
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr. , 29 April 2022
Sec. Nutrigenomics
Volume 9 - 2022 | https://doi.org/10.3389/fnut.2022.868805
 Jerry Maria Sojan1
Jerry Maria Sojan1 Manu Kumar Gundappa2
Manu Kumar Gundappa2 Alessio Carletti3,4
Alessio Carletti3,4 Vasco Gaspar4
Vasco Gaspar4 Paulo Gavaia3,4
Paulo Gavaia3,4 Francesca Maradonna1*
Francesca Maradonna1* Oliana Carnevali1*
Oliana Carnevali1*The micronutrient boron (B) plays a key role during the ossification process as suggested by various in vitro and in vivo studies. To deepen our understanding of the molecular mechanism involved in the osteogenicity of B and its possible interaction with vitamin D3 (VD), wild-type AB zebrafish (Danio rerio) were used for morphometric analysis and transcriptomic analysis in addition to taking advantage of the availability of specific zebrafish osteoblast reporter lines. First, osteoactive concentrations of B, VD, and their combinations were established by morphometric analysis of the opercular bone in alizarin red-stained zebrafish larvae exposed to two selected concentrations of B (10 and 100 ng/ml), one concentration of VD (10 pg/ml), and their respective combinations. Bone formation, as measured by opercular bone growth, was significantly increased in the two combination treatments than VD alone. Subsequently, a transcriptomic approach was adopted to unveil the molecular key regulators involved in the synergy. Clustering of differentially expressed genes revealed enrichment toward bone and skeletal functions in the groups co-treated with B and VD. Downstream analysis confirmed mitogen-activated protein kinase as the most regulated pathway by the synergy groups in addition to transforming growth factor-β signaling, focal adhesion, and calcium signaling. The best-performing synergistic treatment, B at 10 ng/ml and VD at 10 pg/ml, was applied to two zebrafish transgenic lines, Tg(sp7:mCherry) and Tg(bglap:EGFP), at multiple time points to further explore the results of the transcriptomic analysis. The synergistic treatment with B and VD induced enrichment of intermediate (sp7+) osteoblast at 6 and 9 days post fertilization (dpf) and of mature (bglap+) osteoblasts at 15 dpf. The results obtained validate the role of B in VD-dependent control over bone mineralization and can help to widen the spectrum of therapeutic approaches to alleviate pathological conditions caused by VD deficiency by using low concentrations of B as a nutritional additive.
Osteogenesis is a process that can be modulated by several factors, including macro- and micronutrient supplementation. Among the various micronutrients, it has been demonstrated that boron (B) has an important role in the development and maintenance of bone (1–3). B plays a crucial biological role in bone health by modulating the functions of various essential nutrients, including vitamin D3 (VD), calcium (Ca), and phosphorous (P), which are known to affect bone mineralization in both humans and animals (4–10). The presence of B is restricted to the mineral component of skeletal tissues and not to the organic matrix (11). Contrasting results were found in previous studies about the effect of B on the absorption of Ca and P, both essential nutrients for the skeletal formation. Dietary supplementation with B increased Ca and P absorption and balance in wethers and rats (12, 13) and promoted the improvement of the mechanical properties of bone tissue (14–18), while no such effects were observed in barrows (19). However, in vertebrates, B deficiency results in impaired osteogenesis, negatively affecting bone development (20, 21). Supplementation of B resulted in the improvement of bone strength and microstructure in mice (22). This was also observed in ostrich, where low concentrations of B supplemented through water led to increased osteogenesis through bone morphogenetic protein (BMP)-2 regulation (23). In addition, B was found to be able to reduce inflammation correlated with reduced bone mineral density, thereby improving the bone health (24), suggesting its use in the treatment of osteochondrosis (25). B supplementation was also able to enhance the fracture-healing process in rats (26). In addition to the in vivo evidence of the beneficial effects of B in the mineralization process, clear evidence was obtained in vitro as well. In pre-osteoblastic cell line MC3T3-E1, B was found to positively regulate mineralized tissue-associated proteins and messenger RNA (mRNA) expression of genes involved in osteoblastic functions such as osteocalcin (bglap), osteopontin (spp1), and collagen type 1 (27). In the same study, B concentrations of 10 and 100 ng/ml were able to increase in vitro mineralization in MC3T3-E1 cells (27). Similar concentrations were used in human bone marrow mesenchymal stem cells; although cell proliferation was not affected, an increased expression of BMPs and osteocalcin, along with elevation in the activity of alkaline phosphatase, was reported (28). Similarly, osteo-inductive properties of encapsulated B within scaffolds were confirmed in MC3T3-E1 cells (29).
Skeletal diseases such as osteoporosis, osteomalacia, and rickets are a significant medical burden worldwide, and VD deficiency is one of the major and perhaps the most preventable factor leading to bone fragility (30, 31). In a previous study, the supplementation of B in broiler chickens with VD deficiency alleviated symptoms associated with the insufficiency such as disruptions in the mineral metabolism (8). B supplementation further improved the biochemical characteristics such as Ca and P levels, thereby resulting in a healthier bone despite the VD deficiency (8). Similar observations were found in VD-deficient rats where B was able to increase Ca, Mg, and P (32). B interacts with VD probably by compensating the disturbances in the energy-substrate consumption or by enhancing the macro-mineral content of bone, in addition to the possibility of VD-independent regulation of the indices of maturation of cartilage (33). B was also shown to inhibit enzymes that catabolize VD, thereby causing an upregulatory impact on VD status (34). The exact mode of interaction between B and VD is not yet clear and needs further exploration.
Zebrafish (Danio rerio) is an increasingly relevant model for bone-related studies due to the many technical advantages associated with its use (35). In particular, translucent larval stages and cost-effective genetic manipulation translated into an increased availability of fluorescent reporter lines, which are particularly useful for in vivo cellular tracking, allowing the study of the fate and differentiation of specific bone cell types (36). Considering the necessity to provide valid solution to promote bone health, two concentrations of B were supplemented alone and in synergy with VD to zebrafish larvae and the impacts were studied at transcriptional level. Recent advances in next-generation sequencing guaranteed an easy access to RNA-sequencing data, which are being exploited here, and it allows the investigation of the action of compounds of interest on the bone metabolism and defines key pathways modulated by therapeutic interventions. To further validate the effects of B and VD on early skeletal development, Tg(Ola.sp7:mCherry-Eco.NfsB)pd46, and Tg(Ola.bglap:EGFP)hu4008, hereinafter mentioned as Tg(sp7:mCherry) and Tg(bglap:EGFP), respectively. D. rerio lines were chosen for this study as sp7 is a zinc-finger-containing transcription factor expressed in pre-osteoblasts and immature osteoblasts, making it an excellent marker for cell tracking of osteoblastic cells (37), and bglap is a mature osteoblast marker, making it suitable to look into the effects of treatments on osteoblast maturation (36).
The analyzed data provide a greater understanding of the actions of these compounds in modulating the overall transcriptome with particular attention to the exact molecular regulation of skeletal development and time course of expression of sp7 and bglap proteins through transgenic lines. Furthermore, it opens the potential of using micronutrients like B as an additive to enhance the osteogenic efficiency of VD, thereby providing a low-cost solution to tackle the nutritional VD deficiency in biomedicine and aquaculture.
Adult wild-type AB female and male zebrafish specimens reared in the fish facility at Università Politecnica delle Marche (Ancona, Italy) were set up for overnight breeding at 2:1 ratio. The embryos were collected and divided into six groups in triplicates. Treatment concentrations were modified and adapted from previous studies (28, 38):
• Control: control group with ethanol at 0.1%.
• VD: VD group with VD3 (1α,25-dihydroxyvitamin D3; Sigma-Aldrich, USA) at 10 pg/ml.
• B10: B at 10 ng/ml.
• B10VD: B at 10 ng/ml with VD at 10 pg/ml.
• B100: B at 100 ng/ml.
• B100VD: B at 100 ng/ml with VD at 10 pg/ml.
Boric acid (Sigma-Aldrich, Germany) was used to make the concentrations of B across different treatment groups. The larvae were continuously treated via waterborne exposure with the respective compounds until 8 days post fertilization (dpf) and 70% of the water with respective treatments was renewed daily. VD was dissolved in ethanol before using in the treatments; therefore, ethanol (0.1%) was added to the control and two B groups without VD to ensure constant ethanol concentrations in all the groups. The sampling was done at 9 dpf for AR-S staining, image acquisition, and RNA extraction.
AR-S staining is one of the most used methods for studying bone mineralization (39). For fluorescence imaging, larvae (n = 5 per replicate per group) were exposed to an overdose of 300 mg/L MS-222 (ethyl 3-aminobenzoate methane sulfonate; Sigma-Aldrich, USA) and were stained with AR-S (Fluka Chemika, Switzerland) at 0.01% for 15 min. After washing with H2O, stained larvae were placed in a lateral position onto an agarose gel (2%). Images of the stained larvae were taken using a Zeiss Axio Imager M2 fluorescent microscope (Milan, Italy) set with a green light filter (λex = 530–560 nm and λem = 580 nm). Images were acquired using constant parameters and analyzed using ImageJ (version 2.1.0/1.53c) software after splitting the color channels of the RGB images. Eight-bit images were adjusted uniformly for all the images to achieve optimum contrast and brightness for improved visibility of the operculum bone.
Broodstock from the transgenic lines used in our experiments, Tg(sp7:mCherry) and Tg(bglap:EGFP), were maintained in a recirculating water system (Tecniplast, Italy) at the aquatic animal experimental facilities of the Centre of Marine Sciences, Faro, Portugal. Eggs were produced with an in-house breeding program and maintained in static conditions until hatching at 3 dpf. Larvae were then screened with a Leica MZ10F fluorescence stereomicroscope (Leica, Germany) and 400 fish expressing the reporter proteins were selected and randomly distributed into four 300 ml beaker cups (100 fish/beaker) with the respective treatments in water. The selected four experimental groups were
• Control: control group with ethanol at 0.1%.
