- 1Hubei Key Laboratory of Animal Nutrition and Feed Science, Wuhan Polytechnic University, Wuhan, China
- 2State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, Institute of Agro-Product Safety and Nutrition, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 3Institute of Food Sciences, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
Sucralose is a non-nutritive artificial sweetener (NNS) used in foods or beverages to control blood glucose levels and body weight gain. The consumption of NNS has increased in recent years over the world, and many researches have indicated long-term sucralose administration altered the gut microbiome composition of mice. These studies all focus on the US Food and Drug Administration (FDA) defined acceptable daily intake (ADI), approximately 5 mg/kg BW/day for human. In our study, mice were given with T1-4 (0.0003, 0.003, 0.03, and 0.3 mg/mL) of sucralose, respectively, Control group mice were given normal water. In particular, 0.3 mg/mL of sucralose was equal to the ADI (5 mg/kg BW/day). After 16 weeks, all mice were weighted and sacrificed, the liver of each mouse was isolated and weighed, segments of jejunum, ileum and colon were collected for H&E-stained. The contents of jejunum, ileum, cecum and colon were collected for 16S rRNA gene sequencing. The results showed sucralose administration affects the intestinal barrier function evidenced by distinct lymphocyte aggregation in ileum and colon while not change the mice body weight. The 16S rRNA gene sequencing of the mice gut microbiome suggested sucralose administration significantly changed the composition of gut microbiota, especially in T1 and T4 group. For example, a reduction of probiotics abundance (Lachnoclostridium and Lachnospiraceae) was found in cecum of T4 group mice compared with Control group. On the other hand, Allobaculum, which was reported positively correlated with diabetes, was increased in the T1 and T4 group. In addition, the potential pathogens, including Tenacibaculum, Ruegeria, Staphylococcus were also increased in jejunum, ileum and colon by sucralose administration in T1 and T4 group. These new findings indicate that low dose of sucralose (T1) alter gut microbiome in mice, and these adverse health effects are equal to ADI level (T4). Overall, our study provides guidance and suggestions for the use of sucralose in foods and beverages.
Introduction
Global consumption of sugar-free foods is increasing. Non-nutritional sweeteners (NNS) added to beverages and foods are defined as sweetener with higher sweetness and lower calorie content than caloric or nutritional sweeteners (such as sucrose or corn syrup) (1). Sucralose also named trichlorogalactosucrose and TGS, is a NNS, zero-calorie artificial sweetener (2). It is a substitute for chlorinated sugar, and its sweetness is 600 times than sucrose, because of its low production cost, high thermal stability and solubility, sucralose has become an important sugar substitute in foods and beverage (3, 4). US Food and Drug Administration (FDA) defined acceptable daily intake (ADI) approximately 5 mg/kg BW/day for human (5, 6). The adverse health effects of sucralose have been highly argued over the years. For example, a large number of early studies have shown that most of the ingested sucralose will not be absorbed and metabolized by the body, and it will not change with gut peristalsis (7, 8). However, researches have confirmed that sucralose can change the composition of gut microbiome, inhibiting intestinal development, and aggravating HFD-induced hepatic steatosis in adulthood (5, 9).
Gut microbiome refers to the complex community of microorganisms living in the digestive tract of human and animals, its number is about 10 times than our body cells (10). The balance between host and gut microbiome is essential to maintain a healthy gut barrier and optimal immune homeostasis, which helps to prevent the occurrence of diseases (11, 12). Gut microbiome contribute to the metabolic health of the human host, when aberrant, it will cause the pathogenesis of various common metabolic disorders including obesity, type 2 diabetes, non-alcoholic liver disease, cardio-metabolic diseases and malnutrition (13). Related research used fecal samples from Sprague Dawley rats that received artificial sweetener sucralose (1.1%) for 12 weeks, the results show that sucralose administration reduced the total number of anaerobic bacteria, aerobic bacteria, bifidobacteria, Lactobacillus, Bacteroides and Clostridium (14). Uebanso research showed that the abundance of Clostridium flora in the high-dose sucralose group decreased significantly, and the concentrations of butyric acid and bile acid increased in a dose-dependent manner with the intake of sucralose (15). So, sucralose administration significantly altered mice gut microbiome, and reduced the abundance of beneficial bacteria.
Although many studies have deeply explored the impact of sucralose on gut microbiome, most studies were close to the concentration of ADI (5 mg/kg BW/day) (16, 17). In this study, we found low concentration sucralose also significantly altered gut microbiome by setting four concentration gradients, and it might involve in the development of diabetes. It provides a research basis for the adverse effect mechanism of sucralose on human health, and provides guidance and theoretical support for the practical application of artificial sweeteners.
Materials and Methods
Animals and Sampling
Forty specific pathogen-free (SPF) C57BL/6J male mice weaned at the age of 28 days were purchased from SLAC Laboratory Animal Co., Ltd (Shanghai, China). The mice were raised in cages at 25 ± 2°C for 12 h light/dark cycles with free access to water and mouse chow. After acclimatization for 1 week, the mice were weighed and randomly divided into 5 groups and treated for 16 weeks as follows: Control group mice were given distilled water (C, n = 8), Trichlorogalactosucrose (TGS) 1–4 groups mice were given a sucralose solution of 0.0003 g/mL (T1, n = 8), 0.003 mg/mL (T2, n = 8), 0.03 mg/mL (T3, n = 8), 0.3 mg/mL (T4, n = 8) per day. FDA defined ADI for sucralose in humans were 5 mg per kg (body weight) (18), 0.3 mg/ml is equal to a mouse with an average body weight of 0.02 kg, according to the following calculation:
At the end of 16-week study, all mice were weighed individually and euthanized. The liver of each mouse was isolated and weighed. Segments of jejunum, ileum and colon were collected and fixed in 4% paraformaldehyde for H&E-stained. The contents of jejunum, ileum, cecum and colon were collected and stored at −20°C until DNA isolation and 16S rRNA gene sequencing. More detailed bioinformatics methods can be found in a previous study (19).
Histological Staining
The jejunum, ileum, and colon tissue segments (1 cm) were collected for histological staining from 3 mice per group, the tissues were rinsed with PBS, immediately fixed in 4% paraformaldehyde, and then cut into sections (4–5 mm), the H&E staining were used to stain the tissue sections according the methods described by previous study (20).
Histopathological scores were calculated according to the methods described by Ma (21): Epithelial surface loss, crypt destruction, and immune cell infiltration (0: no change, 1: localized and mild, 2: localized and moderate, 3: localized and severe, 4: extensive and moderate, 5: extensive and severe).
DNA Extraction and PCR Amplification
Microbial genomic DNA was extrtacted from each intestinal content according to the manufacturer's instructions (QIAamp DNA Stool Mini Kit QIAGEN, CA). The V4-V5 region of the bacteria 16S ribosomal RNA gene was amplified by PCR (95°C for 2 min, followed by 25 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 5 min) using primers 515 F 5′-barcode- GTGCCAGCMGCCGCGG)-3′ and 907 R 5′-CCGTCAATTCMTTTRAGTTT-3′, where the barcode is an eight-base sequence unique to each sample. The PCR reactions were performed in triplicate using 20 μL mixture which contained 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of forward primer (5 μM), 0.8 μL of reverse primer (5 μM), 0.4 μL of FastPfu Polymerase, 0.2 μL of BSA and 10 ng of template DNA, then add ddH2O to 20 μL. Amplicons were extracted from 2% agarose gels and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, U.S.) according to the manufacturer's instructions and quantified using QuantiFluor™-ST (Promega, U.S.).
