- 1Division of Germplasm Evaluation, Indian Council of Agricultural Research-National Bureau of Plant Genetic Resources, New Delhi, India
- 2Department of Biotechnology, Ministry of Science and Technology, Government of India, New Delhi, India
Linseed or flaxseed is a well-recognized nutritional food with nutraceutical properties owing to high omega-3 fatty acid (α-Linolenic acid), dietary fiber, quality protein, and lignan content. Currently, linseed enjoys the status of a ‘superfood’ and its integration in the food chain as a functional food is evolving continuously as seed constituents are associated with lowering the risk of chronic ailments, such as heart diseases, cancer, diabetes, and rheumatoid arthritis. This crop also receives much attention in the handloom and textile sectors as the world’s coolest fabric linen is made up of its stem fibers which are endowed with unique qualities such as luster, tensile strength, density, bio-degradability, and non-hazardous nature. Worldwide, major linseed growing areas are facing erratic rainfall and temperature patterns affecting flax yield, quality, and response to biotic stresses. Amid such changing climatic regimes and associated future threats, diverse linseed genetic resources would be crucial for developing cultivars with a broad genetic base for sustainable production. Furthermore, linseed is grown across the world in varied agro-climatic conditions; therefore it is vital to develop niche-specific cultivars to cater to diverse needs and keep pace with rising demands globally. Linseed genetic diversity conserved in global genebanks in the form of germplasm collection from natural diversity rich areas is expected to harbor genetic variants and thus form crucial resources for breeding tailored crops to specific culinary and industrial uses. Global genebank collections thus potentially play an important role in supporting sustainable agriculture and food security. Currently, approximately 61,000 germplasm accessions of linseed including 1,127 wild accessions are conserved in genebanks/institutes worldwide. This review analyzes the current status of Linum genetic resources in global genebanks, evaluation for agro-morphological traits, stress tolerance, and nutritional profiling to promote their effective use for sustainable production and nutrition enhancement in our modern diets.
Introduction
Linseed or flax (Linum usitatissimum L.) has been cultivated for seed oil and stem fiber since ancient times across the world. Globally, six countries (Canada, Kazakhstan, Russia, China, USA, and India) are the major producers (1). At present, flax (fiber-type) ranks as the third largest textile crop, whereas linseed (oil-type) ranks the fifth among the oil crops in the world (1). True to the species name ‘usitatissimum’ meaning ‘very useful,’ linseed/flax stem, seeds, and seed oil have a wide range of applications in the preparation of food, nutritional and industrial products. The oil has wide industrial utility in paints and varnishes because of its unique drying properties attributable to its distinctive fatty acid composition (2). Other diverse uses of linseed oil include the manufacturing of hardboards, brake linings, printing ink, linoleum, and soaps. Fiber flax is used in handloom, textiles, and polymeric composites owing to its natural fibers endowed with unique luster and tensile strength (3, 4). The utilization of linseed plants for food, feed, fiber, and value added products has been reviewed comprehensively in the recent past (5, 6).
Linseed has emerged as a well-recognized nutrition-rich food because of its high omega-3 fatty acid (α-linolenic acid; ALA) content, dietary fiber, high quality protein, and lignans. With respect to omega-3 fatty acids, linseed is considered one of the richest plant-based sources and has around 55% ALA of the total fatty acids (7, 8). It also has an impressive omega 6/omega 3 fatty acid ratio of 0.3:1. As the modern-day diet predominates high omega 6 fats, nutritionists recommend a higher intake of essential omega 3 fats in food, which offer tremendous health benefits. Linseed has high fiber content (27.4%) along with high protein content (18.29%) (5). The seeds contain up to 800 times more lignans than other plant foods (9) and are principally composed of secoisolariciresinol diglucoside (SDG) (294–700 mg/100 g) (10–12). ALA together with SDG and dietary fiber has been reported to lower the risk of multiple chronic ailments such as heart diseases, stroke, hormonal disorders, cancer, diabetes, and rheumatoid arthritis (13–16). Currently, cancer is ranked as the second most important cause of death worldwide leading to a heavy global economic toll estimated at $1.16 trillion per annum. Over 2 million new cases were diagnosed globally in the year 2020, of which 24.5% accounted for breast cancer (17). Daily consumption of 25 g ground flax or 50 mg SDG has been found to be associated with a decreased risk of breast cancer, its recurrence, and mortality risk among survivors (10, 18). A survey reported increased intake of flaxseed in regular diets in cancer patients (19) since people prefer to use natural therapies in addition to conventional medical treatments for the management of the disease. The integration of linseed into the food chain as a functional food is evolving continuously, and it is popularly being called a ‘nutritional punch’. Whole seeds, milled powder, extracted oil, and mucilage are extensively used as nutritional additives in the preparation of baked/ready-to-eat cereal products, bars, salad dressings, bread, muffins, sweets, processed meat and spaghetti, etc. (5). Renewed interest has led to an increase in consumer demand for linseed-based products not only for culinary use but also for novel industrial applications, such as geotextiles, biopolymers, and biofuels. However, unpredictable environmental stresses, such as drought, salinity, heat, diseases, and pest pressure, result in huge losses in yield and quality of oil/fiber (20–23). Hence, in the near future, climate change may exert strong pressure on flax breeding programs to adapt cultivars to changing conditions at an accelerating rate. Furthermore, the narrow genetic base of modern cultivars poses a major constraint to achieving sustainable yields to cater to diverse needs. Genebank collections of cultivated and wild species of the genus Linum from natural diversity-rich areas are important sources of genetic diversity for finding new alleles to meet new environmental challenges. Keeping this in view, the present review article is intended to apprise (1) an overview of the origin, domestication, taxonomy, and gene pool; (2) the status of global ex-situ collections of Linum sp., (3) trait-specific accessions identified through germplasm evaluation for agro-morphological traits, nutrition profiling, and biotic and abiotic stress tolerance in target environments to promote the effective use of these materials; (4) enhancement of the use of trait specific germplasm through the application of recent advances in genomic resources, molecular and biotechnological tools; and (5) documentation and access to germplasm/information through a global exchange system to facilitate transboundary flow.
Origin and domestication
Vavilov (24) described two centers of origin based on two distinct morphotypes, linseed (oil) type (short stature, bushy, profusely branched, and small seeded for the seed or oil purpose) originated in south-western Asia comprising India, Afghanistan, and Turkey and the fiber flax type (erect, long, pliable, unbranched stem with few branches restricted to top and bold seeded for fiber purpose) originated in the Mediterranean region including Asia Minor, Egypt, Algeria, Spain, Italy, and Greece. Later, Zeven and de Wet (25) and Damania et al. (26) reported Central Asia as the primary center of origin and the Mediterranean region as the secondary center of origin.
The domestication and subsequent spread of linseed across continents have not been clearly delineated. However, the archaeological evidence suggests that it was domesticated around 8,000 years ago in the Fertile Crescent and consequently spread to Europe and the rest of the world (27). Other dominant regions of diversity include the Indian subcontinent, Abyssinia, and the Mediterranean (24), where the present-day domesticated form, L. usitatissimum, originated from the wild ancestor L. bienne in geographic isolation. An independent domestication event in Central Asia (Indo-Afghan region) resulted in the development and production of fiber varieties as reported by Duk et al. (28). Crops, such as linseed, having multiple utilities may carry variable domestication signatures. Therefore, multiple domestication events have been suggested to be associated with different regions and included lineages that contain oil, fiber, and winter varieties (29). The long-term domestication of stem fiber and seed oil has diversified it into different types, including fiber, oil, and intermediate (dual-purpose) type (30–33). The first flax varieties were reported to be oil type, and later domestication for fiber use was reported, and multiple paths of flax domestication have been suggested (29, 34, 35). Very recently, Guo et al. (36) also confirmed that oil flax is the ancestor of cultivated flax and revealed signatures of artificial selection during the oil-to-fiber type transition.
Taxonomy, gene pool, and interspecific hybridization
Linum is the largest genus in the family Linaceae and consists of nearly 230 species with diverse chromosome numbers ranging from 2n = 16, 18, 30, 36, and 60 or more (37, 38). The scientific names and classifications used for Linum taxa in local, national, or regional floras are not consistent. The Global Biodiversity Information Facility lists only 252 accepted species names and 22 doubtful species names in the genus Linum (39), whereas Turkish and European floras list more species. The database “Plant List” (40) includes 460 scientific plant names of species rank for the genus Linum of which 108 are accepted species names and 229 are unresolved. Many synonyms exist and are used in scientific communications without cross-references. For example, L. bienne and L. angustifolium are considered synonymous in several databases, such as GRIN, Flora Europaea, The Plant List, and NCBI taxonomy; however, genome analysis based on molecular markers showed that L. bienne is a subspecies of L. usitatissimum rather than a separate species (41, 42). Several morphological, cytological, and molecular characterizations have revealed that L. bienne is the wild progenitor of cultivated flax (35, 43–52).
Despite many taxonomic, cytological, and evolutionary studies conducted for the genus Linum, flax genetic resources could not be classified into distinct gene pools as proposed by Harlan and de Wet (53) or modified by Gepts and Papa (54). In addition to classification, there are ambiguities in the identification of species, mainly L. perenne, L. lewisii, L. flavum, and L. africanum; hence, taxonomic revisions had been suggested (55, 56). Jhala et al. (57) also emphasized that species delimitation needs clarification for classification and efficient conservation of genetic diversity in the genus Linum. Thus, the absence of a coherent taxonomic review for reference to communicate interspecific diversity has made Linum systematics and taxonomy unclear. Understanding this requirement, a recent conspectus was presented by Fu (58) using flax as a case, and different flax species were assigned to various gene pools based on the existing literature. L. usitatissimum and L. bienne were categorized under the primary gene pool as they are interfertile and share the same chromosome number. Interspecific crosses between L. usitatissimum and other species, such as L. africanum, L. angustifolium, L. corymbiferum, and L. decumbens, having n = 15 chromosomes have been reported as successful (45). In addition, successful crosses with L. nervosum, L. pallescens, L. africanum, L. corymbiferum, L. decumbens, L. hirsutum, L. floccosum, and L. tenue have been reported in either direction based on crossability studies of cultivated flax with wild species (38, 59). The tertiary gene pool includes all 200 other species of the genus Linum that cannot hybridize with cultivated flax but could be exploited using advanced biotechnological tools (Figure 1). Several Linum species have the potential for beneficial trait introgression such as for lowering the linolenic acid content (L. tenuifolium, L. sulcatum, L. hudsonoides); drought and cold hardiness (L. perenne); resistance to linseed bud fly and Alternaria blight (L. grandiflorum, L. bienne); oil, fiber quality, and yield improvement (L. bienne), number of tillers (L. strictum); resistance to rust (L. grandiflorum, L. bienne, L. africanum, L. creptans, L. flocossum, L. gallicum, L. marginale, L. perenne, L. strictum, L. tenue, L. trigyna L. alpinum, L. corymbiferum, L. hispidum); and medicinal use as purgative (L. cartharticum L.) (48, 57, 60–62). The latest interest has been to explore the cut flower potential of perennial flax (L. austriacum, L. perenne, and L. lewisii) for ornamental use in floral arrangements (62). Thus, despite its huge potential, interspecific hybridization in Linum is hitherto unexplored to a large extent. A well-defined breeding program aided by biotechnological and molecular biology techniques is required to harness the potential of the wild gene pool and support germplasm enhancement.
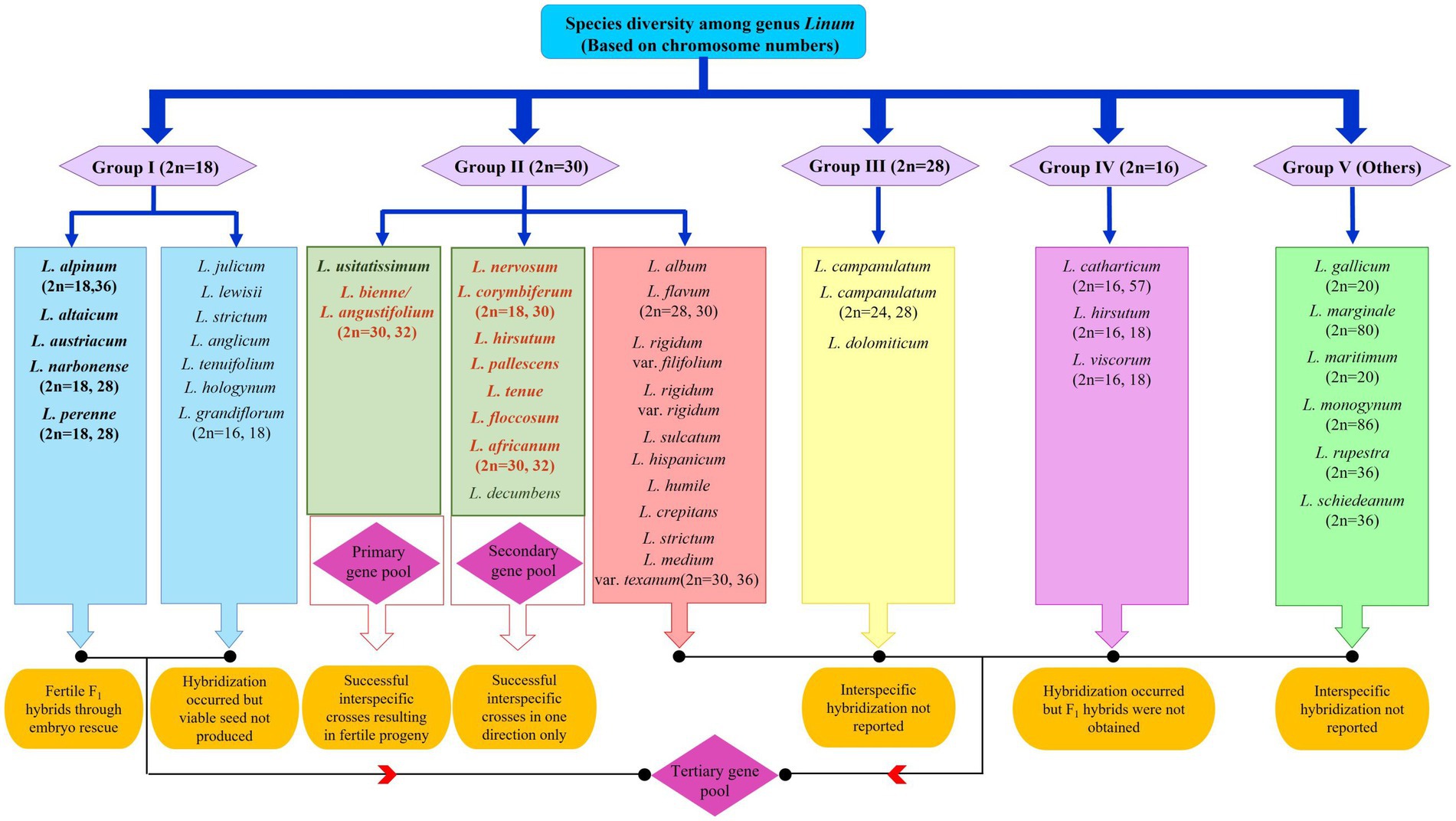
Figure 1. Species diversity in the genus Linum and status of interspecific hybridization among different gene pools. Grouping is based on chromosome numbers as proposed by Gill (37). Bold letters indicate the greatest potential to hybridize with cultivated linseed. The colored font indicates most potential wild species for utilization in breeding programs as donors for specific traits of economic importance such as oil and fiber quality, disease resistance, and abiotic stress tolerance.
