- 1Microbiota Division, Department of Gastroenterology and Hepatology, The First Medical Center, Chinese People's Liberation Army (PLA) General Hospital, Beijing, China
- 2State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, Institute of Food Research, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 3Department of Gastroenterology, Sir Run Run Shaw Hospital, Hangzhou, China
Gut microbiota dysbiosis is closely associated with ulcerative colitis (UC). Prebiotic therapy is a potential approach for UC management especially remission maintaining. Xylo-oligosaccharide (XOS) is an efficient prebiotic with proven health benefits and few side effects. However, the effects of XOS on the gut microbiota of patients with UC have not been investigated previously. The aim of this study was to evaluate the prebiotic effects of XOS on the fecal microbiota of patients with UC in clinical remission using an in vitro fermentation model. Five patients with UC in clinical remission and five healthy volunteers were enrolled in this study. Fresh fecal samples of UC patients were diluted and inoculated in yeast extract, casitone and fatty acid (YCFA) medium alone or with XOS. After fermentation for 48 h, samples were collected for 16S rDNA sequencing to investigate the gut microbiota composition. Differences in the gut microbiota between healthy volunteers and UC patients in clinical remission were detected using original fecal samples. Subsequently, the differences between the YCFA medium alone or with XOS samples were analyzed to illustrate the effects of XOS on the gut microbiota of UC patients. In both principal coordinate analysis (PCoA) and principal component analysis (PCA), the fecal samples of UC patients differed from those of healthy volunteers. Linear discriminant analysis effect size (LEfSe) analysis revealed that the relative abundances of g_Roseburia and g_Lachnospiraceae_ND3007_group were higher in healthy volunteers than in UC patients, while o_Lactobacillales abundance showed the opposite trend (P < 0.05). Wilcoxon rank-sum test bar plot showed that the abundances of g_Eubacterium_halli_group and g_Lachnospiraceae_ND3007_group were higher in the healthy volunteers than in the UC patients (P < 0.05). In addition, in UC patients, the Wilcoxon rank-sum test showed that XOS fermentation promoted the growth of bacterial groups including g_Roseburia, g_Bifidobacterium, and g_Lactobacillus, which is beneficial for recovery of intestinal diseases. These results suggest that XOS can relieve dysbiosis in the feces of UC patients in clinical remission and thus represent a potential prebiotic material for maintaining remission.
Introduction
Ulcerative colitis (UC) is a chronic disease involving recurrent colonic inflammation causing damage to the mucosa or submucosa of colon, and the occurrence of UC is increasing globally (1). Although the etiology and pathogenesis of UC are unclear, previous studies have shown that epithelial barrier integrity disruption, intestinal immunity disorders and in particular, gut microbiota dysbiosis contribute to its development and progression (2). The treatments for UC include controlling the active inflammation and maintaining remission by using amino salicylates, antibiotics, corticosteroids and immunomodulatory drugs. However, these therapies were more likely to bring long-term side effects especially in maintaining remission due to lack of low side effect drugs for long-term use (3). In recent years, modulating the dysbiosis of gut microbiota has been become a new strategy for UC management and shown a fine application prospect (4).
Prebiotics are non-digestible compounds found in many natural foods that selectively stimulate the growth and activity of one or several bacterial groups to produce beneficial effects on the host with few side effects (5). Many clinical trials have been carried out to assess the effects of administering prebiotics such as fructo-oligosaccharide (FOS) for UC treatment and limited benefits have been shown (6, 7). Besides, a review showed that the use of traditional prebiotics including FOS and inulin may be useful especially in IBD patients with low clinical activity of the disease or to maintain remission (8). However, some researches indicated that there was no significant beneficial effect for the use of prebiotics to IBD patients, which may be due to inclusion of IBD patients with high clinical activity or insufficient use of prebiotics (9). Therefore, patient selection for clinical trials is important to the research.
Xylo-oligosaccharide (XOS) is a more efficient prebiotic than traditional prebiotics with proven health benefits via adjusting gut microbiota (10). For example, in vitro and in vivo studies have shown that XOS significantly enriched bifidobacterial populations, which mitigated inflammatory diseases (11, 12). What's more, XOS with bifidobacterium have been proved to alleviate colitis in an animal model (13).
Thus, the aim of this study was to evaluate the prebiotic effect of XOS on the fecal microbiota of UC patients in clinical remission via an in vitro fermentation model (14).
Materials and Methods
Fecal Sample Origins
Five patients with UC in clinical remission diagnosed by colonoscopy and five healthy volunteers, aged between 18 and 60 years, were enrolled in this case-control study. The age and sex of the UC patients and healthy volunteers are matched and details are shown in Supplementary Table 1. The exclusion criteria were as follows: (1) diabetes, cancers or other systemic or serious diseases; (2) pregnancy or lactation; and (3) probiotic, prebiotic or antibiotic agent use within 4 weeks prior to fecal sample collection. Volunteers were provided informed, written consents for collection and research of fecal samples. All the procedures used in the present study were approved by the Ethics Committee of Chinese PLA General Hospital (S2016-130-01).
Fermentation Medium
In brief, the basic growth medium (yeast extract, casitone and fatty acid; YCFA) contained the following compounds: tryptone, 10 g/L; yeast extract, 2.5 g/L; L-cysteine, 1 g/L; NaCl, 0.9 g/L; CaCl2·6H2O, 0.09 g/L; KH2PO4, 0.45 g/L; K2HPO4, 0.45 g/L; MgSO4·7H2O, 0.09 g/L; vitamin I solution, 200 mL; and hemin solution, 2 mL. The vitamin I solution had the following components: vitamin B8, 0.05 mg/mL; vitamin B12, 0.05 mg/mL; 4-aminobenzoique acid, 0.15 mg/mL; vitamin B9, 0.25 mg/mL; and pyridoxamine, 0.75 mg/mL. The hemin solution was 1 mg/mL in 1M sodium hydroxide. XOS (Sigma Aldrich, USA) was added (8 g/L) as the sole carbon source. After adding resazurin (0.1 mg/L), an indicator of anaerobic conditions, the medium was adjusted to pH 6.5, and 5 mL was dispensed into a 10 mL bottle that was flushed with N2 before sterilization in an autoclave.