Library Construction and Sequencing
Purified PCR products were quantified by Qubit®3.0 (Life Invitrogen) and every 24 amplicons whose barcodes were different were mixed equally. The pooled DNA product was used to construct Illumina Pair-End library following Illumina's genomic DNA library preparation procedure. Then the amplicon library was paired-end sequenced (2 × 250) on an Illumina Novaseq platform [Mingke Biotechnology (Hangzhou) Co., Ltd] according to the standard protocols. The original image data files obtained by high-throughput sequencing were converted into Sequenced Reads by Base Calling analysis, the results were stored in FASTQ (referred to as fq) format file, which contains sequence information of reads and their corresponding sequencing quality information.
Statistical Analysis
All statistical analyses were performed by SPSS 23.0 (IBM, New York, NY, United States) using One way ANOVA (22). Data are presented as the mean ± SEM. Results were considered significant when P < 0.05.
Results
Sucralose Administration Did Not Change the Phenotype of Mice
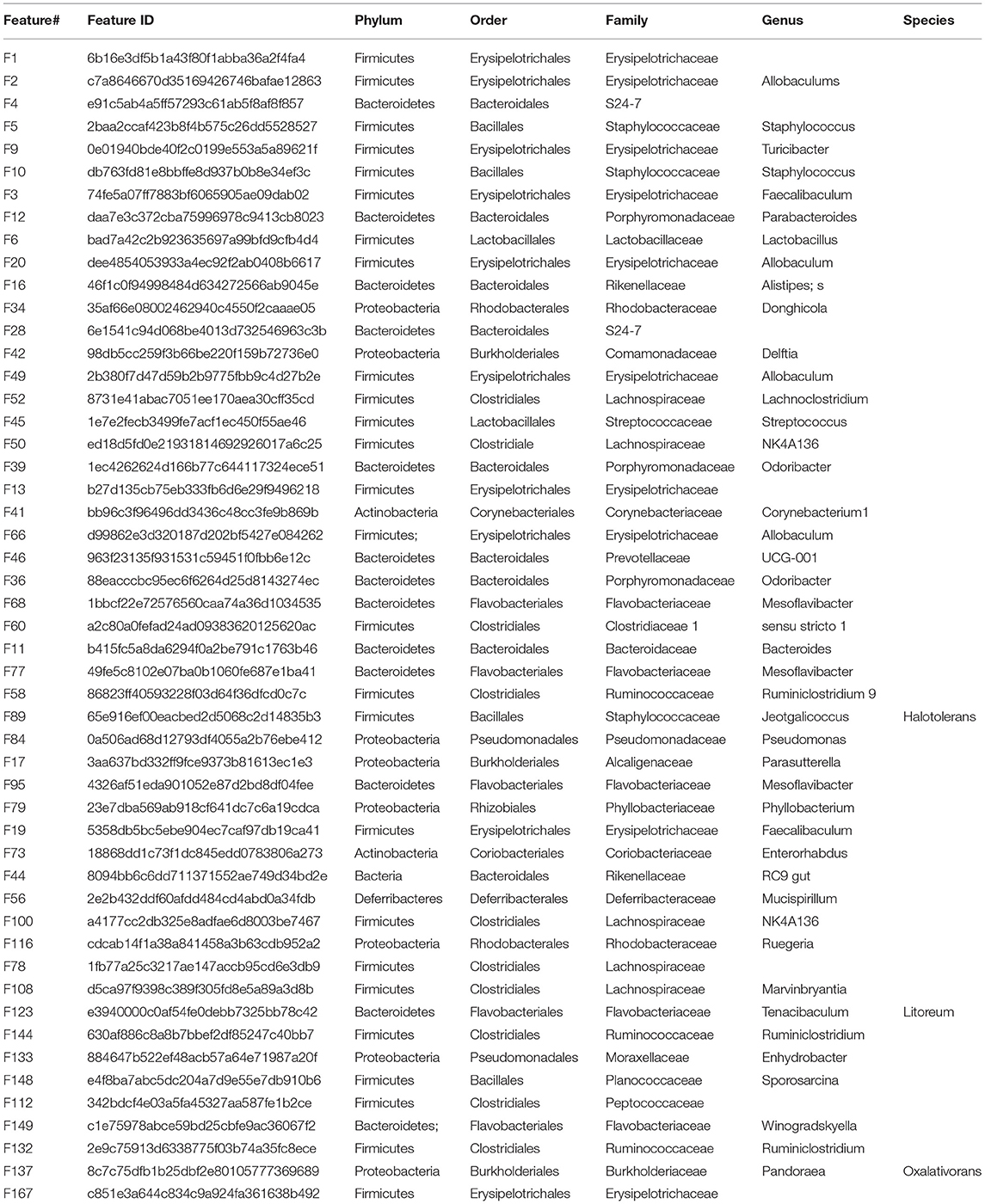
In order to confirm the effect of zero-calorie sucralose on body nutritional absorption, Mice was given with T1-4 (0.0003, 0.003, 0.03, and 0.3 mg/mL) of sucralose in drinking water, respectively. The results showed mice body weight and liver weight were not significant differences between Control group and T1–T4 groups (Figures 1A,B). To observe the effect of sucralose on intestines of mice. We stained mice intestinal tissue segments of each group. Compared with the Control group, the intestinal barrier and goblet cells of T1-4 groups were significantly damaged, and there was distinct lymphocyte aggregation in ileum and colon of T1 group and ileum of T4 group (Figure 1C). According to the scores of H&E staining, T1 and T4 group presented with severe acute colitis, crypt destruction, disappearance of superficial epithelial cells and goblet cells, and the increased infiltration of inflammatory cell (Figure 1D).
