- International Institutes of Medicine, The Fourth Affiliated Hospital of Zhejiang University School of Medicine, Yiwu, Zhejiang, China
The circadian clock governs activity of many physiological processes, thereby playing a pivotal role in human health. Circadian disruption is closely associated with cancer development; in particular, recent discoveries have provided strong evidence supporting specific functions of different molecular clock components in either promoting or inhibiting tumorigenesis. This narrative review aims to summarize the existing data on molecular connections between the clock and cancer. These results along with future efforts pave the road to targeting the circadian clock as a novel pathway for therapeutic intervention. Given the implications of chrono-nutrition interventions such as time-restricted feeding in extending lifespan, chrono-nutrition may have preventive and therapeutic applications for individuals with and at-risk of age-related diseases including cancer.
Introduction
Evolved by most living creatures, the circadian clock allows organisms to organize their behavior and physiology to anticipate diurnal environmental changes. The mammalian circadian machinery is centrally controlled by core clock genes which form two interlocked transcriptional feedback loops. The BMAL1::CLOCK (brain and muscle ARNT-like 1 and CLOCK) heterodimer activates transcription of the clock repressors PER1/2/3 and CRY1/2, which subsequently form a heterodimer, translocate into the nucleus, and inhibit transcriptional activity of BMAL1::CLOCK. In the other loop, RORs and REV-ERBs, which are also targets of BMAL1::CLOCK, feedback to collaboratively regulate BMAL1::CLOCK by stimulating or repressing Bmal1 transcription, respectively (1). Up to 80% of the protein-coding genes exhibit circadian transcription somewhere in the body of primates, highlighting the molecular clock as a crucial mechanism monitoring a broad range of physiological functions, including sleep, feeding, hormone secretion, metabolism, and immune responses (2, 3). Environmental or genetic disruption of circadian homeostasis exacerbates the development of clinically relevant disorders, such as sleep disorders, obesity, diabetes, and cancers, prompting pharmacological manipulation of the clock for novel treatments (4).
The normal circadian rhythms have long been acknowledged as a tumor suppressor, given that epidemiologic studies have suggested night shift work being a carcinogenic factor, particularly for breast cancer (5, 6). Systemic studies also revealed circadian clock genes are frequently dysregulated or mutated across many human cancer types (7–9). Furthermore, the disruption of circadian rhythms is closely associated with cancer incidence, stage, and survival rate (7, 10, 11). While recent discoveries in different cancer systems have confirmed the significant roles of the circadian machinery in cancer biology, studies conducted in specific cancer systems have provided consistent evidence showing that certain central clock molecules may play oncogenic roles (10). Devising strategies for clock-based cancer therapeutics therefore calls for in-depth mechanistic studies to guide cancer-specific treatments for optimal efficacy and safety. This article will discuss the broad spectrum of molecular functions of the circadian clock in driving cancer progression.
Circadian control of cell cycle progression and DNA damage response
The circadian clock and the cell cycle are interconnected biological circuits both frequently dysregulated in cancer (12). Discovering BMAL1 and CLOCK, the forementioned master transcription factors of circadian rhythms, are required for cancer cell proliferation has encouraged experimental and clinical investigations to inform novel anticancer strategies. Downregulating BMAL1 or CLOCK triggers reduced cell proliferation rate in the glioblastoma stem cells (GSCs), hepatocellular carcinoma (HCC) cells, and leukemia stem cells, featured by a symbolic cellular response of cell cycle dysregulation (13–15). BMAL1 or CLOCK inhibition arrests the cell cycle at the S/G2 phase, which is likely associated with the increased expression of p21 and reduced level of CYCLIN B1 (14, 16). Meanwhile, BMAL1/CLOCK knockdown induces the downregulation of the WEE1 gene, which encodes a checkpoint kinase essential for cancer cells’ survival and, consequently, leads to enhanced genome instability and apoptosis (14). Genetic studies demonstrated that transcriptional regulation of WEE1 and p21 cooperatively contributes to cancer cell proliferation promoted by BMAL1::CLOCK (14). Taken together, cell cycle regulation is an important cellular mechanism used by the circadian clock for tumor growth control (Figure 1).
