
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr. , 08 September 2021
Sec. Nutrition and Food Science Technology
Volume 8 - 2021 | https://doi.org/10.3389/fnut.2021.740286
This article is part of the Research Topic Wheat Biofortification to Alleviate Global Malnutrition View all 11 articles
The dietary needs of humans for provitamin A carotenoids arise from their inability to synthesize vitamin A de novo. To improve the status of this essential micronutrient, special attention has been given to biofortification of staple foods, such as wheat grains, which are consumed in large quantities but contain low levels of provitamin A carotenoids. However, there remains an unclear contribution of metabolic genes and homoeologs to the turnover of carotenoids in wheat grains. To better understand carotenoid catabolism in tetraploid wheat, Targeting Induced Local Lesions in Genomes (TILLING) mutants of CCD4, encoding a Carotenoid Cleavage Dioxygenase (CCD) that cleaves carotenoids into smaller apocarotenoid molecules, were isolated and characterized. Our analysis showed that ccd4 mutations co-segregated with Poltergeist-like (pll) mutations in the TILLING mutants of A and B subgenomes, hence the ccd-A4 pll-A, ccd-B4 pll-B, and ccd-A4 ccd-B4 pll-A pll-B mutants were analyzed in this study. Carotenoid profiles are comparable in mature grains of the mutant and control plants, indicating that CCD4 homoeologs do not have a major impact on carotenoid accumulation in grains. However, the neoxanthin content was increased in leaves of ccd-A4 ccd-B4 pll-A pll-B relative to the control. In addition, four unidentified carotenoids showed a unique presence in leaves of ccd-A4 ccd-B4 pll-A pll-B plants. These results suggested that CCD4 homoeologs may contribute to the turnover of neoxanthin and the unidentified carotenoids in leaves. Interestingly, abnormal spike, grain, and seminal root phenotypes were also observed for ccd-A4 pll-A, ccd-B4 pll-B, and ccd-A4 ccd-B4 pll-A pll-B plants, suggesting that CCD4 and/or PLL homoeologs could function toward these traits. Overall, this study not only reveals the role of CCD4 in cleavage of carotenoids in leaves and grains, but also uncovers several critical growth traits that are controlled by CCD4, PLL, or the CCD4-PLL interaction.
Wheat is among the most widely cultivated and consumed staple food crops around the world. Wheat grains are a rich source of starch and proteins for human nutrition (1). In recent years, efforts have been directed toward enhancing the production of provitamin A carotenoids, particularly β-carotene, in wheat grains through breeding and biotechnology for improved vitamin A nutrition (2). However, provitamin A carotenoids produced in wheat grains may be subjected to degradation by carotenoid cleavage dioxygenases (CCDs) that cleave carotenoids (C40) into smaller apocarotenoid molecules (3). Therefore, it is imperative to better understand the activity and function of CCDs in wheat grains for achieving a high level of provitamin A carotenoid accumulation. Previous comparative analyses using CCDs from mouse [β-carotene 15,15′-monooxygenase-1 (BCMO1), BCMO2, and retinal pigment epithelium 65 (RPE65)], Synechocystis sp. PCC 6803 [lignostilbene dioxygenase (ACO)], and maize [9-cis-epoxycarotenoid dioxygenase (VP14)] revealed that CCDs from different kingdoms all contain four conserved histidine residues and additional acidic amino acids for binding of iron through hydrogen bonding, which is essential for the activity of the iron-dependent CCD enzymes (4).
Of the four CCDs (CCD1, CCD4, CCD7, and CCD8) identified in different plant species, CCD7 and CCD8 are dedicated to the biosynthesis of apocarotenoid phytohormones strigolactones, whereas CCD1 and CCD4 contribute to the production of various other apocarotenoid molecules in diverse tissues (5). Our recent study showed that CCD1 and CCD4 homoeologs are differentially expressed in tetraploid and hexaploid wheat grains (6). When assayed for enzyme activity using recombinant proteins, wheat CCD1 homoeologs, but not CCD4 homoeologs, converted β-carotene, lutein, and zeaxanthin to apocarotenoids, suggesting a role of CCD1 homoeologs in carotenoid degradation in wheat grains (6). However, genetic studies with Arabidopsis seeds demonstrated that AtCCD4 plays a major role in degradation of β-carotene during desiccation of Arabidopsis seeds, though AtCCD4 exhibited low in vitro enzyme activity toward β-carotene (7). Additionally, total carotenoids in chrysanthemum petals and the violaxanthin content in potato tubers were increased when CCD4 expression was downregulated via RNA interference (RNAi) in these tissues, suggesting its role in carotenoid cleavage in planta (8, 9). Taking into consideration these reports from wheat and other plants, it remains to be determined whether CCD4 could catalyze carotenoid cleavage reactions in wheat, which could be interrogated through genetic manipulation of CCD4 enzyme activity or gene expression. Emerging evidence has suggested the role of apocarotenoid signaling molecules, other than strigolactones and abscisic acid [ABA; produced by nine-cis-epoxycarotenoid dioxygenases (NCEDs)], in controlling plant growth, development, and interactions with the environment (5). Therefore, it will be important to also understand the function of apocarotenoid molecules generated by CCD4 in wheat.
To investigate the function of CCD4 homoeologs in carotenoid metabolism and plant growth in tetraploid wheat, we isolated Targeting Induced Local Lesions in Genomes (TILLING) mutants of CCD4 homoeologs from a tetraploid wheat mutant library in this study. CCD4 is located near a gene annotated as Poltergeist-like (PLL) on chromosome 6 of tetraploid wheat (Figure 1A). PLL and its homolog Poltergeist (POL) belong to the protein phosphatase type 2C (PP2C) family and contain metal ion-interacting domains with highly conserved amino acid residues (10). The Arabidopsis POL and PLL1 were shown to function in maintenance and differentiation of stem cells as well as additional developmental pathways such as formation of the central vasculature and embryo development (10–15). Although the Arabidopsis pol and pll1 single mutants only displayed weak developmental phenotypes relative to wild-type plants (10, 15), the pol pll1 double mutants were seedling lethal, indicating that both POL and PLL1 are crucial for plant development (13). Besides PLL1, there are four additional PLLs in Arabidopsis: PLL2-PLL5. While the mutant analysis demonstrated that PLL4 and PLL5 regulate leaf morphology, pll2 and pll3 mutants did not exhibit any distinguishable growth phenotypes when compared to wild-type plants, suggesting that PLL2 and PLL3 may not play a developmental role in Arabidopsis (12).
