- 1School of Marine Sciences, Ningbo University, Ningbo, China
- 2Key Laboratory of Aquacultural Biotechnology, Ministry of Education, Ningbo University, Ningbo, China
Microbial community may systematically promote the development of fermentation process of foods. Traditional fermentation is a spontaneous natural process that determines a unique nutritional characteristic of crab paste of Portunus trituberculatus, However, rare information is available regarding the development pattern and metabolic role of bacterial community during the fermentation of crab paste. Here, using a 16S rRNA gene amplicon sequencing technology, we investigated dynamics of bacterial community and its relationship with metabolites during the fermentation of crab paste. The results showed that bacterial community changed dynamically with the fermentation of crab paste which highlighted by consistently decreased α-diversity and overwhelming dominance of Vibrio at the later days of fermentation. Vibrio had a positive correlation with trimethylamine, hypoxanthine, formate, and alanine while a negative correlation with inosine and adenosine diphosphate. In contrast, most of other bacterial indicators had a reverse correlation with these metabolites. Moreover, Vibrio presented an improved function potential in the formation of the significantly increased metabolites. These findings demonstrate that the inexorable rise of Vibrio not only drives the indicator OTUs turnover in the bacterial community but also has incriminated the quality of crab paste from fresh to perished.
1. Introduction
By giving the distinctive flavor and taste, traditional fermentation process has been conducted on a catalog of seafoods such as fish (1, 2), shrimp (3), and crab (4). On one hand, the physiologically active substances such as vitamins (3), amino acids (5), and organic acids (6) are produced during the fermentation of aquatic products. On the other hand, some undesirable substances such as hypoxanthine (Hx) and trimethylamine (TMA) could be simultaneously formed (7). Hx is the main cause of the bitter taste and unpleasant smell of aquatic products (8, 9) whereas TMA may induce cancer (10) and cardiovascular disease (11) if taken excessively. In this regard, fermented seafood quality is a hotspot for food safety, which play a vital role for human health.
Traditional fermentation of seafoods is a spontaneous natural process in which autolytic enzymes and microbes jointly develop a unique nutritional characteristic (12). Among them, microbial community could be adaptive and metabolically diverse and may systematically promote the development of fermentation process (13). Particularly, some studies have explored the microbial metabolic functions in the production of volatile flavor compounds from shrimp paste (12) and fermented mandarin fish (14). Like Hx from adenosine triphosphate (ATP) (15), and TMA from trimethylamine N-oxide (TMAO) (16, 17). Therefore, comprehensively elucidating how the microbial community changes and its relationship with fermented substances over fermentation is fundamental for the seafood quality.
As a popular fermented aquatic product, crab paste is made by directly mixing fresh meat of the swimming crab, Portunus trituberculatus, with seasonings including salt, sugar, monosodium glutamate, and liquor (4). Twenty-eight metabolites such as amino acids, organic acids, and organic bases have been detected in crab paste based on our previous metabolomic work (7). Although some cultural bacteria such as Staphylococcus, Arthrobacter, and Sphingobacterium have been found in crab paste (18–21). Rare information is available regarding the succession of bacterial community in such an evolving, complex mixture of crab paste and its metabolic contribution to fermented substances, which might be important to the quality control of crab paste.
Therefore, in this study, a 16S rRNA gene amplicon sequencing technology was used to analyze the change of bacterial community structure across the fermentation of crab paste. Combining multivariate statistical analysis and PICRUSt2 functional prediction, we aimed to reveal the following: (1) the dynamics of α-diversity and composition of bacterial community with the fermentation of crab paste, (2) the relationships between bacterial taxa and fermented products of crab paste, (3) the functional potential of Vibrio in the formation of fermented products.
2. Materials and methods
2.1. Crab paste processing
Live seven swimming crabs (205.69 ± 17.25 g) were purchased from a crab aquafarm in Ningbo, China and anaesthetized on ice. Each crab was cleaned using tap water and cut into pieces immediately. For crab paste processing, the crab pieces were mixed with 4% sucrose, 2% salt, 1% monosodium glutamate, and 1% white liquor with 40% ethanol and were packaged in an airtight plastic bag for 7 days-fermentation at 4°C. The samples were respectively taken at 1, 3, 5, and 7 days of fermentation and the fresh crab samples were used as control. All samples were stored at −80°C for further analysis.
2.2. DNA extraction, 16S rRNA gene amplification, and illumina sequencing
Genomic DNA was extracted from 0.5 g of crab paste for each sample using the FastDNA Spin kit (MP Biomedicals, USA). The concentration and purity of DNA extracts were measured by a NanoDrop ND-1000 spectrophotometer. The V3–V4 region of 16S rRNA gene was amplified using the primer sets 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) with dual barcodes (21). To reduce the bias during amplification, PCR reactions were performed in triplicate for each sample. Following purification, assessment of fragment size, and quantification, PCR amplicons for each sample were aggregated in equimolar ratios and sequenced with the Illumina MiSeq platform (Illumina, USA) for generating paired end reads. Raw sequence data are available in the NCBI Sequence Read Archive under BigProject PRJNA808815.
2.3. Sequence processing
Paired-end reads were merged using FLASH (22). The merged sequences were quality filtered and processed using the QIIME2 pipeline (23). Following chimera detection using UCHIME (24), the remaining high-quality sequences without chimeras were sorted into the operational taxonomic units (OTUs) with a cutoff of 97% sequence similarity using UCLUST (25). To obtain the taxonomic information, the sequence with the highest abundance and coverage in each OTU was selected for assignment against the Greengenes database (release 13.8) using PyNAST (26). After removing the sequences which cannot be assigned to bacteria, a total of 3,288,164 clean reads (mean 93,948 reads per sample) were detected in the 35 crab paste samples. To avoid unequal sequencing depth, the OTU table was rarefied at 44,350 reads per sample for further analysis.
2.4. Statistical analysis
Shannon index, Richness, and phylogenetic diversity indices were calculated using QIIME. Pielou’s evenness was calculated using the R package “vegan.” Differences in α-diversity indices and bacterial populations were compared using the Kruskal-Wallis test in the “agricolae” package. A non-metric multidimensional scaling (NMDS) and a principal coordinate analysis (PCoA) were jointly used to visualize the differences in bacterial communities of crab paste between each fermentation time point, with three non-parametric multivariate analyses of dissimilarity based on Bray-Curtis distance using the “vegan” package, including MRPP, ANOSIM, and Adonis. To identify indicative bacteria that associated with fermentation time, the indicators at the OTU level were screened using the R “labdsv” package (27). The indicators of dominant bacteria at the OTU level (with average relative abundance > 0.01% in all samples) were screened with a significant difference between groups (p < 0.05) and indicator value (Indval) > 0.5 using the R package “labdsv.” Spearman correlations between indicator OTUs and significantly changed metabolites (7) were calculated and visualized using the R “psych” and “pheatmap” packages, respectively.
To infer the functional potential of Vibrio, the OTU table of Vibrio was used for predicted 16S rRNA gene copy number by PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Observed States, v2.1.0-b) pipeline (28). Here, only genes involved in the metabolism pathways of significantly changed metabolites of crab paste were predicted. Functional annotations of the genes were obtained according to the KEGG database.1 Differences in functional genes were compared using the Kruskal-Wallis test in the “agricolae” package. All analyses and plots were completed in R 4.0.4 (R Foundation for Statistical Computing, Vienna, Austria).