Global ex situ holdings of Linum genetic resources
Earlier reviews on ex situ collections were presented by Maggioni et al. (63) who provided the status of ex situ germplasm of linseed in Europe and Diederichsen (55) for global collections. The N.I. Vavilov Research Institute for Plant Industry (VIR) in St. Petersburg, Russia, and the All-Russian Flax Research Institute (VNIIL) at Torzhok, constitute the largest collections of approximately 6,000 accessions at each institute (55, 64, 65). The Indian National GeneBank (INGB) at the Indian Council of Agricultural Research-National Bureau of Plant Genetic Resources (ICAR-NBPGR), New Delhi, holds about 2,900 accessions (66). In addition, around 2,942 accessions are being maintained by the All India Coordinated Research Project on Linseed, ICAR-Indian Institute of Oilseed Research, Hyderabad, India (https://aicrp.icar.gov.in/linseed/ (67)). The Plant Gene Resources of Canada (PGRC) holds around 3,551 accessions of cultivated and 152 accessions of 25 wild species assembled from 72 countries. Currently, a total of 59,786 germplasm accessions of L. usitatissimum and 1,129 accessions belonging to different wild Linum species are conserved worldwide (Figure 2; Supplementary Table 1).
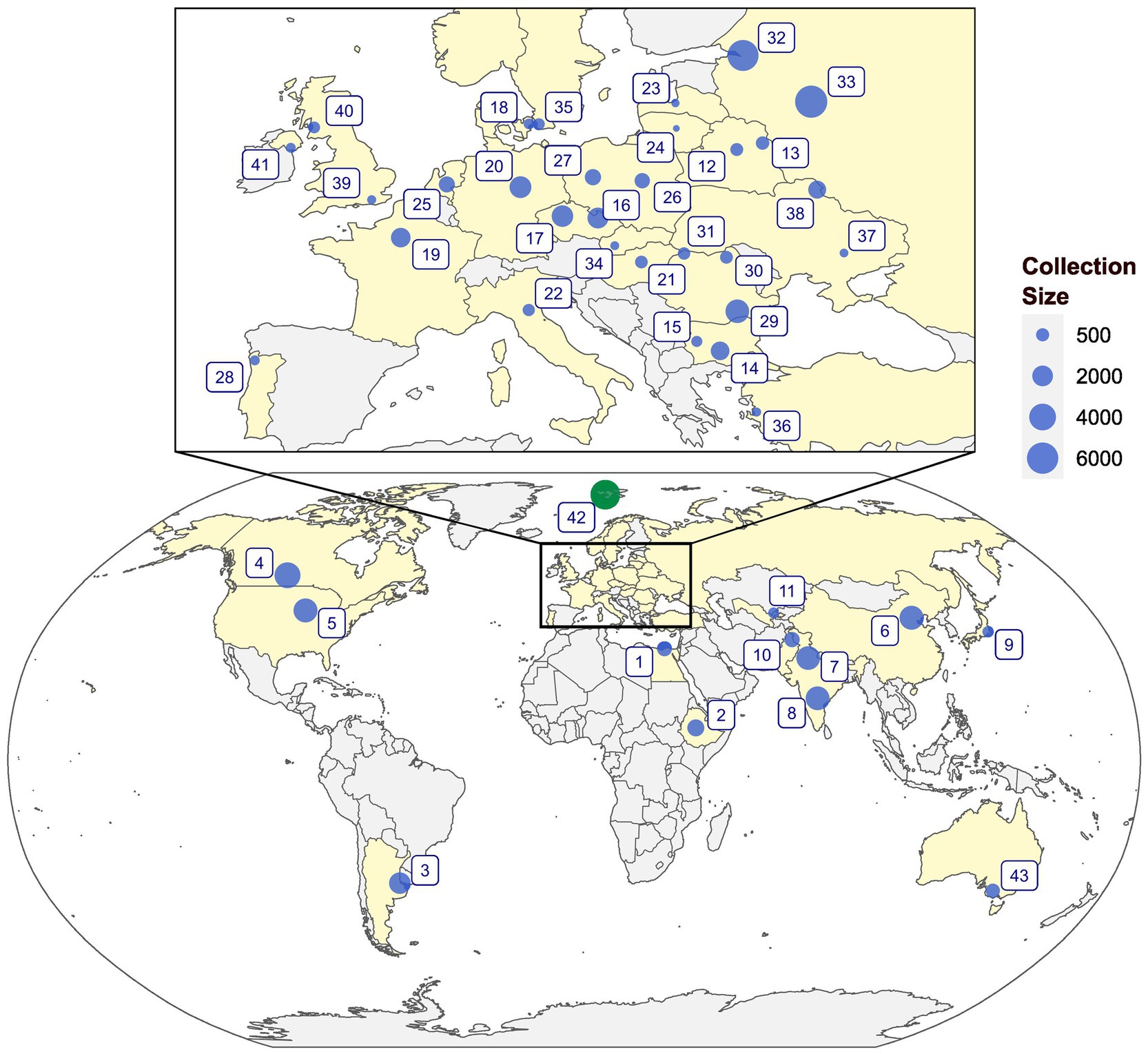
Figure 2. An overview of the global genebanks and institutes holding major collections of Linum genetic resources. 1: NGBGR, Egypt; 2: EBI, Ethiopia; 3: BG-CNIA, Argentina; 4: PGRC, Canada; 5: NCRPIS, United States; 6: ICS-CAAS, China; 7: NGB, ICAR-NBPGR, India; 8: AICRP, ICAR-IIOR, India; 9: NARO, Japan; 10: BCI, Pakistan; 11: UzRIPI, Uzbekistan; 12: NA, Belarus; 13: NA, Belarus; 14: IPGR, Bulgaria; 15: ABI, Bulgaria; 16: AGRITEC, Czechia; 17: CRI, Czechia; 18: NA, Denmark; 19: INRAE-VERSAILLES, France; 20: IPK, Germany; 21: NODiK, Hungary; 22: CREA-CI-BO, Italy; 23: LSFRI, Latvia; 24: LIA, Lithuania; 25: CGN, Netherlands; 26: IHAR, Poland; 27: IWNIRZ, Poland; 28: BPGV-INIAV, Portugal; 29: NARDI, Romania; 30: BRGV, Romania; 31: SCDA Livada, Romania; 32: VIR, Russia; 33: VNIIL, Russia; 34: SVKPIEST, Slovakia; 35: NordGen, Sweden; 36: AARI, Turkey; 37: IOK, Ukraine; 38: IBC, Ukraine; 39: RBGK, United Kingdom; 40: SRUC, United Kingdom; 41: NA, United Kingdom; 42: SGSV, Norway; and 43: AGG, Australia.
Evaluation of Linum germplasm collections
Agro-morphological traits
Many diversity assessment studies were conducted in PGRC flax collection and a wide range of variations for important traits, such as the onset of flowering (37–69 days), plant height (17–130 cm), and thousand seed weight (2.8 to 11.5 g), were reported (68–71). You et al. (72) phenotyped the PGRC flax core collection for agronomic, seed quality, fiber, and disease resistance traits (Pasmo, Powdery mildew, and Fusarium wilt) in multilocation-year environments and reported significant phenotypic variation in both fiber and oil accessions. Similarly, Worku et al. (73) evaluated Ethiopian collection for agronomic traits and Zhuchenko and Rozhmina (65) evaluated flax landraces for fiber quality and enlisted many accessions with superior trait value. In India, agro-morphological characterization and diversity analyses have shown broad range of phenotypic expression and many trait-specific accessions for early flowering, early maturity, oil content, bold seededness, high-test weight, and ALA were identified (5, 74–79). In pale flax, phenotypic diversity was studied by Diederichsen and Hammer (43) in Canadian and Uysal et al. (80) in Turkish germplasm. A high range of variation in pale flax was recorded for traits, such as plant height (38.4–123.3 cm), no. of days until the start of flowering (97–129), days to maturity (178–207), thousand seed weight (1.10–2.70 g), and seed color. Researchers are exploring advanced technologies, such as genomic tools and marker-assisted breeding, to accelerate the development of high-quality linseed varieties with desirable traits.
Germplasm evaluation for nutritional and nutraceutical traits
Biochemical characterization and evaluation of linseed for nutritional traits have been performed around the world. Green and Marshall (81) studied 214 accessions of linseed for the content of seed oil, seed weight, and fatty-acid composition and observed that the oil concentration in flaxseed was in the range of 33.3–46.4%, which positively correlated with high seed weight. Earlier Zimmerman and Klosterman (82) also reported a similar oil content in 1,175 linseed germplasm accessions. Variation in seed oil content, as well as the fatty acid composition of cultivated flax germplasm accessions at PGRC grown in western Canada, was studied by Diederichsen and Fu (68), Diederichsen and Raney (69), Diederichsen et al. (71, 83) in 2,934 germplasm accessions and a min (31.4%) to max (45.7%) oil content was found to be contributed by brown seeded linseed (2730) and yellow seeded accessions (84). Various Indian researchers (5, 76, 77, 79, 85) evaluated small subsets of linseed germplasm for qualitative and quantitative traits and reported oil content between 29.4 and 42.6%. For ALA content, a range of 48.08 to 57.58% was reported by Bayrak et al. (86) in Turkish and Romanian genotypes, while a similar variation (48.9 to 59.9%) was recorded in a Polish flax collection by Silska (87). In the Indian germplasm, the ALA content varied from 39.5% in germplasm accession IC564687 to 57.1% in IC564631 (85) and to as low as 33.14% (88). The proportion of ALA in a huge set of PGRC flax collection (2,243 accessions) was reported to be 39.6 to 66.7% with a mean of 52.6% (55).
Linseed also has high protein content which generally varies from 10.5 to 31% (7, 11). Flaxseed meal is a significant by-product (de-oiled) extracted from the processing of flaxseed and is generally rich in protein content of up to 40%. Protein contentmay vary with the environmental, genetic factors, processing techniques and is negatively correlated with oil content. Oomah et al. (89) evaluated 109 accessions to determine the composition of carbohydrates, protein, and oil content. Until recently, sparse information has been reported on germplasm evaluation for protein content in linseed. Similarly, the evaluation of linseed germplasm for SDG content has been performed to a limited extent only. A varied amount of SDG content ranging from 12.9 to 14.3 mg/g (90, 91) and 11.9–25.9 mg/g in whole linseed (92) has been reported. Thus, genotypic variation had a profound effect on the seed SDG content.
Recently, seed coat mucilage content has been recognized as a new character of interest for industrial applications. Large scale evaluation for mucilage content (assessed as Mucilage indicator value; MIV) was carried out by Diederichsen et al. (83), who screened 1,689 accessions from the PGRC collection and identified potential genetic resources. Canadian germplasm had been reported to have higher MIV (22.1 to 343.4 cSt mL g−1) than North American cultivars (90.6–246.1 cSt mL g−1). The scarcity of information on the chemical composition and functional properties of flax mucilage has limited its use in industries (93). However, recently, linseed mucilage has been recognized as having high technological value as a retardant polymer in pharmaceutical applications as well as in the food industry as a cost-effective natural polymer. Table 1 summarizes the major studies reporting variability for agro-morphological, phenological, fiber, and nutritional traits in flax germplasm and sources of trait-specific superior accessions. The health-related properties of linseed and their rejuvenated importance in animal and human nutrition have stimulated research and breeding programs for novel traits in germplasm collections. The current status of this research area is promising, with several studies identifying linseed varieties with high levels of desirable nutritional and nutraceutical traits. The future of linseed germplasm evaluation for nutritional and nutraceutical traits seems promising with an increasing focus on developing high-quality varieties and functional food products that can provide health benefits beyond basic nutrition.
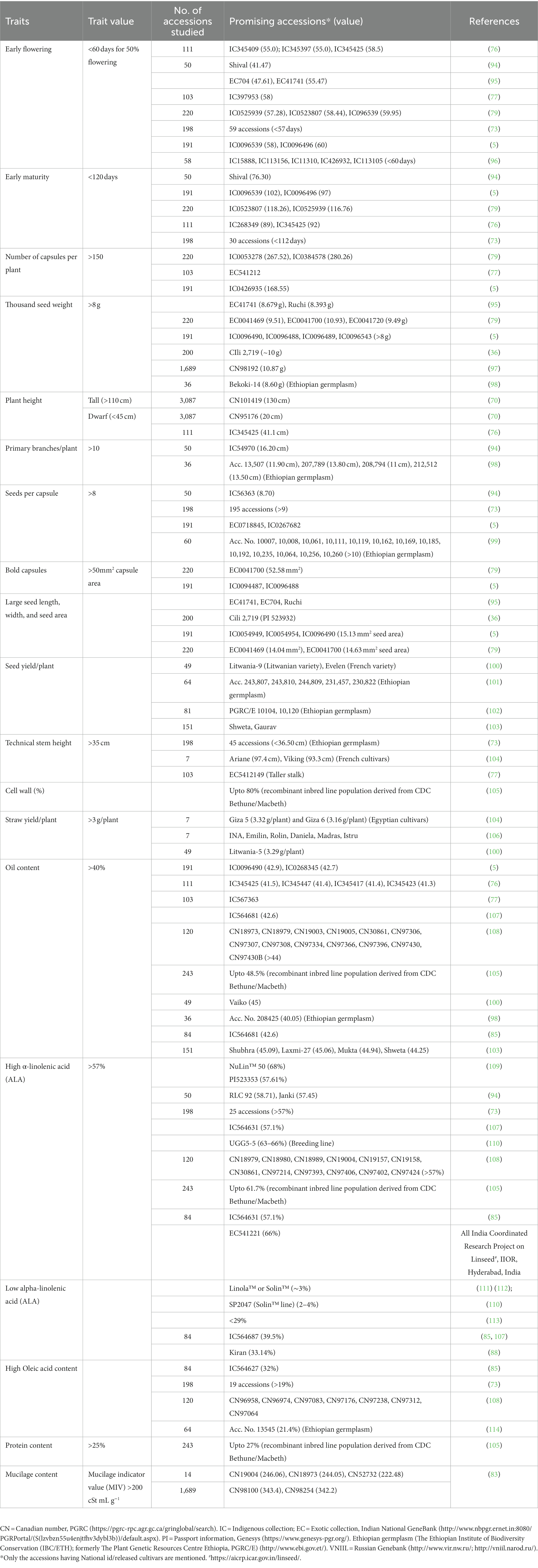
Table 1. Potential accessions identified in linseed and fiber flax germplasm for various agro-morphological and nutritional traits.
Evaluation for major biotic stresses
Among biotic stresses, wilt caused by Fusarium oxysporum f. sp. lini has been recognized as the most devastating disease worldwide resulting in 80 to 100% loss in yield (115, 116). Differential diversity in Fusarium wilt tolerance in flax with higher resistance in accessions from East Asia, moderate resistance in American accessions, lower than average resistance in accessions from Europe and the Indian subcontinent was reported by Diederichsen et al. (117). Although most domesticated varieties are moderately resistant to Fusarium wilt (118, 119), the low genetic diversity of flax varieties and climate change may lead to increased aggressiveness of pathogenic breeds, substantially increasing the risk of disease. Another important fungal pathogen is Alternaria linicola Groves and Skolko, which is mostly prevalent in northwestern Europe and causes seedling blight (120), whereas A. lini Dey is found predominantly in the Indian subcontinent and causes flower and stem blight. Field evaluations under artificial epiphytotic conditions resulted in the identification of either no or very few lines as resistant (84, 121–124).
Powdery mildew, caused by the fungus Oidium lini Skoric, is another important disease of flax, which is reported to cause yield losses between 12–38% in India (125), upto 18% in the United Kingdom (126) and 10–20% in Canada (127). Sources of resistance to powdery mildew have been reported (121, 128, 129) in natural hotspot areas (Table 2). Genes conferring resistance to local O. lini isolates have been identified in Indian flax breeding lines (152, 153) and Canadian and European oil cultivars (127). Recently, several powdery mildew resistance QTLs have been identified (154, 155).