• VD: VD group with VD3 (1α,25-dihydroxyvitamin D3, Sigma-Aldrich, USA) at 10 pg/ml.
• B10: B at 10 ng/ml.
• B10VD: B at 10 ng/ml with VD3 at 10 pg/ml.
Boric acid (USB Corporation, USA) was used to make the required concentrations of B groups. Fish (n = 20) were sampled at four different time points, 6, 9, 12, and 15 dpf, stained with 0.01% AR-S (Sigma-Aldrich, USA) or 0.1% calcein (Sigma-Aldrich, USA) to label mineralized structures and imaged using a Leica MZ10F fluorescence stereomicroscope equipped with a green fluorescence filter (λex = 546/10 nm) and a barrier filter (λem = 590 nm) for Tg(sp7:mCherry) and AR-S stained fish; and with a blue fluorescence filter (λex = 470/40 nm) and a barrier filter (λem = 515 nm) for Tg(bglap:EGFP) and calcein-stained fish. All images were acquired with a DFC7000T color camera (Leica, Germany) according to the following parameters, namely, 24-bit colored image, exposure time 2 s (green channel) and 1 s (red channel), gamma 1.00, image format 1920 × 1440 pixels, and binning 1 × 1. Fluorescence images were processed with ZFBONE macro toolset for Fiji (40).
At 9 dpf, the larvae were sampled using an overdose of MS-222 and stored at −80°C. There were three biological replicates for each experimental group, and each replicate consisted of a pool of seven larvae. Total RNA was extracted from each replicate sample using RNAeasy® Minikit (QIAGEN, Germany) and eluted in 20 μl of molecular grade nuclease-free water. Final RNA concentrations were determined using a nanophotometer (Implen, Germany). Total RNA was treated with DNase (10 IU at 37°C for 10 min; Sigma-Aldrich, USA), and quality was confirmed using gel electrophoresis (1% gel) and stored at −80°C until library preparation for RNA sequencing. iScript cDNA Synthesis Kit (Bio-Rad, USA) was used to perform cDNA synthesis using 1 μg of total RNA and stored at −20°C until further use in real-time PCRs (RT-PCR).
Samples to be used for RNA sequencing were confirmed for concentration using Invitrogen Qubit 3.0 Fluorometer with the RNA assay kit and for integrity using Agilent Tapestation. Illumina TruSeq RNA libraries were prepared by Novogene Ltd. (Cambridge, UK) and sequenced on an Illumina Novaseq6000. All the triplicate samples of each group were sequenced to generate approximately 30 million paired end reads of 150 base pairs (bp) each (Supplementary Table 1). The read data were assessed for its quality using FastQC version 0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were then trimmed using TrimGalore version 0.4.4 (https://github.com/FelixKrueger/TrimGalore) by setting the parameters -q 30, -stringency 5, -length 40. Specifically, reads were trimmed for any adapters, and bases with a Phred score of <25 were trimmed off. Trimmed reads <40 bp were also removed.
The final cleaned up reads were then mapped to the D. rerio reference genome (GRCz11) retrieved from Ensembl genome database. Mapping was performed using STAR aligner (41) with the parameters (–outSAMtype BAM SortedByCoordinate, –outSAMunmappedWithin, –outSAMattributes Standard). Gene-level read count data was generated using featureCounts (42) with the parameters (–primary, -C, -t exon, -g gene_id), and the rest were set to default.
Differential gene expression was performed using DESeq2 1.26.0 (43) within R 3.6.1 (44). Finally, genes with false discovery rate < 0.05 and absolute log2 fold change values (FC) >0.5 were considered as differentially expressed. Principal component analysis (PCA) plots were generated to remove any outlier samples from the data using plotPCA function within DESeq2 1.26.0 followed by hierarchical clustering across samples using the function heatmap.2 within the package gplots 3.0.1.1 (45) to confirm the clustering of replicates. A list of differentially expressed genes (DEGs) was generated for all treatment combinations and concatenated to generate a final list of genes that were differentially expressed in at least one combination. The DEGs were then clustered using partition around medoids (PAM) algorithm (46) into different clusters based on the DESeq2 median ratio normalized expression values across different treatments using the package cluster 2.1.0 (47). The optimum number of clusters was identified using Gap statistic method (48) within the package factoextra 1.0.6 (49). The normalized counts were mean centralized across different treatments within each cluster and visualized using the package ggplot2 3.2.1(50).
Gene set enrichment analysis was performed using Clusterprofiler 3.14.3 package (51). Annotations for D. rerio were retrieved from the package org.Dr.eg.db 3.8.2 (52), and gene set enrichment analysis was performed for DEGs in the clusters individually. Gene Ontology (GO) terms falling under the categories Biological Process (BP), Cellular Component (CC), and Molecular Function (MF) with a p < 0.05 were considered as significant and used for downstream analysis. Enriched GO terms and genes within each cluster were then used to generate bipartite networks using the package ggnetwork 0.5.1 (53) within R. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed against the DEGs present in the three clusters and filtered for p < 0.05 to be considered significant. Bubble plots for enriched KEGG pathways across all the clusters were generated using ggplot2 3.2.1.
RT-PCRs were performed with SYBR green in a CFX thermal cycler (Bio-Rad, Italy) in triplicate as previously described (54). The thermal profile for all reactions was 3 min at 95°C followed by 45 cycles of 20 s at 95°C, 20 s at 60°C, and 20 s at 72°C. Dissociation curve analysis showed a single peak in all the cases. Ribosomal protein L13a (rpl13a) and ribosomal protein, large, P0 (rplp0) were used as the housekeeping genes to standardize the results by eliminating variation in mRNA and cDNA quantity and quality. No amplification product was observed in negative controls and primer-dimer formation was never seen. Data were analyzed using iQ5 Optical System version 2.1 (Bio-Rad), including Genex Macro iQ5 Conversion and Genex Macro iQ5 files. Modification of gene expression between the groups is reported as relative mRNA abundance (arbitrary units). Primers were used at a final concentration of 10 pmol/ml. Primer sequences are listed in Supplementary Table 2.
Data of all groups were normally distributed as assessed using Shapiro-Wilk's test (p > 0.05), and there was homogeneity of variances, as assessed using Levene's test for equality of variances (p > 0.05). The differences between the control and the treatments were tested with a one-way analysis of variance (ANOVA) followed by Tukey's post-hoc test (p < 0.05) for the experiments with the transgenic lines and the image data of AR-S staining. All the tests were performed using R version 3.6.1(44), and plots were generated using ggplot2 3.2.1.
No major differences in opercular bone growth were observed between the two concentrations of B (B10 and B100), whereas an increase in mineralization of the opercula was observed in groups treated with VD with respect to control, B10, and B100. However, both the synergy groups (B10VD and B100VD) showed an increase in mineralized area of the opercular bone when compared to VD treatment (Figure 1A). Quantitative analysis of the integrated pixel density within the operculum area was performed as it adequately proxies the intensity of fluorescence signal, thus providing an index of the density of AR-S staining (i.e., mineralization). Using ImageJ (version 2.1.0/1.53c), the area of the operculum (OpA) and the area of the head (HA) were manually selected and the raw integrated density within the area of the operculum (Opl) was extracted. Opl was then normalized with the area of the head to compensate for differences in size among fish. Normalized pixel density within the operculum (Opl/HA) did not vary among fish treated with ethanol (control) and the two concentrations of B (B10, B100) while VD and both synergy groups significantly increased mineralization of the operculum with respect to the control group and both B concentrations (Figure 1B). Importantly, opercular bone mineralization was significantly increased in fish treated with both synergy groups (B10VD, B100VD) compared to VD in what appeared to be a dose-dependent manner, although no significant differences were found between the synergy groups with different concentrations of B (Figure 1B).