Static Fermentation
Static fermentation was conducted as described previously (15). Fresh fecal samples (0.8 g) were homogenized with 8 mL of 0.1 M anaerobic phosphate-buffered saline (pH 7.0) using an automatic fecal homogenizer (Halo Biotechnology, China) to make 10% (w/v) slurries as soon as the fecal samples arrived at the laboratory.
In addition, 0.5 mL of the fecal slurry was inoculated into a sterilization bottle of 5 mL growth medium and subjected to anaerobic fermentation at 37°C. After 48 h of fermentation, the broth was centrifuged. The precipitate of the broth were stored at −80°C for DNA extraction (16). The original feces of UC patients and healthy volunteers were defined as samples of the U_FAE and N_FAE groups, while the fecal fermentation precipitate samples of UC patients in YCFA and the XOS-containing media were defined as samples of the U_Y and U_XOS groups.
DNA Extraction and Sequencing
Bacterial genomic DNA was extracted from all samples of four groups mentioned above using a QIAamp DNA Stool Mini Kit according to the manufacturer's instructions (Qiagen, Germany). The concentration of extracted DNA was determined by a NanoDrop 2000 (NanoDrop Technologies, USA) and confirmed by 1.0% agar gel electrophoresis. The V3-V4 region of the bacterial 16S rRNA genes was amplified using the barcoded primers 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′). Amplicons were extracted from 2% agarose gels, purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, USA) according to the manufacturer's instructions and quantified using QuantiFluor™-ST (Promega, USA). Purified amplicons were pooled in equimolar amounts and paired-end sequenced (2 × 250) on an Illumina MiSeq platform according to standard protocols performed by Promegene Technology, Shenzhen, China.
Bioinformatics and Statistical Analysis
Sequences were identified by their barcodes using the Quantitative Insights in Microbial Ecology (QIIME) 1.9.1 pipeline. Low-quality sequences were removed before further analysis. Sequences were clustered into operational taxonomic units (OTUs) with a 97% similarity cutoff by Uparse (version 7.0.1090). The OTUs were assigned to taxa using the silva138 (16s bacteria database) and data were analyzed by R package (version 3.3.1). Within-community diversity (α-diversity) indexes (the ACE, Simpson and Shannon indexes) were calculated by wilcoxon rank-sum test. Venn diagram was used to show the number of common and unique species in different groups by R. Community bar diagram and heatmap were used for exhibition of community species composition and abundance by R. β-Diversity was estimated by calculating the Bray-Curtis distance matrix with QIIME and R, and statistically examined by analysis of similarities (ANOSIM). Principal component analysis (PCA) was performed to explore the variance in microbiota composition. Principal coordinate analysis (PCoA) was conducted based on the OTU level using the R package. Statistically significant differences in the relative abundances of taxa in different groups were calculated by using the linear discriminant analysis (LDA) effect size (LEfSe) method. Taxa with LDA results >2 were considered significantly enriched. Comparison differences of species between two groups were executed by wilcoxon rank-sum test in the stats package of R (17).
Results
Sequencing Data
After quality filtering, trimming and data annotation, totals of 359 OTUs and 329 OTUs were identified in the N_FAE and U_FAE groups, respectively, while 320 and 313 OTUs were found in the U_Y and U_XOS groups, respectively. A total of 235 core OTUs were detected in all four groups (Figure 1). We also found that the N_FAE group had the most unique species (52 species), with 7, 4 and 4 unique species in U_FAE, U_Y and U_XOS groups, respectively. Most species in the gut microbiota of UC patients and healthy volunteers could be cultured by static fermentation, which meant that this in vitro simulation system of the gut microbiota was practicable.
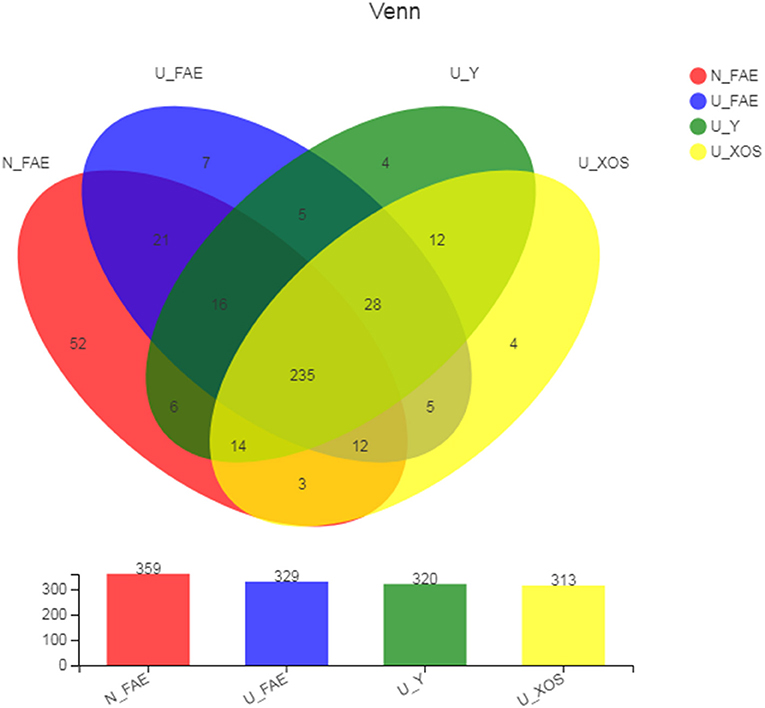
Figure 1. Common and unique OTUs of four groups in the Venn diagram. The OTUs were assigned to taxa using the silva138 (16s bacteria database). N_FAE: original fecal samples of healthy volunteers, n = 5; U_FAE: original fecal samples of UC patients, n = 5; U_Y: fecal fermentation samples in YCFA of UC patients, n = 5; U_XOS: fecal fermentation samples in YCFA+XOS of UC patients, n = 5.
