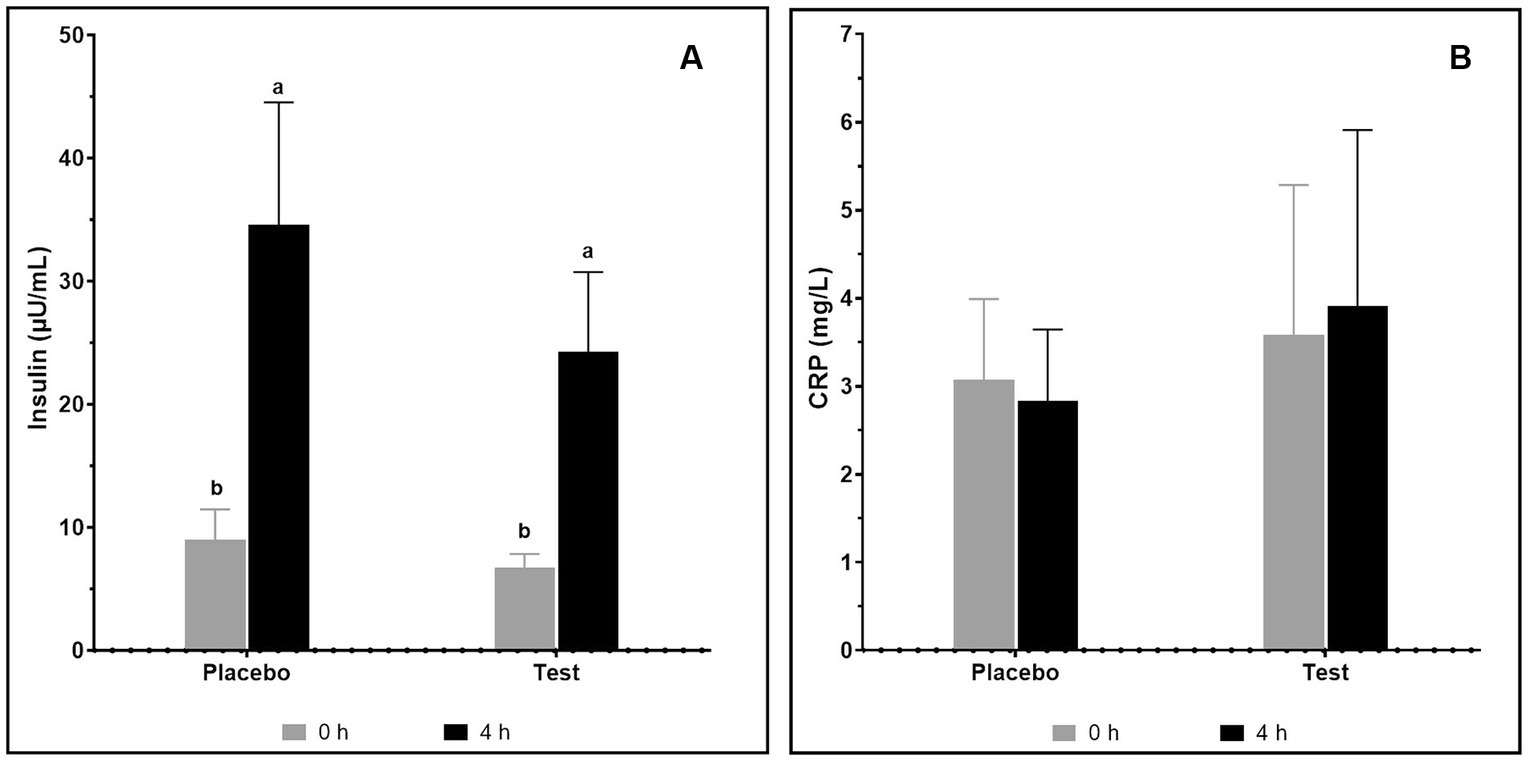
Figure 5. Insulin (A) and CRP (B) on consumption of pea protein (placebo) and pea protein + enzymes-probiotics blend (test) on day 15. Values represented as mean ± standard error. Data were statistically analyzed using the Student’s t-test. Lower letters placed on the bar represent statistical difference at p ≤ 0.05.
The variations in the gut microbiome profile of the test arm after supplementation with pea protein and enzymes-probiotics blend on day 1 and day 15 were assessed by visualizing and characterizing for direct quantitative comparison of abundances. This was followed by alpha and beta diversity measurements, and then determining differentially abundant species across the comparing groups. As whole metagenome sequencing was performed, the profiling of all the microbial taxa (including fungi and viruses) within each sample was done successfully.
