- Genomics Laboratory, Department of Biotechnology and Bioinformatics, Jaypee University of Information Technology, Solan, India
Mycobiota is not only associated with healthy homeostasis in the human gut but also helps to adapt to the environment. Food habits, alcohol consumption, intake of probiotics, and contaminated food with a mycotoxin, often lead to the alteration in the mycobiota composition. Impaired immunity of the host may affect fungal symbiosis leading to mycosis. The human gut adapts to the commensalism fungi belonging to the phylum Ascomycota and Basidiomycota. Diet habits such as plant-or animal-based, phytoestrogens enriched plant products, fat-rich diets also influence the colonization of certain fungal species in the mammalian gut. Food habits or mycotoxin-contaminated food or fungal peptides have an impact on bacterial-fungal interaction and human health. The mycobiota population such as Fusarium, Humicola, Aspergillus, and Candida are altered due to alcohol intake in alcoholic liver disease. The role of associated gut mycobiota due to irregular bowel habits or lifestyle change has been observed in inflammatory bowel disease. In this review, it has been observed that Saccharomyces, Aspergillus, Fusarium, Cladosporium, Candida, and Malassezia were the common genus in the human mycobiota. Therefore, this study focused on how diet habits and alcohol intake, among others., influence mycobiota composition that may affect the human immune system or overall health.
Introduction
The human system harbors many diverse and unculturable species such as bacteria, protozoan, viruses, and fungi, to name a few, which constitute microbiota (1, 2). These species live on and inside the human body, but in comparison with the bacteria, the diversity and abundance of fungi are relatively lower (3). Thus, fungal microbiota or are often less explored (4, 5). The association of the fungal community to human health is a well-known fact (6). The relationships between organisms within a microbial community (i.e., symbiosis) or the unbalance of microbial community composition (i.e., dysbiosis) and their role within a host is an active area of research. Dysbiosis not only permits or promotes the growth of certain fungal species, but is also associated with alteration in internal homeostasis, and influences the systemic immunity of the individuals (7). The alteration of mycobiota composition may depend on types of food intake or due to the recent rise in immune-compromised patients. In addition, fungal communities may also be beneficial in micronutrient extraction, as well as in the production of enzymes and vitamins, to aid digestion (3). Thus, understanding the relationship between host-fungi and food-fungi seems critical for good health.
Recently, there has been a focus on characterizing bacteria in the healthy individual and with disease conditions, with a limited effort on fungal microbiota due to their lower abundance and culturing complications (8). Studies on human microbiota have revealed that predominant mycobiota in the human system belongs to Ascomycota and Basidiomycota phyla with commonly observed genus from Saccharomyces, Aspergillus, Fusarium, Cladosporium, Candida, and Malassezia, among others (8, 9). Based on the recent development, this mini-review addresses the symbiosis or dysbiosis of human mycobiota associated with food habits or other factors (Figure 1) and their impact on human health.
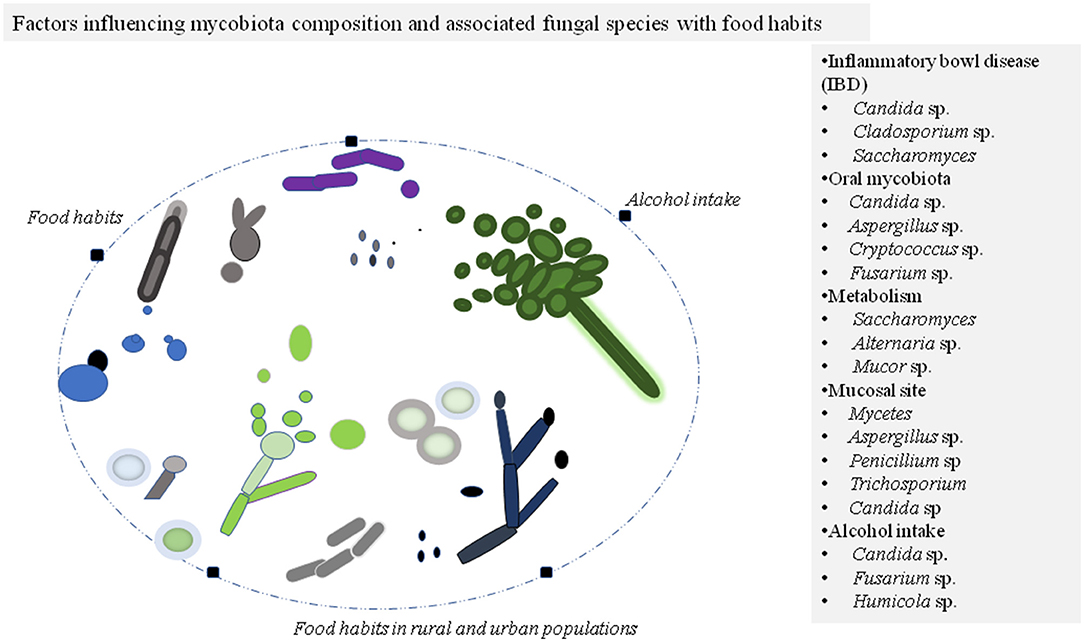
Figure 1. Factors affecting mycobiota composition of the individuals commonly belong to phylum Ascomycota and Basidiomycota. The different morphotypes of fungal species are illustrated in colors for presentation purposes. The modulated fungal species during the disease state and other conditions are listed in the panel.
