
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr. , 16 November 2021
Sec. Nutrition and Microbes
Volume 8 - 2021 | https://doi.org/10.3389/fnut.2021.760316
This article is part of the Research Topic Interactions Between Natural Polysaccharides and Gut Microbiota View all 11 articles
The purpose of the study was to assess the artificial saliva (AS) pH on ruminal fermentation and rumen bacteria community in the rumen simulation technique (RUSITEC) system. The experiment was performed in two treatments (low AS pH vs. high AS pH) with four replicates. The low AS pH was sustained by altering the composition of the AS (NaHCO3 from 9.8 to 1.96 g/L, Na2HPO4 from 9.3 to 1.86 g/L) according to McDougall's method. The diets were supplemented with 16 g basic diets with forage to the concentrate ratio of 50:50. The experiments were conducted over 13-day incubation periods, with 9 days adaption and 4 days sample collection. The results showed low AS pH decreased dry matter (DM) degradability (64.37 vs. 58.67%), organic matter (OM) degradability (64.38 vs. 59.32%), neutral detergent fiber (NDF) degradability (46.87 vs. 39.94%), acid detergent fiber (ADF) degradability (38.16 vs. 31.13%), and crude protein (CP) degradability (70.33 vs. 62.99%), respectively. Compared with the high AS pH, the low AS pH increased the proportion of butyrate (P = 0.008) and decreased the proportion of propionate (P < 0.001). At the bacteria community, the low AS pH increased the abundances of Spirochaetes (P = 0.001) and Synergistetes (P = 0.004) and decreased the Verrucomicrobia abundance (P = 0.004) in solid-associated bacteria. At the genus level, the low AS pH increased the abundance of Lactobacillus (P = 0.050) and decreased the abundance of Schwartzia (P = 0.002) in solid-associated bacteria. The abundances of Prevotellaceae_YAB2003_group (P = 0.040), Schwartzia (P = 0.002), and Ruminobacter (P = 0.043) were lower in the low AS pH group compared with the high AS pH group in liquid-associated bacteria. Low AS pH decreased the number of Ruminococcus albus, Ruminococcus flavefaciens, Fibrobacter succinogenes (P < 0.001) both in the solid- and liquid-associated bacteria, respectively. The results of the present study included three groups of bacteria communities according to the different sensitives to rumen pH: the abundances of Lactobacillus, Succinivibrio, and Prevotella_7 are increased with decreasing AS pH; the amounts of R. albus, R. flavefaciens, F. succinogenes as well as the abundances of Schwartzia and Ruminobacter decreased with the reducing AS pH; the abundances of Selenomonas_1, Rikenellaceae_RC9_gut_group, and Succiniclasticum were not affected by the AS pH in RUSTITEC.
A high grain-based diet has been a common strategy to improve animal performance in ruminant production. However, the fermentable carbohydrate diets can lead to the accumulation of organic acids in the rumen, which results in the reduction of ruminal pH, and increases the risk of subacute rumen acidosis (SARA) (1, 2). SARA was described as the daily average rumen pH between 6.25 and 5.5 (3). The main SARA model was obtained by increasing the dietary proportions of grain or decreasing physically effective fiber (peNDF) content (4, 5). The SARA induction approaches have a different impact on the rumen fermentation and bacterial community because of the different substrates (6). The low dietary peNDF induced SARA usually increased the feed intake of dairy cows (7), and the increased feed intake resulted in increasing the production of volatile fatty acids (VFA) and decreased pH (8). Therefore, the low peNDF induced SARA needs to avoid the impact of different feed intake between the treatments. Decreasing the peNDF intake for ruminants could reduce the chewing time and the amount of saliva secretion (9). The in vitro SARA model that induced in the rumen simulation technique (RUSITEC) system usually by decreasing the buffer capacity of artificial saliva (AS) (10, 11), which could simulate the low peNDF induced SARA. In addition, the RUSITEC system was designed to ensure the identical substrate intake and rumen passage rate during the fermentation that avoids the disturbance of different feed intake and rumen content passage rate of in vivo when the ruminants received different dietary peNDF. Orton et al. (10) decreased the buffer capacity of AS (NaCl from 28 up to 118.5 mmol/L and NaCO3 from 97.0 to 20 mmol/L) decreased pH from 7.0 to 6.0 in the RUSITEC system.
The ruminal pH plays an important role in affecting the communities of rumen bacteria. Li et al. (8) found the low-peNDF diet induced SARA increased the numbers of Fibrobacter succinogenes and Ruminococcus flavefaciens for the dairy goats. The increased feed intake and cellulolytic bacteria were due to the more substrates or particulate surfaces available for these bacteria attachment and proliferation (12). The solid-associated bacteria attached to the feed particles play a key role in fiber digestion, while the liquid-associated bacteria have significant functions in the metabolism of soluble nutrients (13, 14). There is a difference in the bacteria community between the solid and liquid fractions. The rumen bacteria are influenced by the combination of substrate, physical structure, and pH environment. Li et al. (15) demonstrated that the three groups of bacteria communities change under grain-induced SARA: pH-sensitive but substrate insensitive bacteria, pH-insensitive but substrate sensitive bacteria, and bacteria that are both pH and substrate sensitive. However, it is difficult to design and execute experiments in vivo to test this hypothesis. The RUSITEC system is an optional tool in vitro model to simulate the rumen microbial fermentation and could strictly control the effects of substrate and pH independently (10).
Therefore, we hypothesized that the low AS pH would alter the rumen bacteria community, which also lead to the variation of the rumen fermentation. The objectives of this study were to determine the effects of AS pH on the nutrients digestion, rumen fermentation, and ruminal bacteria community.
All the procedures involving animals were carried out in accordance with the Biological Studies Animal Care and Use Committee of Gansu Province, China (2005–2012).
The study was conducted using RUSITEC (Sanshin, Tokyo, Japan) as described by Kajikawa et al. (16). The RUSITEC system contained eight fermenters with a volume of 800 ml each per tank. The inoculum used in the fermenters was obtained from four ruminal fistulated Hu lambs fed two equivalent meals at 07:00 and 19:00 daily in the form of totally mixed ration (TMR) pellets with forage to concentrate of 80:20. The rumen contents were collected through the ruminal fistula before the morning feed and separated into liquid and solid fractions by four layers of cheesecloth. The squeezed solid inoculum (70 g wet weight) was enclosed in a nylon bag (7 × 13 cm, pore size: 100 μm). On the 1st day during fermentation, 400 ml of liquid inoculum was distributed to each fermenter under CO2 flux after mixing with an equal volume of AS, and two bags were placed in the fermenter, one with feed and the other with solid inoculum. After 24 h, the bag with the inoculum was replaced by a new bag with the feed. Subsequently, the bag that included the feed incubated 48 h was replaced by a new feed bag. A continuous infusion of AS at a rate of 600 ml/day was maintained in each fermenter. The fermenters were kept in a water bath at 39°C and slowly moved up and down by an electric motor (five times per minute).
The fermenters were randomly assigned to the two treatments with four replicates of each treatment. The treatment included high AS pH (pH 7.0) or low AS pH (pH 6.0) according to McDougall's method (17) (Table 1). The low AS pH was sustained by decreasing the AS buffer capacity (NaHCO3 from 9.8 to 1.96 g/L and Na2HPO4 from 9.3 to 1.86 g/L). The pH of all the fermenters was recorded at 07:30, 15:30, and 23:30 daily throughout the experiment periods. The diets were supplemented with 16 g basic diets with forage to the concentrate ratio of 50:50 (Table 2). The diets were ground through a 2 mm sieve. The experiment was conducted for 13-day incubation periods, with 9 days adaption and 4 days sample collection.
