
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr. , 23 December 2021
Sec. Nutritional Immunology
Volume 8 - 2021 | https://doi.org/10.3389/fnut.2021.746592
This article is part of the Research Topic Nutrients, Gut Microbiome, and Intestinal Inflammation View all 35 articles
Age-related gut barrier dysfunction and dysbiosis of the gut microbiome play crucial roles in human aging. Dietary methionine restriction (MR) has been reported to extend lifespan and reduce the inflammatory response; however, its protective effects on age-related gut barrier dysfunction remain unclear. Accordingly, we focus on the effects of MR on inflammation and gut function. We found a 3-month methionine-restriction reduced inflammatory factors in the serum of aged mice. Moreover, MR reduced gut permeability in aged mice and increased the levels of the tight junction proteins mRNAs, including those of occludin, claudin-1, and zona occludens-1. MR significantly reduced bacterial endotoxin lipopolysaccharide concentration in aged mice serum. By using 16s rRNA sequencing to analyze microbiome diurnal rhythmicity during 24 h, we found MR moderately recovered the cyclical fluctuations of the gut microbiome which was disrupted in aged mice, leading to time-specific enhancement of the abundance of short-chain fatty acid-producing and lifespan-promoting microbes. Moreover, MR dampened the oscillation of inflammation-related TM7-3 and Staphylococcaceae. In conclusion, the effects of MR on the gut barrier were likely related to alleviation of the oscillations of inflammation-related microbes. MR can enable nutritional intervention against age-related gut barrier dysfunction.
Aging is accompanied by a decline in the functional capacity of body systems, including cognitive, cardiovascular, and physiological health. The gut has been reported to play crucial roles in the aging process (1). Moreover, aging can alter microbial abundance and cause microbiota dysbiosis (2). Loss of gut microbiota diversity can increase chronic low-grade inflammation and reduce cognitive function during aging (3). The gut microbes could produce short-chain fatty acids (SCFAs) from carbohydrate fermentation. It has been reported that Sutterella, Bacteroides, Lactobacillus, Prevotella and Bacteroidales are SCFAs producing microbe (4). Moreover, Desulfovibrionaceae, Staphylococcus, and Ruminococcaceae lipopolysaccharidesare has been reported to be associated with the microbiota inflammatory properties (5, 6). Several studies have revealed that elderly adults and aged mice have lower levels of Firmicutes, Actinobacteria, and SCFA-producing microbiota (e.g., Lachnospiraceae, Faecalibaculum, and Ruminococcaceae) and higher levels of inflammatory gastrointestinal bacteria, such as Proteobacteria, Desulfovibrio, and Staphylococcus, compared with the microbiome of younger adults and mice (7–10). Age-related changes in the gut microbiome are strongly related to intestinal barrier permeability and age-associated inflammation of the host. Christensenella, Akkermansia, and Bifidobacterium, showed potential life span–promoting effects (11–13). Furthermore, Escherichia coli can induce cognitive impairment and colitis, which is increased in the feces of aged mice and elderly adults (14).
Mammalian circadian clock system is composed of suprachiasmatic nucleus (SCN) in the hypothalamus, which acts as the central pacemaker, and other cells/tissues, such as the liver, gut, and muscle, which act as peripheral oscillators (15, 16). Aging has a negative effect on the circadian clock, and animals and humans show impairment of rest-activity rhythms with age. SCN neurons can disrupt the circadian phase, and the amplitude is reduced in peripheral oscillators with age (17, 18). Mutation of circadian clock gene reduces lifespan and increases oxidative damage and neuronal degeneration in Drosophila (19). In addition, disorders of the circadian clock system affect microbial community gene expression, decrease host immune ability and increase the synthesis and transportation of lipopolysaccharide (LPS); these changes are similar to those observed in the gut microbiome of Alzheimer's disease patients (20, 21). Interestingly, the circadian clock is involved in gut microbiota-gut epithelium crosstalk and regulates multiple functions, including intestinal permeability, body composition, and the immune response (22–24). Notably, the abundance of gut bacteria exhibit daily oscillations over a 24-h period (25, 26). The total biomass of the gut microbiome also shows diurnal fluctuations, affecting more than 20% of functional pathways (27). Furthermore, the oscillating pattern of genes in germ-free and wild-type mice was quite different, suggesting that the gut microbiome is associated with host molecular rhythms (28). Similarly, germfree or antibiotic-treated mice exhibit reduced expression of clock and metabolic genes and significantly dampened diurnal histone signaling owing to a lack of histone deacetylase 3 expression (22, 29). The diurnal rhythms of the gut microbiome may contribute to the development of healthy host states. However, whether the circadian rhythm affects the aging process by regulating the gut microbiome remains unclear.
Dietary restriction (DR) extends lifespan in various organisms, including rat, mouse, and fruit fly models, and prevents age-related circadian reprograming in various tissues (30, 31). Methionine is a sulfur-containing essential amino acid that is enriched in animal products. Methionine restriction (MR) can mimic the effects of DR and is effective for suppressing proliferation, increasing longevity, and alleviating inflammation and obesity (32). More specifically, both 80 and 40% MR diets reduce mitochondrial reactive oxygen species generation, which may lead to a healthy lifespan in rodent models (32). In MR-treated mice, the inflammatory responses in the liver and white adipose tissue were found to be significantly downregulated (33). Notably, MR (0.172%) decreases endogenous oxidative molecular damage in the rat brain, further ameliorating aging-related neurodegenerative diseases (34). In addition, a recent study showed that MR improves gut function by regulating the gut microbiome and reducing intestinal permeability in high-fat diet (HFD)-fed mice (35), and feeding of an MR diet for 1 month leads to sex-specific changes in the gut microbiome (36). Moreover, MR time-specifically increases the abundance of SCFA-producing bacteria and decreases the inflammation-related bacteria Desulfovibrionales and Staphylococcaceae (37), and restriction of sulfur amino acids improves gut barrier function and upregulates claudins (38). Taken together, these findings support that MR may alter the gut microbiome in aged mice. However, whether MR affects diurnal fluctuations in the gut microbiome and the potential effects of MR on age-related gut homeostasis remain unknown. Accordingly, in this study, we revealed the effects of MR on age-related inflammation and gut barrier damages.
Fifteen-month-old and two-month-old male C57BL/6J mice (Vital River Laboratory Animal Technology, Beijing, China) were housed under a 12/12-h light-dark cycle (lights on 08:00, lights off 20:00). Fifteen-month-old mice were randomized into two groups after adaptively feeding. Aged group fed a standard chow diet and MR group fed a MR diet for 3 months. Two-month-old mice were fed a standard chow diet. The detailed dietary compositions of these diets are provided in Supplementary Materials. We have obeyed relevant ethical regulations and research were approved in advance by Northwest A&F University (approval no. N81803231).
Total RNA was isolated from frozen colon using TRIzol method, diluted to uniform concentration, and reverse-transcribed into cDNA. RT-qPCR was performed using an UltraSYBR Mixture (Cowin Bio., Jiangsu, China) on a CFX96TM real-time system (Bio-Rad, CA, USA), operated at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. The 2−ΔΔCT method was used to calculated the mRNA expression. Gene-specific mouse primers were used as listed in Supplementary Materials.
Serum samples were obtained by eyeball extirpating under anesthesia. Serum LPS contents were detected by ELISA kits (Xinle Bio., Shanghai, China).
The proximal colons were fixed in paraformaldehyde (4% in PBS, v/v) and embedded in paraffin. For H&E staining, colon sections were cut and stained with hematoxylin and eosin. Three animals per group were used for the assessment of goblet cell numbers. The number of goblet cells on each villus was counted.
For immunofluorescence staining, sections were exposed to Claudin-1 antibodies (Abcam, Ab15098, USA) at 4°C for 10 h, and incubated with biotinylated antibodies. After DAPI staining, the sections were sealed with antifading mounting medium (Solarbio, Beijing, China). Stained sections were observed using an inverted fluorescence microscope (Olympus, Tokyo, Japan).
