- 1School of Life Sciences, Anhui University, Hefei, China
- 2Key Laboratory of Human Microenvironment and Precision Medicine of Anhui Higher Education Institutes, Anhui University, Hefei, China
- 3Health Nutrition, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, Japan
- 4ALMADO Inc., Tokyo, Japan
Inflammatory bowel disease (IBD) is known to be associated with compositional and metabolic changes in the gut microbiota. The aim of this study was to investigate whether dietary eggshell membrane (ESM) improves survival rate or ameliorates gut dysbiosis in a spontaneous IBD model of interleukin-10 knockout (IL10−/−) mice. Female C57BL/6J wild-type (WT) and IL10−/− mice (KO) were fed an AIN-93G basal diet or an ESM diet (KOE) for 19 weeks. Gut microbiota profiles were analyzed via 16S rRNA sequencing, and short-chain fatty acids in cecal content were analyzed with high-performance liquid chromatography. The results demonstrated that ESM supplementation significantly improved the survival rate and body composition in KO mice. Alpha diversity analysis of the microbiota revealed that ESM supplementation significantly increased gut microbial diversity, which was decreased in IL10−/− mice. The Firmicutes/Bacteroidetes ratio was recovered to a normal level by ESM supplementation, suggesting that ESM helps maintain the compositional balance of the gut microbiota. ESM increased relative abundance of commensal bacterial Ruminococcus and Bacteroidales S24-7 and reduced the abundance of the proinflammatory-related bacterium, Enterobacteriaceae. Additionally, ESM supplementation promoted the production of butyrate in cecal contents and downregulated the expression of proinflammatory genes, including interleukin-1β (Il-1β) and tumor necrosis factor-α (Tnf-α) in IL10−/− mice colon, indicating anti-inflammatory functions. These findings suggest that ESM may be used as a beneficial dietary intervention for IBD.
Introduction
Inflammatory bowel disease (IBD) is characterized as a cluster of chronic gut inflammation disorders, including Crohn's disease and ulcerative colitis (1). Despite extensive efforts, the exact underlying mechanisms of IBD have not been clearly elucidated. However, several risk factors, including genetics, immune system, and environmental factors, play important roles in the development of IBD (2). Recently, abnormal microbial colonization of the gastrointestinal tract has been considered a possible pathogenesis of IBD in genetically susceptible individuals (3). Many previous studies have identified a perturbation of gut microbiome in both IBD patients and mouse colitis models (4–6). Compared to chemical-induced colitis models, interleukin-10 knockout (IL-10−/−) mice are often applied in mechanistic studies to examine the pathogeny of spontaneous, immune-mediated, chronic gastrointestinal inflammation (7, 8). Germ-free IL-10−/− mice showed no proof of colitis or related immune system activation, suggesting the critical role of gut microbiota in IBD pathogenesis (9).
Current treatments for IBD mainly rely on anti-inflammatory drugs and surgery to relieve symptomatic complications, which have several side effects, including loss of immunotolerance and drug resistance (10). Administration of prebiotics, probiotics, or synbiotics has been demonstrated to ameliorate some colitis symptoms in both IBD patients and rodent colitis models (7, 11–13). These studies imply that targeting the gut microbiota is an effective strategy for IBD treatment (14).
Eggshell membrane (ESM) is a by-product of the egg process industry and is considered an environmental-burden waste. Our previous study found that dietary ESM exerts biochemical functions that can counter liver injury and fibrosis through modulating the PPARγ-endothelin 1 interaction signaling pathway (15). Moreover, dietary ESM was shown to ameliorate dextran sulfate sodium (DSS)-induced colitis through exerting anti-inflammatory effects and modulating the gut microbiota (16). The DSS-colitis model is widely used to mimic human IBD, mainly because of the massive epithelial injury and wholesale invasion of the lamina propria by gut microbiota, however, this is unlikely to be a crucial mechanism of human IBD (17). IL-10−/− mice display chronic intestinal inflammation through impaired immunoregulatory function of antigen-presenting cells as well as dysfunction of macrophages (17). The efficacy of ESM in IL-10−/− mice with spontaneous colitis has not yet been elucidated, thus we investigated the potential protective role of ESM on the effects of colitis and gut microbiota modulation in IL-10−/− female mice.
Materials and Methods
Animal and Diets
The animal experiment was approved by the Animal Care and Use Committee of the University of Tokyo. Female wild-type (WT) mice and IL-10−/− C57BL/6J mice, aged 9 or 10 weeks, bred in our laboratory were used in this study. The mice were individually housed in cages under a controlled temperature (23 ± 2°C), 12 h light-dark cycle environment (lights on from 08:00 a.m. to 20:00 p.m.) with a relative humidity of 40–60%. The mice were acclimatized for 1 week and then randomly divided into three groups based on their genetic types and diets: WT (WT mice; control diet of AIN93G, n = 10), KO (IL-10−/− mice; control diet, n = 16), KOE (IL-10−/− mice; 8% ESM diet, n = 16) for 19 weeks. The dietary composition is shown in Supplementary Table S1. As the digestibility of ESM is ~46%, the ESM diet was adjusted with cornstarch and casein to maintain the caloric and protein balance. Animals had free access to the experimental diet and tap water.
Sample Collection and Cecal Short-Chain Fatty Acid Analysis
At the end of experimental period, all 12 h-fasted mice were anesthetized using isoflurane before sacrificing. After sacrificing the mice, the tissues and cecal contents were collected, weighed, and stored at −80°C prior to analysis. The concentration of short-chain fatty acids (SCFAs) in cecal contents was measured using ion-exclusion high-performance liquid chromatography (HPLC), as described in a previous study (18). Briefly, 50 mg cecal contents were mixed with 100 μl distilled water and 15 μl 12% (v/v) perchloric acid. After centrifuging the mixture for 10 min at 13,000 × g, the collected supernatants were filtered with a 0.45 μm membrane filter (Cosmonice Filter W; Nacalai Tesque, Kyoto, Japan). The filtered samples were injected into an SIL-30AC autosampler (Shimadzu, Kyoto, Japan) for quantitative analysis.