During the last 4 days of the experiment, the ice water was added around the over flow bottle to terminate fermentation. On days 10 and 11, about 10 ml of fermenter fluid were collected at 0, 3, 6, 9, and 12 h after the morning feed, the ruminal pH was immediately measured with a mobile pH meter (PHB-4, Shanghai Hongyi instrument Limited, Shanghai, China). Then, 5-ml of rumen fluid was preserved with 1 ml of metaphosphoric acid (25% wt/vol) and stored at −20°C for the determination of VFA. On day 12, about 10 ml of ruminal fluid was collected from each fermenter and immediately stored at −80°C to exact bacterial DNA. On day 13, about 20% of solid contents from each nylon bag were frozen at −80°C for the solid phase bacteria DNA extraction. The bag from each vessel on days 10, 11, and 12 was collected, washed one time with 100 ml of artificial saliva, washed with cold water until the outflow was clear, and stored to determine dry matter (DM) disappearance. The DM disappearance was calculated from the loss in weight after oven drying at 65°C for 48 h by using the following equation: DM disappearance (%) = {(g Sample DM – g Residue DM – g Bag DM)/g Sample DM} *100, and the residues were analyzed for DM, organic matter (OM), neutral detergent fiber (NDF), acid detergent fiber (ADF), and crude protein (CP).
The content of DM, ash, and N in the feed and residues were determined according to the Association of Official Analytical Chemists (AOAC) method (18). The DM content was determined by drying at 105°C in a forced-air oven for 4 h. The ash content was determined by complete combustion in a muffle furnace (PrepASH-340, Precisa, Swizerland) at 550°C for 6 h. The N contents of the feed bag were carried out by a protein analyzer (K9840, Hanon Advanced Technology Group Co., Ltd, Jinan, China) according to the Kjeldahl method and CP was calculated as N × 6.25 (18). The NDF and ADF were determined by the method of Van Soest et al. (19).
The thawed rumen fluid samples were centrifuged at 2,500 ×g at 4°C for 5 min, and the supernatants were processed as described by Liang et al. (20). The VFA concentrations were measured with gas chromatography (GC) on a Thermo Fisher Trace 1300 GC system (TRACE 1300, Thermo Scientific, Milan, Italy) as described by Li et al. (15). The GC was fitted with a silica capillary column (DB-FFAP, 30 m × 0.32 mm × 0.25 μm, Agilent Technologies Co., Ltd, Santa Clara, CA, USA), and crotonic acid (1% wt/vol) was used as the internal standard. The injector and detector temperatures were set at 240°C. The following temperature program was used: the temperature was increased from 50 to 190°C at a rate of 25°C/min, and the temperature increased was increased to 200°C at 10°C/min for 5 min. Finally, the temperature was increased to 220°C at a rate of 10°C/min and was held for 5 min. The concentration of lactate was determined by a commercial Lactate Analysis Kit (Nanjing Jiancheng Technology Co., LTD., Nanjing, China).
The DNA of rumen bacteria was extracted by an E.Z.N. A® Bacterial DNA Kit (Omega Bio-Tek, Inc., Norcross, GA, USA) according to the instructions from the manufacturer. The final elution volume was 80 μl, and DNA concentration and purity were measured by an ND-2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The primer design for all the rumen bacteria to amplify was selected on the basis of the published literature (Table 3). The quantitative real-time PCR (qPCR) protocol was described by Liang et al. (20). Each sample contained 1 μl of DNA, 10 μl of SYBR Green (TransGen Biotech, Beijing, China), 0.6 μl of each primer, and 8.6 μl of ddH2O in a final volume of 20 μl. The amplification conditions were as follows: 95°C for 10 s; 40 cycles of 10 s at 95°C; 30 s at 60°C; 72°C for 10 s; and a final cycle at 72°C for 5 min. To obtain melting curve data, the temperature increased in 0.5°C increments from 65 to 95°C. All investigated PCR products had only single melting peaks. The relative abundance of rumen bacteria was expressed as a proportion of total rumen bacterial 16S rRNA according to the equation: relative quantification = 2−(CTctarget−CTctotal bacteria), where CT represents a threshold cycle (26). Before the statistical analysis, the percentage of each microbe target was calculated as (2−ΔCT) × 100, then, the data were log10–scale transformed before the statistical analysis (20). The quantity of each species was expressed as the log10 copy number of 16S rRNA gene copies per milliliter of rumen fluid.
The sequence analysis and bioinformatics were conducted by SMRT Portal (version. 2.7; PacBio, CA, USA). The Lima (version. 1.7.0; PacBio, CA, USA) software was applied to export circular consensus sequencing (CCS) sequences from raw data and perform Barcode identification for the CCS sequences. Then, the chimera was filtered by UCHIME (version. 4.2; Tiburon, CA, USA) software to get the Optimization-CCS (27). We cluster Optimization-CCS sequences to get operational taxonomic units (OTU) by USEARCH (version 10.0; Tiburon, CA, USA) software (28), then get the species classification according to the sequence composition of OTU. The principal coordinate analysis (PCoA) plot of samples according to the distance matrix was obtained to analyze. The Ace, Chao1, Shannon, and Simpson indexes of each sample were statistically calculated by using Mothur (version v.1.30; Mothur, Michigan, USA) to evaluate the alpha diversity at 97% similarity level (29). According to OTU analysis results, a taxonomic analysis was performed with RDP Classifier (version 2.2; RDP Classifier, Michigan, USA) at the taxonomic level of phylum and genus (30). The raw sequencing data were in the Sequence Read Archive (SRA) of NCBI and can be accessed via accession number: PRJNA752826.
The nutrients degradation, fermentation parameters, and rumen bacteria abundances were analyzed by using SPSS software version 17.0 (IBM, Armonk, NY, United States). The independent sample T-test was used to calculate the differences in the results between the high AS pH group and the low AS pH group in this experiment. The effect of time on fermentation variables was used as a repeated measure. The model included the effects of AS pH, time, and their interaction as fixed effects, and individual fermenters as a random effect. The Kruskal–Wallis test was used to test the rumen bacteria in the solid and liquid fraction at the phylum and genus. The significant difference of data was analyzed by Kruskal–Wallis one-way ANOVA analysis. The significance was set as P ≤ 0.05 and the tendencies were considered when 0.05 < P < 0.10.
Decreasing the AS buffer capacity resulted in a reduction in average pH to 6.02 in the low AS group. The effect of AS pH on the nutrients degradabilities is presented in Table 4. The degradabilities of DM, OM, NDF, ADF, and CP were lower in the low AS pH group compared with the high AS pH group (P < 0.001).
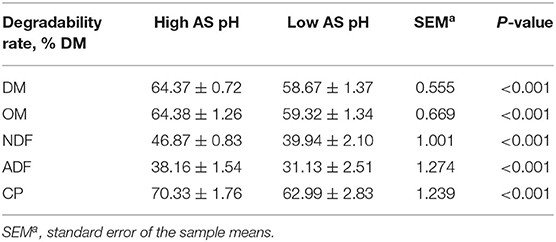
Table 4. Effect of AS pH on the nutrients degradability in the rumen simulation technique (RUSITEC).
The effect of AS pH on VFA in RUSITEC is shown in Table 5. The total concentration of VFA tended to be lower (P = 0.080) in the low AS pH group compared with the high AS pH group. The proportions of acetate, isobutyrate, and the concentration of lactate were not affected by different AS pH (P > 0.05). The low AS pH decreased the proportion of propionate (P < 0.001) and increased the proportions of butyrate (P = 0.008), isovalerate (P = 0.012), valerate (P < 0.001), and the ratio of acetate to propionate (P < 0.001).
