- 1Key Laboratory of Animal Protein Deep Processing Technology of Zhejiang Province, College of Food and Pharmaceutical Sciences, Ningbo University, Ningbo, China
- 2Center for Global Health, School of Public Health, Nanjing Medical University, Nanjing, China
- 3Ordos Agriculture and Animal Husbandry Technology Extension Centre, Ordos, China
- 4Institute of Environmental Research at Greater Bay Area, Guangzhou University, Guangzhou, China
Frequent meat frauds have become a global issue because adulteration risks the food safety, breaches market rules, and even threatens public health. Multiplex PCR is considered to be a simple, fast, and inexpensive technique that can be applied for the identification of meat products in food industries. However, relatively less is known about a multiplex PCR method authenticating seven animal species simultaneously in one reaction due to technological challenge. Through screening new species-specific primers and optimizing PCR system, a heptaplex PCR method was established, which could simultaneously detect seven meat ingredients of camel (128 bp), pigeon (157 bp), chicken (220 bp), duck (272 bp), horse (314 bp), beef (434 bp), and pork (502 bp) in a single-tube reaction. DNA sequencing solidly validated that each set of primers specifically amplified target species from total DNA mixtures of seven meat species. The developed multiplex assay was stable and sensitive enough to detect 0.01–0.025 ng DNA from various meat treatments including raw, boiled, and autoclaved meat samples or target meat content of 0.1% total meat weight, suggesting the suitability of the heptaplex PCR technique for tracing target meats with high accuracy and precision. Most importantly, a market survey validated the availability of this multiplex PCR technique in real-world meat products with a good application foreground.
Introduction
Frequent meat frauds have become a global issue, which perhaps risks the food safety, breaks the rules of the market, and even threatens public health (1, 2). Specifically, since the European horsemeat scandal in 2013, the events regarding ingredient substitution of expensive meat with low-cost ones for extra economic benefits have frequently occurred worldwide (3). The false information on the label will mislead the choices of consumers, resulting in serious religious issues and health problems (4, 5). For example, meat products harboring pork ingredients are not permitted in Islamic countries (6, 7). Similar to soy allergy, certain meat species may trigger allergic reactions, especially for sensitized patients, which may cause a severe health risk of infectious diseases, metabolic disorders, and allergies (8–11). Therefore, a practical technique to identify the animal origin with rapid, sensitive, and accurate characteristics is of great importance (10, 11).
The techniques have been continuously evolved for identifying the origin of meat species in the recent years (2, 12–15). DNA molecules present in every cell and possess high stability, and therefore, DNA-based techniques combined with polymerase chain reaction (PCR) provide more reliable methods in discriminating meat species (13, 16, 17). Both conventional multiplex and real-time PCR techniques are considered to be reliable methods with high sensitivity and specificity in authenticating meat species (18). Real-time PCR techniques are generally used for quantifying the amount of a target sequence in a reaction system. As reported, accurate quantification could only be achieved with a proper reference material, as some ingredients in the recipe might be co-extracted with the DNA and interfere with the quantification process (19, 20). In this regard, multiplex PCR presents a simple, efficient, and inexpensive technique, which are being widely applied for qualitatively authenticating animal origin in food industries (1). Nowadays, this technique remains to be considered as a practical method used for identifying the origin of meat species.
Mitochondrial DNA has high copy number in each cell and possesses strong stability, which has allowed a low detection limit and broad availability in various meat products (21). Extensive studies have found that mitochondrial genes such as cytochrome b, 12S and 16S rRNA, D-loop, ATPase subunits 6 and 8, and NADH dehydrogenase have been broadly targeted for PCR protocols in the identification of meat species (19, 21–24). Here, using mitochondrial DNA sequences retrieved from camel, pigeon, chicken, duck, horse, beef, and pork, seven sets of primers that specifically amplified seven animal species were designed with differential lengths through PCR assays. Simplex and multiplex PCR assays demonstrated the specific, sensitive, and efficient properties of all primers. This study ultimately constructs a heptaplex PCR method, which can simultaneously detect seven ingredients in meat products.
Materials and Methods
DNA Extraction
Fresh pure meat of camel, pigeon, chicken, duck, horse, beef, and pork was obtained from local retailers and markets, which was transported on ice to the laboratory for processing immediately. A total of 67 raw or heat processing of meat balls (10), meat slices (7), kebab (5), sausages (10), cutlets (3), drysaltery (7), jerky (10), steak (4), dry meat stripe (5), braised pigeon (3), and fried pigeon (3) were obtained from markets as well as from online supermarket platform. All samples were stored at −80°C to inhibit DNA degradation. Genomic DNA from meat samples was isolated using the EasyPure® Genomic DNA Kit (Beijing Trans Gen Biotech Co., Ltd., Beijing, China) according to the manufacturer's instructions. DNA concentration was examined by a NanoDrop 2000 spectrophotometer (NanoDrop 2000, UV–Vis spectrophotometer, USA) (25).
