- 1School of Animal Science and Technology, Jiangxi Agricultural University, Nanchang, China
- 2Shenzhen Aohua Group Co., Ltd., Shenzhen, China
- 3Department of Animal, Veterinary and Food Sciences, Aquaculture Research Institute, University of Idaho, Moscow, ID, United States
The intestinal dysfunction induced by high plant protein diets is frequently observed in farmed fish, and probiotics of Bacillus genus were documented to benefit the intestinal health through the modulation of intestinal microbiota without clearness in its underlying mechanism yet. Fusobacteria, Proteobacteria, and Firmicutes were observed to be the dominate phyla, but their proportion differentiated in the intestinal bacterial community of Pengze crucian carp (Carassius auratus var. Pengze) fed different diets in this study. Dietary supplementation of B. cereus and B. subtilis could reshape the intestinal bacterial community altered by high plant protein diets through a notable reduction in opportunistic pathogen Aeromonas together with an increase in Romboutsia and/or Clostridium_sensu_stricto from Firmicutes. Due to the alteration in the composition of bacterial community, Pengze crucian carp exhibited characteristic ecological networks dominated by cooperative interactions. Nevertheless, the increase in Aeromonas intensified the competition within bacterial communities and reduced the number of specialists within ecological network, contributing to the microbial dysbiosis induced by high plant protein diets. Two probiotics diets promoted the cooperation within the intestinal bacterial community and increased the number of specialists preferred to module hubs, and then further improved the homeostasis of the intestinal microbiota. Microbial dysbiosis lead to microbial dysfunction, and microbial lipopolysaccharide biosynthesis was observed to be elevated in high plant protein diets due to the increase in Aeromonas, gram-negative microbe. Probiotics B. cereus and B. subtilis restored the microbial function by elevating their amino acid and carbohydrate metabolism together with the promotion in the synthesis of primary and secondary bile acids. These results suggested that dietary supplementation of probiotics B. cereus and B. subtilis could restore the homeostasis and functions of intestinal microbiota in Pengze crucian carp fed high plant protein diets.
Introduction
Aquaculture industry, one of the fastest growing animal food-producing sectors in agriculture, provides a large amount of high-quality protein to consumers around the world (1, 2). In recent decades, intestinal dysfunction is widely observed in farmed fish due to the excessive use of plant protein in commercial feed, and substantial evidences indicated that the occurrence of intestinal dysfunction is usually accompanied by a dysbiosis of intestinal microbiota (3–5). It is well-accepted that micro-ecological ecosystem formed by symbiotic microbes plays a crucial role in the health status of host through production of digestion-related enzymes, vitamins synthesis, protection from pathogens, and promotion of immune maturation, etc. (6, 7). Hence, regulating the homeostasis of intestinal microbiota would be one effective strategy to maintain the health of farmed fish.
Probiotics, defined as live microorganisms, have attracted more and more attentions for their benefits to the homeostasis of intestinal microbiota in aquatic animal, as well as growth performance, nutrient digestion, antioxidant capacity, and immunity system (8). Bacillus genus serves as one of the most common probiotic species and is widely applied to aquaculture industry (9, 10). Numerous researches have investigated the effect of Bacillus species on the intestinal microbiota of fishes like grass carp (Ctenopharyngodon idellus) (11), turbot (Scophthalmus maximus) (12), common carp (Cyprinus carpio L.) (13), olive flounder (Paralichthys olivaceus) (14), Nile tilapia (Oreochromis niloticus) (15), and tongue sole (Cynoglossus semilaevis) (16). Nevertheless, most of these studies in farmedc fish focus on the effects of Bacillus species on the composition of bacterial community, which is not enough to interpret the modulation of intestinal microbiota homeostasis by probiotics.
The number of species and their abundance are the most basic elements for constructing the bacterial community, whereas the homeostasis of intestinal microbiota depends on complex species-species interactions within the bacterial community (17). Physiological benefits were observed to be associated with probiotics consumption in human and animal model without significant effects on microbial composition, indicating that probiotics promoted the homeostasis of intestinal microbiota rather than altered its composition (18, 19). Trillions of bacteria, residing predominantly in the gastrointestinal tract, interact with each other to accomplish systems functions through the flow of energy, matter, and information (20). Microbes in micro-ecological ecosystem depend on interspecific interactions to form a dynamic ecological network, in which species perform different topological roles due to their ecological niche (20). Previous evidence indicated that B. cereus G19 could affect microbial interactions and increase the number of generalists to improve the intestinal microbiota homeostasis (21). The symbiotic microbes in the intestine are verified to play a crucially important role in the host metabolism, since microbes possess 100-fold genes more than host and can synthesize a large number of enzymes (22). Therefore, intestinal microbiota is considered to be an auxiliary metabolic organ and participate in metabolic process of host, such as amino acid, carbohydrate, energy, and lipid metabolism through provision of fermentation end products (23–26). Relying on complex species-species interactions, the bacterial community maintains its stability in the intestine, and also accomplishes a system metabolic function simultaneously (27, 28).
Pengze crucian carp (Carassius auratus var. Pengze) is a widely farmed freshwater omnivorous fish in China. Previous studies of crucian carp confirmed that high plant protein diets were damaged to the intestinal health (29, 30), while probiotics Bacillus could recover the negative effects and improve growth, antioxidant capability and filet quality (31–34). The studies about Pengze crucian carp have documented that B. cereus (35, 36) and B. subtilis (37) could improve the growth performance and intestinal health status, while the two probiotics on intestinal microbiota was unclear. And thus, the objective of the current study was to investigate the effects of B. cereus and B. subtilis on the intestinal microbiota, and further explore their underlying mechanisms of modulating the intestinal microbiota homeostasis.
Materials and methods
Experimental procedure and sample collection
Juvenile Pengze crucian carp (Carassius auratus var. Pengze) were bought from Fishery Science Research Institute, Jiujiang. Prior to the experiment, the fish were reared in a floating cage (4.0 × 4.0 × 2.0 m) and fed basal diets for 3 weeks to acclimatize to the experiment conditions. After being fasted for 24 h, 180 similar-sized individuals (mean initial weight 12.91 ± 0.02 g) were randomly distributed into nine cages (1.5 × 1.5 × 1.5 m). Each group has three replicates with a density of 20 fish per cage. The dietary ingredient preparation, weighed accurately and mixed thoroughly, basal diets (Control group) made well and dried, were followed by the standard procedure of diets made for the Pengze crucian carp (36, 37). And then, abundant fresh cells of the Bacillus cereus (CD group) and B. subtilis (BS group) obtained through spreading cultivation, and mixed thoroughly with the basal diets at the dose of 1 × 109 CFU/kg by spraying. The dose of two probiotics was chosen in accordance with previous studies (36, 37), and the concentration of probiotics B. cereus and B. subtilis was determine by countess II automated cell counter (Thermo Fisher Scientific, Shanghai, China) before mixing. Fish were fed to apparent satiation three times a day (8: 00, 13: 00, and 16: 00) for 70 days. During the experimental period, water quality conditions were stable (water temperature, 25.5 ± 3.2°C; dissolved oxygen > 6.0 mg L−1; NH4+-N <0.3 mg L−1; -N <0.1 mg L−1, respectively). At the end of the experiment, prior to sampling, experimental fish were anesthetized with 100 mg/L MS222 (Tricaine methanesulfonate, Sigma-Aldrich Co. LLC.). The intestinal content in hindgut from eight Pengze crucian carp in each group was collected and frozen at −80°C until further analysis. The experimental protocols together with Pengze crucian carp handling and sampling have been approved by the Committee on Research Ethics of the Department of Laboratory Animal Science, Jiangxi Agricultural University.