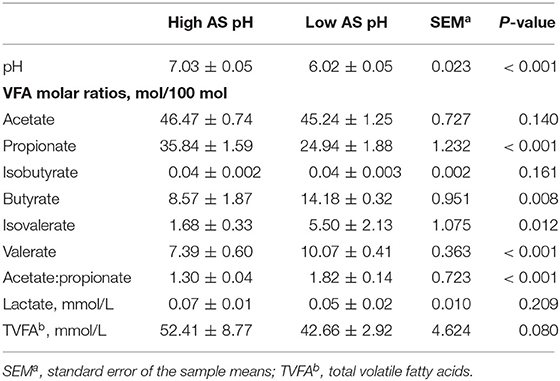
Table 5. Effect of artificial saliva (AS) pH on pH and volatile fatty acids in the rumen simulating fermenter (RUSITEC).
The effect of AS pH on the fermentation parameters at 0, 3, 6, 9, and 12 h after feeding is shown in Supplementary Table 1. An interaction between AS pH and time affected the rumen pH (P = 0.003); the low AS pH had lower rumen pH than high AS pH (P < 0.001), and rumen pH was decreased at 0 and 9 h after feeding (P < 0.001). The proportion of acetate was increased at 0 h and decreased at 3 h after feeding in the high AS pH group, respectively (P = 0.039). The proportion of butyrate was affected by an interaction between the AS pH and time (P = 0.037). The low AS pH had a greater proportion of butyrate than high AS pH (P = 0.002), and the proportion of butyrate was decreased at 0 h and increased at 3 and 9 h between the low and high AS pH group (P < 0.001). The proportion of valerate was higher at 0, 3, 6, and 9 h than at 12 h after feeding in the low AS pH group (P = 0.009).
The effect of AS pH on the α diversity and phylum abundances of the ruminal bacteria are shown in Table 6. The sequence coverage sufficiently met a coverage >97% for all the samples. Across all the samples, a total of 102,043 CCS sequences were obtained, and an average of 6,378 CCS sequences per sample. In total, 94% of CCS sequences were classified at the phyla level and 66% at the genus level. In the solid-associated bacteria, the ACE, Chao1, and Simpson indexes were not affected by AS pH (P > 0.05). However, the Shannon index was greater in the high AS pH group (P = 0.031) compared with the low AS pH group. In the liquid-associated bacteria, the ACE, Chao1, and Shannon indexes were greater (P < 0.05) in the high AS pH group compared with the low AS pH group, whereas the Simpson index tended to be higher (P = 0.098). The ACE, Chao1, Simpson, and Shannon indexes have no difference between solid-associated bacteria and liquid-associated bacteria.
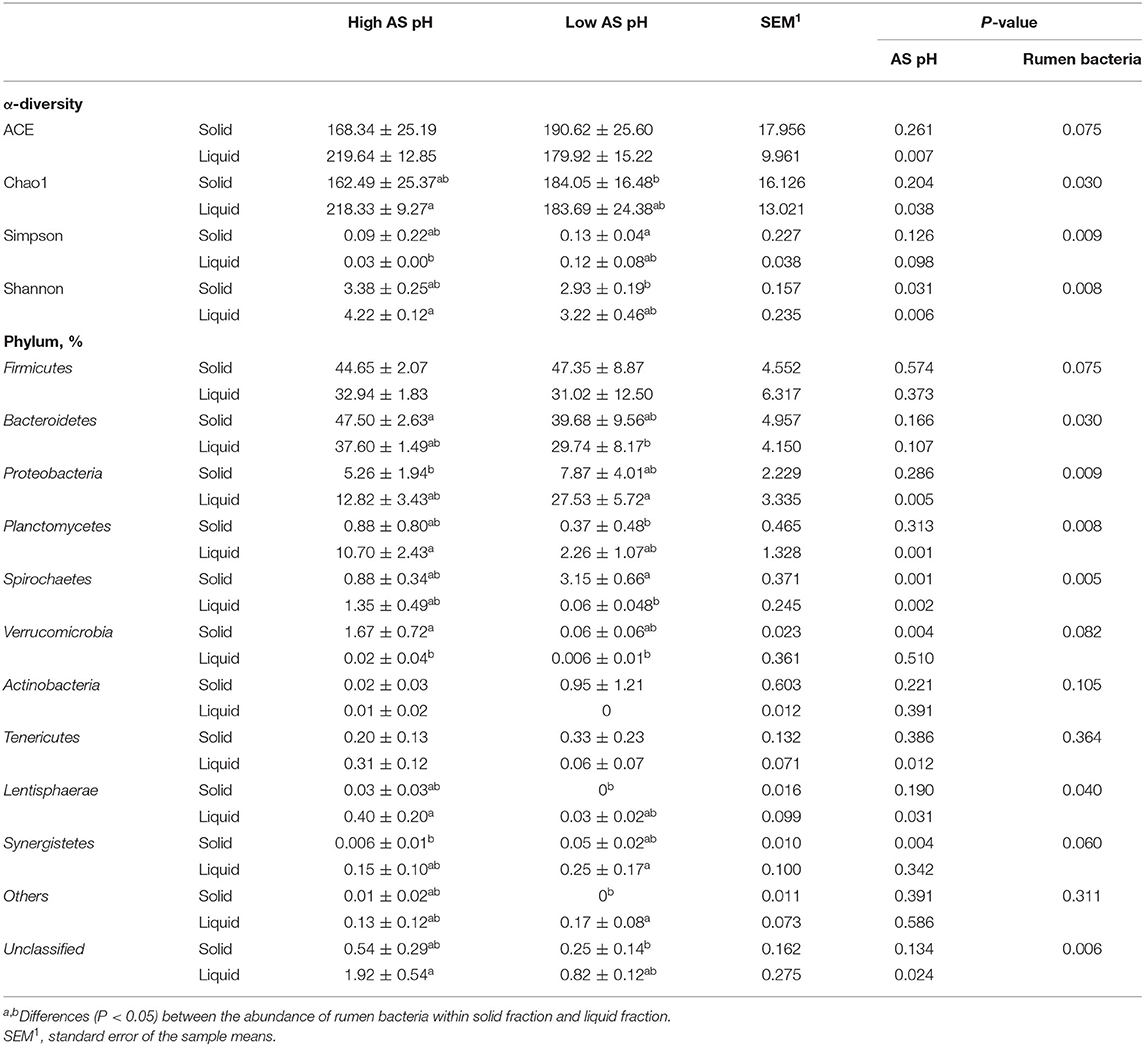
Table 6. Effect of AS pH on the α-diversity of the rumen bacteria and community at phylum level in RUSITEC.
The abundance of Firmicutes and Bacteroidetes was not affected by AS pH treatment (P > 0.05). The low AS pH increased the abundances of Spirochaetes (P = 0.001), Synergistetes (P = 0.004), and decreased the Verrucomicrobia abundance (P = 0.004) in the solid-associated bacteria. In the liquid-associated bacteria, low AS pH increased the abundance of Proteobacteria (P = 0.005) and decreased the abundances of Planctomycetes (P = 0.001), Spirochaetes (P = 0.002), Tenericutes (P = 0.012), and Lentisphaerae (P = 0.031). At the AS pH 6.0, the abundance of Spirochaetes was greater in solid than a liquid fraction. For the liquid and solid fraction, the abundances of Firmicutes, Bacteroidetes, and Proteobacteria were similar in solid fraction compared with the liquid fraction.