The composition of microbial kingdom between day 1 (PP_Pre) and day 15 (PP_Post) of supplementation with pea protein and enzymes-probiotics blend is shown as stacked bar plots (Figure 6A). A slight but not statistically significant variation was observed in the microbial composition, particularly a small decline in the bacterial and viral populations post-consumption of test supplementation. The downtrend in bacterial and viral composition was from 99.37% (PP_Pre) to 99.13% (PP_Post) and 0.11% (PP_Pre) to 0.04% (PP_Post), respectively, after 15 days of supplementation with pea protein and enzymes-probiotics blend. Furthermore, the uptrend in archeal and eukaryotic abundance was from 0.12% (PP_Pre) to 0.33% (PP_Post), and 0.40% (PP_Pre) to 0.49% (PP_Post), respectively, after 15 days of supplementation. The relative abundance of this pattern can also be seen at phylum level (Figure 6B). The 15-day test supplementation showed the possibility of a downtrend in the abundance of Bacteroidetes and Proteobacteria, which may be due to a reduction in the abundance of Holdemanella (Holdemanella porci and Holdemanella biformis) (Figures 6F, 7A,B) and Prevotella copri. There was also a decline in Haemophilus pittmaniae of the phylum Proteobacteria (Figure 7C). The observed uptrend in the abundance of Firmicutes was largely due to an increase in the relative abundance of Roseburia (Roseburia intestinalis, Roseburia hominis, and Roseburia inulinivorans) (Figures 6F, 7D,E).
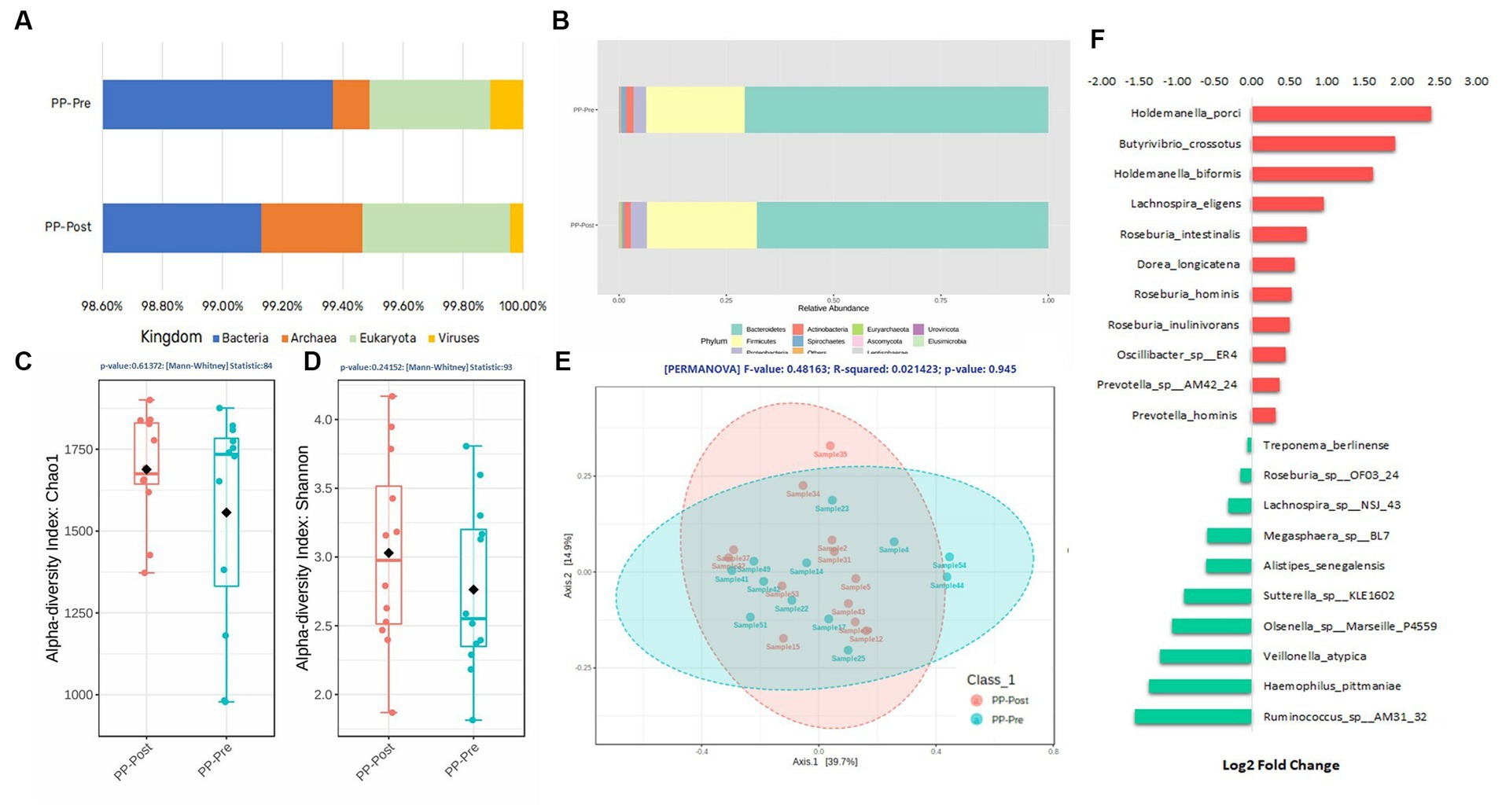
Figure 6. Composition of the gut microbiome after consumption of pea protein + enzymes-probiotics blend on day 1 (PP_Pre) and day 15 (PP_Post). (A) Quantitative comparison of abundances Kingdom level; (B) Quantitative comparison of abundances Phylum level; (C) Alpha Diversity—Chao1; (D) Alpha Diversity—Shannon Index; (E) Beta Diversity—Bray-Curtis distance, PERMANOVA; (F) Log2 Fold Change, of some of the most differentially abundant species.
