
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr., 21 March 2023
Sec. Nutrition and Metabolism
Volume 10 - 2023 | https://doi.org/10.3389/fnut.2023.1128272
 Weili Xu1,2,3†
Weili Xu1,2,3† Weijiao Liu1,2,3†
Weijiao Liu1,2,3† Jinming Li1,2,3†
Jinming Li1,2,3† Xiangzhen Zhu1,2,3
Xiangzhen Zhu1,2,3 Li Wang1,2,3
Li Wang1,2,3 Dongyang Li1,2,3
Dongyang Li1,2,3 Kaixin Zhang1,2,3
Kaixin Zhang1,2,3 Jichao Ji1,2,3
Jichao Ji1,2,3 Xueke Gao1,2,3*
Xueke Gao1,2,3* Junyu Luo1,2,3*
Junyu Luo1,2,3* Jinjie Cui1,2,3*
Jinjie Cui1,2,3*The cotton aphid, Aphis gossypii Glover, is a species of polyphagous aphid with many biotypes, and its host transfer has always been the focus of research on the control of cotton aphid. An important factor affecting aphid specialization is the nutritional association with microbial symbionts that provide the host with nutrients lacking in the diet. We analyzed the microbial composition and biodiversity of reared on zucchini for 10 generations (T1–T10) and cotton as a control (CK), by high-throughput Illumina sequencing of 16S ribosomal RNA genes. The findings showed that the change in plant hosts decreased the richness and variety of microbial species. Regardless of whether the plant host is altered or not, Proteobacteria and Firmicutes are the predominate phyla in cotton-specialized aphid. Additionally, cotton-specialized aphids that live in zucchini had considerably lower relative abundances of non-dominant phyla (Bacteroidetes) than cotton hosts. At the genus level the dominant communities were Buchnera, Acinetobacter, and Arsenophonus. The relative abundance of Buchnera was significantly higher in aphids reared on zucchini than those on cotton, whereas the opposite was observed for Acinetobacter, as well as for some non-dominant communities (Stenotrophomonas, Pseudomons, Flavobacterium, Novosphingobium). Collectively, this study clarifies the dynamic changes of symbiotic bacteria in cotton-specialized aphids reared on zucchini for multiple generations. Among them, Buchnera is crucial for the cotton-specialized aphid to get nutrients during the transfer of the host and has a favorable impact on the colonization of cotton-specialized aphid populations on zucchini hosts. It not only enriches our understanding of the relationship between the bacterial microbiota of aphids and their adaptability to new hosts, zucchini, but also expands the current body of research on the mechanisms underlying the host shifting ability of cotton-specialized aphids.
Aphis gossypii Glover (Hemiptera: Aphididae) is a significant agricultural pest of several crops that causes significant feeding damage and spreads viruses (1). Due to their small size, large base, wide distribution, and complex life history, aphids are able to adapt to most environments, making them an increasingly rampant threat to agricultural and forestry development, and a source of great economic losses (2). As a result, A. gossypii has been a major problem in agricultural production for many years. The migration of A. gossypii is known to be an important factor affecting cotton aphid control. When A. gossypii do not have optimal host plants, they produce winged aphids that migrate to other host plants and use them to supplement their own needs for development (3). Thus, host adaptation is an important condition for the outbreak and spread of A. gossypii, and the study of adaptive mechanisms of A. gossypii regarding host transfer is of great significance for cotton aphid control.
Host specialization refers to the preference of insects to feed and live on specific host plants and the specificity of long-term feeding under natural conditions. While there are some polyphagous insects that can feed on hundreds of different host plants, less than 10% of plant feeding insects can feed on three or more different host plants (4). Most of these insects tend to feed on several closely related plants, or have a strong preference for a single host plant with a specific chemical composition (5). Studies have demonstrated that cotton aphids can produce population differentiation on different plant hosts, and various specific host biotypes have been identified around the world, including cotton and melon (6, 7). In China, based on host plant preferences and complete mitochondrial sequences, A. gossypii has been divided into cotton and cucumber biotypes (8). The two biotypes were unable to form populations after transferring to other plant hosts, and their survival rates drastically dropped, according to host transfer tests (9). However, it has also been shown that specialization is not absolute and that cotton-specialized aphids can migrate to each other through zucchini as bridge hosts. Cotton-specialized aphids can be successfully transferred to cucumber and cotton after several generations of culture on zucchini successfully transferred to cucumber and cotton, with no significant differences from aphids on the original host after transfer (10). However, it is unknown why zucchini can act as an intermediate bridge for host transformation and how cotton-specialized aphids absorb sufficient nutrients during host adaptation.
A very close relationship exists between insects and their endosymbionts, and interactions between A. gossypii and their bacterial communities have received increased attention in recent years. aphid endosymbiotic bacteria include primary endosymbionts, such as Buchnera, and secondary symbionts which include Rickettsia spp, Wolbachia spp, Hamiltonella, Arsenophonus, Acinetobacter, Micrococcus, Regiella insecticola, Serratia, and Pseudomonadaceae. Different symbiotic bacteria have different effects (11–16). Studies have shown that symbiotic bacteria can improve aphids’ heat tolerance and help them acclimate to high temperature environments (17, 18). It also has a significant impact on the aphids’ body color and resistance to diseases and parasitic wasps (19–21). In order for insects to adjust to the new host of nutrient utilization and metabolism, its symbiotic bacteria in the body can not only synthesize host the necessary amino acids, cholesterol, protein, and other nutrients, but also provide host decomposition of host plant cell wall enzymes, can also use the host plant secondary substances for detoxification, and can participate in the nitrogen cycle of the host, turning a host of nitrogen waste into nutrients (22, 23). It has been shown that Aphis craccivora, harboring Arsenophonus bacteria could better utilize Robinia pseudoacacia as a plant host. After inoculating Arsenophonus into an A. craccivora strain that lacked this bacterial endosymbiont and was incapable of colonizing Robinia pseudoacacia, the aphid strain acquired the ability to colonize Robinia pseudoacacia (24).
The research of symbiotic bacteria in cotton-specialized aphids, particularly in host transfer process, has not been documented, despite the fact that symbiotic bacteria of aphids are widely recognized. In this study, cotton-specialized aphids were transferred to and reared on an intermediate zucchini host for 10 generations, and changes in the diversity and abundance of aphid bacterial endosymbionts were assessed by sequencing the V3∼V4 region of the 16S rRNA gene using the Illumina MiSeq platform. We aimed to better understand how cotton-specialized aphids can adapt to cucumber hosts after being reared on an intermediate zucchini host at the level of aphid bacterial endosymbionts. This study contributes to the understanding of the relationship between the endosymbiotic bacterial composition of cotton-specialized aphids and their host specialization and shifting abilities.