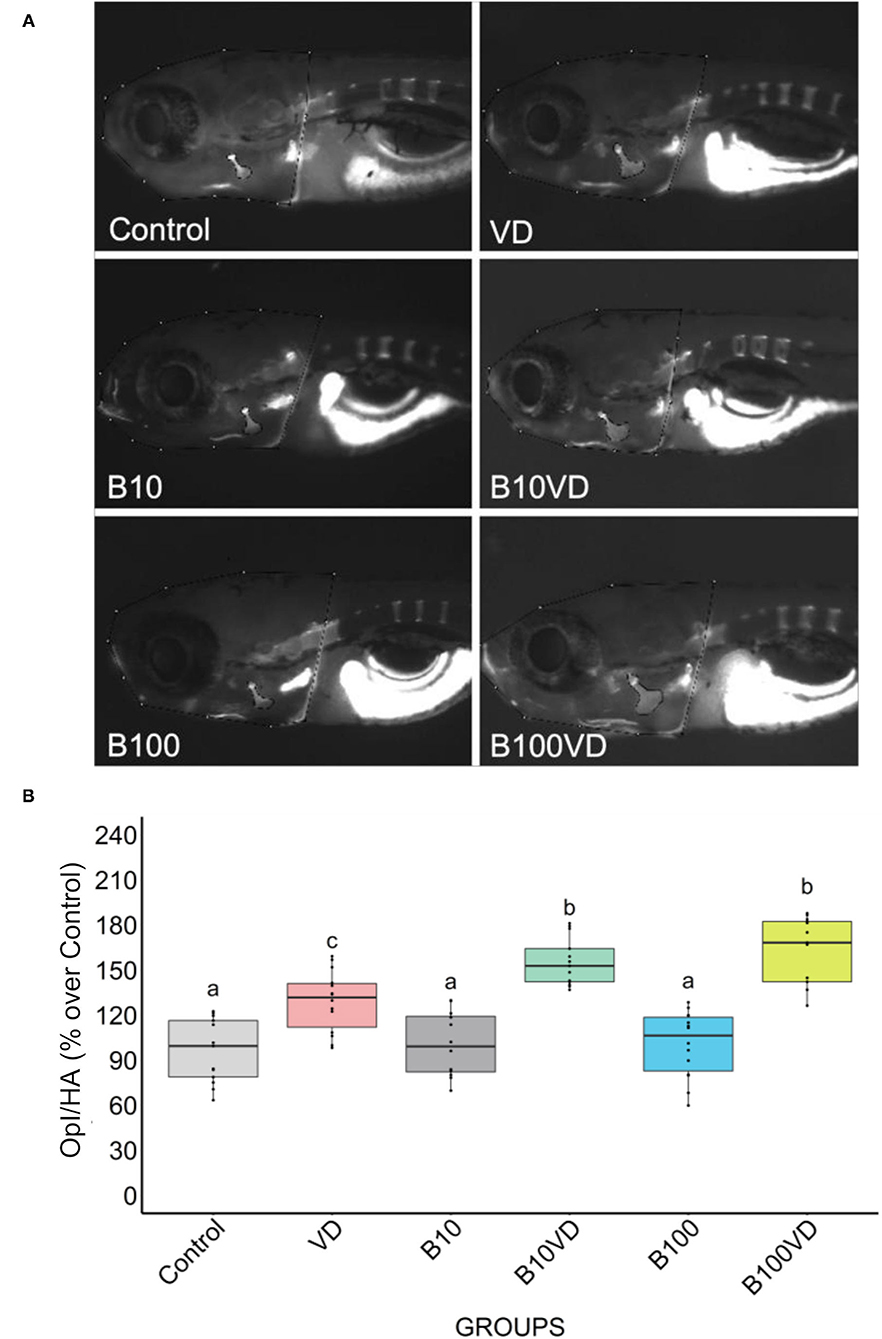
Figure 1. Fluorescence microphotograph of AR-S-stained larvae and ImageJ quantification of the opercular bone mineralization. (A) Mineralized bones stained by AR-S staining at 9 dpf following different treatments. (B) Quantitative analysis of the operculum bone integrated pixel density normalized by head area (Opl/HA) showed as % over control in fish treated with different concentrations of B and VD. Different letters above each graph indicate statistically significant differences among different groups. One-way ANOVA and Tukey's post-hoc test were used, and statistical significance was set at p < 0.05.
Approximately 30 million paired end reads of 150 bp were generated across each sample of RNA in triplicates from each treatment group at 9 dpf. PCA on the read count data stratified the different treatments and replicates into distinct clusters (Figure 2A). Hierarchical clustering of the top 1500 DEGs clustered the different replicates of different treatments together (Figure 2B), confirming uniformity among the replicates. However, control and B10 samples showed an interspersing in both the PCA and hierarchical clustering, indicating very low variability in expression between these groups, which is also consistent with the dimensions of the operculum between the two treatment groups (Figure 1B). Differential expression analysis on all these groups combined resulted in a set of 7,341 genes differentially expressed in at least one of the different contrasts listed in Figure 2C and Supplementary Table 3. Comparing the differential expression of different treatments against control revealed B10VD to be a highly responsive group against control with 1,477 and 724 genes up- and downregulated, respectively (Figure 2C). B10 treatment in contrast just revealed four downregulated and one upregulated gene, showing to be the treatment with the lowest transcriptional response against control (Figure 2C).
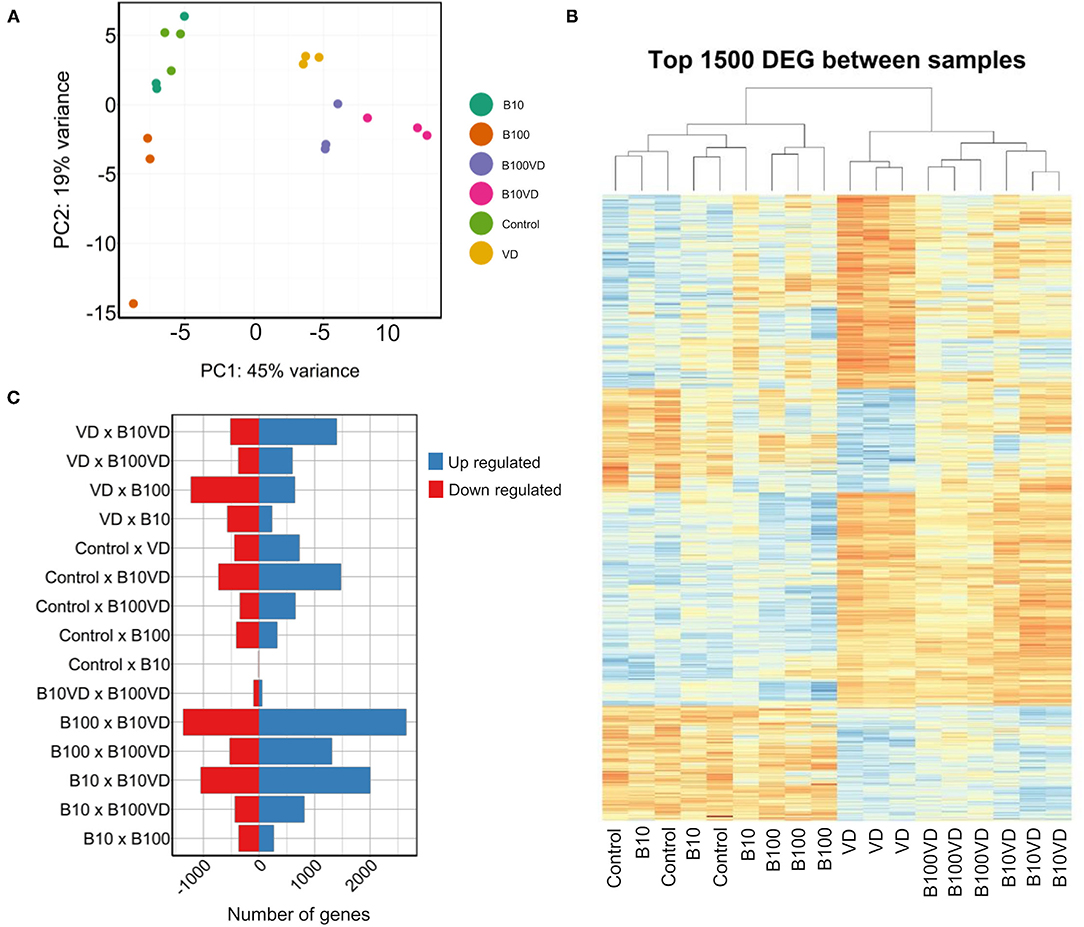
Figure 2. Sample clustering and DEGs across different combinations. (A) PCA of all the samples used in the experiment. (B) Hierarchical clustering of the top 1,500 DEGs across all the samples used in the experiment. (C) Number of upregulated and downregulated genes across different combinations of experimental groups.
Gap statistic method determined three clusters to be optimum for PAM clustering (Supplementary Figure 1). PAM clustering defined three clusters C1, C2, C3 with 2,993, 2,014, and 2,245 genes in each cluster (Figure 3A), respectively. The three clusters revealed distinct expression patterns across different treatments, and normalized expression values in cluster 1 were highest for B100 and lowest for B10VD (Figure 3B). Cluster 2 genes displayed a contrasting expression pattern between VD (decreased) and B10VD (increased) compared to control, while cluster 3 harbored genes showed increased expression in VD, B10VD, B100VD groups compared to the control and other treatment groups.
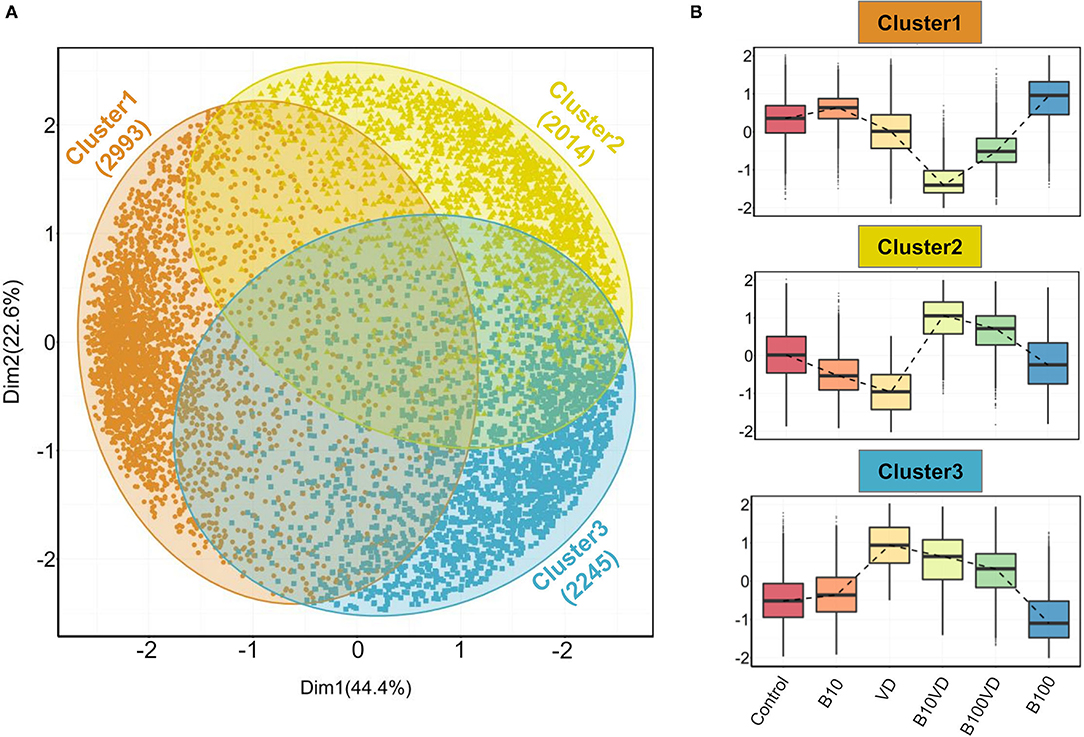
Figure 3. Outputs from PAM clustering of all the DEGs. (A) PCA visualizing the three clusters labeled cluster 1, cluster 2, and cluster 3 (indicated in orange, yellow, and blue colors, respectively) as defined by the PAM clustering. (B) Expression patterns of normalized gene expression values across different treatment groups in each of the three clusters. The expression counts were mean-centered before plotting them as box plots. The dashed lines within each panel connect the median values across different treatment groups.