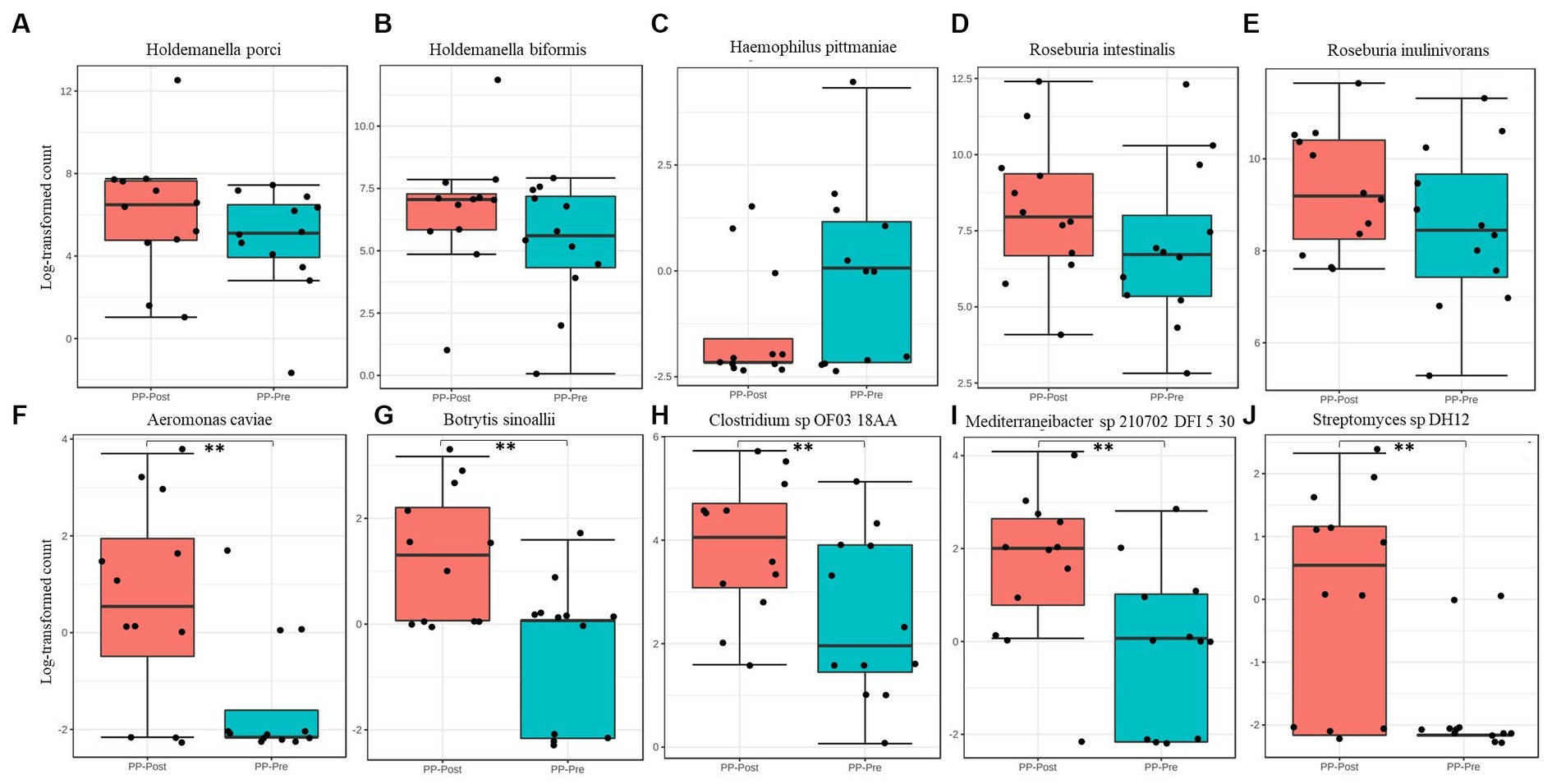
Figure 7. Differential abundance of species (A) Holdemanella porci, (B) Holdemanella biformis, (C) Haemophilus pittmaniae, (D) Roseburia intestinalis, (E) Roseburia inulinivorans, (F) Aeromonas caviae, (G) Botrytis sinoallii, (H) Clostridium sp. OF03 18AA, (I) Mediterraneibacter sp. 210,702 DFI 5 30, and (J) Streptomyces sp. DH12. ** represents significant difference between day 1 (PP_Pre) and day 15 (PP_Post) of supplementation at p ≤ 0.01.