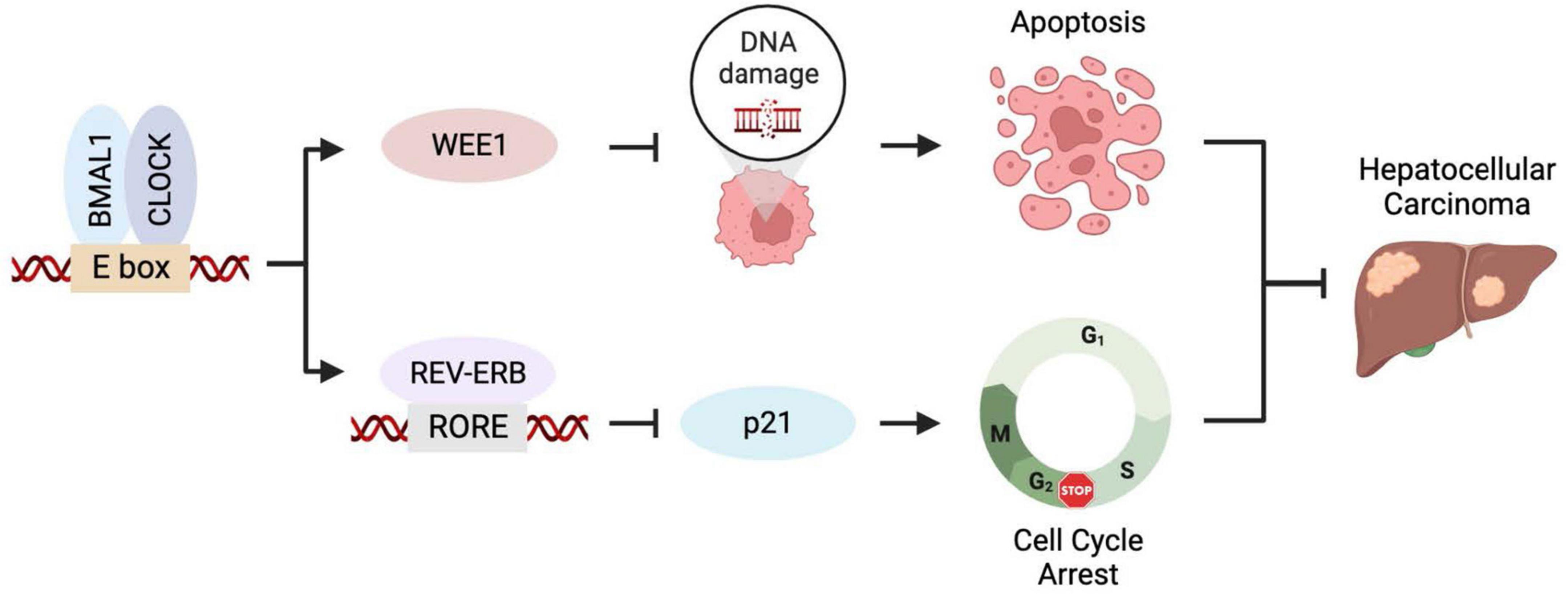
Figure 1. BMAL1::CLOCK promotes HCC growth by controlling cell cycle regulators. BMAL1::CLOCK directly activates the transcription of WEE1 but inhibits p21 expression via regulating the REV-ERBs level. Inhibition of BMAL1::CLOCK downregulates WEE1 leading to mitotic catastrophe and apoptosis and elevates p21 level that causes cell cycle arrest. The two mechanisms collaboratively contribute to the pro-proliferative activity of BMAL1::CLOCK in HCC.
Genome instability resulting from compromised DNA damage responses is a major hallmark of cancer that contributes to cancer initiation and progression (17). DNA damage response activates checkpoints that enforce cell cycle arrest to allow DNA repair before cell division (18) and therefore shares similar regulation by the clock as in the cell cycle pathways. Besides, studies have found that the core clock components may regulate earlier stages of the DNA damage response via interaction with the damaged DNA or molecules involved in DNA damage detection and signaling. For example, the CLOCK protein was reported to relocalize to the DNA lesion sites and participate in the checkpoint activation (19). Consequently, loss of CLOCK or BMAL1 in mice results in enhanced sensitivity to DNA cross-linking agents (20). CRYs are regulated by DNA damage through post-translational modification and subsequently protect genome integrity by modulating ATR activity (21, 22). PERs physically interact with the checkpoint proteins ATM and CHK2, promote DNA damage-induced apoptosis, and protect animals from tumor development (23–25). The clock factor TIMELESS interacts with the DNA damage response kinases CHK1 and ATR and contributes to DNA damage checkpoint response (26). Overall, most circadian clock components are generally positive regulators of the DNA damage response, thereby suppressing cancer development in healthy tissues. Notably, in prostate cancer, the CRY1 expression is elevated by androgen, endowing it with pro-tumorigenic functions to promote DNA repair and cancer cell survival after ionizing radiation treatment (27).
In addition to DNA damage response, the circadian clock machinery strictly controls the activity of DNA damage repair, particularly the nucleotide excision repair (NER) pathway which specifically repairs DNA lesions caused by UV irradiation, cigarette smoke, and the cross-linking agent cisplatin. The underlying mechanisms involve the clock-controlled circadian expression of xeroderma pigmentosum group A (XPA), an essential scaffold protein of the NER machinery, leading to its peak expression at ZT10 (28). Besides, because the NER repair is facilitated by locally activated transcription, DNA damages at rhythmically expressed genes exhibit a circadian repair on the transcribed strand in phase with the rhythmicity of gene expression (29). Another important link between the clock and DNA damage repair is that the circadian-controlled metabolite nicotinamide adenine dinucleotide (NAD+) acts as the substrate for the poly(ADP-Ribose) polymerase 1 (PARP1) and sirtuin enzymes that contribute to genome stability through participation in the base excision repair (BER), homologous recombination (HR), non-homologous end-joining (NHEJ), and NER (30, 31).
Circadian clock regulation of the classical oncogene Myc
The C-MYC proto-oncogene is a multifunctional transcription factor implicated in many aspects of cellular processes, including proliferation, differentiation, metabolism, and apoptosis (32). Multiple lines of evidence indicate that the cellular level of MYC protein is regulated by the circadian clock. For example, in mice, ubiquitin-mediated MYC degradation is enhanced by CRY2, which recruits the SCFFBXL3 E3 ligase to the phosphorylation-modified MYC (33). Consequently, the absence of CRY2 leads to MYC upregulation and enhanced incidence of lymphoma (33). On the other hand, in GSCs, BMAL1 directly binds the promoter region of MYC. Disrupting BMAL1/CLOCK or overexpressing CRY1 decreases MYC levels and impairs GSC proliferation (13). In lung cancer, conversely, Bmal1 works as a tumor suppressor whose loss leads to increased C-MYC expression and enhanced proliferation (34). Taken together, the MYC regulation reflects cancer-specific functions of the core clock components.
