
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr. , 29 September 2022
Sec. Nutrition and Metabolism
Volume 9 - 2022 | https://doi.org/10.3389/fnut.2022.1006253
 Hui Xue1,2
Hui Xue1,2 Yunyun Zhao1,2
Yunyun Zhao1,2 Li Wang1,2
Li Wang1,2 Xiangzhen Zhu1,2
Xiangzhen Zhu1,2 Kaixin Zhang1,2
Kaixin Zhang1,2 Dongyang Li1,2
Dongyang Li1,2 Jichao Ji1,2
Jichao Ji1,2 Lin Niu1,2
Lin Niu1,2 Jinjie Cui1,2*
Jinjie Cui1,2* Junyu Luo1,2*
Junyu Luo1,2* Xueke Gao1,2*
Xueke Gao1,2*The vast majority of parasitoids are capable of precise and meticulous regulation of nutrition and metabolism within the host. An important building block of life, amino acids are critical to the development of parasitoids. To date, research on how parasitoids regulate host amino acid metabolism remains limited. In this study, Aphis gossypii and its dominant parasitoid Binodoxys communis were used as a study system to explore how parasitism may change the regulation of amino acids in A. gossypii with UHPLC-MS/MS and RT-qPCR techniques. Here, for the first 8 h of parasitism the abundance of almost all amino acids in cotton aphids increased, and after 16 h most of the amino acids decreased. An amino acid of parasitic syndrome, the content of Tyr increased gradually after being parasitized. The expression of genes related to amino acid metabolism increased significantly in early stages of parasitism and then significantly decreased gradually. At the same time, the abundance of Buchnera, a cotton aphid specific symbiont increased significantly. Our comprehensive analyses reveal impacts of B. communis on the amino acid regulatory network in cotton aphid from three aspects: amino acid metabolism, gene expression, and bacterial symbionts. Therefore, this research provides an important theoretical basis for parasitoid nutritional regulation in host, which is highly significant as it may inform the artificial reproduction of parasitoids and the biological control of insect pests.
The regulation of host physiological mechanisms by parasitism is a highly complex and refined process which has evolved for thousands and tens of thousands of years. Parasites precisely control host metabolism so that the nutritional environment in the host is more suitable for the growth, development, and nutritional needs of the parasite (1–3). Although the phenomenon of parasitic insects changing host nutritional homeostasis is common (3, 4), its regulatory pathways and mechanisms remain largely unknown.
Parasitic insects, especially parasitic wasps, are representative models for the study of parasitic regulation. The parasitic characteristics of parasitic wasp larvae are very prominent in the regulation of host nutrition. Due to the parasitic characteristics and long-term development and evolution of parasitic wasps, a strong dependence on the host has evolved (5, 6). Normally, parasitic wasps mainly feed on the host’s hemolymph in early stages, and directly feed on the host’s body fat in later stages (7, 8). Studies have shown that the fatty acid composition of the parasitic wasp is very similar to that of the host (9), which may be due to parasitoids lacking an enzyme system for fatty acid synthesis. Therefore in order to meet their physiological and metabolic requirements, parasitic wasps directly use the fatty acids of the host (10). Lysiphlebia japonica directly increases the triglyceride content of cotton aphids to meet the growth and development of cotton aphid larvae by increasing the expression of genes related to the tricarboxylic acid cycle and glycolysis pathway in cotton aphid (11, 12). Furthermore, carbohydrates are important energy stores for organisms, and thus important substances for parasitic wasps to regulate in their hosts. Transcriptome analysis showed that after Microplitis mediator parasitized Helicoverpa armigera, more than 30 genes related to nutrition metabolism pathway were differentially expressed in the hemolymph of H. armigera, of which 20 genes related to glucose metabolism were significantly enriched (13).
Lipids and carbohydrates, as important energy-supplying substances for organisms, are crucial to the development of parasitic wasps, and similarly amino acids, as globally recognized nutritional small-molecules to life, are indispensable and important regulatory substances for mammals (14–18), plants (19, 20), and insects (21). Amino acids are traditionally classified into nutritionally essential (EAA) and non-essential (NEAA) categories according to animal growth or nitrogen balance (22), and play important roles in regulating animal food intake, nutrient metabolism, gene expression, and cell signaling (23–26). It has been reported that parasitoid wasps have a strong dependence on host amino acids due to their lack of some amino acid synthesis functions (27). Aphids are a special example of amino acid metabolism. Aphids feed on plant sap, but this sap is a poor diet and lacks certain essential amino acids, but bacterial symbionts (Buchnera) in host cells synthesize these amino acids providing them for the host (28–30). In this study, Aphis gossypii and its dominant parasitic wasp Binodoxys communis were used as the research system. Exploring the synergistic regulation mechanisms of parasite-host-symbiotic bacteria based on the specific amino acid metabolism patterns of parasitoid parasitism and aphids is both valuable and interesting from ecological, evolutionary, and agricultural perspectives.
At present, there are few studies on the regulation of host amino acid metabolism by parasitoids. The control mechanisms of parasitoids on host amino acid metabolism is remains unknown and may have powerful implications in the field in regards to fundamental questions in ecology and evolution as well as pest regulation. In this study, UHPLC-MS/MS and RT-qPCR techniques were used to detect changes in host amino acid content, related regulatory genes, and host symbiotic bacteria abundances under parasitic conditions to reveal the regulatory patterns and potential regulation mechanisms of parasitoids on host cotton aphids, thus building an important theoretical foundation for how parasitoids affect host metabolism.
With A. gossypii and B. communis as the model research system, A. gossypii reproduced by parthenogenesis, and B. communis took cotton aphid as the host. Aphids were reared on cotton leaves at 26 ± 1°C and 65 ± 5% relative humidity with a 14:10 h light: dark photoperiod. B. communis were reared in laboratory on cotton aphids at 26 ± 1°C with a 14:10 h light: dark photoperiod and 65 ± 5% relative humidity. In order to obtain parasitized cotton aphids, 2-day-old cotton aphids were exposed to the B. communis environment, and parasitized cotton aphids were transferred to a B. communis-free environment after parasitism behavior was observed. In addition, the parasitoid wasps used in the experiment were all mated, and there were 10 female and male parasitoids in each parasitoid environment.
The samples used for amino acid extraction and RT-qPCR in this study were the cotton aphid bodies parasitized by B. communis for 8, 16 h, 1, 2, and 3 day, respectively. The cotton aphid samples that were not parasitized but had the same growth and development were used as the control group. In a sterile environment, the cotton aphids parasitized for 16 h, 1, 2, and 3 day were manually dissected to remove the parasitic wasp larvae, and the control group was also subjected to the same corresponding dissection test. Three biological replicates were set for each stage of treatment and control samples. Cotton aphid samples were snap-frozen in liquid nitrogen immediately after dissection and stored at −80°C.
Samples were weighed (20 mg) and transferred to Eppendorf tubes. After the addition of two small steel balls and 250 μL of extraction solution (precooled at −20°C, acetonitrile-methanol-water, 2:2:1, containing isotopically-labeled internal standard mixture), the samples were vortexed for 30 s, homogenized at 40 Hz for 4 min, and sonicated for 5 min in ice-water bath. The homogenate and sonicate cycle was repeated 3 times, followed by incubation at −40°C for 1 h and centrifugation at 12,000 rpm (RCF = 13,800 (× g), R = 8.6 cm) and 4°C for 15 min. The supernatant was diluted 4 times, vortexed for 30 s to mix, and then transferred to an auto-sampler vial for UHPLC-MS/MS analysis.
The UHPLC separation was carried out using an Agilent 1290 Infinity II series UHPLC System (Agilent Technologies), equipped with a Waters ACQUITY UPLC BEH Amide column (100 × 2.1 mm, 1.7 μm). The mobile phase A was 1% formic acid in water, and the mobile phase B was 1% formic acid in acetonitrile. The column temperature was set at 35°C. The auto-sampler temperature was set at 4°C and the injection volume was 1 μL.
