
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Nutr. , 13 June 2022
Sec. Food Chemistry
Volume 9 - 2022 | https://doi.org/10.3389/fnut.2022.913883
This article is part of the Research Topic Food Rich in Phenolic Compounds and Their Potential to Fight Obesity View all 5 articles
The consumption of phytochemicals, bioactive compounds in fruits and vegetables, has been demonstrated to ameliorate obesity and related metabolic symptoms by regulating specific metabolic pathways. This review summarizes the progress made in our understanding of the potential of phytochemicals as metabolic signals: we discuss herein selected molecular mechanisms which are involved in the occurrence of obesity that may be regulated by phytochemicals. The focus of our review highlights the regulation of transcription factors toll like receptor 4 (TLR4), nuclear factor (erythroid-derived 2)-like 2 (Nrf2), the peroxisome proliferator-activated receptors (PPARs), fat mass and obesity-associated protein (FTO) and regulation of microRNAs (miRNA). In this review, the effect of phytochemicals on signaling pathways involved in obesity were discussed on the basis of their chemical structure, suggesting molecular mechanisms for how phytochemicals may impact these signaling pathways. For example, compounds with an isothiocyanate group or an α, β-unsaturated carbonyl group may interact with the TLR4 signaling pathway. Regarding Nrf2, we examine compounds possessing an α, β-unsaturated carbonyl group which binds covalently with the cysteine thiols of Keap1. Additionally, phytochemical activation of PPARs, FTO and miRNAs were summarized. This information may be of value to better understand how specific phytochemicals interact with specific signaling pathways and help guide the development of new drugs to combat obesity and related metabolic diseases.
Obesity, normally results from an imbalance between energy consumption and energy expenditure, is one of the most prevalent problems challenging public health today. Obesity increases the risk of diseases including type 2 diabetes (1), cardiovascular disease and some types of cancer (2). However, no effective therapy has been developed for the treatment of obesity except for exercise and dietary regimes. Pharmacological treatment has been used for the long-term treatment of severely obese patients. Currently, orlistat, the world's only weight loss drug approved by the US Food and Drug Administration, has been used by some obese people (3). However, its application is limited for the side effects such as diarrhea, and vomiting (4). Therefore, the search for novel food and food compound interventions as a monotherapy or along with existing therapies is a main focus for nutritionists (5).
Phytochemicals, bioactive compounds in fruits and vegetables are produced through primary or secondary metabolism. They may be synthesized to protect plants from a variety of stresses, such as pathogenic attacks, predators, and UV irradiation (6). In recent years, there have been many studies on the regulation of signaling pathways by phytochemicals, most of which aim at the biological activity and physiological function of phytochemicals; however, little attention has been paid to the underlying molecular mechanism and preferred structure of phytochemicals for pathway activation.
In this review, the structure-activity relationship of phytochemicals on obesity and related chronic diseases were discussed. We summarize the progress made in our understanding of the anti-obesity effect of phytochemicals and their mechanisms of action: the effect of phytochemicals on transcription factors and some related molecular pathways. We focus on the transcription factors toll like receptor 4 (TLR4), peroxisome proliferator-activated receptors (PPARs), and transcription factors nuclear factor (erythroid-derived 2)-like 2 (Nrf2). Additionally, the fat mass and obesity-associated protein (FTO), and microRNA (miRNA) regulation of gene expression are also discussed.
Toll-like receptors (TLRs) are a family of pattern-recognition receptors (PRR) that trigger innate immune and inflammatory responses in response to invading microorganisms and non-microbial endogenous molecules (7). TLR4, one of the thirteen TLRs identified in mammals, influences symptoms induced by high fat diet-induced obesity, including insulin resistance (8–10), inflammation (8, 11, 12), and hepatic lipid accumulation (13–15).
TLR4 molecular structure includes a leucine-rich repeat (LRR) domain in the ectodomain, which is involved in the recognition of pathogen-associated molecular patterns (PAMPs), and a Toll/interleukin-1 receptor homology (TIR) domain in the cytoplasm, in which highly conserved cysteine residues are located (16). In addition to this cytoplasmic conserved region, other cysteine residues also reside in the extracellular domain.
Cells of myeloid origin such as monocytes and macrophages exhibit the highest levels of TLR4 expression (17). In liver, TLR4 is expressed by hepatocytes and non-parenchymal cells (NPCs), including liver sinusoidal endothelial cells (LSECs) and Kupffer cells (KCs) (18). Stimulation of TLR4 by ligands such as lipopolysaccharides (LPS), leads to the activation of two downstream signaling pathways: MyD88-dependent and MyD88-independent (TRIF-dependent) signaling pathways. Pro-inflammatory cytokines, such as tumor necrosis factor alpha (TNFα), are typically the major final product of the TLR signaling pathway (Figure 1). TNFα plays an important role in lipid metabolism as well as hepatocyte cell death in the development of obesity (19–23), which promotes lipid accumulation in hepatocytes induces insulin resistance, increases FFA levels, and sustains intracellular lipid retention (19). On the other hand, TNFα promotes hepatic cholesterol accumulation by inducing expression of LDL receptor and by inhibiting efflux of cholesterol (21). TLR4 knockout mice have been reported to protect against insulin resistance induced by high fat diet (HFD) (8). In obese individuals, elevated expression of TLR4 and the adaptor proteins including MyD88, interleukin-1 receptor-associated kinase 1 (IRAK1) and factor receptor-associated factor 6 (TRAF6), was observed to correlate with the elevated expression of TNF-α and interleukin-6 (IL-6) (24, 25).
Phytochemicals that inhibit the activation of TLR4, may ameliorate obesity associated symptoms. It has been well-documented that molecules with the α, β-unsaturated carbonyl groups can react with biological nucleophiles such as a sulfhydryl group (thiol group) by a Michael addition (Figure 2) (26–28). Phytochemicals with α, β-unsaturated carbonyl groups including withaferin A, kaempferide, isoliquiritigenin and curcumin were reported to ameliorate obesity and related metabolic symptoms by the suppression of TLR4.
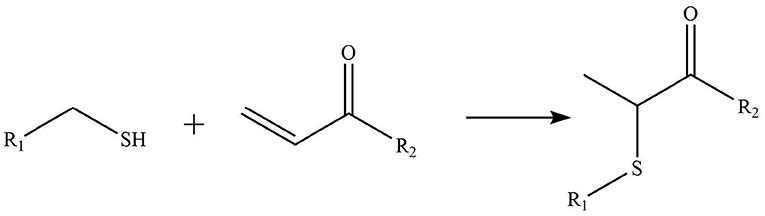
Figure 2. Reaction of thiol groups in the TLR4 signaling pathway with the α,β-unsaturated carbonyl group of phytochemicals.
Withaferin A, extracted from Withania somnifera plant, has been verified to attenuate several metabolic diseases. Mohamad reported that administrate with withaferin A (1.25 mg/kg/d) for 12 weeks protected against high-fat diet induced obesity through reducing hepatic mRNA expressions of TLR4, NF-κB, TNF-α), chemokine (C-C motif) ligand-receptor, and cyclooxygenase 2 (COX2) (29).
Tang and colleagues found that obesity, glycolipid metabolism disorder, inflammation, and oxidative stress were effectively alleviated by kaempferide treatment for 16 weeks in HFD mice, and the beneficial effects of kaempferide may be associated with inhibition of the TLR4/IκBα/NF-κB signaling pathways (30).
Isoliquiritigenin is a flavonoid derived from Glycyrrhiza uralensis, which was proved to improve HFD-induced adipose tissue fibrosis with decreased expression of TLR4 (31).
The diarylheptanoid curcumin is a naturally occurring yellow pigment found in the plant curcuma longa. It has been reported that curcumin exhibited anti-inflammatory property by interference with TLR4 and its downstream signaling pathway in HFD-induced obese mice (32–34). The molecular interactions between curcumin and TLR4 including: (1) Curcumin is also known to inhibit the activation of IkB kinase β (IKKβ), which is the main downstream of TLR4. The α, β-unsaturated carbonyl group of curcumin reacts with thiol group containing cysteine residue in the activation loop of IKKβ (35). (2) TLR4 dimerization is blocked by curcumin: the α, β-unsaturated carbonyl group of curcumin interacts with free thiol groups in cysteine residues in extracellular and cytoplasmic domains of TLR4. Therefore, curcumin can inhibit LPS-induced activation of both MyD88- and TRIF-dependent pathways of TLR4, and results in the inhibition of both NF-kB and interferon regulatory factor 3 (IRF3) (36).
On the other hand, phytochemicals such as resveratrol, epigallocatechin-3-gallate (EGCG), quercetin, luteolin, and analogs of luteolin, do not inhibit TLR4 dimerization. Instead, they inhibit TLR4 signaling by specifically inhibiting TANK binding kinase 1 (TBK1) kinase activity and consequently downregulate the expression of TBK1-targeted genes, including TNF-α, and IL-6 (37). Nevertheless, a clear understanding of the molecular determinants of TBK1 regulation and substrate selection has not been achieved. TBK1 activity can be regulated by phosphorylation of the serine 172 residue within the kinase activation loop (38, 39). Future work is warranted to investigate how specifically these phytochemicals regulated TBK1 kinase activity.
PPARs belong to a subfamily of the nuclear receptor superfamily of ligand-inducible transcription factors (40). Three PPAR subtypes including PPARα, PPARβ (also known as PPARδ), and PPARγ, have been identified (41, 42). Peroxisome proliferator-activated receptors control the expression of genes involved in adipogenesis, lipid metabolism, inflammation, and the maintenance of metabolic homeostasis, as summarized in Figure 3 (43).
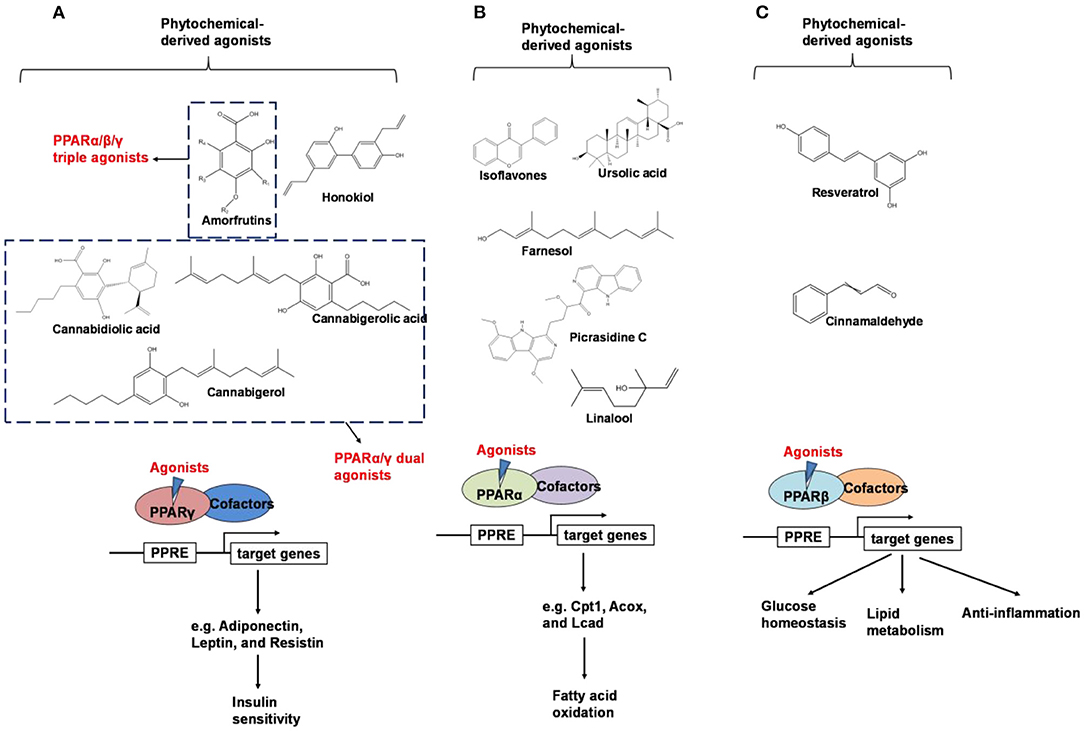
Figure 3. An overview of phytochemical-derived agonists and the biological function of PPARs. (A) Agonists of PPARγ. (B) Agonists of PPARα. (C) Agonists of PPARβ.