α-Diversity Analysis
Species richness and diversity can be evaluated by measuring α-diversity, for which four indexes are commonly used: ACE and Sobs represent species richness indexes, while Shannon and Simpson represent species diversity indexes. We found that all four indexes in patients with UC in remission were lower than those in healthy volunteers, but these differences were not significant (Figure 2). Moreover, α-diversity decreased after fermentation, especially in the U_XOS group (although again, this was not significant, P > 0.05). This result revealed that growth of some bacteria may be inhibited by fermentation, while some bacteria showed excessive growth, especially with XOS. In general, XOS fermentation could not ameliorate the reduced α-diversity of the gut microbiota in patients with UC.
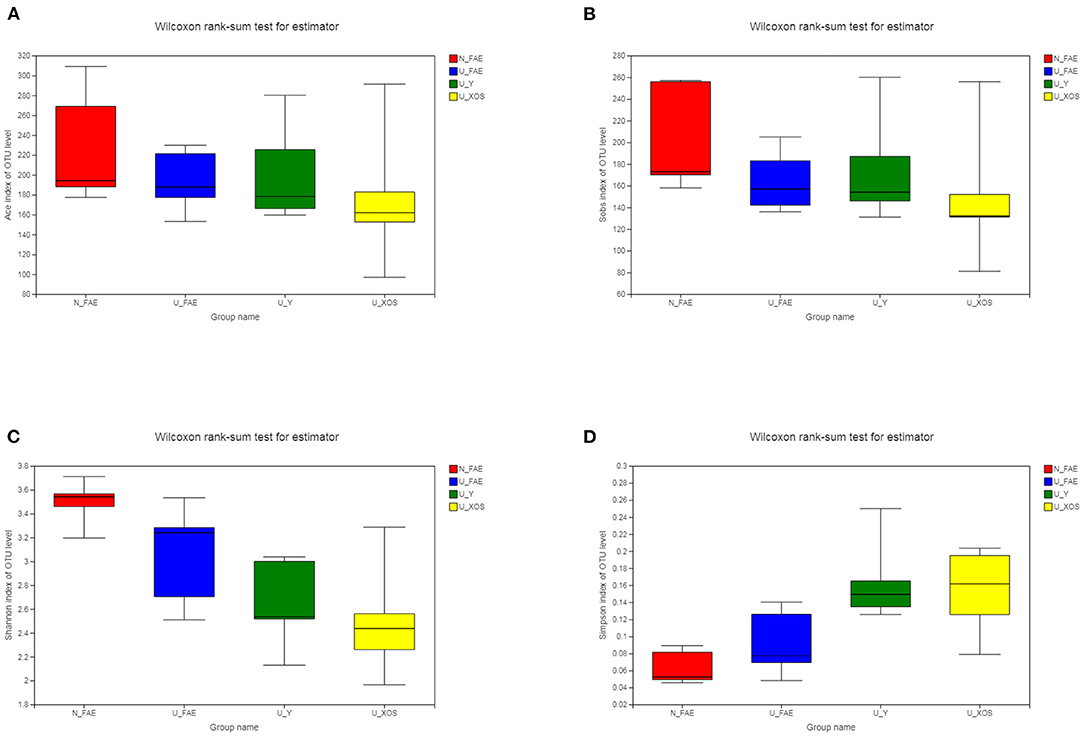
Figure 2. α-Diversity of the four groups. ACE index (A), Sobs index (B), Shannon index (C), and Simpson index (D) in patients with UC in remission were lower than those in healthy volunteers, but these differences were not significant.
β-Diversity Analysis
β-diversity analysis was performed by calculating the Bray-Curtis distance matrix to reveal differences among the four groups, the significance of which was examined by ANOSIM. According to PCA, the four groups showed obvious separation, and the P-value determined by ANOSIM was 0.001, which meant that the differences among the four groups were statistically significant (Figure 3). More importantly, a similar trend was observed in the PCoA. Analysis of all samples indicated that fermentation had a significant impact on the microbiota composition, as the two FAE groups and two fermentation groups exhibited marked separation.
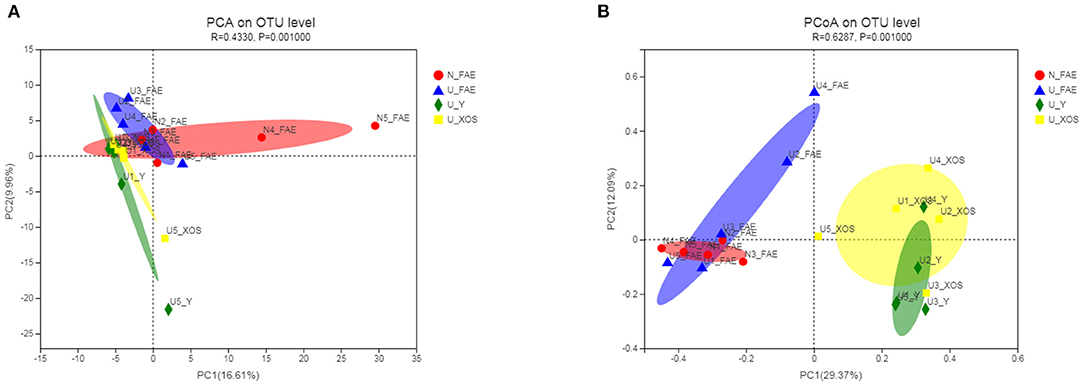
Figure 3. β-diversity of the four groups. According to PCA (A), the four groups showed obvious separation and a similar trend was observed in the PCoA (B).
Community Bar Plot Analysis
Community bar plot analysis was carried out to investigate the microbiota composition in the four groups at the phylum and family levels. At the phylum level, the N_FAE samples were found to contain more Actinobacteria and less Proteobacteria than the U_FAE group. In addition, more Actinobacteria and less Fusobacteria were detected in the U_XOS group than in the U_Y group (Figure 4). At the family level, the relative abundances of Bacteroidaceae and Bifidobacteriaceae were higher, whereas that of Enterobacteriaceae was lower, in the U_FAE group than in the N_FAE group. Furthermore, the U_XOS group differed from the U_Y group in having increased abundances of Bifidobacteriaceae and Selenomonadaceae, and having decreased abundances of Lachnospiraceae and Fusobacteriaceae (Figure 4).

