- 1Microbiome Medicine Center, Department of Laboratory Medicine, Zhujiang Hospital, Southern Medical University, Guangzhou, China
- 2Department of Neonatology, Nanfang Hospital, Southern Medical University, Guangzhou, China
Background: Aminophylline is widely used in the treatment of preterm infants, but it can cause feeding intolerance events, in which gut microbial dysbiosis may have a role. This study aims to investigate the relationship between the gut microbiome of preterm infants treated with aminophylline and the occurrence of feeding intolerance and weight gain rate.
Methods: This study included a cohort of 118 preterm infants. Survival analysis and multivariate Cox regression were used to evaluate the relationship between aminophylline treatment and the occurrence of feeding intolerance. 16S rRNA V4 region gene sequencing was used to characterize the microbiome of fecal samples from the cohort. Linear discriminant analysis effect size was used to analyze the differential abundance of bacteria related to aminophylline treatment. Wilcoxon test, Kruskal–Wallis test, Spearman correlation coefficients and generalized linear mixed models were used to analyze the correlation between the differential bacteria and feeding intolerance events as well as the weight gain.
Results: The results showed that the use of aminophylline could significantly increase the occurrence of feeding intolerance. The relative abundances of Streptococcus and Rothia in the gut microbiome of preterm infants were positively correlated with both the occurrence of feeding intolerance and the use of aminophylline, while the relative abundance of Staphylococcus was negatively correlated. In particular, preterm infants with a lower relative abundance of Rothia were more likely to develop feeding intolerance associated with aminophylline, and this difference existed before the onset of feeding intolerance. Moreover, it took longer for individuals with a lower relative abundance of Streptococcus to reach 2 kg weight. The contribution of Streptococcus to weight gain was greater than that of Bifidobacterium or Lactobacillus.
Conclusion: The gut microbiome in preterm infants treated with aminophylline was characterized by a decrease in Streptococcus and Rothia and an increase in Staphylococcus. These microbes, especially Rothia, were positively correlated with the occurrence of feeding intolerance. Streptococcus but not Bifidobacter likely participated in the weight gain of preterm infants in early life.
Introduction
Aminophylline is widely used in the treatment of critically ill neonates and preterm infants to stimulate the respiratory center, prevent renal injury after hypoxia and ischemia, and relax pulmonary vascular smooth muscle (1, 2). Similar to adults, neonates will experience increased heart rate, increased blood sugar, and increased urine output during the use of aminophylline, and these symptoms are usually mild. However, in the course of clinical application, it was found that some neonates had related gastrointestinal reactions to aminophylline, accompanied by the occurrence of feeding intolerance (FI) events, manifested as vomiting, abdominal distension, gastric retention, and even bloody stools after enteral feeding (3, 4). FI is an important event that affects early enteral nutrition in preterm infants, leading to a prolonged time to full enteral feeding, the occurrence of extrauterine growth retardation, and an increase in parenteral nutrition time (4, 5); FI is also the early manifestation of some severe digestive tract diseases and systemic infections, such as necrotizing enterocolitis and neonatal sepsis (6). The prevention and treatment of FI are of great significance in improving the survival rate of preterm infants.
As a factor closely related to the health of the human digestive tract, the gut microbiome has been reported in recent years to play an important role in early life (7), participate in the occurrence and development of many neonatal diseases, and serve as the key factor in growth and nutrition (8). Preterm infants experience a number of unique challenges to the establishment of their microbiota, the medicine of maternal and neonatal exposure, birth gestational, and the sterile environment of the neonatal intensive care unit (NICU) may all alter the microbiome. The gut microbiome of preterm infants in early life is dominated by opportunistic pathogens (such as Staphylococcus, Streptococcus, and Enterobacter), and the abundance of Bifidobacterium is low. The gut microbiome of preterm infants in this period follows a specific rule (9, 10), under which it is most closely related to age after birth (11) and is greatly influenced by postnatal diseases and clinical intervention (10, 12–14).
Based on the abovementioned clinical phenomena and studies, we hypothesize that the use of aminophylline in early life is associated with specific changes in the gut microbiome and that this association might be related to the occurrence of FI and even to the growth, development and nutrient absorption of preterm infants in early life. Therefore, we designed a prospective observational cohort study, combined with a nested case-control model, to focus on the gut microbiome of preterm infants related to aminophylline treatment for the first time and elucidate its relationship with FI events and early weight gain. The results provide clues for further mechanistic studies.
Materials and Methods
Study Participants and Experimental Design
This study was based on a prospective observational cohort of preterm infants in the NICU of Nanfang Hospital of Southern Medical University. Inclusion criteria included (a) gestational age (GA) <35 weeks and birth weight <2000 g; (b) a transfer to the NICU after birth and survival for >72 hours; and (c) informed consent of family members. The exclusion criteria included the following: (a) patients with digestive tract malformations and inherited metabolic diseases; (b) patients who used methylxanthine drugs other than aminophylline; and (c) patients whose families refused treatment or who had not reached full enteral feeding when transferred to another hospital due to other diseases. According to whether aminophylline was used, the cohort was divided into two groups for comparative analysis. The usage of aminophylline and the age at the time of the FI incident was investigated, and clinical data such as GA, weight, sex mode of delivery, time to full enteral feeding and daily weight were collected. Subjects were matched for the nested case-control design according to the needs of the analysis. The diagnostic criteria for FI are as follows: (a) gastric residual volume exceeding 50% of the previous feeding amount, accompanied by vomiting and/or abdominal distension; or (b) failure of the feeding plan, including a reduction, delay or interruption of enteral feeding (15).
The research was reviewed and approved by the Ethics Committee of Nanfang Hospital of Southern Medical University (NFEC-2021-054). The study was enrolled in the Chinese Clinical Trial Register (trial registration No. ChiCTR2100044469).
Sample Collection and 16S Ribosomal RNA Gene Sequencing
Postnatal fecal samples were collected from cohort members 1–2 times per week until discharge. After being collected from diapers, the fresh fecal samples were stored at −40°C in time, transported back to the laboratory within 3 days, and then stored at −80°C. According to the instructions, the QIAamp Mini Kit was uniformly used to extract DNA from the fecal samples. The V4 hypervariable region of the 16S ribosomal RNA (rRNA) gene was amplified and sequenced using an Illumina iSeq 100 platform.
Processing of Sequence Data
The raw fastq sequence data were demultiplexed into separate paired-end fastq files based on the barcodes using in-house Perl scripts. Amplicon sequence variants (ASVs) were obtained by denoising each sample paired-end fastq file using the R package DADA2 (16) (version 1.6.0). The PyNAST (17) algorithm was used to align representative sequences, and FastTree (18) was used to build a phylogenetic tree. Taxonomic classification was performed with the Ribosomal Database Project classifier (19) against the GreenGenes database (20) (version 13.8). The ASVs from reagent controls were excluded from downstream analysis. Each sample was rarefied to 3,180 sequences, and those samples with fewer than 3,180 sequences were discarded from downstream analysis.
