
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Nutr. , 21 June 2022
Sec. Nutrition and Metabolism
Volume 9 - 2022 | https://doi.org/10.3389/fnut.2022.925267
This article is part of the Research Topic The Mechanism of Plant-derived Polysaccharides Regulating the Obesity and Metabolic Diseases in Humans View all 6 articles
 Haotian Gu1†
Haotian Gu1† Ping Hu1†
Ping Hu1† Yahui Zhao1
Yahui Zhao1 Yaya Liu1
Yaya Liu1 Yi-Ting Wang1
Yi-Ting Wang1 Abdelkareem A. Ahmed2,3
Abdelkareem A. Ahmed2,3 Hao-Yu Liu1*
Hao-Yu Liu1* Demin Cai1*
Demin Cai1*Under the influences of modern lifestyle, metabolic syndromes (MetS), including insulin resistance, obesity, and fatty liver, featuring a worldwide chronic disease, greatly raise the risk of type 2 diabetes, heart disease, and stroke. However, its pathogenesis is still unclear, and there are limited drugs with strong clinical efficacy and specificity. Given the close connection between impaired lipid metabolism and MetS onset, modulating the lipid metabolic genes may provide potential prospects in the development of MetS therapeutics. Nuclear receptors are such druggable transcription factors that translate physiological signals into gene regulation via DNA binding upon ligand activation. Recent studies reveal vital functions of the NRs retinoic acid's receptor-related orphan receptors (RORs), including RORα and RORγ, in the gene regulation in lipid metabolism and MetS. This review focuses on the latest developments in their actions on MetS and related metabolic disorders, which would benefit future clinically therapeutic applications.
Metabolic syndrome (MetS) is a cluster of conditions manifested as insulin resistance, obesity, fatty liver, and hyperlipidemia (1). The number of patients with MetS in the USA is more than one-third of the total population, while over 200 million Chinese adults suffer from it (2). As a complex condition, MetS conditions often occurs together and elevates the risk of type 2 diabetes, prevalence of stroke and myocardial infarction, and mortality rate of cardiovascular diseases, as well as compromise life quality and cause great economic burden (3). Given the primary pathological components in MetS, including impaired glucose and lipid metabolism, the current treatments of healthy lifestyles changes, including improved dietary plans, regular physical activity, reducing stress, and stopping smoking are helping, but not enough (4–6). For the purposes of drug discovery, nuclear receptors (NRs) like peroxisome proliferator-activated receptor (PPARs), liver X receptor (LXRs), REV-ERBs, and retinoic acid receptor-related orphan receptors (RORs) have favorable attributes, with most containing protected internal cavities predisposed for occupation with hydrophobic molecules of volumes typical of drugs. Moreover, these transcription factors control central pathways impacting a wide range of pathophysiological events ranging from cancers to metabolic diseases (7, 8).
The subfamily RORs (α, β, γ) has the typical NR domain structure involving a ligand-binding domain (LBD), along with a highly-conserved DNA-binding domain (DBD), an N-terminal domain, and a hinge domain spacing the DBD and LBD (9, 10). Accumulating evidence reveals that RORs as transcription factors exert the potential therapeutic capacities in numerous MetS-related metabolic diseases (11). In particular, the RORα and RORγ are closely associated with fatty acids and sterol metabolism, and their trans-activities could be driven by a series of sterols and synthetic ligands that function either as agonists or antagonists (12). In contrast, the expression of ROR-β is restricted to the brain and retina, and is commonly associated with retinal neurogenesis, etc. (13). These targetable characteristics provide the possibility of RORs-mediated therapeutic targets of metabolic illnesses. This mini-review aims at summarizing the connections between these orphan NRs and the onset of MetS and related disorders for better understanding of the underlying mechanisms in the process of pathogenesis. To explore the feasibility of using orphan NR ligands in the treatment of metabolic diseases according to published documents will benefit the theoretical strategies for further small-molecule drugs' development and clinical intervention.
