- 1Animal Science and Technology College, Beijing University of Agriculture, Beijing, China
- 2Faculty of Science and Engineering, Centre for Healthy Eating and Food Innovation, Maastricht University - Campus Venlo, Venlo, Netherlands
Approximately 35% of rapeseed meal (RSM) dry matter (DM) are carbohydrates, half of which are water-soluble carbohydrates. The cell wall of rapeseed meal contains arabinan, galactomannan, homogalacturonan, rhamnogalacturonan I, type II arabinogalactan, glucuronoxylan, XXGG-type and XXXG-type xyloglucan, and cellulose. Glycoside hydrolases including in the degradation of RSM carbohydrates are α-L-Arabinofuranosidases (EC 3.2.1.55), endo-α-1,5-L-arabinanases (EC 3.2.1.99), Endo-1,4-β-mannanase (EC 3.2.1.78), β-mannosidase (EC 3.2.1.25), α-galactosidase (EC 3.2.1.22), reducing-end-disaccharide-lyase (pectate disaccharide-lyase) (EC 4.2.2.9), (1 → 4)-6-O-methyl-α-D-galacturonan lyase (pectin lyase) (EC 4.2.2.10), (1 → 4)-α-D-galacturonan reducing-end-trisaccharide-lyase (pectate trisaccharide-lyase) (EC 4.2.2.22), α-1,4-D-galacturonan lyase (pectate lyase) (EC 4.2.2.2), (1 → 4)-α-D-galacturonan glycanohydrolase (endo-polygalacturonase) (EC 3.2.1.15), Rhamnogalacturonan hydrolase, Rhamnogalacturonan lyase (EC 4.2.2.23), Exo-β-1,3-galactanase (EC 3.2.1.145), endo-β-1,6-galactanase (EC 3.2.1.164), Endo-β-1,4-glucanase (EC 3.2.1.4), α-xylosidase (EC 3.2.1.177), β-glucosidase (EC 3.2.1.21) endo-β-1,4-glucanase (EC 3.2.1.4), exo-β-1,4-glucanase (EC 3.2.1.91), and β-glucosidase (EC 3.2.1.21). In conclusion, this review summarizes the chemical and nutritional compositions of RSM, and the microbial degradation of RSM cell wall carbohydrates which are important to allow to develop strategies to improve recalcitrant RSM carbohydrate degradation by the gut microbiota, and eventually to improve animal feed digestibility, feed efficiency, and animal performance.
Introduction
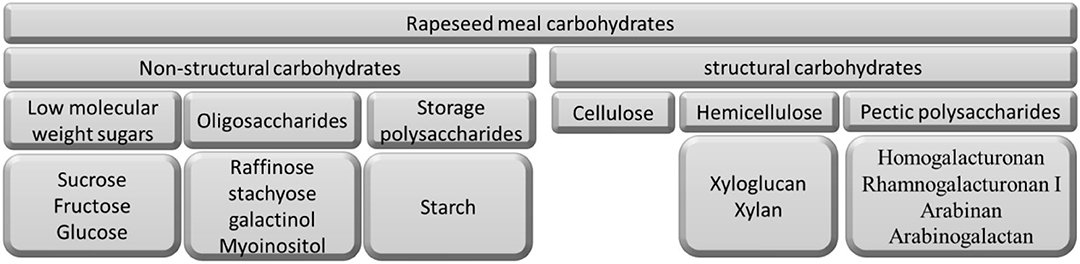
The main ingredients of animal feeds are plant carbohydrates, which consist of more than 70% of dry matter (DM). Plant carbohydrates can be classified as non-structural carbohydrates (low molecular weight sugars, oligosaccharides, and storage polysaccharides) and structural polysaccharides (non-starch polysaccharides) (1). Non-starch polysaccharides (NSP) cannot be broken down by endogenous digestive enzymes, nor be absorbed in the upper gastrointestinal tract (GIT). However, they can be (partly) degraded by the gut microbiota in the large intestine, and absorbed by the host in the form of short-chain fatty acids (SCFA), in this manner contributing to additional energy extraction from the diet. In addition, NSP may affect the digestion of other nutrients by the means of physical hindrance or physiological changes in the gut, such as increased digesta viscosity (2). Thus, it is vital to understand the NSP composition and structure of feed ingredients to improve animal feed efficiency.
Rapeseed meal (RSM), a byproduct of rapeseed oil production, is not only a suitable protein source but also a potential energy source as animal feed. Non-structural carbohydrates make up ~3–5% of the DM. Meal production processes could affect fiber production from RSM. These include expeller pressed (rapeseed oil is physically extracted using heat), cold pressed (rapeseed oil is physically extracted without heat treatment) and solvent extraction (rapeseed oil is extracted from the meal by physical expeller extraction followed by solvent washing). Previous research showed that expeller-pressed meal had more crude fiber compared to the cold-pressed meal (3). Another research demonstrated that solvent-extraction meal had more neutral detergent fiber and acid detergent fiber compared to the cold-pressed meal (4). Non-starch polysaccharides constitute 20–40% of RSM (5, 6), which are represented by pectic polysaccharides (homogalacturonan, rhamnogalacturonan I, arabinan, galactomannan, and arabinogalactan), hemicelluloses (xyloglucan and glucuronoxylan), and cellulose (7). Reports show that the NSP can only be degraded for 3–6% in chickens (5, 8, 9), and around 58–68% in pigs. This is rather low compared to other NSP-rich feed ingredients, such as sugar beet pulp (~85% of NSP is degraded by pigs) (1). Knowledge about the relationship between the chemical structure of NSP and GIT degradation is scant.
Plant cell walls are composed of a primary and a secondary layer, which are both built from polysaccharides, lignin, and protein. Polysaccharides contribute the most to the cell wall composition. The primary cell wall of RSM contains pectin, cellulose, and xyloglucan. The cellulose microfibrils are interlinked with xyloglucan via hydrogen bonds forming a stiff network (10). Pectins are linked to each other and cross-linked with (Hemi)cellulose (11, 12). In the secondary cell wall of RSM, the main carbohydrates are 4-o-methyglucuronoxylan, xyloglucan, and cellulose (13). This review aims to provide an overview of the carbohydrate composition of RSM, as well as the microbial degradation of RSM. Rapeseed derives from several species belonging to the genus Brassicaceae. In the current review, B. napus is discussed.
Nutritional characteristics of RSM carbohydrates
Carbohydrates in rapeseed meal (B. napus) are mainly pectins and (Hemi)celluloses and comprise 35–36% of the RSM dry matter (DM) (7, 14). The common feedstuff analysis shows commercial RSM contains 12.1% crude fiber and 34%nitrogen-freee extract (NFE) (Table 1). Carbohydrates of RSM can be categorized into non-structural carbohydrates and structural carbohydrates: a portion of the NFE is non-structural carbohydrates, whereas the crude fiber and the remainder of the NFE are structural polysaccharides. The composition of the different categories of RSM carbohydrates is displayed in Table 1.
Non-structural carbohydrates
The non-structural carbohydrates in RSM are comprised of low molecular weight sugars, oligosaccharides, and storage polysaccharides (Figure 1).
Low molecular weight sugars
Low molecular weight sugars mainly are sucrose (2.3–2.9%), fructose (0.05–0.16%), and glucose (0.05–0.16%) (15).
Oligosaccharides
The primary oligosaccharides found in RSM are raffinose, stachyose, galactinol, and Myo-inositol. Of those, stachyose has the highest concentration (0.4–0.5%), followed by raffinose (0.05–0.16%), galactinol (0.1%), and Myo-inositol (0.1%) (6).
Storage polysaccharides
Only low concentrations of storage polysaccharides are present in RSM. The primary storage polysaccharide is starch. The starch content of the seed approaches 50% during early development, but starch almost completely disappears when energy stores are converted into oil as the seed matures. Rommi et al. (14) reported that starch concentrations found in the intact seed were the same as dehulled rapeseed press cakes (0.2% of DM), while the starch content of canola meal was up to 2.5% of DM (17). Starch granules are comprised of two main macromolecules: amylose and amylopectin. Amylose is a linear polymer consisting of α-1,4-linked D-glucose units (GU) and includes 500–600 GU in its native form, which can be subdivided into 1–20 chains. Amylopectin is a large branched macromolecule (molecular weight 107-109 kDa), with ~5% of its total linkages consisting of β-1,6-linked GU plus a large number of short α-1,4-linked linear GU chains (12–70 GU) (18).
Structural polysaccharides
The largest part of the RSM carbohydrates is made up of structural polysaccharides (Figure 1). These include cellulose, hemicellulose (xyloglucan and xylan), and pectic polysaccharides (homogalacturonan, rhamnogalacturonan I, arabinan, and arabinogalactan).
Polysaccharides composition
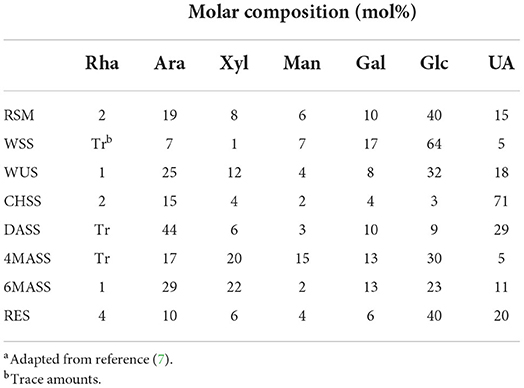
RSM contains 34.5% NSP of its DM, and RSM is high in glucosyl (40 mol%), arabinosyl (19 mol%), and uronyl residues (18 mol%) (7, 19). Water-soluble and water-insoluble carbohydrates comprise 18.7 and 15.8% of the total carbohydrates content of RSM, respectively (Table 1). Water-soluble carbohydrates mainly contain glucosyl (64 mol%) and some galactosyl residues (17 mol%) (7), while water-insoluble carbohydrates are mainly glucosyl (32 mol%), arabinosyl (25 mol%), uronyl (18 mol%), and xylosyl residues (12%) (Table 2).