Ussing chamber assays were performed as previously described (39). Briefly, 1.5 cm colons were unfolded and placed on Ussing chambers (KingTech, Beijing, China). The chamber was separated into two parts by the unfolded tissue. The tissue was exposed to carbogen-gassed Krebs buffers (7.35 g CaCl2·2H2O, 13.67 g NaCl, 7.01 g KCl, 4.2 g NaHCO3, 4.88 g MgCl2·6H2O, 3.96 g glucose, 3.74 g NaH2PO4·2H2O, dissolved in 2 L ddH2O, pH 7.4) at 37°C. For permeability measurements, fluorescein (0.09 g/L) were replaced on the luminal side and the fluorescence intensity of the serosal buffer was determined on the other side.
Fecal samples were collected every 4 h over 24 h before sacrifice through repeated sampling from the same mice after dietary intervention. Total DNA was isolated using an E.Z.N.A. Stool DNA Extraction Kit (Omega, GA, USA), and the V3 to V4 regions of the 16S rRNA gene were amplified using region-specific primers (341_F: 5′-CCTACGGGNGGCWGCAG-3′; and 802_R: 5′-TACNVGGGTATCTAATCC-3′). Raw reads were merged, trimmed, and denoised to construct operational taxonomic units (OTUs) and identified using Usearch (version 7.1). Venn diagram evaluation, β-diversity assessment, principal coordinate analysis (PCoA), and partial least squares-discriminant analysis (PLS-DA) were performed using Qiime2 (2018.11) combined with the R-package vegan and ANCOM2. Taxonomy-based analyses were used to identify significant differences in phylotypes under MR treatment at distinct energy densities. Biomarker-discovery responses to MR were analyzed by linear discriminant analysis (LDA) effect size (LEfSe). Significant biomarkers were detected using the default threshold of α value < 0.05 and LDA score >2.
Gut microbiome data were reported as mean ± SEM, other data were reported as max, min, and median. Significant differences between mean values were determined by one-way ANOVA. For multiple comparisons, Tukey's test was performed using GraphPad Prism 7.0 software. Means were considered significantly different when the p-value was <0.05. JTK_CYCLE (v3.1) was used to determine whether an OTU was cyclical (ADJ. p < 0.05) (40).
Aged mice were fed with an MR diet for 3 months. MR feeding significantly decreased the body weights of aged mice (p < 0.01, Figure 1A). However, MR-fed aged mice consumed more food compared to control diet-fed aged mice (p < 0.01, Figure 1B). Moreover, methionine intake was significantly decreased under MR feeding (p < 0.01, Figure 1C). The inflammatory response during aging is one of the causes of other age-related symptoms. To evaluate the effects of MR on age-induced systemic inflammation, which have been reported to be a risk factor for age-related biological degradation, inflammatory factors were investigated. As shown in Figures 1D,E, interleukin (IL)-1β and tumor necrosis factor (TNF)-α levels in serum were dramatically increased in aged mice, and this effect was reversed in MR-fed aged mice (Figures 1D,E, p < 0.05).
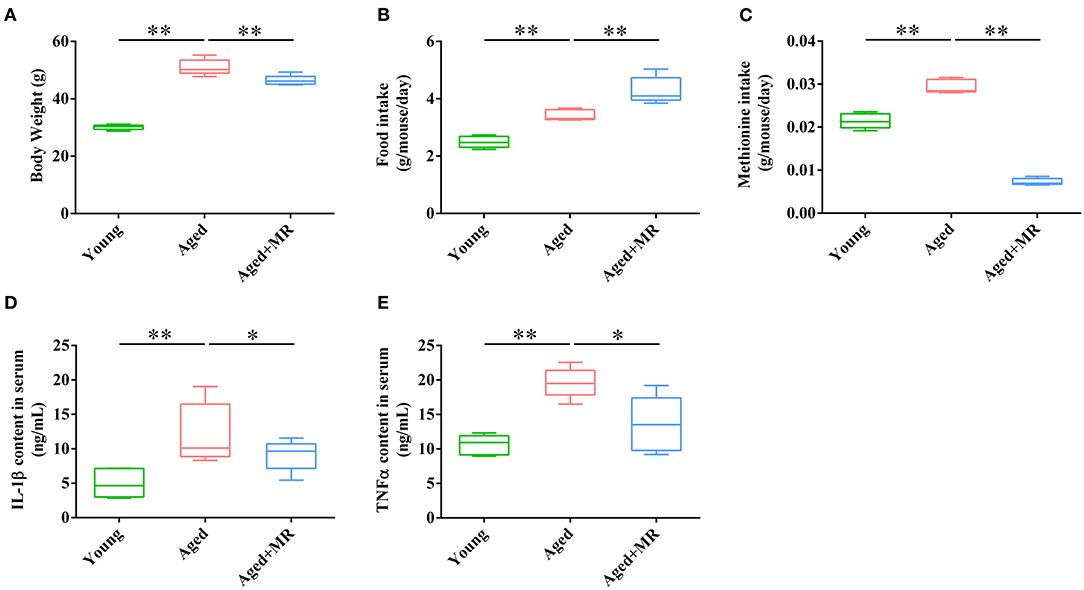
Figure 1. Effects of methionine restriction on the body weight, food intake, and serum cytokines concentrations of aged mice. (A) Body weight; (B) food intake; (C) methionine intake; (D) IL-1β content in serum; (E) TNF-α content in serum. The boxplot elements are defined as following: center line, median; box limits, upper and lower quartiles; whiskers, min to max (n = 5/group). Significant differences between mean values were determined using one-way ANOVA analysis of variance with Tukey's multiple comparison test; *p < 0.05, **p < 0.01.
Intestinal barrier integrity plays a critical role in age-related systemic inflammation. There is no significant difference of muscular thickness between young and aged mice. We found MR increased the villi length, and number of goblet cells in aged mice (p < 0.01, Figures 2A–D). Moreover, immunofluorescence staining for claudin-1 showed that aging decreased the expression of claudin-1, whereas MR increased claudin-1 in aged mice (Figure 2A). Furthermore, we observed that MR prevented gut leakage (Figure 2E). In addition, the expression levels of occludin, claudin-1, and zona occuldens-1 (Zo-1) mRNAs, which encode tight-junction proteins in the gut barrier, were also elevated by MR (p < 0.01, Figures 2F–H). The intestinal barrier is the first defense against LPS-induced inflammatory activation in the body. Consistent with this, LPS content was increased in aged mouse serum, whereas MR reversed the abnormal increase (p < 0.05, Figure 2I). This result showed that MR reduced age-related inflammation, possibly because of its effects on improving gut barrier function.