Bacterial 16S rRNA Gene Sequencing
Bacterial genomic DNA from mouse cecal content was extracted with the QIAamp Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The V3–V4 of the 16S rRNA gene were amplified with the primer set, 341F: 5′-CCTACGGGNGGCWGCAG-3′; and 806R: 5′-GACTACHVGGGTATCTAATCC-3′, and then incorporated with Illumina adapters for subsequent sequencing. Thereafter, the libraries were conducted with a single Illumina MiSeq run (MiSeq Reagent Kit V3, 600 cycles; Illumina, San Diego, CA, USA) according to the manufacturer's instructions.
Bioinformatic Analysis
The collected sequencing data was analyzed with QIIME (v. 1.8.0) software and the method is described in our previous study. First, fast length adjustment of short reads (FLASH) (v.1.2.11) was used to assemble the paired-end reads. Assembled reads with an average Q-value < 25 were filtered out using in-house script. The same number of filter-passed reads was randomly selected from each sample and used for further analysis. The selected reads were then processed using the QIIME pipeline. The high-quality sequences were clustered into operational taxonomic units (OTUs) at 97% sequence similarity, and OTUs were assigned with the Greengenes database (v.13.8). The microbial functionality profiles were predicted using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt, v.2.1.4) to generate the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. The predicted metagenomic data were aligned to the KEGG database, and the differences between groups were compared using STAMP (Welch's t-test, two-sided).
RNA Extraction and Real-Time Quantitative Polymerase Chain Reaction
The colonic mucosa was scraped off gently with a blunt-edged glass slide. Total RNA was extracted from colonic mucosa using TRIzol™ reagent (Ambion® Life Technologies™, Foster City, CA, USA) and RNA Isolation Kit (NucleoSpin RNA II; Macherey Nagel, Düren, German). cDNA was synthesized from extracted total RNA with the PrimeScript™ RT Master Mix (Takara Bio, Tokyo, Japan) according to the manufacturer's instructions. The real-time quantitative polymerase chain reaction (qPCR) was performed using the Thermal Cycler Dice Real Time System TP800 (Takara Bio Inc.). The primer sequences used in the study are described in Supplementary Table S2. The expression level of ribosomal protein lateral stalk subunit P1 (Rplp1) was used as the internal reference for normalization of the target gene mRNA expression.
Statistical Analysis
Data are expressed as mean ± standard error (SE) or boxplots with median, minimum, and maximum values. The normality and equal variances of the data were performed by Shapiro-wilk and Hartley's test firstly. Then, statistical analysis was performed using one-way ANOVA and Tukey-Kramer test. The survival analysis was performed using a Log-rank (Mantel-Cox) test in GraphPad Prism 9 (San Diego, CA). P < 0.05 was considered statistically significant.
Results
ESM Administration Significantly Improved Survival Rate and Body Composition in IL10−/− Mice