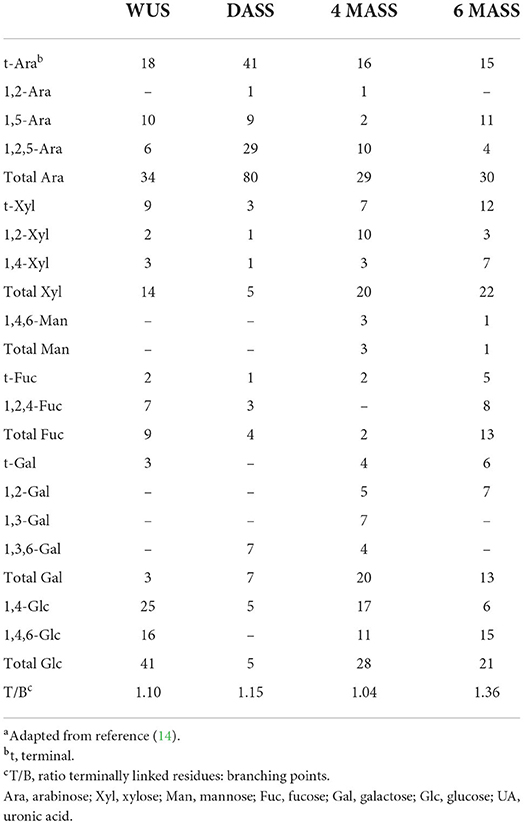
The detailed monosaccharide constituent compositions of the Water unextractable solids (WUS), which were sequentially extracted with (i) chelating agent (ChSS) to release calcium-bound pectins, (ii) dilute alkali (DASS) to release pectins tightly bound to hemicellulose, (iii) 4, and (iv) 6 molar alkali (4 MASS and 6 MASS) to release hemicelluloses, leaving cellulose that remains in the residue (RES), are presented in Table 2. The composition of the glycosidic residues indicates the presence of α-1,5-linked arabinan branched at O-2, galactomannan, homogalacturonan, rhamnogalacturonan I, type II arabinogalactan, glucuronoxylan, XXGG-type and XXXG-type xylo(X)glucan(G), and cellulose in RSM (7, 14, 20). Arabinan consists of a linear backbone of α-L-1,5-linked arabinose (Ara) units with α-1,2-linked or α-1,3-linked Ara units (Figure 2). Galactomannan has a backbone of β-1,4-linked mannose units, substituted with α-1,6-linked galactose units (Figure 1). Homogalacturonan (HG) is the simplest form of pectin, consisting of a linear chain of α-1,4-linked galacturonic acid, and part of its carboxyl groups esterified with acetyl or methyl groups (Figure 2). Rhamnogalacturonan I (RG-I) consists of alternating rhamnose (Rha) and galacturonic acid (GalA) residues [-,2)-α-Rha-(1,4)-α-D-GalA-(1,-], which is highly ramified with single terminal β-D-Gal and/or α-D-Ara at position O-4 or O-3 of the rhamnosyl residues where the α-D-GalA residues are often O-acetyl esterified at O-2 and/or O-3 (Figure 2) (21). Type II arabinogalactan (AG) is composed of a β-1,3-galactan backbone and β-1,6-galactan side chains (Figure 2). The side chains are variably decorated with L-arabinose. Glucuronoxylan (GX) has a linear backbone of β-(1-4)-linked D-xylosyl (Xyl) residues, which can be ramified with acetyl and arabinosyl residues, and some of the Xyl residues are decorated with a single α-D-glucuronic acid (GlcA) or 4-O-methyl-D-glucuronic acid (MeGlcA) residue at O2 (Figure 2) (22, 23). Both XXGG- and XXXG-type xyloglucan (XG) exist in RSM. XXGG (Figure 2) consists of a β-1,4-linked D-glucosyl (Glc) backbone carrying various side chains of D-β-1,2-Gal-D-α-1,6-Xyl and L-α-1,2-Ara-D-β- D-α-1,6-Xyl (24), whereas, XXXG (Figure 2) comprises a β-1,4-linked D-glucosyl backbone carrying continuous side chains of D-α-1,6-Xyl, D-β-1,2-Gal-D-α-1,6-Xyl, and L-α-1,2-Ara-D-β-1,2-Gal-D-α-1,6-Xyl in every 4 residues of D-β-1,4-Glc (24). Cellulose consists of long linear chains of β-1,4-linked D-Glc residues, with a degree of polymerization between 2,000 and 14,000 residues (Figure 2). The backbone of these polysaccharides can also be esterified with methyl-esters, ethyl-esters, and glycosyls (arabinosyl, galactosyl, mannosyl, gucosyl, xylosyl, or frucosyl).
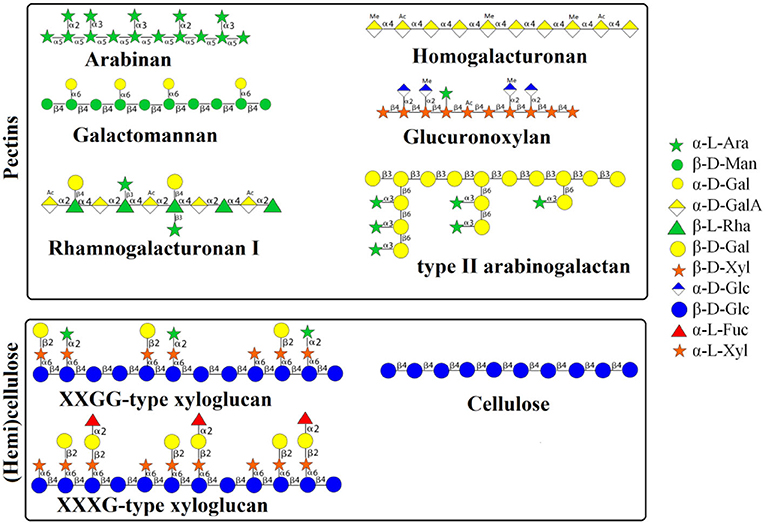
Figure 2. Schematic representation of structures of polysaccharides in RSM. RSM, rapeseed meal; WSS, water soluble solids; WUS, water unextractable solids; ChSS, Chelating Agent Soluble Solids; DASS, Dilute Alkali Soluble Solids; 4 MASS, 4 Molar Alkali Soluble Solids; 6 MASS, 6 Molar Alkali Soluble Solids; RES, residue; Ara, arabinose; Xyl, xylose; Man, mannose; Fuc, fucosyl; Gal, galactose; Glc, glucose; UA, uronic acids.
Except for containing glucosyl residues, RES (Table 2) also has quite some arabinosyl and uronyl residues (10 and 20%, respectively), which indicates that some pectic polysaccharides are tightly associated with cellulose microfibrils. This suggests that the cell wall polysaccharide matrix of rapeseed meal is strongly interlinked. The specific structures of dietary fibers of RSM are still not entirely understood. The main restriction may be the analysis method, but nowadays, the comprehensive microarray polymer profiling (CoMPP) technique is a powerful tool for probing cell wall structure studies (25). The profiles generated by CoMPP provide a global snapshot of cell-wall composition. It cannot only detect the amount of the particular polysaccharides but also their linkage type as discussed in the next section.
Glycosidic linkage type
A covalent link between one carbohydrate molecule and a second carbohydrate molecule is called a glycosidic bond. Glycosidic linkage is the type of bond between two adjacent glycosides in the chain of polysaccharides (26).
The proposed sugar linkage compositions of RSM fractions are shown in Table 3. The data can only be used in a qualitative manner instead of quantitative, due to the poor DMSO solubility of the fractions and the high amount of uronic acids (which are not detected with this method) present in some samples (27).
Microbial degradation of RSM cell wall carbohydrates
Low molecular weight sugars and starch can be 100% digested and/or absorbed, while (oligo- and poly-)saccharides are considered indigestible in the small intestine due to the lack of the necessary enzymes in monogastric animals (8). Indeed, mammalian genomes do not encode most of the enzymes needed to degrade the structural polysaccharides present in plant material. Instead, a complex mutual dependence has developed between the mammalian host and symbiotic gut microorganisms that do possess the ability to access the abundant source of energy in carbohydrates that are indigestible by the host. The gut microbiota has glycoside hydrolases (GH) that can degrade the oligo- and polysaccharides into small oligomers and monosaccharides which are subsequently taken up and fermented.
Glycoside hydrolases
Glycoside hydrolases (GHs) are a vast repertoire of cell wall-degrading enzymes that hydrolyze glycosidic bonds between two or more carbohydrate modules or sugar and a non-sugar moiety within carbohydrates or oligosaccharides (28, 29). GH families widely exist in prokaryotic, eukaryotic, and archaea species. A total of 173 GH families have been identified until now (accessed on May-2022, http://www.cazy.org/Glycoside-Hydrolases.html).
Carbohydrate binding modules (CBMs) are the non-catalytic part of cell-wall-degrading enzymes, and they are attached to the GH catalytic modules. Usually, CBMs have to recognize and bind to the specific polysaccharides first, before the GHs cleave the polysaccharides (30).
Degradation of arabinan
Arabinan can be hydrolyzed by α-L-Arabinofuranosidases (EC 3.2.1.55) and endo-α-1,5-L-arabinanases (EC 3.2.1.99), which are found in GH families 3, 43, 51, 54, and 62, and which release arabinosyl oligomers and L-arabinose (31). A previous study reported that Bacillus subtilis contained a series of arabinan-degrading genes, abnA, abn2, abfA, and abf2, which were induced by arabinose and arabinan, repressed by glucose, and subjected to temporal regulation (32). AbnA and abn2 encode extracellular endo-α-1,5-L-arabinanases belonging to GH43, which hydrolyzes arabinan (branched) and linear α-1,5-L-arabinan and produces arabinose and arabino-oligosaccharides (33). These resulting products, are subsequently transported into the cell by different transport systems. Arabinose enters the cell mainly through the araE permease (34), and the uptake of arabinose oligomers occurs most likely via araNPQ, an ABC-type transporter (35). These products are further digested into the monosaccharide arabinose by two intracellular arabinofuranosidases, abfA and abf2, which are α-L-arabinofuranosidases (EC 3.2.1.55) belonging to GH51. AbfA acts preferentially on (1 → 5) arabinofuranosyl linkages, and in contrast, abf2 is most active on (1 → 2) and (1 → 3) linkages (36). After this, L-arabinose is converted into D-xylulose-5-phosphate, which is further catabolized through the pentose phosphate pathway. The induction mechanism of these genes is mediated through negative control by the key regulator of arabinose metabolism, araR. The transcriptional repression of the abfA and abf2 genes is achieved by a tightly controlled mechanism but the regulation of abnA is more flexible.
The presence of α-L-arabinofuranosidases (Table 4) has also been determined in Bifidobacterium longum subsp. longum (37), Bacteroides thetaiotaomicron VPI-5482 (38), Sulfolobus solfataricus P2 (39), Anoxybacillus kestanbolensis AC26Sari (40), Monoglobus pectinilyticus (41), and Roseburia faecis M72/1 (42). Endo-α-1,5-L-arabinanases (Table 4) are present in Paenibacillus polymyxa (43), Bacillus licheniformis (44), Caldicellulorsiruptor saccharolyticus (45), Bacillus subtilis (36), Bacillus thermodenitrificans (46), Pseudomonas fluorecens subsp. cellulosa (47), Monoglobus pectinilyticus (41), and Roseburia faecis M72/1 (42).
