- 1Department of Health and Exercise Science, Colorado State University, Fort Collins, CO, United States
- 2Center for Healthy Aging, Colorado State University, Fort Collins, CO, United States
Aging is the greatest risk factor most diseases, including cardiovascular disorders, cancers, diabetes, and neurodegeneration, but select nutritional interventions may profoundly reduce the risk for these conditions. These interventions include calorie restriction, intermittent fasting, protein restriction, and reducing intake of certain amino acids. Certain ad libitum diets, including the Mediterranean, Finnish Geriatric Intervention Study to Prevent Cognitive Impairment and Disability, and Okinawan diets also promote healthy aging. Evidence indicates that these dietary strategies influence aging and healthspan by acting on the biological “hallmarks of aging” and especially upstream nutrient sensing pathways. Recent advances in “omics” technologies, including RNA-sequencing (transcriptomics), have increased our understanding of how such nutritional interventions may influence gene expression related to these biological mediators of aging, primarily in pre-clinical studies. However, whether these effects are also reflected in the human transcriptome, which may provide insight on other downstream/related cellular processes with aging, is an emerging topic. Broadly, the investigation of how these nutritional interventions influence the transcriptome may provide novel insight into pathways associated with aging, and potential targets to treat age-associated disease and increase healthspan. Therefore, the purpose of this mini review is to summarize what is known about the transcriptomic effects of key dietary/nutritional interventions in both pre-clinical models and humans, address gaps in the literature, and provide insight into future research directions.
Introduction
Older age is the major risk factor for cardiovascular disorders, cancers, diabetes, sarcopenia, frailty, and neurodegeneration (1). As a result, the “Geroscience” concept has emerged in an attempt to understand the relationships between aging biology and age-related diseases, with the hope that targeting aging itself will increase healthspan (the number of healthy, disease-free years in older age) (2). The mechanisms that drive aging/age-related diseases, termed the “hallmarks of aging,” represent key molecular targets for interventions to improve healthspan (3).
Nutrition has a profound impact on the biology of aging and disease. Several interventions in particular act on most hallmarks of aging to improve cardiometabolic, physical and cognitive health, and to delay age-related disease and increase lifespan. These interventions include calorie restriction (CR), intermittent fasting (IF) (4), alternate day fasting (ADF), prolonged fasting (PF), time-restricted feeding (TRF), protein restriction, and reduced intake of certain amino acids. Specific eating patterns, including the Mediterranean, Okinawan, and FINGER (Finnish Geriatric Intervention Study to Prevent Cognitive Impairment and Disability) diets also improve health (5). Information on how these interventions impact health and lifespan (largely by influencing the hallmarks of aging) has accumulated over several decades (1). However, recent advances in “omics” technologies, including RNA-sequencing, may provide additional, important understanding of the common or distinct biological mechanisms underlying these interventions and yield novel insight into targetable, genomic/cellular pathways associated with aging and disease.
The effects of healthy aging interventions on the transcriptome are not fully understood, mostly due to a lack of sequencing data. However, evidence to date suggests that key transcriptomic pathways involved in aging including glucose homeostasis/insulin signaling, inflammation, oxidative phosphorylation, immune responses, and circadian signaling are similarly influenced by most healthy aging/nutritional interventions (Figure 1 and Table 1). The purpose of this mini review is to detail how healthy aging interventions affect these pathways (and related genes/modulators), and to outline additional pathways/genes that are distinctly modulated with select healthy dietary interventions. While most existing transcriptomic and genetic studies have used pre-clinical models, we also provide insight into how some of these interventions affect the human transcriptome. Finally, we describe gaps in the research, future directions, and goals in the field of dietary interventions and transcriptomics.
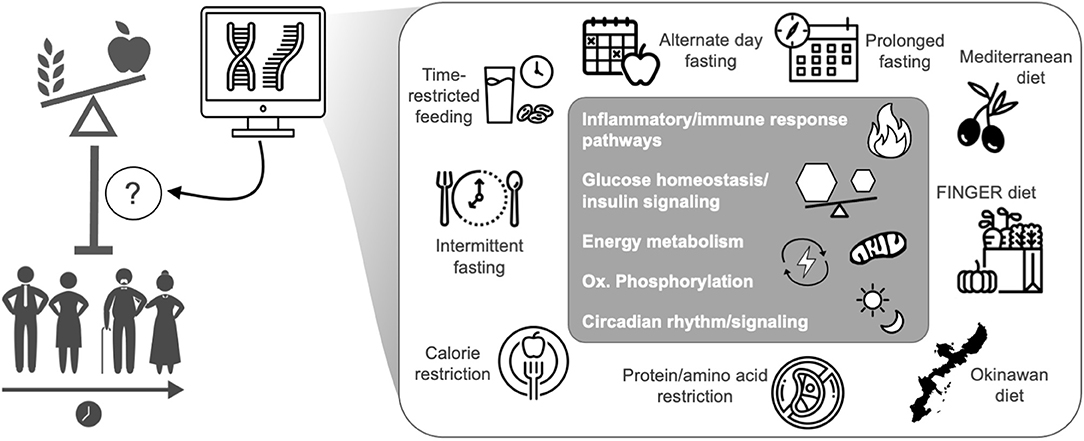
Figure 1. Transcriptomics has increased the understanding of how health-promoting nutritional interventions influence gene expression related to the biological mediators of aging and healthspan, mostly in pre-clinical models. The transcriptomic effects of healthspan-promoting nutritional interventions are still under investigation, but common effects among all interventions are noted in the gray box. Icons from nounproject.com under Creative Commons license.