Illumina sequencing of bacterial 16S rRNA gene
PowerFecal™ DNA Isolation Kit (MoBio Laboratories, Inc.) was used for DNA extraction of digesta samples. Amplification of the 16S rRNA V3-V4 region was performed as described previously with barcoded fusion primers of 341F and 805R (38). High-throughput sequencing was performed using the Illumina HiSeq platform. All of the sequencing data can be found in the Sequence Read Archive (SRA) database at NCBI under accession number PRJNA872491.
Bioinformatics and statistical analysis
The raw sequences were sorted into different samples according to the barcodes by using the BIPES pipeline, followed by a quality-control step remove low-quality amplicon sequences by VSEARCH (39). The clean sequences were then clustered into operational taxonomic units (OTUs) with 99% sequence similarity and annotated using the Ribosomal Database (rdp_16s_v16_sp). A total of 3,056,535 effective sequences and 1,375 OTUs were generated. Alpha diversity and the relative abundance of taxa analyses were calculated by R software v 4.1.3. The Wilcoxon test was used to test the α-diversity index, and the relative abundance of taxa using R software. Principal coordinates analysis (PCoA) based on the Bray-Curtis dissimilarity analyses was employed to visualize bacterial community structure and the difference in bacterial community was calculated by permutational analysis of variance (PERMANOVA) based on Bray–Curtis distance (40).
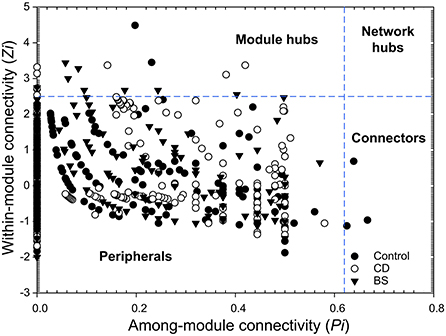
Based on the abundance profiles of individual OTUs, molecular ecological network analysis was performed to evaluate species-species interactions within bacterial community (http://ieg2.ou.edu/MENA). Random matrix theory (RMT)-based approach was used for ecological network construction and topological roles identification (41). The network was visualized using Circos and Cytoscape 3.9.0, respectively. Based on modularity property, each network was separated into modules by the fast-greedy modularity optimization. According to values of within-module connectivity (Zi) and among module connectivity (Pi), the topological roles of different nodes can be categorized into four types: peripherals (Zi ≤ 2.5, Pi ≤ 0.62), connectors (Zi ≤ 2.5, Pi > 0.62), module hubs (Zi > 2.5, Pi ≤ 0.62) and network hubs (Zi > 2.5, Pi > 0.62).
Functional gene and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were predicted using PICRUSt2 software (42) against a Greengenes reference database (Greengenes 13.5). And then, the non-metric multidimensional scaling (NMDS) and analysis of similarity (ANOSIM) were used to evaluate the overall differences in predicted bacterial functional composition related to metabolism based on Bray-Curtis distance at KEGG orthology (KO) level (43). A two-sided Welch's t-test was used to identify significant different metabolic pathways in the two groups by software STAMP (44), with P < 0.05 considered significant.
Results
Diversity and composition of the bacterial community
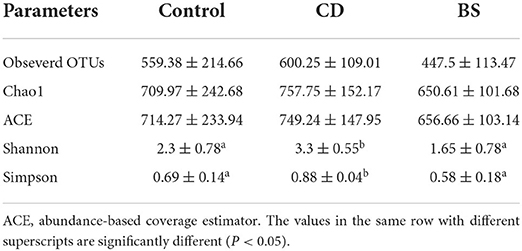
Compared to the Control group, dietary supplementation of B. cereus increased the number of observed OTUs, Chao1, ACE, Shannon, and Simpson in bacterial community of Pengze crucian carp, whilst a significant difference was recorded in Shannon and Simpson (P < 0.05, Table 1). However, no notable difference in α-diversity index mentioned above was observed between BS and Control groups. As shown in Figure 1, the bacterial community of carp was predominated by Fusobacteria (Control: 49.48%; CD: 19.25%; BS: 26.01%), Proteobacteria (Control: 26.16%; CD: 27.63%; BS: 11.60%), and Firmicutes (Control: 20.26%; CD: 47.45%; BS: 60.59%). At class level, fish fed basal diets was mainly rich in Fusobacteriia (49.48%), Gammaproteobacteria (11.86%), Bacilli (11.23%), Alphaproteobacteria (11.15%), and Clostridia (6.50%); Clostridia (28.33%), Fusobacteriia (19.25%), Bacilli (16.58%), Gammaproteobacteria (12.22%), and Alphaproteobacteria (11.23%) took dominate in the intestinal bacterial community in CD group; BS group recorded a high percentage of Clostridia (59.16%), Fusobacteriia (26.01%), Alphaproteobacteria (6.65%), and Gammaproteobacteria (4.15%) in bacterial community (Figure 2).
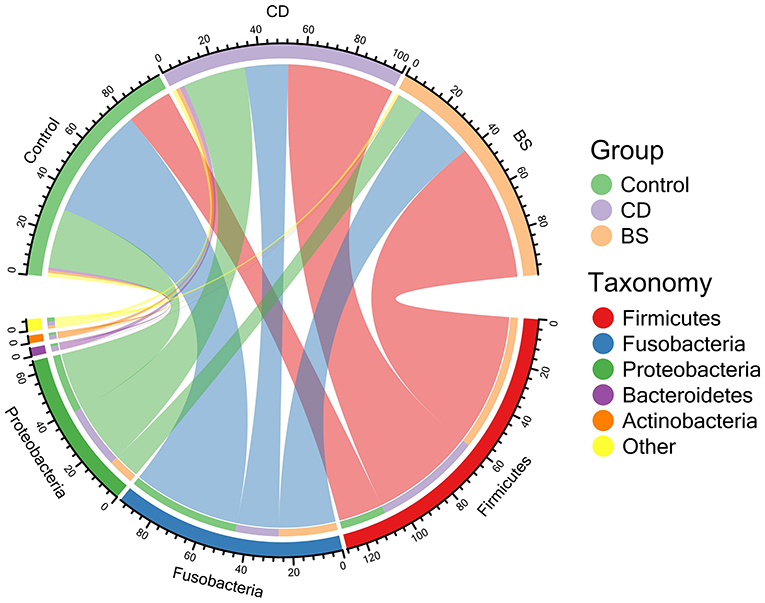
Figure 1. Chord diagram exhibited the relative abundance of bacterial phyla above ≥ a cutoff value of 2%.