Encoded by NR1F1, RORα is widely expressed in tissues like liver, lung, skin, kidney, adipose tissue, etc. (14, 15). RORα's full body knockout mice exhibit improved metabolic outputs on both chow and high fat diets, without overt behavioral dysfunctions (16). The natural RORα spontaneous mutant mice, i.e., the staggerer (sg) mice would be the initial evidence that RORα modulates metabolic processes by showing a lower expression pattern of genes involved in lipid metabolism, including apolipoprotein A-1 (apoA1) and apolipoprotein C-III (apoCIII) (17, 18). Thus, RORαsg/sg mice display a lean body type compared to that of the wild-type mice. The sg mice perform the protective functions against diet-induced MetS and associated complications, such as fatty liver, adipose tissue inflammation, and glucose metabolic abnormalities (19). It is suggested that RORα plays a positive role in metabolic diseases by reducing oxidative stress and inflammation in livers when activated (20). Intriguingly, liver-specific RORα deletion (RORαLKO) in mice shows no significant effects on glucose and lipid metabolism in response to either chow or western-style diet (21). In contrast, RORαLKO mice show the typical MetS including hepatic steatosis, obesity, and IR when supplemented with high fat diet (22). This is attributed to the RORα-mediated PPARγ transcriptional repression by HDAC3 recruitment. In agreement, Kim et al. also demonstrated that RORαLKO mice are more susceptible to high-fat diet-induced non-alcoholic steatohepatitis (NASH) where RORα exerts a beneficial role in mitochondrial quality control by enhancing oxygen consumption rate and the expression of key genes Bnip3 and PGC-1α (23). Same in this RORα deletion model, RORα inhibition induces suppression of hepatic polyploidization and hepatocyte DNA endoreplication to ameliorate the high fat diet (HFD)-triggered NASH, providing another insight of the protective mechanisms of RORα against MetS by maintaining genome integrity (24). In addition, RORα-targeting lysosomal acidification recovery is a new mechanism behind the HFD-induced NASH when RORα is knocked out (25). This is supported by several pieces of evidence, e.g., the lower lysosomal acidity, increased lysosomal translocation of the mechanistic target of rapamycin, accumulated immature cathepsin D, LC3-II, p62/sequestosome 1 (SQSTM1), and upregulated neighbor of BRCA1 gene 1 (NBR1) observed in the RORαLKO mice. Importantly, the reduced key genes enrolled in lysosomal function in the livers of mice with HFD-caused NASH, and patients are activated by RORα. Regarding genes expression, a couple of genes involved in glucolipid metabolism, including phosphoenolpyruvate carboxykinase (Pepck), glucose-6 phosphatase (G6pc), glycerol-3-phosphate acyltransferase (G3pat), and perilipin 2 (Plin2) are downregulated in the RORα−/− mice fed a HFD diet (26–28). In contrast, the sulfotransferase Sult1e1 and Sult2a1, as the sulfotransferase and bile acids sulfonation, are drastically upregulated in the RORα-deficient mice (29, 30). The genome-wide ChIP-seq analysis in the liver reveals that RORα directly binds to the ROR response elements at these glucolipid genes including Pepck1, G6pc, Apoa1, Fasn, and Elovl3 to transcriptionally control their expression (31, 32).
It is worth mentioning that adipose tissues-associated inflammation is tightly linked to the development of MetS and related metabolic disorders (33, 34). RORα modulates insulin sensitivity and contribute to diminished inflammation in the RORα−/− mice (35). Lipid droplets are seen to be reduced in the adipose tissue of RORα−/− mice fed with an HFD, while decreased inflammatory macrophages infiltration and downregulated inflammatory genes are observed (26). Because of the potential role of interleukin-1 receptor antagonist (IL-1rn) in IR, the reduced Il-1rn expression in adipose tissue of RORα−/− mice possibly leads to less susceptibility to MetS (36). Moreover, the neutral cholesterol ester hydrolase 1 (NCEH1), as a direct target of RORα, is required for RORα-mediated lipid droplets deposition in macrophage, and functions via a phorbol myristate acetate-dependent mechanism (37). To further define the potential role of RORα-expressing macrophages in the generation of an aberrant metabolic state, RORαfl/flLysMCre/+ mice, which do not express RORα in myeloid cells, were maintained on an HFD, and metabolic parameters were assessed (38). These mice had significantly impaired weight gain and improved metabolic parameters in comparison to RORαfl/fl control mice. Further analysis of the immune cell populations within white adipose tissue deposits demonstrates a decrease in inflammatory adipose tissue macrophages (ATM). It is noted that endoplasmic reticulum (ER) stress and unfolding protein response (UPR) signaling play a critical role in inflammatory response in obese adipose tissue (39). Liu et al. revealed that RORα is also involved in this process, that RORα is potently induced by inflammatory stimuli in macrophages and is positively regulated by the inflammatory response by activating ER stress and UPR response (40). In contrast to adipose tissue, RORα exhibits the protection against NASH via enhancing M2 polarization in liver macrophages, while myeloid-specific knockout RORα are susceptible to HFD (41). Interestingly, it is also documented that RORα deficiency, which is specifically in macrophages, does not affect the development of IR, obesity, and NASH (42). This suggests a different Cre or Lyz2 copy numbers, but not a specific effect of RORα deletion in macrophages causes the impairments on NASH.