Degradation of galactomannan
Endo-1,4-β-mannanase (EC 3.2.1.78), β-mannosidase (EC 3.2.1.25) and α-galactosidase (EC 3.2.1.22) are involved in the degradation of galactomannan into monosaccharides (48). β-mannanase degrades randomly within the main chain of galactomannans and produces shorter galactomanno-oligosaccharides that can be further hydrolyzed by β-mannosidase and α-galactosidase. β-mannosidase hydrolyses β-1,4-linked mannose residues from the non-reducing end of the galactomanno-oligosaccharides and α-galactosidase hydrolyses terminal α-1,6-linked galactose residues from galactomannans or the galactomanno-oligosaccharides. The gut microbes Bacillus subtilis YH12 (49), Bacillus subtilis TD7 (50), Bacillus subtilis Bs5 (51), Bacillus licheniformis DSM13 (52), Sphingomonas sp. JB13 (53), Sphingobacterium sp. GN25 (54), Klebsiella oxytoca KUB-CW2-3 (55), Enterobacter sp. strain N18 (56), Flavobacterium sp. (57), Pseudomonas cellulosa (47), Monoglobus pectinilyticus (41), and Bacteroides ovatus (58) are reported to have endo-1,4-β-mannanase (Table 4). While Bifidobacterium sp. (59), Bifidobacterium longum subsp. longum NCC2705 (60), Bacteroides ovatus (61, 62), Cellvibrio mixtus (63), Bacteroides thetaiotaomicron (64), Pseudomonas cellulose (47), and Kitasatospora sp. (65) are reported to have β-mannosidase (Table 4). Moreover, Lactobacillus acidophilus NCFM (66), Lactobacillus crispatus ST1 (67), Lactobacillus brevis (68), Bifidobacterium animalis subsp. lactis Bl-04 (66), Arthrobacter sp. C2-2 (69), Bacillus megaterium (70), Dictyoglomus thermophilum (71), Bacillus stearothermophilus NCIM-5146 (72), Bacillus stearothermophilus NUB 3621 (73), Bacteroides ovatus 0038-1 (74), ?ifidobacterium bifidum NCIMB41171 (75), ?ifidobacterium adolescentis DSM20083 (76), ?ifidobacterium breve 203 (77), Clostridium stercorarium (78), and Monoglobus pectinilyticus (41) are reported to have α-galactosidases (Table 4).
Asperigillus niger contains galactomannan-degradation genes, aglA, algB, and algC (encoding α-galactosidases), and mndA (encoding a β-mannosidase). AglA and aglB have been classified into GH27, while aglC has been classified into GH36, and mndA belongs to GH2. The metabolism mechanism of galactomannan is that α-galactosidase (encoded by aglC) and β-mannosidase (encoded by mndA) hydrolyze the galactomannan to the oligosaccharide Gal2Man5, where mndA cleaves single mannose units from the non-reducing end of the substrate until it reaches a galactose side-group (79), and afterward the non-reducing galactose group is hydrolyzed by α-galactosidase (aglA, aglB, or aglC). AglB and mndA play a major role in the degradation of galactomannan in A. niger. The expression of aglA is high on galactose and galactose-containing oligosaccharides, but is fully repressed in the presence of glucose (80). Little is known about genes, and especially their regulation, in members of the gut microbiota.
Degradation of homogalacturonan
Homogalacturonan (HG) can be cleaved by α-1,4-L-galacturonan reducing-end-disaccharide-lyase (pectate disaccharide-lyase) (EC 4.2.2.9), (1 → 4)-6-O-methyl-α-D-galacturonan lyase (pectin lyase) (EC 4.2.2.10), (1 → 4)-α-D-galacturonan reducing-end-trisaccharide-lyase (pectate trisaccharide-lyase) (EC 4.2.2.22), α-1,4-D-galacturonan lyase (pectate lyase) (EC 4.2.2.2), and (1 → 4)-α-D-galacturonan glycanohydrolase (endo-polygalacturonase) (EC 3.2.1.15) (89, 124–128). Pectin lyase (EC 4.2.2.10) provides cleavage of α-1,4-linked D-galacturonan methyl ester to give oligosaccharides with 4-deoxy-6-O-methyl-α-D-galact-4-enuronosyl groups at their non-reducing ends, while α-1,4-D-galacturonan lyase (EC 4.2.2.2) cleaves α-1.4-linked D-galacturonan to give oligosaccharides with 4-deoxy-α-D-galact-4-enuronosyl groups at their non-reducing ends. Afterwards, pectate disaccharide-lyase (EC 4.2.2.9) hydrolyzes these oligosaccharides to (1,4-α-D-galacturonosyl)n − 2 and 4-(4-deoxy-α-D-galact-4-enuronosyl)-D-galacturonate, and (1,4-α-D-galacturonosyl)n − 2 will be cleaved by pectin lyase (EC 4.2.2.10 or EC 2.2.2) again until the disaccharide results. Polygalacturonase (EC 3.2.1.15) randomly hydrolyzes (1 → 4)-α-D-galactosiduronic linkages in pectate and other galacturonans. Pectate disaccharide-lyase (Table 4) has been reported in Caldicellulosiruptor bescii (81), Bacteroides thetaiotaomicron (82), Bacteroides ovatus (83), Bacillus pumilus BK2 (84), Eubacterium eligens, Faecalibacterium prausnitzii (85), and Monoglobus pectinilyticus (41). Pectate lyase (Table 4) has been reported in Caldicellulosiruptor bescii (81), genus Bacillus: Bacillus subtilis, B. licheniformis, B. cereus, B. circulans, B. pasteurii, B. amyloliquefaciens, and B. pumilus (86, 87), Paenibacillus sp. (88), Clostridium cellulovorans (89), Streptomyces thermocarboxydus (90), Bacteroides thetaiotaomicron (82), Bacteroides ovatus (83), Eubacterium eligens, Faecalibacterium prausnitzii (85), and Monoglobus pectinilyticus (41). Endo-polygalacturonase (Table 4) has been reported in Caldicellulosiruptor bescii (81), Bacteroides thetaiotaomicron (82), Bacteroides ovatus (83), Eubacterium eligens, Faecalibacterium prausnitzii (85), Monoglobus pectinilyticus (41), and Bifidobacterium longum subsp. longum (91).
Degradation of rhamnogalacturonan I
Utilization of rhamnogalacturonan I (RG-I) by microbes is mediated by a series of enzymes, which is well-studied in Bacillus subtilis (92). Two main enzymes, a hydrolase, and a lyase are involved in the degradation of the RG-I backbone, whereas a few other enzymes are responsible for the breakdown of the RG-I side chains. Rhamnogalacturonan hydrolase (rhamnogalacturonan α-L-rhamnopyranohydrolase) cleaves α-1,2 linkages between GalA and Rha (129). Rhamnogalacturonan lyase [-L-rhamnopyranosyl-(1 → 4)-alpha-D-galactopyranosyluronate endolyase (EC 4.2.2.23)] cleaves the α-1,4 linkages of RG-I resulting in a double bond in the on-reducing GalA residue (130). Rhamnogalacturonan lyase and hydrolase (Table 4) have been reported in Caldicellulosiruptor bescii (81), Bacteroides thetaiotaomicron (82), Bacteroides ovatus (83), Bacillus subtilis (92), Bacillus licheniformis (93), Cellvibrio japonicus (94), Clostridium cellulolyticum (95), Bacillus licheniformis DSM13 (93), Pseudomonas cellulose (94), Penicillium chrysogenum (96), and Monoglobus pectinilyticus (41).
Degradation of type II arabinogalactan
Exo-β-1,3-galactanase (EC 3.2.1.145) cleaves the β-1,3-galactan backbone of type II arabinogalactan via bypassing the β-1,6-galactan side chains and releasing β-1,6-galactooligosaccharides and their derivatives (131). The β-1,6-galactan side chains are hydrolyzed to β-1,6-galactooligosaccharides of various degrees of polymerization (DP) by endo-β-1,6-galactanase (EC 3.2.1.164) (132, 133). On the other hand, exo-β-1,6-galactobiohydrolase releases β-1,6-galactobiose (β-1,6-Gal2) from the non-reducing terminal end of β-1,6-galactooligosaccharides, and α-L-arabinofuranosidase (EC 3.2.1.55) releases arabinofuranose (Araf) from α-1,3-Araf-substituted β-1,6-galactooligosaccharides (134). Exo-β-1,3-galactanase (Table 4) has been reported in Monoglobus pectinilyticus (41), Bifidobacterium longum subsp. longum (91), Clostridium thermocellum (97), Phanerochaete chrysosporium (98), Sphingomonas sp. (99), Bacteroides thetaiotaomicron (100), Bacteroides ovatus (100), Bacteroides caccae (101), and Bacteroides cellulosilyticus (101). Endo-β-1,6-galactanase (Table 4) had been reported in Streptomyces avermitilis NBRC14893 (102), Bacteroides ovatus (100), Bacteroides caccae (101), Bacteroides cellulosilyticus (101), and Bifidobacterium longum subsp. longum (91). Exo-β-1,6-galactobiohydrolase (Table 4) has been reported in Monoglobus pectinilyticus (41), Bacteroides thetaiotaomicron (101), Streptomyces avermitilis (102), Bacteroides ovatus (100), Bacteroides caccae (101), Bacteroides cellulosilyticus (101), and Bifidobacterium longum subsp. longum (91).
Degradation of glucuronoxylan
Two enzymes, β-(1, 3–5)-D-xylan xylanohydrolase (endo-β-1,4 xylanase) (EC 3.2.1.8) and 1,4-β xylohydrolase (exo-β-1,4 xylanase) (EC 3.2.1.37), are involved in degrading the β-1,4 xylosyl linkages in unsubstituted domains along the xylan backbone of glucuronoxylan (GX) (135, 136). Glucuronoxylanase cleaves glucuronosyl moietes which are substituted as monomeric side chains on the xylan backbone (137). Endo-β-1,4-xylanase (Table 4) has been reported in Pseudomonas boreopolis G22 (103), Bacteroides ovatus (82), Monoglobus pectinilyticus (41), Bacteroides thetaiotaomicron (100), Bacteroides caccae (101), Bacteroides cellulosilyticus (101), Clostridium thermocellum (104), Bacillus subtilis (105), and Streptomyces turgidiscabies (106). Exo-β-1,4 xylanase (Table 4) has been reported in Monoglobus pectinilyticus (41), Luteimicrobium xylanilyticum (107), Amycolatopsis mediterranei (107), Clostridium thermocellum (104), Bacillus subtilis (105), and Streptomyces turgidiscabies (106).
