- 1State Key Laboratory of Biobased Material and Green Papermaking, School of Food Science and Engineering, Qilu University of Technology, Shandong Academy of Sciences, Jinan, China
- 2College of Health Sciences, Shandong University of Traditional Chinese Medicine, Jinan, China
- 3Yucheng People's Hospital, Dezhou, China
- 4College of Natural Resources and Environment, South China Agricultural University, Guangzhou, China
As the essential regulator of intestinal bacterial diversity, probiotics are a potential treatment for chronic high-salt diet (HSD)–induced metabolic dysfunction. Probiotic cells entrapped in microgels have been confirmed as being more effective than free cells in protecting bacteria against unfavorable conditions, that is, enhancing their stress resistance. This study explored the physiological mechanism by which probiotic microgels relieve HSD–induced hepatorenal injury. Herein, Lactobacillus rhamnosus was encapsulated in alginate-chitosan microgels which the percentage of alginate/chitosan was applied 1.5:0.5 (w/w) in this system, and the encapsulation significantly improved the probiotic viability in simulated gastrointestinal conditions. Mice were fed an HSD with L. rhamnosus (SDL) or L. rhamnosus microgels (SDEL). After 8 weeks of administration, dietary sodium was confirmed as inducing the hepatic and renal damages in mice, based on indicators, including serum biomarker levels, histopathological features of tissues, and pro-inflammatory cytokine contents in blood levels. However, the serum levels of urea nitrogen, creatinine, uric acid, glutamic-pyruvic transaminase, glutamic-oxalacetic transaminase, and alkaline phosphatase in the SDL and SDEL-fed mice were significantly lowered compared to the HSD-fed mice, especially in the SDEL group. HSD increased the abundances of Anaeroplasma, Enterorhabdus, Parvibacter, and Bacteroides, while the microgels increased the abundances of Lactobacillus, Bifidobacterium, Mucispirillum, and Faecalibaculum. Significant variations of fecal metabolome were validated for SDEL-treated mice, containing those linked to entero-hepatic circulation (e.g., cholic acid), carbohydrate metabolism (i.e., L-lactic acid), and increased antioxidants including citric acid. Furthermore, the probiotic microgels ameliorated intestinal damage by improving barrier and absorption functions. These results augmented existing knowledge on probiotic application for salt toxicity.
Introduction
Probiotics are defined as living microorganisms that, when consumed in sufficient amounts, can exert beneficial effects on the host (1). For instance, probiotics can alleviate aging-related leaky gut (2), ameliorate autistic-like behaviors (3), reduce complications associated with colorectal cancer surgery (4), promote gastrointestinal symptoms in patients with celiac disease (5), and relieve constipation (6). Notably, healthy benefits can be gained from the oral intake of adequate amounts of living cells in quantities sufficient for survival through the upper gastrointestinal tract to the small intestine to colonize at the mucosal membrane of the colon. However, the viability of probiotics can be reduced during food preservation or handing through oxygen enrichment and/or thermal treatments (7). Encapsulation technology is one of the most effective methods available to improve the viability and stability of probiotics in fermented foods and the human gastrointestinal tract. Several materials have been investigated to encapsulate probiotics, such as polysaccharides (predominantly sodium alginate, chitosan, and starch), proteins, and lipids, which are employed to protect probiotics against the freezing and acid associated with the harsh environments present in commercial manufacturing, preservation, and oral delivery (8). Alginate and chitosan are naturally occurring biopolymers that are finding widespread applications in food and pharmaceutical industry. Alginate is used extensively in food industry as a thickener, emulsifier and as a stabilizer. Chitosan is a potentially useful pharmaceutical material owing to its good biocompatibility and low toxicity (9). Alginate and chitosan, by virtue of their physicochemical properties and mild gelation conditions, have gained the acceptance of researchers as matrices for the purpose of probiotics peroral delivery (10).
Chronic kidney disease (CKD) and renal dysfunction impose a tremendous burden on the healthcare system worldwide. It is widely considered that renal injury is caused by a suboptimal diet in humans, including high-salt diets (HSDs), high-fat diets, and/or excess foods (11–13). The human gastrointestinal tract provides habitat for more than 100 trillion commensal bacteria. Metagenomic analyses have revealed that CKD is highly correlated with intestinal dysbacteriosis, which may be linked to metabolites from the gut microbiota (14, 15). Renal excretory capability is an indispensable role of the host-microbial symbiosis. It enables intestinal absorption of the beneficial microbial metabolites, whereas the kidney removes useless and potentially harmful metabolites (16). This method of interorgan communication, closely processed through small molecule messengers, is regarded as remote sensing and signaling (17). Accumulating evidence from patients with CKD displays strong support for the kidney's role in host-microbial symbiosis (18, 19). Some of microbiota-derived metabolites, including indoxyl sulfate, deoxycholic acid (DCA), and trimethylamine oxide, were confirmed to accumulate in the blood parallel to the cause of the renal dysfunction development and proven to be related to clinical outcomes in patients with CKD (20–22). This paradigm has been coined the gut-kidney axis. Although probiotics have displayed beneficial effects on renal injury through modulating the intestinal microbiota modulation (23, 24), to date, the mechanism and impact of probiotics microgels on salt-induced renal impairment have not yet been elucidated.
Previously, research studies have validated that HSD could cause liver injury and affect the intestinal microbiota in mice, particularly reducing the abundance of Lactobacillus genus in the gut (25, 26). Additionally, oral treatment with Lactobacillus improved salt-induced experimental autoimmune encephalomyelitis and hypertension in mice (27). Considering these previous findings, we hypothesized that L. rhamnosus in microgels could effectively ameliorate salt-induced hepatorenal injury. Accordingly, L. rhamnosus was encapsulated in alginate/chitosan microgels, the physicochemical properties were analyzed, and they were used to treat the HSD mice in this study. After 8 weeks of treatment, the composition of the intestinal microbiota, fecal metabolites, serum biochemical indexes, hepatic and renal morphology, and intestinal function of mice was determined. The current research identifies mechanisms for the beneficial outcomes of L. rhamnosus in microgels on hepatorenal injury, and facilitating the development of probiotic supplements for treating CKD.
Materials and Methods
Bacterial Strain and Culture Conditions
Lactobacillus rhamnosus ATCC 7469 used in this study was obtained from the Guangdong Microbial Culture Collection Center (Guangzhou, China). L. rhamnosus stock solution was preserved in de Man, Rogosa and Sharp (MRS) broth supplemented with 25% glycerol (v/v) at −80°C and incubated aerobically in the fresh MRS broth medium for 12 h at 37°C. Cultures were centrifuged at 8,000 rpm for 10 min at 4°C, washed twice with normal saline solution (0.9% NaCl solution) to obtain cells, resuspended as bacterial cell pellets at 1010 colony-forming units (CFU)/mL and set aside for later use. In this study, the total viable bacteria were enumerated in triplicate using the drop-plate method after 2 days grown at 37°C on an MRS agar plate.
Microencapsulation of L. rhamnosus
Bacterium encapsulation was performed using the method presented by Gao et al. (28) with some modifications. Briefly, sodium alginate solution of 1.5% (w/v) was prepared by being dissolved in normal saline solution and then filtered with a sterile membrane filter (0.22 μm pore size; Millipore, Bedford, MA, USA). Then 1 mL of resuspended L. rhamnosus solution, containing 1010 CFU, was centrifuged at 8,000 rpm for 5 min and suspended in sodium alginate solution. Approximately 1 mL suspension was extruded into the 1.1% (w/v) CaCl2 solution by an 0.5 mm-external diameter needle to shape the calcium alginate beads. The beads were hardened for 30 min and then transferred to 0.5% (w/v) chitosan solution at a beads/solution ratio of 1:10 (v/v) for 20 min to form an alginate-chitosan microcapsule membrane. The beads were then washed with normal saline solution to remove excess chitosan. Charges on the membranes were counteracted by adding a 0.15% alginate solution, coating the bead with another membrane for 20 min. Finally, the microcapsules were collected, washed, and transferred into 0.055 M sodium citrate for 5 min to harvest the microcapsules.
Morphology of L. rhamnosus Microgels
Frozen microgels were freeze-dried at −40°C and 1 MPa in a vacuum dryer (LGJ-10, Songyuan Huaxing Technology Develop Co., Ltd., Beijing, China) for 48 h. Lyophilized L. rhamnosus microgels were mounted on aluminum specimen stubs, sputter-coated with a thin layer of gold, and investigated using scanning electron microscopy (SEM; Carl Zeiss EVO MA15-Smart SEM, Zeiss, Germany) under different magnifications.
In vitro Simulated Digestion of L. rhamnosus Microgels
Simulated intestinal fluid (SIF) and simulated gastric fluid (SGF), which simulate digestive fluids in the gastrointestinal tract, were established as directed by the U.S.P. (https://www.usp.org). Briefly, 1 L SGF (pH 1.2) was prepared to contain 2 g NaCl, 3.2 g pepsin, 7 mL HCl, and deionized H2O. The stock intestinal solution was prepared including 6.8 g KH2PO4, 77 mL NaOH (0.2 M), and 0.5 L deionized H2O. Subsequently, 10 g pancreatin were mixed into the stock intestinal solution, with an adjusted pH of 6.8, and diluted with deionized H2O to 1 L to obtain SIF. Each 1 mL L. rhamnosus solution or the L. rhamnosus microgels (1010 CFU) was mixed separately with 9 mL of SGF, SIF, or 2% bile-salt solution, and digested in an incubator shaker at 37°C. The probiotic viability was determined at 0–60 min, using 10 min intervals. The probiotic amount of the microcapsules is quantified in log 1010 CFU/mL by sonicating 100 μL of the wet capsules in 1 mL of peptone water for 10 s using a Vibra-cell probe sonicator (VC 505, Sonics, USA). Then the viable bacteria were assessed using the drop-plate method after 2 days grown at 37°C on an MRS agar plate (29).
Viability of Probiotics During Long-Time Storage
One-milliliter sample of free or encapsulated cells was mixed with 9 mL of sodium citrate after storing at 4°C for 8 weeks. After vortex mixing, samples were placed on a shaker at 37°C until microgels totally disintegrated. Viable bacteria were calculated by the plate count method on MRS agar in triplicate.
Animal Intervention
Thirty-two male C57BL/6J mice (8-week-old; specific pathogen-free) were purchased from Pengyue Laboratory Animal Technology Co., Ltd. (Jinan, China) with production license number SCXK (Lu) 2019-0003. All mice were kept singly in clean conditions with a 12 h light/dark cycle and ample ventilation, at an ambient temperature of 24°C, and relative humidity of 50–70%. This study, approved by the Animals Ethics Committee of the Experimental Animal Center of Shandong University of Traditional Chinese Medicine (No. SYXKLU20170022, Jinan, China), complied with the EU Directive 2010/63/EU for the care and use of laboratory animals.