Figure 4. Community bar plot analysis of the four groups. (A) Microbiota composition at the phylum level; (B) microbiota composition at the family level.
Identification of Differentially Abundant Taxa
Next, LEfSe analysis was used to identify the differentially abundant taxa between the N_FAE and U_FAE groups. The results showed that the relative abundances of g_Roseburia, g_Fusicatenibacter, g_Lachnospiraceae_ND3007_group, g_Butyricimonas, g_Eubacterium_halli_group, g_Oscillibacter, g_Lachnospiraceae_UCG-010, g_Bilophila, g_Turicibacter and g_Lachnospiraceae_FCS020_group were higher in the N_FAE group, while that of o_Lactobacillales was higher in the U_FAE group (Figure 5A). Subsequently, differences between the proportions of these taxa were detected by the wilcoxon rank-sum test bar plot analysis at the genus level. The abundances of g_Eubacterium_halli_group, g_Lachnospiraceae_ND3007_group, g_Bilophila, g_Turicibacter, and g_Butyricimonas were higher in the N_FAE group (P < 0.05), whereas the abundances of g_Bifidobacterium, g_Lactobacillus, and g_Lactococcus were higher in the U_FAE group, although the difference was not significant (Figures 6A,C). To illustrate the effects of XOS on the gut microbiota of UC patients in clinical remission, LEfSe and difference analyses between the U_Y and U_XOS groups were executed. LEfSe analysis showed that the abundances of g_Lachnoclostridium, g_norank_f_Lachnospiraceae, and g_Flavonifractor were higher in the U_Y group than in the U_XOS group (Figure 5B). A wilcoxon rank-sum test bar plot at the genus level showed that g_Roseburia, g_Fusicatenibacter, g_Bifidobacterium, and g_Lactobacillus were more abundant, whereas g_Oscillibacter, g_Bilophila, and g_Lachnospiraceae_UCG-010 were less abundant, in the U_XOS group than in the U_Y group, but the differences were not significant (Figures 6B,D). In addition, g_Turicibacter was not detected in the U_XOS group or U_Y group, which meant that it could not be cultured in the fermentation environment.
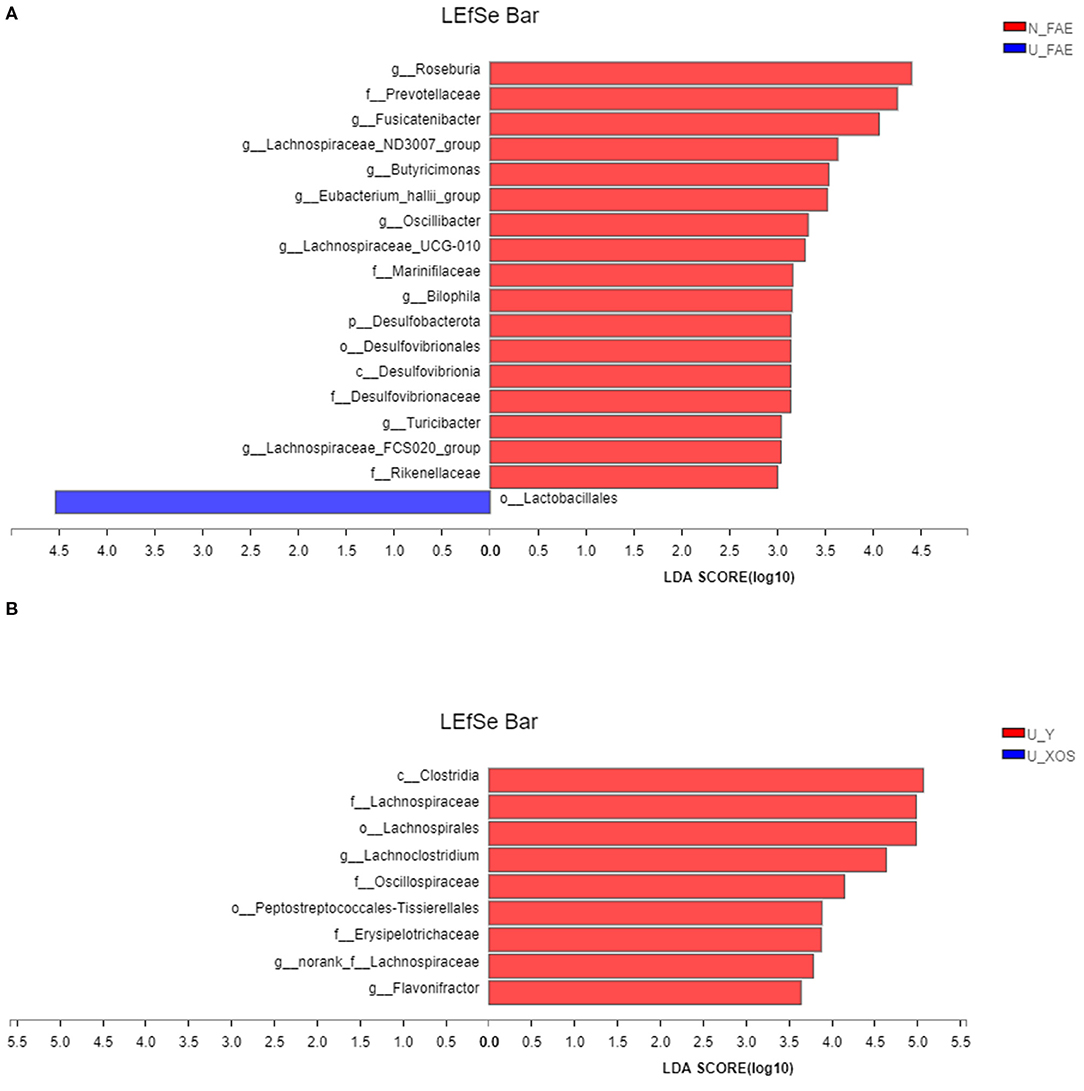
Figure 5. LEfSe analysis to identify the differentially abundant taxa. (A) N_FAE group vs. U_FAE group; (B) U_Y group vs. U_XOS group.
