- 1Co-Innovation Center of Jiangsu Marine Bio-industry Technology, Jiangsu Ocean University, Lianyungang, China
- 2Jiangsu Key Laboratory of Marine Bioresources and Environment/Jiangsu Key Laboratory of Marine Biotechnology, Jiangsu Ocean University, Lianyungang, China
- 3School of Food Science and Engineering, Jiangsu Ocean University, Lianyungang, China
- 4Key Laboratory of Marine Drugs, School of Medicine and Pharmacy, The Ministry of Education of China, Ocean University of China, Qingdao, China
Urolithin A (UA) has received considerable research attention because of its health benefits. However, only a few strains have been reported to produce UA from ellagic acid (EA), and the molecular mechanisms underlying the gut microbiota-mediated transformation of ellagic acid into urolithin A is limited. In the present study, a single strain FUA027 capable of converting ellagic acid into UA in vitro was isolated from the fecal samples. The strain was identified as Enterococcus faecium through the morphological, physiological, biochemical and genetic tests. UA was produced at the beginning of the stationary phase and its levels peaked at 50 h, with the highest concentration being 10.80 μM. The strain Enterococcus faecium FUA027 is the first isolated strain of Enterococcus sp. producing urolithin A from ellagic acid, which may be developed as probiotics and used to explore molecular mechanisms underlying the biotransformation of ellagic acid into UA.
Introduction
A healthy diet not only supports the energy and physiological needs of the body even in the absence of excess food intake but also prevents diseases and positively influences health (1), the role of microbiota in health and diseases is also being highlighted (2). This point is supported by scientific evidence obtained by investigating foods rich in ellagic acid and ellagitannin (3). Ellagic acid is a polyphenol and a natural antioxidant widely found in many vegetables, fruits, nuts, and teas (4, 5). However, their health applications have been limited due to their poor water solubility and low bioavailability (6). Only a small amount of this polyphenol is absorbed by the epithelial cells of the small intestine (7). The first reported urolithins were urolithin A and urolithin B that were found in kidney stones of sheep. Besides, the intestinal flora of human and animals also transforms some of the unabsorbed ellagic acid or ellagitannin into urolithins, mainly including isourolithin, urolithin A, urolithin B, and urolithin C. Urolithins, which are more easily absorbed than their precursors, have been widely investigated and have exhibited various biological activities (4, 8), such as anticancer, antioxidative (9), anti-inflammatory, neuroprotective, antiaging, and anti-obesity activities (10). Significantly, urolithin A can improve mitochondrial function, which is the important characteristic of urolithin A compared with other intestinal metabolites.
Urolithin A does not exist in the natural state, which can be produced by the metabolism of ellagic acid through intestinal microorganisms of human. The bioconversion capacity of ellagitannin/ellagic acid to urolithins varies from person to person (11). People are classified into three urolithin metabotypes on the basis of urolithins metabolized by their intestinal microbiota, namely UM-A, UM-B, and UM-0 (12). UM-A only produces urolithin A; UM-B is characterized by urolithin B and isourolithin A, in addition to a low concentration of urolithin A, whereas UM-0 does not produce these urolithins. Urolithin A was one of the most widely studied urolithins, which reduced triglyceride accumulation in the liver (13). Moreover, it can inhibit inflammatory factors, such as TNF-α, IL-1β, it played an important role in the treatment of various inflammations (13). Urolithin A exhibited stronger anti-inflammatory activity than urolithin B. Urolithin A stimulates mitophagy and improves muscle health in both mice and humans (14, 15), and urolithin A prevents the accumulation of dysfunctional mitochondria to extends the lifespan of the nematode Caenorhabditis elegans (16–18). Moreover, urolithin A was shown to be safe by a clinical study, and it improved exercise capacity in mouse models of age-related decline of muscle function. Based on the bioavailability of urolithin A in muscle, the preventive effect of urolithin A treatment on age-related muscle decline in 16-month-old male C57BL/6J mice fed a high-fat diet (HFD) was evaluated by Ryu, D., et al. From the age of 16 months, the diet was supplemented with 50 mg/kg/d urolithin A for 8 months. Urolithin A treatment resulted in a significant increase in muscle function measured at 22 and 24 months of age compared to control HFD-fed mice (18). Consequently, urolithin metabotypes affect the metabolism and bioactivity of dietary polyphenols including ellagic acid and ellagitannins, thereby making them exhibit different effects on people’s health (17).
Although several studies have demonstrated that the gut microbiota composition is a critical factor in the bioconversion of ellagic acid and ellagitannins to urolithins (19), the mechanism underlying this transformation is unclear. This would help us maximize the biological activitiy of urolithins, especially for the metabotypes UM-0 and UM-B (12, 20). In 2020, Amazentis sold high-purity urolithin A as a nutritional product for improving muscular strength and fighting aging. So far, chemical synthesis is the only method for producing urolithin A for industrial purposes. Compared with chemical synthesis, biosynthesis of urolithin A is an economical and environmentally friendly method (21). Therefore, screening strains metabolizing ellagitannin/ellagic acid to urolithin A and exploring their metabolite regulation are necessary (22). However, intestinal bacteria with the ability to transform ellagic acid into urolithin A have rarely been reported. To date, Gordonibacter urolithinfaciens DSM 27213T = CCUG 64261T, Gordonibacter pamelaeae DSM 19378T = CCUG 55131T (23, 24), Ellagibacter isourolithinifaciens DSM 104140T = CCUG 70284T (12, 25), and Bifidobacterium pseudocatenulatum INIA P815 have been reported to produce urolithins from ellagic acid (26). Among them, only B. pseudocatenulatum INIA P815 showed urolithin A-producing ability (26).
In the present study, a urolithin A-producing strain, designated FUA027, was isolated from human feces. The strain was identified as Enterococcus faecium on the basis of the results of the morphological, physiological, biochemical and genetic tests. The time course of the strain-mediated transformation of ellagic acid into urolithin A was also determined through high-performance liquid chromatography (HPLC) and ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS) (23, 27).
Materials and methods
Chemicals
L-Cysteine hydrochloride, urolithin A, urolithin B, urolithin C, isourolithin A, urolithin M6, and urolithin D were purchased from Dalton Pharma Services (Toronto, Canada). Ellagic acid was purchased from Sigma-Aldrich (St. Louis, MO). MS- or LC-grade solvents, namely methanol, acetonitrile, formic acid, and ethanol were purchased from Merck (Darmstadt, Germany).
Collection of human fecal samples
All 7 volunteers (ages 22–27) with no history of gastrointestinal diseases signed an experimental informed consent form at the Jiangsu Ocean University (Lianyungang, China). The volunteers had not used antibiotics 3 months before the study. After took 0.7 g food/kg body weight/day for 2 consecutive weeks, feces samples were collected at 6:30 AM on the 15th day before they ate any food. Within 10 min, the samples were used for the fermentation experiments described below (24, 28).