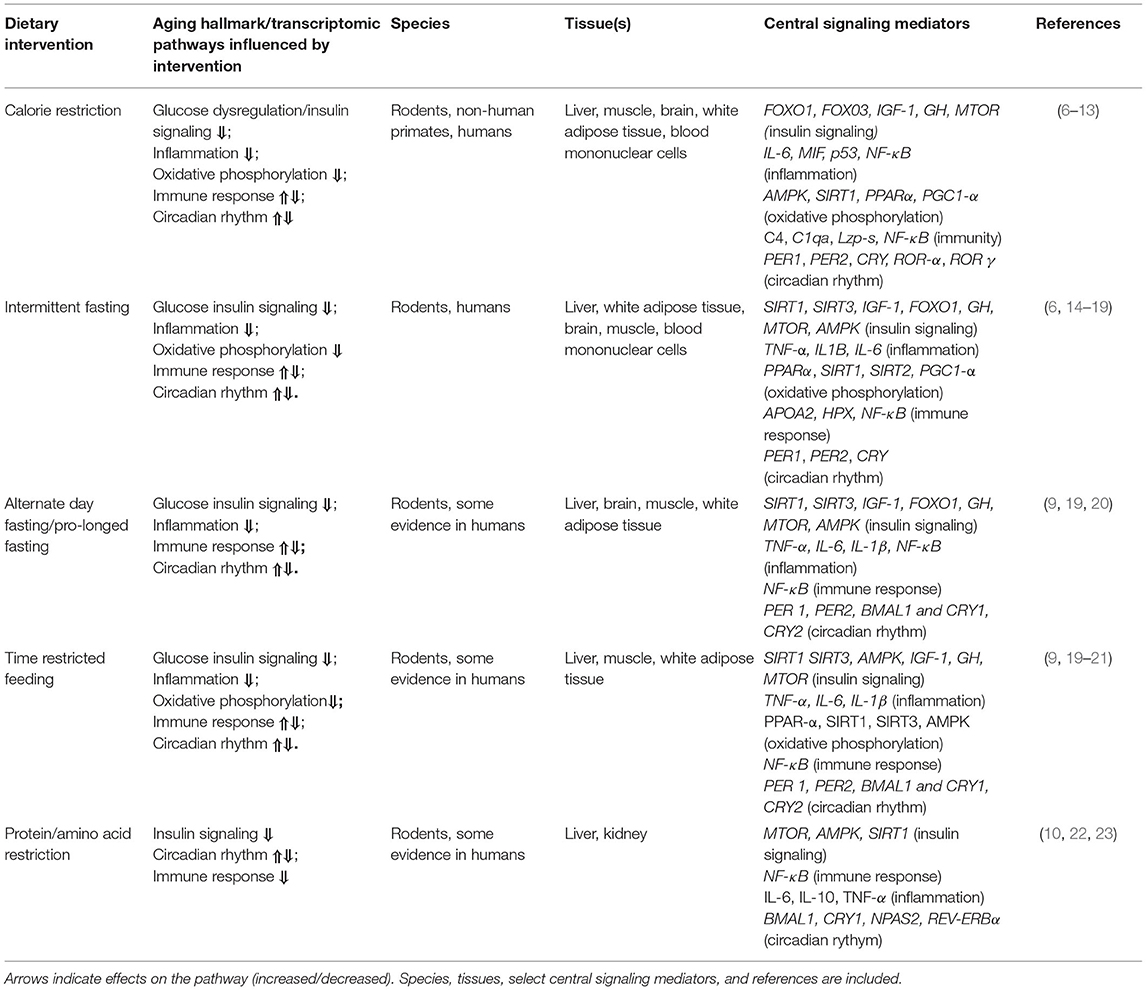
Table 1. Commonalities among the effects of select healthy dietary interventions on biological hallmarks of aging in transcriptomic evidence.
Calorie Restriction
Perhaps the most powerful anti-aging nutritional intervention, CR is defined as a reduction in 10–50% daily caloric intake without malnutrition. CR increases healthspan and lifespan through many mechanisms, including improved glucose homeostasis and mitochondrial health, and by inhibiting all molecular hallmarks of aging (24). The transcriptomic effects of CR in pre-clinical models have been extensively investigated in multiple tissues. Seminal studies showed that long-term CR reverses gene expression changes associated with age-related inflammation and impaired stress responses, DNA-replication/cell cycle defects, oxidative stress, tumorigenesis, and macromolecular damage (11, 25, 26). Additional results indicate that CR largely reduces transcriptomic signatures associated with immune activation, inflammatory signaling, glucose homeostasis, and AMP-activated protein-kinase/insulin-like growth factor 1 (AMPK/IGF-1) signaling (evolutionarily conserved energy sensing pathways). Also, consistent with the observation that glucose homeostasis/insulin signaling are central players in the effects of CR on metabolism, CR changes the expression of key genes affecting energy/metabolism-modulating proteins and insulin secretion (largely reducing the expression of these genes) (6). In addition to glucose homeostasis, other pathways/genes contributing to DNA repair, fatty acid metabolism, and citric acid cycle/oxidative phosphorylation appear to be key transcriptomic mediators of the effects of CR in white adipose tissue and liver in pre-clinical models (27–29).
More recently, the influence of CR on the liver transcriptome in the Rhesus macaque (a close relative to humans sharing 93% DNA homology) was investigated. A two-year CR intervention reduced transcriptome signatures of immune activation and inflammation (a main hallmark of aging) and increased pathways associated with ribosome activity, branched-chain amino acid metabolism, fatty acid degradation, and RNA transport mechanisms (7). These results suggest that Rhesus macaques and mice respond similarly to CR, with main transcriptomic changes in inflammatory pathways, immune function, and fatty acid degradation (related to glucose metabolism) in multiple tissues (30).
In humans, analyses from multiple studies indicates that both short-term (e.g., 4 weeks) and long-term (e.g., 1 year) CR upregulates transcriptome modules associated with stem cell maintenance, blood vessel remodeling, and lipid metabolism in subcutaneous fat tissue (31). Other studies indicate that very long-term (10 years) voluntary CR causes gene expression patterns related to enhanced lipid and glucose metabolism, and downregulation of the insulin/IGF-1 growth pathway (resulting in a younger transcriptional profile) in muscle (a main metabolic tissue), consistent with the pre-clinical mouse and non-human primate data (8).
Intermittent Fasting
IF is an umbrella term for eating patterns (which also include ADF, PF, and TRF) involving an extended time with little or no energy intake (fasting), all of which are associated with improved glucose homeostasis (and reduced insulin) and increased stress resistance (32). It has been suggested that the key benefit of IF comes from fasting periods (rather than reduced total energy intake), which can induce hormesis (responses to moderate stress with downstream protective and anti-aging effects). For example, 11 h/day of restricted feeding (e.g., no calorie intake) reduces liver pathology and extends median lifespan in male mice (33). IF (16 h/day) in mice also results in global transcriptomic changes in the liver (a key metabolic organ) including modulation of pathways contributing to sterol, alcohol, and cholesterol synthesis, and changes in gene regulation modulating circadian rhythm (14). In addition to these effects, IF enhances transcriptional signatures of peroxisome proliferator activator receptor (PPAR) activity, a major mediator of oxidative phosphorylation, highlighting the influence of IF on homeostasis and metabolic function (34). Finally, IF (12–16 h/day) results in global transcriptomic changes in the mouse brain reflecting increased neuroplasticity and reduced inflammation, which parallels the effects of CR on the brain (15).
The transcriptomic effects of IF in humans are still under investigation (studies are underway), but initial results from studies of individual genes indicate that IF (5 days of periodic fasting) increases transcriptomic signatures of nutrient and energy sensing/oxidative phosphorylation gene expression including AMP-activated protein kinase (AMPK), Forkhead box protein 01 (FOXO1), Sirtuin 1 (SIRT1), and Sirtuin 3 (SIRT 3). These genes/proteins are key players in cellular regulation pathways and centrally implicated stress responses, cardiometabolic/systemic health, and longevity (35). Another study (36) confirmed these results with data showing that IF contributes to a marginal increase in SIRT3 gene expression, which was associated with a decrease in plasma insulin levels. While these preliminary results in humans have primarily focused on changes in specific gene expression, the global transcriptomic effects of IF (36-h fast) appear to be similar to CR with changes in gene expression related to fatty acid oxidation, cell cycle and apoptosis pathways, and reductions in key inflammatory genes/pathways (16).