Degradation of xyloglucan
A set of glucanases and glycosidases are involved in cleaving xyloglucan (XG) into monosaccharides by two-step degradation (138, 139). Endo-β-1,4-glucanase (EC 3.2.1.4) hydrolyzes XG into large fragments, which are further degraded into monosaccharides by α-xylosidase (EC 3.2.1.177) and β-glucosidase (EC 3.2.1.21) (140). Endo-β-1,4-glucanase (Table 4) has been reported in Monoglobus pectinilyticus (41), Caldicellulosiruptor kronotskyensis (108), Roseburia sp. (42), Eubacterium rectale group (42), Ruminococcus champanellensis (109), Ruminococcus bromii (110), Ruminiclostridium cellulolyticum (111), and Phaeoacremonium minimum (112). Exo-β-1,4 xylanase (Table 4) has been reported in Monoglobus pectinilyticus (41), Luteimicrobium xylanilyticum (107), Amycolatopsis mediterranei (107), Clostridium thermocellum (104), Bacillus subtilis (105), Streptomyces turgidiscabies (106), Ruminiclostridium cellulolyticum (111), and Phaeoacremonium minimum (112). While α-D-xylosidase (Table 4) has been reported in Sulfolobus solfataricus P2 (39), Talaromyces thermophilus (113), Cellvibrio japonicus (114), Bacteroides thetaiotaomicron (82), Bacteroides ovatus (82), and Monoglobus pectinilyticus (41). Moreover, β-glucosidase (Table 4) has been reported in Bifidobacterium adolescentis (115), Bacteroides ovatus (116), Listeria innocua (117), Streptomyces venezuelae (118), Pyrococcus furiosus (119), Cellvibrio japonicus (114), Caldicellulosiruptor saccharolyticus (120), Microbispora bispora (121), Thermoanaerobacter brockii (122), Thermobifida fusca (103), Pseudomonas sp. (123), Monoglobus pectinilyticus (41), Ruminococcus champanellensis (109), and Ruminococcus bromii (110).
Degradation of cellulose
The enzymes of the cellulase system consist of endo-β-1,4-glucanase (EC 3.2.1.4), exo-β-1,4-glucanase (EC 3.2.1.91), and β-glucosidase (EC 3.2.1.21). Endo-β-1,4-glucanase and exo-β-1,4-glucanase cleave cellulose to cellodextrins and cellobiose, which are then degraded to glucose by β-glucosidase (141, 142). Endo and exo-β-1,4-glucanase (Table 4) have been reported in Monoglobus pectinilyticus (41), Caldicellulosiruptor kronotskyensis (108), Roseburia sp. (42), Eubacterium rectale group (42), Ruminococcus champanellensis (109), Ruminococcus bromii (110), Ruminiclostridium cellulolyticum (111), Paenibacillus sp. (143), and Phaeoacremonium minimum (112). The enzyme β-glucosidase (Table 4) has been reported in Bifidobacterium adolescentis (115), Bacteroides ovatus (116), Listeria innocua (117), Streptomyces venezuelae (118), Pyrococcus furiosus (119), Cellvibrio japonicus (114), Caldicellulosiruptor saccharolyticus (120), Microbispora bispora (121), Thermoanaerobacter brockii (122), Thermobifida fusca (103), Pseudomonas sp. (123), Monoglobus pectinilyticus (41), Ruminococcus champanellensis (109), and Ruminococcus bromii (110).
Processing to increase use of recalcitrant fibers
Our previous studies have already shown that physical processing technologies cannot significantly increase fiber degradability in monogastric animals (19, 144, 145). However, the utilization of enzymes added to feed is a promising method to improve fiber fermentability.
Enzymatic and chemical modification
Supplementation of cell wall degrading enzymes to improve feed efficiency for pigs gets more and more attention from the feed industry nowadays. Supplementation of cell wall degrading enzymes can remove side-chains of polysaccharides in plant cell wall, which make them more accessible to bacterial enzymes (145). In addition, cell wall degrading enzymes are able to reduce the digesta viscosity, which might affect absorption of other nutrients, by cleaving the viscous polysaccharides (e.g., pectin). In the end, carbohydrases can depolymerize polysaccharides to oligosaccharides, which have potential prebiotic effects on the gut microbiota, leading to the health benefits for the host animal.
A previous study showed that pre-processing with sulfuric acid could increase utilization of carbohydrates from lignocellulosic biomass (146). Mild acid-treated rye (together with heat treatment) was reported to improve the release of arabinosyl residues in chickens (147). Alkaline pretreatment of rapeseed meal in the feed improved its fermentation in pigs (7). Therefore, chemical treatment in feed might be a promising method to improve fiber fermentation in pigs.
We have shown recently that RSM processed by enzymatic and chemical treatment led to modulation of the gut microbiota using a newly developed in vitro swine large intestinal model (SLIM) (148). In brief, our studies demonstrated that both enzymatic (cellulase or 2 different pectinases) and chemical (6 N sodium hydroxide) pretreatment on RSM shifted its cell wall polysaccharide structure, subsequently altering microbial community composition and functional profile compared to untreated RSM, and eventually increased fiber degradability as evaluated by SCFA production in SLIM. Moreover, it was validated in pigs, by the mobile nylon bag technique, that cellulase and alkaline treatment on RSM improved the overall degradation of RSM (149–151).
Potential emerging technologies
High hydrostatic pressure, a non-thermal pasteurization technology, is applied to physical modification of chemical structure of compound (e.g., polysaccharides). High hydrostatic pressure could promote the water into interior of the matrix. As a result, high pressure (100–1,000 MPa) leads to the destruction of cell wall matrix (affecting non-covalent bonds, including hydrophobic interactions, hydrogen bonds, and van der Waals forces) and reduce substrate particle size. Research demonstrated that pectin (extracted from potato peel waste) could increase galacturonic acid content and decrease the esterification degree of pectin after high hydrostatic pressure at 200 MPa for 5 min (152). Another study reported that high hydrostatic pressure could significant reduce the molecular weight, degree of esterification and degree of acylation of sugar beet pectin at 250–500 MPa for 30 min (153). These reports have shown that high hydrostatic pressure could modify the polysaccharide structure, which might have a potential application value in improving fiber degradability of RSM in monogastric animals.
Cold plasma (CP) processing is a technology that causing the ionization of neutral gas (argon, helium, and nitrogen) and generating active species (free radicals, electrons, ions), which could induce oxidative damage in the cell wall compounds, such as polysaccharides. Therefore, CP could change structure and properties of polysaccharides. Study showed that CP could increase the viscosity and enhance the emulsion stabilization capacity of xanathan gum (154). Prasertsung et al. (155) reported that CP could degrade starches and cellulose into sugars and glucose (155). Therefore, CP might modify polysaccharides of RSM by inducing oxidative damage in its cell wall polysaccharides.
Conclusions and perspectives
Rapeseed meal is not only a promising protein ingredient but also a potentially energy source for non-ruminant animal diets. RSM contains a high amount of cell wall polysaccharides, even higher when compared to soybean meal commonly used in the feed industry. However, information on degradation and utilization of RSM carbohydrates upon feeding these diets is insufficient, and for this it is essential to understand the characterization and quantification of RSM carbohydrates. For the sake of improving the degradation of RSM carbohydrates, it is necessary to understand the mode of actions of RSM polysaccharide degradation by microbes, discover factors that limit their utilization and absorption in the GIT, and develop strategies to improve efficiency and productivity. The use of exogenous enzymes, carbohydrases, becomes a good solution to improve RSM carbohydrate degradation, and eventually to improve animal digestibility, feed efficiency, and animal performance.
The current review summarized the polysaccharide types of RSM, gut microbiota which could degrade the polysaccharides, and glycoside hydrolases which used by the microorganism. The information offered above could be used to develop novel engineering microbes and enzymic preparations to improve RSM utilization in the future studies. One possible way is to deep mine the genomes of theses microbes for encoded the carbohydrases by bioinformatic tools. What is more, these enzymes can be overexpressed in an expression host and subsequently used to process RSM to investigate its degradability in livestock.
Author contributions
CL prepared the original draft and edited the manuscript. X-LQ and KV critically reviewed the manuscript. All authors listed have approved the final manuscript for publication.
Funding
This research has been partly made possible with the support of the Dutch Province of Limburg, with a grant to the Centre for Healthy Eating and Food Innovation (HEFI) of Maastricht University—Campus Venlo to KV. This study was also supported by the Young Teachers' Research and Innovation Ability Improvement Program of Beijing University of Agriculture (QJKC2022026), Technology Innovation spark action support program of Beijing University of Agriculture, and 2022 Beijing Municipal Education Commission Classification Development Project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Pustjens AM, De Vries S, Bakuwel M, Gruppen H, Gerrits WJ, Kabel MA. Unfermented recalcitrant polysaccharide structures from rapeseed (Brassica napus) meal in pigs. Ind Crops Prod. (2014) 58:271–9. doi: 10.1016/j.indcrop.2014.04.036
2. Kiarie E, Romero LF, Nyachoti CM. The role of added feed enzymes in promoting gut health in swine and poultry. Nutr Res Rev. (2013) 26:71–88. doi: 10.1017/S0954422413000048
3. Leming R, Lember A. Chemical composition of expeller-extracted and cold-pressed rapeseed cake. Agraarteadus. (2005) 16:96–109.
4. Agyekum AK, Woyengo TA. Nutritive value of expeller/cold-pressed canola meal and pre-pressed solvent-extracted carinata meal for broiler chicken. Poult Sci. (2022) 101:101528. doi: 10.1016/j.psj.2021.101528
5. Slominski BA, Campbell LD. Non-starch polysaccharides of canola meal: quantification, digestibility in poultry and potential benefit of dietary enzyme supplementation. J Sci Food Agric. (1990) 53:175–84. doi: 10.1002/jsfa.2740530205
6. Simbaya J, Slominski BA, Rakow G, Campbell LD, Downey RK, Bell JM. Quality characteristics of yellow-seeded Brassica seed meals: protein, carbohydrate, and dietary fiber components. J Agric Food Chem. (1995) 43:2062–6. doi: 10.1021/jf00056a020
7. Pustjens AM, Schols HA, Kabel MA, Gruppen H. Characterisation of cell wall polysaccharides from rapeseed (Brassica napus) meal. Carbohydr Polym. (2013) 98:1650–6. doi: 10.1016/j.carbpol.2013.07.059
8. Slominski BA. Hydrolysis of galactooligosaccharides by commercial preparations of α-galactosidase and β-fruetofuranosidase: potential for use as dietary additives. J Sci Food Agric. (1994) 65:323–30. doi: 10.1002/jsfa.2740650310
9. Meng X, Slominski B. Nutritive values of corn, soybean meal, canola meal, and peas for broiler chickens as affected by a multicarbohydrase preparation of cell wall degrading enzymes. Poult Sci. (2005) 84:1242–51. doi: 10.1093/ps/84.8.1242
10. Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. (1993) 3:1–30. doi: 10.1111/j.1365-313X.1993.tb00007.x
12. Zykwinska A, Thibault JF, Ralet MC. Competitive binding of pectin and xyloglucan with primary cell wall cellulose. Carbohydr Polym. (2008) 74:957–61. doi: 10.1016/j.carbpol.2008.05.004
13. Pustjens AM. Fate of rapeseed meal polysaccharides during digestion in pigs and poultry: effect of processing and enzyme addition. In: Fiber Fermentation in Pigs And Poultry Sense and Nonsense of Its Manipulation. Wageningen: Wageningen University and Research (2013). p. 64.