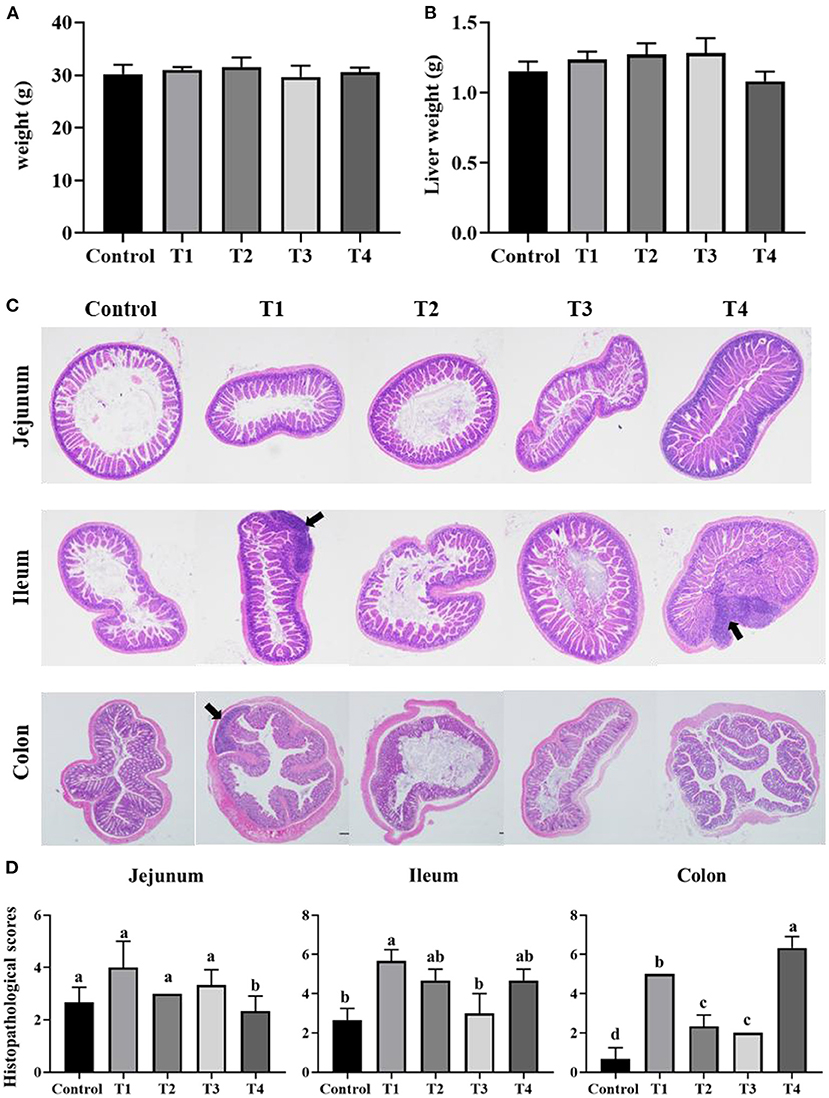
Figure 1. Bar chart showing the mice body weight (A) and liver weight (B). Representative H&E-stained sections from jejunum, ileum, cecum and colon (C), the arrowhead points in the direction where lymphocyte aggregation. Histopathological scores of the H&E staining (D). Data was expressed as mean ± SEM (n = 8) and analyzed by one-way ANOVA analysis. The different superscript letters on the histogram represent a significant difference (D) (P < 0.01).
Sucralose Administration Altered Mice Gut Microbiome
To validate that sucralose administration will change the structure of gut microbiome in mice, the jejunal, ileal, cecal, colonic contents were collected for 16S rRNA gene sequencing. The alpha-diversity indicated that the number of features and Shannon index had an upward trend from the Control group to T1 and T2 groups, and there was a downward trend from T2 group to T3 and T4 group in mice jejunum (Figures 2A,B), ileum (Figures 2C,D), cecum (Figures 2E,F) and colon (Figures 2G,H). The beta-diversity showed significant changes in mice gut microbiome community membership and structure from Control group to T1-4 groups. Especially in T1 group (Figures 3A–D), its clustering is far away from all other groups in jejunum (Figure 3A) ileum (Figure 3B) cecum (Figure 3C) and colon (Figure 3D). In mice jejunum, ileum and cecum, T3 and T4 groups were closer to control group. In colon, T4 group was as far away from the control group as T1 group.
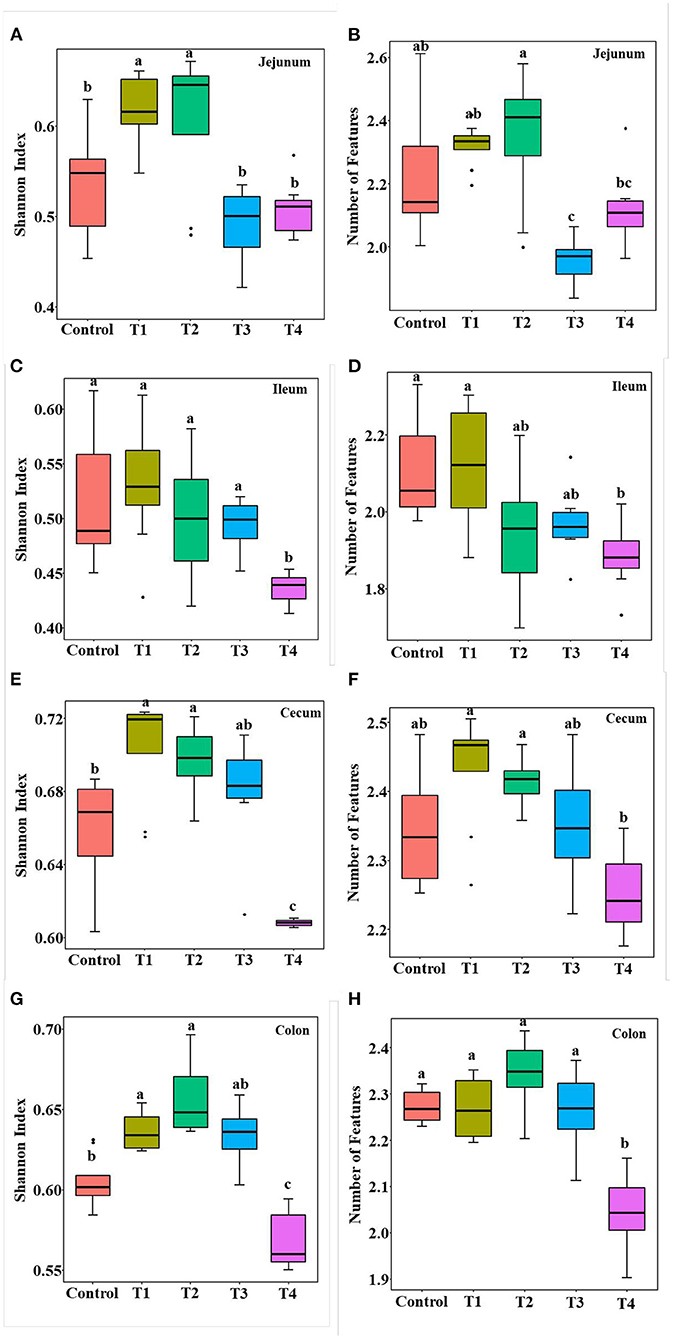
Figure 2. Alpha diversity including Shannon index (A,C,E,G) and the number of features (B,D,F,H) in control group, Trichlorogalactosucrose (TGS) 1–4 (T1, T2, T3, T4) groups in jejunum (A,B), ileum (C,D), cecum (E,F), colon (G,H). Data was processed through log10, and expressed as mean ± SEM (n = 8) and analyzed by one-way ANOVA analysis. The different superscript letters on the boxplot represent a significant difference (P < 0.01).
Figure 3. PCA of the mice gut microbial community composition of the Control and T1-4 group based on the Bray-Curtis distances showed distinct clusters, P-value and R-value were calculated by ANOSIM. The jejunum (A), ileum (B), cecum (C), and colon (D) microbial community structure between the Control group and T1–4 groups were differentiated by colors (red, brown, green, blue, purple, respectively).