Biostatistical Analysis
All subsequent biostatistical analyses were conducted using QIIME (21) (version 1.9.1) software or the R language (version 4.1.0). Kaplan–Meier survival curves were generated and compared by the log-rank test. Multivariate Cox regression analysis was performed to identify significant factors associated with FI. The Shannon index and phylogenetic diversity (PD) whole-tree index were calculated to estimate the α-diversity of the gut microbiome. Assessment of bacterial compositional changes was performed using the MaAsLin2 (22) (version 1.6.0) R package and linear discriminant analysis (LDA) effect size (LEfSe) (23). To better show the relative abundance distribution of bacteria in the figure, the relative abundances were arcsine square root transformed. Two nested case-control cohorts were obtained by 1:1 propensity score matching (PSM) to reduce some common confounding biases. To estimate the effect sizes of different variables, a multivariate generalized linear mixed model (GLMM) was fitted by taking groups with weekly weight gain rate (weekly_WGRs)≥ 0.06 as the dependent variable, some variables of interest as fixed effects, and patients as a random effect in the R package ‘lme4' (version 1.1–27.1).
For comparisons of categorical data, the chi-square test or Fisher's exact test was performed depending on the theoretical frequency. The Wilcoxon rank-sum test and the Kruskal- Wallis test, respectively, were compared of distributions between two groups or between two groups. Correlations between two variables were estimated with Spearman correlation coefficients. The Benjamini–Hochberg false discovery rate (FDR) method was adopted to adjust for multiple comparisons. For all comparisons, a P-value or FDR-corrected P-value <0.05 was considered statistically significant.
Results
Characterization of the Cohort
During the recruitment period, 151 infants from the prospective cohort of preterm infants at Nanfang Hospital of Southern Medical University were screened, and 118 infants enrolled into the study (Figure 1). The recruited infants had an average GA of 31.2±2.5 weeks, an average birth weight of 1.49 ± 0.41 kg, an aminophylline usage rate of 75.42, and a 52.54% incidence of FI (Table 1). A total of 467 fecal samples were collected, and 6,013,140 high-quality sequences were obtained after iSeq sequencing and quality control. The median number of sequences contained in each sample was 11,991 (3,526–43742); after the data were denoised with DADA2, 8086 ASVs were obtained. Next, the samples were flattened to 3,500 sequences, and samples from patients with unknown FI onset times were removed; 7,507 ASVs that remained after these steps were included in the downstream analysis.
The Incidence of FI in Preterm Infants Treated With Aminophylline Was Significantly Higher Than That in the Control Group
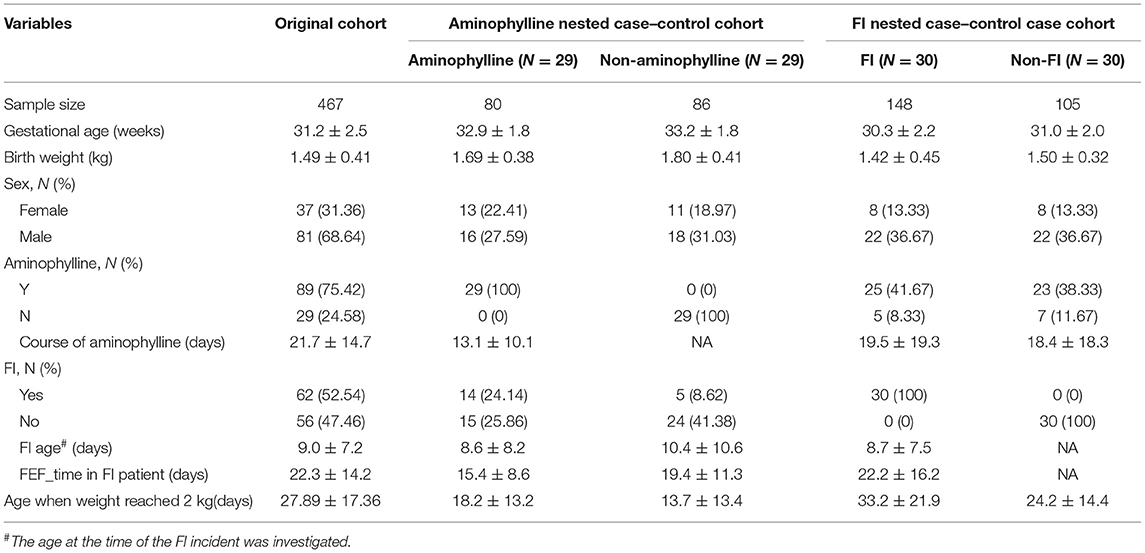
To evaluate the effect of aminophylline treatment on FI events in this study, we used Kaplan–Meier survival analysis to compare aminophylline treatment during the most frequent period of FI events (the first 3 weeks after birth). The results showed that the risk of FI events in the aminophylline treatment group (aminophylline: Y) was significantly increased (Figure 2A). This difference still existed after controlling for factors such as GA, gender and birth weight of the cohort (Figure 2B, P = 0.037). According to whether aminophylline was used, we further established a small nested case-control cohort by PSM of the whole cohort. The nested case-control cohort for aminophylline included a total of 166 samples, collected from 29 patients in the treatment group and 29 patients in the control group. There were no significant differences in GA, birth weight, or sex between the two groups in the aminophylline nested case-control cohort (Table 1). The incidence of FI in the aminophylline treatment group from the nested case-control cohort was 48.28%, while that in the control group was 17.24%, and the difference was still statistically significant (Figure 2C, P =0.012). When the course of aminophylline treatment was further analyzed across all aminophylline-treated populations, it was found that preterm infants with FI events had a longer course of aminophylline than preterm infants who did not (Figure 2D, P < 0.001), and the length of the course was positively correlated with the time to reach full enteral feeding in preterm infants (Figure 2E, ρ = 0.42, P < 0.001).