Cotton (Zhong-mian49) and zucchini (Chunbaiyu) plants were used as the host plants in this study. They were grown in a growth chamber maintained at 26 ± 1°C, 75 ± 5% RH, and under a 14:10 h light/dark cycle. Cotton can be used to feed aphids when it reaches the 4-leaf stage.
Cotton-specialized aphids were collected from the experimental field station of the Institute of Cotton Research, Chinese Academy of Agricultural Sciences (CAAS) (36°5’34.8″N, 114°31’47.19″E) in May 2019. Cotton-specialized aphids were reared in petri dishes (diameter 9 cm) containing 1.8% agar and fresh host plant leaves, kept at 26 ± 1°C, 75 ± 5% relative humidity (20), under a 14:10-h light/dark photoperiod. A. gossypii biotypes were identified based on mitochondrial gene sequence polymorphisms described in the literature (25, 26). The identified cotton biotype aphids were maintained on cotton plants in the laboratory for over 40 generations (10 months) without exposure to any pesticides. Cotton-specialized aphids were transferred to and reared on zucchini leaves for 10 generations, and 3 day old aphids from each generation were collected. Four biological replicates were used for each generation.
The control group (CK) was 3 day old cotton-specialized aphids that had only fed on cotton hosts; treatment groups consisted of 3 day old cotton-specialized aphids reared on the zucchini host from generations 1 to 10 (T1-T10). After collection, the 3 day old aphid samples were flash frozen in liquid nitrogen and stored at -80°C. Bacterial DNA was extracted from the aphid samples using the MagPure Stool DNA KF Kit B (Magen, China) according to the manufacturer’s instructions. DNA purity and concentration were determined via a Microplate Reader and Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Carlsbad, CA, USA), respectively. The quality of the DNA samples was checked by 1% agarose gel electrophoresis.
According to the specificity of the V3∼V4 variable region of the bacterial 16S rRNA gene, the degenerate PCR primers 515F (5’-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) were used to amplify the DNA. Both forward and reverse primers were labeled with Illumina adaptor, pad, and adaptor sequences. PCR amplification was performed in a 50 μL reaction mixture containing 30 ng of DNA template. The PCR cycling conditions were 95°C for 3 min, 30 cycles of 95°C for 45 s, 56°C for 45 s, 72°C for 45 s, and a final extension for 10 min at 72°C. PCR products were purified using Agencourt AMPure XP beads and eluted in elution buffer. PCR libraries were quality checked using the Agilent Technologies 2100 Bioanalyzer (Agilent, USA), and qualified libraries were used for sequencing on the Illumina HiSeq 2500 PE250 platform (BGI, Shenzhen, China) according to protocols described by Gregory Caporaso and Eric J. de Muinck (27, 28), to generate 2 × 250 bp paired-end reads. Sequences obtained in this study have been deposited in GenBank1 under the accession number PRJNA764358.
Raw reads were filtered to remove adapters, poly-N, and low quality (quality score < 20) sequences using the cutadapt v2.6 software. Filtered sequences were assembled into pairs by FLASH (Fast Length Adjustment of Short reads, v1.2.11) software (29), with the minimum overlap length set to 15 bp and the mismatch ratio of overlap regions ≤ 0.1, to obtain tags for the hypervariable region. The tags were clustered into OTUs (Operational Taxonomic Unit) at 97% sequence similarity using scripts in the USEARCH (v7.0.1090) software (30). UCHIME (v4.2.400) (31) was used to perform a reference-based chimera removal according to the gold database (v20110519) to obtain OTU abundance statistics tables for each sample. Finally, OTUs in each sample were classified using Ribosomal Database Project (RDP) Classifier 1.9.1 (32), using a minimum confidence threshold of 0.6 OTUs without annotation results; species that were not included in the analysis project were removed from the annotation results and the remaining OTUs were used for subsequent analysis. The community composition of each sample was calculated at the levels of phylum, class, order, family, genus, and species. Initially, the species abundance in all samples was less than 0.50% and all unclassified species were merged into “Others”. Alpha and beta diversity were estimated by MOTHUR (v1.31.2) (33) and QIIME (v1.80) (34) at the OTU level, respectively. PCoA (Principal coordinate analysis) was used to display differences between the samples based on the beta diversity distance matrix. Linear discriminant analysis (LDA) effect size (LEfSe) was used2 to analyze differences in relative abundance of OTUs between the different treatment groups and the control group. The LDA threshold value was set at >4 for treatment groups T1-T9, while that for treatment group T10 was set at 3.5. LDA score distribution histograms were developed using PERMANOVA to compare the differences in community structure between groups. For tests comparing only two groups, the Wilcoxon rank-sum test was used to show the significance of microbial community composition between different groups. If the number of groups was more than two, the Kruskal-Wallis Hest (kruskal. test in R) was used for multi-sample comparison. P-value <0.05 was used to assess the significance of differences.
From sequences of 16S rRNA gene amplicon libraries from all ten generations (T1–T10) and control group (CK) had a total original reading of 1,641.93 Mbp before quality control (individual raw read numbers ranged from 60,461 to 299,627). A total of 3,106,199 paired reads were generated from 44 samples, with an average of 70,720 reads obtained for each sample (Table 1). After filtering, a total of 3,110,270 high-quality clean tags having an average length of 253 bp were obtained, with an average of 70,595 tags per sample. The percentage of bases with an effective sequencing error rate of <1% ranged from 99.94 to 99.97%. Reads for A. gossypii bacterial communities were produced, and 60 to 100 OTUs were identified (Table 1), with 97% identity cut-offs for each treatment group. A total of 10 phyla, 63 families, and 127 genera were annotated. Based on the observed species index, the sparse curves for all samples were almost saturated and stable, indicating that the 16S rRNA gene sequence was abundant and our analysis had sufficient depth to capture most of the microbial diversity information (Supplementary Figure 1).
