- 1Department of Nutritional Sciences, University of Toronto, Toronto, ON, Canada
- 2Joannah and Brian Lawson Centre for Child Nutrition, University of Toronto, Toronto, ON, Canada
Cranberries have known anti-inflammatory properties, which extend their benefits in the context of several chronic diseases. These benefits highly rely on the polyphenol profile of cranberries, one of few foods rich in A-type proanthocyanidin (PAC). A-type PAC comprises flavan-3-ol subunits with an additional interflavan ether bond in the conformational structure of the molecule, separating them from the more commonly found B-type PAC. PACs with a degree of polymerization higher than three are known to reach the colon intact, where they can be catabolyzed by the gut microbiota and biotransformed into lower molecular weight organic acids that are available for host absorption. Gut microbiota-derived metabolites have garnered much attention in the past decade as mediators of the health effects of parent compounds. Though, the mechanisms underlying this phenomenon remain underexplored. In this review, we highlight emerging evidence that postulates that polyphenols, including ones derived from cranberries, and their metabolites could exert anti-inflammatory effects by modulating host microRNAs. Our review first describes the chemical structure of cranberry PACs and a pathway for how they are biotransformed by the gut microbiota. We then provide a brief overview of the benefits of microbial metabolites of cranberry in the intestinal tract, at homeostasis and in inflammatory conditions. Finally, we discuss the role of microRNAs in intestinal health and in response to cranberry PAC and how they could be used as targets for the maintenance of intestinal homeostasis. Most of this research is pre-clinical and we recognize that conducting clinical trials in this context has been hampered by the lack of reliable biomarkers. Our review discusses the use of miRNA as biomarkers in this context.
1. Introduction
The American Cranberry (Vaccinium macrocarpon) is the most consumed species of cranberry among the four known types in North America [reviewed in Feghali et al. (1)]. For decades, cranberries have attracted attention in the field of nutritional sciences because of their abundant, unique polyphenol profile that renders them as powerful antioxidants and modulators of inflammation (2). Polyphenols are also increasingly regarded as potential prebiotic compounds. This is because polyphenols of higher molecular weight that are too large to be absorbed in the small intestine, are then directed to the colon, where they are catabolized into organic acids by the gut microbiota; these become available for host absorption and thus, could mediate the health effects of polyphenols (3). Cranberry and its polyphenols also influence the composition of the gut microbiota, thereby potentially affecting its function and production of secondary metabolites from various dietary substrates. Both cranberry components and the gut microbiota affect the intestinal gene expression program. In particular, evidence has emerged that these effects manifest transcriptionally as well as post-transcriptionally via microRNAs (miRNAs) (4–6). It is thus possible that cranberry and the microbiota interact to affect host miRNAs and that this underlies downstream effects. The aim of this review is to provide a comprehensive assessment of the reciprocal interaction between dietary cranberry and the gut microbiota and to discuss the role of intestinal miRNAs in this context.
2. Polyphenols in cranberry
2.1. Chemical structure
In cranberry juice, the four main phenolic classes identified include phenolic acids and three flavonoid classes: anthocyanins, flavonols, and flavan-3-ols (7). About 85% (by weight) of the total flavan-3-ols are represented by proanthocyanidins (PACs). Cranberry PACs are typically formed by epicatechin subunits (flavan-3-ol monomers) with an average degree of polymerization (DP) of four to seven, which can extend up to 12 (8, 9). Opposite to most fruits PACs which contain B-type bonds (typically linked through C4 → C8 or C4 → C6, cranberries are among the richest sources of A-type PACs (10), with most PACs containing at least one A-type linkage (11). A-type bonds are doubly linked, containing an additional ether bond between the C2 of the upper unit of one heterocyclic ring, to the oxygen-bearing C5 or C7 on the lower unit awarding the molecule a degree of conformational inflexibility (Figure 1).
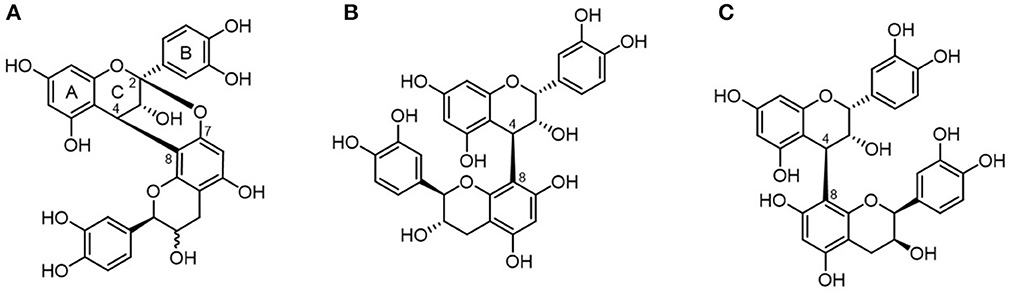
Figure 1. Chemical structure of proanthocyanidins. PACs are differentiated into A-type (A) proanthocyanidin A and B-type (B) proanthocyanidin B1 or (C) proanthocyanidin B2 depending on their interflavanic linkages.
2.2. Consumption, bioavailability and microbial metabolism
2.2.1. Consumption
Representative surveys of the European and American populations have estimated PAC daily intake to range between 95 to 160 mg/day, with marked differences among countries and dietary habits (12, 13). A 240 ml serving of a 27% juice cranberry cocktail contains an average of 38 mg of PAC [reviewed in Blumberg et al. (14)]; this amount corresponds approximately to the recommended daily intake of 36 mg/day that was shown to prevent urinary tract infections [reviewed in Howell et al. (15)]. Clinical studies have altered the composition of cranberry juice to contain more PAC, ranging from 77 to 118 mg per 240 ml serving in doubly concentrated 54% cranberry juice (16, 17), up to 922.2 mg per 240 ml serving in 117% cranberry juice (18). So theoretically, based on a colonic volume estimate of 561 ml (19), the consumption of a serving of PAC-enriched cranberry beverage could result in colonic exposure to PAC in concentrations ranging from 137 μg/ml to as high as 1,644 μg/ml. Colonic PAC is available for microbial metabolism (20) which could ultimately affect the magnitude of exposure across individuals.
2.2.2. Bioavailability
Several studies have reported that cranberry polyphenols and their metabolites are bioavailable to the host. A variety of phenolic acids, flavonoids and their metabolites have been detected in human plasma and urine following acute or prolonged consumption of cranberry juice (17, 21–23). Hydrosoluble PACs are the only PAC molecules available for absorption and become accessible to the enterocyte surface in the small intestine. The absorption of non-soluble PACs depends on their DP, MW, and structure. An in vitro study showed that A-type PACs with lower DP (dimer, trimer and tetramer) can transverse the intestinal monolayer at low rates (24). Intestinal epithelial cells are important sites of PACs modification; these polymers undergo methylation and glucuronidation processes, producing methylated and glycosylated PACs with higher bioavailability (25). Polymers reach the colon intact where they are metabolized by the gut microbiota in a diverse range of metabolites (25, 26), with different hydroxylation profiles and aliphatic side chain lengths, such as phenylacetic, phenylpropionic, and phenylvaleric acids (3, 27, 28), which are available for absorption by colonocytes.
2.3. PACs metabolism by the microbiota
The gut microbiota is an indispensable player for the processing of ingested food. The functional potential of the microbiota is estimated to be encoded in millions of genes (29), many of which are necessary to metabolize otherwise undigestible dietary components that reach the colon. Specifically, in the colon, the backbones of non-absorbed PACs undergo a series of bacterial enzymatic transformations resulting in the generation of less complex compounds. Figure 2 presents a compendium of these reactions that include C-ring cleavage, dihydroxylation and decarboxylation. Once absorbed, the microbial metabolites of PACs are transported through the portal vein to the liver, where they undergo further metabolism by phase II enzymes (30) before being released into systemic circulation. There is currently limited research to identify gut microbial taxa involved in the catabolism of cranberry polyphenols. Different bacterial genera have been reported to initiate the metabolism of flavan-3-ols, including Eubacterium, Flavonifractor, Eggerthella, Lactobacillus, and Enterococcus (20, 31, 32). Clostridium coccoides, Eubacterium spp., Eggerthella lenta, Adlercreutzia equolifaciens, Slackia equolifaciens, and lactobacilli species carry PAZymes (polyphenol-associated enzymes) (33). PACs first undergo partial acid-catalyzed interflavan cleavage to form procyanidin dimers epicatechins units, which are then transformed by colonic bacteria. Eggerthella lenta, Clostridium spp. and Eubacterium sp. are involved in opening the C-rings in epicatechins (34); following C-ring cleavage, Flavonifractor plautii further converts this intermediate into 5-(3′-4′-dihydroxyphenyl)-γ-valerolactone through A-ring breakdown and lactone formation (34, 35) (Figure 2).