Screening of human fecal samples converting ellagic acid into urolithin A in vitro
In total, 10 g of fecal samples were diluted 1/10 (w/v) with anaerobe basal broth (ABB, Oxoid, Basingstoke, England) and homogenized using a stomacher in filter bags (29, 30). 1 ml of filtered fecal suspensions were inoculated into 100 ml of ABB medium supplemented with 20.00 μM ellagic acid and 0.05% L-cysteine hydrochloride and cultured for 5 days in an anaerobic chamber (Don Whitley Scientific Limited, West Yorkshire, England) with an atmosphere consisting of N2/H2/CO2 (80:10:10) at 37°C. Ellagic acid was first dissolved in polyethylene glycol (PEG) 300. The resulting solution was added to ABB medium 20.00 μM. In addition, media without fecal suspension and those without ellagic acid were used as controls. Samples (5 ml) were collected once per day from each culture, extracted with 5 ml C2H3N:H2O:CH2O2 (80:19.9:0.1), and further analyzed using High Performance Liquid Chromatography (HPLC), as explained below. If urolithin A was detected in the broth, it was used to screen urolithin A-producing bacteria. ABB medium (g/L): Peptone 16.0, Yeast extract 7.0, NaCl 5.0, Starch 1.0, Glucose 1.0, C3H3NaO3 1.0, Arginine 1.0, C4H4Na2O4 0.5, L-Cysteine HCl 0.5, NaHCO3 0.4, Fe4(O7P2)3 0.5, C34H32ClFeN4O4 0.005,Vitamin K 0.0005, C2H3NaO2S 0.5, C4H10O2S2 1.0,pH6.8 ± 0.2.
Screening of urolithin A-producing bacteria from the fermentation broth
The urolithin A-containing fermentation broth was diluted to 10–3 to 10–6 with ABB media. An aliquot of the diluted suspension (0.1 ml) was spread on the ABB agar plate, and the plate was incubated at 37°C in an anaerobic chamber under the condition of N2/H2/CO2 (80:10:10, V:V:V). Selected single colonies were inoculated into 50 ml of ABB containing 20 μM ellagic acid (31). After anaerobic incubation at 37°C for 4 days, the broths were extracted with 50 ml C2H3N:H2O:CH2O2 (80:19.9:0.1, V:V:V) and further analyzed through HPLC and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses, explained below. The urolithin A-producing strains were stored in ABB media with 20% (v/v) glycerol at –80°C for further experiments.
Identification and taxonomical studies of strain FUA027
The morphological, physiological, and biochemical properties of strain FUA027 were investigated using previously described methods (32). Genomic DNA was extracted using a DNA extraction kit (TIANamp Bacteria DNA Kit, Beijing Tiangen Biotechnology Co., Ltd.). The 16S rDNA sequence was amplified through a PCR procedure by using the universal primers (27F 5’-AGAGTTTGATCCTGGCTCAG-3’, 1492R 5’-TACGACTTAACCCCAATCGC-3’) (33). The PCR product was purified and sequenced. A similarity search in the EMBL database was performed using the BLAST program (National Center for Biotechnology Information). Sequence alignment was executed using the Clustal W algorithm, and a neighbor-joining phylogenetic tree was constructed using MEGA 11 software.
Growth kinetics and time-course transformation of ellagic acid into urolithin A by strain Enterococcus faecium FUA027
Enterococcus faecium FUA027was streaked to single colonies on an ABB agar plate at 37°C for 6 days. A liquid culture for the inoculum was prepared by transferring a single colony to a tube containing 5 ml ABB by using a sterile inoculation loop. Two milliliters of the inoculum were transferred to 100 ml ABB media containing 20 μM ellagic acid and incubated under anaerobic conditions consisting of N2/H2/CO2 (80:10:10, V:V:V) at 37°C for 7 days. During incubation, 5-ml aliquots were taken and extracted with 5 ml of C2H3N:H2O:CH2O2 (V:V:V, 80:19.9:0.1) for HPLC and UPLC-MS analyses as described below. Similarly, culture samples were collected every 2 h on all 7 days of incubation (34), and the cell density was measured at 600 nm using a microplate reader (SpectraMax 190, Molecular Devices, Sunnyvale, CA, USA), the experiment was repeated three times. Data were subjected to statistical analysis of standard deviation. The quantitative estimates represent mean values ± standard error.
High-performance liquid chromatography analyses
The extracted samples were evaporated to dryness under a N2 stream at room temperature. The resultant extract was dissolved in 500 μl methanol/water (50:50, v/v). A 0.22-μm cellulose acetate filter (Millipore, Madrid, Spain) was used to filter the extract. The HPLC analysis was performed on a ZORBAX SB-C18 column (250 mm × 4.6 mm, 5 μm particle size, Agilent Technologies) at 25°C. The gradient started at 20% A and 80% B, followed by 70% A and 30% B at 15 min, 95% A and 5% B at 20 min, 100% A and 0% B at 21 min, 100% A and 0% B at 23 min, 20% A and 80% B at 24 min, and 20%A and 80% B at 25 min. The flow rate was 1 ml/min, and the column temperature was 25°C. Injection volume was 5 μl for each sample. UV chromatograms were recorded at 305 nm.
Liquid chromatography–mass spectrometry/mass spectrometry analyses
The Q-Exactive LC-MS/MS (Thermo Fisher) was used to analyze urolithin A using a C18 column [(Waters Ltd., MA, USA) ACQUITY UPLC® BEH C18, 2.1 × 50 mm, 1.7 μm; 0.5 ml/min] and HR-ESI-MS scan range from 150.0 to 2,000.0 Da. Sheath gas flow rate, 45 arb; aux gas flow rate; 15 arb; Capillary temp:320°C; S-Lens RF Level 50 T. chromatographed using LC-MS/MS at 30°C. The mobile phase consisted of solvent A (water, 0.1% formic acid) and solvent B (acetonitrile). Separation was performed at a flow rate of 0.5 ml/min with the following gradient elution: 0–10.5 min, a linear gradient runs from 10 to 100% solvent B; 10.5–12.5 min, 100% solvent B; 12.5–12.6 min, a linear gradient runs from 100 to 10% solvent B; 12.6–16.5 min 10% solvent B for re-equilibration. The limits of detection (LODs) were determined based on a signal-to-noise ratio (S/N) of 3 and of 10 for the limit of quantification (LOQ). The LODs: 0.5 μM (EA and urolithin A), 0.2 μM (urolithin C), the dynamic range: 0.5–20 μM. Urolithin A and urolithin C in the fermentation broth were first identified through HPLC and further identified by comparing the molecular mass of the compound obtained with that of a pure standard by using LC-MS/MS (35, 36).