3. Results
3.1. Change of bacterial community during fermentation
After the sequencing of 16S rRNA amplicons, a total of 3,288,164 high-quality sequences from 35 samples were obtained, with an average of 93,948 ± 21,784 reads per sample. Subsequent analysis was performed on a minimum of 44,350 reads per sample after normalization and homogenization.
To explore change in the bacterial community of crab paste during fermentation, changes of bacterial α-diversity indices were investigated. Bacterial α-diversity of crab paste was at the highest level before fermentation (at day 0), then decreased after fermentation (Figure 1). The Shannon index, richness of observed species, and phylogenetic diversity decreased sharply at the first day of fermentation (p < 0.05) while the Pielous’s evenness index showed a significant drop from the third day of fermentation (p < 0.05). Although the bacterial α-diversity showed certain fluctuation at day 3, it finally reached the lowest level at the end of fermentation. Overall, bacterial α-diversity indices of crab paste were all reduced over fermentation time.
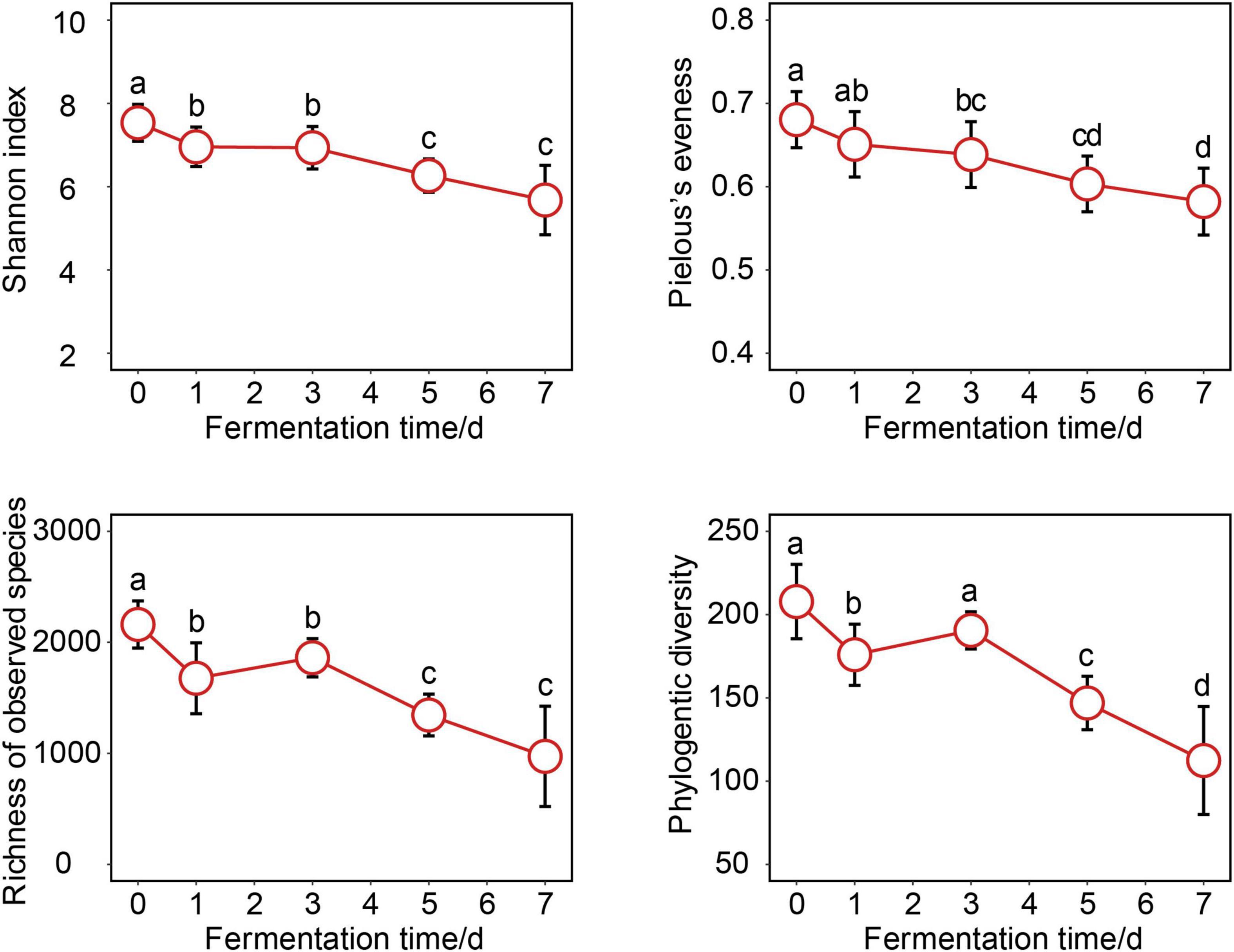
Figure 1. Changes of bacterial α-diversity indices over crab paste fermentation. Data present means ± standard deviation. Different letters indicate significant differences among groups (p < 0.05).
Next, we analyzed whether bacterial community composition of crab paste changed with fermentation time. We found that the bacterial community of crab paste presented a clear succession tendency along the first axis after fermentation if fresh crab samples were excluded (Figures 2A, B). Three non-parametric dissimilarity analyses including MRPP, ANOSIM, and Adonis showed a significant difference between fermentation time points (p < 0.05) except between day 0 and day 1 as well as between day 1 and day 3 (Table 1). These observations indicate that the main variation in the crab paste microbiota occurs at the later period of fermentation.