Food Habits
Dietary intake/choice of food is an important factor that determines microbial composition in the human gut (10). Maintaining the balance among microbes including fungal species in the gut is critical for better gastrointestinal health (11). The human mycobiota diversity is influenced by the diet of an individual, including fermented food products, bread, as well as alcoholic beverages (12). Yeast cells are the most common microorganism for fermented food products, e.g., cheese (13), bread (14), beer (15), and others (16). Other species such as Debaryomyces hansenii, a yeast species, have been detected in salt fermented products and Penicillium roqueforti in blue cheese (17). Studies on the decreased intake of bread and beer have been shown to minimize the amount of Saccharomyces cerevisiae in human stools (18). In a controlled study, it was examined that Penicillium was correlated with a plant-based diet and Candida was enriched in animal-based diet participants (12). To decipher the relation between obesity with diets, the variation in the human gut microbiome in obese was assessed and compared with that of non-obese subjects by ITS-based sequencing (19, 20). The fungal species found in non-obese were Pichia, Candida, Aspergillus, Mucor, and Saccharomyces, S. cerevisiae being the most abundant. No specific changes in fungal species were observed in obese patients, except the absence of a minor phylum Zygomycota. In obese patients, Candida and Penicillium were abundant, while Mucor was lacking. In non-obese patients, the Mucor species suggested being associated with weight loss (19). In other studies in obese subjects, the risk of developing cardiovascular diseases and type 2 diabetes was reported (8). The fat-rich diet has affected mycobiota composition in the gut (e.g., the low abundance of S. cerevisiae) and possibly led to a shift in microbiota composition in mice (20). Furthermore, this has been found in correlation with a low abundance of S. cerevisiae in obese subjects in comparison with the gut of control subjects (19).
To understand the impact of dietary intake on the interaction among fungal and bacterial species, fecal samples from Indian and Japanese adults were subjected for sequencing (21). Indian population having plant-rich polysaccharides in their diet showed the abundance of Prevotella and Candida in comparison with the Japanese population. Therefore, vegetarian or animal-based diet or both pertaining to good health or diseased-state concerning mycobiota needs more research work (22, 23). In particular, fungal species producing toxins, such as aflatoxin, ochratoxin P, zearalenone, patulin, etc. may be carcinogenic or severely harmful to the host (24, 25). These toxins, either from consumed foods or from the resident mycobiota may lead to adverse effects including their impact on host immune response (26, 27). Thus, the mycotoxin produced from the gut mycobiota also needs investigation. These findings presented here provided a general understanding of how diets (plant- or animal-based, fat-rich, or fermented product) might influence the dynamics of gut mycobiota of the host. Thus, consumption of a balanced diet that could manifest the suitable microbial composition in the gut likely to promote good health.
Several veggies, legumes, grains, especially in soy, are structurally and/or functionally similar to mammalian estrogens, called phytoestrogen (28). It can interfere with steroid biosynthesis (29), thus modulating the free circulating endogenous hormones including estrogens. Stimulatory or inhibitory effects on the growth of the fungal communities have been observed by estrogen/17-β-estradiol (30). For instance, it has been reported the stimulation of the growth of Candida albicans by 17-β-estradiol (31). Elevated estrogen levels in the host and occurrence of Candidiasis are often associated (32). The stimulatory effect of 17-β-estradiol allows C. albicans to colonize on mucosal surfaces, including the reproductive tract (33). Overgrowth of C. albicans in the mucosal surfaces leads to Vulvovaginal candidiasis, characterized by scratching and inflammation (34). On the other hand, 17-β-estradiol showed an inhibitory effect on Paracoccidioides, a thermoregulated dimorphic fungus causing systemic mycosis (paracoccidioidomycosis) in Latin America (35). It has been observed that females are about 13–70 times less likely than males to develop the clinical disease and women are resistant to paracoccidioidomycosis (36). Furthermore, 17-β-estradiol has been shown to block/delay the morphogenesis of Paracoccidioides via stress responses (Heat shock proteins), signaling pathways through kinase nodes (37). Thus, endogenous hormone or dietary intake of phytoestrogen may have a profound effect on a few fungal species, and during the disease state, the estrogen level may need manipulation via restricted diet or supplements.
Alcohol Intake
Alcohol is often consumed along with food around the world. Owing to chronic alcohol intake, the composition of bacterial and fungal species is altered. Alcoholic liver disease (ALD) is responsible for half of all cirrhosis deaths. Chronic ethanol intake (up to 8 weeks) increased the fungal richness and diversity among mice. The mycobiota population increased significantly in particular such as Fusarium, Humicola, Aspergillus, and Candida. ALD raised the plasma level of β-glucan in mice (38). It increases the translocation of microbial products of fungal species from the intestinal lumen to the systemic circulation that causes inflammation of the liver. This leads to hepatocyte damage and facilitates the development of ethanol-induced liver disease. However, anti-fungal treatment to mice suppressed fungal overgrowth but also affected liver disease. The distribution of mycobiota was seen in patients with alcoholic hepatitis and compared with that of patients with alcohol use disorder (39). C. albicans was primarily observed in patients with alcoholic hepatitis, while normal patients were tested for Penicillium. Moreover, anti-S. cerevisiae antibodies in serum (ASCA) were measured to determine the systemic immune response to fungal products or fungi. Patients with alcoholic hepatitis had higher levels of ASCA relative to patients with alcohol consumption disorder and non-alcoholic patients. ASCA were also observed in other subjects such as IBD (40, 41) celiac diseases (42), thus antibodies against S. cerevisiae in serum could act as a marker for the inflammatory response (43). Additionally, the fungi-bacteria association has been observed to correlate positively in alcoholic hepatitis patients (44). Thus, screening of fungal species or antibodies against them could be recommended in these subjects to assess the health risk.