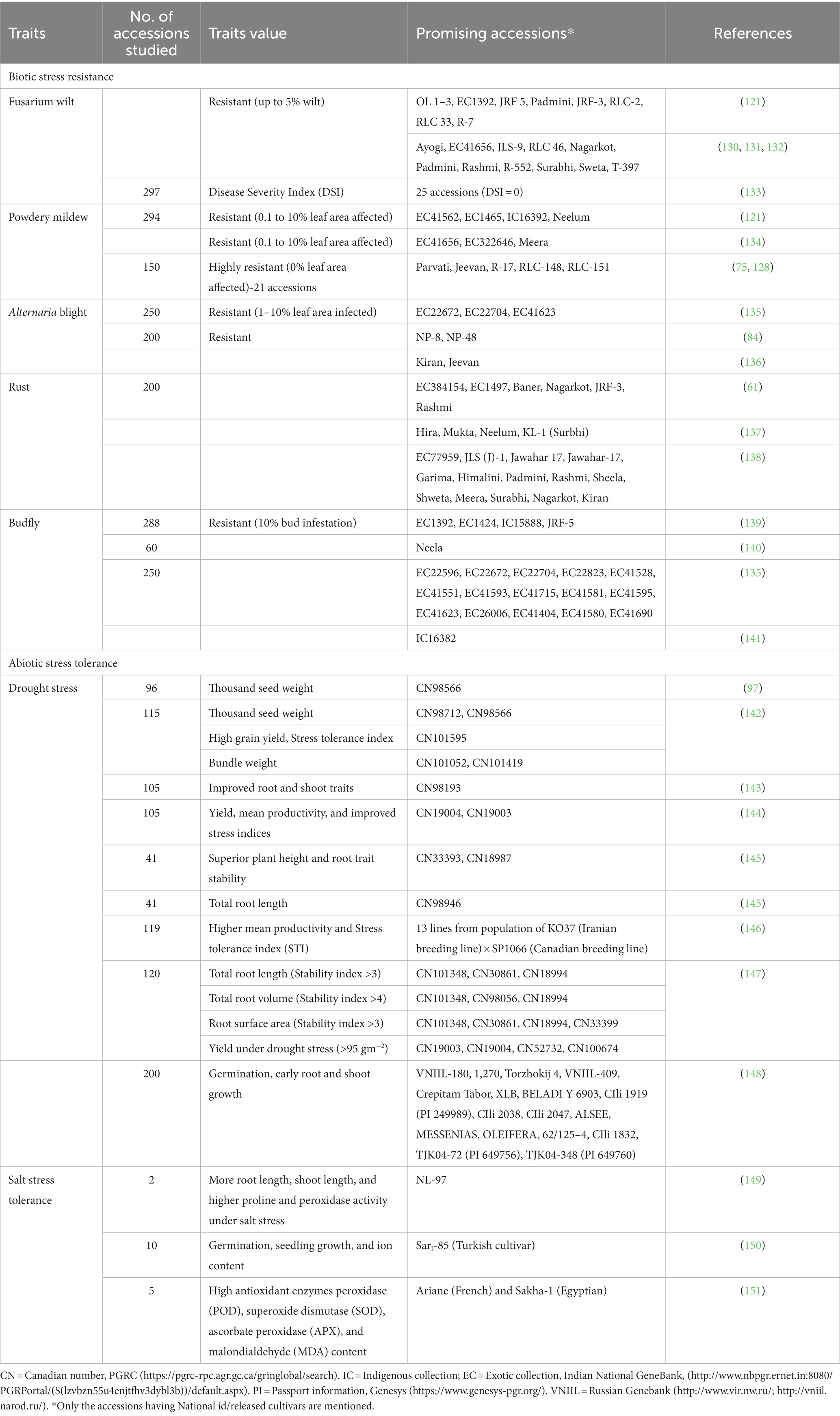
Table 2. Evaluation of linseed germplasm for tolerance to major biotic and abiotic stresses and sources of promising accessions.
In the case of linseed rust, caused by the fungus, Melompsora lini, field evaluations to identify sources of resistance, physiologic races, and inheritance of resistance have been reported (137, 138, 156–159) (Table 2). Misra (160) and Singh et al. (116) reported that during screening of wild Linum species for rust resistance, the species L. africanum, L. augustifolium, L. creptans, L. flocossum, L. gallicum, L. marginale, L. perenne, L. strictum, L. tenue, and L. trigyna were found resistant to all the races, while L. mysorense and L. pallecum were found susceptible.
Among insect pests, defoliators, such as semilooper (Plusia orichalcea Fabr), Lucern caterpillar (Spodoptera exigua Hubn.), tobacco caterpillar (Spodoptera litura Fabr.), and Bihar hairy caterpillar (Spilarctia obliqua Walk.), appear as sporadic and region-specific pests with high incidence in some cropping seasons. Bud fly (Dasyneura lini Barnes) is a key pest of this crop in Asia, particularly India, Pakistan, and Bangladesh, while it appears as a pest of less economic significance for flaxseed/fiber flax in Europe (161). The linseed incidence of Dasyneura lini was first recorded in India by Pruthi and Bhatia (162) and has been reported as the main pest causing yield losses upto 90% in central India (163–165). A few accessions were recorded as resistant (below 10% bud fly infestation score) or moderately resistant germplasm under field conditions in deliberate late planting (139, 165–169) (Table 2). Studies have reported that varietal attributes, such as short flowering periods, flower shape, thin sepals, and higher polyphenol content, render resistance or escape from midges (139, 140, 170, 171). However, limited breeding efforts have been taken to improve this complex polygenic trait.
Evaluation for major abiotic stresses
Drought, salinity, and heat are the major environmental stressors that negatively affect flowering initiation, plant height, seed yield, straw, and fiber yield in linseed. Heller et al. (172) observed that the fiber formation process in the stem is the most intensive in the vegetative growth period till the end of flowering and therefore weather conditions in this period determine the fiber content and quality. Heller and Byczyńska (173) reported a reduction in fiber yield from 35–50% while screening 51 flax cultivars under water limitation to identify superior yielding lines. In most developing countries, linseed is extensively cultivated in rain-fed or moisture-scarce utera conditions and therefore, root system architecture plays an important role in determining water and nutrient acquisition from soil. Linseed has a shallower and less aggressive tap-rooted system compared to other oilseeds such as canola, sunflower, and safflower (174, 175). A deeper and dense root system could extract water more efficiently from deep soil and is thus advantageous, particularly in rainfed production areas (144, 176). Differential performance among flax genotypes under different moisture regimes attributed to variations of their root systems has been reported (97, 177, 178). Linseed is predominantly a rainfed crop in India and therefore Indian genotypes are likely to be more drought tolerant owing to long-time divergent selection under moisture-scarce conditions as also corroborated by several studies (97, 142, 147, 179).
Soil salinization significantly affects the growth and distribution of linseed (151). Only a few studies have reported screening of linseed germplasm against salinity-alkalinity stresses to identify salinity-tolerant lines using germination, biomass and K+/Na+ ratio (149, 150, 180–183), and the underlying biochemical and physiological mechanisms (151) (Table 2). Yu et al. (184) reported differentially expressed genes (DEGs) and saline-alkaline tolerant miRNAs in flax (Lus-miRNAs) for the first time. Wu et al. (185) identified genes that might enhance salt tolerance by increasing root length and improving membrane injury and ion distribution. Lately, Li et al. (148) screened 200 accessions of flax germplasm for salt tolerance and revealed that stress tolerance indices for germination and early root and shoot traits of the oil flax subpopulation were significantly higher than those of the fiber flax subpopulation. They suggested that oil flax may contain more resistance sites related to abiotic stress, and the screening of resistant germplasm and resistance genes should focus more on oil flax.
Heat stress has a significant impact on the climatic adaptation of linseed, particularly superior quality fiber cultivars as it is a cool-season crop. A rise in temperature during flowering and seed filling may lead to necrosis of the ovules, resulting in a reduced seed set (186). Gusta et al. (187) studied genetic variability among flax cultivars in response to temperature stress and reported negative effects on flowering and seed yield. Cross et al. (188) studied the adverse effects of heat stress (>40°C for 5 days) on reproductive organ functioning and reported a decline in boll formation and seed setting although the composition of the oil was not affected. Fofana et al. (189) reported that warmer and drier environmental conditions can cause a lowering of ALA by 5% owing to thermos-sensitivity of the linked enzyme, fatty acid desaturase (FAD2). The molecular mechanism underlying heat stress tolerance in flax is largely unknown. Recently, Saha et al. (20, 190) attempted to describe the role of heat shock factors (HSFs) and DNA hypomethylation with regard to heat stress adaptation in flax.
Genomic resources in linseed
Molecular marker resources in linseed
The PCR-based dominant marker systems, such as RAPD, AFLP, and ISSR, have been employed in flax for linkage mapping and genetic diversity studies since the 1990s until the recent past (99, 191–201). In addition, retrotransposon-based inter-retrotransposon amplified polymorphism (IRAP) markers have been developed in flax and utilized for genetic diversity studies (202). The most preferred PCR-based markers, simple sequence repeats (SSRs) (110, 203), were identified in linseed to the tune of a few hundred until 2011 (50, 51, 204–207). From 2012 onward, there was a substantially higher number of SSRs reported in linseed by different research groups largely due to the advances in next-generation sequencing (NGS) technologies. Cloutier et al. (110) reported 1,164 and 342 SSRs from BAC-end genomic sequences (BESs) and ESTs, respectively. Kale et al. (208) used 454 GS-FLX platform for the sequencing of PCR amplicons and designed 290 SSRs. Furthermore, a reduced representation genome sequencing (RRGS) approach was also used to identify 1,574 SSR loci (209). More recently, 24,375 SSR motifs were identified employing a pseudomolecule-scale genome-wide scan of the flax genome for the development of SSR markers (210). From the identified SSR markers, the polymorphic loci have also been identified in different studies which were used for distinguishing flax and linseed cultivars (206, 207). In terms of functional marker resources, 580 regulatory gene-derived simple sequence repeat (ReG-SSR) markers from transcription factor-coding genes and long non-coding RNAs have been identified (211). In wild species of flax, Linum bienne, 44 microsatellite loci have been identified using genome skimming; of which, 16 have been used for genotyping six L. bienne populations (212). More recently, a few to hundreds of thousands of single nucleotide polymorphic (SNP) marker loci have been unraveled by an array of techniques such as reduced representation sequencing approaches and whole genome resequencing in linseed accessions (36, 78, 143, 213, 214). Detailed information on the major marker resources available in linseed has been shown ( Supplementary Table 2). These resources, especially SNP markers, have enabled the construction of high-density linkage maps, identification of QTLs, and thereby a better understanding of the genetic architecture of several complex traits in linseed (78, 215–219).
Genetic diversity in linseed germplasm elucidated by molecular markers
Different marker systems, such as AFLP, RAPD, ISSR, IRAP, SSR, and SNPs, have been employed in linseed specifically for studying the genetic diversity (33, 79, 195, 197, 198, 202, 220–222). The genetic diversity studies with sufficiently large germplasm accessions (>100) have been considered in this review. The genetic diversity of 708 accessions of cultivated flax and 10 wild species was studied using polymorphic IRAP markers (202). The 708 accessions of cultivated flax comprised 143 landraces, 387 varieties, and 178 breeding lines from 36 countries. The robust 141 reproducible data points per accession were obtained from 10 polymorphic IRAP primers with 52% polymorphism and 0.34 Shannon diversity index. The study showed the highest genetic diversity in wild Linum species (polymorphism: 100% and Jaccard similarity: 0.57), followed by landraces (58%, 0.63), breeding lines (48%, 0.85), and cultivars (50%, 0.81).
Genetic diversity analysis of 168 linseed accessions predominantly of Indian origin was conducted using 50 SSR loci, which unraveled a total of 337 alleles (74). The mean Shannon’s information index for all three populations was 0.23. Similarly, in the case of Ethiopian linseed landraces, IRAP and ISSR markers showed a comparable level of molecular diversity (PIC, 0.16; GD, 0.19) in 203 accessions (222). The genetic diversity of the PGRC flax core collection was studied using 448 microsatellite markers (33). The genetic structure of the core set showed two major groups with six sub-groups having weak population differentiation, weak relatedness (mean = 0.287), and abundant genetic diversity in the total panel (5.32 alleles per locus). The sub-groups were found to have a high proportion of private alleles. Similarly, 350 globally distributed flax genotypes were studied using 6,200 SNP markers, which clustered the 350 accessions into seven sub-populations with moderate genetic diversity (average H = 0.22 and I = 0.34) (221). There was a significant positive correlation (r = 0.30 and p < 0.01) between genetic and geographic distances in the whole collection. Overall, the moderate to low genetic diversity reported in different studies on linseed using varied marker systems are as anticipated and could be accounted for the self-pollinated nature of the crop.
QTL mapping and genome-wide association analysis
In one of the earliest QTL mappings for Fusarium wilt resistance in flax using a doubled-haploid population, two AFLP markers (afB13 and afXR6) were found tightly linked to flax rust resistance (223). A linkage map-based QTL study with 329 SNP and 362 SSR markers using a RILs population of a cross of two Canadian varieties, CDC Bethune and Macbeth, had unraveled 20 QTLs for 14 traits comprising oil, fatty acid, iodine, and protein content as well as fiber quality traits (105). Interestingly, one SSR marker Lu2031 on LG4 (chromosome 4, coordinates: 14489225–14489333) was found to be linked to QTLs of five different traits including cell wall (fiber components), straw weight, seeds per boll, days to maturity, and yield (105, 217). Plant height and technical length are crucial traits for fiber flax. A total of 19 QTLs were identified for both traits following linkage map-based QTL analysis using two RIL populations and a total of 4,497 SNPs were anchored on 15 linkage groups with an average marker density of 2.71 cM (224). A high-density linkage map of flax was constructed using 112 F2 plants and 2,339 specific-locus amplified fragment (SLAF) markers on 15 linkage groups with a total length of 1483.25 cM and a mean distance of 0.63 cM between two adjacent markers (216). The study also helped map 12 QTLs for 6 flax fiber-related traits. In biotic stress, three QTLs have been identified for powdery mildew resistance located on LG1, 7, and 9 accounting for 97% of the phenotypic variation and suggesting a dominant gene action (225). Until 2020, 313 QTLs for quantitative traits comprising seed yield & agronomic traits (155 QTLs), seed quality (75 QTLs), fiber (11 QTLs), and diseases (72 QTLs) have been identified in flax, most of which were mapped on chromosome-scale pseudomolecules (217).
In addition to biparental QTL mapping, genome-wide association studies (GWAS) have been extensively employed in flax for the genetic dissection of complex traits. In the early years of last decades, SSR markers have been used for association mapping of seed weight trait in linseed, which identified five SSR markers associated with the trait (50). Later, considerably high numbers of SNPs identified by reduced representation sequencing and whole genome resequencing approaches have been used in GWAS. The GWAS strategy was reported for genetic dissection of seed weight trait in flax by two independent research groups, unraveling the associated SNP markers and important candidate genes such as PHO1, cytochrome P450, and ubiquitin-proteosome pathway genes (36, 214). GWAS was also employed for genetic dissection of flowering time in linseed on 200 accessions of the Canadian flax core collection using 70,935 curated SNPs by single and multi-locus methods (ML-GWAS). A total of 40 quantitative trait nucleotides (QTNs) associated with 27 QTL were identified for flowering time accounting for 3.06–14.71% of trait variation (215). ML-GWAS was also used on a panel of germplasm accessions of the National Genebank of India for dissecting flowering time, maturity, and plant height trait using 68,925 SNPs identified by genotyping by sequencing. The study identified 53, 30, and 27 stable QTNs for flowering time traits, days to maturity, and plant height, respectively (78). GWAS was also successfully employed for identifying genomic regions associated with pasmo resistance (PR) in flax in a panel of 370 core collections. In total, 10 different statistical models identified a total of 692 unique QTNs associated with 500 putative QTLs from six phenotypic PR datasets. Interestingly, 45 of the identified QTLs spanned 85 resistance gene analogs including a large toll interleukin receptor, nucleotide-binding site, and leucine-rich repeat (TNL) type gene cluster (213). In a similar fashion, GWAS have been deployed in the genetic dissection of other complex traits in linseed including fiber-related traits, fatty acid biosynthesis, mucilage and seed hull content, capsule numbers, branch numbers, and other important agronomic traits (21, 214, 226–228). Detailed information on GWAS in linseed has been presented ( Supplementary Table 3). The identified QTLs/QTNs/associated markers/candidate genes are expected to facilitate linseed researchers for the crop improvement and tailoring niche specific, stress tolerant cultivars through genomics/marker-assisted breeding.