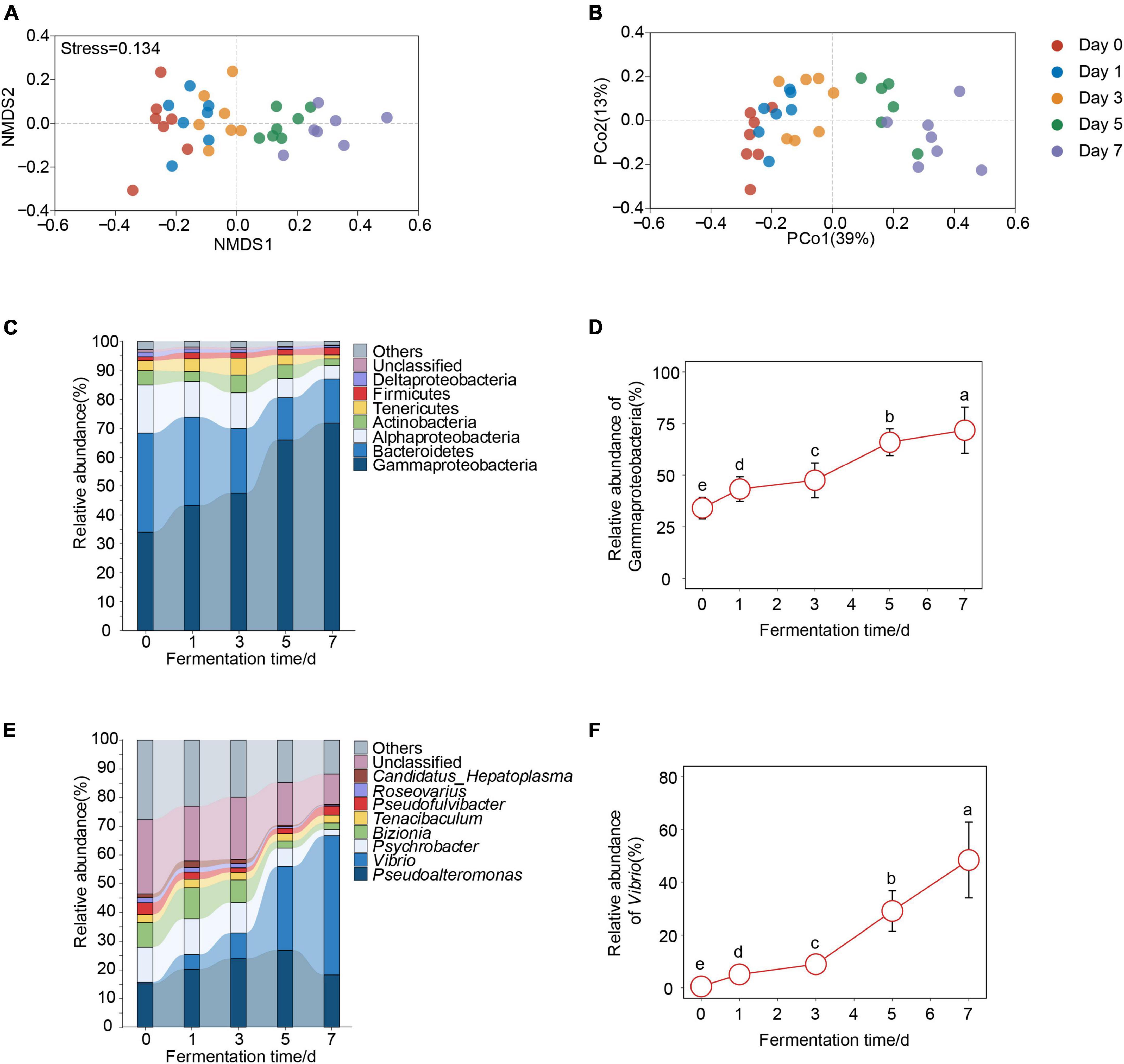
Figure 2. Changes of bacterial communities over crab paste fermentation. Non-metric multidimensional scaling (NMDS) plot (A) and principal coordinate analysis (PCoA) plot (B) based on Bray-Curtis dissimilarity visualizing compositional variations of bacterial communities of crab paste at day 0 (red), day 1 (blue), day 3 (orange), day 5 (green), and day 7 (purple). The relative abundances of bacterial communities at the phyla/proteobacterial classes (C) and genus (E) Level in crab paste during fermentation. Species with relative abundance < 1% were classified as “others”. Significantly increased phyla/proteobacterial classes (D) and genera (F) over crab paste fermentation. Data present means ± standard deviation. Different letters indicate significant differences among groups (p < 0.05).
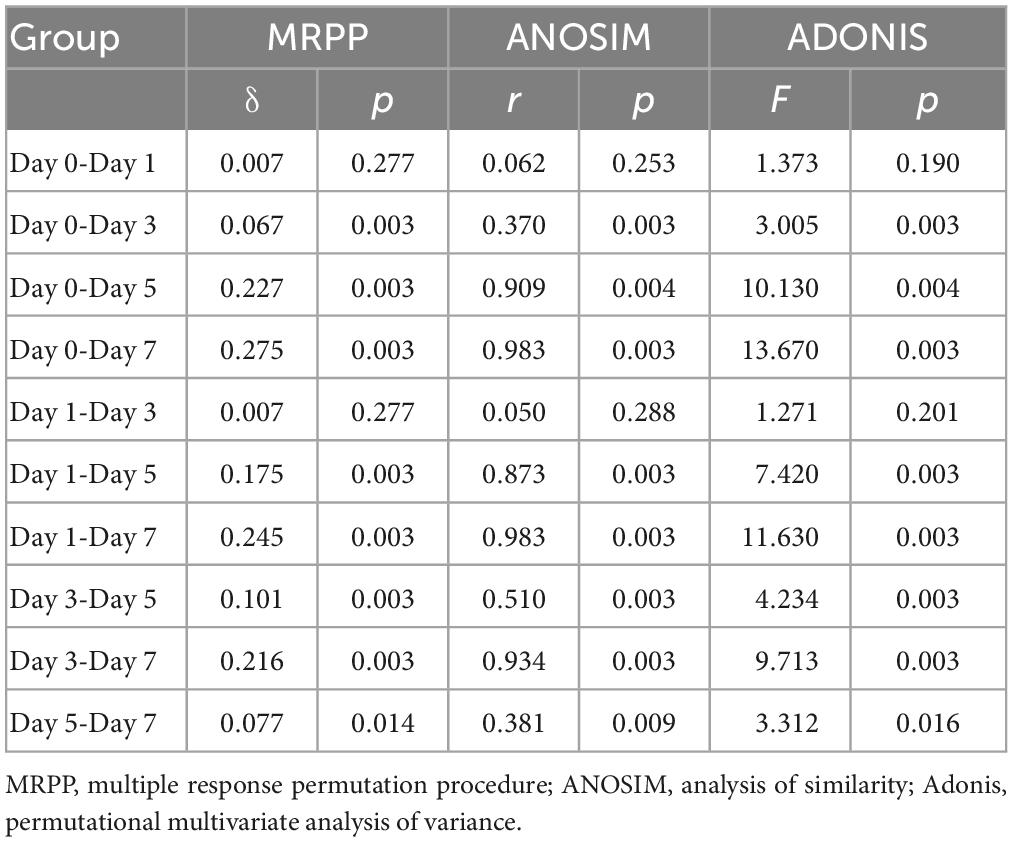
Table 1. Significance tests of the differences in bacterial communities of crab paste between fermentation time points.
The changes in bacterial community composition of crab paste over fermentation time were detectable at the phylum/class level (Figure 2C). Seven dominant phyla/classes including Gammaproteobacteria, Bacteroides, Alphaproteobacteria, Actinobacteria, Tenericutes, Firmicutes, and Deltaproteobacteria all changed across time. Of note, the relative abundance of Gammaproteobacteria highly increased over fermentation time (Figure 2D). Among them, the relative abundance of Gammaproteobacteria continued to increase to the extreme dominance (71.9%) at day 7. In contrast, the relative abundances of Bacteroides, Alphaproteobacteria, and Deltaproteobacteria decreased over fermentation time (Supplementary Figure 1A). The bacterial changes in crab paste over fermentation time were also detectable at the genus level (Figure 2E). The average relative abundance of Vibrio markedly increased throughout the fermentation, reaching up to 48% at day 7 (Figure 2F). In contrast, the relative abundances of Psychrobacter, Bizionia, and Roseovarius significantly decreased over time (Supplementary Figure 1B). Taken together, these observations indicate a substantial rising of Gammaproteobacteria (mainly Vibrio) in the bacterial community of crab paste due to fermentation.
3.2. Indicator OTUs associated with fermentation time
The bacterial assemblages which characterized the discrete bacterial communities of crab paste due to fermentation were further examined. A total of 32 indicator OTUs were screened out at different fermentation time points (Figure 3). In detail, nearly half of the indicator OTUs were most abundant in the fresh crabs (at day 0), such as five OTUs belonging to Flavobacteriaceae (OTU 148, OTU 4657, OTU 6196, OTU 5946, and OTU 1499), three OTUs belonging to Gracilibacteria (OTU 194, OTU 455, and OTU 383), and two OTUs belonging to Mesonia (OTU 4231 and OTU 6680) (Figure 3A). As fermentation proceeded, indicators OTU 344 belonging to Aequorivita viscosa and OTU 338 belonging to Halomonas were most abundant in the crab paste at day 1 (Figure 3B). Further, only one indicator OTU 651 belonging to Brachybacterium presented the highest relative abundance at day 3 and one indicator OTU 329 belonging to Vibrio did at day 5 (Figure 3B). At day 7, 11 OTUs (OTU 35, OTU 28, OTU 26, OTU 24, OTU 7, OTU 140, OTU 14, OTU 275, OTU 77, OTU 8, and OTU 9) belonging to Vibrio dominated in the bacterial community of crab paste, accompanied by OTU 6592 belonging to Gammaproteobacteria (Figure 3A).