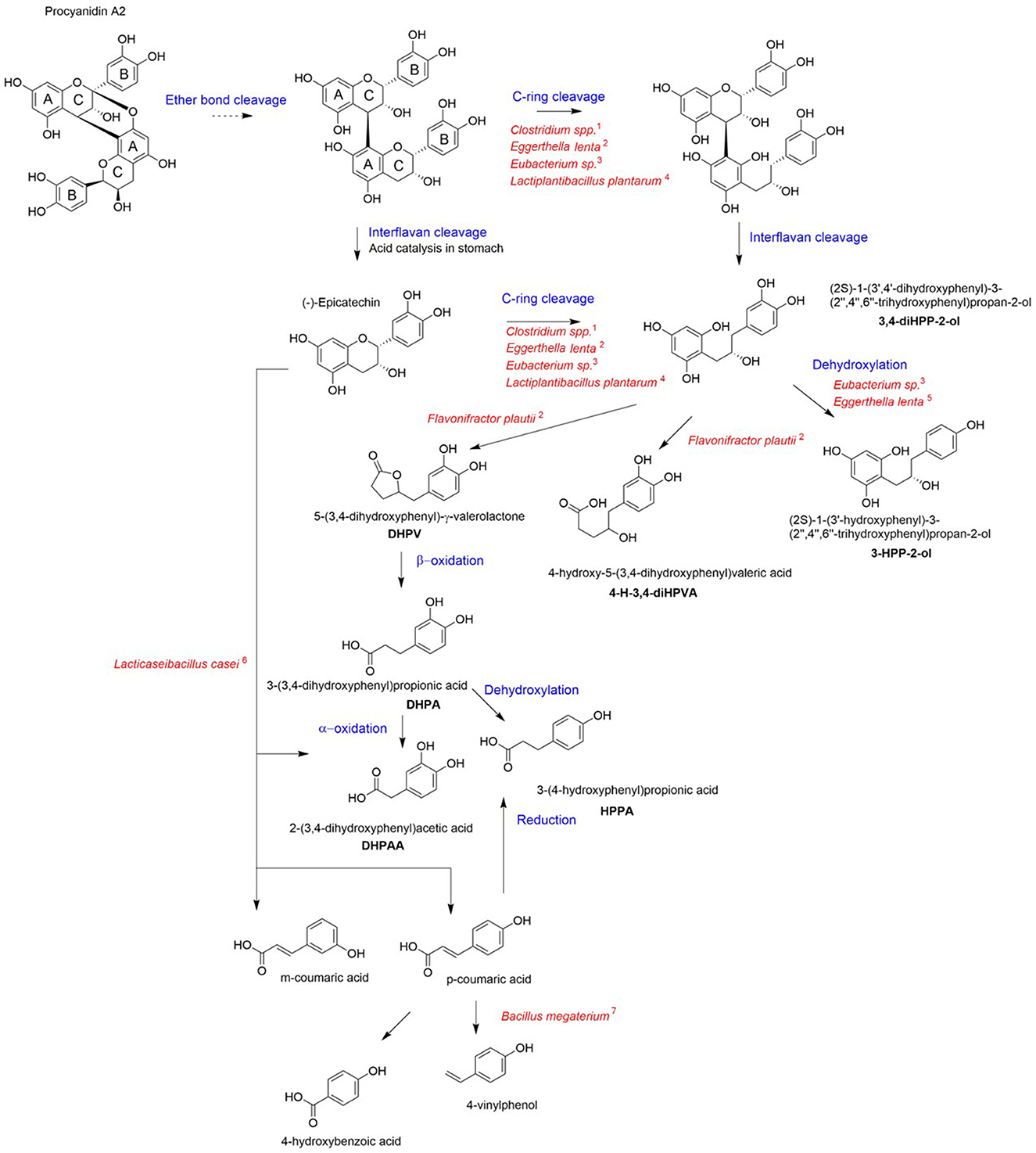
Figure 2. Proposed pathways for cranberry PAC microbial catabolism. Blue indicates the chemical reaction for each step and red corresponds to the bacterial taxa involved in the metabolite formation for each step. The catabolic pathway was generated via integration of published data [1: Corrêa et al. (36); 2: Kutschera et al. (37); 3: Wang et al. (38); 4: Sánchez-Patán et al. (39); 5: Jin and Hattori (40); 6: Li et al. (41); 7: Torres and Rosazza (42)].
In vitro microbiota fermentation of cranberry PACs was shown to produce higher amounts of 2-(3′-hydroxyphenyl)-acetic acid compared to apple (43). In comparison to grape seed polyphenols, cranberry PACs were catabolized to a greater extent and produced distinctly high amounts of 3-(3′,4′-dihydroxyphenyl)-propionic acid, 3-(4-hydroxyphenyl)-propionic acid (HPPA); 3,4-dihydroxyphenylacetic acid (DHPAA), and phenylpropionic acid (20). Both HPPA and DHPAA metabolites were reported to be produced from A-type PAC in vitro, using a pig cecum model (44). Moreover, HPPA was identified as the most highly produced metabolite from the bioconversion of PAC using Lacticaseibacillus rhamnosus (32). Inter-individual variability exists in the production of metabolites from cranberry polyphenols (23, 45), which may relate to bacterial PAZymes polymorphism, (46), host gut microbiota composition and diet (47). Nonetheless, PAC metabolites have been consistently detected in high amounts in human plasma or urine after consumption of cranberry juice. HPPA was identified as the most abundant polyphenol metabolite in plasma of healthy young individuals (23). Similarly, DHPAA has been found in high amounts in the urine of healthy older adults after acute cranberry juice exposure (22), as well as in overweighted adults who received a low-calorie cranberry extract beverage for 8 weeks (17).
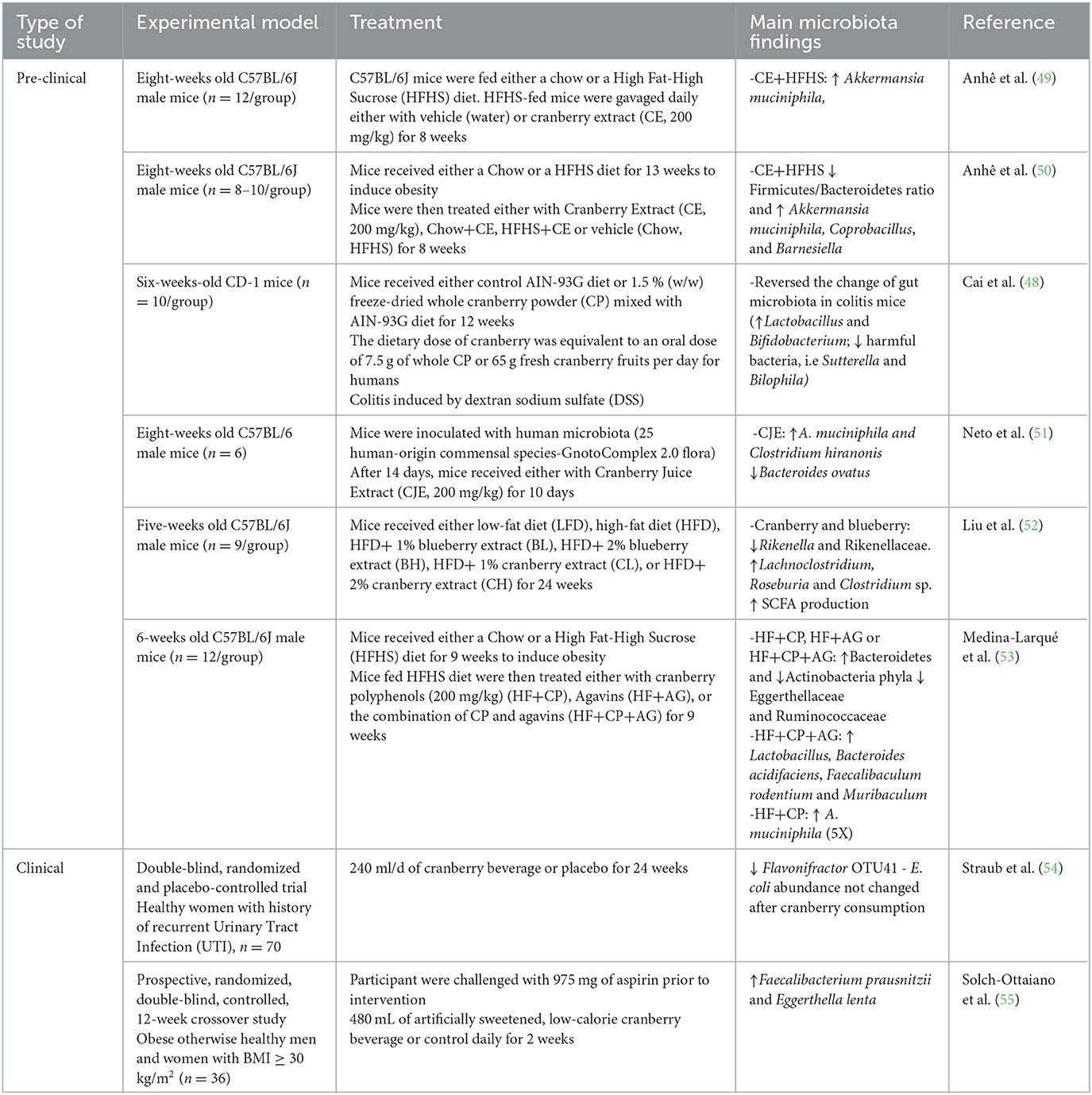
Interestingly, the ingestion of cranberry modifies the composition of the microbiota, as shown in pre-clinical and clinical studies including both health and disease conditions (Table 1). The limited number of studies makes it difficult to identify a common pattern of microbial responses. Though, overall, the effect appears to favor a health-compatible microbial phenotype. This is sometimes accompanied by a measured physiological benefit, such as in a mouse model of colitis where cranberry powder administration reversed the microbiota changes found in inflamed mice and resulted in decreased colitis severity and production of pro-inflammatory cytokines (48). Taken together, these findings suggest that the bi-directional interaction between cranberry and the gut microbiota is an important determinant of cranberry benefits.
3. Cranberry PAC, the microbiota, PAC microbial metabolites and the anti-inflammatory effects of cranberries
In vivo studies that have demonstrated effects of cranberry in models of inflammation have implicated the microbiota in these effects. For example, in a mouse colitis model, whole cranberry decreased inflammatory cytokines in the colonic mucosa (48, 56) and whole cranberry powder was able to counteract the microbiota alterations by restoring Lactobacillus and Bifidobacterium with implications for better repair and protection from cancer (48). In mouse models of diet-induced obesity, PAC-rich extracts improved metabolic parameters (reduced liver triglyceride accumulation, improved insulin sensitivity) during high fat-high sucrose feeding (50) and shifted gut microbiota from a Firmicutes/Ruminococcus enterotype enriched in opportunistic bacteria to an enterotype enriched in Lactobacillus and Akkermansia, while boosting butyrate-producing bacteria (57). Interestingly, polyphenol-degrading bacterial families such as Eggerthellaceae were also favored, suggesting that polyphenol metabolism accompanies microbiota shifts that are favorable to metabolic health. Causal relationships are difficult to demonstrate. Though, studies that investigate the effects of microbial metabolites of cranberry polyphenols would be helpful. Similarly to its parent compound PAC (58), DHPPA was also reported for its anti-inflammatory properties in vitro [reviewed in Mithul Aravind et al. (59)]. Moreover, one in vivo study showed that pre-treatment with DHPPA at a dose of 30 mg/kg caused a significant decrease of abdominal contractions induced by acetic acid administration in rats (60). Interestingly, protective effects of PAC microbial metabolites DHPPA and HPPA were also seen against p-cresol, a toxic compound produced by the gut microbiota from l-tyrosin. Both metabolites prevented p-cresol-induced alterations of cell membrane integrity and cell death in vitro (61). Though, in vivo studies testing the direct effects of PAC metabolites in the context of chronic inflammation are lacking. In terms of the mechanisms that may underlie anti-inflammatory effects of PAC and its metabolites, gene expression regulation has been consistently described. This includes genes regulating the NF-kB pathway and downstream cytokines (58, 59).