Alternate Day Fasting and Prolonged Fasting
ADF and PF are types of IF involving feeding days (on which food is consumed ad libitum), and fasting days on which no calories are consumed. The fasting days can occur every other day or intermittently (e.g., fasting 2 days per week or even up to 21 days once per year, which is more commonly known as PF) (37, 38). Evidence shows that these dietary interventions induce metabolic effects similar to IF, including improving glucose homeostasis and reducing liver triglyceride content in mice (39). In pre-clinical models, these diets flip a “metabolic switch” activating key transcriptional regulators of fatty acid and energy metabolism and inflammation (9, 40), and transcriptional evidence shows that ADF (24-h fast) modulates key processes associated with circadian rhythm, RNA processing, and oxidative metabolism (41).
In humans, a prolonged 10-day fast results in transcriptomic signatures of increased lipolysis/lipid metabolism and reduced activation of pathways related to glycolysis and oxidative phosphorylation. Interestingly, this prolonged fasting period also contributes to the upregulation of inflammatory pathways and macrophage activation in subcutaneous fat tissue (42), reflecting a potential hormesis response and downstream protective mechanisms. Another study showed that short-term fasting is associated with enrichment of transcriptomic pathways involved in fatty acid oxidation, cell cycling and apoptosis, and a decrease in the expression of pro-inflammatory genes in peripheral blood mononuclear cells (16). These apparently divergent results suggest that the protocol, length of fasting, and tissues analyzed are important factors to consider when studying the influence of these dietary interventions on the human transcriptome.
Time-Restricted Feeding
TRF is a another type of IF and allows for eating only during a certain time window in the day (e.g., eating ad libitum for 8 h or less a day in humans) (37). TRF is different from IF in that the daily eating time frame remains constant, which is thought to influence circadian clock pathways. TRF also promotes cardiometabolic/systemic health by maintaining metabolic homeostasis (e.g., improving glucose tolerance and insulin sensitivity, and reducing blood lipid levels) (43). Additional effects of TRF include enhanced mitochondrial health, DNA-repair, and autophagy, all of which coincide with improved glucose metabolism and potentially reduced risk for most age-related diseases (44). In mice, TRF contributes to transcriptomic changes to circadian clock gene expression (increased expression/rhythmicity) and metabolic/nutrient sensing pathways (changes that parallel improved metabolic health and the prevention of fatty liver) (21), all of which are similar to effects seen with IF, ADF, and PF.
There is limited research on the effects of TRF on the human transcriptome/gene expression, but recent evidence suggests that a daily 8-h eating time frame influences genes responsible for amino acid transport in muscle (45), and reduces gene expression signatures related to growth and metabolism. Research on the effects of fasting has recently focused on peripheral/circadian clock genes, with some studies suggesting no universal effect (17) and others suggesting that that TRF in particular influences several genes implicated in circadian rhythm (46). Such changes in circadian clock gene expression might impact healthspan and lifespan, but these links are poorly understood (47).
Protein and Amino Acid Restriction
The restriction of dietary protein and certain amino acids promotes healthspan and lifespan in pre-clinical models (48) and may mimic CR to some extent. This may be an important observation, especially considering that most people may find it difficult to adhere to most calorie deficit interventions, especially in Western societies where there is essentially free access to calorie-dense foods. Long-term reduction of dietary protein intake (through dilution with non-digestible fiber) improves healthspan and lifespan via reduced growth pathway activation and improved mitochondrial function in male and female mice (49), and short-term protein restriction reduces circulating triglycerides via changes in hepatic APOE expression (50). Similarly, reduced intake of branched-chain amino acids (BCAAs; leucine, isoleucine, and valine) also improves health and lifespan in male and female mice by inhibiting mechanistic target of rapamycin (MTOR) nutrient sensing pathway activation, improving glucose homeostasis, and reducing insulin resistance (22). While the aforementioned studies are not transcriptomic studies, the results suggest that protein and amino acid restriction may partly mimic the gene expression changes and metabolic effects seen with fasting or related nutritional interventions.
Protein restriction is known influence key genes associated with circadian rhythm, metabolic signaling (including insulin signaling), and oxidative phosphorylation, in addition to other transcriptional factors/genes linked with healthspan and lifespan extension (51, 52). On the other hand, BCAA intake is associated with transcriptomic effects including changes in cell cycle/metabolic pathways, apoptosis (programmed cell death), p53 activity (cell division), and NF-κB signaling (innate and adaptive immunity) (22). These results highlight the contribution of these diet-induced transcriptomic effects to changes in metabolic/energy-sensing processes, as well as the modulation of inflammation—all of which are associated with aging and age-related disease (53).
Common Transcriptomic Pathways Affected by Most Well-Studied Healthy Dietary Interventions
The common transcriptomic pathways modulated by most health-promoting dietary interventions involve glucose/insulin signaling, inflammation, oxidative phosphorylation, immune responses, and circadian rhythm (Table 1). Based on limited transcriptomic/genetic data, these processes seem to be similarly affected by most interventions, regardless of species or tissue, although specific genes within pathways may be activated/repressed depending on the intervention type (e.g., the duration of CR or fasting period). We also note that some processes like circadian rhythm and immunity are especially complex and may be differentially modulated (e.g., increased or decreased) and highly dependent on length or type of intervention (54). Below, we briefly describe the common biological processes/transcriptomic pathways that are affected by most healthy aging interventions.
Glucose Homeostasis and Insulin Signaling
Metabolic dysfunction is considered a central hallmark of aging, and current transcriptomic data shows that all healthy aging dietary interventions influence pathways involved with glucose homeostasis and insulin signaling. Most evidence suggests that these interventions improve health by suppressing key insulin-related signaling pathways and gene expression. These main genes include Insulin-like growth factor-1 (IGF-1, reduced), Sirtuins (SIRT1 and SIRT3, increased), Growth hormone (GH, increased), Mechanistic target of Rapamycin (MTOR, reduced), and FOXO genes (increased) (6), all of which have key roles in energy sensing, growth and metabolism (5).
Inflammation
Inflammation is considered a key hallmark of aging because it can directly affect all other biological hallmarks of aging (55). Most healthy dietary interventions targeting energy intake reduce transcriptomic evidence of inflammatory activation, and this has been demonstrated in multiple species/tissues. Key inflammatory mediators involved in these pathways include Interluekin-6 (IL-6), Tumor necrosis factor alpha (TNF-α), and Interleukin 1 beta (IL-1β). Importantly, additional health-promoting diets including the FINGER and Mediterranean (described below) also similarly modulate these pro-inflammatory cytokines (and transcriptomic evidence of their activity) in humans (56).