Circadian clock in cancer stem cells
Cancer stem cells (CSCs) are maintained as a subpopulation of tumor cells by self-renewal. Due to this property and their inherent resistance to mainstream treatments, CSCs are key players in tumor relapses (35). As compared to normal neural stem cells, the GSCs are highly sensitive to perturbation of the circadian pathway. For example, the stemness of GSCs was lost upon BMAL1 or CLOCK downregulation, as presented by reduced frequency of sphere formation and decreased expression of core GSC maintenance transcription factors, including SOX2 and OLIG2 (13). Similarly, targeting Clock or Bmal1 also leads to reduced self-renew capacity and differentiation of murine leukemia stem cells (15).
Circadian clock regulation of cancer metabolism
Chronic jet lag may cause spontaneous development of HCC following global liver metabolic dysfunction and increased incidence of non-alcoholic fatty liver disease (NAFLD) (36). This suggests that metabolic disruption plays an important role in HCC development associated with circadian dysfunction. In addition, BMAL1 and CLOCK maintain GSCs by promoting glycolysis and mitochondrial oxidative phosphorylation (OXPHOS), two major mechanisms of energy production for cancer cells, supported by reduced oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) upon BMAL1 or CLOCK knockdown (13). Chromatin binding of BMAL1 is reprogrammed in GSCs to directly regulate multiple metabolic enzymes involved in glycolysis and the TCA cycle which is tightly coupled with OXPHOS (13). In addition, the molecular clock was suggested to modulate the activity of autophagy that plays a vital role in fueling the metabolic demands of cancer cells (37). Overall, the molecular mechanisms of circadian clock-regulated cancer metabolism involve both anabolism and catabolism.
Circadian regulation of tumor microenvironment
The molecular clock has been demonstrated to play a role in tumor microenvironment (TME). For example, BMAL1::CLOCK recruits immune-suppressive microglia cells to the TME of glioblastoma (GBM) via transcriptional activation of the OLFML3 gene (38). Circadian expression of the co-stimulatory molecule CD80 in dendritic cells governs a circadian response of tumor antigen-specific CD8+ T cells to melanoma development (39). The CLOCK protein interacts with HIF-1 to activate VEGF expression, facilitating angiogenesis and metastasis in colorectal cancer (40).
Circadian clock roles in aging
Increasing evidence has linked disruption of the circadian clock function to aging (41). During the aging process, the circadian control declines, resulting in dampened and occasionally shifted oscillations of sleep-week cycle, body temperature, suprachiasmatic nucleus (SCN) activity, hormone release, and plasma glucose levels (41–43). Mice deficient in BMAL1, CLOCK, or PER show reduced life span and premature aging phenotypes, including sarcopenia, osteoporosis, loss of soft tissues, and cataracts (44–47).
Circadian alignment of feeding time combined with >12 h fasting interval (time-restricted feeding, TRF) enhances the lifespan benefits of calorie restriction (CR), indicating that optimizing the phase of circadian gene expression may be a powerful intervention for increasing life span and wellbeing (48). In addition to optimizing the expression of catabolic factors and disease markers, long-term CR and TRF enhance the circadian amplitude of the core clock and output genes (49, 50) and thereby entrain the clock in the SCN or peripheral organs, respectively (51). Interestingly, in both flies and mice, the lifespan extension benefits of CR require core clock genes (52–54). CR induces globally reprogrammed protein acetylation whose cycling organizes the enhancement of circadian oscillations (55). Therefore, reprogramming circadian rhythms holds significant importance for increased longevity achieved by feeding regimens.
Providing molecular mechanisms, deficiency of the BMAL1 gene accelerates aging in both human and cynomolgus monkey mesenchymal progenitor cells (MPCs), attributable to a non-canonical transcription-independent role of BMAL1 in stabilizing heterochromatin which prevents activation of pro-senescence retrotransposons (56). Furthermore, comparative transcriptomic analysis showed that a species’ maximum lifespan negatively correlates with the expression of genes responsible for cellular energy metabolism which are more prone under circadian regulation potentially to avoid persistent high expression (57).
The NCI Annual Plan and Budget Proposal for Fiscal Year 2020 stated that the “greatest risk factor for cancer is advancing age”. The fundamental roles of the clock in aging regulation indicates that anti-aging may be an important mechanism used by the circadian clock in cancer prevention. This concept encourages a healthy circadian rhythm reinforced by chrono-nutrition in everyday practice aiming to combat age-related diseases including cancer.
Circadian disruption in cancer
Transcriptome analysis of patient specimens revealed that a large body of genes cycling in non-cancerous tissues, including core clock genes, may lose oscillations in tumors. For example, serum shock stimulated clear circadian rhythms in the non-tumorigenic breast epithelial cells MCF10A, while the oscillation patterns were not observed in the breast cancer cells MCF7 (7). The CYCLOPS algorism identified quite a number of transcripts cycling in non-cancerous liver samples, including CRY1, PER1, and ARNTL2, lost rhythmic expression in biopsies of human HCC (9). Notably, circadian transcription and metabolism in the liver could be reprogrammed by distal tumors in the lung or breast, highlighting the effects of the tumor macroenvironment on systemic homeostasis of circadian rhythms (58, 59).
Several possible mechanisms can explain circadian disruption during tumorigenesis. C-MYC is a transcription factor constitutively expressed in over 70% of human cancers (32). It has been proposed to disrupt the circadian rhythms in tumor cells by binding an E-box motif element that drives activation of the core clock gene NR1D1 (60). In epigenetic studies, we reported that, relative to normal neural stem cells, the genome-wide BMAL1 occupancy was considerably enhanced in GSCs coupled with reprogrammed chromatin structure (13). The ectopic recruitment of BMAL1 to thousands of additional binding sites drives essential hallmarks of cancer progression in these cells, including ramped-up metabolism and proliferative activity and maintenance of CSC state (13). Thus, cancer cells extensively reprogram the circadian rhythms to render novel oncogenic functions supporting disease physiology. Cell-specific chromatin structure guides deposition of the master circadian clock transcription factors and is thus a pivotal physiological mechanism defining heterogenous circadian rhythms in health and disease (61, 62). Notably, cancer reprogramming of the circadian machinery does not necessarily result in disturbance of the relatively robust central clock oscillation (13, 15).