4. MicroRNAs and intestinal homeostasis
MiRNAs are a class of small (~22 nucleotides in length), non-coding RNAs that are highly conserved across species and regulate gene expression at the post-transcriptional level (62). Thus far, 2,693 miRNAs have been annotated in humans (miRBase release 22.1) (63) and it is estimated that over 60% of human protein-coding genes are targeted by miRNAs (64). Overall, the potential impact of miRNAs is markedly pronounced because one miRNA can have hundreds of targets, while multiple miRNAs can also share the same target. In the intestine, lack of mature miRNA forms results in impaired barrier function and an inflammatory phenotype (65, 66). More recently, evidence has emerged supporting a role for intestinal miRNA in host-microbiota crosstalk. We and others demonstrated that miRNA expression is microbiota-dependent in various intestinal regions (4, 6, 67). Intestinal miRNAs can also be found in the lumen (68, 69), where they may regulate bacterial growth and gene expression (69, 70). Luminal miRNAs can be ultimately recovered in the feces (fecal miRNAs). Intestinal miRNA signatures are increasingly being used to discern between states of health and disease. In IBD, miRNA are aberrantly expressed in tissue biopsies, body fluids and feces of patients with Crohn's disease (CD) and active ulcerative colitis (UC) in comparison to healthy controls [reviewed in James et al. (71)], including elevated miR-301a expression acting as a promotor of intestinal and mucosal inflammation (72, 73). Moreover, fecal miRNA signatures correlate with disease activity (74), and recent studies suggest that they may play a role in IBD pathology by shaping the intestinal microbiota [reviewed in Casado-Bedmar and Viennois (75)]. In fact, IBD-associated miR-199a-5p, miR-1226, miR-548ab, and miR-515–5p were found to affect the proliferation of intestinal E. coli, Fusobacterium nucleatum and segmented filamentous bacteria (76). MiRNAs also play an extensive role in the progression and proliferation of colorectal cancer, they are associated with resistance to chemoradiotherapy and are involved in the regulation of signaling pathways such as the Wnt/β-catenin and ERK-MAPK pathways [reviewed in Zhang et al. (77)]. MiRNA signatures in tumor tissue, cancer-derived exosomes, stool, and plasma have been explored as predictive indicators to identify the best treatment strategies for patients based on factors including predicted chemoresistance or metastatic properties of the cancer [reviewed in To et al. (78)]. More recently, a panel of circulating miRNAs has been identified as potential biomarkers of gastroenterological cancers [reviewed in Hoshino (79)], celiac disease [reviewed in Felli et al. (80)], and other human infectious diseases [reviewed in Ojha et al. (81)].
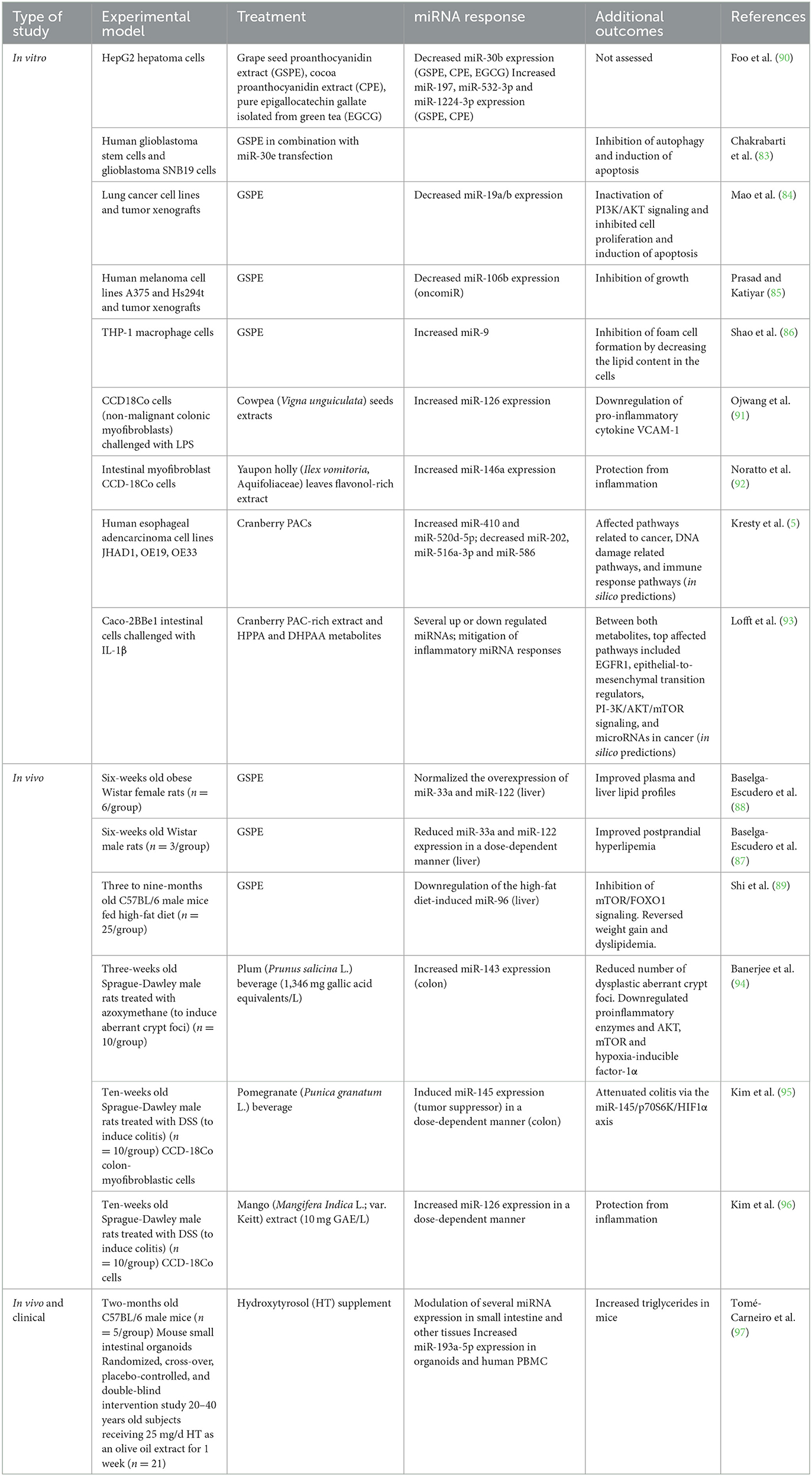
4.1. Dietary polyphenols and microRNA modulation
Dietary compounds modulate host miRNA expression in various organs including the intestine. The impact of dietary polyphenols on miRNA expression have been studied in different in vitro and in vivo models (Table 2). Grapeseed PAC, commonly referred to as grape seed proanthocyanidin extract (GSPE), has been studied more extensively compared to PAC derived from cranberries. There is emerging evidence that the biological effects of GSPE depend on miRNA modulation. In vitro, GSPE treatment resulted in the differential expression of miR-30b, miR-197, miR-532-3p, and miR-1224-3p in HepG2 hepatoma cells (82). In human glioblastoma stem cells, GSPE in combination with miR-30e transfection led to the induction of apoptosis (83). In lung cancer cell lines and tumor xenografts, GSPE decreased miR-19a/b expression, which inactivated PI3K/AKT signaling and inhibited cell proliferation and induced apoptosis (84). GSPE also inhibited growth of human melanoma cell lines by downregulating the oncomiR miR-106b (85). In THP-1 macrophage cells, GSPE inhibited foam cell formation by decreasing the lipid content in the cells and inducing miR-9, which downregulated the expression of ACAT1, encoding for an enzyme that catalyzes intracellular free cholesterol to cholesterol esters (86). In animal models, consumption of GSPE improved plasma and liver lipid profiles, and normalized the overexpression miR-33a and miR-122 in the liver of obese rats (87, 88). Similarly, in C57BL/6 mice fed high-fat diets, GSPE reversed weight gain and dyslipidemia in mice, which was thought to be attributed to the ability of GSPE to downregulate the high-fat diet-induced increase of miR-96 and inhibition of mTOR/FOXO1 signaling (89). Together, these studies suggest that GSPE may interact with specific host miRNAs or modulate their expression to exert a specific biological effect.
Kresty and colleagues were the first to evaluate the ability of cranberry PACs on miRNA expression profiles in esophageal adencarcinoma and its precursor, Barrett's esophagus tissue as well as EAC cell lines (5). Across the samples tested, five miRNAs (miR-410, miR-520d-5p, miR-202, miR-516a-3p and miR-586) were commonly aberrantly expressed, which were implicated in multiple biological processes, including pathways in cancer, DNA damage related pathways, and immune response pathways (5). Remarkably, PACs inversely modulated 10 miRNAs (let-7b, miR-106b, miR-143, miR-199a, miR-215, miR-223, miR-23b, miR-32, miR-543, and miR-7) dysregulated in esophageal adenocarcinoma or Barrett's esophagus tissues, showing that cranberry PAC may in part normalize the expression of miRNAs in esophageal cancer. However, the extent to which cranberry PAC mitigate the aberrant expression of miRNAs in gastrointestinal cancers and other intestinal inflammatory pathologies is not clear. We recently investigated intestinal miRNA signatures in Caco-2BBe1 intestinal cells -a homogeneous clone of Caco-2 cells with a morphology, stability and brush-border skeleton comparable to the human enterocytes (98)- pre-treated with PACs derived from cranberry, metabolites DHPAA, or HPPA and stimulated or not with IL-1β (93). At homeostasis, each treatment was associated with a distinctive miRNA signature, but the two metabolites exerted a partially shared miRNA response. More miRNAs were altered by the metabolites compared to PAC, proposing the notion that the microbial catabolism of PAC initiated by the gut microbiota could facilitate its miRNA-regulatory effects. Thus, the gut microbiota may affect intestinal miRNAs via its metabolites, and this closely echoes previous findings for microbiota-produced SCFAs (99, 100). The gene targets of miRNAs responding to the metabolites were significantly enriched in many pathways relating to cell growth and development and pathways in cancer (93). PAC-responsive miRNAs, miR-1260b and miR-542-5p, were previously found to be modulated in response to genistein (101, 102) and glyceollins (103), respectively. Metabolites-associated and shared miRNAs were miR-1260a, miR-130a-3p, miR-625-5p, miR-6721-5p and miR-20a. The latter was recently reported to be downregulated in response to resveratrol (104).
Interestingly, PAC and DHPAA reversed the expression of 80% and 15% of miRNAs deregulated by IL-1β, respectively (93), these miRNAs may be key mediators of the anti-inflammatory effects associated with cranberry polyphenols and microbial metabolites (105, 106).