The Mice Gut Core Microbiome
To identify the core microbiome in mice gut, Top 51 bacterial features of mice gut core microbiome was obtained by referring the research of Li (23). Most of these features are associated with the phylum Firmicutes (n = 26), Bacteroidetes (n = 14). At the family level, the top three families were Erysipelotrichaceae (n = 10), Lactobacillaceae (n = 6) Staphylococcaceae (n = 4). The top feature was Allobaculums (n = 4) (F2, F20, F49, F66) at genus level. These features sequence and taxonomy are shown in Table 1. Phylogenetic tree analysis indicated the Firmicutes had the highest level of abundance in the phylum, the next was Proteobacteria based on the top 129 genus level (Figure 4).
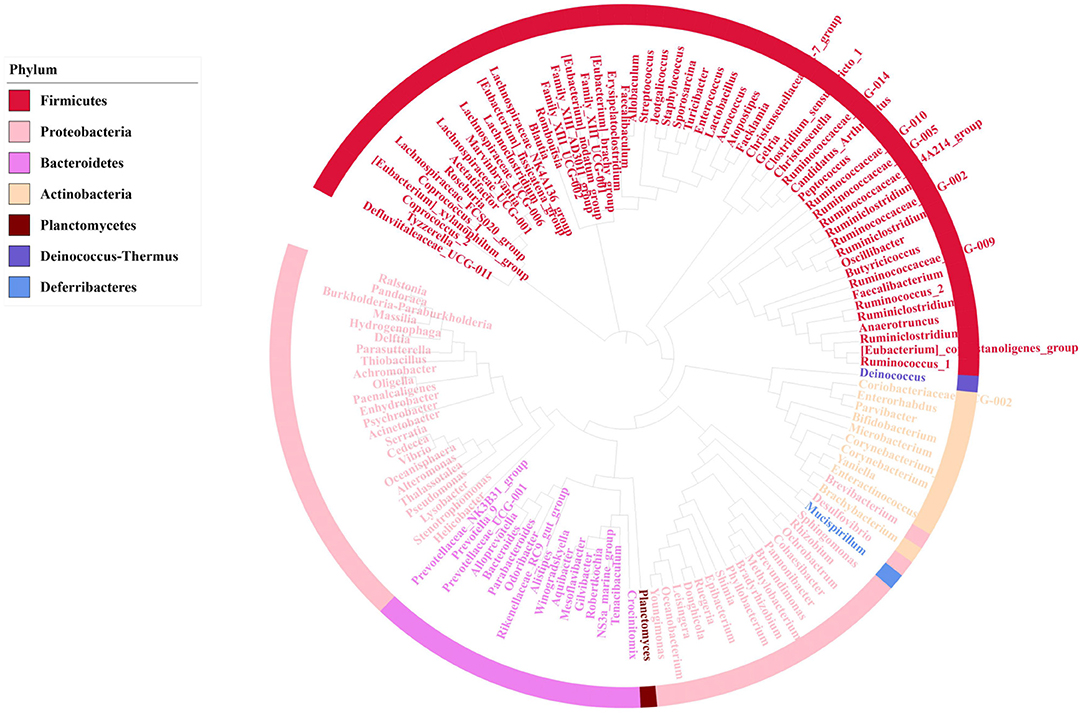
Figure 4. Phylogenetic tree analysis showing top 129 bacterial taxa based on 16S rRNA gene V4-V5 hypervariable regions, after removing 21 features which were uncultured or no rank in genus level. The innermost clades and labels were colored by genus.
We next confirmed the shifts of mice microbiome in different gut segments, top 30 most abundant bacterial features were shown on the bar chart (Figure 5). Among top 5 taxa, in jejunum and cecum, compared with Control group, Firmicutes-Allobaculums (F2) had an upward trend in T1 and T4 group, and it significantly rose in ileum and cecum (P < 0.05). In the colon, the T4 group had a significant increase in Firmicutes-Allobaculums (F49 and F66) compared with the Control group. Firmicutes-Staphylococcus (F5) increased significantly in T1 group compared with Control group in four different gut segments (P < 0.05) (Figure 5).
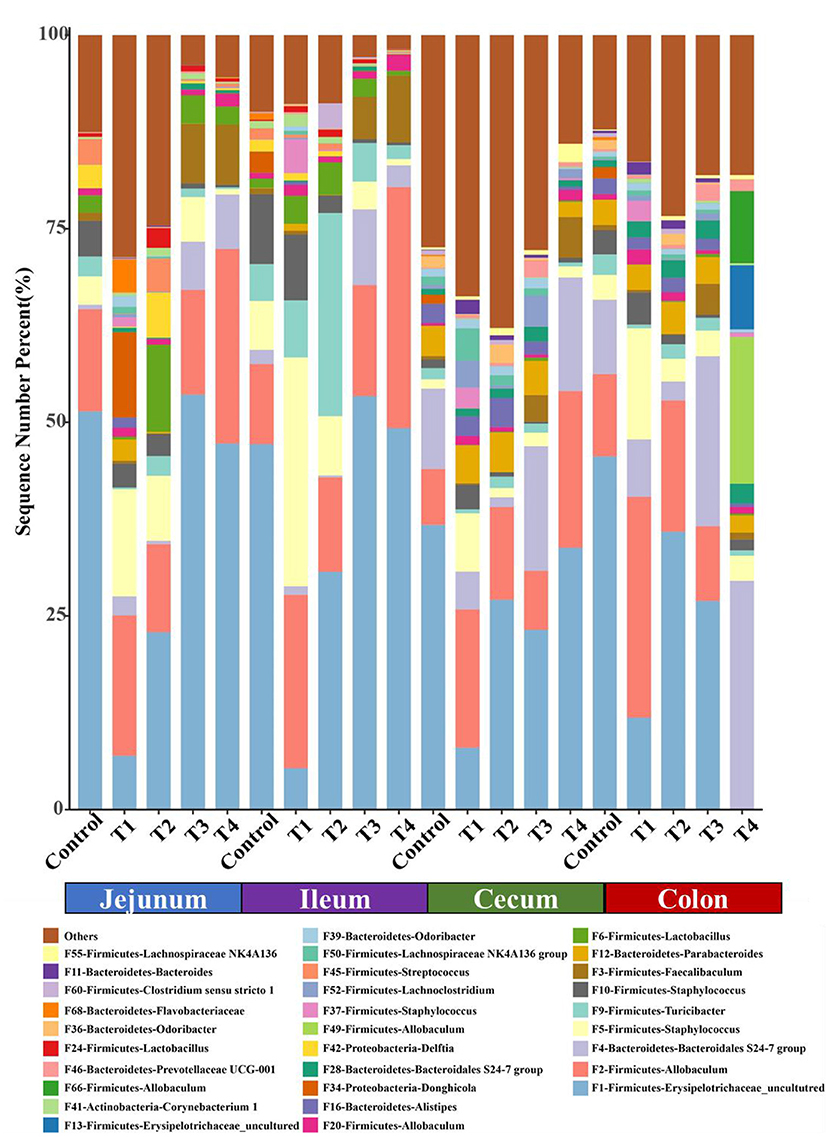
Figure 5. The top 30 features in Control group and T1–4 groups of jejunum, ileum, cecum and colon microbiome in mice. Each color indicates the relative abundance of a bacterial taxon on the bar chart.