Whole genome sequence
In 2012, a de novo assembly of the flax genome sequence of the cultivar CDC Bethune was made available, having an estimated 81% genome coverage from 302 Mb non-redundant sequences (229). However, there was a misassembly observed in several regions at the genome level. This genome assembly was further improved using the BioNano genome (BNG) optical map of CDC Bethune in 2018 (230). The refined scaffold sequences and validation of the BAC-based physical map were performed followed by further scaffolding of BNG contigs to the super BNG contigs. These super BNG contigs were then assigned to the 15 flaxseed chromosomes with the help of the genetic maps. These pseudomolecules constituted total 316 Mb sequences covering 97% of the annotated genes (230). These resources were pivotal in localizing the earlier identified and new QTLs/markers for important traits to the chromosome-scale pseudomolecules (78, 215, 217). More recently, the genome of fiber flax cultivar Atlant was sequenced using the Oxford Nanopore and Illumina platforms reporting the complete assembly with a total length of 361.7 Mb (N50 = 350 kb) and 97.40% completeness (231). Furthermore, a chromosome-scale high-quality genome sequence of another fiber flax cultivar YY5 is reported with HiFi and Hi-C technology, which helped substantially improved the earlier assembly of fiber flax (232). Besides these, scaffold-level genome assemblies of the other three cultivars including linseed ‘Longya-10’, fiber purpose cultivar ‘Heiya-14’, and pale seed color flax are also available (233).
The plastid genome of L. usitatissimum was sequenced and assembled showing a typical circular DNA molecule of 156,721 bp length (234). The assembled plastid genome showed a total of 109 unique genes, 2 pseudogenes, 176 SSRs, 20 tandem repeats, and 39 dispersed repeats.
These whole genome sequence assemblies would be valuable resources for further fine mapping of causal genetic variants, precise positioning of markers, and thereby in genomics/marker-assisted breeding in linseed.
Effective utilization of Linum genetic resources and trait information
Linum core collections
The large size of germplasm collections conserved at genebanks poses restrictions for the evaluation, utilization, and maintenance of these resources (235). Therefore, the concept of “core collections” (containing approximately 5–10% of the whole collection) was introduced while preserving the maximum genetic diversity of the entire collection with minimum repetitiveness to promote utilization (236, 237). Diverse germplasm in the form of the core collection has stimulated the research to have more and important insights into the genetic variability conserved in huge collections in genebanks. Approximately 10,000 genetically diverse flax accessions in the form of core collections have been maintained in global genebanks (55). Initially, small collections representing donors for specific quality traits and disease resistance were assembled by Kutuzova (238) and Brutch (64) at The N.I. Vavilov Institute (St. Petersburg). Later van Soest and Bas (239) developed a core set of 84 accessions from 506 fiber flax accessions at The Centre for Genetic Resources at Wageningen in the Netherlands. Diederichsen et al. (70) assembled a PGRC flax core collection comprising 407 accessions encompassing both the oil and fiber morphotypes from a world collection of around 3,500 accessions. This core set has been characterized phenotypically for agronomic, fiber quality, and disease resistance traits (72) and to identify the candidate genes and QTLs for seed quality traits and drought tolerance (33, 50, 142, 147), thus contributing to the efficient utilization of genetic resources.
Documentation and access to germplasm and trait information
Documentation of data on germplasm accessions and their characterization and evaluation is of pivotal importance to facilitate the sustainable use of genetic resources. World Information and Early Warning System (WIEWS) maintained by the Plant Genetic Resources of Food and Agriculture Organization, Rome, Italy hosts information on major flax collections.1 Based on the recommendations by IPGRI (240), most of the genebanks have developed their own documentation systems, wherein the passport data are linked to germplasm accessions. These data are often accessible to the users through genebank databases/websites ( Supplementary Table 1). Figshare repository (Federal Research Center for Bast Fiber Crops, Russia) holds data on quantitative phenotypes of flax.2 PGRC has provided full access to the passport, characterization, and evaluation of data of the whole flax collection along with photographs of each accession of the core set.3 For European flax collections, the passport data is available in International Flax Database.4 The PGR information of Indian linseed collection has been duly documented by ICAR-NBPGR in user-friendly databases such as PGR Portal and mobile/desktop apps Genebank, PGRMap (http://www.nbpgr.ernet.in/PGR_Databases.aspx). The evaluated germplasm and trait-specific accessions have been published through catalogs, workshop proceedings, and notification of germplasm registration (241–246). The Inventory of elite germplasm/genetic stocks is available at the NBPGR website.5 The catalogs listing 50 linseed accessions as donors for specific traits including disease resistance (238) and fiber characterization data of 250 flax accessions (64) were published by the Vavilov Institute. The comprehensive dataset of flax accessions from the Russian Federal Research Center for Bast Fiber has been recently published by Rozhmina et al. (247). FIBexDB is a database based on transcriptomic data for flax where the expression pattern for genes and co-expression network are available and pairwise comparison can be made (248).
Exchange of germplasm and information
Universal accessibility to information through unified record maintenance in the system is essential to promote the wider use of genetic resources in breeding programs. With the aid of modern information technology tools, a well-developed global network system through inter-operatable data sets is in place now to facilitate the exchange of information and trait discovery. An overview of the evaluation of Linum genebank collections through comprehensive phenomics and genomics for the identification of germplasm traits relevant to climate resilience and sustainability is presented in Figure 3. Global genebank systems and databases facilitate the exchange of germplasm and the flow of information for enhanced utilization. Institutional, regional, and global platforms provide inter- operatable data sets and create customized data analysis tools to address the needs of end users and different stake holders (e.g., genebank managers, curators, researchers, breeders, students, and farmers). Accession-level information on plant genetic resources secured in genebanks can be retrieved through FAO WIEWS.6 To ease the search of accession level information, a transition from individual genebank website access to multi-institutional/regional portals, such as the European Plant Genetic Resources Search catalogue (EURISCO; http://eurisco.ecpgr.org), the CGIAR system Wide Information Network for Genetic Resources (SINGER; http://www.singer.cgiar.org/), and the Germplasm Resources Information Network (GRIN; http://www.ars-grin.gov/) of United states Department of Agriculture (USDA), has helped a lot. On a global scale, GENESYS (https://www.genesys-pgr.org/) portal integrates data from the website as a single data entry point for users to mine genetic diversity and order germplasm of interest. Currently, it contains information on around 50% of the global accessions; of which, around 24,555 accessions belong to Linum sp., and this information is continuously evolving over time (249).
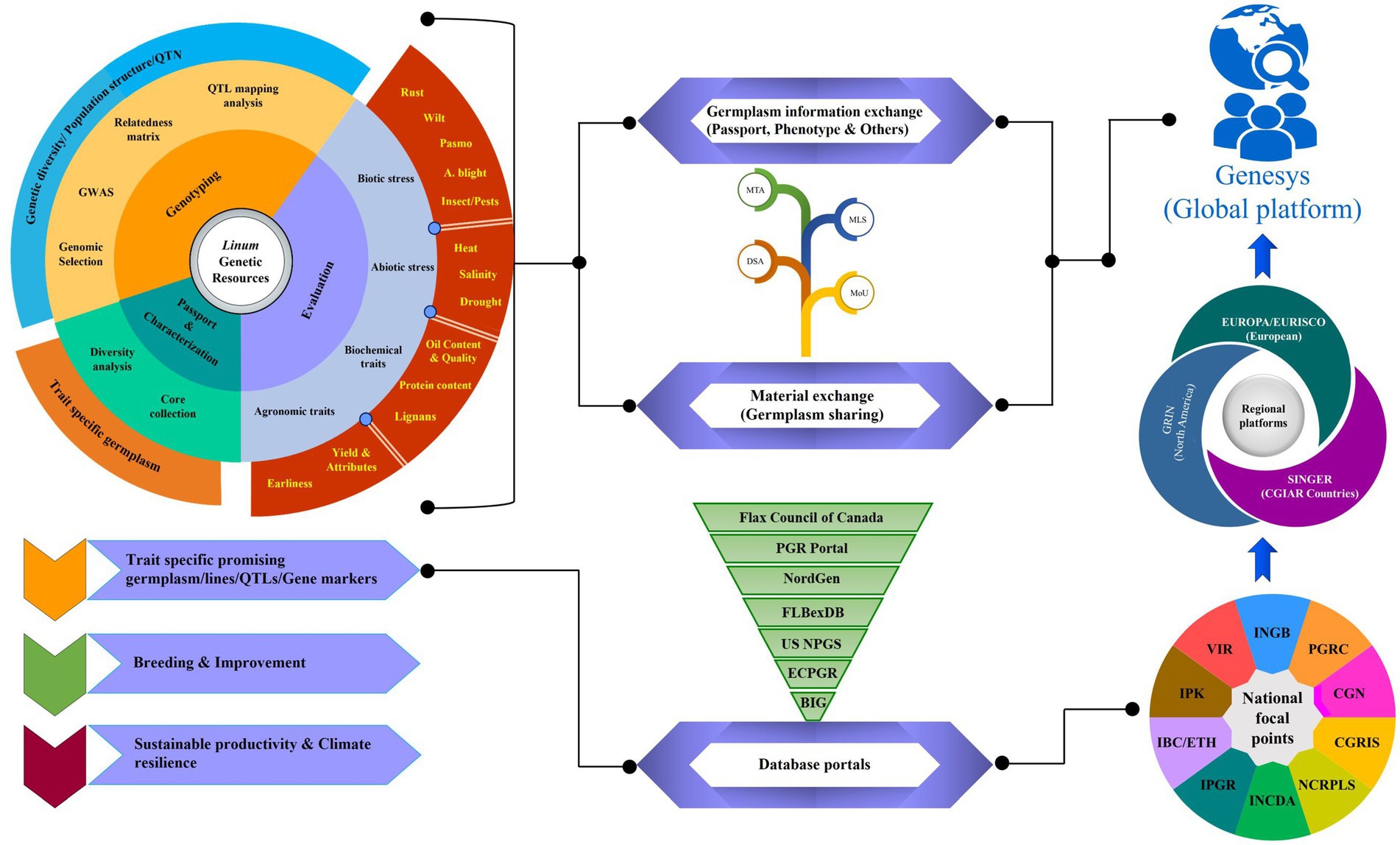
Figure 3. A comprehensive overview of the global network for evaluation, conservation, and exchange of Linum genetic resources. The requisite information on conserved genetic resources in genebanks at the national, regional, and global levels is accessible at the respective focal point. Some of the genebanks have been transitioned and progressed from individual institute-level limited operations to multi-institutional, regional, as well as global platforms for access to genetic resources and information. CGIAR, Consultative Group on International Agricultural Research; SINGER, System-wide Information Network for Genetic Resources; EURISCO, European Plant Genetic Resources Search Catalogue; GRIN, Germplasm Resources Information Network; ECPGR, European Central Crop Databases; BIG, Germany’s Federal Information System on Genetic Resources; US NPGS, US National Plant Germplasm System; NordGen, Nordic Genetic Resources Centre; FIBexDB, Plant Fiber Expression Database; INGB-Indian National Genebank; CGN, Centre for Genetic Resources; the Netherlands; VIR, N.I. Vavilov Research Institute of Plant Industry; INCDA, Research Institute for Cereals and Industrial Crops; IBC/ETH, The Ethiopian Institute of Biodiversity Conservation; IPK, The Leibniz Institute of Plant Genetics and Crop Plant Research; Gatersleben; Germany; NCRPLS, North Central Regional Plant Introduction Station; USA; CGRIS, Institute of Crop Germplasm Resources; China; PGRC, Plant Gene Resources of Canada; Saskatoon; MLS, Multilateral System; SMTA, Standard Material Transfer Agreement; MoU, Memorandum of Understanding; DSA, Data Sharing Agreement.
Conclusion and future perspectives
Germplasm evaluation is a critical aspect of linseed research, as it involves the identification and characterization of genetic resources for the improvement of the crop. Research in linseed germplasm evaluation has been focused on the identification and characterization of diverse germplasm collections for traits, such as oil content, fatty acid composition, seed size, and resistance to biotic and abiotic stresses. This has been accomplished through the use of molecular markers, biochemical analysis, and field evaluations. The present work attempts to review and systematize the existing scientific knowledge about Linum genetic resources with respect to the present status of the collection, evaluation, and utilization of major traits of economic importance. The conservation and systematic evaluation of Linum genetic diversity is essential to overcome environmental challenges and produce resilient cultivars. Amid the alarming rate of climate change, potential genetic resources from diversity rich and environmentally adapted areas need to be identified for sustainable production. Many studies have reported value-rich and trait-specific promising linseed accession from India, especially high salt and drought tolerant genotypes (68, 142, 147, 148, 179). Therefore, the identification of promising donors for economically important traits and novel molecular tags/genes for important traits in the Indian material will add new information to the domain of knowledge, as of now, only PGRC flax core germplasm has been extensively evaluated for various traits. Moreover, several reports have indicated a narrow genetic base in Canadian linseed cultivars (196, 197, 204), which is an impediment to further breeding progress. In addition, the scarce availability of compatible wild species to incorporate novel variation and the limited molecular breeding have hampered flax yield and quality improvements, thus limiting the competitiveness of the species. Owing to these facts, conventional breeding could result in very low and slow yield gains in linseed. Hence, the utilization of advanced genomic tools to identify key genes and pathways associated with important traits can facilitate the development of molecular breeding approaches for targeted futuristic improvement. Furthermore, the integration of omics technologies in future research and breeding should be emphasized to ensure sustainability in yield and climate resilience. In view of these facts, a mega initiative ‘Leveraging genetic resources of linseed through comprehensive phenotyping and genotyping approaches’ under the Mission program on ‘Minor oilseed of Indian origin’ is being taken up presently in India with financial support from the Department of Biotechnology, Govt. of India, wherein whole INGB linseed collection is being phenotyped and genotyped extensively for key agronomic, quality traits, and major biotic and abiotic stresses. The access to evaluation data, genome-wide availability of molecular markers, and QTLs/QTNs/SNPs/candidate genes underlying important traits will ensure further genetic gains in linseed through markers/genomics-assisted and/or haplotype-based breeding.
Author contributions
VK, AK, SK and KS conceptualized the theme. VK wrote the initial draft and edited the manuscript. MS, DW, KG, SL, BT, and JA helped in the preparation of manuscript in the respective area of expertise. SY and JA helped in the preparation of figures and tables and editing of the manuscript. AK, KS and VK reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by funding for the project (No. BT/Ag/Network/Linseed/2019–20) from Department of Biotechnology (DBT), Government of India.