The beta diversities of bacteria communities within different AS pH for each fraction were calculated and visualized through the two-dimensional PCoA analysis using the binary-Jaccard (Figure 1). A significant difference between the bacterial communities in the AS pH treatment was noted. Both principal components accounted for 34.79% (PC1) and 27.78% (PC2) of the explained variance.
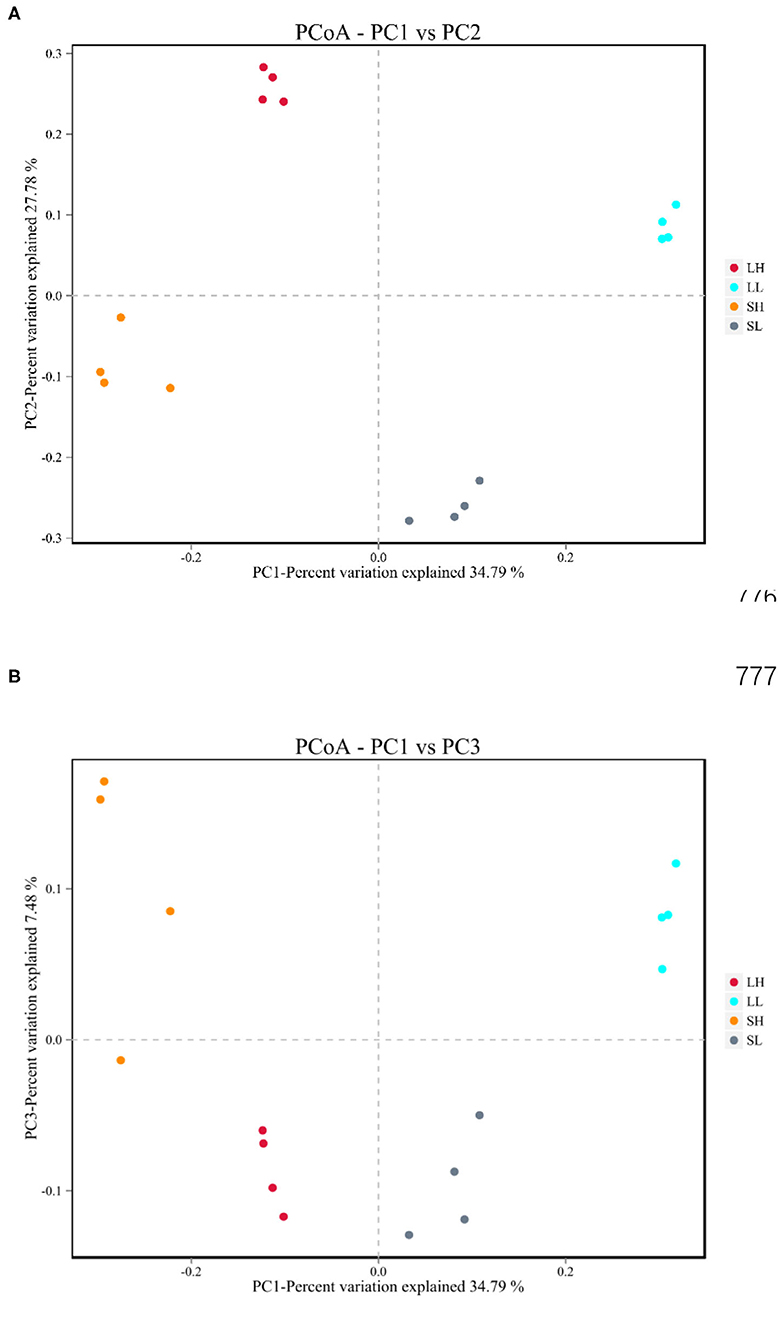
Figure 1. Effects of artificial saliva (AS) pH on the β diversity in the rumen bacteria (A,B) in RUSITEC. LH, the high artificial saliva (AS) pH in the liquid-associated bacteria; LL, the low AS pH in the liquid-associated bacteria; SH, the high AS pH in the solid-associated bacteria; SL, the low AS pH in the solid-associated bacteria.
The ruminal bacteria with abundances >1% at the genus level were presented in Table 7. The low AS pH increased the abundance of Lactobacillus (P = 0.050) and decreased the abundances of Schwartzia (P = 0.002) in solid-associated bacteria compared with the high AS pH group. In the solid-associated bacteria, the abundance of Succinivibrio tended to be greater (P = 0.059) in the low AS pH group compared with the high AS pH group, whereas the abundance of Ruminobacter tended to be lower (P = 0.086). In the liquid-associated bacteria, the abundances of Prevotellaceae_YAB2003_group (P = 0.040), Schwartzia (P = 0.002), and Ruminobacter (P = 0.043) were lower in the low AS pH group compared with the high AS pH group. However, the abundances of Succinivibrio (P < 0.001) and Prevotella_1 (P = 0.001) were higher in the low AS pH treatment compared with the high AS pH group. At the AS pH 7.0 and 6.0, the abundance of Prevotellaceae_YAB2003_group was greater in a solid fraction than a liquid fraction (P < 0.05). The abundance of Prevotella_1 was decreased in a solid fraction when the AS pH was 6.0 (P = 0.008), while the abundance of Prevotella_7 was increased (P = 0.007).
The effect of AS pH on the number of rumen bacteria is presented in Table 8. The low AS pH decreased the number of Ruminococcus albus, R. flavefaciens, F. succinogenes (P < 0.001) both in the solid- and liquid-associated bacteria, respectively. The low AS pH tended to increase the amount of Prevotella brevis (P = 0.091) in liquid-associated bacteria. The low AS pH decreased the amounts of Selenomonas ruminantium in solid-associated bacteria (P = 0.022) and tended to decrease in liquid-associated bacteria (P = 0.065). The number of S. ruminantium was increased in solid fractions both in high AS pH and low AS pH (P < 0.001). At the high AS pH, the amounts of P. brevis, Butyrivibrio fibrisolvens, and total bacteria were increased in solid fraction compared with the liquid fraction (P < 0.05). The number of F. succinogenes was greater in solid fraction than liquid fraction at the low AS pH (P < 0.001).
The rumen pH is the most monitored parameter for SARA diagnosis. According to the severity of SARA, the average daily pH threshold was 5.50–6.25 (3). In the current study, decreasing the AS buffer capacity resulted in an average pH of 6.02. The ruminal pH was an important factor that affect the degradation of NDF and OM degradation in the rumen (31). In our study, low AS pH decreased the degradabilities of DM, OM, NDF, and ADF. These results are consistent with the previous reports that the digestion rates of DM and NDF were reduced with the decreasing AS pH (7.0 vs. 4.9) in vitro (32). The decreased digestibilties of NDF and ADF at the low AS pH are mainly attributed to the reduction of cellulolytic bacteria populations and the ability of cellulolytic bacteria to attach to the feed particles (33).