Food Habits in Rural and Urban Populations
The fungal composition of our gut is largely affected by the consumption of packed food such as dairy products, meat products, and frozen vegetables in urban environments vs. fresh food products in local rural environments (45). To understand the influence of the environment on fungal composition in humans, a cohort of 151 Amerindians living in remote communities (French, Guinea) was studied at 4-year intervals. They showed a rich diversity of fungi relative to people living in western cultures. People living in industrial areas have C. albicans as a predominant species in their intestines (11). Furthermore, Candida krusei and S. cerevisiae were found to be abundant in remote communities. C. albicans were more common in females and crowded areas. C. krusei and S. cerevisiae were known to be associated with foodborne by plants and water. It was proposed that there may be cross-transmission of the strains of C. albicans between humans and animals (11). A wide diversity of fungi has been found in the human gastrointestinal (GI) tract and detected in stool samples. Fungi found in stools are highly affected by food or the oral cavity. S. cerevisiae-free diet renders this strain undetectable in stools. The level of C. albicans in the stool has correlated with the cleaning of teeth (18). The study of Sun et al. (46) also studied the fecal mycobiome using metagenomic sequencing to associate fungal communities in the rural and urban populations of China. In brief, S. cerevisiae was more abundant while Candida dubliniensis was low in abundance in urban compared with rural population suggesting that distinct food habits in the shaping of gut mycobiota composition. The enrichment of fungal species in the gut of these populations may modulate the metabolism of the host to confer good health need more such studies.
Impact of Mycobiota Composition on Human Health
The mycobiota is a part of human microbiota and plays important role in regulating innate and adaptive immune homeostasis (47–49). While the mycobiota composition of the host may have an independent effect on the microbial environment of the intestine and immune development (50), the balance of microbial communities in providing immunity to protect from invading pathogens to the immune dysregulation is critical for health benefits to human (47, 51). The dynamics of fungal species, such as Malassezia, Candida, Aspergillus, Cladosporium, Saccharomyces, and Penicillium, among others, are reported in various health conditions (Table 1). Therefore, mycobiota associated with gastrointestinal diseases were discussed here.
Gut Mycobiota and Associated Diseases
The chronic and excessive intestinal inflammation due to the resident or foreign antigens causes inflammatory bowel disease (IBD) (67). It is categorized into two main clinical conditions, namely Crohn's disease (CD) and ulcerative colitis (UC), which are due to the activation of the immune response against certain microbiota in the gut influenced by environmental factors (7, 68). It is characterized by inflammation of the gastrointestinal tract, altered bowel habits, and abdominal pain. The fungal diversity of the gastrointestinal tract is contributed by many species, including Penicillium, Candida, Aspergillus, Saccharomyces, Cryptococcus, and Malassezia (57). In pediatric subjects, stool samples were collected from IBD patients and healthy subjects and were characterized by deep sequencing of rRNA gene segments specific to the fungal domain. Pichia jidanii, Candida species, and Torula yeast were abundant in IBD patients. The healthy subjects showed Clostridium cladosporioides dominance (56). In IBD patients, particularly CD was screened with antibodies against S. cerevisiae, suggesting an inappropriate immune response to this fungus (57). In addition, the study of Jain et al. (69) showed the colonization of D. hansenii, a yeast species, at intestinal wounds of mice and inflamed mucosal tissue of CD human subjects, and mucosal healing dysfunction. Thus, inflammation in the gut either because of yeast colonization and/or immune suppressive therapy needs more studies. Genetic variation such as polymorphism in CLEC7A, a gene encoding for Dectin-1, can induce UC in mammals due to a weak immune response against fungi (55). Dectin-1, a pattern recognition receptor for β-glucan, is involved in adaptive immune response via T-helper cells, and the polymorphism in the receptor makes the host susceptible to invasive infections (70). The work of Tang et al. (71) used Dectin-1 deficient mice to demonstrate that Dectin-1 regulates regulatory T (Treg) cell differentiation under the influence of microbiota composition, implying the role of Dectin-1 in intestinal immunity. Chitooligosaccharide (COS), a derivative of chitosan, possesses antimicrobial activity. The alteration in the diversity of mycobiota was observed in colorectal cancer (CRC) patients before and after COS induction. In addition, it has been associated with decreased symptoms of colitis-induced CRC (72). The data showed that the abundance of two phyla in intestinal mycobiota: Ascomycota and Basidiomycota (73).
Fungal species such as Candida and Aspergillus release mycotoxins into non-neuronal tissues that pass through the bloodstream. These toxins target astrocytes and oligodendrocytes, as a result of which the blood-brain barrier is weakened followed by degradation of myelin (74). The above studies showed mycobiota plays a critical in modulating the metabolism of host and intestinal immunity. It was suggestive that the intestinal environment could be managed by modulating the mycobiota composition to achieve a healthy gut environment.