The regulation of gene transcription is identical in all three PPAR subtypes. Upon ligand binding, PPARs form heterodimers with retinoid X receptor (RXR). The PPAR-RXR heterodimer translocate to the nucleus, where it binds to peroxisome proliferator response elements (PPREs) in the promoter region of the target genes. The transcription process is then initiated, since a conformational change in PPAR-RXR complexes cause the dissociation of co-repressors and recruitment of transcriptional cofactors, while in the absence of a bound ligand, the heterodimer remains bound to the promoter region of its target genes in complex with co-repressors and associated histone deacetylases and chromatin-modifying enzymes, inhibiting the activation of target genes (44, 45).
PPARγ is predominantly expressed in the adipose tissue and plays a central role in lipid and glucose metabolism (46). PPARγ is activated by binding with small lipophilic ligands, mainly fatty acids. Synthetic PPARγ agonists, such as thiazolidinediones (TZDs), which were first reported as insulin-sensitizing drugs in the early 1980s (47), and have been widely used as a therapeutic compound in the treatment of type 2 diabetes, but their underlying mechanism remain unclear until the middle 1990s, when scientists found TDZs were ligands for PPARγ (48).TZDs-induced activation of PPARγ regulates the production and secretion of adipokines, including adiponectin, leptin, and resistin, which impact insulin sensitivity through endocrine signaling pathways (49–51).
Aside from the availability of agonists, the transcriptional activity of PPARγ is also regulated by its phosphorylation status (52). The ability of ligand to suppress Ser273 phosphorylation is well-correlated with their anti-diabetic effectiveness. TZDs inhibit the Cdk5-mediated Ser273 phosphorylation of PPARγ in adipose tissue (52), which in turn, down-regulate expression of genes involved in obesity, including adipsin, a fat-cell-selective gene, and adiponectin, an insulin-sensitizing adipokine (53). PPARγ ligands with poor agonistic activity but potent anti-diabetic effects were revealed to be strong inhibitors of the PPARγ phosphorylation by Cdk5 (54). Consequently, suppression of Ser273 phosphorylation of PPARγ was suggested as a promising approach for development of a new generation of anti-diabetic agents (53).
Activation of another set of genes leads to side effects of PPARγ activation. For instance, TZDs induce the expression of genes involved in adipocyte differentiation and fatty acid storage, such as adipocyte fatty acid binding protein (aP2) (55), phosphoenolpyruvate carboxykinase (PEPCK) (56), and lipoprotein lipase (LPL) (57). It has been reported that ectopic expression of PPARγ in non-adipogenic cells converts them into adipocytes (55), whereas PPARγ knockout mice are unable to develop adipose tissue (58). TZDs promote fat accumulation in type 2 diabetic patients is an example of these undesirable clinical side effects (59).
TZDs was fallen into disuse for their side effects and adverse events in recent years. Water retention was one of the serious clinical events due to TZDs, as well as edema and heart failure (60). The use of TDZs in diabetes clinic visits fell sharply as the side effects reported repeatedly, from 41% in 2005 to 16% in 2012 (61). Therefore, the use of selective PPARγ modulators is a potential way to prevent the side effects of some PPARγ agonists. Different binding modes influence the recruitment of coactivators and elicit a lower transactivation potential of the receptor, which results in the maintenance of antidiabetic activity while minimizing potential side effects of PPARγ modulators (62). The PPARγ ligand binding domain (LBD) consists of 13 α-helices, H1–H12 and H2', and one β-sheet region. Full agonists stabilize H12 by forming hydrogen bonds with the side chains of Ser 289, His 323, His 449, and Tyr 473. However, selective PPARγ modulators stabilize the β-sheet through a hydrogen bond with Ser 342 and helix H3 by hydrophobic interactions (63, 64), while other selective PPARγ modulators also display weaker interactions with the residues that stabilize H12 (65).
Therefore, inhibition of phosphorylation of Ser 273 in PPARγ, as well as the use of selective PPARγ modulators are two preferred therapeutic strategies for improving insulin sensitivity while preventing adipogenesis (53, 66).
Weidner et al. (66) found that amorfrutin, a family of isoprenoid-substituted benzoic acid derivatives from edible parts of Glycyrrhiza foetida and Amorpha fruticosa, possesses powerful antidiabetic effect. In diet-induced obese and db/db mice, amorfrutin treatment strongly improved insulin resistance, decreased plasma triglycerides and inflammatory parameters without concomitant increase of lipid accumulation or other unwanted side effects such as hepatoxicity (66). It might because that amorfrutins bind to and modulate PPARγ, which results in selective gene expression and physiological profiles markedly different from activation by synthetic TZDs. On the other hand, amorfrutins block HFD-induced PPARγ Ser273 phosphorylation in mouse adipocytes, leading to dysregulation of a large number of genes whose expression is altered in obesity (52, 66). Other natural products, such as honokiol, a lignan isolated from the bark, seed cones, and leaves of trees belonging to the genus Magnolia was also reported to improve metabolic parameters in diabetic animal models, with reduced side effects in comparison to TZDs agonists (54, 67).
PPARα is widely expressed in tissues with high fatty acid catabolic activity, including adipose tissue, heart, muscle, liver, kidney, and intestine (68). PPARα is also found to be widely expressed in the digestive tract and hippocampus (69). Activation of PPARα improves insulin resistance, promotes fatty acid catabolism, and inhibits transcription of genes related to the inflammatory response (70).
Several endogenous PPARα ligands have been proposed, including phospholipid 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine, fatty acids such as palmitic acid, oleic acid, linoleic acid, arachidonic acid, oleoylethanolamide, 3-hydroxy-(2,2)-dimethyl butyrate, hexadecanamide, and 9-octadecenamide (69, 71, 72). Synthetic PPARα agonists such as fibrates lower triglyceride levels and raise high density lipoprotein (HDL), and are used to treat severe hypertriglyceridemia (73). PPARα knockout mice display a fatty liver phenotype (74, 75). Overexpression of PPARα improves glucose tolerance in diet-induced obese mice (76).
Shay and Banz (77) suggested the effect of soy intake on lipid metabolism may be due to isoflavones, the major bioactive compound in soy, acting as activator of PPARα. Linalool, aroma and flavors in most herbal essential oils and teas, was found to act as a direct ligand of PPARα and reduce plasma TG levels, and the reduction was markedly attenuated by silencing PPARα expression (43). Ursolic acid, a natural triterpene compound found in various fruits and vegetables, is a PPARα agonist and was found to control the expression of genes related to lipid metabolism (78, 79). Goto et al. (78) reported that farnesol, a natural organic compound which is an acyclic sesquiterpene alcohol, mediated improvement of obesity-associated metabolic disorders through a PPARα-dependent manner. Picrasidine C, a dimeric β-carboline-type alkaloid isolated from the root of Picrasma quassioides, was identified as a selective PPARα agonist by binding with PPARα LBD forming hydrogen bonds with Cys276 and Thr279, therefore exhibiting potential in treating hyperlipidemia, atherosclerosis, and hypercholesterolemia (80).
As reviewed by Rigano et al., structurally related compounds may have robust differences on binding affinity to PPARα. For instance, daidzein or formononetin slightly activated PPARα, while the metabolite 6-hydroxydaidzein exerted a much higher agonistic PPARα activity. Similarly, 3′-hydroxygenistein exhibited a more potent on PPARα activation than its precursor genistein. Biochanin A, differing from genistein only by methylation of the 4'OH group, was robustly more potent than its precursor. In contrast, the metabolites dihydrogenistein and dihydrodaidzein did not transactivate PPARα (69, 81). The pentacyclic triterpene oleanolic acid was found to stimulate PPARα activation in keratinocytes while the closely related ursolic acid, differing only by the methylation pattern on ring E, failed to exhibit this activity (82). The reason for the differences is not well-understood: it may be related to an increase in the bioavailability of the molecules, an improved interaction with the binding site, different abilities to recruit coactivators or co-repressors, and/or cross-activation of other nuclear receptors (69).
As a phosphoprotein, PPARα can also be activated through post-translational modification. The phosphorylation sites in PPARα include Ser12, Ser21, Ser179, and Ser230 (83–87). The increased transactivation of PPARα may occur via decreased co-repressor interaction, such as NCoR, or increased interaction with co-activators, such as Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) (88). Since PGC-1α is one of the major transcription factors of non-shivering thermogenesis (89), phytochemicals that potential to activate its co-activator PPARα draws a lot of attention to induce thermogenesis in rodent models. Li et al. (90) reported that tyrosol, one of the main polyphenolic compounds in extra virgin olive oil, acted as a ligand which binds with PPARα, and increased its downstream genes expression, such as uncoupling protein 1, iodothyronine deiodinase 2 and PGC-1α, resulted in reduced body weight gain in HFD-induced obese mice.
Peroxisome proliferator-activated receptor β (PPARβ) is expressed ubiquitously (91). PPARβ, also a lipid ligand-inducible transcription factor, regulates lipid metabolism and glucose homeostasis. PPARβ is also shown to suppress the activities of several transcription factors, including NFκB, and activator protein 1, thus regulating anti-inflammatory cellular responses (92, 93). Compared to non-obese subjects, obese patients exhibited reduced PPARβ expression in both the subcutaneous and visceral adipose tissues (94). Previous reports have shown that PPARβ agonists, such as GW501516, ameliorates insulin resistance by increasing the expression of the insulin receptor, of significance for treatment of insulin resistance in patients with type 2 diabetes mellitus (95). Consistently, administered with the PPARβ agonist MBX-8025 for 8 weeks, overweight patients presented favorable trends in the body fat percentage, lean body mass and waist circumference (96).
Resveratrol is a natural stilbene found in grapes and red wine. Tsukamoto et al. (97) demonstrated that resveratrol treatment activated PPARβ in bovine arterial endothelial cells. Qin et al. (98) showed that resveratrol was able to regulate PPARβ expression in retinal pigment epithelial cells in a dose-dependent manner. In addition, Lu et al. suggest that the treatment of resveratrol, modulated adipokine expression and improves insulin sensitivity in adipocytes by down-regulating PPARβ (99).
Since the ligand binding domains among PPARs are 60–70% identical (100), phytochemicals may act as dual agonists for PPARs or as pan agonists. For example, D'Aniello et al. identified cannabigerolic acid, cannabidiolic acid and cannabigerol from C. sativa as PPARα/γ dual agonists (101). Amorfrutin has binding affinities for PPARα, PPARβ, and PPARγ (66).
Nuclear factor erythroid 2-related factor 2 (Nrf2), a transcription factor in the basic leucine zipper (bZIP) superfamily, is an attractive target for obesity and metabolic syndrome treatment and prevention. Dual roles of Nrf2 in obesity and related metabolic diseases, including the regulation of antioxidant defenses and hepatic fatty acid metabolism have been reported (102, 103).
Nrf2 behaves as a cellular redox status sensor and under normal circumstances, most Nrf2 is sequestered, bound to the cytoskeletal-anchoring protein Kelch-like ECH-associated protein 1 (Keap1), a cysteine rich, homodimeric zinc-finger protein in the cytosol. Keap1 acts as a substrate adaptor protein for the cullin 3-containing E3-ligase complex and targets Nrf2 for ubiquitination, which leads to proteasome mediated degradation of Nrf2. During the exposure of electrophilic or oxidative stressors, Nrf2 releases from sequestration as stressors interact with cysteine thiols of Keap1, followed by translocation to the nucleus. Nrf2 dimerizes with small Maf proteins or other leucine zipper proteins, binds to antioxidant response elements (ARE) in the promoter or enhancer of target genes, and thus regulates genes involved in the protection against oxidative/electrophilic stress (104, 105). These target genes include NAD(P)H dehydrogenase, quinone 1(Nqo1), glutathione S-transferase (Gst), and heme oxygenase-1 (Ho-1) (103). Electrophilic phytochemicals, such as curcumin and sulforaphane, possess α,β-unsaturated carbonyl groups which interact with thiol groups in cysteine residues of the Keap1 protein, releasing Nrf2 into the nucleus, thus activating downstream genes (106).
Human Keap1 has at least 25 reactive thiols, most of which are found in the IVR (intervening linker region) redox-sensitive region (107, 108). Specially, seven critical cysteine residues, including Cys151, Cys257, Cys273, Cys288, Cys297, Cys434, and Cys613 (Figure 4A), are responsible for sensing of alkenals and redox signals, which are required for the stimulation of Nrf2 (109). Not all phytochemicals preferably interact with the same region of Keap1 protein. Luo et al. found that for isoliquiritigenin, the top reactive thiols on human Keap1 were cysteine residues Cys151 and Cys266. However, for xanthohumol (a prenylated flavonoid isolated from hops) and 10-shogaol (pungent constituents of ginger), top reactive thiols on Keap1 were Cys151, Cys319, Cys613 and Cys151, Cys257, Cys368, respectively (110). The different interactions between these electrophiles with Keap1 might be due to their unique structures and reactivities (Figure 4B). It is imperative to investigate the reason for these differences.
