
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr., 10 May 2024
Sec. Nutrition and Microbes
Volume 11 - 2024 | https://doi.org/10.3389/fnut.2024.1304045
This article is part of the Research TopicExploring Dietary Polysaccharides: Molecular Insights into Gastrointestinal Health and the Brain-Gut AxisView all 6 articles
 Karley K. Mahalak1*
Karley K. Mahalak1* LinShu Liu1
LinShu Liu1 Jamshed Bobokalonov1,2
Jamshed Bobokalonov1,2 Adrienne B. Narrowe1
Adrienne B. Narrowe1 Jenni Firrman1
Jenni Firrman1 Kyle Bittinger3,4
Kyle Bittinger3,4 Weiming Hu3
Weiming Hu3 Steven M. Jones3
Steven M. Jones3 Ahmed M. Moustafa3,4
Ahmed M. Moustafa3,4Introduction: Studies have shown that a diet high in fiber and prebiotics has a positive impact on human health due largely to the fermentation of these compounds by the gut microbiota. One underutilized source of fiber may be rice bran, a waste product of rice processing that is used most frequently as an additive to livestock feed but may be a good source of fibers and other phenolic compounds as a human diet supplement. Previous studies focused on specific compounds extracted from rice bran showed that soluble fibers extracted from rice bran can improve glucose response and reduce weight gain in mouse models. However, less is known about changes in the human gut microbiota in response to regular rice bran consumption.
Methods: In this study, we used a Simulator of the Human Intestinal Microbial Ecology (SHIME®) to cultivate the human gut microbiota of 3 different donors in conditions containing either soluble or insoluble fiber fractions from rice bran. Using 16S rRNA amplicon sequencing and targeted metabolomics via Gas Chromatography-Mass Spectrometry, we explored how gut microbial communities developed provided different supplemental fiber sources.
Results: We found that insoluble and soluble fiber fractions increased short-chain fatty acid production, indicating that both fractions were fermented. However, there were differences in response between donors, for example the gut microbiota from donor 1 increased acetic acid production with both fiber types compared with control; whereas for donors 2 and 3, butanoic acid production increased with ISF and SF supplementation. Both soluble and insoluble rice bran fractions increased the abundance of Bifidobacterium and Lachnospiraceae taxa.
Discussion: Overall, analysis of the effect of soluble and insoluble rice bran fractions on the human in vitro gut microbiota and the metabolites produced revealed individually variant responses to these prebiotics.
Regular intake of dietary fiber is beneficial for human health due in part to reduced risks of cardiovascular disease, diabetes, colorectal cancers, and more (1–5). Dietary fibers that are found in plant-based foods and cereals including wheat, oat, millet, and rice are of particular interest due to their reported beneficial properties (6, 7). Dietary fibers remain unchanged through the digestive tract until they reach the colon, where they are then fermented by the gut microbiome (8). This fermentation process promotes a healthy mucosal barrier in the colon and an increase in the production of short-chain fatty acids (SCFAs), among other beneficial health effects (9). SCFAs absorbed by the human cells lining the intestine as an energy source, and contribute to overall health by functioning as carbon sources for the colonocytes (butyrate) and liver cells (acetic acid and propanoic acid) (10). Many studies have shown the beneficial effects of SCFAs on the body, including anti-inflammatory, anti-obesity, and immunoregulatory effects (10, 11). The ability of dietary fibers to aid against metabolic and other disorders is often attributed to their ability to regulate digestion, absorption of nutrients, and metabolism (2).
It is important to distinguish between dietary fiber types when looking at their effects on health. Two main fiber types, categorized as soluble and insoluble fibers, are important components of a healthy diet. Soluble fibers are quickly fermented by the gut microbiota in the colon, can increase satiety, and reduce the speed of movement through the large intestine (1, 12). Conversely, insoluble fibers increase the rate of passage through the large intestine and contribute to fecal bulking, however, there is less fermentation associated with insoluble fibers in the colon (13). While soluble fibers are well known for their fermentability by the gut microbiota, and consequently how they increase the production of SCFAs by the gut microbiota, less is known about insoluble fibers. In mouse studies, insoluble fibers have been shown to reduce weight gain and cholesterol, as well as increase SCFA production and modify the abundance of particular gut microbial community members (14). Insoluble fibers from bamboo have also been shown to increase SCFA production after 24 h of in vitro batch fermentation (15). Insoluble fibers extracted from soy husk and subjected to 48 h of in vitro batch fermentation by human fecal samples significantly changed the gut microbial community composition at the phylum level (16). Taken together, these findings indicate that insoluble fiber impacts the gut microbiota in a more meaningful way than was previously thought.
Rice is a staple food source around the world, with consumption increasing in the Americas due to the globalization of food (17). Unprocessed, whole-grain rice is considered healthier, as it contains more fibers and polyphenols, which are associated with the health benefits of rice and other whole-grain cereals. The milling of rice removes the outer layer of the rice grain, which is termed rice bran. Due to its high fat content, particularly free fatty acids, and the lipase enzyme, rice bran requires processing before use in human consumption to ensure it does not become rancid, an intensive process called stabilization (18). Due to the necessity of this process for human consumption, rice bran is often used instead as livestock feed as a bulking agent (19). However, if there are suitably notable beneficial health effects of rice bran extracts on gut health, there could be increased interest in its use as a food supplement.
Previous studies on the effect of rice bran components on the human gut microbiome have demonstrated that both of the soluble fiber components increased SCFA production in vitro in a short-term batch fermentation model. In those studies, rice bran modified the microbial community composition but did not increase the abundance of Bifidobacterium and Lactobacillus species, which did increase with other fiber treatments, such as fructooligosaccharides (20, 21). In vivo studies using dietary interventions with mouse models showed that phenolics found in the fibers from rice bran had antihyperglycemic effects, and in an irritable bowel disease (IBD) model, fermented rice bran helped alleviate symptoms of IBD (22–24).
The beneficial effects of dietary fiber have been studied in relation to overall health as well as gut health for many years. However, there are few studies directly comparing fiber types from the same plant on the gut microbiota. To explore the effect of portions of rice bran that compose soluble fiber vs. those that compose insoluble fiber on the gut microbiota, we used an in vitro simulator of the human intestinal microbial ecosystem (SHIME®), mimicking the gut microbiota of three different healthy, adult donors. Genomic and metabolomic analyses were performed to determine differential effects between soluble and insoluble fiber rice bran components as part of a regular diet.
The SHIME® platform was operated following the manufacturer’s guidelines (Prodigest, Belgium) as described previously (25). The defined medium (DM) used in the bioreactors for this experiment was adult MSHIME media with added starch purchased from Prodigest (Ghent, Belgium) (25). Soluble (SF) and Insoluble fractions (ISF) from stabilized Rice Bran were procured from RiceBran Technologies, (Texas, USA). The media used in the control was supplemented with 5 g/L of either SF, ISF, or itself as the control. The DM plus ISF was sonicated for 10 min prior to being autoclaved. Pancreatic Juice (PJ) was made from 12.5 g/L NaHCO3 (Sigma-Aldrich, Saint Louis, MO), 6 g/L Bile Salts (BD, Franklin Lakes, NJ), and 0.9 g/L pancreatin (Sigma-Aldrich, Saint Louis, MO). Mucin-agar was prepared using 5% type II porcine mucin (Sigma-Aldrich) and 1% bacterial agar in sterile MilliQ water as described previously (25).
Screened, healthy, homogenized human fecal samples were obtained from OpenBiome (Cambridge, Massachusetts, USA) as described previously (25, 26). In brief, all 3 donor samples were harvested from 3 separate Americans with an average Body Mass Index (BMI), between the ages of 21 and 45 years of age, who had been free of antibiotics for at least 1 year and consumed a typical Western diet. After testing negative for pathogens, fecal samples were homogenized in a glycerol buffer solution to a 10% final concentration and stored at −80°C. Donors used for this study were selected at random from a group of consumers meeting these criteria.
The SHIME® (Prodigest, Ghent, Belgium) was set up and run similarly to what was described previously (27) with slight modifications. As illustrated in Figure 1, the bioreactors were split into sets of 3, for a total of 3 digestive systems per experimental run. For each set of digestive systems, there were bioreactors simulating the stomach/small intestine, the proximal colon (PC) and the distal colon (DC). The DM and PJ were pumped separately into the stomach/small intestine reactors, and then subsequently into the PC, the DC, and then to waste. Regions are differentiated by volume and pH control, where the stomach (140 mL) was held at pH of 1.9–2.1, the small intestine (200 mL) at a pH of 6.5–7.1, the PC (500 mL) at a pH of 5.6–5.9, and the DC (800 mL) at a pH of 6.6–6.9. Each PC and DC has a luminal (liquid) and mucosal (mucosal agar beads) phase as described previously (25). Inoculum from each donor was tested separately under control, SF, and ISF conditions. Each time, the experiment was performed for 2 weeks to establish community stability, and then 1 additional week for analysis. Luminal and mucosal samples from the cultivars were taken at regular intervals throughout the experiment and stored in a − 80°C freezer prior to analysis.