Bacterial Taxa Differentially Represented in Mice Gut Microbiome
Mice gut bacterial features were analyzed by using LEfSe (24), the abundance of these significantly different features were shown on the heat map. In jejunum (Figure 6A), Bacteroidetes-Tenacibaculum (F123) and Proteobacteria-Ruegeria (F116) had a significantly increase in T1 group compared with other groups (Control, T2, T3, T4). In ileum (Figure 6C), Firmicutes-Allobaculum (F2) in T1 and T4 group were significantly higher than other group (Control, T2, T3), Firmicutes-Staphylococcus (F5, F37, F10) and Actinobacteria-Corynebacterium 1 (F41) had a significantly increase in T1 group compared with other groups (Control, T2, T3, T4). In cecum (Figure 6B), Firmicutes-Lachnoclostridium (F134) and Firmicutes-Lachnospiraceae UCG-006(F156) in Control group were significantly higher than T1 and T4 group. In colon (Figure 6D), after removing these features which were uncultured or no rank in genus level, we found the Firmicutes-Allobaculum (F20) in T1 group and Firmicutes-Allobaculum (F66, F49) in T4 group increased significantly than other groups (Control, T2, T3).
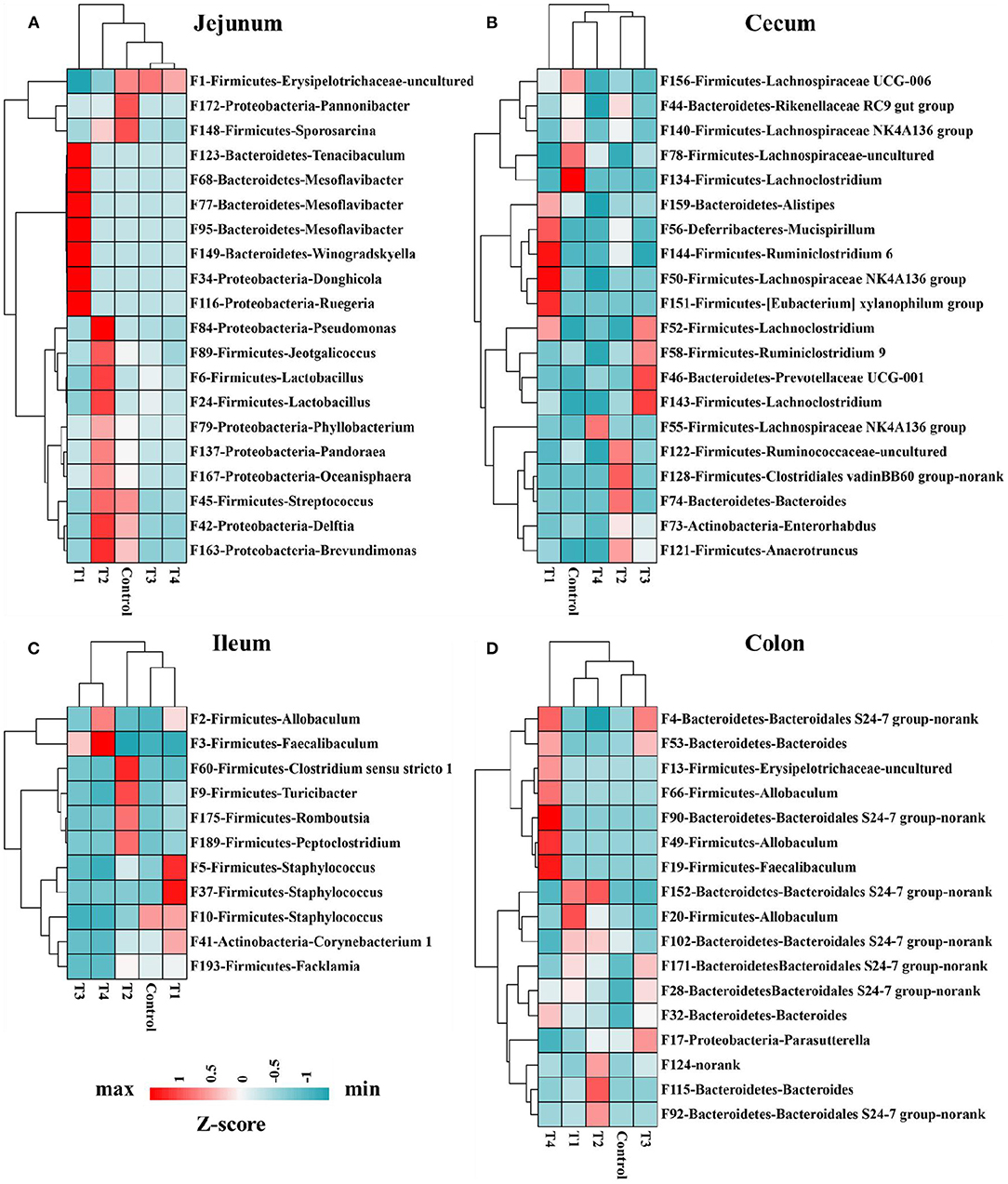
Figure 6. Heat map indicated 68 bacterial taxa were identified by LEfSe (LDA > 3) in mice jejunum (n = 20) (A), ileum (n = 11) (C), cecum (n = 20) (B), and colon (n = 17) (D) microbiome. The top 1,000 features were used for LEfSe analysis. Heat map shows the average relative abundances on a Z-score.
Discussion
Sweetness, whether provided by sugar or artificial sweeteners, enhances human appetite and reduce stress, so it is the preference of most people (25, 26). Besides, sucralose is one of the most consumed NNS in the world, since entering the food and beverage market (27), Recent research had shown that sucralose was used instead of sugar to reduce calorie and blood sugar intake (28). However, its effect on human health has been always controversial in recent years. Thus, in the present study, we examined the effects of sucralose (0.0003, 0.003, 0.03, and 0.3 mg/mL) on mice weight. Finally, we found that sucralose administration did not change the phenotype of mice, including the body weight and liver weight. Resent research showing the body weight remained constant by short-term sucralose consumption in human (16). Azad searched Medline, Embase and Cochrane Library for randomized controlled trials that evaluated interventions for NNS, NNS administration had no significant effect on BMI in 1,003 participants (29). This all consistent with our study, and confirms that sucralose as a zero-calorie sweetener does not provide energy to the body.