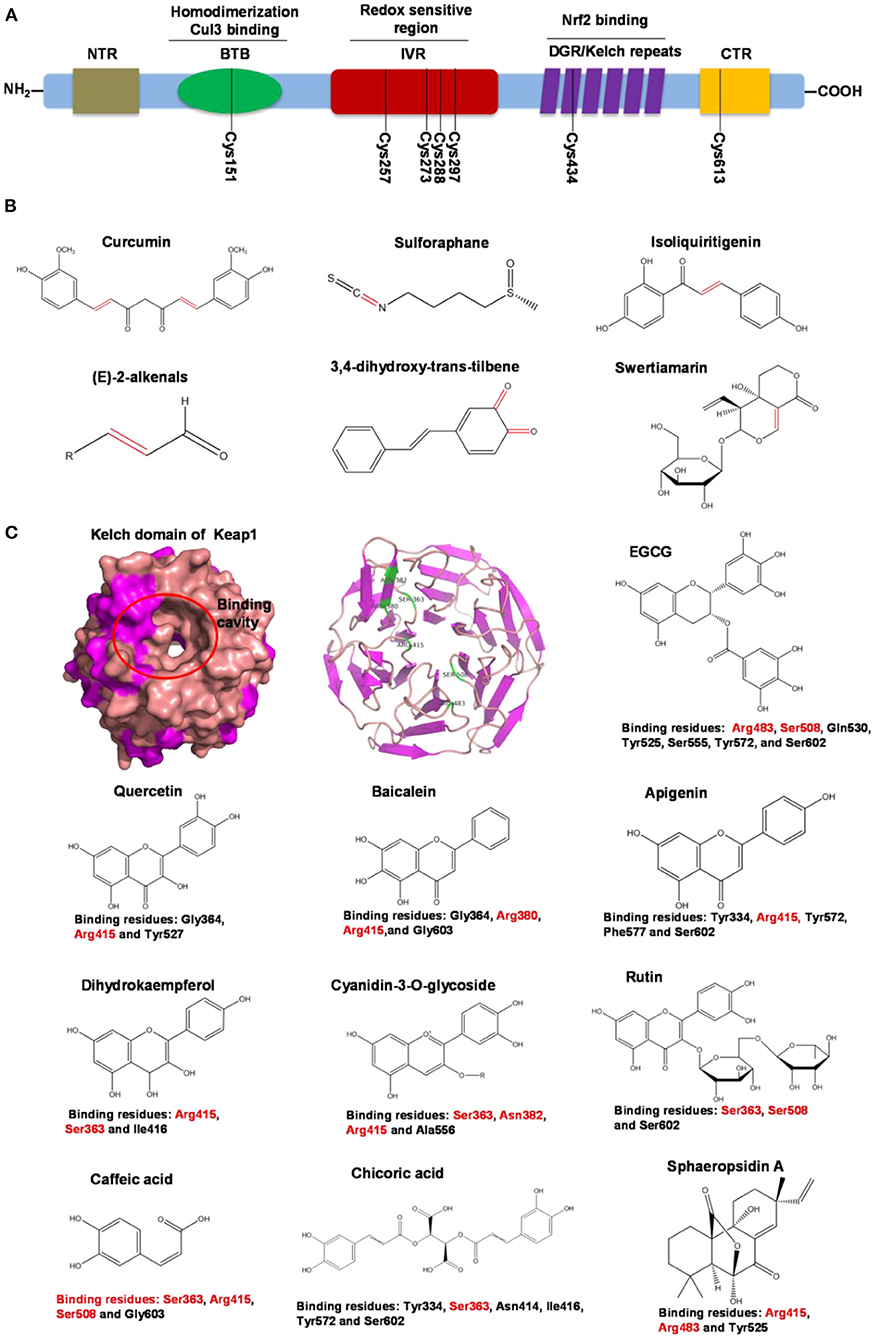
Figure 4. Summary of Keap1 protein structure and binding to phytochemicals. (A) Domain structures of Keap1 protein. Seven cysteine residues (Cys151, Cys257, Cys273, Cys288, Cys297, Cys434, and Cys613) are responsible for covalent binding with phytochemicals to activate Nrf2. (B) Phytochemicals with α, β-unsaturated carbonyl groups having highly potent binding with cysteine residues of Keap1 through covalent modification. (C) Non-covalent binding cavity and residues of Kelch domain of Keap1 with phytochemicals. The reactive unsaturated carbonyl group of phytochemicals and potential non-covalent binding residues of Kelch domain of Keap1 are highlighted using red font.
Notably, electrophilic Nrf2 activators might be not highly selective for Keap1 over other cytoplasmic ubiquitous cysteines due to their action involving the covalent reaction with cysteine thiols, which leads to non-specific effects by perturbing multiple targets except for Keap1 (111). Alternatively, direct and non-covalent disrupting of Keap1-Nrf2 protein-protein interaction (PPI) has emerged as a rationale for selective activation of Nrf2. The C-terminal Kelch domain of Keap1 is responsible for its binding with Nrf2 (109). Among Kelch domain residues, three highly conserved arginine residues (Arg380, Arg415, and Arg483) and other serine, glutamate and asparagine residues (Ser363, Ser508, and Asn382) play important roles in Keap1 binding to Nrf2 (112). Numerous common natural phytochemicals (quercetin, EGCG, baicalein, caffeic acid, sphaeropsidin A, dihydrokaempferol, rutin, apigenin, and anthocyanins) can bind to Kelch domain through the formation of non-covalent bonds (H-bond, π-π/H-π interaction, hydrophobic effects, van der Waals and ionic bonds) with the specific residues involving Tyr334, Ser363, Gly364, Asn382, Arg415, Arg483, Ser508, Tyr525, Tyr527, Gln530, Ser555, Ala556, Tyr572, Phe577, Ser602, and Gly603, thereby directly disrupting Keap1-Nrf2 interaction to activate Nrf2 (Figure 4C) (106, 113–119). However, these interactions of the Kelch domain of Nrf2 with phytochemicals were predicted based on molecular docking, which should be further compared and verified by using direct biomolecular interaction techniques, such as isothermal titration calorimetry, surface plasmon resonance, and biolayer interferometry.
Besides activating antioxidant genes, Nrf2 is also known to participate in the regulation of hepatic fatty acid metabolism in rodents, as a negative regulator of genes that promote hepatosteatosis (120). Nrf2 has been reported to directly impact the regulation of genes including ATP citrate lyase (Acly), acetyl-CoA carboxylase 1 (Acaca), fatty acid synthase (Fasn) and fatty acid elongase 6 (Elovl6) (104, 121). Hepatic lipogenesis is negatively regulated by Nrf2 in mice administrated a high fat diet (122, 123).
Activation of Nrf2 pathway via phytochemical supplementation has been found to protect mice from obesity. In a study by Okada et al. (124) sulforaphane supplementation suppressed oxidative stress and hepatic fibrosis in mice induced by MCD diet. Sulforaphane also activates lipolysis by transcriptionally regulating genes related to lipid metabolism, including adipose triglyceride lipase (Atgl) and hormone-sensitive lipase (Hsl) (125). Nagata et al. (126), revealed the anti-obesity effect of glucoraphanin, precursor of sulforaphane, in HFD-fed mice, and found that 0.3% w/w glucoraphanin for oral administration for 14 weeks significantly reduced body weight, alleviated hepatic steatosis and improved insulin sensitivity in wild type mice but not in Nrf2 KO mice. They further investigated that whole body energy expenditure was increased, accompanied with over expression of Ucp1 in WAT, but these results were not found in Nrf2 KO mice. Thus, stimulation of energy metabolism by Nfr2 activation was considered effective to combat obesity. Other natural phytochemicals (i.e., sesamol, curcumin, Garcina cambogia, timosaponin) that can activate Nrf2 are potential candidates to prevent obesity and improve metabolic disease via Nrf2 pathway (127–130).
Interestingly, mice deficient in Nrf2 were protected from high fat diet induced obesity, including improved glucose tolerance, reduced hepatic triglyceride content and decreased liver weight (131, 132). Chartoumpekis et al. (133) found that Nrf2 knockout mice displayed decreased fat mass in association with small adipocytes and are resistant to diet-induced obesity. Xu et al. (134) found that enhanced Nrf2 activity induced insulin resistance in leptin-deficient mice. The mechanisms proposed include interaction of Nrf2 with other pathways (Fibroblast Growth Factor 21, or lipid synthesis enzymes) mainly in liver and white adipose tissue (135).
Despite contrary findings that both Nrf2 gain or loss of function may protect from obesity, this likely happens through distinct mechanisms/pathways. A detailed assessment of obesity in mouse models with Nrf2 deletion or overexpression is warranted to determine the molecular pathways underlying the positive and negative effect of Nrf2.
Fat mass and obesity-associated protein (FTO) is a member of the Fe (II)- and oxoglutarate-dependent AlkB dioxygenase family, and is known for the strong association of the multiple single-nucleotide polymorphisms located in its intron 1 with risk of obesity (136). FTO knockout or loss-of-function mutations lead to reduced body weight, and its overexpression contributes to obesity (137). The mechanism of FTO-induced obesity is summarized in Figure 5A: FTO expression in the brain positively regulates food intake through neuropeptide Y (NPY) and ghrelin (137, 138). FTO in the adipose tissue has been reported to control the expression of uncoupling protein 1, a mitochondrial inner membrane proton channel linked to thermogenesis, through FOXO1 (139). Adipogenesis is promoted by FTO by targeting Atg5- and Atg7-mediated autophagy as well as adipogenic regulator factor Runx1t1 (140, 141). The role of FTO in controlling thermogenesis and adipogenesis in the adipose tissue largely depends on its regulation of m6A mRNA methylation, which is the most abundant mRNA modification in mammals and is involved in various biological processes including obesity and obesity associated-metabolic disorders (139, 142).
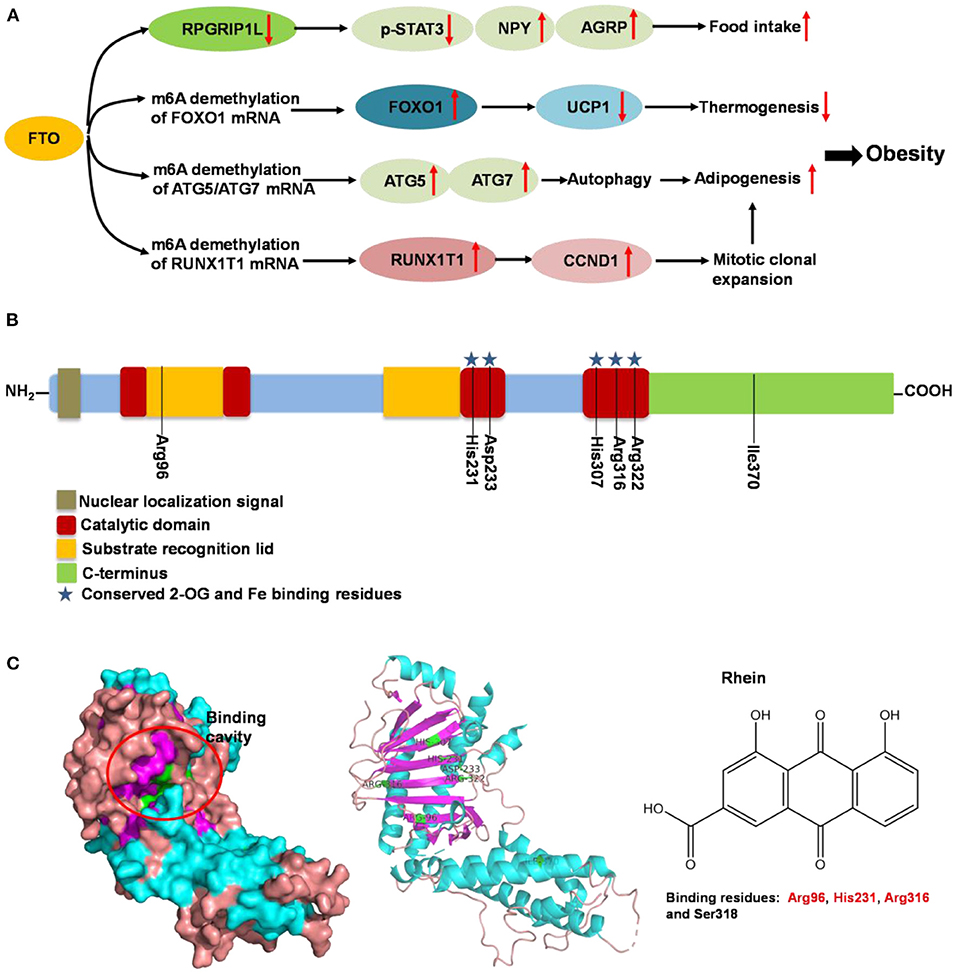
Figure 5. Summary of molecular mechanism of FTO-induced obesity, FTO protein structure and its binding with rhein. (A) Mechanism of FTO-induced obesity through regulating food intake, thermogenesis and adipogenesis. (B) Domain structures of FTO protein. Seven amino acid residues (Arg96, His231, Asp233, His307, Arg316, Arg322, and Ile370) are responsible for FTO activity. (C) Binding cavity and residues of FTO protein with rhein. The potential binding residues of FTO are highlighted using red font.