Reduction in the rumen pH lower than 6.0 has a negative impact on the amount of cellulolytic bacteria (R. albus, R. R. flavefaciens, F. succinogenes, and B. fibrisolvens) in the rumen (15, 34). As expected, the low AS pH decreased the amount of ruminal R. albus, R. flavefaciens, F. succinogenes, and B. fibrisolvens in the solid- and liquid-associated bacteria with the identical substrates chemical compositions. However, Li et al. (8) found that the ruminal cellulolytic bacteria, such as the amounts of F. succinogenes and R. flavefaciens were increased when the dairy goats experience the low-peNDF diet induced SARA. Khafipour et al. (6) found the mild grain-induced SARA increased the populations of R. albus and R. flavefaciens. The result of cellulolytic bacteria was not consistent between the low peNDF-induced SARA with the high grained-induced SARA in vivo (6). In the present study, we intended to stimulate the low peNDF induced SARA by decreasing the AS pH in Rustitec, which is similar to the decreased saliva secretion when the ruminants received the low peNDF diets in vivo. The increased cellulolytic bacteria when the cows or goats received the low peNDF diets (decreased the roughage particle size without changing the roughage to concentrate ratio) mainly attributed to the increased surface area for microbial attachment (8, 35–37). However, the particle size of feed was identical between the treatments, and the changes of cellulolytic bacteria only response to the different AS pH in the present study. Therefore, the results of the present study indicated that the low AS pH indeed decreased the number of cellulolytic bacteria when the substrate was identical in the Rustitec. The rumen bacteria abundance of the solid fraction is significantly higher than that of the liquid fraction (38). The amounts of B. fibrisolvens and F. succinogenes, total bacteria were enriched in a solid fraction in our study. This result was in accordance with De Mulder et al. (14), who identified that cellulolytic bacteria are prevalent in the solid fraction.
The ruminal genus of Prevotella.app is considered to be associated with starch degradation and growth well at low pH conditions (6, 39). In our study, the amount of P. brevis was not affected by AS pH in the solid-associated bacteria, but the number of P. brevis in the liquid-associated bacteria tended to be increased in the low AS pH group.
In the current study, the degradability of CP decreased in the low AS pH group. Several studies have indicated that the low AS pH decreased or unaffected CP degradability (32, 40). The plant proteins were integrated within non-protein polymers, such as polysaccharides, which may limit the access of proteolytic bacteria to the substrate (41). It is possible that the low AS pH decreased ruminal cellulolytic activity and led to a reduction in the CP degradability due to the limitation of the access of proteases to their matrix (8).
In our experiment, the total concentration of VFA was reduced in the low AS pH treatment. This result was in accordance with Jiao et al. (42), who found that the total concentration of VFA (42.66 vs. 52.41 mmol/L) declined when pH was at 5.8 compared with the pH 6.5 in vitro. The declined total VFA concentration in the present study is mainly attributed to the decreased OM degradability. In our study, the proportion of acetate was increased at 0 h and decreased at 3 h after feeding in the high AS pH group. Because the fermenters are opened to supply new nylon bags with feed; this operation exposes the cellulolytic bacteria to oxygen and inhibits the activity (43). The decrease AS pH reduced the proportion of propionate in our experiment. The results were consistent with Strobel and Russell (44) found the concentration of propionate from starch fermentation (2.9 vs. 1.1 mM) decreased when the pH decreased from 6.7 to 5.8. The previous studies reported the amylolytic bacteria to produce amounts of propionate, but many cellulolytic bacteria generate a large amount of succinate, an intermediate that is eventually converted to propionate (45). The decreased proportion of propionate was because the low ruminal pH inhibited the succinate conversion to propionate. The lower molar ratio of the propionate in the low AS pH group also resulted in a higher acetate to propionate compared with the high AS pH group. These results are different from Cardozo et al. (46), who reported the ratio of acetate to propionate was lower when pH was decreased from 7.0 to 5.5 because the high-grain diets decreased the acetate production and increased the propionate production in the rumen.
In the present study, the proportion of butyrate was greater in the low AS pH group compared with the high AS pH group. The results were in accordance with Esdale and Satter (47), who reported that the butyrate production was higher at pH 5.6 compared with at pH 6.2 in vitro. In addition, Shriver et al. (48) found butyrate production increased as pH was decreased from 6.2 to 5.8 in vivo. Calsamiglia et al. (32) identified that the concentration of butyrate was only affected by the changes of pH and not affected by diet compositions in vitro. The results could be associated with the increasing abundance of Prevotellaceae (e.g., Prevotella_1) in the liquid-associated bacteria that resulted in the increased butyrate production in vitro (49). In the current study, the proportion of butyrate was decreased at 0 h and increased at 3, 6, and 9 h after feeding. At 2 h after the start of incubation, the16S rDNA copy numbers of amylolytic bacteria attached to the grain were increased (50), which may promote butyrate production at 3, 6, and 9 h after feeding.
The concentrations of isovalerate and valerate in the rumen were related to the protein degradation and fermentation of branched-chain AA (51). The isovalerate and valerate are also considered as stimulating factors that enhanced the growth of cellulolytic bacteria (52). In the current study, the low AS pH increased the proportion of isovalerate and valerate, and the proportion of valerate was higher at 0, 3, 6, and 9 h than at 12 h after feeding. It had a low pH, which inhibited the growth of cellulolytic bacteria. The RUSITEC system fermenters were opened when the bags were replaced by the new nylon bags with feed; this operation exposed cellulolytic bacteria to oxygen and inhibited the activity of cellulolytic bacteria (43). This therefore would have resulted in the accumulation of valerate and isovalerate in the fermenters and increased at 3, 6, 9, and 12 h after feeding.
Using the sequence and bioinformatics analysis, we obtained 6,378 CCS sequences on average for each sample with good coverage (>97.0%). In accordance with our hypothesis, both the microbial α-diversity and β-diversity were affected by AS pH treatment. Meanwhile, most of the alpha diversity indices (except Simpson index) decreased with the low AS pH in the liquid-associated bacteria, suggesting that the low pH significantly decreased the activity and number of ruminal bacteria. These results are in agreement with Shen et al. (53), who reported the reduction of pH decreased the bacteria alpha diversity. In addition, the PCoA analysis also showed that the bacterial communities of the high AS pH and low AS pH clustered separately, indicating their distinct bacterial compositions in the rumen. These results were similar to the founding by Li et al. (15) who found the bacterial compositions were different between the sheep with high rumen pH and low rumen pH with identical feed composition. Interestingly, the ACE, Chao1, Simpson, and Shannon indexes have no difference between solid fractions and liquid fractions in our study. Because the solid- and liquid-associated bacteria do not have differences in the taxonomic composition but can be distinguished based on the relative abundance of species (14).
In the present study, the relative abundances of ruminal Firmicutes and Bacteroidetes were not affected by AS pH. The Firmicutes are predominantly composed of Gram-positive bacteria in the rumen, which are metabolically capable of utilizing the fermentable carbohydrates (54). Previous studies showed that feeding high-grain diets for cattle increased the abundance of ruminal Firmicutes (55). However, AS pH did not affect the abundance of Firmicutes in our study suggests that the Firmicutes were pH-insensitive bacteria. The Bacteroidetes are the most abundant Gram-negative bacteria found in the anaerobic communities of the rumen, and low pH resulted in the death and lysis of Gram-negative bacteria (15, 54). However, the abundance of Bacteroidetes was not affected by AS pH in the present study. Although the Bacteroidetes were not different in statistics, the value of Bacteroidetes decreased in the low pH group (47.50 vs. 39.68% in solid-associated bacteria and 37.60 vs. 29.74% in liquid-associated bacteria). Wang et al. (56) reported that feeding high-concentrate diets decreased the ruminal pH and increased the abundance of Proteobacteria in the rumen for cows. Furthermore, the low AS pH increased the abundance of Proteobacteria in the liquid-associated bacteria in this experiment. This result suggests that the phylum of Proteobacteria can tolerate the low pH condition. For the liquid and solid fraction, the abundances of Firmicutes, Bacteroidetes, and Proteobacteria were similar in solid fraction compared with the liquid fraction in our study. It is possible that the fermenters were moved up and down by an electric motor, and promoted the exchange of rumen bacteria in solid fraction and liquid fraction. The Spirochaetes commonly fermented xylan and pectin in feed (57). After 6 h fermentation, the Spirochaetes phyla became abundant in the forage-adherent community (58). The Spirochaetes was greater in low pH conditions in the solid-associated bacteria, which was also greater in high AS pH in the liquid-associated bacteria in the current study. And the abundance of Spirochaetes was greater in a solid fraction than a liquid fraction. These results indicated that Spirochaetes tend to colonize in the solid phase in the rumen.