Mycobiota and Impact on Host Immune System
Fungi are also part of oral microbial communities along with bacterial species and these microbes enter to human gut through ingested foodstuff (75). The fungal mycobiome has been explored in the oral cavity from healthy individuals and reported a total of 101 species, 74 cultivable, and 11 non-cultivable (58). Candida was followed by Cladosporium, Aurobasidium, Saccharomyces, Aspergillus, Fusarium, and Cryptococcus, among the most common species. Four of these species, namely Candida, Aspergillus, Fusarium, and Cryptococcus, are pathogenic to immunocompromised humans when present in abundance (76–79). In the oral microenvironment, C. albicans associates with Streptococcus species to promote bacterial colonization (80), and synergistic interaction between fungal-bacterial species contribute to the development of multi-microbial biofilm (81–83). Thus, interspecies interaction and microbial biofilm formation in the oral cavity surfaces need more such studies, consequently, enable better strategies to treat oral diseases. In rural areas or unhygienic environments, human beings are often exposed to fungal spores that disturb the mycobiota composition under healthy conditions (84). In adverse conditions, inhaled fungal spores cause severe health deterioration (7). In another study, the mouth and lung fungal microbiota was compared between healthy individuals and recipients of lung transplants (60). Candida species were found in the oral wash of lung transplant recipients because of antibiotic therapy and immunosuppressant. There was limited fungal ITS amplification in the bronchoalveolar lavage of healthy individuals, whereas detectable fungi of Candida, Aspergillus, or Cryptococcus were present in lung transplant recipients (59). Thus, balanced oral microbial communities are required to be a healthy oral system. If microorganisms are altered may enhance the risk of oral diseases.
The human immunity system is affected by microbiota composition, and secondary metabolites and/or peptides produced by them (85, 86). To understand the role of the fungal component in modulating immunity, when the Influenza A virus was infected in antibiotic-treated mice, they were more vulnerable to colitis-induced dextran sodium sulfate (DSS) and displayed decreased CD8+ T cells. The protective immunity resumes adequately when commensal fungal species, such as S. cerevisiae or C. albicans was administered. It was concluded that the immune response may have been produced by mannan, an abundant component of the fungal cell wall. It was therefore noted that the function of the immunocyte was dependent on the composition of the mycobiota in the organism (87). Mucosal and systemic fungal infections cause CD4+ T cells to respond to induce Th17 or Th1 cells (47). During the dysbiosis of gut microbiota, the CD4+ T cells dysfunction has been observed in childhood atopic subjects (88). C. albicans and Aspergillus fumigatus are pathogenic fungi and elicit antigen-presenting cells to produce effector T-helper cells against these microorganisms (47, 89–91). On the other hand, S. cerevisiae, a non-pathogenic yeast, and A. fumigatus were observed inducing both the Th1 and Th17 subsets of CD4+ T cells (92, 93). Thus, pathogenic or non-pathogenic fungal spores can promote T-cell response. However, systemic antibodies in humans against major inducers such as C. albicans showed protection against disseminated C. albicans or Candida auris (94). Furthermore, the production of CARD9-dependent antibodies repertoire in shaping host immunity implicated the role of fungal communities in the human gut (94). Therefore, certain mycobiota composition, their interactions, and/or fungal cell wall components in the microbial environment allow individuals to may remain healthy. On the other hand, dysbiosis or the alteration of mycobiota structure and/or function may occur as a consequence of infection, e.g., COVID-19. The study of Zuo et al. (95) observed a significant change in the fecal mycobiota in COVID-19 patients in comparison with control subjects. These patients showed an increased abundance of opportunistic fungal pathogens, such as C. albicans, C. auris, Aspergillus flavus, and Aspergillus niger, in their fecal samples. Thus, impaired immunity during the infection and delay in recovery of immunity allows opportunistic pathogens to colonize in the host. Additionally, patients recovered from the COVID-19 infection but probably not their immune system since there were still shown infections with black fungi/mucormycosis with high mortality (96). Therefore, microbial symbiosis or balance of bacteria-fungi-virus communities in the host in shaping innate or acquired immunity to the host presents future research opportunities.
Conclusion and Perspectives
Microbial micro-environment in the human system and its physiological diversity, or the factors that influence its colonization inside the body, is still at a primitive stage. The dynamics of fungal species belonging to Malassezia, Candida, Aspergillus, Cladosporium, Saccharomyces, and Penicillium, among others, were among the most common genera associated with symbiosis or dysbiosis in humans. Morphological and metabolic changes in these species are essential for survival within hosts and need better understanding. The bacterial predominance in the oral cavity or intestines to achieve optimal health and the role of fungi in gastro-intestinal ecology that fungi influence bacterial activities through various interactions needs thorough investigation. Moreover, exploration of genes encoding proteins produced by fungal species metabolizing certain substrates could result in food recommendations. Furthermore, the identification of coordinated expression of protein from the resident microbes in the gut microenvironment may assist our understanding of the relationship between bacteria-fungi or host-microbe. In vitro or in vivo experiments should be performed to identify human microbial communities for a normal healthy condition and those associated with the disease. In particular, it is possible to examine the microbiome composition of individuals with neurological disorders to assess their diet and lifestyle, which promote healthy brain activities and actions. Overall, a sound understanding of the biological properties of the mycobiota could increase our quality of life in coordination with diet, lifestyle, and climate.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
I am thankful to the Department of Biotechnology and Bioinformatics, Jaypee University of Information Technology, Solan, HP for providing facilities.