Following the determination of the FTO crystal structure by Chai's group in 2010 (Figure 5B) (143), FTO is widely viewed as an attractive biological target; potentially a small-molecule inhibitor specifically targeting FTO could be developed for the treatment of metabolic disorders such as obesity and diabetes. Some synthetic compounds, such as CHTB, FB23, and diacerein, occupying the αKG and/or substrate binding site have been identified as potent inhibitors of FTO, potentially regulating obesity (144, 145). Peng et al. (139) used a structure-based hierarchical virtual screening approach to identify potential FTO inhibitors from 1,323 FDA-approved drugs, and found entacapone as a chemical inhibitor of FTO which directly binds to FTO and inhibits its demethylation activity, mediating metabolic regulation through FOXO1. Development of FTO-specific inhibitors from natural phytochemicals is a promising strategy to avoid the side effects of synthetic drugs for treatment of obesity and other chronic diseases (146). The natural phytochemical Rhein was identified as the first FTO inhibitor that disrupts FTO activity by directly binding to the catalytic domain, which blocks access to the ssRNA substrate (Figure 5C) (147), and rhein remarkably suppresses adipogenesis in the stage-specific and dose-dependent manners (https://pubmed.ncbi.nlm.nih.gov/34790688/). But, side-by-side comparison analysis revealed that the rhein treatment and FTO knockdown triggered the differential gene regulatory patterns, though both resulting in impaired adipocyte formation, suggesting separate regulation of global m6A pattern and adipogenesis mediated by rhein (148). So, the interrelation of rhein-FTO- adipogenesis remain elusive. Epigallocatechin gallate has been shown to inhibit adipogenesis by regulating FTO expression in an mRNA m6A-dependent manner (149). Baicalin has been reported to ameliorate high-fat diet-induced obesity and hepatic steatosis through carnitine palmitoyltransferase 1 (CPT1) (150), of which mRNA serves as a potential substrate of FTO for m6A modification, predicted by m6A-Atlas (151). Zhong et al. (152, 153), reported that Angelica sinensis alleviated HFD-induce obesity through altering expression of FTO gene. Betaine was also found to decrease FTO expression and improved m6A methylation in adipose tissue of wild-type mice with high-fat diet, resulting in decreased final body weight and improved glucose tolerance (154). In another human trial included 214 participants revealed that, Garcinia cambogia supplement with 1,000 mg/day for 6 months significantly reduced body weight and improved serum lipid profile, but these beneficial effects were hampered by polymorphisms of FTO gene (155). Suggesting that, FTO may at least partially involved in this anti-obesity process.
The miRNAs are a class of regulatory RNAs. They are small non-coding RNA molecules, averaging 22 ribonucleotides in length, and may repress gene expression post-transcriptionally by binding to untranslated regions and coding sequences of target mRNAs (156). The class of RNA was first discovered in Caenorhabditis elegans in 1993, and then identified in vertebrates and plants (157).
The miRNA coding sequences are transcribed by RNA polymerase II to yield primary miRNAs (pri-miRNAs) in the nucleus, and further processed by Drosha RNAse III endonuclease and microprocessor complex subunit DGCR8 to release precursor miRNAs (pre-miRNAs), which are about 60–70 ribonucleotides long. The pre-miRNAs are transported into the cytoplasm by Exportin-5 and cleaved by Dicer complex, another RNase III enzyme, to generate miRNA/miRNA double stranded molecules (158). Upon separation of the two strands, the guide strand binds to an Argonaute (Ago) protein and is integrated into the RNA-induced silencing complex (RISC), where it targets and binds to the 3′-untranslated region (3′UTR) of target mRNAs via base pair complementarity (159). This binding leads to degradation or translational repression of target mRNAs (160).
Thousands of different miRNAs have been identified, and miRNAs are now recognized as one of the most abundant classes of gene-regulatory molecules in multicellular organisms (161). Computational and experimental studies have shown that some miRNAs play a role in lipid metabolism and glucose homeostasis, therefore, may be influencing the pathogenesis of metabolic diseases (162).
MicroRNA-122 (miR-122), an abundant liver-specific miRNA, accounts for approximately 70% of total miRNAs in the liver. microRNA-122 is the first miRNA to be linked recently to fat and cholesterol metabolism, suggesting it as a therapeutic target for metabolic diseases (163, 164).
Temporary miR-122 inhibition resulted in reduced plasma cholesterol levels in both normal and diet-induced obese mice (163, 165), by down-regulating hepatic gene expression including HMG-CoA reductase and phosphomevalonate kinase (PMVK), which are involved in cholesterol biosynthesis (163). Consistently, a study of non-human primates demonstrated that miR-122 inhibition caused a dose-dependent decrease in plasma cholesterol, indicating a therapeutic potential in the treatment of hypercholesterolemia in humans (166). A study from Wang et al. found that in young adults, elevated circulating miR-122 is positively associated with obesity and insulin resistance (167).
Excessive retention of triglyceride within hepatocytes was observed in miR-122-knockout mice. Hsu et al. found that both liver-specific and germline miR-122 knockout mice exhibited increased hepatic triglyceride accumulation and progressive steatohepatitis, which was not seen in the mice with temporary miR-122 inhibition (168). The underlying mechanism may be due to the up-regulation of triglyceride biosynthesis related gene expression in the liver: 1-acylglycerol-3-phosphate O-acyltransferase 1 (Agpat1) and monoacylglycerol O-acyltransferase 1 (Mogat1), (168) and the reduction of microsomal TG transfer protein (Mttp) (169), which normally functions to enhance the rate of lipid transfer between vesicles (170).
Phytochemicals which modulate the expression of miR-122 possess the potential to regulate lipid metabolism associated with obesity and metabolic syndrome. Epigallocatechin-3-gallate was found to decrease miR-122 expression. Using 1H NMR spectroscopy, EGCG was observed to bind directly to miR-122 through an interaction with all of the rings of the EGCG molecule (171). However, resveratrol was found to bind directly to miR-122 primarily through an A ring interaction, which increases the expression of miR-122. It is hypothesized that the size and chemical structure of these molecules deferentially influenced the expression of miRNA, which may account for the opposite effects of resveratrol and EGCG on miRNA modulation (171).
Other phytochemicals were reported as modulators of miRNA levels. Polyphenols in acai and red muscadine grape protect human umbilical vascular endothelial cells from glucose and lipopolysaccharide-induced inflammation, partly acting through the upregulation of miR-126 expression (172). Zerumbone, a phytochemical isolated from the subtropical Zingiberaceae family, was reported to reverse high-fat diet-induced adiposity by restoring AMPK-regulated lipogenesis and the regulation of miRNA-146b mediated adipogenesis (173). Fisetin, which present in fruits and vegetables such as strawberries, apple, cucumber, persimmon, grape and onion, was shown to suppresses the expression of hepatic miR-378, a miR located in intron of the peroxisome proliferator-activated receptor gammacoactivator-1 beta (PGC-1β), resulting in preventing obesity and hepatic lipid accumulation induced by high fat diet in mice (174). Polyphenols derived from Hibiscus sabdariffa were reported to regulate the expression of miR-103, miR-107 and miR-122 and attenuated weight gain, liver steatosis and insulin resistance in hyperlipidemic mice (175).
Emerging data suggest that phytochemicals alter the expression of miRNAs involved in regulation of cancer pathobiology by modulating the expression of miRNAs through mechanisms including epigenetic, transcriptional, and miRNA processing (176). However, relatively little is known about how phytochemicals regulate miRNAs related to obesity and metabolic syndrome (177). It is hypothesized that similar mechanisms might be involved, although more investigation is needed. Since each mammalian miRNA may regulate a large number of target genes, it can be that several different miRNAs can act synergistically at multiple target sites of a single Mrna (178). Thus, it is imperative to manipulate multiple candidate miRNAs in different combinations rather than changing one miRNA at a time for a full functional characterization of their regulatory impact.
Plenty number of phytochemicals have been reported as candidates for the management of obesity. However, the role of the structure of phytochemicals in specific signaling pathways remains unclear. In this review, we discussed the regulation of phytochemicals on five different signaling pathways involved in obesity and related symptoms (Table 1). In particular, we discussed specific molecular structures required for regulation to occur. We suggest that, in the future, more attention should be paid to the preliminary research on the correlation between structures of phytochemical and specific target molecular using accessible methods, such as molecular docking by computer or in vitro kinetic measurements. Although the current evidence is limited, we suggest that in some cases, the optimal molecular structure for a specific pathway to become activated or regulated may ultimately be deduced.
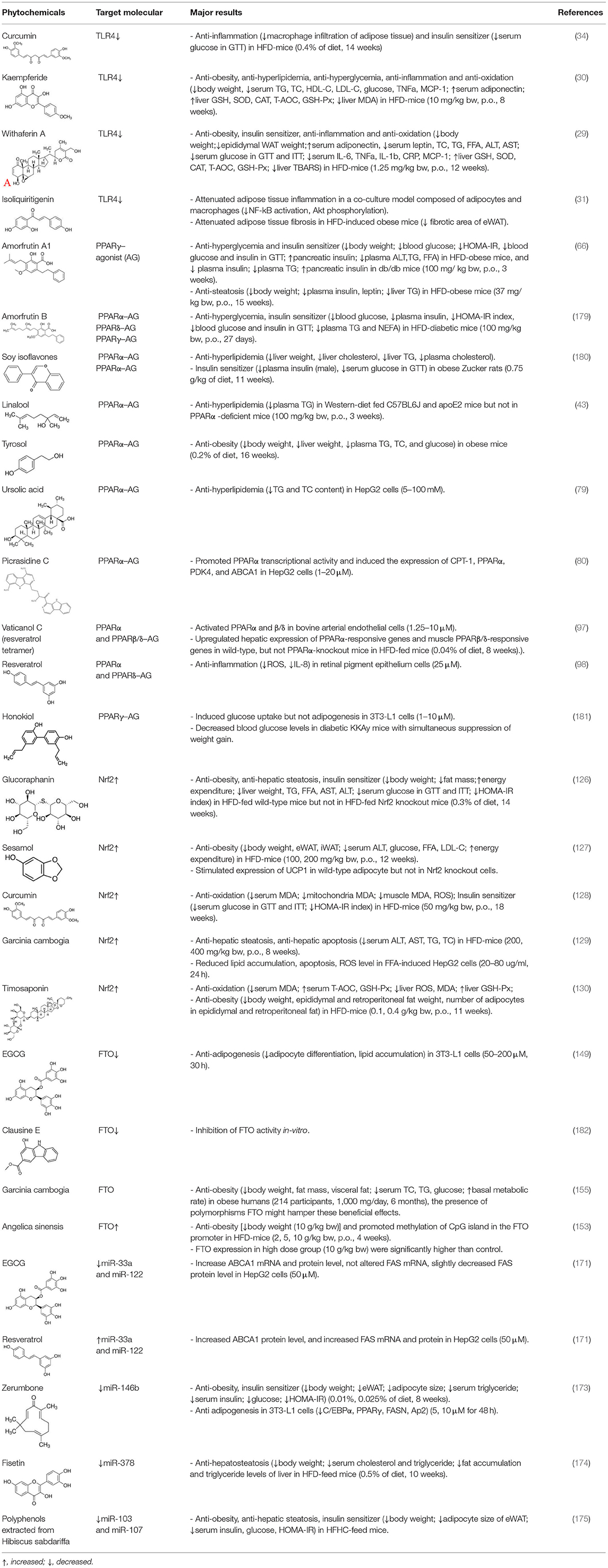
Table 1. Phytochemicals targeting specific signaling pathway act against obesity and related symptoms.