Results and discussion
Screening of human fecal samples converting ellagic acid into urolithin A in vitro
Urolithins were identified through a direct comparison of the retention times with those of pure standards (10). The retention times of standards were analyzed by the HPLC at absorbance of 305 nm. The results revealed that urolithin A was produced by three human fecal samples, which indicates that three volunteers were metabotype A (Figure 1) (the concentration of urolithin A: volunteer L, 2.15 μM; volunteer X, 0.56 μM; volunteer H, 3.32 μM). A similar result was reported by Cortés-Martín et al., indicating that aging is a key factor that influences the distribution of urolithin metabotypes (11).
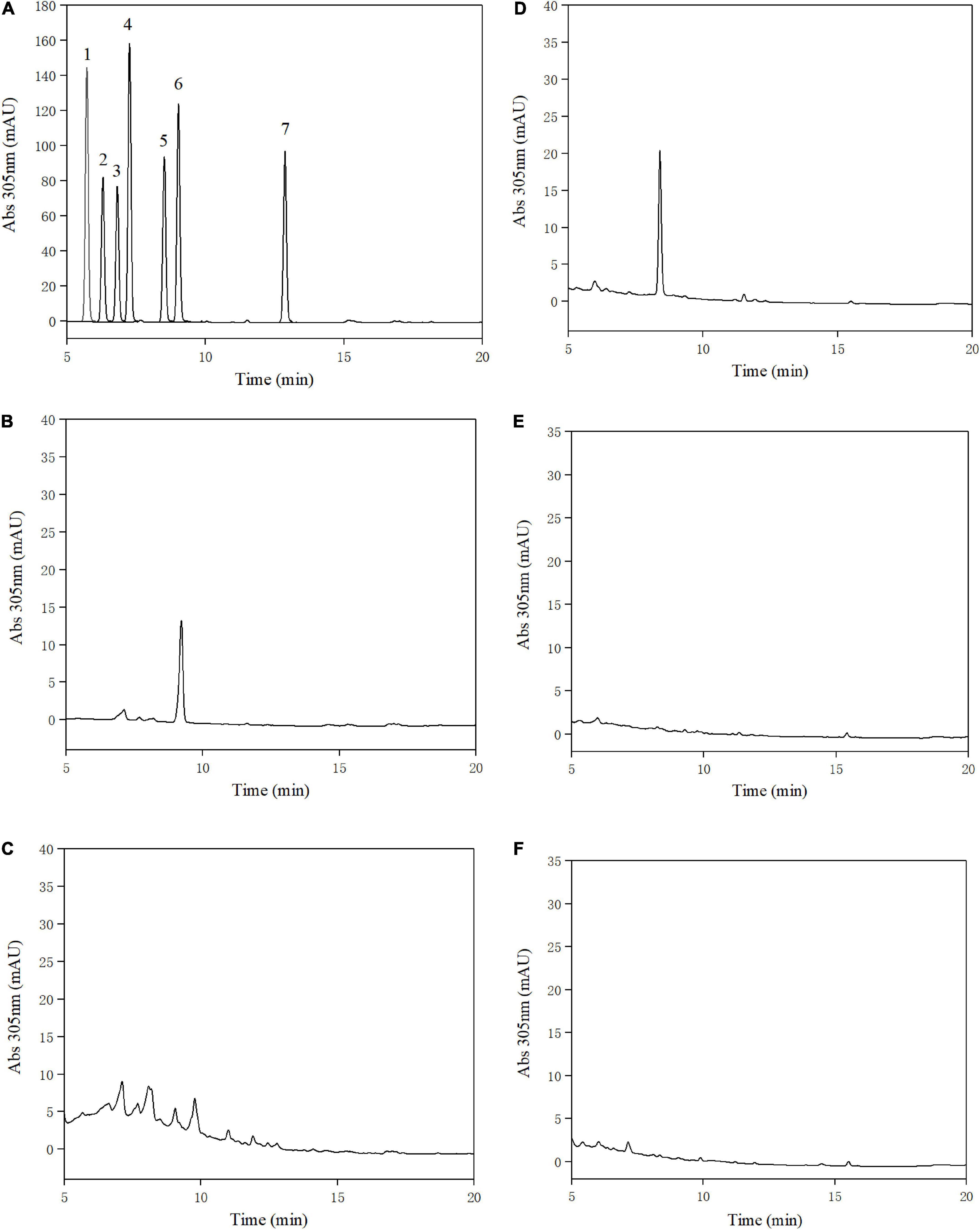
Figure 1. High-performance liquid chromatography (HPLC) analysis of urolithins obtained and standard samples at 305 nm. Standard samples of urolithins: 1. urolithin D, 2. ellagic acid, 3. urolithin M6, 4. urolithin C, 5. isourolithin A, 6. Urolithin A, 7. urolithin B (A), fecal samples of volunteer L (B), volunteer X (C), volunteer H (D), medium without fecal suspension (E), medium without ellagic acid (F).
Screening of urolithin A-producing bacteria
A total of 395 strains were screened for urolithin A production. According to the results of HPLC analysis (Figure 2) and LC-MS/MS (Figure 3), only FUA027 strain can convert ellagic acid into urolithin A under anaerobic conditions. Several reports have shown that the specific intestinal microflora of some people can transform ellagic acid/ellagitannins consumed into urolithin A (37). The species or composition of gut flora that produce urolithin A has not, however, been investigated. Although urolithin A was found in humans nearly 40 years ago, the pathway for its transformation remains unknown (23). Therefore, to completely comprehend the molecular mechanism of strains or flora underlying the transformation of ellagic acid into urolithins, strains capable of producing urolithin A should be screened and investigated (38). Moreover, probiotics must be developed to increase the urolithin A production efficiency of low-yield individuals (37, 39).