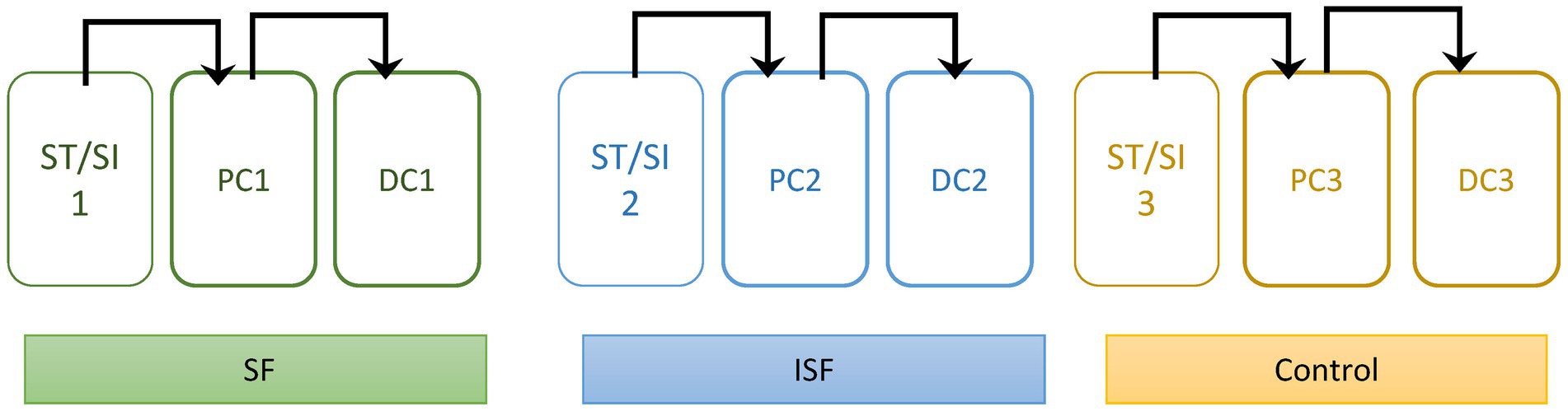
Figure 1. Diagram of Experimental set-up. Each colon has a stomach/small intestine reactor (ST/SI), a proximal colon (PC), and a distal colon (DC). Each colon was also given different treatments: soluble fraction of rice bran (SF), insoluble fraction of rice bran (ISF), and the control. Contents move sequentially through the colon as shown by the arrows.
DNA was extracted and analyzed at the Microbiome Center, Children’s Hospital of Philadelphia (CHOP) from bacterial pellets obtained at each sampling timepoint using the PowerSoil Pro DNA extraction kit (Qiagen, Hilden, Germany) as described previously (28). DNA was then analyzed using 16S rRNA-tagged gene sequencing. An Illumina Miseq (San Diego, CA, USA) was used to perform the sequencing using a 2 × 250 bp reagent kit following manufacturer guidelines. Libraries were generated as described previously, targeting the V4 region of the 16S rRNA gene (28). The negative controls used were extraction blanks and DNA-free water with the positive control being known amounts of 16S rRNA gene fragments.
Analysis and sample preparation was performed as described previously (29). In brief, samples were collected and stored in −80°C, then thawed immediately prior to analysis. After sample preparation, SCFAs were analyzed using GC/MS Shimadzu QP2010 Ultra (Shizmadzu, Columbia, MD, USA) equipped with a Stabilwax-DA column, 30 m, 0.25 mm ID, 0.25 μm (Restek Corporation, Bellefonte, PA, USA). 1 μL of each sample was injected in a 1:20 split mode at 260°C, with an interface temperature of 280°C and an ion source temperature of 220°C. A standard stock solution with a concentration of 5 mg/mL of each of the following: acetic acid, butanoic acid, 2-methylbutanoic acid, valeric acid, propionic acid, isobutanoic acid, hexanoic acid, heptanoic acid, 2-methylvaleric acid, 3-methylvaleric acid, 4-methylvaleric acid, 2-methylvutyric acid, and isovaleric acid was used as described previously (29). The data were acquired with a mass range of m/z 25–375 in full scan mode.
Sequence data were processed using QIIME2 (30). Read pairs were processed to identify amplicon sequence variants with DADA2 (31). Taxonomic assignments were generated by comparison to the Greengenes reference database (32), using the naive Bayes classifier implemented in scikit-bio (33). A phylogenetic tree was inferred from the sequence data using MAFFT (34). Similarity between samples was assessed by weighted and unweighted UniFrac distance (35, 36), as well as percent shared species (Jaccard index) and Bray-Curtis distance. PICRUSt2 was used to perform functional estimation based on the 16S rRNA community profiles (37).
Data files from QIIME were analyzed in the R environment for statistical computing, using the QIIMER library, which we developed.1 Global differences in bacterial community composition were visualized using Principal Coordinates Analysis. Community-level differences between sample groups were assessed using the PERMANOVA test, which allows sample-sample distances to be applied to an ANOVA-like framework (38). Statistical analysis and visualizations were done using R (v.4.1.3) (39) and the packages tidyverse (40) and ggplot2 (41). Differential estimated enzyme abundance (EC) was calculated using MaAsLin2 (42).
The ability of both soluble (SF) and insoluble (ISF) rice bran supplementation to elicit changes to the gut microbiota was tested using three donors via the SHIME platform. This platform was specifically selected because it allowed us to examine changes to both the proximal (PC) and distal (DC) colons while simultaneously mimicking both the luminal and mucosal communities for each donor. Following stabilization of the SHIME communities (2 weeks) (25), samples were taken for genomic analysis using 16S rRNA tagged sequencing to evaluate changes in community structure. Due to high interindividual variation of the gut microbiota, analyses are shown separated by donor, the region of the gut, and the type of community.
First, we considered richness, or the number of unique members of the community. By this measure, as seen in Figure 2A, both the luminal and mucosal communities showed broadly similar patterns of response within each donor, but these patterns varied across donors. Using this measure, we found a significant difference between the SF and ISF treatments, which was particularly strong in the proximal region of the colon with donors 1 and 3. Donor #2, however, did not show a significant difference between any of the conditions. Shannon’s diversity index (Figure 2B) was used to explore the heterogeneity of the microbial community, and we found again that the communities from donor #2 displayed a different pattern than donors 1 and 3. In donors 1 and 3, we found that supplementation with ISF increased diversity of the PC microbial communities to levels higher than that of either SF or the control. However, in the DC luminal communities, ISF supplementation did not affect microbial diversity whereas supplementation with SF caused a decrease in diversity. In the mucosal phase, however, we observed a decrease in diversity with ISF and SF supplementation compared with the control. The final measure of alpha diversity used to analyze these microbial communities was Faith’s phylogenetic diversity (Figure 2C), used to look for changes according to phylogeny and not by taxon alone. Using this measure we found a similar pattern, that donor 2 showed no significant changes in diversity associated with supplementation, whereas donors 1 and 3 showed an increase in diversity with ISF, especially in the PC. Similarly to the other alpha diversity measures discussed, in the DC, we observed a decrease in diversity with SF supplementation compared with control.
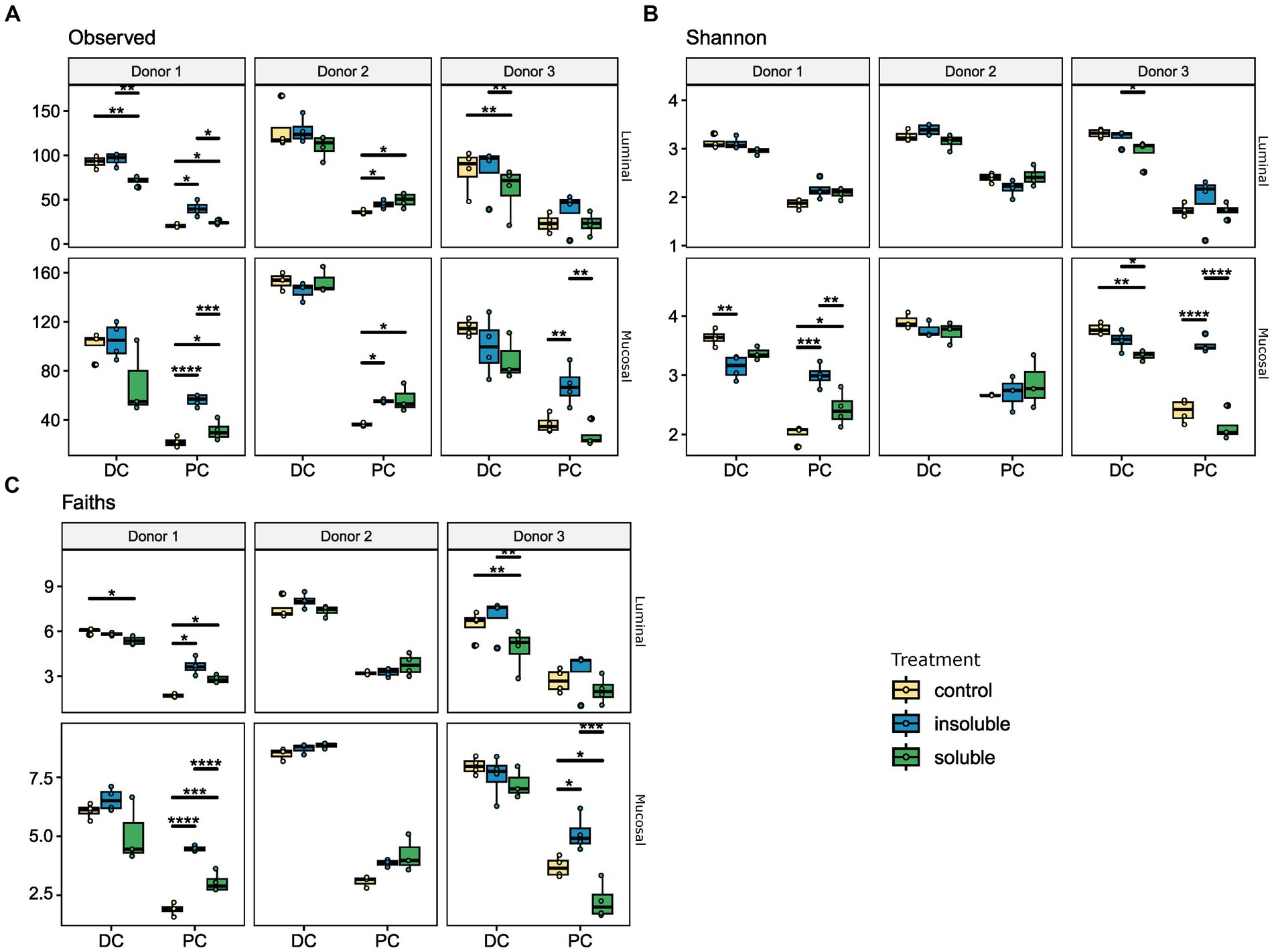
Figure 2. Alpha diversity analysis reveals significant changes with ISF and SF treatment. Donors 1–3, and luminal/mucosal phases are separated for this analysis. For all measures, PC and DC differ significantly. (A) Number of observed ASVs, for luminal and mucosal samples, ISF and SF differ significantly; (B) Shannon’s diversity index, for mucosal samples SF and ISF differ significantly; (C) Faith’s phylogenetic diversity, the luminal phase ISF differs significantly from control and SF, for mucosal phase ISF is significantly different from control. Significance was determined using Tukey’s multiple comparison of means. * = q < 0.05; ** = q < 0.01; *** = q < 0.001; **** = q < 0.0001.