Immune Response
Long periods of calorie restriction or periodic fasting may impair immune system pathways, while short bouts of fasting are emerging as potential interventions for immune system repair and maintenance (10). There is evidence that most healthy aging interventions influence immune responses to a certain degree, mostly by modulating NF-κB, a transcriptional factor which plays critical roles in immune response and cytokine production (38). Some evidence suggests that CR reduces additional transcripts (57) involved in immune function including Complement component 4 (C4) and Complement C1q (C1q), but whether these genes are also influenced by the other interventions remains to be determined.
Oxidative Phosphorylation
All healthy nutritional interventions act on the mitochondria, and consequently, pathways/genes contributing to cellular respiration. Most evidence suggests that these interventions reduce oxidative phosphorylation gene expression (58) consequently attenuating the production of reactive oxygen species (ROS; a key contributor to aging/age-related disease). Increased expression/activity of mitochondrial biogenesis and turnover pathways with healthy nutritional interventions may also contribute to greater mitochondrial efficiency (despite reductions in oxidative phosphorylation). Key genes associated with these pathways are mostly energy sensing in nature and include the Sirtuins, Peroxisome proliferator activated-receptor α (PPAR-α), and AMPK (59).
Circadian Rhythm
Most physiological and metabolic processes are regulated by circadian clock genes that are responsible for the synchronization of biological processes within an organism (60). Recently, it has been suggested that circadian rhythm dictated by peripheral clock genes can be re-synchronized (thus resulting in positive health benefits) under CR or PF (61). Key modulators of circadian rhythm pathways affected by most nutritional interventions include the Period circadian regulators (PER1 and PER2) and the Cryptochromes (CRY) (61).
Exploration of the Transcriptomic Effects of Additional Health-Promoting Diets
Most of the well-studied interventions above are calorie or macronutrient restrictive. It is therefore important to understand how other health-promoting ad libitum nutritional interventions influence the genome. Indeed, emerging data show that several healthy dietary/nutritional interventions or patterns act on the transcriptome to affect processes associated with aging, age-related disease, and longevity.
The Finnish Geriatric Intervention Study to Prevent Cognitive Impairment and Disability (FINGER), Mediterranean, and Okinawan diets all improve health in humans. These diets are characterized by ample fruit and vegetable consumption and low-to-moderate protein intake (with limited red meat consumption), low-to-moderate fat consumption (healthy fats including olive oils and avocado), and moderate-to-high carbohydrate consumption (whole grains, potatoes) (62, 63). The FINGER diet also includes cognitive function-enhancing exercises and a weekly physical activity routine (64).
The most well-studied of these diets, in terms of transcriptomic impact, is the Mediterranean diet. The Mediterranean diet elicits a transcriptomic response in humans including reduced inflammatory/inflammation response gene expression (65) (possibly due to a high consumption of monounsaturated fatty acids and olive oils) and enhanced circadian clock transcript levels (66). Other studies show that the Mediterranean diet affects transcriptomic pathways involved with cardiovascular health (e.g., atherosclerosis and hypertension) including lipid and cholesterol metabolism, which appears to be a main benefit of this diet (67). The transcriptomic effects of an Okinawan or FINGER dietary intervention remain to be determined; however, based on the above observations that the Mediterranean diet may have transcriptomic effects similar to those of CR/fasting, profiling the influence of these other diets on the transcriptome could be critical for identifying the ideal, non-fasting dietary pattern to promote healthy aging.
Research Gaps and Future Directions
As described above and in Figure 1 and Table 1, the available evidence suggests that the common transcriptomic effects of healthspan-promoting nutritional interventions include favorable modulation of gene expression patterns related to inflammatory signaling, immune function, energy sensing/glucose homeostasis, oxidative phosphorylation (including mitochondrial function), and circadian regulation. Importantly, most of this evidence is based on: (1) pre-clinical studies with some limited/emerging clinical data; and (2) various experimental and methodological approaches, including gene expression/qPCR panels, microarrays, and most recently RNA-sequencing (see individual references for details). More research is needed to increase our understanding of human-specific transcriptomic effects of these interventions, and to leverage newer, comprehensive RNA-seq approaches—ideally in “multi-omics” studies (e.g., RNA-seq combined with proteomics) that may provide further mechanistic insight. In addition to these general future directions, below (and in Figure 2) we outline several key issues that require further investigation in this area.
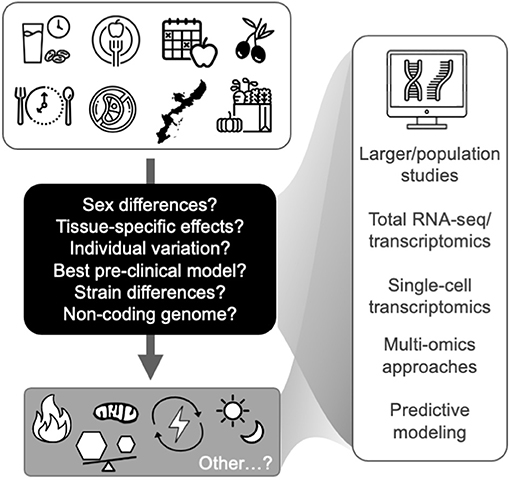
Figure 2. Future studies are needed to identify key transcriptomic modules that relate to the hallmarks of aging (bottom gray box) and represent targets for healthy aging in humans. Future directions and key issues (shown in the black box and at right) in transcriptomic studies of healthspan-promoting nutritional interventions are also noted. Icons from nounproject.com under Creative Commons license.
Suitability of Rodent Models
It is generally accepted that common responses to the most robust healthspan-promoting nutritional interventions are shared among mice and non-human primates, (68). However, it remains to be determined whether mice are a good translational model for studying the transcriptomic impact of nutritional interventions as they relate to humans. With CR (perhaps the most studied and robust nutritional intervention) for example, global plasma biomarker changes with CR in mice mimic those seen with fasting in humans (69), but recent research shows more divergent transcriptome effects of certain healthy nutritional interventions in mice and humans. For instance, an increase in stem cell maintenance and vascularization pathway activation is associated with CR in human subcutaneous white adipose tissue (WAT), but in mouse epididymal WAT these processes are downregulated with CR (31).