14. Rommi K, Hakala TK, Holopainen U, Nordlund E, Poutanen K, Lantto R. Effect of enzyme-aided cell wall disintegration on protein extractability from intact and dehulled rapeseed (Brassica rapa L. and Brassica napus L) press cakes. J Agric Food Chem. (2014) 62:7989–97. doi: 10.1021/jf501802e
15. Bell J. Nutrients and toxicants in rapeseed meal: a review. J Anim Sci. (1984) 58:996–1010. doi: 10.2527/jas1984.584996x
16. Nair J, Penner GB, Yu P, Lardner H, Mcallister T, Damiran D, et al. Evaluation of canola meal derived from Brassica juncea and Brassica napus seed as an energy source for feedlot steers. Can J Anim Sci. (2015) 95:599–607. doi: 10.4141/cjas-2015-055
17. Slominski B, Campbell L. The carbohydrate content of yellow-seeded canola. In: Proceedings of the 8th International Rapeseed Congress. Sasakatoon (1991). p. 9–11.
18. Gallant D, Bouchet B, Buleon A, Perez S. Physical characteristics of starch granules and susceptibility to enzymatic degradation. Eur J Clin Nutr. (1992) 46:S3–16.
19. De Vries S, Pustjens A, Kabel M, Kwakkel R, Gerrits W. Effects of processing technologies and pectolytic enzymes on degradability of nonstarch polysaccharides from rapeseed meal in broilers. Poult Sci. (2014) 93:589–98. doi: 10.3382/ps.2013-03476
20. Ghosh P, Ghosal P, Thakur S, Lerouge P, Loutelier-Bourhis C, Driouich A, et al. Cell wall polysaccharides of Brassica campestris seed cake: isolation and structural features. Carbohydr Polym. (2004) 57:7–13. doi: 10.1016/j.carbpol.2004.01.012
21. Colquhoun IJ, De Ruiter GA, Schols HA, Voragen AG. Identification by NMR spectroscopy of oligosaccharides obtained by treatment of the hairy regions of apple pectin with rhamnogalacturonase. Carbohydr Res. (1990) 206:131–44. doi: 10.1016/0008-6215(90)84012-J
22. Ebringerová A, Heinze T. Xylan and xylan derivatives-biopolymers with valuable properties, 1. Naturally occurring xylans structures, isolation procedures and properties. Macromol Rapid Commun. (2000) 21:542–56. doi: 10.1002/1521-3927(20000601)21:9<542::AID-MARC542>3.0.CO;2-7
23. Peña MJ, Zhong R, Zhou G-K, Richardson EA, O'neill MA, Darvill AG, et al. Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell. (2007) 19:549–63. doi: 10.1105/tpc.106.049320
24. Larsbrink J, Rogers TE, Hemsworth GR, Mckee LS, Tauzin AS, Spadiut O, et al. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature. (2014) 506:498–502. doi: 10.1038/nature12907
25. Kračun SK, Fangel JU, Rydahl MG, Pedersen HL, Vidal-Melgosa S, Willats WGT. Carbohydrate microarray technology applied to high-throughput mapping of plant cell wall glycans using comprehensive microarray polymer profiling (CoMPP). High-throughput glycomics and glycoproteomics. Methods Protoc. (2017) 147–65. doi: 10.1007/978-1-4939-6493-2_12
26. Forsee WT, Springfield JD, Schutzbach J. Effect of phospholipids on alpha-1, 2-mannosidase activity. J Biol Chem. (1982) 257:9963–7. doi: 10.1016/S0021-9258(18)33971-1
27. Hilz H, Bakx EJ, Schols HA, Voragen AG. Cell wall polysaccharides in black currants and bilberries-characterisation in berries, juice, and press cake. Carbohydr Polym. (2005) 59:477–88. doi: 10.1016/j.carbpol.2004.11.002
28. Flint HJ, Bayer EA. Plant cell wall breakdown by anaerobic microorganisms from the mammalian digestive tract. Ann N Y Acad Sci. (2008) 1125:280–8. doi: 10.1196/annals.1419.022
29. Van Den Brink J, De Vries RP. Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol. (2011) 91:1477. doi: 10.1007/s00253-011-3473-2
30. Crouch LI, Labourel A, Walton PH, Davies GJ, Gilbert HJ. The contribution of non-catalytic carbohydrate binding modules to the activity of lytic polysaccharide monooxygenases. J Biol Chem. (2016) 291:7439–49. doi: 10.1074/jbc.M115.702365
31. Cartmell A, Mckee LS, Pena MJ, Larsbrink J, Brumer H, Kaneko S, et al. The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases. J Biol Chem. (2011) 286:15483–95. doi: 10.1074/jbc.M110.215962
32. Raposo MP, Inacio JM, Mota LJ, De Sa-Nogueira I. Transcriptional regulation of genes encoding arabinan-degrading enzymes in Bacillus subtilis. J Bacteriol Res. (2004) 186:1287–96. doi: 10.1128/JB.186.5.1287-1296.2004
33. Leal TF, De Sa-Nogueira I. Purification, characterization and functional analysis of an endo-arabinanase (AbnA) from Bacillus subtilis. FEMS Microbiol Lett. (2004) 241:41–8. doi: 10.1016/j.femsle.2004.10.003
34. Sá-Nogueira I, Ramos SS. Cloning, functional analysis, and transcriptional regulation of the Bacillus subtilis araE gene involved in L-arabinose utilization. J Bacteriol Res. (1997) 179:7705–11. doi: 10.1128/jb.179.24.7705-7711.1997
35. Sá-Nogueira I, Nogueira TV, Soares S, De Lencastre H. The Bacillus subtilis L-arabinose (ara) operon: nucleotide sequence, genetic organization and expression. Microbiology. (1997) 143:957–69. doi: 10.1099/00221287-143-3-957
36. Inacio JM, De Sa-Nogueira I. Characterization of abn2 (yxiA), encoding a Bacillus subtilis GH43 arabinanase, Abn2, and its role in arabino-polysaccharide degradation. J Bacteriol. (2008) 190:4272–80. doi: 10.1128/JB.00162-08
37. Komeno M, Hayamizu H, Fujita K, Ashida H. Two novel α-l-Arabinofuranosidases from Bifidobacterium longum subsp. longum belonging to glycoside hydrolase family 43 cooperatively degrade arabinan. Appl Environ Microbiol. (2019) 85:e02582–18. doi: 10.1128/AEM.02582-18
38. Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA. (2009) 106:5859–64. doi: 10.1073/pnas.0901529106
39. Morana A, Paris O, Maurelli L, Rossi M, Cannio R. Gene cloning and expression in Escherichia coli of a bi-functional β-D-xylosidase/α-L-arabinosidase from Sulfolobus solfataricus involved in xylan degradation. Extremophiles. (2007) 11:123–32. doi: 10.1007/s00792-006-0020-7
40. Canakci S, Kacagan M, Inan K, Belduz AO, Saha BC. Cloning, purification, and characterization of a thermostable α-L-arabinofuranosidase from Anoxybacillus kestanbolensis AC26Sari. Appl Microbiol Biotechnol. (2008) 81:61. doi: 10.1007/s00253-008-1584-1
41. Kim CC, Healey GR, Kelly WJ, Patchett ML, Jordens Z, Tannock GW, et al. Genomic insights from Monoglobus pectinilyticus: a pectin-degrading specialist bacterium in the human colon. ISME J. (2019) 13:1437–56. doi: 10.1038/s41396-019-0363-6
42. Sheridan PO, Martin JC, Lawley TD, Browne HP, Harris HM, Bernalier-Donadille A, et al. Polysaccharide utilization loci and nutritional specialization in a dominant group of butyrate-producing human colonic Firmicutes. Microbial Genom. (2016) 2:e000043. doi: 10.1099/mgen.0.000043
43. Wang S, Yang Y, Zhang J, Sun J, Matsukawa S, Xie J, et al. Characterization of abnZ2 (yxiA1) and abnZ3 (yxiA3) in Paenibacillus polymyxa, encoding two novel endo-1,5-α-l-arabinanases. Bioresourc Bioprocess. (2014) 1:14. doi: 10.1186/s40643-014-0014-8
44. Seo E-S, Lim Y-R, Kim Y-S, Park C-S, Oh D-K. Characterization of a recombinant endo-1, 5-α-l-arabinanase from the isolated bacterium Bacillus licheniformis. Biotechnol Bioprocess Eng. (2010) 15:590–4. doi: 10.1007/s12257-009-3138-5
45. Hong M-R, Park C-S, Oh D-K. Characterization of a thermostable endo-1, 5-α-L-arabinanase from Caldicellulorsiruptor saccharolyticus. Biotechnol Lett. (2009) 31:1439. doi: 10.1007/s10529-009-0019-0
46. Takao M, Akiyama K, Sakai T. Purification and characterization of thermostable endo-1, 5-α-L-arabinase from a strain of Bacillus thermodenitrificans. Appl Environ Microbiol. (2002) 68:1639–46. doi: 10.1128/AEM.68.4.1639-1646.2002
47. Hogg D, Woo E-J, Bolam DN, Mckie VA, Gilbert HJ, Pickersgill RW. Crystal structure of mannanase 26A from Pseudomonas cellulosa and analysis of residues involved in substrate binding. J Biol Chem. (2001) 276:31186–92. doi: 10.1074/jbc.M010290200
48. Ademark P, De Vries RP, Hägglund P, Stålbrand H, Visser J. Cloning and characterization of Aspergillus niger genes encoding an α-galactosidase and a β-mannosidase involved in galactomannan degradation. Eur J Biochem. (2001) 268:2982–90. doi: 10.1046/j.1432-1327.2001.02188.x
49. Liu H-X, Gong J-S, Li H, Lu Z-M, Li H, Qian J-Y, et al. Biochemical characterization and cloning of an endo-1, 4-β-mannanase from Bacillus subtilis YH12 with unusually broad substrate profile. Process Biochem. (2015) 50:712–21. doi: 10.1016/j.procbio.2015.02.011
50. Li C-Y, Liu F-F, Ye J, Liu J-F, Yang S-Z, Zhang H-Z, et al. A low-temperature active endo-β-1, 4-mannanase from Bacillus subtilis TD7 and its gene expression in Escherichia coli. Appl Environ Biotechnol. (2018) 3:17–25. doi: 10.26789/AEB.2018.02.004
51. Huang JL, Bao LX, Zou HY, Che SG, Wang GX. High-level production of a cold-active B-mannanase from Bacillus subtilis Bs5 and its molecular cloning and expression. Mol Genet Microbiol Virol. (2012) 27:147–53. doi: 10.3103/S0891416812040039