Next, beta diversity was evaluated using the weighted UniFrac distance metric and portrayed via principal coordinate analysis (PCoA). In Figure 3, these results were separated by colon region (PC vs. DC) and phase (luminal vs. mucosal). Similarly, to the alpha diversity data, we found that in most cases, donor 2 communities were the most distinct from the other two donors and treatment, indicating that donor is the most important factor in response to supplementation of fiber. In the distal lumen and distal mucosa plots, communities were highly similar, with donor 3 showing the most divergence in the distal mucosal community. The PC communities were more diverse compared with the DC communities with the most changes between treatments found in the proximal mucosal communities. Statistical analysis performed using PERMANOVA found that supplementation with both SF and ISF caused significant changes (q < 0.05) in beta diversity compared with control but were not significantly different from each other. Overall, this analysis shows that the largest difference in microbial diversity for these communities is dependent on the donor rather than the type of supplement.
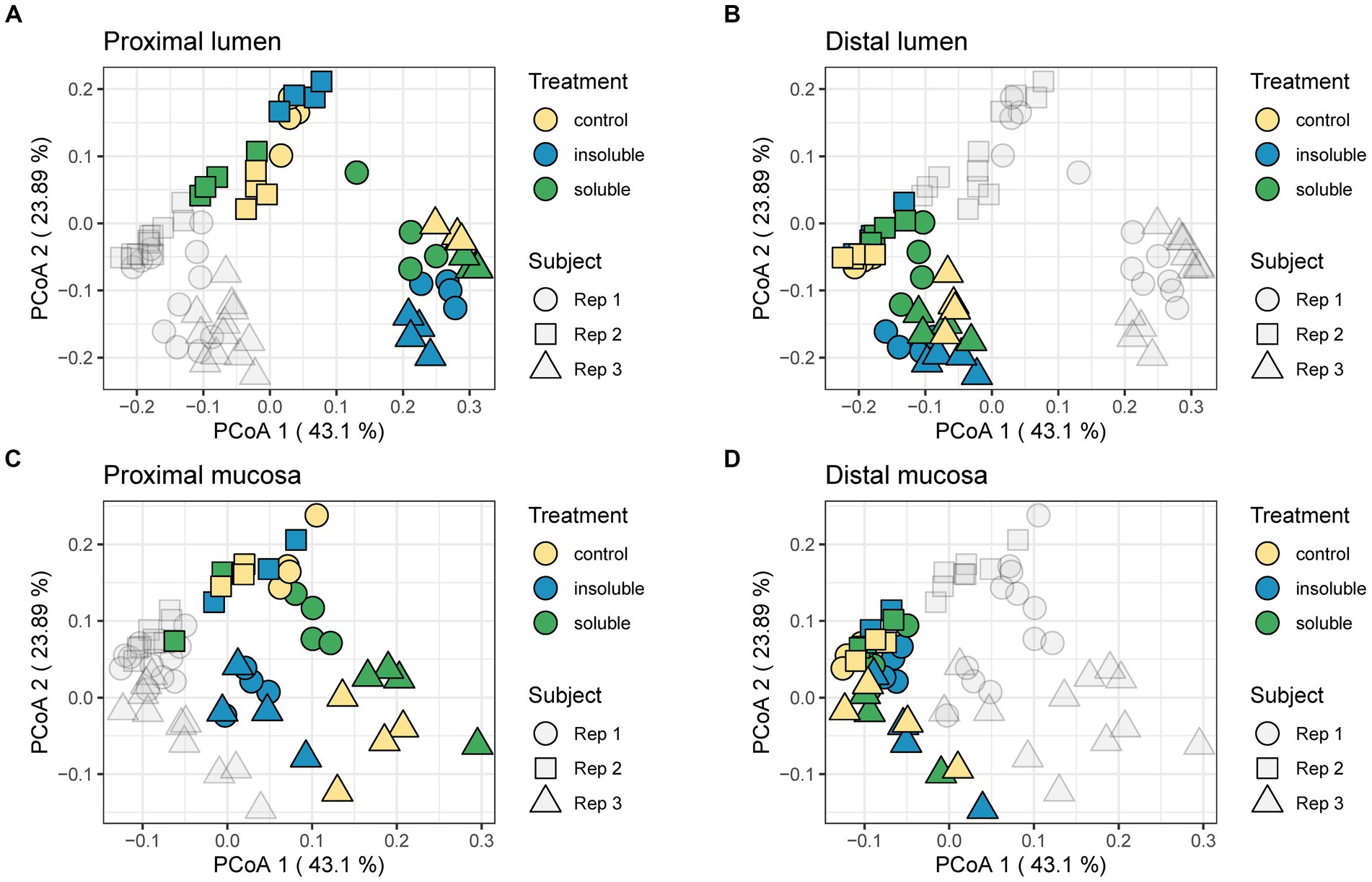
Figure 3. Weighted UniFrac distance shows shifts in the community. (A) Proximal lumen; (B) Distal lumen; (C) Proximal mucosa; (D) Distal mucosa. PERMANOVA analysis was used to determine significance. Control treatment differed significantly from ISF (F = 1.0078; q < 0.05) and SF (F = 0.6998; q < 0.05); ISF and SF did not differ significantly from each other regardless of region.
Although the results of UniFrac distances demonstrated that both SF and ISF elicited shifts in the microbial communities to the same extent, it did not provide information on which taxa were responding to these supplements. Using sequencing data from above, changes to the addition of SF and ISF were determined based on the relative abundance of the microbiota at the phylum level. Similar to the results of alpha diversity and UniFrac distances, supplementation with SF and ISF caused donor-dependent changes to the taxonomic structure at the phylum level (Figure 4). At this level, we found that both treatments caused a notable but not statistically significant increase in the Actinobacteria in the donor 1 communities, as well as a significant (q < 0.0001) increase in Fusobacteria in the DC in the lumen. This increase in Fusobacteria was also apparent in donor 3, but only with SF treatment (q < 0.05). In the mucosal phylum level analysis (Figure 4B), there is a similar pattern, though this part of the gut microbial community is more highly dominated by the Firmicutes than the luminal counterpart.
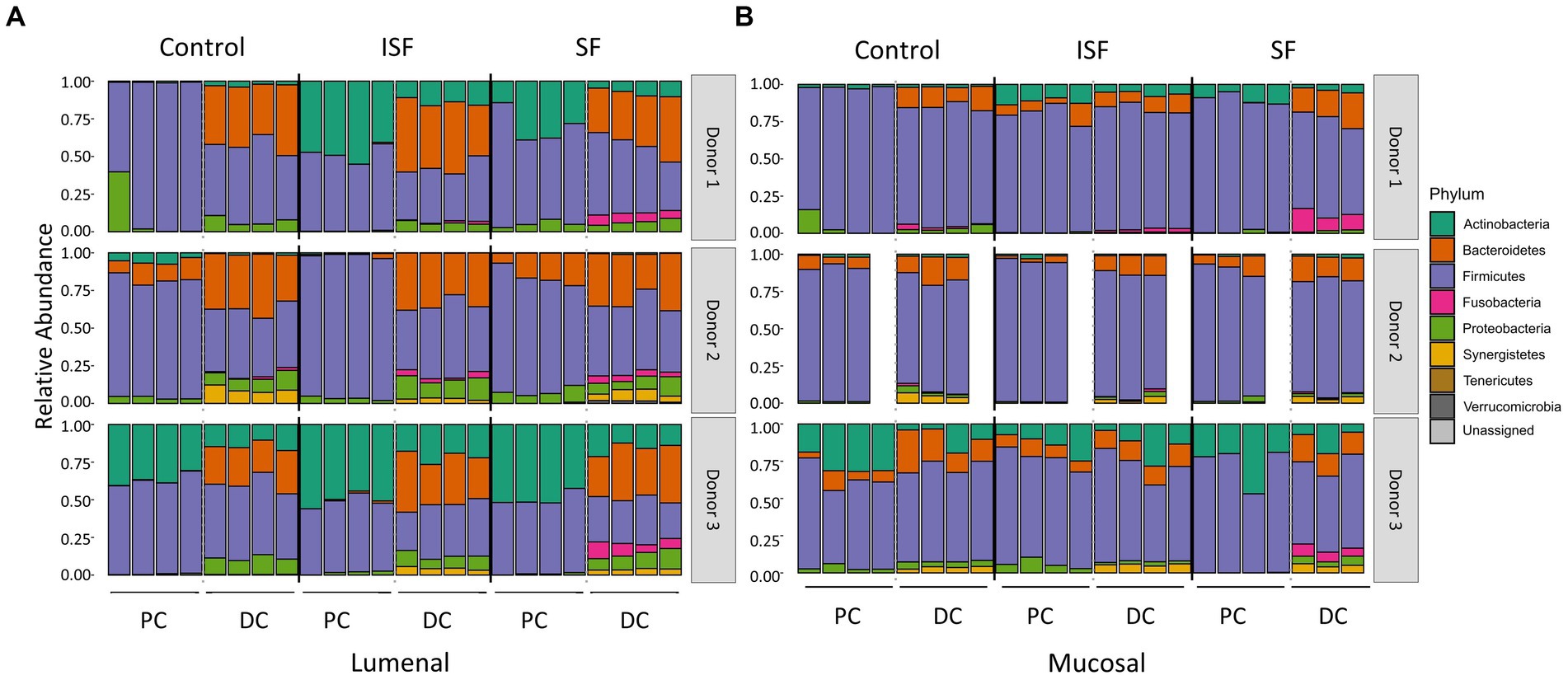
Figure 4. Phyla relative abundance of the luminal and mucosal communities. (A) Luminal community; (B) mucosal community. Statistical significance (q < 0.05) was determined via ANOVA.