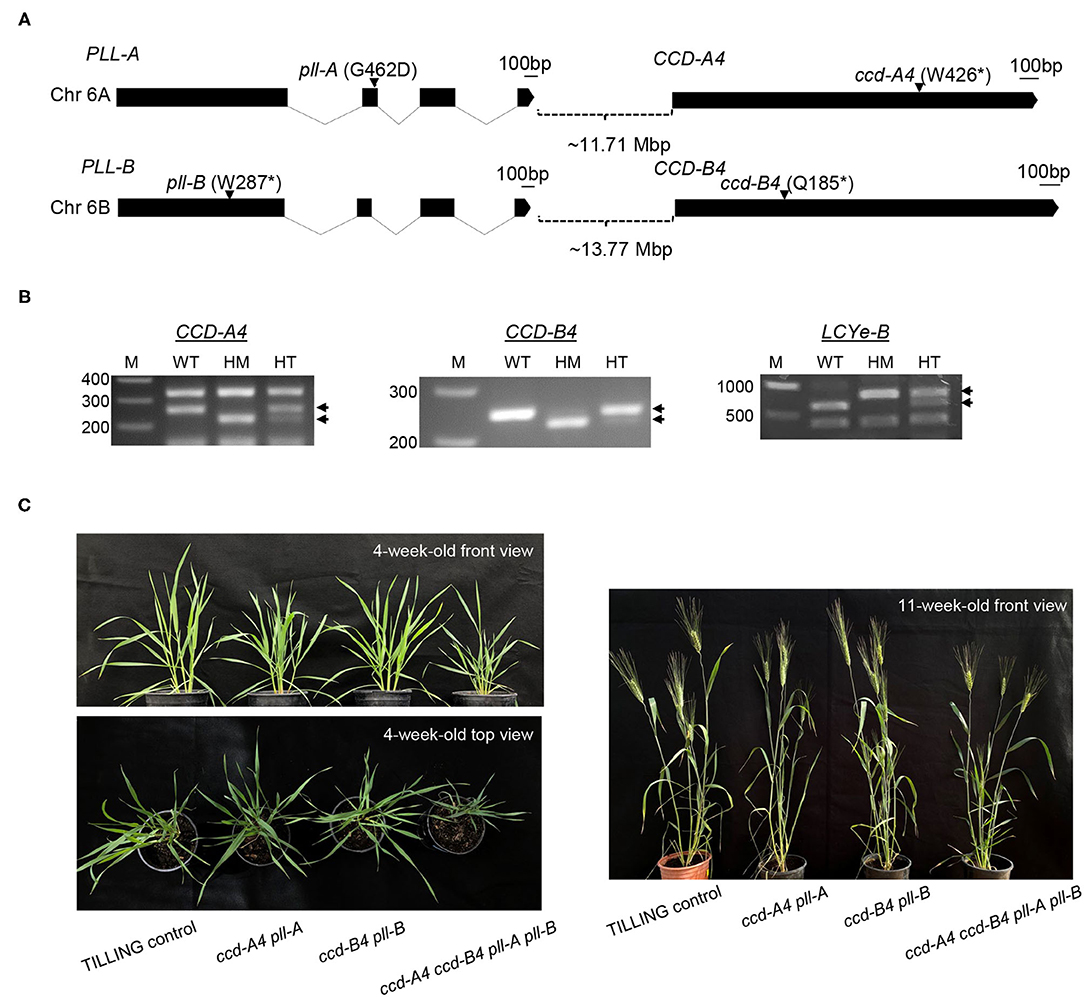
Figure 1. Tetraploid wheat TILLING mutants of CCD-A4, CCD-B4, PLL-A, and PLL-B. (A) Exon-intron diagrams of CCD-A4, CCD-B4, PLL-A, and PLL-B. Exons and introns are shown by black boxes and solid lines, respectively. The black bar denotes 100 bp. Locations of mutations are indicated with arrows. Mutant alleles of CCD4 and PLL homoeologs are linked and co-segregated in progenies of ccd-A4 and ccd-B4 mutants. Chr, chromosome; Mbp, megabase pair; *stop codon. (B) Cleaved Amplified Polymorphic Sequences (CAPS) and derived CAPS (dCAPS) markers for genotyping of wild-type and mutant CCD-A4, CCD-B4, and LCYe-B alleles. The PCR products were digested with HphI (CCD-A4 primer pair), RsaI (CCD-B4 primer pair), and DdeI (LCYe-B primer pair). Diagnostic bands for each marker are indicated with arrows. WT, homozygous wild type for the target gene; HM, homozygous mutant; HT, heterozygous mutant; M, DNA size marker. (C) Images of 4-week-old and 11-week-old TILLING control as well as ccd-A4 pll-A, ccd-B4 pll-B, and ccd-A4 ccd-B4 pll-A pll-B mutant plants.
Our analysis showed that the ccd4 TILLING mutant lines isolated in this study also contain mutations in PLL and that the ccd4 mutations are linked to the pll mutations in both A and B subgenomes in the backcrossed (BCed) progenies. Because CCD4 and PLL are closely located in the same chromosomal region, there is a low frequency of recombination between the two genes during meiosis and they are inherited together in the next generation. As such, the double (ccd-A4 pll-A and ccd-B4 pll-B) and quadruple (ccd-A4 ccd-B4 pll-A pll-B) mutants were analyzed in this study for biochemical and growth phenotypes and agronomic traits.
Wheat seeds were surface-sterilized using 1% (w/v) sodium hypochlorite solution containing 0.1% (v/v) Triton X-100, and rinsed with running water for at least 3 times. The sterilized seeds were placed on two layers of damp germination paper (Hoffman Manufacturing, Inc., Corvallis, OR) in a petri dish and stored at 4°C for 3 d to synchronize germination. The cold-treated seeds were subsequently moved to room temperature (~22°C) and germinated in the dark for 2–4 d to allow root development. The seedlings were then transplanted in soil and grown in a climate-controlled greenhouse (~22°C) under long-day conditions (16-h light/8-h dark). For leaf carotenoid and gene expression analyses, the fourth leaf counted from the top on the primary tiller of 4-week-old plants was collected, frozen in liquid nitrogen, and stored at −80°C until analysis. For grain carotenoid, spike, and grain yield analyses, spikes were collected from wheat plants at the harvest-ready stage and dried at room temperature (~22°C) for 1 week before the measurements. Grains were hand-cleaned to remove dry husks, and those harvested from the same plant were pooled and considered as one biological replicate. For grain carotenoid analysis, 25 whole grains were randomly sampled from each biological replicate and used for extraction.
Mutants of CCD-A4 and CCD-B4 (i.e., ccd-A4 and ccd-B4) were identified from the exome-sequenced TILLING mutant library of tetraploid wheat cv. Kronos (https://dubcovskylab.ucdavis.edu/wheat_blast) using the respective DNA sequences as queries. To reduce the additional mutations caused by ethyl methanesulfonate (EMS), the M4 ccd-A4 (line T4-0842) and ccd-B4 (line T4-3179) mutants were BCed to the wild-type parental plant Kronos for one generation. The BC1 ccd-A4 and ccd-B4 mutant plants were then intercrossed and the heterozygous progenies harboring ccd-A4 and ccd-B4 mutant alleles were chosen for self-pollination. From the segregating population of the self-pollinated plants, mutants that are homozygous for ccd-A4 or ccd-B4, or both ccd-A4 and ccd-B4 were selected. TILLING controls are plants containing wild-type CCD-A4 and CCD-B4 alleles, and have a mutational load similar to that of ccd-A4, ccd-B4, and ccd-A4 ccd-B4 mutants; TILLING controls were also selected from the segregating population. Cleaved Amplified Polymorphic Sequences (CAPS) and derived CAPS (dCAPS) markers were designed for CCD-A4 and CCD-B4 and used in the genotyping analysis (Supplementary Table 1).
A mutated lycopene ε-cyclase-B (LCYe-B) allele is also present in the M4 ccd-B4 plant (line T4-3179), but segregated independently from the ccd-B4 mutation when BCed to the wild-type Kronos plants. TILLING control and the homozygous BC1 ccd-A4, ccd-B4, and ccd-A4 ccd-B4 mutants were inspected for LCYe-B alleles using a CAPS marker (Supplementary Table 1); all of the above-mentioned genotypes contain the wild-type LCYe-B allele and were used in this study. The PLL homoeologs were amplified from the ccd-A4, ccd-B4, ccd-A4 ccd-B4 mutants and TILLING control and subjected to DNA sequencing (primer pairs are listed in Supplementary Table 2). This confirmed that the ccd-A4 mutant harbored pll-A and PLL-B alleles, the ccd-B4 mutant contained PLL-A and pll-B alleles, and the ccd-A4 ccd-B4 mutant carried pll-A and pll-B alleles. TILLING control plants were verified to possess only wild-type CCD-A4, CCD-B4, PLL-A, and PLL-B alleles. Because the ccd4 mutants also contain homozygous pll mutations, they were therefore designated the ccd-A4 pll-A, ccd-B4 pll-B, and ccd-A4 ccd-B4 pll-A pll-B mutants.