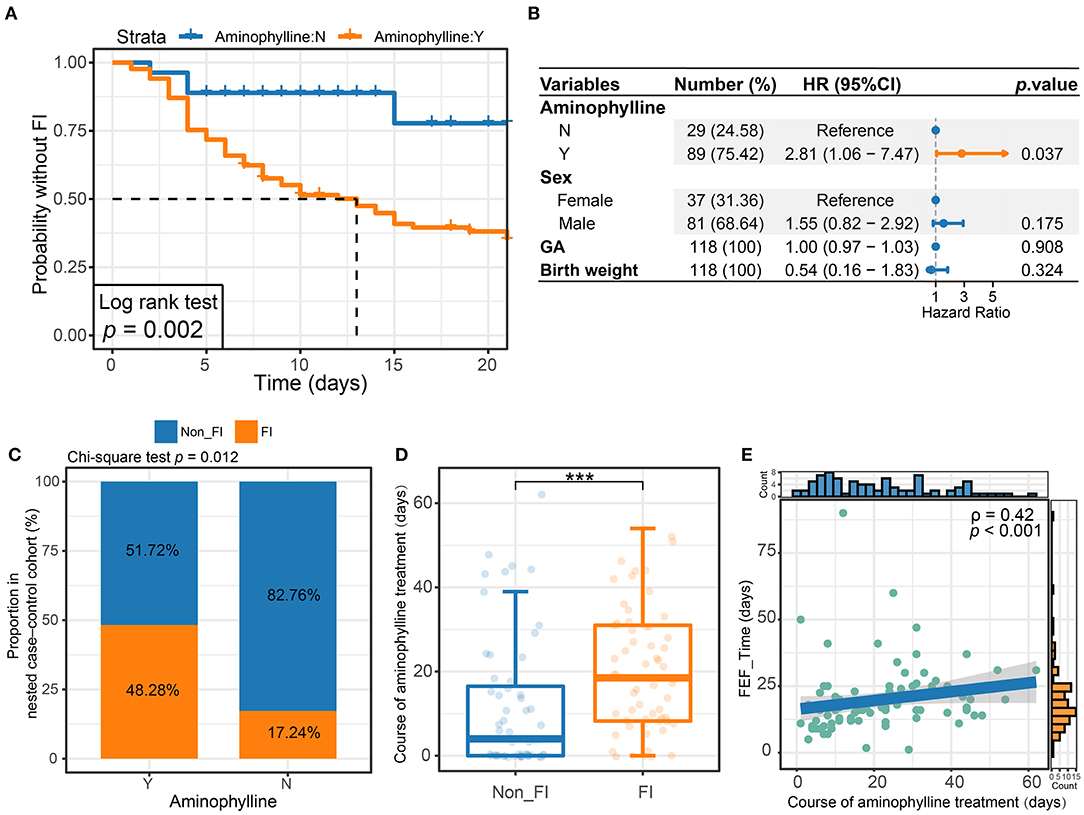
Figure 2. The relationship between aminophylline treatment and FI events. (A) Kaplan–Meier survival analysis of FI between groups treated with aminophylline. The group treated with aminophylline had a higher FI risk (log rank P = 0.002). (B) Forest plot showing the results of multivariate Cox regression analyses. The points and horizontal lines correspond to the adjusted hazard ratios and 95% confidence intervals. Orange indicates statistical significance (P < 0.05). (C) Stacked bar plot showing that the proportions of FI were different between groups treated with aminophylline in the aminophylline nested case-control cohort. (D) Boxplot showing the course of aminophylline in patients with FI and without FI. ***, P < 0.001. (E) Scatter plot showing the correlation between time to full enteral feeding (FEF_Time) and the course of aminophylline in patients taking aminophylline. The histograms above and on the right show the distribution of the course of aminophylline and FEF_Time, respectively. Spearman correlation coefficients (ρ values) and P-values are shown in the top left corner.
Preterm Infants Treated With Aminophylline Showed Characteristic Changes in the Gut Microbiome, Which Were Related to FI
We sequenced and identified the fecal samples of the preterm infants after birth by 16S rRNA sequencing. The gut microbiome of preterm infants in this study was mainly composed of Firmicutes and Proteobacteria, among which the bacterial genera with the highest relative abundance were Enterococcus, Streptococcus, Staphylococcus, Klebsiella and Acinetobacter. However, Bifidobacterium, Escherichia and other genera had low relative abundance, and a few individuals were characterized by an absolute dominance of a single genus of gut microbes (Figure 3A). When the cohort was divided according to the use or non-use of aminophylline, it was found that the relative abundances of Streptococcus, Staphylococcus, Klebsiella and Escherichia in the two groups differed (Figure 3B).
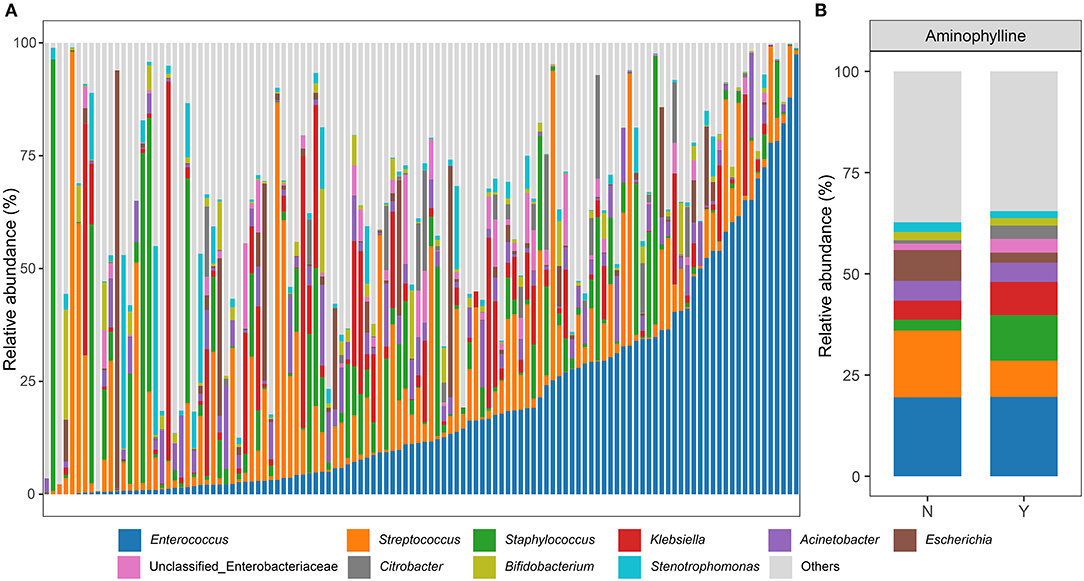
Figure 3. Genus-level microbiome composition in patients. (A) Relative abundances of the top 10 abundant genera are displayed. Each bar represents the mean abundance of all samples of a patient. (B) The relative abundances at the genus level between patients who received aminophylline and those who did not.
To further clarify whether there was a significant difference in the microbiome between the aminophylline treatment group and the control group, we conducted LEfSe analysis, compared the microbiome composition of the two groups at the genus level, and estimated the change in the relative abundance of microbes at the genus level combined with MaAsLin2 (FDR < 0.25) (Figures 4A,B). The results showed that Rothia (β = −1.689675, P = 0.000159), Staphylococcus (β = 1.428663, P = 0.010141) and Streptococcus (β = −1.478244, P = 0.011750) had significant differences in relative abundance between the gut microbiomes of the two groups. Therefore, we focused on the relationship between these three bacteria and aminophylline treatment. Using the Wilcoxon test, we found that the relative abundances of Rothia (P < 0.001) (Figure 4C) and Streptococcus (P < 0.001) (Figure 4D) in the treatment group decreased significantly; however, Staphylococcus (P < 0.001) (Figure 4E) increased significantly, and these differences were still significant after considering the influence of postmenstrual age on the evolution of the microbiome in the GLMM (Rothia, P = 0.007; Streptococcus, P = 0.002; Staphylococcus, P = 0.0455). At the same time, the relative abundances of Rothia (ρ =-0.12, P = 0.02) and Streptococcus (ρ = 0.13, P = 0.01) in the gut microbiome of preterm infants who took aminophylline were negatively correlated with the course of treatment, while Staphylococcus (ρ =-0.17, P < 0.001) was positively correlated with it (Figures 4F–H).