Next, we examined the abundances of three bacterial families (Bifidobacteriaceae, Lachnospiraceae, and Lactobacillaceae) that are known to contribute to SCFA production and fermentation of fibers and are likely to have overlapping roles in metabolism (Figure 5, statistical significance shown in Supplementary Table S1). We observed that Bifidobacteriaceae is a key member of the community in donors 1 & 3 and is significantly increased with ISF and SF compared with control in the lumen for donor 1 (q < 0.0005, q < 0.05, respectively). In the mucosa, Bifidobacteriaceae increased significantly with SF and ISF treatment, Lachnospiraceae decreased significantly with ISF and SF treatment (q < 0.0005; q < 0.05), and Lactobacillaceae (q < 0.0005) decreased significantly with SF treatment in the PC of donor 1. Donor 2 exhibited a significant decrease in Bifidobacterium with for ISF (q < 0.005) and SF (q < 0.0005) treatment in both the luminal and mucosal community PCs, though the overall Bifidobacterium content in the gut microbiome is low in donor 2. Donor 2 had a larger abundance of both Lachospiraceae and Lactobacillaceae than the other two donors but did not experience a significant change in abundance of either of these families with either SF or ISF treatment. Donor 3 did not exhibit significant changes in Bifidobacterium abundance based on supplement type, however, donor 3 did have a substantial abundance of Bifidobacteriaceae in both the lumen and the mucosa, similar to donor 1. Lactobacillaceae and Lachnospiraceae did increase significantly (q < 0.0005) with ISF treatment in the luminal and mucosal phases of donor 3’s PC, but it is also worth noting that these families have low abundance in donor 3 overall.
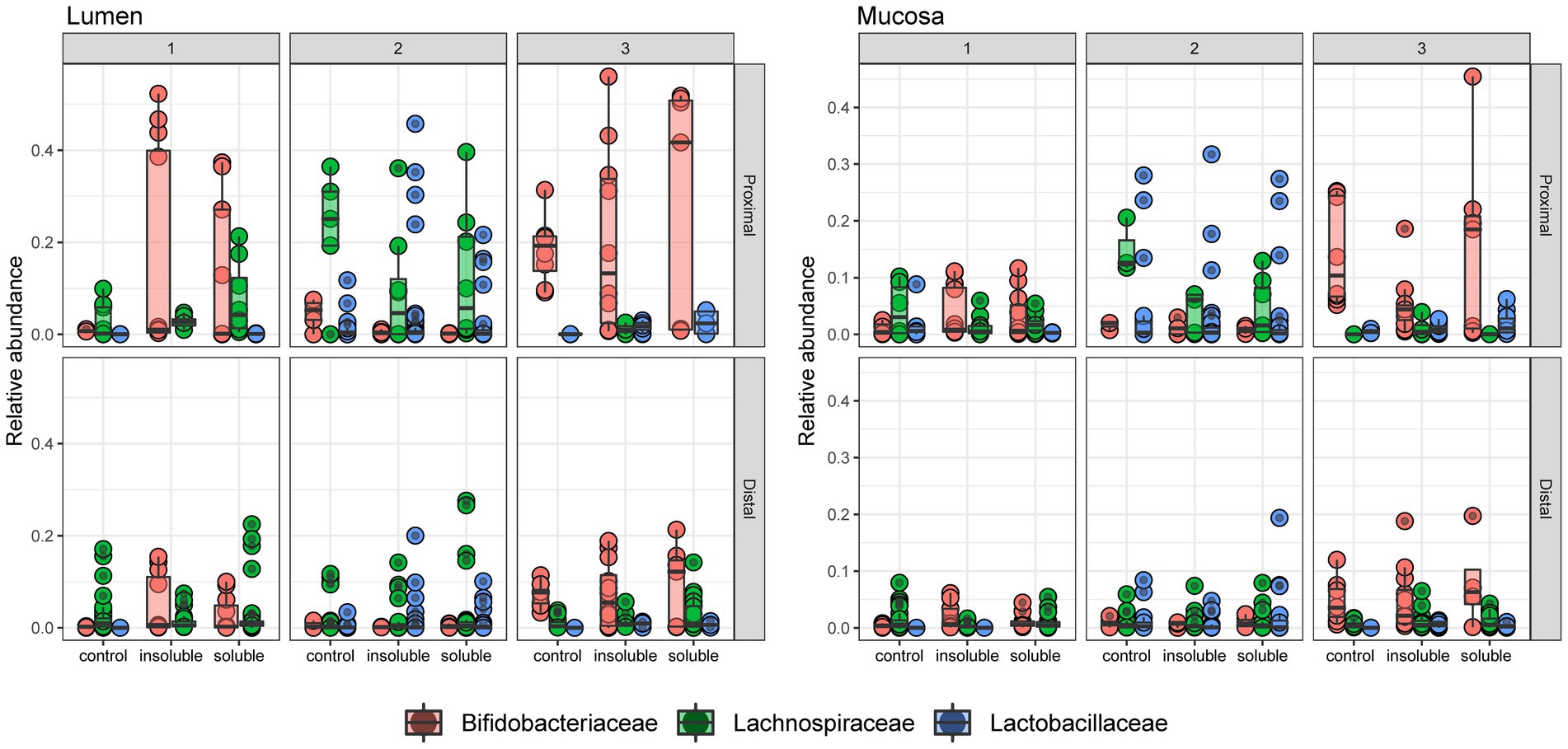
Figure 5. Relative abundance of 3 key taxa at the family level. Relative abundance of Bifidobacteriaceae, Lachnospiraceae, and Lactobacillaceae in each community. Significance determined via Tukey’s multiple comparison of means and shown in Supplementary Table S1.
This data was also used to look more closely at specific taxa known to ferment dietary fibers or that showed significant changes in response to these different supplementation options (Figure 6; Supplementary Table S2). As expected, changes seen at this level are donor-, region-, and phase-dependent. Bifidobacterium (Figure 6A) significantly increased in relative abundance with ISF in the PC regions for donor in the luminal phase (q < 0.0005) and the mucosal phase (q < 0.0005), with a smaller, non-significant increase in the DC as well. In the mucosal phase, ISF decreased in relative abundance of Bifidobacterium in comparison with control for donor 3, however this change was not statistically significant. Treatment with SF also increased Bifidobacterium in the luminal phase of the PC in donor 1. Conversely, ISF (q < 0.005) and SF (q < 0.0005) both caused a significant decrease in Bifidobacterium abundance in the PC of donor 2, however the starting abundance of Bifidobacterium was already low.
