
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr. , 24 August 2022
Sec. Food Chemistry
Volume 9 - 2022 | https://doi.org/10.3389/fnut.2022.990380
This article is part of the Research Topic Flavor Chemistry of Food: Mechanism, Interaction, New Advances View all 24 articles
Zygosaccharomyces rouxii was a highly salt-tolerant yeast, playing an important role in soy sauce fermentation. Previous studies reported that Z. rouxii under salt treatment produces better fermented food. However, the detailed change of main flavor substance was not clear. In this study, the physiological and transcriptomic analyses of Z. rouxii under salt treatment was investigated. The results revealed the high salt tolerance of Z. rouxii. Analysis of physiological data showed that the proportion of unsaturated fatty acids was significantly increased with the increment of salt concentrations. The analysis of organic acids showed that the content of succinic acid was significantly higher in the salt-treated Z. rouxii while oxalic acid was only identified at the 18% salt concentration-treated group. Results of volatile substances analysis showed that concentrations of 3-methyl-1-butanol and phenylethyl alcohol were significantly increased with the increment of salt concentrations. A comparison of transcriptome data showed that the genes involved in the TCA cycle and the linoleic acid synthesis process exhibited different expressions, which is consistent with the results of physiological data. This study helps to understand the change of main flavor substance of Z. rouxii under salt treatment and guide their applications in the high salt liquid state fermentation of the soy sauce.
Zygosaccharomyces rouxii belongs to the genus Zygosaccharomyces, which plays an important part in the manufacture of traditional fermented foods, such as soy sauce and miso. It mainly participates in consuming the sugars and amino acids of the soybeans and wheat during soy sauce fermentation. Apart from catalyzing the glucose conversion into ethanol, Z. rouxii is also capable of releasing higher alcohols from free amino acids via the Ehrlich pathway and produces a sweet and caramel type of flavor (1). The main flavor substances are two 4-hydroxyfuranones derivatives, 4-hydroxy-2,5-dimethyl-3(2H)-furanone (HDMF) and 4-hydroxy-2(or 5)-ethyl-5(or 2)-methyl-3(2H)-furanone (HEMF), the latter is also a potent antioxidant and has anticarcinogenic effects (2–4). In addition, it was also reported that the quality of fermented foods was improved and the fermentation process could be accelerated by the addition of Z. rouxii as a starter culture (5).
The volatile and non-volatile substances, such as organic acids, alcohols, and amino acids, contribute to the nutrition, unique taste, and aroma of fermented foods. Organic acids are one of the crucial compounds present in fermented products that not only have a huge effect on the sour taste but several of which also have other flavor attributes including a bitter or salty taste (6). Additionally, organic acids also affect the pH of the fermented products, which further affects the product quality, such as flavor stability (7, 8). Succinic acid, derived from the fermentation of glucose, is generally found in various fermented foods such as soy sauce, seasonings, and miso. All of these fermentation products have very distinct and marked flavors, which might attribute in part to a flavor enhancement by the small amounts of succinic acid. This would imply that succinic acid may have vital effects on various flavors that cannot be replicated with other food organic acids (9). As a major inhibitory and volatile compound, acetic acid can cause intracellular acidification or cell death and inhibit cell metabolism that acts to stress microbiota, finally leading to the reduction of yeast fermentative performance (10, 11). Intriguingly, acetic acid is generally used in foods as a flavor enhancer and flavoring agent. In addition to ethanol, alcohols, and organic acids, aldehydes, the main aroma-active compounds, ketones, esters, furans, phenols, and pyrazines were known as critical characteristic flavor substances of fermented foods produced via Z. rouxii (12).
Zygosaccharomyces rouxii inevitably suffer from multiple abiotic stresses covering osmotic, acidic, oxidative, temperature, and alcohol stresses during the process of fermented food production. Especially the high salt concentration triggers osmotic stress resulting in structural and physiological damage of cells, and thus negatively affects the growth and survival of organisms (13, 14). Previous studies have reported that Z. rouxii harbor a good tolerance toward high concentrations of salt, and the related physiological, transcriptomic, and metabolomics analysis of the salt tolerance mechanism of Z. rouxii have been comprehensively elucidated (5, 15, 16). Even though the high salinity environment leads to the injury cell and the low metabolic activity of Z. rouxii, on the other hand, a characteristic of Z. rouxii fermentation under that condition was also observed in the past several decades, accelerating and enhancing the flavor formation of fermented foods (17, 18). In addition, Z. rouxii is capable of slowly fermenting the sugars and amino acids present in high salty, typically in 18–20% NaCl environment, releasing flavors vital to the quality during soy sauce fermentation (19). Considerable investigations have been made regarding the enhancing and accelerating flavor formation of fermented foods by Z. rouxii under the high salinity environment in the last several decades. Nevertheless, to the best of our knowledge, little progress has been made on the detailed mechanism of enhancing and accelerating flavor formation by Z. rouxii under the above-mentioned abiotic stress. In the post-genomic era, with the advance of the omics approaches such as metabolomics, genomics, transcriptomics, and proteomics for flavor materials component and ratio analyses, a comprehensive understanding of the mechanisms, processes and environmental stress responses might obtain about the functions of Z. rouxii on fermented foods (15, 16, 20). These may also help to understand the genetic, biochemical, and physiological constraints on the metabolic processes of Z. rouxii in a high salinity environment.
High-salt condition is the main environmental factor in the high salt liquid state fermentation of the soy sauce (18–20% salt concentration), and Z. Rouxii would inevitably be subject to high-salinity stress. The present study aims to delineate the related mechanism of flavor formation by Z. rouxii under a high salinity environment. Based on physiological and transcriptome analysis, the salt tolerance response of Z. rouxii including growth performance, changes in organic acids content, intracellular fatty acid, and volatile substances, as well as the overall transcription levels of key genes were investigated. Results of this study not only contribute to further completing the mechanism of flavor formation of Z. rouxii under high salt stress, but also might be beneficial to enhance and accelerate flavor formation, and thus ultimately improve its fermentation characteristics and guide their applications in the production of traditional fermented foods.
The Z. rouxii CICC 32899 strain, purchased from China Center of Industrial Culture Collection, was used in this study. It was grown statically in a YPD medium (yeast paste 10 g/L, protein 20 g/L, glucose 20 g/L, pH 6.0) at 30°C for 24 h for activating strain. To observe the growth of Z. rouxii at different salt concentrations, the activated Z. rouxii was inoculated in an YPD medium treated with different salt concentrations (0, 6, 12, and 18%) for 3 days, each sample was performed in triplicate.