Design of Seven Species-Specific Primer Pairs
Mitochondrial genes were selected as targets for designing primers according to high divergence and conservation within the animal species (23). As seen in Table 1, sequences of cytochrome c oxidase subunit III of camel (GenBank accession no. MH109991.1), NADH dehydrogenase subunit 6 of pigeon (KP168712.1), D-loop of chicken (MK163565.1), ATPase subunit 6 of duck (MK770342.1), NADH dehydrogenase subunit 4 of horse (MN187574.1), cytochrome c oxidase subunit II of beef (MN714195.1), and 16S rRNA gene of pork (KJ746666.1) were retrieved from the National Center of Biotechnology Information (NCBI) database. The conservative and variable regions among animal species were examined by MEGA6 alignment tool. Combined Oligo 7.0 with BLAST programs (www.ncbi.nlm.nih.gov/blast/), new primers were designed according to physical parameters of melting temperature, cross-reactivity, self-complementarity, and secondary structures (7). Primers were synthesized by Shanghai Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China). To detect the mismatch present in the target and non-target species, all primer pairs were in silico screened against 14 land animals, camel (Camelus bactrianus), pigeon (Columba livia), chicken (Gallus gallus), duck (Anas platyrhynchos), horse (Equus caballus), cattle (Bos taurus), pig (Sus scrofa), turkey (Meleagris gallopavo), goose (Anser cygnoides), sheep (Ovis aries), ostrich (Struthio camelus), dog (Canis lupus), rabbit (Oryctolagus cuniculus), cat (Felis catus), and 3 aquatic species, small yellow croaker (Larimichthys polyactis), black carp (Mylopharyngodon piceus), and tuna (Thunnus orientalis) using ClustalW software. Specificity of primer pairs was examined by simplex PCR assays, respectively.
Simplex and Multiplex PCR Assays
Polymerase chain reaction assays were performed as previously described (25). For simplex PCR, PCR amplification was achieved using EasyTaq® DNA Polymerase Kit (Beijing TransGen Biotech Co., Ltd., Beijing, China). PCR system was composed of 2.5 μl of 10 x EasyTaq® Buffer, 2 μl of 2.5 mM dNTPs, 0.5 μl of EasyTaq DNA polymerase, 0.5 μl of 10 μm each primer, genomic DNA of each species, and refilled ddH2O to 25 μl in a single reaction. The reaction was elicited by a 5-min denaturation at 94°C, followed by 34 cycles (94°C for 30 s, 63°C for 30 s, and 72°C for 45 s) and a final elongation at 72°C for 5 min. Using the same PCR amplification condition as that of simplex PCR, we employed a set of seven species-specific primers and corresponding genomic DNA as templates to develop a heptaplex PCR assay. For multiplex PCR, PCR system included 2.5 μl EasyTaq® Buffer (10 x), 2 μl dNTPs (2.5 mm), 0.5–1 μl EasyTaq DNA Polymerase (5 units μl−1), 0.5 μl each primer of seven species (10 μM), 1 μl genomic DNA of each species at the indicated concentrations from 10 to 0.1 ng μl−1, and refilled ddH2O to 25 μl. All PCR fragments were amplified using T100™ Thermal Cycler (Bio-Rad, Germany) and analyzed through 4% agarose gels using 4S GelRed Nucleic Acid Stain, which were visualized by Gel DocTM XR+ System with Image LabTM Software (BIO-RAD) (26).
Sequencing of PCR Products
Each PCR product was purified from the gel with DiaSpin DNA Gel Extraction Kit (Shanghai Sangon Biological Engineering Technology & Services Co., Ltd., Shanghai, China) and cloned into a pEASY®-T5 Zero Cloning vector (TransGen Biotech Co., Ltd., Beijing, China) according to the manufacturer's protocol. Each plasmid DNA was extracted using a SanPrep Column Plasmid Mini-Preps Kit (Shanghai Sangon Biological Engineering Technology & Services Co., Ltd., Shanghai, China). PCR amplification with vector primers M13F (5'-GTAAAACGACGGCCAGT-3') and M13R (5'- CAGGAAACAGCTATGAC-3') was carried out using the template of plasmid DNA and then sequenced by an automated DNA sequencer (Applied Biosystems, Foster City, CA, USA). Determination of DNA base composition of the sequences was accomplished by a BLAST search against the NCBI nucleotide database.
Evaluation of Primers' Specificity, Sensitivity, and Reproducibility
The specificity of each primer pair was assessed by using template DNA extracted from all species (camel, pigeon, chicken, duck, horse, cattle, pork, turkey, goose, sheep, rabbit, ostrich, dog, cat, small yellow croaker, tuna, and black carp). For the preliminary phase of the experiment, simplex and multiplex PCR assays were individually performed using the template DNA isolated from raw animal species. For sensitivity test, a series of PCR assays were performed using serial dilutions of the premixed genomic DNA templates of all target species in one reaction. A total of ten concentrations of the target templates ranging from 10 ng to 0.01 ng were used for PCR amplification. To further determine the sensitivity of model mixtures, each raw meat tissue of pork, horse, duck, chicken, camel, and pigeon was weighed at 0.1, 0.25, 0.5, 1, 2.5, 5, 10, and 15 of total weight, respectively. Then, all meat weighed at the same proportion were gathered with beef together and then homogenized separately using different triturators to avoid cross-contaminations. The dynamic range and the limit of detection were determined through 4% agarose gel analysis. For reproducibility assay, raw meat samples were boiled (97–99°C, 30 min) and microwave-cooked (750 W, 10 min) and then used for DNA isolation. Similar to sensitivity test, PCR amplification was carried out and the reproducibility was analyzed by agarose gel analysis (27).