The bacteria genus of Lactobacria was suitable for growth at pH 6.0 (59). The previous studies indicated that the increased non-fiber carbohydrate for ruminant promoted the growth of amylolytic and other starch-digesting bacterial species, such as Lactobacillus (60–62). In the current study, the relative abundance of Lactobacillus was increased when the AS pH decreased. Wang et al. (63) reported that the abundance of ruminal Lactobacillus was increased when cows intake the SARA diet. These studies indicated that Lactobacillus affected not only the dietary compositions but also the magnitude of pH. The Prevotella species were essential to hemicellulose degradation in the rumen, and Prevotella_1 and Prevotellaceae_YAB2003 (Bactenroidetes) were identified to have the ability to degrade hemicellulose or xylan in vivo (64, 65). In the current study, the low AS pH increased the abundances of Prevotella_1, and decreased the abundances of Prevotellaceae_YAB2003, and Prevotella_7 in the liquid-associated bacteria. These results indicate the sensitivity of Prevotella strains to AS pH was inconsistent. Similarly, the abundance of Prevotellaceae_YAB2003_group was greater in a solid fraction than a liquid fraction. The abundance of Prevotella_1 was decreased in solid fractions at the low AS pH, whereas the abundance of Prevotella_7 was increased. The Prevotellaceae comprises up to 40% of the community in the liquid samples, and ruminal Prevotella is non-cellulolytic but has a broad saccharolytic and proteolytic potential (14, 66). The abundances of Prevotellaceae_YAB2003_group and Prevotella_1 were increased in liquid fraction, which primarily consumed the soluble nutrients.
The function of Succinivibrio produced succinate, the precursor of propionate (67). In this research, the related abundance of Succinivibrio was higher in the low AS pH group compared with low AS pH, whereas the proportion of propionate was decreased. It should be presumed that the conversion of succinate to propionate acid was inhibited by low pH and producing less propionate. In addition, the Schwartzia fermented succinate and produced propionate (68). The Schwartzia abundance decreased in the low AS pH group, which was coordinated with the results of propionate in this study. The low AS pH decreased the abundance of Ruminobacter (Firmicutes) in this current study. Wang et al. (69) reported that the cow intake high-forage diets increased the ruminal Ruminobacter abundance. Mu et al. (70) found that fed a high grain-diet induce cow SARA increased the abundance of Ruminobacter. These results indicated that the growth of Ruminobacter in the rumen was affected by the combination of pH and diet compositions.
The nutrients degradabilities were decreased by reducing AS pH in the present study. The reduction of AS pH increased the proportion of butyrate, valerate, and isovalerate and decreased the proportion of propionate. The results of the present study indicated the three groups of bacteria communities according to the different sensitives to rumen pH: the abundances of Lactobacillus, Succinivibrio, Prevotella_7 are increased with decreasing AS pH; the amounts of R. albus, R. flavefaciens, F. succinogenes as well as the abundances of Schwartzia and Ruminobacter decreased with reducing AS pH; the abundances of Selenomonas_1, Rikenellaceae_RC9_gut_group, and Succiniclasticum were not affected by AS pH in Rustitec. In addition, the effect of the interaction of rumen pH and diets on the rumen bacteria community should be further investigated.
The data for this study can be found in the NCBI database (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA752826).
The animal study was reviewed and approved by the Biological Studies Animal Care and Use Committee of Gansu Province, China (2005–12). Written informed consent was obtained from the owners for the participation of their animals in this study.
ToG collected the sample, analyzed the data, and drafted the manuscript. TaG, YC, LG, FaL, and GY collected the sample. FeL presented the idea of this manuscript, supported the funding, analyzed the conclusions, and revised the manuscript. All authors contributed to the article and approved the submitted version.
The research was financially supported by the National Natural Science Foundation of China, grant number (No. 32072754) and the Natural Science Foundation of Gansu Province (20JR5RA299), China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The author thanks GY of the Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, who kindly assisted on the experiment site.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.760316/full#supplementary-material
1. Kleen JL, Hooijer GA, Rehage J, Noordhuizen JP. Subacute ruminal acidosis (SARA): A review. J Vet Med. (2003) 50:406–14. doi: 10.1046/j.1439-0442.2003.00569.x
2. Seddik H, Xu L, Wang Y, Mao SY. A rapid shift to high-grain diet results in dynamic changes in rumen epimural microbiome in sheep. Animal. (2018) 13:1–9. doi: 10.1017/S1751731118003269
3. Sauvant D, Meschy F, Mertens D. Les composantes de l'acidose ruminale et les effets acidogènes des rations. Prod Anim. (1999) 12:49–60. doi: 10.20870/productions-animales.1999.12.1.3854
4. Kleen JL, Hooijer GA, Rehage J, Noordhuizen JP. Subacute ruminal aacidosis in Dutch dairy herds. Vet Rec. (2009) 164:681–3. doi: 10.1136/vr.164.22.681
5. Colman E, Khafipour E, Vlaeminck B, De Baets B, Plaizier JC, Fievez V. Grain-based versus alfalfa-based subacute ruminal acidosis induction experiments: similarities and differences between changes in milk fatty acids. J. Dairy Sci. (2013) 96:4100–11. doi: 10.3168/jds.2012-6109
6. Khafipour E, Li SC, Plaizier JC, Krause DO. Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl Environ Microb. (2009) 75:7115–24. doi: 10.1128/AEM.00739-09
7. Lechartier C, Peyraud JL. The effects of forage proportion and rapidly degradable dry matter from concentrate on ruminal digestion in dairy cows fed corn silage-based diets with fixed neutral detergent fiber and starch contents. J Dairy Sci. (2010) 93:666–81. doi: 10.3168/jds.2009-2349
8. Li F, Li Z, Li S, Ferguson JD, Cao Y, Yao J, et al. Effect of dietary physically effective fiber on ruminal fermentation and the fatty acid profile of milk in dairy goats. J Dairy Sci. (2014) 97:2281–90. doi: 10.3168/jds.2013-6895
9. Park JH, Kim KH, Park PJ, Jeon BT, Oh MR, Jang SY, et al. Effects of physically effective neutral detergent fibre content on dry-matter intake, digestibility and chewing activity in beef cattle fed total mixed ration. Anim Prod Sci. (2014) 55:166. doi: 10.1071/AN14241
10. Orton T, Rohn K, Breves G, Brede M. Alterations in fermentation parameters during and after induction of a subacute rumen acidosis in the rumen simulation technique. J Anim Physiol An N. (2020) 104:1678–89. doi: 10.1111/jpn.13412
11. Brede M, Orton T, Pinior B, Roch FF, Dzieciol M, Zwirzitz B, et al. PacBio and Illumina MiSeq amplicon sequencing confirm full recovery of the bacterial community after subacute ruminal acidosis challenge in the Rusitec system. Front Microbiol. (2020) 11:1813. doi: 10.3389/fmicb.2020.01813
12. Wanapat M, Cherdthong A. Use of real-time PCR technique in studying rumen cellulolytic bacteria population as affected by level of roughage in swamp buffalo. Curr Microbiol. (2009) 58:294–9. doi: 10.1007/s00284-008-9322-6
13. McAllister TA, Bae HD, Jones GA, Cheng KJ. Microbial attachment and feed digestion in the rumen. J Anim Sci. (1994) 72:3004–18. doi: 10.2527/1994.72113004x
14. De Mulder T, Goossens K, Peiren N, Vandaele L, Haegeman A, De Tender C, et al. Exploring the methanogen and bacterial communities of rumen environments: solid adherent, fluid and epimural. Fems Microbiol Ecol. (2016) 93:fiw251. doi: 10.1093/femsec/fiw251
15. Li F, Wang Z, Dong C, Li F, Wang W, Yuan F, et al. Rumen bacteria communities and performances of fattening lambs with a lower or greater subacute ruminal acidosis risk. Front Microbiol. (2017) 8:2506. doi: 10.3389/fmicb.2017.02506
16. Kajikawa H, Jin H, Terada F, Suga T. Operation and Characteristics of Newly Improved and Marketable Artificial Rumen (Rusitec). Japan: Memoirs of National Institute of Livestock and Grassland Science (2003).