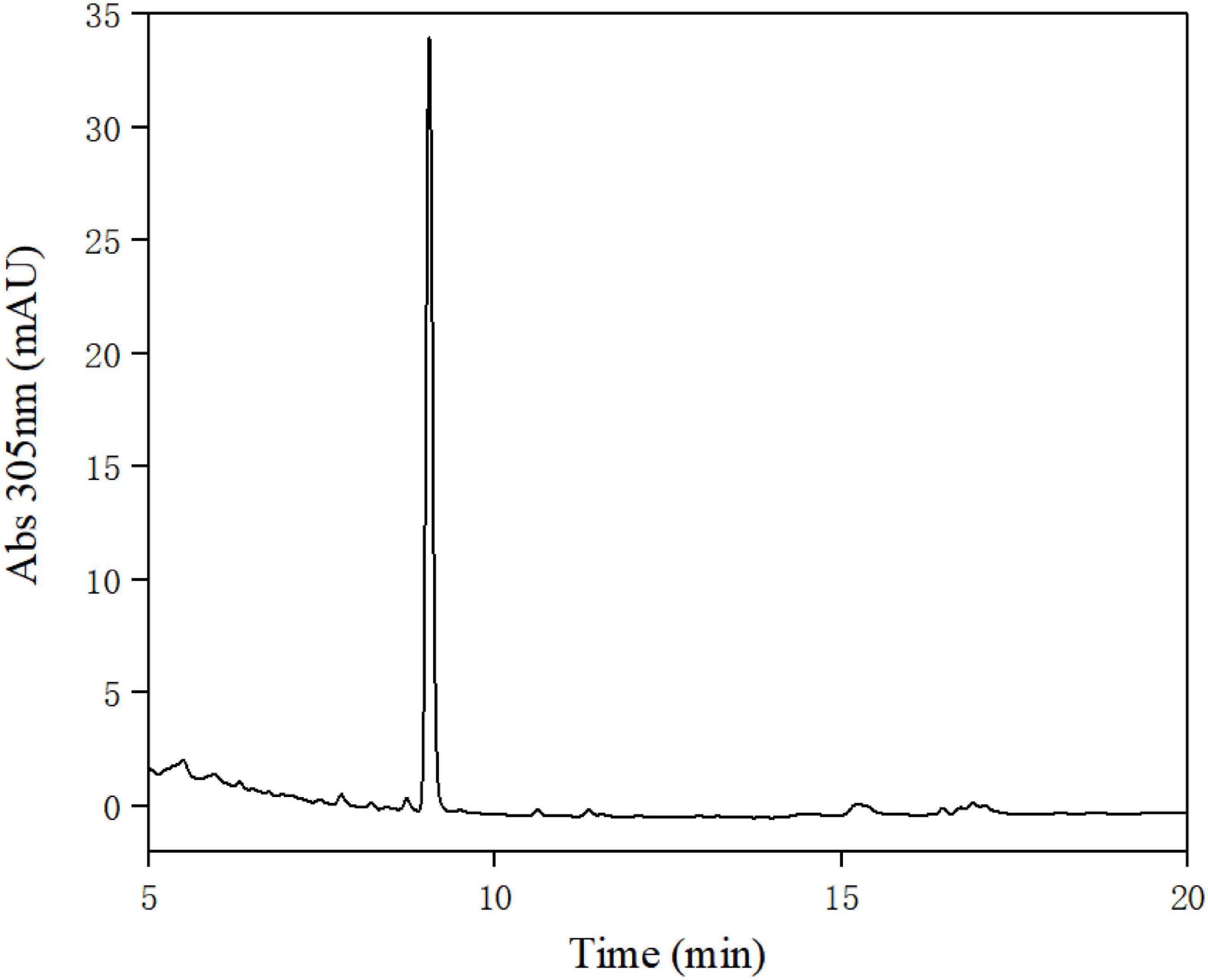
Figure 2. High-performance liquid chromatography (HPLC) analysis of the extract of the fermentation broth of strain FUA027.
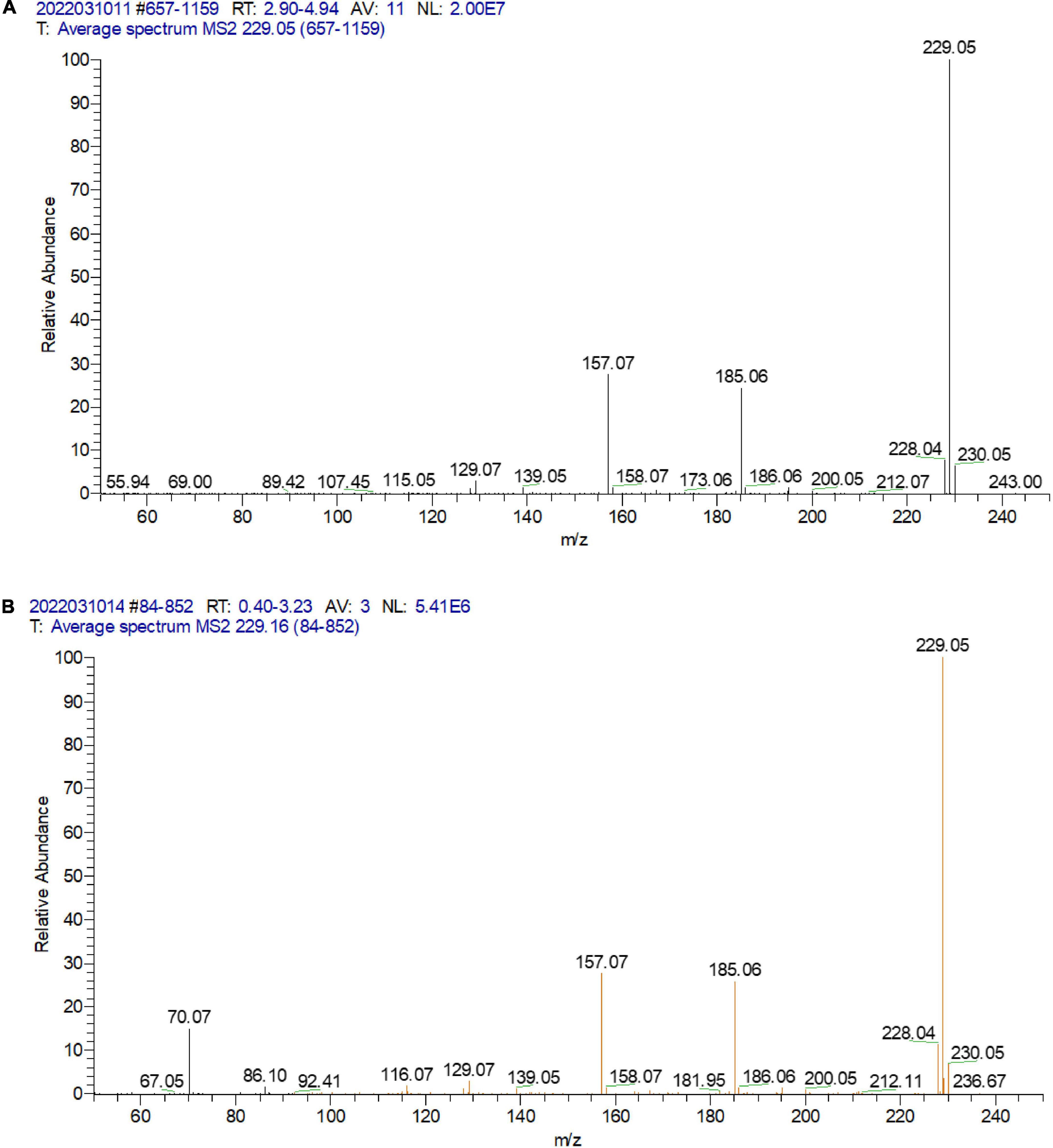
Figure 3. LC-MS/MS analysis of the standard urolithin A (A) and the urolithin A of the extract of the fermentation broth of strain FUA027 (B).