Acknowledgments
The authors thank the Director, ICAR-National Bureau of Plant Genetic Resources (NBPGR), New Delhi, India, and the Head, Division of Germplasm Evaluation, ICAR-NBPGR for guidance and support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2023.1165580/full#supplementary-material
Footnotes
1. ^ http://apps3.fao.org/wiews/
2. ^ https://figshare.com/s/86a68ecfacf6872ef239
4. ^ http://www.ecpgr.cgiar.org/database/crops/flax
5. ^ http://www.nbpgr.ernet.in:8080/registration/InventoryofGermplasm.aspx
References
1. FAO (2022) https://www.fao.org/faostat/en/#data/QCL [Accessed 5 June, 2022].
2. Przybylski, R . Flax oil and high linolenic oils In: Bailey’s industrial oil and fat products (2005). 281–301. doi: 10.1002/047167849x.bio010
3. Foulk, JA, Fuqua, MA, Ulven, CA, and Alcock, MM. Flax fiber quality and influence on interfacial properties of composites. Int J Sustain Eng. (2010) 3:17–24. doi: 10.1080/19397030903348710
4. Yan, L, Chow, N, and Jayaraman, K. Flax fiber and its composites- a review. Compos Part B Eng. (2014) 56:296–317. doi: 10.1016/j.compositesb.2013.08.014
5. Kaur, V, Kumar, S, Yadav, R, Wankhede, DP, Aravind, J, Radhamani, J, et al. Analysis of genetic diversity in Indian and exotic linseed germplasm and identification of trait specific superior accessions. J Environ Biol. (2018) 39:702–9. doi: 10.22438/jeb/39/5/MRN-849
6. Zuk, M, Richter, D, Matuła, J, and Szopa, J. Linseed, the multipurpose plant. Ind Crop Prod. (2015) 75:165–77. doi: 10.1016/j.indcrop.2015.05.005
7. Rabetafika, HN, Van Remoortel, V, Danthine, S, Paquot, M, and Blecker, C. Flaxseed proteins: food uses and health benefits. Int J Food Sci Technol. (2011) 46:221–8. doi: 10.1111/j.1365-2621.2010.02477.x
8. Rubilar, M, Gutiérrez, C, Verdugo, M, Shene, C, and Sineiro, J. Flaxseed as a source of functional ingredients. J Plant Nutr Soil Sci. (2010) 10:373–7. doi: 10.4067/S0718-95162010000100010
9. Westcott, NA, and Muir, AD. Flax seed lignan in disease prevention and health promotion. Phytochem Rev. (2003) 2:401–17. doi: 10.1023/B:PHYT.0000046174.97809.b6
10. De Silva, SF, and Alcorn, J. Flaxseed lignans as important dietary polyphenols for cancer prevention and treatment: chemistry, pharmacokinetics, and molecular targets. Pharmaceuticals. (2019) 12:68. doi: 10.3390/ph12020068
12. Touré, A, and Xueming, X. Flaxseed lignans: source, biosynthesis, metabolism, antioxidant activity, bio-active components, and health benefits. Compr Rev Food Sci Food Saf. (2010) 9:261–9. doi: 10.1111/j.1541-4337.2009.00105.x
13. Adolphe, JL, Whiting, SJ, Juurlink, BH, Thorpe, LU, and Alcorn, J. Health effects with consumption of the flax lignan secoisolariciresinol diglucoside. Br J Nutr. (2010) 103:929–38. doi: 10.1017/S0007114509992753
14. Kajla, P, Sharma, A, and Sood, DR. Flaxseed-a potential functional food source. J Food Sci Technol. (2015) 52:1857–71. doi: 10.1007/s13197-014-1293-y
15. Tarpila, A, Wennberg, T, and Tarpila, S. Flaxseed as a functional food. Curr Top Nutraceutical Res. (2005) 3:167–88. doi: 10.18231/j.ijnmhs.2020.008
16. Westcott, ND, and Paton, D. Complex containing lignan, phenolic and aliphatic substances from flax process for preparing. PubChem Patent Summary. (2001) 6:264–853.
17. GLOBOCAN (2020) Available at: https://gco.iarc.fr/today/home [Accessed 29 July, 2022].
18. Flower, G, Fritz, H, Balneaves, LG, Verma, S, Skidmore, B, Fernandes, R, et al. Flax and breast cancer: a systematic review. Integr Cancer Ther. (2014) 13:181–92. doi: 10.1177/1534735413502076
19. Boucher, BA, Cotterchio, M, Curca, IA, Kreiger, N, Harris, SA, Kirsh, VA, et al. Intake of phytoestrogen foods and supplements among women recently diagnosed with breast cancer in Ontario. Canada Nutr Cancer. (2012) 64:695–703. doi: 10.1080/01635581.2012.687426
20. Saha, D, Mukherjee, P, Dutta, S, Meena, K, Sarkar, SK, Mandal, AB, et al. Genomic insights into HSFs as candidate genes for high-temperature stress adaptation and gene editing with minimal off-target effects in flax. Sci Rep. (2019) 9:5581. doi: 10.1038/s41598-019-41936-1
21. Yadav, B, Kaur, V, Narayan, OP, Yadav, SK, Kumar, A, and Wankhede, DP. Integrated omics approaches for flax improvement under abiotic and biotic stress: current status and future prospects. Front Plant Sci. (2022) 13:931275. doi: 10.3389/fpls.2022.931275
22. Zare, S, Mirlohi, A, Saeidi, G, Sabzalian, MR, and Ataii, E. Water stress intensified the relation of seed color with lignan content and seed yield components in flax (Linum usitatissimum L.). Sci Rep. (2021) 11:23958. doi: 10.1038/s41598-021-02604-5
23. Saha, D, Shaw, AK, Datta, S, and Mitra, J. Evolution and functional diversity of abiotic stress-responsive NAC transcription factor genes in Linum usitatissimum L. Environ Exp Bot. (2021) 188:104512. doi: 10.1016/j.envexpbot.2021.104512
24. Vavilov, NI . The origin, variation, immunity and breeding of cultivated plants. Chron Bot. (1951) 13:1–366. doi: 10.2134/agronj1952.00021962004400020016x
25. Zeven, AC, and JMJ, D-W. Dictionary of cultivated plants and their regions of diversity. Wagenigen: The Netherlands: Centre for Agricultural Publishing and Documentation (1982).
26. Damania, A, Valkoun, J, Willcox, G, and Qualset, CO. (1998). The origin of agriculture and crop domestication.
27. van Zeist, W, and Bakker-Heeres, JAH. Evidence for linseed cultivation before 6,000 B.C. J Archaeol Sci. (1975) 2:215–9. doi: 10.1016/0305-4403(75)90059-X
28. Duk, M, Kanapin, A, Rozhmina, T, Bankin, M, Surkova, S, Samsonova, A, et al. The genetic landscape of fiber flax. Front Plant Sci. (2021) 12:764612. doi: 10.3389/fpls.2021.764612
29. Fu, YB . Genetic evidence for early flax domestication with capsular dehiscence. Genet Resour Crop Evol. (2011) 58:1119–28. doi: 10.1007/s10722-010-9650-9
30. Diederichsen, A, and Ulrich, A. Variability in stem fibre content and its association with other characteristics in 1177 flax (Linum usitatissimum L.) genebank accessions. Ind Crop Prod. (2009) 30:33–9. doi: 10.1016/j.indcrop.2009.01.002
31. Liu, FH, Chen, X, Long, B, Shuai, RY, and Long, CL. Historical and botanical evidence of distribution, cultivation and utilization of Linum usitatissimum L. (flax) in China. Veget Hist Archaeobot. (2011) 20:561–6. doi: 10.1007/s00334-011-0311-5
32. Povkhova, LV, Melnikova, NV, Rozhmina, TA, Novakovskiy, RO, Pushkova, EN, Dvorianinova, EM, et al. Genes associated with the flax plant type (oil or fiber) identified based on genome and transcriptome sequencing data. Plan Theory. (2021) 10:2616. doi: 10.3390/plants10122616
33. Soto-Cerda, BJ, Diederichsen, A, Ragupathy, R, and Cloutier, S. Genetic characterization of a core collection of flax (Linum usitatissimum L.) suitable for association mapping studies and evidence of divergent selection between fiber and linseed types. BMC Plant Biol. (2013) 13:78. doi: 10.1186/1471-2229-13-78
34. Allaby, RG, Peterson, GW, Andrew, DM, and Fu, YB. Evidence of the domestication history of flax (Linum usitatissimum) from genetic diversity of the Sad2 locus. Theor Appl Genet. (2005) 112:58–65. doi: 10.1007/s00122-005-0103-3
35. Fu, YB, and Allaby, RG. Phylogenetic network of Linum species as revealed by non-coding chloroplast DNA sequences. Genet Resour Crop Evol. (2010) 57:667–77. doi: 10.1007/s10722-009-9502-7
36. Guo, D, Jiang, H, Yan, W, Yang, L, Ye, J, Wang, Y, et al. Resequencing 200 flax cultivated accessions identifies candidate genes related to seed size and weight and reveals signatures of artificial selection. Front Plant Sci. (2020) 10:1682. doi: 10.3389/fpls.2019.01682
38. Seetharam, A . Interspecific hybridization in Linum. Euphytica. (1972) 21:489–95. doi: 10.1007/BF00039344
39. GBIF (2022) Global biodiversity information facility. Available at: http://www.gbif.org/ (Accessed November 13, 2022).
40. Royal botanic gardens Kew and Missouri Botanical Garden (2022) The plant list, a working list of all plant species. Available at: http://www.theplantlist.org/ (Accessed November 13, 2022).
41. Muravenko, OV, Lemesh, VA, Samatadze, TE, Amosova, AV, Grushetskaya, ZE, Popov, KV, et al. Genome comparisons with chromosomal and molecular markers for three closely related flax species and their hybrids. Russ J Genet. (2003) 39:414–21. doi: 10.1023/A:1023309831454
43. Diederichsen, A, and Hammer, K. Variation of cultivated flax (Linum usitatissimum L. subsp. usitatissimum) and its wild progenitor pale flax (subsp. angustifolium (Huds.) Thell.). Genet Resour Crop Evol. (1995) 42:263–72. doi: 10.1007/BF02431261
44. Fu, YB, and Peterson, GW. Characterization of expressed sequence tag derived simple sequence repeat markers for 17 Linum species. Botany. (2010) 88:537–43. doi: 10.1139/B10-019
45. Gill, KS, and Yermanos, DM. Cytogenetic studies on the genus Linum I. hybrids among taxa with 15 as the haploid chromosome number. Crop Sci. (1967) 7:623–7. doi: 10.2135/cropsci1967.0011183X000700060021x
46. Gill, KS, and Yermanos, DM. Cytogenetic studies on the genus Linum II. Hybrids among taxa with nine as the haploid chromosome number. Crop Sci. (1967) 7:627–31. doi: 10.2135/cropsci1967.0011183X000700060022x
47. Soto-Cerda, BJ, Carrasco, RJ, Aravena, GA, Urbina, HA, and Navarro, CS. Identifying novel polymorphic microsatellites from cultivated flax (Linum usitatissimum L.) following data mining. Plant Mol Biol Report. (2011) 29:753–9. doi: 10.1007/s11105-010-0270-5
48. Soto-Cerda, BJ, Diederichsen, A, Duguid, S, Booker, H, Rowland, G, and Cloutier, S. The potential of pale flax as a source of useful genetic variation for cultivated flax revealed through molecular diversity and association analyses. Mol Breed. (2014) 34:2091–107. doi: 10.1007/s11032-014-0165-5
49. Soto-Cerda, BJ, Duguid, S, Booker, H, Rowland, G, Diederichsen, A, and Cloutier, S. Genomic regions underlying agronomic traits in linseed (Linum usitatissimum L.) as revealed by association mapping. J Integr Plant Biol. (2014) 56:75–87. doi: 10.1111/jipb.12118
50. Soto-Cerda, BJ, Duguid, S, Booker, H, Rowland, G, Diederichsen, A, and Cloutier, S. Association mapping of seed quality traits using the Canadian flax (Linum usitatissimum L.) core collection. Theor Appl Genet. (2014) 127:881–96. doi: 10.1007/s00122-014-2264-4
51. Soto-Cerda, BJ, Saavedra, HU, Navarro, NC, and Ortega, PM. Characterization of novel genic SSR markers in Linum usitatissimum (L.) and their transferability across eleven Linum species. Electron J Biotechnol. (2011) 14:4. doi: 10.2225/vol14-issue2-fulltext-6
52. Tammes, T . The genetics of the genus Linum In: JP Lotsy and WA Goddijn, editors. Bibliographia Genetica. The Netherlands: Martinus Nijhoff (1928). 1–34.
53. Harlan, JR, and de Wet, JMJ. Toward a rational classification of cultivated plants. Taxon. (1971) 20:509–17. doi: 10.2307/1218252
54. Gepts, P, and Papa, R. Possible effects of (trans) gene flow from crops on the genetic diversity from landraces and wild relatives. Environ Biosaf Res. (2003) 2:89–103. doi: 10.1051/ebr:2003009
55. Diederichsen, A . Ex situ collections of cultivated flax (Linum usitatissimum L.) and other species of the genus Linum L. Genet Resour Crop Evol. (2007) 54:661–78. doi: 10.1007/s10722-006-9119-z
56. Diederichsen, A . A taxonomic view on genetic resources in the genus Linum L. for flax breeding In: CA Cullis , editor. Plant genetics and genomics: Crops and models. Springer (2019). 23:1–15
57. Jhala, AJ, Hall, LM, and Hall, JC. Potential hybridization of flax with weedy and wild relatives: An avenue for movement of engineered genes? Crop Sci. (2008) 48:825–40. doi: 10.2135/cropsci2007.09.0497
58. Fu, YB . A molecular view of flax gene pool In: C Cullis , editor. Plant genetics and genomics: Crops and models. Springer (2019). 16–36.
59. Bari, G, and Godward, MBE. Interspecific crosses in Linum. Euphytica. (1970) 19:443–6. doi: 10.1007/BF01902918
60. Hegi, G . Illustrierte Flora von Mitteleuropa, illustrated flora of Central Europe. Munchen: Lehmanns Verlag (1925). 5 p 3-38.
61. Kumar, A, Ekka, S, and Tripathi, UK. Evaluation of linseed germplasm for resistance against rust (Melampsora lini). J Oilseeds Res. (2016) 33:56–61.
62. Tork, DG, Anderson, NO, Wyse, DL, and Betts, KJ. Perennial flax: a potential cut flower crop. HortScience. (2022) 57:221–30. doi: 10.21273/HORTSCI16098-21
63. Maggioni, L, Pavelek, M, van Soest, LJM, and Lipman, E. (2002). “Flax genetic resources in Europe” in Ad hoc meeting: Prague, Czech Republic. International Plant Genetic Resources Institute, Rome, Italy.
64. Brutch, NB . The flax genetic resources collection held at the N.I. Vavilov institute, Russian Federation In: Flax genetic resources. IPGRI, Maccarese Rome: Europe (2002). 61–5.
65. Zhuchenko, AA, and Rozhmina, TA. Mobilization of flax genetic resources In: VILAR and VNIIL. Russia: Starica (2000)
66. Kaur, V, Yadav, R, and Wankhede, DP. Linseed genetic resources for climate change intervention and future breeding. J Appl Nat Sci. (2017) 9:1112–8. doi: 10.31018/jans.v9i2.1331
67. FAO . The second report on the state of the world’s plant genetic resources for food and agriculture. Rome (2010).
68. Diederichsen, A, and Fu, Y. Phenotypic and molecular (RAPD) differentiation of four infraspecific groups of cultivated flax (Linum usitatissimum L. subsp. usitatissimum). Genet Resour Crop Evol. (2006) 53:77–90. doi: 10.1007/s10722-004-0579-8
69. Diederichsen, A, and Raney, JP. Seed colour, seed weight and seed oil content in Linum usitatissimum accessions held by plant gene resources of Canada. Plant Breed. (2006) 125:372–7. doi: 10.1111/j.1439-0523.2006.01231.x
70. Diederichsen, A, Kusters, PM, Kessler, D, Bainas, Z, and Gugel, RK. Assembling a core collection from the flax world collection maintained by plant gene resources of Canada. Genet Resour Crop Evol. (2013) 60:1479–85. doi: 10.1007/s10722-012-9936-1
71. Diederichsen, A, Raney, JP, Fu, YB, and Richards, KW. (2002). “Diversity in the flax collection at plant gene resources of Canada” in Proceedings of the 59th Flax Institute of the United States. Doublewood Inn, Fargo, North Dakota, USA. Flax Institute, Fargo, USA. pp. 138–143.