Figure 3 exhibited a significant difference in microbial species among three groups. Fish fed high plant protein diets had a higher average relative abundance of Aeromonas (8.39%, P < 0.05) and Cetobacterium (46.49%, P < 0.01) compared to BS group (2.39 and 24.00%, respectively), whilst Control group was higher than CD group (17.08%) in the value for Cetobacterium (P < 0.01). CD group recorded a higher average relative abundance of Aeromonas (6.65%, P < 0.05) and Clostridium_sensu_stricto (5.45%, P < 0.01) compared to BS group (2.39 and 1.92%, respectively). A lowest relative abundance of Romboutsia was observed in Control group (2.20%), which was dramatically different from that in CD (22.65%) and BS (56.40%) groups (P < 0.01); BS group was higher than CD group (P < 0.01) in this respect simultaneously. Strikingly, principal coordinates analysis (PCoA) displayed a clear separation of bacterial community among three groups at OTU level, and significant differences among each other were further confirmed by PERMANOVA (P < 0.001, Figure 4).
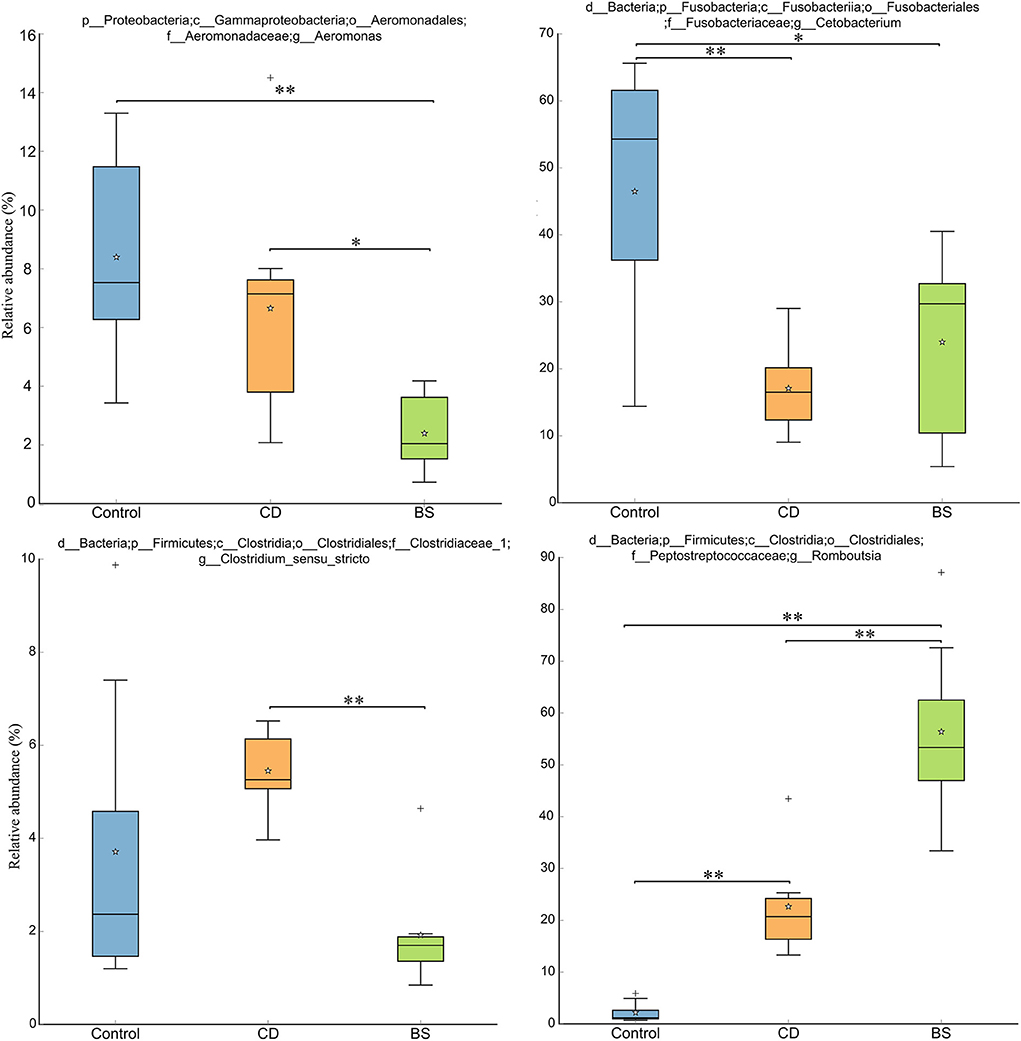
Figure 3. Box plots showing significant variations of relative abundances of intestinal microbiota. **P < 0.01, *P < 0.05.
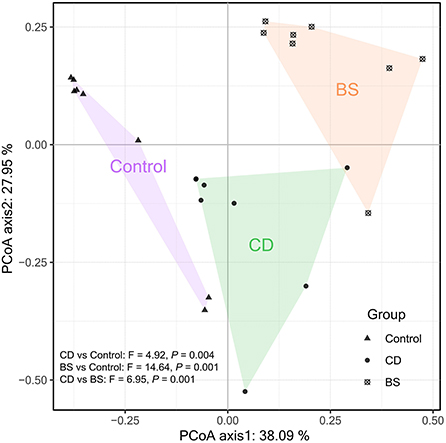
Figure 4. Principal coordinates analysis (PCoA) plot based on Bray-Curtis dissimilarity visualizing dissimilarities in the intestinal bacterial community.
Ecological network analysis
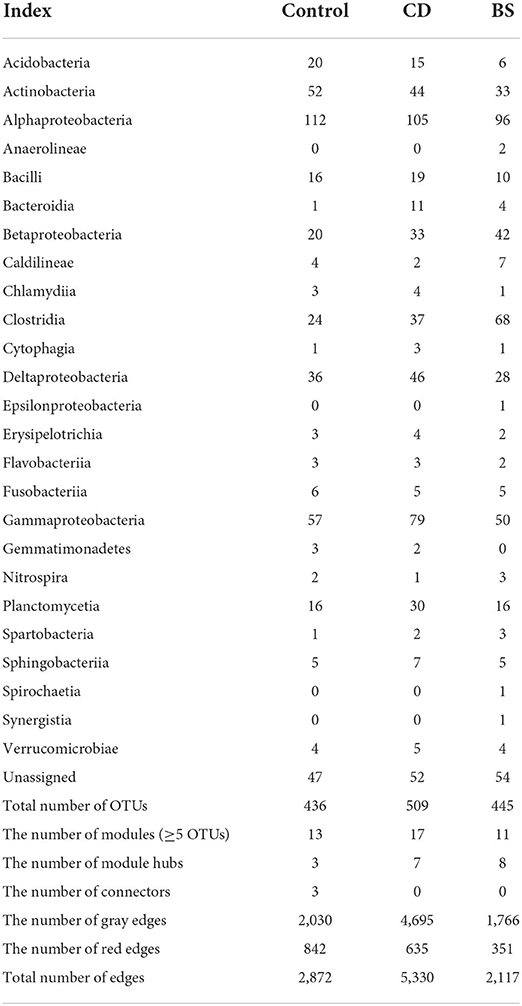
Circos plot described interactions across different species within bacterial community, and 26 different bacterial classes were observed among three groups (Figure 5A). OTUs from Alphaproteobacteria, Gammaproteobacteria, Clostridia, Actinobacteria, Deltaproteobacteria, Betaproteobacteria, and Planctomycetia were recorded to take dominate in the ecological networks within bacterial community (Table 2). Within Control network, there were 436 OTUs and 2,872 edges including 2,030 gray edges (positive interactions) and 842 red edges (negative interactions) between two OTUs. Total of 509 OTUs and 5,330 edges (gray edges: 4,695; red edges: 635) were observed in CD network, and BS network consisted of 445 OTUs and 2,117 edges (gray edges: 1,766; red edges: 351).