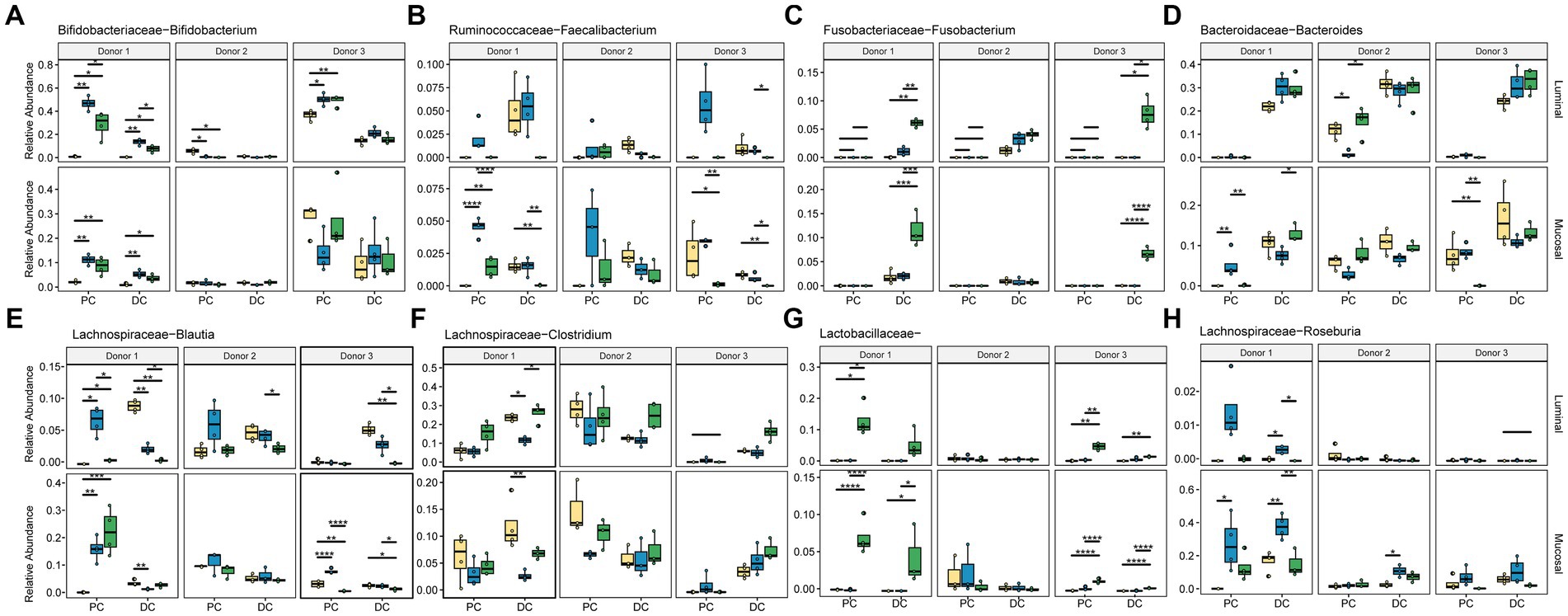
Figure 6. Relative abundance of significant taxa. Colors are as found in previous figures: yellow indicates control, blue indicates ISF, green indicates SF. Significance was determined using ANOVA and is shown in Supplementary Table S2. (A) Bifidobacteriaceae Bifidobacterium; (B) Ruminococcaceae Faecalibacterium; (C) Fusobacteriaceae Fusobacterium; (D) Bacteroidaceae Bacteroides; (E) Lachnospiraceae Blautia; (F) Lachnospiraceae Clostridium; (G) Lactobacillaceae; (H) Lachnospiraceae Roseburia. * = q < 0.05; ** = q < 0.01; *** = q < 0.001; **** = q < 0.0001.
Faecalibacterium (Figure 6B) relative abundance significantly increased with ISF treatment only in donors 1 and 3 which was evident in the luminal PC. SF, on the other hand, significantly increased the abundance of Fusobacterium in the DC of donor 1 in both the luminal and mucosal phases (Figure 6C). The relative abundance of Bacteroides (Figure 6D), one of the more abundant members of the gut microbiota, did not change significantly with either fiber treatment.
The Lachnospiraceae family had the most members that changed significantly with fiber supplementation, including Blautia, and Clostridium. Blautia (Figure 6E) was overall more abundant in the PC but increased in abundance with ISF treatment in the luminal phase (q < 0.005) and mucosal phase (q < 0.05) for donor 1. With SF treatment, Blautia saw an increase with the PC of donor 1 (q < 0.05) and the DC of donor 3 (q < 0.05). Clostridium (Figure 6F) abundance significantly increased with ISF treatment for donor 1 in the DC, luminal and mucosal phases (q < 0.05), and it increased abundance with SF treatment in the DC of donor 3, luminal and mucosal phases (q < 0.0005). Roseburia, on the other hand, appears to reside only in the mucosal phase of the colons, and did not change in a statistically significant way. For this taxon (Figure 6H), we found that treatment with ISF increased abundance over control and SF in both the PC and the DC with donor 1, but in the other two donors was mostly present in the DC. Donor 1 also showed an increased in abundance of Roseburia, but only in the PC, and again this change was not significant. Another category, the otherwise un-categorized Lactobacillaceae genera (Figure 6G), showed significant increases in abundance with SF treatment, however, this is largely the case with donor 1 and donor 3 (q < 0.0005) in the PC. However, for donor 1 there was a smaller, but still significant increase in the DC as well (q < 0.05). Overall, our data demonstrates that responses to these rice bran supplements are highly dependent on individual taxa and donor communities.
To understand how the gut microbiota changed functionally due to supplementation, we performed GC–MS/MS to analyze the concentration of SCFAs for each donor under each condition (Figure 7; Supplementary Table S3). As expected, overall concentrations of SCFAs increased in the DC compared with the PC regardless of treatment type. Interestingly, donors 2 and 3 had similar responses to treatment with respect to SCFA concentrations, whereas the response of donor 1 was much more unique. For example, acetic acid increased with both ISF and SF supplementation for donor 1 (q < 0.0005) but decreased for donors 2 and 3 (q < 0.0005). Propanoic acid decreased in response to ISF in the PC of donor 1 (q < 0.0005) and the DC (q < 0.05) of donor 3 but increased for donor 2 in both the PC and DC (q < 0.0005). Both treatment groups increased butanoic acid for donors 2 and 3 in the PC, though only SF with donor 3 showed a significant change (q < 0.0005) but SF treatment decreased butanoic acid in concentration for donor 1 (q < 0005). The concentration of pentanoic acid was very low in donor 1 but was present in the DC of donors 2 and 3. Supplementation with SF decreased pentanoic acid in both donors, but only significantly so with donor 3 (q < 0.05). ISF caused an increase in pentanoic acid concentration in donor 2, (though this change was not significant) while decreasing the concentration in donor 3 (q < 0.05). Branched-chain short-chain fatty acids (BCSCFAs) 3-methylbutanoic acid, 2-methylpropanoic acid, and 2-methylbutanoic acid had similar responses to the treatments. In donor 1, these compounds were only present in the DC, and both rice bran supplements increased the concentration of all three BCSCFAs (q < 0.0005). In donor 2, SF decreased the concentrations of the 3 BCSCFAs in both the PC and the DC, whereas 3-methylbutanoic acid and 2-methylpropanoic acid increased in the PC with ISF supplementation, however these changes were not statistically significant. With donor 3, all 3 BCSCFAs increased in response to ISF in both the PC and the DC (significance shown in Supplementary Table S3), whereas the SF treatment remained the same as control in the PC but slightly decreased in the DC. This data, along with that of the microbial community diversity and taxon abundance confirms that the response of the gut microbiota to SF and ISF supplementation depends largely on the native microbial community.
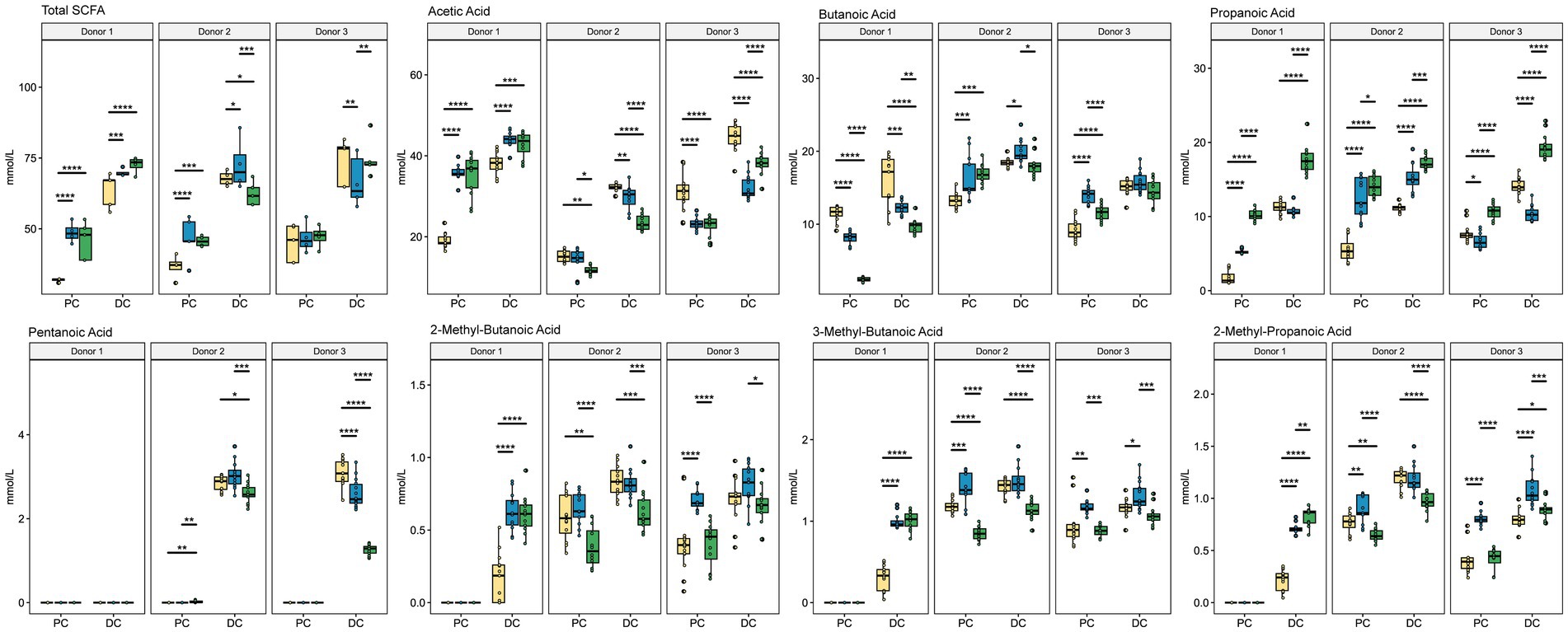
Figure 7. SCFA Concentration. Colors are as found in previous figures: yellow indicates control, blue indicates ISF, and green indicates SF. Significance was determined using Tukey’s multiple comparisons of means (q < 0.05) shown in Supplementary Table S3. * = q < 0.05; ** = q < 0.01; *** = q < 0.001; **** = q < 0.0001.