During the 19-week experimental period, there was no significant difference in the total food intake among the experimental groups. After the 19-week treatment, the survival rate was significantly lower in the KO group than in the WT group (P < 0.01); however, ESM supplementation significantly improved the survival rate of IL10−/− mice (Figure 1A). From the 8- to 19-week treatment period, body weight was significantly lower in the KO group than in the WT group. ESM treatment largely reversed the body weight loss in IL10−/− mice (Figure 1B). At necropsy, KO mice showed reduced liver, visceral fat, and gastrocnemius muscle weights, which was mitigated by ESM supplementation (Figures 1C–E). The cecal content weight in KOE group mice was higher than that in WT mice (Figure 1F). The mice colon length was not significantly different among the experimental groups (Figure 1G). However, the mice colon weight was significantly elevated in the KO group compared to the WT group (Figure 1H). Accordingly, the ratio of the colon weight (mg) to length (cm), an indirect indicator of edema and inflammation, increased in the KO group compared to the WT group, which was slightly alleviated by ESM supplementation (Figure 1I).
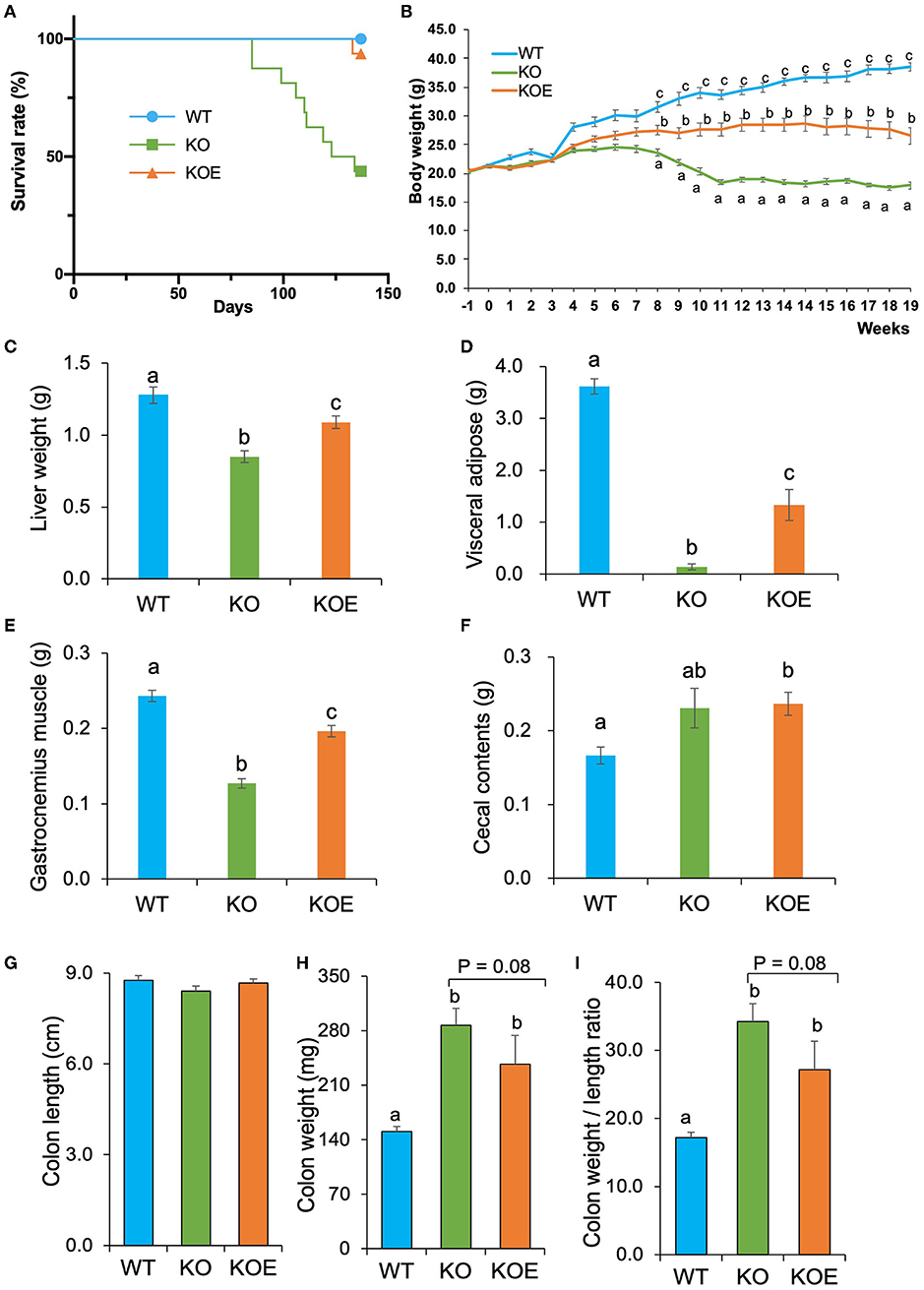
Figure 1. Effect of eggshell membrane (ESM) supplementation on the survival rate and body composition in IL10−/− mice. (A) Survival rate in wild-type (WT) mice fed with control diet or IL10−/− mice fed with control diet (KO group) or ESM supplemental diet (KOE group) for 19 weeks. During the treatment period, body weight (B) was measured for each group. After 19-week treatment, liver weight (C), visceral fat pad weight (D), gastrocnemius muscle weight (E), cecal content weight (F), colon length (G), colon weight (H), and colon weight to length ratio (I) were calculated (WT: n = 8, KO and KOE: n = 7, respectively). Data are presented as mean ± standard error. Superscript with different letters indicate significant difference at P < 0.05.
ESM Administration Modulated the Composition of Gut Microbiota in IL10−/− Mice
To analyze the gut microbiota, a total of 28,313 filter-passed high-quality reads per sample were extracted for further QIIME pipeline analysis. Alpha diversity analysis showed that Shannon and Simpson indices were lower in the KO group than in the WT group (Figures 2A,B). However, Shannon index-based diversity was recovered by ESM supplementation. Principal component analysis of unweighted UniFrac analysis showed a distinct difference in the gut bacterial profiles between the WT and KO groups (Figure 2C). The KOE samples were clustered between the WT and KO groups.
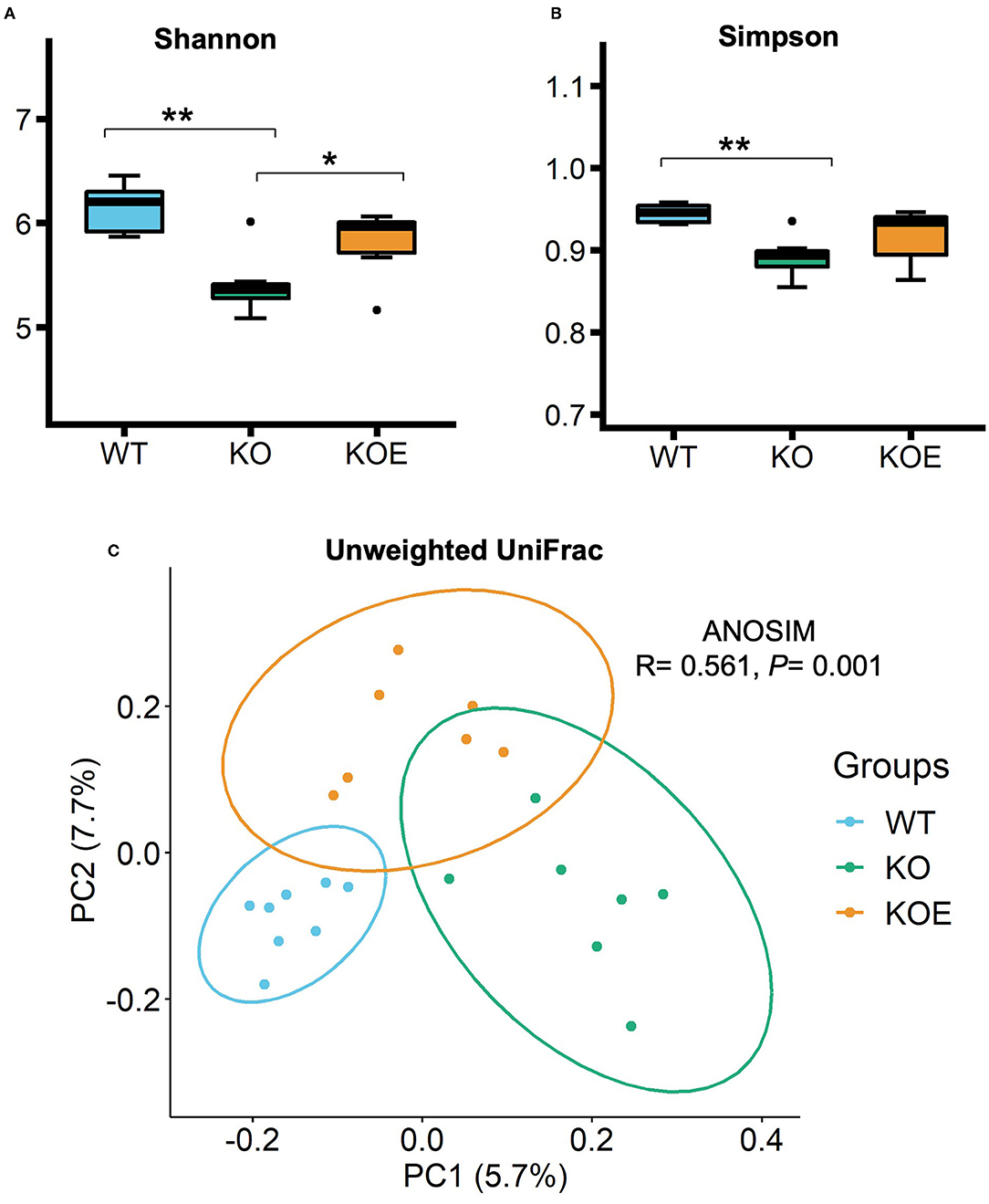
Figure 2. The α- and β-diversities of gut bacteria in mice of treatment groups [wild-type (WT): n = 8, IL10−/− (KO) and KO with eggshell membrane supplementation (KOE): n = 7, respectively]. After the 19-week treatment, total bacterial DNA in cecal contents were extracted for 16S rRNA gene sequencing. Gut bacterial diversity and richness within samples were measured by (A) Shannon and (B) Simpson indices. The principal component analysis (PCoA) of Unweighted UniFrac (C) and ANOSIM analysis were conducted to compare the gut bacterial profiles among the three groups. Statistical analysis of α-diversity data was performed by ANOVA and the Tukey-Kramer test. *P < 0.05, **P < 0.01. The dots (•) in the boxplots indicate outliers.