Identification of the urolithin A-producing strain FUA027
The cells of FUA027 strain were gram-positive and rod-shaped (Figure 4). Simultaneously, based on the comparison of the results of physiological and biochemical measurements with the identification standards (32, 40, 41), strain FUA027 was preliminarily identified as E. faecium (Table 1). The 16S rDNA sequence was submitted to GenBank (Accession No.: OM670243). The phylogenetic tree, which was constructed for comparing the 16S rDNA sequences, suggested that strain FUA027 belongs to the genus Enterococcus (Figure 5). The closest relative of this strain was strain ATCC 19434T with a valid published name of E. faecium (99.36% similarity). Therefore, strain FUA027 was identified as E. faecium.
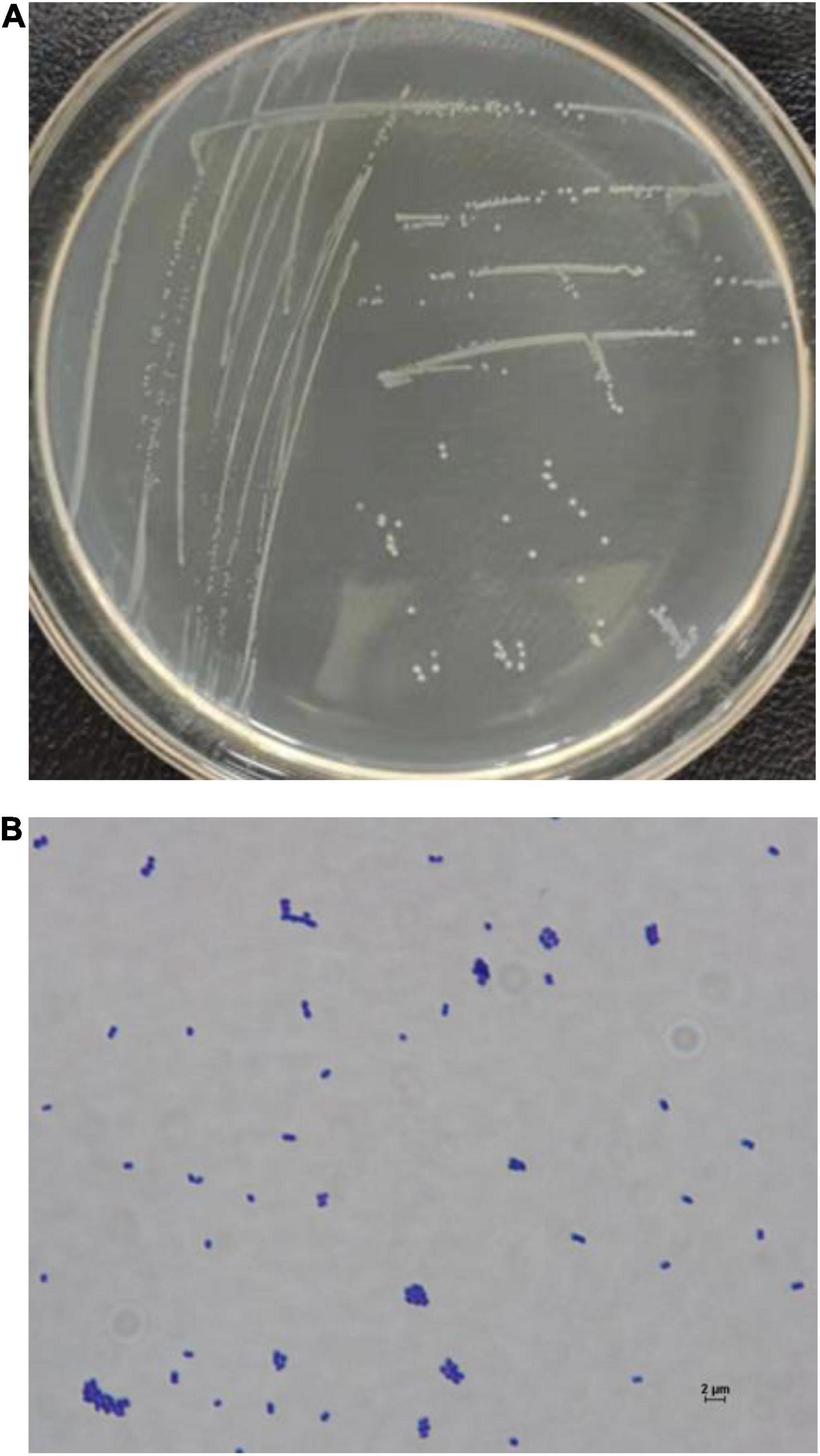
Figure 4. The colony morphology of the strain FUA027 on the ABB plate (A), light microscopic picture (40 × 100 magnification) (B).

Figure 5. Phylogenetic tree of strain FUA027 within the genus Enterococcus based on 16S rDNA sequences. The branching pattern was generated by using the maximum-likelihood method. Based on 100 replications, bootstrap percentages >50% are shown. The bar below the tree represents 1 nucleotide changes per 1,000 nucleotides.
A balance of intestinal flora is beneficial for the health of organisms, conducive to biological activity (7). E. faecium is a part of the normal flora of human and animal intestines. When E. faecium, harmful intestinal bacteria in humans (Escherichia coli), and human intestinal probiotics (Bifidobacterium) were cultured, E. faecium was found to inhibit these harmful bacteria along with Bifidobacterium. E. faecium rapidly adheres to the intestinal mucosa after entering the intestine, forming an intestinal barrier (42, 43) and inducing the production of cytokines, interferons, and interleukins to enhance the body’s nonspecific immunity and improve disease resistance. Thus, E. faecium can be developed into probiotics (40, 44). The strain FUA027 was deposited in China General Microbiological Culture Collection Center (CGMCC) with accession number 24964.
Growth kinetics and time-course transformation of ellagic acid into urolithin A by Enterococcus faecium FUA027
The growth curves of strain FUA027 and its associated transformation of ellagic acid into urolithin A are presented in Figure 6. The lag phase was 5.67 ± 0.78 h, and the growth rate was 0.26 ± 0.04/h in the presence of ellagic acid. Ellagic acid catabolism and urolithin formation occur during the early stage of the mid-logarithmic growth phase of E. faecium FUA027. The first metabolite produced after incubating ellagic acid with E. faecium FUA027 was urolithin C (thihydroxy-urolithin), which was observed at 28 h and whose levels peaked at 40 h, with the maximum concentration being 1.54 μM. Then, the urolithin C levels declined progressively as urolithin A was formed (Figure 6). Urolithin A was detected after 40 h of incubation, and a maximum urolithin A concentration of 10.80 μM was attained at 50 h. Then, the urolithin A levels decreased gradually (Figure 6). Ellagic acid (20.00 μM) was finally converted to urolithin A (6.44 μM). The urolithin A levels decreased gradually is a phenomenon that can’t be fully explained. Further research is necessary to better understand the reasons of phenomenon.