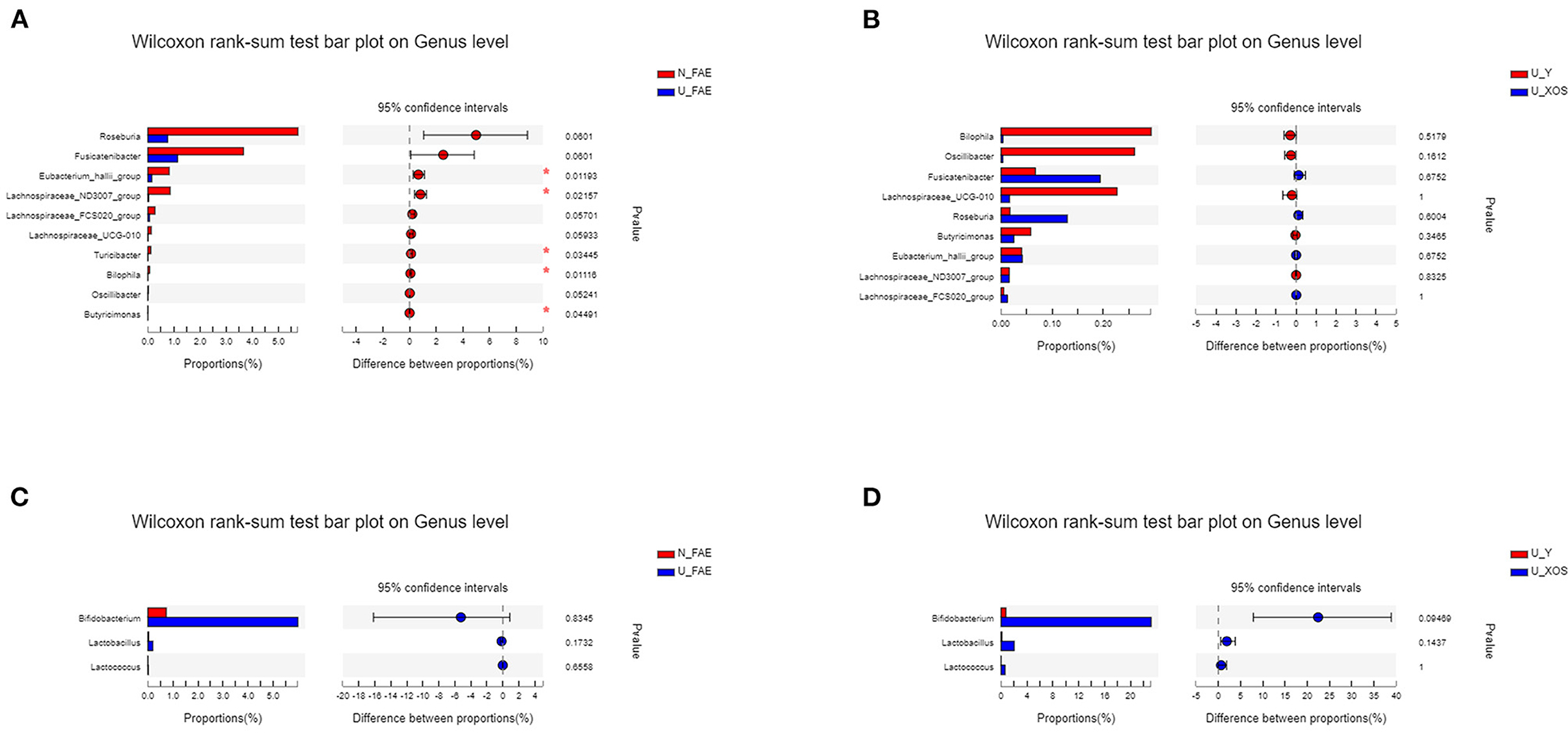
Figure 6. Differential abundance analysis to identify the differentially abundant taxa by the wilcoxon rank-sum test. (A) N_FAE group vs. U_FAE group; (B) U_Y group vs U_XOS group; (C) proportions of Bifidobacterium, Lactobacillus and Lactobacillus in the two original fecal groups; (D) proportions of Bifidobacterium, Lactobacillus and Lactobacillus in the two fermentation groups.
Top-Species Identification and Phylogenetic Tree for Every Sample in the Four Groups
A hierarchical clustering tree was constructed to illustrate the microbiota composition and show the cluster relationship of every sample (Figure 7A). In this analysis, we found that samples of the N_FAE, U_FAE, U_Y, and U_XOS groups separated well. Community heatmap analysis at the genus level revealed the top 20 taxa of the four groups and the differentially abundant taxa, such as g_Roseburia and g_Bifidobacterium. In addition, a phylogenetic tree was drawn to illustrate the evolutionary relationship of the top 20 species via a community heatmap (Figure 7B).