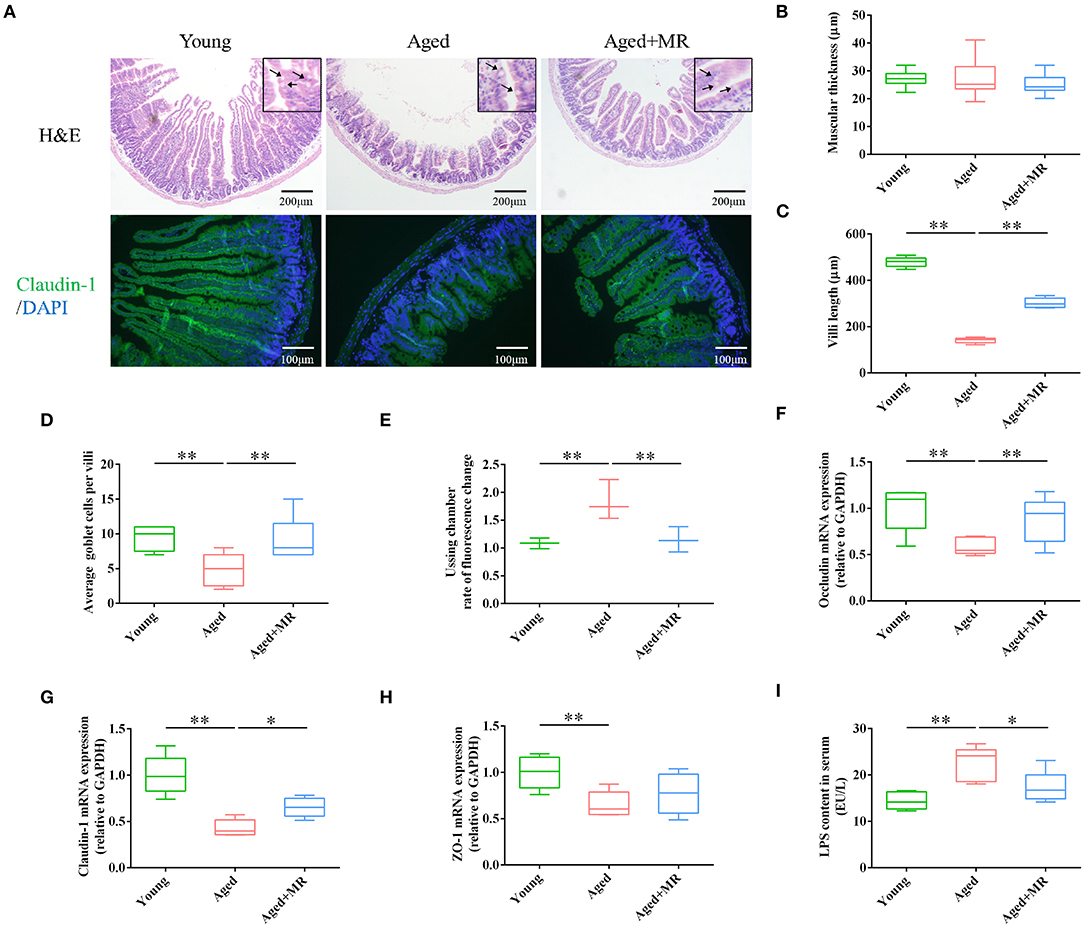
Figure 2. Effects of methionine restriction on the gut barrier integrity of aged mice. (A) Representative images of hematoxylin and eosin staining and immunofluorescence staining of claudin-1/DAPI in mouse colon tissues (n = 3/group). Arrows point to goblet cells. (B) Muscular thickness; (C) villus length; (D) average goblet cell per villi; (E) rate of fluorescence changes during Ussing chamber analysis (n = 3/group). (F–H) mRNA expression of occludin, claudin-1, and Zo-1; loading control: Gapdh (n = 5/group). (I) LPS content in serum. (n = 5/group). The boxplot elements are defined as following: center line, median; box limits, upper and lower quartiles; whiskers, min to max. Significant differences between mean values were determined using one-way ANOVA analysis of variance with Tukey's multiple comparison test; *p < 0.05, **p < 0.01.
We identified 1102 OTUs in mice fecal of 3 conditions. Venn diagram showed that microbial composition of mice in each group was significantly changed (Figure 3A). As shown in Figures 3B,C, the gut microbiome was distinctly different between the young and aged groups, as demonstrated by PCA and PLS-DA. Moreover, aging significantly increased the observed OTUs, Chao index, and ACE index compared with that in young mice (p < 0.01, Figures 3D–G). However, MR increased the Simpson index in aged mice (p < 0.01, Figure 3H). MR affected the relative abundance of gut microbiome. Specifically, compared with the young group, the proportion of Bacteroidetes decreased and the proportions of Firmicutes and Proteobacteria increased in the aged group. After MR treatment, an increase in the proportion of Bacteroidetes with a decrease in Firmicutes was observed (Figure 3I). LEfSe analysis suggested that Akkermansia, Verrucomicrobiaceae, and Bacteroidaceae were universal markers in the young group; Erysipelotrichaceae, Staphylococcus, and Helicobacteraceae were dominant microbes in the aged group; and Prevotella, Bacteroidales, Desulfovibrionales, and Bifidobacteriales were aged + MR-specific markers (Figures 3J,K).
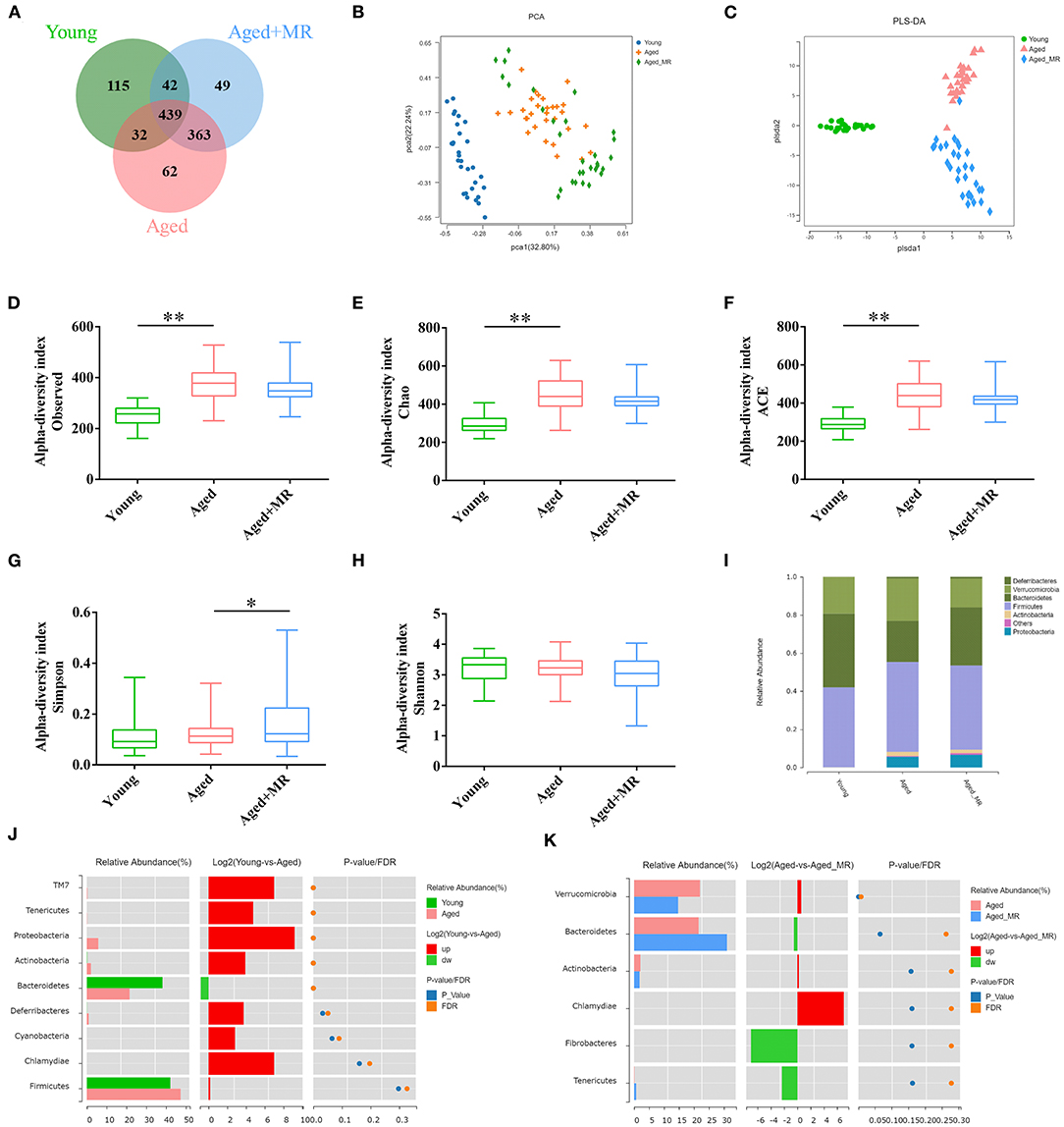
Figure 3. Effects of methionine restriction on the gut microbiota composition of aged mice. (A) Venn diagram displays the number of shared and unique OTUs among Young, Aged and Aged+MR. (B) Principal coordinate analysis (PCoA) and (C) Partial least squares discrimination analysis (PLS-DA) indicated the beta-diversity of gut microbiome.; (D) Observed species index, (E) Chao index, (F) Ace index, (G) Simpson index, and (H) Shannon index indicated the alpha-diversity of gut microbiome. The boxplot elements are defined as following: center line, median; box limits, upper and lower quartiles; whiskers, min to max. Significant differences between mean values were determined using one-way ANOVA analysis of variance with Tukey's multiple comparison test; *p < 0.05, **p < 0.01. (I) Gut microbiome composition at the phylum level. Differential analysis of the gut microbiome between (J) young and (K) aged + MR groups.