Chrononutrition and circadian medicine in cancer treatment
As narrated above, maintaining a healthy circadian rhythm through consistent daily sleep patterns and diet increases resilience in fighting cancer. In parallel, efforts have been made to identify chemicals that can strengthen our circadian rhythms aiming to overcome circadian clock dampening associated with aging. For example, the small molecule Nobiletin, a naturally synthesized compound enriched in citrus peels, enhances the circadian clock amplitude and protects against metabolic diseases by agonizing the ROR nuclear receptors (63).
In addition to disease prevention, exciting approaches for the circadian clock management of diseases have been developed to limit eating windows, modify drug scheduling, or target clock components. Independent epidemiological studies discovered that the risk of breast cancer and prostate cancer was significantly associated with late night eating behavior (64–66). In mouse orthotopic models, TRF markedly inhibits tumor initiation, progression, and metastasis of obesity-driven postmenopausal breast cancer accompanied by restored circadian gene expression in the tumor suggesting that TRF might suppress tumorigenesis by regulating tumor clock (67). Similarly, in a transgenic MMTV-PyMT model of spontaneous breast cancer, TRF prevented the procarcinogenic effects of high-fat diet (68). These studies support the beneficial role of chrono-nutrition in cancer management and have encouraged clinical trials to prescribe TRF for the treatment of cancer in humans (ClinicalTrials.gov, identifier NCT05259410).
Clinically actionable genes strongly correlate with clock genes in transcriptional expression, suggesting that circadian drug efficacy should be necessarily investigated and considered (3, 7). Synchronizing drug delivery with a patient’s biological clock, known as chronomedicine, has been clinically validated for the improvement of drug efficacy in pathological conditions such as asthma, hypertension, and cardiovascular disease (69). Extending the boundary of chronotherapy further to the surgical procedures, patients who received cardiac surgery in the afternoon and subsequently suffered from major heart damage were half the patients who underwent the same surgery in the morning (70).
In cancer clinical trials, nevertheless, the application of chronochemotherapy has produced inconsistent results within different clinical trials and patient populations. For example, chronochemotherapy benefits observed in a group of 31 women with ovarian cancer (71) were not replicated in a larger study (72). In a retrospective study reviewing 166 glioblastoma patients, Damato et al. found that patients taking morning temozolomide exhibited longer overall survival compared to evening, especially in MGMT-methylated patients (73). However, when the same group conducted a small feasibility study, they found that among 35 patients on different dosing schedules there was no significant difference in either adverse side effects or survival (74). A phase III trial of 564 colorectal cancer patients from 10 European countries receiving chronomodulated infusion of fluorouracil, leucovorin, and oxaliplatin revealed no benefit of the chronotherapy to the whole study population; however, compared with the conventional treatment group, the risk of death was decreased by 25% for men but increased by 38% for women treated with chronotherapy (75). These confusing clinical trials exemplify the heterogeneity of the circadian rhythms in cancer patients, warranting an aforehand definition of the circadian phase potentially through evaluation of appropriate blood-based biomarkers.
By contrast, the emerging fundamental roles of core clock components in cancer biology have urged developing and examining small molecules directly targeting the clock proteins. Encouragingly, existing clock-targeting small molecules have exhibited specific tumor growth inhibition in glioblastoma and other cancer types (10, 13, 37). Considering the broad roles of the core clock components in managing specific cancers, it is favorable for future endeavors to develop cancer-specific strategies with the goal of improving standard first-line regimens. A better understanding of the clock-cancer interactions will inform blood- or tissue-based biomarkers for molecular stratification of cancer patients that may show differential sensitivity to clock-targeting treatments.
Conclusion
The molecular clock is emerging as a fundamental regulator of cancer development and prognosis. Here we review recent endeavors that collectively reveal the clock’s multifaceted roles in different aspects of cancer development (Figure 2). Further mechanistic investigation in various cancer systems and pathways assisted by clinical and omics data collected from cancer patients will expand the functional landscape of the circadian clock aiming to guide pharmacological strategies.