Good’s coverage, which estimates the percentage of the total species represented in a sample, averaged 99.96% thereby suggesting that the majority of microbial species present in A. gossypii were included in this study (Table 1). Alpha diversity analysis was used to determine the complexity of species diversity of each sample through several indices, including observed species, the diversity index of Ace, and Chao 1, all of which showed that there were significant differences in Shannon and Simpson indexes among the control and treatment groups (P < 0.05), demonstrating that microbial diversity and evenness decreased among treatments (Figure 1A). Petal plots showed that there were 44 common OTUs in the control (CK) and all treatment (T1-T10) groups, and T7 had the most OTUs. In contrast to the 8 unique OTUs in the control (CK) group, T6 and T8 had the least number and no unique OTUs (Figure 1B). In the PCoA analyses based on species level OTU, points that are close together represent samples that were highly similar in their community composition. Representative T1, T5, and T10 were selected from the treatment group (T1-T10) for comparison with CK and the results showed that the bacterial communities of CK and the selected treatment groups were separated (Figures 1C, D), and that difference between were statistically significant (P < 0.001).
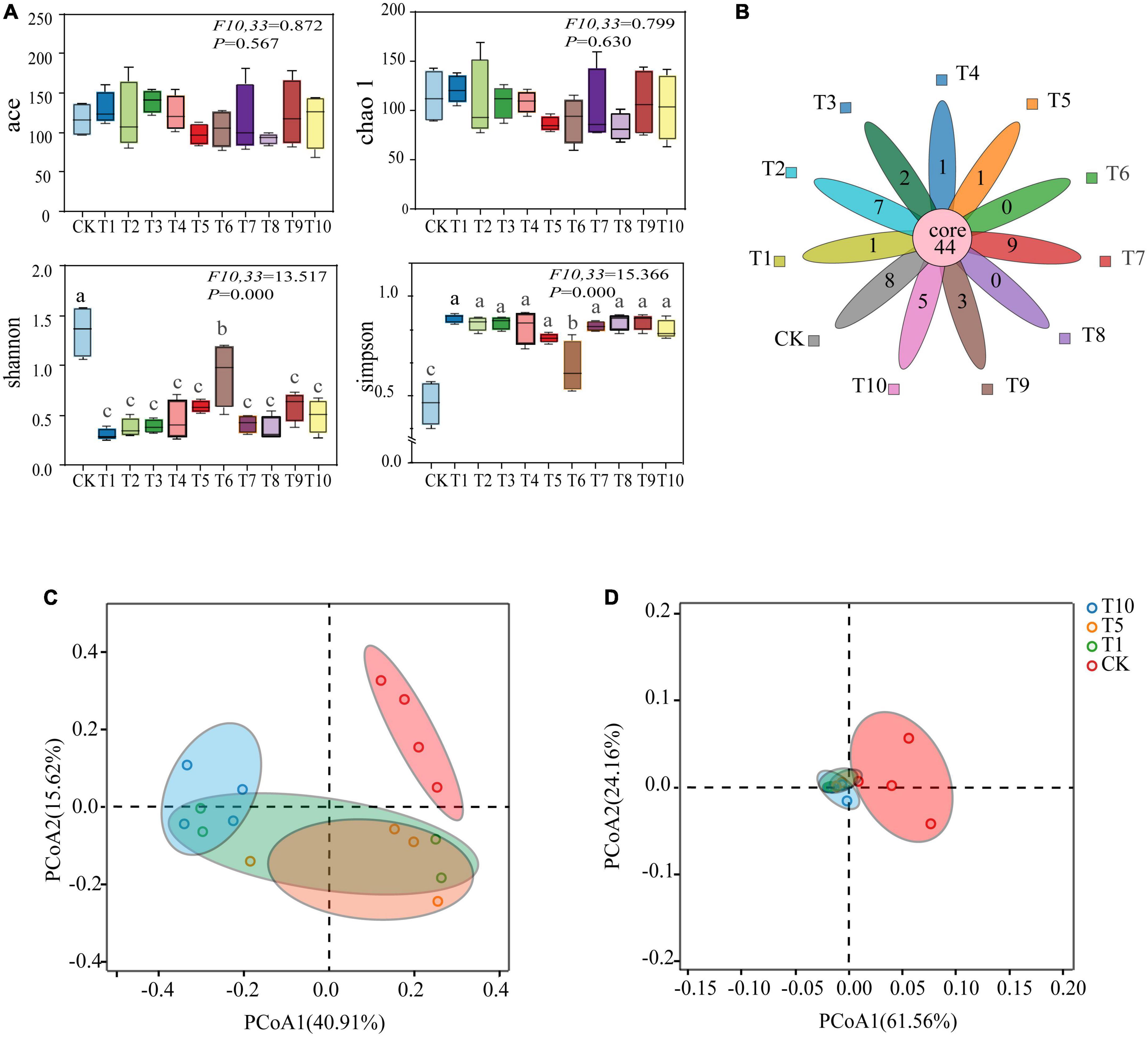
Figure 1. Sequencing analysis of 16S rDNA gene amplicons of cotton-specialized aphids with diversity indices. (A) Boxplot of α-diversity measured by the four indexs. (B) Core-Pan OTU presentation of the common and unique OTUs of all samples in the petal diagram. (C) Unweighted UniFrac metrics were used to determine pairwise distances among all of the samples. (D) Wighted UniFrac metrics were used to determine pairwise distances among all of the samples.
To better understand differences between the microbial communities in A. gossypii aphids before and after host transfer, we classified the microbial communities of the control (CK) and treatment (T1-T10) groups at the phylum level (Supplementary Table 1 and Figure 2A). Among them, the dominant phyla were Proteobacteria, Firmicutes, and Bacteroidetes, with the abundance of Proteobacteria being highest regardless of host transfer (average 99.03% of all samples). The relative abundance of Proteobacteria was lowest in the T6 group (97.31%) and highest in the T1 group (99.70%). There were no significant differences in relative abundance between the treatment and control groups, or between the individual treatment groups (T1-T10). As for Firmicutes, the relative abundance fluctuated steadily in groups T1-T4 and was mostly consistent with CK (0.05%), however the abundance began to shift in group T5 (0.14%) and reached its maximum in group T6 (1.97%) which was significantly different (P < 0.05) than the other groups. The relative abundance of Firmicutes in groups T7-T10 was slightly higher than CK. Compared with CK (1.39%), the mean relative abundance of Bacteroidetes was significantly (P < 0.05) lower in groups T1-T10 but there was no significant difference between the individual treatment groups.