An Agilent 6,460 triple quadrupole mass spectrometer (Agilent Technologies), equipped with an AJS electrospray ionization (AJS-ESI) interface, was used for assay development. Standard ion source parameters were: capillary voltage = + 4,000/-3500 V, Nozzle Voltage = + 500/-500 V, gas (N2) temperature = 300°C, gas (N2) flow = 5 L/min, sheath gas (N2) temperature = 250°C, sheath gas flow = 11 L/min, nebulizer = 45 psi. The MRM parameters for each of the targeted analytes were optimized using flow injection analysis, by injecting the standard solutions of the individual analytes, into the API source of the mass spectrometer. Several of the most sensitive transitions were used in the MRM scan mode to optimize the collision energy for each Q1/Q3 pair. Among the optimized MRM transitions per analyte, the Q1/Q3 pairs with the highest sensitivity and selectivity were selected as the “quantifier” for quantitative monitoring. The additional transitions acted as “qualifiers” for the purpose of verifying the identity of the target analytes. Agilent MassHunter Work Station Software (B.08.00, Agilent Technologies) was employed for MRM data acquisition and processing.
A single peak was filtered to retain only peak area data with no more than 50% null in a single group or no more than 50% in all groups. The sorted data were analyzed by PCA (principal component analysis) and OPLS-DA (orthogonal projections to latent structures-discriminant analysis) using SIMCA (V16.0.2, Sartorius Stedim Data Analytics AB, Umea, Sweden). Differences between treatment means were statistically analyzed in SPSS for Windows (SPSS 20.0, Chicago, IL, USA) with the Student’s t-test.
The samples were soaked in 75% ethanol for 30 s and then rinsed 3 times with ddH2O. DNA was extracted using the Fast DNA Spin Kit for SOIL (MP Biomedicals, Santa Ana, CA, USA). The concentration and purity of extracted DNA were detected by NanoDrop 2000C (Thermo Fisher Scientific, USA), and the DNA quality was measured with 1.5% agarose gel electrophoresis. Total RNA was isolated from samples using the TRIZOL total RNA isolation system (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. RNA quality was measured on 1.5% agarose gels and quantified using a Nano-Drop 2000 (Thermo Scientific, Wilmington, DE, USA). cDNA was synthesized from 1 μg of total RNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Dalian, China), according to the manufacturer’s instructions. cDNA was used for RT-qPCR assay and DNA was used to detect bacterial copy number.
RT-qPCR was performed with the StepOnePlus™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using TransStart® Top Green qPCR SuperMix (Yeasen Biotech, Shanghai, China) in 10 μl reaction volume. And the protocol was performed: 95°C for 3 min followed 40 cycles of 95°C for 5 s and 60°C for 30 s. Dimethyladenosine transferase (GenBank: KM507111) and peptidyl-prolyl cis-trans isomerase (GenBank: KF018924) were used as reference genes for gene expression normalization (11). Related gene-specific primers are shown in Supplementary Table 1. Gene expression data were analyzed using the 2–ΔΔ Ct method (31). Differences between treatment means were statistically analyzed in SPSS for Windows (SPSS 20.0, Chicago, IL, USA) with the Student’s t-test.
To estimate the Buchnera copy number, “absolute” real-time qPCR was conducted on DNA extracted from parasitized and non-parasitized aphids at 8, 16 h, 1, 2, and 3 day after parasitism (three biological replicates per treatment). The copy number of the target gene (F:5′-AGTATCGTAGAGGGAGGTA-3′, R:5′-CTTTCGCCACAGGTATTC-3′) was calculated according to the standard curve after dilution of the target gene sequence cloned in a Peasy-T1 cloning vector (TransGen Biotech, China). Using the StepOnePlus™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) the following protocol was performed: 95°C for 3 min followed 40 cycles of 95°C for 5 s and 60°C for 30 s. Working in 10 μl volume using TransStart® Top Green qPCR SuperMix (Yeasen Biotech, Shanghai, China). The corresponding standard curve for each reaction plate was calculated accordingly and three technical replicates were used for each sample. Differences in copy number between samples were statistically analyzed with Student’s t-test using SPSS 20.0.
The quality parameters of PCA model R2X are all greater than 0.5 and the samples are basically in 95% confidence interval, therefore, the current model may be reliably used to characterize the metabolic differences between samples (Figure 1). Parasitized and unparasitized cotton aphids were visibly grouped separately in the same period, although there was a cross phenomenon in different developmental stages, but the amino acid metabolisms between the treatment (parasitized) and control (unparasitized) groups differed significantly (Figure 1).
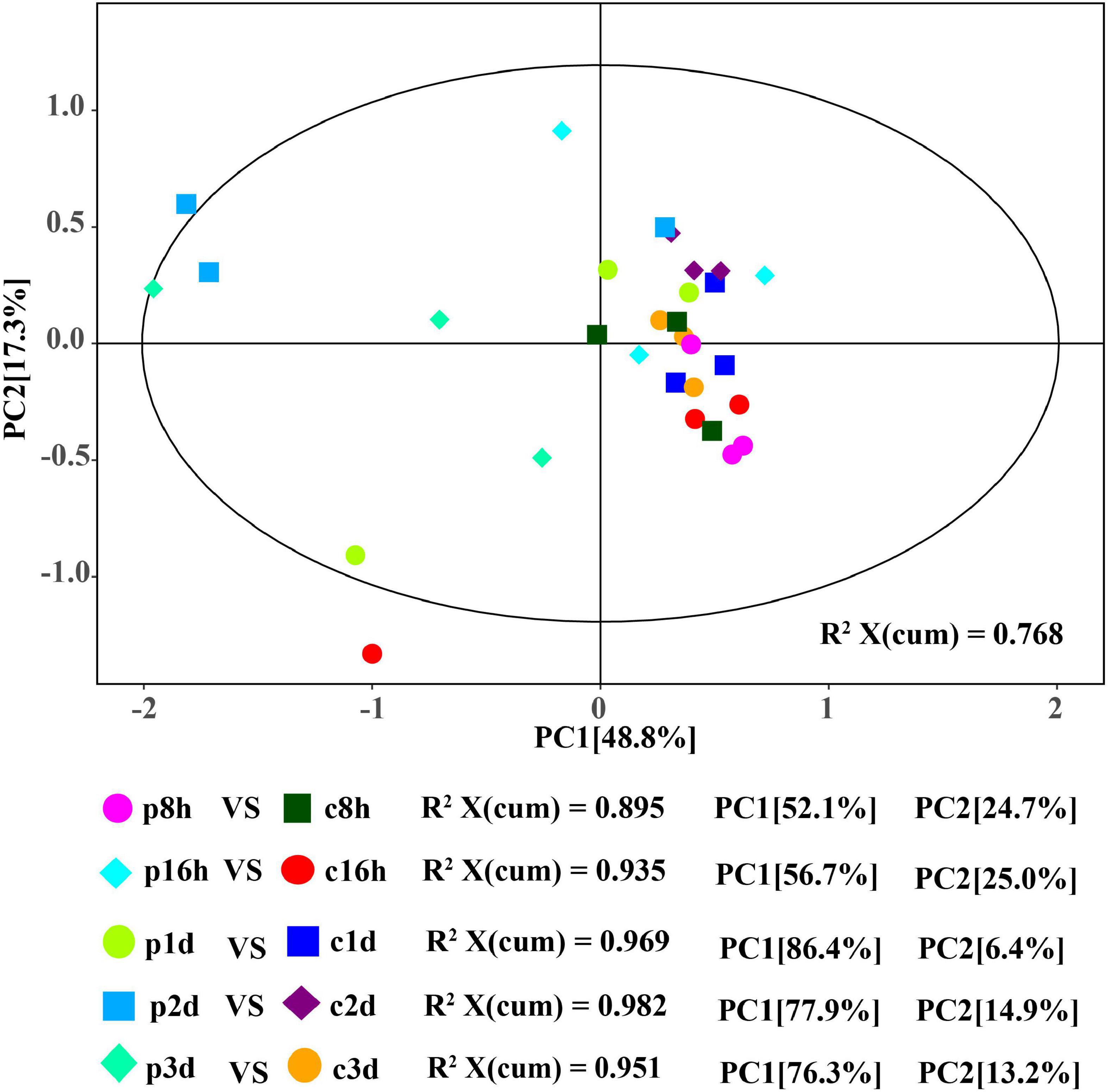
Figure 1. Score plot of the PCA models applied to parasitized and control aphids. p8h, p16h, p1d, p2d, and p3d indicate parasitized cotton aphids for 8, 16 h, 1, 2, and 3 days, respectively. Similarly, c8h, c16h, c1d, c2d, and c3d represent the above-treated control groups, respectively.