The measurement of alpha and beta diversity confirmed minor variations in diversities, but there were no statistically significant differences among the diversity measures on days 1 and 15 of the supplementation with pea protein and enzymes-probiotics blend. The Chao1 diversity displayed an increasing trend in the mean species richness in day 15 samples as compared to day 1 samples, with the diversity becoming more similar across all samples (lower deviation across samples) (Figure 6C). Furthermore, the Shannon diversity measure also showed increasing trend in diversities of day 15 samples with almost similar evenness as compared to day 1 samples (Figure 6D). The beta diversity measure by Bray-Curtis distance had not established any statistically significant difference between day 1 and day 15 of supplementation, even though it displayed clustering between the groups, represented by the ellipses (Figure 6E).
Though there were no statistically significant variations in microbiome profile at the higher taxonomic levels, we observed several specific changes at species level that were statistically, potentially, and functionally significant. Based on the consensus approach employed with five different DA tools, we could establish a total of five species to be significantly differentially abundant (p < 0.05) between day 1 and day 15. The comparative abundance plots of Aeromonas caviae, Botrytis sinoallii, Clostridium sp. OF03 18AA, Mediterraneibacter sp. 210,702 DFI 5 30, and Streptomyces sp. DH12 are displayed in Figures 7F-J.
The present clinical trial was designed to evaluate the effect of the enzymes-probiotics blend on pea protein digestibility, amino acid bioavailability, and gut microbiota after regular consumption of high protein. Furthermore, the safety of the supplement was studied in terms of the adverse events, an alteration in serum insulin and CRP levels, as well as the changes in fecal and urine nitrogen levels.
Test supplementation showed an uptrend in the rate of amino acid absorption compared with the placebo on day 15, but not on day 1. The inability of the supplement to demonstrate positive effect on the amino acid absorption on day 1 illustrated that the external enzyme supplementation alone could not enhance the digestion and absorption of amino acids from the pea protein, but the probiotics aid the process. Consumption of the supplement for 15 consecutive days demonstrated the likelihood of augmenting the absorption of total amino acids (AUC) and maximum concentration (Cmax), as well as the tendency to reduce the time required to reach the maximum concentration (Tmax) in the blood. These results illustrated the possibility of enzymes-probiotics blend to positively alter the pea protein digestion and absorption. Previously, our in vitro study had showed that the co-ingestion of Pepzyme AG (containing acid proteases) with protein (whey, pea, and collagen) improved the protein digestion (13).
Probiotics are known to help digestion of protein by promoting healthy intestinal flora. They regulate and influence intestinal bacteria related to protein digestion. Some probiotics could also secrete proteolytic enzymes capable of hydrolyzing proteins and can also induce host digestive proteases and peptidases. They also reduce the production of toxic metabolites by controlling or inhibiting the fermentation of harmful proteins. Furthermore, probiotics also elevate the absorption of smaller peptides and amino acids by altering the abilities of the epithelium lining and promoting transport (20). The role of Bacillus coagulans, Bacillus subtilis, Lactobacillus acidophilus, and Lactobacillus plantarum in enhancing protein digestion has been well documented (18, 31–34). Previously, Jager et al. (18) documented the ability of probiotics to enhance the digestion of pea proteins. Garcia et al. (21) firstly hydrolyzed the pea proteins with digestive proteases and then fermented them using Lactobacillus plantarum, leading to production of smaller peptides which could be easily absorbed in gastrointestinal track. Furthermore, the enzymes-probiotics blend has been shown to boost the nutrient digestibility and to stabilize the microbial environment, thus promoting healthy gut in Nile tilapia (35). It is worth noticing that the digestion potential of probiotics could be augmented by blending them with exogenous proteases. In the current trial, 15-day supplementation of protein along with the enzymes-probiotics blend demonstrated an uptrend in rate of amino acid absorption and total amino acid absorption compared with the only protein supplementation. This could be possibly owed to the role of enzymes in digestion of protein and role of probiotics in regulating and influencing gut microbiota related to proteolysis.