In addition to issues related to divergent mouse-human transcriptome effects, some commonly used mouse strains do not respond well to CR (e.g., they exhibit reduced lifespan) and/or other interventions. Most research on nutritional interventions and the transcriptome has utilized C57/B6 mice, a homogenous, inbred strain. However, It appears that CR results in greater lifespan extension in non-inbred mice when compared to inbred mice and may not be as effective in certain genetic backgrounds/strains (70). The underlying mechanisms are still under investigation, but some evidence shows that certain mouse strains exhibit completely opposite transcriptional responses to CR compared with other strains (e.g., BALBC, DBA), and these effects are tissue-specific (31). As such, when examining the influence of healthy nutritional interventions on the rodent transcriptome, it is important to consider the genetic strain/background and tissue(s) before drawing conclusions. This point parallels the need to consider genetic variability in humans (71) when considering the role and transcriptomic profiles of healthy aging interventions in aging/age-related disease. Indeed, it may be necessary to use transcriptomics and/or other omics approaches to develop “precision” dietary strategies tailored to individual people.
Tissues for Studying the Effects of Nutritional Interventions on the Transcriptome
Most pre-clinical nutritional intervention studies have largely focused on transcriptomic changes in liver and fat, and to a lesser extent, in brain and muscle. Liver, fat, and muscle are key metabolic tissues that are centrally implicated in the transcriptomic and metabolic responses to healthy aging nutritional interventions (72), and it appears that the responses to certain dietary interventions are different depending on type of tissue analyzed. Some transcriptomic pathways, like the insulin/IGF1 signaling response, seem to be equally affected by fasting across multiple tissues in rodents (73). However, the major metabolic tissues/organs of the body have vastly different metabolic rates (74) and may therefore respond to nutritional interventions differently in other gene expression pathways. As an example, AMPK, a key energy sensor and metabolism regulator, is activated by fasting in metabolic tissues like fat, liver, and muscle. However, the activation/response of AMPK (and related downstream mechanisms) to fasting in other tissues (e.g., brain, kidney) may vary (75). Current and future research should aim to determine transcriptomic overlap and differences of healthy nutritional interventions in multiple tissues, as this will broaden the scope of targetable mechanisms in tissue-specific diseases.
Sex-Specific Effects of Healthy Nutritional Interventions on the Aging Transcriptome
Male and female mice respond differently to anti-aging nutritional interventions (76). Key differences between sexes in response to nutritional interventions could be due to amino acid metabolism in the liver (77) or the ability of female mice to use increased energy from fat (through β-oxidation) during food restriction (78). Male and female mice also respond differently to varying intervention regimens (e.g., 20 vs. 40% CR) with different cardiometabolic and lifespan responses (24). Some recent RNA-seq studies have addressed these differences and found sex-specific responses in gene expression patterns with fasting and amino-acid restriction (79). Future research should continue to focus on such similarities/differences in transcriptomic responses to healthy aging nutritional interventions in both male and female mice, which may be key for understanding clinical applications.
The Non-coding Genome and Healthy Aging Nutritional Interventions
The non-coding genome (e.g., non-coding RNAs, microRNAs) is often ignored in transcriptomic studies on aging. However, certain non-coding transcripts are emerging as key players in aging and disease, and could be critical in understanding the transcriptomic effects of nutritional interventions. For example, microRNAs, which negatively regulate gene expression of their targets at post-transcriptional levels, may be intricately involved in aging gene expression pathways and the hallmarks of aging (e.g., genome stability) (80). Some non-coding/microRNAs are influenced by CR to modify chromatin, which may partly explain how this intervention regulates gene expression during aging (81). Even less commonly investigated, non-coding repetitive elements (RE) make up more than 60% of the genome (82), and it has been suggested that some of these RE can become active during aging, perhaps due to decreased chromatin stability (83). Indeed, there is a global increase in RE transcripts in model organisms and humans with aging (84), and some of these RE may be associated with certain hallmarks of aging, including inflammation, oxidative stress, and cellular senescence (85, 86). Interestingly, CR is associated with a global reduction in RE transcripts in liver, which further supports the idea that global RE transcript dysregulation may be an important transcriptional hallmark of aging and healthy nutritional interventions (28). The influence of other nutritional interventions on RE with aging remains to be determined, but could be a promising area of future investigation.
Conclusions