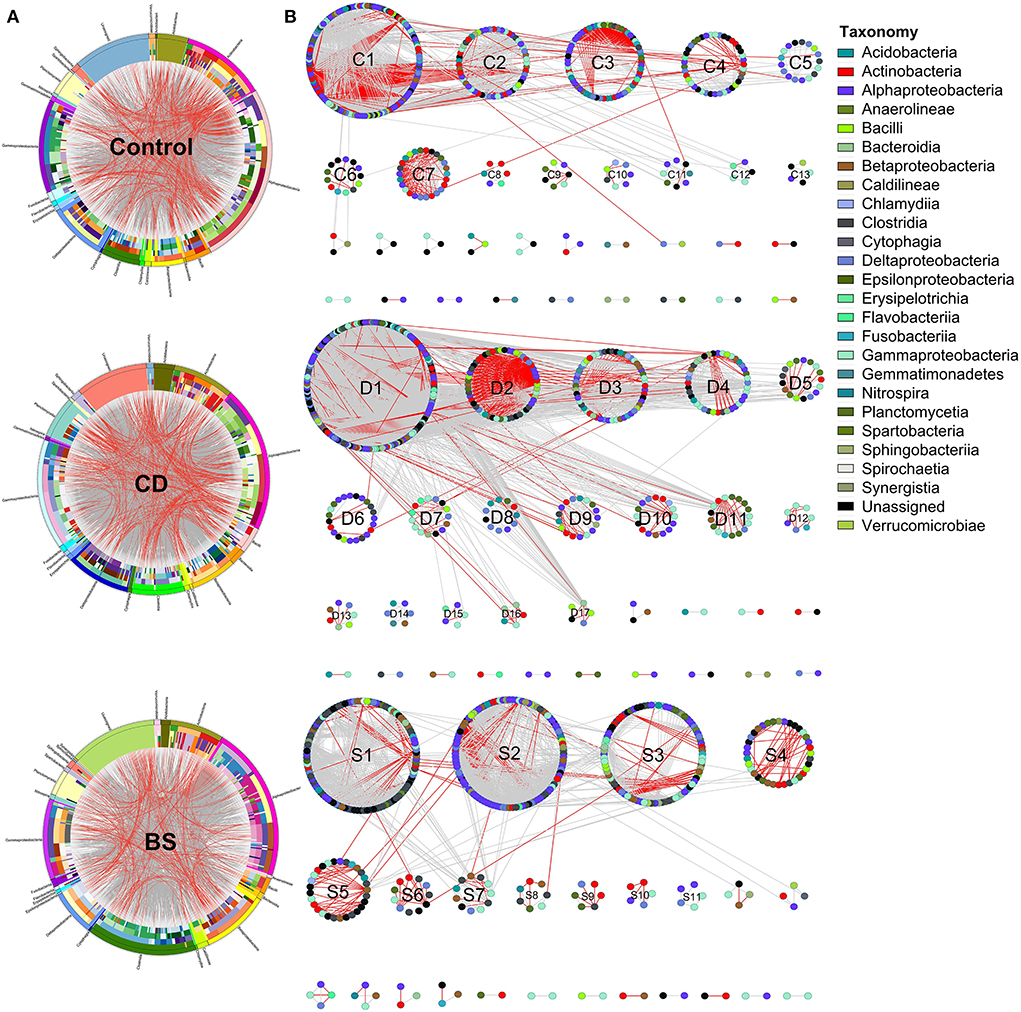
Figure 5. Circular plot (A) and ecological network (B) descriptions of the interspecific interactions within bacterial community. The width of the bars represents the abundance of each taxon. The bands with different colors demonstrate the source of different genera. The taxonomic levels were class, order, family, genera, and species from the outside to the inside of the circle, respectively. Each node in network graph indicates one OTU. Colors of the nodes indicate different major classes. The edges (gray edge, positive interaction and red edge, negative interaction) inside the circle and ecological network represent the interactions between species.
Different separate modules were observed in three ecological networks (Figure 5B). The bacterial ecological network in Control group had 13 submodules (≥ 5 nodes), among which four submodules recorded more than 30 nodes including C1 (141 OTUs), C2 (53 OTUs), C3 (63 OTUs), and C4 (41 OTUs); 17 submodules with more than five nodes were observed in CD network, in which submodules such as D1 (182 OTUs), D2 (54 OTUs), D3 (54 OTUs), and D4 (40 OTUs) were four biggest submodules; BS network possessed 11 submodules (≥5 nodes), among which S1 (141 OTUs), S2 (53 OTUs), S3 (63 OTUs), and S3 (41 OTUs) contained more than 30 nodes. Positive interactions took dominate in these three networks, whereas many red edges were observed between C1 and C2 submodules.
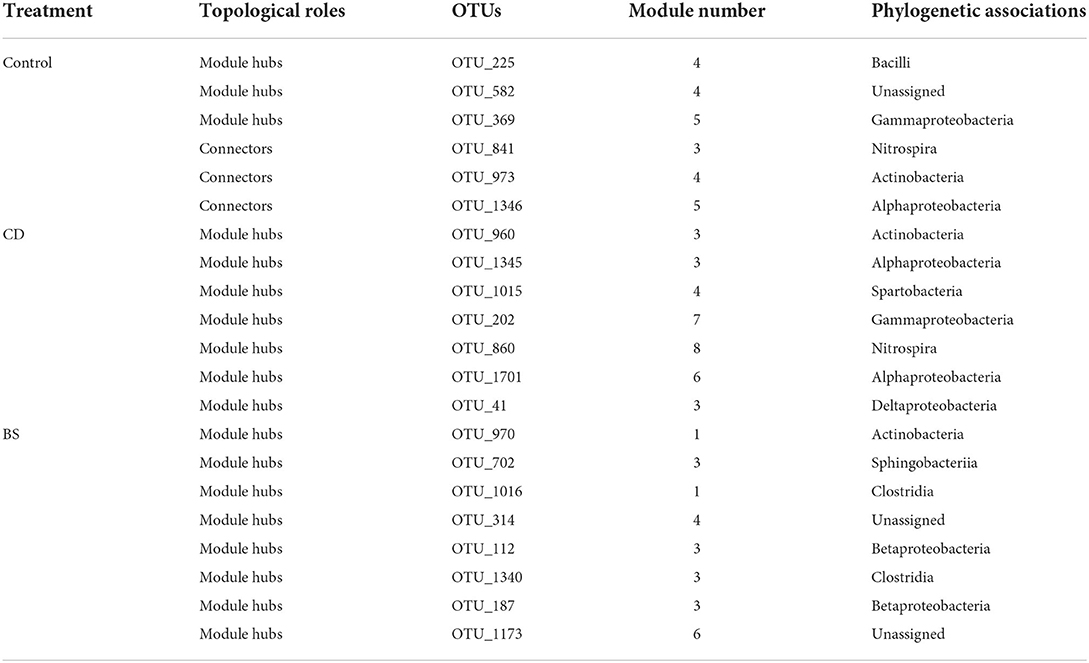
The species performed different topological roles in the ecological network, in which most of the nodes were peripherals and several nodes performed as module hubs or connectors (Figure 6). Control network 3 module hubs and 3 connectors mainly from submodules C3 (one OTU from Nitrospira), C4 (three OTUs from Bacilli, Actinobacteria, and Unassigned bacteria), and C5 (two OTUs from Alphaproteobacteria and Gammaproteobacteria); only 7 module hubs were observed in D3 (three OTUs from Actinobacteria, Alphaproteobacteria, and Gammaproteobacteria), D4 (one OTU from Spartobacteria), D6 (one OTU from Alphaproteobacteria), D7 (one OTU from Gammaproteobacteria), D8 (one OTU from Nitrospira) from CD network; similarly, 7 module hubs were found in S1 (two OTUs from Actinobacteria and Clostridia), S3 (four OTUs from Betaproteobacteria, Clostridia, and Sphingobacteriia), S4 (one OTU from Unassigned bacteria), and S6 one OTU from Unassigned bacteria) from BS network (Table 3).