The RORγ is another key player in the modulation of MetS-related metabolic diseases. In this regard, insulin sensitivity and glucose tolerance were observed along with decreased glucose content in the liver of RORγ−/− mice (43–45). This may be attributed to the suppression of hepatic gluconeogenesis and blunted transformation to glucose from pyruvate to glucose (43, 45). Indeed, ectopic RORγ activation in the RORγ−/− mice or RORγ-deficient primary hepatocytes facilitates the glucose production recovery by improving the rate of gluconeogenesis (43). Consistent with the output, key genes involved in glucolipid metabolism, including G6pc, Pklr, Gck, Glut2, Gckr, Pparδ, Insig2a, Elovl3, and Sult1e1, are remarkably upregulated compared to those of RORγ−/− mice (30, 43, 44, 46). Given that Insig2a and Elovl3 are critical for insulin signaling and directly driven by RORγ, the restoration of these genes by exogenous RORγ over-expression support the RORγ-mediated MetS by ameliorating IR (47, 48). A genome-wide cistrome analysis of mouse liver reveals that RORγ directly binds to promoters and/or the enhancers of genes enrolled in gluconeogenesis and glycolysis, e.g., G6pc, Pepck, Gckr, Pklr, Glut2, Gck, Gys2, Pcx, and Klf15 (44, 49). Thus, the downregulated transcripts of these genes in the RORγ−/− mice are accompanied by impaired glucose generation. Importantly, the Insig2 repression is an indicator of activated lipogenesis in an SREBP1-dependent negative feedback loop; loss of RORγ in mice will disrupt the dynamic balance of lipid metabolism (48, 49). This would result in reduced contents of triglycerides in liver and blood with enhanced hepatic lipogenic gene expressions in RORγ-deficient mice when fed with HFD.
It is noted that cholesterol and bile acids were also reduced in liver and blood when Insig2 expression was lower in the RORγ null mice (46, 49). Indeed, RORγ modulates MetS more likely via regulating sterols' metabolism. The early finding of RORγ-mediated cholesterol metabolism should be attributed to the RORγ isoform 2, RORγt, which is mainly expressed in immune cells and is required for Th17 differentiation (12, 50). During this process, the key genes involved in cholesterol dynamic metabolism, including Hmgxs1, Hmgcr, Lss, Dhcr24, Fdft1, Cyp7a, Cyp27a1, Abca1, and Abcg1, were all affected (51). Given that squalene synthesis of mevalonate pathway is critical for cholesterol production, squalene supplementation is shown to improve RORγ-mediated transactivation in in vitro validation experiment. It is, therefore, expected that this transactivation was inhibited when the key gene Fdft1 enrolled in squalene biosynthesis was deficient (52). In mammals, RORγ-controlling sterols' metabolism is intrinsically associated with the typical transcription factors like sterol regulatory element binding protein 2 (SREBP2) and liver X receptor (53–56). The increased intermediates like zymosterol and desmosterol involved in SREBP2-enhanced cholesterol biosynthesis via upregulating the genes, which encode the rate-limiting enzymes involving HMGCR and HMGCS, have been demonstrated to activate the sensitivity to RORγ (53, 54, 57). It is documented that RORγ directly binds to cholesterol biosynthetic genes, and positively regulates these genes over SREBP2 in a breast cancer cell line (58, 59). Intriguingly, RORγ is found to share the binding peaks with SREBP2 and determines the SREBP2 recruitments on the target genes. In the in vivo model using piglets, ChIP-seq analysis provides the proof that hepatic RORγ exhibited strong enrichments on the cholesterol biosynthesis genes at the enhancer region (60). In the liver organoids, RORγ agonists, such as desmosterol and a commercial drug SR0987, displayed drastical restoration of reduced cholesterol content, along with biosynthetic genes caused by a time-restricted feeding (61). Moreover, ectopic RORγ overexpression performed similar effects with those of the agonists possibly via epigenetically recruited histone active marks H3K27ac. A recent study also revealed RORγ as an essential mediator in facilitating the interaction of PPARα and SREBP2 and in controlling the cholesterol biosynthesis genes transcription (62). This new complex of “PPARα-RORγ-SREBP2” explored a new mechanism on cholesterol reprogramming in a context and cell-type-specific manner, which would be beneficial for the invention of therapeutic strategy for MetS and related metabolic diseases. Indeed, PPARα plays a novel role in MetS because of the changed functions induced by RORγ in different scenarios, suggesting the complicated crosstalk among these NRs (62, 63). Given the fact that LXRs modulate the cholesterol intake and efflux by the key enzymes such as LDLR and ABCA1/G1, it is suggested that a diminished RORγ availability is associated with decreased cholesterol production in response to LXR activation (53, 55, 64). Recent studies provide a pivotal clue that the derivative of lithocholic acids (LCA), 3-oxoLCA, is a novel inhibitor of RORγ (65, 66). Because the cholic acids are synthesized from cholesterol and are mainly modulated by nuclear receptor FXR, it is likely that RORγ is enrolled in the regulation of cholesterol-bile acid circulation and, maybe, a new mechanism to maintain cholesterol homeostasis against MetS. It is worth addressing that circadian rhythm exerts critical roles in MetS and RORα/γ are primary modulators of clock genes expression. This RORα/γ-driven circadian clock gene regulation is therefore another type of events controlling the biological balance against MetS and related disorders (7, 67).