To investigate the stability of the relative abundance of gut microbes over the course of a day, we use JTK_CYCLE to define the rhythmicity of each OTU. Venn diagram analysis showed the distribution of OTUs that were considered to be rhythmic, with an ADJ. p-value <0.05, in each group (Figure 4A). Moreover, the gut microbiome of aged mice had the most rhythmic OTUs, accounting for 17.48% of the total number of OTUs. However, the gut microbiome of young mice and MR-fed aged mice ranged from 11 to 12% (Figure 4B). In OTUs with oscillations, peaks appeared at different times. In the young group, the cycling OTUs showed peaks distributed over a day (Figure 4C). The peaks of these OTUs changed in the feces of aged mice. However, these alterations were alleviated by MRD. The peaks of these OTUs were shifted rather than dramatically disturbed compared with those in young mice (Figure 4C). These data indicated that the composition of the gut microbiome changed over time and that MR could have different influences on the gut microbiome at different time points.
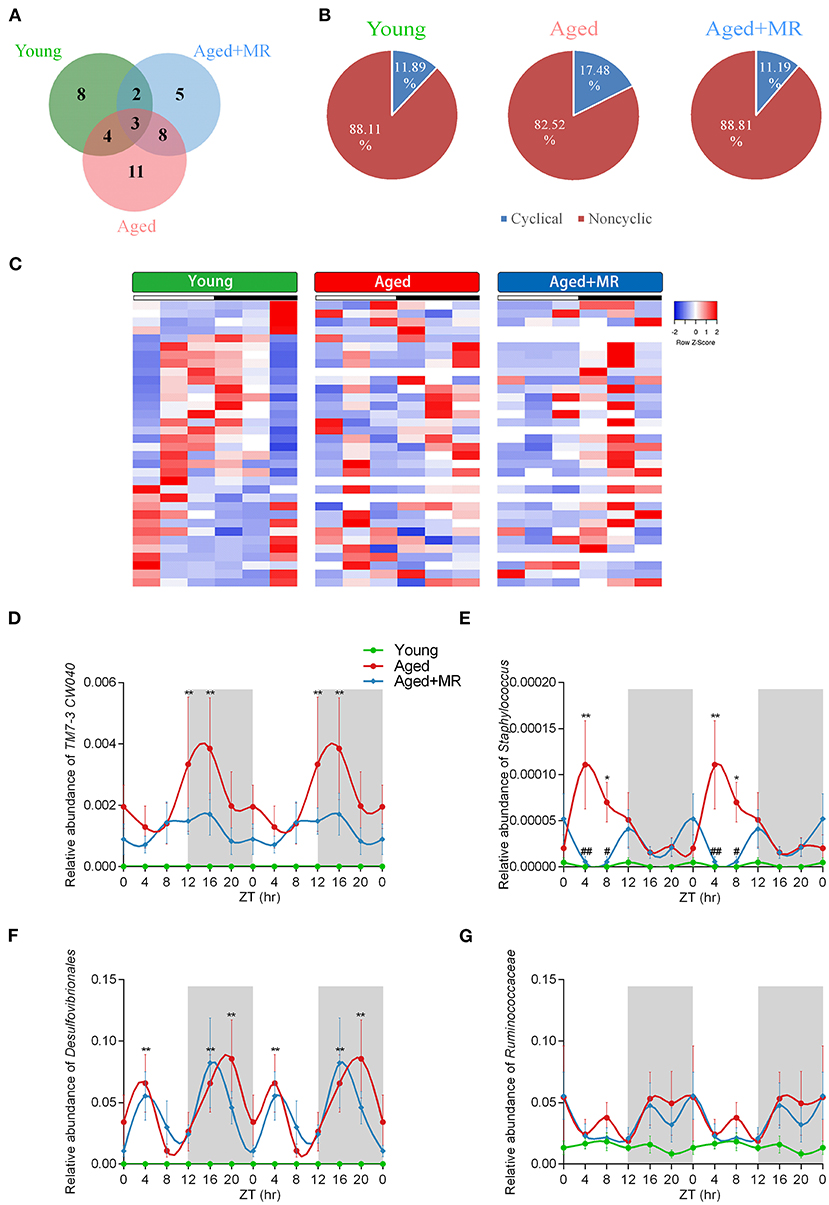
Figure 4. Effects of methionine restriction on diurnal rhythms of the gut microbiome in aged mice. (A) Venn diagrams displays the number of shared and unique cycling OTUs. (B) Proportion of cycling and non-cycling OTUs. (C) Heatmap of cycling OTUs organized by peak, from the young group. (D–G) Diurnal patterns of relative abundances of TM7-3 CW040, Staphylococcus, Desulfovibrionales, and Ruminococcaceae over 24 h. Data are double-plotted for clear visualization. Data are presented as means ± SEMs; n = 5/time point. *p < 0.05, **p < 0.01, vs. the young group; #p < 0.05, ##p < 0.01, vs. the aged group. Significant differences between means were determined using one-way ANOVA analysis of variance with Tukey's multiple-comparison test.
The results of analyses of inflammation-related microbiome, involving TM7-3 CW040, Staphylococcaceae, Desulfovibrionales, and Ruminococcaceae, showed that the aged group exhibited dramatic increases in TM7-3 CW040, Staphylococcaceae, and Desulfovibrionales compared with the young mice. No significant differences in Ruminococcaceae were detected between the young and aged groups. The relative abundance of Staphylococcaceae was lower in the aged + MR group than in the aged group at ZT4 and ZT8. Moreover, the aged group showed disordered cyclical oscillations of Staphylococcaceae, Desulfovibrionales, and Ruminococcaceae (ADJ. p = 0.18, 0.08, and 0.05, respectively). After MR feeding, the aged + MR group exhibited similar cyclical fluctuations in Staphylococcaceae, Desulfovibrionales, and Ruminococcaceae as the young group (Figures 4D–G).
To consider circadian effects, the family, genus, and species levels of the gut microbiome were evaluated. Specific differences in the relative abundances of these organisms are shown in Figure 5A, including SCFA-producing microbes (e.g., Prevotella, Bacteroidales, Bacteroides, Lachnospiraceae, and Sutterella) and potential life span-promoting microbes (e.g., Escherichia coli, Akkermansia, and Bifidobacterium), from the light to the dark phase. The aged + MR group showed a marked increase in the relative abundance of Prevotella compared with the aged group, particularly at ZT0 and ZT4. Moreover, the young group exhibited higher relative abundances of Bacteroidales, Bacteroides, and Lachnospiraceae than the aged groups, but had a lower relative abundance of Sutterella. A previous study demonstrated that aging could alter the Sutterella to Barneseilla ratio (41). Additionally, we found that MR significantly increased the relative abundance of Bacteroidales at ZT0. Although Lachnospiraceae has been found to increase SCFA levels, this microbe is also associated with obesity. Our findings showed that MR slightly decreased the relative abundance of Lachnospiraceae in the aged mice. In addition, in the young and aged + MR groups, Prevotella, Bacteroides, Lachnospiraceae, Escherichia coli, and Bifidobacterium exhibited similar cyclical oscillation, whereas the cyclical fluctuations of Bacteroides and Bifidobacterium were disrupted (ADJ. p = 0.17 and 0.08, respectively) compared with those in the young and aged + MR groups (Figure 5B). These results together indicated that MR enhanced the relative abundances of SCFA-producing and life span-promoting microbiomes and restored the diurnal fluctuations of specific microbiomes in aged mice.