The GenBank accession numbers of the selected proteins are: Synechocystis sp. PCC 6803 lignostilbene dioxygenase, BAA18428; maize VP14, AAB62181; tetraploid wheat CCD-A4, KU975448; tetraploid wheat CCD-B4, KU975449; AtPLL1, NP_181078; AtPOL, NP_850463; tetraploid wheat PLL-A, XP_037447847 (this sequence from Triticum dicoccoides is identical to PLL-A from T. turgidum); tetraploid wheat PLL-B, XP_037453069 (this sequence from T. dicoccoides is identical to PLL-B from T. turgidum). The protein sequences were aligned using Clustal Omega (16) and the alignment diagrams were prepared using BoxShade (https://embnet.vital-it.ch/software/BOX_form.html).
Leaves and mature whole grains were ground into fine powder in liquid nitrogen using a mortar and pestle. Total carotenoids were extracted from ~50 mg leaves and ~300 mg whole grain flour using the methods described in (6) and (17), respectively. Carotenoids extracted from grain flour were saponified using 2 M KOH [dissolved in methanol containing 0.01% (w/v) butylated hydroxytoluene]. Following saponification in dark for 30 min, equal volumes of diethyl ether and H2O were added for phase separation. Re-extraction of carotenoids from the water phase, pooling and washing the diethyl ether layers, and drying and re-dissolving of carotenoid residues were carried out as described (17). Ten microliter of leaf or grain carotenoid extract (resuspended in ethyl acetate) was injected on a reverse-phase HPLC column and analyzed using a previously established gradient (18).
Total RNA was extracted from ~50 mg of ground leaves using a cetyltrimethylammonium bromide (CTAB)-based method (19). After treatment with RNase-free DNase I (Fermentas, Glen Burnie, MD), reverse transcription was carried out using the iScript™ Advanced cDNA Synthesis Kit (BioRad, Hercules, CA) following the manufacturer's instructions. For each qPCR reaction, 0.1 μl first-strand cDNA (equivalent to 6.25 ng total RNA) was used as template for amplification with the iTaq™ Universal SYBR® Green Supermix (BioRad). Primers used for the real-time qPCR analysis of CCD-A4 and CCD-B4, as well as the reference genes Ta2291 and Ta54227, were as previously described (6).
For analysis of spike and grain traits, nine plants of TILLING control or each of the mutant genotypes were assessed for the total number of spikes per plant. The number of spikelets on each spike was counted and averaged for all spikes in the same plant to be considered as one biological replicate. To determine the length of primary spike, the spike on the first tiller of the plant was measured for the distance between the base and the top of the spike without the awn. The total number of grains was counted for each plant. Grain weight was then calculated by dividing the weight of all grains harvested from a plant by the number of grains for that plant. Images of grains were captured using an Epson V600 scanner (Epson, Los Alamitos, CA) with the crease side down. The grain length and width were determined using ImageJ (20) and averaged for all grains collected from each plant.
For analysis of seminal root traits, one sterilized and cold-treated wheat seed was placed in between a piece of germination paper and a clear sheet protector with the crease side of the seed oriented toward the germination paper and the embryo end pointing downwards. The seeds were grown vertically at the room temperature (~22°C) either entirely in the dark or under long-day conditions (16-h light/8-h dark) with a light intensity of 120 μmol m−2 s−1. After germinating at room temperature for 3 d, the seedlings were scanned using an Epson V600 scanner (Epson) and images saved as TIFF files. Coleoptiles and seminal roots were hand-traced in the images and the trait parameters were determined using ImageJ (20).
One-way Analysis of Variation (ANOVA) followed by Tukey's Honestly Significant Difference test were performed for the carotenoid quantification, gene expression, spike, grain, and seminal root data using JMP (SAS Institute, Cary, NC).
By searching an exome-sequenced tetraploid wheat TILLING mutant library (21), 78 and 109 lines were identified that contain mutations in the open reading frame (ORF) of CCD-A4 and CCD-B4, respectively. Of these mutations, three led to premature stop codons in CCD-A4: W426* in line T4-0842, W441* in line T4-2477, and Q628* in line T4-0594, and one in CCD-B4: Q181* in line T4-3179. Lines T4-0842 (W426*) and T4-3179 (Q181*) were used in this study and designated as ccd-A4 and ccd-B4. The W426* mutation in ccd-A4 truncated 203 amino acids out of the 628 amino acids of CCD-A4. The Q181* mutation in ccd-B4 led to a CCD-B4 protein missing 452 amino acids from the C-terminus (Figure 1A). These truncated CCD-A4 and CCD-B4 proteins do not contain the entire four-His and three-Glu iron coordination system essential for CCD enzyme activity, suggesting that they are loss-of-function mutations (Supplementary Figure 1) (22).
The exome sequences of ccd-A4 and ccd-B4 were also analyzed for possible mutations in other carotenoid metabolic genes, including lycopene β-cyclase (LCYb), LCYe, β-carotene hydroxylase 1 (HYD1), HYD2, and CCD1. The absence of mutations was verified for all of these gene homoeologs (i.e., both A and B subgenomes) except for LCYe-B; ccd-B4 (line T4-3179 at the M4 generation) also contained a mutation in LCYe-B. The ccd-A4 and ccd-B4 mutants were each BCed to the wild-type parental line Kronos for one generation. The LCYe-B/lcye-B (chromosome 3) and CCD-B4/ccd-B4 (chromosome 6) alleles segregated independently during BC as they are located on different chromosomes. We confirmed that all the ccd4 mutants used in this study (at the BC1 generation) only contain the wild-type LCYe-B alleles according to genotyping analysis using a CAPS marker (Figure 1B). The BC1 ccd-A4 and ccd-B4 mutants were crossed and the progenies genotyped to select the ccd-A4 ccd-B4 double mutants using CAPS and dCAPS markers (Figure 1B).
Genes surrounding CCD4 on chromosome 6 were also inspected for mutations using the exome sequences of ccd-A4 and ccd-B4. Both ccd-A4 and ccd-B4 mutants carried mutated PLL alleles (Figure 1A) and we confirmed that pll mutations were linked with ccd4 mutations in the BC1 ccd4 mutants by DNA sequencing (data not shown). The mutation of PLL-A led to an amino acid substitution (G462D) and the mutation of PLL-B led to a truncated PLL-B (W287*) protein (Figure 1A). These mutated proteins miss either a critical amino acid in a highly conserved region (pll-A) or essential functional components of protein phosphatases (pll-B), suggesting that they are likely loss-of-function mutations (Supplementary Figure 2). Taken together from the mutant analysis, the ccd-A4, ccd-B4, and ccd-A4 ccd-B4 mutants used in this study are indeed ccd-A4 pll-A, ccd-B4 pll-B, and ccd-A4 ccd-B4 pll-A pll-B, respectively. In addition to these mutant genotypes, TILLING control plants that are wild-type for the CCD-A4, CCD-B4, PLL-A, and PLL-B alleles, and carry a similar mutational load as the mutants were analyzed in parallel with the mutants.
To understand the impact of ccd4 mutations on carotenoid accumulation, total carotenoids were extracted from leaves and mature whole grains of TILLING control and the mutant plants and analyzed on high performance liquid chromatography (HPLC) (Figure 2; Tables 1, 2). In leaves, a small but significant increase was observed in neoxanthin levels in ccd-B4 pll-B and ccd-A4 ccd-B4 pll-A pll-B, in violaxanthin levels in ccd-A4 pll-A and ccd-B4 pll-B, in lutein levels in ccd-B4 pll-B, and in total carotenoids in ccd-B4 pll-B and ccd-A4 ccd-B4 pll-A pll-B, relative to TILLING control (Table 1). There was no significant change in the β-carotene content between TILLING control and the mutants (Table 1). Although accumulated at very low levels, two unidentified peaks eluted at 8.89 min (peak 1) and 10.01 min (peak 2) showed differential accumulation only in ccd-A4 ccd-B4 pll-A pll-B (Figure 2B; Supplementary Table 1). Additionally, two unidentified peaks eluted at 13.28 min (peak 3) and 13.39 min (peak 4) were present in the leaf carotenoid extracts of ccd-A4 ccd-B4 pll-A pll-B but absent in other genotypes (Figure 2B; Supplementary Table 3). Peaks 1–4 appear to be carotenoid molecules as they possess the characteristic three-peak absorption profiles for carotenoids (Figure 2C). In mature whole grains, lutein, β-carotene, and total carotenoids were not significantly different among TILLING control and the mutant genotypes (Table 2).