In order to obtain more reliable information about the correlation between the differences in metabolites between control and treatment groups, orthogonal variables in amino acids that were not related to the classification variables by OPLS-DA analysis were filtered, and non-orthogonal variables and orthogonal variables were analyzed separately. The quality of the OPLS-DA model was tested by 7-fold cross validation. The R2X, R2Y, and Q2 of each comparison group were all greater than 0.739, 0.915, and 0.593, respectively (Table 1). The OPLS-DA score map can effectively distinguish the experimental group from the control groups (Supplementary Figures 1A–F), indicating that the current model can effectively distinguish samples from the parasitic and non-parasitic groups. The original model R2Y’s score approaches 1 through the permutation test for 200 random changes in the order of categorical variables, indicating that the established model conforms to the real situation of the sample data. The intercept of the regression line of Q2 and the vertical axis is less than 0. At the same time, with the gradual decrease in permutation retention, the proportion of the permuted Y variable increases, and the Q of the random model gradually decreases. Therefore, the original model is robust and there is no overfitting phenomenon (Supplementary Figures 1G–K).
The content changes of all detected EAAs in each parasitic stage showed that the total EAAs of cotton aphids increased significantly at 8 h, began to decrease significantly at 16 h, and continued to trend downward 1–3 day (Figure 2). The heat map in Figure 3A summarize the log2(Fold change) in the essential amino acid content in cotton aphids in different stages of parasitism. The abundances of essential amino acids in cotton aphids were almost all greater at 8 h, and most of the essential amino acids showed a downward trend from 16 h (Figure 3). Only the abundance of the amino acid 1Mhis decreased in parasitized aphids at 8 h, and the decrease reached a significant level at the 2 day timepoint (Figure 3B). The amino acid His also decreased significantly in the 2 day timepoint (Figure 3C). Furthermore, the content of amino acid Phe decreased significantly in the 16 h timepoint, but showed an upward trend in the 3 day timepoint (Figure 3D). The overall analysis showed that only the quantities of Arg and Lys were significantly increased in different stages of parasitism (Figures 3E,F). Thr decreased significantly at 16 h (Figure 3G). Changes in 3His, Met, Try and Val did not reach significant levels (Figures 3H–K).
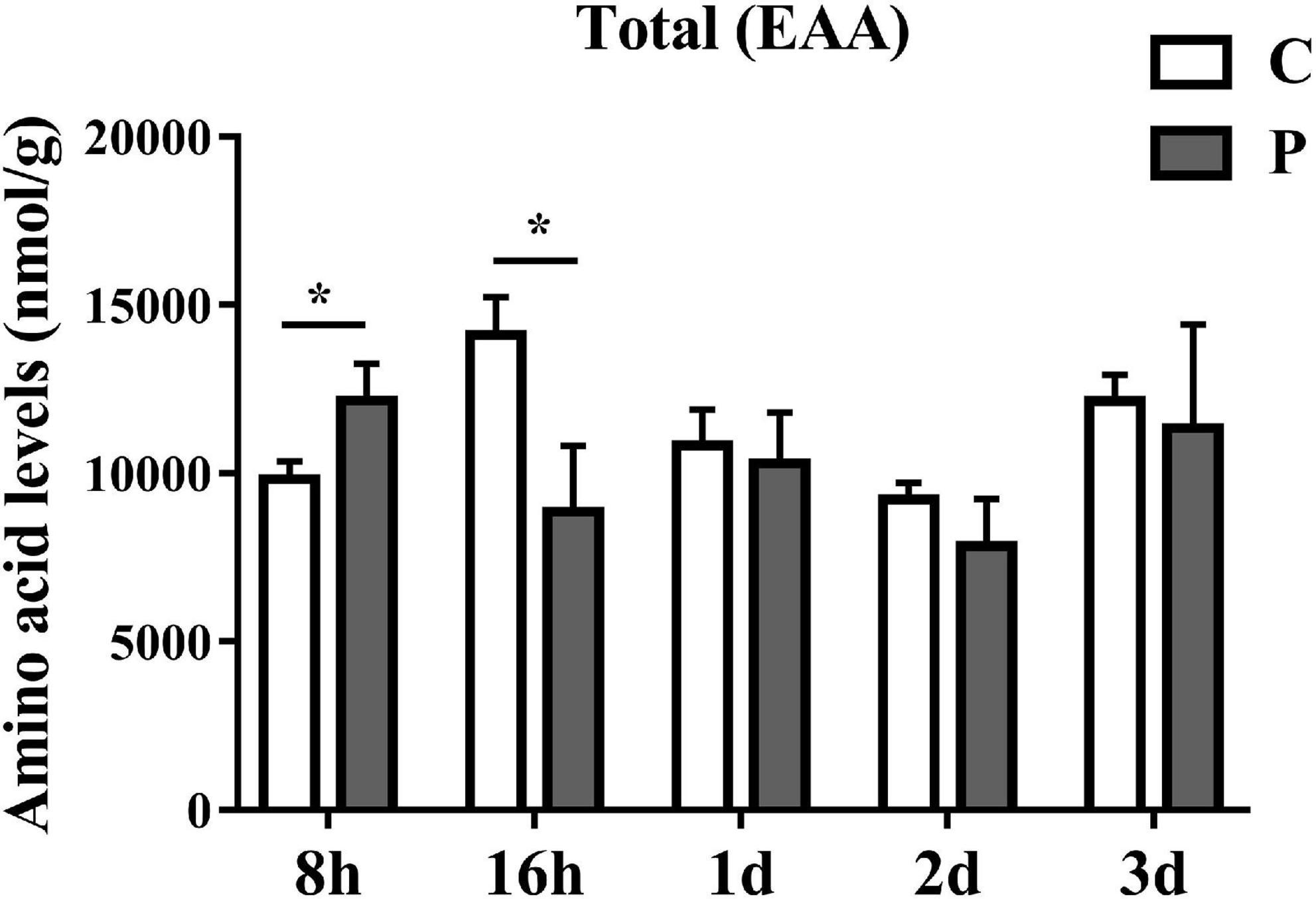
Figure 2. Parasitism causes changes in the content of total EAA (Essential Amino Acids) in cotton aphid. Statistical significance (Student’s t-test): *P < 0.05; **P < 0.01; ***P < 0.001.
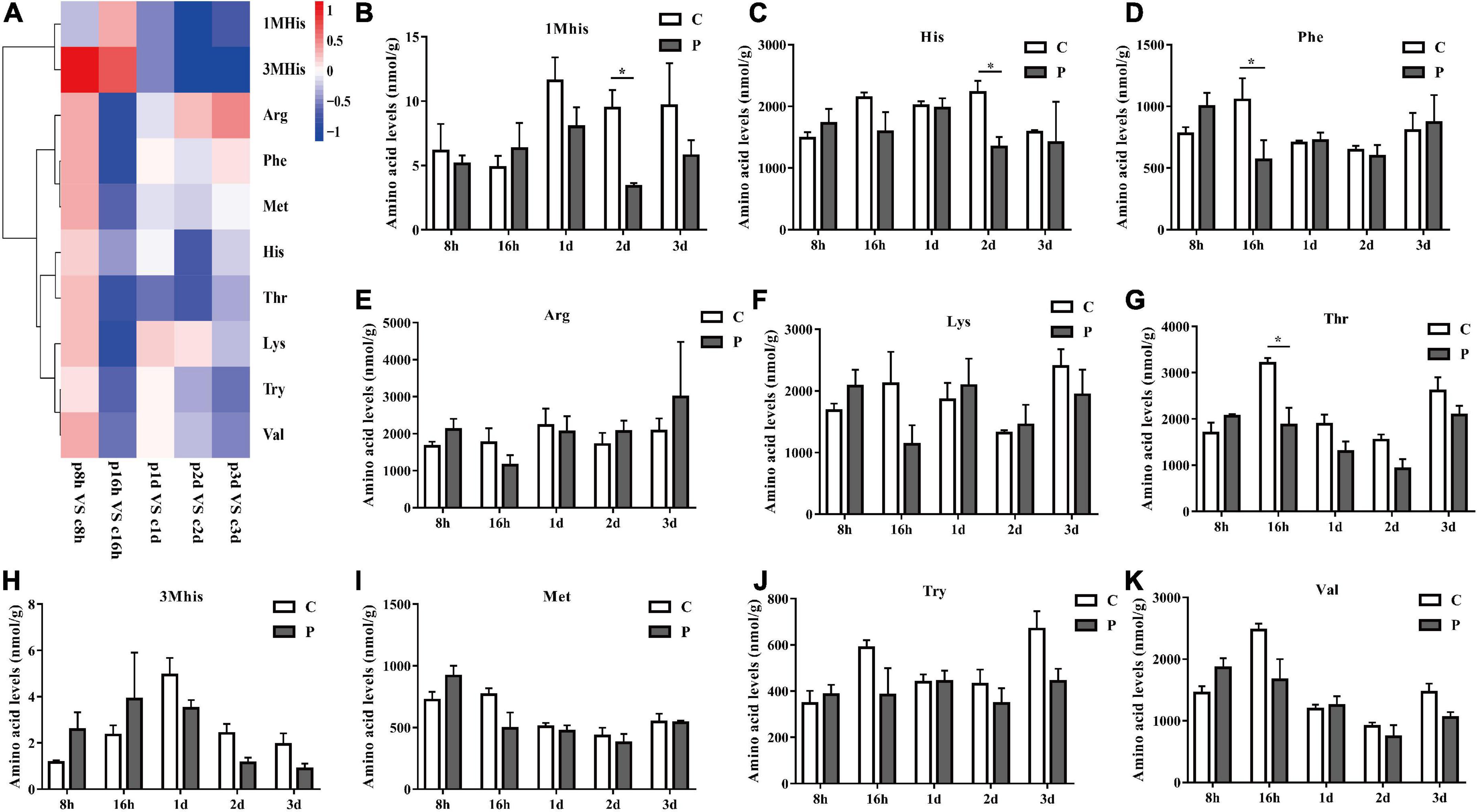
Figure 3. Parasitism leads to the change of EAA content of cotton aphid. Heatmap (A) and column (B–K) display of changes in EAA. 1Mhis, 1-Methyl-L-histidine; 3Mhis, 3-Methyl-L-histidine; C, Non-parasitized cotton aphids; P, Parasitized cotton aphids. The log2(Fold change) of amino acid content change is represented by heat map color scale, red indicates up-regulation and blue indicates down-regulation. Statistical significance (Student’s t-test): *P < 0.05; **P < 0.01; ***P < 0.001.