Analysis of community structure and SCFAs portrayed high interindividual variability in the response of the gut microbiota to both SF and ISF. As each microbiome is distinct, this variability was somewhat expected, however, we hypothesized that while the taxa and functional response may have been dependent on the donors’ starting community structure, genetic pathways enriched due to SF and ISF would be similar and not driven by interindividual variability. Therefore, PICRUSt2 was used to predict the presence of genes that may be responsible for cellulose metabolism within these gut microbial communities. A portion of this metabolic pathway is shown in Figure 8, illustrating enzymes whose abundance was significantly altered by either SF or ISF treatment. The taxa relevant to each enzyme are shown in adjacent heatmaps. In this figure, Lachnospiraceae is a major contributor to the enzymes involved in the conversion of cellulose to cellobiose, and for some of the Lachnospiraceae family members, they are only associated with specific treatment types. For example, Coprococcus is only present with control and ISF use and Roseburia is present with both ISF and SF use, but not present for the control for cellulase (EC3.2.1.4).
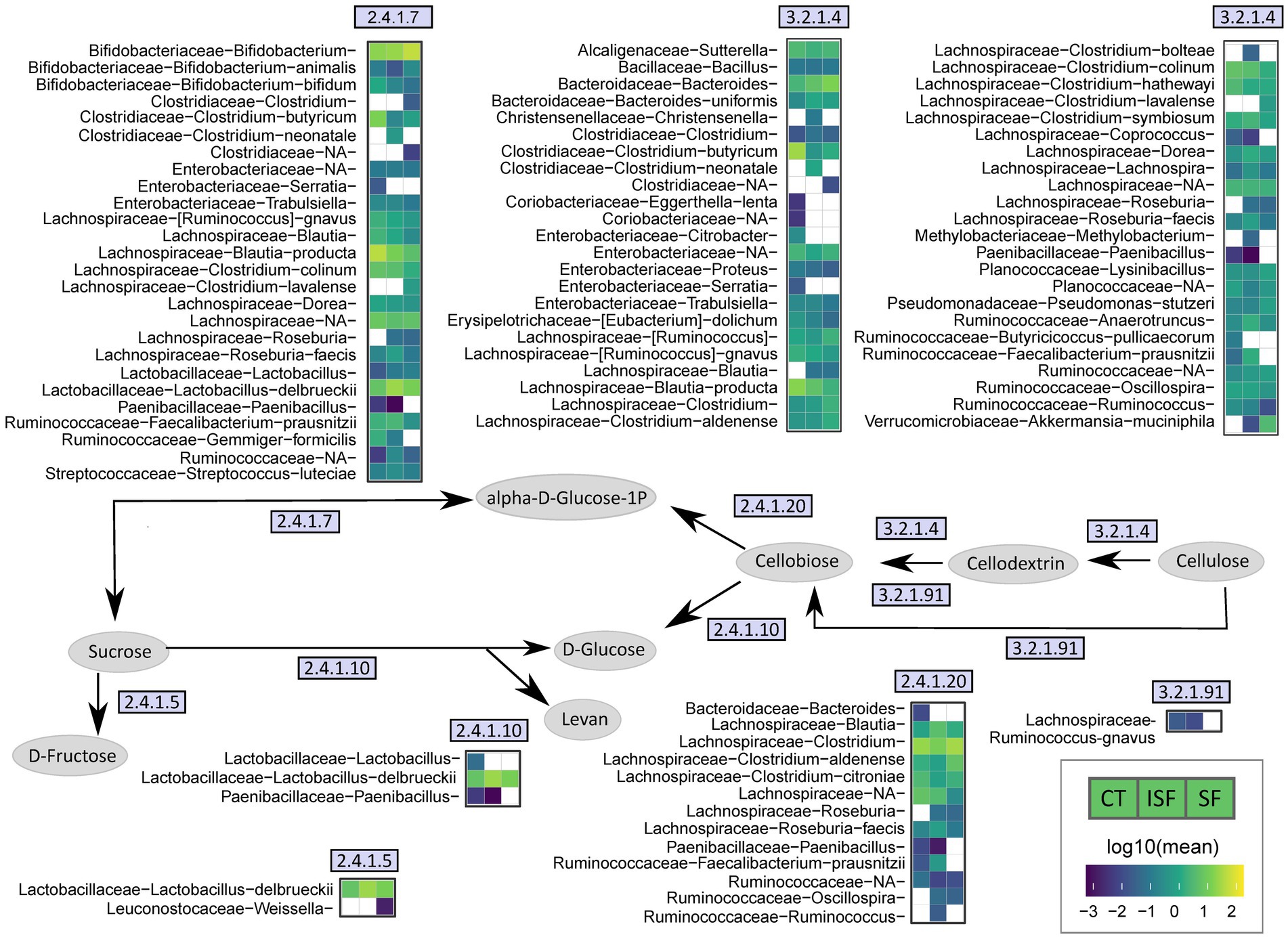
Figure 8. Simplified cellulose metabolism pathway. This figure illustrates a part of the cellulose metabolism pathway by the gut microbiota, using EC numbers to identify genes associated with enzymes involved in the process that are significantly changed with either ISF or SF supplementation. Heatmaps demonstrate specific taxa that are associated with each enzyme and may contribute to its activity.
Lactobacillus delbrueckii is associated with sucrose phosphorylase (EC 2.4.1.7), levansucrase (EC 2.4.1.10), and dextransucrase (EC 2.4.1.5), and has a greater abundance with ISF and SF compared with control. Bifidobacterium appears to be more represented with SF than with control or ISF with sucrose phosphorylase (EC 2.4.1.7). From our relative abundance data shown above, we know that Bifidobacterium has a greater presence with donors 1 and 3, while Lactobacillus delbrueckii has a greater abundance with donor 2 (Figure 5). This indicates that Bifidobacterium and Lactobacillus delbrueckii may have overlapping roles in starch metabolism, and that this functional redundancy is likely what allows for gut microbiota from different donors to ferment the same dietary fibers.
Continued industrialization and westernization of communities around the world has led to an increase in gut health disorders, including Crohn’s disease and other irritable bowel diseases, as well as related problems such as obesity, diabetes, and autoimmune disorders (43, 44). Many of these changes are thought to be due to the increase in consumption of processed foods with a reduction of whole foods in the diet, especially high-fiber, nutrient-dense vegetables, and whole grains. These dietary changes that have occurred over time have gradually lessened the diversity of the average person’s gut microbial community, a change that continues with each subsequent generation (45). In this study, we used by-products of rice processing in the form of rice bran fractionated into soluble and insoluble parts, largely fibers, to find how these supplements can shape the gut microbiota over time. We used the SHIME® platform to determine how the same starting inoculum from 3 donors responded to growth media supplemented with either fraction of the rice bran fiber compared with the control.
Through genomic analysis, this study found that supplementation of both SF and ISF caused changes in the gut microbial community but that the nature and direction of these changes varied by donor. Surprisingly, we observed a greater increase in alpha diversity with ISF than with SF compared with control. While the common understanding is that soluble fiber is easily fermented by the gut microbiota while insoluble fiber is not, our findings that the ISF fractions change the gut microbiota are not unheard of considering other recent research looking into the effects of insoluble fiber from other staple crops on the gut microbiota (14, 16). In previous work, researchers found that insoluble fiber from soybeans increased the diversity of the mouse gut microbiota, the relative abundance of specific taxa, and the production of SCFAs, indicating that fermentation of these insoluble fibers was indeed occurring (14). In an in vitro study using human feces as the starting material, other researchers found that the use of insoluble fibers from soy husks modified the gut microbiota via an increase in alpha diversity and an increase in specific taxa, with a notable increase in Bifidobacterium (16). In this study, we observed a similar increase in alpha diversity with ISF, however, changes in Bifidobacterium abundance were more variable and were highly dependent on donor, community type, and colon region (PC or DC). Bifidobacterium are an integral part of a healthy gut microbiota, and members of the genus are frequently used as probiotics and as part of functional foods to improve gut health (46). Here, we found that for Donors 1 and 3, Bifidobacterium was either unchanged or increased in both the PC and DC regions of the lumen, however, Bifidobacterium abundances were higher in the PC than the DC, suggesting that their preferred substrates are available in the PC and have likely been depleted before reaching the DC. This pattern holds in the mucosal for donors 1 and 3 except for the PC of donor 3, where Bifidobacterium abundances were lower with treatment compared to controls, however, it is important to note that as these are relative abundances, these decreases could reflect an increase in other taxa, rather than a decrease in Bifidobacteria. Interestingly, Bifidobacteriaceae was almost completely absent in donor 2 and appeared to be replaced by the members of the Lachnospiraceae. For donor 2, which did not have a large community presence of Bifidobacterium, we did find that other family members also involved in fiber metabolism are likely to be involved, most notably Lactobacillus delbrueckii, which has been shown previously to be involved in inulin metabolism via the ability to uptake inulin and degrade it as a source of carbon for growth (47). L. delbrueckii is a probiotic strain frequently used in culturing yogurt, which may suggest different dietary habits of the donors used in this study (48).
Another taxon that increased significantly with ISF treatment in our study was Faecalibacterium, which increased significantly in abundance in the PC for all donors. Species within Faecalibacterium are associated with a lower incidence of IBD and other diseases related to colonic inflammation and have been demonstrated to produce SCFAs, including acetate and butyrate (49). The abundance of Faecalibacterium has been significantly increased in previous studies with the use of other dietary fibers, like inulin in human studies (50). The increase of this genus with ISF treatment may contribute to the increase in acetic acid and butanoic acid seen in some of our donors as well. These findings, that similar members of the gut microbiota are involved in degradation of additional fiber types, like those found in rice bran, highlight that there is functional redundancy integral to the gut microbiota. This is likely what allows for beneficial effects from an increase in dietary fiber for most individuals, despite individual differences in the gut microbial community.