72. You, FM, Jia, G, Xiao, J, Duguid, SD, Rashid, KY, Booker, HM, et al. Genetic variability of 27 traits in a core collection of flax (Linum usitatissimum L.). front. Plant Sci. (2017) 8:1636. doi: 10.3389/fpls.2017.01636
73. Worku, N, Heslop-Harrison, JS, and Adugna, W. Diversity in 198 Ethiopian linseed (Linum usitatissimum) accessions based on morphological characterization and seed oil characteristics. Genet Resour Crop Evol. (2015) 62:1037–53. doi: 10.1007/s10722-014-0207-1
74. Chandrawati, SN, Kumar, R, Kumar, S, Singh, PK, Yadav, VK, et al. Genetic diversity, population structure and association analysis in linseed (Linum usitatissimum L.). Physiol Mol Biol Plants. (2017) 23:207–19. doi: 10.1007/s12298-016-0408-5
75. Dhirhi, N, and Mehta, N. Estimation of genetic variability and correlation in F2 segregating generation in linseed (Linum usitatisimum L.). Plant Arch. (2019) 19:475–84.
76. Dikshit, N, and Sivaraj, N. Analysis of agro-morphological diversity and oil content in Indian linseed germplasm. Grasas Aceites. (2015) 66:e060. doi: 10.3989/gya.0689141
77. Nizar, MA, and Mulani, RM. Genetic diversity in indigenous and exotic linseed germplasm (Linum usitatissimum L.). Electron. J Plant Breed. (2015) 6:848–54.
78. Saroha, A, Pal, D, Gomashe, SS, Akash, KV, Ujjainwal, S, et al. Identification of QTNs associated with flowering time, maturity, and plant height traits in Linum usitatissimum L. using genome-wide association study. Front Genet. (2022) 13:811924. doi: 10.3389/fgene.2022.811924
79. Saroha, A, Pal, D, Kaur, V, Kumar, S, Bartwal, A, Aravind, J, et al. Agro-morphological variability and genetic diversity in linseed (Linum usitatissimum L.) germplasm accessions with emphasis on flowering and maturity time. Genet Resour Crop Evol. (2022) 69:315–33. doi: 10.1007/s10722-021-01231-3
80. Uysal, H, Kurt, O, Fu, YB, Diederichsen, A, and Kusters, P. Variation in phenotypic characters of pale flax (Linum bienne mill.) from Turkey. Genet Resour Crop Evol. (2012) 59:19–30. doi: 10.1007/s10722-011-9663-z
81. Green, AG, and Marshall, DR. Variation for oil quantity and quality in linseed (Linum usitatissimum). Aust J Agric Res. (1981) 32:599–607. doi: 10.1071/AR9810599
82. Zimmerman, DC, and Klosterman, HJ. The distribution of fatty acids in linseed oil from the world collection of flax varieties. Proc North Dakota Acad Sci. (1959) 13:71–5.
83. Diederichsen, A, Raney, JP, and Duguid, SD. Variation of mucilage in flax seed and its relationship with other seed characters. Crop Sci. (2006) 46:365–71. doi: 10.2135/cropsci2005.0146
84. Kumar, N, and Tripathi, UK. Screening of linseed germplasm for resistance against Alternaria spp. cause blight disease in linseed (Linum usitatissimum L.). J Pharmacogn Phytochem. (2018) 7:1389–92.
85. Sivaraj, N, Sunil, N, Pandravada, SR, Kamala, V, Babu, A, Vinod, K, et al. Variability in linseed (Linum usitatissimum) germplasm collections from peninsular India with special reference to seed traits and fatty acid composition. Indian J Agric Sci. (2012) 82:102–5.
86. Bayrak, A, Kiralan, M, Ipek, A, Arslan, N, Cosge, B, and Khawar, KM. Fatty acid compositions of linseed (Linum usitatissimum L.) genotypes of different origin cultivated in Turkey. Biotechnol Biotechnol Equip. (2010) 2:1836–42. doi: 10.2478/V10133-010-0034-2
87. Silska, G . Genetic resources of flax (Linum usitatissimum L.) as very rich source of α-linolenic acid. Herba Pol. (2017) 63:26–33. doi: 10.1515/hepo-2017-0022
88. Pali, V, and Mehta, N. Evaluation of oil content and fatty acid compositions of flax (Linum usitatissimum L.) varieties of India. J Agric Sci. (2014) 6:198–207. doi: 10.5539/jas.v6n9p198
89. Oomah, BD, Kenaschuk, EO, Cui, W, and Mazza, G. Variation in the composition of water-soluble polysaccharides in flaxseed. J Agric Food Chem. (1995) 43:1484–8. doi: 10.1021/jf00054a013
90. Madhusudhan, B . Potential benefits of flaxseed in health and disease - a perspective. Agric Conspec Sci. (2009) 74:67–72.
92. Eliasson, C, Kamal-Eldin, A, Andersson, R, and Aman, P. High-performance liquid chromatographic analysis of secoisolariciresinol diglucoside and hydroxycinnamic acid glucosides in flaxseed by alkaline extraction. J Chromatogr A. (2003) 1012:151–9. doi: 10.1016/s0021-9673(03)01136-1
93. Kaewmanee, T, Bagnasco, L, Benjakul, S, Lanteri, S, Morelli, CF, Speranza, G, et al. Characterisation of mucilages extracted from seven Italian cultivars of flax. Food Chem. (2014) 148:60–9. doi: 10.1016/j.foodchem.2013.10.022
94. Kumar, M, Patel, M, Chauhan, R, Tank, C, and Solanki, S. Delineating multivariate divergence, heritability, trait association and identification of superior omega-3-fatty acid specific genotypes in linseed (Linum usitatissimum L.). Genetika. (2021) 53:825–36. doi: 10.2298/GENSR2102825K
95. Hussain, ME, Goyal, VK, Paul, PJ, Yadav, Y, Jha, UC, and Moitra, PK. Assessment of genetic variability, diversity, and identification of promising lines in linseed germplasm for harnessing genetic gain in central plain of the Indian subcontinent. J Plant Breed Crop Sci. (2022) 14:12–20. doi: 10.5897/JPBCS2021.0990
96. Chandrawati, SN, Yadav, VK, Kumar, R, Kumar, S, and Yadav, HK. Genetic variability and interrelationship among morphological and yield traits in linseed (Linum usitatissimum L.). Genetika. (2016) 48:881–92. doi: 10.2298/GENSR1603881C
97. Diederichsen, A, Rozhmina, TA, Zhuchenko, AA, and Richards, KW. Screening for broad adaptation in 96 flax (Linum usitatissimum L.) accessions under dry and warm conditions in Canada and Russia. PGR Newsl. (2006) 146:9–16.
98. Dabalo, DY, Singh, BCS, and Weyessa, B. Genetic variability and association of characters in linseed (Linum usitatissimum L.) plant grown in Central Ethiopia region. Saudi. J Biol Sci. (2020) 27:2192–206. doi: 10.1016/j.sjbs.2020.06.043
99. Adugna, W, Labuschagne, MT, and Viljoen, CD. The use of morphological and AFLP markers in diversity analysis of linseed. Biodivers Conserv. (2006) 15:3193–205. doi: 10.1007/s10531-005-6970-8
100. Bakry, AB, Ibrahim, OM, Abd El-Fattah Elewa, T, and El-Karamany, MF. Performance assessment of some flax (Linum usitatissimum L.) varieties using cluster analysis under sandy soil conditions. Agric Sci. (2014) 5:677–86. doi: 10.4236/as.2014.58071
101. Ali, MF, Mekbib, F, and Wakjira, A. Morphological diversity of Ethiopian linseed (Linum usitatissimum L.) landrace accessions and non-native cultivars. J Plant Breed Genet. (2014) 2:115–24.
102. Tadesse, T, Parven, A, Singh, H, and Weyessa, B. Estimates of variability and heritability in linseed germplasm. Int J Sustain Crop Prod. (2010) 5:8–16.
103. Siddiqui, A, Shukla, SK, Rastogi, A, Bhargava, A, Niranjan, A, and Lehri, A. Relationship among phenotypic and quality traits in indigenous and exotic accessions of linseed. Pesq Agropec Bras. (2016) 51:1964–72. doi: 10.1590/s0100-204x2016001200007
104. El-Hariri, DM, Al-Kordy, MA, Hassanein, MS, and Ahmed, MA. Partition of photosynthates and energy production in different flax cultivars. J Nat Fibers. (2005) 1:1–15. doi: 10.1300/J395v01n04_01
105. Kumar, S, You, FM, Duguid, S, Booker, H, Rowland, G, and Cloutier, S. QTL for fatty acid composition and yield in linseed (Linum usitatissimum L.). Theor Appl Genet. (2015) 128:965–84. doi: 10.1007/s00122-015-2483-3
106. Ottai, MES, Al-Kordy, MAA, Hussein, RM, and Hassanein, MS. Genetic diversity among Romanian fiber flax varieties under Egyptian conditions. Aust J Basic Appl Sci. (2012) 6:162–8.
107. Biradar, SA, Ajithkumar, K, Rajanna, B, Savitha, AS, Shubha, GV, Shankergoud, I, et al. Prospects and challenges in linseed (Linum usitatissimum L.) production: a review. J Oilseeds Res. (2016) 33:1–13.
108. Thambugala, D, Duguid, S, Loewen, E, Rowland, G, Booker, H, You, FM, et al. Genetic variation of six desaturase genes in flax and their impact on fatty acid composition. Theor Appl Genet. (2013) 126:2627–41. doi: 10.1007/s00122-013-2161-2
109. Friedt, W, Bickert, C, and Schaub, H. In vitro breeding of high linolenic, doubled haploid lines of linseed (Linum usitatissimum L.) via androgenesis. Plant Breed. (1995) 114:322–6. doi: 10.1111/J.1439-0523.1995.TB01242.X
110. Cloutier, S, Ragupathy, R, Miranda, E, Radovanovic, N, Reimer, E, Walichnowski, A, et al. Integrated consensus genetic and physical maps of flax (Linum usitatissimum L.). Theor Appl Genet. (2012) 125:1783–95. doi: 10.1007/s00122-012-1953-0
111. Green, AG . Genetic control of polyunsaturated fatty acid biosynthesis in flax (Linum usitatissimum) seed oil. Theor Appl Genet. (1986) 72:654–61. doi: 10.1007/BF00289004
112. Rowland, GG . An EMS-induced low-linolenic-acid mutant in McGregor flax (Linum usitatissimum L.). Can J Plant Sci. (1991) 71:393–6. doi: 10.4141/cjps91-054
113. Green, AG, and Marshall, AR. Isolation of induced mutants in linseed (Linum usitatissimum L.) having reduced linolenic acid content. Euphytica. (1984) 33:321–8. doi: 10.1007/BF00021128
114. Fikere, M, Mekbib, F, and Wakjira, A. Seed oil diversity of Ethiopian linseed (Linum usitatissimum L.) landraces accessions and some exotic cultivars. Afr J Biochem Res. (2013) 7:76–85. doi: 10.5897/AJBR.9000214
115. Dmitriev, AA, Krasnov, GS, Rozhmina, TA, Novakovskiy, RO, Snezhkina, AV, Fedorova, MS, et al. Differential gene expression in response to fusarium oxysporum infection in resistant and susceptible genotypes of flax (Linum usitatissimum L.). BMC Plant Biol. (2017) 17:253. doi: 10.1186/s12870-017-1192-2
116. Singh, J, Singh, PK, and Srivastava, RL. Diseases of linseed (Linum usitatissium L.) in India and their management a review. J Oilseeds Res. (2017) 34:52–69.
117. Diederichsen, A, Rozhmina, T, and Kudrjavceva, L. Variation patterns within 153 flax (Linum usitatissimum L.) genebank accessions based on evaluation for resistance to fusarium wilt, anthracnose and pasmo. Plant Genet Resour. (2008) 6:22–32. doi: 10.1017/S1479262108913897
118. Rozhmina, TA . Identification of genes for resistance to fusarium wilt in flax, breeding seed production agrotechnology, economics and primary processing of flax. (2002) 30:48–52. Torzhok, Russia
119. Rozhmina, TA, and Loshakova, NI. Samples of spinning and oil flax (Linum usitatissimum L.). sources of effective genes for resistance to Fusarium wilt and its dependence on temperature. Agric Biol. (2016) 51:310–7.
120. Evans, N, Mcroberts, N, Hitchcock, D, and Marshall, G. Assessing linseed (Linum usitatissimum) resistance to Alternaria linicola using a detached cotyledon assay. Ann Appl Biol. (1997) 130:537–47. doi: 10.1111/j.1744-7348.1995.tb06671.x
121. Dash, J, Naik, BS, and Mohapatra, UB. Field screening of linseed genotypes for resistance to powdery mildew (Oidium lini Skoric) in the north central plateau zone of Odisha. Int J Adv Res. (2016) 4:961–2. doi: 10.21474/IJAR01/204
122. Naik, BS, and Sahoo, KC. Survey of diseases of linseed in Mayurbhanj district of Odisha. Int J Adv Res. (2016) 4:1407–10. doi: 10.21474/IJAR01/235
123. Pandey, MK, Singh, R, Singh, A, and Mishra, BK. Influence of dates of sowing, disease incidence and crop yield against Alternaria blight of linseed. J Pharmacogn Phytochem. (2019) 8:1459–61.
124. Singh, RB, Singh, AK, and Srivastava, RK. Assessment of yield losses due to Alternaria blight of linseed. J Oilseeds Res. (2003) 20:168–9.
125. Pandey, RN, and Mishra, DP. Assessment of yield loss due to powdery mildew of linseed. Indian Bot Rep. (1992) 11:62–4.
126. Beale, RE . Studies of resistance in linseed cultivars to Oidium lini and Botrytis cinerea. In: RJ Fraud Williams , editor. Production and protection of linseed. Aspects of applied biology series, association of applied biologists. Wellesbourne, Warwick, UK: Horticulture Research International (1991). 85–90.
127. Rashid, K, and Duguid, S. Inheritance of resistance to powdery mildew in flax. Can J Plant Pathol. (2005) 27:404–9. doi: 10.1080/07060660509507239
128. Dhirhi, N, Mehta, N, and Singh, S. Screening of powdery mildew tolerance in linseed (Linum usitatisimum L.). J Plant Dev Sci. (2017) 9:153–6.
129. Dutt, S, Chahal, AS, Lobana, KS, and Singh, M. Evaluation of genetic stock of linseed (Linum usitatissimum L.) against powdery mildew in Punjab. J Res. (1975) 12:6–8.
130. Kamthan, KP, Shukla, AK, and Misra, DP. Independent genetic resistance to wilt, rust and powdery mildew in linseed I. J Agric Biol Sci. (1991) 7:56–60.
131. Kishore, R, Pandey, M, Tripathi, UK, and Singh, J. Evaluation of elite genotypes of linseed against Fusarium wilt. Indian Phytopathol. (2011) 64:209.
132. Singh, RB, and Singh, RN. Date of sowing and varieties for root-rot and wilt complex in linseed (Linum usitatissimum L.). Indian J Agric Sci. (2011) 81:287–9.