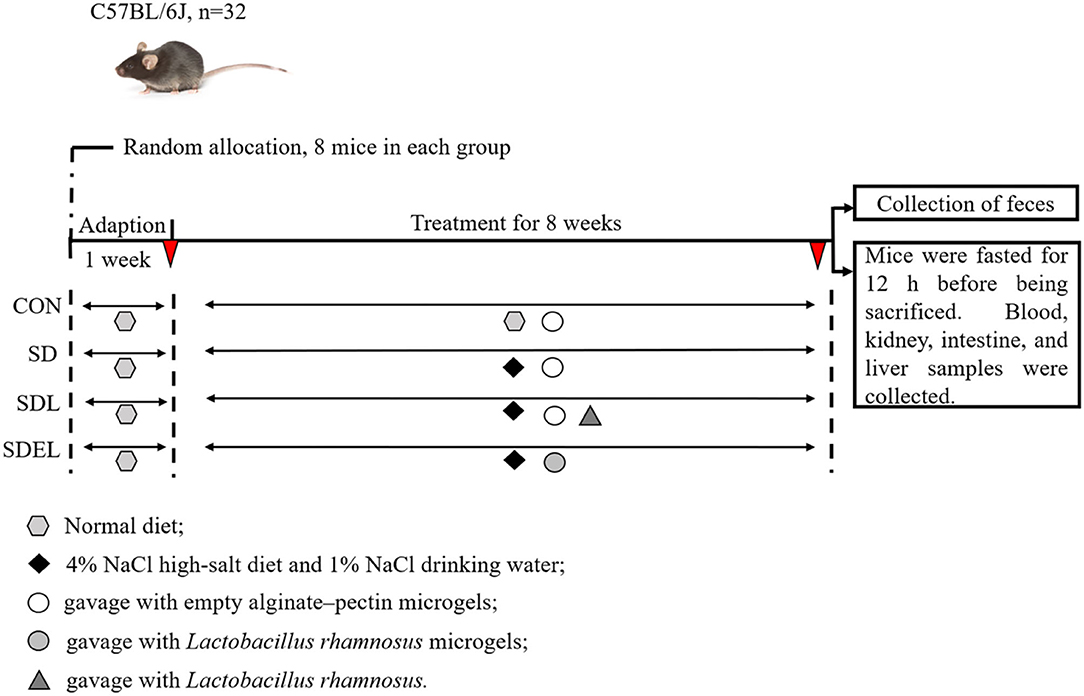
Mice were given a basic rodent-chow pellet diet and water ad libitum during the first week to enable acclimatization to the environment, followed by an 8-week administered diet (Figure 1). The animals were stochastically divided into four groups, each containing eight mice: CON group fed with a standard diet; SD group fed with 4% NaCl diet; SDL group fed with 4% NaCl diet and orally treated with 200 μL fresh L. rhamnosus suspensions (2 × 109 CFU); and SDEL group fed with 4% NaCl diet and orally treated with 200 μL fresh L. rhamnosus microgels (2 × 109 CFU) suspensions. Meanwhile, the mice in the CON, SD, and SDL groups were orally treated with the same volumes (200 μL) of the empty alginate-chitosan microgels suspensions. Furthermore, potable water for the SD, SDL, and SDEL mice was supplemented with 1% NaCl. All diets were sterilized by the 25 kGy γ-irradiation and prepared following the American Institute of Nutrition (AIN)-93G purified diet standard to satisfy the nutritional requirements for laboratory rodents. Fodder and potable water for the experimental animals were changed daily, and extra fodder was stored at −20°C. The animals were weighed weekly over the 8 weeks of diet intervention experiments.
Sample Collection
The sampling methods were carried out essentially as described previously (30). At the end of the experimental period, the mice were not fed in the evening, and the next day, fresh feces and serum were collected, and the mice were sacrificed using cervical dislocation. After rinsing three times with sterile normal saline, renal, hepatic, colonic and small intestinal tissues were fixed using fresh 4% paraformaldehyde solution. After sectioning, these tissues were assessed with hematoxylin and eosin (H&E) staining to observe the pathological changes under a microscope. In addition, the kidney and liver tissues were investigated by Masson staining to evaluate the histology of organ fibrosis, and the pathological scores of kidney and liver tissues were calculated per the method of Fang et al. (31).
Gut Microbiota Profiling
The purity of DNA was isolated from fecal samples by following the QIAamp DNA stool mini kit protocol (Qiagen, GmbH, Hilden, Germany). Processing of separated DNAs, Illumina MiSeq amplicon sequencing, and procedures for library generation were performed according to the general method described previously (30). The V4–V5 regions of the bacterial 16S rRNA gene sequences were amplified using the system of polymerase chain reaction (PCR) containing the specific primers (515F: 5′-GTGCCAGCMGCCGCGG-3′ and 907R: 5′-CCGTCAATTCMTTTGAGTTT-3′) and a HiFi Hot Start Ready Mix (KAPA Biosystems, Woburn, MA, USA). Amplicons from the second PCR round were purified, pooled, and paired-end (PE) sequenced.
The raw Illumina paired-end reads in FASTQ format were quality trimmed using Trimmomatic software (32) to detect and cut off ambiguous bases and then matched using FLASH software (33) following the parameters as previously reported (25). The Quantitative Insights into Microbial Ecology (QIIME, version 1.8.0) pipeline was employed to process sequence data with 75% of the bases showing quality scores above 20 (base-calling accuracy of 99%) (34). All effective tags were clustered into operational taxonomic units (OTUs) at a 97% stringency threshold using the workflow provided by the QIIME wrappers (35). All representative reads were identified and classified into taxonomic levels using a nucleotide Basic Local Alignment Search Tool comparison against the Greengenes (16S rRNA) database and the classifier tool of the Ribosomal Database Project II Classifier with a confidence score threshold of 70% (36).
Fecal Metabolomic Profiling
In brief, fecal pellets were spiked with 20 μL of an L-2-chlorophenylalanine solution (1 mg/mL in distilled water) as an internal standard and the metabolites were extracted with chloroform and methanol. After homogenization and bath sonication, the supernatant liquid was transferred into a fresh glass vial. The microbial-host co-metabolites were analyzed using an Agilent 7890A GC system (Agilent Technologies, Santa Clara, CA, USA) coupled to a Pegasus HT time-of-flight mass spectrometry (Leco, Saint Joseph, MI, USA) following the parameters as previously reported (26).
Analysis of Serum Biochemical Indexes and Intestinal Function
Biochemical indicators in serum, including blood urea nitrogen (BUN), creatinine (CRE), uric acid (UA), glucose (GLC), glutamic-pyruvic transaminase (ALT), glutamic-oxalacetic transaminase (AST), alkaline phosphatase (ALP), and triglycerides (TG), were evaluated on an automatic biochemical analyzer (Au680, Beckman Coulter, Inc., Brea, CA, USA). The serum interleukin (IL)-17a and IL-22 levels were measured using enzyme-linked immunosorbent assay (ELISA) kits (Gersion Bio-Technology Co., Ltd., Beijing, China) according to the manufacturer's protocol.
To assess the intestinal barrier function, the serum levels of D-lactate (D-LA) and diamine oxidase (DAO) were analyzed using the respective ELISA kits (Gersion Bio-Technology Co., Ltd.) according to the manufacturer's protocol. The morphological development of the small intestine, including crypt depth and villus height, were measured using Image Pro-Plus version 6.0 software (Media Cybernetics, Inc., Rockville, MD, USA) to analyze the absorptive function. The pH value of the colonic contents was determined as previously described (26).
Data Analysis
All results are represented as the mean ± standard deviation (SD) of replicate tests or as the median with interquartile ranges. As appropriate, the t-tests were performed to analyze the independent measurements using SPSS version 22.0 software (SPSS Inc., Chicago, IL, USA). One-way analysis of variance was used to analyze the differences between groups; multiple groups were compared with the Turkey post-hoc test. Significance was established at P < 0.05. All data analyses were run in R-statistical software (version 2.13.0; Free Software Foundation, Boston, MA, USA). Multivariate analyses [principal co-ordinates analysis (PCoA)] was performed using the Simca-P 14.1 software package (Umetrics, Umea, Sweden).
Results
Characterization of L. rhamnosus Microgels in vitro
In this section, SEM was employed to inspect the morphology of the L. rhamnosus microgels (Figure 2A). The SEM results show that the freeze-dried microgels were regularly olive-shaped, and the surfaces of the microgels appeared shrunken and wrinkled. Small cracks and high porosity could be found at the rough surface of microgels under higher magnification, consistent with a previous finding (37). The viability of bacterial cells is decreased by acidic and alkaline digestive fluids, enzymes, and bile salts in the host's gastrointestinal tract. We evaluated the effect of encapsulation of cell viability under simulated gastrointestinal environments to mimic these harsh conditions and calculate the survival rates of free or encapsulated probiotic cells. The results indicated that in both free and encapsulated cells, the viability of bacterial cells was reduced in simulated in vivo conditions. The viability of free probiotic cells was lost after 30 min exposure in both SGF and SIF (Figures 2B,C). The effect of encapsulation on L. rhamnosus viability in long-term storage was evaluated by storing freeze-dried samples at 4°C for 8 weeks to simulate commercial storage. The viability of free or encapsulated probiotic cells did not significantly differ at the first and second week in storage (P > 0.05). Subsequently, the viability of free probiotic cells was significantly decreased than that of encapsulated probiotic cells at the third week to 8 week (P < 0.001; Figure 2E).