Results
Specificity Assays of the Designed Primers
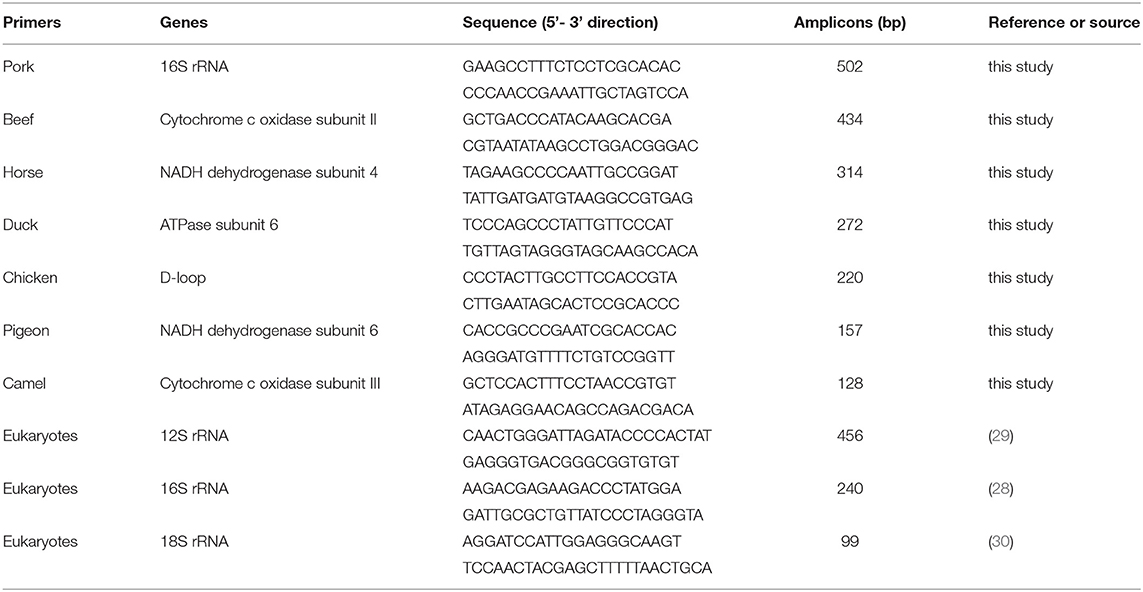
To obtain species-specific primers, many candidate primers for each species were checked. Using the primer pairs in Table 1, PCR amplification produced the only band with target meat species but not non-target species after the analysis of gel electrophoresis. As shown in Figure 1A, PCR products showed differential bands with the predicted size of 128, 157, 220, 272, 314, 434, and 502 bp for camel, pigeon, chicken, duck, horse, beef, and pork species, respectively. To ensure the quality of genomic DNA templates, three sets of universal eukaryotic primers were selected as positive controls in a single PCR, which targets 18S rRNA, 16S rRNA, and 12S rRNA genes with the predicted size of 99-bp, 240-bp, and 456-bp PCR fragments in all meat species (28–30). In our study, all meat samples generated the target PCR bands with intensities similar to each other (Figure 1B), suggesting the good quality of genomic DNA existing in meat resource. Using genomic DNA of a single meat species as the template, PCR product was obtained just using all primer mixtures of seven meat species, but not that of six non-target primer pairs without target counterpart (Figure 1C). In addition, species-specific primer pair generated PCR fragments with template DNA mixture of seven but not six meat species excluding target one through PCR amplification (Figure 1D), suggesting that the primer pair can specifically amplify target species. To make the conclusion more solid, the 128, 157, 220, 272, 314, 434, and 502-bp amplicons in Figure 1D were cloned and then sequenced. As expected, DNA sequencing validated the accurate amplification of camel, pigeon, chicken, duck, horse, cattle, and pork by a BLAST search against the NCBI nucleotide database. In addition, the specificity of all primer pairs was validated against 17 animal species which indicated through PCR analysis (data not shown). Collectively, this assay demonstrates that new primers are highly specific and are suitable for the authentication of meat species in real-world foodstuffs.
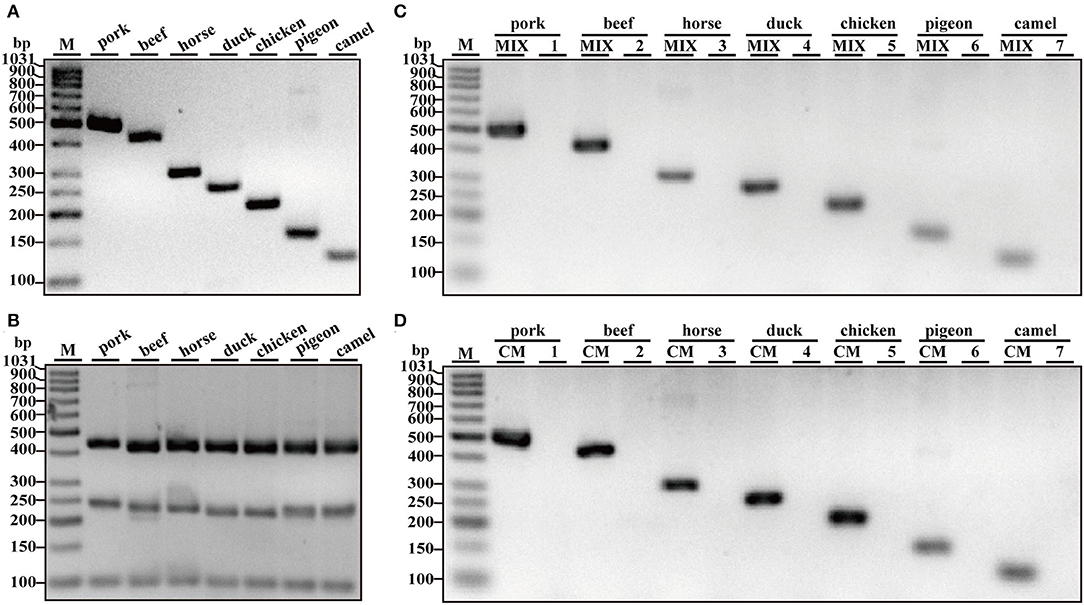
Figure 1. Verification of primer specificity with conventional simplex PCR. (A) Simplex PCR detection using species-specific primers for camel, pigeon, chicken, duck, horse, beef, and pork origin and respective genomic DNA as a template. (B) PCR amplification with premixed universal primers of eukaryotic 12S rRNA, 16S rRNA, and 18S rRNA genes for each meat species, respectively. (C) PCR amplification using individual template DNA from camel, pigeon, chicken, duck, horse, beef, and pork species. MIX, a mixture of seven primer pairs of camel, pigeon, chicken, duck, horse, beef, and pork species; 1–7, a mixture of six primer pairs of six nontarget species. (D) PCR amplification with species-specific primers for camel, pigeon, chicken, duck, horse, beef, and pork species. CM, a complete mixture of seven species including camel, pigeon, chicken, duck, horse, beef, and pork; 1–7, a complete DNA mixture of six meat species except target species. Lane M is ladder DNA.