The overall trends in changes of total non-essential amino acids (NEAAs) content were similar to that of EAAs. Parasitism led to a significant increase in total NEAAs content at 8 h and a significant decrease at 16 h (Figure 4). The overall NEAAs trends in abundance are clearly visualized through heat map analysis (Figure 5A). The contents of amino acids Asp, β-Ala, Ala, Glu, Cit, Gln, and Asn decreased significantly (Figures 5B–H), but the content of Tyr increased significantly with as length of parasitism increased (Figure 5I). Tyr is also the key amino acid to reflect a parasitic effect. Orn increased 2.1 times significantly at 3 day (Figure 5J), and Pro content had an observable increasing trend but did not reach statistical significance (Figure 5K). Gly increased significantly at 8 h (Figure 5L), while Hyp and Ser had no significant change (Figures 5M,N).
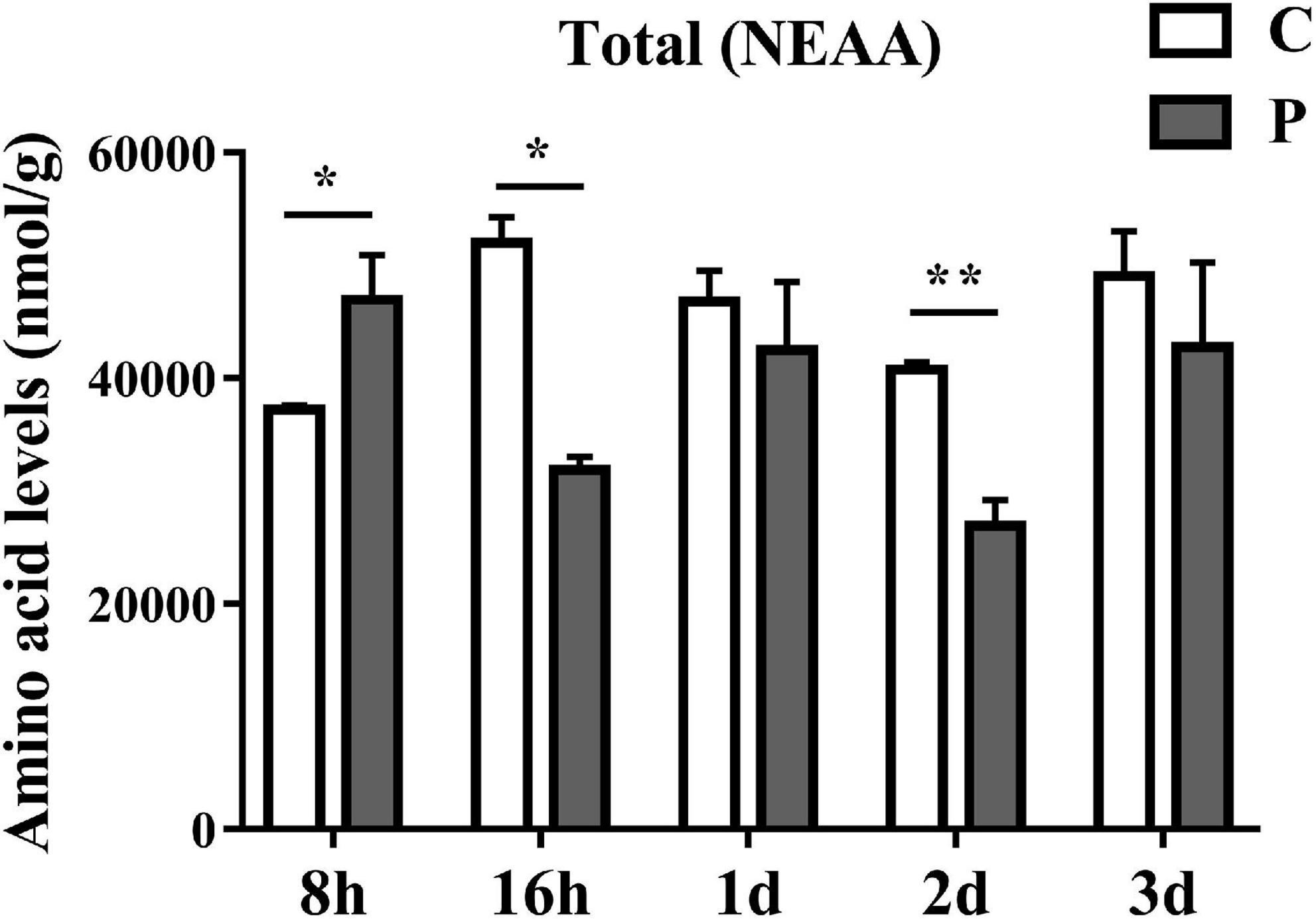
Figure 4. Parasitism causes changes in the content of total NEAA (Non-Essential Amino Acids) in cotton aphid. C, Non-parasitized cotton aphids; P, Parasitized cotton aphids. Statistical significance (Student’s t-test): *P < 0.05; **P < 0.01; ***P < 0.001.