17. McDougall EI. Studies on ruminant saliva. I. the composition and output of sheep's saliva. Biochem J. (1948) 43:99–109. doi: 10.1042/bj0430099
18. Association of Official Analytical Chemists (AOAC). Official Methods of Analysis, 16th ed. Washington, DC: AOAC International (1997).
19. Van Soest PJ, Robertson JB, Lewis BA. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci. (1991) 74:3583–97. doi: 10.3168/jds.S0022-0302(91)78551-2
20. Liang YS, Li GZ, Li XY, Lu JY, Li FD, Tang DF, et al. Growth performance, rumen fermentation, bacteria composition, and gene expressions involved in intracellular pH regulation of rumen epithelium in finishing hu lambs differing in residual feed intake phenotype. J Anim Sci. (2017) 95:1727–38. doi: 10.2527/jas.2016.1134
21. Tajima K, Aminov RI, Nagamine T, Matsui H, Nakamura M, Benno Y. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl Environ Microbe. (2001) 67:2766–74. doi: 10.1128/AEM.67.6.2766-2774.2001
22. Denman SE, McSweeney CS. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. Fems Microbiol Ecol. (2006) 58:572–82. doi: 10.1111/j.1574-6941.2006.00190.x
23. Stevenson DM, Weimer PJ. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biot. (2007) 75:165–74. doi: 10.1007/s00253-006-0802-y
24. Koike S, Kobayashi Y. Development and use of competitive PCR assays for the rumen cellulolytic bacteria: Fibrobacter succinogenes, Ruminococus albus and Ruminococcus flavefaciens. Fems Microbiol Lett. (2001) 204:361–6. doi: 10.1111/j.1574-6968.2001.tb10911.x
25. Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology-Sgm. (2002) 148:257–66. doi: 10.1099/00221287-148-1-257
26. Chen XL, Wang JK, Wu YM, Liu JX. Effects of chemical treatments of rice straw on rumen fermentation characteristics, fibrolytic enzyme activities and populations of liquid- and solid-associated ruminal microbes in vitro. Anim Feed Sci Tech. (2008) 141:1–14. doi: 10.1016/j.anifeedsci.2007.04.006
27. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. (2011) 27:2194–200. doi: 10.1093/bioinformatics/btr381
28. Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. (2013) 10:996–8. doi: 10.1038/nmeth.2604
29. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing MOTHUR: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb. (2009) 75:7537–41. doi: 10.1128/AEM.01541-09
30. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb. (2007) 73:5261–7. doi: 10.1128/AEM.00062-07
31. Mould FL, Ørskov ER. Manipulation of rumen fluid pH and its influence on cellulolysis in sacco, dry matter degradation and the rumen microflora of sheep offered either hay or concentrate. Anim Feed Sci Tech. (1983) 10:1–14. doi: 10.1016/0377-8401(83)90002-0
32. Calsamiglia S, Cardozo P, Ferret WA, Bach A. Changes in rumen microbial fermentation are due to a combined effect of type of diet and pH. J Anim Sci. (2008) 86:702–11. doi: 10.2527/jas.2007-0146
33. Sung HG, Kobayashi Y, Chang J, Ha A, Hwang IH. Low ruminal pH reduces dietary fiber digestion via reduced microbial attachment. Asian Austr J Anim. (2007) 20:200–7. doi: 10.5713/ajas.2007.200
34. Nagaraja TG, Titgemeyer EC. Ruminal acidosis in beef cattle: the current microbiological and nutritional outlook. J Dairy Sci. (2007) 90:E17–38. doi: 10.3168/jds.2006-478
35. Miron J, Ben-Ghedalia D, Morrison M. Invited Review: adhesion mechanisms of rumen cellulolytic bacteria. J Dairy Sci. (2001) 84:1294–309. doi: 10.3168/jds.S0022-0302(01)70159-2
36. Bhandari SK, Ominski KH, Wittenberg KM, Plaizier JC. Effects of chop length of alfalfa and corn silage on milk production and rumen fermentation of dairy cows. J Dairy Sci. (2007) 90:2355–66. doi: 10.3168/jds.2006-609
37. Zebeli Q, Tafaj M, Junck B, Oelschlaeger V, Ametaj BN, Drochner W. Evaluation of the response of ruminal fermentation and activities of nonstarch polysaccharide-degrading enzymes to particle length of corn silage in dairy cows. J Dairy Sci. (2008) 91:2388–98. doi: 10.3168/jds.2007-0810
38. Pei C, Mao S, Cheng Y, Zhu W. Diversity, abundance and novel 16S rRNA gene sequences of methanogens in rumen liquid, solid and epithelium fractions of Jinnan cattle. Animal. (2010) 4:20–9. doi: 10.1017/S1751731109990681
39. Matsui H, Ogata K, Tajima K, Nakamura M, Nagamine T, Aminov RI, et al. Phenotypic characterization of polysaccharidases produced by four prevotella type strains. Curr Microbiol. (2000) 41:45–9. doi: 10.1007/s002840010089
40. Cerrato-Sánchez M, Calsamiglia S, Ferret A. Effects of patterns of suboptimal pH on rumen fermentation in a dual-flow continuous culture system. J Dairy Sci. (2007) 90:4368–77. doi: 10.3168/jds.2006-804
41. Wallace RJ, Onodera R, Cotta MA. Metabolism of Nitrogen-Containing Compounds. Dordrecht: Springer (1997). doi: 10.1007/978-94-009-1453-7_7
42. Jiao P, Wei C, Sun Y, Xie X, Zhang Y, Wang S, et al. Screening of live yeast and yeast derivatives for their impact of strain and dose on in vitro ruminal fermentation and microbial profiles with varying media pH levels in high-forage beef cattle diet. J Sci Food Agr. (2019) 99:6751–60. doi: 10.1002/jsfa.9957
43. Gizzi G, Zanchi R, Sciaraffia F. Comparison of microbiological and fermentation parameters obtained with an improved rumen in vitro technique with those obtained in vivo. Anim Feed Sci Tech. (1998) 73:291–305. doi: 10.1016/S0377-8401(98)00150-3
44. Strobel HJ, Russell JB. Effect of pH and energy spilling on bacterial protein synthesis by carbohydrate-limited cultures of mixed rumen bacteria. J Dairy Sci. (1986) 69:2941–7. doi: 10.3168/jds.S0022-0302(86)80750-0
46. Cardozo PW, Calsamiglia S, Ferret A, Kamel C. Screening for the effects of natural plant extracts at different pH on in vitro rumen microbial fermentation of a high-concentrate diet for beef cattle. J Anim Sci. (2005) 83:2572–9. doi: 10.2527/2005.83112572x
47. Esdale WJ, Satter LD. Manipulation of ruminal fermentation. IV. Effect of altering ruminal pH on volatile fatty acid production. J Dairy Sci. (1972) 55:964–70. doi: 10.3168/jds.S0022-0302(72)85603-0
48. Shriver BJ, Hoover WH, Sargent JP, Crawford RJ, Thayne WV. Fermentation of a high concentrate diet as affected by ruminal pH and digesta flow. J Dairy Sci. (1986) 69:413–9. doi: 10.3168/jds.S0022-0302(86)80419-2