Taxonomic analysis revealed that all treatment groups contained four dominant phyla: Firmicutes, Bacteroidetes, Proteobacteria, and Verrucomicrobia (Figures 3A,B). At the phylum level, the abundance of Bacteroidetes was enriched in KO mice and reversed in KOE mice (Figure 3B). The abundance of Verrucomicrobia was decreased in KO mice but recovered by ESM supplementation. The ratio of Firmicutes to Bacteroidetes was significantly lowered in the KO group compared to the WT group, which was almost completely restored by ESM treatment (Figure 3C). At the genus level, commensal bacteria, such as Ruminococcus, were significantly reduced in the KO group but recovered in KOE, compared to the WT group. The family Bacteroidales S24-7 also followed this trend (Figure 3D). In contrast, Bacteroides, unclassified Enterobacteriaceae, and Blautia were significantly enriched in the KO group over the WT and KOE groups (Figure 3D). Collectively, these data indicated that ESM treatment markedly modulated the composition of gut bacteria in IL10−/− mice, resulting in a more similar bacterial structure to WT mice.
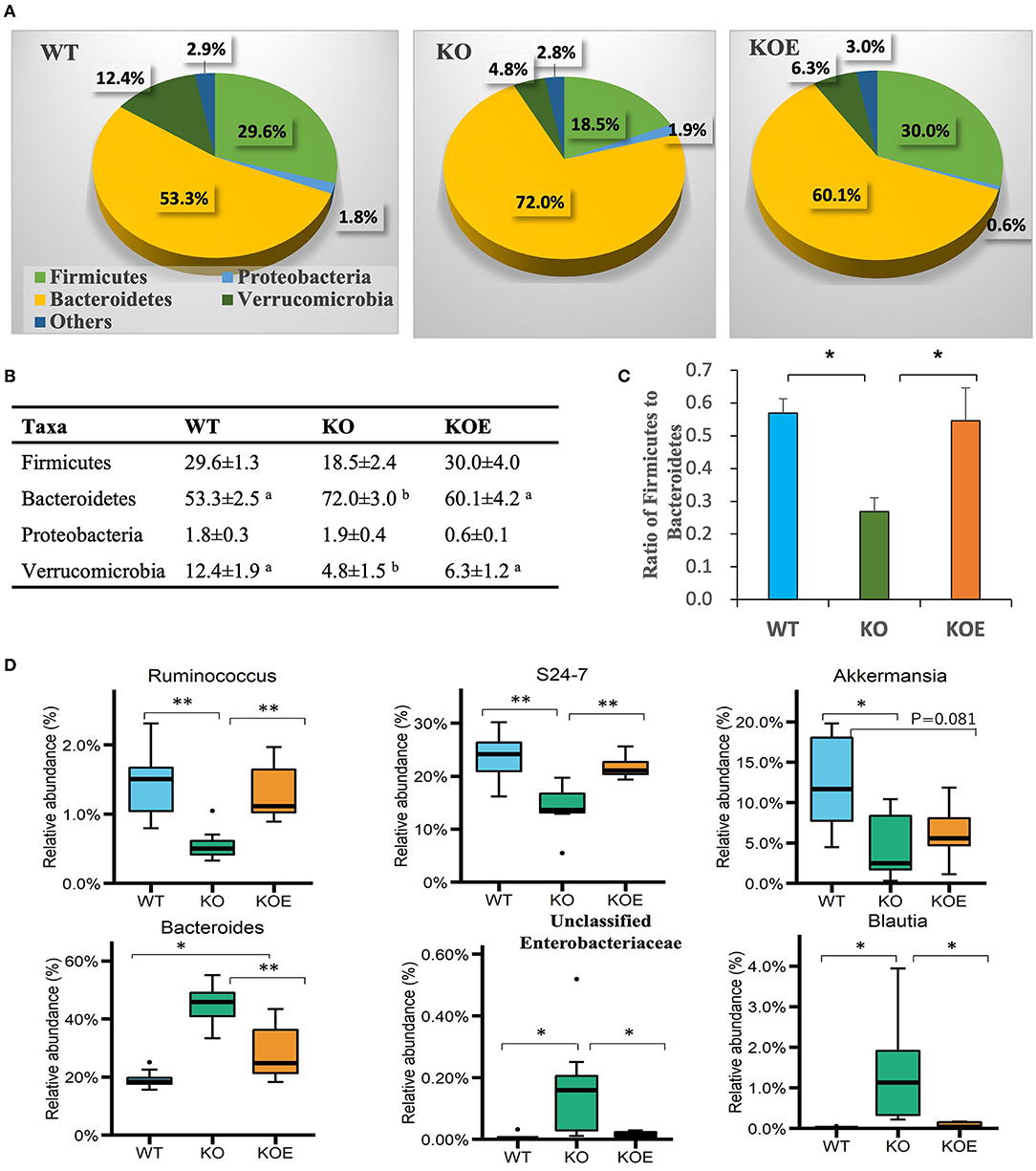
Figure 3. Effect of eggshell membrane (ESM) on gut bacterial composition in IL10−/− (KO) mice [wild-type (WT): n = 8, KO and KO with ESM supplementation (KOE): n = 7, respectively]. Relative abundance of phyla is demonstrated in the pie graphs (A) and table (B). (C) The ratio of Firmicutes to Bacteroidetes in the three groups. (D) The abundance of gut bacterial taxa at the genus level. Different superscript letters indicate significant differences at P < 0.05. *P < 0.05, **P < 0.01. The dots (•) in the boxplots indicate outliers.