Z-YD and TL: conceptualization. XL: writing—original draft preparation. BZ and LZ: writing—review and editing. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of P.R. China (Grant No. 82160168), Shuangqian Project of Scientific and Technological Innovation of High-end Talents-Natural Science, Jiangxi Province (Grant No. jxsq2020101063), and Natural Science Foundation of Jiangxi Province (Grant No. 20212BAB216075).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
TG, triglyceride; TC, total cholesterol; HDLC, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; MCP-1, monocyte chemoattractant protein-1; GSH, glutathione; SOD, superoxide dismutase; CAT, catalase; T-AOC, total antioxidant capacity; GSH-Px, glutathione peroxidase; MDA, malonaldehyde; WAT, white adipose tissue; FFA, free fatty acid; ALT, alanine transaminase; AST, aspartate aminotransferase; IL-1b, interleukin-1b; GTT, glucose tolerance test; ITT, insulin tolerance test; CRP, C-reactive protein; TBARS, thiobarbituric acid reactive substance; eWAT, epididymal white adipose tissue; HOMA-IR, homeostasis model assessment of insulin resistance; NEFA, non-esterified fatty acid; CPT-1, carnitine palmitoyl transferase 1; PDK-4, pyruvate dehydrogenase kinase-4; ABCA1, ATP-binding cassette transporter A1; ROS, reactive oxygen species; IL-8, interleukin-8; FAS, fatty acid synthase; C/EBPα, CCAAT-enhancer binding protein alpha; Ap2, adaptor protein 2.
1. Hoare D, Bussooa A, Neale S, Mirzai N, Mercer J. The future of cardiovascular stents: bioresorbable and integrated biosensor technology. Adv Sci. (2019) 6:1900856. doi: 10.1002/advs.201900856
2. Chatterjee S, Khunti K, Davies MJ. Type 2 diabetes. Lancet. (2017) 389:2239–51. doi: 10.1016/S0140-6736(17)30058-2
3. Gadde KM, Martin CK, Berthoud HR, Heymsfield SB. Obesity: pathophysiology and management. J Am Coll Cardiol. (2018) 71:69–84. doi: 10.1016/j.jacc.2017.11.011
4. Konopelniuk VV, Goloborodko II, Ishchuk TV, Synelnyk TB, Ostapchenko LI, Spivak MY, et al. Efficacy of Fenugreek-based bionanocomposite on renal dysfunction and endogenous intoxication in high-calorie diet-induced obesity rat model-comparative study. EPMA J. (2017) 8:377–90. doi: 10.1007/s13167-017-0098-2
5. Konstantinidi M, Koutelidakis AE. Functional foods and bioactive compounds: a review of its possible role on weight management and obesity's metabolic consequences. Medicines. (2019) 6:94. doi: 10.3390/medicines6030094
6. Molyneux RJ, Lee ST, Gardner DR, Panter KE, James LF. Phytochemicals: the good, the bad and the ugly? Phytochemistry. (2007) 68:2973–85. doi: 10.1016/j.phytochem.2007.09.004
7. Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. (2011) 30:16–34. doi: 10.3109/08830185.2010.529976
8. Jia L, Vianna CR, Fukuda M, Berglund ED, Liu C, Tao C, et al. Hepatocyte Toll-like receptor 4 regulates obesity-induced inflammation and insulin resistance. Nat Commun. (2014) 5:3878. doi: 10.1038/ncomms4878
9. Benomar Y, Amine H, Crépin D, Al Rifai S, Riffault L, Gertler A, et al. Central resistin/TLR4 impairs adiponectin signaling, contributing to insulin and FGF21 resistance. Diabetes. (2016) 65:913–26. doi: 10.2337/db15-1029
10. Ji Y, Sun S, Shrestha N, Darragh LB, Shirakawa J, Xing Y, et al. Toll-like receptors TLR2 and TLR4 block the replication of pancreatic β cells in diet-induced obesity. Nat Immunol. (2019) 20:677–86. doi: 10.1038/s41590-019-0396-z
11. Marcelo R, Philip C. Obesity, inflammation, toll-like receptor 4 and fatty acids. Nutrients. (2018) 10:432. doi: 10.3390/nu10040432
12. Degirmenci I, Ozbayer C, Kebapci MN, Kurt H, Colak E, Gunes HV. Common variants of genes encoding TLR4 and TLR4 pathway members TIRAP and IRAK1 are effective on MCP1, IL6, IL1β, and TNFα levels in type 2 diabetes and insulin resistance. Inflamm Res. (2019) 68:801–814. doi: 10.1007/s00011-019-01263-7
13. Rivera CA, Adegboyega P, van Rooijen N, Tagalicud A, Allman M, Wallace M. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. J Hepatol. (2007) 47:571–9. doi: 10.1016/j.jhep.2007.04.019
14. Spruss A, Kanuri G, Wagnerberger S, Haub S, Bischoff SC, Bergheim I. Toll-like receptor 4 is involved in the development of fructose-induced hepatic steatosis in mice. Hepatology. (2009) 50:1094–104. doi: 10.1002/hep.23122
15. Wu HM, Ni XX, Xu QY, Wang Q, Li XY, Hua J. Regulation of lipid-induced macrophage polarization through modulating peroxisome proliferator-activated receptor-gamma activity affects hepatic lipid metabolism via a Toll-like receptor 4/NF-κB signaling pathway. J Gastroenterol Hepatol. (2020) 35:1998–2008. doi: 10.1111/jgh.15025
16. Shirey KA, Lai W, Brown LJ, Blanco JCG, Beadenkopf R, Wang Y, et al. Select targeting of intracellular Toll-interleukin-1 receptor resistance domains for protection against influenza-induced disease. Innate Immun. (2020) 26:26–34. doi: 10.1177/1753425919846281
17. Vaure C, Liu Y. A comparative review of toll-like receptor 4 expression and functionality in different animal species. Front Immunol. (2014) 5:316. doi: 10.3389/fimmu.2014.00316
18. Guo J, Friedman SL. Toll-like receptor 4 signaling in liver injury and hepatic fibrogenesis. Fibrogenesis Tissue Repair. (2010) 3:21. doi: 10.1186/1755-1536-3-21
19. Tomita K, Tamiya G, Ando S, Ohsumi K, Chiyo T, Mizutani A, et al. Tumour necrosis factor alpha signalling through activation of Kupffer cells plays an essential role in liver fibrosis of non-alcoholic steatohepatitis in mice. Gut. (2006) 55:415–24. doi: 10.1136/gut.2005.071118
20. Cawthorn WP, Sethi JK. TNF-alpha and adipocyte biology. FEBS Lett. (2008) 582:117–31. doi: 10.1016/j.febslet.2007.11.051
21. Ma KL, Ruan XZ, Powis SH, Chen Y, Moorhead JF, Varghese Z. Inflammatory stress exacerbates lipid accumulation in hepatic cells and fatty livers of apolipoprotein E knockout mice. Hepatology. (2008) 48:770–81. doi: 10.1002/hep.22423
22. Marí M, Caballero F, Colell A, Morales A, Caballeria J, Fernandez A, et al. Mitochondrial free cholesterol loading sensitizes to TNF- and Fas-mediated steatohepatitis. Cell Metab. (2006) 4:185–98. doi: 10.1016/j.cmet.2006.07.006
23. Zhang W, Kudo H, Kawai K, Fujisaka S, Usui I, Sugiyama T, et al. Tumor necrosis factor-alpha accelerates apoptosis of steatotic hepatocytes from a murine model of non-alcoholic fatty liver disease. Biochem Biophys Res Commun. (2010) 391:1731–6. doi: 10.1016/j.bbrc.2009.12.144
24. Ahmad R, Al-Mass A, Atizado V, Al-Hubail A, Al-Ghimlas F, Al-Arouj M, et al. Elevated expression of the toll like receptors 2 and 4 in obese individuals: its significance for obesity-induced inflammation. J Inflamm. (2012) 9:48. doi: 10.1186/1476-9255-9-48
25. Xu M, Ge C, Qin Y, Gu T, Lv J, Wang S, et al. Activated TNF-α/RIPK3 signaling is involved in prolonged high fat diet-stimulated hepatic inflammation and lipid accumulation: inhibition by dietary fisetin intervention. Food Funct. (2019) 10:1302–1316. doi: 10.1039/C8FO01615A
26. Rungeler P, Castro V, Mora G, Goren N, Vichnewski W, Pahl HL, et al. Inhibition of transcription factor NF-kappaB by sesquiterpene lactones: a proposed molecular mechanism of action. Bioorg Med Chem. (1999) 7:2343–52. doi: 10.1016/S0968-0896(99)00195-9
27. Dinkova-Kostova AT, Massiah MA, Bozak RE, Hicks RJ, Talalay P. Potency of Michael reaction acceptors as inducers of enzymes that protect against carcinogenesis depends on their reactivity with sulfhydryl groups. Proc Natl Acad Sci USA. (2001) 98:3404–9. doi: 10.1073/pnas.051632198
28. Siedle B, García-Piñeres AJ, Murillo R, Schulte-Mönting J, Castro V, Rüngeler P, et al. Quantitative structure-activity relationship of sesquiterpene lactones as inhibitors of the transcription factor NF-kappaB. J Med Chem. (2004) 47:6042–54. doi: 10.1021/jm049937r
29. Abu Bakar MH, Azmi MN, Shariff KA, Tan JS. Withaferin A protects against high-fat diet-induced obesity via attenuation of oxidative stress, inflammation, and insulin resistance. Appl Biochem Biotechnol. (2019) 188:241–59. doi: 10.1007/s12010-018-2920-2
30. Tang H, Zeng Q, Ren N, Wei Y, He Q, Chen M, et al. Kaempferide improves oxidative stress and inflammation by inhibiting the TLR4/IκBα/NF-κB pathway in obese mice. Iran J Basic Med Sci. (2021) 24:493–8. doi: 10.22038/ijbms.2021.52690.11892
31. Watanabe Y, Nagai Y, Honda H, Okamoto N, Yamamoto S, Hamashima T, et al. Isoliquiritigenin attenuates adipose tissue inflammation in vitro and adipose tissue fibrosis through inhibition of innate immune responses in mice. Sci Rep. (2016) 6:23097. doi: 10.1038/srep23097
32. Yu S, Wang X, He X, Wang Y, Gao S, Ren L, et al. Curcumin exerts anti-inflammatory and antioxidative properties in 1-methyl-4-phenylpyridinium ion (MPP(+))-stimulated mesencephalic astrocytes by interference with TLR4 and downstream signaling pathway. Cell Stress Chaperones. (2016) 21:697–705. doi: 10.1007/s12192-016-0695-3
33. Patel SS, Acharya A, Ray RS, Agrawal R, Raghuwanshi R, Jain P. Cellular and molecular mechanisms of curcumin in prevention and treatment of disease. Crit Rev Food Sci Nutr. (2020) 60:887–939. doi: 10.1080/10408398.2018.1552244
34. Islam T, Koboziev I, Albracht-Schulte K, Mistretta B, Scoggin S, Yosofvand M, et al. Curcumin reduces adipose tissue inflammation and alters gut microbiota in diet-induced obese male mice. Mol Nutr Food Res. (2021) 65:e2100274. doi: 10.1002/mnfr.202100274
35. Miura K, Seki E, Ohnishi H, Brenner DA. Role of toll-like receptors and their downstream molecules in the development of nonalcoholic Fatty liver disease. Gastroenterol Res Pract. (2010) 2010:362847. doi: 10.1155/2010/362847
36. Youn HS, Saitoh SI, Miyake K, Hwang DH. Inhibition of homodimerization of Toll-like receptor 4 by curcumin. Biochem Pharmacol. (2006) 72:62–9. doi: 10.1016/j.bcp.2006.03.022
37. Zhao L, Lee JY, Hwang DH. Inhibition of pattern recognition receptor-mediated inflammation by bioactive phytochemicals. Nutr Rev. (2011) 69:310–20. doi: 10.1111/j.1753-4887.2011.00394.x
38. Clark K, Peggie M, Plater L, Sorcek RJ, Young ER, Madwed JB, et al. Novel cross-talk within the IKK family controls innate immunity. Biochem J. (2011) 434:93–104. doi: 10.1042/BJ20101701
39. Ma X, Helgason E, Phung QT, Quan CL, Iyer RS, Lee MW, et al. Molecular basis of Tank-binding kinase 1 activation by transautophosphorylation. Proc Natl Acad Sci USA. (2012) 109:9378–83. doi: 10.1073/pnas.1121552109
40. Laudet V, Hänni C, Coll J, Catzeflis F, Stéhelin D. Evolution of the nuclear receptor gene superfamily. EMBO J. (1992) 11:1003–13. doi: 10.1002/j.1460-2075.1992.tb05139.x
41. Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. (1990) 347:645–50. doi: 10.1038/347645a0
42. Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W. Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell. (1992) 68:879–87. doi: 10.1016/0092-8674(92)90031-7
43. Jun HJ, Lee JH, Kim J, Jia Y, Kim KH, Hwang KY, et al. Linalool is a PPARα ligand that reduces plasma TG levels and rewires the hepatic transcriptome and plasma metabolome. J Lipid Res. (2014) 55:1098–110. doi: 10.1194/jlr.M045807
44. Gearing KL, Göttlicher M, Teboul M, Widmark E, Gustafsson JA. Interaction of the peroxisome-proliferator-activated receptor and retinoid X receptor. Proc Natl Acad Sci USA. (1993) 90:1440–4. doi: 10.1073/pnas.90.4.1440
45. Yu S, Reddy JK. Transcription coactivators for peroxisome proliferator-activated receptors. Biochim Biophys Acta. (2007) 1771:936–51. doi: 10.1016/j.bbalip.2007.01.008
46. Zieleniak A, Wójcik M, Wozniak LA. Structure and physiological functions of the human peroxisome proliferator-activated receptor gamma. Arch Immunol Ther Exp. (2008) 56:331–45. doi: 10.1007/s00005-008-0037-y
47. Fujita T, Sugiyama Y, Taketomi S, Sohda T, Kawamatsu Y, Iwatsuka H, et al. Reduction of insulin resistance in obese and/or diabetic animals by 5-[4-(1-methylcyclohexylmethoxy)benzyl]-thiazolidine-2,4-dione (ADD-3878, U-63,287, ciglitazone), a new antidiabetic agent. Diabetes. (1983) 32:804–10. doi: 10.2337/diab.32.9.804
48. Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma). J Biol Chem. (1995) 270:12953–6. doi: 10.1074/jbc.270.22.12953
49. Choi SS, Park J, Choi JH. Revisiting PPARγ as a target for the treatment of metabolic disorders. BMB Rep. (2014) 47:599–608. doi: 10.5483/BMBRep.2014.47.11.174
50. Wu L, Guo C, Wu J. Therapeutic potential of PPARγ natural agonists in liver diseases. J Cell Mol Med. (2020) 24:2736–48. doi: 10.1111/jcmm.15028
51. Ahsan W. The journey of thiazolidinediones as modulators of PPARs for the management of diabetes: a current perspective. Curr Pharm Design. (2019) 25:2540–54. doi: 10.2174/1381612825666190716094852
52. Choi JH, Banks AS, Estall JL, Kajimura S, Boström P, Laznik D, et al. Obesity-linked phosphorylation of PPARγ by cdk5 is a direct target of the anti-diabetic PPARγ ligands. Nature. (2010) 466:451. doi: 10.1038/nature09291
53. Jones D. Potential remains for PPAR-targeted drugs. Nat Rev Drug Discov. (2010) 9:668–9. doi: 10.1038/nrd3271
54. Wang L, Waltenberger B, Pferschy-Wenzig EM, Blunder M, Liu X, Malainer C, et al. Natural product agonists of peroxisome proliferator-activated receptor gamma (PPARγ): a review. Biochem Pharmacol. (2014) 92:73–89. doi: 10.1016/j.bcp.2014.07.018
55. Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. (1994) 8:1224–34. doi: 10.1101/gad.8.10.1224
56. Tontonoz P. PPARγ2 regulates adipose expression of the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. (1995) 15:351–7. doi: 10.1128/MCB.15.1.351
57. Schoonjans K, Peinado-Onsurbe J, Lefebvre AM, Heyman RA, Briggs M, Deeb S, et al. PPARalpha and PPARgamma activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. (1996) 15:5336–48. doi: 10.1002/j.1460-2075.1996.tb00918.x
58. Wang Q, Tang J, Jiang S, Huang Z, Song A, Hou S, et al. Inhibition of PPARγ, adipogenesis and insulin sensitivity by MAGED1. J Endocrinol. (2018) 239:167–80. doi: 10.1530/JOE-18-0349
59. Rosen CJ. Revisiting the rosiglitazone story–lessons learned. N Engl J Med. (2010) 363:803–6. doi: 10.1056/NEJMp1008233
60. Nesto RW, Bell D, Bonow RO, Fonseca V, Grundy SM, Horton ES, et al. Thiazolidinedione use, fluid retention, and congestive heart failure: a consensus statement from the American Heart Association and American Diabetes Association. Diabetes Care. (2004) 27:256–63. doi: 10.2337/diacare.27.1.256
61. Turner LW, Nartey D, Stafford RS, Singh S, Alexander GC. Ambulatory treatment of type 2 diabetes in the U.S., 1997-2012. Diabetes Care. (2014) 37:985–92. doi: 10.2337/dc13-2097
62. Lavecchia A, Cerchia C. Selective PPARγ modulators for Type 2 diabetes treatment: how far have we come and what does the future hold? Future Med Chem. (2018) 10:703–5. doi: 10.4155/fmc-2018-0021
63. Bruning JB, Chalmers MJ, Prasad S, Busby SA, Kamenecka TM, He Y, et al. Partial agonists activate PPARgamma using a helix 12 independent mechanism. Structure. (2007) 15:1258–71. doi: 10.1016/j.str.2007.07.014
64. Yasmin S, Capone F, Laghezza A, Piaz FD, Loiodice F, Vijayan V, et al. Novel benzylidene thiazolidinedione derivatives as partial PPARγ agonists and their antidiabetic effects on type 2 diabetes. Sci Rep. (2017) 7:14453. doi: 10.1038/s41598-017-14776-0
65. Montanari R, Saccoccia F, Scotti E, Crestani M, Godio C, Gilardi F, et al. Crystal structure of the peroxisome proliferator-activated receptor gamma (PPARgamma) ligand binding domain complexed with a novel partial agonist: a new region of the hydrophobic pocket could be exploited for drug design. J Med Chem. (2008) 51:7768–76. doi: 10.1021/jm800733h
66. Weidner C, de Groot JC, Prasad A, Freiwald A, Quedenau C, Kliem M, et al. Amorfrutins are potent antidiabetic dietary natural products. Proc Natl Acad Sci USA. (2012) 109:7257–62. doi: 10.1073/pnas.1116971109
67. Pirat C, Farce A, Lebègue N, Renault N, Furman C, Millet R, et al. Targeting peroxisome proliferator-activated receptors (PPARs): development of modulators. J Med Chem. (2012) 55:4027–61. doi: 10.1021/jm101360s
68. Lefebvre P, Chinetti G, Fruchart JC, Staels B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J Clin Invest. (2006) 116:571–80. doi: 10.1172/JCI27989
69. Rigano D, Sirignano C, Taglialatela-Scafati O. The potential of natural products for targeting PPARα. Acta Pharm Sin B. (2017) 7:427–38. doi: 10.1016/j.apsb.2017.05.005
70. Videla LA, Pettinelli P. Misregulation of PPAR functioning and its pathogenic consequences associated with nonalcoholic fatty liver disease in human obesity. PPAR Res. (2012) 2012:107434. doi: 10.1155/2012/107434
71. Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, et al. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. (2009) 138:476–88. doi: 10.1016/j.cell.2009.05.036
72. Roy A, Kundu M, Jana M, Mishra RK, Yung Y, Luan CH, et al. Identification and characterization of PPARα ligands in the hippocampus. Nat Chem Biol. (2016) 12:1075–83. doi: 10.1038/nchembio.2204
73. Fruchart JC, Brewer HB, Jr, Leitersdorf E. Consensus for the use of fibrates in the treatment of dyslipoproteinemia and coronary heart disease. Fibrate Consensus Group. Am J Cardiol. (1998) 81:912–7.
74. Chen X, Acquaah-Mensah GK, Denning KL, Peterson JM, Wang K, Denvir J, et al. High-fat diet induces fibrosis in mice lacking CYP2A5 and PPARα: a new model for steatohepatitis-associated fibrosis. Am J Physiol Gastrointest Liver Physiol. (2020) 319:G626–35. doi: 10.1152/ajpgi.00213.2020
75. Montagner A, Polizzi A, Fouché E, Ducheix S, Lippi Y, Lasserre F, et al. Liver PPARα is crucial for whole-body fatty acid homeostasis and is protective against NAFLD. Gut. (2016) 65:1202–14. doi: 10.1136/gutjnl-2015-310798
76. Hogh KL, Uy CE, Asadi A, Baker RK, Riedel MJ, Gray SL. Overexpression of peroxisome proliferator-activated receptor α in pancreatic β-cells improves glucose tolerance in diet-induced obese mice. Exp Physiol. (2013) 98:564–75. doi: 10.1113/expphysiol.2012.068734
77. Shay NF, Banz WJ. Regulation of gene transcription by botanicals: novel regulatory mechanisms. Annu Rev Nutr. (2005) 25:297–315. doi: 10.1146/annurev.nutr.25.050304.092639
78. Goto T, Takahashi N, Hirai S, Kawada T. Various terpenoids derived from herbal and dietary plants function as PPAR modulators and regulate carbohydrate and lipid metabolism. PPAR Res. (2010) 2010:483958. doi: 10.1155/2010/483958
79. Jia Y, Bhuiyan MJ, Jun HJ, Lee JH, Hoang MH, Lee HJ, et al. Ursolic acid is a PPAR-α agonist that regulates hepatic lipid metabolism. Bioorg Med Chem Lett. (2011) 21:5876–80. doi: 10.1016/j.bmcl.2011.07.095
80. Zhao S, Kanno Y, Li W, Sasaki T, Zhang X, Wang J, et al. Identification of Picrasidine C as a Subtype-Selective PPARα Agonist. J Nat Prod. (2016) 79:3127–33. doi: 10.1021/acs.jnatprod.6b00883
81. Mueller M, Hobiger S, Jungbauer A. Red clover extract: a source for substances that activate peroxisome proliferator-activated receptor alpha and ameliorate the cytokine secretion profile of lipopolysaccharide-stimulated macrophages. Menopause. (2010) 17:379–87. doi: 10.1097/gme.0b013e3181c94617
82. Lee HK, Nam GW, Kim SH, Lee SH. Phytocomponents of triterpenoids, oleanolic acid and ursolic acid, regulated differently the processing of epidermal keratinocytes via PPAR-alpha pathway. Exp Dermatol. (2006) 15:66–73. doi: 10.1111/j.0906-6705.2005.00386.x
83. Barger PM, Browning AC, Garner AN, Kelly DP. p38 mitogen-activated protein kinase activates peroxisome proliferator-activated receptor alpha: a potential role in the cardiac metabolic stress response. J Biol Chem. (2001) 276:44495–501. doi: 10.1074/jbc.M105945200
84. Juge-Aubry CE, Hammar E, Siegrist-Kaiser C, Pernin A, Takeshita A, Chin WW, et al. Regulation of the transcriptional activity of the peroxisome proliferator-activated receptor alpha by phosphorylation of a ligand-independent trans-activating domain. J Biol Chem. (1999) 274:10505–10. doi: 10.1074/jbc.274.15.10505
85. Compe E, Drané P, Laurent C, Diderich K, Braun C, Hoeijmakers JH, et al. Dysregulation of the peroxisome proliferator-activated receptor target genes by XPD mutations. Mol Cell Biol. (2005) 25:6065–76. doi: 10.1128/MCB.25.14.6065-6076.2005
86. Blanquart C, Mansouri R, Paumelle R, Fruchart JC, Staels B, Glineur C. The protein kinase C signaling pathway regulates a molecular switch between transactivation and transrepression activity of the peroxisome proliferator-activated receptor alpha. Mol Endocrinol. (2004) 18:1906–18. doi: 10.1210/me.2003-0327
87. Kaupang S, Hansen TV. The PPAR Ω pocket: renewed opportunities for drug development. PPAR Res. (2020) 2020:1–21. doi: 10.1155/2020/9657380
88. Brunmeir R, Xu F. Functional regulation of PPARs through post-translational modifications. Int J Mol Sci. (2018) 19:1738. doi: 10.3390/ijms19061738
89. Ma X, Xu L, Alberobello AT, Gavrilova O, Bagattin A, Skarulis M, et al. Celastrol protects against obesity and metabolic dysfunction through activation of a HSF1-PGC1α transcriptional axis. Cell Metab. (2015) 22:695–708. doi: 10.1016/j.cmet.2015.08.005
90. Li X, Wei T, Li J, Yuan Y, Wu M, Chen F, et al. Tyrosol ameliorates the symptoms of obesity, promotes adipose thermogenesis, and modulates the composition of gut microbiota in HFD fed mice. Mol Nutr Food Res. (2022) e2101015. doi: 10.1002/mnfr.202101015. [Epub ahead of print].