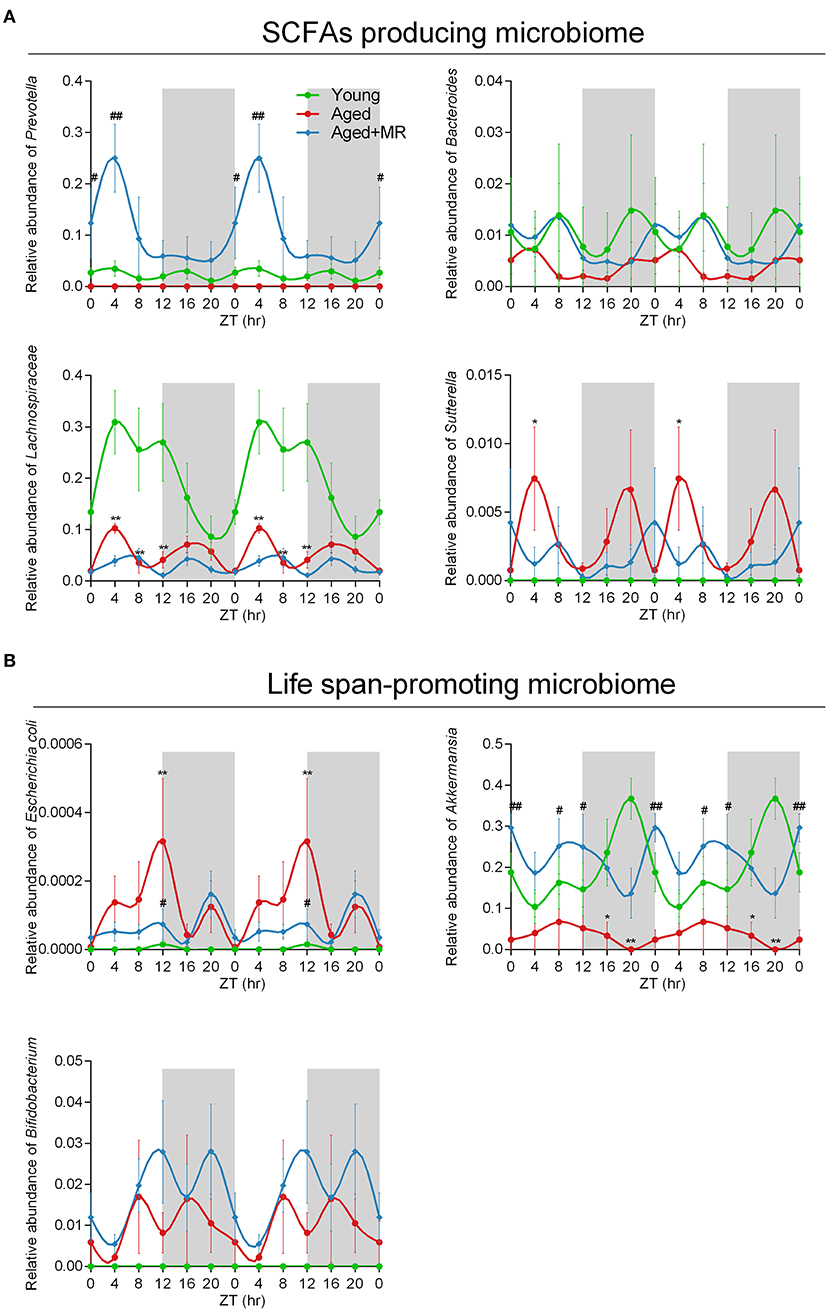
Figure 5. Methionine restriction increased the abundances of SCFA-producing and life span-promoting microbes. (A) Diurnal patterns of the relative abundances of Prevotella, Bacteroides, Lachnospiraceae, and Sutterella over 24 h. (B) Diurnal patterns of the relative abundances of Escherichia coli, Akkermansia, and Bifidobacterium over 24 h. Data are presented as means ± SEMs; n = 5/time point. *p < 0.05, **p < 0.01, vs. the young group; #p < 0.05, ##p < 0.01, vs. the aged group. Significant differences between means were determined using one-way ANOVA analysis of variance with Tukey's multiple-comparison test.
To clarify the correlations among the characteristic indicators, tight junction proteins, and inflammatory factors, Pearson's correlation analysis was performed (Figure 6). The results showed that characteristic indicators and specific rhythm microbiomes were highly correlated. More specifically, Met intake was positively correlated with serum TNF-α levels (Pearson's coefficient = 0.83). Importantly, the correlation of oscillation characteristic indicators and abundance in the light period (ZT4, ZT8, and ZT12) was slightly higher than that in the dark period (ZT16, ZT20, and ZT0).
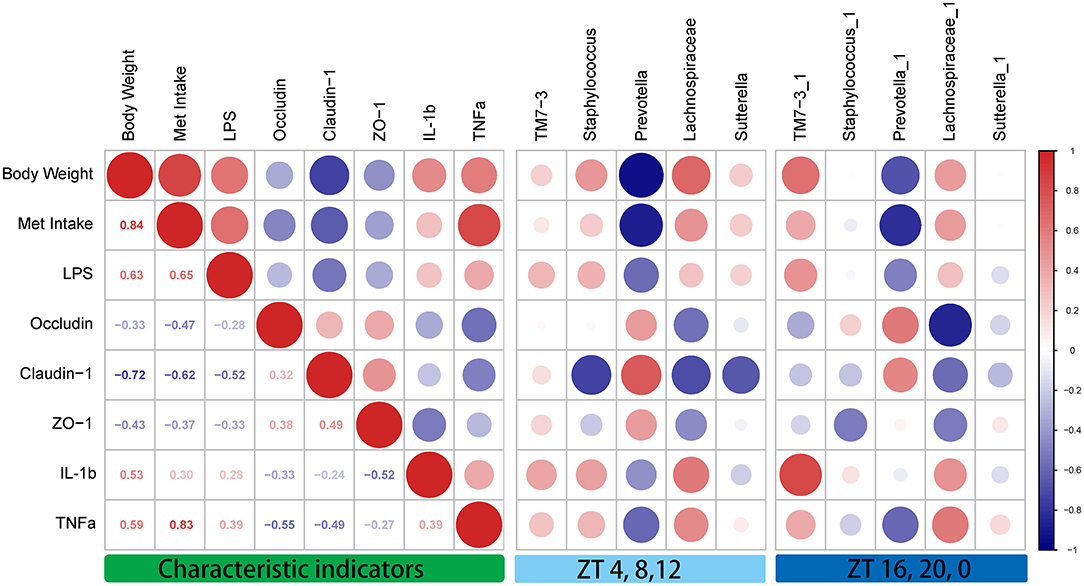
Figure 6. Correlation analysis between the characteristic indicators and relative abundances of the gut microbiome during daytime (ZT4, ZT8, and ZT12) and nighttime (ZT16, ZT20, and ZT0). Pearson's correlation analysis was performed for body weight, methionine intake, inflammatory factors, and relative abundance of the gut microbiome. Red, positive correlation; blue, negative correlation.