The concentrations of SCFAs in mice cecal contents were quantified using a HPLC system. The level of butyrate, which is considered a beneficial bacterial metabolite in the intestines, was significantly increased in the KOE group over the KO group (Figure 4A). The levels of other SCFAs, including acetate, propionate, isobutyrate, and valerate, were not altered by ESM treatment (Figure 4A). The concentration of isovalerate was significantly lower in the KOE group than the KO group. To understand the relationship between specific bacterial taxa and cecal SCFAs, a Pearson-correlation heatmap was generated by selecting taxa at the family level and the levels of cecal SCFAs (Figure 4B). Particularly, a strong positive correlation was observed between Ruminococcaceae, Oscillospira, Clostridiales, and Bacteroidales S24-7 and the level of cecal butyrate, respectively. Bacteroides showed a positive correlation with isovalerate levels and a negative correlation with acetate and butyrate levels.
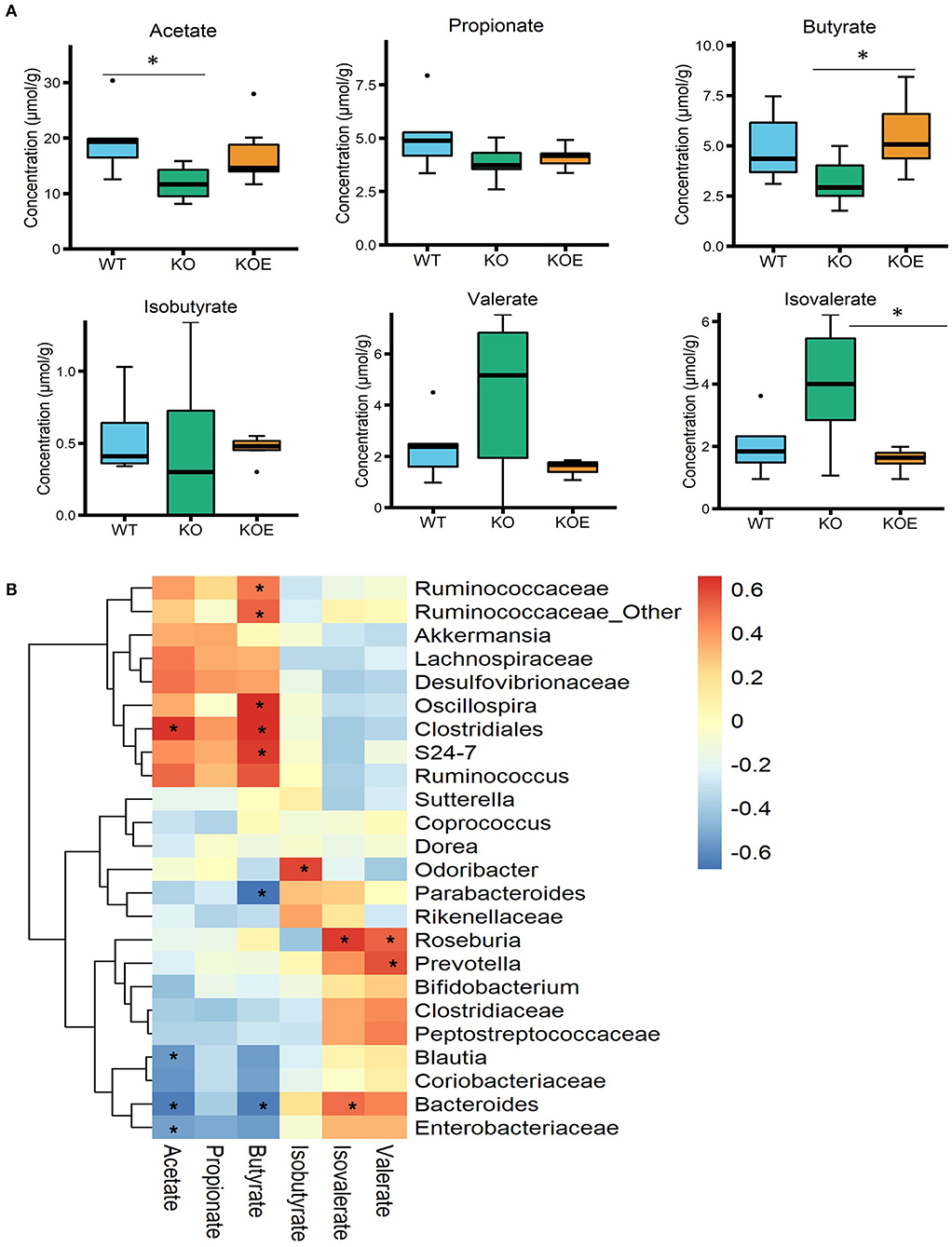
Figure 4. Effects of eggshell membrane (ESM) on the (A) levels of short-chain fatty acids (SCFAs) in cecal contents [wild-type (WT): n = 8, IL10−/− (KO) and KO with ESM supplementation (KOE): n = 7, respectively]. (B) The heatmap showing the Pearson correlation between SCFAs and the relative abundance of the selected taxa at the family level. *P < 0.05. The dots (•) in the boxplots indicate outliers.
To obtain a better understanding of the influence of ESM supplementation on gut bacteria, PICRUSt analysis was conducted to predict the bacterial gene functional profiles based on the 16S rRNA gene sequences. There are 28 KEGG pathways associated with gut microbiota that were significantly altered in the KO group compared to the WT group (Supplementary Figure S1A). ESM supplementation affected 21 KEGG pathways when compared to the KO group. Among them, 14 pathways (marked with *) that were altered in the KO group were recovered by ESM treatment (Supplementary Figure S1B).
ESM Inhibited the Augmented Proinflammatory Gene Expression in IL10−/− Mice
To further investigate the molecular mechanisms responsible for the observed beneficial effects of ESM on disease indices of IL10-KO mice, we analyzed relative gene expression in the mouse colonic mucosa with qPCR. The result showed the relative mRNA levels of Tnf-α and Il1β were dramatically upregulated in IL10−/− mice, however, the gene expression was significantly inhibited by ESM treatment (Figure 5). The expression of the other genes related to intestinal homeostatic functions, including Il17, Tgf-β1, Tlr4, and Apc, were not significantly altered by ESM supplementation.