52. Songsiriritthigul C, Buranabanyat B, Haltrich D, Yamabhai M. Efficient recombinant expression and secretion of a thermostable GH26 mannan endo-1, 4-β-mannosidase from Bacillus licheniformis in Escherichia coli. Microb Cell Factories. (2010) 9:20. doi: 10.1186/1475-2859-9-20
53. Zhou J, Zhang R, Gao Y, Li J, Tang X, Mu Y, et al. Novel low-temperature-active, salt-tolerant and proteases-resistant endo-1, 4-β-mannanase from a new Sphingomonas strain. J Biosci Bioeng. (2012) 113:568–74. doi: 10.1016/j.jbiosc.2011.12.011
54. Zhang R, Zhou J, Gao Y, Guan Y, Li J, Tang X, et al. Molecular and biochemical characterizations of a new low-temperature active mannanase. Folia Microbiol. (2015) 60:483–92. doi: 10.1007/s12223-015-0391-1
55. Pongsapipatana N, Damrongteerapap P, Chantorn S, Sintuprapa W, Keawsompong S, Nitisinprasert S. Molecular cloning of kman coding for mannanase from Klebsiella oxytoca KUB-CW2-3 and its hybrid mannanase characters. Enzyme Microb Technol. (2016) 89:39–51. doi: 10.1016/j.enzmictec.2016.03.005
56. You J, Liu J-F, Yang S-Z, Mu B-Z. Low-temperature-active and salt-tolerant β-mannanase from a newly isolated Enterobacter sp. strain N18. J Biosci Bioeng. (2016) 121:140–6. doi: 10.1016/j.jbiosc.2015.06.001
57. Zakaria MM, Ashiuchi M, Yamamoto S, Yagi T. Optimization for β-mannanase production of a psychrophilic bacterium, Flavobacterium sp. Biosci Biotechnol Biochem. (1998) 62:655–60. doi: 10.1271/bbb.62.655
58. Reddy SK, Bågenholm V, Pudlo NA, Bouraoui H, Koropatkin NM, Martens EC, et al. A β-mannan utilization locus in Bacteroides ovatus involves a GH36 α-galactosidase active on galactomannans. FEBS Lett. (2016) 590:2106–18. doi: 10.1002/1873-3468.12250
59. Cordeiro RL, Pirolla RaS, Persinoti GF, Gozzo FC, De Giuseppe PO, Murakami MT. N-glycan utilization by bifidobacterium gut symbionts involves a specialist β-mannosidase. J Mol Biol. (2019) 431:732–47. doi: 10.1016/j.jmb.2018.12.017
60. Sonnenburg JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. (2006) 4:e413. doi: 10.1371/journal.pbio.0040413
61. Krishnaswamyreddy S. galactmannan Degradation by Fungi and Gut Bacteria: Structural Enzymology and Fine-Tuned Substrate Specifcity. Lund: Lund University (2016).
62. Bågenholm V, Reddy SK, Bouraoui H, Morrill J, Kulcinskaja E, Bahr CM, et al. Galactomannan catabolism conferred by a polysaccharide utilization locus of Bacteroides ovatus enzyme synergy and crystal structure of a β-mannanase J Biol Chem. (2017) 292:229–43. doi: 10.1074/jbc.M116.746438
63. Dias FM, Vincent F, Pell G, Prates JA, Centeno MS, Tailford LE, et al. Insights into the molecular determinants of substrate specificity in glycoside hydrolase family 5 revealed by the crystal structure and kinetics of Cellvibrio mixtus mannosidase 5A. J Biol Chem. (2004) 279:25517–26. doi: 10.1074/jbc.M401647200
64. Tailford LE, Money VA, Smith NL, Dumon C, Davies GJ, Gilbert HJ. Mannose foraging by Bacteroides thetaiotaomicron structure and specificity of the β-mannosidase, BtMan2A. J Biol Chem. (2007) 282:11291–9. doi: 10.1074/jbc.M610964200
65. Rahmani N, Kashiwagi N, Lee J, Niimi-Nakamura S, Matsumoto H, Kahar P, et al. Mannan endo-1,4-β-mannosidase from Kitasatospora sp. isolated in Indonesia and its potential for production of mannooligosaccharides from mannan polymers. AMB Express. (2017) 7:100. doi: 10.1186/s13568-017-0401-6
66. Hachem MA, Fredslund F, Andersen JM, Jonsgaard Larsen R, Majumder A, Ejby M, et al. Raffinose family oligosaccharide utilisation by probiotic bacteria: insight into substrate recognition, molecular architecture and diversity of GH36 α-galactosidases. Biocatal Biotransformat. (2012) 30:316–25. doi: 10.3109/10242422.2012.674717
67. Ojala T, Kuparinen V, Koskinen JP, Alatalo E, Holm L, Auvinen P, et al. Genome sequence of Lactobacillus crispatus ST1. J Bacteriol Res. (2010) 192:3547–8. doi: 10.1128/JB.00399-10
68. Comfort DA, Bobrov KS, Ivanen DR, Shabalin KA, Harris JM, Kulminskaya AA, et al. Biochemical analysis of Thermotoga maritima GH36 α-galactosidase (Tm GalA) confirms the mechanistic commonality of clan GH-D glycoside hydrolases. Biochemistry. (2007) 46:3319–30. doi: 10.1021/bi061521n
69. Skalova T, Dohnálek J, Spiwok V, Lipovová P, Vondráčková E, Petroková H, et al. Cold-active β-galactosidase from Arthrobacter sp. C2-2 forms compact 660 kDa hexamers: crystal structure at 19 Å resolution. J Mol Biol. (2005) 353:282–94. doi: 10.1016/j.jmb.2005.08.028
70. Huang Y, Zhang H, Ben P, Duan Y, Lu M, Li Z, et al. Characterization of a novel GH36 α-galactosidase from Bacillus megaterium and its application in degradation of raffinose family oligosaccharides. Int J Biol Macromol. (2018) 108:98–104. doi: 10.1016/j.ijbiomac.2017.11.154
71. Schröder C, Janzer V-A, Schirrmacher G, Claren J, Antranikian G. Characterization of two novel heat-active α-galactosidases from thermophilic bacteria. Extremophiles. (2017) 21:85–94. doi: 10.1007/s00792-016-0885-z
72. Gote M, Khan M, Gokhale D, Bastawde K, Khire J. Purification, characterization and substrate specificity of thermostable α-galactosidase from Bacillus stearothermophilus (NCIM-5146). Process Biochem. (2006) 41:1311–7. doi: 10.1016/j.procbio.2006.01.003
73. Lisowska B. Genomic Analysis and Metabolic Modelling of Geobacillus thermoglucosidasius NCIMB 11955. Bath: University of Bath (2016).
74. Gherardini F, Babcock M, Salyers AA. Purification and characterization of two alpha-galactosidases associated with catabolism of guar gum and other alpha-galactosides by Bacteroides ovatus. J Bacteriol Res. (1985) 161:500–6. doi: 10.1128/jb.161.2.500-506.1985
75. Goulas T, Goulas A, Tzortzis G, Gibson GR. A novel α-galactosidase from ?ifidobacterium bifidum with transgalactosylating properties: gene molecular cloning and heterologous expression. Appl Microbiol Biotechnol. (2009) 82:471–7. doi: 10.1007/s00253-008-1750-5
76. Van Laere K, Hartemink R, Beldman G, Pitson S, Dijkema C, Schols H, et al. Transglycosidase activity of Bifidobacterium adolescentis DSM 20083 α-galactosidase. Appl Microbiol Biotechnol. (1999) 52:681–8. doi: 10.1007/s002530051579
77. Zhao H, Lu L, Xiao M, Wang Q, Lu Y, Liu C, et al. Cloning and characterization of a novel α-galactosidase from Bifidobacterium breve 203 capable of synthesizing Gal-α-1, 4 linkage. FEMS Microbiol Lett285. (2008) 278–83. doi: 10.1111/j.1574-6968.2008.01246.x
78. Kimura T, Sakka K, Ohmiya K. Cloning, sequencing, and expression of the gene encoding the Clostridium stercorarium α-Galactosidase Aga36A in Escherichia coli. Biosci Biotechnol Biochem. (2003) 67:2160–6. doi: 10.1271/bbb.67.2160
79. Ademark P, Lundqvist J, Hägglund P, Tenkanen M, Torto N, Tjerneld F, et al. Hydrolytic properties of a β-mannosidase purified from Aspergillus niger. J Biotechnol. (1999) 75:281–9. doi: 10.1016/S0168-1656(99)00172-8
80. De Vries RP, Van Den Broeck HC, Dekkers E, Manzanares P, De Graaff LH, Visser J. Differential expression of three α-galactosidase genes and a single β-galactosidase gene from Aspergillus niger. Appl Environ Microbiol. (1999) 65:2453–60. doi: 10.1128/AEM.65.6.2453-2460.1999
81. Hamouda HI, Ali N, Su H, Feng J, Lu M, Li F-L. Exploration of two pectate lyases from Caldicellulosiruptor bescii reveals that the CBM66 module has a crucial role in pectic biomass degradation. Appl Environ Microbiol. (2020) 86:e00787–20. doi: 10.1128/AEM.00787-20
82. Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, Mcnulty NP, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. (2011) 9:e1001221. doi: 10.1371/journal.pbio.1001221
83. Luis AS, Martens EC. Interrogating gut bacterial genomes for discovery of novel carbohydrate degrading enzymes. Curr Opin Chem Biol. (2018) 47:126–33. doi: 10.1016/j.cbpa.2018.09.012
84. Klug-Santner BG, Schnitzhofer W, Vršanská M, Weber J, Agrawal PB, Nierstrasz VA, et al. Purification and characterization of a new bioscouring pectate lyase from Bacillus pumilus BK2. J Biotechnol. (2006) 121:390–401. doi: 10.1016/j.jbiotec.2005.07.019
85. Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJ, Garcia-Gil LJ, Flint HJ. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl Environ Microbiol. (2012) 78:420–8. doi: 10.1128/AEM.06858-11
86. Hugouvieux-Cotte-Pattat N, Condemine G, Shevchik VE. Bacterial pectate lyases, structural and functional diversity. Environ Microbiol Rep. (2014) 6:427–40. doi: 10.1111/1758-2229.12166
87. Bekli S, Aktas B, Gencer D, Aslim B. Biochemical and molecular characterizations of a novel pH-and temperature-stable pectate lyase from Bacillus amyloliquefaciens S6 for industrial application. Mol Biotechnol. (2019) 61:681–93. doi: 10.1007/s12033-019-00194-2
88. Yaniv O, Jindou S, Frolow F, Lamed R, Bayer EA. A simple method for determining specificity of carbohydrate-binding modules for purified and crude insoluble polysaccharide substrates. In: Biomass Conversion. Totowa, NJ: Humana Press (2012). p. 101–7.