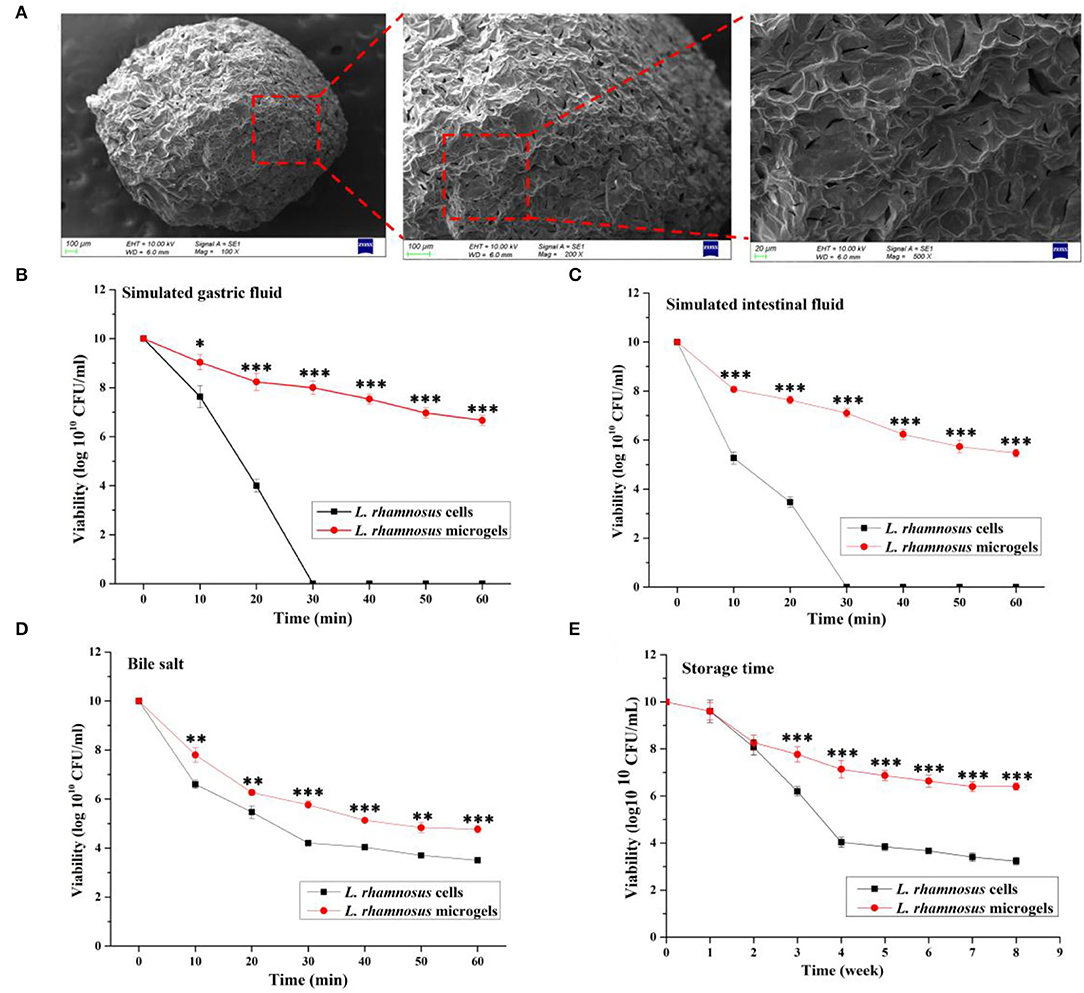
Figure 2. Morphology and characteristics of L. rhamnosus microgels. (A) The freeze-dried L. rhamnosus microgels were characterized using scanning electron microscope. The viability of L. rhamnosus microgels in a simulated gastrointestinal environment was separately assessed in (B) simulated gastric fluid, (C) simulated intestinal fluid, and (D) bile salt. (E) Anti-inactivation of L. rhamnosus microgels were analyzed in 8 weeks storage. Values are presented as the mean ± SD; n = 3. *P < 0.05, **P < 0.01, ***P < 0.001 vs. L. rhamnosus microgels.
Interestingly, the viability of the probiotics encapsulated in alginate/chitosan microgels was greatly improved, with higher resistance to gastric fluids. The viability of L. rhamnosus in microgels was 6.66 ± 0.21 and 5.47 ± 0.15 log1010 CFU/mL at 60 min exposure in SGF and SIF, respectively (Figures 2B,C). The viability of L. rhamnosus in microgels (4.77 ± 0.15 log1010 CFU/mL) exposed to 2% bile salt for 60 min was significantly increased over that of the free probiotic cells (3.50 ± 0.10 log1010 CFU/mL) (P < 0.05, Figure 2D). Therefore, the L. rhamnosus cells were successfully encapsulated in alginate/chitosan microgels, and the microgels improved the viability and resistance of bacterial cells in simulated gastrointestinal environments.
Analysis of Hepatorenal Injury
After 8 weeks of supplementation, biomarkers of renal function (serum BUN, CRE, and UA contents) and liver injury (serum AST, ALT, and ALP activities) were measured. The results showed that the serum levels of BUN (Figure 3E), CRE (Figure 3F) and UA (Figure 3G) in the SD group were significantly increased compared with those in the CON group (P < 0.05), confirming that dietary salt-induced the renal dysfunction occurred in the SD-fed mice. Likewise, the serum ALT (Figure 3H), AST (Figure 3I), and ALP (Figure 3J) activities in the SD group were significantly increased compared with those in the CON group (P < 0.05), confirming that impaired liver function existed in the SD-fed mice. Additionally, the serum concentration of glycolipid (including GLC and TG) and inflammatory factors (e.g., IL-17a and IL-22) in the SD group were considerably increased compared with those of the CON group (P < 0.05, Figures 3K–N). The treatment of the SD-fed mice with L. rhamnosus or its microgels significantly reduced all of the serum biochemical parameters levels. Notably, the BUN, CRE, AST, GLC, TG, IL-17a, and IL-22 levels in the SDEL were significantly decreased compared with those in the SDL group (P < 0.05). Finally, there were no statistically significant differences in the mice's body weights among the four groups throughout the experimental period (P > 0.05; Supplementary Figure 1).
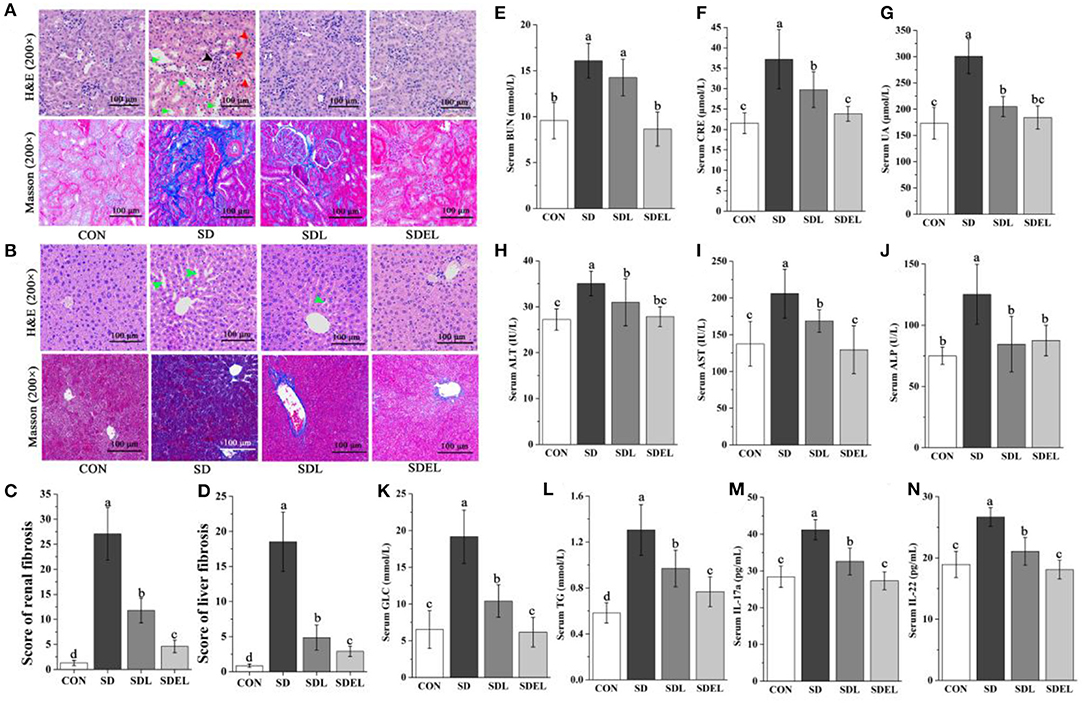
Figure 3. L. rhamnosus and L. rhamnosus microgels administration attenuated HSD-induced hepatorenal injury. (A) Representative kidney sections with H&E and Masson staining (200× magnification). The black arrow indicates vacuolations and swelling of glomerular cells, red arrows indicate interstitial angiectasis hyperemia, and green arrows indicate edema in renal interstitial spaces. (B) Representative liver sections with H&E and Masson staining (200× magnification). Green arrows indicate the loss of cellular boundaries around the central vein of hepatocytes. Compare (C) the renal fibrosis and (D) the liver fibrosis scores in various groups. Serum levels of (E) BUN, (F) CRE, (G) UA, (H) ALT, (I) AST, (J) ALP, (K) GLC, (L) TG, (M) IL-17a, and (N) IL-22 were determined in different groups. Data are expressed as mean ± SD; n = 8. Different letters were significantly different (P < 0.05).
We used H&E staining to examine the histological changes of pathological tissue morphology and glomerular volume, as well as Masson staining to show the degree of fibrosis (Figures 3A,C). The severity of the tissue lesions varied among the four groups. The CON group showed structural integrality in renal tissue cells, normal appearance of the hepatic lobular structure and central vein, and no changes in glomerular morphology. In the SD group, the renal tissues appeared to have undergone some definitive pathological changes, including vacuolations and swelling of glomerular cells (black arrow), interstitial angiectasis hyperemia (red arrows), edema in renal interstitial spaces (green arrows), and severe renal tubule interstitial fibrosis. Treatment of SDL and SDEL mice could significantly curtail these adverse changes in the SD group. In particular, following SDEL intervention, swelling of the glomerular cells was alleviated. The nuclei were typical, and interstitial angiectasis hyperemia decreased. In cross-sections, the appearances of the kidney, glomerulus, and renal tubule structure were regular, and renal tubule interstitial fibrosis levels were restored significantly.
The H&E and Masson staining methods were also employed to observe the micromorphology of the liver tissues (Figures 3B,D). In the CON group, the liver showed a typical structure, with the nucleus located in the middle of the hepatic cells, no apoptosis, and hepatocytes arranged as cords and distributed uniformly. In the SD group, the hepatic cords were dissociated loosely and randomly, and there was neutrophilic granulocyte infiltration in hepatocytes, loss of cellular boundaries around the central vein of hepatocytes (green arrows), and prominent fibrosis in the liver tissue. As expected, the hepatic injury was effectively ameliorated to varying extents after 8 weeks of treatment with L. rhamnosus or L. rhamnosus microgels. In comparison with SDL-fed mice, the liver cells of the SDEL-fed mice were more evenly arranged with clear nuclei, no swollen cytoplasmic vesicular, and no fibrosis in liver tissues. Briefly, treatment with L. rhamnosus microgels significantly mitigated the hepatorenal injury of SD-fed mice.