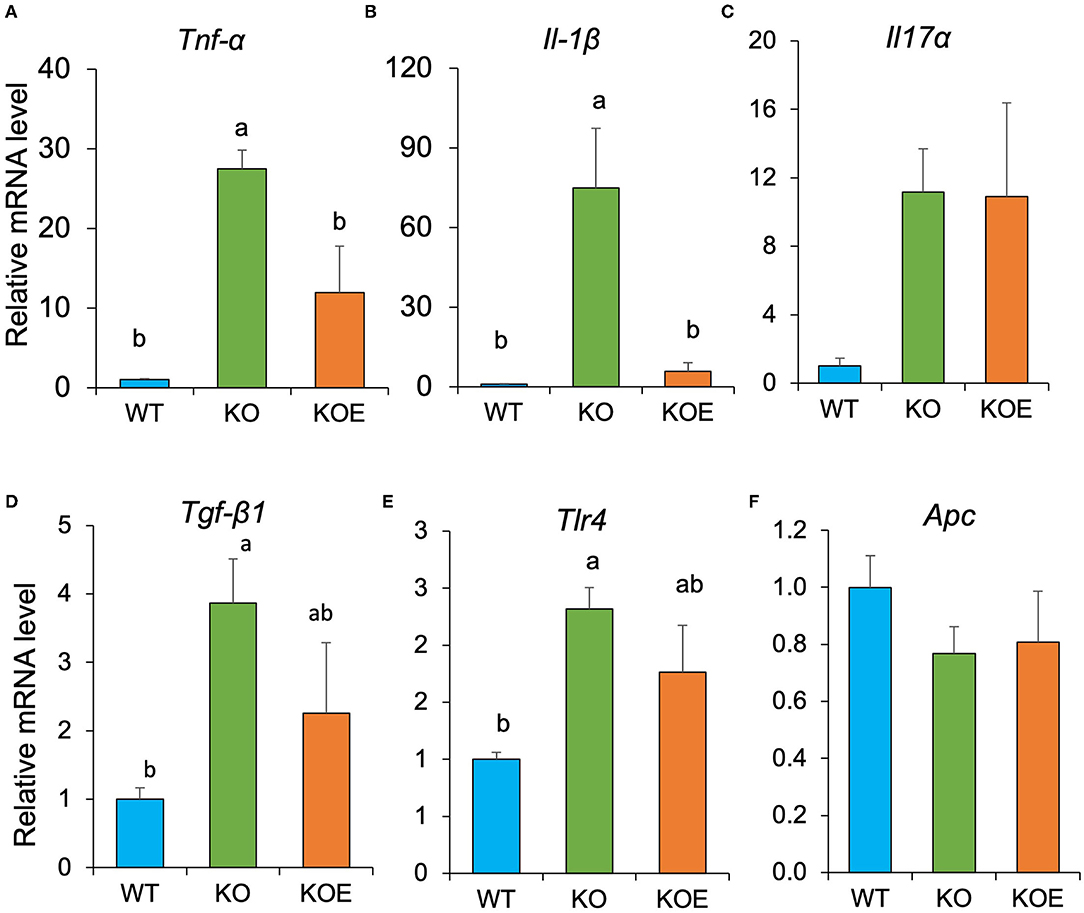
Figure 5. Eggshell membrane (ESM) modulates relative gene expression of several proinflammatory cytokines relative to RPLP1 in the mouse colonic mucosa. After the 19-week treatment, relative mRNA expression of Tnf-α (A), Il1β (B), Il17a (C), Tgf-β1 (D), Tlr4 (E), and Apc (F) in colonic mucosa were examined using quantitative real-time polymerase chain reaction (qPCR). Data are presented as mean ± standard error. Statistical analysis was performed by one-way ANOVA and the Tukey-Kramer test. Superscript with different letters indicate significant difference at P < 0.05.
Discussion
Eggshell membrane (ESM), primarily composed of collagen-like proteins, is considered as a safety and novel functional dietary ingredient. In this study, IL-10−/− mice, as a spontaneous colitis model, were used to explore the effect of ESM on colitis. The survival rate and body weight loss were ameliorated following ESM treatment in the IL-10−/− mice. Coincidently, the reduced tissue weights in the liver, visceral adipose, and gastrocnemius muscles were reversed following ESM treatment by the end of experimental period. These findings suggested that the colitis mouse model was successfully constructed, and ESM supplementation was effective in ameliorating some symptoms of colitis.
The alteration of gut bacterial composition is considered to play a critical role in the onset and progression of IBD. Therefore, we examined the effects of ESM on the composition of gut bacteria via 16S rRNA gene sequencing. Our results indicated that bacterial diversity was reduced in IL-10−/− mice, but this was reversed by ESM supplementation. Moreover, the gut microbiota of the three groups was clustered into different groups, where the KOE group was positioned between the WT and KO groups. These results suggest that the gut dysbiosis observed in colitis mouse model might be improved by ESM treatment. Firmicutes and Bacteroidetes were dominant in the mice intestine according to the taxonomic analysis. The Firmicutes / Bacteroidetes (F/B) ratio was reduced in IL-10−/− mice but was recovered to a normal level (similar to WT) following ESM treatment. The F/B ratio is generally considered to play an important role in maintaining regular gut homeostasis. The altered F/B ratio is a marker of dysbiosis, whereby an increase is normally observed with obesity, and a decrease is associated with IBD (19). However, an increased or unchanged F/B ratio was reported in other previous studies in IL-10−/− mice (7, 20). This disparity may be attributed to the difference in housing conditions, maturity or other environmental factors.
At the genus level, Ruminococcus was significantly decreased in the KO group, but recovered in KOE mice, compared to the WT group. The family Bacteroidales S24-7 followed the same trend. The symbiont of the genus Ruminococcus often inhabits both the animal and human gastrointestinal tract, and can use fermentable carbohydrates for growth. For instance, Ruminococcus bromii has the ability to ferment resistant starch and subsequently contribute dramatically to butyrate production in the gut (21). Consistently, our results showed that treatment with ESM enhanced the production of butyrate, which is positively associated with the elevated level of Ruminococcaceae. Meanwhile, members of the family S24-7, recently renamed as Muribaculaceae, are dominant in the murine gut microbiota and have been identified in the gastrointestinal tract of other animals (22). The abundance of S24-7 has been shown to decrease in mice fed with a high-fat diet but to increase in those fed with dietary fibers (23, 24). In addition, the abundance of S24-7 recovered to a higher level following treatment-induced colitis remission in a mouse model (25). In our study, S24-7 was found to have a positive correlation with cecal butyrate. These results imply that S24-7 may play a critical role in the amelioration of colitis by treatment with ESM.