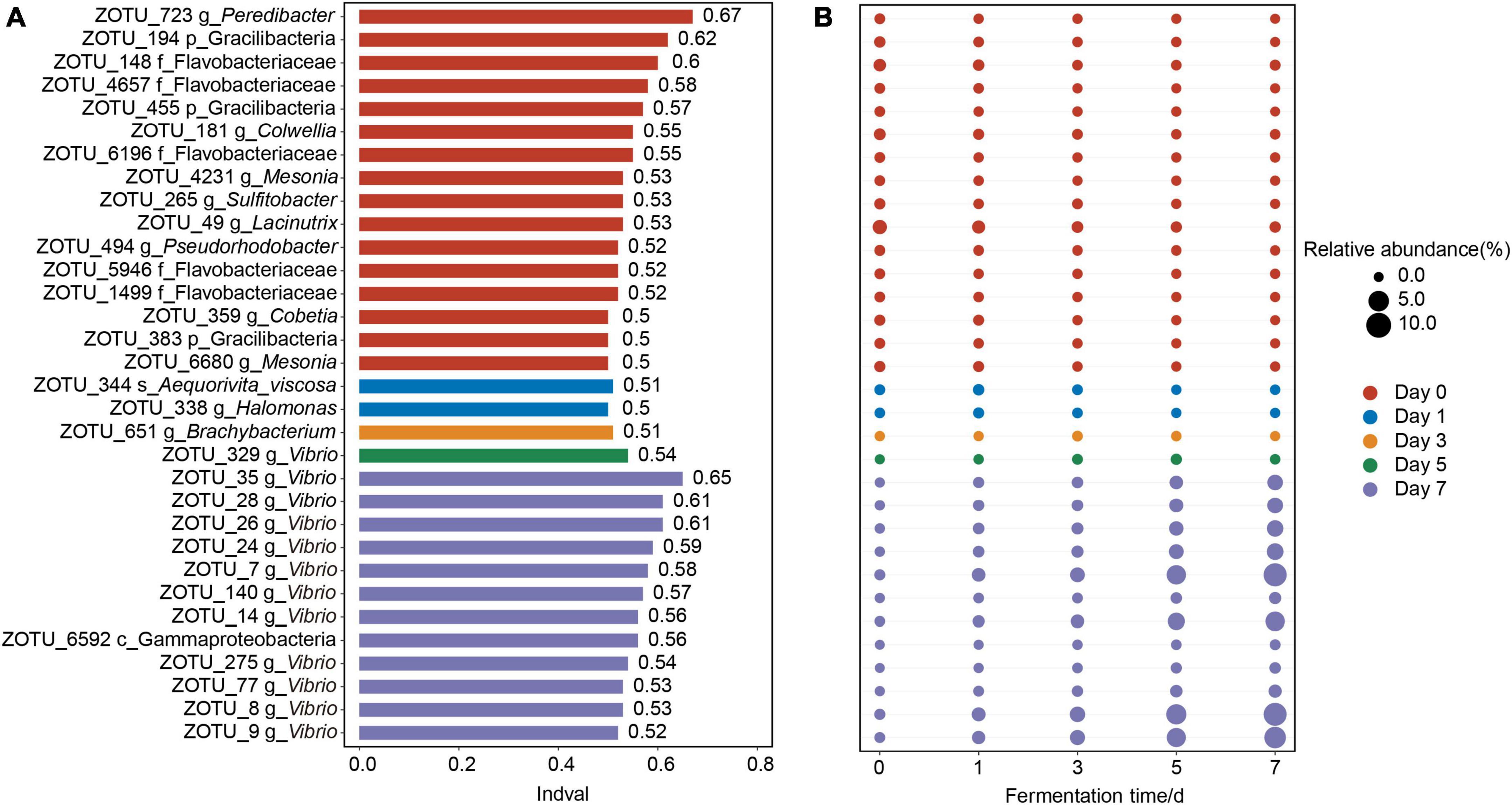
Figure 3. The 32 indicator operational taxonomic units (OTUs) values (A) and relative abundances (B) of crab paste at day 0 (red), day 1 (blue), day 3 (orange), day 5 (green), and day 7 (purple). The length of the bar in plot (A) represents Indval of one indicator OTU. The diameters of the circles in plot (B) are proportional to the relative abundances of the OTUs, with the red, blue, orange, green, and purple circles indicating the peak relative abundances at five fermentation time points, respectively.
3.3. Relationships between indicator OTUs and metabolites
To understand how bacteria relate to metabolites during crab paste fermentation, we analyzed the relationships between indicator OTUs and significantly changed metabolites based on our previous metabolomic results (7). As expected, the heatmap shows a close correlation between bacteria and metabolites with red denoting positive correlation, whereas blue denoting negative correlation (Figure 4). Notably, almost all of indicator OTUs could be classified into two clusters according to the taxonomic information: cluster I (not Vibrio) and cluster II (Vibrio) except OTU6592 which could not be assigned to genus due to the limited 16S rRNA gene information. The cluster I was complex during the first three days of fermentation while cluster II tended to be simple and highlighted by one remarkable indicator OTU turnover since the fifth day of fermentation. Specifically, 12 Vibrio OTUs overwhelmingly dominated in cluster II with one OTU emerging at day 5. Moreover, the two clusters presented a complementary correlation with metabolites. In detail, most of OTUs in cluster I showed positive correlations with inosine, adenosine diphosphate (ADP), taurine, and 2-pyridinemethanol, whereas negative correlations with formate, Hx, and TMA. In contrast, most of OTUs in cluster II presented reverse correlations with metabolites which highlighted by positive correlations with alanine, formate, Hx, and TMA while negative correlations with lactate, inosine, ADP, taurine, trigonelline, and 2-pyridinemethanol. These observations indicate that the succession of bacterial community may play a vital role in the metabolite formation of crab paste during fermentation.