Tremendous advances have been made in studying the underlying mechanisms in the pathogenesis of MetS and related metabolic diseases in recent decades. However, the pivotal targetable controllers are still being explored to find a therapeutic strategy. An increasing number of reports have provided evidence that RORα/γ exert numerous actions on lipid metabolism of MetS, as summarized in Figure 1. Given these NR transcriptional factors governing the development and progress of MetS, further studies are needed to understand the precise molecular mechanisms behind their crosstalk, and how alteration of NR function affects these interactions and MetS pathologies. Importantly, unlike the established master regulator of several metabolic pathways, RORα/γ exhibit diversity and plasticity of RORs functions and actions in a context and subtype-specific manner. These explorations of untapped RORs' features are worthy of further investigations on their biological importance. Elucidating the remaining intricacies will subsequently afford viable approaches for treating MetS. Even though current efforts to target the MetS-related pathogenesis have failed to develop efficient therapeutic agents, researchers in this field have gained substantial insights from the cases of those that have been available for use by targeting modulators and drivers like RORs in animal models of disease for the past. Furthermore, identification of endogenous high-affinity or synthetic ligands of RORs α/γ will not only increase our understanding of their actions on MetS and related disorders, but also probe the therapeutic potential of these receptors as bona fide drug targets for the treatments.
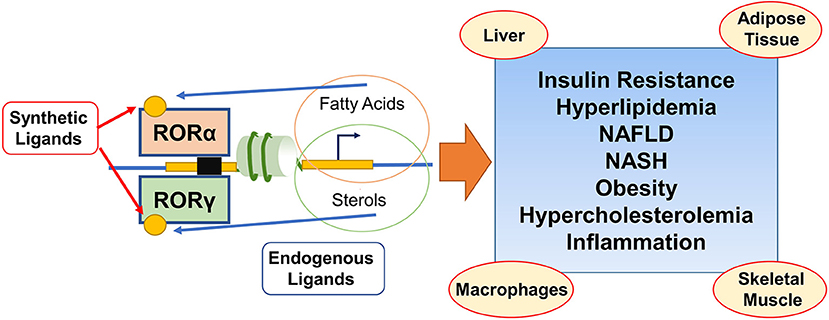
Figure 1. Schematic illustration depicting that coordinated actions of RORα/γ on lipid and sterol metabolic programming during the development of the metabolic syndrome and related disorders. Both RORα and RORγ regulate the target genes involved in lipid/sterol metabolism through ROR response elements and are exciting modulators in metabolic syndrome and related disorders in multiple tissues. RORs (ant)agonists from endogenous or synthetic generation, act as the potential prosects in the treatment of insulin resistance, hyperlipidemia, NAFLD, NASH, obesity, hypercholesterolemia, and inflammation.
DC and H-YL: conceptualization. HG, PH, YZ, YL, and Y-TW: writing original draft. PH, H-YL, AA, and DC: review, editing, and resources. All authors contributed to the article and approved the submitted version.