The growth of Z. rouxii was mainly reflected by the OD values of Z. rouxii. Z. rouxii had a maximum absorption at 600 nm, thus this wavelength was chosen to determine the OD values. All samples were collected for 12, 24, 48, 60, and 72 h, and then the bacterial broth was diluted in a ratio of 1:3 for determination the OD values. It was determined by the UV spectrophotometer with a wavelength of 600 nm, deionized water as a control and the diluted bacteria solution as samples. The OD values are equal to the OD read multiplied by the dilution multiple. Each sample was performed in triplicate.
Lipids were extracted from cell homogenates and methylated by following previously described methods (21). Specifically, Z. rouxii mycelia were filtered, collected, washed with distilled water, and freeze dried. After that, the mycelia of each group were powdered and weighed. The same weight of mycelia powder was subjected to lipid extraction. The lipid extracts were incubated in chloroform with 2% H2SO4–MeOH solution at 70°C for 2 h to obtain fatty acid methyl esters (FAMEs). The FAME components were separated and analyzed by QP2010 gas chromatography-mass spectrometry (GC-MS) (Shimazjin, Kyoto, Japan). The system was equipped with Supelco SP-2340 fused silica capillary column (30 m × 0.25 mm i.d., with a film thickness of 0.2 μm; Bellefonte, PA, United States). FAMEs were identified by comparing their mass spectra with a spectrum database. FA peaks were identified based on comparisons of their retention times to the external standards or similarity search. The relative amounts of individual FA components were performed by using the peak area of the most intensive ion of each peak, and the percentage of UFAs was also calculated (21).
Same treated samples were also used to determine the volatile substances via GC-MS based on Benucci et al. method with some changes (22). It analyzed by QP2010 gas chromatography-mass spectrometry (GC-MS) (Shimazjin, Kyoto, Japan). The system was equipped with Supelco SP-2340 fused silica capillary column (30 m × 0.25 mm i.d., with a film thickness of 0.25 μm; Bellefonte, PA, United States).
After 3 days of liquid culture, Z. rouxii mycelium was filtered with a 0.45 μm membrane, and then the content of organic acids was determined for the following analysis. Briefly, the mycelia were lyophilized, vacuum freeze dried to a constant weight, and ground to powder. Two grams of dry powder were weighted from each sample was used to perform High Performance Liquid Chromatography (HPLC) detection. HPLC assay was conducted on Waters Alliance e2695 HPLC (Milford, MA, United States) using a UV detector set at 210 nm equipped with Aminex HPX-87H (Bio-Rad, CA, United States) column (300 × 7.8 mm, 9 μm). The analysis conditions were as follows: the mobile phase was 0.065% H3PO4 with isocratic elution at flow rate of 0.6 ml min–1, and 10 μl injection sample volume. A standard organic acids curve was generated using tartaric acid, propanedioic acid, acetic acid, citric acid, succinic acid, malic acid, and oxalic acid standard (Sigma-Aldrich, Burlington, MA, United States). These yield of organic acids was calculated using the detected peak area according to the standard curve.
After obtaining the cell samples, RNA extraction was performed using the fungal RNA kit (Omega Bio-Tek, Norcross, GA, United States). The RNA concentrations were detected by using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, United States), and the RNA integrity values were analyzed with a Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, United States). To ensure reliability and reproducibility, equal quantities of RNA of each pool from three individual cultures were used for cDNA library construction. Afterward, the mRNA was enriched from pooled total RNA using oligo (dT) magnetic beads and then digested into short pieces with fragmentation buffer at 94°C for 5 min and then reversely transcripted into cDNA using random hexamer primers. Using these mRNA fragments as templates, the first-strand cDNA was synthesized, followed by second-strand cDNA synthesis by DNA polymerase I and RNase H (23). The cDNA fragments were purified using QIAquick PCR extraction kit, end repaired, and ligated to Illumina sequencing adapters to create the cDNA library. After that the libraries were sequenced by the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, United States). The obtained raw reads containing adapters or low-quality bases will be further filtered by our previous criterion to get clean reads, and thus, removed (21). Moreover, the Bowtie2 software was used to remove reads that mapped to ribosome RNA (rRNA) database to get the final clean reads, which were further employed for assembly and transcriptome analysis (24). RNA-seq data of Z. rouxii under distinct salt stress were deposited in the NCBI/SRA database, under the BioProject accession number PRJNA837122.
The obtained clean-read datasets were aligned to Z. rouxii reference genome using HISAT2 (25). The RSEM software was used to quantify gene abundances, and the quantification of gene expression level was normalized using the FPKM (Fragments Per Kilobase of transcript perMillion mapped reads) method (26, 27). Differentially expressed genes (DEGs) across samples were identified using DESeq2 v1.18.1 on R package (version 3.4.2). The log2 (fold change) over 1 and false discovery rate (FDR) within 0.05 were set as the threshold for significant DEGs (28). Besides, the identified DEGs were carried out into hierarchical clustering, with the KEGG pathway enrichment analysis (29).
To validate the transcriptional level results from RNA-seq data analysis, six genes including elongase 1–4, D9D, and D12D genes which are involved in the linoleic acid biosynthesis in Z. rouxii, were selected for real-time RT-PCR validation. Total RNA was extracted with E.Z.N.A. Fungal RNA Kit (Omega Bio-Tek, Norcross, GA, United States) according to the protocols of the manufacturer. Real-time RT-PCR was performed according to our previous work (30). GAPDH served as the reference gene for the normalization of the target gene expression and to correct for variation between samples. Primers used for the candidate genes were designed based on the Illumina sequencing data by using Primer Premier 5 and listed in Supplementary Table 1. The comparative 2–ΔΔCT method was employed to calculate the relative expression between the target genes.
All the data obtained in this experiment are presented as the mean of three replicates. Data from the same period were evaluated by two-way nested analysis of variance (ANOVA), followed by the least significant difference test (LSD) for mean comparison. All statistical analysis was performed with SAS 9.20 software (SAS Institute Inc., Cary, NC, United States) at a p < 0.05.
To investigate the effects of salt stress on the growth performance of Z. rouxii, a preliminary study of different concentrations of salt treatments was conducted (Figure 1). Z. rouxii grew well in the control condition, and the maximum biomass yield (OD 600) reached to 2.1. With the salt concentrations increasing from 0 to 18%, the growth of Z. rouxii was gradually inhibited, and the highest OD 600 decreased from 2.1 to 1.2. This result suggested a great effect of salt stress on the growth performance of Z. rouxii. Of note, the similar growth performance of Z. rouxii was detected at 6 and 12% salt concentration, which illustrated both of these salt concentrations harbor the analogous effect on Z. rouxii growth. Moreover, the almost same maximum OD 600 was identified at 0, 6, and 12% salt concentrations, elucidating the high salt tolerance of Z. rouxii. The Z. rouxii grew slowly and almost stagnant during the period from 60 to 72 h, the samples thus cultured for 72 h were selected for the follow-up analysis.