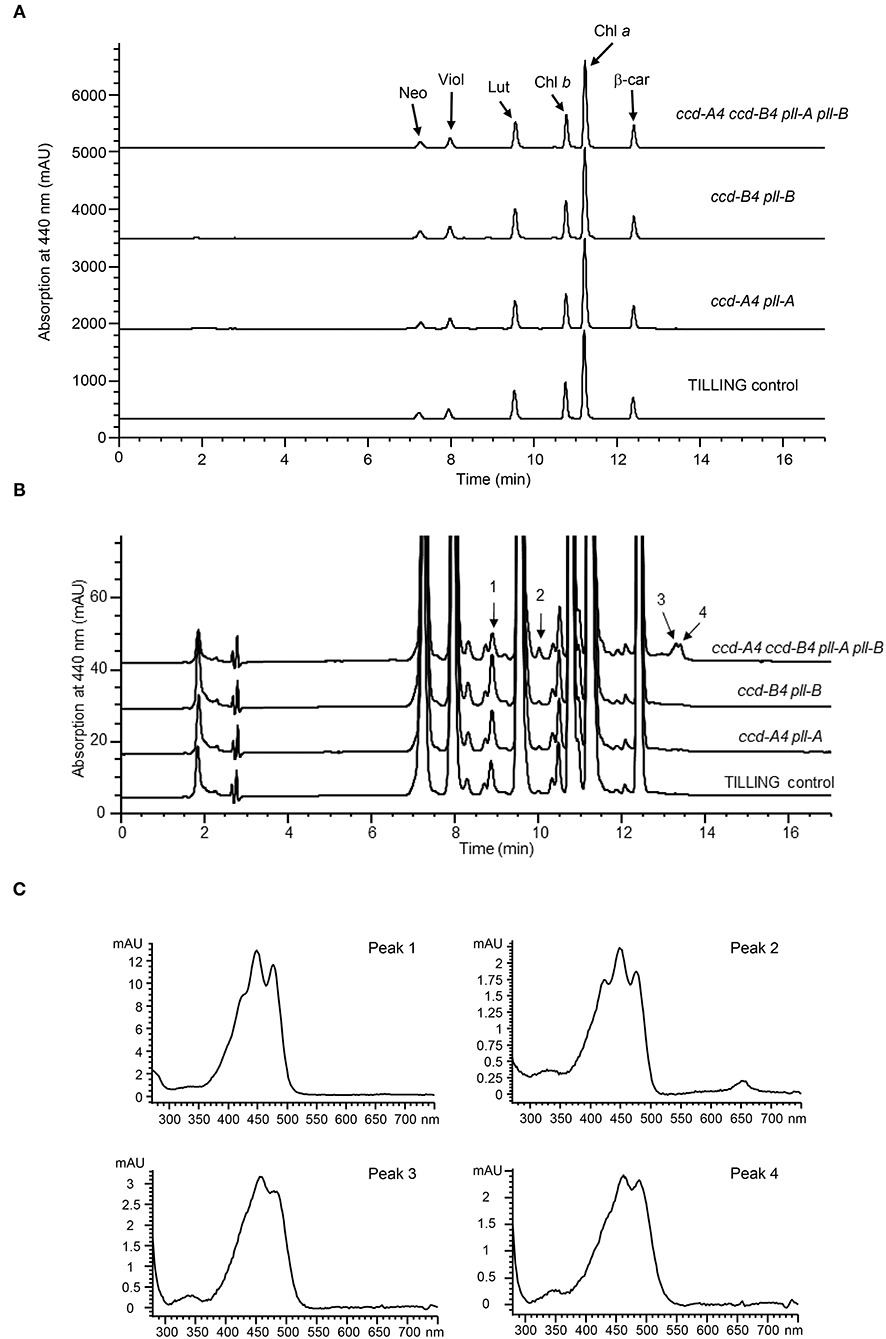
Figure 2. HPLC analysis of carotenoid profiles in leaves of TILLING control and mutant plants. (A) HPLC chromatograms of TILLING control, ccd-A4 pll-A, ccd-B4 pll-B, and ccd-A4 ccd-B4 pll-A pll-B. Neo, neoxanthin; Viol, violaxanthin; Lut, lutein; Chl b, chlorophyll b; Chl a, chlorophyll a; β-car, β-carotene. (B) A zoomed view of HPLC chromatograms showing peaks that are differentially accumulated in ccd-A4 ccd-B4 pll-A pll-B and TILLING control. (C) Absorption spectra of peaks 1–4.
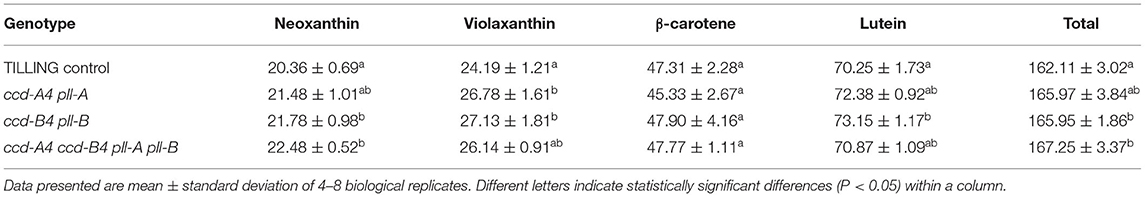
Table 1. Carotenoids (mmol mol−1 chlorophylls a + b) in leaves of 4-week-old TILLING control and mutant plants.
To examine whether the mutation of one CCD4 homoeolog may cause changes in the expression of the other CCD4 homoeolog, transcript levels of CCD-A4 and CCD-B4 in leaves of the mutant and TILLING control plants were determined using real-time qPCR (Figure 3). Expression levels of the mutated CCD-A4 and CCD-B4 alleles were only about 30% of the wild-type alleles in TILLING control, suggesting that the mutations not only led to premature stop codons, but also reduced the stability of the respective mRNAs (Figure 3). While the expression of CCD-B4 in ccd-A4 pll-A maintained at a level comparable to that in TILLING control, the expression of CCD-A4 was slightly higher in ccd-B4 pll-B than TILLING control, suggesting that the ccd-B4 mutation may induce CCD-A4 expression (Figure 3).
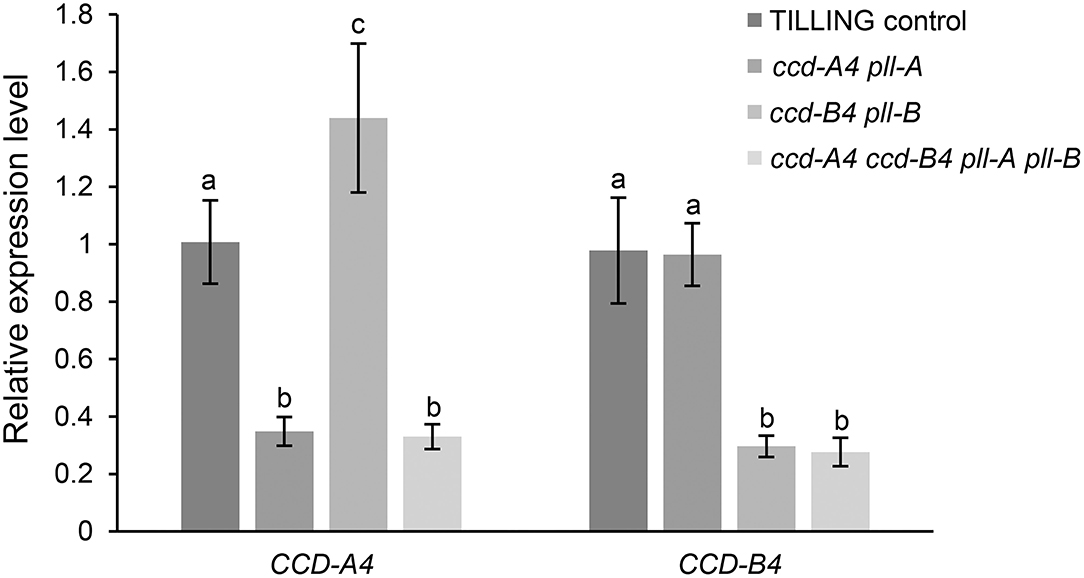
Figure 3. Expression levels of CCD-A4 and CCD-B4 in leaves of TILLING control and mutant plants. Data presented are mean ± standard deviation of 4-8 biological replicates. Different letters indicate significantly different expression (P < 0.05) for CCD-A4 or CCD-B4 in different genotypes.