Nutrition has a profound impact on aging/age-related disease and lifespan. The transcriptomic effects of healthy nutritional interventions are still under investigation, but glucose regulation/insulin sensitivity (growth/metabolic homeostasis), oxidative phosphorylation, inflammatory/immune function, and circadian pathways appear to be key mediators of healthspan that are modulated by these interventions at the transcriptome level in multiple metabolic tissues and species. In order to identify additional genes/transcriptomic modules that may represent targets for promoting healthy aging, future transcriptome/multi-omic studies are needed to address key issues (e.g., differential sex and tissue responses, more comprehensive genomic profiling) and characterize the effects of more diets (e.g., Okinawan)—ideally in human subjects.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work is supported by the National Institute on Aging and National Institutes of Health (AG060302; TL and F32AG069361; DW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Partridge L, Deelen J, Slagboom PE. Facing up to the global challenges of ageing. Nature. (2018) 561:45–56. doi: 10.1038/s41586-018-0457-8
2. Sierra F, Kohanski R. Geroscience and the trans-NIH Geroscience Interest Group, GSIG. Geroscience. (2017) 39:1–5. doi: 10.1007/s11357-016-9954-6
3. López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. (2013) 153:1194–217. doi: 10.1016/j.cell.2013.05.039
4. Armenise C, Lefebvre G, Carayol J, Bonnel S, Bolton J, Di Cara A, et al. Transcriptome profiling from adipose tissue during a low-calorie diet reveals predictors of weight and glycemic outcomes in obese, non-diabetic subjects. Am J Clin Nutr. (2017) 106:736–46. doi: 10.3945/ajcn.117.156216
5. Wahl D, Cogger VC, Solon-Biet SM, Waern RV, Gokarn R, Pulpitel T, et al. Nutritional strategies to optimise cognitive function in the aging brain. Ageing Res Rev. (2016) 31:80–92. doi: 10.1016/j.arr.2016.06.006
6. Derous D, Mitchell SE, Green CL, Wang Y, Han JDJ, Chen L, et al. The effects of graded levels of calorie restriction: X. Transcriptomic responses of epididymal adipose tissue. J Gerontol A Biol Sci Med Sci. (2018) 73:279–88. doi: 10.1093/gerona/glx101
7. Rhoads TW, Burhans MS, Chen VB, Hutchins PD, Rush MJP, Clark JP, et al. Caloric restriction engages hepatic RNA processing mechanisms in rhesus monkeys. Cell Metab. (2018) 27:677–88.e5. doi: 10.1016/j.cmet.2018.01.014
8. Mercken EM, Crosby SD, Lamming DW, JeBailey L, Krzysik-Walker S, Villareal DT, et al. Calorie restriction in humans inhibits the PI3K/AKT pathway and induces a younger transcription profile. Aging Cell. (2013) 12:645–51. doi: 10.1111/acel.12088
9. Anton SD, Moehl K, Donahoo WT, Marosi K, Lee SA, Mainous AG, et al. Flipping the metabolic switch: understanding and applying the health benefits of fasting. Obesity. (2018) 26:254–68. doi: 10.1002/oby.22065
10. Longo VD, Cortellino S. Fasting, dietary restriction, and immunosenescence. J Allergy Clin Immunol. (2020) 146:1002–4. doi: 10.1016/j.jaci.2020.07.035
11. Swindell WR. Genes and gene expression modules associated with caloric restriction and aging in the laboratory mouse. BMC Genomics. (2009) 10:585. doi: 10.1186/1471-2164-10-585
12. Kim SE, Mori R, Shimokawa I. Does calorie restriction modulate inflammaging via FoxO transcription factors? Nutrients. (2020) 12:1959. doi: 10.3390/nu12071959
13. López-Lluch G, Navas P. Calorie restriction as an intervention in ageing. J Physiol. (2016) 594:2043–60. doi: 10.1113/JP270543
14. Ng GY, Kang SW, Kim J, Alli-Shaik A, Baik SH, Jo DG, et al. Genome-wide transcriptome analysis reveals intermittent fasting-induced metabolic rewiring in the liver. Dose Response. (2019) 17. doi: 10.1177/1559325819876780
15. Kim J, Kang SW, Mallilankaraman K, Baik SH, Lim JC, Balaganapathy P, et al. Transcriptome analysis reveals intermittent fasting-induced genetic changes in ischemic stroke. Hum Mol Genet. (2018) 27:1497–513. doi: 10.1093/hmg/ddy057
16. Elliott RM, de Roos B, Duthie SJ, Bouwman FG, Rubio-Aliaga I, Crosley LK, et al. Transcriptome analysis of peripheral blood mononuclear cells in human subjects following a 36 h fast provides evidence of effects on genes regulating inflammation, apoptosis and energy metabolism. Genes Nutr. (2014) 9:432. doi: 10.1007/s12263-014-0432-4
17. Zhao L, Hutchison AT, Wittert GA, Thompson CH, Lange K, Liu B, et al. Intermittent fasting does not uniformly impact genes involved in circadian regulation in women with obesity. Obesity. (2020) 28(Suppl. 1):S63–7. doi: 10.1002/oby.22775
18. Straus DS, Takemoto CD. Effect of fasting on insulin-like growth factor-I (IGF-I) and growth hormone receptor mRNA levels and IGF-I gene transcription in rat liver. Mol Endocrinol. (1990) 4:91–100. doi: 10.1210/mend-4-1-91
19. Mattson MP, Arumugam TV. Hallmarks of brain aging: adaptive and pathological modification by metabolic states. Cell Metab. (2018) 27:1176–99. doi: 10.1016/j.cmet.2018.05.011
20. Longo VD, Panda S. Fasting, circadian rhythms, and time-restricted feeding in healthy lifespan. Cell Metab. (2016) 23:1048–59. doi: 10.1016/j.cmet.2016.06.001
21. Chaix A, Lin T, Le HD, Chang MW, Panda S. Time-restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metabolism. (2019) 29:303–19.e4. doi: 10.1016/j.cmet.2018.08.004
22. Solon-Biet SM, Cogger VC, Pulpitel T, Wahl D, Clark X, Bagley E, et al. Branched chain amino acids impact health and lifespan indirectly via amino acid balance and appetite control. Nat Metab. (2019) 1:532–45. doi: 10.1038/s42255-019-0059-2
23. Oishi K, Uchida D, Itoh N. Low-carbohydrate, high-protein diet affects rhythmic expression of gluconeogenic regulatory and circadian clock genes in mouse peripheral tissues. Chronobiol Int. (2012) 29:799–809. doi: 10.3109/07420528.2012.699127
24. Mitchell SJ, Madrigal-Matute J, Scheibye-Knudsen M, Fang E, Aon M, González-Reyes JA, et al. Effects of sex, strain, and energy intake on hallmarks of aging in mice. Cell Metab. (2016) 23:1093–112. doi: 10.1016/j.cmet.2016.05.027
25. Cao SX, Dhahbi JM, Mote PL, Spindler SR. Genomic profiling of short- and long-term caloric restriction effects in the liver of aging mice. Proc Nat Acad Sci. (2001) 98:10630–5. doi: 10.1073/pnas.191313598
26. Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. (1999) 285:1390–3. doi: 10.1126/science.285.5432.1390