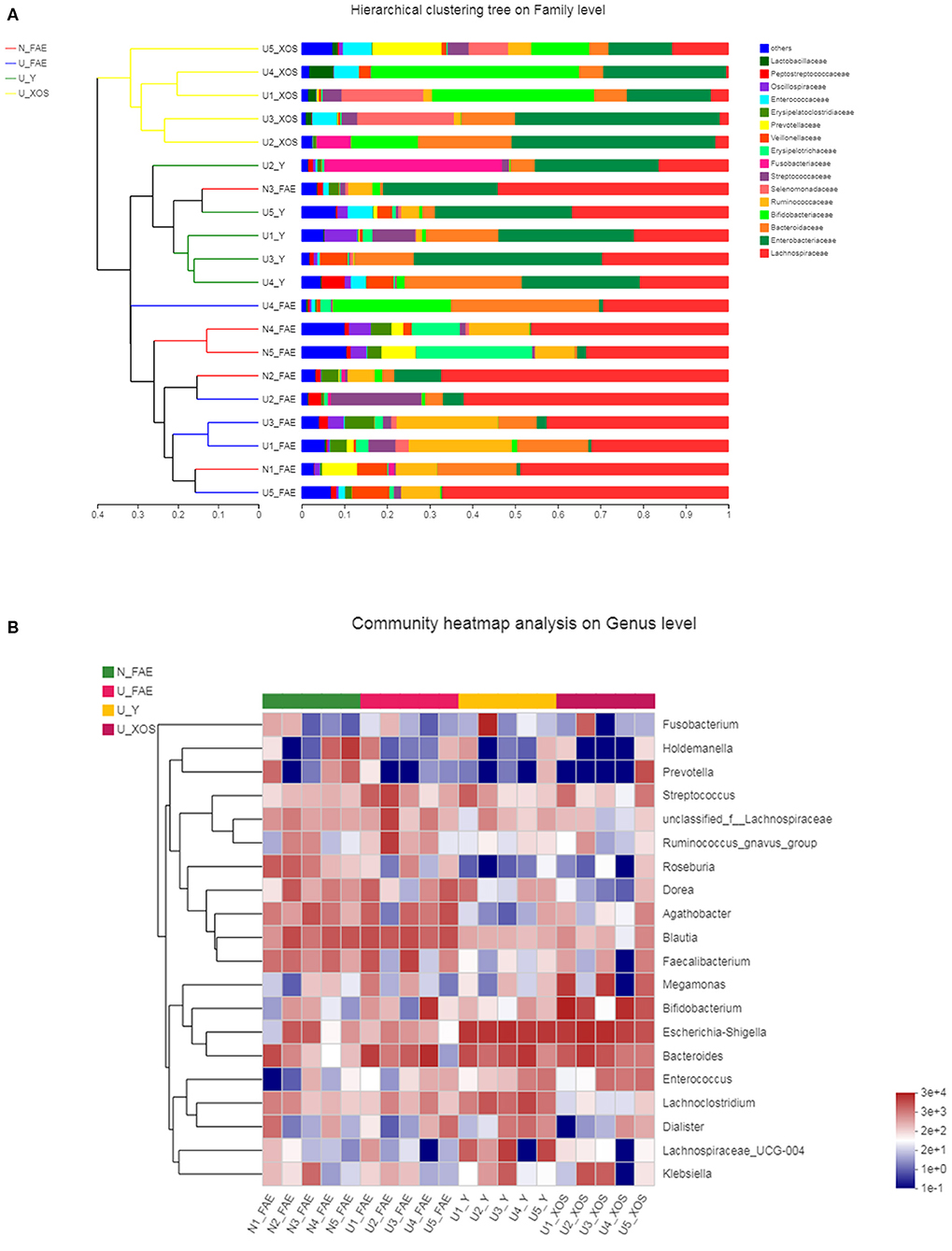
Figure 7. Top-species analysis of the four groups. (A) Hierarchical clustering tree on family level; (B) community heatmap analysis on genus level.
Discussion
UC is characterized by colonic inflammation, remission and relapse, the etiology and pathogenesis of this intractable disease are still unclear (1). Increasing evidence has shown that gut microbiota dysbiosis plays an important role in UC development and progression (2). In addition, the interplay between immune system disorders and gut microbiota dysbiosis has attracted the interest of researchers. New therapeutic methods, such as fecal microbiota transplantation (FMT), have been proven to be safer and more efficacious therapeutic approach for microbiota-associated diseases (18, 19).
Another potential way to modulate or maintain gut microbiota composition is the application of prebiotics, which are defined as substances that selectively stimulate the growth of beneficial microbes, particularly administered in combination with their target group of beneficial microbes (20). To date, many studies have assessed the preventive or severity-reducing effects of prebiotics in UC patients (6, 7). While some studies have shown that prebiotics are beneficial in inducing remission, others have shown no significant influence on UC. The discrepancy may be associated with disease status, individual gut microbiota differences and prebiotic type. Thus, the search for efficient prebiotics is critical.
XOS is one of the most efficient prebiotics for proliferation of Bifidobacterium (10), which produces a variety of organic acids and inhibits the growth of harmful bacteria via changing the gut microbiota composition. Generally, the best models for investigation of prebiotic effects on intestinal microbiota were in vivo testing involving use of humans and animals (21). However, in vivo human trials deserve strict ethical constraints and in vivo animal trials often give results that are not reproducible in humans (22). Furthermore, in vivo testing is often expensive and time consuming, usually with limited study numbers while in vitro fermentation models can be characterized by inoculation of single or multiple ingredients with fecal microbiota in the meantime (23).
Therefore, in the present study, we evaluated the prebiotic effect of XOS on the fecal microbiota of patients with UC in clinical remission using an in vitro fermentation model. The results indicated that the feces of healthy volunteers had more unique species and most species in the gut microbiota of UC patients could be cultured by a static fermentation model, which meant that this simulation model of the gut microbiota was practicable. The species richness and diversity indexes in patients with UC in remission were lower than those in healthy volunteers, but the differences were not significant, which implied that the gut microbiota may be recovered after disease remission. Moreover, the α-diversity decreased after fermentation, especially in the U_XOS group, but the difference was not significant (Figure 2). This revealed that some bacteria could not be cultured, while some were overgrown, especially with XOS fermentation, which did not significantly change the α-diversity. In general, XOS fermentation could not improve the α-diversity of the gut microbiota in patients with UC in remission. PCA showed that the four groups were obviously separated, which meant that the differences among the four groups were significant. More importantly, PCoA showed results similar to those of PCA, but the fecal samples of UC patients were more different from the fermentation samples than from N_FAE, which meant that the gut microbiota community composition was clearly changed by fermentation.
To illustrate the effects of XOS on the gut microbiota of UC patients in clinical remission, wilcoxon rank-sum test bar plots at the genus level were used, and results showed that g_Roseburia, g_Fusicatenibacter, g_Bifidobacterium, and g_Lactobacillus were more abundant, and g_Oscillibacter, g_Bilophila, and g_Lachnospiraceae_UCG-010 were less abundant, in the U_XOS group than in the U_Y group.