Figure 4. Differences in the microbiome between the aminophylline-treated group and the control group. (A) A cladogram of the LEfSe analysis shows the significantly differentially abundant bacteria between the group that received aminophylline and the group that did not. Taxonomic levels are represented by rings with phyla in the innermost ring and genera in the outermost ring. The significantly different taxa are colored by different colors representing the two groups (LDA score >2), and the yellow dots represent the taxa without significant differences. (B) The changes in the relative abundance of genera between the groups with and without aminophylline treatment, estimated using MaAsLin2 by taking the non-aminophylline group as a reference. Each bar represents the coefficient estimated by MaAsLin2, and the depth of the color represents the P-value (only genera with a P-value <0.05 are displayed). (C–E) Boxplots and jittered points show the abundance distribution of Rothia (C), Staphylococcus (D) and Streptococcus (E) between groups treated with aminophylline. Each point represents a sample. ***, P < 0.001. (F–H) Scatter plots showing the correlation between the abundances of Rothia (F), Staphylococcus (G) and Streptococcus (H) and the course of aminophylline in patients taking aminophylline. The histograms above and on the right show the distribution of the course of aminophylline and abundances, respectively. Spearman correlation coefficients (ρvalues) and P-values are shown in the top left corner.
In addition to finding that the characteristics of the microbiome were related to the treatment of aminophylline, we also found that when FI occurred in preterm infants, the microbiome also showed certain predictable characteristics. The α-diversity (PD whole tree, Shannon) of this group of preterm infants gradually decreased in the following three stages: before onset, within 1 week after onset, and 1 week after onset (Figures 5A,B), and the differences among groups were statistically significant, indicating that there were significant changes in the microbiome before and after the onset of FI. To explore whether these changes were associated with aminophylline-related microbiome characteristics, we also compared three aminophylline-related bacteria between the FI group and the non-FI group. The results showed that the relative abundances of Rothia (P < 0.001) and Streptococcus (P = 0.02307) also showed a significant downward trend in the FI group, while that of Staphylococcus (P = 0.007105) showed a significant increase (Figures 5C–E). These characteristics are consistent with the changes in three bacteria in the aminophylline treatment group. When a further comparison was made between the samples before the onset in the FI group and those before the corresponding age (taking the median age of 6 days in the FI group) in the non-FI group, it was found that the relative abundance of Rothia decreased significantly before the onset of FI (Figure 5F, P = 0.04576).
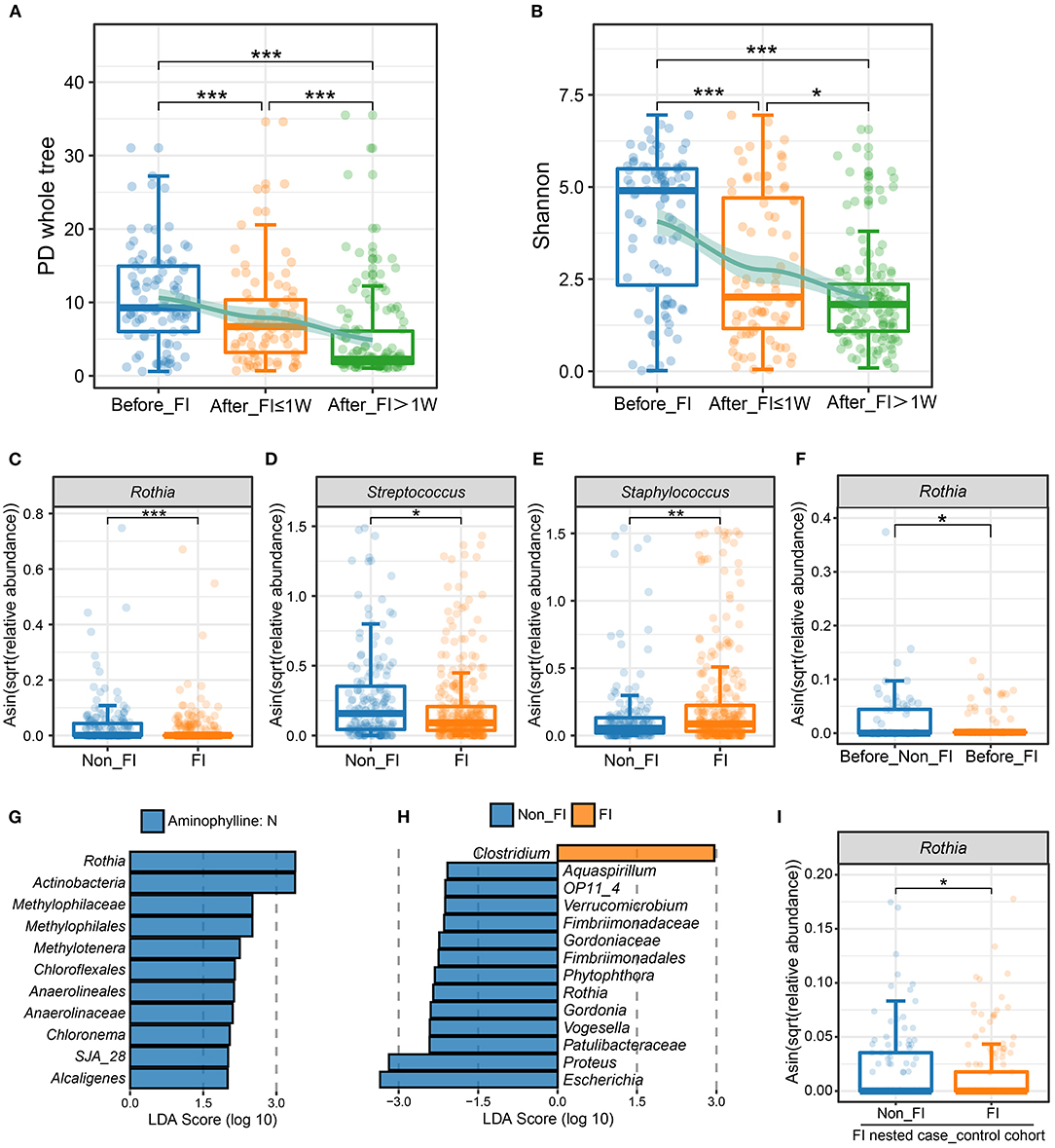
Figure 5. Differences in the microbiome between patients with FI and without FI. (A,B) Comparison of the PD whole-tree index (A) and Shannon index (B) among different FI stage groups. The jittered scatter plot represents the α-diversity index of each sample. The curves on the top layers of the graphs were fitted with the LOESS method. *, FDR < 0.05; **, FDR < 0.01; ***, FDR < 0.001. (C–E) Boxplots and jittered points show the abundance distribution of Rothia (C), Streptococcus (D) and Staphylococcus (E) between the non-FI and FI groups. Each point represents a sample. *, P < 0.05; **, P < 0.01; ***: P < 0.001. (F) Boxplot and jittered points show the abundance distribution of Rothia between the before-FI (samples before the FI onset) and before-non-FI (samples before 6 days age) groups. Each point represents a sample. *, P < 0.05. (G) Significantly differentially abundant bacterial taxa between FI patients with and without aminophylline, estimated by LEfSe. Each bar represents the LDA score for a specific taxon. The color of a bar indicates that the taxon was enriched in the corresponding group. (H) Significantly different bacterial taxa between the non-FI and FI groups among patients treated with aminophylline. (I) Comparison of Rothia abundance between the non-FI and FI groups in the FI nested case-control cohort. *, P < 0.05.