27. Wheatley KE, Nogueira LM, Perkins SN, Hursting SD. Differential effects of calorie restriction and exercise on the adipose transcriptome in diet-induced obese mice. J Obes. (2011) 2011:265417. doi: 10.1155/2011/265417
28. Wahl D, Cavalier AN, Smith M, Seals DR, LaRocca TJ. Healthy aging interventions reduce repetitive element transcripts. J Gerontol A Biol Sci Med Sci. (2020) 76:805–10. doi: 10.1101/2020.06.25.172023
29. Mutch DM, Pers TH, Temanni MR, Pelloux V, Marquez-Quiñones A, Holst C, et al. A distinct adipose tissue gene expression response to caloric restriction predicts 6-mo weight maintenance in obese subjects. Am J Clin Nutr. (2011) 94:1399–409. doi: 10.3945/ajcn.110.006858
30. Lopaschuk GD. Fatty acid oxidation and its relation with insulin resistance and associated disorders. Ann Nutr Metabol. (2016) 68(Suppl. 3):15–20. doi: 10.1159/000448357
31. Swindell WR, List EO, Berryman DE, Kopchick JJ. Transcriptional profiling identifies strain-specific effects of caloric restriction and opposite responses in human and mouse white adipose tissue. Aging. (2018) 10:701–46. doi: 10.18632/aging.101424
32. de Cabo R, Mattson MP. Effects of intermittent fasting on health, aging, and disease. N Engl J Med. (2019) 381:2541–51. doi: 10.1056/NEJMra1905136
33. Mitchell SJ, Bernier M, Mattison JA, Aon MA, Kaiser TA, Anson RM, et al. Daily fasting improves health and survival in male mice independent of diet composition and calories. Cell Metab. (2019) 29:221–8.e3. doi: 10.1016/j.cmet.2018.08.011
34. Tyagi S, Gupta P, Saini AS, Kaushal C, Sharma S. The peroxisome proliferator-activated receptor: a family of nuclear receptors role in various diseases. J Adv Pharm Technol Res. (2011) 2:236–40. doi: 10.4103/2231-4040.90879
35. Lilja S, Stoll C, Krammer U, Hippe B, Duszka K, Debebe T, et al. Five days periodic fasting elevates levels of longevity related christensenella and sirtuin expression in humans. Int J Mol Sci. (2021) 22:2331. doi: 10.3390/ijms22052331
36. Wegman MP, Guo MH, Bennion DM, Shankar MN, Chrzanowski SM, Goldberg LA, et al. Practicality of intermittent fasting in humans and its effect on oxidative stress and genes related to aging and metabolism. Rejuvenation Res. (2015) 18:162–72. doi: 10.1089/rej.2014.1624
37. Rynders CA, Thomas EA, Zaman A, Pan Z, Catenacci VA, Melanson EL. Effectiveness of intermittent fasting and time-restricted feeding compared to continuous energy restriction for weight loss. Nutrients. (2019) 11:2442. doi: 10.3390/nu11102442
38. Longo VD, Di Tano M, Mattson MP, Guidi N. Intermittent and periodic fasting, longevity and disease. Nature Aging. (2021) 1:47–59. doi: 10.1038/s43587-020-00013-3
39. Henderson CG, Turner DL, Swoap SJ. Health effects of alternate day fasting versus pair-fed caloric restriction in diet-induced obese C57Bl/6J male mice. Front Physiol. (2021) 12:641532. doi: 10.3389/fphys.2021.641532
40. Sokolović M, Sokolović A, Wehkamp D, Ver Loren van Themaat E, de Waart DR, Gilhuijs-Pederson LA, et al. The transcriptomic signature of fasting murine liver. BMC Genomics. (2008) 9:528. doi: 10.1186/1471-2164-9-528
41. Kinouchi K, Magnan C, Ceglia N, Liu Y, Cervantes M, Pastore N, et al. Fasting imparts a switch to alternative daily pathways in liver and muscle. Cell Rep. (2018) 25:3299–314.e6. doi: 10.1016/j.celrep.2018.11.077
42. Fazeli PK, Zhang Y, O'Keefe J, Pesaresi T, Lun M, Lawney B, et al. Prolonged fasting drives a program of metabolic inflammation in human adipose tissue. Mol Metab. (2020) 42:101082. doi: 10.1016/j.molmet.2020.101082
43. McAllister MJ, Pigg BL, Renteria LI, Waldman HS. Time-restricted feeding improves markers of cardiometabolic health in physically active college-age men: a 4-week randomized pre-post pilot study. Nutr Res. (2020) 75:32–43. doi: 10.1016/j.nutres.2019.12.001
44. Mattson MP, Longo VD, Harvie M. Impact of intermittent fasting on health and disease processes. Ageing Res Rev. (2017) 39:46–58. doi: 10.1016/j.arr.2016.10.005
45. Lundell LS, Parr EB, Devlin BL, Ingerslev LR, Altintaş A, Sato S, et al. Time-restricted feeding alters lipid and amino acid metabolite rhythmicity without perturbing clock gene expression. Nat Commun. (2020) 11:4643. doi: 10.1038/s41467-020-18412-w
46. Jamshed H, Beyl RA, Della Manna DL, Yang ES, Ravussin E, Peterson CM. Early time-restricted feeding improves 24-hour glucose levels and affects markers of the circadian clock, aging, and autophagy in humans. Nutrients. (2019) 11:1234. doi: 10.3390/nu11061234
47. Giebultowicz JM. Circadian regulation of metabolism and healthspan in Drosophila. Free Radic Biol Med. (2018) 119:62–8. doi: 10.1016/j.freeradbiomed.2017.12.025
48. Yu D, Richardson NE, Green CL, Spicer AB, Murphy ME, Flores V, et al. The adverse metabolic effects of branched-chain amino acids are mediated by isoleucine and valine. Cell Metab. (2021) 33:905–22.e6. doi: 10.1016/j.cmet.2021.03.025
49. Solon-Biet SM, McMahon AC, Ballard JWO, Ruohonen K, Wu LE, Cogger VC, et al. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. (2020) 31:654. doi: 10.1016/j.cmet.2020.01.010
50. Treviño-Villarreal JH, Reynolds JS, Bartelt A, Langston PK, MacArthur MR, Arduini A, et al. Dietary protein restriction reduces circulating VLDL triglyceride levels via CREBH-APOA5–dependent and –independent mechanisms. JCI Insight. (2018) 3:e99470. doi: 10.1172/jci.insight.99470
51. Wahl D, Solon-Biet SM, Wang QP, Wali JA, Pulpitel T, Clark X, et al. Comparing the effects of low-protein and high-carbohydrate diets and caloric restriction on brain aging in mice. Cell Rep. (2018) 25:2234–43.e6. doi: 10.1016/j.celrep.2018.10.070
52. Gokarn R, Solon-Biet SM, Cogger VC, Cooney GJ, Wahl D, McMahon AC, et al. Long-term dietary macronutrients and hepatic gene expression in aging mice. J Gerontol A Biol Sci Med Sci. (2018) 73:1618–25. doi: 10.1093/gerona/gly065