The family with the most significant changes in abundance with either SF or ISF supplementation was the Lachnospiraceae family. This family is a major contributor to the production of SCFAs, greater production of which is associated with better health outcomes (51–53). However, the impact of various players within Lachnospiraceae on human health is more controversial. Blautia and Roseburia, two genera that increased with ISF and SF compared with control, are major SCFA producers and have been previously demonstrated as beneficial for their anti-inflammatory properties and their contribution to the health of the immune system (54, 55).
The fermentation of dietary fibers by the gut microbiota produces SCFAs as microbial metabolites that are important for gut and overall health. For example, acetic acid has been shown to alleviate aging symptoms in gut health of mice (56). The production of butanoic acid in particular is known to benefit tight-junction formation in the gut, and therefore increase barrier integrity of the gut (57). Butanoic acid is also linked to the production of mucus by the goblet cells of the large intestine (58). Propanoic acid has been shown to have anti-inflammatory effects, to improve insulin sensitivity, and improve satiety (59). Our study demonstrated increases in SCFA production, suggesting broader trends, however many of these changes were subject to interindividual variability. For example, the reactors from all donors had significantly increased propionate concentrations in the SF relative to controls in both the PC and DC, while the propionate response to ISF was mixed. Acetic acid production displayed a more pronounced donor effect, with significant increases in acetic acid production in ISF and SF relative to controls in donor 1 reactors, with the opposite effect in donor 3 reactions. Interestingly in the case of donor 3 reactors, the acetic acid production in the control samples was already among the highest measured concentrations. Similarly, for butanoic acid, donors 2 and 3 had increases or stable concentrations of butanoic acid in the supplemented samples relative to controls with an opposite trend for donor 1 where the control samples had the highest concentrations. As with acetic acid, in those communities where the concentrations were highest in the control samples, it’s possible that those communities were already predisposed to certain pathways. The extent of inter-individual variability in gut microbiota is well known and likely underlies the complex SCFA results shown here. This variability can be seen in taxa known to be associated with SCFA production and mirrors the changes in the population of SCFA producing members of the gut microbiota. Combined with our results, we interpret the increase in these families to indicate that supplementation with both SF and ISF may have beneficial effects on the gut microbiota.
Overall, this study determined that both soluble and insoluble fractions of fiber from rice bran elicited a microbial response, however, the nature of the response varied among the donor and colon regions. This is likely due to the different initial states of the donors’ microbial communities and agrees with the well-known phenomenon of a high level of variability among individuals and the gut microbiome (60). These nuanced variations are important to consider and underpin the growing fields of personalized nutrition and personalized medicine and underscores the need for fine-grained sampling across donors and colon regions and additional such studies with larger cohorts, however, overall, these results suggest a potential use of fiber from rice bran as a functional food which merits continued exploration.
The metabolite analysis data is available as supplementary material, found in Supplemental Table 4. The original sequencing contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1032541/.
Ethical approval was not required for the studies involving humans because fecal samples were purchased from Openbiome, and acquired from a by-product of routine care or industry. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
KM: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. LL: Conceptualization, Investigation, Writing – review & editing. JB: Conceptualization, Investigation, Methodology, Writing – review & editing. AN: Conceptualization, Data curation, Formal analysis, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. JF: Investigation, Methodology, Writing – review & editing. KB: Data curation, Formal analysis, Writing – review & editing. WH: Data curation, Formal analysis, Writing – review & editing. SJ: Investigation, Methodology, Writing – review & editing. AM: Investigation, Methodology, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the USDA In-House Project 8072-41000-108-00-D, “In Vitro Human Intestinal Microbial Ecosystems: Effect of Diet.” This research used resources provided by the SCINet project of the USDA Agricultural Research Service, ARS project number 0500-00093-001-11-D.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture (USDA). USDA is an equal opportunity provider and employer.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2024.1304045/full#supplementary-material
Supplementary Table 4 | Gas-Chromatography-Mass Spectrometry analysis of short-chain fatty acid content raw data in mmol/L. RB1/2/3 indicates donors 1, 2, 3; UC indicates proximal colon, DC indicates distal colon.
1. Gidley, MJ, and Yakubov, GE. Functional categorisation of dietary fibre in foods: beyond ‘soluble’vs ‘insoluble’. Trends Food Sci. (2019) 86:563–8. doi: 10.1016/j.tifs.2018.12.006
2. Anderson, JW, Baird, P, Davis, RH Jr, Ferreri, S, Knudtson, M, Koraym, A, et al. Health benefits of dietary fiber. Nutr Rev. (2009) 67:188–205. doi: 10.1111/j.1753-4887.2009.00189.x
3. Chuang, S-C, Norat, T, Murphy, N, Olsen, A, Tjønneland, A, Overvad, K, et al. Fiber intake and total and cause-specific mortality in the European prospective investigation into Cancer and nutrition cohort. Am J Clin Nutr. (2012) 96:164–74. doi: 10.3945/ajcn.111.028415
4. Threapleton, DE, Greenwood, DC, Evans, CE, Cleghorn, CL, Nykjaer, C, Woodhead, C, et al. Dietary fibre intake and risk of cardiovascular disease: systematic review and meta-analysis. BMJ. (2013) 347:6879. doi: 10.1136/bmj.f6879
5. He, Y, Wang, B, Wen, L, Wang, F, Yu, H, Chen, D, et al. Effects of dietary fiber on human health. Food Sci Hum Wellness. (2022) 11:1–10. doi: 10.1016/j.fshw.2021.07.001
6. NPV, P, and Joye, IJ. Dietary fibre from whole grains and their benefits on metabolic health. Nutrients. (2020) 12:3045. doi: 10.3390/nu12103045
7. Baniwal, P, Mehra, R, Kumar, N, Sharma, S, and Kumar, S. Cereals: functional constituents and its health benefits. Pharm Innov. (2021) 10:343–9. doi: 10.22271/tpi.2021.v10.i2e.5681
8. Slavin, JL. Position of the American dietetic association: position of the American dietetic association: health implications of dietary Fiber. J Am Diet Assoc. (2008) 108:1716–31. doi: 10.1016/j.jada.2008.08.007
9. Flint, HJ, Scott, KP, and Louis, P. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. (2012) 9:577–89. doi: 10.1038/nrgastro.2012.156
10. Xiong, R-G, Zhou, D-D, Wu, S-X, Huang, S-Y, Saimaiti, A, Yang, Z-J, et al. Health benefits and side effects of short-chain fatty acids. Food Secur. (2022) 11:2863. doi: 10.3390/foods11182863
11. De Vadder, F, Kovatcheva-Datchary, P, Goncalves, D, Vinera, J, Zitoun, C, Duchampt, A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. (2014) 156:84–96. doi: 10.1016/j.cell.2013.12.016
12. Lentle, RG, and Janssen, PWM. Manipulating digestion with foods designed to change the physical characteristics of Digesta. Crit Rev Food Sci Nutr. (2010) 50:130–45. doi: 10.1080/10408390802248726
13. Ozyurt, VH, and Ötles, S. Effect of food processing on the physicochemical properties of dietary fibre. Acta Sci Pol Technol Aliment. (2016) 15:233–45. doi: 10.17306/J.AFS.2016.3.23
14. Wang, B, Yu, H, He, Y, Wen, L, Gu, J, Wang, X, et al. Effect of soybean insoluble dietary fiber on prevention of obesity in high-fat diet fed mice via regulation of the gut microbiota. Food Funct. (2021) 12:7923–37. doi: 10.1039/D1FO00078K
15. Ge, Q, Li, H-q, Zheng, Z-y, Yang, K, Li, P, Xiao, Z-q, et al. In vitro fecal fermentation characteristics of bamboo insoluble dietary fiber and its impacts on human gut microbiota. Food Res Int. (2022) 156:111173. doi: 10.1016/j.foodres.2022.111173
16. Yang, L, Zhao, Y, Huang, J, Zhang, H, Lin, Q, Han, L, et al. Insoluble dietary fiber from soy hulls regulates the gut microbiota in vitro and increases the abundance of bifidobacteriales and lactobacillales. J Food Sci Technol. (2020) 57:152–62. doi: 10.1007/s13197-019-04041-9
17. Muthayya, S, Sugimoto, JD, Montgomery, S, and Maberly, GF. An overview of global rice production, supply, trade, and consumption. Ann N Y Acad Sci. (2014) 1324:7–14. doi: 10.1111/nyas.12540