49. Esquivel-Elizondo SR, Ilhan ZE, Garcia-Pena EI, Krajmalnik-Brown R, Beiko RG. Insights into butyrate production in a controlled fermentation system via gene predictions. mSystems. (2017) 2:e00051–e00017. doi: 10.1128/mSystems.00051-17
50. Kozakai K, Nakamura T, Kobayashi Y, Tanigawa T, Osaka I, Kawamoto S, et al. Effect of mechanical processing of corn silage on in vitro ruminal fermentation, and in situ bacterial colonization and dry matter degradation. Can J Anim Sci. (2007) 87:259–67. doi: 10.4141/A06-028
51. Martínez ME, Ranilla MJ, Tejido ML, Saro C, Carro MD. Comparison of fermentation of diets of variable composition and microbial populations in the rumen of sheep and Rusitec fermenters. II. Protozoa population and diversity of bacterial communities. J Dairy Sci. (2010) 93:3699–712. doi: 10.3168/jds.2009-2934
52. Hespell RB, Cotta MA. Degradation and utilization by Butyrivibrio-Fibrisolvens H17c of xylans with different chemical and physical-properties. Appl Environ Microb. (1995) 61:3042–50. doi: 10.1128/aem.61.8.3042-3050.1995
53. Shen H, Xu Z, Shen Z, Lu Z. The regulation of ruminal short-chain fatty acids on the functions of rumen barriers. Front Physiol. (2019) 10:1305. doi: 10.3389/fphys.2019.01305
54. Huo W, Zhu W, Mao S. Impact of subacute ruminal acidosis on the diversity of liquid and solid-associated bacteria in the rumen of goats. World J Microbiol Biotechnol. (2014) 30:669–80. doi: 10.1007/s11274-013-1489-8
55. Sato S. Pathophysiological evaluation of subacute ruminal acidosis (SARA) by continuous ruminal pH monitoring. Anim Sci J. (2016) 87:168–77. doi: 10.1111/asj.12415
56. Wang LJ, Zhang GN, Li Y, Zhang YG. Effects of high forage/concentrate diet on volatile fatty acid production and the microorganisms involved in VFA production in cow rumen. Animals. (2020) 10:12. doi: 10.3390/ani10020223
57. Paster BJ, Canale-Parola E. Physiological diversity of rumen spirochetes. Appl Environ Microbiol. (1982) 43:686–93. doi: 10.1128/aem.43.3.686-693.1982
58. Liu J, Zhang M, Xue C, Zhu W, Mao S. Characterization and comparison of the temporal dynamics of ruminal bacterial microbiota colonizing rice straw and alfalfa hay within ruminants. J Dairy Sci. (2016) 99:9668–81. doi: 10.3168/jds.2016-11398
59. Mackie RI, Gilchrist FM. Changes in lactate-producing and lactate-utilizing bacteria in relation to pH in the rumen of sheep during stepwise adaptation to a high-concentrate diet. Appl Environ Microbiol. (1979) 38:422–30. doi: 10.1128/aem.38.3.422-430.1979
60. Goad DW, Goad CL, Nagaraja TG. Ruminal microbial and fermentative changes associated with experimentally induced subacute acidosis in steers. J Anim Sci. (1998) 76:234–41. doi: 10.2527/1998.761234x
61. Metzler-Zebeli BU, Schmitz-Esser S, Klevenhusen F, Podstatzky-Lichtenstein L, Wagner M, Zebeli Q. Grain-rich diets differently alter ruminal and colonic abundance of microbial populations and lipopolysaccharide in goats. Anaerobe. (2013) 20:65–73. doi: 10.1016/j.anaerobe.2013.02.005
62. Mickdam E, Khiaosa-ard R, Metzler-Zebeli BU, Klevenhusen F, Chizzola R, Zebeli Q. Rumen microbial abundance and fermentation profile during severe subacute ruminal acidosis and its modulation by plant derived alkaloids in vitro. Anaerobe. (2016) 39:4–13. doi: 10.1016/j.anaerobe.2016.02.002
63. Wang H, Pan X, Wang C, Wang M, Yu L. Effects of different dietary concentrate to forage ratio and thiamine supplementation on the rumen fermentation and ruminal bacterial community in dairy cows. Anim Prod Sci. (2015) 55:189–93. doi: 10.1071/AN14523
64. Emerson EL, Weimer PJ. Fermentation of model hemicelluloses by Prevotella Strains and B. Fibrisolvens in pure culture and in ruminal enrichment cultures. Appl Microbiol Biot. (2017) 101:4269–78. doi: 10.1007/s00253-017-8150-7
65. Wang B, Ma PM, Diao QY, Tu Y. Saponin-induced shifts in the rumen microbiome and metabolome of young cattle. Front Microbio. (2019) 10:356. doi: 10.3389/fmicb.2019.00356
66. Avgustin G, Wallace RJ, Flint HJ. Phenotypic diversity among ruminal isolates of Prevotella ruminicola: proposal of P. brevis sp. nov., Prevotella bryantii sp. nov., and Prevotella albensis sp. nov. and redefinition of Prevotella ruminicola. Int J Syst Bacteriol. (1997) 47:284–8. doi: 10.1099/00207713-47-2-284
67. Elolimy AA, Arroyo JM, Batistel F, Iakiviak MA, Loor JJ. Association of residual feed intake with abundance of ruminal bacteria and biopolymer hydrolyzing enzyme activities during the periportal period and early lactation in Holstein dairy cows. J Anim Sci. (2018) 9:43. doi: 10.1186/s40104-018-0258-9
68. Vangylswyk NO, Hippe H, Rainey FA. Schwartzia Succinivorans Gen. Nov., Sp. Nov., another ruminal bacterium utilizing succinate as the sole energy source. Int J Syst Bacteriol. (1997) 47:155–9. doi: 10.1099/00207713-47-1-155
69. Wang L, Li Y, Zhang Y, Wang L. The effects of different concentrate-to-forage ratio diets on rumen bacterial microbiota and the structures of Holstein cows during the feeding cycle. Animals. (2020) 10:957. doi: 10.3390/ani10060957
70. Mu YY, Qi WP, Zhang T, Zhang JY, Mao SY. Gene function adjustment for carbohydrate metabolism and enrichment of rumen microbiota with antibiotic resistance genes during subacute rumen acidosis induced by a high-grain diet in lactating dairy cows. J Dairy Sci. (2021) 104:2087–105. doi: 10.3168/jds.2020-19118
Keywords: artificial saliva, rumen bacteria, rumen pH, in vitro, ruminant
Citation: Guo T, Guo T, Cao Y, Guo L, Li F, Li F and Yang G (2021) Changes in the Fermentation and Bacterial Community by Artificial Saliva pH in RUSITEC System. Front. Nutr. 8:760316. doi: 10.3389/fnut.2021.760316
Received: 18 August 2021; Accepted: 29 September 2021;
Published: 16 November 2021.
Edited by:
Ren-You Gan, Institute of Urban Agriculture, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Anusorn Cherdthong, Khon Kaen University, ThailandCopyright © 2021 Guo, Guo, Cao, Guo, Li, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Li, bGZlaUBsenUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.