References
1. Backhed F, Fraser CM, Ringel Y, Sanders ME, Sartor RB, Sherman PM, et al. Defining a healthy human gut microbiome: current concepts, future directions, clinical applications. Cell Host Microbe. (2012) 12:611–22. doi: 10.1016/j.chom.2012.10.012
2. Hmpc, H. M. P. C. Structure, function and diversity of the healthy human microbiome. Nature. (2012) 486:207–14. doi: 10.1038/nature11234
3. Huffnagle GB, Noverr MC. The emerging world of the fungal microbiome. Trends Microbiol. (2013) 21:334–41. doi: 10.1016/j.tim.2013.04.002
4. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. (2010) 464:59–65. doi: 10.1038/nature08821
5. Lynch MD, Neufeld JD. Ecology and exploration of the rare biosphere. Nat Rev Microbiol. (2015) 13:217–29. doi: 10.1038/nrmicro3400
6. Wampach L, Heintz-Buschart A, Hogan A, Muller EEL, Narayanasamy S, Laczny CC, et al. Colonization and succession within the human gut microbiome by archaea, bacteria, and microeukaryotes during the first year of life. Front Microbiol. (2017) 8:738. doi: 10.3389/fmicb.2017.00738
7. Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. (2014) 14:405–416. doi: 10.1038/nri3684
8. Lai GC, Tan TG, Pavelka N. The mammalian mycobiome: A complex system in a dynamic relationship with the host. Wiley Interdiscip Rev Syst Biol Med. (2019) 11:e1438. doi: 10.1002/wsbm.1438
9. Wheeler ML, Limon JJ, Underhill DM. Immunity to commensal fungi: detente and disease. Annu Rev Pathol. (2017) 12:359–85. doi: 10.1146/annurev-pathol-052016-100342
10. Graf D, Di Cagno R, Fak F, Flint HJ, Nyman M, Saarela M, et al. Contribution of diet to the composition of the human gut microbiota. Microb Ecol Health Dis. (2015) 26:26164. doi: 10.3402/mehd.v26.26164
11. Angebault C, Djossou F, Abelanet S, Permal E, Ben Soltana M, Diancourt L, et al. Candida albicans is not always the preferential yeast colonizing humans: a study in Wayampi Amerindians. J Infect Dis. (2013) 208:1705–16. doi: 10.1093/infdis/jit389
12. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. (2014) 505:559–63. doi: 10.1038/nature12820
13. Frohlich-Wyder MT, Arias-Roth E, Jakob E. Cheese yeasts. Yeast. (2019) 36:129–41. doi: 10.1002/yea.3368
14. Heitmann M, Zannini E, Arendt E. Impact of Saccharomyces cerevisiae metabolites produced during fermentation on bread quality parameters: a review. Crit Rev Food Sci Nutr. (2018) 58:1152–64. doi: 10.1080/10408398.2016.1244153
15. Alba-Lois L, Segal-Kischinevzky C. Yeast fermentation and the making of beer and wine. Nat Educ. (2010) 3:17
16. Nielsen J. Yeast systems biology: model organism and cell factory. Biotechnol J. (2019) 14:e1800421. doi: 10.1002/biot.201800421
17. Hallen-Adams HE, Kachman SD, Kim J, Legge RM, Martínez I. Fungi inhabiting the healthy human gastrointestinal tract: a diverse and dynamic community. Fungal Ecol. (2015) 15:9–17. doi: 10.1016/j.funeco.2015.01.006
18. Auchtung TA, Fofanova TY, Stewart CJ, Nash AK, Wong MC, Gesell JR, et al. Investigating colonization of the healthy adult gastrointestinal tract by fungi. mSphere. (2018) 3:e00092-18. doi: 10.1128/mSphere.00092-18
19. Mar Rodriguez M, Perez D, Javier Chaves F, Esteve E, Marin-Garcia P, Xifra G, et al. Obesity changes the human gut mycobiome. Sci Rep. (2015) 5:14600. doi: 10.1038/srep14600
20. Heisel T, Montassier E, Johnson A, Al-Ghalith G, Lin YW, Wei LN, et al. High-fat diet changes fungal microbiomes and interkingdom relationships in the murine gut. mSphere. (2017) 2:e00351-17. doi: 10.1128/mSphere.00351-17
21. Pareek S, Kurakawa T, Das B, Motooka D, Nakaya S, Rongsen-Chandola T, et al. Comparison of Japanese and Indian intestinal microbiota shows diet-dependent interaction between bacteria and fungi. NPJ Biofilms Microbiomes. (2019) 5:37. doi: 10.1038/s41522-019-0110-9
22. Hoffman SR, Stallings SF, Bessinger RC, Brooks GT. Differences between health and ethical vegetarians. Strength of conviction, nutrition knowledge, dietary restriction, and duration of adherence. Appetite. (2013) 65:139–44. doi: 10.1016/j.appet.2013.02.009