The results of this in-vitro study were consistent with previously published data showing stimulation of Bifidobacterium enrichment by XOS (24). Bifidobacterium, a classic probiotic, has obvious effects on the alleviation of UC symptoms and inflammation, and is widely used in the management of UC (25–27). However, the abundances of Bifidobacterium, Lactococcus and Lactobacillus were higher (no significant) in UC patients in remission than in healthy volunteers, which may be explained by prior intake and colonization by probiotics. Moreover, Roseburia, a butyrate-producing bacterium, significantly decreased in patients with IBD in clinical remission and increased obviously by XOS in this study. A previous study showed Roseburia had an anti-inflammatory effect on dextran sulfate sodium (DSS)-induced colitis (28) and alleviated colitis by maintaining the Treg/Th17 balance in an experimental colitis model (29). Another study demonstrated that Roseburia flagellin inhibited activation of the NLRP3 inflammasome and pyroptosis via miR-223-3p/NLRP3 signaling in macrophages (30). All these reveal the importance of Roseburia in UC development and management. Furthermore, in patients with ulcerative colitis in the central European part of Russia, there was a significant decrease in the genus of Fusicatenibacte. The current research showed that the supplementation of XOS could increase the level of Fusicatenibacter, which may be beneficial for the UC treatment (31). Additionally, the number of Lactococcus and Lactobacillus, increased under the supplementation of XOS (Figure 6). It is known that representatives of Lactococcus and Lactobacillus have therapeutic properties such as improvement of normal microbiota, prevention of infectious diseases and food allergies, modulation of innate and adaptive immune response (32). All these results indicated that XOS could promote the growth of some probiotics in the gut microbiota of UC patients in remission, which may be conducive to the alleviation of inflammation. But due to the limited sample size and lack of clinical trials, the clinical effect of XOS in UC patients still needs to be proven in the future.
In conclusion, our research indicates that this fermentation model provides a convenient in-vitro method for screening effective prebiotics for UC patients. The findings also suggest that XOS has the potential to relieve dysbiosis in UC patients in clinical remission, thus XOS may represent a potential prebiotic for UC management. The sample size of our study is small and further in-vivo studies are needed to verify the clinical effects of XOS.
Data Availability Statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA787766.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Chinese PLA General Hospital (S2016-130-01). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YY and XW: conceptualization. ZoL and ZhL: methodology, formal analysis, and writing-original draft preparation. LZ and ND: resources. GS and LP: writing-review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Ministry of Science and Technology of the People's Republic of China (2015AA020701).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.778542/full#supplementary-material
References
1. Agrawal M, Spencer EA, Colombel JF, Ungaro RC. Approach to the management of recently diagnosed inflammatory bowel disease patients: a user's guide for adult and pediatric gastroenterologists. Gastroenterology. (2021) 161:47–65. doi: 10.1053/j.gastro.2021.04.063
2. Glassner KL, Abraham BP, Quigley EMM. The microbiome and inflammatory bowel disease. J Allergy Clin Immunol. (2020) 145:16–27. doi: 10.1016/j.jaci.2019.11.003
3. De Souza HSP, Fiocchi C, Iliopoulos D. The IBD interactome: an integrated view of aetiology, pathogenesis and therapy. Nat Rev Gastroenterol Hepatol. (2017) 14:739–49. doi: 10.1038/nrgastro.2017.110
4. Derikx LA, Dieleman LA, Hoentjen F. Probiotics and prebiotics in ulcerative colitis. Best Pract Res Clin Gastroenterol. (2016) 30:55–71. doi: 10.1016/j.bpg.2016.02.005
5. Jadhav P, Jiang Y, Jarr K, Layton C, Ashouri JF, Sinha SR. Efficacy of dietary supplements in inflammatory bowel disease and related autoimmune diseases. Nutrients. (2020) 12:2156. doi: 10.3390/nu12072156
6. Casellas F, Borruel N, Torrejón A, Varela E, Antolin M, Guarner F, et al. Oral oligofructose-enriched inulin supplementation in acute ulcerative colitis is well tolerated and associated with lowered faecal calprotectin. Aliment Pharmacol Ther. (2007) 25:1061–7. doi: 10.1111/j.1365-2036.2007.03288.x
7. Furrie E, Macfarlane S, Kennedy A, Cummings JH, Walsh SV, O'neil DA, et al. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut. (2005) 54:242–9. doi: 10.1136/gut.2004.044834
8. Zhang XF, Guan XX, Tang YJ, Sun JF, Wang XK, Wang WD, et al. Clinical effects and gut microbiota changes of using probiotics, prebiotics or synbiotics in inflammatory bowel disease: a systematic review and meta-analysis. Eur J Nutr. (2021) 60:2855–75. doi: 10.1007/s00394-021-02503-5
9. Akutko K, Stawarski A. Probiotics, prebiotics and synbiotics in inflammatory bowel diseases. J Clin Med. (2021) 10:2466. doi: 10.3390/jcm10112466
10. Broekaert WF, Courtin CM, Verbeke K, Van De WT, Verstraete W, Delcour JA. Prebiotic and other health-related effects of cereal-derived arabinoxylans, arabinoxylan-oligosaccharides, and xylooligosaccharides. Crit Rev Food Sci Nutr. (2011) 51:178–94. doi: 10.1080/10408390903044768