In contrast, Bacteroides, unclassified Enterobacteriaceae, and Blautia were significantly higher in the KO group than in WT or KOE groups. The family Enterobacteriaceae contains symbiotic bacteria and several well-known pathogens, such as Escherichia coli, Klebsiella, and Shigella. An increased abundance of Enterobacteriaceae has been reported in both IBD patients and mouse colitis models (26, 27). Consistently, our previous study showed that ESM treatment decreased the abundance of Enterobacteriaceae in DSS-induced IBD model mice (16). The proinflammatory properties of Enterobacteriaceae have been well studied, but the mechanism by which Enterobacteriaceae is enriched in IBD is not fully clear. Meanwhile, the commensal bacterium Blautia is a dominant genus in the gut microbiota that plays a critical role in maintaining gut environmental balance and preventing gut inflammation through regulation of intestinal regulatory T cells and production of SCFAs (28). Decreased levels of Blautia have been found in patients with Crohn's disease or colorectal cancer (29, 30). However, several studies found higher levels of Blautia in the feces of IBD patients and ulcerative colitis than in healthy individuals (31, 32). Given these conflicting results, it will be interesting to explore the role of Blautia in the etiology of colitis. Due to the current shortcomings of 16S rRNA gene sequencing for gut microbiota, this study could not identify the gut bacterial taxa at the species or strain levels. Therefore, more in-depth studies should be performed to investigate the detailed roles of the gut microbiota, especially at the species or even strain levels.
Through the analysis of relative gene expression in the mouse colonic mucosa with qPCR, we confirmed aggravated colon inflammation through the increased expression of proinflammatory gene mRNA, such as Tnf-α and Il1β. However, ESM supplementation ameliorated the colon inflammatory state by downregulating these proinflammatory genes. Previous studies have showed that butyrate exerts anti-inflammatory effects by activating GPR41 and GPR43 and inhibiting NFκB activation (33, 34). Together, these results indicated the possibility that ESM ameliorates the colon inflammatory state through the modulation of gut microbiota and its metabolites. Further study is required to confirm this possibility.
Conclusions
In conclusion, the current study demonstrated the effectiveness of ESM supplementation in improving the survival rate and ameliorating the reduction of body weight, body fat, and muscle volume in IL10−/− mice. Furthermore, ESM supplementation improved the imbalanced gut microbiota composition, as indicated by the higher bacterial diversity and richness, the increased abundance of commensal gut bacteria (Ruminococcus and Bacteroidales S24-7), and the decreased level of potentially harmful bacteria (Enterobacteriaceae) in IL10−/− mice. Moreover, ESM supplementation promoted the production of butyrate in the gut and downregulated proinflammatory cytokine gene expression in the colon. These findings suggest that ESM could be used as a dietary intervention in colitis treatment.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found below: DDBJ; DRA013317.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee of the University of Tokyo.
Author Contributions
HJ, HK, and YH: conceptualization. HJ: methodology, supervision, and project administration. YY: formal analysis, writing original draft preparation, and visualization. WL, KF, and XL: data curation. HJ and HK: writing review and editing and funding acquisition. YH: resources. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded in part by Grant-in-Aid, grant number 18K11095 from the Japan Society for the Promotion of Science (JSPS).
Conflict of Interest
YH is working as a researcher in ALMADO Inc. in Tokyo.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.895665/full#supplementary-material
References
1. Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet. (2007) 369:1627–40. doi: 10.1016/S0140-6736(07)60750-8
2. Molodecky NA, Kaplan GG. Environmental risk factors for inflammatory bowel disease. Gastroenterol Hepatol. (2010) 6:339–46.
3. Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. (2012) 9:599–608. doi: 10.1038/nrgastro.2012.152
4. Maharshak N, Packey CD, Ellermann M, Manick S, Siddle JP, Young Huh E, et al. Altered enteric microbiota ecology in interleukin 10-deficient mice during development and progression of intestinal inflammation. Gut Microbes. (2013) 4:316–24. doi: 10.4161/gmic.25486
5. Håkansson, Tormo-Badia N, Baridi A, Xu J, Molin G, Hagslätt ML, et al. Immunological alteration and changes of gut microbiota after dextran sulfate sodium (DSS) administration in mice. Clin Exp Med. (2015) 15:107–20. doi: 10.1007/s10238-013-0270-5
6. Santoru ML, Piras C, Murgia A, Palmas V, Camboni T, Liggi S, et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep. (2017) 7:9523. doi: 10.1038/s41598-017-10034-5
7. Chen H, Xia Y, Zhu S, Yang J, Yao J, Di J, et al. Lactobacillus plantarum LP-Onlly alters the gut flora and attenuates colitis by inducing microbiome alteration in interleukin-10 knockout mice. Mol Med Rep. (2017) 16:5979–85. doi: 10.3892/mmr.2017.7351
8. Büchler G, Wos-Oxley ML, Smoczek A, Zschemisch NH, Neumann D, Pieper DH, et al. Strain-specific colitis susceptibility in IL10-deficient mice depends on complex gut microbiota-host interactions. Inflamm Bowel Dis. (2012) 18:943–54. doi: 10.1002/ibd.21895
9. Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. (1998) 66:5224–31. doi: 10.1128/IAI.66.11.5224-5231.1998
10. Triantafillidis JK, Merikas E, Georgopoulos F. Current and emerging drugs for the treatment of inflammatory bowel disease. Drug Des Devel Ther. (2011) 5:185–210. doi: 10.2147/DDDT.S11290