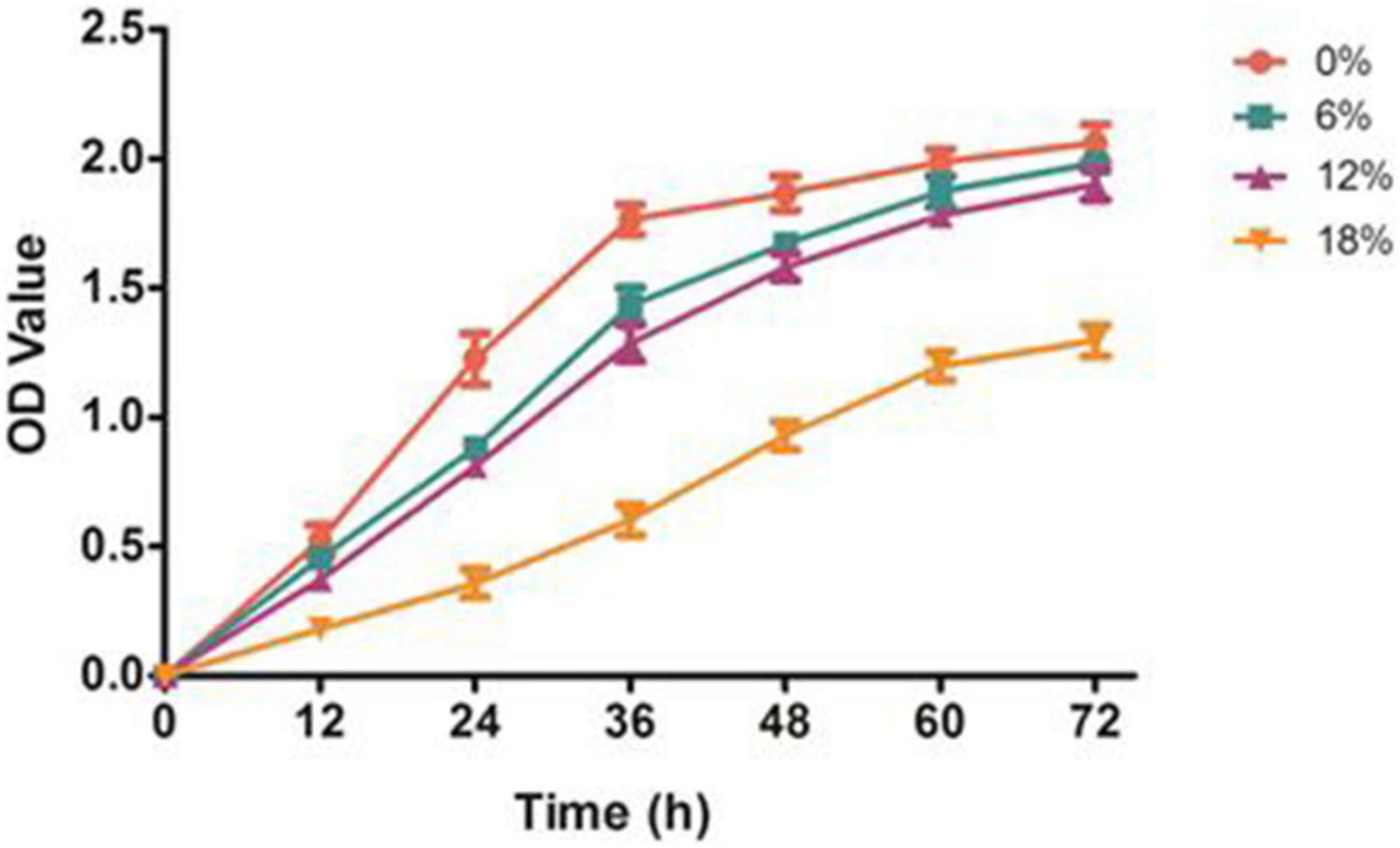
Figure 1. Effect of salt stress on the growth of Z. rouxii. 0, 6, 12, and 18% represent different salt concentrations, respectively.
Previous studies have documented that changes of the membrane lipid composition have impacts on the membrane fluidity and then affect the stress resistance of yeast (31, 32). Z. rouxii inevitably encountered salt stress in the manufacture of high salty foods. However, the effect of salt stress on the fermented food produced by Z. rouxii is poorly understood. In the present study, the fatty acid content profiles of Z. rouxii grown under salt stress and control conditions were qualitative and quantitative determinations by GC-MS. Our results revealed that the unsaturation of Z. rouxii increases with increasing salt concentrations (Supplementary Figure 1). In addition, the main composition of unsaturated and saturated fatty acid was also present on the Figure 2. The composition of fatty acid in Z. rouxii cells was dominated by C18 and C16, of them, the proportion of oleic acid (C18:1) was significantly increased with the increment of salt concentrations. Moreover, the proportion of linoleic acid was slightly increased, while palmitoleic acid was hardly increased at different salt concentrations. As for saturated fatty acid, palmitic acid and stearic acid were decreased significantly, while myristic acid and arachidic acid was slightly decreased (Figure 2).
Volatile substances are essential compositions of flavor in fermented food (33). Here, GC-MS was applied to determine the volatile compounds present in the extracts of Z. rouxii. A total of 98, 91, 94, and, 90 volatile compounds was identified at 0, 6, 12, and 18% salt-treated group, of which, 33 volatiles were found commonly to all Z. rouxii (Supplementary Table 2). Alcohols and acids made more contributions to the total volatiles in comparison with esters, furans, aldehydes, phenols, and pyrazines. Furthermore, the main seven volatile compounds were identified as the major volatile compounds in Z. rouxii due to their higher concentration, such as ethanol, 3-methyl-1-butanol, and phenylethyl alcohol (Figure 3). Among them, the relative contents of ethanol, 2-ethylhexanol, and 2, 4-dimethylbenzaldehyde were decreased with the increment of salt concentration. In contrast, the relative contents of two edible spices, 3-methyl-1-butanol and phenylethyl alcohol were increased, whereas 2-methyl-1-propanol was kept steady under distinct salt stress. Noteworthily, phenethyl acetate, a food flavoring agent, was not detected in the control group.