23. Suhr MJ, Hallen-Adams HE. The human gut mycobiome: pitfalls and potentials–a mycologist's perspective. Mycologia. (2015) 107:1057–73. doi: 10.3852/15-147
24. Shankar J, Tiwari S, Shishodia SK, Gangwar M, Hoda S, Thakur R, et al. Molecular insights into development and virulence determinants of aspergilli: a proteomic perspective. Front Cell Infect Microbiol. (2018) 8:180. doi: 10.3389/fcimb.2018.00180
25. Brand B, Stoye NM, Guilherme MDS, Nguyen VTT, Baumgaertner JC, Schuffler A, et al. Identification of patulin from penicillium coprobium as a toxin for enteric neurons. Molecules. (2019) 24:2776. doi: 10.3390/molecules24152776
26. Shankar J. An overview of toxins in Aspergillus associated with pathogenesis. Int. J. LifeSc. Bt and Pharm. Res. (2013) 2:16–31
27. Adeyeye SAO. Fungal mycotoxins in foods: a review. Cogent Food Agric. (2016) 2:1213127. doi: 10.1080/23311932.2016.1213127
28. Patisaul HB, Jefferson W. The pros and cons of phytoestrogens. Front Neuroendocrinol. (2010) 31:400–19. doi: 10.1016/j.yfrne.2010.03.003
29. Adlercreutz H, Mousavi Y, Clark J, Hockerstedt K, Hamalainen E, Wahala K, et al. Dietary phytoestrogens and cancer: in vitro and in vivo studies. J Steroid Biochem Mol Biol. (1992) 41:331–7. doi: 10.1016/0960-0760(92)90359-Q
30. Clemons KV, Shankar J, Stevens DA. Mycologic endocrinology. Adv Exp Med Biol. (2016) 874:337–63. doi: 10.1007/978-3-319-20215-0_16
31. Cheng G, Yeater KM, Hoyer LL. Cellular and molecular biology of Candida albicans estrogen response. Eukaryot Cell. (2006) 5:180–91. doi: 10.1128/EC.5.1.180-191.2006
32. Cotch MF, Hillier SL, Gibbs RS, Eschenbach DA. Epidemiology and outcomes associated with moderate to heavy Candida colonization during pregnancy. Vaginal Infections and Prematurity Study Group. Am J Obstet Gynecol. (1998) 178:374–80. doi: 10.1016/S0002-9378(98)80028-8
33. Sobel JD, Faro S, Force RW, Foxman B, Ledger WJ, Nyirjesy PR, et al. Vulvovaginal candidiasis: epidemiologic, diagnostic, therapeutic considerations. Am J Obstet Gynecol. (1998) 178:203–11. doi: 10.1016/S0002-9378(98)80001-X
34. Bradford LL, Ravel J. The vaginal mycobiome: a contemporary perspective on fungi in women's health and diseases. Virulence. (2017) 8:342–51. doi: 10.1080/21505594.2016.1237332
35. Restrepo A. The ecology of Paracoccidioides brasiliensis: a puzzle still unsolved. Sabouraudia. (1985) 23:323–34. doi: 10.1080/00362178585380481
36. Shankar J, Restrepo A, Clemons KV, Stevens DA. Hormones and the resistance of women to paracoccidioidomycosis. Clin Microbiol Rev. (2011) 24:296–313. doi: 10.1128/CMR.00062-10
37. Shankar J, Wu TD, Clemons KV, Monteiro JP, Mirels LF, Stevens DA. Influence of 17beta-estradiol on gene expression of Paracoccidioides during mycelia-to-yeast transition. PLoS One. (2011) 6:e28402. doi: 10.1371/journal.pone.0028402
38. Yang AM, Inamine T, Hochrath K, Chen P, Wang L, Llorente C, et al. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Invest. (2017) 127:2829–1. doi: 10.1172/JCI90562
39. Lang S, Duan Y, Liu J, Torralba MG, Kuelbs C, Ventura-Cots M, et al. Intestinal fungal dysbiosis and systemic immune response to fungi in patients with alcoholic hepatitis. Hepatology. (2020) 71:522–38. doi: 10.1002/hep.30832
40. Vermeire S, Peeters M, Vlietinck R, Joossens S, Den Hond E, Bulteel V, et al. Anti-Saccharomyces cerevisiae antibodies (ASCA), phenotypes of IBD, and intestinal permeability: a study in IBD families. Inflamm Bowel Dis. (2001) 7:8–15. doi: 10.1097/00054725-200102000-00002
41. Israeli E, Grotto I, Gilburd B, Balicer RD, Goldin E, Wiik A, et al. Anti-Saccharomyces cerevisiae and antineutrophil cytoplasmic antibodies as predictors of inflammatory bowel disease. Gut. (2005) 54:1232–6. doi: 10.1136/gut.2004.060228
42. Granito A, Muratori L, Muratori P, Guidi M, Lenzi M, Bianchi FB, et al. Anti-saccharomyces cerevisiae antibodies (ASCA) in coeliac disease. Gut. (2006) 55:296.