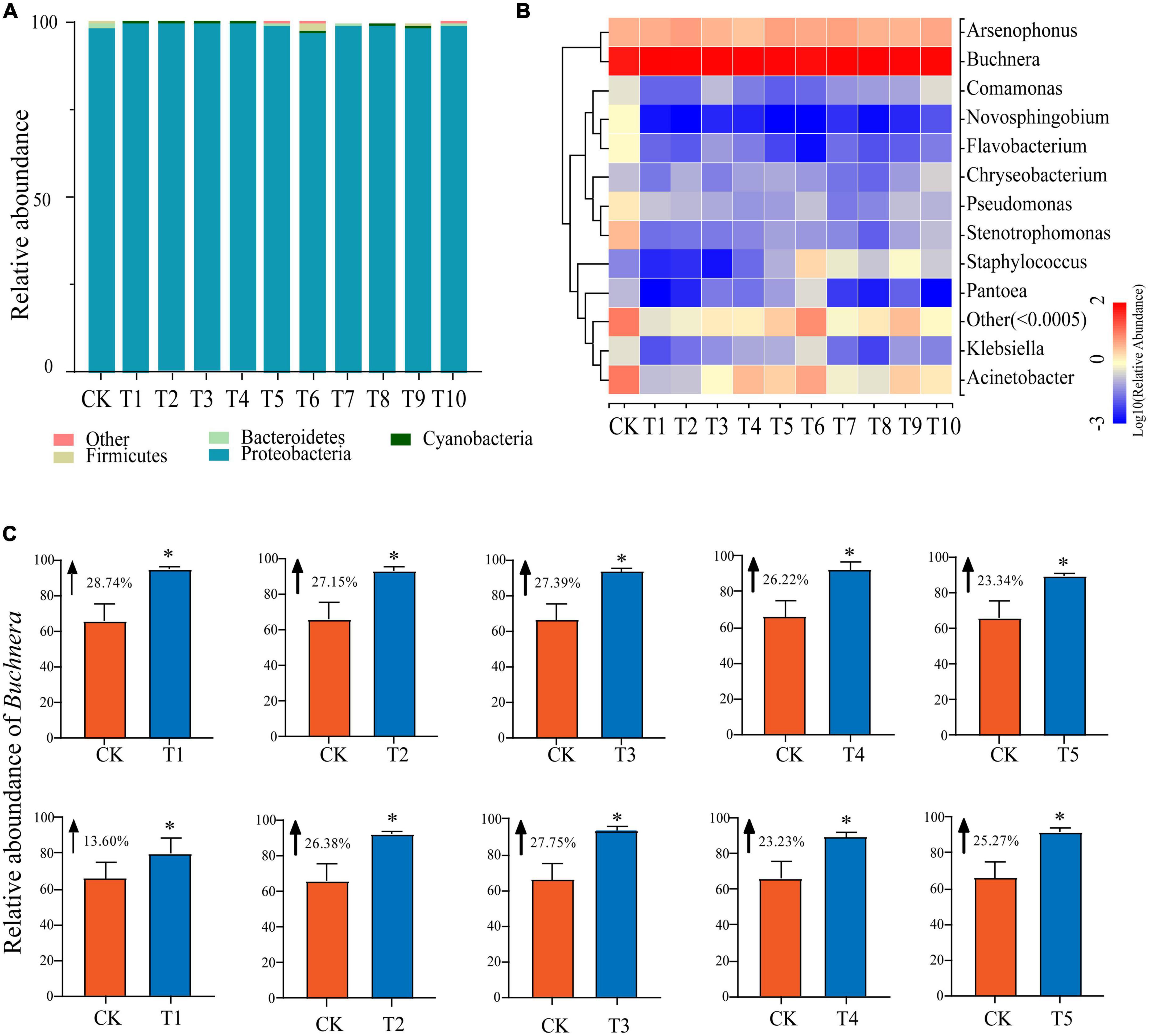
Figure 2. Changes in relative abundance of symbiotic bacteria in cotton-specialized aphids fed on zucchini for 1 to 10 generations and the control group (CK). (A) Relative abundance of symbiotic bacteria in cotton-specialized aphids fed on zucchini for 1 to 10 generations and the control group (CK) at the phylum level. (B) Relative abundance of symbiotic bacteria in cotton-specialized aphids fed on zucchini for 1 to 10 generations and the control group (CK) at the genus level. (C) Transfer to the Zucchini host significantly affected the relative abundance of Buchnera in the microbial communities of cotton-specialized aphids. Pairwise comparison between the treatment group and CK, the data is mean ± SD. *P < 0.05.
At the genus level, the most abundant genera (relative abundance > 1.00%) in the CK group were Buchnera (66.08%), Acinetobacter (11.28%), Arsenophonus (3.95%), Stenotrophomonas (3.41%), Pseudomonas (1.42%), Flavobactera (1.03%), and Novosphingobium (1.02%) (Supplementary Table 2 and Figure 2B). Compared with CK, the top three most abundant genera in the treatment groups were Buchnera, Acinetobacter, and Arsenophonus. Excluding Buchnera and Arsenophonus, the abundance of other symbiotic bacteria decreased significantly (P < 0.05) after plant host transfer, and the abundance of Flavobacterium and Novosphingobium approached 0. There were no significant differences between individual treatment groups. The relative richness of Arsenophonus fluctuated significantly among T1-T10 treatment groups, among which T5 (5.33%) was the largest and T4 (2.78%) was significantly smaller than CK (3.95%) (P < 0.05). Buchnera, as the dominant genera in CK, persisted in the treatment groups (T1-T10). In addition, all treatment groups (T1-T10) were significantly higher than the control group CK (66.08%) (P < 0.05), and reached the highest value at T1 (94.82%) (Figure 2C).
LEfSe analysis was performed to screen for biomarkers in aphid bacterial communities (Figure 3). A different number of biomarkers were identified in the treatment group compared with the control (LDA > 4, only T10 LDA > 3.5). It was found that as the number of progenies of the aphid population increased, the specific microbial biomarkers decreased in each generation at specific LDA thresholds until only Buchnera was found in the T9 and T10 generations of aphids. And Buchnera was found in the whole generation T1-T10 of zucchini hosts. These results suggest that Buchnera may play an important role in the adaptation of cotton aphids to the intermediate zucchini host.