This work was supported by the Postgraduate Research & Practice Innovation Program of Jiangsu Province (SJCX21_1619), National Natural Science Foundation of China (32002243), Natural Science Foundation of Jiangsu Province (BK20200932), Natural Science Foundation of the Higher Education Institutions of Jiangsu Province (20KJB230001), the Jiangsu Agricultural Science And Technology Innovation Fund [CX(21)2014 and CX(21)3125], and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Panov A, Mayorov VI, Dikalov S. Metabolic Syndrome and beta-Oxidation of long-chain fatty acids in the brain, heart, and kidney mitochondria. Int J Mol Sci. (2022) 23:4047. doi: 10.3390/ijms23074047
2. Miao R, Fang X, Wei J, Wu H, Wang X, Tian J. Akt: A potential drug target for metabolic syndrome. Front Physiol. (2022) 13:822333. doi: 10.3389/fphys.2022.822333
3. Coronati M, Baratta F, Pastori D, Ferro D, Angelico F, Del Ben M. Added fructose in non-alcoholic fatty liver disease and in metabolic syndrome: a narrative review. Nutrients. (2022) 14:1127. doi: 10.3390/nu14061127
4. Biccire FG, Bucci T, Menichelli D, Cammisotto V, Pignatelli P, Carnevale R, et al. Mediterranean diet: a tool to break the relationship of atrial fibrillation with the metabolic syndrome and non-alcoholic fatty liver disease. Nutrients. (2022) 14:1260. doi: 10.3390/nu14061260
5. Tang Q, Li Y, He J. MANF: an emerging therapeutic target for metabolic diseases. Trends Endocrinol Metab. (2022) 33:236–46. doi: 10.1016/j.tem.2022.01.001
6. Bungau SG, Tit DM, Vesa CM, Abid A, Szilagyi DV, Radu AF, et al. Non-conventional therapeutical approaches to acne vulgaris related to its association with metabolic disorders. Eur J Pharmacol. (2022) 923:174936. doi: 10.1016/j.ejphar.2022.174936
7. Solt LA, Kojetin DJ, Burris TP. The REV-ERBs and RORs: molecular links between circadian rhythms and lipid homeostasis. Future Med Chem. (2011) 3:623–38. doi: 10.4155/fmc.11.9
8. Marin R, Diaz M. Estrogen Interactions With lipid rafts related to neuroprotection. Impact of brain ageing and menopause. Front Neurosci. (2018) 12:128. doi: 10.3389/fnins.2018.00128
9. Frigo DE, Bondesson M, Williams C. Nuclear receptors: from molecular mechanisms to therapeutics. Essays Biochem. (2021) 65:847–56. doi: 10.1042/EBC20210020
10. Cai D, Liu H, Zhao R. Nuclear receptors in hepatic glucose and lipid metabolism during neonatal and adult life. Curr Protein Pept Sci. (2017) 18:548–61. doi: 10.2174/1389203717666160627081751
11. Marciano DP, Chang MR, Corzo CA, Goswami D, Lam VQ, Pascal BD, et al. The therapeutic potential of nuclear receptor modulators for treatment of metabolic disorders: PPARgamma, RORs, and Rev-erbs. Cell Metab. (2014) 19:193–208. doi: 10.1016/j.cmet.2013.12.009
12. Jetten AM, Kang HS, Takeda Y. Retinoic acid-related orphan receptors alpha and gamma: key regulators of lipid/glucose metabolism, inflammation, and insulin sensitivity. Front Endocrinol. (2013) 4:1. doi: 10.3389/fendo.2013.00001
13. Byun H, Lee HL, Liu H, Forrest D, Rudenko A, Kim IJ. Rorbeta regulates selective axon-target innervation in the mammalian midbrain. Development. (2019) 146:dev171926. doi: 10.1242/dev.171926
14. Hamilton BA, Frankel WN, Kerrebrock AW, Hawkins TL, FitzHugh W, Kusumi K, et al. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature. (1996) 379:736–9. doi: 10.1038/379736a0
15. Steinmayr M, Andre E, Conquet F, Rondi-Reig L, Delhaye-Bouchaud N, Auclair N, et al. staggerer phenotype in retinoid-related orphan receptor alpha-deficient mice. Proc Natl Acad Sci USA. (1998) 95:3960–5. doi: 10.1073/pnas.95.7.3960
16. Lau P, Tuong ZK, Wang SC, Fitzsimmons RL, Goode JM, Thomas GP, et al. Roralpha deficiency and decreased adiposity are associated with induction of thermogenic gene expression in subcutaneous white adipose and brown adipose tissue. Am J Physiol Endocrinol Metab. (2015) 308:E159–71. doi: 10.1152/ajpendo.00056.2014