Gene function enrichment analysis for all the DEGs across each cluster revealed distinct functional enrichment for GO terms and KEGG analysis. Cluster 1 was enriched for a total of 47 GO terms across BP, MF, and CC categories with a very high representation of terms involved in catabolic processes (GO:0019941, GO:0010499, GO:0043632, GO:0006511) and proteasome assembly (GO:0043248) (details of GO terms are given in Supplementary Table 4). A total of 117 GO terms were enriched in cluster 2, and there was no overrepresentation of GO terms contributing to a particular function across all the three GO categories (Supplementary Table 4). Cluster 1 and cluster 2 revealed no significant enrichment for functions or processes related to skeletal system. Interestingly, cluster 3 stood out among the three with a high enrichment for GO terms involved in bone and skeleton system functioning (Supplementary Table 4). In cluster 3, a total of 44 GO terms were enriched in the BP category out of which 23 GO terms were involved in processes, leading to bone and skeletal system functioning, such as extracellular matrix (ECM) organization (GO:0030198; p = 1.29e−16) and skeletal system development (GO:0001501; p = 0.000504). In addition, within cluster 3, GO terms involved in skeletogenesis were again enriched in the CC (<10 terms) and MF (<5 terms) category (Supplementary Table 4); however, the key focus was diverted toward BP considering more than half of the enriched processes were related to bone and skeletal system. The bipartite network showing interactions of genes and enriched GO terms (BP) across the clusters displayed minimum overlap between enriched GO terms across three different clusters (Figure 4). This further indicates that genes showing distinct expression profiles across the three clusters are in fact enriched for distinct functions. Also, many genes in cluster 3 are shared across multiple GO terms with bone and skeletal system functions as highlighted in Figure 4, which otherwise is not observed in the other two clusters.
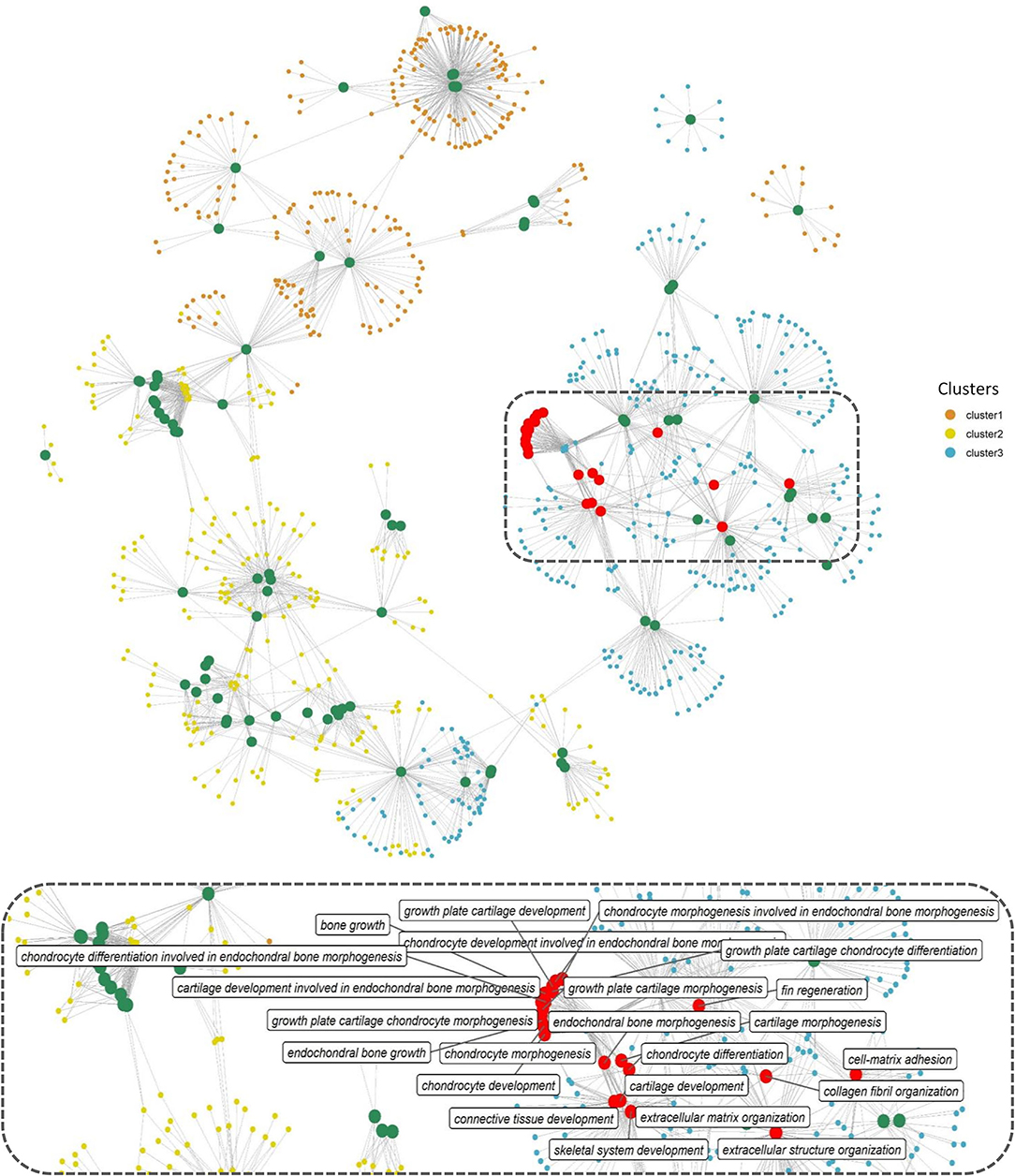
Figure 4. Network representation of enriched GO terms (BP) performed against the DEGs across three different clusters (Supplementary Table 4), the large-sized green and red-colored nodes indicate enriched GO terms, the red nodes in particular highlight GO terms in bone and skeleton system functioning, the region within the dashed rectangle is zoomed at the bottom of the network to highlight the nomenclature of key GO terms. Each small node of orange, green, and blue colors indicates a gene contributing to the enriched GO term across different clusters.
We further investigated the strong enrichment for skeletal GO terms (BP) by generating a quantitative matrix of the number of genes falling within each GO term across three clusters (Figure 5A). Increased number of genes involved in skeletal functions within cluster 3 correlates well with the strong enrichment in GO terms. KEGG analysis revealed cluster 1 to be enriched for 11 KEGG pathways, while cluster 2 displayed enrichment for only one pathway, phototransduction, involving 21 genes (Figure 5B; Supplementary Table 5). Cluster 3, which showed a strong bias toward skeletal functions in GO analysis, was enriched for five KEGG pathways, out of which focal adhesion (dre04510), ECM-receptor interaction (dre04512), and regulation of actin cytoskeleton (dre04810) contribute toward bone/skeletal functions (Figure 5B). Heatmaps showing normalized expression values of genes falling within the key enriched bone and skeletal-related GO terms such as skeletal system development (GO:0001501), ECM organization (GO:0030198), bone growth (GO:0098868), endochondral bone growth (GO:0003416), endochondral bone morphogenesis (GO:0060350), and chondrocyte differentiation involved in endochondral bone morphogenesis (GO:0003413) revealed an increased expression in the B10VD group compared to all other groups (Figure 5C). There is a bias toward increased expression of skeletal system genes in B10VD despite the overall expression pattern of cluster 3 showing the highest expression for the VD group (Figures 3A, 5C).