Analysis of Fecal Microbiota
After removing unqualified sequences, the CON, SD, SDL, and SDEL groups yielded more than 51,309, 69,761, 71,065, and 71,732 effective tags, respectively. Species were annotated with representative 16S rRNA gene sequence fragments. The effective sequences of all samples qualified OTUs were clustered based on ≥97% sequence identity. The box-and-whisker plot (Figure 4A) shows the relative abundances of seven major phyla analyzed to be significantly different (P < 0.05) among the CON, SD, SDL, and SDEL groups, as follows: Bacteroidetes, Actinobacteria, Epsilonbacteraeotat, Deferribacteres, Acidobacteria, Verrucomicrobia, and Patescibacteria. Linear discriminant analysis effect size (LEfSe) identified the differentially abundant bacterial genera that were differentially abundant in the four groups. At the family level, Eggerthellaceae, Bacteroidaceae, Rikenellaceae, Clostridiaceae_1, and Anaeroplasmataceae, as the biomarkers, were more abundant in the SD group. In contrast, Bifidobacteriaceae, Muribaculaceae, Deferribacteraceae, Lactobacillaceae, Alcaligenaceae, and Helicobacteraceae were more abundant in the SDEL group (Figure 4B). At the genus level, Anaeroplasma, Enterorhabdus, Lactococcus, Parvibacter, Tyzzerella, Lachnoclostridium, and Bacteroides, as the biomarker, were more abundant in the SD group, while Lactobacillus, Bifidobacterium, Parasutterella, Helicobacter, Mucispirillum, Faecalibaculum, Muribaculum, and Acetatifactor were more abundant in the SDEL group (Figure 4C). The α-diversity—which consists of richness estimates (i.e., Chao 1 index) and diversity values (including Shannon and Simpson indices)—of the microbial community were measured. The richness estimates of the microbial community in the SD, SDL, and SDEL groups was significantly higher than that of the CON group, while the diversity values of the microbial community in CON and SD groups was lower than that of the SDEL group (Figure 4D). PCoA indicated the bacterial diversity among the four groups; 27.79% of the total variance was attributable to the three principal components (PC1, PC2, and PC3), which were stable and reliable. The chord diagram reveals the top 25 abundant genera in the four groups (Figure 4F), showing that Ambiguous, Alistipes, and Bacteroides were the three predominant taxa in all the groups, accounting for 2.51–24.25% of the total OTUs. The differential abundance among the four groups showed that the L. rhamnosus microgels caused variation, for instance, in the levels of hepatorenal injury and the structure of the mouse gut microbiome. The discrepancy of functional profiles between different groups predicted by PICRUST (38). The results showed the overall differences in KEEG abundances among the microbiomes and revealed that biosynthesis of other secondary metabolites, metabolic diseases, immune system diseases, glycan biosynthesis and metabolism, transport and catabolism, excretory system, metabolism of other amino acids, metabolism of cofactors and vitamins, carbohydrate metabolism, and lipid metabolism in the SDEL group was significantly lower than that in the SD group (Figure 4G; P < 0.005).
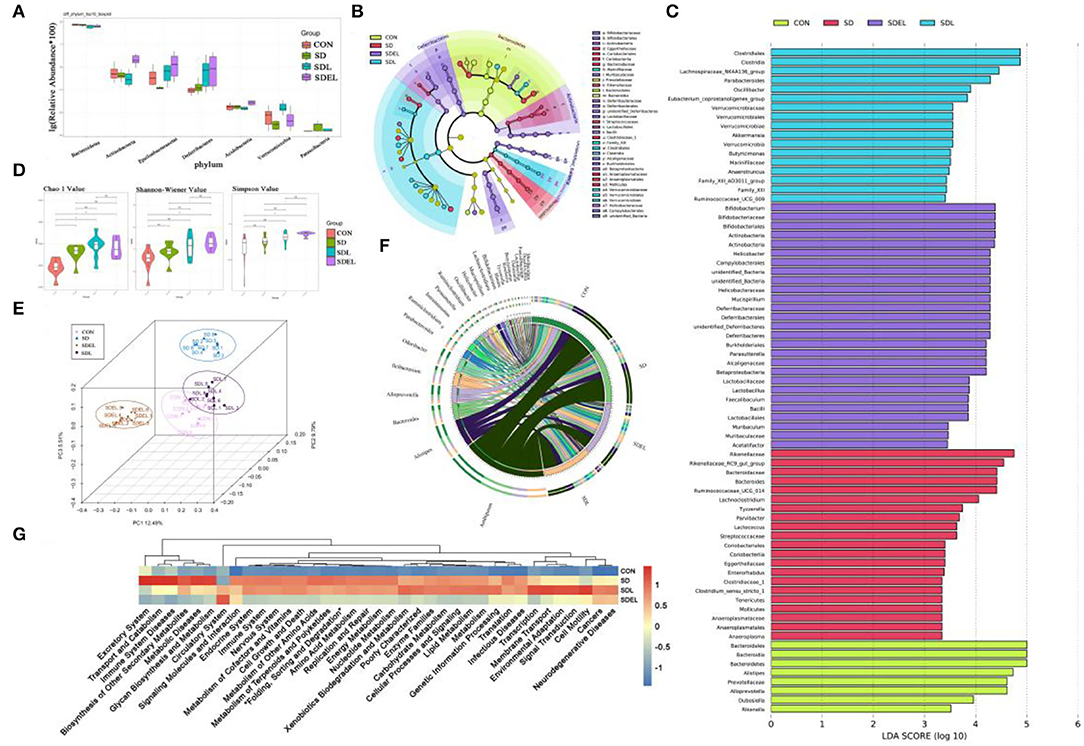
Figure 4. (A) Relative abundance of seven phyla in the CON, SD, SDL, and SDEL groups: Bacteroidetes, Actinobacteria, Epsilonbacteraeotat, Deferribacteres, Acidobacteria, Verrucomicrobia, and Patescibacteria. (B) Cladogram of linear discriminant analysis effect size (LEfSe) analysis identified the most differentially abundant taxa of gut microbiota in the different groups. (C) Histogram of LDA scores by comparing inter-group variance at the relative abundance level by LEfSe analysis. (D) Microbial richness estimates (Chao index) and diversity indices (Shannon–Wiener and Simpson) in the different group at the 8-week point. (E) PCoA shows separation of the fecal bacterial composition of the different groups. (F) The chord diagram reveals the four groups' top 25 abundant genera (abundance >0.1%). (G) Microbial community functions predicted by PICRUSt of the different groups.
Metabolic Functionality
Fecal metabolic profiling is a functional readout of gut microbiota that enables comprehension of the possible mechanisms underlying how the microbiota influences host health (39). A total of 592 peaks were identified, and 468 metabolites persisted after eliminating background noise using the interquartile range denoising method. Missing raw data values were generated using half of the minimum value. PCoA (Figure 5A) illustrates the data distribution and the separation in the metabolic compositions of the different groups. Specifically, 69.20% of the total variance resulted from PC1 PC2, and PC3, which were stable and reliable. The variables by which the content differed between the SD and SDEL groups are shown in the form of a volcano plot (Figure 5B), wherein each dot represents a metabolite: red dots represent upregulated metabolites, blue dots represent downregulated metabolites, and green dots represent metabolites with non-significant differences (P > 0.05). However, the volcano graph was complex since the various variables were included. Consequently, 70 differential metabolites between the SD and SDEL groups were identified based on the variable importance in the projection (VIP) value (>1.0) of the orthogonal partial least squares discriminant analysis and the Student's t-test P-value (< 0.05). The heatmap visualization (Figure 5C) was used to show differential metabolites, thus refining the analysis. A total of 58 metabolites, such as cholic acid, glucose, uridine, kynurenic acid, urocanic acid, hippuric acid, and L-tyrosine, exhibited significantly decreased abundance in the feces of SDEL-treated mice, whereas the abundance of 12 metabolites, such as L-lactic acid, 3-hydroxybutyric acid, saccharic acid, aspartate, citric acid, and β-tocopherol was significantly increased compared to levels in the feces of SD-fed mice. In particular, the abundance of L-lactic acid in the SDEL group increased the most, more 3.65 times than that of the SD group (Supplementary Table 1).
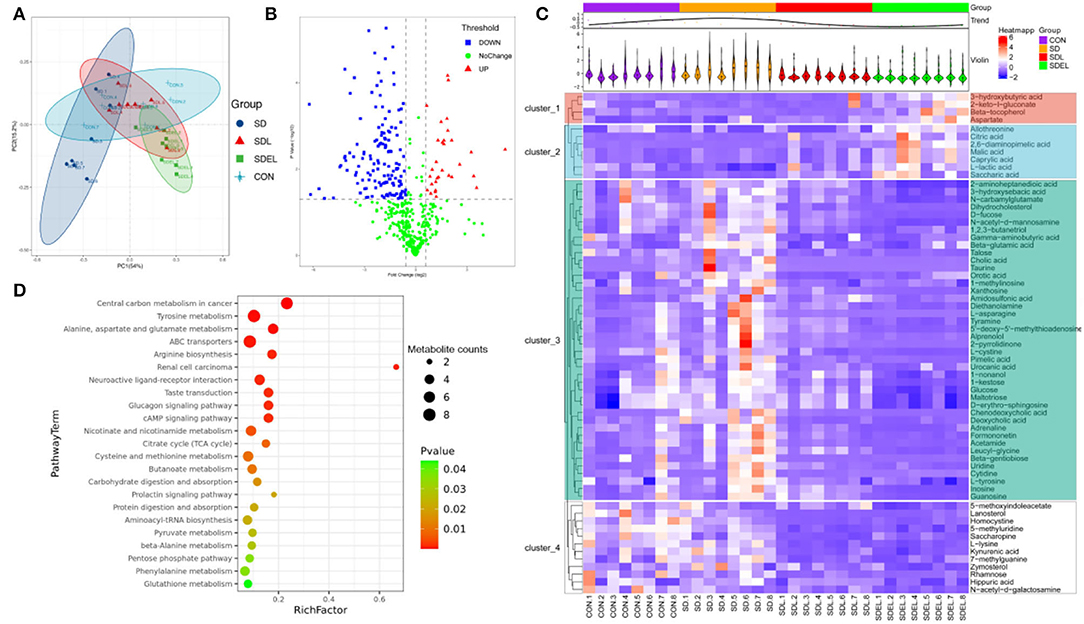
Figure 5. Fecal metabolic profiling of the CON, SD, SDL, and SDEL groups. (A) Scatterplot of PCoA scores in different groups. (B) Volcano plot showing the differential variables between the SD and SDEL groups. Each metabolite is represented by a dot, where the red dots indicate up-regulation, the blue dots indicate down-regulation, and the green dots indicate no statistical difference. (C) Heatmap of 70 metabolites at normalized levels differentially abundant (P < 0.05) among the three groups. The dendrogram shows the distances of the metabolites based on their relative abundances. The normalized abundance values are depicted visually from red to blue, representing the highest and lowest abundances. (D) Enriched KEGG pathways in the SD group compared with the SDEL group. The statistical significance values (P < 0.05) are depicted visually from red to green, representing the most and least differences, respectively. The dot sizes on the vertical axis represent the metabolite counts in the metabolic pathways.