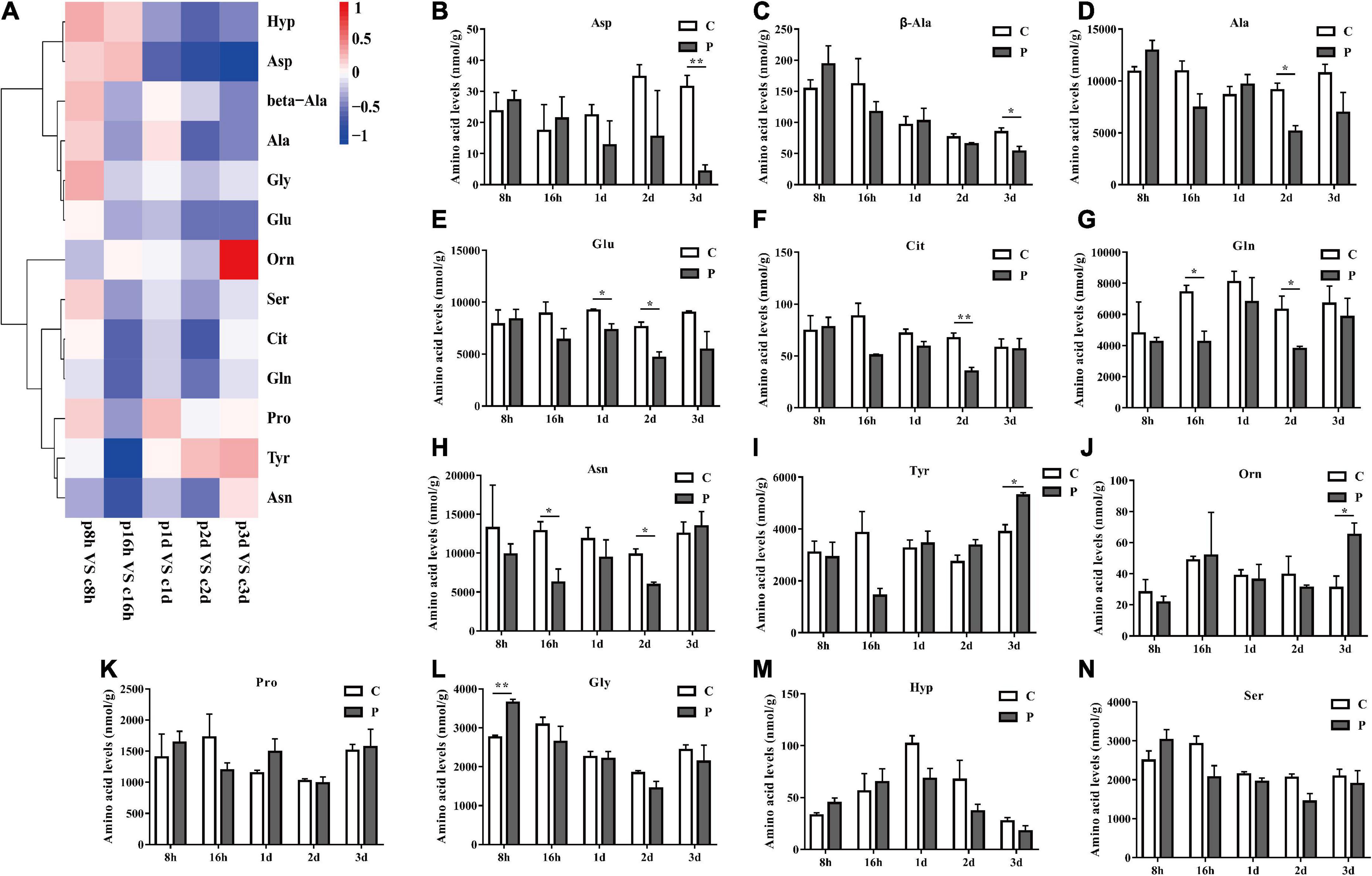
Figure 5. Parasitism leads to changes in NEAA content of cotton aphid. Heatmap (A) and column (B–N) display of changes in NEAA. C, Non-parasitized cotton aphids; P, Parasitized cotton aphids. The log2(Fold change) of amino acid content change is represented by heat map color scale, red indicates up-regulation and blue indicates down-regulation. Statistical significance (Student’s t-test): *P < 0.05; **P < 0.01; ***P < 0.001.
According to the difference of amino acid metabolism level caused by parasitism, we detected 8 key genes related to amino acid metabolism. The expression of amino acid transporter and glutamate synthase genes increased by 1.25 and 1.55 times at 8 h, 1.46 times and 3.51 times at 16 h, decreased after 16 h, and reached statistically significant levels at 3 day (Figures 6A,B). The expression levels of alanine aminotransferase, aspartate aminotransferase, and ribosomal protein S6 kinase genes were consistent, all decreasing at 8 h and increasing significantly at 16 h, and decreasing to a significant level at 3 day (Figures 6C–E). There was no significant difference in the expression levels of ornithine aminotransferase between 1 and 3 day, but the expression also significantly increased at 16 h (Figure 6F). There was no significant change in the expression levels of alanine-glyoxylate aminotransferase and target of rapamycin (Figures 6G,H). We grouped the aforementioned genes into metabolic pathways, including a heatmap representation of the fold change in gene expression, clearly showing the regulatory relationships between amino acid metabolism and synthesis, as well as the changes in pathway genes that utilize amino acids for protein synthesis (Figure 6I).
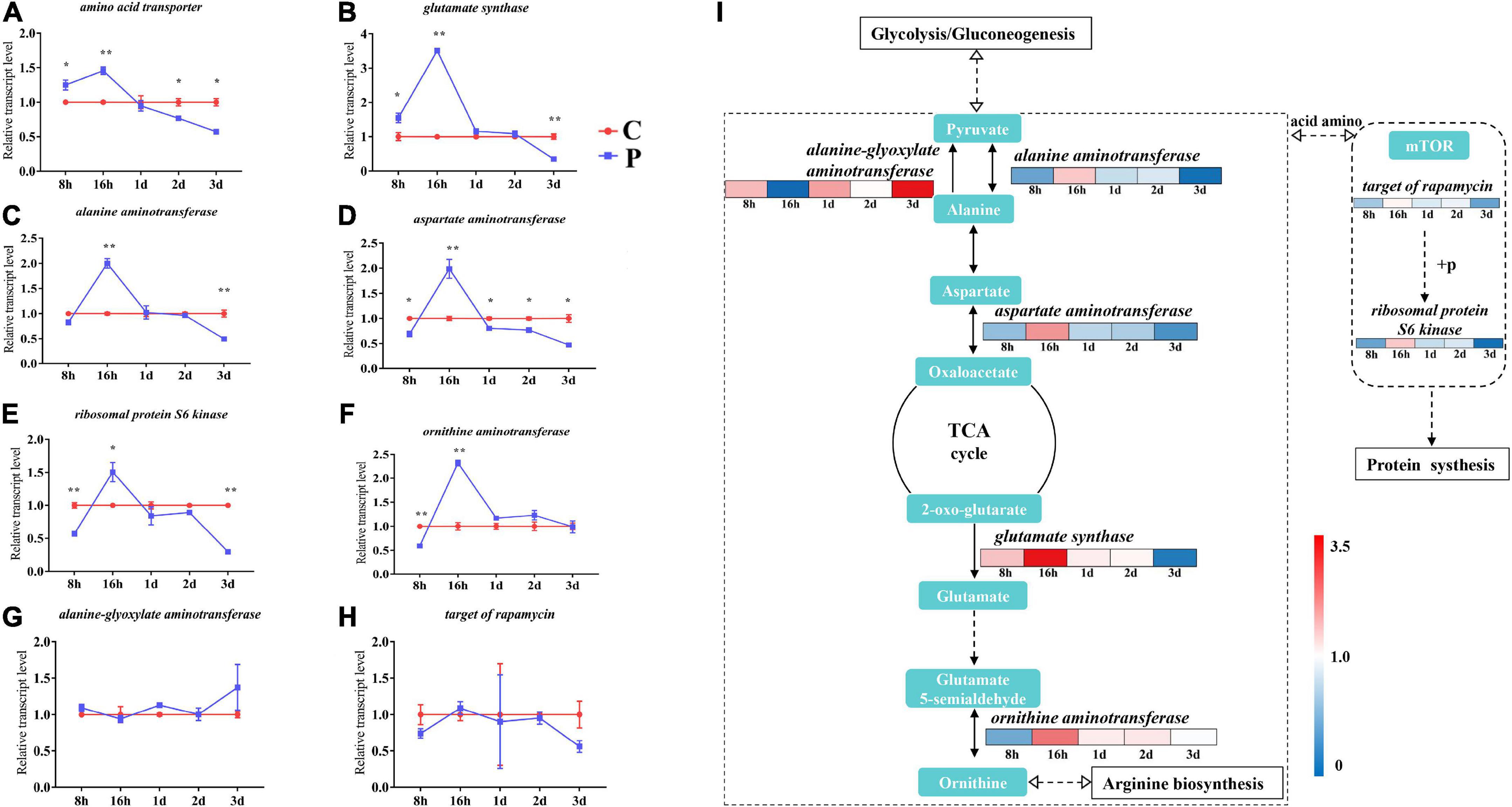
Figure 6. Changes in expression of amino acid metabolism genes in cotton aphid. Line chart (A–B) and signaling pathway (I) display of gene expression. C, Non-parasitized cotton aphids; P, Parasitized cotton aphids. The fold change in gene expression in the pathway is represented by the color scale, red indicates up-regulation and blue indicates down-regulation. Statistical significance (Student’s t-test): *P < 0.05; **P < 0.01; ***P < 0.001.
As an obligate symbiotic bacteria in cotton aphid, Buchnera plays a key role in regulating the supply of amino acids. We detected the changes in the abundance of Buchnera bacteria measured with on copy number in cotton aphids at different stages of parasitism, and found that the abundance of Buchnera increased significantly at 8, 16 h and 2 day, which increased by 1.26, 1.63, and 1.50 times, respectively. During the 1 day period, although the difference of Buchnera abundance was not significant, it increased 1.3 times (Figure 7).