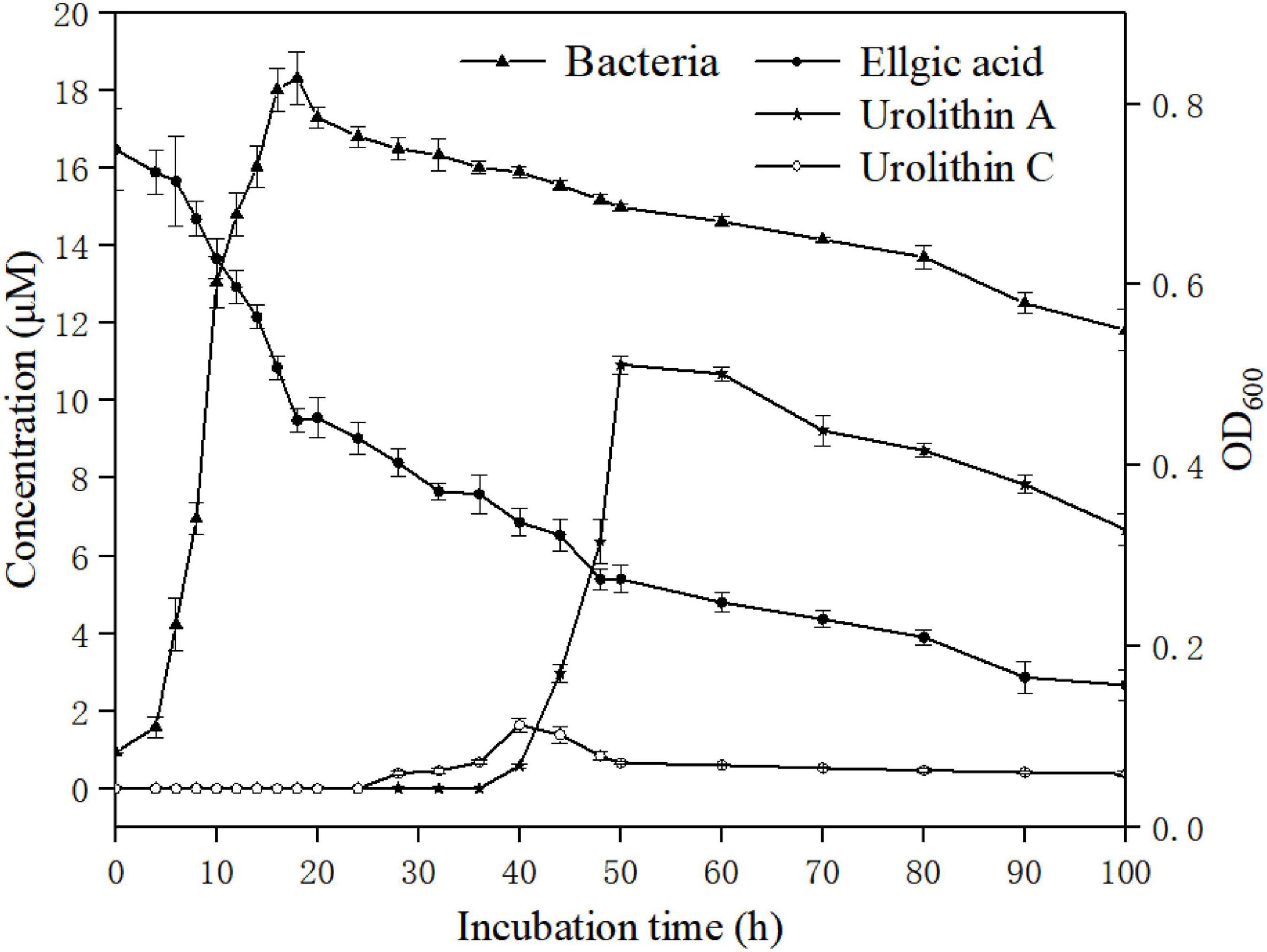
Figure 6. Bacterial growth and time course of urolithin production by the strain FUA027. Ellgic acid (solid circles); Urolithin A (solid stars); Urolithin C (open circles); Bacterial growth curve (solid triangles).
To date, four urolithin-producing strains have been reported; however, they differ from one another in ellagic acid biotransformation processes. Gordonibacter was the first reported genus to transform ellagic acid into urolithins, producing urolithin M5, urolithin M6, and urolithin C. Ellagic acid catabolism and urolithin formation occurred during the stationary growth phase of G. urolithinfaciens and G. pamelaeae. Urolithin C was the end product of ellagic acid transformation by the two strains (23, 24). Ellagic acid and urolithin C catabolism also occurred during the stationary growth phase in E. isourolithinifaciens, but isourolithin A was the end product of ellagic acid catabolism (25). The end products of ellagic acid catabolism by strain B. pseudocatenulatum INIA P815 were urolithins A and B. The transition from ellagic acid to the intermediary urolithins was rapid; however, only urolithins A and B, but not the intermediary urolithins, were detected (26). Similar results have been reported by García-Villalba et al. (34). The conversion rate of urolithin A in a single bacterium (Bifidobacterium pseudocatenulatum INIA P815) reported in the literature is less than 2% (26). Compared with the reported strain, the conversion rate of urolithin A in a single bacterium (E. faecium FUA027) is 32.2%, the conversion rate was higher than the reported strain significantly. FUA027 is the great candidate strain to study the mechanism underlying the biotransformation of ellagic acid into urolithin A.
Conclusion
The transformation of ellagic acid into urolithin A by a single bacterial strain is described herein. This is the first description of an E. faecium strain capable of converting ellagic acid into urolithin A in vitro. The strain E. faecium FUA027 could potentially be useful for the development of next-generation probiotics, functional foods, and food complements and be used to explore the regulation of their transcription.
Data availability statement
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
XZ: conceptualization, methodology, software, and writing – original draft. YF: resources and supervision. GY: conceptualization and resources. XH: data curation and supervision. YH: software and validation. MX and YZ: investigation. FH: formal analysis. SL: methodology, supervision, and project administration. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions, the Key Natural Science Foundation of the Jiangsu Higher Education Institutions of China (20KJA550001), the National Natural Science Foundation of Jiangsu Ocean University (KQ20041 and KQ20052), and Project “333” of Jiangsu Province, Open-end Funds of Jiangsu Key Laboratory of Marine Bioresources and Environment (SH20191204).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Cena H, Calder PC. Defining a healthy diet: evidence for the role of contemporary dietary patterns in health and disease. Nutrients. (2020) 12:334–48. doi: 10.3390/nu12020334
2. Hou K, Wu ZX, Chen XY, Wang JQ, Zhang D, Xiao C, et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. (2022) 7:135.