When grown in the greenhouse under long-day conditions, ccd-A4 ccd-B4 pll-A pll-B plants were apparently smaller than ccd-A4 pll-A, ccd-B4 pll-B, and TILLING control plants at both early (4-week-old, prior to the emergence of spikes) and late (11-week-old, physiological maturity) developmental stages (Figure 1C). To evaluate the effect of ccd4 and pll mutations on plant performance and grain quality, several agronomic traits related to grain yield were determined that include the number of spikes per plant, number of spikelets per spike, grain number per plant, grain weight, and grain size (length and width) (Figure 4). When evaluated using tissues collected at the harvest-ready stage of mature plants, ccd-A4 ccd-B4 pll-A pll-B has 120% more spikes per plant than TILLING control, and ~50% more spikes than ccd-A4 pll-A and ccd-B4 pll-B (Figure 4A). However, ccd-A4 ccd-B4 pll-A pll-B has 40% less spikelets per spike than TILLING control, and ~30% less spikelets per spike than ccd-A4 pll-A and ccd-B4 pll-B (Figure 4B). As a result of the reduced number of spikelets on the spike, the primary spike in ccd-A4 ccd-B4 pll-A pll-B was ~12% shorter compared with other genotypes (Figures 4C,D).
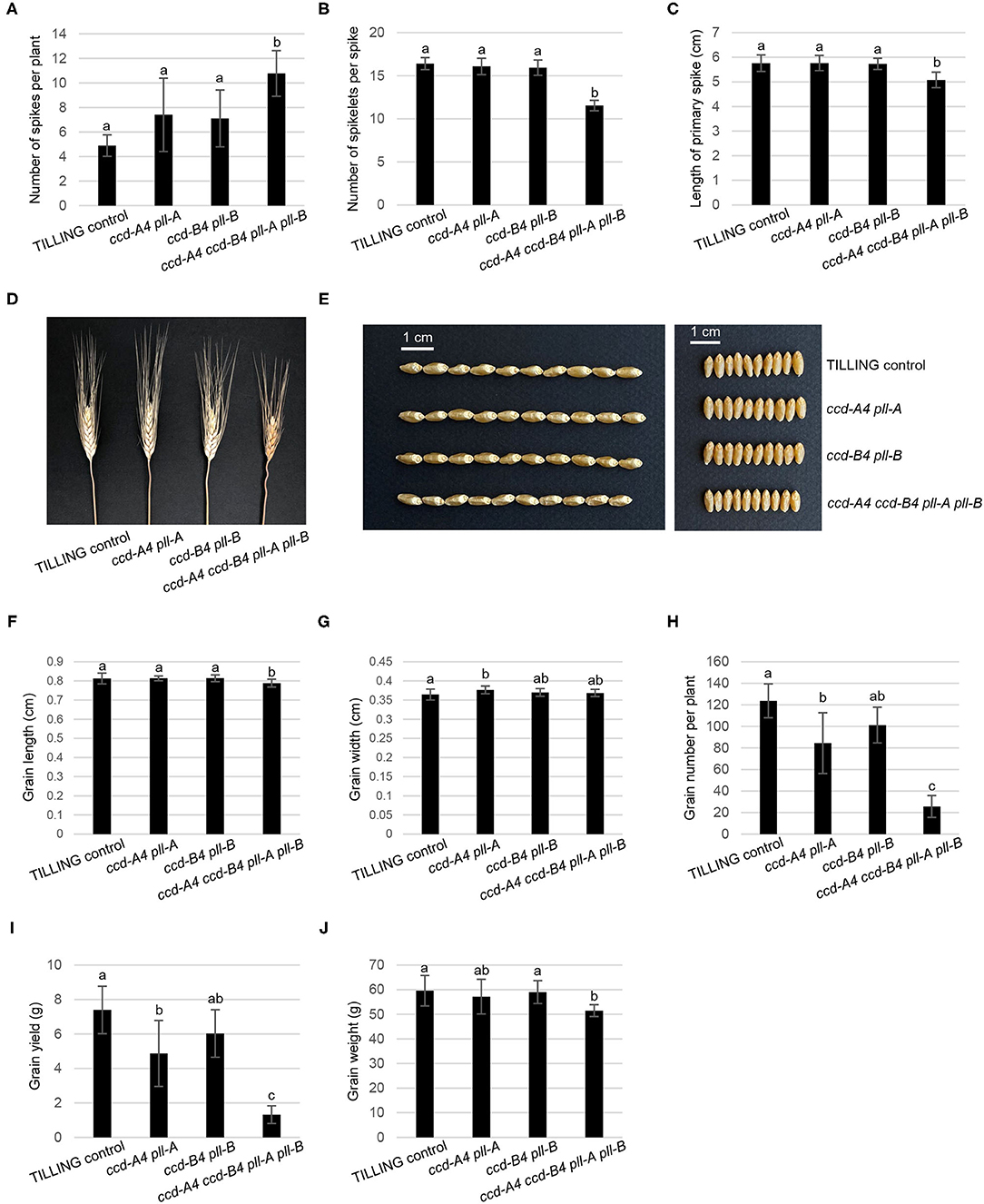
Figure 4. Spike and grain phenotyping of TILLING control and mutant plants. (A) Number of spikes per plant. (B) Number of spikelets per spike. (C) Length of primary spike. (D) Image of TILLING control and mutant spikes. (E) Images of TILLING control and mutant grains. (F) Grain length. (G) Grain width. (H) Grain number per plant. (I) Grain yield. (J) Grain weight. The mean ± standard deviation of 9 biological replicates are shown. Different letters denote significantly different (P < 0.05) for each spike or grain trait.
Besides the visibly different spike phenotypes (Figure 4D), grains harvested from ccd-A4 ccd-B4 pll-A pll-B appeared to be slightly smaller than those from TILLING control and other mutants (Figure 4E). When quantified, ccd-A4 ccd-B4 pll-A pll-B grains were 3% shorter than TILLING control and other mutants, whereas grain width was comparable for grains of all genotypes (Figures 4F,G). On the other hand, the number of grains per plant and grain yield were more drastically reduced for ccd-A4 ccd-B4 pll-A pll-B relative to TILLING control, ccd-A4 pll-A, and ccd-B4 pll-B, suggesting that the ccd-A4 ccd-B4 pll-A pll-B mutant has reduced fertility (Figures 4H,I). Indeed, we observed that spikes on the lateral tillers of ccd-A4 ccd-B4 pll-A pll-B were sterile despite its large number of lateral tillers (data not shown). Consistent with the relatively smaller grains, the average weight of grains was also decreased by ~11% in ccd-A4 ccd-B4 pll-A pll-B compared to TILLING control and other mutants (Figure 4J).
In mature plants at harvest, ccd-A4 ccd-B4 pll-A pll-B plants possess a largely reduced root volume, which may lead to less biomass production and yield as the root system is responsible for taking up water and mineral nutrients from the soil (Figure 5A). Wheat plants contain both seminal roots that develop from the radical and nodal (aka. crown or adventitious) roots that develop from nodes of the stem. To understand whether the ccd4 and/or pll mutations affect root growth, seminal root traits of seedlings were analyzed for 3-day-old seedlings grown in dark or long-day conditions, as both lighting schemes have been used in wheat seed germination (Figures 5, 6). A total of seven seminal root traits were evaluated, including network width, network depth, network width to depth ratio, convex hull area (the area of the smallest convex polygon to enclose the root system), number of seminal roots, seminal root length, and seminal root angle (the angle between the outermost seminal roots). Coleoptile lengths of the seedlings were also measured.