In this paper, the protective effects of MR on age-related systemic inflammation were investigated. The mechanism may involve protecting the gut barrier and reducing LPS, consistent with the results of reshaping the gut microbiome (Figure 7). We also found that intestinal tight junction proteins were upregulated and the age-related rhythmic disturbance of intestinal bacteria was alleviated by MR.
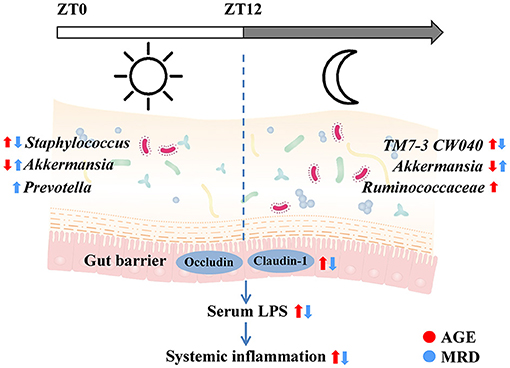
Figure 7. Methionine restriction alleviated systemic inflammation in aged mice by reshaping the gut microbiome, improving gut barrier function, and reducing serum lipopolysaccharide.
The effects of MR on lifespan extension have been reported in various animal models (32). Recent studies have shown that MR can alleviate age-related bone density reduction, cognitive dysfunction, systemic inflammation, and oxidative stress (35, 42–44). Moreover, during aging, intestinal function gradually degrades, which may facilitate other age-related degenerative processes. Aging leads to aging of intestinal cells, damage to intestinal barrier function, and reduction of intestinal tight junction protein levels, which in turn promotes the degradation of overall body function (3, 45, 46). Additionally, improving aging-related intestinal barrier damage can extend lifespan (46, 47). Mechanistically, MR can promote the production of glutathione and regulate the production of H2S by activating the nutritional response signal fibroblast growth factor 21, thereby prolonging lifespan and reducing inflammation (48, 49). However, some studies have shown that the anti-inflammatory effects of MR are partly independent of these mechanisms (50). Furthermore, the regulatory effects of MR on the intestinal function of aging mice have not been clearly studied. In an HFD-induced obese mouse model, an MRD reshaped the gut microbiome, reduced inflammatory factors, and ultimately alleviated metabolic syndrome in obese mice (35, 37). MR alleviates fat accumulation and inflammatory responses by reshaping gut microbiome of 8-month-old mice (51). In young mice, dietary methionine supplementation alters one-carbon metabolism and DNA methylation in the proximal jejunum, and alters the normal gut physiology (52). Increasing or decreasing the level of methionine in the diet will cause changes in the composition of gut microbes, with some differences between males and females (36). Consistent with this, in our study, dramatic changes in gut microbiome composition were found in MR-treated 18-month-old mice, with a lower level of inflammatory factors.
Intestinal barrier integrity is necessary for maintaining the stability of the body. Gut microbial disruption leads to compromised gut barrier function and sustained systemic inflammation (53). Microbiome-host interactions are related to the inflammatory factors production in humans (54), and LPS produced by the gut microbiota accelerates inflammation and aging in mice (55). In the current study, we found that MR reduced serum LPS levels in aged mice and that LPS levels were correlated with Met intake. In addition, the increase in LPS associated with age further activates the intestinal inflammatory factors TNF-α and IL-1β, triggering a systemic inflammatory response. In another study in our group, an increase in LPS caused by aging was found to exacerbate the cognitive impairment associated with aging (56). Furthermore, damage to the intestinal barrier function and activation of inflammatory factors have been used as triggers or biomarkers for Crohn's disease, Parkinson's disease, and metabolic syndrome (57–59). We found that MR reduced the levels of serum LPS and inflammatory factors in aged mice, which may be related to enhancement of the integrity of the intestinal barrier and upregulation of tight junction proteins by MR. The results of correlation analysis also supported this speculation.
The gut microbiome has a circadian rhythm similar to that of organisms, and this rhythm plays critical roles in the biological functions of the host, including digestion, absorption, and metabolism (60, 61). Feeding time and food composition have been shown to affect the diurnal rhythm of the gut microbiome (62, 63). In our study, MR affected the diurnal rhythms of some specific intestinal bacteria, particularly those related to inflammation-associated, SCFA-producing, and lifespan-promoting microbes. The relative abundance of TM-7 has been reported to be increased in the intestine and oral cavity of patients with inflammatory bowel disease (64, 65). In addition, the relative abundances of Desulfovibrionaceae and Staphylococcaceae, related to the activation of host inflammatory factors, are down-regulated by MR. In particular, Desulfovibrio and other sulfate-reducing bacteria, hydrogen sulfide producer, are related to inflammatory response (66). Sutterella, Bacteroides, Lactobacillus, Prevotella, and Bacteroidales are associated with SCFA production (4). Akkermansia improves the colonic mucus thickness decline and attenuates immune activation in aged mice (67). Bifidobacterium and Lactobacillus have been reported to impair brain function and further lead to neurodegenerative disorders (68). We found that MR increased the relative abundance of Staphylococcaceae during the daytime and decrease the relative abundance of TM7-3 CW040 at night in aged mice. MR has no difference in up-regulating the relative abundance of Akkermansia between a day.
This study has potential limitations. Firstly, the synchronism between gut microbiota and gut barrier function upon MR diets would be estimated if there are serum LPS concentrations with different time points. Secondly, there may be commensal bacteria that become opportunistic pathogens only under certain conditions. The classification of inflammation-related, SCFA producing and age-related microbes in presented study may not be applicable to all pathologies. Metagenomics sequencing could provide more precise information on how gut microbiota affects host functions. Thirdly, we only chose a single dose of Met-restricted diet commonly used in MR studies. The effect of different Met intakes on the gut microbiome is not clear. A wider range of Met-restricted level will reveal a clearly correlation between Met intake and the relative abundance of gut bacteria. Moreover, feeding time was not monitored in this study, which was proved to have great influence on the diurnal rhythm of the gut microbiome. The relationship between dietary methionine content and the relative abundance of gut microbiome on aging and aging-related gut dysfunction are under further investigation.
In this study, we found that MR alleviated systemic inflammation, improved gut barrier function, and reshaped the gut microbiome. The effects of MR on Staphylococcus, TM7-3 CW040, and Prevotella varied during the 24-h observation period. These findings provide important insights into the application of MR for prevention of age-related gut barrier damage and inflammation-related diseases.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA747265.
The animal study was reviewed and approved by Animal Studies Committee at Northwest A&F University.
BR, LW, AM, and YL performed the experiments and analyzed the data. ZL and XL designed the study. BR, LW, and ZL wrote the paper. BR, AM, and YL prepared the figures. All authors discussed the results and commented on the paper.