The significantly different metabolites occurring in the SD and SDEL groups were assigned based on metabolic pathways present in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. This analysis suggested that the differentially abundant metabolites in the SDEL group were all associated with central carbon metabolism in cancer, ABC transporters, arginine biosynthesis, tyrosine, alanine, aspartate and glutamate metabolism as well as renal cell carcinoma pathways (P < 0.01, frequency-distance relationship correction; Figure 4D).
Integration of Among Metabolites, Bacteria, and Clinical Chemistry Data
Pearson's correlation analysis was employed to determine the correlations among gut microbiota differentially metabolites, and serum biochemical parameters. The resulting metabolic association heatmap (Figure 6) displays positive and negative correlations among the identified genera, the metabolite levels, and the clinical chemistry data. The dominant genera in SD group, including Anaeroplasma, Enterorhabdus, Lactococcus, Parvibacter, Tyzzerella, Lachnoclostridium, and Bacteroides, displayed strong positive correlations with D-erythro-sphingosine, glucose, kynurenic acid, maltotriose, N-acetyl-d-mannosamine, L-lysine, 5-methyluridine, 2-aminoheptanedioic acid, N-carbamylglutamate, deoxycholic acid, 3-hydroxysebacic acid, and all of the serum biochemical parameters. The dominant genera in the SDEL group, including Lactobacillus, Bifidobacterium, Parasutterella, Helicobacter, Mucispirillum, Faecalibaculum, Muribaculum, and Acetatifactor, displayed strong positive correlations with L-lactic acid, 3-hydroxybutyric acid, and allothreonine. Overall, L. rhamnosus microgels impacted the bacterial community and substantially altered the fecal metabolic profiling, affecting the host health.
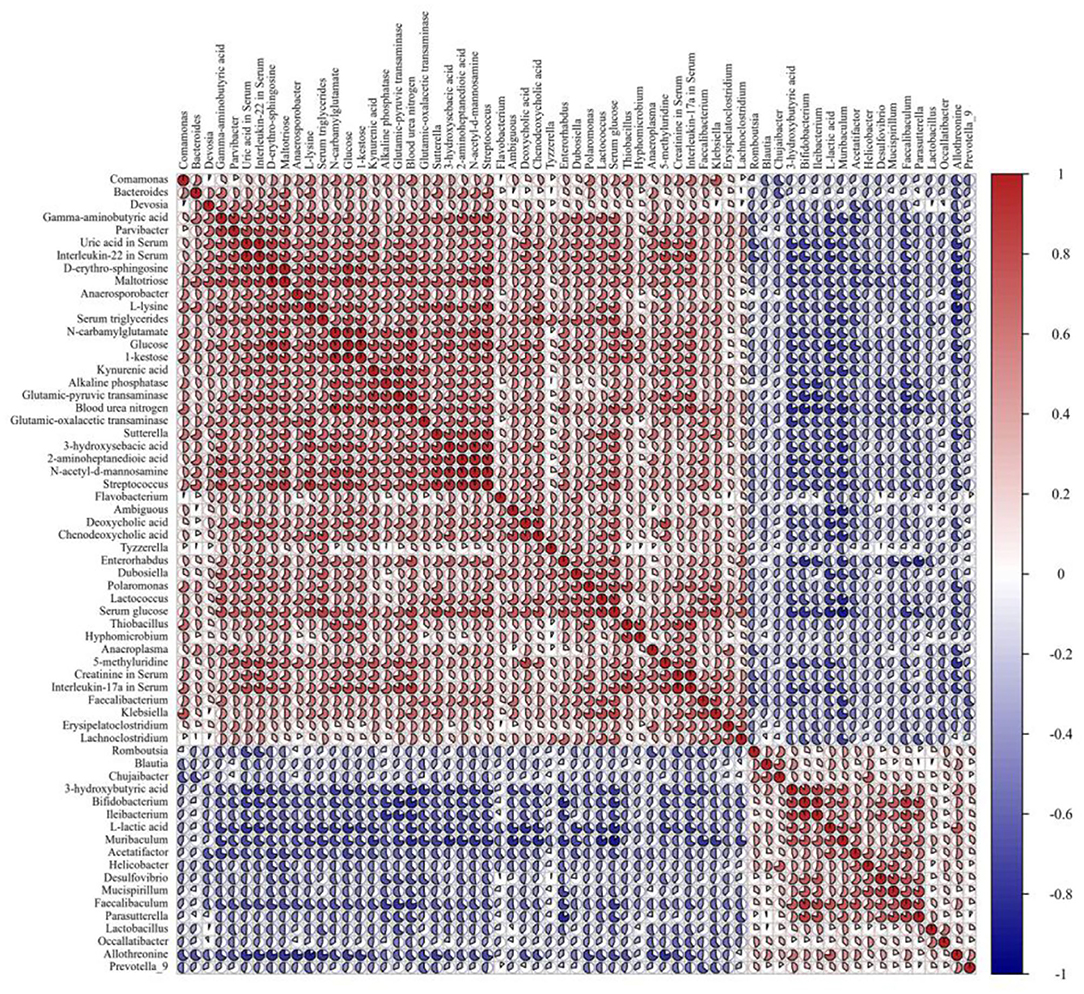
Figure 6. The Pearson correlation analysis among the 36 gut bacteria (P < 0.05), 17 fecal metabolites (P < 0.001), and 10 clinical chemistry data (P < 0.05) in the SD group compared with the SDEL group. R-values, the sector area, are depicted from blue to red, where red represents positive correlations and blue negative correlations.
Intestinal Function and Morphology
The colonic barrier function and the small intestinal absorption functions were investigated in the CON-, SD-, SDL-, and SDEL-fed mice. Serum DAO activity and D-LA concentration were assessed to evaluate the effects of SDEL treatment on the intestinal permeability of SD-fed mice. Serum levels of DAO and D-LA were significantly higher in the SD group compared to the CON group (P < 0.05), indicating impaired colonic barrier function in the SD-fed mice (Figures 7A,B). In contrast, the levels of DAO and D-LA were significantly decreased in the SDL and SDEL groups compared to the SD group (P < 0.05), indicating reduced colonic barrier function following treatment of L. rhamnosus and its microgels. However, the DAO and D-LA levels in the SDEL group were significantly lower than the SDL group (P < 0.05), suggesting that L. rhamnosus microgels significantly reduced intestinal permeability in SDEL-fed mice. The pH of the colonic contents of the SD group (8.56 ± 0.24) was significantly different to that of the CON (7.54 ± 0.28), SDL (7.81 ± 0.28), and SDEL (7.31 ± 0.32) groups (P < 0.05). There was no statistically significant difference in pH value between the CON and SDEL groups (P > 0.05; Figure 7C). Furthermore, the villus height (V), crypt depth (C), and the V/C ratio in the small intestine were analyzed to evaluate the function of small intestinal absorption. The V/C ratio in the SDEL group was significantly higher than that in the CON, SD, and SDL groups (P < 0.05; Figure 7D), indicating that the L. rhamnosus microgels significantly restored the normal function of small intestinal absorption.
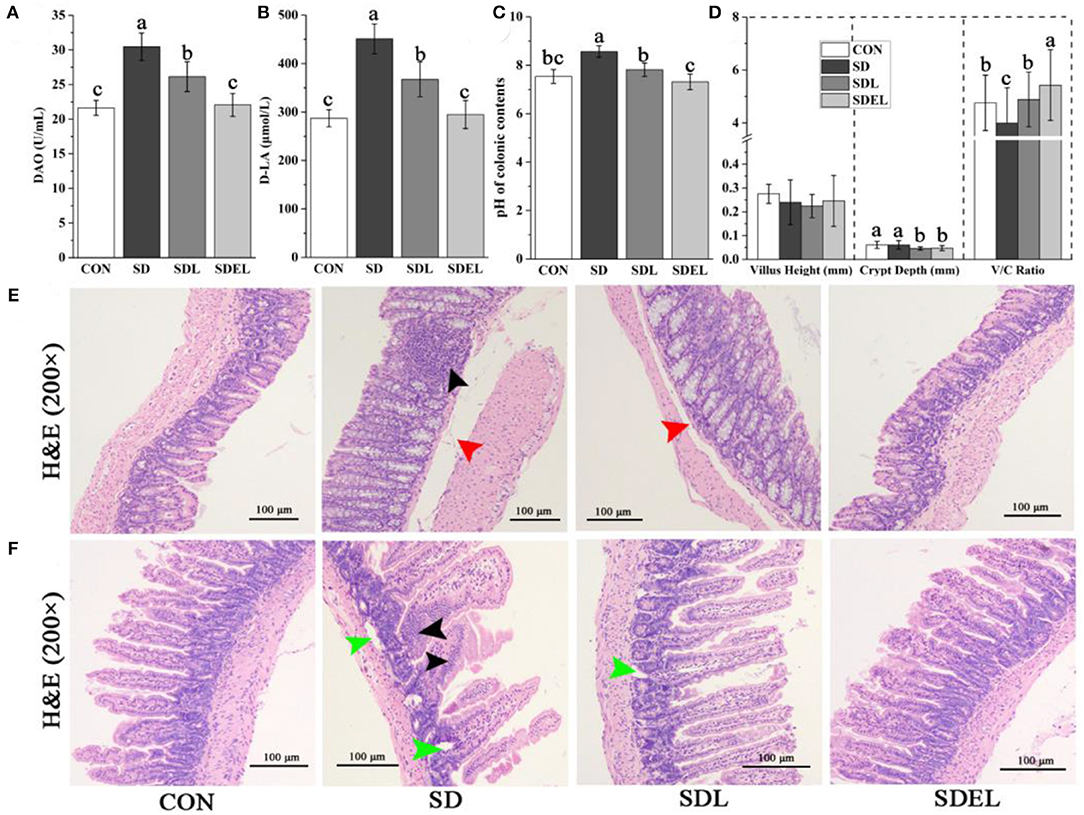
Figure 7. The colonic and small intestinal functions of the CON-, SD, SDL, and SDEL-fed mice. (A) Serum DAO activities and (B) D-LA concentrations indicate mice's colonic barrier function. (C) The pH of the colonic contents of the mice in different groups. (D) The villus height (V, mm), crypt depth (C, mm), and V/C ratio, which indicate the absorption function of the small intestine of the mice in different groups. (E) Colonic photomicrographs (100× magnification) of the mice in different groups. The black arrow indicates inflammatory cell infiltration; red arrows indicate edema in the submucosal layer. (F) Photomicrographs (100× magnification) of small intestinal villi of the mice in different groups. Black arrows and green arrows indicate infiltration of inflammatory cells and necrotized local muscularis. Data are expressed as mean ± SD; n = 8. Different letters were significantly different (P < 0.05).