133. Kanapin, A, Bankin, M, Rozhmina, T, Samsonova, A, and Samsonova, M. Genomic regions associated with Fusarium wilt resistance in flax. Int J Mol Sci. (2021) 22:12383. doi: 10.3390/ijms222212383
134. Ajithkumar, K, Biradar, SA, and Rajanna, B. Screening of linseed germplasms for resistance against powdery mildew caused by Oidium lini Skoric. J Mycopathol Res. (2015) 53:247–51.
135. Ramakant, CMP, and Singh, RB. Screening of linseed genotypes against Alternaria blight and bud fly. Crop Res. (2008) 35:124–7.
136. Singh, RB, Singh, HK, and Parmar, A. Yield loss assessment due to Alternaria blight and its management in linseed. Pak J Biol Sci. (2014) 17:511–6. doi: 10.3923/pjbs.2014.511.516
137. Sinha, JN, Singh, AP, and Kumar, P. Reaction of linseed germplasm to rust (Melampsora lini). J Appl Biol. (1993) 3:80–2.
139. Pal, R, and Malik, YP. Histological basis of resistance in linseed against bud fly (Dasyneura lini Barnes) in Central Uttar Pradesh. Int J Agri Invent. (2020) 5:25–30. doi: 10.46492/IJAI/2020.5.1.3
140. Ekka, RK, Mandal, S, Meena, RS, and Padamshali, S. Some physical basis of host plant resistance in linseed (Linum usitatissimum L.), against bud fly, Dasyneuralini (Barnes). J Pharmacogn Phytochem. (2018) 7:2010–5.
142. Sertse, D, You, FM, Ravichandran, S, Soto-Cerda, BJ, Duguid, S, and Cloutier, S. Loci harboring genes with important role in drought and related abiotic stress responses in flax revealed by multiple GWAS models. Theor Appl Genet. (2021) 134:191–212. doi: 10.1007/s00122-020-03691-0
143. Sertse, D, You, FM, Ravichandran, S, and Cloutier, S. The complex genetic architecture of early root and shoot traits in flax revealed by genome-wide association analyses. Front Plant Sci. (2019) 10:1483. doi: 10.3389/fpls.2019.01483
144. Soto-Cerda, BJ, Cloutier, S, Gajardo, HA, Aravena, G, and Quian, R. Identifying drought-resilient flax genotypes and related-candidate genes based on stress indices, root traits and selective sweep. Euphytica. (2019) 215:41–16. doi: 10.1007/s10681-019-2362-0
145. Soto-Cerda, BJ, Cloutier, S, Gajardo, HA, Aravena, G, Quian, R, and You, FM. Drought response of flax accessions and identification of quantitative trait nucleotides (QTNs) governing agronomic and root traits by genome-wide association analysis. Mol Breed. (2020) 40:1–24. doi: 10.1007/s11032-019-1096-y
146. Asgarinia, P, Mirlohi, A, Saeidi, G, Mirik, MAA, Gheysari, M, and Razavi, VS. Selection criteria for assessing drought tolerance in a segregating population of flax (Linum usitatissimum L.). Can J Plant Sci. (2017) 97:424–37. doi: 10.1139/cjps-2016-0140
147. Soto-Cerda, BJ, Larama, G, Gajardo, H, Inostroza-Blancheteau, C, Cloutier, S, Fofana, B, et al. Integrating multi-locus genome-wide association studies with transcriptomic data to identify genetic loci underlying adult root traits responses to drought stress in flax (Linum usitatissimum L.). Environ Exp Bot. (2022) 202:105019. doi: 10.1016/j.envexpbot.2022.105019
148. Li, X, Guo, D, Xue, M, Li, G, Yan, Q, and Jiang, H. Genome-wide association study of salt tolerance at the seed germination stage in flax (Linum usitatissimum L.). Genes. (2022) 13:486. doi: 10.3390/genes13030486
149. Patil, N, Datir, S, and Shah, P. Salt-induced physiological and biochemical changes in two varieties of Linum usitatissimum L. Int J Curr Microbiol App Sci. (2015) 4:296–304.
150. Kaya, MD, Day, S, Cikili, Y, and Arslan, N. Classification of some linseed (Linum usitatissimum L.) genotypes for salinity tolerance using germination, seedling growth, and ion content. Chil J Agric Res. (2012) 72:27–32. doi: 10.4067/s0718-58392012000100005
151. El-Beltagi, HS, Salama, ZA, and El Hariri, DM. Some biochemical markers for evaluation of flax cultivars under salt stress conditions. J Nat Fibers. (2008) 5:316–30. doi: 10.1080/15440470802252487
152. Basandrai, D, and Basandrai, AK. Genetics of field resistance to powdery mildew in flax. Indian Phytopathol. (2000) 53:224–6.
153. Singh, BM, and Saharan, GS. Inheritance of resistance to Oidium lini Skoric in linseed (Linum usitatissimum L.). Euphytica. (1979) 28:531–2. doi: 10.1007/BF00056614
154. Asgarinia, P, Cloutier, S, Duguid, S, Rashid, K, Mirlohi, A, Banik, M, et al. Mapping quantitative trait loci for powdery mildew resistance in flax (Linum usitatissimum L.). Crop Sci. (2013) 53:2462–72. doi: 10.2135/cropsci2013.05.0298
155. Speck, A, Trouvé, JP, Enjalbert, J, Geffroy, V, Joets, J, and Moreau, L. Genetic architecture of powdery mildew resistance revealed by a genome-wide association study of a worldwide collection of flax (Linum usitatissimum L.). Front Plant Sci. (2022) 13:871633. doi: 10.3389/fpls.2022.871633
156. Goray, SC, Khosla, HK, Upadhyaya, YM, Naik, SL, and Mandoli, SL. Inheritance of wilt resistance in linseed. Indian J Agric Sci. (1987) 57:625–7. doi: 10.1093/oxfordjournals.jhered.a111281
157. Misra, DP, and Pandey, RN. Linkage of resgenes for powdery-mildew and rust resistance in linseed. Indian J Agric Sci. (1981) 51:559–62.
158. Misra, DP, and Shukla, AK. Genetic identity for rust resistance in some linseeds. Indian J Agric Sci. (1981) 27:444–52. doi: 10.1177/0019556119810209
159. Saharan, GS, and Singh, BM. Inheritance of resistance in linseed to rust caused by Melampsora lini under field conditions. Indian J Mycol Pl Pathol. (1980) 10:136–41.
160. Misra, DP . Rust resistance in the species and varieties of Linum against Indian physiological races of linseed rust. J Oilseeds Res. (1964) 8:277–9.
161. Biswas, GC, and Das, GP. Insect and mite pests diversity in the oilseed crops ecosystems in Bangladesh. Bangladesh J Zool. (2011) 39:235–44. doi: 10.3329/bjz.v39i2.10594
162. Pruthi, HS, and Bhatia, HL. A new cecidomyiidae pest of linseed in India. Indian J Agric Sci. (1937) 7:797–808.
163. Malik, YP . Estimation of economic threshold level for bud fly, Dasyneura lini Barnes in linseed, Linum usitatissimum L. J Oilseeds Res. (2005) 22:100–2.
164. Malik, YP . Yield losses due to bud fly, Dasyneura lini Barnes in linseed. J Oilseeds Res. (2006) 23:363.
165. Malik, YP, Husain, K, Singh, SV, and Srivastava, RL. Development of management module for bud fly, Dasyneura lini in linseed. Indian J Entomol. (2000) 62:260–9.
166. Gupta, MP . Field screening of linseed, Linum usitatissimum Linn. Genotypes against the incidence of budfly, Dasyneura lini Barnes and blight, Alternaria lini Dey. J Oilseeds Res. (2004) 21:108–11.
167. Pal, R, and Malik, YP. Screening of linseed germplasm against bud fly Dasyneura lini (Barnes) under field condition. Int J Pure Appl Biosci. (2018) 6:196–201. doi: 10.18782/2320-7051.6542
168. Pal, RK, and Singh, R. Screening of linseed germplasms against bud fly, Dasyneura lini Barnes. Int J Plant Prot. (2010) 3:410–1.
169. Reddy, N, Maheshwari, JJ, Patil, PV, Ghorapade, PB, and Gohokar, RT. Simplified triple test cross analysis for bud fly resistance and yield components in linseed, Linum usitatissimum L. J Oilseed. (2009) 26:166–8.
170. Gupta, AK . Biophysical basis of resistance in linseed against bud fly, Dasyneura lini (Barnes). Indian J Entomol. (2015) 77:345–52. doi: 10.5958/0974-8172.2015.00073.5
171. Sood, NK, and Pathak, SC. Thickness of sepal: a factor for resistance in linseed against Dasyneura lini Barnes. Indian J Plant Prot. (1990) 52:28–30.
172. Heller, K, Konczewicz, W, Byczyńska, M, Łukaszewska, N, and Praczyk, M. (2008). “The effect of fiber flax growing technologies on plants ontogenesis and cultivars yielding capacity” in Conference on flax and other bast plants. Saskatoon: Saskatchewan, Canada. pp. 315–325.
173. Heller, K, and Byczyńska, M. The impact of environmental factors and applied agronomy on quantitative and qualitative traits of flax fiber. J Nat Fibers. (2015) 12:26–38. doi: 10.1080/15440478.2013.879088
174. Hall, LM, Booker, H, Siloto, RMP, Jhala, AJ, and Weselake, RJ. Flax (Linum usitatissimum L.) In: T McKeon, D Hayes, D Hildebrand, and R Weselake, editors. J.Industrial oil crops. AOCS Press, New York: Elsevier (2016). 157–94.
175. Kar, G, Kumar, A, and Martha, M. Water use efficiency and crop coefficients of dry season oilseed crops. Agric Water Manag. (2007) 87:73–82. doi: 10.1016/j.agwat.2006.06.002
176. Dash, P, Cao, Y, Jailani, A, Gupta, P, Venglat, P, Xiang, D, et al. Genome-wide analysis of drought induced gene expression changes in flax (Linum usitatissimum). GM Crops Food. (2014) 5:106–19. doi: 10.4161/gmcr.29742
177. Foster, R, Pooni, H, and Mackay, I. The impact of water deprivation on the performance of Linum usitatissimum cultivars. J Genet Breed. (1998) 52:63–71.
178. Lisson, S, and Mendham, N. Agronomic studies of flax (Linum usitatissimum L.) in South-Eastern Australia. Aust J Exp Agric. (2000) 40:1101–12. doi: 10.1071/EA00059
179. Diederichsen, A, and Raney, JP. Pure-lining of flax (Linum usitatissimum L.) genebank accessions for efficiently exploiting and assessing seed character diversity. Euphytica. (2008) 164:255–73. doi: 10.1007/s10681-008-9725-2
180. EL Afry, MM, EL, OSA, EL, KESA, and EL, YGSA. Exogenous application of ascorbic acid for alleviation the adverse effects of salinity stress in flax (Linum usitatissimum L.). Middle East J. (2018) 7:716–39.
181. Guo, R, Zhou, J, Hao, W, Gu, F, Liu, Q, Li, H, et al. Germination, growth, chlorophyll fluorescence and ionic balance in linseed seedlings subjected to saline and alkaline stresses. Plant Prod Sci. (2014) 17:20–31. doi: 10.1626/pps.17.20
182. Nasri, N, Maatallah, S, Saidi, I, and Lachâal, M. Influence of salinity on germination, seedling growth, ion content and acid phosphatase activities of Linum usitatissimum L. J Anim Plant Sci. (2017) 27:517–21.
183. Qayyum, MA, Bashir, F, Maqbool, MM, Ali, A, Bashir, S, and Abbas, Q. Implications of saline water irrigation for linseed on seed germination, seedling survival and growth potential. Sarhad J Agric. (2019) 35:1289–97. doi: 10.17582/journal.sja/2019/35.4.1289.1297
184. Yu, Y, Wu, G, Yuan, H, Cheng, L, Zhao, D, Huang, W, et al. Identification and characterization of miRNAs and targets in flax (Linum usitatissimum) under saline, alkaline, and saline-alkaline stresses. BMC Plant Biol. (2016) 16:124. doi: 10.1186/s12870-016-0808-2
185. Wu, J, Zhao, Q, Wu, G, Yuan, H, Ma, Y, Lin, H, et al. Comprehensive analysis of differentially expressed unigenes under NaCl stress in flax (Linum usitatissimum L.) using RNA-Seq. Int J Mol Sci. (2019) 20:369. doi: 10.3390/ijms20020369
186. Kraft, JM, Kommedahl, T, and Linck, AL. Histological study of malformation in flaxseed after exposure to 31°C. Bot Gaz. (1963) 125:367–71. doi: 10.1086/336221
187. Gusta, LV, O’Conner, BJ, and Bhatty, RS. Flax (Linum usitatissimum L.) response to chilling and heat stress on flowering and seed yield. Can J Plant Sci. (1997) 77:97–9. doi: 10.4141/P95-205
188. Cross, RH, Mckay, SAB, McHughen, AG, and Bonham-Smith, PC. Heat-stress effects on reproduction and seed set in Linum usitatissimum L. (flax). Plant Cell Environ. (2003) 26:1013–20. doi: 10.1046/j.1365-3040.2003.01006.x
189. Fofana, B, Cloutier, S, Duguid, S, Ching, J, and Rampitsch, C. Gene expression of stearoyl-ACP desaturase and D12 fatty acid desaturase 2 is modulated during seed development of flax (Linum usitatissimum). Lipids. (2006) 41:705–20. doi: 10.1007/s11745-006-5021-x
190. Saha, D, Shaw, AK, Datta, S, Mitra, J, and Kar, G. DNA hypomethylation is the plausible driver of heat stress adaptation in Linum usitatissimum. Physiol Plant. (2022) 174:e13689. doi: 10.1111/ppl.13689
191. Aldrich, J, and Cullis, CA. RAPD analysis in flax: optimization of yield and reproducibility using klenTaq1 DNA polymerase, Chelex 100, and gel purification of genomic DNA. Plant Mol Biol Report. (1993) 11:128–14. doi: 10.1007/BF02670471
192. Cullis, CA, Oh, TJ, and Gorman, MB. (1995), “Genetic mapping in flax (Linum usitatissimum)” in Proc. 3rd Meeting Int. Flax Breed Res Group. St. Vale´ ryencaux. pp. 161–169.
193. Cullis, CA, Swami, S, and Song, Y. RAPD polymorphisms detected between flax genotrophs. Plant Mol Biol. (1999) 41:795–800. doi: 10.1023/a:1006385606163
194. Everaert, I, Riek, JD, Loose, MD, Waes, JV, and Bockstaele, EV. Most similar variety grouping for distinctness evaluation of flax and linseed (Linum usitatissimum L.) varieties by means of AFLP and morphological data. Plant Var Seeds. (2001) 14:69–87.