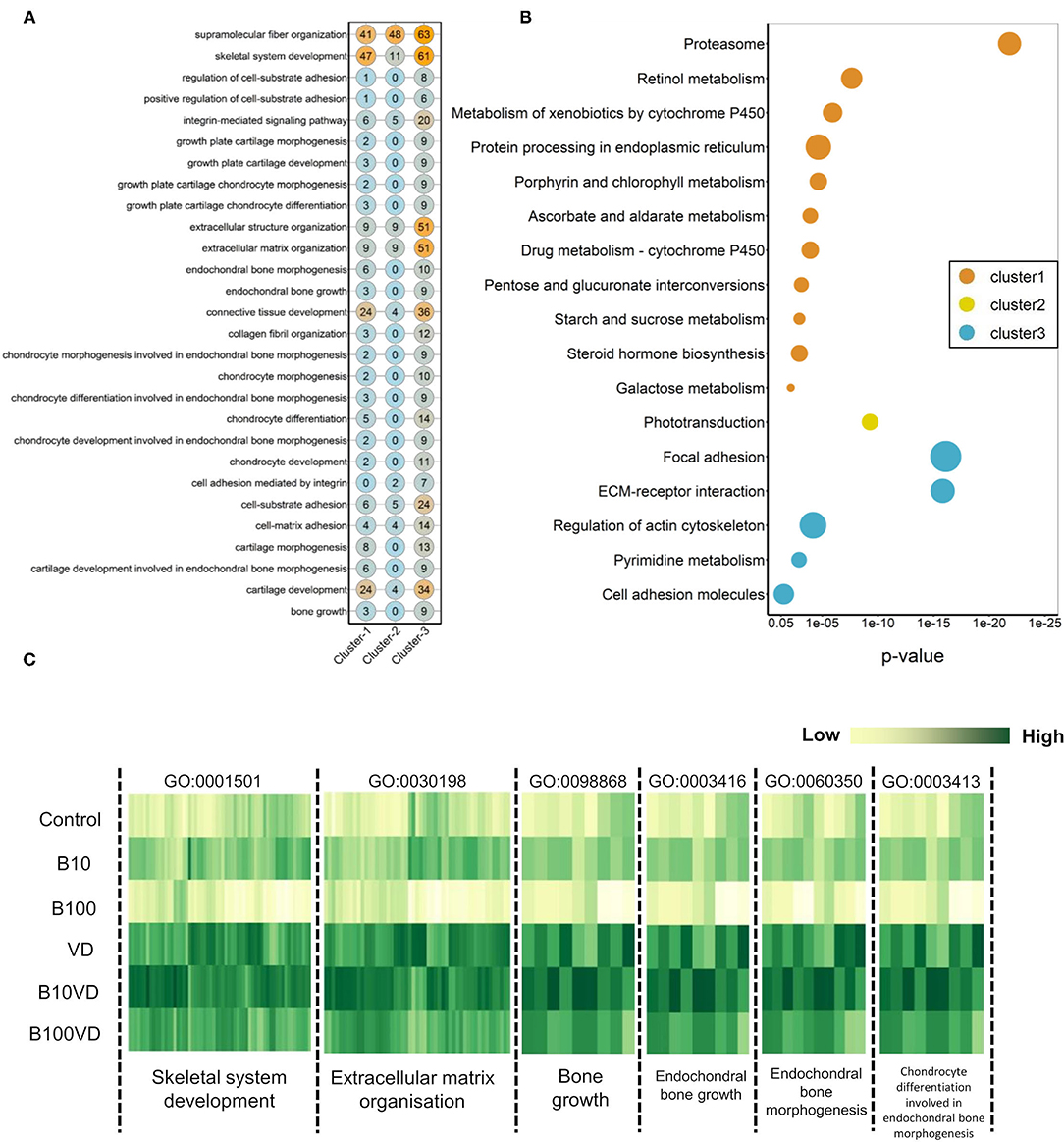
Figure 5. (A) Matrix visualizing the number of DEGs falling under enriched GO terms involved in skeletal system development. Y-axis highlights the key enriched GO terms, and X-axis describes the cluster number. The numbers within each circle indicate the total genes falling within that GO term in each cluster. (B) Bubble plot visualizing KEGG pathways significantly enriched (p < 0.05) across the three different clusters. Color of the bubbles indicates the cluster they fall into, and size of the bubble indicates the number of genes. (C) Heatmap visualizing normalized expression values of genes contributing to enriched skeletal GO terms in the cluster 3. Y-axis highlights the different treatment groups, and X-axis describes the enriched GO terms.
A total of 354 genes related to bone and skeletal system were identified from the 28 GO terms listed in Figure 5A. In comparison to control, 101 DEGs for VD, 126 DEGs for B10VD, and 64 DEGs for B100VD were found. Since we observed more DEGs with B10VD, which is the synergy group with less concentration of B, downstream analysis was focused on B10VD alone. A total of 55 genes (3 downregulated and 52 upregulated) were commonly differentially expressed for B10VD and VD compared to control. In addition, 71 DEGs were unique for B10VD and 46 were specific for VD. Some important genes like col1a1a and ucmab were common DEGs among B10VD and VD, whereas dcn was upregulated only in B10VD. The osteoclast marker gene ctsk (cathepsin K), on the contrary, was found to be upregulated only in VD and not in B10VD.
We further decided to investigate synergy by exploring the KEGG pathways that were not enriched in the current analysis but were key for bone and skeletal development in zebrafish. KEGG pathway maps for mitogen-activated protein kinase (MAPK) (dre04010), transforming growth factor beta (TGF-β) (dre04350), focal adhesion (dre0510), WNT signaling pathway (dre04310), and Ca signaling (dre04020) were generated incorporating the log FC data for B10VD, VD treatments in contrast to control (Supplementary Figure 2). Expression patterns of the genes from these pathways further confirmed B10VD synergy to be more effective in supporting bone and skeletal development. Key candidate genes from these pathways such as cacn3b, egfra, mapk14b, mras, ppp3cca, and rps6ka3b were confirmed for their upregulation in B10VD than VD or B100VD (Supplementary Table 3; Supplementary Figure 2).
Given their importance for osteogenesis and mineralogenesis, eight marker genes (cacn3b, dusp2, egfra, hspb1, mapk14b, mrasb, ppp3cca, rps6ka3b) involved in the MAPK pathway were selected for validation of the transcriptomic data by RT-PCR. As shown in Supplementary Figure 3, the relative FC in RT-PCR was consistent with RNA-Seq results, suggesting that the transcript identification and quantification were extremely consistent between the two techniques. Most genes from RNA-Seq analysis were in good accordance with the expression intensities by RT-PCR, although the result was dissimilar for mapk14b in the VD group, probably due to the difference in sensitivity of each technique.
Since B10VD is the synergy group with a lower concentration of B, yet with more DEGs, to further investigate the synergy effect at various stages of skeletal development, additional analysis was performed at multiple time points using two zebrafish fluorescent reporter lines, one expressing mCherry under the control of the medaka (Oryzias latipes) sp7 (osterix) promoter, the other expressing EGFP downstream to the promoter of medaka bglap (osteocalcin) (Figures 6A,B). Areas of the operculum and sp7+ and bglap+ areas, showing early and mature osteoblasts, respectively, were measured (Figures 6C,D). Synergy group (B10VD) exhibits the largest mineralized area of the operculum as well as the largest sp7+ and bglap+ areas at all the time points analyzed (Figures 6C,D). The sp7+ area normalized with the head area (sp7+A/HA) was significantly higher in VD and B10VD with respect to control at 6 dpf, whereas at 9 dpf only the synergy group showed a significantly higher fluorescence signal with respect to control. At 12 and 15 dpf, B10VD remained the group with the largest sp7+ area, but significant differences were only found with the B group at 12 dpf. No differences between the control and B groups were detected throughout the experiment (Figure 6C). GFP fluorescence marking mature osteoblasts normalized with head area (bglap+A/HA) showed a significant increase for the synergy group compared to every other group at 15 dpf. Importantly, this time point was characterized by the strongest signal among all end points studied. Synergy group (B10VD) also showed a significantly larger bglap+ area than control at every time point studied, whereas VD showed a larger bglap+ area than control at all time points except 15 dpf. Synergy group also showed a significantly stronger signal compared to VD or B alone at 15 dpf. In accordance with what was observed for sp7 expression pattern, no differences were detected between control and B at any of the end points evaluated. Except VD, all other groups showed a general increase of bglap+ area from 6 to 15 dpf (Figure 6C).
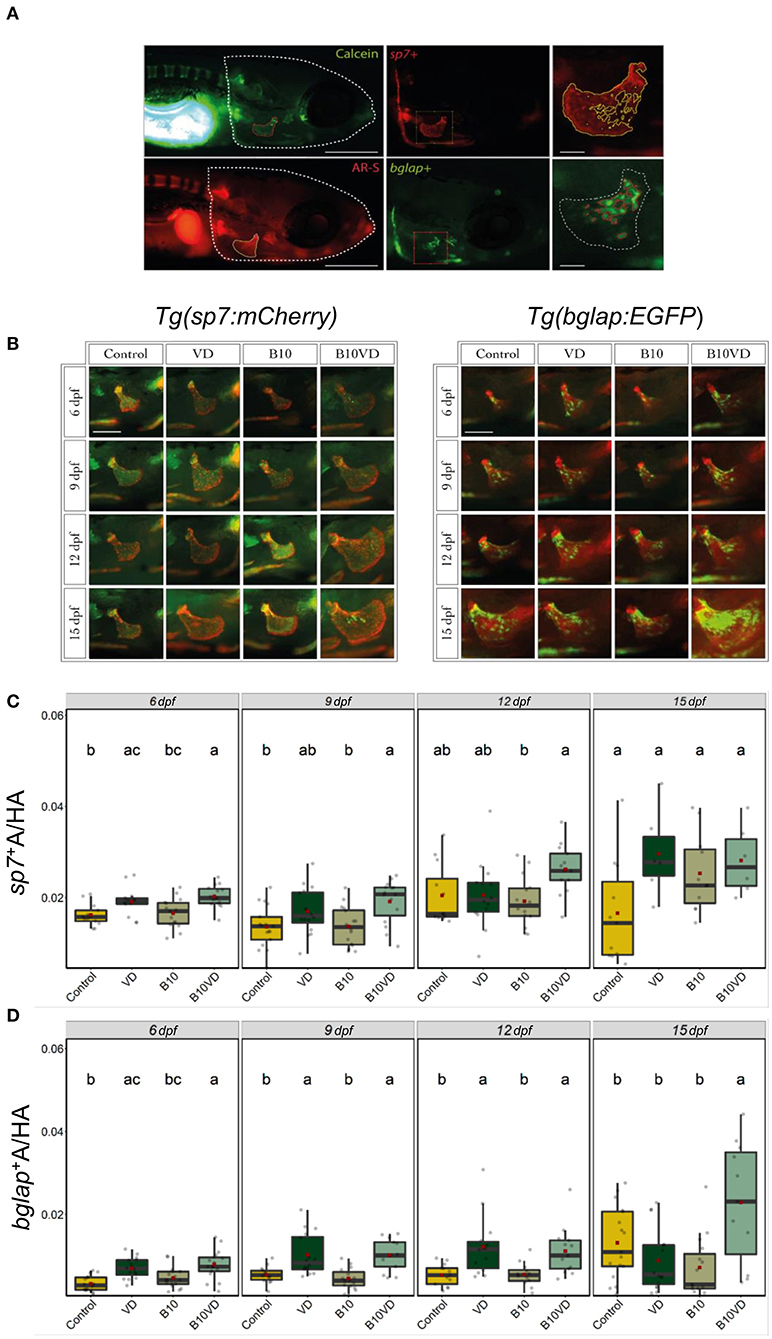
Figure 6. Early and mature osteoblasts respectively tracked by the fluorescence expression of Tg(sp7:mCherry) and Tg(bglap:EGFP). Four groups of treatment are ethanol at 0.1% (Control), VD group with VD3 at 10 pg/ml (VD), B at 10 ng/ml (B10), B at 10 ng/ml with VD3 at 10 pg/ml (B10VD). (A) An example of how the areas of the operculum and sp7+ and bglap+ were measured (big scale bar = 0.35 mm; small bar = 0.05 mm). (B) A table with merged pictures of operculum from the sp7:mCherry line stained with calcein and the bglap:EGFP line stained with AR-S (scale bar = 0.17 mm). (C) The sp7+ area inside the operculum was normalized with the total head area at time points 6, 9, 12, and 15 dpf. (D) The bglap+ area inside the operculum was normalized with the total head area at time points 6, 9, 12, and 15 dpf. One-way ANOVA and Tukey's post-hoc test were used. Data are presented as means ± SD, and different letters represent statistical significance at p < 0.05.