The microbiota profiling data displayed an increasing trend in the abundance of Archaea, and specifically that of Methanobrevibacter smithii belonging to the phylum Euryarchaeota, post-test supplementation. This was interesting, as a previous study with whey protein supplementation reported an increase in Archaea diversity (36). M. smithii is highly prevalent microbes of the gut (37). In an earlier study, the presence of M. smithii had promoted calories intake from the diet in an animal model (38). The potential increase in the methanogenic archaea observed after supplementation of test for 15 days needs further detailed study with respect to the role of the Archaea and its functional impact. The supplementation also showed tendency to reduce the viral abundance and increase the abundance of Eukaryota (mainly fungi). At the phylum level, there was a downtrend in the abundance of Bacteroidetes and Proteobacteria and an uptrend in the Firmicutes (Roseburia intestinalis and Roseburia inulinivorans) on day 15. This can be functionally important as Roseburia intestinalis being one of the keystone species that influences the structure of the microbial communities (39), while Roseburia inulinivorans being one of the primary species associated with recovery of post-antibiotic exposure (40). On the other hand, pathogens like Haemophilus pittmaniae, belonging to phylum Proteobacteria, showed a tendency to reduce their abundance post-supplementation. However, none of these species were significantly differentially abundant across the groups. The comparative abundance of five species, namely Aeromonas caviae, Botrytis sinoallii, Clostridium sp. OF03 18AA, Mediterraneibacter sp. 210,702 DFI 5 30, and Streptomyces sp. DH12, was estimated to be significantly different in the test on day 15 by at least 2 Log2Folds across the groups. Most of these microbiome species have been identified and characterized within the gut microbiome of humans recently. For instance, Clostridium sp. OF03 18AA and Mediterraneibacter sp. 210,702 DFI 5 30, both belonging to the phylum Firmicutes, are relatively new and comes under the group of uncharacterized isolates of Clostridium (41) and Mediterraneibacter, respectively (42). Hence, their increase in abundance or its functional association post-supplementation of pea protein and enzymes-probiotics blend needs further evaluation. Of the five species, Streptomyces sp. DH12 was perhaps the one with most potential to have a positive impact on the gut microbiome, with broad-spectrum antibacterial activity toward Gram-positive and Gram-negative pathogens, based on its genomic characterization (43). This was estimated to have increased by ~3-fold after supplementation with pea protein and an enzymes-probiotics blend. Though the statistically significant differences were not observed among the test and placebo group at phylum level of gut microbial population, the study provided inkling of possible positive effect which supplement might offer on long-term consumption.
Enzymes-probiotics supplementation was found to be safe in this trial as no adverse effects were recorded. Furthermore, it did not negatively affect the metabolism as shown by unaltered nitrogen levels of urine and feces as well as CRP on the initial and final day of the trial in both the placebo and test arms. This confirms the safety of the supplementation. The level of serum insulin was increased after consumption of pea protein in placebo as well as in test arm on day 15. Thondre et al. (44) has already documented the role of pea protein on the secretion of insulin. Though statistically insignificant, low levels of serum insulin in the test over the placebo arm may be attributed to the fixed time-point (0 h and 4 h) analysis. A potential decline in the Tmax after 15-day test supplementation might be owing to earlier insulin spike in the blood. The time kinetics for the serum insulin levels after protein supplementation would definitely help to shed more light on the observed results. Although an increase in protein digestibility was seen in test arm, no statistically significant difference was found between test and placebo arms. The variation in the digestive processes and amino acid metabolism of each individual and relatively small subject size making it difficult to justify the results statistically.
The placebo-controlled, double-blinded, randomized, and crossover study conducted on 15 healthy subjects demonstrated the possible effect of the enzymes-probiotics blend (Pepzyme Pro) on the digestion of pea protein and gut health. Supplementation of the enzymes-probiotics blend along with the pea protein for 15 consecutive days provided an inkling of a possible uptrend in the rate of amino acid absorption, total amino acid absorption (AUC), Cmax, and a downtrend in the Tmax compared with the placebo arm. Supplementation did not show statistically significant variations in microbiome profile at the higher taxonomic levels; expect changes observed with respect to Archaea and a few uncharacterized species. A detailed study related to the gut microbiome might be useful to characterize those species. The small sample size and metabolic variations had affected the statistical significance of the data. A study on a large population would be useful to shed more light on the details.