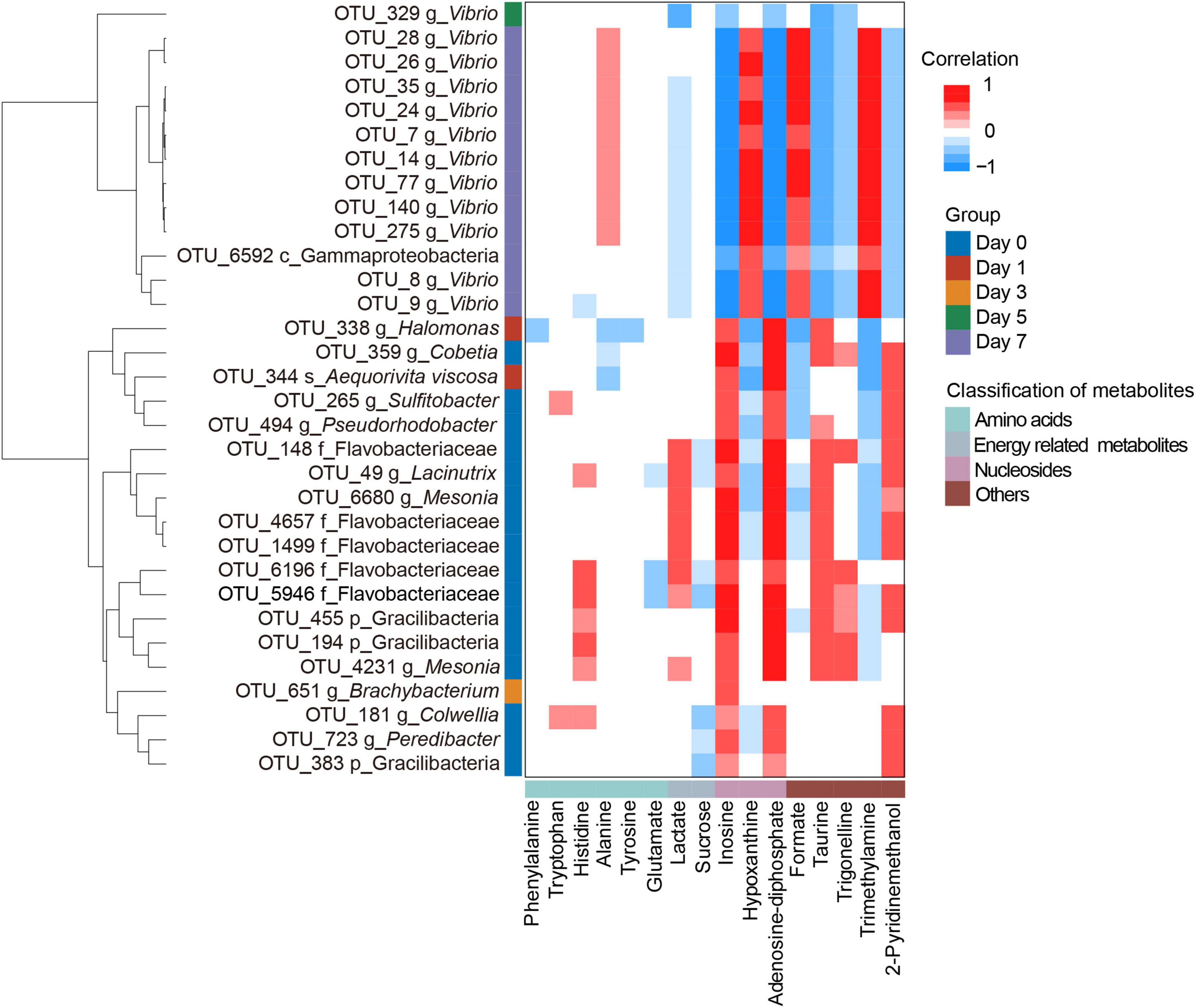
Figure 4. Clustering heatmap showing the Spearman’s correlations between the indicative bacterial operational taxonomic units (OTUs) and the significantly changed metabolites, with red and blue indicating significant positive and negative correlations, respectively.
3.4. Vibrio function prediction analysis
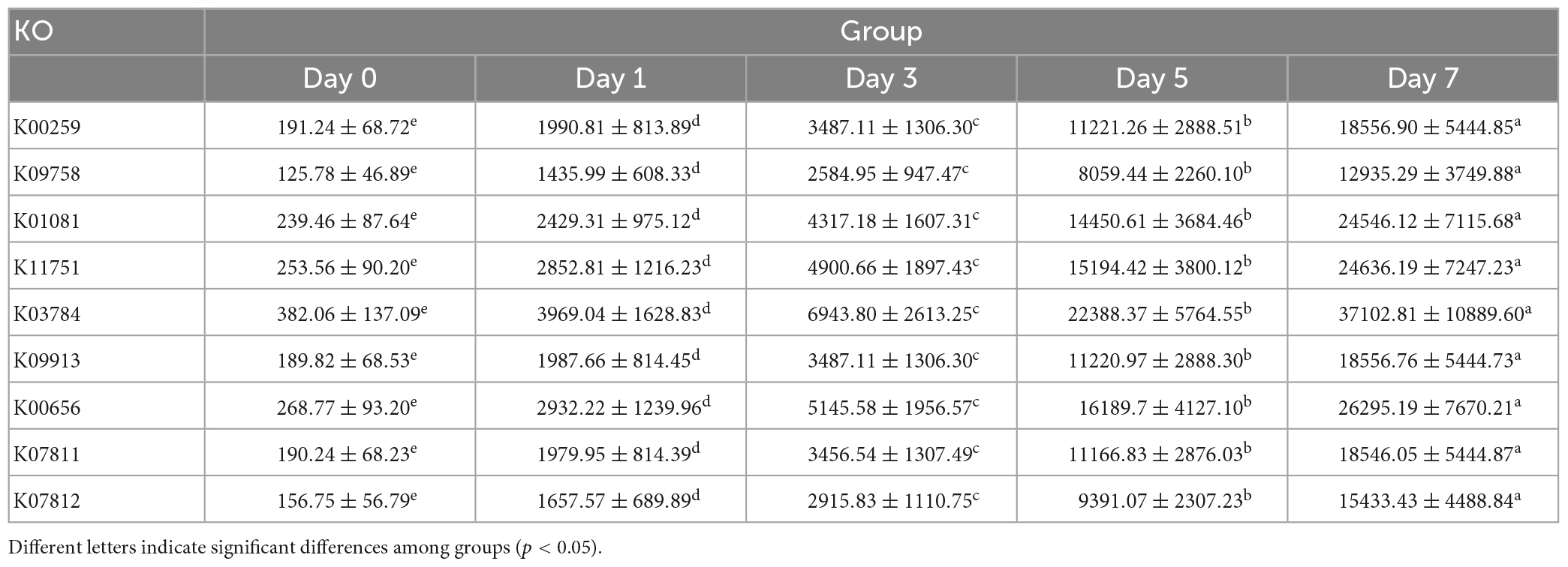
Given the predominance of Vibrio in the bacterial community due to fermentation, large differences in functional profiles of the genus Vibrio between before and after fermentation were predicted by PICRUSt2. We found a significant increase in the abundances of nine predicted function genes of Vibrio (p < 0.05) (Table 2 and Figure 5), which were involved in the metabolic pathways of significantly changed metabolites of crab paste during fermentation (7). For example, the abundances of torA and torZ encoding trimethylamine-N-oxide reductase increased approximately 98-fold at day 7 compared to those at day 0. These results indicate that Vibrio may play a critical role in the metabolite changes of crab paste during fermentation.