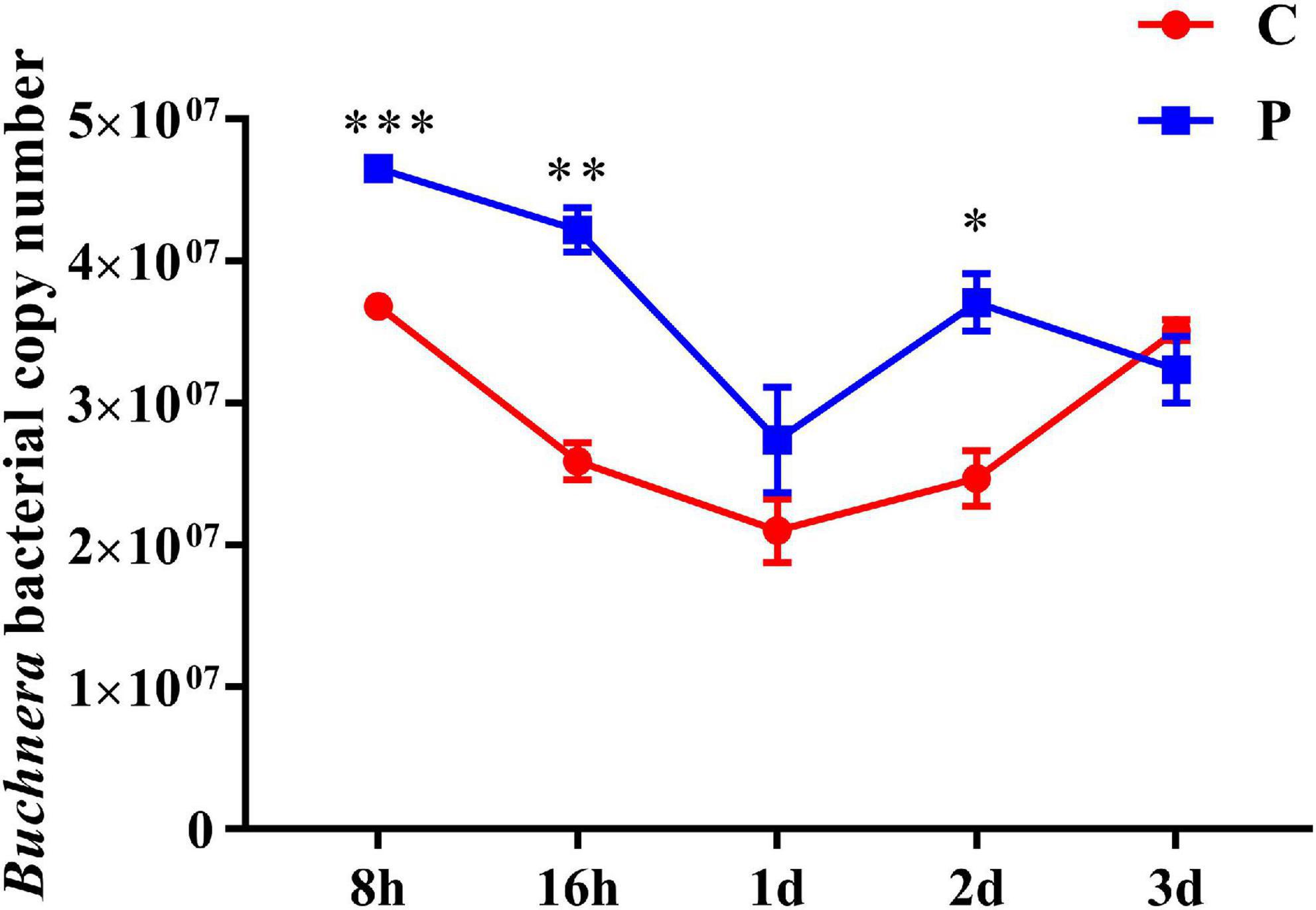
Figure 7. Changes in abundance of symbiotic bacteria Buchnera in cotton aphid. C, Non-parasitized cotton aphids; P, Parasitized cotton aphids. Statistical significance (Student’s t-test): *P < 0.05; **P < 0.01; ***P < 0.001.
Parasites are ubiquitous in nature, and nutrient utilization is one of the most important strategies of parasitism (32). Parasitoid wasps create suitable host nutritional adaptations by regulating host nutrient metabolism (32). Amino acids are important nutrients for organisms, and understanding the regulation of parasitoids’ amino acid metabolism in the host is helpful to deeply understand the nutritional requirements of parasitoids, and is of great significance to the breeding and biological control of parasitoids. In this study, the amino acid content of parasitized and unparasitized cotton aphids at different developmental stages were analyzed with PCA and OPLS-DA. The differences between these samples were clearly distinguished, indicating that parasitism significantly affects the amino acid metabolism of cotton aphids, and that B. communis has a certain regulatory effects on the nutrition of cotton aphids.
Previous in vitro studies have demonstrated that the host provides some amino acids that are of considerable importance for the nutritional and metabolic adaptation of parasitoid wasps (33, 34). For example, there is currently evidence that the larval stage of Aphidius ervi is able to directly ingest the host’s sugars and amino acids through the body surface and gut. These nutrients are efficiently absorbed by the larval epidermis, but the rate of transport gradually decreases over the course of parasitization (33). With the decrease in the amino acid transport rate to the parasitoid wasp, the activity level of the host amino acid transport gene was likewise lowered, which was also confirmed by our findings here with B. communis, another endoparasitic wasp. In this study, the content of almost all amino acids in cotton aphids parasitized for 8 h increased, and this temporary increase was accompanied by a decrease in amino acid content. The most direct explanation for the change of amino acid content is related to the rate of parasitoid nutrient absorption and the activity of absorbed proteins (34). B. communis in the earliest stage of development at 8 h has no ability to absorb substances and lacks a digestive tract (35). B. communis has a faster rate of amino acid absorption in the early stage of larval development, for example, the content of some amino acids is greatly reduced at 16 h after parasitization. This indicates that parasitic wasp larvae begin to absorb or feed on some amino acids for physiological metabolism, and that the rate of absorption may be greater than cotton aphid’s ability to synthesize these molecules. The content of 1Mhis, Phe, His, Thr, Ala, Glu, Cit, and Gln decreased significantly from 16 h to 2 day, but there was no statistically significant difference in the later stages of development. Similarly, previous studies have shown that parasitoid larvae absorption rates of amino acids decrease in the later stage of development (33). Perhaps B. communis needs some amino acids to supplement in the later stage of growth and development. In regards to the nutritional regulation of parasitic wasps on the host, the TOR (target of rapamycin)-S6K (ribosomal protein S6 kinase) pathway was found to be inhibited, and the transcriptional translation process of cotton aphid may be decreased (36), which could meet the needs of parasitoids by reducing the consumption of amino acids (27).
As an amino acid with late and statistically significant parasitic effects, tyrosine is indispensable for the growth and development of endoparasitoid larvae (37). Tyrosine is an aromatic amino acid, which is synthesized in large quantities by phenylalanine through an aromatic shuttle mediated by symbiotic bacteria (38). Phenylalanine, as the only derivative precursor of tyrosine, decreased significantly at 16 h in our study, probably because tyrosine also decreased significantly at 16 h. In order to supplement the supply of tyrosine, a large amount of phenylalanine is needed to be involved in the formation of tyrosine. Another explanation is that the immune response of insects to biological attacks involves the extensive use of tyrosine through the phenoloxidase pathway (39), which may also be a means for parasitic wasps to regulate host tyrosine content. At present, the lack of lipid synthesis ability of parasitoid wasps has always been the focus of current research (40, 41). The N-acetylaspartate (NAA) pathway, which is involved in the production of aspartic acid, forms acetic acid, which is the only prerequisite for lipid synthesis (42). Furthermore, the NAA pathway is also an important regulatory pathway for lipid metabolism (43, 44). The change of aspartate content of cotton aphid was consistent with the trend in aspartate aminotransferase gene abundance. At the same time, aspartate aminotransferase is a part of malate-aspartate shuttle in human body, which is involved in gluconeogenesis in liver, kidney and glycerol in adipose tissue (45), which therefore suggests that parasitoids may use aspartic acid as an important regulator of lipid synthesis and thus regulate the lipid metabolism of cotton aphids. Based on this hypothesis, more verification experiments are needed to prove it.