43. Seibold F. ASCA: genetic marker, predictor of disease, or marker of a response to an environmental antigen? Gut. (2005) 54:1212–3. doi: 10.1136/gut.2005.065227
44. Gao B, Zhang X, Schnabl B. Fungi-bacteria correlation in alcoholic hepatitis patients. Toxins (Basel). (2021) 13:143. doi: 10.3390/toxins13020143
45. Strati F, Di Paola M, Stefanini I, Albanese D, Rizzetto L, Lionetti P, et al. Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front Microbiol. (2016) 7:1227. doi: 10.3389/fmicb.2016.01227
46. Sun Y, Zuo T, Cheung CP, Gu W, Wan Y, Zhang F, et al. Population-level configurations of gut mycobiome across 6 ethnicities in urban and rural China. Gastroenterology. (2021) 160:272–86.e11. doi: 10.1053/j.gastro.2020.09.014
47. Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. (2012) 3:4–14. doi: 10.4161/gmic.19320
48. Iliev ID, Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat Rev Immunol. (2017) 17:635–46. doi: 10.1038/nri.2017.55
49. Li XV, Leonardi I, Iliev ID. Gut mycobiota in immunity and inflammatory disease. Immunity. (2019) 50:1365–79. doi: 10.1016/j.immuni.2019.05.023
50. Van Tilburg Bernardes E, Pettersen VK, Gutierrez MW, Laforest-Lapointe I, Jendzjowsky NG, Cavin JB, et al. Intestinal fungi are causally implicated in microbiome assembly and immune development in mice. Nat Commun. (2020) 11:2577. doi: 10.1038/s41467-020-16431-1
51. Perez JC. Fungi of the human gut microbiota: roles and significance. Int J Med Microbiol. (2021) 311:151490. doi: 10.1016/j.ijmm.2021.151490
52. Ma X, Li C, Hou J, Gu Y. Isolation and identification of culturable fungi from the genitals and semen of healthy giant pandas (Ailuropoda melanoleuca). BMC Vet Res. (2017) 13:344. doi: 10.1186/s12917-017-1231-0
53. Yano J, Noverr MC, Fidel PL Jr. Cytokines in the host response to Candida vaginitis: identifying a role for non-classical immune mediators, S100 alarmins. Cytokine. (2012) 58:118–28. doi: 10.1016/j.cyto.2011.11.021
54. Kalia N, Singh J, Kaur M. Microbiota in vaginal health and pathogenesis of recurrent vulvovaginal infections: a critical review. Ann Clin Microbiol Antimicrob. (2020) 19:5. doi: 10.1186/s12941-020-0347-4
55. Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. (2012) 336:1314–7. doi: 10.1126/science.1221789
56. Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, et al. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. (2015) 21:1948–56. doi: 10.1097/MIB.0000000000000454
57. Richard ML, Lamas B, Liguori G, Hoffmann TW, Sokol H. Gut fungal microbiota: the Yin and Yang of inflammatory bowel disease. Inflamm Bowel Dis. (2015) 21:656–65. doi: 10.1097/MIB.0000000000000261
58. Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. (2010) 6:e1000713. doi: 10.1371/journal.ppat.1000713
59. Chabe M, Aliouat-Denis CM, Delhaes L, Aliouat El M, Viscogliosi E, Dei-Cas E. Pneumocystis: from a doubtful unique entity to a group of highly diversified fungal species. FEMS Yeast Res. (2011) 11:2–17. doi: 10.1111/j.1567-1364.2010.00698.x
60. Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. (2012) 186:536–45. doi: 10.1164/rccm.201204-0693OC
61. Black PN, Udy AA, Brodie SM. Sensitivity to fungal allergens is a risk factor for life-threatening asthma. Allergy. (2000) 55:501–4. doi: 10.1034/j.1398-9995.2000.00293.x
62. Tillie-Leblond I, Tonnel AB. Allergic bronchopulmonary aspergillosis. Allergy. (2005) 60:1004–13. doi: 10.1111/j.1398-9995.2005.00887.x
63. Bond SL, Workentine M, Hundt J, Gilkerson JR, Leguillette R. Effects of nebulized dexamethasone on the respiratory microbiota and mycobiota and relative equine herpesvirus-1, 2, 4, 5 in an equine model of asthma. J Vet Intern Med. (2020) 34:307–21. doi: 10.1111/jvim.15671
64. Manning SC, Holman M. Further evidence for allergic pathophysiology in allergic fungal sinusitis. Laryngoscope. (1998) 108:1485–96. doi: 10.1097/00005537-199810000-00012
65. Montone KT, Livolsi VA, Feldman MD, Palmer J, Chiu AG, Lanza DC, et al. Fungal rhinosinusitis: a retrospective microbiologic and pathologic review of 400 patients at a single University medical center. Int J Otolaryngol. (2012) 2012:684835. doi: 10.1155/2012/684835
66. Woolnough KF, Richardson M, Newby C, Craner M, Bourne M, Monteiro W, et al. The relationship between biomarkers of fungal allergy and lung damage in asthma. Clin Exp Allergy. (2017) 47:48–56. doi: 10.1111/cea.12848
67. Lee SH, Kwon JE, Cho ML. Immunological pathogenesis of inflammatory bowel disease. Intest Res. (2018) 16:26–42. doi: 10.5217/ir.2018.16.1.26
68. Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, et al. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe. (2016) 19:865–73. doi: 10.1016/j.chom.2016.05.003
69. Jain U, Ver Heul AM, Xiong S, Gregory MH, Demers EG, Kern JT, et al. Debaryomyces is enriched in Crohn's disease intestinal tissue and impairs healing in mice. Science. (2021) 371:1154–9. doi: 10.1126/science.abd0919
70. Thakur R, Shankar J. Comprehensive in-silico analysis of high-risk non-synonymous SNPs in dectin-1 gene of human and their impact on protein structure. Curr Pharmacogenomics Person Med. (2017) 15:144–55. doi: 10.2174/1875692116666180115142706
71. Tang C, Kamiya T, Liu Y, Kadoki M, Kakuta S, Oshima K, et al. Inhibition of dectin-1 signaling ameliorates colitis by inducing lactobacillus-mediated regulatory T cell expansion in the intestine. Cell Host Microbe. (2015) 18:183–97. doi: 10.1016/j.chom.2015.07.003
72. Wu M, Li J, An Y, Li P, Xiong W, Yan D, et al. Chitooligosaccharides prevents the development of colitis-associated colorectal cancer by modulating the intestinal microbiota and mycobiota. Front Microbiol. (2019) 10:2101. doi: 10.3389/fmicb.2019.02101
73. Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. (2017) 152:327–39.e24. doi: 10.1053/j.gastro.2016.10.012
74. Daneman R, Prat A. The blood-brain barrier. Cold Spring Harb Perspect Biol. (2015) 7:a020412. doi: 10.1101/cshperspect.a020412
75. Marsh PD, Zaura E. Dental biofilm: ecological interactions in health and disease. J Clin Periodontol. (2017) 44 (Suppl. 18):S12–S22. doi: 10.1111/jcpe.12679
77. Shankar J, Nigam S, Saxena S, Madan T, Sarma PU. Identification and assignment of function to the genes of Aspergillus fumigatus expressed at 37 degrees C. J Eukaryot Microbiol. (2004) 51:428–32. doi: 10.1111/j.1550-7408.2004.tb00390.x
78. Dinardo AR, Schmidt D, Mitchell A, Kaufman Y, Tweardy DJ. First description of oral cryptococcus neoformans causing osteomyelitis of the mandible, manubrium and third rib with associated soft tissue abscesses in an immunocompetent host. Clin Microbiol Case Rep. (2015) 1:017.