Functional predictions of intestinal microbiota with PICRUSt2
To understand the bacterial function of the Pengze crucian carp, 7,341 KEGG orthology groups (KOs) were obtained through PICRUSt2 in this study (Figure 7A). The bacterial functional composition was clustered to three groups, and a significant difference between each other was further confirmed through ANOSIM (P < 0.05). KEGG functional categories related to metabolic function were further analyzed including Amino acid metabolism, Carbohydrate metabolism, Digestive system, Energy metabolism, Glycan biosynthesis and metabolism, Lipid metabolism, and Protein families: metabolism. There were 24 dramatically different metabolic pathways and one protein family observed between CD and Control groups (P < 0.05). Compared to Control group, the bacterial community in CD group possessed 18 enriched metabolic pathways involved in amino acid metabolism (three pathways), carbohydrate metabolism (four pathways), glycan biosynthesis and metabolism (five pathways), and lipid metabolism (six pathways, Figure 7B). Figure 7C exhibited 42 metabolic pathways and seven protein families between CD and BS groups, and microbial function related to amino acid and lipid metabolism was more active in CD group. A total of 27 significantly different metabolic pathways and seven protein families was recorded between BS and Control groups (P < 0.05, Figure 7D), and BS group was rich in five pathways from amino acid metabolism, two pathways from carbohydrate metabolism, two pathways from energy metabolism, three pathways from lipid metabolism, and four protein families related to metabolism.
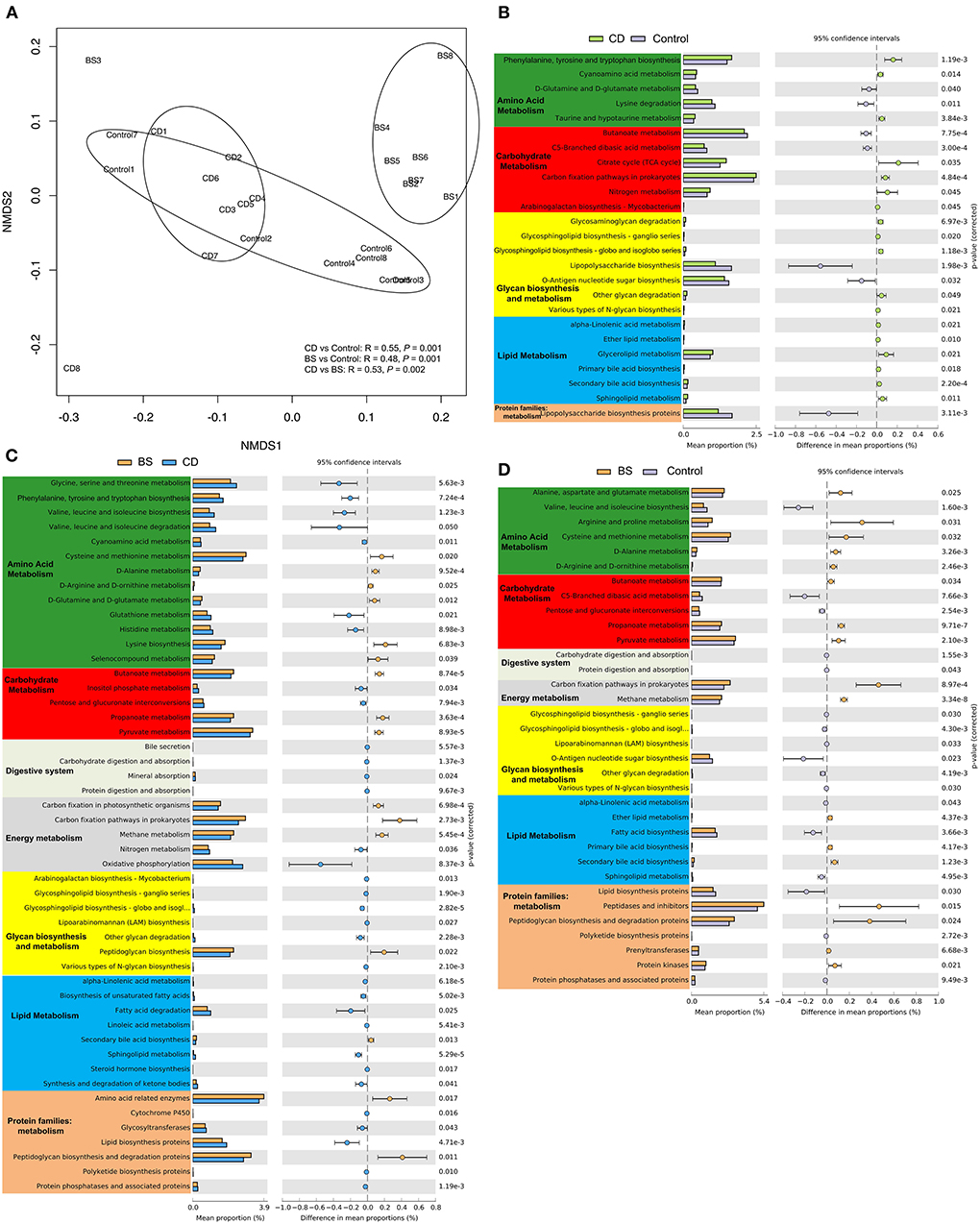
Figure 7. Non-metric multidimensional scaling (NMDS) plot visualizing bacterial functional community dissimilarities using Bray-Curtis distance (A). Differentially abundant KEGG pathways between CD and Control (B), BS and CD (C), or BS and Control (D) groups by STAMP.