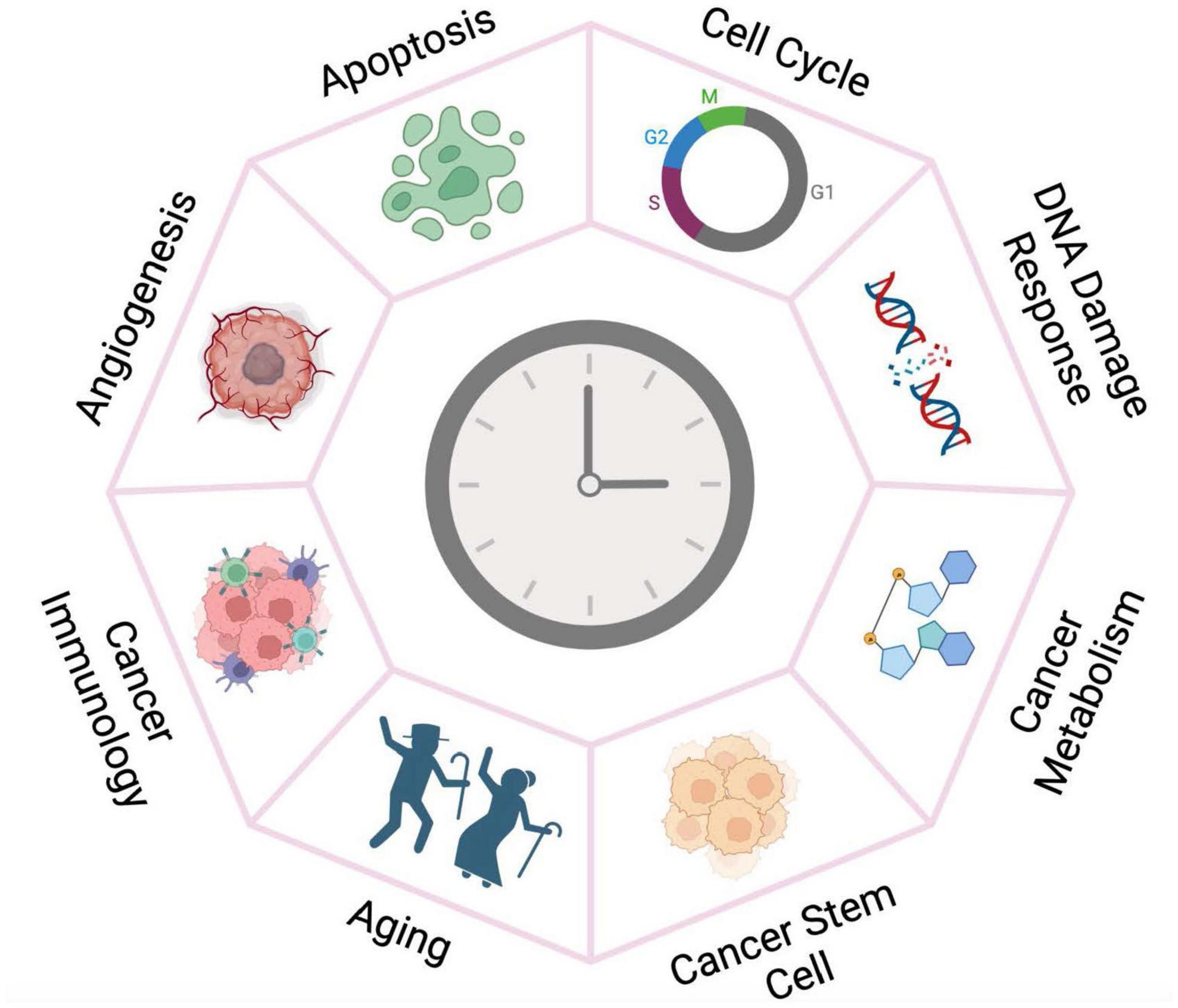
Figure 2. The circadian clock plays versatile roles in cancer development. The circadian clock machinery is reprogrammed by oncogenic pathways and chromatin remodeling in cancer tissues, thereby gaining fundamental functions in cancer development. Targeting the clock may therefore have more deleterious impact on the growth of tumor tissue than for the normal cells.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Takahashi J. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. (2017) 18:164–79. doi: 10.1038/nrg.2016.150
2. Mure L, Le H, Benegiamo G, Chang M, Rios L, Jillani N, et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science. (2018) 359:eaao0318. doi: 10.1126/science.aao0318
3. Zhang R, Lahens N, Ballance H, Hughes M, Hogenesch J. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci U S A. (2014) 111:16219–24. doi: 10.1073/pnas.1408886111
4. Rasmussen E, Takahashi J, Green C. Time to target the circadian clock for drug discovery. Trends Biochem Sci. (2022) 47:745–58. doi: 10.1016/j.tibs.2022.04.009
5. Straif K, Baan R, Grosse Y, Secretan B, Ghissassi F, Bouvard V, et al. Carcinogenicity of shift-work, painting, and fire-fighting. Lancet Oncol. (2007) 8:1065–6. doi: 10.1016/S1470-2045(07)70373-X
6. Burki T. Night shift work and breast cancer. Lancet Oncol. (2019) 20:e352. doi: 10.1016/S1470-2045(19)30383-3
7. Ye Y, Xiang Y, Ozguc F, Kim Y, Liu C, Park P, et al. The genomic landscape and pharmacogenomic interactions of clock genes in cancer chronotherapy. Cells. (2018) 6:314–28.e2. doi: 10.1016/j.cels.2018.01.013
8. Morales-Santana S, Morell S, Leon J, Carazo-Gallego A, Jimenez-Lopez J, Morell M. An overview of the polymorphisms of circadian genes associated with endocrine cancer. Front Endocrinol. (2019) 10:104. doi: 10.3389/fendo.2019.00104
9. Anafi R, Francey L, Hogenesch J, Kim J. CYCLOPS reveals human transcriptional rhythms in health and disease. Proc Natl Acad Sci U S A. (2017) 114:5312–7. doi: 10.1073/pnas.1619320114
10. Battaglin F, Chan P, Pan Y, Soni S, Qu M, Spiller E, et al. Clocking cancer: The circadian clock as a target in cancer therapy. Oncogene. (2021) 40:3187–200. doi: 10.1038/s41388-021-01778-6
11. Battaglin F, Cao S, Millstein J, Puccini A, Tokunaga R, Naseem M, et al. Polymorphism in the circadian clock pathway to predict outcome in patients (pts) with metastatic colorectal cancer (mCRC): Data from TRIBE and FIRE-3 phase III trials. J Clin Oncol. (2018) 36:3576. doi: 10.1200/JCO.2018.36.15_suppl.3576
12. Gaucher J, Montellier E, Sassone-Corsi P. Molecular cogs: Interplay between circadian clock and cell cycle. Trends Cell Biol. (2018) 28:368–79. doi: 10.1016/j.tcb.2018.01.006
13. Dong Z, Zhang G, Qu M, Gimple R, Wu Q, Qiu Z, et al. Targeting glioblastoma stem cells through disruption of the circadian clock. Cancer Discov. (2019) 9:1556–73. doi: 10.1158/2159-8290.CD-19-0215
14. Qu M, Zhang G, Qu H, Vu A, Wu R, Tsukamoto H, et al. Circadian regulator BMAL1::CLOCK promotes cell proliferation in hepatocellular carcinoma by controlling apoptosis and cell cycle. Proc Natl Acad Sci U S A. (2023) 120:e2214829120. doi: 10.1073/pnas.2214829120
15. Puram R, Kowalczyk M, de Boer C, Schneider R, Miller P, McConkey M, et al. Core circadian clock genes regulate leukemia stem cells in AML. Cell. (2016) 165:303–16. doi: 10.1016/j.cell.2016.03.015
16. Farshadi E, Yan J, Leclere P, Goldbeter A, Chaves I, van der Horst G. The positive circadian regulators CLOCK and BMAL1 control G2/M cell cycle transition through Cyclin B1. Cell Cycle. (2019) 18:16–33. doi: 10.1080/15384101.2018.1558638
17. Negrini S, Gorgoulis V, Halazonetis T. Genomic instability — an evolving hallmark of cancer. Nat Rev Mol Cell Biol. (2010) 11:220–8. doi: 10.1038/nrm2858
18. Pilié P, Tang C, Mills G, Yap T. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol. (2019) 16:81–104. doi: 10.1038/s41571-018-0114-z
19. Cotta-Ramusino C, McDonald E, Hurov K, Sowa M, Harper J, Elledge S. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science. (2011) 332:1313–7. doi: 10.1126/science.1203430