Morphometric assessment of the mineralizing opercular bone in early-stage zebrafish larvae was previously described as a suitable tool for the screening of osteogenic compounds (38, 55). The pro-osteogenic properties of VD are known since a long time but the possible synergistic actions of VD with micronutrients to promote bone mineralization are understood less. Here we tested two concentrations of B, one concentration of VD and their respective combinations, and observed the induction of osteogenic effect as observed by increased mineralized operculum areas in both the synergy groups compared to the supplementation of VD alone but even more pronounced effect when compared to the control and the B treatments alone. Following this preliminary screening, and with the scope of exploring the molecular determinants of the observed phenotype, we analyzed the transcriptome of the treated larvae to obtain a large multivariate dataset of DEGs with particular attention given to pathways associated with the skeletal system. The PAM clustering approach was previously proven to be appropriate to cluster and define expression patterns among the DEGs (56) and the same was observed here, where a dominance of skeletal- or bone-related pathways was observed in one out of the three clusters. This cluster of interest (C3) had a specific pattern for groups with VD, where VD and its synergy groups stayed in the high-expression side, whereas control and groups with B stayed in the low-expression side. Interestingly, when looking at the expression patterns of genes contributing to enriched skeletal function GO terms, only B10VD showed higher expression than the VD group, suggesting a potential role that the synergy between VD and B could be playing in inducing the pro-mineralogenic effect observed. In addition, transcriptomic responses showed that more genes with positive effects on the skeleton were highly expressed in the combination group of VD with a lower concentration of B (B10VD) than the other synergy group with a higher concentration of B (B100VD). Thus, on a molecular level, 10 ng/ml B with VD showed the strongest positive effect on metabolic pathways associated with skeletal development.
Among B10VD and VD, there were common DEGs, including col1a1a, expressed in developing bony elements and ectoderm, and ucmab, which play a pivotal role in zebrafish skeletal development (57, 58). Some important genes were expressed only for B10VD such as dcn, which encodes for an important ECM glycoprotein that has a role in regulation of bone mass by modulating TGF-β activity (59). The most affected pathway was observed to be the MAPK signaling, where many genes were found to be overexpressed in the synergy groups compared to VD. RSK2 (encoded by rps6ka3a gene) was shown to be highly upregulated in the B10VD group relative to B100VD and VD groups, which is particularly relevant, given its importance for osteoblast differentiation and function (60). It is in fact involved at the distal end of the MAPK pathway, where it plays a key role in bone turnover by phosphorylating different substrates, such as cAMP response element-binding protein, which is a known inducer of osteoblast differentiation and c-Fos, which is an osteoclast differentiation inducer (61–63). C-Fos was found to be downregulated in both VD and B10VD, but osteoclast marker gene ctsk was upregulated only in VD. This indicates that the synergy group possibly has a reduced VD-induced osteoclast formation through VD receptors in the nuclei as found in previous studies (64). The synergy groups also showed significant upregulation for p38, which is involved in the runx2 activation (65), giving further evidence to the pro-osteogenic effect of B and VD synergy in the induction of osteoblast differentiation. Fgfr4 receptor, a gene in the ERK signaling pathway that is involved in the runx2 transcriptional activation, was also found to be highly upregulated in the synergy groups, pointing out at a possibility of runx2 regulation, and thereby to be involved in the differentiation and proliferation of osteoblasts (66, 67). Although runx2 was not one of the DEGs observed here and that could be due to the specific skeletal development stage of the larvae under study. Fgfr4, being the receptor for fgf6, also plays a pivotal role in osteoblast and osteoclast differentiation (68). The calcineurin, which is part of multiple bone-related pathways such as MAPK, WNT, and Ca signaling, was found to be upregulated in the B10VD synergy group. Calcineurin is known to be expressed in osteoblasts and enhances the differentiation of osteoblasts, thereby increasing bone formation (69). The focal adhesion pathway was also regulated in both the synergy groups where fibronectin 1 (fn1) was highly upregulated. fn1, through activation of the WNT pathway, stimulates osteoblast differentiation and ECM mineralization as observed in previous in vitro studies, and it is known to be produced by osteoblasts during bone generation (70, 71). Additionally, fn1 was found to play a role in osteoblast compaction through fibronectin fibrillogenesis cell-mediated matrix assembly, which is crucial for the bone ECM mineralization mediated by osteoblast (72, 73). In the KEGG analysis of the TGF-β/BMP pathway, smad4 was largely upregulated in the B10VD group. Proteins belonging to the Smad family are associated with the BMP pathway, whose activation is paramount for bone mineralization and osteoblast differentiation (74, 75). The transcriptomic analysis clearly evidenced the upregulation of genes involved in bone formation by osteoblast differentiation, ECM formation, and mineralization in the synergy groups, particularly in B10VD, with a higher effect than VD treated alone.
From the combined morphological and transcriptomic data, we decided to further investigate the synergy of VD with B at a lower concentration, 10 ng/ml. To explore the effect of the B-VD synergy on bone development in a time-dependent manner, cellular dynamics of intermediate and late osteoblasts, as labeled by sp7 and bglap expression, respectively, were verified. Sp7 is a highly conserved, zinc finger-containing transcription factor essential within the stepwise genetic program regulating osteoblast differentiation. It is required for the activation of a repertoire of genes directly associated to osteoblast maturity and bone ECM formation and mineralization such as bglap, spp1, col10a1a/b, and sparc (76, 77). In zebrafish, sp7 expression is considered to be labeling intermediate stages of osteoblast differentiation and tends to be downregulated in mature osteoblasts (76). It is first observed at the time of the onset of the primary cranial ossified structures (36 hpf), with its expression pattern perfectly co-localizes with the whole mineralized domain of the bone (76). In contrast, Osteocalcin (Bglap) is a small osteoblast-secreted protein well accepted as a marker of mature and matrix-secreting osteoblasts (78). Plenty of evidence suggests that VD exerts a direct stimulating effect on osteoblasts proliferation and differentiation through the VD receptor in humans and other mammals (79). Similarly, it has been previously observed in zebrafish that treatment with VD is able to increase the expression of sp7 and blglap at very early stages of skeletal development (6 dpf) (38). Accordingly, in this study a significant increase in the area of sp7+ cells within the opercular bone was observed for VD alone and when in combination with B at early stages of opercular development (6 dpf), but only the synergy group increased the amount of intermediate osteoblast (sp7+) populating the operculum at 9 dpf. Similarly, when looking at more advanced (bglap+) osteoblasts stages, both VD alone and synergy group increased bglap+ osteoblasts in early stages (6, 9, and 12 dpf), but the synergy group extended the induction of mature osteoblast up to 15 dpf. As a result, larger mineralized opercular bone was observed in the synergy group at 15 dpf. Overall, B administered alone was not able to affect the intermediate or mature osteoblast populations and the effect was observed only in synergy with VD.
In conclusion, our study indicates that B may be able to potentiate the osteoblast-stimulating effect of VD or exert its pro-osteogenic effect in a VD-dependent manner. Our transcriptome analysis suggests that these effects could be through the induction of molecular programs involving the activation of MAPK and TGF-β/BMP pathways. These findings could lead to a promising approach in developing medicines to alleviate VD deficiency by synergically adding micronutrients along with VD to increase the overall efficiency of the treatment. In a world where improving bone health has become a matter of crucial importance, there is a high translational value for the findings of this study in the development of prophylactic measures focusing on improving VD supplementation efficiency in nutrition. Future studies should explore other outcomes of the combinatorial treatments such as its effects on stress or immunity to have a complete overview.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: All the sequence data can be accessed through NCBI Bioproject - PRJNA796753.
The experiment involving wildtype zebrafish were conducted in accordance with the Italian law on animal experimentation and were approved by the Ethics Committee of the Università Politecnica delle Marche, Ancona, Italy and by the Italian Ministry of Health (Aut. No. 583/2020-PR). All the experimental procedures involving transgenic zebrafish followed the EU Directive 2010/63/EU and Portuguese legislation - Decreto-Lei 113/2013 - for animal experimentation and welfare. Animal handling and experiments were performed by qualified operators accredited by the Portuguese Direção-Geral de Alimentação e Veterinária (DGAV) under authorization no. 012769/2021.