Sensitivity Assays of Heptaplex PCR
After the specificity analysis of each set of primers, a heptaplex PCR system was developed using seven pairs of species-specific primers. To analyze the dynamic range as well as the limit of detection (LOD), multiplex PCR assays were performed with serial dilutions of template DNA of each target species ranging from 10 ng to 0.01 ng in per PCR. As shown in Figure 2A, top bands of five meat species (pork, beef, horse, duck, and chicken) were obviously observed at all indicated DNA concentrations ranging from 0.01 to10 ng DNA. In comparison, pigeon and camel had relatively weak bands under the condition of 0.01 and 0.25 ng template DNA, especially 0.01 ng DNA; however, they seemed to be recognized.
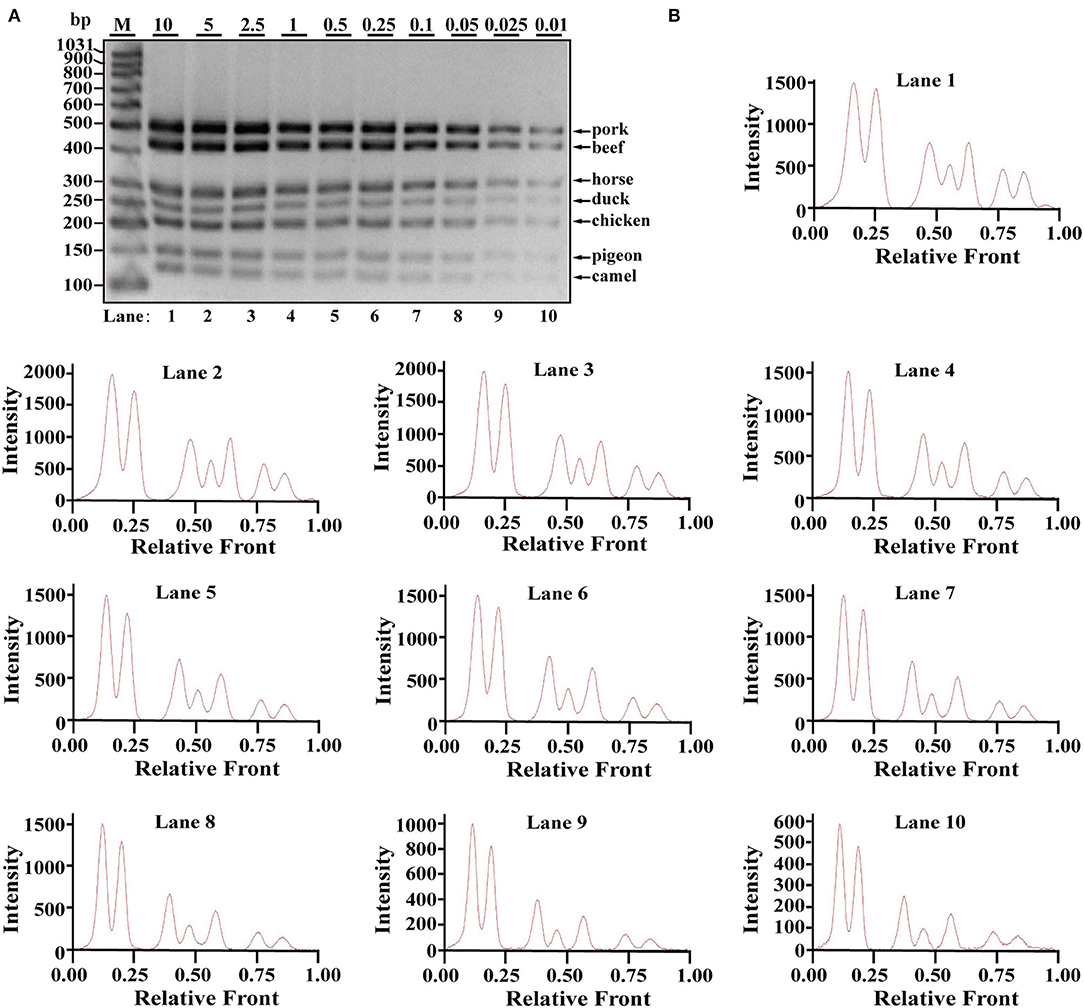
Figure 2. Validation of the sensitivity of multiplex PCR assay. (A) Gel image of PCR fragments amplified by multiplex PCR using premixed DNA templates of seven species (10, 5, 2.5, 1, 0.5, 0.25, 0.1, 0.05, 0.025, and 0.01 ng) with species-specific primers of seven meat species in a single PCR. (B) The corresponding electropherogram of gel image represented pork, beef, horse, duck, chicken, pigeon, and camel in each lane. Lanes 1–10 are presented with labels (10, 5, 2.5, 1, 0.5, 0.25, 0.1, 0.05, 0.025, and 0.01) in (A). The value of number at the horizontal line means the relative position of peaks distant from the top of agarose gel. The value of number at the vertical line means the fluorescent intensity of DNA-bound dyes using 4S GelRed nucleic acid stain. Lane M is ladder DNA.