Previously, Falabella et al. (37) proposed that bacterial endosymbionts affect the early growth and development of parasitic wasp larvae in aphids, and that the growth and reproduction of aphids depends on the obligate symbiotic bacterium Buchnera which provides essential amino acids (46, 47). Interestingly, parasitic wasps do not let go of the regulation of every detail of the host in order to satisfy their own growth and development. Other studies have proposed that parasitoids can alter the host’s microbial environment to increase favorable conditions for their colonization, and that parasitoids can adapt and even modulate the host’s microbial composition (48). Our results confirm the previous evidence that the parasitism of B. communis leads to an increase in the abundance of Buchnera in cotton aphids, which likely further supplements the biosynthesis of carbohydrates, lipids, and coenzymes in parasitic wasps by ensuring a steady supply of amino acids from the host (49, 50). Studies have shown that the symbiotic bacterial composition of parasitic wasp larvae is similar to that of host aphids, and that in particular the abundance of Buchnera is higher in parasitic wasp larvae (49). The shifts in Buchnera abundance in cotton aphid were no longer statistically significant in late stages of parasitization, perhaps in part because the bacterial composition of the parasitoid wasp was highly similar to that of the host. The Buchnera symbiont in the cotton aphid in the early stages of parasitism is most likely acquired by the parasitoid, and the parasitoid’s dependence on the host Buchnera gradually decreases in the later stage of development. Further investigation and experimentation is needed to confirm this hypothesis.
Parasitic wasps, as important biological controls natural enemy insects of pests, are critical to pest management (51). This research further clarifies the nutritional regulation and nutritional requirements of parasitoids, providing a strong theoretical basis for practical and inexpensive management of pests through the large-scale artificial reproduction of parasitoids.
Binodoxys communis has a significant effect on amino acid metabolism of A. gossypii, involving associated gene expression changes and changes in abundance of symbionts associated with amino acid metabolism. This study provides an important theoretical basis for parasitic wasps to regulate the metabolism of the host A. gossypii, and provides a key reference for guiding parasitoid breeding and biological control of pests.
The data of this study can be provided directly by the corresponding author for further inquiries.
JC, JL, and XG conceived and designed the research. HX conducted the experiments and wrote the manuscript. YZ, LW, DL, XZ, and KZ contributed the new reagents and analytical tools. HX, YZ, JJ, and LN analyzed the data. All authors contributed to the study conception and design, read and approved the manuscript.
This research was supported by the National Natural Science Foundation of China (Grant no. 32001919) and Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences.
We thank for the support of experimental technology and equipment provided by Cotton Research Institute of Chinese Academy of Agricultural Sciences.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.1006253/full#supplementary-material
PCA, principal components analysis; OPLS-DA, orthogonal projections to latent structures-discriminant analysis; 1Mhis, 1-Methyl-L-histidine; 3Mhis, 3-Methyl-L-histidine; Phe, Phenylalanine; Arg, Arginine; His, Histidine; Met, Methionine; Thr, Threonine; Try, Tryptophan; Val, Valine; Lys, Lysine; Hyp, 4-Hydroxyproline; β -Ala, beta-Alanine; Gly, Glycine; Ala, Alanine; Asp, Aspartic acid; Cit, Citrulline; Glu, Glutamic acid; Gln, Glutamine; Orn, Ornithine; Pro, Proline; Ser, Serine; Tyr, Tyrosine; Asn, Asparagine.
1. Wang Y, Wu X, Wang Z, Chen T, Zhou S, Chen J, et al. Symbiotic bracovirus of a parasite manipulates host lipid metabolism via tachykinin signaling. PLoS Pathog. (2021) 17:e1009365. doi: 10.1371/journal.ppat.1009365
2. Zhang J, Wang F, Yuan B, Yang L, Yang Y, Fang Q, et al. A novel cripavirus of an ectoparasitoid wasp increases pupal duration and fecundity of the wasp’s Drosophila melanogaster host. ISME J. (2021) 15:3239–57. doi: 10.1038/s41396-021-01005-w
3. Becchimanzi A, Avolio M, Di Lelio I, Marinelli A, Varricchio P, Grimaldi A, et al. Host regulation by the ectophagous parasitoid wasp Bracon nigricans. J Insect Physiol. (2017) 101:73–81. doi: 10.1016/j.jinsphys.2017.07.002
4. Gao X, Luo J, Zhu X, Wang L, Ji J, Zhang L, et al. Growth and fatty acid metabolism of Aphis gossypii parasitized by the parasitic wasp Lysiphlebia japonica. J Agric Food Chem. (2019) 67:8756–65. doi: 10.1021/acs.jafc.9b02084
5. Pennacchio F, Strand MR. Evolution of developmental strategies in parasitic hymenoptera. Annu Rev Entomol. (2006) 51:233–58. doi: 10.1146/annurev.ento.51.110104.151029
6. Mrinalini, Siebert AL, Wright J, Martinson E, Wheeler D, Werren JH. Parasitoid venom induces metabolic cascades in fly hosts. Metabolomics. (2015) 11:350–66. doi: 10.1007/s11306-014-0697-z
7. Nakamatsu Y, Fujii S, Tanaka T. Larvae of an endoparasitoid, Cotesia kariyai (Hymenoptera: Braconidae), feed on the host fat body directly in the second stadium with the help of teratocytes. J Insect Physiol. (2002) 48:1041–52. doi: 10.1016/s0022-1910(02)00192-0
8. Salvador G, Cônsoli FL. Changes in the hemolymph and fat body metabolites of Diatraea saccharalis (Fabricius) (Lepidoptera: Crambidae) parasitized by Cotesia flavipes (Cameron) (Hymenoptera: Braconidae). Biol Control. (2008) 45:103–10.
9. Nakamatsu Y, Tanaka T. Venom of Euplectrus separatae causes hyperlipidemia by lysis of host fat body cells. J Insect Physiol. (2004) 50:267–75. doi: 10.1016/j.jinsphys.2003.12.005
10. Visser B, Ellers J. Lack of lipogenesis in parasitoids: a review of physiological mechanisms and evolutionary implications. J Insect Physiol. (2008) 54:1315–22. doi: 10.1016/j.jinsphys.2008.07.014
11. Zhang S, Luo JY, Lv LM, Wang CY, Li CH, Zhu XZ, et al. Effects of Lysiphlebia japonica (Ashmead) on cotton-melon aphid Aphis gossypii Glover lipid synthesis. Insect Mol Biol. (2015) 24:348–57. doi: 10.1111/imb.12162
12. Gao X, Zhang S, Luo J, Lü L, Zhang L, Cui J. Lipidomics and RNA-Seq study of lipid regulation in Aphis gossypii parasitized by Lysiphlebia japonica. Sci Rep. (2017) 7:1364. doi: 10.1038/s41598-017-01546-1
13. Lin Z, Wang RJ, Cheng Y, Du J, Volovych O, Han LB, et al. Insights into the venom protein components of Microplitis mediator, an endoparasitoid wasp. Insect Biochem Mol Biol. (2019) 105:33–42. doi: 10.1016/j.ibmb.2018.12.013
14. Young VR, Borgonha S. Adult human amino acid requirements. Curr Opin Clin Nutr Metab Care. (1999) 2:39–45. doi: 10.1097/00075197-199901000-00008
15. Ryan PJ, Riechman SE, Fluckey JD, Wu G. Interorgan metabolism of amino acids in human health and disease. Adv Exp Med Biol. (2021) 1332:129–49. doi: 10.1007/978-3-030-74180-8_8
16. Baker DH. Animal models of human amino acid responses. J Nutr. (2004) 134:1646s–50s. doi: 10.1093/jn/134.6.1646S
17. Bruhat A, Cherasse Y, Chaveroux C, Maurin AC, Jousse C, Fafournoux P. Amino acids as regulators of gene expression in mammals: molecular mechanisms. Biofactors. (2009) 35:249–57. doi: 10.1002/biof.40
18. Ji X, Hou C, Gao Y, Xue Y, Yan Y, Guo X. Metagenomic analysis of gut microbiota modulatory effects of jujube (Ziziphus jujuba Mill.) polysaccharides in a colorectal cancer mouse model. Food Funct. (2020) 11:163–73.