17. Kim EJ, Yoon YS, Hong S, Son HY, Na TY, Lee MH, et al. Retinoic acid receptor-related orphan receptor alpha-induced activation of adenosine monophosphate-activated protein kinase results in attenuation of hepatic steatosis. Hepatology. (2012) 55:1379–88. doi: 10.1002/hep.25529
18. Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J Biol Chem. (2008) 283:18411–21. doi: 10.1074/jbc.M710526200
19. Zhao YC, Xu LW, Ding S, Ji QQ, Lin N, He Q, et al. Nuclear receptor retinoid-related orphan receptor alpha deficiency exacerbates high-fat diet-induced cardiac dysfunction despite improving metabolic abnormality. Biochim Biophys Acta Mol Basis Dis. (2017) 1863:1991–2000. doi: 10.1016/j.bbadis.2016.10.029
20. Han YH, Kim HJ, Kim EJ, Kim KS, Hong S, Park HG, et al. RORalpha decreases oxidative stress through the induction of SOD2 and GPx1 expression and thereby protects against nonalcoholic steatohepatitis in mice. Antioxid Redox Signal. (2014) 21:2083–94. doi: 10.1089/ars.2013.5655
21. Molinaro A, Caesar R, L'Homme L, Koh A, Stahlman M, Staels B, et al. Liver-specific RORalpha deletion does not affect the metabolic susceptibility to western style diet feeding. Mol Metab. (2019) 23:82–7. doi: 10.1016/j.molmet.2019.02.010
22. Kim K, Boo K, Yu YS, Oh SK, Kim H, Jeon Y, et al. RORalpha controls hepatic lipid homeostasis via negative regulation of PPARgamma transcriptional network. Nat Commun. (2017) 8:162. doi: 10.1038/s41467-017-00215-1
23. Kim HJ, Han YH, Na H, Kim JY, Kim T, Kim HJ, et al. Liver-specific deletion of RORalpha aggravates diet-induced nonalcoholic steatohepatitis by inducing mitochondrial dysfunction. Sci Rep. (2017) 7:16041. doi: 10.1038/s41598-017-16077-y
24. Kim JY, Yang IS, Kim HJ, Yoon JY, Han YH, Seong JK, et al. RORalpha contributes to the maintenance of genome ploidy in the liver of mice with diet-induced nonalcoholic steatohepatitis. Am J Physiol Endocrinol Metab. (2022) 322:E118–31. doi: 10.1152/ajpendo.00309.2021
25. Kim HJ, Han YH, Kim JY, Lee MO. RORalpha enhances lysosomal acidification and autophagic flux in the hepatocytes. Hepatol Commun. (2021) 5:2121–38. doi: 10.1002/hep4.1785
26. Kang HS, Okamoto K, Takeda Y, Beak JY, Gerrish K, Bortner CD, et al. Transcriptional profiling reveals a role for RORalpha in regulating gene expression in obesity-associated inflammation and hepatic steatosis. Physiol Genomics. (2011) 43:818–28. doi: 10.1152/physiolgenomics.00206.2010
27. Chauvet C, Vanhoutteghem A, Duhem C, Saint-Auret G, Bois-Joyeux B, Djian P, et al. Control of gene expression by the retinoic acid-related orphan receptor alpha in HepG2 human hepatoma cells. PLoS ONE. (2011) 6:e22545. doi: 10.1371/journal.pone.0022545
28. Wang Y, Solt LA, Burris TP. Regulation of FGF21 expression and secretion by retinoic acid receptor-related orphan receptor alpha. J Biol Chem. (2010) 285:15668–73. doi: 10.1074/jbc.M110.102160
29. Ou Z, Shi X, Gilroy RK, Kirisci L, Romkes M, Lynch C, et al. Regulation of the human hydroxysteroid sulfotransferase (SULT2A1) by RORalpha and RORgamma and its potential relevance to human liver diseases. Mol Endocrinol. (2013) 27:106–15. doi: 10.1210/me.2012-1145
30. Kang HS, Angers M, Beak JY, Wu X, Gimble JM, Wada T, et al. Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol Genomics. (2007) 31:281–94. doi: 10.1152/physiolgenomics.00098.2007
31. Wang R, Campbell S, Amir M, Mosure SA, Bassette MA, Eliason A, et al. Genetic and pharmacological inhibition of the nuclear receptor RORalpha regulates TH17 driven inflammatory disorders. Nat Commun. (2021) 12:76. doi: 10.1038/s41467-020-20385-9
32. Han YH, Kim HJ, Choi H, Lee S, Lee MO. RORalpha autoregulates its transcription via MLL4-associated enhancer remodeling in the liver. Life Sci. (2020) 256:118007. doi: 10.1016/j.lfs.2020.118007
33. Nishimura S, Manabe I, Nagasaki M, Eto K, Yamashita H, Ohsugi M, et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. (2009) 15:914–20. doi: 10.1038/nm.1964
34. Hotamisligil GS. Inflammation and metabolic disorders. Nature. (2006) 444:860–7. doi: 10.1038/nature05485