Using Image LabTM Software, the electropherograms were drawn based on the bands. The visible bands were matched with intact peak patterns, whereas weak bands were equipped with defective peak patterns. As can be seen from Figure 2B, the fluorescent intensities were gradually decreased from lanes 1 to 10 along with reduced content of genomic DNA template, reflecting their reduced PCR products. In accordance with gel view, electropherograms showed intact peak patterns of pork, beef, horse, duck, and chicken in lines 1–10. By contrast, lower intensities of camel and pigeon were found in all lines, whereas defective peak pattern of both camel and pigeon was present in line 10. Collectively, it was postulated that LOD of heptaplex PCR method was 0.01 ng DNA for pork, beef, horse, duck, and chicken, whereas it was 0.025 ng DNA for pigeon and camel. In addition, model beef adulterated with six meat tissues of pork, horse, duck, chicken, camel, and pigeon at 0.1, 0.25, 0.5, 1, 2.5, 5, 10, or 15% of total weight was individually used for genomic DNA extraction. Using the multiplex PCR method, the specific amplicons for each species were clearly displayed even at target meat percentages of 0.1% (Figures 3A,B).
Figure 3. Validation of the sensitivity of multiplex PCR assay. (A) Gel image of PCR fragments amplified by multiplex PCR using model mixtures of pork, horse, duck, chicken, pigeon, and camel added to beef at 15, 10, 5, 2.5, 1, 0.5, 0.25, and 0.1% of total weight with species-specific primers of seven meat species in a single PCR. (B) The corresponding electropherogram of gel image represented pork, beef, horse, duck, chicken, pigeon, and camel in each lane. Lanes 1–8 are presented with labels (15, 10, 5, 2.5, 1, 0.5, 0.25, and 0.1%) in (A). The value of number at the horizontal line means the relative position of peaks distant from the top of agarose gel. The value of number at the vertical line means the fluorescent intensity of DNA-bound dyes using 4S GelRed nucleic acid stain. Lane M is ladder DNA.
Reproducibility Assay of Heptaplex PCR in Heat Processing Meat
To examine the efficiency of each set of primers for detecting thermally processed meat, both boiled and microwave-cooked treatments were adopted to process raw meat samples. After genomic DNA isolated from boiled meat samples, heptaplex PCR was carried out and the products were analyzed through 4% agarose gels. As shown in Supplementary Figures S1A, S1B, seven bands of seven meat species were obviously observed at the range of 0.01–10 ng DNA, and intact peak patterns of seven species were found in lines 1–10, suggesting that LOD of heptaplex PCR method was 0.01 ng DNA for pork, beef, horse, duck, chicken, pigeon, and camel under the condition of boiled treatment. Using genomic DNA isolated from microwave-cooked meat samples, top bands of five meat species (pork, beef, horse, duck, and chicken) were obviously observed at the range of 0.025–10 ng DNA, and intact peak patterns of pork, beef, horse, duck, and chicken were found in lines 1–9 (Supplementary Figures S2A, S2B). Combined the data obtained from both boiled and microwave-cooked treatments, the threshold value for discrimination of heat processing meat was about 0.01–0.025 ng DNA. The results suggest that new primers are suitable for authenticating animal species in real-world meat products.
Application of Multiplex PCR Assays on Commercial Foodstuffs
Typical cases of intentional meat adulteration involve the substitution or addition of animal ingredients not declared on the label of the products. A total of 67 commercial samples of pork, beef, horse, pigeon, and camel were purchased for the identification of real-world meat products through this heptaplex PCR technique. As summarized in Table 2, most of meat samples contained the identical ingredients as labeled without any contamination. However, some samples that declared to be 100% pure meat content were found to be substituted with other meat ingredients. As illustrated, 5 of 15 (33.3%) pork samples, 6 of 15 (40.0%) beef samples, 3 of 12 (25%) horse samples, 1 of 10 (10%) pigeon samples, and 3 of 15 (20%) camel samples contained some meat ingredients unlisted. This survey unmasked that cheap meat of chicken, duck, and pork are likely to frequently used as a substitute ingredient for red meat. Most importantly, the survey further validated the efficiency of heptaplex PCR technique in authenticating commonly consumed meat species.
Discussion
Food frauds are the topical issues that must be addressed due to quality and safety purposes and to maintain consumers' trust (31). Nowadays, adulteration practice has been ingeniously done with the morphological and physical characteristics similar to pure meat (6). Multiplex PCR assays have some advantages in discriminating animal origins, because they are easily accomplished through simple agarose gel analysis and dramatically minimizes the cost (32–34). As shown in Supplementary Table S1, much information is available on recently published multiplex PCR methods. While the detection number of meat origin mainly focuses on two to six species in one reaction, relatively less is known on the identification of more meat species in a single PCR. Although some studies have authenticated more than ten animal species, they are achieved by two-tube multiplex PCR assays (7, 27). Accordingly, it is still lack of reliable, low-cost, and high-throughput detection methods for supervising more species origin of meat. This study has set up a multiplex PCR method for simultaneous identification of seven meat species including ruminant, poultry, and pork materials.