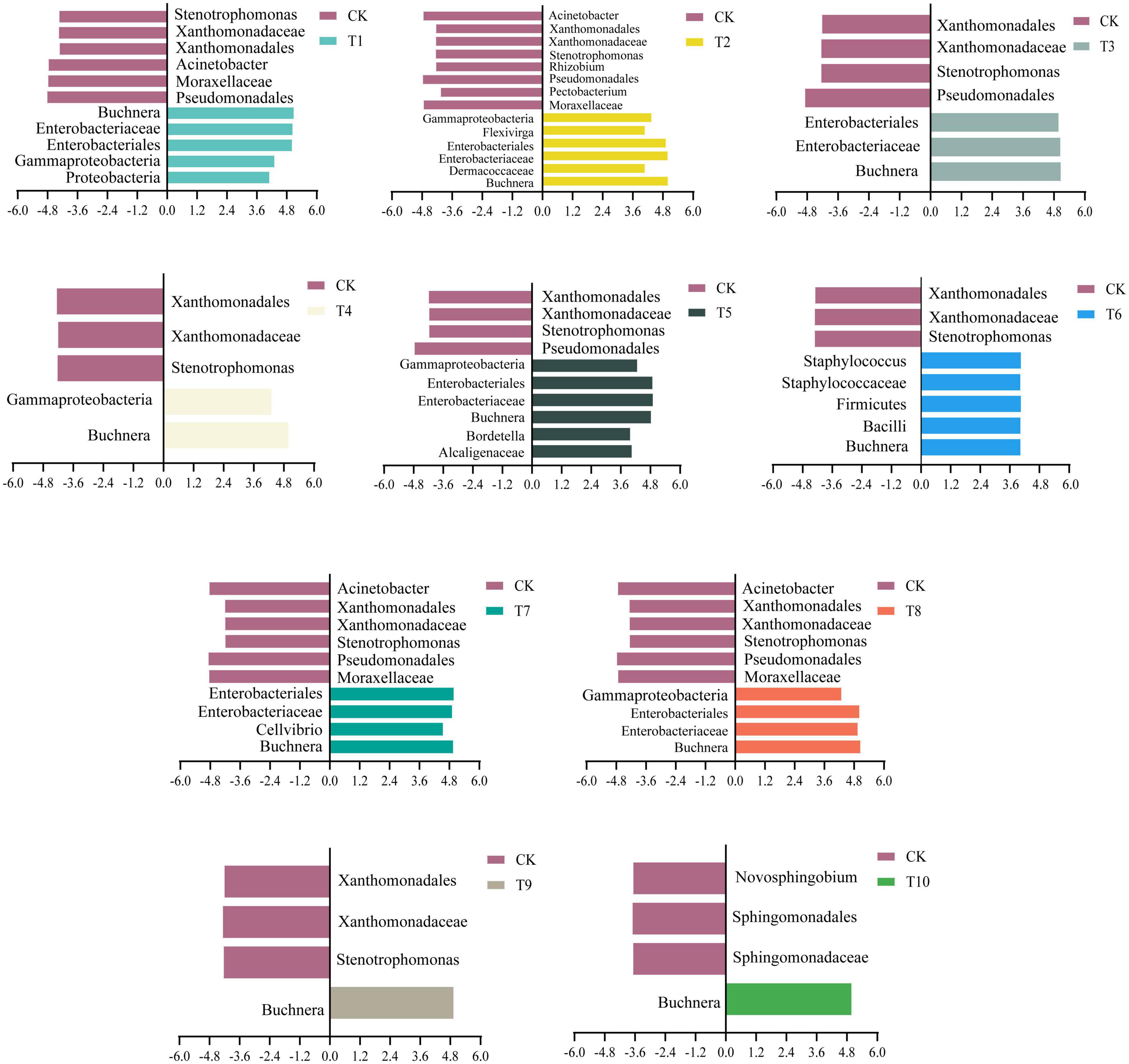
Figure 3. Comparison of biomarkers of different generations of cotton-specialized aphids on cotton host (CK) and transferred to zucchini host (T1-T10).
Biotypes represent excesses in the evolutionary process of species. In insects, most biotypes are classified according to the ability to use different hosts (34). However, the presence of an intermediate host, zucchini, suggests that specialization is not absolute and that some endosymbiotic bacteria provide essential substances to compensate for the nutritional maladjustment of the host’s diet (8). Therefore, this study reveals for the first time the transformation mechanism of cotton specialized aphids using zucchini as an intermediate host from the perspective of symbiotic bacteria (Figure 4). The results showed that the presence of Buchner’s provided an important role for the transformation of aphids in plant hosts.
It is well known that almost all insects contain symbiotic bacteria, which form mutually beneficial relationships with their aphid hosts in different ways. Hemiptera cannot produce cholesterol, vital amino acids, or vitamins, and the nutrients they take in from plant sap are insufficient and imbalanced, according to studies on the roles of symbiotic bacteria (23). While Primary symbiotic bacteria would play their role to provide nutrients. However a large number of studies have shown that Secondary symbiotic bacteria can also affect the growth and development, reproductive ability, and environmental adaptation of their host insects (35, 36). According to the research by Sasaki et al., the symbiotic yeast bacteria in Nilaparvatalugens, the primary pest of rice, have high activity of uricase, which may use the metabolic waste (uric acid) of the host to produce amino acids needed for Nilaparvatalugens’ growth and development (37).
In our study, the microbiota of A. gossypii was dominated by a few bacterial taxa both before and after host transition. At the phylum level, Proteobacteria and Firmicutes were the dominant taxa, with the abundance of Proteobacteria being highest at 99.09%, which is similar to that found in other hemipteran insects (38, 39) (Figure 2A). Firmicutes and Proteobacteria are essential to maintaining the growth and development of insects, aiding in the metabolism of secondary metabolites from host plants. Bacteroidetes are often the dominant bacterial phylum in the intestinal microbe communities of many insects such as the pea aphid in order Hemiptera and the longhorn beetle in Coleoptera, which can degrade plant cell wall polysaccharides (40, 41). The third major community in this study’s cotton aphids was the Bacteroidetes, and when zucchini was used as the host plant, the population’s richness drastically fell. Therefore, it may be assumed that the decline in Bacteroidetes may make it difficult for aphids to efficiently enter the cell wall for effective nutrition while feeding on zucchini. Actinobacteria can promote the growth and development, and improve the resilience of their insect hosts (42, 43). The lack of actinomycetes in aphids that we discovered in this research, leads us to believe that the aphids’ immunity may have been diminished as a result of the transition in plant hosts, and also that the change in host may have increased aphid maladaptation. In addition, the species diversity of the microbial community in cotton-specialized aphids decreased significantly after plant host transfer. Such results are consistent with the results of Mottal et al. who showed a low diversity of symbiotic bacteria in environmentally sensitive honey bees, i.e., the lower the diversity of symbiotic bacteria, the weaker the resistance (44, 45).