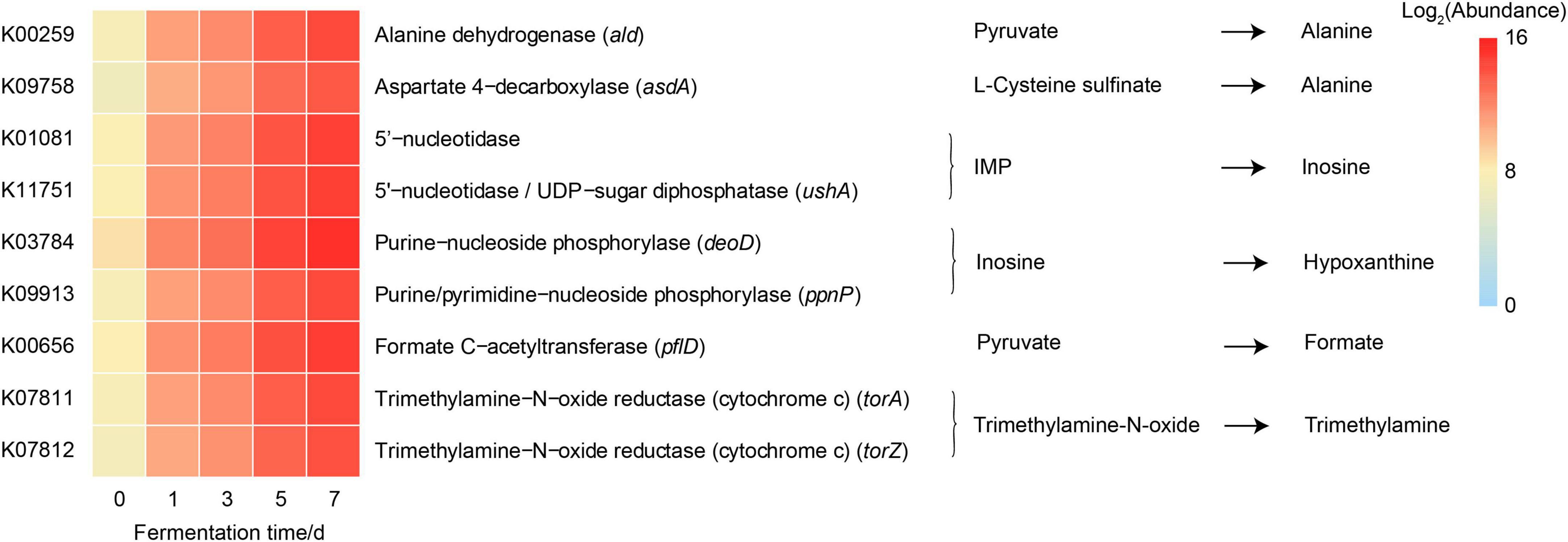
Figure 5. Heatmap showing the abundances of predicted function genes of Vibrio involved in the metabolic pathways of significantly changed metabolites of crab paste during fermentation. The data of gene abundances were log2 transformed.
4. Discussion
Microorganisms impart crab paste a unique flavor and nutritional composition, but they may also lead to spoilage (18–20). Understanding the dynamic changes in the bacterial community may help in the quality control in seafood fermentation. Our microbial study across 7-day fermentation of crab paste allowed an in-depth look into the dynamics of a developing fermented seafood ecosystem. We observed a consistent decrease in bacterial α-diversity over fermentation (Figure 1). The decrease of bacterial α-diversity due to fermentation has been observed in fermented tempeh (29), kimchi (30), and fish (2). The reason why the great loss of bacterial α-diversity during fermentation may be the overabundance of Gamaproteobacteria which inexorably rising from the fifth day of fermentation (Figures 2C, D). Specifically, merely the relative abundance of Gamaproteobacteria (mainly Vibrio) continuously increased with fermentation which accompanied by a substantial decrease in the relative abundances of other phyla. Notably, only Vibrio became predominant in the later days of crab paste fermentation (Figures 2E, F, 3A, B), which is consistent with the change characteristics of gut bacteria in the diseased swimming crab (31) and shrimp (32). Vibrio comprises about > 100 species (33). some Vibrio species such as V. harveyi, V. alginolyticus, and V. parahaemolyticus form pathogenic or symbiotic relationships with marine animals (34–37). This genus seemly had a robust competitive ability and physiological adaption in crab paste environment with high sugar, high salt, and high liquor contents. Such a characteristic change of bacterial community highlighted by an inexorable rise of Vibrio may drive the evolution of metabolomic profile of crab paste during fermentation as observed before (7).
Our correlation analysis and functional prediction can confirm this assertion. As we expected, the results of correlation analysis strongly support a vital role of Vibrio in the formation of TMA, Hx, formate, and alanine in the crab paste during fermentation (Figure 4). TMA is mainly responsible for the fishy odor and often associated with decomposition of aquatic animals, therefore this simple amine is regarded as well-known fish freshness index (38). An excessive intake of TMA will lead to a TMA poisoning (39) and it has been identified as a uremic toxin (40). TMA has been found to be a product of various types of bacteria such as Escherichia, Shewanella, Photobacterium, and Vibrio via the reduction of TMAO (16, 41–44). Of note, in this study, two genes K07811 (torA) and K07812 (torZ) encoding two isozymes of TMAO reductase (TorA and TorZ) were predicted in the genome of Vibrio. TMAO reductase is capable to reduce TMAO to TMA. Moreover, the abundances of these two genes were largely enhanced with the fermentation of crab paste (Figure 5). As such, we speculate that Vibrio contributes most to the TMA formation by TMAO reductase system and drives the spoilage of crab paste.
Moreover, Vibrio positively correlated to Hx but negatively correlated to ADP and inosine (Figure 4). These three compounds are intermediate metabolites in the pathway of ATP catabolism as observed in the postmortem muscle of the mud crab Scylla paramamosain (15, 45). Among them, inosine and Hx are two important determinants of various nucleotide freshness indicators such as K value, Ki value, and H value (46–48). It is generally believed that the breakdown from ATP to inosine monophosphate (IMP) results from autolysis enzyme action, while the further degradation of IMP to inosine and Hx probably results from microbial action (49). In this regard, the production of inosine and Hx may attribute to bacterial metabolism. Moreover, we predicted the genes including K01081 and ushA (K11751) which encoding 5′-nucleotidase/UDP-sugar diphosphatase involved in the conversion of IMP to inosine in Vibrio. We also predicted the genes including deoD (K03784) and ppnP (K09913) which encoding purine-nucleoside phosphorylase and purine-pyrimidine-nucleoside phosphorylase involved in the degradation of inosine to Hx in this genus. Notably, the abundances of these four genes were all highly elevated with the fermentation of crab paste (Figure 5). These observations strongly imply the crucial functional potentials of Vibrio in the ATP catabolism. Previous evidence supports this notion in which Vibrio can secrete extracellular hydrolases decomposing inosine to Hx (50). Given the importance of inosine and Hx in calculating the nucleotide indices, Vibrio is further inferred as an important driver in the spoilage of crab paste from the perspective of nucleotide freshness indicator.
In addition, our results suggest that Vibrio may contribute to the formate and alanine production in the crab paste (Figures 4, 5). This is in line with earlier observations in which formate and alanine can be produced from glucose and other carbohydrates under the action of Vibrio (51, 52). Although formate and alanine are not regarded as the freshness index of seafood, these observations indicate a more diverse metabolic potential of Vibrio during the fermentation of crab paste.
Collectively, these findings combined with our previous metabolomic findings demonstrate that the succession of bacterial community highlighted by the rise of Vibrio during the fermentation of crab paste could be Darwinism encouraging survival of the fittest by requiring extinction of the unfit. Such a change in microbial ecology indeed correlates with a changed metabolic activity of bacteria, which subsequently driving the quality switch from fresh to spoilt crab paste.
5. Conclusion
This study revealed a dynamic change of bacterial community with the fermentation of crab paste which highlighted by consistently decreased α-diversity and overwhelming dominance of Vibrio at the later days of fermentation. Vibrio had a positive correlation with TMA, Hx, formate, and alanine while a negative correlation with inosine and ADP. In contrast, most of other bacterial indicators had a reverse correlation with these metabolites. Moreover, Vibrio presented an improved function potential in the formation of the significantly increased metabolites. Collectively, these findings combined with our previous metabolomic findings demonstrate that the inexorable rise of Vibrio not only drives the indicator OTUs turnover in the bacterial community but also is implicated in the quality switch from fresh to spoilt crab paste. In fact, natural microbial fermentation can be a double-edged sword. How to control the microbial fermentation of seafood with great precision merits need to be studied in future.