89. Tamaru Y, Doi RH. Pectate lyase A, an enzymatic subunit of the Clostridium cellulovorans cellulosome. PNAS. (2001) 98:4125–9. doi: 10.1073/pnas.071045598
90. Tonouchi A, Hara Y, Umehara R, Sanuki T, Fukusawa T, Miyairi K. Cloning of the gene encoding an endo-acting pectate lyase from Streptomyces thermocarboxydus. Biosci Biotechnol Biochem. (2010) 74, 433–6. doi: 10.1271/bbb.90693
91. Fujita K, Sakaguchi T, Sakamoto A, Shimokawa M, Kitahara K. Bifidobacterium longum subsp. longum exo-β-1, 3-galactanase, an enzyme for the degradation of type II arabinogalactan. Appl Environ Microbiol. (2014) 80:4577–84. doi: 10.1128/AEM.00802-14
92. Ochiai A, Itoh T, Kawamata A, Hashimoto W, Murata K. Plant cell wall degradation by saprophytic Bacillus subtilis strains: gene clusters responsible for rhamnogalacturonan depolymerization. Appl Environ Microbiol. (2007) 73:3803–13. doi: 10.1128/AEM.00147-07
93. Silva IR, Larsen DM, Meyer AS, Mikkelsen JD. Identification, expression, and characterization of a novel bacterial RGI Lyase enzyme for the production of bio-functional fibers. Enzyme Microb Technol. (2011) 49:160–6. doi: 10.1016/j.enzmictec.2011.04.015
94. Mckie VA, Vincken J-P, Voragen AG, Van Den Broek LA, Stimson E, Gilbert HJ. A new family of rhamnogalacturonan lyases contains an enzyme that binds to cellulose. Biochem J. (2001) 355:167–77. doi: 10.1042/bj3550167
95. Pagès S, Valette O, Abdou L, Bélaïch A, Bélaïch J-P. A rhamnogalacturonan lyase in the Clostridium cellulolyticum cellulosome. J Bacteriol Res. (2003) 185:4727–33. doi: 10.1128/JB.185.16.4727-4733.2003
96. Kunishige Y, Iwai M, Nakazawa M, Ueda M, Tada T, Nishimura S, et al. Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26. FEBS Lett. (2018) 592:1378–88. doi: 10.1002/1873-3468.13034
97. Ichinose H, Kuno A, Kotake T, Yoshida M, Sakka K, Hirabayashi J, et al. Characterization of an exo-β-1, 3-galactanase from Clostridium thermocellum. Appl Environ Microbiol. (2006) 72:3515–23. doi: 10.1128/AEM.72.5.3515-3523.2006
98. Ichinose H, Yoshida M, Kotake T, Kuno A, Igarashi K, Tsumuraya Y, et al. An exo-β-1, 3-galactanase having a novel β-1, 3-galactan-binding module from Phanerochaete chrysosporium. J Biol Chem. (2005) 280:25820–9. doi: 10.1074/jbc.M501024200
99. Sakamoto T, Tanaka H, Nishimura Y, Ishimaru M, Kasai N. Characterization of an exo-β-1, 3-d-galactanase from Sphingomonas sp. 24T and its application to structural analysis of larch wood arabinogalactan. Appl Microbiol Biotechnol. (2011) 90:1701–10. doi: 10.1007/s00253-011-3219-1
100. Fujita K, Sasaki Y, Kitahara K. Degradation of plant arabinogalactan proteins by intestinal bacteria: characteristics and functions of the enzymes involved. Appl Microbiol Biotechnol. (2019) 103:7451–7. doi: 10.1007/s00253-019-10049-0
101. Cartmell A, Muñoz-Muñoz J, Briggs JA, Ndeh DA, Lowe EC, Baslé A, et al. A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation. Nat Microbiol. (2018) 3:1314–26. doi: 10.1038/s41564-018-0258-8
102. Ichinose H, Kotake T, Tsumuraya Y, Kaneko S. Characterization of an endo-β-1, 6-galactanase from Streptomyces avermitilis NBRC14893. Appl Environ Microbiol. (2008) 74:2379–83. doi: 10.1128/AEM.01733-07
103. Chen S, Wilson DB. Proteomic and transcriptomic analysis of extracellular proteins and mRNA levels in Thermobifida fusca grown on cellobiose and glucose. J Bacteriol Res. (2007) 189:6260–5. doi: 10.1128/JB.00584-07
104. Sharma K, Fontes CM, Najmudin S, Goyal A. Molecular organization and protein stability of the Clostridium thermocellum glucuronoxylan endo-β-1, 4-xylanase of family 30 glycoside hydrolase in solution. J Struct Biol. (2019) 206:335–44. doi: 10.1016/j.jsb.2019.04.005
105. St. John FJ, Rice JD, Preston JF. Characterization of XynC from Bacillus subtilis subsp. subtilis strain 168 and analysis of its role in depolymerization of glucuronoxylan. J Bacteriol Res. (2006) 188:8617–26. doi: 10.1128/JB.01283-06
106. Maehara T, Yagi H, Sato T, Ohnishi-Kameyama M, Fujimoto Z, Kamino K, et al. GH30 glucuronoxylan-specific xylanase from Streptomyces turgidiscabies C56. Appl Environ Microbiol. (2018) 84:e01850–17. doi: 10.1128/AEM.01850-17
107. Lee SH, Lee MJ, Cho H-Y, Lee JS, Rhee YH, Shin D-H, et al. Genetic and functional characterization of a novel GH10 endo-β-1, 4-xylanase with a ricin-type β-trefoil domain-like domain from Luteimicrobium xylanilyticum HY-24. Int J Biol Macromol. (2018) 106:620–8. doi: 10.1016/j.ijbiomac.2017.08.063
108. Conway JM, Pierce WS, Le JH, Harper GW, Wright JH, Tucker AL, et al. Multidomain, surface layer-associated glycoside hydrolases contribute to plant polysaccharide degradation by Caldicellulosiruptor species. J Biol Chem. (2016) 291:6732–47. doi: 10.1074/jbc.M115.707810
109. Moraïs S, David YB, Bensoussan L, Duncan SH, Koropatkin NM, Martens EC, et al. Enzymatic profiling of cellulosomal enzymes from the human gut bacterium, R uminococcus champanellensis, reveals a fine-tuned system for cohesin-dockerin recognition. Environ Microbiol. (2016) 18:542–56. doi: 10.1111/1462-2920.13047
110. Ze X, David YB, Laverde-Gomez JA, Dassa B, Sheridan PO, Duncan SH, et al. Unique organization of extracellular amylases into amylosomes in the resistant starch-utilizing human colonic Firmicutes bacterium Ruminococcus bromii. MBio. (2015) 6:e01058–15. doi: 10.1128/mBio.01058-15
111. Ravachol J, De Philip P, Borne R, Mansuelle P, Maté MJ, Perret S, et al. Mechanisms involved in xyloglucan catabolism by the cellulosome-producing bacterium Ruminiclostridium cellulolyticum. Sci Rep. (2016) 6:1–17. doi: 10.1038/srep22770
112. Matsuzawa T, Kameyama A, Yaoi K. A novel isoprimeverose-producing enzyme from Phaeoacremonium minimum is active with low concentrations of xyloglucan oligosaccharides. FEBS Open Bio. (2019) 9:92–100. doi: 10.1002/2211-5463.12549
113. Guerfali M, Gargouri A, Belghith H. Catalytic properties of Talaromyces thermophilus α-l-arabinofuranosidase and its synergistic action with immobilized endo-β-1, 4-xylanase. J Mol Catal. (2011) 68:192–9. doi: 10.1016/j.molcatb.2010.11.003
114. Larsbrink J, Izumi A, Ibatullin FM, Nakhai A, Gilbert HJ, Davies GJ, et al. Structural enzymatic characterization of a glycoside hydrolase family 31 α-xylosidase from Cellvibrio japonicus involved in xyloglucan saccharification. Biochem J. (2011) 436:567–80. doi: 10.1042/BJ20110299
115. Florindo RN, Souza VP, Manzine LR, Camilo CM, Marana SR, Polikarpov I, et al. Structural and biochemical characterization of a GH3 β-glucosidase from the probiotic bacteria Bifidobacterium adolescentis. Biochimie. (2018) 148:107–15. doi: 10.1016/j.biochi.2018.03.007
116. Hemsworth GR, Thompson AJ, Stepper J, Sobala ŁF, Coyle T, Larsbrink J, et al. Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut. Open Bio. (2016) 6:160142. doi: 10.1098/rsob.160142
117. Nakajima M, Yoshida R, Miyanaga A, Abe K, Takahashi Y, Sugimoto N, et al. Functional and structural analysis of a β-glucosidase involved in β-1, 2-glucan metabolism in Listeria innocua. PLoS ONE. (2016) 11:e0148870. doi: 10.1371/journal.pone.0148870
118. Zmudka MW, Thoden JB, Holden HM. The structure of DesR from Streptomyces venezuelae, a β-glucosidase involved in macrolide activation. Protein Sci. (2013) 22:883–92. doi: 10.1002/pro.2204
119. Cota J, Corrêa TL, Damasio AR, Diogo JA, Hoffmam ZB, Garcia W, et al. Comparative analysis of three hyperthermophilic GH1 and GH3 family members with industrial potential. N Biotechnol. (2015) 32:13–20. doi: 10.1016/j.nbt.2014.07.009
120. Hong M-R, Kim Y-S, Park C-S, Lee J-K, Kim Y-S, Oh D-K. Characterization of a recombinant β-glucosidase from the thermophilic bacterium Caldicellulosiruptor saccharolyticus. J Biosci Bioeng. (2009) 108:36–40. doi: 10.1016/j.jbiosc.2009.02.014
121. Wright RM, Yablonsky MD, Shalita ZP, Goyal A, Eveleigh D. Cloning, characterization, and nucleotide sequence of a gene encoding Microbispora bispora BglB, a thermostable beta-glucosidase expressed in Escherichia coli. Appl Environ Microbiol. (1992) 58:3455–65. doi: 10.1128/aem.58.11.3455-3465.1992
122. Breves R, Bronnenmeier K, Wild N, Lottspeich F, Staudenbauer WL, Hofemeister J. Genes encoding two different beta-glucosidases of Thermoanaerobacter brockii are clustered in a common operon. Appl Environ Microbiol. (1997) 63:3902–10. doi: 10.1128/aem.63.10.3902-3910.1997