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/; PRJNA1020557.
The studies involving humans were approved by Institutional ethics committee Charak Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
AR: Conceptualization, Methodology, Visualization, Writing – review & editing. TG: Data curation, Formal analysis, Visualization, Writing – original draft. DD: Data curation, Formal analysis, Writing – review & editing. GK: Data curation, Formal analysis, Writing – original draft. SJ: Conceptualization, Data curation, Formal analysis, Methodology, Visualization, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This clinical study received funding from Advanced Enzymes Technologies. The funder had role in study design, decision to publish, and preparation of the manuscript but actual conduct of the study and data collection.
The authors gratefully acknowledge Specialty Enzymes & Probiotics for providing the supplements and thank the ProClin Research team for study conduct.
AR, SJ, and TG are paid employees of Advanced Enzyme Technologies, which had funded this clinical study and has a corporate affiliation with Specialty Enzymes and Probiotics. Specialty Enzymes and Probiotics had no role in study design and actual conduct of the study. DD is a director and GK is a paid employee of Leucine Rich Bio Pvt. Ltd.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Karlund, A , Gómez-Gallego, C , Turpeinen, A , Palo-Oja, O , El-Nezami, H , and Kolehmainen, M . Protein supplements and their relation with nutrition, microbiota composition and health: is more protein always better for sportspeople? Nutrients. (2019) 11:829. doi: 10.3390/nu11040829
2. Frestedt, JL , Zenk, JL , Kuskowski, MA , Ward, LS , and Bastian, ED . A whey-protein supplement increases fat loss and spares lean muscle in obese subjects: a randomized human clinical study. Nutr Metabol. (2008) 5:1–7. doi: 10.1186/1743-7075-5-8
3. Herring, C , Bazer, F , Johnson, G , and Wu, G . Impacts of maternal dietary protein intake on fetal survival, growth, and development. Exp Biol Med. (2018) 243:525–33. doi: 10.1177/1535370218758275
4. Hertzler, S , Lieblein-Boff, J , Weiler, M , and Allgeier, C . Plant proteins: assessing their nutritional quality and effects on health and physical function. Nutrients. (2020) 12:3704. doi: 10.3390/nu12123704
5. Langyan, S , Yadava, P , Khan, FN , Dar, ZA , Singh, R , and Kumar, A . Sustaining protein nutrition through plant-based foods. Front Nutr. (2022) 8:1237. doi: 10.3389/fnut.2021.772573
6. Gorissen, S , Crombag, J , Senden, J , Waterval, W , Bierau, J , Verdijk, L, et al. Protein content and amino acid composition of commercially available plant-based protein isolates. Amino Acids. (2018) 50:1685–95. doi: 10.1007/s00726-018-2640-5
7. Guillin, F , Gaudichon, C , Guérin-Deremaux, L , Lefranc-Millot, C , Airinei, G , Khodorova, N, et al. Real ileal amino acid digestibility of pea protein compared to casein in healthy humans: a randomized trial. Am J Clin Nutr. (2021) 115:353–63. doi: 10.1093/ajcn/nqab354
8. Lu, ZX , He, JF , Zhang, YC , and Bing, DJ . Composition, physicochemical properties of pea protein and its application in functional foods. Crit Rev Food Sci Nutr. (2020) 60:2593–605. doi: 10.1080/10408398.2019.1651248
9. Ge, J , Sun, C , Corke, H , Gul, K , Gan, R , and Fang, Y . The health benefits, functional properties, modifications, and applications of pea (Pisum sativum L.) protein: current status, challenges, and perspectives. Compr Rev Food Sci Food Saf. (2020) 19:1835–76. doi: 10.1111/1541-4337.12573
10. Lonnie, M , Laurie, I , Myers, M , Horgan, G , Russell, W , and Johnstone, A . Exploring health-promoting attributes of plant proteins as a functional ingredient for the food sector: a systematic review of human interventional studies. Nutrients. (2020) 12:2291. doi: 10.3390/nu12082291
11. Kumar, M , Tomar, M , Punia, S , Dhakane-Lad, J , Dhumal, S , Changan, S, et al. Plant-based proteins and their multifaceted industrial applications. LWT. (2022) 154:112620. doi: 10.1016/j.lwt.2021.112620
12. Awosika, T , and Aluko, RE . Enzymatic pea protein hydrolysates are active trypsin and chymotrypsin inhibitors. Foods. (2019) 8:200. doi: 10.3390/foods8060200