A limited number of studies have investigated the effects of polyphenol metabolites on different biological outcomes and the role of miRNAs as putative mediators. In a study comparing the anti-inflammatory effects of quercetin with its two metabolites isorhamnetin and quercetin-3-glucuronide in vitro, pre-incubation of murine macrophage cells with quercetin or its metabolites quercetin-3-glucuronide and isorhamnetin, prior to LPS stimulation decreased mRNA and protein levels of TNF-α, decreased Il1b, Il6 gene expression, and macrophage inflammatory protein-1α, and thereby NF-κB activation in comparison to LPS stimulation alone. Interestingly, quercetin and its metabolite isorhamnetin reversed the LPS-induced upregulation of miR-155, a miRNA identified as a modulator of inflammatory responses (107). When analyzing the effect of trans-resveratrol and its metabolites, trans-resveratrol-3-O-sulfate, trans-resveratrol-3′-O-glucuronide, and trans-resveratrol-4′-O-glucuronide on the expression of adipogenic transcription factors and miRNAs in 3T3-L1 murine adipocytes, Eseberri et al. (108) found that of the nine miRNAs analyzed, only miR-155 responded to different treatments. Trans-resveratrol and its metabolites trans-resveratrol-3′-O-glucuronide, and trans-resveratrol-4′-O-glucuronide significantly upregulated the expression of miR-155, which led to a decrease of CCAAT enhancer-binding protein-ß gene expression (108). In both studies, the authors mentioned the hepatic and intestinal metabolism of quercetin or trans-resveratrol in general however there was no mention of the role of the gut microbiota. The microbiota and miRNAs may interact to control host gene expression [reviewed in Malmuthuge and Guan (109)], however, the interplay existing between gut microbiota and miRNAs in the presence of dietary polyphenols is still not completely elucidated.
4.2. Crosstalk between polyphenols, miRNAs and microbiota: a clinical perspective
An emerging body of evidence suggests the presence of a tripartite association between cranberry polyphenols, gut microbiota, and miRNAs (Figure 3). These interactions are likely to at least partially underlie the beneficial health effects of cranberries.
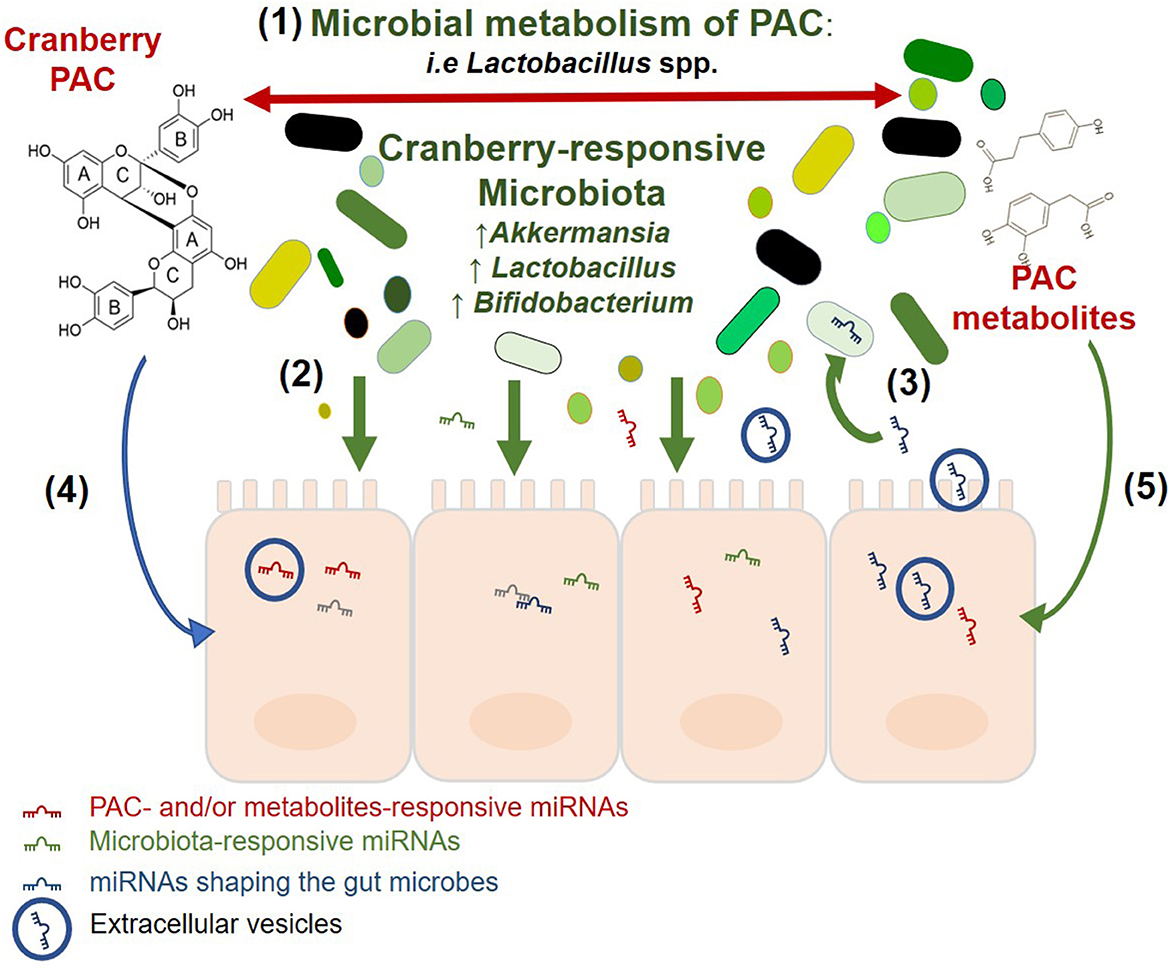
Figure 3. Potential association between cranberry PAC, gut microbiota, and miRNAs. Visualization of the relationships between PAC, microbiota and miRNA in the intestinal ecosystem, based on the available literature. The two-way interaction between cranberry and the gut microbiota as described in the text and reviewed by Prasain and Barnes (110). Gut Microbiota affect the host intestinal response and miRNA signature as previously reported by Dalmasso et al. (4), Singh et al. (6), Peck et al. (67). Intestinal miRNAs may enter resident bacteria, regulate their growth, and shape the gut microbiota [Liu et al. (69), Liu et al. (70)], and Cranberry PAC and their microbial metabolites alter the expression of intestinal miRNAs Lofft et al. (93).
4.2.1. Cranberry and gut microbiota
The relationship between cranberry polyphenols and gut microorganisms relies on the microbial metabolism of polyphenols into smaller molecules and bioactive components that can either be involved in trophic interactions or further metabolized by other microbial taxa that possess PAZymes. This cascade of microbial activities modifies polyphenol bioavailability and, as a consequence, the intestinal niche. Cranberry extracts and polyphenols were previously reported to significantly modify the abundance of beneficial bacteria such as Lactobacillus and Bifidobacterium as well as A. muciniphila (48–50, 52, 111). A. muciniphila has been suggested as a next-generation probiotic in the context of obesity (112) and diabetes (113, 114). This is aligned with the understanding that cranberry may also exert prebiotic effects, by conferring health beneficial effects to the host via the modulation of gut microbiota. Microbial taxa and their metabolites trigger host responses, including via gene expression regulation. These effects may be post-transcriptional and include miRNAs. In vitro, intestinal miRNAs are modulated by cranberry polyphenols and their microbial metabolites (93); intestinal miRNAs also respond to bacterial taxa such as Lactobacillus (115, 116) and Bifidobacterium (117), which were identified as cranberry-responsive bacteria (48, 118). In tandem, while the gut microbiota affects the expression of miRNA, luminal miRNAs have been shown to affect members of the microbiota (69). Interestingly, the mucin-degrading and cranberry-responsive A. muciniphila was found to be targeted by miR-30d, resulting in increased abundance of the bacterium (70). MiR-30d is a member of the miR-30 family which also includes miR-30b and miR-30e, previously reported to be significantly differently expressed in response to GSPE treatment (82, 83). Cranberry polyphenols and their microbial metabolites may mediate prebiotic effects by modulating intestinal miRNAs.
4.2.2. Cranberry and host responses
On the host side, along with microbiota-dependent modulation of the intestinal response, the polyphenolic compounds interact with cell membranes, changing their structure and function. For decades, polyphenols have been considered simple antioxidant molecules capable of interacting with host cells. However, this fact has been questioned (119) and different forms of interactions between host and polyphenols have been suggested (120). It is recognized that the function of polyphenolic compounds is attributed to their ability to interact with membrane surfaces through hydrophobic, electrostatic, or covalent binding [reviewed in Hendrich (121), Reis and De Freitas (122)]. These interactions with cell membranes may be the basis by which polyphenolic compounds exert their beneficial effects on the host (123–125)]. They may also depend on the nature and function of the proteins involved, enzymatic activities, binding sites, and the interaction between specific sites in DNA and transcription factors. Previous studies reported the impact of polyphenols on molecular signal transduction pathways such as cell migration/proliferation, metabolic disorders, oxidative stress as well as inflammation cascades (126). As described above, the beneficial effects of polyphenols also involve the regulation of cell signaling (127, 128), since they act as regulation factors at the transcriptional (gene expression) (58, 129), and posttranscriptional (microRNAs) levels that affect different processes such as cell growth and apoptosis (130). Many in vitro and in vivo studies support the capacity of polyphenols to modulate cell functionality through the modification of gene expression and protein levels (58, 129, 131) or by an epigenetic mechanism involving miRNAs (5). An emerging body of evidence indicates that miRNAs serve as mediators in regulating polyphenols′ beneficial effects [reviewed in Milenkovic (132), Corrêa et al. (133), Majidinia et al. (134)] and targeting miRNAs could be a novel strategy for inflammation and disease.
4.2.3. Clinical perspective
In clinical practice, the number of intervention studies of cranberry has increased in recent years. Many of these studies focused on the beneficial effects of cranberries as a whole food primarily on outcomes related to UTIs and H. pylori infections. Only few studies have explored the microbial response to cranberry intervention. The studies are inconsistent in terms of populations studied and the cranberry products administered, which were mainly cranberry juice or powders, the equivalent dose of polyphenols and the duration of intervention and detection, as well as biomarkers used, either for health outcomes or for microbial metabolism of the cranberry polyphenols. To the best of our knowledge, no study has explored the miRNA-mediated effect of cranberries in a clinical setting. The use of miRNAs may open new avenues in clinical studies. In particular, the study of miRNA signatures in response to cranberry and/or its components in various human samples, such as plasma, urine or feces, in health and disease, may help identifying new biomarkers of administration and response. For example, Seo and colleagues analyzed miRNA signatures in mice exposed to a high-fat and high-fructose diet (HFrD) containing or not 5% polyphenol-rich wine grape seed flour (GSF) (135). MiR-129-5p was identified as significantly affected by GSF in both blood and feces and strongly correlated with biomarkers of obesity such as body weight gain, liver weight, adipose tissue weight, triglyceride, total cholesterol, and HDL (135). The study identified for the first time a potential biomarker for monitoring grape seed flour efficacy.