123. Mckie VA, Black GW, Millward-Sadler SJ, Hazlewood GP, Laurie JI, Gilbert HJ. Arabinanase A from Pseudomonas fluorescens subsp. cellulosa exhibits both an endo-and an exo-mode of action. Biochem J. (1997) 323:547–55. doi: 10.1042/bj3230547
124. Nagel CW, Vaughn RH. The degradation of oligogalacturonides by the polygalacturonase of Bacillus polymyxa. Arch Biochem Biophys. (1961) 94:328–32. doi: 10.1016/0003-9861(61)90047-9
125. Edstrom R, Phaff H. Purification and certain properties of pectin trans= eliminase from Aspergihs fonsecaeus. J Biol Chem. (1963) 239:2403–8. doi: 10.1016/S0021-9258(18)93866-4
126. Nasuno S, Starr M. Polygalacturonic acid trans-eliminase of Xanthomonas campestris. Biochem J. (1967) 104:178. doi: 10.1042/bj1040178
127. Mayans O, Scott M, Connerton I, Gravesen T, Benen J, Visser J, et al. Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases. Structure. (1997) 5:677–89. doi: 10.1016/S0969-2126(97)00222-0
128. Shevchik VE, Kester HC, Benen JA, Visser J, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Characterization of the exopolygalacturonate lyase PelX of Erwinia chrysanthemi 3937. J Bacteriol Res. (1999) 181:1652–63. doi: 10.1128/JB.181.5.1652-1663.1999
129. Mutter M, Beldman G, Schols HA, Voragen A. Rhamnogalacturonan [alpha]-L-rhamnopyranohydrolase (a novel enzyme specific for the terminal nonreducing rhamnosyl unit in rhamnogalacturonan regions of pectin). Plant Physiol. (1994) 106:241–50. doi: 10.1104/pp.106.1.241
130. Mutter M, Colquhoun IJ, Schols HA, Beldman G, Voragen A. Rhamnogalacturonase B from Aspergillus aculeatus is a rhamnogalacturonan [alpha]-L-rhamnopyranosyl-(1->4)-[alpha]-D-galactopyranosyluronide lyase. Plant Physiol. (1996) 110:73–7. doi: 10.1104/pp.110.1.73
131. Fujita K, Takashi Y, Obuchi E, Kitahara K, Suganuma T. Characterization of a novel β-l-Arabinofuranosidase in Bifidobacterium longum functional elucidation of a DUF1680 protein family member. J Biol Chem. (2014) 289:5240–9. doi: 10.1074/jbc.M113.528711
132. St John FJ, González JM, Pozharski E. Consolidation of glycosyl hydrolase family 30: a dual domain 4/7 hydrolase family consisting of two structurally distinct groups. FEBS Lett. (2010) 584:4435–41. doi: 10.1016/j.febslet.2010.09.051
133. Aspeborg H, Coutinho PM, Wang Y, Brumer H, Henrissat B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol Biol. (2012) 12:186. doi: 10.1186/1471-2148-12-186
134. Fujita K, Sakamoto A, Kaneko S, Kotake T, Tsumuraya Y, Kitahara K. Degradative enzymes for type II arabinogalactan side chains in Bifidobacterium longum subsp. longum. Appl Microbiol Biotechnol. (2019) 103:1299–310. doi: 10.1007/s00253-018-9566-4
135. Takenishi S, Tsujisaka Y. On the modes of action of three xylanases produced by a strain of Aspergillus niger van Tieghem. Agric Biol Chem. (1975) 39:2315–23. doi: 10.1271/bbb1961.39.2315
136. Comtat J, Joseleau J-P. Mode of action of a xylanase and its significance for the structural investigation of the branched L-arabino-D-glucurono-D-xylan from redwood (Sequoia sempervirens). Carbohydr Res. (1981) 95:101–12. doi: 10.1016/S0008-6215(00)85299-4
137. Nishitani K, Nevins D. Glucuronoxylan xylanohydrolase. A unique xylanase with the requirement for appendant glucuronosyl units. J Biol Chem. (1991) 266:6539–43. doi: 10.1016/S0021-9258(18)38151-1
138. Koyama T, Hayashi T, Kato Y, Matsuda K. Degradation of xyloglucan by wall-bound enzymes from soybean tissue I. Occurrence of xyloglucan-degrading enzymes in soybean cell wall. Plant Cell Physiol. (1981) 22:1191–8.
139. Koyama T, Hayashi T, Kato Y, Matsuda K. Degradation of xyloglucan by wall-bound enzymes from soybean tissue II: degradation of the fragment heptasaccharide from xyloglucan and the characteristic action pattern of the α-D-xylosidase in the enzyme system. Plant Cell Physiol. (1983) 24:155–62. doi: 10.1093/pcp/24.2.155
140. Hayashi T. Xyloglucans in the primary cell wall. Annu Rev Plant Biol. (1989) 40:139–68. doi: 10.1146/annurev.pp.40.060189.001035
141. Béguin P, Aubert J-P. The biological degradation of cellulose. FEMS Microbiol Rev. (1994) 13:25–58. doi: 10.1111/j.1574-6976.1994.tb00033.x
142. Teeri TT. Crystalline cellulose degradation: new insight into the function of cellobiohydrolases. Trends Biotechnol. (1997) 15:160–7. doi: 10.1016/S0167-7799(97)01032-9
143. Adlakha N, Sawant S, Anil A, Lali A, Yazdani SS. Specific fusion of β-1, 4-endoglucanase and β-1, 4-glucosidase enhances cellulolytic activity and helps in channeling of intermediates. Appl Environ Microbiol. (2012) 78:7447–54. doi: 10.1128/AEM.01386-12
144. De Vries S, Pustjens A, Schols H, Hendriks W, Gerrits W. Improving digestive utilization of fiber-rich feedstuffs in pigs and poultry by processing and enzyme technologies: a review. Anim Feed Sci Technol. (2012) 178:123–38. doi: 10.1016/j.anifeedsci.2012.10.004
145. De Vries S, Pustjens AM, Kabel MA, Salazar-Villanea S, Hendriks WH, Gerrits WJ. Processing technologies and cell wall degrading enzymes to improve nutritional value of dried distillers grain with solubles for animal feed: an in vitro digestion study. J Agric Food Chem. (2013) 61:8821–8. doi: 10.1021/jf4019855
146. Liu Z-S, Wu X-L, Kida K, Tang Y-Q. Corn stover saccharification with concentrated sulfuric acid: effects of saccharification conditions on sugar recovery and by-product generation. Bioresour Technol. (2012) 119:224–33. doi: 10.1016/j.biortech.2012.05.107
147. Teitge D, Campbell G, Classen H, Thacker P. Heat pretreatment as a means of improving the response to dietary pentosanase in chicks fed rye. Can J Anim Sci. (1991) 71:507–13. doi: 10.4141/cjas91-060
148. Long C, De Vries S, Venema K. Polysaccharide source altered ecological network, functional profile, and short-chain fatty acid production in a porcine gut microbiota. Benef Microbes. (2020) 11:591–610. doi: 10.3920/BM2020.0006
149. Long C, De Vries S, Venema K. Differently pre-treated rapeseed meals affect in vitro swine gut microbiota composition. Front Microbiol. (2020) 11:570985. doi: 10.3389/fmicb.2020.570985
150. Long C, Rosch C, De Vries S, Schols H, Venema K. Cellulase and alkaline treatment improve intestinal microbial degradation of recalcitrant fibers of rapeseed meal in pigs. J Agric Food Chem. (2020) 68:11011–25. doi: 10.1021/acs.jafc.0c03618
151. Long C, Venema K. Pretreatment of rapeseed meal increases its recalcitrant fiber fermentation and alters the microbial community in an in vitro model of swine large intestine. Front Microbiol. (2020) 11:588264. doi: 10.3389/fmicb.2020.588264
152. Xie F, Zhang W, Lan X, Gong S, Wu J, Wang Z. Effects of high hydrostatic pressure and high pressure homogenization processing on characteristics of potato peel waste pectin. Carbohydr Polym. (2018) 196:474–82. doi: 10.1016/j.carbpol.2018.05.061
153. Peng X-Y, Mu T-H, Zhang M, Sun H-N, Chen J-W, Yu M. Effects of pH and high hydrostatic pressure on the structural and rheological properties of sugar beet pectin. Food Hydrocoll. (2016) 60:161–9. doi: 10.1016/j.foodhyd.2016.03.025
154. Bulbul V, Bhushette PR, Zambare RS, Deshmukh R, Annapure US. Effect of cold plasma treatment on Xanthan gum properties. Polym Test. (2019) 79:106056. doi: 10.1016/j.polymertesting.2019.106056
Keywords: rapeseed meal, animal performance, cell wall polysaccharides, carbohydrates, gut microbial degradation
Citation: Long C, Qi X-L and Venema K (2022) Chemical and nutritional characteristics, and microbial degradation of rapeseed meal recalcitrant carbohydrates: A review. Front. Nutr. 9:948302. doi: 10.3389/fnut.2022.948302
Received: 19 May 2022; Accepted: 31 August 2022;
Published: 28 September 2022.
Edited by:
Shabir Ahmad Mir, Government College for Women, Srinagar, IndiaReviewed by:
Fang Geng, Chengdu University, ChinaManzoor Ahmad Shah, Government College for Women Anantnag, India
Mudasir Bashir Mir, Govt. Degree College Baramulla, India
Copyright © 2022 Long, Qi and Venema. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Koen Venema, ay52ZW5lbWFAbWFhc3RyaWNodHVuaXZlcnNpdHkubmw=
 Cheng Long
Cheng Long Xiao-Long Qi1
Xiao-Long Qi1