35. Cook DN, Kang HS, Jetten AM. Retinoic acid-related orphan receptors (RORs): regulatory functions in immunity, development, circadian rhythm, and metabolism. Nucl Receptor Res. (2015) 2:101178. doi: 10.11131/2015/101185
36. Somm E, Henrichot E, Pernin A, Juge-Aubry CE, Muzzin P, Dayer JM, et al. Decreased fat mass in interleukin-1 receptor antagonist-deficient mice: impact on adipogenesis, food intake, and energy expenditure. Diabetes. (2005) 54:3503–9. doi: 10.2337/diabetes.54.12.3503
37. Matsuoka H, Tokunaga R, Katayama M, Hosoda Y, Miya K, Sumi K, et al. Retinoic acid receptor-related orphan receptor alpha reduces lipid droplets by upregulating neutral cholesterol ester hydrolase 1 in macrophages. BMC Mol Cell Biol. (2020) 21:32. doi: 10.1186/s12860-020-00276-z
38. Hams E, Roberts J, Bermingham R, Hogan AE, O'Shea D, O'Neill L, et al. Role for retinoic acid-related orphan receptor alpha (RORalpha) expressing macrophages in diet-induced obesity. Front Immunol. (2020) 11:1966. doi: 10.3389/fimmu.2020.01966
39. Saltiel AR, Olefsky JM. Inflammatory mechanisms linking obesity and metabolic disease. J Clin Invest. (2017) 127:1–4. doi: 10.1172/JCI92035
40. Liu Y, Chen Y, Zhang J, Liu Y, Zhang Y, Su Z. Retinoic acid receptor-related orphan receptor alpha stimulates adipose tissue inflammation by modulating endoplasmic reticulum stress. J Biol Chem. (2017) 292:13959–69. doi: 10.1074/jbc.M117.782391
41. Han YH, Kim HJ, Na H, Nam MW, Kim JY, Kim JS, et al. RORalpha induces KLF4-mediated M2 polarization in the liver macrophages that protect against nonalcoholic steatohepatitis. Cell Rep. (2017) 20:124–35. doi: 10.1016/j.celrep.2017.06.017
42. L'Homme L, Sermikli BP, Molendi-Coste O, Fleury S, Quemener S, Le Maitre M, et al. Deletion of the nuclear receptor RORalpha in macrophages does not modify the development of obesity, insulin resistance and NASH. Sci Rep. (2020) 10:21095. doi: 10.1038/s41598-020-77858-6
43. Takeda Y, Kang HS, Freudenberg J, DeGraff LM, Jothi R, Jetten AM. Retinoic acid-related orphan receptor gamma (RORgamma): a novel participant in the diurnal regulation of hepatic gluconeogenesis and insulin sensitivity. PLoS Genet. (2014) 10:e1004331. doi: 10.1371/journal.pgen.1004331
44. Takeda Y, Kang HS, Lih FB, Jiang H, Blaner WS, Jetten AM. Retinoid acid-related orphan receptor gamma, RORgamma, participates in diurnal transcriptional regulation of lipid metabolic genes. Nucleic Acids Res. (2014) 42:10448–59. doi: 10.1093/nar/gku766
45. Meissburger B, Ukropec J, Roeder E, Beaton N, Geiger M, Teupser D, et al. Adipogenesis and insulin sensitivity in obesity are regulated by retinoid-related orphan receptor gamma. EMBO Mol Med. (2011) 3:637–51. doi: 10.1002/emmm.201100172
46. Takeda Y, Jothi R, Birault V, Jetten AM. RORgamma directly regulates the circadian expression of clock genes and downstream targets in vivo. Nucleic Acids Res. (2012) 40:8519–35. doi: 10.1093/nar/gks630
47. Raichur S, Lau P, Staels B, Muscat GE. Retinoid-related orphan receptor gamma regulates several genes that control metabolism in skeletal muscle cells: links to modulation of reactive oxygen species production. J Mol Endocrinol. (2007) 39:29–44. doi: 10.1677/jme.1.00010
48. Zou H, Yang N, Zhang X, Chen HW. RORgamma is a context-specific master regulator of cholesterol biosynthesis and an emerging therapeutic target in cancer and autoimmune diseases. Biochem Pharmacol. (2022) 196:114725. doi: 10.1016/j.bcp.2021.114725
49. Zhang Y, Papazyan R, Damle M, Fang B, Jager J, Feng D, et al. The hepatic circadian clock fine-tunes the lipogenic response to feeding through RORalpha/gamma. Genes Dev. (2017) 31:1202–11. doi: 10.1101/gad.302323.117
50. Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. (2009) 7:e003. doi: 10.1621/nrs.07003
51. Hu X, Wang Y, Hao LY, Liu X, Lesch CA, Sanchez BM, et al. Sterol metabolism controls T(H)17 differentiation by generating endogenous RORgamma agonists. Nat Chem Biol. (2015) 11:141–7. doi: 10.1038/nchembio.1714