Data availability statement
The original contributions presented in this study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA808815.
Ethics statement
The animal study was reviewed and approved by Animal Care and Use Committee of Ningbo University.
Author contributions
T-HX: conceptualization, validation, methodology, data curation, formal analysis, software, and writing—original draft. CS, C-KM, and C-LW: supervision and writing—review and editing. Y-FY: conceptualization, validation, methodology, data curation, formal analysis, funding acquisition, supervision, and writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Key Scientific and Technological Grant of Zhejiang for Breeding New Agricultural Varieties (2021C02069-6), 2025 Technological Innovation for Ningbo (2019B10010), China Agriculture Research System of MOF and MARA, and K. C. Wong Magna Fund in Ningbo University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2023.1092573/full#supplementary-material
Footnotes
References
1. Yang Z, Liu S, Lv J, Sun Z, Xu W, Ji C, et al. Microbial succession and the changes of flavor and aroma in Chouguiyu, a traditional Chinese fermented fish. Food Biosci. (2020) 37:100725. doi: 10.1016/j.fbio.2020.100725
2. Li C, Zhao Y, Wang Y, Li L, Huang J, Yang X, et al. Contribution of microbial community to flavor formation in tilapia sausage during fermentation with Pediococcus pentosaceus. LWT Food Sci Technol. (2022) 154:112628. doi: 10.1016/j.lwt.2021.112628
3. Afifah D, Dewi U, Anggraeni R, Tsani A, Widyastuti N, Fulyani F, et al. Antioxidant activity, microbiological quality, and acceptability of spontaneously fermented shrimp sausage (Litopenaeus vannamei). J Food Qual. (2022) 2022:1–8. doi: 10.1155/2022/5553432
4. Ye Y, Zhang L, Tang H, Yan X. Survey of nutrients and quality assessment of crab paste by 1H NMR spectroscopy and multivariate data analysis. Chin Sci Bull. (2012) 57:3353–62. doi: 10.1007/s11434-012-5119-x
5. Kleekayai T, Pinitklang S, Laohakunjit N, Suntornsuk W. Volatile components and sensory characteristics of Thai traditional fermented shrimp pastes during fermentation periods. J Food Sci Technol. (2016) 53:1399–410. doi: 10.1007/s13197-015-2142-3
6. Shim K, Mok J, Jeong Y, Park K, Jang M. Effect of organic acids on the formation of biogenic amines in fermented anchovy sauce comprising raw anchovy materials with different levels of freshness. J Food Sci Technol. (2022) 59:703–14. doi: 10.1007/s13197-021-05065-w
7. Chen D, Ye Y, Chen J, Yan X. Evolution of metabolomics profile of crab paste during fermentation. Food Chem. (2016) 192:886–92. doi: 10.1016/j.foodchem.2015.07.098
8. Özogul F, Özden Ö, Özoğul Y, Erkan N. The effects of gamma-irradiation on the nucleotide degradation compounds in sea bass (Dicentrarchus labrax) stored in ice. Food Chem. (2010) 122:789–94. doi: 10.1016/j.foodchem.2010.03.054
9. Dervisevic M, Custiuc E, Çevik E, Durmus Z, Şenel M, Durmus A. Electrochemical biosensor based on REGO/Fe3O4 bionanocomposite interface for xanthine detection in fish sample. Food Control. (2015) 57:402–10. doi: 10.1016/j.foodcont.2015.05.001
10. Bulushi I, Poole S, Deeth H, Dykes G. Biogenic amines in fish: Roles in intoxication, spoilage, and nitrosamine formation—a review. Crit Rev Food Sci Nutr. (2009) 49:369–77. doi: 10.1080/10408390802067514
11. He S, Jiang H, Zhuo C, Jiang W. Trimethylamine/Trimethylamine-N-oxide as a key between diet and cardiovascular diseases. Cardiovasc Toxicol. (2021) 21:593–604. doi: 10.1007/s12012-021-09656-z
12. Lv X, Li Y, Cui T, Sun M, Bai F, Li X, et al. Bacterial community succession and volatile compound changes during fermentation of shrimp paste from Chinese Jinzhou region. LWT Food Sci Technol. (2020) 122:108998. doi: 10.1016/j.lwt.2019.108998
13. Bao R, Liu S, Ji C, Liang H, Yang S, Yan X, et al. Shortening fermentation period and quality improvement of fermented fish, chouguiyu, by co-inoculation of Lactococcus lactis M10 and Weissella cibaria M3. Front Microbiol. (2018) 9:3003. doi: 10.3389/fmicb.2018.03003
14. Shen Y, Wu Y, Wang Y, Li L, Li C, Zhao Y, et al. Contribution of autochthonous microbiota succession to flavor formation during Chinese fermented mandarin fish (Siniperca chuatsi). Food Chem. (2021) 348:129107. doi: 10.1016/j.foodchem.2021.129107
15. Lin W, He Y, Shi C, Mu C, Wang C, Li R, et al. ATP catabolism and bacterial succession in postmortem tissues of mud crab (Scylla paramamosain) and their roles in freshness. Food Res Int. (2022) 155:110992. doi: 10.1016/j.foodres.2022.110992
16. Dos Santos J, Iobbi-Nivol C, Couillault C, Giordano G, Méjean V. Molecular analysis of the trimethylamine N -oxide (TMAO) reductase respiratory system from a Shewanella species. J Mol Biol. (1998) 284:421–33. doi: 10.1006/jmbi.1998.2155
17. Kim K, Lee S, Chun B, Jeong S, Jeon C. Tetragenococcus halophilus MJ4 as a starter culture for repressing biogenic amine (cadaverine) formation during saeu-jeot (salted shrimp) fermentation. Food Microbiol. (2019) 82:465–73. doi: 10.1016/j.fm.2019.02.017
18. Ma C, Xu Z, Yang X. Studies on microbiological quality and safety characteristics of bottled crab paste in cold storage. J Shanghai Fish Univ. (2008) 2:222–6.
19. Zhang C, Chen Y, Quan J, Xu J, Su X, Zhang C, et al. Bacterial diversity of salted mudsnail and crab paste. Food Sci. (2011) 32:185–8.
20. Huang J, Chen Y, Quan J, Zhang C, Li Y, Qiu D, et al. Isolation of identification of cultivable microorganisms in bottled crab paste and salted mudsnail. Food Sci. (2011) 32:217–20.