With regard to cranberry, some miRNAs that were found to respond to PACs, have previously been proposed as fecal or serum biomarkers of gastrointestinal diseases. For example, miR-1260b and miR-542-5p were previously found to be upregulated in CRC and other cancers and suggested to be used as predictive markers (136–139). Interestingly, these miRNAs were found to be downregulated in response to PAC (93). Of the cranberry metabolites-responsive miRNAs in the intestine, miR-130a-3p, found to be upregulated in response to HPPA and DHPAA, is also identified as an anti-tumor miRNA (140). In a similar pattern, the HPPA- and DHPAA-induced miRNA, miR-625-5p, is known to serve as a tumor suppressor and its overexpression has been shown to inhibit the proliferation of gastric cancers (141). These findings suggest that selected miRNAs could be used as potential biomarkers to monitor cranberry intervention efficacy or as a target to mitigate or prevent disease.
Author contributions
AT, ZL, and EC conceived the review and drafted the manuscript. AT, ZL and BL-I conducted the literature search and constructed the tables. AT and BL-I created the figures. EC supervised this work. All authors edited, revised, read, and approved the final manuscript.
Funding
Research in the laboratory of EMC was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) (Grant # RGPIN-2019-06100) and the Joannah and Brian Lawson Center for Child Nutrition at the University of Toronto. EMC was the recipient of the Lawson Family Chair in Microbiome Nutrition Research. ZL was the recipient of an NSERC Canada Graduate Scholarship for Master's (CGSM).
Conflict of interest
EC has received research support from Ocean Spray Cranberries and Lallemand Health Solutions; and has received consultant fees or speaker and travel support from Danone and Lallemand Health Solutions.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Feghali K, Feldman M, La VD, Santos J, Grenier D. Cranberry proanthocyanidins: natural weapons against periodontal diseases. J Agric Food Chem. (2012) 60:5728–35. doi: 10.1021/jf203304v
2. Yahfoufi N, Alsadi N, Jambi M, Matar C. The immunomodulatory and anti-inflammatory role of polyphenols. Nutrients. (2018) 10. doi: 10.3390/nu10111618
3. Déprez S, Brezillon C, Rabot S, Philippe C, Mila I, Lapierre C, et al. Polymeric proanthocyanidins are catabolized by human colonic microflora into low-molecular-weight phenolic acids. J Nutr. (2000) 130:2733–8. doi: 10.1093/jn/130.11.2733
4. Dalmasso G, Nguyen HT, Yan Y, Laroui H, Charania MA, Ayyadurai S, et al. Microbiota modulate host gene expression via microRNAs. PLoS ONE. (2011) 6:e19293. doi: 10.1371/journal.pone.0019293
5. Kresty LA, Clarke J, Ezell K, Exum A, Howell AB, Guettouche T, et al. MicroRNA alterations in Barrett's esophagus, esophageal adenocarcinoma, and esophageal adenocarcinoma cell lines following cranberry extract treatment: Insights for chemoprevention. J Carcinog. (2011) 10:34. doi: 10.4103/1477-3163.91110
6. Singh N, Shirdel EA, Waldron L, Zhang RH, Jurisica I, Comelli EM, et al. The murine caecal microRNA signature depends on the presence of the endogenous microbiota. Int J Biol Sci. (2012) 8:171–86. doi: 10.7150/ijbs.8.171
7. Duarte S, Gregoire S, Singh AP, Vorsa N, Schaich K, Bowen WH, et al. Inhibitory effects of cranberry polyphenols on formation and acidogenicity of Streptococcus mutans biofilms. FEMS Microbiol Lett. (2006) 257:50–6. doi: 10.1111/j.1574-6968.2006.00147.x
8. Foo LY, Lu Y, Howell AB, Vorsa N. The structure of cranberry proanthocyanidins which inhibit adherence of uropathogenic P-fimbriated Escherichia coli in vitro. Phytochemistry. (2000) 54:173–81. doi: 10.1016/S0031-9422(99)00573-7
9. Neto CC, Krueger CG, Lamoureaux TL, Kondo M, Vaisberg AJ, Hurta RA, et al. MALDI-TOF MS characterization of proanthocyanidins from cranberry fruit (Vaccinium macrocarpon) that inhibit tumor cell growth and matrix metalloproteinase expression in vitro. J Sci Food Agric. (2006) 86:18–25. doi: 10.1002/jsfa.2347
10. Gu L, Kelm MA, Hammerstone JF, Beecher G, Holden J, Haytowitz D, et al. Screening of foods containing proanthocyanidins and their structural characterization using LC-MS/MS and thiolytic degradation. J Agric Food Chem. (2003) 51:7513–21. doi: 10.1021/jf034815d
11. Feliciano RP, Meudt JJ, Shanmuganayagam D, Krueger CG, Reed JD. Ratio of “A-type” to “B-type” proanthocyanidin interflavan bonds affects extra-intestinal pathogenic Escherichia coli invasion of gut epithelial cells. J Agric Food Chem. (2014) 62:3919–25. doi: 10.1021/jf403839a
12. Wang Y, Chung SJ, Song WO, Chun OK. Estimation of daily proanthocyanidin intake and major food sources in the U.S. diet. J Nutr. (2011) 141:447–2. doi: 10.3945/jn.110.133900
13. Vogiatzoglou A, Mulligan AA, Luben RN, Lentjes MA, Heiss C, Kelm M, et al. Assessment of the dietary intake of total flavan-3-ols, monomeric flavan-3-ols, proanthocyanidins and theaflavins in the European Union. Br J Nutr. (2014) 111:1463–73. doi: 10.1017/S0007114513003930
14. Blumberg JB, Camesano TA, Cassidy A, Kris-Etherton P, Howell A, Manach C, et al. Cranberries and their bioactive constituents in human health. Adv Nutr. (2013) 4:618–32. doi: 10.3945/an.113.004473
15. Howell AB, Dreyfus JF, Chughtai B. Differences in urinary bacterial anti-adhesion activity after intake of cranberry dietary supplements with soluble versus insoluble proanthocyanidins. J Diet Suppl. (2021) 1–18. doi: 10.1080/19390211.2021.1908480
16. Novotny JA, Baer DJ, Khoo C, Gebauer SK, Charron CS. Cranberry juice consumption lowers markers of cardiometabolic risk, including blood pressure and circulating C-reactive protein, triglyceride, and glucose concentrations in adults. J Nutr. (2015) 145:1185–93. doi: 10.3945/jn.114.203190
17. Chew B, Mathison B, Kimble L, Mckay D, Kaspar K, Khoo C, et al. Chronic consumption of a low calorie, high polyphenol cranberry beverage attenuates inflammation and improves glucoregulation and HDL cholesterol in healthy overweight humans: a randomized controlled trial. Eur J Nutr. (2018). doi: 10.1007/s00394-018-1643-z
18. Rodriguez-Mateos A, Feliciano RP, Boeres A, Weber T, Dos Santos CN, Ventura MR, et al. Cranberry (poly)phenol metabolites correlate with improvements in vascular function: a double-blind, randomized, controlled, dose-response, crossover study. Mol Nutr Food Res. (2016) 60:2130–40. doi: 10.1002/mnfr.201600250
19. Pritchard SE, Marciani L, Garsed KC, Hoad CL, Thongborisute W, Roberts E, et al. Fasting and postprandial volumes of the undisturbed colon: normal values and changes in diarrhea-predominant irritable bowel syndrome measured using serial MRI. Neurogastroenterol Motil. (2014) 26:124–30. doi: 10.1111/nmo.12243
20. Sánchez-Patán F, Barroso E, Van De Wiele T, Jiménez-Girón A, Martín-Alvarez PJ, Moreno-Arribas MV, et al. Comparative in vitro fermentations of cranberry and grape seed polyphenols with colonic microbiota. Food Chem. (2015) 183:273–82. doi: 10.1016/j.foodchem.2015.03.061
21. Zampariello CA, Mckay DL, Dolnikowski GG, Blumberg JB, Chen CYO. Determination of Cranberry Proanthocyanidin A2 in Human Plasma and Urine Using LC-MS/MS. (2012) 124–8. doi: 10.1096/fasebj.26.1_supplement.124.8
22. Mckay DL, Chen CY, Zampariello CA, Blumberg JB. Flavonoids and phenolic acids from cranberry juice are bioavailable and bioactive in healthy older adults. Food Chem. (2015) 168:233–40. doi: 10.1016/j.foodchem.2014.07.062
23. Feliciano RP, Mills CE, Istas G, Heiss C, Rodriguez-Mateos A. Absorption, metabolism and excretion of cranberry (poly)phenols in humans: A dose response study and assessment of inter-individual variability. Nutrients. (2017) 9. doi: 10.3390/nu9030268
24. Ou K, Percival SS, Zou T, Khoo C, Gu L. Transport of cranberry A-type procyanidin dimers, trimers, and tetramers across monolayers of human intestinal epithelial Caco-2 cells. J Agric Food Chem. (2012) 60:1390–6. doi: 10.1021/jf2040912
25. Raab T, Barron D, Vera FA, Crespy V, Oliveira M, Williamson G, et al. Catechin glucosides: occurrence, synthesis, and stability. J Agric Food Chem. (2010) 58:2138–49. doi: 10.1021/jf9034095
26. Wu T, Grootaert C, Voorspoels S, Jacobs G, Pitart J, Kamiloglu S, et al. Aronia (Aronia melanocarpa) phenolics bioavailability in a combined in vitro digestion/Caco-2 cell model is structure and colon region dependent. J Funct Foods. (2017) 38:128–39. doi: 10.1016/j.jff.2017.09.008
27. Cortés-Martín A, Selma MV, Espín JC, García-Villalba R. The human metabolism of nuts proanthocyanidins does not reveal urinary metabolites consistent with distinctive gut microbiota metabotypes. Mol Nutr Food Res. (2019) 63:e1800819. doi: 10.1002/mnfr.201800819
28. Pereira-Caro G, Gaillet S, Ordóñez JL, Mena P, Bresciani L, Bindon KA, et al. Bioavailability of red wine and grape seed proanthocyanidins in rats. Food Funct. (2020) 11:3986–4001. doi: 10.1039/D0FO00350F
29. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. (2010) 464:59–65. doi: 10.1038/nature08821