Due to the high homology existing in meat species, proper target genes are required for establishing the multiple PCR system. Mitochondrial DNA sequence has variable region with intraspecific and interspecific polymorphism in animal cells, which is highly suitable for the discrimination of closely related animal species (35). Besides, mitochondrial DNA sequences have multiple copies within the ring structure, which is more stable during meat processing. Therefore, the polymorphism site of mitochondrial sequences was selected as a target region for designing species-specific primers in meat source. Because multiple targets can be simultaneously detected by a multiplex PCR method in a single/assay platform, species-specific primers should be analyzed, screened, and optimized to eliminate interaction among animal species and thus ruled out the cross-reactivity in meat resource. Based on this, many candidates of species-specific primers were designed throughout target mitochondrial DNA sequences and ultimately determined seven sets of species-specific primers for camel (128 bp), pigeon (157 bp), chicken (220 bp), duck (272 bp), horse (314 bp), beef (434 bp), and pork (502 bp). The specificity test validates that all primers are specific to each own species without cross-reactivity with other 16 animal species indicated. In addition, DNA sequencing further validates the specificity of species-specific primers. Notably, different lengths of multiple amplicons as well as the competition among PCR amplification modules may result in different PCR efficiencies and, consequently, affect LOD of multiplex PCR methods. As reported, the detection limits of multiplex PCR assays vary from 1 pg to 0.32 ng (Supplementary Table S1). Here, LOD of this multiplex PCR technique is about 0.01–0.025 ng DNA in various meat samples of raw, boiled, and microwave-cooked meat. To further evaluate the availability of multiplex PCR in real-world foodstuffs, model mixtures were constructed by adding each meat sample of pork, horse, duck, chicken, camel, and pigeon weighed at 0.1 to 15% of total weight to beef. The proposed multiplex PCR method can clearly detect even at target meat percentages of 0.1% (Figure 3), whereas target meat content can be availably detected at percentages of 0.01 to 9.1% total weight (Supplementary Table S1). The data suggest that this method is adequate for meat authentication.
The demand for beef is increasing worldwide due its nutritional value of protein, amino acids, and trace elements. A high incidence of beef substitution and mislabeling is prevailing for monetary benefits. Hence, detection of beef adulteration is indispensable for safeguarding consumer rights and food safety. Application of multiplex PCR assay on commercial meat products revealed some interesting and shocking findings that cheap poultry and pork meats are frequently adulterated to meat products (Table 2). In accordance with previous studies, meat resource is frequently adulterated with cheap- or poor-quality meat such as chicken, duck, and pork, which easily escapes visual detection (36–38). Perhaps, economic benefits are a critical factor for the substitution of expensive and high-quality meat with inferior and low-cost ones, and therefore, the amounts of adulterated ingredients should be easily detected. In this regard, these traces such as 0.1% adulterant may be mixed unintentionally with other origins from either the place of origin or at processing level. Identification of animal origin can ensure the authenticity and traceability of meat products, protecting consumers' health and complying with religious faith. However, serially diluted DNA from raw meats could not be used as a reference for DNA isolated from commercial meat samples, as DNA co-extraction might occur due to the presence of inclusion in the recipe and interferes with the quantification process. Therefore, multiplex PCR assays perhaps fail to determine the percentage of adulteration of a real-world meat product. In addition, less is known about how the magnitude of adulteration poses health threats, especially for sensitized patients. It is necessary to deal with the challenging issues in the future study. Nonetheless, this study provides a developed heptaplex PCR assay showing a reliable, efficient, and sensitive detection method for the discrimination of meat species origin in actual adulteration event.
Conclusions
This study provides a heptaplex PCR technique for the identification of meat products mislabeling prevailing in real-world foodstuffs, which achieves simultaneous detection of seven animal species of camel, pigeon, chicken, duck, horse, beef, and pork species. Meat origin can be availably detected at the concentration of 0.01–0.025 ng DNA or target meat content of 0.1% total meat weight in one-tube reaction system, indicating that this assay has qualified for the authentication of species origin of meat in real-world foodstuffs. The availability of the assay has been further confirmed by the findings obtained from application of multiplex PCR on commercial meat products. Collectively, this multiplex PCR technique could be more broadly used for the identification of species origin of meat in foodstuffs through the analysis of simple agarose gel.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
DP, ZC, and JH: conception and design of the investigation and work. SZ, GZ, HZ, XZh, XZe, and ZW: completion of the experiments. SZ, GZ, ZC, JH, QL, and DP: evaluation and analysis of the results. SZ, GZ, ZC, and JH: manuscript writing. SZ, GZ, HZ XZh, XZe, ZW, DP, JH, QL, and ZC: final approval of manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Natural Science Foundation of China (NSFC) (31901668), the Natural Science Foundation of Zhejiang Province of China (LY22C200002), the Scientific Research Fund of Zhejiang Provincial Education Department (Y201940932), the Natural Science Foundation of Ningbo (2019A610436 and 2021J108), and the School Research Project in Ningbo University (XYL19011).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.890537/full#supplementary-material