The intestinal barrier is composed of physical barrier (intestinal epithelium and mucus elements) (30), immunologic (immune cells) (31), and microbial community (32). Intestinal barrier regulates the two-way flow of water, ions and macromolecules between the lumen and the host (33). Epithelial barrier dysfunction has been reported in a variety of intestinal diseases, including inflammatory bowel disease (IBD) and ulcerative colitis (34). With the administration of sucralose, T1-4groups intestinal epithelial barrier was destroyed, as evidenced by the lymphocyte aggregation, especially in T1 and T4 groups. Resent study indicated that high concentrations of sucralose [10 mmol, the public may consume generous sweetener in the diet to achieve up to 10 mmol exposure to sweeteners (35)] induced apoptosis and cell death of intestinal epithelial cells, low concentrations of sucralose (0.1 mmol) down-regulated cell surface claudin 3 (36). Dai et al. (5) research showed that maternal sucralose administration significantly inhibited intestinal development and destroyed the intestinal barrier function in 3-week-old offspring. MUC2 is one of the important products of goblet cells and is closely related to the formation of the mucus layer. Research showed that compared with the control group, the production of MUC2 was significantly decreased in the sucralose group (5). After 6 months of sucralose administration, the genes related to LPS synthesis increased significantly (37). The relative mRNA expression levels of proinflammatory factors, including IL-1β, IFN-γ, and TNF-α were significantly higher in sucralose group than those in control group in colon (5). In our study, sucralose administration induced lymphocyte aggregation, which may lead to the increase of inflammatory factors. These revealed that sucralose administration might disrupt intestinal barrier function, the sucralose concentration and the effect on intestinal barrier of these studies all consistent with T4 group in our study, more interestingly, we also found the intestinal barrier was significantly damaged in group T1.
Gut microbiome is also an important part of the intestinal barrier (38), it is a complex and dynamic system, intestinal imbalanced states or even unhealthy stable states will develop, or potentially lead to diseases, including IBD, Nonalcoholic fatty liver disease (NALFD) and Irritable Bowel Syndrome (IBS) (39, 40). In this study, we demonstrate low dose of sucralose alter gut microbiome in mice by using 16S rRNA gene sequencing, T1-4 groups mice accessed 0.0003 g/mL, 0.003 mg/mL, 0.03 mg/mL, 0.3 mg/mL sucralose for 16 weeks, 0.1 mg/ml of sucralose solution was FDA acceptable daily intake (18). The results showed the number of features and Shannon index had an upward trend in T1 group and a downward trend in T4 group compared with Control group, Beta-diversity indicated T1 group was distinct from other groups, especially Control group. Sánchez-Tapia research found 1.5% (1.5 mg/mL) concentration of sucralose led to the lowest α-diversity in rats gut microbiota, its PCoA analysis revealed that gut microbiota was differentially shifted by sucralose (41). Many previous studies had shown that ADI (0.1 mg/ml) of sucralose significantly altered mice gut microbiome (37, 42). These results were consistent with T4 group in our study, however, the new finding in our research was T1 (0.0003 mg/mL) group, like T4 (0.3 mg/mL), also altered mice gut microbiome.
Core microbiome is essential to understand its function in the gut, it has been well- researched in different species (43, 44). Generally, a core microbiome indicates common bacterial present in all or most (e.g., >90%) of the communities in the host (45). In this study, a total of 51 core microbiome members of mice were identified in five groups. Most of these features are associated with the phylum Firmicutes (n = 26), Bacteroidetes (n = 14). Research have shown 2.5% sucralose treatment group increased the Firmicutes in phylum level (46). In our study, we found the top feature was Allobaculums (n = 4) (F2, F20, F49, F66) at genus level. Besides, Allobaculums of T1 and T4 group were significantly higher in mice jejunum, ileum and colon. Many research had shown Allobaculums significantly increased in diabetes model group compared with normal group (47, 48). However, whether sucralose administration will induce diabetes by altering gut microbiota, it requires further research.
Sucralose intake associated bacterial features were identified by using LEfSe, an algorithm that not only analyze statistical significance but also biological consistency. The results showing, in jejunum, Bacteroidetes-Tenacibaculum (F123) and Proteobacteria-Ruegeria (F116) significantly increased in T1 group compared with Control group. Tenacibaculum is a genus of gram negative, filamentous bacteria, related to the disease (tenacibaculosis) existing in aquaculture farms all over the world (49). Rubio-Portillo research identified Ruegeria OUT was associated with tissue necrosis in their hosts (50). In ileum, Firmicutes-Staphylococcus (F5, F37, F10) and Actinobacteria-Corynebacterium 1 (F41) significantly increased in T1 group compared with Control groups. The representative species of Staphylococcus are Staphylococcus aureus, it is a pathogen that usually colonizes the human anterior nostrils. This pathogen is one of main causes of life-threatening bloodstream infections, as sepsis and endocarditis (51). In cecum, Firmicutes-Lachnoclostridium (F134) and Firmicutes-Lachnospiraceae UCG-006 (F156) significantly decreased in T4 group than Control group. Lachnoclostridium was significantly up-regulated after treatment of obesity and inflammatory bowel disease (IBD) (52, 53). Lachnospiraceae is the major producers of short-chain fatty acids (SCFA), and is significantly related with enhanced gut barrier function (54, 55). In colon, Firmicutes-Allobaculum (F20) in T1 group and Firmicutes-Allobaculum (F66, F49) in T4 group increased significantly than other groups (Control, T2, T3). Allobaculum is not only positive related to diabetes (47, 48), but also related to ileal RORγT and IL-17 levels (56), induced susceptibility to autoimmune encephalitis (57), increased the expansion of inflammatory T helper 17 cells in gut (58). So, low dose of sucralose (T1, 0.0003 g/mL) consumption significantly altered mice gut microbiome, it might contribute to the increased expression of pro-inflammatory, these changes same as most previous study had indicated sucralose intake at human ADI (T4, 0.03 mg/mL) altered the gut microbiome in mice (5, 18, 41, 46).
Conclusion
Overall, our study demonstrated sucralose administration did not change mice body weight, but low dose of sucralose (0.0003 mg/mL) significantly altered mice gut microbiome, including the increases of Tenacibaculum, Ruegeria, Staphylococcus and Allobaculum in genus level in mice jejunum, ileum and colon. The decrease of Lachnoclostridium and Lachnospiraceae in cecum of T4 group mice. Although the sucralose of ADI (0.3 mg/mL) level also altered the gut microbiome in mice, the human daily intake of sucralose is usually lower than this concentration. We should focus on the low dose of sucralose administration in human. Finally, our research is limited to the effect of low-dose sucralose on the gut microbiome of mice, and the relevance to human metabolic diseases warrant further investigation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA787401.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Zhejiang Academy of Agricultural Sciences.
Author Contributions
ZZ, YX, YR, and JL designed the experiment. ZZ, LM, HP, and XW conducted the animal experiments. ZZ, YX, LM, YR, and JL wrote and revised the manuscript. ZZ, YX, LM, YR, and JL did experimental analysis, collected, and analyzed the data. All authors reviewed the manuscript and contributed to the article and approved the submitted version.