3. Tomas-Barberan FA, Garcia-Villalba R, Gonzalez-Sarrias A, Tomas- Selma MV, Espín JC. Ellagic acid metabolism by human gut microbiota: consistent observation of three urolithin phenotypes in intervention trials, independent of food source, age, and health status. J. Agric. Food Chem. (2014) 62:6535–8. doi: 10.1021/jf5024615
4. Toney AM, Fox D, Chaidez V, Ramer-Tait AE, Chung S. Immunomodulatory role of urolithin a on metabolic diseases. Biomedicines. (2021) 9:192–210. doi: 10.3390/biomedicines9020192
5. García-Villalba R, Beltran D, Frutos MD, Selma MV, Espín JC, Tomás-Barberán FA. Metabolism of different dietary phenolic compounds by the urolithin producing humangut bacteria Gordonibacter urolithinfaciens and Ellagibacter isourolithinifaciens. Food Funct. (2020) 2020:7012–22. doi: 10.1039/d0fo01649g
6. Alfei S, Marengo B, Zuccari G. Oxidative stress, antioxidant capabilities, and bioavailability: ellagic acid or urolithins? Antioxidants. (2020) 9:707.
7. Zhang M, Cui S, Mao B, Zhang Q, Zhao J, Zhang H, et al. Ellagic acid and intestinal microflora metabolite urolithin: a review on its sources, metabolic distribution, health benefits, and biotransformation. Crit. Rev. Food Sci. Nutr. (2022) 2022:21–3. doi: 10.1080/10408398.2022.2036693
8. Avila-Galvez MA, Gimenez-Bastida JA, Gonzalez-Sarrias A, Espín JC. Tissue deconjugation of urolithin a glucuronide to free urolithin a in systemic inflammation. Food Funct. (2019) 10:3135–41. doi: 10.1039/c9fo00298g
9. Djedjibegovic J, Marjanovic A, Panieri E. Ellagic acid-derived urolithins as modulators of oxidative stress. Oxid. Med. Cell. Longev. (2020) 2020:5194508. doi: 10.1155/2020/5194508
10. García-Villalba R, Gimenez-Bastida JA, Cortés-Martín A, Ávila-Gálvez MÁ, Tomás-Barberán FA, Selma MV, et al. Urolithins: a comprehensive update on their metabolism, bioactivity, and associated gut microbiota. Mol. Nutr. Food Res. (2022) 2022:e2101019. doi: 10.1002/mnfr.202101019
11. Cortes-Martin A, Garcia-Villalba R, Gonzalez-Sarrias A, Romo-Vaquero M, Loria-Kohen V, Ramírez-de-Molina A, et al. The gut microbiota urolithin metabotypes revisited: the human metabolism of ellagic acid is mainly determined by aging. Food Funct. (2018) 9:4100–6. doi: 10.1039/c8fo00956b
12. Selma MV, Beltran D, Luna MC, Romo-Vaquero M, García-Villalba R, Mira A, et al. Isolation of human intestinal bacteria capable of producing the bioactive metabolite isourolithin a from ellagic acid. Front. Microbiol. (2017) 8:1521. doi: 10.3389/fmicb.2017.01521
13. Romo-Vaquero M, Cortes-Martin A, Loria-Kohen V, Ramírez-de-Molina A, García-Mantrana I, Collado MC, et al. Deciphering the human gut microbiome of urolithin metabotypes: association with enterotypes and potential cardiometabolic health implications. Mol. Nutr. Food Res. (2019) 63:e1800958. doi: 10.1002/mnfr.201800958
14. Yang J, Guo Y, Henning SM, Chan B, Long J, Zhong J, et al. Ellagic acid and its microbial metabolite urolithin a alleviate diet-induced insulin resistance in mice. Mol. Nutr. Food Res. (2020) 64:e2000091. doi: 10.1002/mnfr.202000091
15. Singh A, D’Amico D, Andreux PA, Fouassier AM, Blanco-Bose W, Evans M, et al. Urolithin a improves muscle strength, exercise performance, and biomarkers of mitochondrial health in a randomized trial in middle-aged adults. Cell Rep. Med. (2022) 3:100633. doi: 10.1016/j.xcrm.2022.100633
16. Kang I, Buckner T, Shay NF, Gu L, Chung S. Improvements in metabolic health with consumption of ellagic acid and subsequent conversion into urolithins: evidence and mechanisms. Adv. Nutr. (2016) 7:961–72. doi: 10.3945/an.116.012575
17. Andreux PA, Blanco-Bose W, Ryu D, Burdet F, Ibberson M, Aebischer P, et al. The mitophagy activator urolithin a is safe and induces a molecular signature of improved mitochondrial and cellular health in humans. Nat. Metab. (2019) 1:595–603. doi: 10.1038/s42255-019-0073-4
18. Ryu D, Mouchiroud L, Andreux PA, Katsyuba E, Moullan N, Nicolet-Dit-Félix AA, et al. Urolithin a induces mitophagy and prolongs lifespan in c. elegans and increases muscle function in rodents. Nat. Med. (2016) 22:879–88. doi: 10.1038/nm.4132
19. D’Amico D, Andreux PA, Valdes P, Singh A, Rinsch C, Auwerx J. Impact of the natural compound urolithin a on health, disease, and aging. Trends Mol. Med. (2021) 27:687–99. doi: 10.1016/j.molmed.2021.04.009
20. DaSilva NA, Nahar PP, Ma H, Eid A, Wei Z, Meschwitz S, et al. Pomegranate ellagitannin-gut microbial-derived metabolites, urolithins, inhibit neuroinflammation in vitro. Nutr. Neurosci. (2017) 22:185–95. doi: 10.1080/1028415X.2017.1360558
21. Evtyugin DD, Magina S, Evtuguin DV. Recent advances in the production and applications of ellagic acid and its derivatives. a review. Molecules. (2020) 25:2745. doi: 10.3390/molecules25122745
22. Espin JC, Gonzalez-Sarrias A, Tomas-Barberan FA. The gut microbiota: a key factor in the therapeutic effects of (poly) phenols. Biochem. Pharmacol. (2017) 139:82–93. doi: 10.1016/j.bcp.2017.04.033
23. Selma MV, Beltran D, Garcia-Villalba R, Espín JC, Tomás-Barberán FA. Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter species. Food Funct. (2014) 5:1779–84. doi: 10.1039/c4fo00092g
24. Selma MV, Tomas-Barberan FA, Beltran D, García-Villalba R, Espín JC. Gordonibacter urolithinfaciens sp. nov., a urolithin-producing bacterium isolated from the human gut. Int. J. Syst. Evol. Microbiol. (2014) 64:2346–52. doi: 10.1099/ijs.0.055095-0
25. Beltran D, Romo-Vaquero M, Espin JC. Ellagibacter isourolithinifaciens gen. nov., sp. nov., a new member of the family Eggerthellaceae, isolated from human gut. Int. J. Syst. Evol. Microbiol. (2018) 68:1707–12. doi: 10.1099/ijsem.0.002735
26. Gaya P, Peirotén Á, Medina M. Bifidobacterium pseudocatenulatum INIA P815: the first bacterium able to produce urolithins a and b from ellagic acid. J. Funct. Foods. (2018) 45:95–9.