References
1. Li YC, Liu SY, Meng FB, Liu DY, Zhang Y, Wang W, et al. Comparative review and the recent progress in detection technologies of meat product adulteration. Compr Rev Food Sci F. (2020) 19:2256–96. doi: 10.1111/1541-4337.12579
2. Chung SM, Hellberg RS. Effects of poor sanitation procedures on cross-contamination of animal species in ground meat products. Food Control. (2020) 109:106927. doi: 10.1016/j.foodcont.2019.106927
3. Li TT, Wang JS, Wang ZY, Qiao L, Liu R, Li SS, et al. Quantitative determination of mutton adulteration with single-copy nuclear genes by real-time PCR. Food Chem. (2021) 344:128622. doi: 10.1016/j.foodchem.2020.128622
4. Li JC Li JP, Liu RX, Wei YX, Wang SW. Identification of eleven meat species in foodstuff by a hexaplex real-time PCR with melting curve analysis. Food Control. (2021) 121:107599. doi: 10.1016/j.foodcont.2020.107599
5. Nugraha WS, Chen D, Yang S-H. The effect of a Halal label and label size on purchasing intent for non-Muslim consumers. J. Retail. Consum. Serv. (2022) 65:102873. doi: 10.1016/j.jretconser.2021.102873
6. Li TT, Jalbani YM, Zhang GL, Zhao ZY, Wang ZY, Zhao XY, et al. Detection of goat meat adulteration by real-time PCR based on a reference primer. Food Chem. (2019) 277:554–7. doi: 10.1016/j.foodchem.2018.11.009
7. Li JC Li JP, Xu SG, Xiong SY, Yang JN, Chen X, et al. A rapid and reliable multiplex PCR assay for simultaneous detection of fourteen animal species in two tubes. Food Chem. (2019) 295:395–402. doi: 10.1016/j.foodchem.2019.05.112
8. Tanabe S, Miyauchi E, Muneshige A, Mio K, Sato C, Sato M, et al. Method of detecting pork in foods for verifying allergen labeling and for identifying hidden pork ingredients in processed foods. Biosci Biotech Bioch. (2007) 71:1663–7. doi: 10.1271/bbb.70075
9. Wang J, He ZY, Raghavan V. Soybean allergy: characteristics, mechanisms, detection and its reduction through novel food processing techniques. Crit Rev Food Sci. (2022). doi: 10.1080/10408398.2022.2029345
10. Holzhauser T, Johnson P, Hindley JP, O'Connor G, Chan CH, Costa J, et al. Are current analytical methods suitable to verify VITAL(R) 20/30 allergen reference doses for EU allergens in foods? Food Chem Toxicol. (2020) 145:111709. doi: 10.1016/j.fct.2020.111709
11. Wilson JM, Platts-Mills TAE. Red meat allergy in children and adults. Curr Opin Allergy Cl. (2019) 19:229–35. doi: 10.1097/Aci.0000000000000523
12. da Costa PA, Cobuccio L, Mainali D, Rault M, Cavin C. Rapid analysis of food raw materials adulteration using laser direct infrared spectroscopy and imaging. Food Control. (2020) 113:107114. doi: 10.1016/j.foodcont.2020.107114
13. Mansouri M, Khalilzadeh B, Barzegari A, Shoeibi S, Isildak S, Bargahi N, et al. Design a highly specific sequence for electrochemical evaluation of meat adulteration in cooked sausages. Biosens Bioelectron. (2020) 150:111916. doi: 10.1016/j.bios.2019.111916
14. Barbin DF, Badaro AT, Honorato DCB Ida EY, Shimokomaki M. Identification of turkey meat and processed products using near infrared spectroscopy. Food Control. (2020) 107:106816. doi: 10.1016/j.foodcont.2019.106816
15. Leng T, Li F, Xiong LA, Xiong Q, Zhu MT, Chen Y. Quantitative detection of binary and ternary adulteration of minced beef meat with pork and duck meat by NIR combined with chemometrics. Food Control. (2020) 113:107203. doi: 10.1016/j.foodcont.2020.107203
16. Mokhtar NFK, El Sheikha AF, Azmi NI, Mustafa S. Potential authentication of various meat-based products using simple and efficient DNA extraction method. J Sci Food Agr. (2020) 100:1687–93. doi: 10.1002/jsfa.10183
17. Amaral JS, Santos CG, Melo VS, Costa J, Oliveira MBPP, Mafra I. Identification of duck, partridge, pheasant, quail, chicken and turkey meats by species-specific PCR assays to assess the authenticity of traditional game meat Alheira sausages. Food Control. (2015) 47:190–5. doi: 10.1016/j.foodcont.2014.07.009
18. Liu GQ, Luo JX, Xu WL Li CD, Guo YS, Guo L. Improved triplex real-time PCR with endogenous control for synchronous identification of DNA from chicken, duck, and goose meat. Food Sci Nutr. (2021) 9:3130–41. doi: 10.1002/fsn3.2272
19. Thanakiatkrai P, Dechnakarin J, Ngasaman R, Kitpipit T. Direct pentaplex PCR assay: An adjunct panel for meat species identification in Asian food products. Food Chem. (2019) 271:767–72. doi: 10.1016/j.foodchem.2018.07.143
20. Kitpipit T, Sittichan K, Thanakiatkrai P. Direct-multiplex PCR assay for meat species identification in food products. Food Chem. (2014) 163:77–82. doi: 10.1016/j.foodchem.2014.04.062
21. Fajardo V, Gonzalez I, Rojas M, Garcia T, Martin R. A review of current PCR-based methodologies for the authentication of meats from game animal species. Trends Food Sci Tech. (2010) 21:408–21. doi: 10.1016/j.tifs.2010.06.002
22. Kumar A, Kumar RR, Sharma BD, Gokulakrishnan P, Mendiratta SK, Sharma D. Identification of species origin of meat and meat products on the DNA basis: a review. Crit Rev Food Sci. (2015) 55:1340–51. doi: 10.1080/10408398.2012.693978
23. Ali ME, Razzak MA, Abd Hamid SB, Rahman MM, Al Amin M, Abd Rashid NR, et al. Multiplex PCR assay for the detection of five meat species forbidden in Islamic foods. Food Chem. (2015) 177:214–24. doi: 10.1016/j.foodchem.2014.12.098
24. Galal-Khallaf A. Multiplex PCR and 12S rRNA gene sequencing for detection of meat adulteration: a case study in the Egyptian markets. Gene. (2021) 764:145062. doi: 10.1016/j.gene.2020.145062
25. Cai ZD, Zhou S, Liu QQ, Ma H, Yuan XY, Gao JQ, et al. A simple and reliable single tube septuple PCR assay for simultaneous identification of seven meat species. Foods. (2021) 10:1083.