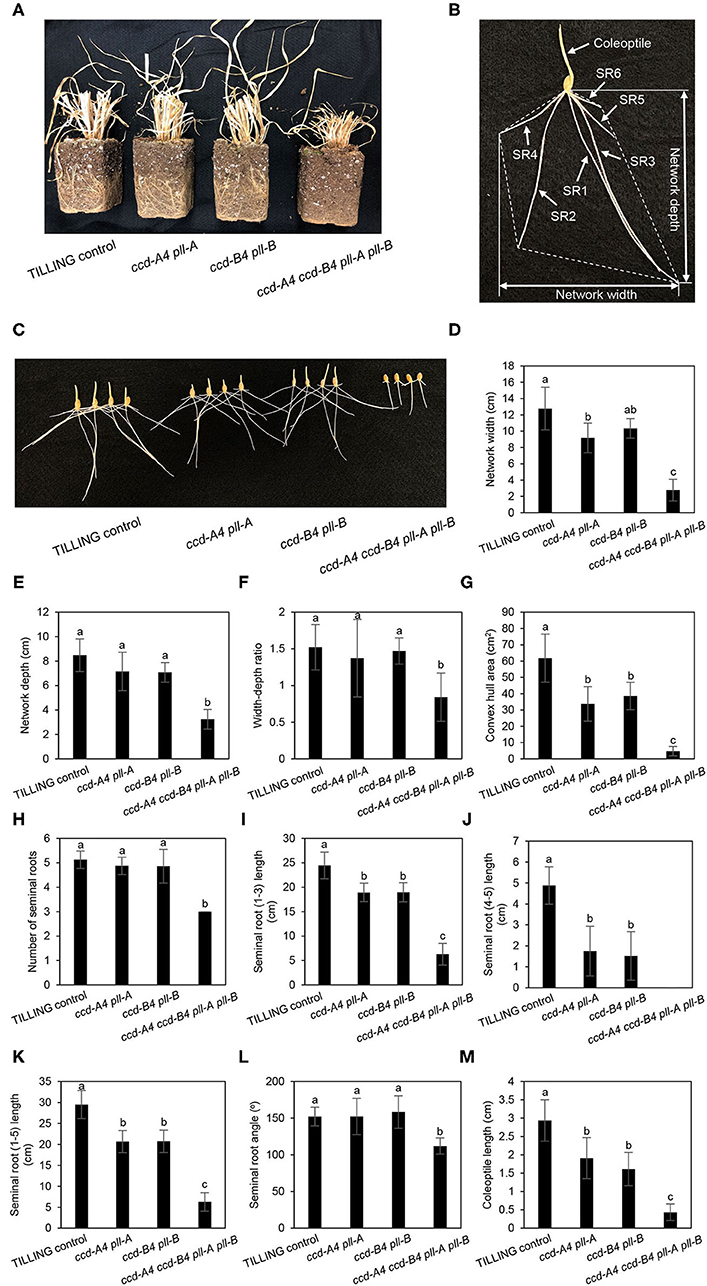
Figure 5. Seminal root phenotyping of TILLING control and mutant seedlings grown in the dark. (A) Overview of root growth for TILLING control and mutant plants at harvest. (B) Diagram showing analysis of wheat seminal root parameters. The convex hull area refers to the smallest area covered by a convex polygon containing the root system and is delineated with dotted lines. SR, seminal root. (C) Image of 3-day-old seedlings grown in the dark. (D–L) Represent different seminal root traits. (M) Coleoptile length. The mean ± standard deviation of 11 seedlings are shown. Different letters denote significantly different (P < 0.05) for each seminal root trait or coleoptile length.
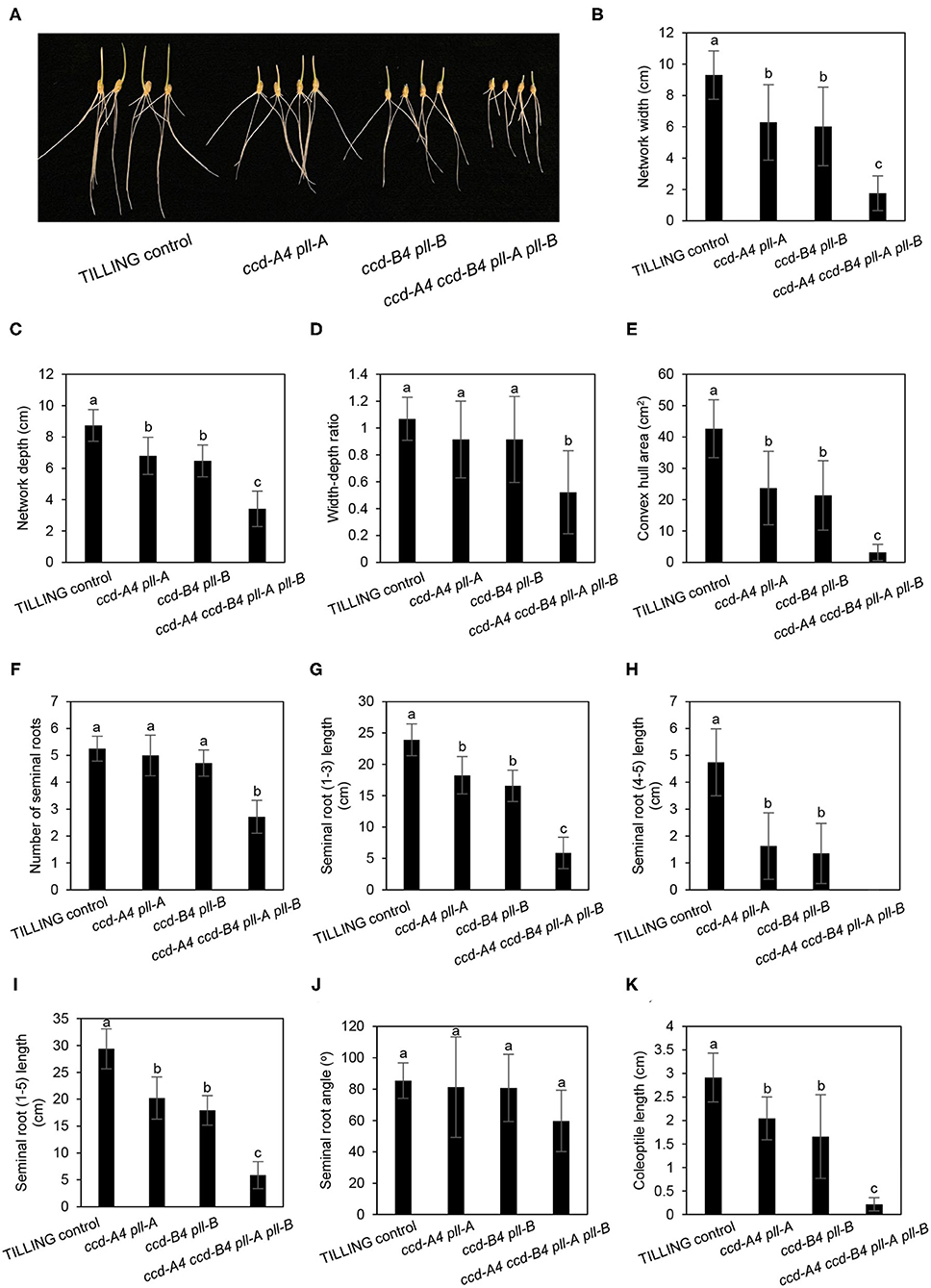
Figure 6. Seminal root phenotyping of TILLING control and mutant seedlings grown under long-day conditions. (A) Image of 3-day-old seedlings grown under long-day conditions. (B–J) Represent different seminal root traits. (K) Coleoptile length. The mean ± standard deviation of 11 seedlings are shown. Different letters denote significantly different (P < 0.05) for each seminal root trait or coleoptile length.