30. Monagas M, Urpi-Sarda M, Sánchez-Patán F, Llorach R, Garrido I, Gómez-Cordovés C, et al. Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food Funct. (2010) 1:233–53. doi: 10.1039/c0fo00132e
31. Ozdal T, Sela DA, Xiao J, Boyacioglu D, Chen F, Capanoglu E, et al. The Reciprocal interactions between polyphenols and gut microbiota and effects on bioaccessibility. Nutrients. (2016) 8:78. doi: 10.3390/nu8020078
32. Rupasinghe HPV, Parmar I, Neir SV. Biotransformation of cranberry proanthocyanidins to probiotic metabolites by Lactobacillus rhamnosus enhances their anticancer activity in HepG2 cells in vitro. Oxid Med Cell Longev. (2019) 2019:4750795. doi: 10.1155/2019/4750795
33. Rodríguez-Daza MC, Pulido-Mateos EC, Lupien-Meilleur J, Guyonnet D, Desjardins Y, Roy D, et al. Polyphenol-mediated gut microbiota modulation: toward prebiotics and further. Frontiers in Nutrition. (2021) 8:9456. doi: 10.3389/fnut.2021.689456
34. Lee CC, Kim JH, Kim JS, Oh YS, Han SM, Park JHY, et al. 5.-(3′,4′-Dihydroxyphenyl-γ-valerolactone), a major microbial metabolite of proanthocyanidin, attenuates THP-1 monocyte-endothelial adhesion. Int J Mol Sci. (2017) 18:1363. doi: 10.3390/ijms18071363
35. Pasinetti GM, Singh R, Westfall S, Herman F, Faith J, Ho L, et al. The Role of the gut microbiota in the metabolism of polyphenols as characterized by gnotobiotic mice. J Alzheimers Dis. (2018) 63:409–21. doi: 10.3233/JAD-171151
36. Corrêa TA, Rogero MM, Hassimotto NM, Lajolo FM. The two-way polyphenols-microbiota interactions and their effects on obesity and related metabolic diseases. Front Nutri. (2019) 6:188. doi: 10.3389/fnut.2019.00188
37. Kutschera M, Engst W, Blaut M, Braune A. Isolation of catechin-converting human intestinal bacteria. J Appl Microbiol. (2011) 111:165–75. doi: 10.1111/j.1365-2672.2011.05025.x
38. Wang LQ, Meselhy MR, Li Y, Nakamura N, Min BS, Qin GW, et al. The heterocyclic ring fission and dehydroxylation of catechins and related compounds by Eubacterium sp. strain SDG-2. A human intestinal bacterium. Chem Pharm Bull. (2001) 49:1640–3. doi: 10.1248/cpb.49.1640
39. Sánchez-Patán F, Tabasco R, Monagas M, Requena T, Peláez C, Moreno-Arribas MV, et al. Capability of Lactobacillus plantarum IFPL935 to catabolize Flavan-3-ol compounds and complex phenolic extracts. J Agric Food Chem. (2012) 60:7142–51. doi: 10.1021/jf3006867
40. Jin JS, Hattori M. Isolation and characterization of a human intestinal bacterium Eggerthella sp. CAT-1 capable of cleaving the C-ring of (+)-catechin and (-)-epicatechin followed by p-dehydroxylation of the B-ring. Biol Pharm Bull. (2012) 35:2252–6. doi: 10.1248/bpb.b12-00726
41. Li S, Chen L, Yang T, Wu Q, Lv Z, Xie B, et al. Increasing antioxidant activity of procyanidin extracts from the pericarp of Litchi chinensis processing waste by two probiotic bacteria bioconversions. J Agric Food Chem. (2013) 61:2506–12. doi: 10.1021/jf305213e
42. Torres YTJL, Rosazza JP. Microbial transformations of p-coumaric acid by Bacillus megaterium and Curvularia lunata. J Nat Prod. (2001) 64:1408–14. doi: 10.1021/np010238g
43. Ou K, Sarnoski P, Schneider KR, Song K, Khoo C, Gu L, et al. Microbial catabolism of procyanidins by human gut microbiota. Mol Nutr Food Res. (2014) 58:2196–205. doi: 10.1002/mnfr.201400243
44. Engemann A, Hübner F, Rzeppa S, Humpf HU. Intestinal metabolism of two A-type procyanidins using the pig cecum model: detailed structure elucidation of unknown catabolites with Fourier transform mass spectrometry (FTMS). J Agric Food Chem. (2012) 60:749–57. doi: 10.1021/jf203927g
45. Manach C, Williamson G, Morand C, Scalbert A, Rémésy C. Bioavailability and bioefficacy of polyphenols in humans. I Review of 97 bioavailability studies. Am J Clin Nutr 81. (2005) 230S−42S. doi: 10.1093/ajcn/81.1.230S
46. Cortés-Martín A, Selma MV, Tomás-Barberán FA, González-Sarrías A, Espín JC. Where to look into the puzzle of polyphenols and health? The postbiotics and gut microbiota associated with human metabotypes. Mol Nutr Food Res. (2020) 64:e1900952. doi: 10.1002/mnfr.201900952
47. Han Y, Xiao H. Whole food-based approaches to modulating gut microbiota and associated diseases. Annu Rev Food Sci Technol. (2020) 11:119–43. doi: 10.1146/annurev-food-111519-014337
48. Cai X, Han Y, Gu M, Song M, Wu X, Li Z, et al. Dietary cranberry suppressed colonic inflammation and alleviated gut microbiota dysbiosis in dextran sodium sulfate-treated mice. Food Funct. (2019) 10:6331–41. doi: 10.1039/C9FO01537J
49. Anhê FF, Roy D, Pilon G, Dudonné S, Matamoros S, Varin TV, et al. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut. (2015) 64:872–83. doi: 10.1136/gutjnl-2014-307142
50. Anhê FF, Nachbar RT, Varin TV, Vilela V, Dudonné S, Pilon G, et al. A polyphenol-rich cranberry extract reverses insulin resistance and hepatic steatosis independently of body weight loss. Mol Metab. (2017) 6:1563–73. doi: 10.1016/j.molmet.2017.10.003
51. Neto CC, Mortzfeld BM, Turbitt JR, Bhattarai SK, Yeliseyev V, Dibenedetto N, et al. Proanthocyanidin-enriched cranberry extract induces resilient bacterial community dynamics in a gnotobiotic mouse model. Microb Cell. (2021) 8:131–42. doi: 10.15698/mic2021.06.752
52. Liu J, Hao W, He Z, Kwek E, Zhu H, Ma N, et al. Blueberry and cranberry anthocyanin extracts reduce bodyweight and modulate gut microbiota in C57BL/6 J mice fed with a high-fat diet. Eur J Nutr. (2021) 60:2735–46. doi: 10.1007/s00394-020-02446-3
53. Medina-Larqué AS, Rodríguez-Daza MC, Roquim M, Dudonné S, Pilon G, Levy É, et al. Cranberry polyphenols and agave agavins impact gut immune response and microbiota composition while improving gut barrier function, inflammation, and glucose metabolism in mice fed an obesogenic diet. Front Immunol. (2022) 13:871080. doi: 10.3389/fimmu.2022.871080
54. Straub TJ, Chou WC, Manson AL, Schreiber HL, Walker BJ, Desjardins CA, et al. Limited effects of long-term daily cranberry consumption on the gut microbiome in a placebo-controlled study of women with recurrent urinary tract infections. BMC Microbiol. (2021) 21:53. doi: 10.1186/s12866-021-02106-4
55. Solch-Ottaiano RJ, Judkins TC, Matott SH, Mcdermott CE, Nieves C, Wang Y, et al. High polyphenolic cranberry beverage alters specific fecal microbiota but not gut permeability following aspirin challenge in healthy obese adults: a randomized, double-blind, crossover trial. J Funct Foods. (2022) 99:105332. doi: 10.1016/j.jff.2022.105332
56. Monk JM, Lepp D, Zhang CP, Wu W, Zarepoor L, Lu JT, et al. Diets enriched with cranberry beans alter the microbiota and mitigate colitis severity and associated inflammation. J Nutr Biochem. (2016) 28:129–39. doi: 10.1016/j.jnutbio.2015.10.014
57. Rodríguez-Daza MC, Roquim M, Dudonné S, Pilon G, Levy E, Marette A, et al. Berry polyphenols and fibers modulate distinct microbial metabolic functions and gut microbiota enterotype-like clustering in obese mice. Front Microbiol. (2020) 11:2032. doi: 10.3389/fmicb.2020.02032
58. Hannon DB, Thompson JT, Khoo C, Juturu V, Vanden Heuvel JP. Effects of cranberry extracts on gene expression in THP-1 cells. Food Sci Nutr. (2017) 5:148–59. doi: 10.1002/fsn3.374
59. Mithul Aravind S, Wichienchot S, Tsao R, Ramakrishnan S, Chakkaravarthi S. Role of dietary polyphenols on gut microbiota, their metabolites and health benefits. Food Res Int. (2021) 142:110189. doi: 10.1016/j.foodres.2021.110189
60. Larrosa M, Luceri C, Vivoli E, Pagliuca C, Lodovici M, Moneti G, et al. Polyphenol metabolites from colonic microbiota exert anti-inflammatory activity on different inflammation models. Mol Nutr Food Res. (2009) 53:1044–54. doi: 10.1002/mnfr.200800446
61. Wong X, Carrasco-Pozo C, Escobar E, Navarrete P, Blachier F, Andriamihaja M, et al. Deleterious effect of p-cresol on human colonic epithelial cells prevented by proanthocyanidin-containing polyphenol extracts from fruits and proanthocyanidin bacterial metabolites. J Agric Food Chem. (2016) 64:3574–83. doi: 10.1021/acs.jafc.6b00656
62. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
63. Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. (2019) 47:D155–62. doi: 10.1093/nar/gky1141
64. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. (2009) 19:92–105. doi: 10.1101/gr.082701.108
65. Mckenna LB, Schug J, Vourekas A, Mckenna JB, Bramswig NC, Friedman JR, et al. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. (2010) 139:1654–64. doi: 10.1053/j.gastro.2010.07.040
66. Biton M, Levin A, Slyper M, Alkalay I, Horwitz E, Mor H, et al. Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat Immunol. (2011) 12:239–46. doi: 10.1038/ni.1994
67. Peck BCE, Mah AT, Pitman WA, Ding S, Lund PK, Sethupathy P, et al. Functional transcriptomics in diverse intestinal epithelial cell types reveals robust microRNA sensitivity in intestinal stem cells to microbial status. J Biol Chem. (2017) 292:2586–600. doi: 10.1074/jbc.M116.770099
68. Koga Y, Yasunaga M, Takahashi A, Kuroda J, Moriya Y, Akasu T, et al. MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening. Cancer Prev Res. (2010) 3:1435–42. doi: 10.1158/1940-6207.CAPR-10-0036