Discussion
Overuse of plant protein in diets was proved to induce the intestinal disorder together with notably negative effects on the intestinal microbiota of farmed fish (3, 5). Substantial evidences have verified that the alteration in diversity of intestinal bacterial community has been associated with the growth performance of aquatic animal (45), and a reduction in the microbial α-diversity together with a lower growth was found in fish fed with a high soybean meal diet compared to high fish meal diets (5). As we know, the probiotics could improve host health through the modulation of intestinal microbiota. Hence, the objective of this study was to evaluate the function of the two probiotics. The present study indicated that B. cereus could dramatically elevate the microbial α-diversity in terms of the increase in Shannon and Simpson index. The positive effect of B. cereus on the growth performance of Pengze crucian carp observed in our previous studies (35, 36). Although B. subtilis supplementation exhibited no significant influence on the microbial α-diversity, a notable alteration in the microbial composition was observed in Pengze crucian carp, as well in carp fed with B. cereus. Firmicutes, Proteobacteria, and Fusobacteria were the dominant phyla colonizing the intestine of farmed fish, and evidences have suggested that high level of plant protein could decrease the abundance of Firmicutes together with an increase in the proportion of Fusobacteria and Proteobacteria in crucian carp (46). Here, Pengze crucian carp fed high plant protein diets also exhibited a high proportion of Fusobacteria and Proteobacteria due to a significant increase of Aeromonas and Cetobacterium. Specially, Aeromonas species from Gammaproteobacteria, gram-negative microbes, are reported as an opportunistic pathogen and have been isolated from wound fish (47, 48). Accordingly, the significant increase in Aeromonas indicated the negative effects of high plant protein diets on the intestinal microbiota. However, after consuming probiotics, the abundance of Firmicutes was restored through the increase of Clostridium_sensu_stricto and/or Romboutsia ratios in this study, confirming the benefits of B. cereus and B. subtilis on the intestinal microbiota of Pengze crucian carp.
The interspecific interactions enable the intestinal microbiota to form an ecological network, through which the micro-ecological ecosystem can maintain its dynamic homeostasis in host (17). It is well-known that the interspecific interactions present in bacterial community may be due to the species performing similar or complementary function (49). Here, a high cooperation (positive interactions) was shown in bacterial community from middle intestine of Pengze crucian carp fed different diets. According to the ecological theory of r/K selection, r-strategy species are considered to be representative community occupying a nutrient-rich environment, characterized by low competition, high capacity of nutrient utilization, and high growth rates (50). The middle intestine is one place full of various nutrients, creating a nutrient-rich living environment for a r-strategy bacterial community in Pengze crucian carp. Previous evidences have revealed that the cooperation-dominated community would be more stable since cooperative interactions are more robust to population perturbations in spatial condition, while competitive interactions, on the other hand, are susceptible to disruption (51, 52). Nevertheless, due to the significant increase in opportunistic pathogen Aeromonas, a relatively high competition (negative interactions) was displayed in the intestinal bacterial community of carp fed high plant protein diets. Because of the variation in bacterial community, the carp exhibited characteristic submodules in network, in which the dominant microbiome was the major component. From the ecological viewpoint, peripherals may represent specialists, whereas connectors and module hubs may be related to generalists, and network hubs are super-generalists (53). The generalists played by species act as structural and functional keystone and play a crucial role in maintaining the property of network, so increasing the number of connectors and module hubs can promote the network's stability. Here, the number of connectors and module hubs was observed in carp fed probiotics diets, and similar results was observed in sea cucumber (21). Took together, probiotics B. cereus and B. subtilis could improve intestinal microbiota homeostasis of Pengze crucian carp by enhancing the cooperation within bacterial community and increasing the number of generalists in ecological network.
The microbial fermentation is one of most important capacities of intestinal microbiota to participate in host metabolism by secreting digestive enzymes, which are observed to vary among microbial species (20, 22, 26). Therefore, the alteration in the microbial composition caused a notable variation in the microbial metabolic function in the intestine of carp in present study. Accordingly, high plant protein diets disturbed the stability of intestinal microbiota and induced microbial dysfunctions, contributing to the inhibition in the growth performance of Pengze crucian carp (29, 35, 36). Meanwhile, due to the increase of opportunistic pathogen Aeromonas, the microbial function related to lipopolysaccharide biosynthesis was dramatically increased in carp fed high plant protein diets, and this was maybe one important reason for the occurrence of intestinal inflammation induced by plant protein since lipopolysaccharide could trigger TLR4-mediated inflammatory pathway (54). As expected, probiotics B. cereus and B. subtilis could restore the microbial function, and affect host protein metabolism by elevating the amino acid metabolism of microbial community as well in carbohydrate metabolism, due to the significant increase in Clostridia ratio from Firmicutes. Clostridia has been proved to participate in amino acid metabolism and degrade polysaccharides (55–57). Moreover, the current study revealed that dietary two Bacillus probiotics could affect the lipid metabolism of carp by promoting the synthesis of primary and secondary bile acids in enteric cavity. It is widely accepted that microbial fermentation processes depended on the consortium of microbial community through microbe–microbe interactions (27, 58), and thus microbial dysbiosis would lead to metabolic dysfunction (59). Hence, dietary high plant protein diets induced microbial dysfunction, and Bacillus supplementation diets could improve the homeostasis of intestinal microbiota and recover the microbial function in Pengze crucian carp.
In conclusion, though altering the microbial composition and affecting species-species interactions and microbial topological roles in the ecological network performed by intestinal bacterial community, probiotics B. cereus and B. subtilis could recover the microbial dysbiosis and dysfunction induced by high plant protein diets.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA872491.
Ethics statement
The animal study was reviewed and approved by Committee on Research Ethics of the Department of Laboratory Animal Science, Jiangxi Agricultural University.
Author contributions
MP designed the experiments and supervised the manuscript. JL and PF carried out the animal experiment and sample analysis with the help of XY and wrote the manuscript. VK revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by Double Thousand Program of Jiangxi Province (2019) and Excellent Youth Cultivation Project of National Natural Science Foundation of China (20202ZDB01010).