27. Espin JC, Larrosa M, Garcia-Conesa MT. Biological significance of urolithins, the gut microbial ellagic acid-derived metabolites: the evidence so far. Evid-based complement. Alternat Med. (2013) 2013:697–715. doi: 10.1155/2013/270418
28. Yang L, Hou K, Zhang B, Ouyang C, Lin A, Xu S. Preservation of the fecal samples at ambient temperature for microbiota analysis with a cost-effective and reliable stabilizer EffcGut. Sci Total Environ. (2020) 741:140423. doi: 10.1016/j.scitotenv.2020.140423
29. Danylec N, Gobl A, Stoll DA, Hetzer B, Kulling SE, Huch M, et al. Rubneribacter badeniensis gen. nov., sp. nov. and Enteroscipio rubneri gen. nov., sp. nov., new members of the Eggerthellaceae isolated from human faeces. Int. J. Syst. Evol. Microbiol. (2018) 68:1533–40. doi: 10.1099/ijsem.0.002705
30. Maini Rekdal V, Bess EN, Bisanz JE, Turnbaugh PJ, Balskus EP. Discovery and inhibition of an interspecies gut bacterial pathway for levodopa metabolism. Science. (2019) 364:1055. doi: 10.1126/science.aau6323
31. Feria-Gervasio D, Tottey W, Gaci N, Alric M, Cardot JM, Peyret P, et al. Three-stage continuous culture system with a self-generated anaerobia to study the regionalized metabolism of the human gut microbiota. J. Microbiol. Methods. (2014) 96:111–8. doi: 10.1016/j.mimet.2013.11.015
32. Bergey DH. Genus trichodesmium. In: Holt JG editor. Bergey’s manual of determinative bacteriology. Philadelphia, PA: Williams & Wilkins (1994). p. 395–6.
33. Ayyash M, Abushelaibi A, Al-Mahadin S. In-vitro investigation into probiotic characterisation of streptococcus and enterococcus isolated from camel milk. LWT. (2018) 87:478–87. doi: 10.1016/j.lwt.2017.09.019
34. Garcia-Villalba R, Beltran D, Espin JC, Selma MV, Tomás-Barberán FA. Time course production of urolithins from ellagic acid by human gut microbiota. J. Agric. Food Chem. (2013) 61:8797–806. doi: 10.1021/jf402498b
35. González-Sarrías A, García-Villalba R, Núñez-Sánchez M. Identifying the limits for ellagic acid bioavailability: a crossover pharmacokinetic study in healthy volunteers after consumption of pomegranate extracts. J. Funct. Foods. (2015) 19:225–35. doi: 10.1016/j.jff.2015.09.019
36. Tomas-Barberan F, Garcia-Villalba R, Quartieri A. In vitro transformation of chlorogenic acid by human gut microbiota. Mol. Nutr. Food Res. (2014) 58:1122–31. doi: 10.1002/mnfr.201300441
37. Tomas-Barberan FA, Gonzalez-Sarrias A, Garcia-Villalba R, Núñez-Sánchez MA, Selma MV, García-Conesa MT, et al. Urolithins, the rescue of “old” metabolites to understand a “new” concept: metabotypes as a nexus among phenolic metabolism, microbiota dysbiosis, and host health status. Mol. Nutr. Food Res. (2017) 61:1–36. doi: 10.1002/mnfr.201500901
38. Watanabe H, Kishino S, Kudoh M, Yamamoto H, Ogawa J. Evaluation of electron-transferring cofactor mediating enzyme systems involved in urolithin dehydroxylation in Gordonibacter urolithinfaciens DSM 27213. J. Biosci. Bioeng. (2020) 129:552–7. doi: 10.1016/j.jbiosc.2019.11.014
39. Selma MV, Romo-Vaquero M, Garcia-Villalba R, González-Sarrías A, Tomás-Barberán FA, Espín JC, et al. The human gut microbial ecology associated with overweight and obesity determines ellagic acid metabolism. Food Funct. (2016) 7:1769–74. doi: 10.1039/c5fo01100k
40. Shi Y, Zhai M, Li J, Li B. Evaluation of safety and probiotic properties of a strain of Enterococcus faecium isolated from chicken bile. Food Sci. Technol. (2020) 57:578–87. doi: 10.1007/s13197-019-04089-7
41. Dosuky AS, Elsayed TR, Yousef ET, Barakat OS, Nasr NF. Isolation, identification, and application of lactic acid-producing bacteria using salted cheese whey substrate and immobilized cells technology. J. Genet. Eng. Biotechnol. (2022) 20:26. doi: 10.1186/s43141-022-00316-5
42. Singh R, Chandrashekharappa S, Bodduluri SR. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat. Commun. (2019) 10:89. doi: 10.1038/s41467-018-07859-7
43. Suez J, Zmora N, Segal E, Elinav E. The pros, cons, and many unknowns of probiotics. Nat. Med. (2019) 25:716–29. doi: 10.1038/s41591-019-0439-x
Keywords: ellagic acid, urolithin A, Enterococcus faecium, novel probiotic, metabolism
Citation: Zhang X, Fang Y, Yang G, Hou X, Hai Y, Xia M, He F, Zhao Y and Liu S (2022) Isolation and characterization of a novel human intestinal Enterococcus faecium FUA027 capable of producing urolithin A from ellagic acid. Front. Nutr. 9:1039697. doi: 10.3389/fnut.2022.1039697
Received: 08 September 2022; Accepted: 26 October 2022;
Published: 09 November 2022.
Edited by:
Guangxu Ren, Institute of Food and Nutrition Development (CAAS), ChinaReviewed by:
Peijun Tian, Jiangnan University, ChinaKaijian Hou, Shantou University, China
Liang Zhao, China Agricultural University, China
Copyright © 2022 Zhang, Fang, Yang, Hou, Hai, Xia, He, Zhao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shu Liu, amRsaXVzaHVAMTYzLmNvbQ==
 Xiaomeng Zhang
Xiaomeng Zhang Yaowei Fang
Yaowei Fang Guang Yang1,2,3
Guang Yang1,2,3 Shu Liu
Shu Liu