This work was financially supported by the National Natural Science Foundation of China (Nos. 32072214 and 81871118).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Taylor & Francis Editing Services (www.tandfeditingservices.cn) for its linguistic assistance during the preparation of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.746592/full#supplementary-material
1. Branca JJ, Gulisano M, Nicoletti C. Intestinal epithelial barrier functions in ageing. Ageing Res Rev. (2019) 54:100938. doi: 10.1016/j.arr.2019.100938
2. Bana B, Cabreiro F. The microbiome and aging. Annu Rev Genet. (2019) 53:239–61. doi: 10.1146/annurev-genet-112618-043650
3. O'Toole PW, Jeffery IB. Gut microbiota and aging. Science. (2015) 350:1214–5. doi: 10.1126/science.aac8469
4. Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. (2018) 359:1151–6. doi: 10.1126/science.aao5774
5. Zhang-Sun W, Augusto LA, Zhao L, Caroff M. Desulfovibrio desulfuricans isolates from the gut of a single individual: structural and biological lipid A characterization. FEBS Lett. (2015) 589:165–71. doi: 10.1016/j.febslet.2014.11.042
6. Sun L, Bao L, Phurbu D, Qiao S, Sun S, Perma Y, et al. Amelioration of metabolic disorders by a mushroom-derived polyphenols correlates with the reduction of Ruminococcaceae in gut of DIO mice. Food Sci Human Wellness. (2021) 10:442–51. doi: 10.1016/j.fshw.2021.04.006
7. D'Amato A, Mannelli LDC, Lucarini E, Man AL, Le Gall G, Branca JJ, et al. Faecal microbiota transplant from aged donor mice affects spatial learning and memory via modulating hippocampal synaptic plasticity-and neurotransmission-related proteins in young recipients. Microbiome. (2020) 8:1–19. doi: 10.1186/s40168-020-00914-w
8. Fransen F, van Beek AA, Borghuis T, Aidy SE, Hugenholtz F., van der Gaast–de Jongh C, et al. Aged gut microbiota contributes to systemical inflammaging after transfer to germ-free mice. Front Immunol. (2017) 8:1385. doi: 10.3389/fimmu.2017.01385
9. Xu C, Zhu H, Qiu P. Aging progression of human gut microbiota. BMC Microbiol. (2019) 19:1–10. doi: 10.1186/s12866-019-1616-2
10. Loubinoux J, Bronowicki J-P, Pereira IA, Mougenel J-L, Le Faou AE. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol Ecol. (2002) 40:107–12. doi: 10.1111/j.1574-6941.2002.tb00942.x
11. Biagi E, Franceschi C, Rampelli S, Severgnini M, Ostan R, Turroni S, et al. Gut microbiota and extreme longevity. Curr Biol. (2016) 26:1480–5. doi: 10.1016/j.cub.2016.04.016
12. Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi J, Verschoor CP, et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. (2017) 21:455–66. e4. doi: 10.1016/j.chom.2017.03.002
13. Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE. (2010) 5:e10667. doi: 10.1371/journal.pone.0010667
14. Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci. (2011) 108:4586–91. doi: 10.1073/pnas.1000097107
15. Paschos GK, FitzGerald GA. Circadian clocks and metabolism: implications for microbiome and aging. Trends Genet. (2017) 33:760–9. doi: 10.1016/j.tig.2017.07.010
16. Chaix A, Manoogian EN, Melkani GC, Panda S. Time-restricted eating to prevent and manage chronic metabolic diseases. Annu Rev Nutr. (2019) 39:291–315. doi: 10.1146/annurev-nutr-082018-124320
17. Farajnia S, Michel S, Deboer T., Tjebbe vanderLeest H, Houben T, Rohling JH, et al. Evidence for neuronal desynchrony in the aged suprachiasmatic nucleus clock. J Neurosci. (2012) 32:5891–9. doi: 10.1523/JNEUROSCI.0469-12.2012
18. Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Effects of aging on central and peripheral mammalian clocks. Proc Nat Acad Sci. (2002) 99:10801–6. doi: 10.1073/pnas.152318499
19. Krishnan N, Kretzschmar D, Rakshit K, Chow E, Giebultowicz JM. The circadian clock gene period extends healthspan in aging Drosophila melanogaster. Aging. (2009) 1:937. doi: 10.18632/aging.100103
20. Deaver JA, Eum SY, Toborek M. Circadian disruption changes gut microbiome taxa and functional gene composition. Front Microbiol. (2018) 9:737. doi: 10.3389/fmicb.2018.00737
21. Liu P, Wu L, Peng G, Han Y, Tang R, Ge J, et al. Altered microbiomes distinguish Alzheimer's disease from amnestic mild cognitive impairment and health in a Chinese cohort. Brain Behav Immun. (2019) 80:633–43. doi: 10.1016/j.bbi.2019.05.008
22. Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. (2013) 153:812–27. doi: 10.1016/j.cell.2013.04.020
23. Summa KC, Voigt RM, Forsyth CB, Shaikh M, Cavanaugh K, Tang Y, et al. Disruption of the circadian clock in mice increases intestinal permeability and promotes alcohol-induced hepatic pathology and inflammation. PLoS ONE. (2013) 8:e67102. doi: 10.1371/journal.pone.0067102
24. Wang Y, Kuang Z, Yu X, Ruhn KA, Kubo M, Hooper LV. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science. (2017) 357:912–6. doi: 10.1126/science.aan0677
25. Frazier K, Chang EB. Intersection of the gut microbiome and circadian rhythms in metabolism. Trends Endocrinol Metab. (2020) 31:25–36. doi: 10.1016/j.tem.2019.08.013
26. Parkar SG, Kalsbeek A, Cheeseman JF. Potential role for the gut microbiota in modulating host circadian rhythms and metabolic health. Microorganisms. (2019) 7:41. doi: 10.3390/microorganisms7020041
27. Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. (2014) 159:514–29. doi: 10.1016/j.cell.2014.09.048
28. Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. (2015) 17:681–9. doi: 10.1016/j.chom.2015.03.006
29. Kuang Z, Wang Y, Li Y, Ye C, Ruhn KA, Behrendt CL, et al. The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science. (2019) 365:1428–34. doi: 10.1126/science.aaw3134
30. Fontana L, Partridge L, Longo VD. Extending healthy life span—from yeast to humans. Science. (2010) 328:321–6. doi: 10.1126/science.1172539
31. Solanas G, Peixoto FO, Perdiguero E, Jardí M, Ruiz-Bonilla V, Datta D, et al. Aged stem cells reprogram their daily rhythmic functions to adapt to stress. Cell. (2017) 170:678–92. e20. doi: 10.1016/j.cell.2017.07.035
32. Lee BC, Kaya A, Gladyshev VN. Methionine restriction and lifespan control. Ann N Y Acad Sci. (2016) 1363:116. doi: 10.1111/nyas.12973
33. Ghosh S, Wanders D, Stone KP, Van NT, Cortez CC, Gettys TW, et al. systems biology analysis of the unique and overlapping transcriptional responses to caloric restriction and dietary methionine restriction in rats. FASEB J. (2014) 28:2577–90. doi: 10.1096/fj.14-249458
34. Naudi A, Caro P, Jové M, Gómez J, Boada J, Ayala V, et al. Methionine restriction decreases endogenous oxidative molecular damage and increases mitochondrial biogenesis and uncoupling protein 4 in rat brain. Rejuvenation Res. (2007) 10:473–84. doi: 10.1089/rej.2007.0538
35. Yang Y, Zhang Y, Xu Y, Luo T, Ge Y, Jiang Y, et al. Dietary methionine restriction improves the gut microbiota and reduces intestinal permeability and inflammation in high-fat-fed mice. Food Funct. (2019) 10:5952–68. doi: 10.1039/C9FO00766K
36. Wallis KF, Melnyk SB, Miousse IR. Sex-specific effects of dietary methionine restriction on the intestinal microbiome. Nutrients. (2020) 12:781. doi: 10.3390/nu12030781
37. Wang L, Ren B, Hui Y, Chu C, Zhao Z, Zhang Y, et al. Methionine restriction regulates cognitive function in high-fat diet-fed mice: roles of diurnal rhythms of SCFAs producing-and inflammation-related microbes. Mol Nutr Food Res. (2020) 64:2000190. doi: 10.1002/mnfr.202000190