21. Herlemann D, Labrenz M, Jürgens K, Bertilsson S, Waniek J, Andersson A. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. (2011) 5:1571–9. doi: 10.1038/ismej.2011.41
22. Magoč T, Salzberg S. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. (2011) 27:2957–63. doi: 10.1093/bioinformatics/btr507
23. Callahan B, McMurdie P, Rosen M, Han A, Johnson A, Holmes S. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. (2016) 13:581–3. doi: 10.1038/nmeth.3869
24. Edgar R, Haas B, Clemente J, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics (2011) 27:2194–200. doi: 10.1093/bioinformatics/btr381
25. Edgar R. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. (2010) 26:2460–1. doi: 10.1093/bioinformatics/btq461
26. DeSantis T, Hugenholtz P, Larsen N, Rojas M, Brodie E, Keller K, et al. Greengenes, a Chimera-Checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. (2006) 72:5069–72. doi: 10.1128/AEM.03006-05
27. Dufrêne M, Legendre P. Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol Monogr. (1997) 67:345–66. doi: 10.1890/0012-96151997067[0345:SAAIST]2.0.CO;2
28. Wang Y, Wang K, Huang L, Dong P, Wang S, Chen H, et al. Fine-scale succession patterns and assembly mechanisms of bacterial community of Litopenaeus vannamei larvae across the developmental cycle. Microbiome (2020) 8:106. doi: 10.1186/s40168-020-00879-w
29. Chen Y, Li P, He W, Liao L, Xia B, Jiang L, et al. Analysis of microbial community and the characterization of Aspergillus flavus in Liuyang Douchi during fermentation. LWT Food Sci Technol. (2022) 154:112567. doi: 10.1016/j.lwt.2021.112567
30. Yang Y, Fan Y, Li T, Yang Y, Zeng F, Wang H, et al. Microbial composition and correlation between microbiota and quality-related physiochemical characteristics in chongqing radish paocai. Food Chem. (2022) 369:130897. doi: 10.1016/j.foodchem.2021.130897
31. Xia M, Pei F, Mu C, Ye Y, Wang C. Disruption of bacterial balance in the gut of Portunus trituberculatus induced by Vibrio alginolyticus infection. J Ocean Limnol. (2018) 36:1891–8. doi: 10.1007/s00343-018-7121-x
32. Hou D, Zhou R, Zeng S, Wei D, Deng X, Xing C, et al. Intestine bacterial community composition of shrimp varies under low- and high-salinity culture conditions. Front Microbiol. (2020) 11:589164. doi: 10.3389/fmicb.2020.589164
33. Baker-Austin C, Oliver J, Alam M, Ali A, Waldor M, Qadri F, et al. Vibrio spp. infections. Nat Rev Dis Primers. (2018) 4:1–19. doi: 10.1038/s41572-018-0005-8
34. Liu Q, Li H, Wang Q, Liu P, Dai F, Li J. Identification and phylogenetic analysis of a strain of Vibrio alginolyticus, a pathogen in Portunus trituberculatus with toothpaste disease. Mar Freshw Res. (2007) 4:9–13.
35. Yildiz F, Visick K. Vibrio biofilms: So much the same yet so different. Trends Microbiol. (2009) 17:109–18. doi: 10.1016/j.tim.2008.12.004
36. Yan B, Qin G, Bao Z, Zhang X, Bi K, Qin L. Isolation and identification of Vibrio parahaemolyticus from diseased Portunus trituberculatus L. Mar Sci Bull. (2010) 29:560–6.
37. Zhang X, Bai X, Yan B, Bi K, Qin L. Vibrio harveyi as a causative agent of mass mortalities of megalopa in the seed production of swimming crab Portunus trituberculatus. Aquacult Int. (2014) 22:661–72. doi: 10.1007/s10499-013-9695-9
38. Prabhakar P, Vatsa S, Srivastav P, Pathak SS. A comprehensive review on freshness of fish and assessment: Analytical methods and recent innovations. Food Res Int. (2020) 133:109157. doi: 10.1016/j.foodres.2020.109157
39. Anthoni U, Christophersen C, Gram L, Nielsen N, Nielsen P. Poisonings from flesh of the Greenland shark Somniosus microcephalus may be due to trimethylamine. Toxicon. (1991) 29:1205–12. doi: 10.1016/0041-0101(91)90193-U
40. Duranton F, Cohen G, De Smet R, Rodriguez M, Jankowski J, Vanholder R, et al. Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol. (2012) 23:1258–70. doi: 10.1681/ASN.2011121175
41. Gram L, Huss H. Microbiological spoilage of fish and fish products. Int J Food Microbiol. (1996) 33:121–37. doi: 10.1016/0168-1605(96)01134-8
42. Santini C. A novel Sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. (1998) 17:101–12. doi: 10.1093/emboj/17.1.101
43. Dunn A, Stabb E. Genetic analysis of Trimethylamine N -Oxide reductases in the light organ symbiont Vibrio fischeri ES114. J Bacteriol. (2008) 190:5814–23. doi: 10.1128/JB.00227-08
44. Proctor L, Gunsalus R. Anaerobic respiratory growth of Vibrio harveyi, Vibrio fischeri and Photobacterium leiognathi with trimethylamine N-oxide, nitrate and fumarate: Ecological implications. Environ Microbiol. (2000) 2:399–406. doi: 10.1046/j.1462-2920.2000.00121.x
45. Zhang L, Guo L, Mu C, Ye Y, Wang C. Postmortem metabolite profile changes of mud crab (Scylla paramamosain) under different storage conditions. J Ocean Univ China. (2021) 20:608–18. doi: 10.1007/s11802-021-4558-x
46. Song Y, Luo Y, You J, Shen H, Hu S. Biochemical, sensory and microbiological attributes of bream (Megalobrama amblycephala) during partial freezing and chilled storage. J Sci Food Agric. (2012) 92:197–202. doi: 10.1002/jsfa.4572
47. Hong H, Regenstein J, Luo Y. The importance of ATP-related compounds for the freshness and flavor of post-mortem fish and shellfish muscle: A review. Crit Rev Food Sci Nutr. (2017) 57:1787–98. doi: 10.1080/10408398.2014.1001489
48. Garg D, Singh M, Verma N, Monika. Review on recent advances in fabrication of enzymatic and chemical sensors for hypoxanthine. Food Chem. (2022) 375:131839. doi: 10.1016/j.foodchem.2021.131839
49. Jia S, Liu Y, Zhuang S, Sun X, Li Y, Hong H, et al. Effect of ε-polylysine and ice storage on microbiota composition and quality of Pacific white shrimp (Litopenaeus vannamei) stored at 0 °C. Food Microbiol. (2019) 83:27–35. doi: 10.1016/j.fm.2019.04.007
50. Cai S, Cheng H, Pang H, Jian J, Wu Z. AcfA is an essential regulator for pathogenesis of fish pathogen Vibrio alginolyticus. Vet Microbiol. (2018) 213:35–41. doi: 10.1016/j.vetmic.2017.11.016
51. Shieh W, Chen A, Chiu H. Vibrio aerogenes sp. nov., a facultatively anaerobic marine bacterium that ferments glucose with gas production. Int J Syst Evol Microbiol. (2000) 50:321–9. doi: 10.1099/00207713-50-1-321
Keywords: crab paste, swimming crab, fermentation, Vibrio, bacterial community
Citation: Xiong T-H, Shi C, Mu C-K, Wang C-L and Ye Y-F (2023) Rise and metabolic roles of Vibrio during the fermentation of crab paste. Front. Nutr. 10:1092573. doi: 10.3389/fnut.2023.1092573
Received: 08 November 2022; Accepted: 09 February 2023;
Published: 24 February 2023.
Edited by:
Jinxuan Cao, Beijing Technology and Business University, ChinaReviewed by:
Wen-Liang Xiang, Xihua University, ChinaLi Li, Ocean University of China, China
Changle Qi, Huzhou University, China
Copyright © 2023 Xiong, Shi, Mu, Wang and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang-Fang Ye,  eWV5YW5nZmFuZ0BuYnUuZWR1LmNu
eWV5YW5nZmFuZ0BuYnUuZWR1LmNu
 Tian-Han Xiong
Tian-Han Xiong Ce Shi
Ce Shi Chang-Kao Mu
Chang-Kao Mu Chun-Lin Wang
Chun-Lin Wang Yang-Fang Ye
Yang-Fang Ye