Yeast bacteria can produce a greater diversity of organic acids during their metabolisms, such as lactic acid, lemon acid, malic acid, and succinic acid (29). Here, the profile of organic acids produced by Z. rouxii during fermentation at different salt concentrations was determined. The content of seven main organic acids has distinct degree changes under salt stress compared with the control group (Figure 4). Among them, the content of tartaric acid, acetic acid, and citrate acid was decreased significantly with the increment of salt concentrations. In contrast, no significantly increased content of malonic acid and D-malic acid was identified with the salt treatment, with the exception of 18% salt concentration, of which the content of malonic acid was decreased significantly. In addition, succinic acid was not detected in the control group and its content significantly increased with the increment of salt concentrations while oxalic acid was only identified in the 18% salt concentration-treated group.
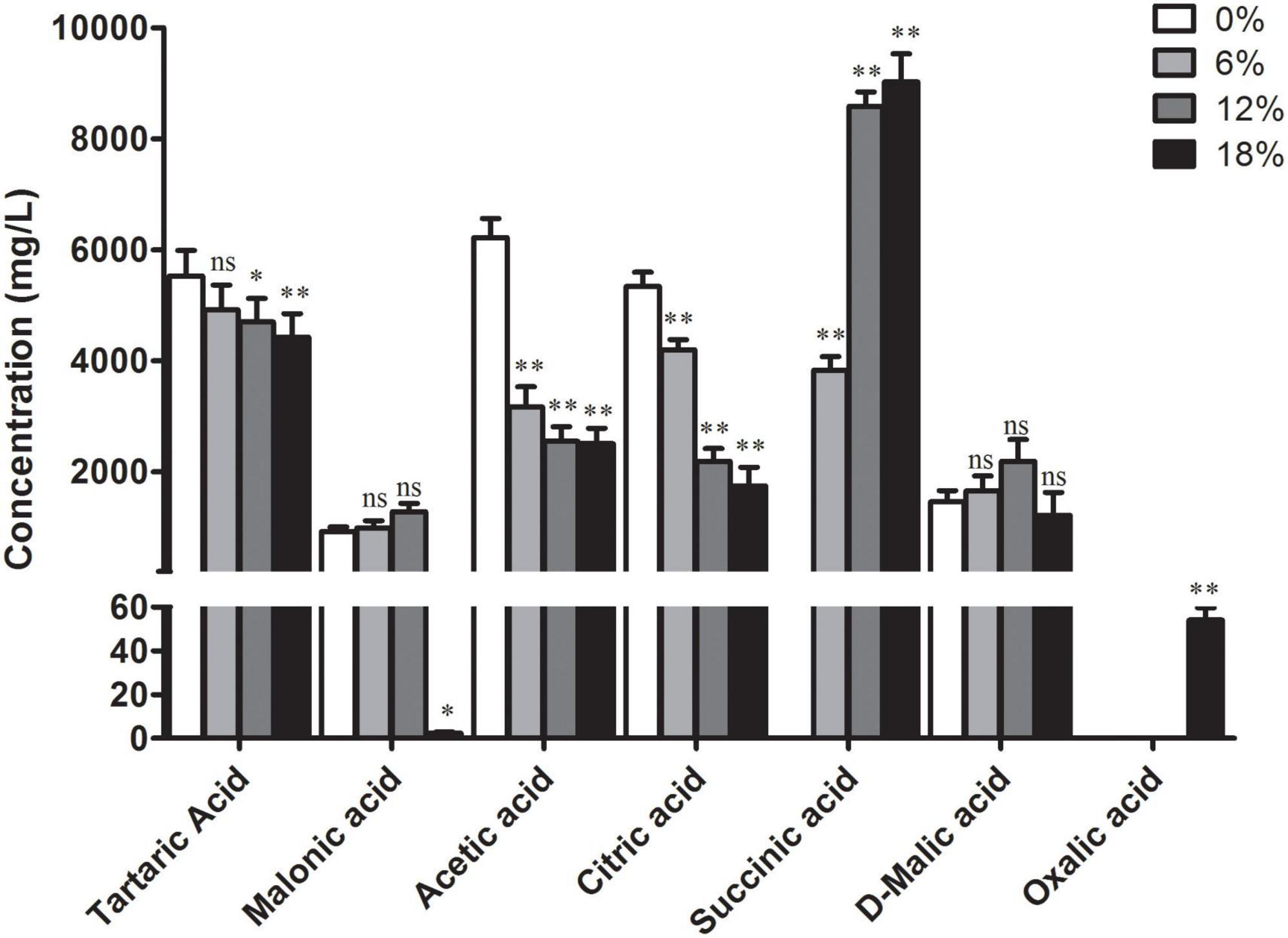
Figure 4. Intracellular content of seven organic acids in Z. rouxii under different salt concentrations, 0, 6, 12, and 18% represent different salt concentrations, respectively. The bars represent the average (±SE) of biological repeats. nsNo significant difference; *p < 0.05; **p < 0.01.
To investigate the environmental stress response of Z. rouxii induced by salt, a transcriptome analysis based on RNA sequencing of Z. rouxii that was subjected to distinct salt concentrations were performed. This resulted into the generation of 43.84, 43.16, 45.05, and 45.66 million clean reads per library, respectively (Table 1). The GC content for all treatments was approximately 43%, and the % ≥Q30 (99.9% accuracy of bases) was greater than 94.4% for all samples, elucidating a good quality of the sequencing data. Moreover, the clean reads were aligned to Z. rouxii genome sequence, and more than 83% of the clean reads for each sample were uniquely mapped to the genome. A summary of the RNA-seq sequencing was shown in Table 1.
To search genes with altered expression levels treated by different salt concentrations, the overall transcription levels of genes was quantified by RPKM metrics. Based on the global transcriptional changes from normalizing the DEG data, a total of 2,575 genes showed altered expression levels in the three salinity treatment groups, as compared to the control. There were 543, 659, and 1,373 DEGs that was identified in the control group and three salinity treatment groups, respectively (Figure 5A). Among them, 232, 173, and 685 genes expression being up-regulated at three salt concentrations compared to control groups, separately, whereas 311, 486, and 688 genes were down-regulated. To better present the effect of salt stress on Z. rouxii, a Venn diagram of DEG distribution at these groups was constructed for follow-up analysis due to their significant differences (Figure 5B). It was observed that only 153 DEGs were commonly shared among the distinct salt concentrations. It is noteworthy that the number of specific DEGs between WT-vs-NaCl-18 groups was significantly higher than that of the other two groups, revealing the involvement of complex developmental events of Z. rouxii cells under 18% salinity treatments (Figure 5B).