52. Santori FR, Huang P, van de Pavert SA, Douglass EF Jr., Leaver DJ, Haubrich BA, et al. Identification of natural RORgamma ligands that regulate the development of lymphoid cells. Cell Metab. (2015) 21:286–98. doi: 10.1016/j.cmet.2015.01.004
53. Bovenga F, Sabbà C, Moschetta A. Uncoupling nuclear receptor LXR and cholesterol metabolism in cancer. Cell Metab. (2015) 21:517–26. doi: 10.1016/j.cmet.2015.03.002
54. Shimano H, Sato R. SREBP-regulated lipid metabolism: convergent physiology - divergent pathophysiology. Nat Rev Endocrinol. (2017) 13:710–30. doi: 10.1038/nrendo.2017.91
55. Mutemberezi V, Guillemot-Legris O, Muccioli GG. Oxysterols: From cholesterol metabolites to key mediators. Prog Lipid Res. (2016) 64:152–69. doi: 10.1016/j.plipres.2016.09.002
56. Cai D, Yuan M, Liu H, Pan S, Ma W, Hong J, et al. Maternal betaine supplementation throughout gestation and lactation modifies hepatic cholesterol metabolic genes in weaning piglets via AMPK/LXR-mediated pathway and histone modification. Nutrients. (2016) 8:646. doi: 10.3390/nu8100646
57. Liu HY, Gu H, Qu H, Bao W, Li Y, Cai D. Aberrant cholesterol metabolic genes regulation in a negative feedback loop induced by an alphacoronavirus. Front Nutr. (2022) 9:870680. doi: 10.3389/fnut.2022.870680
58. Cai D, Zhang X, Chen HW. A master regulator of cholesterol biosynthesis constitutes a therapeutic liability of triple negative breast cancer. Mol Cell Oncol. (2020) 7:1701362. doi: 10.1080/23723556.2019.1701362
59. Cai D, Wang J, Gao B, Li J, Wu F, Zou JX, et al. RORgamma is a targetable master regulator of cholesterol biosynthesis in a cancer subtype. Nat Commun. (2019) 10:4621. doi: 10.1038/s41467-019-12529-3
60. Li K, Li H, Zhang K, Zhang J, Hu P, Li Y, et al. Orphan nuclear receptor rorgamma modulates the genome-wide binding of the cholesterol metabolic genes during mycotoxin-induced liver Injury. Nutrients. (2021) 13:2539. doi: 10.3390/nu13082539
61. Zhang K, Li H, Xin Z, Li Y, Wang X, Hu Y, et al. Time-restricted feeding downregulates cholesterol biosynthesis program via RORgamma-mediated chromatin modification in porcine liver organoids. J Anim Sci Biotechnol. (2020) 11:106. doi: 10.1186/s40104-020-00511-9
62. Liu HY, Hu P, Li Y, Sun MA, Qu H, Zong Q, et al. Targeted inhibition of PPARalpha ameliorates CLA-induced hypercholesterolemia via hepatic cholesterol biosynthesis reprogramming. Liver Int. (2022) 42:1449–66. doi: 10.1111/liv.15199
63. Cai D, Li Y, Zhang K, Zhou B, Guo F, Holm L, et al. Co-option of PPARalpha in the regulation of lipogenesis and fatty acid oxidation in CLA-induced hepatic steatosis. J Cell Physiol. (2021) 236:4387–402. doi: 10.1002/jcp.30157
64. Villa GR, Hulce JJ, Zanca C, Bi J, Ikegami S, Cahill GL, et al. An LXR-cholesterol axis creates a metabolic co-dependency for brain cancers. Cancer Cell. (2016) 30:683–93. doi: 10.1016/j.ccell.2016.09.008
65. Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, et al. Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature. (2020) 577:410–5. doi: 10.1038/s41586-019-1865-0
66. Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature. (2019) 576:143–8. doi: 10.1038/s41586-019-1785-z
Keywords: RORα/γ, metabolic syndromes, sterols, insulin resistance, lipids
Citation: Gu H, Hu P, Zhao Y, Liu Y, Wang Y-T, Ahmed AA, Liu H-Y and Cai D (2022) Nuclear Receptor RORα/γ: Exciting Modulators in Metabolic Syndrome and Related Disorders. Front. Nutr. 9:925267. doi: 10.3389/fnut.2022.925267
Received: 21 April 2022; Accepted: 18 May 2022;
Published: 21 June 2022.
Edited by:
Guiguo Zhang, Shandong Agricultural University, ChinaReviewed by:
Mziwenkosi Nhlanhla Mthiyane, North-West University, South AfricaCopyright © 2022 Gu, Hu, Zhao, Liu, Wang, Ahmed, Liu and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao-Yu Liu, aGFveXUubGl1MDUyM0BvdXRsb29rLmNvbQ==; Demin Cai, ZGVtaW5jYWlAeXp1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.