20. Gorbacheva V, Kondratov R, Zhang R, Cherukuri S, Gudkov A, Takahashi J, et al. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc Natl Acad Sci U S A. (2005) 102:3407–12. doi: 10.1073/pnas.0409897102
21. Kang T, Leem S. Modulation of ATR-mediated DNA damage checkpoint response by cryptochrome 1. Nucl Acids Res. (2014) 42:gku094. doi: 10.1093/nar/gku094
22. Papp S, Huber A, Jordan S, Kriebs A, Nguyen M, Moresco J, et al. DNA damage shifts circadian clock time via Hausp-dependent Cry1 stabilization. eLife Sci. (2015) 4:e04883. doi: 10.7554/eLife.04883
23. Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler H. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. (2006) 22:375–82. doi: 10.1016/j.molcel.2006.03.038
24. Im J, Jung B, Kim S, Lee K, Lee J. Per3, a circadian gene, is required for Chk2 activation in human cells. FEBS Lett. (2010) 584:4731–4. doi: 10.1016/j.febslet.2010.11.003
25. Fu L, Pelicano H, Liu J, Huang P, Lee C. The Circadian gene period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. (2002) 111:41–50. doi: 10.1016/S0092-8674(02)00961-3
26. Ünsal-Kaçmaz K, Mullen T, Kaufmann W, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. (2005) 25:3109–16. doi: 10.1128/MCB.25.8.3109-3116.2005
27. Shafi A, McNair C, McCann J, Alshalalfa M, Shostak A, Severson T, et al. The circadian cryptochrome, CRY1, is a pro-tumorigenic factor that rhythmically modulates DNA repair. Nat Commun. (2021) 12:401. doi: 10.1038/s41467-020-20513-5
28. Kang T, Lindsey-Boltz L, Reardon J, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc Natl Acad Sci U S A. (2010) 107:4890–5. doi: 10.1073/pnas.0915085107
29. Yang Y, Adebali O, Wu G, Selby C, Chiou Y, Rashid N, et al. Cisplatin-DNA adduct repair of transcribed genes is controlled by two circadian programs in mouse tissues. Proc Natl Acad Sci U S A. (2018) 115:E4777–85. doi: 10.1073/pnas.1804493115
30. Peek C, Affinati A, Ramsey K, Kuo H, Yu W, Sena L, et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science. (2013) 342:1243417. doi: 10.1126/science.1243417
31. Ruszkiewicz J, Bürkle A, Mangerich A. Fueling genome maintenance: On the versatile roles of NAD+ in preserving DNA integrity. J Biol Chem. (2022) 298:102037. doi: 10.1016/j.jbc.2022.102037
32. Madden S, de Araujo A, Gerhardt M, Fairlie D, Mason J. Taking the Myc out of cancer: Toward therapeutic strategies to directly inhibit c-Myc. Mol Cancer. (2021) 20:3. doi: 10.1186/s12943-020-01291-6
33. Huber A, Papp S, Chan A, Henriksson E, Jordan S, Kriebs A, et al. CRY2 and FBXL3 cooperatively degrade c-MYC. Mol Cell. (2016) 64:774–89. doi: 10.1016/j.molcel.2016.10.012
34. Papagiannakopoulos T, Bauer M, Davidson S, Heimann M, Subbaraj L, Bhutkar A, et al. Circadian rhythm disruption promotes lung tumorigenesis. Cell Metab. (2016) 24:324–31. doi: 10.1016/j.cmet.2016.07.001
35. Walcher L, Kistenmacher A, Suo H, Kitte R, Dluczek S, Strauß A, et al. Cancer stem cells—origins and biomarkers: Perspectives for targeted personalized therapies. Front Immunol. (2020) 11:1280. doi: 10.3389/fimmu.2020.01280
36. Kettner N, Voicu H, Finegold M, Coarfa C, Sreekumar A, Putluri N, et al. Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell. (2016) 30:909–24. doi: 10.1016/j.ccell.2016.10.007
37. Sulli G, Rommel A, Wang X, Kolar M, Puca F, Saghatelian A, et al. Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature. (2018) 553:351. doi: 10.1038/nature25170
38. Chen P, Hsu W, Chang A, Tan Z, Lan Z, Zhou A, et al. Circadian regulator CLOCK recruits immune-suppressive microglia into the GBM tumor microenvironment. Cancer Discov. (2020) 10:371–81. doi: 10.1158/2159-8290.CD-19-0400
39. Wang C, Barnoud C, Cenerenti M, Sun M, Caffa I, Kizil B, et al. Dendritic cells direct circadian anti-tumour immune responses. Nature. (2023) 614:136–43. doi: 10.1038/s41586-022-05605-0
40. Wang Y, Sun N, Lu C, Bei Y, Qian R, Hua L. Upregulation of circadian gene ‘hClock’ contribution to metastasis of colorectal cancer. Int J Oncol. (2017) 50:2191–9. doi: 10.3892/ijo.2017.3987
41. Hood S, Amir S. The aging clock: Circadian rhythms and later life. J Clin Investig. (2017) 127:437–46. doi: 10.1172/JCI90328