In both the CON- and SDEL-fed mice, colonic sections showed typical structures and boundaries of the mucosal, submucosal, muscularis, and serous layers. The mucosal glands were abundant, goblet cell structures were observed, and the bowel wall thickness was normal (Figure 7E). Conversely, in the colonic tissues of SD-fed mice, edema was found in the submucosal layer with an expanded gap (red arrow) and inflammatory cell infiltration (black arrow) was present between the mucosal glands (Figure 7E). The submucosa was slightly edematous (red arrow) in the colonic tissues of SDL-treated mice (Figure 7E). Furthermore, in both the CON and SDEL groups, the small intestine's submucosal, mucous, muscularis, and serous layers showed typical structures and boundaries (Figure 7F). However, infiltration of inflammatory cells (black arrows) and necrotized local muscularis (green arrows) into the small intestine was observed in the SD group. In the SDL-treated mice, the local muscularis had necrotized (green arrow; Figure 7F).
Discussion
Excessive sodium intake has been identified as a risk factor for CKDs, the leading proponent of premature death worldwide (40). Recently studies have reported the association of salt-induced non-communicable diseases, such as CKDs (14), hepatic steatosis (25), and hypertension (26), with gut microbial dysbiosis. As the microbial habitat, the colon is the principal organ responsible for sodium homeostasis, and colonic epithelial cells help protect mammals against salt overload (41). The beneficial effects of probiotics have been confirmed in combating renal dysfunction by regulating the microbial community (24). Previous studies (42, 43) have confirmed that gavage with only L. rhamnosus can reversed the hepatic inflammation and slight renal injury associated with hyperuricaemia by regulating gut microbiota. However, the acidity of the gastrointestinal tract is a harsh environment that limits the mucoadhesion and colonization of alien probiotics. To date, microcapsules are still an ideal candidate to protect probiotics from the environmental stresses in food and medical industries (44). Zhuge et al. (45) delivered that L. salivarius in microgels could boost viability during transit through the gastrointestinal organs and attenuate liver injury by reducing inflammation and intestinal permeability. Zhao et al. (46) confirmed that probiotic (specifically, Lactobacillus and Bacillus subtilis) microcapsules could reduce inflammation, improve fat metabolism, and restore intestinal barrier functions, thus contributing to the treatment of metabolic syndrome in vivo. Nevertheless, the mechanism and effects of probiotics microgels on salt-induced hepatorenal failure have not been investigated. As Lactobacillus abundance is reduced in the gut by increased dietary sodium intake (26, 27), a correlation between Lactobacillus abundance and clinical outcomes of people consuming high salt diets is particularly interesting. The aims of the current study included encapsulating L. rhamnosus species in alginate and chitosan to prepare the microencapsulation for probiotic protection and evaluate the effects of L. rhamnosus microgels on salt-induced hepatorenal injury.
Both sodium alginate and chitosan are polyelectrolytes. Alginate extracted from brown seaweed (Phaeophyceae) is widely used for drug delivery due to its reliable water-holding capacity and heat stability. As linear copolymers composed of alternating sequences of β-D-mannuronic and α-L-guluronic acid residues (47), alginate will combine with multivalent cations (for example, Ca2+) to form gels or gentle matrices that can trap sensitive substances such as living probiotic bacteria (48, 49). Interestingly, an essential property of alginate is that they form insoluble gels formed at low pH conditions, but become soluble at neutral or higher pHs due to the ionically cross-linked carboxylate groups in the uronate blocks. This feature affords advantages in using of alginate as a support for probiotic delivery, avoiding the solubilization of gels in the gastrointestinal tract and providing adequate protection of bacteria within the acidic environment. Furthermore, the solubility of alginate at the gastrointestinal pH facilitates the release of viable cells into the gut (47). Alginate gel's mechanical and physicochemical properties can be further improved by forming an electrostatic complex membrane with a polycation, such as chitosan (50). Previous studies successfully encapsulated L. plantarum, Escherichia coli, and B. longum (28, 51, 52) into microgels containing alginate and chitosan. In the current study, similar strategies were used to prepare L. rhamnosus microgels using alginate and chitosan as the wall materials. The resulting effective gastric acid and bile acid resistances in vitro (Figure 2) shows uniformity in the reported encapsulation effects with previous studies (29, 53). The probiotic microgels had a defensive and reparative function in HSD-induced hepatorenal injury (Figures 3A–D).
Intestinal bacteria are either directly or indirectly involved in regulating the occurrence and development of hepatorenal injury through microbiome metabolite–host interactions. The structure of gut microbiota and its metabolites differed significantly between the CON and SD groups (Figures 4, 5). The α-diversity of bacterial communities were not confirmed, however, when comparing the CON- and SD-fed mice using the analyses of Shannon-Wiener and Simpson values (Figure 4D). Nonetheless, current study results showed that the SD group's microbiota was particularly enriched with the Parvibacter, Campylobacter, and Marvinbryantia genera. In contrast, other genera decreased, for example, Lactobacillus, Oscillibacter, Roseburia, and Blautia, compared with the CON group (Figures 4C,F), which is consistent with previous studies (25, 27). The abundance of Lactobacillus increased to varying degrees in the feces of SDL- and SDEL-treated mice and is particularly elevated raised to the highest abundance in the SDEL group (Figures 4C,F). Such outcomes confirm that alginate-chitosan encapsulation could exhibit superior protection of L. rhamnosus viability in vivo and ultimately colonization in the gastrointestinal tract.
Concomitantly, the well-known probiotic taxa, including Bifidobacterium, Faecalibaculum, and Mucispirillum, were observed in the SDEL group with more abundance (Figure 4C). These species had play a pivotal role in protection against CKD (54), colitis (55), and colorectal cancer (56), respectively. The enriched probiotics in the SDEL group were perhaps due to a lower pH value in the colonic environment of the SDEL-fed mice (Figure 7C), which stimulates probiotics growth, inhibits potential pathogenic growth, and modulates the immune system (57). As the metabolite of microbial fermentation, lactic acid is a chiral molecule that exists as L-lactic acid and D-lactic acid. L-lactic acid can be directly involved in human metabolism without any side effects. However, because mammalian tissues lack the enzyme metabolizing D-lactic acid, it has been reported that excessive levels of D-lactic acid may cause metabolic disorders and even acidosis (58). A previous study described L. rhamnosus as a homofermenter that solely produces L-lactic acid (59). L-lactic acid has displayed physiological and pathological functions in vivo, such as affecting of Clostridium difficile infections (60), a neuroprotective function (61), and regulating the lipid metabolism (62). In the present study, the abundance of L-lactic acid in the SDEL group, significantly increased by an average of 1.30 times more than in the SDL group (Figure 5C and Supplementary Table 1), which infers that L. rhamnosus may be more abundant than in the SDL group.
Additionally, microbial production of 3-hydroxybutyric acid has many potential applications as an intermediate for the synthesizing antibiotics, vitamins, aromatics, and pheromones in the host (63). 2-keto-L-gluconate (2-KLG), as a critical intermediate in vitamin C synthesis, could decrease the virulence of bacteria in vivo (64). Besides, the gut microbiota can convert primary bile acids—digestive chemicals produced by the liver—to secondary bile acids, including toxic DCA. DCA products pass into the liver through the hepatic portal vein and impair liver dysfunction (65). In the animal models, this metabolite of circulating bile acids was elevated in CKD and can induce vascular mineralization and osteogenic differentiation (66). In this study, the fecal metabolome results showed increased abundances of 3-hydroxybutyric acid and 2-KLG in the fecal samples of SDEL-treated mice. In contrast, the levels of cholic acid and DCA were significantly lowered compared with the feces of SD-treated mice (Figure 5C and Supplementary Table 1). Treatment of L. rhamnosus microgels led to the increased abundances of probiotic species including Lactobacillus, Bifidobacterium, Mucispirillum, and Faecalibaculum, disclosing strong positive correlations with L-lactic acid and 3-hydroxybutyric acid levels (Figure 6), which may exert beneficial effects on the hepatorenal health of the host. Furthermore, metabolites related to the central carbon metabolism in cancer, ABC transporters, arginine biosynthesis, tyrosine, alanine, aspartate and glutamate metabolism, and renal cell carcinoma pathways were significantly changed in the SDEL group relative to the SD group (Figure 5D). Accumulating evidence suggests that proteins associated with the central carbon metabolism in cancer (67), ABC transporters (68), and arginine biosynthesis (69) pathways are involved in the hepatorenal injury.
HSDs can decrease the Lactobacillus abundance in the murine gut, stimulate intestinal Th17 cells to respond, promote IL-17a production by intestinal epithelial cells, and induce inflammatory reactions (26, 70, 71). Meanwhile, IL-17a can govern the expression, pro-inflammatory effects, and tissue-protective properties of IL-22, and the presence or absence of IL-17a regulates the pathological vs. tissue-protective functions of IL-22 (72). This study confirmed that the intestinal Lactobacillus abundance significantly decreased and serum IL-17a and IL-22 levels significantly increased in SD-fed mice, while the L. rhamnosus microgels significantly increased intestinal Lactobacillus abundance and lowered the IL-17a and IL-22 levels in mice (Figures 3M,N, 4C). The intestinal functions, such as the colonic barrier function and the small intestine's absorption function, are highly associated with salt-induced non-communicable diseases (25, 26). Improving intestinal functions is a potential treatment for liver and kidney diseases (12, 73). With a focus on the microenvironment, it can be seen that L. rhamnosus microgels enhanced the colonic barrier function (Figures 7A,B) and the absorption function of the small intestine (Figure 7D), lowered the pH value of colonic contents (Figure 7C), and maintained the normal intestinal structures (Figures 7E,F) in the SD-fed mice. These actions indicated the positive effects of L. rhamnosus microgels in mitigating and repairing hepatorenal injury induced by high dietary salt levels.
Conclusions and Perspectives
In conclusion, the encapsulation of L. rhamnosus into microgels loaded with alginate and chitosan shows excellent outcomes in enhancing probiotic viability in simulated gastrointestinal transit while also attenuating salt-induced hepatorenal injury through regulating the intestinal microbiota and metabolite profiles in mice. The data from the current study confirms the clinical potential of L. rhamnosus microgels in modulating and repairing hepatorenal and intestinal functions, gut microbiota and metabolism, and host immunity. Nevertheless, the mouse's average daily sodium intake was significantly higher than their tolerance, which likely does not occur in humans for extended periods (74). Human hepatorenal injury results from excessive salt consumption and other clinical and environmental factors over long periods (75). Thus, the effects of L. rhamnosus microgels on ameliorating clinical hepatorenal injury requires further clinical investigation to understand the entire scope of L. rhamnosus microgel treatments.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI; PRJNA516991.
Ethics Statement
The animal study was reviewed and approved by Shandong University of Traditional Chinese Medicine (No. SYXKLU20170022, Jinan, China). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
ZZ and BC designed the research. ZZ, XC, ML, JL, WL, and ZC conducted the research. ZZ and XC wrote the manuscript and analyzed the data. BC and BY provided experimental equipment. All authors read and approved the final manuscript.