For dark-grown seedlings, the network width, depth, width to depth ratio, convex hull area, and angle of seminal roots were 26–80% reduced in ccd-A4 ccd-B4 pll-A pll-B relative to TILLING control (Figures 5D–G). While the reduced seminal root angle was due to a lack of seminal roots 4 and 5 in ccd-A4 ccd-B4 pll-A pll-B seedlings after 3 d of germination, the reduced network depth and width resulted from a slower growth of seminal roots 1 (depth) and 2 and 3 (width) in this mutant (Figures 5D,E,L). In fact, the emergence of the fourth seminal root was only observed in very few 11-day-old ccd-A4 ccd-B4 pll-A pll-B seedlings, indicating that the initiation of the fourth seminal root was severely delayed in this mutant (Supplementary Figure 3). The combined absence of seminal roots 4 and 5 and a slow growth of seminal roots 1–3 also led to the reduced network width-depth ratio and convex hull area in 3-day-old ccd-A4 ccd-B4 pll-A pll-B seedlings. The total length of seminal roots #1-3 of ccd-A4 ccd-B4 pll-A pll-B was 75% shorter than that of TILLING control (Figure 5I). Although the 3-day-old ccd-A4 pll-A and ccd-B4 pll-B seedlings possessed 5 seminal roots, their total seminal root lengths were ~30% shorter than TILLING control (Figures 5I–K). The coleoptile lengths correlated with seminal root lengths (seminal roots 1–5) in all mutant genotypes with ccd-A4 pll-A and ccd-B4 pll-B showing 35–45% reduced length, and ccd-A4 ccd-B4 pll-A pll-B 85% reduced length compared to TILLING control (Figure 5M).
For three-day-old seedlings grown under long-day conditions, the network width, depth, width to depth ratio, convex hull area, and angle of seminal roots in ccd-A4 ccd-B4 pll-A pll-B were largely decreased relative to TILLING control. These observed root phenotypes are similar to those observed for the dark-grown seedlings (Figures 6A–E). When compared to TILLING control, the total seminal root length was reduced by 80% in ccd-A4 ccd-B4 pll-A pll-B (containing only seminal roots 1–3), 31% in ccd-A4 pll-A, and 39% in ccd-B4 pll-B (Figures 6I–K). The coleoptile lengths of ccd-A4 pll-A, ccd-B4 pll-B, and ccd-A4 ccd-B4 pll-A pll-B were 30, 43, and 92% decreased relative to TILLING control, respectively (Figure 6K). Interestingly, the long-day-grown seedlings generally exhibited narrower network width, smaller convex hull area, and more shallow seminal root angles than dark-grown seedlings (Figures 5, 6).
In this study, we isolated, generated, and characterized tetraploid wheat TILLING mutants of ccd-A4 pll-A, ccd-B4 pll-B, and ccd-A4 ccd-B4 pll-A pll-B. The carotenoid content was comparable in grains of TILLING control and the mutants, indicating that CCD4 homoeologs do not play a major role in carotenoid turnover in grains, and therefore their activities do not need to be modified for provitamin A biofortification in tetraploid wheat grains. On the other hand, the moderately increased accumulation of neoxanthin in leaves of ccd-B4 pll-B and ccd-A4 ccd-B4 pll-A pll-B, violaxanthin in leaves of ccd-A4 pll-A and ccd-B4 pll-B, and lutein in leaves ccd-B4 pll-B, suggests a potential role of CCD-A4 and CCD-B4 in turnover of these xanthophylls in this tissue (Table 1). By contrast, β-carotene levels were not significantly changed in ccd-A4 pll-A, ccd-B4 pll-B, and ccd-A4 ccd-B4 pll-A pll-B relative to TILLING control in leaves and whole grains (Tables 1, 2), which is consistent with the results of in vitro enzyme assays where incubating recombinant CCD-A4 and CCD-B4 proteins with β-carotene did not yield any products (6). It should be noted that the catalytic activity of CCD-A4/CCD-B4 toward neoxanthin and violaxanthin was not examined in the enzyme assays previously (6).
Interestingly, four unidentified carotenoid peaks showed unique accumulation in leaves of ccd-A4 ccd-B4 pll-A pll-B, suggests that these carotenoid molecules could be potential substrates for CCD-A4 and CCD-B4 in planta. It also indicates that manipulation of CCD4 activities may lead to accumulation of carotenoids that are not normally present in wild-type plants (Figure 2; Supplementary Table 3). Furthermore, two carotenoids (peaks 3 and 4) are only present in ccd-A4 ccd-B4 pll-A pll-B leaves, suggesting that CCD-A4 and CCD-B4 may have overlapping activities toward these two carotenoids and can compensate for each other's missing activity in ccd-A4 pll-A and ccd-B4 pll-B leaves. This notion of overlapping activities is also supported by the observation that CCD-A4 expression was induced when CCD-B4 function was abolished in ccd-B4 pll-B (Figure 3).
Besides analyzing the function of CCD4 homoeologs, the presence of pll-A and pll-B mutations in the TILLING mutants analyzed in this study also provides an opportunity for examining the role of PLL in wheat plants. Although six putative PLL genes were identified in the wheat genome, none of them have been functionally characterized (23). The ccd-A4 pll-A, ccd-B4 pll-B, and ccd-A4 ccd-B4 pll-A pll-B mutants displayed changes in several traits that contribute to wheat plant biomass and yield, including seminal root initiation and architecture, spike fertility, and grain size (Figures 4–6). These growth phenotypes have not been reported in the ccd4 or pol/pll mutants characterized in Arabidopsis or other plant species. Conversely, the tetraploid wheat ccd-A4 ccd-B4 pll-A pll-B mutant does not exhibit the defective meristem and vasculature development phenotypes reported for the Arabidopsis pol/pll mutants (Figure 1).
Among the growth and agronomic traits altered by ccd4 and/or pll mutations, mutations in the A or B subgenome homoeologs of CCD4 and PLL alone already had a significant impact on the convex hull area of seminal roots, the seminal root length (1-3, 4-5), and the coleoptile length; these mutant phenotypes were exacerbated in ccd-A4 ccd-B4 pll-A pll-B where both A and B subgenome homoeologs were knocked out (Figures 5, 6). This suggests that the A and B subgenome homoeologs of CCD4 and/or PLL play distinct roles in controlling the above-mentioned traits. However, for other traits measured, such as number of spikelets per spike, grain yield, and seminal root initiation, significant differences were only observed in ccd-A4 ccd-B4 pll-A pll-B, suggesting that CCD4 and/or PLL homoeologs are functionally redundant for these traits.
Taken together, functional characterization of the ccd-A4 pll-A, ccd-B4 pll-B, and ccd-A4 ccd-B4 pll-A pll-B TILLING mutants uncovered the function of CCD4 in carotenoid accumulation in leaves and grains of tetraploid wheat. Additionally, the mutant analysis revealed that CCD4 and/or PLL homoeologs affect key seminal root, grain, and spike traits—traits that are important for not only wheat yield but also human nutrition as wheat grains are a critical dietary source of starch and proteins for human consumption. While the linked ccd4 and pll mutations in the TILLING mutants pose challenges to discerning the role of CCD4, PLL or the interaction of CCD4 and PLL in controlling the plant growth traits, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) (CRISPR/Cas9) gene editing lines of CCD4 and PLL are currently being generated that mutate each gene individually. These CRISPR/Cas9-induced ccd4 and pll mutant lines will help dissect the function of CCD4 and PLL in plant growth, which will have broad implications in improving wheat yield and nutrient content.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
SY and LT designed the experiments, analyzed the data, and wrote the manuscript. SY performed the experiments. Both authors contributed to the article and approved the submitted version.