91. Grygiel-Górniak B. Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications-a review. Nutr J. (2014) 13:1–10. doi: 10.1186/1475-2891-13-17
92. Tan NS, Vázquez-Carrera M, Montagner A, Sng MK, Guillou H, Wahli W. Transcriptional control of physiological and pathological processes by the nuclear receptor PPARβ/δ. Prog Lipid Res. (2016) 64:98–122. doi: 10.1016/j.plipres.2016.09.001
93. Vázquez-Carrera M. Unraveling the Effects of PPARβ/δ on Insulin Resistance and Cardiovascular Disease. Trends Endocrinol Metab. (2016) 27:319–34. doi: 10.1016/j.tem.2016.02.008
94. Bortolotto JW, Margis R, Ferreira AC, Padoin AV, Mottin CC, Guaragna RM. Adipose tissue distribution and quantification of PPARbeta/delta and PPARgamma1-3 mRNAs: discordant gene expression in subcutaneous, retroperitoneal and visceral adipose tissue of morbidly obese patients. Obes Surg. (2007) 17:934–40. doi: 10.1007/s11695-007-9172-5
95. Kim WJ, Lee W, Jung Y, Jang HJ, Kim YK, Kim SN. PPARβ/δ agonist GW501516 inhibits TNFα-induced repression of adiponectin and insulin receptor in 3T3-L1 adipocytes. Biochem Biophys Res Commun. (2019) 510:621–8. doi: 10.1016/j.bbrc.2019.02.013
96. Bays HE, Schwartz S, Littlejohn T, 3rd Kerzner B, Krauss RM, Karpf DB, et al. MBX-8025, a novel peroxisome proliferator receptor-delta agonist: lipid and other metabolic effects in dyslipidemic overweight patients treated with and without atorvastatin. J Clin Endocrinol Metab. (2011) 96:2889–97. doi: 10.1210/jc.2011-1061
97. Tsukamoto T, Nakata R, Tamura E, Kosuge Y, Kariya A, Katsukawa M, et al. Vaticanol C, a resveratrol tetramer, activates PPARalpha and PPARbeta/delta in vitro and in vivo. Nutr Metab. (2010) 7:46. doi: 10.1186/1743-7075-7-46
98. Qin S, Lu Y, Rodrigues GA. Resveratrol protects RPE cells from sodium iodate by modulating PPARα and PPARδ. Exp Eye Res. (2014) 118:100–8. doi: 10.1016/j.exer.2013.11.010
99. Kang L, Heng W, Yuan A, Baolin L, Fang H. Resveratrol modulates adipokine expression and improves insulin sensitivity in adipocytes: relative to inhibition of inflammatory responses. Biochimie. (2010) 92:789–96. doi: 10.1016/j.biochi.2010.02.024
100. Cronet P, Petersen JF, Folmer R, Blomberg N, Sjöblom K, Karlsson U, et al. Structure of the PPARalpha and -gamma ligand binding domain in complex with AZ 242; ligand selectivity and agonist activation in the PPAR family. Structure. (2001) 9:699–706. doi: 10.1016/S0969-2126(01)00634-7
101. D'Aniello E, Fellous T, Iannotti FA, Gentile A, Allarà M, Balestrieri F, et al. Identification and characterization of phytocannabinoids as novel dual PPARα/γ agonists by a computational and in vitro experimental approach. Biochim Biophys Acta Gen Subj. (2019) 1863:586–97. doi: 10.1016/j.bbagen.2019.01.002
102. Chambel SS, Santos-Gonçalves A, Duarte TL. The dual role of Nrf2 in nonalcoholic fatty liver disease: regulation of antioxidant defenses and hepatic lipid metabolism. Biomed Res Int. (2015) 2015:597134. doi: 10.1155/2015/597134
103. Chartoumpekis DV, Kensler TW. New player on an old field; the keap1/Nrf2 pathway as a target for treatment of type 2 diabetes and metabolic syndrome. Curr Diabetes Rev. (2013) 9:137–45. doi: 10.2174/1573399811309020005
104. Wang C, Cui Y, Li C, Zhang Y, Xu S, Li X, et al. Nrf2 deletion causes “benign” simple steatosis to develop into nonalcoholic steatohepatitis in mice fed a high-fat diet. Lipids Health Dis. (2013) 12:165. doi: 10.1186/1476-511X-12-165
105. Tanaka Y, Ikeda T, Yamamoto K, Ogawa H, Kamisako T. Dysregulated expression of fatty acid oxidation enzymes and iron-regulatory genes in livers of Nrf2-null mice. J Gastroenterol Hepatol. (2012) 27:1711–7. doi: 10.1111/j.1440-1746.2012.07180.x
106. Shin JW, Chun K-S, Kim D-H, Kim S-J, Kim SH, Cho N-C, et al. Curcumin induces stabilization of Nrf2 protein through Keap1 cysteine modification. Biochem Pharmacol. (2020) 173:113820. doi: 10.1016/j.bcp.2020.113820
107. Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. (2004) 24:8477–86. doi: 10.1128/MCB.24.19.8477-8486.2004
108. Stefanson AL, Bakovic M. Dietary regulation of Keap1/Nrf2/ARE pathway: focus on plant-derived compounds and trace minerals. Nutrients. (2014) 6:3777–801. doi: 10.3390/nu6093777
109. Kopacz A, Kloska D, Forman HJ, Jozkowicz A, Grochot-Przeczek A. Beyond repression of Nrf2: an update on Keap1. Free Radic Biol Med. (2020) 157:63–74. doi: 10.1016/j.freeradbiomed.2020.03.023
110. Luo Y, Eggler AL, Liu D, Liu G, Mesecar AD, van Breemen RB. Sites of alkylation of human Keap1 by natural chemoprevention agents. J Am Soc Mass Spectrom. (2007) 18:2226–32. doi: 10.1016/j.jasms.2007.09.015
111. Mou Y, Wen S, Li Y-X, Gao X-X, Zhang X, Jiang Z-Y. Recent progress in Keap1-Nrf2 protein-protein interaction inhibitors. Eur J Med Chem. (2020) 202:112532. doi: 10.1016/j.ejmech.2020.112532
112. Madden SK, Itzhaki LS. Structural and mechanistic insights into the Keap1-Nrf2 system as a route to drug discovery. Biochim Biophys Acta Proteins Proteomics. (2020) 1868:140405. doi: 10.1016/j.bbapap.2020.140405
113. Tanigawa S, Fujii M, Hou D-X. Action of Nrf2 and Keap1 in ARE-mediated NQO1 expression by quercetin. Free Radic Biol Med. (2007) 42:1690–703. doi: 10.1016/j.freeradbiomed.2007.02.017
114. Pang C, Zheng Z, Shi L, Sheng Y, Wei H, Wang Z, et al. Caffeic acid prevents acetaminophen-induced liver injury by activating the Keap1-Nrf2 antioxidative defense system. Free Radic Biol Med. (2016) 91:236–46. doi: 10.1016/j.freeradbiomed.2015.12.024
115. Sun W, Liu X, Zhang H, Song Y, Li T, Liu X, et al. Epigallocatechin gallate upregulates NRF2 to prevent diabetic nephropathy via disabling KEAP1. Free Radic Biol Med. (2017) 108:840–57. doi: 10.1016/j.freeradbiomed.2017.04.365
116. Shi L, Hao Z, Zhang S, Wei M, Lu B, Wang Z, et al. Baicalein and baicalin alleviate acetaminophen-induced liver injury by activating Nrf2 antioxidative pathway: the involvement of ERK1/2 and PKC. Biochem Pharmacol. (2018) 150:9–23. doi: 10.1016/j.bcp.2018.01.026
117. Yang M, Jiang Z-h, Li C-g, Zhu Y-j, Li Z, Tang Y-z, et al. Apigenin prevents metabolic syndrome in high-fructose diet-fed mice by Keap1-Nrf2 pathway. Biomed Pharmacother. (2018) 105:1283–90. doi: 10.1016/j.biopha.2018.06.108
118. Li K, Zhang M, Chen H, Peng J, Jiang F, Shi X, et al. Anthocyanins from black peanut skin protect against UV-B induced keratinocyte cell and skin oxidative damage through activating Nrf 2 signaling. Food Funct. (2019) 10:6815–28. doi: 10.1039/C9FO00706G
119. Li M, Huang W, Jie F, Wang M, Zhong Y, Chen Q, et al. Discovery of Keap1–Nrf2 small–molecule inhibitors from phytochemicals based on molecular docking. Food Chem Toxicol. (2019) 133:110758. doi: 10.1016/j.fct.2019.110758
120. Zambo V, Simon-Szabo L, Szelenyi P, Kereszturi E, Banhegyi G, Csala M. Lipotoxicity in the liver. World J Hepatol. (2013) 5:550–7. doi: 10.4254/wjh.v5.i10.550
121. Meakin PJ, Chowdhry S, Sharma RS, Ashford FB, Walsh SV, McCrimmon RJ, et al. Susceptibility of Nrf2-null mice to steatohepatitis and cirrhosis upon consumption of a high-fat diet is associated with oxidative stress, perturbation of the unfolded protein response, and disturbance in the expression of metabolic enzymes but not with insulin resistance. Mol Cell Biol. (2014) 34:3305–20. doi: 10.1128/MCB.00677-14
122. Tanaka Y, Aleksunes LM, Yeager RL, Gyamfi MA, Esterly N, Guo GL, et al. NF-E2-related factor 2 inhibits lipid accumulation and oxidative stress in mice fed a high-fat diet. J Pharmacol Exp Ther. (2008) 325:655–64. doi: 10.1124/jpet.107.135822
123. Shin S, Wakabayashi J, Yates MS, Wakabayashi N, Dolan PM, Aja S, et al. Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-imidazolide. Eur J Pharmacol. (2009) 620:138–44. doi: 10.1016/j.ejphar.2009.08.022
124. Okada K, Warabi E, Sugimoto H, Horie M, Tokushige K, Ueda T, et al. Nrf2 inhibits hepatic iron accumulation and counteracts oxidative stress-induced liver injury in nutritional steatohepatitis. J Gastroenterol. (2012) 47:924–35. doi: 10.1007/s00535-012-0552-9
125. Lei P, Tian S, Teng C, Huang L, Liu X, Wang J, et al. Sulforaphane improves lipid metabolism by enhancing mitochondrial function and biogenesis in vivo and in vitro. Mol Nutr Food Res. (2019) 63:e1800795. doi: 10.1002/mnfr.201800795
126. Nagata N, Xu L, Kohno S, Ushida Y, Aoki Y, Umeda R, et al. Glucoraphanin ameliorates obesity and insulin resistance through adipose tissue browning and reduction of metabolic endotoxemia in mice. Diabetes. (2017) 66:1222–236. doi: 10.2337/db16-0662
127. Lee DH, Chang SH, Yang DK, Song NJ, Yun UJ, Park KW. Sesamol increases Ucp1 expression in white adipose tissues and stimulates energy expenditure in high-fat diet-fed obese mice. Nutrients. (2020) 12:1459. doi: 10.3390/nu12051459
128. He HJ, Wang GY, Gao Y, Ling WH, Yu ZW, Jin TR. Curcumin attenuates Nrf2 signaling defect, oxidative stress in muscle and glucose intolerance in high fat diet-fed mice. World J Diabetes. (2012) 3:94–104. doi: 10.4239/wjd.v3.i5.94
129. Han JH, Park MH, Myung CS. Garcinia cambogia ameliorates non-alcoholic fatty liver disease by inhibiting oxidative stress-mediated steatosis and apoptosis through NRF2-are activation. Antioxidants. (2021) 10:1226. doi: 10.3390/antiox10081226
130. Liu F, Feng M, Xing J, Zhou X. Timosaponin alleviates oxidative stress in rats with high fat diet-induced obesity via activating Nrf2/HO-1 and inhibiting the NF-κB pathway. Eur J Pharmacol. (2021) 909:174377. doi: 10.1016/j.ejphar.2021.174377
131. Huang J, Tabbi-Anneni I, Gunda V, Wang L. Transcription factor Nrf2 regulates SHP and lipogenic gene expression in hepatic lipid metabolism. Am J Physiol Gastrointest Liver Physiol. (2010) 299:G1211–221. doi: 10.1152/ajpgi.00322.2010
132. Braud L, Pini M, Stec DF, Manin S, Derumeaux G, Stec DE, et al. Increased Sirt1 secreted from visceral white adipose tissue is associated with improved glucose tolerance in obese Nrf2-deficient mice. Redox Biol. (2021) 38:101805. doi: 10.1016/j.redox.2020.101805