26. Yaman BN, Celik PA, Mutlu MB, Cabuk A. A combinational analysis of acidophilic bacterial diversity of an iron-rich environment. Geomicrobiol J. (2020) 37:877–89. doi: 10.1080/01490451.2020.1795320
27. Prusakova OV, Glukhova XA. Afanas'eva GV, Trizna YA, Nazarova LF, Beletsky IP. A simple and sensitive two-tube multiplex PCR assay for simultaneous detection of ten meat species. Meat Sci. (2018) 137:34–40. doi: 10.1016/j.meatsci.2017.10.017
28. Liu WW, Tao J, Xue M, Ji JG, Zhang YH, Zhang LJ, et al. A multiplex PCR method mediated by universal primers for the identification of eight meat ingredients in food products. Eur Food Res Technol. (2019) 245:2385–92. doi: 10.1007/s00217-019-03350-9
29. Vaithiyanathan S, Vishnuraj MR, Reddy GN, Kulkarni VV. Application of DNA technology to check misrepresentation of animal species in illegally sold meat. Biocatal Agric Biote. (2018) 16:564–8. doi: 10.1016/j.bcab.2018.10.012
30. Martin I, Garcia T, Fajardo V, Rojas M, Pegels N, Hernandez PE, et al. SYBR-Green real-time PCR approach for the detection and quantification of pig DNA in feedstuffs. Meat Sci. (2009) 82:252–9. doi: 10.1016/j.meatsci.2009.01.023
31. Safdar M, Junejo Y, Arman K, Abasiyanik MF. A highly sensitive and specific tetraplex PCR assay for soybean, poultry, horse and pork species identification in sausages: development and validation. Meat Sci. (2014) 98:296–300. doi: 10.1016/j.meatsci.2014.06.006
32. Iqbal M, Saleem MS, Imran M, Khan WA, Ashraf K, Zahoor MY, et al. Single tube multiplex PCR assay for the identification of banned meat species. Food Addit Contam B. (2020) 13:284–91. doi: 10.1080/19393210.2020.1778098
33. Wang WJ, Wang XK, Zhang QD, Liu ZH, Zhou X, Liu B, et al. multiplex PCR method for detection of five animal species in processed meat products using novel species-specific nuclear DNA sequences. Eur Food Res Technol. (2020) 246:1351–60. doi: 10.1007/s00217-020-03494-z
34. Mafra I, Ferreira IMPLVO, Oliveira MBPP. Food authentication by PCR-based methods. Eur Food Res Technol. (2008) 227:649–65. doi: 10.1007/s00217-007-0782-x
35. Liu WW, Wang XN, Tao J, Xi BS, Xue M, Sun WP, et al. multiplex PCR assay mediated by universal primers for the detection of adulterated meat in mutton. J Food Protect. (2019) 82:325–30. doi: 10.4315/0362-028x.Jfp-18-302
36. Mane BG, Mendiratta SK, Tiwari AK. Polymerase chain reaction assay for identification of chicken in meat and meat products. Food Chem. (2009) 116:806–10. doi: 10.1016/j.foodchem.2009.03.030
37. Hou B, Meng XR, Zhang LY, Guo JY Li SW, Jin H. Development of a sensitive and specific multiplex PCR method for the simultaneous detection of chicken, duck and goose DNA in meat products. Meat Sci. (2015) 101:90–4. doi: 10.1016/j.meatsci.2014.11.007
Keywords: heptaplex PCR, adulteration, meat species, mitochondrial sequence, commercial foodstuffs
Citation: Zhou S, Zhong G, Zhou H, Zhang X, Zeng X, Wu Z, Pan D, He J, Cai Z and Liu Q (2022) A Heptaplex PCR Assay for Molecular Traceability of Species Origin With High Efficiency and Practicality in Both Raw and Heat Processing Meat Materials. Front. Nutr. 9:890537. doi: 10.3389/fnut.2022.890537
Received: 06 March 2022; Accepted: 09 May 2022;
Published: 23 June 2022.
Edited by:
Hermann Broll, Bundesinstitut für Risikobewertung (BfR), GermanyReviewed by:
Joana Costa, University of Porto, PortugalRené Köppel, Kantonales Labor Zurich, Switzerland
Copyright © 2022 Zhou, Zhong, Zhou, Zhang, Zeng, Wu, Pan, He, Cai and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daodong Pan, ZGFvZG9uZ3BhbkAxNjMuY29t; Jun He, aGVqdW5AbmJ1LmVkdS5jbg==; Zhendong Cai, emhlbmRvbmdjYWlAaG90bWFpbC5jb20=; Y2FpemhlbmRvbmdAbmJ1LmVkdS5jbg==
†These authors have contributed equally to this work
 Song Zhou
Song Zhou Guowei Zhong
Guowei Zhong Hanxiao Zhou1
Hanxiao Zhou1 Xiaoxia Zhang
Xiaoxia Zhang Xiaoqun Zeng
Xiaoqun Zeng Zhen Wu
Zhen Wu Daodong Pan
Daodong Pan Jun He
Jun He Zhendong Cai
Zhendong Cai