Conflict of interest
Author XY was employed by Shenzhen Aohua Group Co., Ltd., Shenzhen, China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ahmad A, Abdullah SRS, Hasan HA, Othman AR, Ismail NI. Aquaculture industry: supply and demand, best practices, effluent and its current issues and treatment technology. J Environ Manage. (2021) 287:112271. doi: 10.1016/j.jenvman.2021.112271
2. Fiorella KJ, Okronipa H, Baker K, Heilpern S. Contemporary aquaculture: implications for human nutrition. Curr Opin Biotechnol. (2021) 70:83–90. doi: 10.1016/j.copbio.2020.11.014
3. Glencross BD, Baily J, Berntssen MH, Hardy R, MacKenzie S, Tocher DR. Risk assessment of the use of alternative animal and plant raw material resources in aquaculture feeds. Rev Aquacult. (2020) 12:703–58. doi: 10.1111/raq.12347
4. Dawood MA. Nutritional immunity of fish intestines: Important insights for sustainable aquaculture. Rev Aquacult. (2021) 13:642–63. doi: 10.1111/raq.12492
5. Zhou Z, Ringø E, Olsen R, Song S. Dietary effects of soybean products on gut microbiota and immunity of aquatic animals: a review. Aquacult Nutr. (2018) 24:644–65. doi: 10.1111/anu.12532
6. Young VB. The intestinal microbiota in health and disease. Curr Opin Gastroen. (2012) 28:63. doi: 10.1097/MOG.0b013e32834d61e9
7. Fava F, Rizzetto L, Tuohy K. Gut microbiota and health: connecting actors across the metabolic system. Proc Nutr Soc. (2019) 78:177–88. doi: 10.1017/S0029665118002719
8. El-Saadony MT, Alagawany M, Patra AK, Kar I, Tiwari R, Dawood MA, et al. The functionality of probiotics in aquaculture: an overview. Fish Shellfish Immunol. (2021) 117:36–52. doi: 10.1016/j.fsi.2021.07.007
9. Kuebutornye FK, Abarike ED, Lu Y, A. review on the application of Bacillus as probiotics in aquaculture. Fish Shellfish Immunol. (2019) 87:820–8. doi: 10.1016/j.fsi.2019.02.010
10. Kawser A, Islam T, Alam MS, Rahman M, Salam MA. Mechanisms of the beneficial effects of probiotic Bacillus spp. in aquaculture. Bacilli Agrobiotechnol. (2022) 453–86. doi: 10.1007/978-3-030-85465-2_20
11. Shi F, Zi Y, Lu Z, Li F, Yang M, Zhan F, et al. Bacillus subtilis H2 modulates immune response, fat metabolism and bacterial flora in the gut of grass carp (Ctenopharyngodon idellus). Fish Shellfish Immunol. (2020) 106:8–20. doi: 10.1016/j.fsi.2020.06.061
12. Ma S, Yu D, Liu Q, Zhao M, Xu C, Yu J. Relationship between immune performance and the dominant intestinal microflora of turbot fed with different Bacillus species. Aquaculture. (2022) 549:737625. doi: 10.1016/j.aquaculture.2021.737625
13. Zhang J, Huang M, Feng J, Chen Y, Li M, Chang X. Effects of dietary Bacillus licheniformis on growth performance, intestinal morphology, intestinal microbiome, and disease resistance in common carp (Cyprinus carpio L). Aquacult Internat. (2021) 29:1343–58. doi: 10.1007/s10499-021-00701-w
14. Niu KM, Khosravi S, Kothari D, Lee WD, Lee BJ, Lim SG, et al. Potential of indigenous Bacillus spp. as probiotic feed supplements in an extruded low-fish-meal diet for juvenile olive flounder, Paralichthys olivaceus. J World Aquacult Soc. (2021) 52:244–61. doi: 10.1111/jwas.12724
15. Wu PS, Liu CH, Hu SY. Probiotic Bacillus safensis NPUST1 administration improves growth performance, gut microbiota, and innate immunity against Streptococcus iniae in Nile tilapia (Oreochromis niloticus). Microorganisms. (2021) 9:2494. doi: 10.3390/microorganisms9122494
16. Wang Y, Wang Q, Xing K, Jiang P, Wang J. Dietary cinnamaldehyde and Bacillus subtilis improve growth performance, digestive enzyme activity, and antioxidant capability and shape intestinal microbiota in tongue sole, Cynoglossus semilaevis. Aquaculture. (2021) 531:735798. doi: 10.1016/j.aquaculture.2020.735798
17. Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: networks, competition, and stability. Science. (2015) 350:663–6. doi: 10.1126/science.aad2602
18. Sanders ME. Probiotics and microbiota composition. BMC Med. (2016) 14:1–3. doi: 10.1186/s12916-016-0629-z
19. Falcinelli S, Picchietti S, Rodiles A, Cossignani L, Merrifield DL, Taddei AR, et al. Lactobacillus rhamnosus lowers zebrafish lipid content by changing gut microbiota and host transcription of genes involved in lipid metabolism. Sci Rep. (2015) 5:1–11. doi: 10.1038/srep09336
20. Montoya JM, Pimm SL, Solé RV. Ecological networks and their fragility. Nature. (2006) 442:259–64. doi: 10.1038/nature04927
21. Yang G, Peng M, Tian X, Dong S. Molecular ecological network analysis reveals the effects of probiotics and florfenicol on intestinal microbiota homeostasis: an example of sea cucumber. Sci Rep. (2017) 7:1–12. doi: 10.1038/s41598-017-05312-1
22. Zhu B, Wang X, Li L. Human gut microbiome: the second genome of human body. Protein Cell. (2010) 1:718–25. doi: 10.1007/s13238-010-0093-z
23. Aron-Wisnewsky J, Warmbrunn MV, Nieuwdorp M, Clément K. Metabolism and metabolic disorders and the microbiome: the intestinal microbiota associated with obesity, lipid metabolism, and metabolic health—pathophysiology and therapeutic strategies. Gastroenterology. (2021) 160:573–99. doi: 10.1053/j.gastro.2020.10.057
24. Portincasa P, Bonfrate L, Vacca M, De Angelis M, Farella I, Lanza E, et al. Gut microbiota and short chain fatty acids: implications in glucose homeostasis. Int J Mol Sci. (2022) 23:1105. doi: 10.3390/ijms23031105
25. Lin R, Liu W, Piao M, Zhu H, A. review of the relationship between the gut microbiota and amino acid metabolism. Amino Acids. (2017) 49:2083–90. doi: 10.1007/s00726-017-2493-3
26. Nieuwdorp M, Gilijamse PW, Pai N, Kaplan LM. Role of the microbiome in energy regulation and metabolism. Gastroenterology. (2014) 146:1525–33. doi: 10.1053/j.gastro.2014.02.008
27. Dearing MD, Kohl KD. Beyond fermentation: other important services provided to endothermic herbivores by their gut microbiota. Integr Comp Biol. (2017) 57:723–31. doi: 10.1093/icb/icx020
28. Yang G, Jian SQ, Cao H, Wen C, Hu B, Peng M, et al. Changes in microbiota along the intestine of grass carp (Ctenopharyngodon idella): community, interspecific interactions, and functions. Aquaculture. (2019) 498:151–61. doi: 10.1016/j.aquaculture.2018.08.062
29. Fang L, Wang Q, Guo X, Pan X, Li X. Effects of dietary sodium butyrate on growth performance, antioxidant capacity, intestinal histomorphology and immune response in juvenile Pengze crucian carp (Carassius auratus Pengze). Aquacult Rep. (2021) 21:100828. doi: 10.1016/j.aqrep.2021.100828
30. Zhu R, Li L, Li M, Yu Z, Wang H, Wu L. The effects of substituting fish meal with soy protein concentrate on growth performance, antioxidant capacity and intestinal histology in juvenile golden crucian carp, Cyprinus carpio × Carassius auratus. Aquacult Rep. (2020) 18:100435. doi: 10.1016/j.aqrep.2020.100435