79. Mutalik VS, Bissonnette C, Kalmar JR, Mcnamara KK. Unique oral presentations of deep fungal infections: a report of four cases. Head Neck Pathol. (2020) 15:682–90. doi: 10.1007/s12105-020-01217-0
80. Diaz PI, Xie Z, Sobue T, Thompson A, Biyikoglu B, Ricker A, et al. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun. (2012) 80:620–32. doi: 10.1128/IAI.05896-11
81. Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. (2007) 71:653–70. doi: 10.1128/MMBR.00024-07
82. Bamford CV, D'mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun. (2009) 77:3696–704. doi: 10.1128/IAI.00438-09
83. Koo H, Andes DR, Krysan DJ. Candida-streptococcal interactions in biofilm-associated oral diseases. PLoS Pathog. (2018) 14:e1007342. doi: 10.1371/journal.ppat.1007342
84. Baxi SN, Portnoy JM, Larenas-Linnemann D, Phipatanakul W. Exposure and health effects of fungi on humans. J Allergy Clin Immunol Pract. (2016) 4:396–404. doi: 10.1016/j.jaip.2016.01.008
85. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. (2016) 16:341–52. doi: 10.1038/nri.2016.42
86. Man AWC, Zhou Y, Xia N, Li H. Involvement of gut microbiota, microbial metabolites and interaction with polyphenol in host immunometabolism. Nutrients. (2020) 12:3054. doi: 10.3390/nu12103054
87. Jiang TT, Shao TY, Ang WXG, Kinder JM, Turner LH, Pham G, et al. Commensal fungi recapitulate the protective benefits of intestinal bacteria. Cell Host Microbe. (2017) 22:809–16.e04. doi: 10.1016/j.chom.2017.10.013
88. Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. (2016) 22:1187–91. doi: 10.1038/nm.4176
89. Chai LY, Van De Veerdonk F, Marijnissen RJ, Cheng SC, Khoo AL, Hectors M, et al. Anti-Aspergillus human host defence relies on type 1 T helper (Th1), rather than type 17 T helper (Th17), cellular immunity. Immunology. (2010) 130:46–54. doi: 10.1111/j.1365-2567.2009.03211.x
90. Thakur R, Anand R, Tiwari S, Singh AP, Tiwary BN, Shankar J. Cytokines induce effector T-helper cells during invasive aspergillosis; what we have learned about T-helper cells? Front Microbiol. (2015) 6:429. doi: 10.3389/fmicb.2015.00429
91. Bacher P, Hohnstein T, Beerbaum E, Rocker M, Blango MG, Kaufmann S, et al. Human anti-fungal Th17 immunity and pathology rely on cross-reactivity against Candida albicans. Cell. (2019) 176:1340–55.e15. doi: 10.1016/j.cell.2019.01.041
92. Rizzetto L, Buschow SI, Beltrame L, Figdor CG, Schierer S, Schuler G, et al. The modular nature of dendritic cell responses to commensal and pathogenic fungi. PLoS One. (2012) 7:e42430. doi: 10.1371/journal.pone.0042430
93. Shankar J, Cerqueira GC, Wortman JR, Clemons KV, Stevens DA. RNA-seq profile reveals Th-1 and Th-17-type of immune responses in mice infected systemically with aspergillus fumigatus. Mycopathologia. (2018) 183:645–58. doi: 10.1007/s11046-018-0254-9
94. Doron I, Leonardi I, Li XV, Fiers WD, Semon A, Bialt-Decelie M, et al. Human gut mycobiota tune immunity via CARD9-dependent induction of anti-fungal IgG antibodies. Cell. (2021) 184:1017–31.e14. doi: 10.1016/j.cell.2021.01.016
95. Zuo T, Zhan H, Zhang F, Liu Q, Tso EYK, Lui GCY, et al. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastroenterology. (2020) 159:1302–10.e5. doi: 10.1053/j.gastro.2020.06.048
Keywords: mycotoxin, phytoestrogens, alcohol intake, gut mycobiome, diet habit, COVID-19, mucormycosis
Citation: Shankar J (2021) Food Habit Associated Mycobiota Composition and Their Impact on Human Health. Front. Nutr. 8:773577. doi: 10.3389/fnut.2021.773577
Received: 10 September 2021; Accepted: 19 October 2021;
Published: 22 November 2021.
Edited by:
Changhong Liu, Hefei University of Technology, ChinaReviewed by:
Zijian Zhang, Baylor College of Medicine, United StatesLikui Feng, The Rockefeller University, United States
Copyright © 2021 Shankar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jata Shankar, amF0YV9zQHlhaG9vLmNvbQ==; amF0YS5zaGFua2FyQGp1aXQuYWMuaW4=; orcid.org/0000-0003-4993-9580
 Jata Shankar
Jata Shankar