Considering the various causes of FI events, to further explore the other bacteria that were involved in FI events related to aminophylline treatment, we also compared the gut microbiomes of all preterm infants with FI events according to whether aminophylline was used, intending to explore the microbiomic characteristics that correspond to FI events related to aminophylline (Figure 5G). At the same time, the gut microbiomes of preterm infants with FI events and those without FI events were compared among all the preterm infants that were treated with aminophylline, screening for bacteria associated with both aminophylline treatment and FI incidence from another perspective (Figure 5H). The results of the two comparisons suggested that the common element was Rothia.
As many basic features of preterm infants may affect the onset of FI, we attempted to further confirm whether the reduced relative abundance of Rothia was an important factor that influenced the occurrence of FI in aminophylline users. In this prospective cohort, we adopted the PSM method, according to whether FI occurred, and matched a group of nested case-control cohorts to the greatest extent for verification (Table 1). A total of 253 samples were included in this small cohort, with the disease group and the control group containing 30 cases each. Aminophylline use was consistent between the two groups, and there was no significant difference in GA, birth weight, or gender. Comparing the differences in Rothia between the two groups again, the results showed that the abundance of Rothia in the FI group after matching was still lower, and this difference was unrelated to GA and body weight (Figure 5I, P = 0.04978).
The Relationship Between Aminophylline-Related Microbiome Features and the Rate of Weight Gain in Preterm Infants
Since (i) both aminophylline treatment and an associated FI event may affect weight gain in preterm infants, and (ii) the results discussed in previous sections suggested that aminophylline treatment was strongly associated with the characteristics of the microbiome, we further investigated weight gain during hospitalization of all preterm infants in this cohort. The age (day of life) when the bodyweight reached 2 kg and the weekly_WGR were used as indicators to evaluate the relationship between some bacteria and the weight gain of preterm infants.
First, we counted the age at which each preterm infant in the cohort reached a bodyweight of 2 kg. If an infant's weight had not reached 2 kg at the time of discharge, the weight gain would be estimated to be 15 g/kg per day. This indicator reflects the speed of weight gain to a certain extent. Statistically, among preterm infants receiving aminophylline, as the duration of medication increased, the age at which the preterm infants reached a weight of 2 kg increased correspondingly (Figure 6A, ρ = 0.46, P < 0.001). To explore whether this effect was related to the characteristics of the microbiome after aminophylline treatment, we performed GLMM with age at 2 kg as the dependent variable and the relative abundance of bacteria, birth weight, GA, and sex as independent variables; we found that the relative abundance of Streptococcus in the gut microbiome of preterm infants was negatively correlated with their age reaching a weight of 2 kg after excluding the influence of other factors (Figure 6B, β = 0.2, P = 0.006), while Rothia (Figure 6C), β = 0.06, P = 0.47), Staphylococcus (Figure 6D, β =-0.02, P = 0.73), Bifidobacterium (Figure 6E, β =-0.09, P = 0.18) and Lactobacillus (Figure 6F, β =-0.13, P = 0.08) were not significantly different.
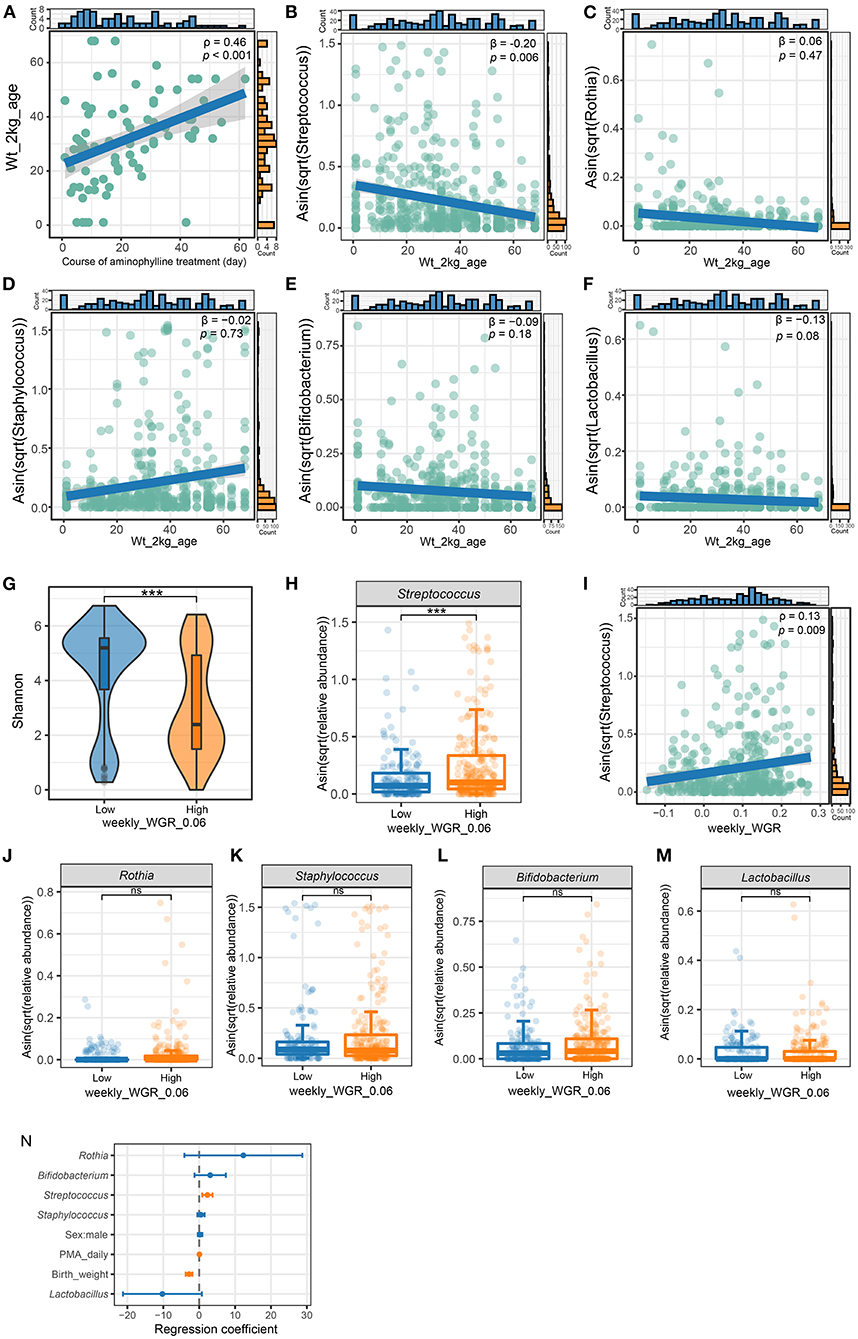
Figure 6. The relationship of weight gain with microbiome and aminophylline. (A) The correlation between age in days when the patient's weight reached two kilograms and the course of aminophylline in patients taking aminophylline. (B–F) The correlation between age in days when the patient's weight reached two kilograms and the abundance of Streptococcus (B), Rothia (C), Staphylococcus (D), Bifidobacterium (E) and Lactobacillus (F). β is the regression coefficient for Streptococcus and age in days when the patient's weight reached two kilograms after adjusting covariates (gestational age, sex, and birth weight). (G) Comparison of the Shannon index between groups with weekly_WGR ≥ 0.06. (H) Comparison of the abundances of Streptococcus. ***: P < 0.001. (I) The correlation between Streptococcus and weekly_WGR for each sample collected. (J–M) Comparison of the abundances of Rothia (J), Staphylococcus (K), Bifidobacterium (L) and Lactobacillus (M) between groups with weekly_WGR ≥ 0.06. ***, P < 0.001; ns, no statistical significance. (N) multivariate GLMM plots to visualize the model result, showing the effect size for each independent variable by taking groups with weekly_WGR ≥ 0.06 as the dependent variable. The points and horizontal lines correspond to the adjusted GLMM coefficients and 95% confidence intervals. Orange significant indicates statistical significance (P < 0.05).