18. Gul, K, Yousuf, B, Singh, A, Singh, P, and Wani, AA. Rice bran: nutritional values and its emerging potential for development of functional food—a review. Bioact Carbohydr Diet Fibre. (2015) 6:24–30. doi: 10.1016/j.bcdf.2015.06.002
19. Goffman, FD, Pinson, S, and Bergman, C. Genetic diversity for lipid content and fatty acid profile in rice bran. J Am Oil Chem Soc. (2003) 80:485–90. doi: 10.1007/s11746-003-0725-x
20. Pham, T, Teoh, K, Savary, BJ, Chen, M-H, McClung, A, and Lee, S-O. In vitro fermentation patterns of Rice bran components by human gut microbiota. Nutrients. (2017) 9:1237. doi: 10.3390/nu9111237
21. Mahalak, KK, Firrman, J, Narrowe, AB, Hu, W, Jones, SM, Bittinger, K, et al. Fructooligosaccharides (FOS) differentially modifies the in vitro gut microbiota in an age-dependent manner. Front Nutr. (2023) 9:1058910. doi: 10.3389/fnut.2022.1058910
22. Yao, W, Gong, Y, Li, L, Hu, X, and You, L. The effects of dietary fibers from rice bran and wheat bran on gut microbiota: an overview. Food Chem X. (2022) 13:100252. doi: 10.1016/j.fochx.2022.100252
23. Zhang, X, Dong, L, Jia, X, Liu, L, Chi, J, Huang, F, et al. Bound phenolics ensure the antihyperglycemic effect of rice bran dietary fiber in db/db mice via activating the insulin signaling pathway in skeletal muscle and altering gut microbiota. J Agric Food Chem. (2020) 68:4387–98. doi: 10.1021/acs.jafc.0c00584
24. Shibayama, J, Kuda, T, Shikano, A, Fukunaga, M, Takahashi, H, Kimura, B, et al. Effects of rice bran and fermented rice bran suspensions on caecal microbiota in dextran sodium sulphate-induced inflammatory bowel disease model mice. Food Biosci. (2018) 25:8–14. doi: 10.1016/j.fbio.2018.07.002
25. Liu, L, Firrman, J, Tanes, C, Bittinger, K, Thomas-Gahring, A, Wu, GD, et al. Establishing a mucosal gut microbial community in vitro using an artificial simulator. PLoS One. (2018) 13:e0197692. doi: 10.1371/journal.pone.0197692
26. Firrman, J, Liu, L, Van den Abbeele, P, Tanes, C, Bittinger, K, and Tomasula, P. Applying advanced in vitro culturing technology to study the human gut microbiota. J Vis Exp. (2019) 144:59054. doi: 10.3791/59054
27. Mahalak, KK, Firrman, J, Lee, J-J, Bittinger, K, Nuñez, A, Mattei, LM, et al. Triclosan has a robust, yet reversible impact on human gut microbial composition in vitro. PLoS One. (2020) 15:e0234046. doi: 10.1371/journal.pone.0234046
28. Mahalak, KK, Firrman, J, Bobokalonov, J, Narrowe, AB, and Bittinger, K. Persistence of the Probiotic Lacticaseibacillus rhamnosus Strain GG (LGG) in an In Vitro Model of the Gut Microbiome. Int J Mol Sci. (2022) 23:12973. doi: 10.3390/ijms232112973
29. Mahalak, KK, Bobokalonov, J, Firrman, J, Williams, R, Evans, B, Fanelli, B, et al. Analysis of the ability of capsaicin to modulate the human gut microbiota in vitro. Nutrients. (2022) 14:1283. doi: 10.3390/nu14061283
30. Bolyen, E, Rideout, JR, Dillon, MR, Bokulich, NA, Abnet, CC, Al-Ghalith, GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. (2019) 37:852–7. doi: 10.1038/s41587-019-0209-9
31. Callahan, BJ, McMurdie, PJ, Rosen, MJ, Han, AW, Johnson, AJA, and Holmes, SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. (2016) 13:581–3. doi: 10.1038/nmeth.3869
32. McDonald, D, Price, MN, Goodrich, J, Nawrocki, EP, DeSantis, TZ, Probst, A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. (2012) 6:610–8. doi: 10.1038/ismej.2011.139
33. Bokulich, NA, Kaehler, BD, Rideout, JR, Dillon, M, Bolyen, E, Knight, R, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. (2018) 6:1–17. doi: 10.1186/s40168-018-0470-z
34. Katoh, K, and Standley, DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. (2013) 30:772–80. doi: 10.1093/molbev/mst010
35. Lozupone, C, and Knight, R. Uni Frac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. (2005) 71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005
36. Lozupone, CA, Hamady, M, Kelley, ST, and Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. (2007) 73:1576–85. doi: 10.1128/AEM.01996-06
37. Douglas, GM, Maffei, VJ, Zaneveld, JR, Yurgel, SN, Brown, JR, Taylor, CM, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. (2020) 38:685–8. doi: 10.1038/s41587-020-0548-6
38. Anderson, MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. (2001) 26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
39. Team, R. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing (2022).
40. Wickham, H, Averick, M, Bryan, J, Chang, W, McGowan, L, François, R, et al. Welcome to the Tidyverse. J Open Source Softw. (2019) 10:1686. doi: 10.21105/joss.01686
42. Mallick, H, Rahnavard, A, McIver, LJ, Ma, S, Zhang, Y, Nguyen, LH, et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput Biol. (2021) 17:e1009442. doi: 10.1371/journal.pcbi.1009442
43. Sonnenburg, ED, and Sonnenburg, JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. (2014) 20:779–86. doi: 10.1016/j.cmet.2014.07.003
44. Jew, S, Abu Mweis, SS, and Jones, PJ. Evolution of the human diet: linking our ancestral diet to modern functional foods as a means of chronic disease prevention. J Med Food. (2009) 12:925–34. doi: 10.1089/jmf.2008.0268
45. Sonnenburg, ED, Smits, SA, Tikhonov, M, Higginbottom, SK, Wingreen, NS, and Sonnenburg, JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. (2016) 529:212–5. doi: 10.1038/nature16504
46. O'Callaghan, A, and Van Sinderen, D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol. (2016) 7:925. doi: 10.3389/fmicb.2016.00925
47. Tsujikawa, Y, Ishikawa, S, Sakane, I, Yoshida, K-i, and Osawa, R. Identification of genes encoding a novel ABC transporter in Lactobacillus delbrueckii for inulin polymers uptake. Sci Rep. (2021) 11:16007. doi: 10.1038/s41598-021-95356-1
48. Lesniewska, V, Rowland, I, Cani, PD, Neyrinck, AM, Delzenne, NM, Naughton, PJJA, et al. Effect on components of the intestinal microflora and plasma neuropeptide levels of feeding Lactobacillus delbrueckii, Bifidobacterium lactis, and inulin to adult and elderly rats. Appl Environ Microbiol. (2006) 72:6533–8. doi: 10.1128/AEM.00915-06
49. Ferreira-Halder, CV, AVS, F, and Andrade, SS. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract Res Clin Gastroenterol. (2017) 31:643–8. doi: 10.1016/j.bpg.2017.09.011
50. Ramirez-Farias, C, Slezak, K, Fuller, Z, Duncan, A, and Holtrop, G. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr. (2008) 101:541–50. doi: 10.1017/S0007114508019880
51. Biddle, A, Stewart, L, Blanchard, J, and Leschine, SJD. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity. (2013) 5:627–40. doi: 10.3390/d5030627
52. Wong, J, Piceno, YM, DeSantis, TZ, Pahl, M, and Andersen, GL. Vaziri NDJAjon. Am J Nephrol. (2014) 39:230–7. doi: 10.1159/000360010
53. Vacca, M, Celano, G, Calabrese, FM, Portincasa, P, Gobbetti, M, and De Angelis, MJM. The controversial role of human gut lachnospiraceae. Microorganisms. (2020) 8:573. doi: 10.3390/microorganisms8040573
54. La Rosa, SL, Leth, ML, Michalak, L, Hansen, ME, Pudlo, NA, Glowacki, R, et al. The human gut Firmicute Roseburia intestinalis is a primary degrader of dietary β-mannans. Nat Commun. (2019) 10:905. doi: 10.1038/s41467-019-08812-y
55. Koh, A, De Vadder, F, Kovatcheva-Datchary, P, and Bäckhed, FJC. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. (2016) 165:1332–45. doi: 10.1016/j.cell.2016.05.041
56. Ma, J, Liu, Z, Gao, X, Bao, Y, Hong, Y, He, X, et al. Gut microbiota remodeling improves natural aging-related disorders through Akkermansia muciniphila and its derived acetic acid. Pharmacol Res. (2023) 189:106687. doi: 10.1016/j.phrs.2023.106687
57. Blaak, E, Canfora, E, Theis, S, Frost, G, Groen, A, Mithieux, G, et al. Short chain fatty acids in human gut and metabolic health. Benefic Microbes. (2020) 11:411–55. doi: 10.3920/BM2020.0057
58. Jung, T-H, Park, JH, Jeon, W-M, and Han, K-S. Butyrate modulates bacterial adherence on LS174T human colorectal cells by stimulating mucin secretion and MAPK signaling pathway. Nutr Res Pract. (2015) 9:343–9. doi: 10.4162/nrp.2015.9.4.343
59. SaH, A-L, Peppelenbosch, MP, Roelofsen, H, Vonk, RJ, and Venema, K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim Biophys Acta. (2010) 1801:1175–83. doi: 10.1016/j.bbalip.2010.07.007
Keywords: gut microbiota, rice bran, Bifidobacterium, short-chain fatty acids, prebiotics
Citation: Mahalak KK, Liu L, Bobokalonov J, Narrowe AB, Firrman J, Bittinger K, Hu W, Jones SM and Moustafa AM (2024) Supplementation with soluble or insoluble rice-bran fibers increases short-chain fatty acid producing bacteria in the gut microbiota in vitro. Front. Nutr. 11:1304045. doi: 10.3389/fnut.2024.1304045
Received: 28 September 2023; Accepted: 11 April 2024;
Published: 10 May 2024.
Edited by:
Carlos Gómez-Gallego, University of Eastern Finland, FinlandReviewed by:
Iman Zarei, University of Eastern Finland, FinlandCopyright © 2024 Mahalak, Liu, Bobokalonov, Narrowe, Firrman, Bittinger, Hu, Jones and Moustafa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karley K. Mahalak, S2FybGV5Lk1haGFsYWtAdXNkYS5nb3Y=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.