19. Hildebrandt TM, Nunes Nesi A, Araujo WL, Braun HP. Amino acid catabolism in plants. Mol Plant. (2015) 8:1563–79. doi: 10.1016/j.molp.2015.09.005
20. Heinemann B, Hildebrandt TM. The role of amino acid metabolism in signaling and metabolic adaptation to stress-induced energy deficiency in plants. J Exp Bot. (2021) 72:4634–45. doi: 10.1093/jxb/erab182
21. Hoedjes KM, Rodrigues MA, Flatt T. Amino acid modulation of lifespan and reproduction in Drosophila. Curr Opin Insect Sci. (2017) 23:118–22. doi: 10.1016/j.cois.2017.07.005
22. Wu G. Amino acids: metabolism, functions, and nutrition. Amino Acids. (2009) 37:1–17. doi: 10.1007/s00726-009-0269-0
23. Wu G, Bazer FW, Dai Z, Li D, Wang J, Wu Z. Amino acid nutrition in animals: protein synthesis and beyond. Annu Rev Anim Biosci. (2014) 2:387–417. doi: 10.1146/annurev-animal-022513-114113
24. Wang J, Wu Z, Li D, Li N, Dindot SV, Satterfield MC, et al. Nutrition, epigenetics, and metabolic syndrome. Antioxid Redox Signal. (2012) 17:282–301. doi: 10.1089/ars.2011.4381
25. Jobgen W, Fu WJ, Gao H, Li P, Meininger CJ, Smith SB, et al. High fat feeding and dietary L-arginine supplementation differentially regulate gene expression in rat white adipose tissue. Amino Acids. (2009) 37:187–98.
26. Wang J, Chen L, Li P, Li X, Zhou H, Wang F, et al. Gene expression is altered in piglet small intestine by weaning and dietary glutamine supplementation. J Nutr. (2008) 138:1025–32. doi: 10.1093/jn/138.6.1025
27. Ye X, Xiong S, Teng Z, Yang Y, Wang J, Yu K, et al. Genome of the parasitoid wasp Cotesia chilonis sheds light on amino acid resource exploitation. BMC Biol. (2022) 20:118. doi: 10.1186/s12915-022-01313-3
28. Price DR, Feng H, Baker JD, Bavan S, Luetje CW, Wilson AC. Aphid amino acid transporter regulates glutamine supply to intracellular bacterial symbionts. Proc Natl Acad Sci USA. (2014) 111:320–5. doi: 10.1073/pnas.1306068111
29. James EB, Feng H, Wilson ACC. mTOR complex 1 implicated in aphid/Buchnera host/symbiont integration. G3. (2018) 8:3083–91. doi: 10.1534/g3.118.200398
30. Hansen AK, Moran NA. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA. (2011) 108:2849–54. doi: 10.1073/pnas.1013465108
31. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. (2001) 29:e45. doi: 10.1093/nar/29.9.e45
32. Pennacchio F, Caccia S, Digilio MC. Host regulation and nutritional exploitation by parasitic wasps. Curr Opin Insect Sci. (2014) 6:74–9. doi: 10.1016/j.cois.2014.09.018
33. Giordana B, Milani A, Grimaldi A, Farneti R, Casartelli M, Ambrosecchio MR, et al. Absorption of sugars and amino acids by the epidermis of Aphidius ervi larvae. J Insect Physiol. (2003) 49:1115–24. doi: 10.1016/j.jinsphys.2003.08.010
34. Caccia S, Leonardi MG, Casartelli M, Grimaldi A, de Eguileor M, Pennacchio F, et al. Nutrient absorption by Aphidius ervi larvae. J Insect Physiol. (2005) 51:1183–92. doi: 10.1016/j.jinsphys.2005.06.010
35. de Eguileor M, Grimaldi A, Tettamanti G, Valvassori R, Leonardi MG, Giordana B, et al. Larval anatomy and structure of absorbing epithelia in the aphid parasitoid Aphidius ervi Haliday (Hymenoptera, Braconidae). Arthropod Struct Dev. (2001) 30:27–37. doi: 10.1016/s1467-8039(01)00017-2
36. Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. (2012) 441:1–21. doi: 10.1042/bj20110892
37. Falabella P, Tremblay E, Pennacchio F. Host regulation by the aphid parasitoid Aphidius ervi: the role of teratocytes. Entomol Exp Appl. (2000) 97:1–9.
38. Liadouze I, Febvay G, Guillaud J, Bonnot G. Effect of diet on the free amino acid pools of symbiotic and aposymbiotic pea aphids, Acyrthosiphon pisum. J Insect Physiol. (1995) 41:33–40.
39. Shelby KS, Webb BA. Polydnavirus-mediated suppression of insect immunity. J Insect Physiol. (1999) 45:507–14. doi: 10.1016/s0022-1910(98)00144-9
40. Visser B, Le Lann C, den Blanken FJ, Harvey JA, van Alphen JJ, Ellers J. Loss of lipid synthesis as an evolutionary consequence of a parasitic lifestyle. Proc Natl Acad Sci USA. (2010) 107:8677–82. doi: 10.1073/pnas.1001744107
41. Lammers M, Kraaijeveld K, Mariën J, Ellers J. Gene expression changes associated with the evolutionary loss of a metabolic trait: lack of lipogenesis in parasitoids. BMC Genomics. (2019) 20:309. doi: 10.1186/s12864-019-5673-6
42. Huber K, Hofer DC, Trefely S, Pelzmann HJ, Madreiter-Sokolowski C, Duta-Mare M, et al. N-acetylaspartate pathway is nutrient responsive and coordinates lipid and energy metabolism in brown adipocytes. Biochim Biophys Acta Mol Cell Res. (2019) 1866:337–48. doi: 10.1016/j.bbamcr.2018.08.017
43. Pessentheiner AR, Pelzmann HJ, Walenta E, Schweiger M, Groschner LN, Graier WF, et al. NAT8L (N-acetyltransferase 8-like) accelerates lipid turnover and increases energy expenditure in brown adipocytes. J Biol Chem. (2013) 288:36040–51. doi: 10.1074/jbc.M113.491324
44. Prokesch A, Pelzmann HJ, Pessentheiner AR, Huber K, Madreiter-Sokolowski CT, Drougard A, et al. N-acetylaspartate catabolism determines cytosolic acetyl-CoA levels and histone acetylation in brown adipocytes. Sci Rep. (2016) 6:23723. doi: 10.1038/srep23723
45. Otto-Şlusarczyk D, Graboń W, Mielczarek-Puta M. Aspartate aminotransferase-key enzyme in the human systemic metabolism. Postepy Hig Med Dosw (Online). (2016) 70:219–30. doi: 10.5604/17322693.1197373
46. Douglas AE. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol. (1998) 43:17–37. doi: 10.1146/annurev.ento.43.1.17
47. Feng H, Edwards N, Anderson CMH, Althaus M, Duncan RP, Hsu YC, et al. Trading amino acids at the aphid-Buchnera symbiotic interface. Proc Natl Acad Sci USA. (2019) 116:16003–11. doi: 10.1073/pnas.1906223116
48. Dheilly NM, Poulin R, Thomas F. Biological warfare: microorganisms as drivers of host-parasite interactions. Infect Genet Evol. (2015) 34:251–9. doi: 10.1016/j.meegid.2015.05.027
49. Gao X, Niu R, Zhu X, Wang L, Ji J, Niu L, et al. Characterization and comparison of the bacterial microbiota of Lysiphlebia japonica parasitioid wasps and their aphid host Aphis gosypii. Pest Manag Sci. (2021) 77:2710–8. doi: 10.1002/ps.6299
50. Gao X, Luo J, Lü L, Zhang L, Zhang S, Cui J. RNA-Seq and UHPLC-Q-TOF/MS based lipidomics study in Lysiphlebia japonica. Sci Rep. (2018) 8:7802. doi: 10.1038/s41598-018-26139-4
Keywords: parasitoids, amino acids, cotton aphids, host-microbe interactions, host-parasitoid interactions
Citation: Xue H, Zhao Y, Wang L, Zhu X, Zhang K, Li D, Ji J, Niu L, Cui J, Luo J and Gao X (2022) Regulation of amino acid metabolism in Aphis gossypii parasitized by Binodoxys communis. Front. Nutr. 9:1006253. doi: 10.3389/fnut.2022.1006253
Received: 29 July 2022; Accepted: 08 September 2022;
Published: 29 September 2022.
Edited by:
Xiaolong Ji, Zhengzhou University of Light Industry, ChinaReviewed by:
Ya-Nan Zhang, Huaibei Normal University, ChinaCopyright © 2022 Xue, Zhao, Wang, Zhu, Zhang, Li, Ji, Niu, Cui, Luo and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinjie Cui, YXljdWlqaW5qaWVAMTYzLmNvbQ==; Junyu Luo, bHVvanVueXUxODE4QDEyNi5jb20=; Xueke Gao, MTUwMzYxMzgzODlAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.