At the genus level, a variety of microbial communities were detected in the aphids, and there were significant differences in microbial communities between aphids feeding on cotton and zucchini hosts. The dominant bacteria in the microbial communities of cotton-specialized aphids fed on zucchini mainly consisted of primary symbiotic bacteria Buchnera and secondary symbiotic bacteria Arsenophonus and Acinetobacter (Supplementary Table 2 and Figure 2B). Studies have shown that only 11 of the 20 amino acids required for aphids to develop can be produced in the body, with the other nine coming from plants. The fitness of cotton aphids will be diminished if one of the required amino acids is lacking or the host plant’s supply is very low, which prevents the plant from providing what the aphids need to grow and develop (22). Douglas discovered that Buchnera in pea aphid may give a range of important amino acids to pea aphid, enabling the host to overcome the nitrogen nutritional barrier of plants (46, 47). By examining the whole genome sequence of the pea aphid, Consortium et al. discovered that Buchnera and the aphids are closely related in terms of nutrient metabolism because they share metabolic pathways for certain important amino acids (48, 49). Our study found that as the dominant species present throughout the host shifting process, the relative abundance of Buchnera significantly increased in cotton-specialized aphids that had been reared on zucchini for 10 generations compared with those reared on cotton (CK). This confirms the important role of Buchnera in host transfer, i.e., when aphids encounter host plants that are not very favorable to them, they increase the symbiotic bacteria. We speculate that it exercises a specific function to regulate the metabolism of nutrients to improve adaptation. It also confirms that endosymbiotic bacteria of aphids mediated by intermediate plant hosts can alter aphid specialization, and the specialization is not a dead-end in the evolution of feeding habit.
Studies have also found that the secondary symbiotic bacteria Arsenophonus plays an important role in resisting natural enemies, expanding the host range, and regulating the reproduction of its aphid host. Infection with of the soybean aphid with Arsenophonus bacteria can help improve the aphid’s ability to adapt to different plant hosts, and promoted growth of the aphid population (50). Acinetobacter was involved in many life activities of aphid host, including food digestion, nitrogen transformation and assimilation, and degradation of harmful substances (51, 52). Other endosymbiotic bacteria, including Stenotrophom, Pseudomons, Flavobacterium, and Novosphingobium, were likewise considerably reduced in the cotton aphid feeding on zucchini, and even their relative richness was close to 0. These further demonstrated the reduction in species variety caused by the transfer of cotton-specific cotton aphid to the host of zucchini. The same study also pointed out that there are complex relationships based on resources, survival niches, and interactions between aphids and bacterial symbionts, known as the metabolic tug of war (53). Arsenophonus infection may have an impact on Buchnera richness, which rose in correlation with Arsenophonus richness (54), whereas Hamiltonian exhibited the reverse connection (25). None of these interactions, nevertheless, were discovered in our analysis.
In summary, endosymbiotic bacteria, particularly Buchnera, played a significant role in nutrition absorption during the transplantation of cotton-specialized aphid to intermediate host zucchini, while other secondary symbiotic bacteria influenced probably more the immunological aspects of cotton-specialized aphid. In other words, the cotton-specialized aphid was not fully adaptation to the intermediate host, zucchini, and it could only improve its adaptation by expanding its own Buchnera. This behavior accelerates the spread of aphids and increases their damage, while avoiding the problem of direct transfer of cotton aphids to poorly adapted hosts such as cucumber due to nutritional deficiencies. However, in general, identifying the significance of intermediate hosts and symbiotic bacteria is only the first step in controlling cotton aphids and in understanding the transformation mechanisms of cotton aphids, and more research is needed to understand the transformation mechanisms of symbiotic bacteria.
To our knowledge, this is the first study of the dynamics of endosymbiotic bacteria in specialized aphids when zucchini act as intermediate hosts. We examined the changes of endosymbiotic bacteria before and after the transfer of aphids from cotton to zucchini host. It was found that the intermediate zucchini host affected bacterial species richness of cotton aphid microbial communities, but did not affect the structural composition of bacterial species. In addition, the dominant bacterial species in A. gossypii were Buchnera and Arsenophonus. In particular, the changes in Buchnera abundance were the most obvious. Buchnera was not only the dominant population before host transformation, but also significantly increased after A. gossypii were transferred to zucchini. This phenomenon confirms the importance of Buchnera in the host transfer process of cotton-specialized aphids. Therefore, we speculate that Buchnera may contribute to aphid-altered specialization through zucchini in a nutrient-providing manner. Our study reveals a complex relationship between symbiotic bacteria in the host transfer ability of A. gossypii and provides a theoretical basis for the mechanism of A. gossypii specialization type alteration.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA764358 (SRA).
XG, JC, and JLu conceived and designed the experiments. WX and WL analyzed the data. DL, XZ, LW, JJ, and XG evaluated the conclusions. WX, JLi, and WL wrote the manuscript. All authors read and approved the final manuscript.
This study was funded by National Key R&D Program of China, 2022YFD1400300, Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences, and Supported by China Agriculture Research System.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2023.1128272/full#supplementary-material
Supplementary Figure 1 | Rarefaction curves based on species abundance data.