To clarify the relationship between weight gain and Streptococcus, we further recorded the ages of preterm infants in weeks at the times when 437 fecal samples were collected from the cohort, and we cross-referenced the samples with the weekly_WGR of preterm infants at this age. Then, according to whether the corresponding weekly_WGR was lower than 6% (converted from the standard of daily weight gain <10 g/kg), all samples were divided into high and low groups. The results of microbiome analysis showed that there were significant differences in α-diversity between these two groups (Figure 6G). The faster the weekly_WGR, the lower the diversity (Shannon, P = 2.13e−08). The relative abundance of Streptococcus was significantly different between the two types of samples (Figure 6H, P < 0.001). Regardless of whether the samples came from the same preterm infant, the relative abundance of Streptococcus was lower in the samples with a slower weekly_WGR (low group), and Streptococcus was positively correlated with weekly_WGR (Figure 6I, ρ = 0.13, P = 0.009). Similarly, the relative abundances of Rothia (P = 0.54), Staphylococcus (P = 0.87), Bifidobacterium (P = 0.07) and Lactobacillus (P = 0.24) were not significantly different between the two groups (Figures 6J–M). Taking a weekly_WGR ≥ 0.06 as the dependent variable and including birth weight, postmenstrual age, gender and other common factors that would affect weight as independent variables, we constructed a GLMM to show the degree of influence of each factor on the weekly_WGR. The model showed that the contribution of Streptococcus's relative abundance to weekly_WGR ≥ 6% was second only to that of postmenstrual age and greater than that of birth weight, with no corresponding contribution from other bacteria (Figure 6N).
Discussion
In recent years, many studies have reported that specific drugs may affect the gut microbiome (24) and may also affect the host through microbial metabolism (25–27). In the course of studying this cohort of 118 preterm infants, we found the following interesting links between aminophylline, FI events, and the gut microbiome.
First, the preterm infants treated with aminophylline not only had a significantly higher incidence of FI events (Figure 2) but also exhibited characteristic changes in the gut microbiome (Figures 3, 4). Aminophylline is a type of methylxanthine drug that acts as a nonselective adenosine receptor antagonist to provide renal protection in hypoxic-ischemic conditions (28) and is also often used to prevent apnea in preterm infants by inhibiting phosphodiesterase (1, 29). Adverse gastrointestinal reactions to aminophylline have been reported frequently (3, 4), but no researchers have investigated whether the drug leads to changes in the gut microbiome. In this study, the duration of aminophylline use was found not only to be associated with an increased incidence of FI in preterm infants (Figures 2A–D) but also to delay enteral nutrition in preterm infants (Figure 2E). Compared with the control group, the gut microbiome of preterm infants treated with aminophylline was characterized by lower relative abundances of Rothia and Streptococcus and a higher relative abundance of Staphylococcus (Figures 4C–E). Moreover, this trend became more pronounced as the duration of the medication course increased (Figures 4F–H).
Second, the three bacteria also showed the same regularity in the FI onset population as in the aminophylline-treated population, especially Rothia, which was closely associated with aminophylline-related FI events (Figures 5G,H). The analysis showed that even in preterm infants with similar aminophylline use, FI was more likely to occur in those with a lower relative abundance of Rothia (Figure 5I), and this difference existed before the onset of FI (Figure 5F). Rothia, a butyrate-producing bacterium, has been found to colonize the human oral cavity and is also part of the characteristic microbiome carried in the gut of vaginally delivered neonates (30). A cohort study reported that it is one of the most characteristic symbiotic bacteria associated with breast milk and the infant's gut (31); research in another cohort also showed that this genus may mediate a protective effect against asthma through the metabolite butyrate (32), reducing chronic airway inflammation (33). While breastfeeding was an effective method to reduce the incidence of FI (34–36), it was speculated that a low relative abundance of Rothia may be a disadvantage during feeding in the aminophylline-treated preterm infant population.