53. Tilstra JS, Clauson CL, Niedernhofer LJ, Robbins PD. NF-κB in aging and disease. Aging Dis. (2011) 2:449–65.
54. Patel SA, Velingkaar N, Makwana K, Chaudhari A, Kondratov R. Calorie restriction regulates circadian clock gene expression through BMAL1 dependent and independent mechanisms. Sci Rep. (2016) 6:25970. doi: 10.1038/srep25970
55. Wahl D, Cavalier AN, LaRocca TJ. Novel strategies for healthy brain aging. Exerc Sport Sci Rev. (2021) 49:115–25. doi: 10.1249/JES.0000000000000242
56. Tsigalou C, Konstantinidis T, Paraschaki A, Stavropoulou E, Voidarou C, Bezirtzoglou E. Mediterranean diet as a tool to combat inflammation and chronic diseases. An overview. Biomedicines. (2020) 8:201. doi: 10.3390/biomedicines8070201
57. Brink TC, Regenbrecht C, Demetrius L, Lehrach H, Adjaye J. Activation of the immune response is a key feature of aging in mice. Biogerontology. (2009) 10:721. doi: 10.1007/s10522-009-9219-1
58. Lanza IR, Zabielski P, Klaus KA, Morse DM, Heppelmann CJ, Bergen HR, et al. Chronic caloric restriction preserves mitochondrial function in senescence without increasing mitochondrial biogenesis. Cell Metabol. (2012) 16:777–88. doi: 10.1016/j.cmet.2012.11.003
59. Wahl D, Solon-Biet SM, Cogger VC, Fontana L, Simpson SJ, Le Couteur DG, et al. Aging, lifestyle, and dementia. Neurobiol Dis. (2019) 130:104481. doi: 10.1016/j.nbd.2019.104481
60. Astafev AA, Patel SA, Kondratov RV. Calorie restriction effects on circadian rhythms in gene expression are sex dependent. Sci Rep. (2017) 7:9716. doi: 10.1038/s41598-017-09289-9
61. García-Gaytán AC, Miranda-Anaya M, Turrubiate I, López-De Portugal L, Bocanegra-Botello GN, López-Islas A, et al. Synchronization of the circadian clock by time-restricted feeding with progressive increasing calorie intake. Resemblances and differences regarding a sustained hypocaloric restriction. Sci Rep. (2020) 10:10036. doi: 10.1038/s41598-020-66538-0
62. Wahl D, Coogan SC, Solon-Biet SM, de Cabo R, Haran JB, Raubenheimer D, et al. Cognitive and behavioral evaluation of nutritional interventions in rodent models of brain aging and dementia. Clin Interv Aging. (2017) 12:1419–28. doi: 10.2147/CIA.S145247
63. Le Couteur DG, Solon-Biet S, Wahl D, Cogger VC, Willcox BJ, Willcox DC, et al. New horizons: dietary protein, ageing and the Okinawan ratio. Age Ageing. (2016) 45:443–7. doi: 10.1093/ageing/afw069
64. Ngandu T, Lehtisalo J, Solomon A, Levälahti E, Ahtiluoto S, Antikainen R, et al. A 2 year multidomain intervention of diet, exercise, cognitive training, and vascular risk monitoring versus control to prevent cognitive decline in at-risk elderly people (FINGER): a randomised controlled trial. Lancet. (2015) 385:2255–63. doi: 10.1016/S0140-6736(15)60461-5
65. Marlow G, Ellett S, Ferguson IR, Zhu S, Karunasinghe N, Jesuthasan AC, et al. Transcriptomics to study the effect of a Mediterranean-inspired diet on inflammation in Crohn's disease patients. Hum Genomics. (2013) 7:24. doi: 10.1186/1479-7364-7-24
66. Herrera-Marcos LV, Lou-Bonafonte JM, Arnal C, Navarro MA, Osada J. Transcriptomics and the Mediterranean diet: a systematic review. Nutrients. (2017) 9:472. doi: 10.3390/nu9050472
67. The Psi. In vivo transcriptomic profile after a Mediterranean diet in high–cardiovascular risk patients: a randomized controlled trial. Am J Clin Nutr. (2013) 98:845–53. doi: 10.3945/ajcn.113.060582
68. Anderson RM, Shanmuganayagam D, Weindruch R. Caloric restriction and aging: studies in mice and monkeys. Toxicol Pathol. (2009) 37:47–51. doi: 10.1177/0192623308329476
69. Mahoney LB, Denny CA, Seyfried TN. Caloric restriction in C57BL/6J mice mimics therapeutic fasting in humans. Lipids Health Dis. (2006) 5:13. doi: 10.1186/1476-511X-5-13
70. Swindell WR. Dietary restriction in rats and mice: a meta-analysis and review of the evidence for genotype-dependent effects on lifespan. Ageing Res Rev. (2012) 11:254–70. doi: 10.1016/j.arr.2011.12.006
71. Wheeler HE, Kim SK. Genetics and genomics of human ageing. Philos Trans R Soc Lond B Biol Sci. (2011) 366:43–50. doi: 10.1098/rstb.2010.0259
72. Lundbom J. Adipose tissue and liver. J Appl Physiol. (2018) 124:162–7. doi: 10.1152/japplphysiol.00399.2017
73. Sharma N, Castorena CM, Cartee GD. Tissue-specific responses of IGF-1/insulin and mTOR signaling in calorie restricted rats. PLoS ONE. (2012) 7:e38835. doi: 10.1371/journal.pone.0038835
74. Shirley MK, Arthurs OJ, Seunarine KK, Cole TJ, Eaton S, Williams JE, et al. Metabolic rate of major organs and tissues in young adult South Asian women. Eur J Clin Nutr. (2019) 73:1164–71. doi: 10.1038/s41430-018-0362-0
75. Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. (2011) 13:1016–23. doi: 10.1038/ncb2329
76. Austad SN, Bartke A. Sex differences in longevity and in responses to anti-aging interventions: a mini-review. Gerontology. (2015) 62:40–6. doi: 10.1159/000381472
77. Della Torre S, Mitro N, Meda C, Lolli F, Pedretti S, Barcella M, et al. Short-term fasting reveals amino acid metabolism as a major sex-discriminating factor in the liver. Cell Metab. (2018) 28:256–67.e5. doi: 10.1016/j.cmet.2018.05.021
78. Freire T, Senior AM, Perks R, Pulpitel T, Clark X, Brandon AE, et al. Sex-specific metabolic responses to 6 hours of fasting during the active phase in young mice. J Physiol. (2020) 598:2081–92. doi: 10.1113/JP278806
79. Tyshkovskiy A, Bozaykut P, Borodinova AA, Gerashchenko MV, Ables GP, Garratt M, et al. Identification and application of gene expression signatures associated with lifespan extension. Cell Metab. (2019) 30:573–93.e8. doi: 10.1016/j.cmet.2019.06.018
80. Jung HJ, Suh Y. MicroRNA in aging: from discovery to biology. Curr Genomics. (2012) 13:548–57. doi: 10.2174/138920212803251436
81. Green CD, Huang Y, Dou X, Yang L, Liu Y, Han J-DJ. Impact of dietary interventions on non-coding RNA networks and mRNAs encoding chromatin-related factors. Cell Rep. (2017) 18:2957–68. doi: 10.1016/j.celrep.2017.03.001
82. Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, et al. Ten things you should know about transposable elements. Genome Biol. (2018) 19:199. doi: 10.1186/s13059-018-1577-z
83. Goodier JL. Restricting retrotransposons: a review. Mobile DNA. (2016) 7:16. doi: 10.1186/s13100-016-0070-z
84. LaRocca TJ, Cavalier AN, Wahl D. Repetitive elements as a transcriptomic marker of aging: evidence in multiple datasets and models. Aging Cell. (2020) 19:e13167. doi: 10.1111/acel.13167
85. De Cecco M, Ito T, Petrashen AP, Elias AE, Skvir NJ, Criscione SW, et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature. (2019) 566:73–8. doi: 10.1038/s41586-018-0784-9
Keywords: aging, RNA-Seq - RNA sequencing, transcriptome, healthspan, nutrition
Citation: Wahl D and LaRocca TJ (2021) Transcriptomic Effects of Healthspan-Promoting Dietary Interventions: Current Evidence and Future Directions. Front. Nutr. 8:712129. doi: 10.3389/fnut.2021.712129
Received: 19 May 2021; Accepted: 14 July 2021;
Published: 10 August 2021.
Edited by:
Sergio Davinelli, University of Molise, ItalyReviewed by:
Andreas Zimmermann, University of Graz, AustriaGiulia Accardi, University of Palermo, Italy
Copyright © 2021 Wahl and LaRocca. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas J. LaRocca, VG9tLkxhUm9jY2FAY29sb3N0YXRlLmVkdQ==
†These authors have contributed equally to this work
 Devin Wahl
Devin Wahl Thomas J. LaRocca
Thomas J. LaRocca