69. Liu S, Cunha Da, Rezende AP, Cialic RM, Wei R., Bry Z. R., et al. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe. (2016) 19:32–43. doi: 10.1016/j.chom.2015.12.005
70. Liu S, Rezende RM, Moreira TG, Tankou SK, Cox LM, Wu M, et al. Oral administration of miR-30d from feces of MS patients suppresses MS-like symptoms in mice by expanding Akkermansia muciniphila. Cell Host Microbe. (2019) 26:779–94. doi: 10.1016/j.chom.2019.10.008
71. James JP, Riis LB, Malham M, Høgdall E, Langholz E, Nielsen BS, et al. MicroRNA biomarkers in IBD—Differential diagnosis and prediction of colitis-associated cancer. Int J Mol Sci. (2020) 21:7893. doi: 10.3390/ijms21217893
72. He C, Shi Y, Wu R, Sun M, Fang L, Wu W, et al. miR-301a promotes intestinal mucosal inflammation through induction of IL-17A and TNF-α in IBD. Gut. (2016) 65:1938–50. doi: 10.1136/gutjnl-2015-309389
73. He C, Yu T, Shi Y, Ma C, Yang W, Fang L, et al. MicroRNA 301A promotes intestinal inflammation and colitis-associated cancer development by inhibiting BTG1. Gastroenterology. (2017) 152:1434–48. doi: 10.1053/j.gastro.2017.01.049
74. Schönauen K, Le N, Von Arnim U, Schulz C, Malfertheiner P, Link A, et al. Circulating and fecal microRNAs as biomarkers for inflammatory bowel diseases. Inflamm Bowel Dis. (2018) 24:1547–57. doi: 10.1093/ibd/izy046
75. Casado-Bedmar M, Viennois E. MicroRNA and gut microbiota: tiny but mighty—novel insights into their cross-talk in inflammatory bowel disease pathogenesis and therapeutics. J Crohn's Colitis. (2021). doi: 10.1093/ecco-jcc/jjab223
76. Ji Y, Li X, Zhu Y, Li N, Zhang N, Niu M, et al. Faecal microRNA as a biomarker of the activity and prognosis of inflammatory bowel diseases. Biochem Biophys Res Commun. (2018) 503:2443–50. doi: 10.1016/j.bbrc.2018.06.174
77. Zhang N, Hu X, Du Y, Du J. The role of miRNAs in colorectal cancer progression and chemoradiotherapy. Biomedicine and Pharmacotherapy. (2021) 134:111099. doi: 10.1016/j.biopha.2020.111099
78. To KK, Tong CW, Wu M, Cho WC. MicroRNAs in the prognosis and therapy of colorectal cancer: from bench to bedside. World J Gastroenterol. (2018) 24:2949–73. doi: 10.3748/wjg.v24.i27.2949
79. Hoshino I. The usefulness of microRNA in urine and saliva as a biomarker of gastroenterological cancer. Int J Clin Oncol. (2021) 26:1431–40. doi: 10.1007/s10147-021-01911-1
80. Felli C, Baldassarre A, Uva P, Alisi A, Cangelosi D, Ancinelli M, et al. Circulating microRNAs as novel non-invasive biomarkers of paediatric celiac disease and adherence to gluten-free diet. eBioMedicine. (2022) 76:103851. doi: 10.1016/j.ebiom.2022.103851
81. Ojha R, Nandani R, Pandey RK, Mishra A, Prajapati VK. Emerging role of circulating microRNA in the diagnosis of human infectious diseases. J Cell Physiol. (2019) 234:1030–43. doi: 10.1002/jcp.27127
82. Arola-Arnal A, Bladé C. Proanthocyanidins modulate microRNA expression in human HepG2 cells. PLoS One. (2011) 6:e25982. doi: 10.1371/journal.pone.0025982
83. Chakrabarti M, Klionsky DJ, Ray SK. miR-30e Blocks autophagy and acts synergistically with proanthocyanidin for inhibition of AVEN and BIRC6 to increase apoptosis in glioblastoma stem cells and glioblastoma SNB19 cells. PLoS ONE. (2016) 11:e0158537. doi: 10.1371/journal.pone.0158537
84. Mao JT, Xue B, Smoake J, Lu QY, Park H, Henning SM, et al. MicroRNA-19a/b mediates grape seed procyanidin extract-induced anti-neoplastic effects against lung cancer. J Nutr Biochem. (2016) 34:118–25. doi: 10.1016/j.jnutbio.2016.05.003
85. Prasad R, Katiyar SK. Down-regulation of miRNA-106b inhibits growth of melanoma cells by promoting G1-phase cell cycle arrest and reactivation of p21/WAF1/Cip1 protein. Oncotarget. (2014) 5:10636–49. doi: 10.18632/oncotarget.2527
86. Shao D, Di Y, Lian Z, Zhu B, Xu X, Guo D, et al. Grape seed proanthocyanidins suppressed macrophage foam cell formation by miRNA-9 via targeting ACAT1 in THP-1 cells. Food Funct. (2020) 11:1258–69. doi: 10.1039/C9FO02352F
87. Baselga-Escudero L, Blade C, Ribas-Latre A, Casanova E, Salvadó MJ, Arola L, et al. Chronic supplementation of proanthocyanidins reduces postprandial lipemia and liver miR-33a and miR-122 levels in a dose-dependent manner in healthy rats. J Nutr Biochem. (2014) 25:151–6. doi: 10.1016/j.jnutbio.2013.09.014
88. Baselga-Escudero L, Pascual-Serrano A, Ribas-Latre A, Casanova E, Salvadó MJ, Arola L, et al. Long-term supplementation with a low dose of proanthocyanidins normalized liver miR-33a and miR-122 levels in high-fat diet-induced obese rats. Nutr Res. (2015) 35:337–45. doi: 10.1016/j.nutres.2015.02.008
89. Shi Y, Jia M, Xu L, Fang Z, Wu W, Zhang Q, et al. miR-96 and autophagy are involved in the beneficial effect of grape seed proanthocyanidins against high-fat-diet-induced dyslipidemia in mice. Phytother Res. (2019) 33:1222–32. doi: 10.1002/ptr.6318
90. Foo LY, Lu Y, Howell AB, Vorsa N. A-Type proanthocyanidin trimers from cranberry that inhibit adherence of uropathogenic P-fimbriated Escherichia coli. J Nat Prod. (2000) 63:1225–8. doi: 10.1021/np000128u
91. Ojwang LO, Banerjee N, Noratto GD, Angel-Morales G, Hachibamba T, Awika JM, et al. Polyphenolic extracts from cowpea (Vigna unguiculata) protect colonic myofibroblasts (CCD18Co cells) from lipopolysaccharide (LPS)-induced inflammation–modulation of microRNA 126. Food Funct. (2015) 6:146–54. doi: 10.1039/C4FO00459K
92. Noratto GD, Kim Y, Talcott ST, Mertens-Talcott SU. Flavonol-rich fractions of yaupon holly leaves (Ilex vomitoria, Aquifoliaceae) induce microRNA-146a and have anti-inflammatory and chemopreventive effects in intestinal myofibroblast CCD-18Co cells. Fitoterapia. (2011) 82:557–69. doi: 10.1016/j.fitote.2011.01.013
93. Lofft Z., Taibi A., Massara P., Tokar T., Paetau-Robinson I., Khoo C., et al. (2022). Cranberry proanthocyanidin and its microbial metabolite 3, 4.-Dihydroxyphenylacetic Acid, but Not 3-(4-Hydroxyphenyl)-Propionic Acid, partially reverse pro-inflammatory microRNA responses in human intestinal epithelial cells. Mol Nutr Food Res. 3:e2100853. doi: 10.1002/mnfr.202100853
94. Banerjee N, Kim H, Talcott ST, Turner ND, Byrne DH, Mertens-Talcott SU, et al. Plum polyphenols inhibit colorectal aberrant crypt foci formation in rats: potential role of the miR-143/protein kinase B/mammalian target of rapamycin axis. Nutr Res. (2016) 36:1105–13. doi: 10.1016/j.nutres.2016.06.008
95. Kim H, Banerjee N, Sirven MA, Minamoto Y, Markel ME, Suchodolski JS, et al. Pomegranate polyphenolics reduce inflammation and ulceration in intestinal colitis-involvement of the miR-145/p70S6K1/HIF1α axis in vivo and in vitro. J Nutr Biochem. (2017) 43:107–15. doi: 10.1016/j.jnutbio.2017.02.005
96. Kim H, Banerjee N, Barnes RC, Pfent CM, Talcott ST, Dashwood RH, et al. Mango polyphenolics reduce inflammation in intestinal colitis-involvement of the miR-126/PI3K/AKT/mTOR axis in vitro and in vivo. Mol Carcinog. (2017) 56:197–207. doi: 10.1002/mc.22484
97. Tomé-Carneiro J, Crespo MC, Iglesias-Gutierrez E, Martín R, Gil-Zamorano J, Tomas-Zapico C, et al. Hydroxytyrosol supplementation modulates the expression of miRNAs in rodents and in humans. J Nutr Biochem. (2016) 34:146–55. doi: 10.1016/j.jnutbio.2016.05.009
98. Peterson MD, Mooseker MS. Characterization of the enterocyte-like brush border cytoskeleton of the C2BBe clones of the human intestinal cell line, Caco-2. J Cell Sci. (1992) 102:581–600. doi: 10.1242/jcs.102.3.581
99. Hu S, Dong TS, Dalal SR, Wu F, Bissonnette M, Kwon JH, et al. The microbe-derived short chain fatty acid butyrate targets miRNA-dependent p21 gene expression in human colon cancer. PLoS ONE. (2011) 6:e16221. doi: 10.1371/journal.pone.0016221
100. Hu S, Liu L, Chang EB, Wang JY, Raufman JP. Butyrate inhibits pro-proliferative miR-92a by diminishing c-Myc-induced miR-17-92a cluster transcription in human colon cancer cells. Mol Cancer. (2015) 14:180. doi: 10.1186/s12943-015-0450-x
101. Hirata H, Ueno K, Nakajima K, Tabatabai ZL, Hinoda Y, Ishii N, et al. Genistein downregulates onco-miR-1260b and inhibits Wnt-signalling in renal cancer cells. Br J Cancer. (2013) 108:2070–8. doi: 10.1038/bjc.2013.173
102. Hirata H, Hinoda Y, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, et al. Genistein downregulates onco-miR-1260b and upregulates sFRP1 and Smad4 via demethylation and histone modification in prostate cancer cells. Br J Cancer. (2014) 110:1645–54. doi: 10.1038/bjc.2014.48
103. Rhodes LV, Tilghman SL, Boue SM, Wang S, Khalili H, Muir SE, et al. Glyceollins as novel targeted therapeutic for the treatment of triple-negative breast cancer. Oncol Lett. (2012) 3:163–71. doi: 10.3892/ol.2011.460
104. Ohishi T, Hayakawa S, Miyoshi N. Involvement of microRNA modifications in anticancer effects of major polyphenols from green tea, coffee, wine, and curry. Crit Rev Food Sci Nutr. (2022) 1–32. doi: 10.1080/10408398.2022.2038540
105. Li S, Liu Z, Fang XD, Wang XY, Fei BY. MicroRNA (miR)-597-5p inhibits colon cancer cell migration and invasion by targeting FOS-like antigen 2. Front Oncol. (2019) 9:495. doi: 10.3389/fonc.2019.00495