13. Jadhav, S , Gaonkar, T , and Rathi, A . In vitro gastrointestinal digestion of proteins in the presence of enzyme supplements: details of antioxidant and antidiabetic properties. LWT. (2021) 147:111650. doi: 10.1016/j.lwt.2021.111650
14. Minevich, J , Olson, MA , Mannion, JP , Boublik, JH , McPherson, JO , Lowery, RP, et al. Digestive enzymes reduce quality differences between plant and animal proteins: a double-blind crossover study. J Int Soc Sports Nutr. (2015) 12:P26. doi: 10.1186/1550-2783-12-S1-P26
15. Oben, J , Kothari, S , and Anderson, M . An open label study to determine the effects of an oral proteolytic enzyme system on whey protein concentrate metabolism in healthy males. J Int Society Sports Nutr. (2008) 5:10. doi: 10.1186/1550-2783-5-10
16. Trehan, A , Verma, MK , Maheshwari, S , Khanna, T , Kumari, P , and Singh, H . An open-label clinical study to determine the effect of enhanced absorption formula (MB EnzymePro®) on the bioavailability of whey protein in healthy male subjects. J Food Process Technol. (2020) 11:820. doi: 10.35248/2157-7110.20.11.820
17. Townsend, J , Morimune, J , Jones, M , Beuning, C , Haase, A , Boot, C, et al. The effect of ProHydrolase® on the amino acid and intramuscular anabolic signalling response to resistance exercise in trained males. Sports. (2020) 8:13. doi: 10.3390/sports8020013
18. Jager, R , Zaragoza, J , Purpura, M , Iametti, S , Marengo, M , and Tinsley, G . Probiotic administration increases amino acid absorption from plant protein: a placebo-controlled, randomized, double-blind, multicenter, crossover study. Probiotics Antimicrob Proteins. (2020) 12:1330–9. doi: 10.1007/s12602-020-09656-5
19. Hemarajata, P , and Versalovic, J . Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Ther Adv Gastroenterol. (2012) 6:39–51. doi: 10.1177/1756283x12459294
20. Wang, J , and Ji, H . Influence of probiotics on dietary protein digestion and utilization in the gastrointestinal tract. Curr Protein Pept Sci. (2019) 20:125–31. doi: 10.2174/1389203719666180517100339
21. Garcia Arteaga, V , Demand, V , Kern, K , Strube, A , Szardenings, M , Muranyi, I, et al. Enzymatic hydrolysis and fermentation of pea protein isolate and its effects on antigenic proteins, functional properties, and sensory profile. Foods. (2022) 11:118. doi: 10.3390/foods11010118
22. McMurdie, PJ , and Holmes, S . Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. (2013) 8:e61217. doi: 10.1371/journal.pone.0061217
23. McMurdie, PJ , and Holmes, S . Shiny-phyloseq: web application for interactive microbiome analysis with provenance tracking. Bioinformatics. (2015) 31:282–3. doi: 10.1093/bioinformatics/btu616
24. Calle, ML . Statistical analysis of metagenomics data. Genomics Inform. (2019) 17:e6. doi: 10.5808/GI.2019.17.1.e6
25. Paulson, JN , Pop, M , and Bravo, HC . metagenomeSeq: statistical analysis for sparse high-throughput sequencing. Bioconductor Package. (2013) 1:191. doi: 10.18129/B9.bioc.metagenomeSeq
26. Paulson, JN , Talukder, H , and Corrada, BH . Longitudinal differential abundance analysis of microbial marker-gene surveys using smoothing splines. BioRxiv. (2017):099457. doi: 10.1101/099457
27. Paulson, JN , Stine, OC , Bravo, HC , and Pop, M . Differential abundance analysis for microbial marker-gene surveys. Nat Methods. (2013) 10:1200–2. doi: 10.1038/nmeth.2658
28. Robinson, MD , McCarthy, DJ , and Smyth, GK . edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. (2010) 26:139–40. doi: 10.1093/bioinformatics/btp616
29. Love, MI , Huber, W , and Anders, S . Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
30. Segata, N , Izard, J , Waldron, L , Gevers, D , Miropolsky, L , Garrett, WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. (2011) 12:R60. doi: 10.1186/gb-2011-12-6-r60
31. Kumar, VV , Sudha, KM , Bennur, S , and Dhanasekar, KR . A prospective, randomized, open-label, placebo-controlled comparative study of Bacillus coagulans GBI-30, 6086 with digestive enzymes in improving indigestion in geriatric population. J Family Med Prim Care. (2020) 9:1108–12. doi: 10.4103/jfmpc.jfmpc_922_19