11. Finegold SM, Li Z, Summanen PH, Downes J, Thames G, Corbett K, et al. Xylooligosaccharide increases bifidobacteria but not lactobacilli in human gut microbiota. Food Funct. (2014) 5:436–45. doi: 10.1039/c3fo60348b
12. Li Z, Summanen PH, Komoriya T, Finegold SM. In vitro study of the prebiotic xylooligosaccharide (XOS) on the growth of Bifidobacterium spp and Lactobacillus spp. Int J Food Sci Nutr. (2015) 66:919–22. doi: 10.3109/09637486.2015.1064869
13. Sheng K, He S, Sun M, Zhang G, Kong X, Wang J, et al. Synbiotic supplementation containing Bifidobacterium infantis and xylooligosaccharides alleviates dextran sulfate sodium-induced ulcerative colitis. Food Funct. (2020) 11:3964–74. doi: 10.1039/D0FO00518E
14. Payne AN, Zihler A, Chassard C, Lacroix C. Advances and perspectives in in vitro human gut fermentation modeling. Trends Biotechnol. (2012) 30:17–25. doi: 10.1016/j.tibtech.2011.06.011
15. Wu QQ, Pi XE, Liu W, Chen HH, Yin YS, Yu HWD, et al. Fermentation properties of isomaltooligosaccharides are affected by human fecal enterotypes. Anaerobe. (2017) 48:206–14. doi: 10.1016/j.anaerobe.2017.08.016
16. Chen JK, Pi XE, Liu W, Ding QF, Wang X, Jia WG, et al. Age-related changes of microbiota in midlife associated with reduced saccharolytic potential: an in vitro study. BMC Microbiol. (2021) 21:47. doi: 10.1186/s12866-021-02103-7
17. Song EJ, Lee ES, Nam YD. Progress of analytical tools and techniques for human gut microbiome research. J Microbiol. (2018), 56:693–705. doi: 10.1007/s12275-018-8238-5
18. Ooijevaar RE, Terveer EM, Verspaget HW, Kuijper EJ, Keller JJ. Clinical application and potential of fecal microbiota transplantation. Annu Rev Med. (2019) 70:335–51. doi: 10.1146/annurev-med-111717-122956
19. Wang YZ, Ren RR, Sun G, Peng LH, Tian YP, Yang YS. Pilot study of cytokine changes evaluation after fecal microbiota transplantation in patients with ulcerative colitis. Int Immunopharmacol. (2020) 85:106661. doi: 10.1016/j.intimp.2020.106661
20. Laurell A, Sjöberg K. Prebiotics and synbiotics in ulcerative colitis. Scand J Gastroenterol. (2017) 52:477–85. doi: 10.1080/00365521.2016.1263680
21. Moon JS, Li L, Bang J, Han NS. Application of in vitro gut fermentation models to food components: a review. Food Sci Biotechnol. (2016) 25:1–7. doi: 10.1007/s10068-016-0091-x
22. Hoshi N, Inoue J, Sasaki D, Sasaki K. The Kobe University Human Intestinal Microbiota Model for gut intervention studies. Appl Microbiol Biotechnol. (2021) 105:2625–32. doi: 10.1007/s00253-021-11217-x
23. Nissen L, Casciano F, Gianotti A. Intestinal fermentation in vitro models to study food-induced gut microbiota shift: an updated review. FEMS Microbiol Lett. (2020) 367:fnaa097. doi: 10.1093/femsle/fnaa097
24. Berger K, Burleigh S, Lindahl M, Bhattacharya A, Patil P, Stålbrand H, et al. Xylooligosaccharides increase bifidobacteria and lachnospiraceae in mice on a high-fat diet, with a concomitant increase in short-chain fatty acids, especially butyric acid. J Agric Food Chem. (2021) 69:3617–25. doi: 10.1021/acs.jafc.0c06279
25. Koretz RL. Probiotics in gastroenterology: how pro is the evidence in adults? Am J Gastroenterol. (2018) 113:1125–36. doi: 10.1038/s41395-018-0138-0
26. Zhang CC, Zhao Y, Jiang JC, Yu LL, Tian FW, Zhao JX, et al. Identification of the key characteristics of Bifidobacterium longum strains for the alleviation of ulcerative colitis. Food Funct. (2021) 12:3476–92. doi: 10.1039/D1FO00017A
27. Wang W, Chen LP, Zhou R, Wang XB, Song L, Huang S, et al. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol. (2014) 52:398–406. doi: 10.1128/JCM.01500-13
28. Luo WW, Shen ZH, Deng MZ, Li XY, Tan B, Xiao MW, et al. Roseburia intestinalis supernatant ameliorates colitis induced in mice by regulating the immune response. Mol Med Rep. (2019) 20:1007–16. doi: 10.3892/mmr.2019.10327
29. Shen ZH, Zhu CX, Quan YS, Yang JM, Yuan W, Yang ZY, et al. Insights into Roseburia intestinalis which alleviates experimental colitis pathology by inducing anti-inflammatory responses. J Gastroenterol Hepatol. (2018) 33:1751–60. doi: 10.1111/jgh.14144
30. Wu X, Pan SY, Luo WW, Shen ZH, Meng XR, Xiao MW, et al. Roseburia intestinalis-derived flagellin ameliorates colitisby targeting miR-223-3p-mediated activation of NLRP3 inflammasome and pyroptosis. Mol Med Rep. (2020) 22:2695–704. doi: 10.3892/mmr.2020.11351
31. Gryaznova MV, Solodskikh SA, Panevina AV, Syromyatnikov MY, Dvoretskaya YD, Sviridova TN, et al. Study of microbiome changes in patients with ulcerative colitis in the Central European part of Russia. Heliyon. (2021) 7:e06432. doi: 10.1016/j.heliyon.2021.e06432
Keywords: ulcerative colitis, xylo-oligosaccharides, in vitro fermentation, gut microbiota, 16S rDNA sequencing
Citation: Li Z, Li Z, Zhu L, Dai N, Sun G, Peng L, Wang X and Yang Y (2021) Effects of Xylo-Oligosaccharide on the Gut Microbiota of Patients With Ulcerative Colitis in Clinical Remission. Front. Nutr. 8:778542. doi: 10.3389/fnut.2021.778542
Received: 17 September 2021; Accepted: 08 December 2021;
Published: 28 December 2021.
Edited by:
Francisco José Pérez-Cano, University of Barcelona, SpainCopyright © 2021 Li, Li, Zhu, Dai, Sun, Peng, Wang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Wang, eHh3dzEwMUBzaW5hLmNvbQ==; Yunsheng Yang, c3VubnlkZGNAcGxhZ2gub3Jn
†These authors have contributed equally to this work and share first authorship
 Zongwei Li
Zongwei Li Zhengpeng Li1,2†
Zhengpeng Li1,2† Gang Sun
Gang Sun Lihua Peng
Lihua Peng Xin Wang
Xin Wang Yunsheng Yang
Yunsheng Yang