Funding
This work received support from the National Natural Science Foundation of Shandong Province (Grant No. ZR2020QC237), National Undergraduate Innovation and Entrepreneurship Training Program in China (Grant No. S202010431032), National Key Research & Development Program in China (Grant No. 2019YFD1002704), and National Natural Science Foundation of China (Grant No. 81803864).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.872808/full#supplementary-material
Abbreviations
CKD, chronic kidney disease; CFU, colony-forming units; CON, control; DCA, deoxycholic acid; HSD, high-salt diet; SDL, HSD with Lactobacillus rhamnosus cells; SDEL, HSD plus L. rhamnosus microgels; SIF, simulated intestinal fluid; SGF, simulated gastric fluid; D-LAD-lactate, DAO, diamine oxidase; IL-17a, interleukin-17a; IL-22, interleukin-22; OUT, operational taxonomic units; ELISA, enzyme-linked immunosorbent assay; BUN, blood urea nitrogen; CRE, creatinine; UA, uric acid; GLC, glucose; ALT, glutamic-pyruvic transaminase; AST, glutamic-oxalacetic transaminase; ALP, alkaline phosphatase; TG, triglycerides; LEfSe, linear discriminant analysis effect size; PCoA, principal co-ordinates analysis.
References
1. Hill C, Guarner F, Reid G, Gibson G, Merenstein D, Pot B, et al. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. (2014) 11:506–14. doi: 10.1038/nrgastro.2014.66
2. Ahmadi S, Wang S, Nagpal R, Wang B, Jain S, Razazan A, et al. A human-origin probiotic cocktail ameliorates aging-related leaky gut and inflammation via modulating microbiota-taurine-tight junction axis. JCI Insight. (2020) 5:e132055. doi: 10.1172/jci.insight.132055
3. Kong Q, Wang B, Tian P, Li X, Zhao J, Zhang H, et al. Daily intake of Lactobacillus alleviates autistic symptoms by ameliorating the 5-hydroxytryptamine metabolic disorder in autistic rats during weaning and sexual maturation. Food Funct. (2021) 12:2591–604. doi: 10.1039/D0FO02375B
4. Darbandi A, Mirshekar M, Shariati A, Moghadam MT, Lohrasbi V, Asadolahi P, et al. The effects of probiotics on reducing the colorectal cancer surgery complications: a periodic review during 2007–2017. Clin Nutr. (2020) 39:2358–67. doi: 10.1016/j.clnu.2019.11.008
5. Seiler CL, Kiflen M, Stefanolo JP, Bai JC, Bercik P, Kelly CP, et al. Probiotics for celiac disease: a systematic review and meta-analysis of randomized controlled trials. Off J Am Coll Gastroenterol. (2020) 115:1584–95. doi: 10.14309/ajg.0000000000000749
6. Tan AH, Lim S-Y, Wei H, Lim J, Low C, Chong C, et al. Probiotics for constipation in parkinson disease: a randomized placebo-controlled study. Neurology. (2020) 96:772–82. doi: 10.1212/WNL.0000000000010998
7. Sanders M, Marco M. Food formats for effective delivery of probiotics. Annu Rev Food Sci Technol. (2010) 1:65–85. doi: 10.1146/annurev.food.080708.100743
8. Terpou A, Papadaki A, Lappa IK, Kachrimanidou V, Bosnea LA, Kopsahelis N. Probiotics in food systems: significance and emerging strategies towards improved viability and delivery of enhanced beneficial value. Nutrients. (2019) 11:1591. doi: 10.3390/nu11071591
9. George M, Abraham TE. Polyionic hydrocolloids for the intestinal delivery of protein drugs: alginate and chitosan — a review. J Control Release. (2006) 114:1–14. doi: 10.1016/j.jconrel.2006.04.017
10. Ta LP, Bujna E, Kun S, Charalampopoulos D, Khutoryanskiy VV. Electrosprayed mucoadhesive alginate-chitosan microcapsules for gastrointestinal delivery of probiotics. Int J Pharm. (2021) 597:120342. doi: 10.1016/j.ijpharm.2021.120342
11. Jobin K, Müller DN, Jantsch J, Kurts C. Sodium and its manifold impact on our immune system. Trends Immunol. (2021) 42:469–79. doi: 10.1016/j.it.2021.04.002
12. Snelson M, Tan SM, Clarke RE, de Pasquale C, Thallas-Bonke V, Nguyen T-V, et al. Processed foods drive intestinal barrier permeability and microvascular diseases. Sci Adv. (2021) 7:eabe4841. doi: 10.1126/sciadv.abe4841
13. Sun Y, Ge X, Li X, He J, Wei X, Du J, et al. High-fat diet promotes renal injury by inducing oxidative stress and mitochondrial dysfunction. Cell Death Dis. (2020) 11:914. doi: 10.1038/s41419-020-03122-4
14. Lobel L, Cao Y, Glickman J, Garrett W. Diet post-translationally modifies the gut microbial proteome to modulate renal function. Science. (2020) 369:1518–24. doi: 10.1126/science.abb3763
15. Yang J, Kim CJ, Go YS, Lee HY, Kim M-G, Oh SW, et al. Intestinal microbiota control acute kidney injury severity by immune modulation. Kidney Int. (2020) 98:932–46. doi: 10.1016/j.kint.2020.04.048
16. Jansen J, Jansen K, Neven E, Poesen R, Othman A, van Mil A, et al. Remote sensing and signaling in kidney proximal tubules stimulates gut microbiome-derived organic anion secretion. Proc Natl Acad Sci USA. (2019) 116:16105–10. doi: 10.1073/pnas.1821809116
17. Nigam SK, Bush KT. Uraemic syndrome of chronic kidney disease: altered remote sensing and signalling. Nat Rev Nephrol. (2019) 15:301–16. doi: 10.1038/s41581-019-0111-1
18. Poesen R, Windey K, Neven E, Kuypers D, De Preter V, Augustijns P, et al. The influence of CKD on colonic microbial metabolism. J Am Soc Nephrol. (2016) 27:1389–99. doi: 10.1681/ASN.2015030279
19. Vaziri ND, Wong J, Pahl M, Piceno YM, Yuan J, DeSantis TZ, et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. (2013) 83:308–15. doi: 10.1038/ki.2012.345
20. Barreto FC, Barreto DV, Liabeuf S, Meert N, Glorieux G, Temmar M, et al. Serum indoxyl sulfate is associated with vascular disease and mortality in chronic kidney disease patients. Clin J Am Soc Nephrol. (2009) 4:1551–8. doi: 10.2215/CJN.03980609
21. Meijers BK, Claes K, Bammens B, de Loor H, Viaene L, Verbeke K, et al. p-Cresol and cardiovascular risk in mild-to-moderate kidney disease. J Am Soc Nephrol. (2010) 5:1182–9. doi: 10.2215/CJN.07971109
22. Zhao J, Zhang QL, Shen JH, Wang K, Liu J. Magnesium lithospermate B improves the gut microbiome and bile acid metabolic profiles in a mouse model of diabetic nephropathy. Acta Pharmacol Sin. (2019) 40:507–513. doi: 10.1038/s41401-018-0029-3
23. Hsu C-N, Lin Y-J, Hou C-Y, Tain Y-L. Maternal administration of probiotic or prebiotic prevents male adult rat offspring against developmental programming of hypertension induced by high fructose consumption in pregnancy and lactation. Nutrients. (2018) 10:1229. doi: 10.3390/nu10091229
24. Wanchai K, Yasom S, Tunapong W, Chunchai T, Eaimworawuthikul S, Thiennimitr P, et al. Probiotic Lactobacillus paracasei HII01 protects rats against obese-insulin resistance-induced kidney injury and impaired renal organic anion transporter 3 (Oat3) function. Clin Sci. (2018) 132: 1545–63. doi: 10.1042/CS20180148
25. Chen X, Zhang Z, Cui B, Jiang A, Tao H, Cheng S, et al. Combination of chronic alcohol consumption and high-salt intake elicits gut microbial alterations and liver steatosis in mice. J Agric Food Chem. (2020) 68:1750–9. doi: 10.1021/acs.jafc.9b07368
26. Zhang Z, Zhao J, Tian C, Chen X, Li H, Wei X, et al. Targeting the gut microbiota to investigate the mechanism of lactulose in negating the effects of a high-salt diet on hypertension. Mol Nutr Food Res. (2019) 63:e1800941. doi: 10.1002/mnfr.201800941
27. Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature. (2017) 551:585–9. doi: 10.1038/nature24628
28. Gao M, Song H, Zheng H, Ren Y, Li S, Liu X, et al. Culture of low density E. coli cells in alginate–chitosan microcapsules facilitates stress resistance by up-regulating luxS/AI-2 system. Carbohydr Polym. (2016) 141:160–5. doi: 10.1016/j.carbpol.2016.01.011
29. Cheow WS, Hadinoto K. Biofilm-like Lactobacillus rhamnosus probiotics encapsulated in alginate and carrageenan microcapsules exhibiting enhanced thermotolerance and freeze-drying resistance. Biomacromolecules. (2013) 14:3214–22. doi: 10.1021/bm400853d
30. Zhang Z, Liu H, Yu B, Tao H, Li J, Wu Z, et al. Lycium barbarum polysaccharide attenuates myocardial injury in high-fat diet-fed mice through manipulating the gut microbiome and fecal metabolome. Food Res Int. (2020) 138:109778. doi: 10.1016/j.foodres.2020.109778
31. Fang H, Yu L, You D, Peng N, Guo W, Wang J, et al. In vivo therapeutic effects and mechanisms of hydroxyasiaticoside combined with praziquantel in the treatment of schistosomiasis induced hepatic fibrosis. Front Bioeng Biotechnol. (2020) 8:613784. doi: 10.3389/fbioe.2020.613784
32. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
33. Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs enables high-throughput genome editing. Nat Biotechnol. (2012) 30:460–5. doi: 10.1038/nbt.2170
34. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. (2010) 7:335–6. doi: 10.1038/nmeth.f.303
35. Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. (2013) 10:996–8. doi: 10.1038/nmeth.2604
36. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. (2007) 73:5261–7. doi: 10.1128/AEM.00062-07
37. Acosta N, Aranaz I, Heras A. Tramadol release from a delivery system based on alginate-chitosan microcapsules. Macromol Biosci. (2003) 3:546–51. doi: 10.1002/mabi.200300009
38. Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. (2013) 31:814–21. doi: 10.1038/nbt.2676
39. Zierer J, Jackson MA, Kastenmüller G, Mangino M, Long T, Telenti A, et al. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet. (2018) 50:790–5. doi: 10.1038/s41588-018-0135-7
40. Borrelli S, Provenzano M, Gagliardi I, Ashour M, Liberti M, Nicola L, et al. Sodium intake and chronic kidney disease. Int J Mol Sci. (2020) 21:4744. doi: 10.3390/ijms21134744
41. Lienhard D, Lauterburg M, Escher G, Frey FJ, Frey BM. High salt intake down-regulates colonic mineralocorticoid receptors, epithelial sodium channels and 11β-hydroxysteroid dehydrogenase type 2. PLoS ONE. (2012) 7:e37898. doi: 10.1371/journal.pone.0037898
42. Ni C, Li X, Wang L, Li X, Zhao J, Zhang H, et al. Lactic acid bacteria strains relieve hyperuricaemia by suppressing xanthine oxidase activity via a short-chain fatty acid-dependent mechanism. Food Funct. (2021) 12:7054–67. doi: 10.1039/D1FO00198A
43. Wang G, Jiao T, Xu Y, Li D, Si Q, Hao J, et al. ifidobacterium adolescentis and Lactobacillus rhamnosus alleviate non-alcoholic fatty liver disease induced by a high-fat, high-cholesterol diet through modulation of different gut microbiota-dependent pathways. Food Funct. (2020) 11:6115–27. doi: 10.1039/C9FO02905B
44. Razavi S, Janfaza S, Tasnim N, Gibson DL, Hoorfar M. Microencapsulating polymers for probiotics delivery systems: preparation, characterization, and applications. Food Hydrocoll. (2021) 120:106882. doi: 10.1016/j.foodhyd.2021.106882
45. Zhuge A, Li B, Yuan Y, Lv L, Li Y, Wu J, et al. Lactobacillus salivarius LI01 encapsulated in alginate-pectin microgels ameliorates D-galactosamine-induced acute liver injury in rats. Appl Microbiol Biotechnol. (2020) 104:7437–55. doi: 10.1007/s00253-020-10749-y
46. Zhao C, Zhu Y, Kong B, Huang Y, Yan D, Tan H, et al. Dual-core prebiotic microcapsule encapsulating probiotics for metabolic syndrome. ACS Appl Mater Interf. (2020) 12:42586–94. doi: 10.1021/acsami.0c13518
47. Le T, Millette M, Mateescu MA, Lacroix M. Modified alginate and chitosan for lactic acid bacteria immobilization. Biotechnol Appl Biochem. (2004) 39(Pt 3):347–54. doi: 10.1042/BA20030158
48. Krasaekoopt W, Bhandari B, Deeth H. Evaluation of encapsulation techniques of probiotics for yoghurt. Int Dairy J. (2003) 13:3–13. doi: 10.1016/S0958-6946(02)00155-3
49. Tønnesen H, Karlsen J. Alginate in drug delivery systems. Drug Dev Ind Pharm. (2002) 28:621–30. doi: 10.1081/DDC-120003853
50. Muzzarelli C, Stanic V, Gobbi L, Tosi G, Muzzarelli RAA. Spray-drying of solutions containing chitosan together with polyuronans and characterisation of the microspheres. Carbohydr Polym. (2004) 57:73–82. doi: 10.1016/j.carbpol.2004.04.002
51. Yang X, Li J, Shi G, Zeng M, Liu Z. Improving 3-phenyllactic acid production of Lactobacillus plantarum AB-1 by enhancing its quorum-sensing capacity. J Food Sci Technol. (2019) 56:2605–10. doi: 10.1007/s13197-019-03746-1
52. Yeung T, Ücok EF, Tiani K, McClements D, Sela D. Microencapsulation in alginate and chitosan microgels to enhance viability of Bifidobacterium longum for oral delivery. Front Microbiol. (2016) 7:494. doi: 10.3389/fmicb.2016.00494
53. Chávarri M, Marañón I, Ares R, Ibáñez FC, Marzo F, Villarán M, et al. Microencapsulation of a probiotic and prebiotic in alginate-chitosan capsules improves survival in simulated gastro-intestinal conditions. Int J Food Microbiol. (2010) 142:185–9. doi: 10.1016/j.ijfoodmicro.2010.06.022
54. Wang X, Yang S, Li S, Zhao L, Hao Y, Qin J, et al. Aberrant gut microbiota alters host metabolome and impacts renal failure in humans and rodents. Gut. (2020) 69:2131–42. doi: 10.1136/gutjnl-2019-319766
55. Herp S, Brugiroux S, Garzetti D, Ring D, Jochum L, Beutler M, et al. Mucispirillum schaedleri antagonizes Salmonella virulence to protect mice against colitis. Cell Host Microbe. (2019) 25:681–94. doi: 10.1016/j.chom.2019.03.004
56. Zagato E, Pozzi C, Bertocchi A, Schioppa T, Saccheri F, Guglietta S, et al. Endogenous murine microbiota member Faecalibaculum rodentium and its human homologue protect from intestinal tumour growth. Nat Microbiol. (2020) 5:511–24. doi: 10.1038/s41564-019-0649-5
57. Roberfroid M, Gibson G, Hoyles L, McCartney A, Rastall R, Rowland I, et al. Prebiotic effects: metabolic and health benefits. Br J Nutr. (2010) 104:S1–63. doi: 10.1017/S0007114510003363
58. Zhang J, Gong G, Wang X, Zhang H, Tian W. Positive selection on D-lactate dehydrogenases of Lactobacillus delbrueckii subspecies bulgaricus. IET Syst Biol. (2015) 9:172–9. doi: 10.1049/iet-syb.2014.0056
59. Wang Y, Li Y, Pei X, Yu L, Feng Y. Genome-shuffling improved acid tolerance and L-lactic acid volumetric productivity in Lactobacillus rhamnosus. J Biotechnol. (2007) 129:510–5. doi: 10.1016/j.jbiotec.2007.01.011
60. Kolling GL, Wu M, Warren CA, Durmaz E, Klaenhammer TR, Timko MP, et al. Lactic acid production by Streptococcus thermophilus alters Clostridium difficile infection and in vitro toxin A production. Gut Microbes. (2012) 3:523–9. doi: 10.4161/gmic.21757
61. Llorente-Folch I, Rueda C, Perez I, Satrústegui J, Pardo B. L-Lactate-mediated neuroprotection against glutamate-induced excitotoxicity requires Aralar/Agc1. J Neurosci. (2016) 36:4443–56. doi: 10.1523/JNEUROSCI.3691-15.2016
62. Shan B, Ai Z, Zeng S, Song Y, Song J, Zeng Q, et al. Gut microbiome-derived lactate promotes to anxiety-like behaviors through GPR81 receptor-mediated lipid metabolism pathway. Psychoneuroendocrinology. (2020) 117:104699. doi: 10.1016/j.psyneuen.2020.104699
63. Liu Q, Ouyang S-P, Chung A, Wu Q, Chen G-Q. Microbial production of R-3-hydroxybutyric acid by recombinant E. coli harboring genes of phbA, phbB, and tesB. Appl Microbiol Biotechnol. (2007) 76:811–8. doi: 10.1007/s00253-007-1063-0
64. Hu FZ, Król JE, Tsai CHS, Eutsey RA, Hiller LN, Sen B, et al. Deletion of genes involved in the ketogluconate metabolism, Entner-Doudoroff pathway, and glucose dehydrogenase increase local and invasive virulence phenotypes in Streptococcus pneumoniae. LoS ONE. (2019) 14:e0209688. doi: 10.1371/journal.pone.0209688
66. Jovanovich A, Isakova T, Block G, Stubbs J, Smits G, Chonchol M, et al. Deoxycholic acid, a metabolite of circulating bile acids, and coronary artery vascular calcification in CKD. Am J Kidney Dis. (2017) 71:27–34. doi: 10.1053/j.ajkd.2017.06.017
67. Sayin VI, LeBoeuf SE, Singh SX, Davidson SM, Biancur D, Guzelhan BS, et al. Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. Elife. (2017) 6:e28083. doi: 10.7554/eLife.28083.018
68. Stanton B. ABC transporters in liver, kidney, and intestine. Kidney Int. (2002) 62:1520–1. doi: 10.1046/j.1523-1755.2002.t01-8-00644.x
69. Varga ZV, Erdelyi K, Paloczi J, Cinar R, Zsengeller ZK, Jourdan T, et al. Disruption of renal arginine metabolism promotes kidney injury in hepatorenal syndrome in mice. Hepatology. (2018) 68:1519–33. doi: 10.1002/hep.29915
70. Miranda P, De Palma G, Serkis V, Lu J, Louis-Auguste M, McCarville J, et al. High salt diet exacerbates colitis in mice by decreasing Lactobacillus levels and butyrate production. Microbiome. (2018) 6:57. doi: 10.1186/s40168-018-0433-4
71. Wei Y, Lu C, Chen J, Cui G, Wang L, Yu T, et al. High salt diet stimulates gut Th17 response and exacerbates TNBS-induced colitis in mice. Oncotarget. (2016) 8:70–82. doi: 10.18632/oncotarget.13783
72. Sonnenberg G, Nair M, Kirn T, Zaph C, Fouser L, Artis D. Pathological versus protective functions of IL-22 in airway inflammation are regulated by IL-17a. J Exp Med. (2010) 207:1293–305. doi: 10.1084/jem.20092054
73. Zhang Z, Chen X, Cui B. Modulation of the fecal microbiome and metabolome by resistant dextrin ameliorates hepatic steatosis and mitochondrial abnormalities in mice. Food Funct. (2021) 12:4504–18. doi: 10.1039/D1FO00249J
74. Relman DA. Microbiota: a high-pressure situation for bacteria. Nature. (2017) 551:571–2. doi: 10.1038/nature24760
Keywords: microgels, high-salt diet, gut microbiome, probiotic, hepatorenal injury
Citation: Zhang Z, Liu J, Li M, Yang B, Liu W, Chu Z, Cui B and Chen X (2022) Lactobacillus rhamnosus Encapsulated in Alginate/Chitosan Microgels Manipulates the Gut Microbiome to Ameliorate Salt-Induced Hepatorenal Injury. Front. Nutr. 9:872808. doi: 10.3389/fnut.2022.872808
Received: 10 February 2022; Accepted: 23 March 2022;
Published: 14 April 2022.
Edited by:
Guifang Tian, Agricultural University of Hebei, ChinaReviewed by:
Xiaojun Huang, Nanchang University, ChinaBabak Haghshenas, Kermanshah University of Medical Sciences, Iran
Copyright © 2022 Zhang, Liu, Li, Yang, Liu, Chu, Cui and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zheng Zhang, emhlbmd6aGFuZzMyNEAxNjMuY29t; Bo Cui, Y3VpYm9wYXBlckAxNjMuY29t; Xiao Chen, NjAyMjAwMjFAc2R1dGNtLmVkdS5jbg==
 Zheng Zhang
Zheng Zhang Jiajian Liu1
Jiajian Liu1 Mengjie Li
Mengjie Li Zhuangzhuang Chu
Zhuangzhuang Chu Bo Cui
Bo Cui Xiao Chen
Xiao Chen