Funding
This work was financially supported by the State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agroproducts, Grant/Award Number: 2010DS700124-ZZ2017, the Open Project of Hubei Key Laboratory of Animal Nutrition and Feed Science (No. 201806), and the National Natural Science Foundation of China (31972999).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Carocho M, Morales P, Ferreira I. Sweeteners as food additives in the XXI century: a review of what is known, and what is to come. Food Chem Toxicol. (2017) 107:302–17. doi: 10.1016/j.fct.2017.06.046
2. Magnuson BA, Carakostas MC, Moore NH, Poulos SP, Renwick AG. Biological fate of low-calorie sweeteners. Nutr Rev. (2016) 74:670–89. doi: 10.1093/nutrit/nuw032
3. AlDeeb OA, Mahgoub H, Foda NH. Sucralose. Profiles Drug Subst Excip Relat Methodol. (2013) 38:423–62. doi: 10.1016/B978-0-12-407691-4.00010-1
4. Martyn D, Darch M, Roberts A, Lee HY, Yaqiong Tian T, Kaburagi N, et al. Low-/no-calorie sweeteners: a review of global intakes. Nutrients. (2018) 10:357. doi: 10.3390/nu10030357
5. Dai X, Guo Z, Chen D, Li L, Song X, Liu T, et al. Maternal sucralose intake alters gut microbiota of offspring and exacerbates hepatic steatosis in adulthood. Gut Microbes. (2020) 11:1043–63. doi: 10.1080/19490976.2020.1738187
6. Olivier-Van Stichelen S, Rother KI, Hanover JA. Maternal exposure to non-nutritive sweeteners impacts progeny's metabolism and microbiome. Front Microbiol. (2019) 10:1360. doi: 10.3389/fmicb.2019.01360
7. Roberts A, Renwick AG, Sims J, Snodin DJ. Sucralose metabolism and pharmacokinetics in man. Food Chem Toxicol. (2000) 38:S31–41. doi: 10.1016/S0278-6915(00)00026-0
8. Sylvetsky AC, Welsh JA, Brown RJ, Vos MB. Low-calorie sweetener consumption is increasing in the United States. Am J Clin Nutr. (2012) 96:640–6. doi: 10.3945/ajcn.112.034751
9. Uebanso T, Ohnishi A, Kitayama R, Yoshimoto A, Nakahashi M, Shimohata T, et al. Effects of low-dose non-caloric sweetener consumption on gut microbiota in mice. Nutrients. (2017) 9:560. doi: 10.3390/nu9060560
10. Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. (1977) 31:107–33. doi: 10.1146/annurev.mi.31.100177.000543
11. Gomaa EZ. Human gut microbiota/microbiome in health and diseases: a review. Antonie Van Leeuwenhoek. (2020) 113:2019–40. doi: 10.1007/s10482-020-01474-7
12. Fassarella M, Blaak EE, Penders J, Nauta A, Smidt H, Zoetendal EG. Gut microbiome stability and resilience: elucidating the response to perturbations in order to modulate gut health. Gut. (2021) 70:595–605. doi: 10.1136/gutjnl-2020-321747
13. Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. (2021) 19:55–71. doi: 10.1038/s41579-020-0433-9
14. Abou-Donia MB, El-Masry EM, Abdel-Rahman AA, McLendon RE, Schiffman SS. Splenda alters gut microflora and increases intestinal p-glycoprotein and cytochrome p-450 in male rats. J Toxicol Environ Health A. (2008) 71:1415–29. doi: 10.1080/15287390802328630
15. Uebanso T, Kano S, Yoshimoto A, Naito C, Shimohata T, Mawatari K, et al. Effects of consuming xylitol on gut microbiota and lipid metabolism in mice. Nutrients. (2017) 9:756. doi: 10.3390/nu9070756
16. Thomson P, Santibañez R, Aguirre C, Galgani JE, Garrido D. Short-term impact of sucralose consumption on the metabolic response and gut microbiome of healthy adults. Br J Nutr. (2019) 122:856–62. doi: 10.1017/S0007114519001570
17. Ahmad SY, Friel J, Mackay D. The effects of non-nutritive artificial sweeteners, aspartame and sucralose, on the gut microbiome in healthy adults: secondary outcomes of a randomized double-blinded crossover clinical trial. Nutrients. (2020) 12:3408. doi: 10.3390/nu12113408
18. Suez J, Korem T, Zeevi D, Zilberman-Schapira G, Thaiss CA, Maza O, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. (2014) 514:181–6. doi: 10.1038/nature13793
19. Zheng Z, Lyu W, Ren Y, Li X, Zhao S, Yang H, et al. Allobaculum Involves in the modulation of intestinal ANGPTLT4 expression in mice treated by high-fat diet. Front Nutr. (2021) 8:690138. doi: 10.3389/fnut.2021.690138
20. Yang H, Xiang Y, Robinson K, Wang J, Zhang G, Zhao J, et al. Gut microbiota is a major contributor to adiposity in pigs. Front Microbiol. (2018) 9:3045. doi: 10.3389/fmicb.2018.03045
21. Ma L, Ni L, Yang T, Mao P, Huang X, Luo Y, et al. Preventive and therapeutic spermidine treatment attenuates acute colitis in mice. J Agric Food Chem. (2021) 69:1864–76. doi: 10.1021/acs.jafc.0c07095
22. Xiao Y, Wu C, Li K, Gui G, Zhang G, Yang H. Association of growth rate with hormone levels and myogenic gene expression profile in broilers. J Anim Sci Biotechnol. (2017) 8:43. doi: 10.1186/s40104-017-0170-8
23. Li Y, Wang X, Wang XQ, Wang J, Zhao J. Life-long dynamics of the swine gut microbiome and their implications in probiotics development and food safety. Gut Microbes. (2020) 11:1824–32. doi: 10.1080/19490976.2020.1773748
24. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. (2011) 12:R60. doi: 10.1186/gb-2011-12-6-r60
25. Jacques A, Chaaya N, Beecher K, Ali SA, Belmer A, Bartlett S. The impact of sugar consumption on stress driven, emotional and addictive behaviors. Neurosci Biobehav Rev. (2019) 103:178–99. doi: 10.1016/j.neubiorev.2019.05.021
26. O'Connor D, Pang M, Castelnuovo G, Finlayson G, Blaak E, Gibbons C, et al. A rational review on the effects of sweeteners and sweetness enhancers on appetite, food reward and metabolic/adiposity outcomes in adults. Food Funct. (2021) 12:442–65. doi: 10.1039/D0FO02424D
27. Schiffman SS, Rother KI. Sucralose, a synthetic organochlorine sweetener: overview of biological issues. J Toxicol Environ Health B Crit Rev. (2013) 16:399–451. doi: 10.1080/10937404.2013.842523
28. Nadolsky KZ, COUNTERPOINT. Artificial sweeteners for obesity-better than sugary alternatives potentially a solution. Endocr Pract. (2021) 27:1056–61. doi: 10.1016/j.eprac.2021.06.013
29. Azad MB, Abou-Setta AM, Chauhan BF, Rabbani R, Lys J, Copstein L, et al. Nonnutritive sweeteners and cardiometabolic health: a systematic review and meta-analysis of randomized controlled trials and prospective cohort studies. Cmaj. (2017) 189:E929–39. doi: 10.1503/cmaj.161390
30. Luissint AC, Parkos CA, Nusrat A. Inflammation and the intestinal barrier: leukocyte-epithelial cell interactions, cell junction remodeling, and mucosal repair. Gastroenterology. (2016) 151:616–32. doi: 10.1053/j.gastro.2016.07.008
31. Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. (2014) 14:667–85. doi: 10.1038/nri3738
32. Chopyk DM, Grakoui A. Contribution of the intestinal microbiome and gut barrier to hepatic disorders. Gastroenterology. (2020) 159:849–63. doi: 10.1053/j.gastro.2020.04.077
33. Odenwald MA, Turner JR. The intestinal epithelial barrier: a therapeutic target? Nat Rev Gastroenterol Hepatol. (2017) 14:9–21. doi: 10.1038/nrgastro.2016.169
34. Torres J, Petralia F, Sato T, Wang P, Telesco SE, Choung RS, et al. Serum biomarkers identify patients who will develop inflammatory bowel diseases up to 5 years before diagnosis. Gastroenterology. (2020) 159:96–104. doi: 10.1053/j.gastro.2020.03.007
35. Gardner C, Wylie-Rosett J, Gidding SS, Steffen LM, Johnson RK, Reader D, et al. Nonnutritive sweeteners: current use and health perspectives: a scientific statement from the american heart association and the american diabetes association. Diabetes Care. (2012) 35:1798–808. doi: 10.2337/dc12-9002
36. Shil A, Olusanya O, Ghufoor Z, Forson B, Marks J, Chichger H. Artificial sweeteners disrupt tight junctions and barrier function in the intestinal epithelium through activation of the sweet taste receptor, T1R3. Nutrients. (2020) 12:1862. doi: 10.3390/nu12061862
37. Bian X, Chi L, Gao B, Tu P, Ru H, Lu K. Gut microbiome response to sucralose and its potential role in inducing liver inflammation in mice. Front Physiol. (2017) 8:487. doi: 10.3389/fphys.2017.00487
38. Allam-Ndoul B, Castonguay-Paradis S, Veilleux A. Gut microbiota and intestinal trans-epithelial permeability. Int J Mol Sci. (2020) 21:6402. doi: 10.3390/ijms21176402
39. Sharpton SR, Schnabl B, Knight R, Loomba R. Current concepts, opportunities, and challenges of gut microbiome-based personalized medicine in nonalcoholic fatty liver disease. Cell Metab. (2021) 33:21–32. doi: 10.1016/j.cmet.2020.11.010
40. Dixit K, Chaudhari D, Dhotre D, Shouche Y, Saroj S. Restoration of dysbiotic human gut microbiome for homeostasis. Life Sci. (2021) 278:119622. doi: 10.1016/j.lfs.2021.119622
41. Sánchez-Tapia M, Miller AW, Granados-Portillo O, Tovar AR, Torres N. The development of metabolic endotoxemia is dependent on the type of sweetener and the presence of saturated fat in the diet. Gut Microbes. (2020) 12:1801301. doi: 10.1080/19490976.2020.1801301
42. Ruiz-Ojeda FJ, Plaza-Díaz J, Sáez-Lara MJ, Gil A. Effects of sweeteners on the gut microbiota: a review of experimental studies and clinical trials. Adv Nutr. (2019) 10:S31–48. doi: 10.1093/advances/nmy037
43. Wang X, Tsai T, Deng F, Wei X, Chai J, Knapp J, et al. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome. (2019) 7:109. doi: 10.1186/s40168-019-0721-7
44. Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. (2012) 488:86–90. doi: 10.1038/nature11237
45. Hamady M, Knight R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. (2009) 19:1141–52. doi: 10.1101/gr.085464.108
46. Wang QP, Browman D, Herzog H, Neely GG. Non-nutritive sweeteners possess a bacteriostatic effect and alter gut microbiota in mice. PLoS ONE. (2018) 13:e0199080. doi: 10.1371/journal.pone.0199080
47. Jia L, Li D, Feng N, Shamoon M, Sun Z, Ding L, et al. Anti-diabetic Effects of Clostridium butyricum CGMCC0313.1 through promoting the growth of gut butyrate-producing bacteria in type 2 diabetic mice. Sci Rep. (2017) 7:7046. doi: 10.1038/s41598-017-07335-0
48. Shen D, Lu Y, Tian S, Ma S, Sun J, Hu Q, et al. Effects of L-arabinose by hypoglycemic and modulating gut microbiome in a high-fat diet- and streptozotocin-induced mouse model of type 2 diabetes mellitus. J Food Biochem. (2021) 45:e13991. doi: 10.1111/jfbc.13991
49. Nowlan JP, Lumsden JS, Russell S. Advancements in characterizing tenacibaculum infections in Canada. Pathogens. (2020) 9:1029. doi: 10.3390/pathogens9121029
50. Rubio-Portillo E, Ramos-Espla AA, Anton J. Shifts in marine invertebrate bacterial assemblages associated with tissue necrosis during a heatwave. Coral Reefs. (2021) 40:395–404. doi: 10.1007/s00338-021-02075-0
51. Kwiecinski JM, Horswill AR. Staphylococcus aureus bloodstream infections: pathogenesis and regulatory mechanisms. Curr Opin Microbiol. (2020) 53:51–60. doi: 10.1016/j.mib.2020.02.005
52. Wang P, Li D, Ke W, Liang D, Hu X, Chen F. Resveratrol-induced gut microbiota reduces obesity in high-fat diet-fed mice. Int J Obes (Lond). (2020) 44:213–25. doi: 10.1038/s41366-019-0332-1
53. Zhang W, Zou G, Li B, Du X, Sun Z, Sun Y, et al. Fecal microbiota transplantation (FMT) alleviates experimental colitis in mice by gut microbiota regulation. J Microbiol Biotechnol. (2020) 30:1132–41. doi: 10.4014/jmb.2002.02044
54. Ma L, Ni Y, Wang Z, Tu W, Ni L, Zhuge F, et al. Spermidine improves gut barrier integrity and gut microbiota function in diet-induced obese mice. Gut Microbes. (2020) 12:1–19. doi: 10.1080/19490976.2020.1832857
55. Vacca M, Celano G, Calabrese FM, Portincasa P, Gobbetti M, De Angelis M. The controversial role of human gut lachnospiraceae. Microorganisms. (2020) 8:573. doi: 10.3390/microorganisms8040573
56. Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. (2014) 158:705–21. doi: 10.1016/j.cell.2014.05.052
57. Miyauchi E, Kim SW, Suda W, Kawasumi M, Onawa S, Taguchi-Atarashi N, et al. Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature. (2020) 585:102–6. doi: 10.1038/s41586-020-2634-9
Keywords: sucralose, low dose, gut microbiome, mice, intestinal barrier
Citation: Zheng Z, Xiao Y, Ma L, Lyu W, Peng H, Wang X, Ren Y and Li J (2022) Low Dose of Sucralose Alter Gut Microbiome in Mice. Front. Nutr. 9:848392. doi: 10.3389/fnut.2022.848392
Received: 04 January 2022; Accepted: 02 February 2022;
Published: 25 February 2022.
Edited by:
Tingtao Chen, Nanchang University, ChinaReviewed by:
Andrew William Moran, University of Liverpool, United KingdomNa Li, Beijing University of Chinese Medicine, China
Copyright © 2022 Zheng, Xiao, Ma, Lyu, Peng, Wang, Ren and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Ren, cmFuZWUxOTc0QDE2My5jb20=; Jinjun Li, bGlqaW5qdW5AemFhcy5hYy5jbg==
†These authors share first authorship
 Zibin Zheng
Zibin Zheng Yingping Xiao
Yingping Xiao Lingyan Ma2
Lingyan Ma2 Wentao Lyu
Wentao Lyu Jinjun Li
Jinjun Li