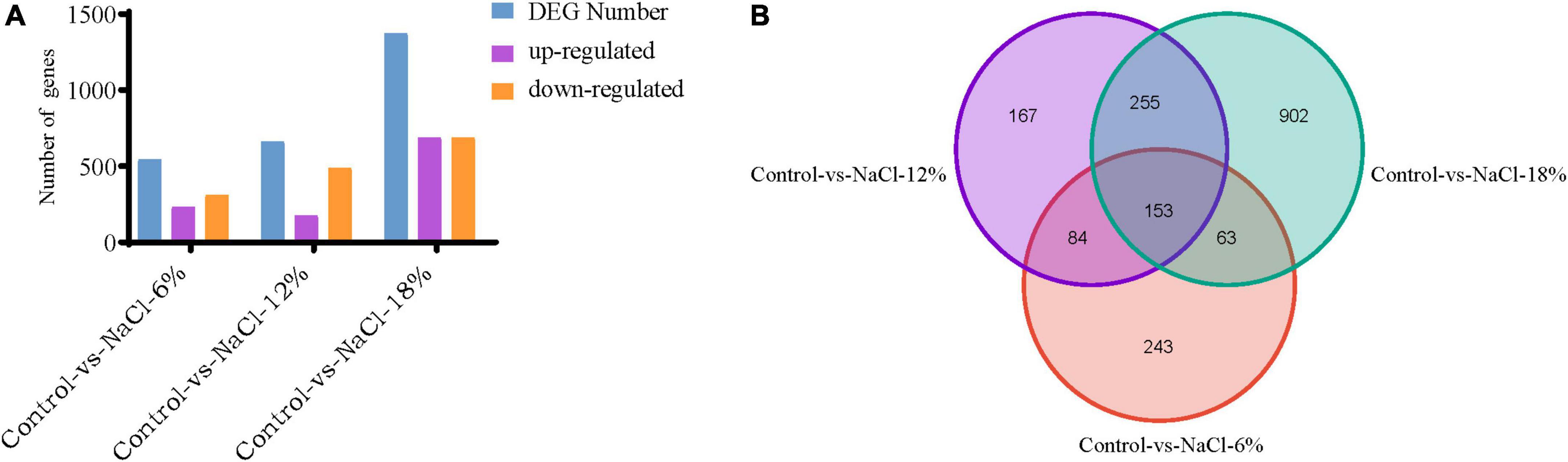
Figure 5. Distribution of differently expressed genes (DEGs) identified in the three samples. (A) Distribution of DEGs in the control group as compared to the salt-treated group. (B) Venn diagram exhibiting DEG distribution in three samples. NaCl-6%, -12%, and -18% represent the salt concentration of 6, 12 and 18% group, respectively.
The citrate cycle (TCA cycle, Krebs cycle) is a crucial aerobic pathway involved in for the final steps of the oxidation of carbohydrates and fatty acids. Hence, differentially expressed genes involved in the TCA cycle was investigated according to the KEGG enrichment analysis. A total of 42 DEGs encodings 10 enzymes were identified to be involved in different steps in carbohydrate metabolism (Figure 6). In addition to phosphoenol-pyruvate carboxykinase, fumarase, and aconitase, both of which are encoded by two DEGs, the remaining enzymes responsible for TCA cycle are encoded by four to eight DEGs, such as succinate dehydrogenase (eight DEGs). The expression level of a majority of these DEGs was up-regulated obviously under 6% salt concentration treatment, and only a partly of DEGs was up-regulated at 12%, while the remaining DEGs were down-regulated or harbor no remarkable change. Intriguingly, almost all of the enzyme-encoding genes identified in the TCA cycle was significantly down-regulated under 18% salinity treatment, for example, succinate dehydrogenase and aconitase gene, and with special attention paid to several malate dehydrogenase and pyruvate dehydrogenase gene, which possessed a high expression level.
Linoleic acid, a principal essential fatty acid, is also a critical structural component of cell membranes and has effect on cell membrane properties like fluidity, flexibility and permeability. These are six crucial enzymes involved in linoleic acid biosynthesis pathway, including elongase 1–4, two rate-limiting enzymes delta 9 fatty acid desaturases (D9D) and delta 12 fatty acid desaturases (D12D). According to the KEGG pathway analysis, 14 genes responsible for linoleic acid biosynthesis in Z. rouxii under salt stress were identified (Figure 7). Just three genes were up-regulated under 6% salinity treatment, the remaining gene were down-regulated or hardly changed, and the same number up-regulated and down-regulated genes were observed under 12% salinity treatment. Additionally, the great majority of genes encoding linoleic acid biosynthesis-related key enzymes were up-regulated under 18% salinity treatment. It has to be noted that four D9D and D12D genes were significant up-regulated under 6 and 18% salinity treatments. This result illustrated that high salt stress is beneficial to unsaturated fatty acid accumulation, which was consistent to the result of the proportion of unsaturated fatty acid determined before. Given that these six genes with notable expression levels in the distinct salt stress, the activity of D9D and D12D genes was enhanced at periods corresponding to linoleic acid production, implying a close correlation between salt stress and transcriptional regulation. This result was further supported by the relative expression levels of gene encoding elongase and D12D using qRT-PCR (Supplementary Figure 2).
Accelerating and enhancing the flavor formation of fermented foods was a paramount pursuit of industrial production. The industrial important microbes, Z. rouxii, was observed this characteristic of fermentation under high salinity condition over the past few decades, and related molecular mechanism was being explored. Nevertheless, no convincing evidence or strongly explanation was obtained regarding the related molecular mechanism of accelerating and enhancing the flavor formation by Z. rouxii fermentation under high salinity condition. Herein, we performed a physiological and transcriptomic analysis to reveal the main flavor substance of Z. rouxii during fermentation under high-salt treatment conditions.