32. Lin, J , Zeng, Q , Zhang, C , Song, K , Lu, K , Wang, L, et al. Effects of Bacillus subtilis supplementation in soybean meal-based diets on growth performance, diet digestibility and gut health in bullfrog Lithobates catesbeianus. Aquac Rep. (2020) 16:100273. doi: 10.1016/j.aqrep.2020.100273
33. Pescuma, M , Hébert, E , Mozzi, F , and Valdez, G . Hydrolysis of whey proteins by Lactobacillus acidophilus, Streptococcus thermophilus and Lactobacillus delbrueckii ssp. bulgaricus grown in a chemically defined medium. J Appl Microbiol. (2007) 103:1738–46. doi: 10.1111/j.1365-2672.2007.03404.x
34. Pranoto, Y , Anggrahini, S , and Efendi, Z . Effect of natural and Lactobacillus plantarum fermentation on in-vitro protein and starch digestibilities of sorghum flour. Food Biosci. (2013) 2:46–52. doi: 10.1016/j.fbio.2013.04.001
35. Maas, RM , Verdegem, MC , Debnath, S , Marchal, L , and Schrama, JW . Effect of enzymes (phytase and xylanase), probiotics (B. amyloliquefaciens) and their combination on growth performance and nutrient utilisation in Nile tilapia. Aquaculture. (2021) 533:736226. doi: 10.1016/j.aquaculture.2020.736226
36. Cronin, O , Barton, W , Skuse, P , Penney, NC , Garcia-Perez, I , Murphy, EF, et al. A prospective metagenomic and metabolomic analysis of the impact of exercise and/or whey protein supplementation on the gut microbiome of sedentary adults. MSystems. (2018) 3:e00044–18. doi: 10.1128/mSystems.00044-18
37. Gaci, N , Borrel, G , Tottey, W , O'Toole, PW , and Brugère, JF . Archaea and the human gut: new beginning of an old story. World J Gastroenterol. (2014) 20:16062–78. doi: 10.3748/wjg.v20.i43.16062
38. Samuel, BS , and Gordon, JI . A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A. (2006) 103:10011–6. doi: 10.1073/pnas.060218710
39. Fisher, CK , and Mehta, P . Identifying keystone species in the human gut microbiome from metagenomic timeseries using sparse linear regression. PLoS One. (2014) 9:e102451. doi: 10.1371/journal.pone.0102451
40. Chng, KR , Ghosh, TS , Tan, YH , Nandi, T , Lee, IR , Ng, AH, et al. Metagenome-wide association analysis identifies microbial determinants of post-antibiotic ecological recovery in the gut. Nat Ecol Evol. (2020) 4:1256–67. doi: 10.1038/s41559-020-1236-0
41. Onate, FP , Roume, H , and Almeida, M . Recovery of metagenome-assembled genomes from a human fecal sample with pacific biosciences high-fidelity sequencing. Microbiol Resour Announc. (2022) 11:e00250–22. doi: 10.1128/mra.00250-22
42. Schneider, D , Zühlke, D , Poehlein, A , Riedel, K , and Daniel, R . Metagenome-assembled genome sequences from different wastewater treatment stages in Germany. Microbiol Resour Announc. (2021) 10:e00504–21. doi: 10.1128/MRA.00504-21
43. Jiao, J , Paterson, J , Busche, T , Rückert, C , Kalinowski, J , Harwani, D, et al. Draft genome sequence of Streptomyces sp. strain DH-12, a soilborne isolate from the Thar Desert with broad-spectrum antibacterial activity. Genome Announc. (2018) 6:e00108–18. doi: 10.1128/genomea.00108-18
44. Thondre, P , Achebe, I , Sampson, A , Maher, T , Guérin-Deremaux, L , Lefranc-Millot, C, et al. Co-ingestion of NUTRALYS® pea protein and a high-carbohydrate beverage influences the glycaemic, insulinaemic, glucose-dependent insulinotropic polypeptide (GIP) and glucagon-like peptide-1 (GLP-1) responses: preliminary results of a randomised controlled trial. Eur J Nutr. (2021) 60:3085–93. doi: 10.1007/s00394-021-02481-8
Keywords: amino acids, absorption, pea protein, digestion, gut microbiota, enzymes, probiotics
Citation: Rathi A, Gaonkar T, Dhar D, Kallapura G and Jadhav S (2024) Study of amino acids absorption and gut microbiome on consumption of pea protein blended with enzymes-probiotics supplement. Front. Nutr. 11:1307734. doi: 10.3389/fnut.2024.1307734
Received: 05 October 2023; Accepted: 02 January 2024;
Published: 23 January 2024.
Edited by:
Oana Lelia Pop, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaReviewed by:
Emiliano Salvucci, National Scientific and Technical Research Council (CONICET), ArgentinaCopyright © 2024 Rathi, Gaonkar, Dhar, Kallapura and Jadhav. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Swati Jadhav, c3dhdGlAYWR2YW5jZWRlbnp5bWVzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.