38. Skrovanek S, Valenzano M, Mullin JM. Restriction of sulfur-containing amino acids alters claudin composition and improves tight junction barrier function. Am J Physiol Regulat Integr Compar Physiol. (2007) 293:R1046–55. doi: 10.1152/ajpregu.00072.2007
39. Liu Z, Dai X, Zhang H, Shi R, Hui Y, Jin X, et al. Gut microbiota mediates intermittent-fasting alleviation of diabetes-induced cognitive impairment. Nat Commun. (2020) 11:1–14. doi: 10.1038/s41467-020-14676-4
40. Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. (2010) 25:372–80. doi: 10.1177/0748730410379711
41. Siddharth J, Chakrabarti A, Pannerec A, Karaz S, Morin-Rivron D, Masoodi M, et al. Aging and sarcopenia associate with specific interactions between gut microbes, serum biomarkers and host physiology in rats. Aging. (2017) 9:1698. doi: 10.18632/aging.101262
42. Ouattara A, Cooke D, Gopalakrishnan R. Huang T-h, Ables GP. Methionine restriction alters bone morphology and affects osteoblast differentiation. Bone Rep. (2016) 5:33–42. doi: 10.1016/j.bonr.2016.02.002
43. Ren B, Wang L, Shi L, Jin X, Liu Y, Liu RH, et al. Methionine restriction alleviates age-associated cognitive decline via fibroblast growth factor 21. Redox Biol. (2021) 41:101940. doi: 10.1016/j.redox.2021.101940
44. Sanchez-Roman I, Barja G. Regulation of longevity and oxidative stress by nutritional interventions: role of methionine restriction. Exp Gerontol. (2013) 48:1030–42. doi: 10.1016/j.exger.2013.02.021
45. Ghosh P, Swanson L, Sayed IM, Mittal Y, Lim BB, Ibeawuchi S-R, et al. The stress polarity signaling (SPS) pathway serves as a marker and a target in the leaky gut barrier: implications in aging and cancer. Life Sci Alliance. (2020) 3:e201900481. doi: 10.26508/lsa.201900481
46. Ahmadi S, Wang S, Nagpal R, Wang B, Jain S, Razazan A, et al. A human-origin probiotic cocktail ameliorates aging-related leaky gut and inflammation via modulating the microbiota/taurine/tight junction axis. JCI insight. (2020) 5:e132055. doi: 10.1172/jci.insight.132055
47. Kühn F, Adiliaghdam F, Cavallaro PM, Hamarneh SR, Tsurumi A, Hoda RS, et al. Intestinal alkaline phosphatase targets the gut barrier to prevent aging. JCI insight. (2020) 5:e134049. doi: 10.1172/jci.insight.134049
48. Richie JP Jr, Leutzinger Y, Parthasarathy S, Maixoy V, Orentreich N, Zimmerman JA. Methionine restriction increases blood glutathione and longevity in F344 rats. FASEB J. (1994) 8:1302–7. doi: 10.1096/fasebj.8.15.8001743
49. Choi K-M, Kim S, Kim S, Lee HM, Kaya A, Chun B-H, et al. Sulfate assimilation regulates hydrogen sulfide production independent of lifespan and reactive oxygen species under methionine restriction condition in yeast. Aging. (2019) 11:4254. doi: 10.18632/aging.102050
50. Sharma S, Dixon T, Jung S, Graff EC, Forney LA, Gettys TW, et al. Dietary methionine restriction reduces inflammation independent of FGF21 action. Obesity. (2019) 27:1305–13. doi: 10.1002/oby.22534
51. Wu G, Shi Y, Han L, Feng C, Ge Y, Yu Y, et al. Dietary methionine restriction ameliorated fat accumulation, systemic inflammation, and increased energy metabolism by altering gut microbiota in middle-aged mice administered different fat diets. J Agric Food Chem. (2020) 68:7745–56. doi: 10.1021/acs.jafc.0c02965
52. Miousse IR, Pathak R, Garg S, Skinner CM, Melnyk S, Pavliv O, et al. Short-term dietary methionine supplementation affects one-carbon metabolism and DNA methylation in the mouse gut and leads to altered microbiome profiles, barrier function, gene expression and histomorphology. Genes Nutr. (2017) 12:1–13. doi: 10.1186/s12263-017-0576-0
53. Banerjee S, Sindberg G, Wang F, Meng J, Sharma U, Zhang L, et al. Opioid-induced gut microbial disruption and bile dysregulation leads to gut barrier compromise and sustained systemic inflammation. Mucosal Immunol. (2016) 9:1418–28. doi: 10.1038/mi.2016.9
54. Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. (2016) 167:1125–36.e8. doi: 10.1016/j.cell.2016.10.020
55. Kim K-A, Jeong J-J, Yoo S-Y, Kim D-H. Gut microbiota lipopolysaccharide accelerates inflamm-aging in mice. BMC Microbiol. (2016) 16:1–9. doi: 10.1186/s12866-016-0625-7
56. Ren B, Yuan T, Zhang X, Wang L, Pan J, Liu Y, et al. Protective effects of sesamol on systemic inflammation and cognitive impairment in aging mice. J Agric Food Chem. (2020) 68:3099–111. doi: 10.1021/acs.jafc.9b07598
57. Dai X, Wang B. Role of gut barrier function in the pathogenesis of nonalcoholic fatty liver disease. Gastroenterology research and practice. (2015) 2015:287348. doi: 10.1155/2015/287348
58. Suenaert P, Bulteel V, Lemmens L, Noman M, Geypens B, Van Assche G, et al. Anti-tumor necrosis factor treatment restores the gut barrier in Crohn's disease. Am J Gastroenterol. (2002) 97:2000–4. doi: 10.1111/j.1572-0241.2002.05914.x
59. Aho VT, Houser MC, Pereira PA, Chang J, Rudi K, Paulin L, et al. Relationships of gut microbiota, short-chain fatty acids, inflammation, and the gut barrier in Parkinson's disease. Mol Neurodegener. (2021) 16:1–14. doi: 10.1186/s13024-021-00427-6
60. Voigt R, Forsyth C, Green S, Engen P, Keshavarzian A. Circadian rhythm and the gut microbiome. Int Rev Neurobiol. (2016) 131:193–205. doi: 10.1016/bs.irn.2016.07.002
61. Liang X, FitzGerald GA. Timing the microbes: the circadian rhythm of the gut microbiome. J Biol Rhythms. (2017) 32:505–15. doi: 10.1177/0748730417729066
62. Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. (2014) 20:1006–17. doi: 10.1016/j.cmet.2014.11.008
63. Zeb F, Wu X, Chen L, Fatima S., Haq I-u, Chen A, et al. Effect of time-restricted feeding on metabolic risk and circadian rhythm associated with gut microbiome in healthy males. Br J Nutr. (2020) 123:1216–26. doi: 10.1017/S0007114519003428
64. Kuehbacher T, Rehman A, Lepage P, Hellmig S, Fölsch UR, Schreiber S, et al. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol. (2008) 57:1569–76. doi: 10.1099/jmm.0.47719-0
65. Bor B, Bedree J, Shi W, McLean J, He X. Saccharibacteria (TM7) in the human oral microbiome. J Dent Res. (2019) 98:500–9. doi: 10.1177/0022034519831671
66. Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10–/– mice. Nature. (2012) 487:104–8. doi: 10.1038/nature11225
67. Van Der Lugt B, Van Beek AA, Aalvink S, Meijer B, Sovran B, Vermeij WP, et al. Akkermansia muciniphila ameliorates the age-related decline in colonic mucus thickness and attenuates immune activation in accelerated aging Ercc1–/Δ7 mice. Immun Ageing. (2019) 16:1–17. doi: 10.1186/s12979-019-0145-z
Keywords: methionine restriction, aging, microbiome diurnal rhythmicity, gut barrier, inflammation
Citation: Ren B, Wang L, Mulati A, Liu Y, Liu Z and Liu X (2021) Methionine Restriction Improves Gut Barrier Function by Reshaping Diurnal Rhythms of Inflammation-Related Microbes in Aged Mice. Front. Nutr. 8:746592. doi: 10.3389/fnut.2021.746592
Received: 24 July 2021; Accepted: 03 December 2021;
Published: 23 December 2021.
Edited by:
Guoxun Chen, The University of Tennessee, United StatesReviewed by:
Jian Tan, The University of Sydney, AustraliaCopyright © 2021 Ren, Wang, Mulati, Liu, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuebo Liu, eHVlYm9saXVAbndzdWFmLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.