42. Hofman M, Swaab D. Living by the clock: The circadian pacemaker in older people. Ageing Res Rev. (2006) 5:33–51. doi: 10.1016/j.arr.2005.07.001
43. Chen C, Logan R, Ma T, Lewis D, Tseng G, Sibille E, et al. Effects of aging on circadian patterns of gene expression in the human prefrontal cortex. Proc Natl Acad Sci U S A. (2016) 113:206–11. doi: 10.1073/pnas.1508249112
44. Lee C. The circadian clock and tumor suppression by mammalian period genes. In: Young M editor. Methods in enzymology. Circadian rhythms. Cambridge, MA: Academic Press (2005). p. 852–61. doi: 10.1016/S0076-6879(05)93045-0
45. Kondratov R, Kondratova A, Gorbacheva V, Vykhovanets O, Antoch M. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. (2006) 20:1868–73. doi: 10.1101/gad.1432206
46. Dubrovsky Y, Samsa W, Kondratov R. Deficiency of circadian protein CLOCK reduces lifespan and increases age-related cataract development in mice. Aging. (2010) 2:936–44. doi: 10.18632/aging.100241
47. Yang G, Chen L, Grant G, Paschos G, Song W, Musiek E, et al. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med. (2016) 8:324ra16. doi: 10.1126/scitranslmed.aad3305
48. Acosta-Rodríguez V, Rijo-Ferreira F, Izumo M, Xu P, Wight-Carter M, Green C, et al. Circadian alignment of early onset caloric restriction promotes longevity in male C57BL/6J mice. Science. (2022) 376:1192–202. doi: 10.1126/science.abk0297
49. Patel S, Velingkaar N, Makwana K, Chaudhari A, Kondratov R. Calorie restriction regulates circadian clock gene expression through BMAL1 dependent and independent mechanisms. Sci Rep. (2016) 6:25970. doi: 10.1038/srep25970
50. Sherman H, Frumin I, Gutman R, Chapnik N, Lorentz A, Meylan J, et al. Long-term restricted feeding alters circadian expression and reduces the level of inflammatory and disease markers. J Cell Mol Med. (2011) 15:2745–59. doi: 10.1111/j.1582-4934.2010.01160.x
51. Chaudhari A, Gupta R, Makwana K, Kondratov R. Circadian clocks, diets and aging. Nutr Healthy Aging. (2017) 4:101–12. doi: 10.3233/NHA-160006
52. Katewa S, Akagi K, Bose N, Rakshit K, Camarella T, Zheng X, et al. Peripheral circadian clocks mediate dietary restriction-dependent changes in lifespan and fat metabolism in Drosophila. Cell Metab. (2016) 23:143–54. doi: 10.1016/j.cmet.2015.10.014
53. Patel S, Chaudhari A, Gupta R, Velingkaar N, Kondratov R. Circadian clocks govern calorie restriction—mediated life span extension through BMAL1- and IGF-1-dependent mechanisms. FASEB J. (2016) 30:1634–42. doi: 10.1096/fj.15-282475
54. Ulgherait M, Chen A, McAllister S, Kim H, Delventhal R, Wayne C, et al. Circadian regulation of mitochondrial uncoupling and lifespan. Nat Commun. (2020) 11:1927. doi: 10.1038/s41467-020-15617-x
55. Sato S, Solanas G, Peixoto F, Bee L, Symeonidi A, Schmidt M, et al. Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell. (2017) 170:664–77.e11. doi: 10.1016/j.cell.2017.07.042
56. Liang C, Ke Q, Liu Z, Ren J, Zhang W, Hu J, et al. BMAL1 moonlighting as a gatekeeper for LINE1 repression and cellular senescence in primates. Nucleic Acids Res. (2022) 50:3323–47. doi: 10.1093/nar/gkac146
57. Lu J, Simon M, Zhao Y, Ablaeva J, Corson N, Choi Y, et al. Comparative transcriptomics reveals circadian and pluripotency networks as two pillars of longevity regulation. Cell Metab. (2022) 34:836–56.e5. doi: 10.1016/j.cmet.2022.04.011
58. Hojo H, Enya S, Arai M, Suzuki Y, Nojiri T, Kangawa K, et al. Remote reprogramming of hepatic circadian transcriptome by breast cancer. Oncotarget. (2017) 8:34128–40. doi: 10.18632/oncotarget.16699
59. Masri S, Papagiannakopoulos T, Kinouchi K, Liu Y, Cervantes M, Baldi P, et al. Lung adenocarcinoma distally rewires hepatic circadian homeostasis. Cell. (2016) 165:896–909. doi: 10.1016/j.cell.2016.04.039
60. Altman B, Hsieh A, Sengupta A, Krishnanaiah S, Stine Z, Walton Z, et al. MYC disrupts the circadian clock and metabolism in cancer cells. Cell Metab. (2015) 22:1009–19. doi: 10.1016/j.cmet.2015.09.003
61. Qu M, Duffy T, Hirota T, Kay S. Nuclear receptor HNF4A transrepresses CLOCK:BMAL1 and modulates tissue-specific circadian networks. Proc Natl Acad Sci U S A. (2018) 115:E12305–12. doi: 10.1073/pnas.1816411115