195. Fu, Y, Guerin, S, Peterson, GW, Diederichsen, A, Rowland, GG, and Richards, KW. RAPD analysis of genetic variability of regenerated seeds in the Canadian flax cultivar CDC normandy. Seed Sci Technol. (2003) 31:207–11. doi: 10.15258/sst.2003.31.1.22
196. Fu, YB, Diederichsen, A, Richards, KW, and Peterson, G. Genetic diversity within a range of cultivars and landraces of flax (Linum usitatissimum L) as revealed by RAPDs. Genet Resour Crop Evol. (2002) 49:167–74. doi: 10.1023/A:1014716031095
197. Fu, YB, Rowland, GG, Duguid, SD, and Richards, K. RAPD analysis of 54 north American flax cultivars. Crop Sci. (2003) 43:1510–5. doi: 10.2135/cropsci2003.1510
198. Kumari, A, Paul, S, and Sharma, V. Genetic diversity analysis using RAPD and ISSR markers revealed discrete genetic makeup in relation to fibre and oil content in Linum usitatissimum L. genotypes. Nucleus. (2018) 61:45–53. doi: 10.1007/s13237-017-0206-7
199. Rajwade, AV, Arora, RS, Kadoo, NY, Harsulkar, AM, Ghorpade, PB, and Gupta, VS. Relatedness of Indian flax genotypes (Linum usitatissimum L.): an inter-simple sequence repeat (ISSR) primer assay. Mol Biotechnol. (2010) 45:161–70. doi: 10.1007/s12033-010-9256-7
200. Uysal, H, Fu, YB, Kurt, O, Peterson, GW, Diederichsen, A, and Kusters, P. Genetic diversity of cultivated flax (Linum usitatissimum L.) and its wild progenitor pale flax (Linum bienne mill.) as revealed by ISSR markers. Genet Resour Crop Evol. (2010) 57:1109–19. doi: 10.1007/s10722-010-9551-y
201. Wiesnerova, D, and Wiesner, I. ISSR-based clustering of cultivated flax germplasm is statistically correlated to thousand seed mass. Mol Biotechnol. (2004) 26:207–14. doi: 10.1385/MB:26:3:207
202. Smykal, P, Kerteszova, NB, Kalendar, R, Corander, J, Schulman, AH, and Pavelek, M. Genetic diversity of cultivated flax (Linum usitatissimum L.) germplasm assessed by retrotransposon-based markers. Theor Appl Genet. (2011) 122:1385–97. doi: 10.1007/s00122-011-1539-2
203. Kumari, R, Wankhede, DP, Bajpai, A, Maurya, A, Prasad, K, Gautam, D, et al. Genome wide identification and characterization of microsatellite markers in black pepper (Piper nigrum): a valuable resource for boosting genomics applications. PLoS One. (2019) 14:e0226002. doi: 10.1371/journal.pone.0226002
204. Cloutier, S, Niu, Z, Datla, R, and Duguid, S. Development and analysis of EST-SSRs for flax (Linum usitatissimum L.). Theor Appl Genet. (2009) 119:53–64. doi: 10.1007/s00122-009-1016-3
205. Deng, X, Long, S, He, D, Li, X, Wang, Y, Hao, D, et al. Isolation and characterization of polymorphic microsatellite markers from flax (Linum usitatissimum L.). Afr J Biotechnol. (2011) 10:734–9.
206. Deng, X, Long, S, He, D, Li, X, Wang, Y, Liu, J, et al. Development and characterization of polymorphic microsatellite markers in Linum usitatissimum. J Plant Res. (2010) 123:119–23. doi: 10.1007/s10265-009-0271-3
207. Roose-Amsaleg, C, Cariou-Pham, E, Vautrin, DA, Tavernier, R, and Solignac, M. Polymorphic microsatellite loci in Linum usitatissimum. Mol Ecol Notes. (2006) 6:796–9. doi: 10.1111/j.1471-8286.2006.01348.x
208. Kale, SM, Pardeshi, VC, Kadoo, NY, Ghorpade, PB, Jana, MM, and Gupta, VS. Development of genomic simple sequence repeat markers for linseed using next-generation sequencing technology. Mol Breed. (2012) 30:597–606. doi: 10.1007/s11032-011-9648-9
209. Wu, J, Zhao, Q, Wu, G, Zhang, S, and Jiang, T. Development of novel SSR markers for flax (Linum usitatissimum L.) using reduced-representation genome sequencing. Front. Plant Sci. (2017) 7:2016. doi: 10.3389/fpls.2016.02018
210. Pan, G, Chen, A, Li, J, Huang, T, Chang, L, Zhao, L, et al. Genome-wide development of simple sequence repeats database for flax (Linum usitatissimum L.) and its use for genetic diversity assessment. Genet Resour Crop Evol. (2020) 67:865–74. doi: 10.1007/s10722-020-00882-y
211. Saha, D, Rana, RS, Das, S, Datta, S, Mitra, J, Cloutier, SJ, et al. Genome-wide regulatory gene-derived SSRs reveal genetic differentiation and population structure in fiber flax genotypes. J Appl Genet. (2019) 60:13–25. doi: 10.1007/s13353-018-0476-z
212. Landoni, B, Viruel, J, Gómez, R, Allaby, RG, Brennan, AC, Picó, FX, et al. Microsatellite marker development in the crop wild relative Linum bienne using genome skimming. Appl Plant Sci. (2020) 8:e11349. doi: 10.1002/aps3.11349
213. He, L, Xiao, J, Rashid, KY, Yao, Z, Li, P, Jia, G, et al. Genome-wide association studies for pasmo resistance in flax (Linum usitatissimum L.). Front Plant Sci. (2018) 9:1982. doi: 10.3389/fpls.2018.01982
214. Xie, D, Dai, Z, Yang, Z, Sun, J, Zhao, D, Yang, X, et al. Genome-wide association study identifying candidate genes influencing important agronomic traits of flax (Linum usitatissimum L.) using SLAF-seq. Front Plant Sci. (2018) 8:8–2232. doi: 10.3389/fpls.2017.02232
215. Soto-Cerda, BJ, Aravena, G, and Cloutier, S. Genetic dissection of flowering time in flax (Linum usitatissimum L.) through single- and multi-locus genome-wide association studies. Mol Gen Genomics. (2021) 296:877–91. doi: 10.1007/s00438-021-01785-y
216. Wu, J, Zhao, Q, Zhang, L, Li, S, Ma, Y, Pan, L, et al. QTL mapping of fiber-related traits based on a high-density genetic map in flax (Linum usitatissimum L.). Front Plant Sci. (2018) 9:9–885. doi: 10.3389/fpls.2018.00885
217. You, FM, and Cloutier, S. Mapping quantitative trait loci onto chromosome-scale pseudomolecules in flax. Methods Protoc. (2020) 3:28. doi: 10.3390/mps3020028
218. You, FM, Rashid, KY, Zheng, C, Khan, N, Li, P, Xiao, J, et al. Insights into the genetic architecture and genomic prediction of powdery mildew resistance in flax (Linum usitatissimum L.). Int J Mol Sci. (2022) 23:4960. doi: 10.3390/ijms23094960
219. You, FM, Xiao, J, Li, P, Yao, Z, Jia, G, He, L, et al. Genome-wide association study and selection signatures detect genomic regions associated with seed yield and oil quality in flax. Int J Mol Sci. (2018) 19:2303. doi: 10.3390/ijms19082303
220. Chandrawati, MR, Singh, PK, Ranade, SA, and Yadav, HK. Diversity analysis in Indian genotypes of linseed (Linum usitatissimum L.) using AFLP markers. Gene. (2014) 549:171–8. doi: 10.1016/j.gene.2014.07.067
221. Hoque, A, Fiedler, JD, and Rahman, M. Genetic diversity analysis of a flax (Linum usitatissimum L.) global collection. BMC Genomics. (2020) 21:557. doi: 10.1186/s12864-020-06922-2
222. Mhiret, WN, and Heslop-Harrison, JS. Biodiversity in Ethiopian linseed (Linum usitatissimum L.): molecular characterization of landraces and some wild species. Genet Resour Crop Evol. (2018) 65:1603–14. doi: 10.1007/s10722-018-0636-3
223. Spielmeyer, W, Green, AG, Bittisnich, D, Mendham, N, and Lagudah, ES. Identification of quantitative trait loci contributing to fusarium wilt resistance on an AFLP linkage map of flax (Linum usitatissimum). Theor Appl Genet. (1998) 97:633–41. doi: 10.1007/s001220050939
224. Zhang, J, Long, Y, Wang, L, Dang, Z, Zhang, T, Song, X, et al. Consensus genetic linkage map construction and QTL mapping for plant height-related traits in linseed flax (Linum usitatissimum L.). BMC Plant Biol. (2018) 18:160. doi: 10.1186/s12870-018-1366-6
225. Wang, YH, Wu, DH, Huang, JH, Tsao, SJ, Hwu, KK, and Lo, HF. Mapping quantitative trait loci for fruit traits and powdery mildew resistance in melon (Cucumis melo). Bot Stud. (2016) 57:19. doi: 10.1186/s40529-016-0130-1
226. Akhmetshina, AO, Strygina, KV, Khlestkina, EK, Porokhovinova, EA, and Brutch, NB. High-throughput sequencing techniques to flax genetics and breeding. Ecol Gen. (2020) 18:103–24. doi: 10.17816/ecogen16126
227. Soto-Cerda, BJ, Cloutier, S, Quian, R, Gajardo, HA, Olivos, M, and You, FM. Genome-wide association analysis of mucilage and hull content in flax (Linum usitatissimum L.) seeds. Int J Mol Sci. (2018) 19:2870. doi: 10.3390/ijms19102870
228. Xie, D, Da, Z, Yang, Z, Tang, Q, Sun, J, Yang, X, et al. Genomic variations and association study of agronomic traits in flax. BMC Genomics. (2018) 19:512. doi: 10.1186/s12864-018-4899-z
229. Wang, Z, Hobson, N, Galindo, L, Zhu, S, Shi, D, McDill, J, et al. The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J. (2012) 72:461–73. doi: 10.1111/j.1365-313x.2012.05093.x
230. You, FM, Xiao, J, Li, P, Yao, Z, Jia, G, He, L, et al. Chromosome-scale pseudomolecules refined by optical, physical, and genetic maps in flax. Plant J. (2018) 95:371–84. doi: 10.1111/tpj.13944
231. Dmitriev, AA, Pushkova, EN, Novakovskiy, RO, Beniaminov, AD, Rozhmina, TA, Zhuchenko, AA, et al. Genome sequencing of fiber flax cultivar Atlant using oxford nanopore and illumina platforms. Front Genet. (2021) 11:590282. doi: 10.3389/fgene.2020.590282
232. Sa, R, Yi, L, Siqin, B, An, M, Bao, H, Song, X, et al. Chromosome-level genome assembly and annotation of the fiber flax (Linum usitatissimum) genome. Front Genet. (2021) 12:735690. doi: 10.3389/fgene.2021.735690
233. Zhang, J, Qi, Y, Wang, L, Wang, L, Yan, X, Dang, Z, et al. Genomic comparison and population diversity analysis provide insights into the domestication and improvement of flax. iScience. (2020) 23:100967. doi: 10.1016/j.isci.2020.100967
234. de Santana, LA, Pacheco, TG, Santos, KGD, Vieira, LDN, Guerra, MP, Nodari, RO, et al. The Linum usitatissimum L. plastome reveals atypical structural evolution, new editing sites, and the phylogenetic position of Linaceae within Malpighiales. Plant Cell Rep. (2018) 37:307–28. doi: 10.1007/s00299-017-2231-z
235. Kaur, V, Aravind, J, Manju, JSR, Kumari, J, Panwar, BS, et al. Phenotypic characterization, genetic diversity assessment in 6,778 accessions of barley (Hordeum vulgare L. ssp. vulgare) germplasm conserved in National Genebank of India and development of a core set. Front Plant Sci. (2022) 13:771920. doi: 10.3389/fpls.2022.771920
236. Brown, AHD . Core collections: a practical approach to genetic resources management. Genome. (1989) 31:818–24. doi: 10.1139/g89-144
237. Frankel, OH . Genetic perspectives of germplasm conservation In: In genetic manipulation: Impact on man and society. Cambridge: Cambridge University Press (1984). 161–70.
238. Kutuzova . Catalogue of the world collection at the VIR, donors of economically important characters for breeding of fiber flax, vol. 714. St. Petersburg: VIR (2000). 50 p.
239. Van Soest, LJM, and Bas, N. Current status of the CGN Linum collection In: Flax genetic resources in Europe. Maccarese Rome: IPGRI (2002). 44–8.
240. Painting, KA, Perry, MC, Dennin, RA, and Ayad, WG. Guidebook for genetic resources documentation, a self-teaching approach to the understanding, analysis and development of genetic resources documentation. Rome: International Plant Genetic Resources Institute (1993).
241. Gupta, K, Archak, S, Pradheep, K, Kumar, S, Jacob, SR, Tyagi, V, et al. ICAR-NBPGR: bridging science and service In: ICAR-National Bureau of plant genetic resources. India: New Delhi (2012-2018). 74.
242. Kak, A, and Gupta, V. (2015) Inventory of registered crop germplasm (2015–2017). ICAR-National Bureau of Plant Genetic Resources, Pusa Campus, Indian Council of Agricultural Research, New Delhi, India. pp. 74.
243. Kak, A, and Tyagi, RK. (2017). Inventory of registered crop germplasm (2010–2014). ICAR-National Bureau of Plant Genetic Resources, Pusa Campus, Indian Council of Agricultural Research, New Delhi, India. pp. 103.
244. Kak, A, Srinivasan, K, and Sharma, SK. (2009). Plant germplasm registration National Bureau of Plant Genetic Resources, Pusa campus, Indian Council of Agricultural Research, New Delhi, India. pp. 75.
245. Srivastava, RL, Singh, K, Singh, J, and Kerkhi, SA. Catalogue on linseed germplasm Project coordinating unit (linseed). Kanpur, UP, India: CSAUA&T (2010)
246. Verma, VD, Mandel, S, Patel, DP, Loknathan, TR, Singh, B, Sapra, RL, et al. Evaluation of linseed germplasm. National Bureau of plant genetic resources. Akola, Maharashtra, India: Regional Station (1993)
247. Rozhmina, T, Mikhail, B, Anastasia, S, Alexander, K, and Maria, S. A comprehensive dataset of flax (Linum uitatissimum L.) phenotypes. Data Br. (2021) 37:107224. doi: 10.1016/j.dib.2021.107224
248. Mokshina, N, Gorshkov, O, Takasaki, H, Onodera, H, Sakamoto, S, Gorshkova, T, et al. FIBexDB: a new online transcriptome platform to analyze development of plant cellulosic fibers. New Phytol. (2021) 231:512–5. doi: 10.1111/nph.17405
249. GENESYS (2023) Available at: https://www.genesys-pgr.org/ [Accessed 31 January, 2023].
Keywords: characterisation, genetic resources, genebank collections, genomics, linseed, flax, nutrition enrichment
Citation: Kaur V, Singh M, Wankhede DP, Gupta K, Langyan S, Aravind J, Thangavel B, Yadav SK, Kalia S, Singh K and Kumar A (2023) Diversity of Linum genetic resources in global genebanks: from agro-morphological characterisation to novel genomic technologies – a review. Front. Nutr. 10:1165580. doi: 10.3389/fnut.2023.1165580
Edited by:
Miroslav Griga, Agritec (Czech Republic), CzechiaReviewed by:
Petr Smýkal, PalackýUniversity in Olomouc, CzechiaRoberto Rodríguez Ramírez, Instituto Tecnológico de Sonora (ITSON), Mexico
Copyright © 2023 Kaur, Singh, Wankhede, Gupta, Langyan, Aravind, Thangavel, Yadav, Kalia, Singh and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vikender Kaur, dmlrZW5kZXIua2F1ckBpY2FyLmdvdi5pbg==
†Present addresses
Boopathi Thangavel, Crop Protection, Indian Council of Agricultural Research-Indian Institute of Oilseeds Research, Hyderabad, Telangana, India
Kuldeep Singh, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Patancheru, Telangana, India
 Vikender Kaur
Vikender Kaur Mamta Singh
Mamta Singh Dhammaprakash Pandhari Wankhede
Dhammaprakash Pandhari Wankhede Kavita Gupta
Kavita Gupta Sapna Langyan
Sapna Langyan Jayaraman Aravind
Jayaraman Aravind Boopathi Thangavel
Boopathi Thangavel Shashank Kumar Yadav
Shashank Kumar Yadav Sanjay Kalia
Sanjay Kalia Kuldeep Singh
Kuldeep Singh Ashok Kumar
Ashok Kumar