1. Biddle PG, Tinsley TW. Some effects of poplar mosaic virus on the growth of poplar trees. New Phytol. (1971) 70:67–75. doi: 10.1111/j.1469-8137.1971.tb02511.x
2. Futuyma DJ, Gould F. Associations of plants and insects in deciduous forest. Ecol Monogr. (1979) 49:33–50. doi: 10.2307/1942571
3. Wu W, Liang XL, Zhao HY, Xu TT, Liu XD. Special plant species determines diet breadth of phytophagous insects: a study on host plant expansion of the host-specialized Aphis gossypii Glover. PLoS One. (2013) 8:e60832. doi: 10.1371/journal.pone.0060832
4. Carletto J, Lombaert E, Chavigny P, Brévault T, Vanlerberghe-Masutti F. Ecological specialization of the aphid Aphis gossypii Glover on cultivated host plants. Mol Ecol. (2015) 18:2198–212. doi: 10.1111/j.1365-294X.2009.04190.x
5. Cui L, Yuan H, Wang Q, Wang Q, Rui C. Sublethal effects of the novel cis-nitromethylene neonicotinoid cycloxaprid on the cotton aphid Aphis gossypii Glover (Hemiptera: Aphididae). Sci Rep. (2018) 8:8915. doi: 10.1038/s41598-018-27035-7
6. Adriaan GJ, Tigges WT, De Vrijer PWF. Host races of Aphis gossypii (Homoptera: Aphididae) on cucumber and chrysanthemum. Environ Entomol. (1994) 5:1235–40. doi: 10.1093/ee/23.5.1235
7. Wang L, Zhang S, Luo JY, Wang CY, Lv LM, Zhu XZ, et al. Identification of Aphis gossypii Glover (Hemiptera: Aphididae) biotypes from different host plants in North China. PLoS One. (2016) 11:e0146345. doi: 10.1371/journal.pone.0146345
8. Wang L, Zhang S, Luo JY, Wang CY, Lv LM, Zhu XZ, et al. Host choice of different host biotypes of cotton Aphid and preliminary analysis of the mechanism. Cotton Sci. (2017) 3:292–300. doi: 10.11963/1002-7807.wlcjj.20170330
9. Wang YM, Zhang PF, Chen J. Host-preference biotypes of the cotton aphid, Aphis gossypii Glover and the behavioral mechanism in their formation. Acta Entomol Sin. (2004) 47:760–7. doi: 10.3321/j.issn:0454-6296.2004.06.012
10. Hu DW, Zhang S, Luo JY, Lu LM, Cui JJ, Zhang X. An example of host plant expansion of host-specialized Aphis gossypii Glover in the field. PLoS One. (2017) 12:e0177981. doi: 10.1371/journal.pone.0177981
11. Tsuchida T, Koga R, Horikawa M, Tsunoda T, Maoka T, Matsumoto S, et al. Symbiotic bacterium modifies aphid body color. Science. (2010) 330:1102–4. doi: 10.1126/science.1195463
12. Tong L, Xiao JH, Xu ZH, Murphy RW, Huang DW. A possibly new Rickettsia-like genus symbiont is found in Chinese wheat pest aphid, Sitobion miscanthi (Hemiptera: Aphididae). J Invertebr Pathol. (2011) 106:418–21. doi: 10.1016/j.jip.2010.12.003
13. Vorburger C, Gehrer L, Rodriguez P. A strain of the bacterial symbiont Regiella insecticola protects aphids against parasitoids. Biol Lett. (2010) 6:109–11. doi: 10.1098/rsbl.2009.0642
14. Vorburger C, Ganesanandamoorthy P, Kwiatkowski M. Comparing constitutive and induced costs of symbiont-conferred resistance to parasitoids in aphids. Ecol Evol. (2013) 3:706–13. doi: 10.1002/ece3.491
15. Lukasik P, Guo H, Asch MV, Henry LM, Godfray H, Ferrari J, et al. Horizontal transfer of facultative endosymbionts is limited by host relatedness. Evolution. (2015) 69:2757–66. doi: 10.1111/evo.12767
16. Lukasik P, Dawid MA, Ferrari J, Godfray HJ. The diversity and fitness effects of infection with facultative endosymbionts in the grain aphid, Sitobion avenae. Oecologia. (2013) 173:985–96. doi: 10.1007/s00442-013-2660-5
17. Montllor CB, Maxmen A, Purcell A. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol. (2002) 27:189–95. doi: 10.1111/jeb.12705
18. Burke G, Fiehn O, Moran N. Effects of facultative symbionts and heat stress on the metabolome of pea aphids. ISME J. (2010) 4:242–52. doi: 10.1038/ismej.2009.114
19. Oliver K, Russell J, Moran N, Hunter M. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci USA. (2003) 100:1803–7. doi: 10.1073/pnas.0335320100
20. Frago E, Mala M, Weldegergis BT, Yang C, Mclean A, Godfray H, et al. Symbionts protect aphids from parasitic wasps by attenuating herbivore-induced plant volatiles. Nat Commun. (2017) 8:1860. doi: 10.1038/s41467-017-01935-0
21. Ferrari J, Darby AC, Daniell TJ, Godfray HC, Douglas A. Linking the bacterial community in pea aphids with host-plant use and natural enemy resistance. Ecol Entomol. (2004) 29:60–5. doi: 10.1111/j.1365-2311.2004.00574.x
22. Douglas A. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol. (1998) 43:17–37. doi: 10.1146/annurev.ento.43.1.17
23. Wilkinson TL, Adams D, Minto LB, Douglas AE. The impact of host plant on the abundance and function of symbiotic bacteria in an aphid. J Exp Biol. (2001) 204:3027–38. doi: 10.1242/jeb.204.17.3027
24. Wagner SM, Martinez AJ, Ruan YM, Kim KL, Lenhart PA, Dehnel AC, et al. Facultative endosymbionts mediate dietary breadth in a polyphagous herbivore. Funct Ecol. (2015) 29:1402–10. doi: 10.1111/1365-2435.12459
25. Zhang S, Luo JY, Wang L, Wang CY, Lv LM, Zhang LJ, et al. The biotypes and host shifts of cotton-melon aphids Aphis gossypii in Northern China. J Integr Agr. (2018) 17:2066–73. doi: 10.1016/0022-1910(95)00086-0
26. Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. (2012) 6:1621–4. doi: 10.1038/ismej.2012.8
27. Muinck EJ, Trosvik P, Gilfillan GD, Hov JR, Sundaram AY. A novel ultra high-throughput 16S rRNA gene amplicon sequencing library preparation method for the Illumina HiSeq platform. Microbiome. (2017) 5:68. doi: 10.1186/s40168-017-0279-1
28. Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. (2011) 27:2957–63. doi: 10.1093/bioinformatics/btr507
29. Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. (2013) 10:996–8. doi: 10.1038/nmeth.2604
30. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. (2011) 27:2194–200. doi: 10.1093/bioinformatics/btr381
31. Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. (2014) 42:633–42. doi: 10.1093/nar/gkt1244
32. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. (2009) 75:7537–41. doi: 10.1128/AEM.01541-09
33. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. (2010) 7:335–6. doi: 10.1038/nmeth.f.303
34. Zhang YC, Lei HX, Miao NH, Liu XD. Comparative transcriptional analysis of the host-specialized aphids Aphis gossypii (Hemiptera: Aphididae). J Econ Entomol. (2017) 110:702–10. doi: 10.1093/jee/tox029
35. Douglas A. The microbial dimension in insect nutritional ecology. Funct Ecol. (2009) 23:38–47. doi: 10.1371/journal.pone.0170332
36. Oliver KM, Degnan PH, Burke GR, Moran N. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol. (2010) 55:247–66. doi: 10.1146/annurev-ento-112408-085305
37. Sasaki T, Kawamura M, Ishikawa H. Nitrogen recycling in the brown planthopper, Nilaparvata lugens: involvement of yeast-like endosymbionts in uric acid metabolism. J Insect Physiol. (1996) 42:125–9. doi: 10.1016/0022-1910(95)00086-0
38. Husseneder C, Park JS, Howells A, Tikhe CV, Davis JA. Bacteria associated with Piezodorus guildinii (Hemiptera: Pentatomidae), with special reference to those transmitted by feeding. Environ Entomol. (2016) 1:159–66. doi: 10.1093/ee/nvw112
39. Li L, Majid A. Metagenomic 16S rDNA amplicon data of microbial diversity of guts of fully fed tropical bed bugs, Cimex hemipterus (F.) (Hemiptera: Cimicidae). Data Brief. (2020) 30:105575. doi: 10.1016/j.dib.2020.105575
40. Adams AS, Jordan MS, Adams SM, Suen G, Goodwin LA, Davenport KW, et al. Cellulose-degrading bacteria associated with the invasive woodwasp Sirex noctilio. ISME J. (2011) 5:1323–31. doi: 10.1038/ismej.2011.14
41. Engel P, Martinson VG, Moran NA. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci USA. (2012) 109:11002–7. doi: 10.1073/pnas.1202970109
42. Dillon R, Charnley K. Mutualism between the desert locust Schistocerca gregaria and its gut microbiota. Res Microbiol. (2002) 153:503–9. doi: 10.1016/s0923-2508(02)01361-x
43. Qin S, Feng WW, Zhang YJ, Wang TT, Xiong YW, Xing K. Diversity of bacterial microbiota of coastal halophyte limonium sinense and amelioration of salinity stress damage by symbiotic plant growth-promoting Actinobacterium Glutamicibacter halophytocola KLBMP 5180. Appl Environ Microbiol. (2018) 84:e01533–18. doi: 10.1128/AEM.01533-18
44. Sheng P, Huang S, Wang Q, Wang A, Zhang H. Isolation, screening, and optimization of the fermentation conditions of highly cellulolytic bacteria from the hindgut of Holotrichia parallela larvae (Coleoptera: Scarabaeidae). Appl Biochem Biotech. (2012) 167:270–84. doi: 10.1007/s12010-012-9670-3
45. Motta EVS, Raymann K, Moran NA. Glyphosate perturbs the gut microbiota of honey bees. Proc Natl Acad Sci USA. (2018) 115:10305–10. doi: 10.1073/pnas.1803880115
46. Wilkinson TL, Koga R, Fukatsu T. Role of host nutrition in symbiont regulation: impact of dietary nitrogen on proliferation of obligate and facultative bacterial endosymbionts of the pea aphid Acyrthosiphon pisum. Appl Environ Microbiol. (2007) 73:1362–6. doi: 10.1128/AEM.01211-06
47. Hardie J, Leckstein P, Biology M. Antibiotics, primary symbionts and wing polyphenism in three aphid species. Insect Biochem Mol Biol. (2007) 37:886–90. doi: 10.1016/j.ibmb.2007.05.005
48. Consortium I, Calevro F, Charles H, Colella S, Sagot M. Genome sequence of the pea Aphid Acyrthosiphon pisum. PLoS Biol. (2010) 8:e1000313. doi: 10.1371/journal.pbio.1000313
49. Ji, X, Peng B, Ding H, Cui B, Nie H, Yan Y. Purification, structure and biological activity of Pumpkin Polysaccharides: a review. Food Rev Int. (2021) 1904973. doi: 10.1080/87559129.2021.1904973
50. Wulff JA, White JA. The endosymbiont Arsenophonus provides a general benefit to Soybean Aphid (Hemiptera: Aphididae) regardless of host plant resistance (Rag). Environ Entomol. (2015) 44:574–81. doi: 10.1093/ee/nvv031
51. Briones-Roblero CI, Rodríguez-Díaz R, Santiago-Cruz JA, Zúñiga G, Rivera-Orduña FN. Degradation capacities of bacteria and yeasts isolated from the gut of Dendroctonus rhizophagus (Curculionidae: Scolytinae). Folia Microbiol. (2017) 62:1–9. doi: 10.1007/s12223-016-0469-4
52. Mason CJ, Lowe-Power TM, Rubert-Nason KF, Lindroth RL, Raffa KF. Interactions between bacteria and aspen defense chemicals at the phyllosphere - herbivore interface. J Chem Ecol. (2016) 42:193–201. doi: 10.1007/s10886-016-0677-z
53. Smith TE, Moran N. Coordination of host and symbiont gene expression reveals a metabolic tug-of-war between aphids and Buchnera. Proc Natl Acad Sci USA. (2020) 117:2113–21. doi: 10.1073/pnas.1916748117
Keywords: Aphis gossypii, Buchnera, bacterial communities, host specialization, zucchini
Citation: Xu W, Liu W, Li J, Zhu X, Wang L, Li D, Zhang K, Ji J, Gao X, Luo J and Cui J (2023) Buchnera breaks the specialization of the cotton-specialized aphid (Aphis gossypii) by providing nutrition through zucchini. Front. Nutr. 10:1128272. doi: 10.3389/fnut.2023.1128272
Received: 20 December 2022; Accepted: 28 February 2023;
Published: 21 March 2023.
Edited by:
Xiaolong Ji, Zhengzhou University of Light Industry, ChinaCopyright © 2023 Xu, Liu, Li, Zhu, Wang, Li, Zhang, Ji, Gao, Luo and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinjie Cui, YXljdWlqaW5qaWVAMTYzLmNvbQ==; Junyu Luo, bHVvanVueXUxODE4QDEyNi5jb20=; Xueke Gao, MTUwMzYxMzgzODlAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.