This work was funded by USDA-NIFA (2017-67013-26164 to LT). SY received support from a China Scholarship Council Scholarship, the Henry A. Jastro Research Award, and the UC Davis, Department of Plant Sciences Graduate Research Fellowship.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Dr. Jorge Dubcovsky at University of California, Davis for providing the wheat TILLING mutant materials. We also thank Cody Bekkering for critical reading of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.740286/full#supplementary-material
Supplementary Figure 1. Multiple sequence alignment of tetraploid wheat CCD-A4 and CCD-B4 as well as maize VP14 and Synechocystis sp. PCC 6803 ACO proteins. The conserved amino acids essential for carotenoid cleavage dioxygenase enzyme activities are indicated with asterisks. The mutated amino acids in CCD-A4 and CCD-B4 are indicated with blue and green arrows, respectively. VP14, 9-cis-epoxycarotenoid dioxygenase; ACO, lignostilbene dioxygenase.
Supplementary Figure 2. Multiple sequence alignment of tetraploid wheat PLL-A and PLL-B as well as Arabidopsis POL and PLL1 proteins. The metal interacting domains and conserved amino acids are underlined and indicated with asterisks, respectively. The mutated amino acids in PLL-A and PLL-B are pointed with blue and green arrows, respectively. POL, Poltergeist; PLL, Poltergeist-like.
Supplementary Figure 3. Images of 11-day-old ccd-A4 ccd-B4 pll-A pll-B and TILLING control seedlings grown in the dark. The white arrow points to the emerged seminal root 4 (SR4) in one of the 11-day-old ccd-A4 ccd-B4 pll-A pll-B seedlings. The white triangles indicate the places where root growth of TILLING control seedlings was constrained by the size of seed pouches.
Supplementary Table 1. Primers and restriction enzymes used in the genotyping analysis.
Supplementary Table 2. Primers used for amplification of the PLL homoeologs.
Supplementary Table 3. Areas (peak area mg−1 fresh weight) of peaks 1–4 integrated in the HPLC analysis shown in Figure 2B.
1. Hazard B, Trafford K, Lovegrove A, Griffiths S, Uauy C, Shewry P. Strategies to improve wheat for human health. Nat Food. (2020) 1:475–80. doi: 10.1038/s43016-020-0134-6
2. Yu S, Tian L. Breeding major cereal grains through the lens of nutrition sensitivity. Mol Plant. (2018) 11:23–30. doi: 10.1016/j.molp.2017.08.006
3. Moreno JC, Mi J, Alagoz Y, Al-Babili S. Plant apocarotenoids: from retrograde signaling to interspecific communication. Plant J. (2021) 105:351–75. doi: 10.1111/tpj.15102
4. Poliakov E, Gentleman S, Cunningham FX, Jr., Miller-Ihli NJ, Redmond TM. Key role of conserved histidines in recombinant mouse beta-carotene 15,15'-monooxygenase-1 activity. J Biol Chem. (2005) 280:29217–23. doi: 10.1074/jbc.M500409200
5. Felemban A, Braguy J, Zurbriggen MD, Al-Babili S. Apocarotenoids involved in plant development and stress response. Front Plant Sci. (2019) 10:1168. doi: 10.3389/fpls.2019.01168
6. Qin X, Fischer K, Yu S, Dubcovsky J, Tian L. Distinct expression and function of carotenoid metabolic genes and homoeologs in developing wheat grains. BMC Plant Biol. (2016) 16:155. doi: 10.1186/s12870-016-0848-7
7. Gonzalez-Jorge S, Ha S, Magallanes-Lundback M, Gilliland L, Zhou A, Lipka A, et al. Carotenoid cleavage dioxygenase4 is a negative regulator of β-carotene content in Arabidopsis seeds. Plant Cell. (2013) 25:4812–26. doi: 10.1105/tpc.113.119677
8. Campbell R, Ducreux LJM, Morris WL, Morris JA, Suttle JC, Ramsay G, et al. The metabolic and developmental roles of carotenoid cleavage dioxygenase4 from potato. Plant Physiol. (2010) 154:656–64. doi: 10.1104/pp.110.158733
9. Ohmiya A, Kishimoto S, Aida R, Yoshioka S, Sumitomo K. Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. (2006) 142:1193–201. doi: 10.1104/pp.106.087130
10. Yu LP, Miller AK, Clark SE. POLTERGEIST encodes a protein phosphatase 2C that regulates CLAVATA pathways controlling stem cell identity at Arabidopsis shoot and flower meristems. Curr Biol. (2003) 13:179–88. doi: 10.1016/S0960-9822(03)00042-3
11. Gagne JM, Clark SE. The protein phosphatases POL and PLL1 are signaling intermediates for multiple pathways in Arabidopsis. Plant Signal Behav. (2007) 2:245–6. doi: 10.4161/psb.2.4.3863
12. Song SK, Clark SE. POL and related phosphatases are dosage-sensitive regulators of meristem and organ development in Arabidopsis. Dev Biol. (2005) 285:272–84. doi: 10.1016/j.ydbio.2005.06.020
13. Song SK, Hofhuis H, Lee MM, Clark SE. Key divisions in the early arabidopsis embryo require POL and PLL1 phosphatases to establish the root stem cell organizer and vascular axis. Dev Cell. (2008) 15:98–109. doi: 10.1016/j.devcel.2008.05.008
14. Song SK, Lee MM, Clark SE. POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for arabidopsis shoot and floral stem cells. Development. (2006) 133:4691–8. doi: 10.1242/dev.02652
15. Yu LP, Simon EJ, Trotochaud AE, Clark SE. POLTERGEIST functions to regulate meristem development downstream of the CLAVATA loci. Development. (2000) 127:1661–70. doi: 10.1242/dev.127.8.1661
16. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. (2011) 7:539. doi: 10.1038/msb.2011.75
17. Dong OX, Yu S, Jain R, Zhang N, Duong PQ, Butler C, et al. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat Commun. (2020) 11:1178. doi: 10.1038/s41467-020-14981-y
18. Qin X, Zhang W, Dubcovsky J, Tian L. Cloning and comparative analysis of carotenoid (-hydroxylase genes provides new insights into carotenoid metabolism in tetraploid (Triticum turgidum ssp. durum) and hexaploid (Triticum aestivum) wheat grains. Plant Mol Biol. (2012) 80:631–46. doi: 10.1007/s11103-012-9972-4
19. Jaakola L, Pirttilä A, Halonen M, Hohtola A. Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol Biotechnol. (2001) 19:201–3. doi: 10.1385/MB:19:2:201
20. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. (2012) 9:671–5. doi: 10.1038/nmeth.2089
21. Krasileva KV, Vasquez-Gross HA, Howell T, Bailey P, Paraiso F, Clissold L, et al. Uncovering hidden variation in polyploid wheat. Proc Natl Acad Sci USA. (2017) 114:E913–21. doi: 10.1073/pnas.1619268114
22. Sui X, Zhang J, Golczak M, Palczewski K, Kiser PD. Key residues for catalytic function and metal coordination in a carotenoid cleavage dioxygenase. J Biol Chem. (2016) 291:19401–12. doi: 10.1074/jbc.M116.744912
Keywords: wheat, carotenoid cleavage dioxygenase, ccd4, poltergeist-like, TILLING
Citation: Yu S and Tian L (2021) Assessing the Role of Carotenoid Cleavage Dioxygenase 4 Homoeologs in Carotenoid Accumulation and Plant Growth in Tetraploid Wheat. Front. Nutr. 8:740286. doi: 10.3389/fnut.2021.740286
Received: 12 July 2021; Accepted: 18 August 2021;
Published: 08 September 2021.
Edited by:
Alexander Arthur Theodore Johnson, The University of Melbourne, AustraliaReviewed by:
Yueliang Zhao, Shanghai Ocean University, ChinaCopyright © 2021 Yu and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Tian, bHRpYW5AdWNkYXZpcy5lZHU=; orcid.org/0000-0001-6461-6072
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.