31. Xu Y, Li Y, Xue M, Xiao Z, Fan Y, Zeng L, et al. Effects of dietary Enterococcus faecalis YFI-G720 on the growth, immunity, serum biochemical, intestinal morphology, intestinal microbiota, and disease resistance of crucian carp (Carassius auratus). Fishes. (2022) 7:18. doi: 10.3390/fishes7010018
32. Yi Y, Zhang Z, Zhao F, Liu H, Yu L, Zha J, et al. Probiotic potential of Bacillus velezensis JW: antimicrobial activity against fish pathogenic bacteria and immune enhancement effects on Carassius auratus. Fish Shellfish Immunol. (2018) 78:322–30. doi: 10.1016/j.fsi.2018.04.055
33. Zhang DX, Kang YH, Zhan S, Zhao ZL, Jin SN, Chen C, et al. Effect of Bacillus velezensis on Aeromonas veronii-induced intestinal mucosal barrier function damage and inflammation in crucian carp (Carassius auratus). Front Microbiol. (2019) 10:2663. doi: 10.3389/fmicb.2019.02663
34. Ahire J, Mokashe N, Chaudhari B. Effect of dietary probiotic Lactobacillus helveticus on growth performance, antioxidant levels, and absorption of essential trace elements in goldfish (Carassius auratus). Probiotics Antimicrob Proteins. (2019) 11:559–68. doi: 10.1007/s12602-018-9428-5
35. Yang G, Shen K, Yu R, Wu Q, Yan Q, Chen W, et al. Probiotic (Bacillus cereus) enhanced growth of Pengze crucian carp concurrent with modulating the antioxidant defense response and exerting beneficial impacts on inflammatory response via Nrf2 activation. Aquaculture. (2020) 529:735691. doi: 10.1016/j.aquaculture.2020.735691
36. Yang G, Cao H, Jiang W, Hu B, Jian S, Wen C, et al. Dietary supplementation of Bacillus cereus as probiotics in Pengze crucian carp (Carassius auratus var. Pengze): effects on growth performance, fillet quality, serum biochemical parameters and intestinal histology. Aquacult Res. (2019) 50:2207–17. doi: 10.1111/are.14102
37. Cao H, Yu R, Zhang Y, Hu B, Jian S, Wen C, et al. Effects of dietary supplementation with β-glucan and Bacillus subtilis on growth, fillet quality, immune capacity, and antioxidant status of Pengze crucian carp (Carassius auratus var. Pengze) Aquaculture. (2019) 508:106–12. doi: 10.1016/j.aquaculture.2019.04.064
38. Yang G, Xu Z, Tian X, Dong S, Peng M. Intestinal microbiota and immune related genes in sea cucumber (Apostichopus japonicus) response to dietary β-glucan supplementation. Biochem Biophys Res Commun. (2015) 458:98–103. doi: 10.1016/j.bbrc.2015.01.074
39. Rognes T, Flouri T, Nichols B, Quince C, Mahé F, VSEARCH. a versatile open source tool for metagenomics. PeerJ. (2016) 4:e2584. doi: 10.7717/peerj.2584
40. Anderson MJ. Permutational Multivariate Analysis of Variance. Auckland: Department of Statistics, University of Auckland (2005). p. 32–46.
41. Deng Y, Jiang Y-H, Yang Y, He Z, Luo F, Zhou J. Molecular ecological network analyses. BMC Bioinf. (2012) 13:1–20. doi: 10.1186/1471-2105-13-113
42. Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. (2020) 38:685–8. doi: 10.1038/s41587-020-0548-6
43. Clarke KR. Non-parametric multivariate analyses of changes in community structure. Austral Ecol. (1993) 18:117–43. doi: 10.1111/j.1442-9993.1993.tb00438.x
44. Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP statistical analysis of taxonomic and functional profiles. Bioinformatics. (2014) 30:3123–4. doi: 10.1093/bioinformatics/btu494
45. Infante-Villamil S, Huerlimann R, Jerry DR. Microbiome diversity and dysbiosis in aquaculture. Rev Aquacult. (2021) 13:1077–96. doi: 10.1111/raq.12513
46. Liu X, Han B, Xu J, Zhu J, Hu J, Wan W, et al. Replacement of fishmeal with soybean meal affects the growth performance, digestive enzymes, intestinal microbiota and immunity of Carassius auratus gibelio♀ × Cyprinus carpio♂. Aquacult Rep. (2020) 18:100472. doi: 10.1016/j.aqrep.2020.100472
47. Fečkaninová A, Koščová J, Mudronová D, Popelka P, Toropilova J. The use of probiotic bacteria against Aeromonas infections in salmonid aquaculture. Aquaculture. (2017) 469:1–8. doi: 10.1016/j.aquaculture.2016.11.042
48. Igbinosa IH, Beshiru A, Odjadjare EE, Ateba CN, Igbinosa EO. Pathogenic potentials of Aeromonas species isolated from aquaculture and abattoir environments. Microb Pathogen. (2017) 107:185–92. doi: 10.1016/j.micpath.2017.03.037
49. Zhou J, Deng Y, Luo F, He Z, Yang Y. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. MBio. (2011) 2:e00122–11. doi: 10.1128/mBio.00122-11
50. Wilson EO, MacArthur RH. The Theory of Island Biogeography. Princeton, NJ: Princeton University Press (1967).
51. Pande S, Kaftan F, Lang S, Svatoš A, Germerodt S, Kost C. Privatization of cooperative benefits stabilizes mutualistic cross-feeding interactions in spatially structured environments. ISME J. (2016) 10:1413–23. doi: 10.1038/ismej.2015.212
52. Ren X, Murray RM, editors. Cooperation enhances robustness of coexistence in spatially structured consortia. In: 2019 18th European Control Conference (ECC). Naples: IEEE (2019).
53. Olesen JM, Bascompte J, Dupont YL, Jordano P. The modularity of pollination networks. PNAS. (2007) 104:19891–6. doi: 10.1073/pnas.0706375104
54. Rhee SH. Lipopolysaccharide: basic biochemistry, intracellular signaling, and physiological impacts in the gut. Intestinal Res. (2014) 12:90. doi: 10.5217/ir.2014.12.2.90
55. Elsden SR, Hilton MG. Amino acid utilization patterns in clostridial taxonomy. Arch Microbiol. (1979) 123:137–41. doi: 10.1007/BF00446812
56. Dai ZL, Wu G, Zhu WY. Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front Biosci-Landmark. (2011) 16:1768–86. doi: 10.2741/3820
57. Mitchell WJ. Physiology of carbohydrate to solvent conversion by clostridia. Adv Microb Physiol. (1997) 39:31–130. doi: 10.1016/S0065-2911(08)60015-6
58. Smid EJ, Lacroix C. Microbe–microbe interactions in mixed culture food fermentations. Curr Opin Biotechnol. (2013) 24:148–54. doi: 10.1016/j.copbio.2012.11.007
Keywords: plant protein, Pengze crucian carp, intestinal microbiota, ecological network, microbial function
Citation: Li J, Fang P, Yi X, Kumar V and Peng M (2022) Probiotics Bacillus cereus and B. subtilis reshape the intestinal microbiota of Pengze crucian carp (Carassius auratus var. Pengze) fed with high plant protein diets. Front. Nutr. 9:1027641. doi: 10.3389/fnut.2022.1027641
Received: 25 August 2022; Accepted: 03 October 2022;
Published: 19 October 2022.
Edited by:
Tingtao Chen, Nanchang University, ChinaReviewed by:
Meiling Zhang, East China Normal University, ChinaXiaoting Zheng, Ghent University, Belgium
Junming Deng, Guangdong Ocean University, China
Copyright © 2022 Li, Fang, Yi, Kumar and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mo Peng, cGVuZ21vd2VsbEBqeGF1LmVkdS5jbg==
 Jiamin Li
Jiamin Li Peng Fang1
Peng Fang1 Vikas Kumar
Vikas Kumar Mo Peng
Mo Peng