133. Chartoumpekis DV, Palliyaguru DL, Wakabayashi N, Fazzari M, Khoo NK, Schopfer FJ, et al. Nrf2 deletion from adipocytes, but not hepatocytes, potentiates systemic metabolic dysfunction after long-term high-fat diet-induced obesity in mice. Am J Physiol Endocrinol Metab. (2018) 315:E180–95. doi: 10.1152/ajpendo.00311.2017
134. Xu J, Kulkarni SR, Donepudi AC, More VR, Slitt AL. Enhanced Nrf2 activity worsens insulin resistance, impairs lipid accumulation in adipose tissue, and increases hepatic steatosis in leptin-deficient mice. Diabetes. (2012) 61:3208–18. doi: 10.2337/db11-1716
135. Chartoumpekis DV, Kensler TW. New player on an old field; the Keap1/Nrf2 pathway as a target for treatment of type 2 diabetes and metabolic syndrome. Curr Diabetes Rev. (2013) 9:137–45. doi: 10.2174/157339913805076490
136. Deng X, Su R, Stanford S, Chen J. Critical enzymatic functions of FTO in obesity and cancer. Front Endocrinol. (2018) 9:396. doi: 10.3389/fendo.2018.00396
137. Church C, Moir L, McMurray F, Girard C, Banks GT, Teboul L, et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet. (2010) 42:1086–92. doi: 10.1038/ng.713
138. Karra E, O'Daly OG, Choudhury AI, Yousseif A, Millership S, Neary MT, et al. A link between FTO, ghrelin, and impaired brain food-cue responsivity. J Clin Investig. (2013) 123:3539–51. doi: 10.1172/JCI44403
139. Peng S, Xiao W, Ju D, Sun B, Hou N, Liu Q, et al. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci Transl Med. (2019) 11:eaau7116. doi: 10.1126/scitranslmed.aau7116
140. Wang X, Wu R, Liu Y, Zhao Y, Bi Z, Yao Y, et al. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. (2020) 16:1221–35. doi: 10.1080/15548627.2019.1659617
141. Zhao X, Yang Y, Sun B-F, Shi Y, Yang X, Xiao W, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. (2014) 24:1403–19. doi: 10.1038/cr.2014.151
142. Wu J, Frazier K, Zhang J, Gan Z, Wang T, Zhong X. Emerging role of m6A RNA methylation in nutritional physiology and metabolism. Obes Rev. (2020) 21:e12942. doi: 10.1111/obr.12942
143. Han Z, Niu T, Chang J, Lei X, Zhao M, Wang Q, et al. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature. (2010) 464:1205–09. doi: 10.1038/nature08921
144. Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. (2019) 35:677–91. e10. doi: 10.1016/j.ccell.2019.03.006
145. Wang R, Han Z, Liu B, Zhou B, Wang N, Jiang Q, et al. Identification of natural compound radicicol as a potent FTO inhibitor. Mol Pharm. (2018) 15:4092–8. doi: 10.1021/acs.molpharmaceut.8b00522
146. Gu J, Xu J, You Q, Guo X. Recent developments of small molecules targeting RNA m6A modulators. Eur J Med Chem. (2020) 196:112325. doi: 10.1016/j.ejmech.2020.112325
147. Chen B, Ye F, Yu L, Jia G, Huang X, Zhang X, et al. Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J Am Chem Soc. (2012) 134:17963–71. doi: 10.1021/ja3064149
148. Huang L, Zhang J, Zhu X, Mi X, Li Q, Gao J, et al. The phytochemical rhein mediates M(6)A-independent suppression of adipocyte differentiation. Front Nutr. (2021) 8:756803. doi: 10.3389/fnut.2021.756803
149. Wu R, Yao Y, Jiang Q, Cai M, Liu Q, Wang Y, et al. Epigallocatechin gallate targets FTO and inhibits adipogenesis in an mRNA m6A-YTHDF2-dependent manner. Int J Obes. (2018) 42:1378–88. doi: 10.1038/s41366-018-0082-5
150. Dai J, Liang K, Zhao S, Jia W, Liu Y, Wu H, et al. Chemoproteomics reveals baicalin activates hepatic CPT1 to ameliorate diet-induced obesity and hepatic steatosis. Proc Natl Acad Sci. (2018) 115:E5896–905. doi: 10.1073/pnas.1801745115
151. Tang Y, Chen K, Song B, Ma J, Wu X, Xu Q, et al. m6A-Atlas: a comprehensive knowledgebase for unraveling the N6-methyladenosine (m6A) epitranscriptome. Nucleic Acids Res. (2020) 49:D134–43. doi: 10.1093/nar/gkaa692
152. Zhong T, Zhang H, Duan X, Hu J, Wang L, Li L, et al. Anti-obesity effect of radix Angelica sinensis and candidate causative genes in transcriptome analyses of adipose tissues in high-fat diet-induced mice. Gene. (2017) 599:92–8. doi: 10.1016/j.gene.2016.11.017
153. Zhong T, Duan XY, Zhang H, Li L, Zhang HP, Niu L. Angelica sinensis suppresses body weight gain and alters expression of the FTO gene in high-fat-diet induced obese mice. Biomed Res Int. (2017) 2017:6280972. doi: 10.1155/2017/6280972
154. Zhou X, Chen J, Chen J, Wu W, Wang X, Wang Y. The beneficial effects of betaine on dysfunctional adipose tissue and N6-methyladenosine mRNA methylation requires the AMP-activated protein kinase α1 subunit. J Nutr Biochem. (2015) 26:1678–84. doi: 10.1016/j.jnutbio.2015.08.014
155. Maia-Landim A, Ramírez JM, Lancho C, Poblador MS, Lancho JL. Long-term effects of Garcinia cambogia/Glucomannan on weight loss in people with obesity, PLIN4, FTO and Trp64Arg polymorphisms. BMC Complement Altern Med. (2018) 18:26. doi: 10.1186/s12906-018-2099-7
156. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
157. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. (1993) 75:843–54. doi: 10.1016/0092-8674(93)90529-Y
158. Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. (2010) 38:323–32. doi: 10.1016/j.molcel.2010.03.013
159. Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. (2005) 123:631–40. doi: 10.1016/j.cell.2005.10.022
160. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. (2014) 15:509–24. doi: 10.1038/nrm3838
161. Landrier JF, Derghal A, Mounien L. MicroRNAs in obesity and related metabolic disorders. Cells. (2019) 8:859. doi: 10.3390/cells8080859
162. Xie H, Sun L, Lodish HF. Targeting microRNAs in obesity. Expert Opin Ther Targets. (2009) 13:1227. doi: 10.1517/14728220903190707
163. Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. (2006) 3:87–98. doi: 10.1016/j.cmet.2006.01.005
164. Gao P, Sun L, He X, Cao Y, Zhang H. MicroRNAs and the Warburg Effect: new players in an old arena. Curr Gene Ther. (2012) 4:285–91. doi: 10.2174/156652312802083620
165. Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs'. Nature. (2005) 438:685–9. doi: 10.1038/nature04303
166. Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. (2008) 452:896–9. doi: 10.1038/nature06783
167. Wang R, Hong J, Cao Y, Shi J, Gu W, Ning G, et al. Elevated circulating microRNA-122 is associated with obesity and insulin resistance in young adults. Eur J Endocrinol. (2015) 172:291–300. doi: 10.1530/EJE-14-0867
168. Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. (2012) 122:2871–83. doi: 10.1172/JCI63539
169. Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. (2012) 122:2884–97. doi: 10.1172/JCI63455
170. Hussain MM, Shi J, Dreizen P. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J Lipid Res. (2003) 44:22–32. doi: 10.1194/jlr.R200014-JLR200
171. Baselga-Escudero L, Blade C, Ribas-Latre A, Casanova E, Suárez M, Torres JL, et al. Resveratrol and EGCG bind directly and distinctively to miR-33a and miR-122 and modulate divergently their levels in hepatic cells. Nucleic Acids Res. (2014) 42:882–92. doi: 10.1093/nar/gkt1011
172. Noratto GD, Angel-Morales G, Talcott ST, Mertens-Talcott SU. Polyphenolics from açaí (Euterpe oleracea Mart.) and red muscadine grape (Vitis rotundifolia) protect human umbilical vascular Endothelial cells (HUVEC) from glucose- and lipopolysaccharide (LPS)-induced inflammation and target microRNA-126. J Agric Food Chem. (2011) 59:7999–8012. doi: 10.1021/jf201056x
173. Ahn J, Lee H, Jung CH, Choi WH, Ha TY. Zerumbone ameliorates high-fat diet-induced adiposity by restoring AMPK-regulated lipogenesis and microRNA-146b/SIRT1-mediated adipogenesis. Oncotarget. (2017) 8:36984–95. doi: 10.18632/oncotarget.16974
174. Jeon TI, Park JW, Ahn J, Jung CH, Ha TY. Fisetin protects against hepatosteatosis in mice by inhibiting miR-378. Mol Nutr Food Res. (2013) 57:1931–7. doi: 10.1002/mnfr.201300071
175. Joven J, Espinel E, Rull A, Aragonès G, Rodríguez-Gallego E, Camps J, et al. Plant-derived polyphenols regulate expression of miRNA paralogs miR-103/107 and miR-122 and prevent diet-induced fatty liver disease in hyperlipidemic mice. Biochim Biophys Acta. (2012) 1820:894–9. doi: 10.1016/j.bbagen.2012.03.020
176. Srivastava SK, Arora S, Averett C, Singh S, Singh AP. Modulation of microRNAs by phytochemicals in cancer: underlying mechanisms and translational significance. Biomed Res Int. (2015) 2015:848710. doi: 10.1155/2015/848710
177. Kang H. MicroRNA-mediated health-promoting effects of phytochemicals. Int J Mol Sci. (2019) 20:2535. doi: 10.3390/ijms20102535
178. Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. (2005) 37:495–500. doi: 10.1038/ng1536
179. Weidner C, Wowro SJ, Freiwald A, Kawamoto K, Witzke A, Kliem M, et al. Amorfrutin B is an efficient natural peroxisome proliferator-activated receptor gamma (PPARγ) agonist with potent glucose-lowering properties. Diabetologia. (2013) 56:1802–12. doi: 10.1007/s00125-013-2920-2
180. Mezei O, Banz WJ, Steger RW, Peluso MR, Winters TA, Shay N. Soy isoflavones exert antidiabetic and hypolipidemic effects through the PPAR pathways in obese Zucker rats and murine RAW 264.7 cells. J Nutr. (2003) 133:1238–43. doi: 10.1093/jn/133.5.1238
181. Atanasov AG, Wang JN, Gu SP, Bu J, Kramer MP, Baumgartner L, et al. Honokiol: a non-adipogenic PPARγ agonist from nature. Biochim Biophys Acta. (2013) 1830:4813–9. doi: 10.1016/j.bbagen.2013.06.021
Keywords: phytochemicals, obesity, transcription factors, structure, metabolic signals
Citation: Li X, Zheng L, Zhang B, Deng Z-Y and Luo T (2022) The Structure Basis of Phytochemicals as Metabolic Signals for Combating Obesity. Front. Nutr. 9:913883. doi: 10.3389/fnut.2022.913883
Received: 06 April 2022; Accepted: 23 May 2022;
Published: 13 June 2022.
Edited by:
Pengfei Xu, University of Pittsburgh, United StatesReviewed by:
Rui Fan, Peking University, ChinaCopyright © 2022 Li, Zheng, Zhang, Deng and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ting Luo, dGluZy5sdW9AbmN1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.