62. Qu M, Qu H, Jia Z, Kay S. HNF4A defines tissue-specific circadian rhythms by beaconing BMAL1::CLOCK chromatin binding and shaping the rhythmic chromatin landscape. Nat Commun. (2021) 12:6350. doi: 10.1038/s41467-021-26567-3
63. He B, Nohara K, Park N, Park Y, Guillory B, Zhao Z, et al. The small molecule nobiletin targets the molecular oscillator to enhance circadian rhythms and protect against metabolic syndrome. Cell Metab. (2016) 23:610–21. doi: 10.1016/j.cmet.2016.03.007
64. Marinac C, Nelson S, Breen C, Hartman S, Natarajan L, Pierce J, et al. Prolonged nightly fasting and breast cancer prognosis. JAMA Oncol. (2016) 2:1049–55. doi: 10.1001/jamaoncol.2016.0164
65. Li M, Tse L, Chan W, Kwok C, Leung S, Wu C, et al. Nighttime eating and breast cancer among Chinese women in Hong Kong. Breast Cancer Res. (2017) 19:31. doi: 10.1186/s13058-017-0821-x
66. Kogevinas M, Espinosa A, Castelló A, Gómez-Acebo I, Guevara M, Martin V, et al. Effect of mistimed eating patterns on breast and prostate cancer risk (MCC-Spain Study). Int J Cancer. (2018) 143:2380–9. doi: 10.1002/ijc.31649
67. Das M, Ellies L, Kumar D, Sauceda C, Oberg A, Gross E, et al. Time-restricted feeding normalizes hyperinsulinemia to inhibit breast cancer in obese postmenopausal mouse models. Nat Commun. (2021) 12:565. doi: 10.1038/s41467-020-20743-7
68. Sundaram S, Yan L. Time-restricted feeding mitigates high-fat diet-enhanced mammary tumorigenesis in MMTV-PyMT mice. Nutr Res. (2018) 59:72–9. doi: 10.1016/j.nutres.2018.07.014
69. Sancar A, Van Gelder R. Clocks, cancer, and chronochemotherapy. Science. (2021) 371:eabb0738. doi: 10.1126/science.abb0738
70. Montaigne D, Marechal X, Modine T, Coisne A, Mouton S, Fayad G, et al. Daytime variation of perioperative myocardial injury in cardiac surgery and its prevention by Rev-Erbα antagonism: A single-centre propensity-matched cohort study and a randomised study. Lancet. (2018) 391:59–69. doi: 10.1016/S0140-6736(17)32132-3
71. Hrushesky W. Circadian timing of cancer chemotherapy. Science. (1985) 228:73–5. doi: 10.1126/science.3883493
72. Lévi F, Benavides M, Chevelle C, Saunier F, Bailleul F, Misset J, et al. Chemotherapy of advanced ovarian cancer with 4’-O-tetrahydropyranyl doxorubicin and cisplatin: A randomized phase II trial with an evaluation of circadian timing and dose-intensity. J Clin Oncol. (1990) 8:705–14. doi: 10.1200/JCO.1990.8.4.705
73. Damato A, Luo J, Katumba R, Talcott G, Rubin J, Herzog E, et al. Temozolomide chronotherapy in patients with glioblastoma: A retrospective single-institute study. Neurooncol Adv. (2021) 3:vdab041. doi: 10.1093/noajnl/vdab041
74. Damato A, Katumba R, Luo J, Atluri H, Talcott G, Govindan A, et al. A randomized feasibility study evaluating temozolomide circadian medicine in patients with glioma. Neurooncol Pract. (2022) 9:193–200. doi: 10.1093/nop/npac003
75. Giacchetti S, Bjarnason G, Garufi C, Genet D, Iacobelli S, Tampellini M, et al. Phase III trial comparing 4-day chronomodulated therapy versus 2-day conventional delivery of fluorouracil, leucovorin, and oxaliplatin as first-line chemotherapy of metastatic colorectal cancer: The European organisation for research and treatment of cancer chronotherapy group. J Clin Oncol. (2016) 24. doi: 10.1200/JCO.2006.06.1440
Keywords: circadian rhythm, cancer, clock-targeting, chronomedicine, chronotherapy
Citation: Qu M (2023) Molecular crosstalk between circadian clock and cancer and therapeutic implications. Front. Nutr. 10:1143001. doi: 10.3389/fnut.2023.1143001
Received: 12 January 2023; Accepted: 14 February 2023;
Published: 02 March 2023.
Edited by:
Reza Rastmanesh, The Nutrition Society, United KingdomReviewed by:
Fabiola Hernández Rosas, Universidad Autónoma de Querétaro, MexicoLorena Aguilar Arnal, Institute of Biomedical Research, National Autonomous University of Mexico, Mexico
Monika Lesicka, Nofer Institute of Occupational Medicine, Poland
Copyright © 2023 Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meng Qu, bWVuZ3F1QHpqdS5lZHUuY24=
 Meng Qu
Meng Qu