11. Ishikawa H, Matsumoto S, Ohashi Y, Imaoka A, Setoyama H, Umesaki Y, et al. Beneficial effects of probiotic Bifidobacterium and galacto-oligosaccharide in patients with ulcerative colitis: a randomized controlled study. Digestion. (2011) 84:128–33. doi: 10.1159/000322977
12. Kato K, Mizuno S, Umesaki Y, Ishii Y, Sugitani M, Imaoka A, et al. Randomized placebo-controlled trial assessing the effect of bifidobacteria-fermented milk on active ulcerative colitis. Aliment Pharmacol Ther. (2004) 22:56–63. doi: 10.1111/j.1365-2036.2004.02268.x
13. Wang H, Xue Y, Zhang H, Huang Y, Yang G, Du M, et al. Dietary grape seed extract ameliorates symptoms of inflammatory bowel disease in IL10-deficient mice. Mol Nutr Food Res. (2013) 57:2253–7. doi: 10.1002/mnfr.201300146
14. Khan I, Ullah N, Zha L, Bai Y, Khan A, Zhao T, et al. Alteration of gut microbiota in inflammatory bowel disease (IBD): Cause or consequence? IBD treatment targeting the gut microbiome. Pathogens. (2019) 8:126. doi: 10.3390/pathogens8030126
15. Jia H, Aw W, Saito K, Hanate M, Hasebe Y, Kato H. Eggshell membrane ameliorates hepatic fibrogenesis in human C3A cells and rats through changes in PPARγ-Endothelin 1 signaling. Sci Rep. (2014) 306:L361–71. doi: 10.1038/srep07473
16. Jia H, Hanate M, Aw W, Itoh H, Saito K, Kobayashi S, et al. Eggshell membrane powder ameliorates intestinal inflammation by facilitating the restitution of epithelial injury and alleviating microbial dysbiosis. Sci Rep. (2017) 7:43993. doi: 10.1038/srep43993
17. Kiesler P, Fuss IJ, Strober W. Experimental models of inflammatory bowel diseases. Cell Mol Gastroenterol Hepatol. (2015) 1:154–70. doi: 10.1016/j.jcmgh.2015.01.006
18. Ramli NS, Jia H, Sekine A, Lyu W, Furukawa K, Saito K, et al. Eggshell membrane powder lowers plasma triglyceride and liver total cholesterol by modulating gut microbiota and accelerating lipid metabolism in high-fat diet-fed mice. Food Sci Nutr. (2020) 8:2512–23. doi: 10.1002/fsn3.1545
19. Stojanov S, Berlec A, Štrukelj B. The influence of probiotics on the firmicutes/bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms. (2020) 8:1–16. doi: 10.3390/microorganisms8111715
20. Son HJ, Kim N, Song C-H, Nam RH, Choi SI, Kim JS, et al. Sex-related alterations of gut microbiota in the C57BL/6 mouse model of inflammatory bowel disease. J Cancer Prev. (2019) 24:173–82. doi: 10.15430/JCP.2019.24.3.173
21. Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. (2012) 6:1535–43. doi: 10.1038/ismej.2012.4
22. Lagkouvardos I, Lesker TR, Hitch TCA, Gálvez EJ, Smit N, Neuhaus K, et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome. (2019) 7:28. doi: 10.1186/s40168-019-0637-2
23. Yang Y, Fukui R, Jia H, Kato H. Amaranth supplementation improves hepatic lipid dysmetabolism and modulates gut microbiota in mice fed a high-fat diet. Foods. (2021) 10:1259. doi: 10.3390/foods10061259
24. Zhu L, Qin S, Zhai S, Gao Y, Li L. Inulin with different degrees of polymerization modulates composition of intestinal microbiota in mice. FEMS Microbiol Lett. (2017) 364:fnx075. doi: 10.1093/femsle/fnx075
25. Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, Michaud M, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. (2014) 8:1403–17. doi: 10.1038/ismej.2014.3
26. Yang I, Eibach D, Kops F, Brenneke B, Woltemate S, Schulze J, et al. Intestinal microbiota composition of interleukin-10 deficient C57BL/6J mice and susceptibility to helicobacter hepaticus-induced colitis. PLoS ONE. (2013) 8:e70783. doi: 10.1371/journal.pone.0070783
27. Litvak Y, Byndloss MX, Tsolis RM, Bäumler AJ. Dysbiotic proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr Opin Microbiol. (2017) 39:1–6. doi: 10.1016/j.mib.2017.07.003
28. Liu X, Mao B, Gu J, Wu J, Cui S, Wang G, et al. Blautia—a new functional genus with potential probiotic properties? Gut Microbes. (2021) 13:1–21. doi: 10.1080/19490976.2021.1875796
29. Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE. (2012) 7:e39743. doi: 10.1371/journal.pone.0039743
30. Chen L, Wang W, Zhou R, Ng SC, Li J, Huang M, et al. Characteristics of fecal and mucosa-associated microbiota in chinese patients with inflammatory bowel disease. Medicine. (2014) 93:e51. doi: 10.1097/MD.0000000000000051
31. Nishino K, Nishida A, Inoue R, Kawada Y, Ohno M, Sakai S, et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J Gastroenterol. (2018) 53:95–106. doi: 10.1007/s00535-017-1384-4
32. Rajilić-Stojanović M, Biagi E, Heilig HGHJ, Kajander K, Kekkonen RA, Tims S, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. (2011) 141:1792–801. doi: 10.1053/j.gastro.2011.07.043
33. Chang P V., Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci. (2014) 111:2247–52. doi: 10.1073/pnas.1322269111
Keywords: inflammatory bowel disease, eggshell membrane, gut microbiota, IL10, colitis
Citation: Yang Y, Jia H, Lyu W, Furukawa K, Li X, Hasebe Y and Kato H (2022) Dietary Eggshell Membrane Powder Improves Survival Rate and Ameliorates Gut Dysbiosis in Interleukin-10 Knockout Mice. Front. Nutr. 9:895665. doi: 10.3389/fnut.2022.895665
Received: 14 March 2022; Accepted: 14 April 2022;
Published: 19 May 2022.
Edited by:
Carlos Gómez-Gallego, University of Eastern Finland, FinlandReviewed by:
Jing Wang, Feed Research Institute (CAAS), ChinaManuela M. Santos, Université de Montréal, Canada
Copyright © 2022 Yang, Jia, Lyu, Furukawa, Li, Hasebe and Kato. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huijuan Jia, YWtha2Vpa2VuQGcuZWNjLnUtdG9reW8uYWMuanA=; Hisanori Kato, YWthdG9xQGcuZWNjLnUtdG9reW8uYWMuanA=
 Yongshou Yang
Yongshou Yang Huijuan Jia
Huijuan Jia Weida Lyu3
Weida Lyu3 Kyohei Furukawa
Kyohei Furukawa