Finally, we also observed that Streptococcus but not Bifidobacterium was involved in weight gain in the early life of preterm infants (Figures 6B,E,F,N), while the diversity of the microbiome decreased with increasing weight gain (Figure 6G). The gut microbiome in early life is closely related to the host's energy balance (37, 38), while Bifidobacterium and Lactobacillus are predominant in the gut of healthy full-term newborns (39, 40), and they are considered to be related to growth and play an important role in early life (41–44). However, multiple studies have found that Enterobacteriaceae, Enterococcus, Staphylococcus, and Streptococcus are the main components of the gut microbiome of preterm infants in early life, (9, 11, 45), and our cohort did show that the relative abundances of these bacteria were high, while the relative abundances of Bifidobacterium and Lactobacillus were low (Figure 3). Interestingly, we found that individuals with a low relative abundance of Streptococcus took longer to reach a bodyweight of 2 kg, but each time they entered a stage of increased Streptococcus abundance, their rate of weight gain increased accordingly (Figures 6H,I). This is consistent with other researchers' observations of a positive correlation between Streptococcus and body mass index (46). However, Bifidobacterium and Lactobacillus were not only low in relative abundance but also unassociated with early weight gain in preterm infants (Figures 6E,F,L–N), which may be due to their low total abundance and inability to contribute to early growth. Although some species of Streptococcus, such as Streptococcus agalactiae, are high-risk factors for infection in preterm infants, Streptococcus itself is one of the common components of the microbiome in early life (47) and is also one of the core microbes from breast milk (31). The role of Streptococcus in the early growth of preterm infants merits further exploration.
Our study was not yet able to show a clear causal association among aminophylline treatment, gut microbiome, FI events, and weight gain; however, for the preterm infant population, our study demonstrated that Streptococcus, Staphylococcus, and especially Rothia were associated with aminophylline treatment and FI events. More importantly, we found that Streptococcus was strongly associated with weight gain in the early life of preterm infants. Despite observing these novel and interesting clues for the first time, our study has some limitations. First, the overall sample size was small. Second, we merely identified associations among changes in the microbiome, aminophylline treatment, associated FI events and weight gain; whether there is truly a causal relationship is a topic that awaits further study in a larger cohort.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA803308.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Nanfang Hospital of Southern Medical University. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author Contributions
WS and HZ contributed to the conception and design of the study. WS, CZ, and YL collected all the materials. WS wrote the first draft of the manuscript. WQ and QL performed the statistical analysis and wrote sections of the manuscript. HZ and WH revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by the National Natural Science Foundation of China (grant No. 81925062 to HZ). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the reviewers and the editor for their helpful comments on this manuscript.
References
1. Shivakumar M, Nayak K, Lewis LES, Kamath A, Purkayastha J. Acute hemodynamic effects of methylxanthine therapy in preterm neonates: Effect of variations in subgroups. J Trop Pediatr. (2019) 65:264–72. doi: 10.1093/tropej/fmy044
2. Armanian AM, Badiee Z, Afghari R, Salehimehr N, Hassanzade A, Sheikhzadeh S, et al. Prophylactic aminophylline for prevention of apnea at higher-risk preterm neonates. Iran Red Crescent Med J. (2014) 16:e12559. doi: 10.5812/ircmj.12559
3. Xiaoyan H, Yanmei C, Zailing L. Clinical features and risk factors of feeding intolerance in premature infants. Chin J Perinat Med. (2020):182–7.
4. Chen Q, Fang JB, Peng WT. Risk prediction of feeding intolerance in preterm infants. Sichuan Da Xue Xue Bao Yi Xue Ban. (2016) 47:749–54.
5. Tang Jun. Feeding intolerance in premature infants: an important clinical problem. Chin J Perinat Med. (2020):177–81.
6. Zhang LP, Lei XP, Luo LJ, Dong WB. Risk factors for necrotizing enterocolitis in very preterm infants: a case-control study in southwest China. J Matern Fetal Neonatal Med. (2019) 32:896–901. doi: 10.1080/14767058.2017.1395011
7. McDonald B, McCoy KD. Maternal microbiota in pregnancy and early life. Science. (2019) 365:984–5. doi: 10.1126/science.aay0618
8. Vuong HE, Pronovost GN, Williams DW, Coley EJL, Siegler EL, Qiu A, et al. The maternal microbiome modulates fetal neurodevelopment in mice. Nature. (2020) 586:281–6. doi: 10.1038/s41586-020-2745-3
9. Rao C, Coyte KZ, Bainter W, Geha RS, Martin CR, Rakoff-Nahoum S. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature. (2021) 591:633–8. doi: 10.1038/s41586-021-03241-8
10. Hill CJ, Lynch DB, Murphy K, Ulaszewska M, Jeffery IB, O'Shea CA, et al. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome. (2017) 5:4. doi: 10.1186/s40168-016-0213-y
11. Shen W, Qiu W, Liu Y, Liao W, Ma Y, He Y, et al. Postnatal age is strongly correlated with the early development of the gut microbiome in preterm infants. Transl Pediatr. (2021) 10:2313–24. doi: 10.21037/tp-21-367
12. Rozé JC, Ancel PY, Marchand-Martin L, Rousseau C, Montassier E, Monot C, et al. Assessment of neonatal intensive care unit practices and preterm newborn gut microbiota and 2-year neurodevelopmental outcomes. JAMA Netw Open. (2020) 3:e2018119. doi: 10.1001/jamanetworkopen.2020.18119
13. Gibson MK, Wang B, Ahmadi S, Burnham CA, Tarr PI, Warner BB, et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbiol. (2016) 1:16024. doi: 10.1038/nmicrobiol.2016.24
14. Li X, Stokholm J, Brejnrod A, Vestergaard GA, Russel J, Trivedi U, et al. The infant gut resistome associates with E. coli, environmental exposures, gut microbiome maturity, and asthma-associated bacterial composition. Cell Host Microbe. (2021) 29:975–87.e4. doi: 10.1016/j.chom.2021.03.017
15. Evidence-Based Medicine Group, Neonatologist Society Chinese Medical, Doctor Association. Clinical guidelines for the diagnosis and treatment of feeding intolerance in preterm infants. Zhongguo Dang Dai Er Ke Za Zhi. (2020) 22:1047–55.
16. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. (2016) 13:581–3. doi: 10.1038/nmeth.3869
17. Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. (2010) 26:266–7. doi: 10.1093/bioinformatics/btp636
18. Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. (2010) 5:e9490. doi: 10.1371/journal.pone.0009490
19. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. (2007) 73:5261–7. doi: 10.1128/AEM.00062-07
20. DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. (2006) 72:5069–72. doi: 10.1128/AEM.03006-05
21. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. (2010) 7:335–6. doi: 10.1038/nmeth.f.303
22. Mallick H, Rahnavard A, McIver LJ, Ma S, Zhang Y, Nguyen LH, et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput Biol. (2021) 17:e1009442. doi: 10.1371/journal.pcbi.1009442
23. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. (2011) 12:R60. doi: 10.1186/gb-2011-12-6-r60
24. Forslund SK, Chakaroun R, Zimmermann-Kogadeeva M, Markó L, Aron-Wisnewsky J, Nielsen T, et al. Combinatorial, additive and dose-dependent drug-microbiome associations. Nature. (2021) 600:500–5. doi: 10.1038/s41586-021-04177-9
25. Tremblay A, Xu X, Colee J, Tompkins TA. Efficacy of a multi-strain probiotic formulation in pediatric populations: a comprehensive review of clinical studies. Nutrients. (2021) 13:1908. doi: 10.3390/nu13061908
26. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. (2018) 359:97–103. doi: 10.1126/science.aan4236
27. Sorboni SG, Moghaddam HS, Jafarzadeh-Esfehani R, Soleimanpour S. A comprehensive review on the role of the gut microbiome in human neurological disorders Clin Microbiol Rev. (2022) 35:e0033820. doi: 10.1128/CMR.00338-20
28. Frymoyer A, Van Meurs KP, Drover DR, Klawitter J, Christians U, Chock VY. Theophylline dosing and pharmacokinetics for renal protection in neonates with hypoxic-ischemic encephalopathy undergoing therapeutic hypothermia. Pediatr Res. (2020) 88:871–7. doi: 10.1038/s41390-020-01140-8
29. Shivakumar M, Jayashree P, Najih M, Lewis LES, Bhat Y R, Kamath A. Shashikala -. comparative efficacy and safety of caffeine and aminophylline for apnea of prematurity in preterm (=34 weeks) neonates: a randomized controlled trial. Indian Pediatr. (2017) 54:279–83. doi: 10.1007/s13312-017-1088-0
30. Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. (2015) 17:690–703. doi: 10.1016/j.chom.2015.04.004
31. Fehr K, Moossavi S, Sbihi H, Boutin RCT, Bode L, Robertson B, et al. Breastmilk feeding practices are associated with the co-occurrence of bacteria in mothers' milk and the infant gut: the CHILD Cohort Study. Cell Host Microbe. (2020) 28:285–97.e4. doi: 10.1016/j.chom.2020.06.009
32. Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. (2015) 7:307ra152. doi: 10.1126/scitranslmed.aab2271
33. Rigauts C, Aizawa J, Taylor S, Rogers GB, Govaerts M, Cos P, et al. Rothia mucilaginosa is an anti-inflammatory bacterium in the respiratory tract of patients with chronic lung disease. Eur Respir J. (2021) 59:2101293. doi: 10.1183/13993003.01293-2021
34. Ford SL, Lohmann P, Preidis GA, Gordon PS, O'Donnell A, Hagan J, et al. Improved feeding tolerance and growth are linked to increased gut microbial community diversity in very-low-birth-weight infants fed mother's own milk compared with donor breast milk. Am J Clin Nutr. (2019) 109:1088–97. doi: 10.1093/ajcn/nqz006
35. Autran CA, Kellman BP, Kim JH, Asztalos E, Blood AB, Spence ECH, et al. Human milk oligosaccharide composition predicts risk of necrotising enterocolitis in preterm infants. Gut. (2018) 67:1064–70. doi: 10.1136/gutjnl-2016-312819
36. Niño DF, Sodhi CP, Hackam DJ. Necrotizing enterocolitis: new insights into pathogenesis and mechanisms. Nat Rev Gastroenterol Hepatol. (2016) 13:590–600. doi: 10.1038/nrgastro.2016.119
37. Vael C, Verhulst SL, Nelen V, Goossens H, Desager KN. Intestinal microflora and body mass index during the first three years of life: an observational study. Gut Pathog. (2011) 3:8. doi: 10.1186/1757-4749-3-8
38. Kozyrskyj AL, Tun HM, Bridgman SL. The Gut Microbiome and Control of Weight Gain. In: Freemark, M. editor. Pediatric Obesity. Contemporary Endocrinology. Cham: Humana Press (2018). doi: 10.1007/978-3-319-68192-4_4
39. Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. (2019) 574:117–21. doi: 10.1038/s41586-019-1560-1
40. DeWeerdt S. How baby's first microbes could be crucial to future health. Nature. (2018) 555:S18–9. doi: 10.1038/d41586-018-02480-6
41. Uzan-Yulzari A, Turta O, Belogolovski A, Ziv O, Kunz C, Perschbacher S, et al. Neonatal antibiotic exposure impairs child growth during the first six years of life by perturbing intestinal microbial colonization. Nat Commun. (2021) 12:443. doi: 10.1038/s41467-020-20495-4
42. Saturio S, Nogacka AM, Alvarado-Jasso GM, Salazar N, de Los Reyes-Gavilán CG, Gueimonde M, et al. Role of bifidobacteria on infant health. Microorganisms. (2021) 9:2415. doi: 10.3390/microorganisms9122415
43. Schwarzer M, Makki K, Storelli G, Machuca-Gayet I, Srutkova D, Hermanova P, et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science. (2016) 351:854–7. doi: 10.1126/science.aad8588
44. Robertson RC, Manges AR, Finlay BB, Prendergast AJ. The human microbiome and child growth - first 1000 days and beyond. Trends Microbiol. (2019) 27:131–47. doi: 10.1016/j.tim.2018.09.008
45. Wandro S, Osborne S, Enriquez C, Bixby C, Arrieta A, Whiteson K. The microbiome and metabolome of preterm infant stool are personalized and not driven by health outcomes, including necrotizing enterocolitis and late-onset sepsis. mSphere. (2018) 3:e00104–18. doi: 10.1128/mSphere.00104-18
46. Korpela K, Zijlmans MA, Kuitunen M, Kukkonen K, Savilahti E, Salonen A, et al. Childhood BMI in relation to microbiota in infancy and lifetime antibiotic use. Microbiome. (2017) 5:26. doi: 10.1186/s40168-017-0245-y
Keywords: aminophylline, gut microbiome, feeding intolerance, preterm infants, weight gain, Streptococcus, Rothia
Citation: Shen W, Qiu W, Lin Q, Zeng C, Liu Y, Huang W and Zhou H (2022) The Gut Microbiome of Preterm Infants Treated With Aminophylline Is Closely Related to the Occurrence of Feeding Intolerance and the Weight Gain. Front. Nutr. 9:905839. doi: 10.3389/fnut.2022.905839
Received: 28 March 2022; Accepted: 29 April 2022;
Published: 27 May 2022.
Edited by:
Zhenbo Xu, South China University of Technology, ChinaReviewed by:
Julio Plaza-Diaz, Children's Hospital of Eastern Ontario (CHEO), CanadaChenhong Zhang, Shanghai Jiao Tong University, China
Tingtao Chen, Nanchang University, China
Copyright © 2022 Shen, Qiu, Lin, Zeng, Liu, Huang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongwei Zhou, YmlvZGVncmFkYXRpb25AZ21haWwuY29t; Weimin Huang, aHdtbmV0QDIxY24uY29t
†These authors have contributed equally to this work and share first authorship
 Wei Shen
Wei Shen Wen Qiu
Wen Qiu Qi Lin1†
Qi Lin1† Hongwei Zhou
Hongwei Zhou