106. Liu C, Yang J, Wu H, Li J. Downregulated miR-585-3p promotes cell growth and proliferation in colon cancer by upregulating PSME3. Onco Targets Ther. (2019) 12:6525–34. doi: 10.2147/OTT.S203175
107. Boesch-Saadatmandi C, Loboda A, Wagner AE, Stachurska A, Jozkowicz A, Dulak J, et al. Effect of quercetin and its metabolites isorhamnetin and quercetin-3-glucuronide on inflammatory gene expression: role of miR-155. J Nutr Biochem. (2011) 22:293–9. doi: 10.1016/j.jnutbio.2010.02.008
108. Eseberri I, Lasa A, Miranda J, Gracia A, Portillo MP. Potential miRNA involvement in the anti-adipogenic effect of resveratrol and its metabolites. PLoS ONE. (2017) 12:e0184875. doi: 10.1371/journal.pone.0184875
109. Malmuthuge N, Guan LL. Non-coding RNAs: regulatory molecules of host–microbiome crosstalk. Trends Microbiol. (2021) 29:713–24. doi: 10.1016/j.tim.2020.12.003
110. Prasain JK, Barnes S. Cranberry polyphenols-gut microbiota interactions and potential health benefits: an updated review. Food Frontiers. (2020) 1:459–64. doi: 10.1002/fft2.56
111. Roopchand DE, Carmody RN, Kuhn P, Moskal K, Rojas-Silva P, Turnbaugh PJ, et al. Dietary polyphenols promote growth of the gut bacterium Akkermansia muciniphila and attenuate high-fat diet-induced metabolic syndrome. Diabetes. (2015) 64:2847–58. doi: 10.2337/db14-1916
112. Abuqwider JN, Mauriello G, Altamimi M. Akkermansia muciniphila, a new generation of beneficial microbiota in modulating obesity: a systematic review. Microorganisms. (2021) 9:1098. doi: 10.3390/microorganisms9051098
113. Shih CT, Yeh YT, Lin CC, Yang LY, Chiang CP. Akkermansia muciniphila is negatively correlated with hemoglobin A1c in refractory diabetes. Microorganisms. (2020) 8:1360. doi: 10.3390/microorganisms8091360
114. Zhang J, Ni Y, Qian L, Fang Q, Zheng T, Zhang M, et al. Decreased abundance of Akkermansia muciniphila leads to the impairment of insulin secretion and glucose homeostasis in lean type 2 diabetes. Adv Sci. (2021) 8:2100536. doi: 10.1002/advs.202100536
115. Wang Q, Sun Q, Wang J, Qiu X, Qi R, Huang J, et al. Identification of differentially expressed miRNAs after Lactobacillus reuteri treatment in the ileum mucosa of piglets. Genes Genomics. (2020) 42:1327–38. doi: 10.1007/s13258-020-00998-6
116. Wang Q, Sun Q, Wang J, Qiu X, Qi R, Huang J, et al. Lactobacillus plantarum 299v changes miRNA expression in the intestines of piglets and leads to downregulation of LITAF by regulating ssc-miR-450a. Probiot Antimicrob Prot. (2021) 13:1093–105. doi: 10.1007/s12602-021-09743-1
117. Taibi A, Singh N, Chen J, Arioli S, Guglielmetti S, Comelli EM, et al. Time- and strain-specific downregulation of intestinal EPAS1 via miR-148a by Bifidobacterium bifidum. Mol Nutri Food Res. (2017) 61:1600596. doi: 10.1002/mnfr.201600596
118. Özcan E, Rozycki MR, Sela DA. Cranberry proanthocyanidins and dietary oligosaccharides synergistically modulate Lactobacillus plantarum physiology. Microorganisms. (2021) 9. doi: 10.3390/microorganisms9030656
119. Hollman PCH, Cassidy A, Comte B, Heinonen M, Richelle M, Richling E, et al. The biological relevance of direct antioxidant effects of polyphenols for cardiovascular health in humans is not established. J Nutr. (2011) 141:989. doi: 10.3945/jn.110.131490
120. Fraga CG, Galleano M, Verstraeten SV, Oteiza PI. Basic biochemical mechanisms behind the health benefits of polyphenols. Mol Aspects Med. (2010) 31:435–45. doi: 10.1016/j.mam.2010.09.006
121. Hendrich AB. Flavonoid-membrane interactions: possible consequences for biological effects of some polyphenolic compounds. Acta Pharmacol Sin. (2006) 27:27–40. doi: 10.1111/j.1745-7254.2006.00238.x
122. Reis A, De Freitas V. When polyphenols meet lipids: Challenges in membrane biophysics and opportunities in epithelial lipidomics. Food Chem. (2020) 333:127509. doi: 10.1016/j.foodchem.2020.127509
123. Tarahovsky YS. Plant polyphenols in cell-cell interaction and communication. Plant Signal Behav. (2008) 3:609–11. doi: 10.4161/psb.3.8.6359
124. Brittes J, Lúcio M, Nunes C, Lima JLFC, Reis S. Effects of resveratrol on membrane biophysical properties: relevance for its pharmacological effects. Chem Phys Lipids. (2010) 163:747–54. doi: 10.1016/j.chemphyslip.2010.07.004
125. Tsuchiya H. Structure-dependent membrane interaction of flavonoids associated with their bioactivity. Food Chem. (2010) 120:1089–96. doi: 10.1016/j.foodchem.2009.11.057
126. Upadhyay S, Dixit M. Role of polyphenols and other phytochemicals on molecular signaling. Oxid Med Cell Longev. (2015) 2015:504253. doi: 10.1155/2015/504253
127. Singh AP, Lange TS, Kim KK, Brard L, Horan T, Moore RG, et al. Purified cranberry proanthocyanidines (PAC-1A) cause pro-apoptotic signaling, ROS generation, cyclophosphamide retention and cytotoxicity in high-risk neuroblastoma cells. Int J Oncol. (2012) 40:99–108.
128. Shi L, Zhang X, Liu X, Jiang Y, Deng Y, Liu J, et al. Cranberry (Vacinium macrocarpon) phytochemicals inhibit hepatic stellate cell activation and liver fibrosis. Food Bioscience. (2021) 42:101176. doi: 10.1016/j.fbio.2021.101176
129. Kresty LA, Howell AB, Baird M. Cranberry proanthocyanidins mediate growth arrest of lung cancer cells through modulation of gene expression and rapid induction of apoptosis. Molecules. (2011) 16:2375–90. doi: 10.3390/molecules16032375
130. Lombardo Bedran TB, Morin MP, Palomari Spolidorio D, Grenier D. Black tea extract and its theaflavin derivatives inhibit the growth of periodontopathogens and modulate Interleukin-8 and β-Defensin secretion in oral epithelial cells. PLoS ONE. (2015) 10:e0143158. doi: 10.1371/journal.pone.0143158
131. Tipton DA, Christian J, Blumer A. Effects of cranberry components on IL-1β-stimulated production of IL-6, IL-8 and VEGF by human TMJ synovial fibroblasts. Arch Oral Biol. (2016) 68:88–96. doi: 10.1016/j.archoralbio.2016.04.005
132. Milenkovic D, Jude B, Morand C. miRNA as molecular target of polyphenols underlying their biological effects. Free Rad Biol Med. (2013) 64:40–51. doi: 10.1016/j.freeradbiomed.2013.05.046
133. Corrêa, Ta F, Rogero MM. Polyphenols regulating microRNAs and inflammation biomarkers in obesity. Nutrition. (2019) 59:150–7. doi: 10.1016/j.nut.2018.08.010
134. Majidinia M, Karimian A, Alemi F, Yousefi B, Safa A. Targeting miRNAs by polyphenols: novel therapeutic strategy for aging. Biochem Pharmacol. (2020) 173:113688. doi: 10.1016/j.bcp.2019.113688
135. Seo KH, Yokoyama W, Kim H. Comparison of polyphenol-rich wine grape seed flour-regulated fecal and blood microRNAs in high-fat, high-fructose diet-induced obese mice. J Funct Foods. (2020) 73:104147. doi: 10.1016/j.jff.2020.104147
136. Liu DR, Guan QL, Gao MT, Jiang L, Kang HX. miR-1260b is a potential prognostic biomarker in colorectal cancer. Med Sci Monit. (2016) 22:2417–23. doi: 10.12659/MSM.898733
137. Zhao J, Cao J, Zhou L, Du Y, Zhang X, Yang B, et al. MiR-1260b inhibitor enhances the chemosensitivity of colorectal cancer cells to fluorouracil by targeting PDCD4/IGF1. Oncol Lett. (2018) 16:5131–9. doi: 10.3892/ol.2018.9307
138. Zhu QN, Renaud H, Guo Y. Bioinformatics-based identification of miR-542-5p as a predictive biomarker in breast cancer therapy. Hereditas. (2018) 155:17. doi: 10.1186/s41065-018-0055-7
139. Silva CM, Barros-Filho MC, Wong DV, Mello JB, Nobre LM, Wanderley CW, et al. Circulating let-7e-5p, miR-106a-5p, miR-28-3p, and miR-542-5p as a promising microRNA signature for the detection of colorectal cancer. Cancers. (2021) 13: 1493. doi: 10.3390/cancers13071493
140. Song GL, Xiao M, Wan XY, Deng J, Ling JD, Tian YG, et al. MiR-130a-3p suppresses colorectal cancer growth by targeting Wnt family member 1 (WNT1). Bioengineered. (2021) 12:8407–18. doi: 10.1080/21655979.2021.1977556
141. Cui N, Sun Q, Liu H, Li L, Guo X, Shi Y, et al. Long non-coding RNA LINC00511 regulates the expression of microRNA-625-5p and activates signal transducers and activators of transcription 3 (STAT3) to accelerate the progression of gastric cancer. Bioengineered. (2021) 12:2915–27. doi: 10.1080/21655979.2021.1940611
Keywords: cranberry, polyphenols, proanthocyanidin, intestinal microbiota, intestinal health, inflammation, microRNA
Citation: Taibi A, Lofft Z, Laytouni-Imbriaco B and Comelli EM (2023) The role of intestinal microbiota and microRNAs in the anti-inflammatory effects of cranberry: from pre-clinical to clinical studies. Front. Nutr. 10:1092342. doi: 10.3389/fnut.2023.1092342
Received: 07 November 2022; Accepted: 05 May 2023;
Published: 23 May 2023.
Edited by:
Mirko Marino, University of Milan, ItalyReviewed by:
Francesco Maria Calabrese, University of Bari Aldo Moro, ItalyTomas Meroño, University of Barcelona, Spain
Copyright © 2023 Taibi, Lofft, Laytouni-Imbriaco and Comelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elena Maria Comelli, ZWxlbmEuY29tZWxsaUB1dG9yb250by5jYQ==
†These authors share first authorship
 Amel Taibi
Amel Taibi Zoe Lofft
Zoe Lofft Bianca Laytouni-Imbriaco
Bianca Laytouni-Imbriaco Elena Maria Comelli
Elena Maria Comelli