JS: conceptualization, methodology, data acquisition and analysis, writing—original draft, and writing—review and editing. MG: data analysis, methodology, and writing—review and editing. AC: data acquisition and analysis, methodology, and writing—review and editing. VG: data acquisition and analysis. PG: supervision and writing—review and editing. FM: conceptualization, supervision, and writing—review and editing. OC: conceptualization, supervision, writing—review and editing, and funding acquisition. All authors contributed to the article and approved the submitted version.
This work is part of a project that has received funding from the European Union's Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie Grant Agreement No. 766347 to OC and PG. Further funding was received through Portuguese national funds from FCT - Foundation for Science and Technology through projects UIDB/04326/2020, UIDP/04326/2020, and LA/P/0101/2020.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Dr. Luca Marisaldi for the valuable comments on the study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.868805/full#supplementary-material
1. McCoy H, Irwin A, Kenney MA, Williams L. Univ of A. Effects of boron supplements on bones from rats fed calcium and magnesium deficient diets. FASEB J. (1991) 5:5. Available online at: https://www.osti.gov/biblio/5213312 (accessed December 17, 2019).
2. Gallardo-Williams MT, Maronpot RR, Turner CH, Johnson CS, Harris MW, Jayo MJ, et al. Effects of boric acid supplementation on bone histomorphometry, metabolism, and biomechanical properties in aged female F-344 rats. Biol Trace Elem Res. (2003) 93:155–69. doi: 10.1385/BTER:93:1-3:155
3. Nielsen FH. Dietary fat composition modifies the effect of boron on bone characteristics and plasma lipids in rats. BioFactors. (2004) 20:161–71. doi: 10.1002/biof.5520200305
4. Nielsen FH. Studies on the relationship between boron and magnesium which possibly affects the formation and maintenance of bones. Magnes Trace Elem. (1990) 9:61–9.
5. Elliot MA, Edwards HM. Studies to determine whether an interaction exists among boron, calcium, and cholecalciferol on the skeletal development of broiler chickens. Poult Sci. (1992) 71:677–90. doi: 10.3382/ps.0710677
6. Meacham SL, Taper LJ, Volpe SL. Effect of boron supplementation on blood and urinary calcium, magnesium, and phosphorus, and urinary boron in athletic and sedentary women. Am J Clin Nutr. (1995) 61:341–5. doi: 10.1093/ajcn/61.2.341
7. Naghii MR, Samman S. The effect of boron on plasma testosterone and plasma lipids in rats. Nutrition Research. (1997) 17:523–31. doi: 10.1016/S0271-5317(97)00017-1
8. Kurtoglu V, Kurtoglu F, Coşkun B. Effects of boron supplementation of adequate and inadequate vitamin D3-containing diet on performance and serum biochemical characters of broiler chickens. Res Vet Sci. (2001) 71:183–7. doi: 10.1053/rvsc.2001.0517
9. Devirian TA, Volpe SL. The physiological effects of dietary boron. Crit Rev Food Sci Nutr. (2003) 43:219–31. doi: 10.1080/10408690390826491
10. Naghii MR, Torkaman G, Mofid M. Effects of boron and calcium supplementation on mechanical properties of bone in rats. BioFactors. (2006) 28:195–201. doi: 10.1002/biof.5520280306
11. Jugdaohsingh R, Pedro LD, Watson A, Powell JJ. Silicon and boron differ in their localization and loading in bone. Bone Reports. (2015) 1:9–15. doi: 10.1016/j.bonr.2014.10.002
12. Brown TF, McCormick ME, Morris DR, Zeringue LK. Effects of dietary boron on mineral balance in sheep. Nutr Res. (1989) 9:503–12. doi: 10.1016/S0271-5317(89)80175-7
13. Hegsted M, Keenan MJ, Siver F, Wozniak P. Effect of boron on vitamin D deficient rats. Biol Trace Elem Res. (1991) 28:243–55. doi: 10.1007/BF02990471
14. Rossi AF, Miles RD, Damron BL, Flunker LK. Effects of dietary boron supplementation on broilers. Poult Sci. (1993) 72:2124–30. doi: 10.3382/ps.0722124
15. Chapin RE, Ku WW, Kenney MA, Mccoy H, Gladen B, Wine RN, et al. The effects of dietary boron on bone strength in rats. Toxicol Sci. (1997) 35:205–15. doi: 10.1093/toxsci/35.2.205
16. Chapin RE, Ku WW, Kenney MA, McCoy H. The effects of dietary boric acid on bone strength in rats. Biol Trace Elem Res. (1998) 66:395–9. doi: 10.1007/BF02783150
17. Wilson JH, Ruszler PL. Effects of boron on growing pullets. Biol Trace Elem Res. (1997) 56:287–94. doi: 10.1007/BF02785300
18. Wilson JH, Ruszler PL. Long term effects of boron on layer bone strength and production parameters. Br Poult Sci. (1998) 39:11–5. doi: 10.1080/00071669889312
19. Armstrong TA, Spears JW. Effect of dietary boron on growth performance, calcium and phosphorus metabolism, and bone mechanical properties in growing barrows. J Anim Sci. (2001) 79:3120–7. doi: 10.2527/2001.79123120x
20. Gorustovich AA, Steimetz T, Nielsen FH, Guglielmotti MB. A histomorphometric study of alveolar bone modelling and remodelling in mice fed a boron-deficient diet. Arch Oral Biol. (2008) 53:677–82. doi: 10.1016/j.archoralbio.2008.01.011
21. Nielsen FH, Stoecker BJ. Boron and fish oil have different beneficial effects on strength and trabecular microarchitecture of bone. J Trace Elem Med Biol. (2009) 23:195–203. doi: 10.1016/j.jtemb.2009.03.003
22. Dessordi R, Spirlandeli AL, Zamarioli A, Volpon JB, Navarro AM. Boron supplementation improves bone health of non-obese diabetic mice. J Trace Elem Med Biol. (2017) 39:169–75. doi: 10.1016/j.jtemb.2016.09.011
23. Zhu, Ansari AR, Xiao k Wang W, Wang L Qiu W, Zheng X Song H Liu H, Zhong J et al. Boron supplementation promotes osteogenesis of tibia by regulating the bone morphogenetic protein-2 expression in african ostrich chicks. Biol Trace Elem Res. (2020) 199:1544–55. doi: 10.1007/s12011-020-02258-w
24. Scorei ID, Scorei RI. Calcium fructoborate helps control inflammation associated with diminished bone health. Biol Trace Elem Res. (2013) 155:315–21. doi: 10.1007/s12011-013-9800-y
25. Johnson EW, Jayroe LM. Prevention Treatment of Osteochondrosis in Animals Humans. (2009). Available online at: https://patents.google.com/patent/US20090214500A1/en (accessed January 3, 2021).
26. Gölge UH, Kaymaz B, Arpaci R, Kömürcü E, Göksel F, Güven M, et al. Effects of boric acid on fracture healing: an experimental study. Biol Trace Elem Res. (2015) 167:264–71. doi: 10.1007/s12011-015-0326-3
27. Hakki SS, Bozkurt BS, Hakki EE. Boron regulates mineralized tissue-associated proteins in osteoblasts (MC3T3-E1). J Trace Elem Med Biol. (2010) 24:243–50. doi: 10.1016/j.jtemb.2010.03.003
28. Ying X, Cheng S, Wang W, Lin Z, Chen Q, Zhang W, et al. Effect of boron on osteogenic differentiation of human bone marrow stromal cells. Biol Trace Elem Res. (2011) 144:306–15. doi: 10.1007/s12011-011-9094-x
29. Gümüşderelioglu M, Tunçay EÖ, Kaynak G, Demirtaş TT, Aydin ST, Hakki SS. Encapsulated boron as an osteoinductive agent for bone scaffolds. J Trace Elem Med Biol. (2015) 31:120–8. doi: 10.1016/j.jtemb.2015.03.008
30. Bussell ME. Improving bone health: addressing the burden through an integrated approach. Aging Clin Exp Res. (2021) 33:2777–86. doi: 10.1007/s40520-021-01971-3
31. Kanis JA, Norton N, Harvey NC, Jacobson T, Johansson H, Lorentzon M, et al. 2021: a new scorecard for osteoporosis in Europe. Arch Osteoporos. (2021) 16:82. doi: 10.1007/s11657-020-00871-9
32. Dupre JN, Keenan MJ, Hegsted M, Brudevold AM. Effects of dietary boron in rats fed a vitamin D-deficient diet. Environ Health Perspect. (1994) 102:55–8. doi: 10.1289/ehp.94102s755
33. Hunt CD, Herbel JL, Idso JP. Dietary boron modifies the effects of vitamin D3 nutrition on indices of energy substrate utilization and mineral metabolism in the chick. J Bone Mineral Res. (1994) 9:171–82. doi: 10.1002/jbmr.5650090206
34. Miljkovic D, Miljkovic N, McCarty MF. Up-regulatory impact of boron on vitamin D function—does it reflect inhibition of 24-hydroxylase? Med Hypotheses. (2004) 63:1054–6. doi: 10.1016/j.mehy.2003.12.053
35. Fernández I, Gavaia PJ, Laizé V, Cancela ML. Fish as a model to assess chemical toxicity in bone. Aquat Toxicol. (2018) 194:208–26. doi: 10.1016/j.aquatox.2017.11.015
36. Valenti MT, Marchetto G, Mottes M, Dalle Carbonare L. Zebrafish: a suitable tool for the study of cell signaling in bone. Cells. (2020) 9:1911. doi: 10.3390/cells9081911
37. Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. (2002) 108:17–29. doi: 10.1016/S0092-8674(01)00622-5
38. Tarasco M, Laizé V, Cardeira J, Cancela ML, Gavaia PJ. The zebrafish operculum: a powerful system to assess osteogenic bioactivities of molecules with pharmacological and toxicological relevance. Comp Biochem Physiol Part C: Toxicol Pharmacol. (2017) 197:45–52. doi: 10.1016/j.cbpc.2017.04.006
39. Bensimon-Brito A, Cardeira J, Dionísio G, Huysseune A, Cancela ML, Witten PE. Revisiting in vivo staining with alizarin red S—a valuable approach to analyse zebrafish skeletal mineralization during development and regeneration. BMC Dev Biol. (2016) 16:2. doi: 10.1186/s12861-016-0102-4
40. Tarasco M, Cordelières FP, Cancela ML, Laizé V. ZFBONE An ImageJ toolset for semi-automatic analysis of zebrafish bone structures. Bone. (2020) 138:115480. doi: 10.1016/j.bone.2020.115480
41. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. Ultrafast universal RNA-seq aligner. Bioinformatics. (2013) 29:15–21. doi: 10.1093/bioinformatics/bts635
42. Liao Y, Smyth GK, Shi W. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. (2014) 30:923–30. doi: 10.1093/bioinformatics/btt656
43. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
44. R Core Team. R: A Language Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing (2019). Available online at: https://www. R-project. org. (accessed December 24, 2021).
45. Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, et al. gplots: Various R Programming Tools for Plotting Data. (2020). Available online at: https://CRAN.R-project.org/package=gplots (accessed December 12, 2021).
46. Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. New York: Wiley (2009).
47. Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K. cluster: Cluster Analysis Basics and Extensions. R package version 210. Berlin: Springer (2019).
48. Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. J R Stat Soc Ser B Stat Methodol. (2001) 63:411–23. doi: 10.1111/1467-9868.00293
49. Kassambara A, Mundt F. Factoextra: Extract Visualize the Results of Multivariate Data Analyses (R Package Version 1.0. 6). (2020). Available online at: https://CRAN.R-project.org/package=factoextra.
51. Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. (2012) 16:284–7. doi: 10.1089/omi.2011.0118
53. Briatte F. ggnetwork: Geometries to Plot Networks with'ggplot2'. R package version 05, 1 (2016).
54. Carnevali O, Notarstefano V, Olivotto I, Graziano M, Gallo P, Di Marco Pisciottano I, et al. Dietary administration of EDC mixtures: a focus on fish lipid metabolism. Aquat Toxicol. (2017) 185:95–104. doi: 10.1016/j.aquatox.2017.02.007
55. Bergen DJM, Kague E, Hammond CL. Zebrafish as an emerging model for osteoporosis: a primary testing platform for screening new osteo-active compounds. Front Endocrinol. (2019) 10:6. doi: 10.3389/fendo.2019.00006
56. Martínez R, Esteve-Codina A, Herrero-Nogareda L, Ortiz-Villanueva E, Barata C, Tauler R, et al. Dose-dependent transcriptomic responses of zebrafish eleutheroembryos to Bisphenol A. Environ Pollut. (2018) 243:988–97. doi: 10.1016/j.envpol.2018.09.043
57. Gistelinck C, Gioia R, Gagliardi A, Tonelli F, Marchese L, Bianchi L, et al. Zebrafish collagen type i: molecular and biochemical characterization of the major structural protein in bone and skin. Sci Rep. (2016) 6:21540. doi: 10.1038/srep21540
58. Neacsu CD, Grosch M, Tejada M, Winterpacht A, Paulsson M, Wagener R, et al. Ucmaa (Grp-2) is required for zebrafish skeletal development. Evidence for a functional role of its glutamate γ-carboxylation. Matrix Biol. (2011) 30:369–78. doi: 10.1016/j.matbio.2011.07.002
59. Bi Y, Stuelten CH, Kilts T, Wadhwa S, Iozzo RV, Robey PG, et al. Extracellular matrix proteoglycans control the fate of bone marrow stromal cells*. J Biol Chem. (2005) 280:30481–9. doi: 10.1074/jbc.M500573200
60. Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology: implication for coffin-lowry syndrome. Cell. (2004) 117:387–98. doi: 10.1016/S0092-8674(04)00344-7
61. Nottmeier C, Decker MG, Luther J, von Kroge S, Kahl-Nieke B, Amling M, et al. Accelerated tooth movement in Rsk2-deficient mice with impaired cementum formation. Int J Oral Sci. (2020) 12:1–8. doi: 10.1038/s41368-020-00102-4
62. Wang Z-Q, Ovitt C, Grigoriadis AE, Möhle-Steinlein U, Rüther U, Wagner EF. Bone and haematopoietic defects in mice lacking c-fos. Nature. (1992) 360:741–5. doi: 10.1038/360741a0
63. Xing J, Ginty DD, Greenberg ME. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science. (1996) 273:959–63. doi: 10.1126/science.273.5277.959
64. Suda T, Ueno Y, Fujii K, Shinki T. Vitamin D and bone. J Cell Biochem. (2003) 88:259–66. doi: 10.1002/jcb.10331
65. Lee K-S, Hong S-H, Bae S-C. Both the Smad and p38 MAPK pathways play a crucial role in Runx2 expression following induction by transforming growth factor-β and bone morphogenetic protein. Oncogene. (2002) 21:7156–63. doi: 10.1038/sj.onc.1205937
66. Cool S, Jackson R, Pincus P, Dickinson I, Nurcombe V. Fibroblast growth factor receptor 4 (FGFR4) expression in newborn murine calvaria and primary osteoblast cultures. Int J Dev Biol. (2002) 46:519–23.
67. Kawane T, Qin X, Jiang Q, Miyazaki T, Komori H, Yoshida CA, et al. Runx2 is required for the proliferation of osteoblast progenitors and induces proliferation by regulating Fgfr2 and Fgfr3. Sci Rep. (2018) 8:13551. doi: 10.1038/s41598-018-31853-0
68. Pawlikowski B, Vogler TO, Gadek K, Olwin BB. Regulation of skeletal muscle stem cells by fibroblast growth factors. Dev Dyn. (2017) 246:359–67. doi: 10.1002/dvdy.24495
69. Sun L, Blair HC, Peng Y, Zaidi N, Adebanjo OA, Wu XB, et al. Calcineurin regulates bone formation by the osteoblast. PNAS. (2005) 102:17130–5. doi: 10.1073/pnas.0508480102
70. Bentmann A, Kawelke N, Moss D, Zentgraf H, Bala Y, Berger I, et al. Circulating fibronectin affects bone matrix, whereas osteoblast fibronectin modulates osteoblast function. J Bone Mineral Res. (2010) 25:706–15. doi: 10.1359/jbmr.091011
71. Yang C, Wang C, Zhou J, Liang Q, He F, Li F, et al. Fibronectin 1 activates WNT/β-catenin signaling to induce osteogenic differentiation via integrin β1 interaction. Lab Investig. (2020) 100:1494–502. doi: 10.1038/s41374-020-0451-2
72. Brunner M, Millon-Frémillon A, Chevalier G, Nakchbandi IA, Mosher D, Block MR, et al. Osteoblast mineralization requires β1 integrin/ICAP-1–dependent fibronectin deposition. J Cell Biol. (2011) 194:307–22. doi: 10.1083/jcb.201007108
73. Sens C, Huck K, Pettera S, Uebel S, Wabnitz G, Moser M, et al. Fibronectins containing extradomain A or B enhance osteoblast differentiation via distinct integrins. J Biol Chem. (2017) 292:7745–60. doi: 10.1074/jbc.M116.739987
74. Chen G, Deng C, Li Y-P. TGF-β and BMP Signaling in osteoblast differentiation and bone formation. Int J Biol Sci. (2012) 8:272–88. doi: 10.7150/ijbs.2929
75. Karner CM, Lee S-Y, Long F. Bmp induces osteoblast differentiation through both Smad4 and mTORC1 Signaling. Mol Cell Biol. (2017) 37:e00253-16. doi: 10.1128/MCB.00253-16
76. Liu Q, Li M, Wang S, Xiao Z, Xiong Y, Wang G. Recent advances of osterix transcription factor in osteoblast differentiation and bone formation. Front Cell Dev Biol. (2020) 8:1575. doi: 10.3389/fcell.2020.601224
77. Niu P, Zhong Z, Wang M, Huang G, Xu S, Hou Y, et al. Zinc finger transcription factor Sp7/Osterix acts on bone formation and regulates col10a1a expression in zebrafish. Sci Bull. (2017) 62:174–84. doi: 10.1016/j.scib.2017.01.009
78. Rutkovskiy A, Stensløkken K-O, Vaage IJ. Osteoblast differentiation at a glance. Med Sci Monit Basic Res. (2016) 22:95–106. doi: 10.12659/MSMBR.901142
Keywords: bone, boron, vitamin D, micronutrients, zebrafish, RNA-Seq, transgenic lines, cell differentiation
Citation: Sojan JM, Gundappa MK, Carletti A, Gaspar V, Gavaia P, Maradonna F and Carnevali O (2022) Zebrafish as a Model to Unveil the Pro-Osteogenic Effects of Boron-Vitamin D3 Synergism. Front. Nutr. 9:868805. doi: 10.3389/fnut.2022.868805
Received: 03 February 2022; Accepted: 02 March 2022;
Published: 29 April 2022.
Edited by:
Yan Huang, University of Arkansas, United StatesReviewed by:
Yang He, Baylor College of Medicine, United StatesCopyright © 2022 Sojan, Gundappa, Carletti, Gaspar, Gavaia, Maradonna and Carnevali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oliana Carnevali, by5jYXJuZXZhbGlAc3RhZmYudW5pdnBtLml0; Francesca Maradonna, Zi5tYXJhZG9ubmFAc3RhZmYudW5pdnBtLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.