High salinity environment represents a stress induced high osmotic perturbation to fungi cells due to the excess salt disturbs osmotic potential and results in metabolic toxicity. In the present study, the growth performance of Z. rouxii was significantly decreased by salt stress, especially in high salt concentration (Figure 1), and this result was analogous to the study of Wang et al. (15). Meanwhile, the number of DEGs reached a peak in the 18% salt treatment group (1,373) while the number was reduced at 6 and 12% salinity (543 and 659), indicating a larger degree of change in gene expressions in the high salt group than that in the low salt treatment (Figure 5). The similar change of gene expressions was also observed in our previous studies on Aspergillus oryzae (21). Further more, the characteristic of Z. rouxii fermentation under hyper-osmotic condition, accelerating and enhancing the flavor formation of fermented foods, thus require a strong salt tolerance of Z. rouxii cells for the long-term fermentation. Previous researches pointed out that several compatible solutes such as carbohydrates, intracellular glycerol, amino acids, and trehalose were accumulated under salt stress, which can be used as osmotic protectants against stress-induced perturbations (15, 34). Aside from the accumulation of these osmotic protectants, changes in the membrane composition and properties represent an essential factor in the adaptation to high salinity environment (35). It is reported that Z. rouxii cells responded to the changes in lipid composition in the high NaCl condition, especially the increment of phospholipids with high oleic acid contents and decreases in triacylglycerol and linoleic acid (36). Moreover, this change can also affect the leakage of glycerol, and keep from the influx of sodium ions under that conditions (37). Herein, the unsaturation of fatty acids in Z. rouxii has a positive correlation with salt concentrations, and the main composition of unsaturated fatty acids was palmitoleic acid, oleic acid, and linoleic acid (Figure 2 and Supplementary Figure 1). These results suggested that Z. rouxii produces more unsaturated fatty acids under salt stress, keeps the structure and function of the cells and helps them adapt to the hypertonic environment. Similar results were also found in Frankia strains, A. oryzae, and halophilic yeast-like melanized fungi (23, 38, 39). Additionally, the relative expression levels of genes related to linoleic acid biosynthesis in Z. rouxii under salt stress further supported the conclusion that salinity stress led to an increase in fatty acid unsaturation to maintain the osmotic balance of cells with this condition. In fact, in the case of exposure of Z. rouxii cells to salinity stress, 7 genes encoding elongase 1, 2, and 4 (from C16:0 to C18:0) were up-regulated (Figure 7). However, the content of C16:0 and C18:0 were both decreased under salt stress. According to the expression level of D9D and D12D, we may know that large amounts of C18:0 were biosynthesized into oleic acid and linoleic acid. Noteworthily, the potential health benefits of unsaturated fatty acids have attracted attention recently, and evidence is mounting on the role that several unsaturated fatty acids might play in the primary and secondary prevention of cardiovascular disease (40).
The volatile and non-volatile substances, such as organic acids, alcohols, and amino acids, have a considerable contribution to the main aroma of fermented foods. Through the analysis of volatile substances of Z. rouxii under salt stress, it is observed that 3-methyl-1-butanol and phenylethyl alcohol were increased with the increment of salt concentrations (Figure 3). 3-Methyl-1-butanol has apple brandy aroma and spicy flavor and is usually use as food spices, mainly for the preparation of apple and banana flavor. It is reported found in over 230 natural sources including apple, apricot, banana, sour cherry, cheese and so on (41). Phenylethyl alcohol usually existed in rose, carnation, hyacinth, Aleppo pine, orange blossom, and other organisms. It has a pleasant floral odor and used as an additive in cigarettes and also used as a preservative in soaps (42). Additionally, it is observed that phenethyl acetate was specially observed in the salt-treat Z. rouxii. Phenethyl acetate occurs in a number of essential oils and is a volatile aroma component of many fruits and alcoholic beverages. Phenylethyl acetate is a colorless liquid with a fine rose scent and a secondary, sweet, honey note. It is used in perfumery as a modifier of phenylethyl alcohol, for example, in rose and lilac compositions. In addition, it is used in a large number of aromas, in keeping with its natural occurrence (43). Hence, we speculate that these volatile substances are the main substance enhancing the flavor formation in Z. rouxii fermentation under salinity environment.
Further more, distinct degree changes of organic acids content in Z. rouxii was observed under salt treatment (Figure 4). The influence of salinity on the content of organic acids varies according to the microorganism and fermented production. The total levels of organic acids were elevated by salinity during the grapevine fermentation, which increased the production of tartaric and malic acids (44). In the less salt-sensitive cv. Korona, the concentration of total organic acids remained fairly constant under salinity environment. In the salt-sensitive cv. Elsanta, the contents of total organic acids, especially citric acid, increased significantly due to salt stress (45). The majority of organic acids were derived from glucose through the citrate cycle. Transcriptome analysis revealed that the pathway of the citrate cycle was activated corresponding to the content of organic acids, of which a majority of related DEGs were significantly up-regulated under salt treatment (Figure 6). The decrease of some organic acids, such as citric acid, may be due to the more conversion of downstream product. From the results, the content of succinic acid was significant higher in the salt-treated Z. rouxii while oxalic acid was only identified at the 18% salt concentration-treated group. Accordingly, we speculate that the content of glucose would be decreased under salt treatment, which need to be confirmed by experiment. Succinic acid is used as a dietary supplement for symptoms related to menopause such as hot flashes and irritability and also used as a flavoring agent for food and beverages (46). The FDA has granted succinic acid with the GRAS status (Generally Recognized as Safe Substance). These results suggested that succinic acid and oxalic acid may endow Z. rouxii under salt concentrations with special flavor.
In summary, we successfully characterized the transcriptomic profiles and the change of main flavor substance of Z. rouxii under salt treatment. Z. rouxii exhibited a good tolerance toward high concentrations of salt. Analysis of physiological data showed a accumulation of unsaturated fatty acids, succinic acid and oxalic acid. In addition, volatile substances analysis showed that concentrations of 3-methyl-1-butanol and phenylethyl alcohol were significantly increased with the increment of salt concentrations. Transcriptomic analysis showed an up-regulated expression of the genes related to TCA cycle and the linoleic acid synthesis. This study helps to understand the change of main flavor substance of Z. rouxii under salt treatment and guide their applications in the high salt liquid state fermentation of the soy sauce.
The RNA-seq data presented in the study are deposited in the NCBI/SRA repository (www.ncbi.nlm.nih.gov/sra), accession number: PRJNA837122.
BH and MA conceived and designed the experiments. BG and RP performed the experiments. GL and YL analyzed the data. RW contributed to reagents, materials, analysis tools, and revised the manuscript. GL and RP wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by the Natural Science Foundation of Jiangxi Province (Grant Nos. 20202BABL203043 and 20212BAB215005) and Graduate Education Reform Project (KSDYJG-2021-21).
Thank to Yayi Tu and Bin Zeng for the critical review of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.990380/full#supplementary-material
Supplementary Figure 1 | Proportion of unsaturated and saturated fatty acids content in Z. rouxii at different salt concentrations.
Supplementary Figure 2 | Relative expression level of six genes encoding crucial enzymes involved in linoleic acid biosynthesis pathway in Z. rouxii under different salt concentrations, 0, 6, 12, and 18% represent different salt concentrations, respectively. Gene 10103, 5047, 217, and 5506 are encoding elongase and gene 5111 and 1576 are encoding D12D, separately. The bars represent the average (±SE) of biological repeats. ns, *, **, and *** indicate statistically significant differences between the control and salt-treated groups (the least significant difference test): ns, no significant difference; *p < 0.05; **p < 0.01; ***p < 0.001.
Supplementary Table 1 | qRT-PCR primers used in this study.
Supplementary Table 2 | Volatile compounds identified in this study.
1. van der Sluis C, Wolken WA, Giuseppin ML, Tramper J, Wijffels RH. Effect of threonine, cystathionine, and the branched-chain amino acids on the metabolism of Zygosaccharomyces rouxii*. Enzyme Microb Technol. (2000) 26:292–300. doi: 10.1016/s0141-0229(99)00165-9
2. Sasaki M, Nunomura N, Matsudo T. Biosynthesis of 4-hydroxy-2(or 5)-ethyl-5(or 2)-methyl-3(2H)-furanone by yeasts. J Agric Food Chem. (1991) 39:934–8. doi: 10.1021/jf00005a027
3. Hecquet L, Sancelme M, Bolte J, Demuynck C. Biosynthesis of 4-hydroxy-2,5-dimethyl-3(2H)-furanone by Zygosaccharomyces rouxii. J Agric Food Chem. (1996) 44:1357–60. doi: 10.1021/jf950435j
4. Kataoka S. Functional effects of Japanese style fermented soy sauce (shoyu) and its components. J Biosci Bioeng. (2005) 100:227–34. doi: 10.1263/jbb.100.227
5. Dakal TC, Solieri L, Giudici P. Adaptive response and tolerance to sugar and salt stress in the food yeast Zygosaccharomyces rouxii. Int J Food Microbiol. (2014) 185:140–57. doi: 10.1016/j.ijfoodmicro.2014.05.015
6. Whiting GC. Organic acid metabolism of yeasts during fermentation of alcoholic beverages-a review. J Inst Brew. (1976) 82:84–92. doi: 10.1002/j.2050-0416.1976.tb03731.x
7. Kaneda H, Takashio M, Tamaki T, Osawa T. Influence of PH on flavour staling during beer storage. J Inst Brew. (1997) 103:21–3. doi: 10.1002/j.2050-0416.1997.tb00932.x
8. Li G, Liu F. Changes in organic acids during beer fermentation. J Am Soc Brew Chem. (2015) 73:275–9. doi: 10.1094/ASBCJ-2015-0509-01
9. Featherstone S. Ingredients used in the preparation of canned foods. In: Featherstone S editor. A Complete Course in Canning and Related Processes. Oxford: Woodhead Publishing (2015). p. 147–211.
10. Sousa MJ, Ludovico P, Rodrigues F, Leão C, Côrte-Real M. “Stress and cell death in yeast induced by acetic acid,” In: Bubulya P Editor. Cell Metabolism – Cell Homeostasis and Stress Response. Princes Gate Court: InTech (2012). doi: 10.5772/27726
11. Chaves SR, Rego A, Martins VM, Santos-Pereira C, Sousa MJ, Côrte-Real M. Regulation of cell death induced by acetic acid in yeasts. Front Cell Dev Biol. (2021) 9:642375. doi: 10.3389/fcell.2021.642375
12. Wah TT, Walaisri S, Assavanig A, Niamsiri N, Lertsiri S. Co-culturing of Pichia guilliermondii enhanced volatile flavor compound formation by Zygosaccharomyces rouxii in the model system of Thai soy sauce fermentation. Int J Food Microbiol. (2013) 160:282–9. doi: 10.1016/j.ijfoodmicro.2012.10.022
13. Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev. (1989) 53:121–47. doi: 10.1128/mr.53.1.121-147.1989
14. Prasad J, McJarrow P, Gopal P. Heat and osmotic stress responses of probiotic Lactobacillus rhamnosus HN001 (DR20) in relation to viability after drying. Appl Environ Microbiol. (2003) 69:917–25. doi: 10.1128/aem.69.2.917-925.2003
15. Wang D, Hao Z, Zhao J, Jin Y, Huang J, Zhou R, et al. Comparative physiological and transcriptomic analyses reveal salt tolerance mechanisms of Zygosaccharomyces rouxii. Process Biochem. (2019) 82:59–67. doi: 10.1016/j.procbio.2019.04.009
16. Wang D, Mi T, Huang J, Zhou R, Jin Y, Wu C. Metabolomics analysis of salt tolerance of Zygosaccharomyces rouxii and guided exogenous fatty acid addition for improved salt tolerance. J Sci Food Agric. (2022) 5:11975. doi: 10.1002/jsfa.11975
17. van der Sluis C, Tramper J, Wijffels RH. Enhancing and accelerating flavour formation by salt-tolerant yeasts in Japanese soy-sauce processes. Trends Food Sci Technol. (2001) 12:322–7. doi: 10.1016/S0924-2244(01)00094-2
18. Liu B, Wang X, Zhao J, Qin L, Shi L, Zhou T, et al. Effects of salinity on the synthesis of 3-methylthiopropanol, 2-phenylethanol, and isoamyl acetate in Zygosaccharomyces rouxii and Z. rouxii 3-2. Bioprocess Biosyst Eng. (2020) 43:831–8. doi: 10.1007/s00449-019-02279-3
19. Hayashida Y, Nishimura K, Slaughter JC. The influence of mash pre-aging on the development of the flavour-active compound, 4-hydroxy-2(or5)-ethyl-5(or2)-methyl-3(2H)-furanone (HEMF), during soy sauce fermentation. Int J Food Sci Technol. (2003) 32:11–4. doi: 10.1046/j.1365-2621.1997.00378.x
20. Solieri L, Vezzani V, Cassanelli S, Dakal TC, Pazzini J, Giudici P. Differential hypersaline stress response in Zygosaccharomyces rouxii complex yeasts: a physiological and transcriptional study. FEMS Yeast Res. (2016) 16:fow063. doi: 10.1093/femsyr/fow063
21. He B, Hu Z, Ma L, Li H, Ai M, Han J, et al. Transcriptome analysis of different growth stages of Aspergillus oryzae reveals dynamic changes of distinct classes of genes during growth. BMC Microbiol. (2018) 18:12. doi: 10.1186/s12866-018-1158-z
22. Benucci I, Cecchi T, Lombardelli C, Maresca D, Mauriello G, Esti M. Novel microencapsulated yeast for the primary fermentation of green beer: kinetic behavior, volatiles and sensory profile. Food Chem. (2021) 340:127900. doi: 10.1016/j.foodchem.2020.127900
23. He B, Ma L, Hu Z, Li H, Ai M, Long C, et al. Deep sequencing analysis of transcriptomes in Aspergillus oryzae in response to salinity stress. Appl Microbiol Biotechnol. (2018) 102:897–906. doi: 10.1007/s00253-017-8603-z
24. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. (2012) 9:357–9. doi: 10.1038/nmeth.1923
25. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. (2015) 12:357–60. doi: 10.1038/nmeth.3317
26. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. (2008) 5:621–8. doi: 10.1038/nmeth.1226
27. Dewey CN, Bo L. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. (2011) 12:323–323.
28. Wang D, Chen H, Yang H, Yao S, Wu C. Incorporation of exogenous fatty acids enhances the salt tolerance of food yeast Zygosaccharomyces rouxii. J Agric Food Chem. (2021) 69:10301–10. doi: 10.1021/acs.jafc.1c03896
29. Zou M, Zhu X, Li X, Zeng X. Changes in lipids distribution and fatty acid composition during soy sauce production. Food Sci Nutr. (2019) 7:764–72. doi: 10.1002/fsn3.922
30. Lv G, Xu Y, Tu Y, Cheng X, Zeng B, Huang J, et al. Effects of nitrogen and phosphorus limitation on fatty acid contents in Aspergillus oryzae. Front Microbiol. (2021) 12:739569. doi: 10.3389/fmicb.2021.739569
31. Rodríguez-Vargas S, Sánchez-García A, Martínez-Rivas JM, Prieto JA, Randez-Gil F. Fluidization of membrane lipids enhances the tolerance of Saccharomyces cerevisiae to freezing and salt stress. Appl Environ Microbiol. (2007) 73:110–6. doi: 10.1128/aem.01360-06
32. Vázquez J, Grillitsch K, Daum G, Mas A, Beltran G, Torija MJ. The role of the membrane lipid composition in the oxidative stress tolerance of different wine yeasts. Food Microbiol. (2019) 78:143–54. doi: 10.1016/j.fm.2018.10.001
33. Shukla S, Choi TB, Park HK, Kim M, Lee IK, Kim JK. Determination of non-volatile and volatile organic acids in Korean traditional fermented soybean paste (Doenjang). Food Chem Toxicol. (2010) 48:2005–10. doi: 10.1016/j.fct.2010.04.034
34. Wood JM, Bremer E, Csonka LN, Kraemer R, Poolman B, van der Heide T, et al. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp Biochem Physiol A Mol Integr Physiol. (2001) 130:437–60. doi: 10.1016/S1095-6433(01)00442-1
35. Russell NJ, Evans RI, ter Steeg PF, Hellemons J, Verheul A, Abee T. Membranes as a target for stress adaptation. Int J Food Microbiol. (1995) 28:255–61. doi: 10.1016/0168-1605(95)00061-5
36. Watanabe Y, Takakuwa M. Change of lipid composition of Zygosaccharomyces rouxii after transfer to high sodium chloride culture medium. J Ferment Technol. (1987) 65:365–9. doi: 10.1016/0385-6380(87)90131-2
37. Yoshikawa S, Mitsui N, Chikara K-I, Hashimoto H, Shimosaka M, Okazaki M. Effect of salt stress on plasma membrane permeability and lipid saturation in the salt-tolerant yeast Zygosaccharomyces rouxii. J Ferment Bioeng. (1995) 80:131–5. doi: 10.1016/0922-338X(95)93207-Z
38. Turk M, Méjanelle L, Šentjurc M, Grimalt JO, Gunde-Cimerman N, Plemenitaš A. Salt-induced changes in lipid composition and membrane fluidity of halophilic yeast-like melanized fungi. Extremophiles. (2004) 8:53–61. doi: 10.1007/s00792-003-0360-5
39. Srivastava A, Singh SS, Mishra AK. Modulation in fatty acid composition influences salinity stress tolerance in Frankia strains. Ann Microbiol. (2014) 64:1315–23. doi: 10.1007/s13213-013-0775-x
40. Abdelhamid AS, Brown TJ, Brainard JS, Biswas P, Thorpe GC, Moore HJ, et al. Omega-3 fatty acids for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst Rev. (2020) 3:CD003177. doi: 10.1002/14651858.CD003177.pub5
41. Kaminarides S, Stamou P, Massouras T. Changes of organic acids, volatile aroma compounds and sensory characteristics of Halloumi cheese kept in brine. Food Chem. (2007) 100:219–25. doi: 10.1016/j.foodchem.2005.09.039
42. Ghosh S, Kebaara BW, Atkin AL, Nickerson KW. Regulation of aromatic alcohol production in Candida albicans. Appl Environ Microbiol. (2008) 74:7211–8. doi: 10.1128/aem.01614-08
43. Trindade de Carvalho B, Holt S, Souffriau B, Lopes Brandão R, Foulquié-Moreno MR, Thevelein JM. Identification of novel alleles conferring superior production of rose flavor phenylethyl acetate using polygenic analysis in yeast. mBio. (2017) 8:e01173–7. doi: 10.1128/mBio.01173-17
44. Li X, Wang C, Li X, Yao Y, Hao Y. Modifications of Kyoho grape berry quality under long-term NaCl treatment. Food Chem. (2013) 139:931–7. doi: 10.1016/j.foodchem.2013.02.038
45. Keutgen A, Pawelzik E. Quality and nutritional value of strawberry fruit under long term salt stress. Food Chem. (2008) 107:1413–20.
Keywords: Zygosaccharomyces rouxii, salt treatment, transcriptomics, flavor substance, physiological
Citation: Pei R, Lv G, Guo B, Li Y, Ai M, He B and Wan R (2022) Physiological and transcriptomic analyses revealed the change of main flavor substance of Zygosaccharomyces rouxii under salt treatment. Front. Nutr. 9:990380. doi: 10.3389/fnut.2022.990380
Received: 09 July 2022; Accepted: 01 August 2022;
Published: 24 August 2022.
Edited by:
Mingquan Huang, Beijing Technology and Business University, ChinaReviewed by:
Georgi Kostov, University of Food Technologies, BulgariaCopyright © 2022 Pei, Lv, Guo, Li, Ai, He and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingqiang Ai, YW1xMTk4M0BxcS5jb20=; Bin He, aGViaW4ubGlAZm94bWFpbC5jb20=; Runlan Wan, d2FucnVubGFuQHN3bXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.