
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr. , 19 May 2021
Sec. Nutrition and Microbes
Volume 8 - 2021 | https://doi.org/10.3389/fnut.2021.690138
This article is part of the Research Topic Gut Microbial Response to Host Metabolic Phenotypes: Volume 1 View all 24 articles
Increasing studies have shown that obesity is the primary cause of cardiovascular diseases, non-alcoholic fatty liver diseases, type 2 diabetes, and a variety of cancers. The dysfunction of gut microbiota was proved to result in obesity. Recent research indicated ANGPTL4 was a key regulator in lipid metabolism and a circulating medium for gut microbiota and fat deposition. The present study was conducted to investigate the alteration of gut microbiota and ANGPTL4 expression in the gastrointestinal tract of mice treated by the high-fat diet. Ten C57BL/6J mice were randomly allocated to two groups and fed with a high-fat diet (HFD) containing 60% fat or a normal-fat diet (Control) containing 10% fat. The segments of ileum and colon were collected for the determination of ANGPTL4 expression by RT-qPCR and immunohistochemical analysis while the ileal and colonic contents were collected for 16S rRNA gene sequencing. The results showed HFD significantly increased mice body weight, epididymal fat weight, perirenal fat weight, liver weight, and the lipid content in the liver (P < 0.05). The relative expression of ANGPTL4 and the ANGPTL4-positive cells in the ileum and colon of mice was significantly increased by HFD treatment. Furthermore, 16S rRNA gene sequencing of the ileal and colonic microbiota suggested that HFD treatment changed the composition of the gut microbiota. The ratio of Firmicutes to Bacteroidetes and the abundance of Allobaculum was significantly higher in the HFD group than in the Control group while the abundance of Adlercreutzia, Bifidobacterium, Prevotellaceae UCG-001, and Ruminococcus was significantly decreased. Interestingly, the abundance of Allobaculum was positively correlated with the expression of ANGPTL4. These findings provide a theoretical foundation for the development of strategies to control the obesity and related diseases by the regulation of ANGPTL4 and gut microbiota.
The epidemic rise in obesity has chronically challenged human health, performance and quality of life with affecting more than 2 billion people in the world and being related to diabetes, cancers and cardiovascular diseases (1, 2). Gut microbiota is composed of numerous bacteria that contribute to nutrition absorption and energy homeostasis (3, 4). Increasing evidences indicate that gut microbiota directly participates in obesity and many other metabolic diseases (4, 5). A recent study have shown that obesity could be induced by the high-fat diet (HFD) (6). Meanwhile, the dysregulation in the composition and metabolic functions of gut microbiota would promote the development of obesity (7). The microbiota transplantation from girls with or without obesity to mice showed a close relationship between gut microbiota and obesity (8). Furthermore, the significant higher abundance ratio of Firmicutes to Bacteroidetes is generally regarded as a marker signal of obesity (9). Therefore, gut microbiota is closely related to host lipid metabolism, the disorder of which alters the composition of gut microbiota. The dysfunctional gut microbiota would further affects the host lipid metabolism in turn (10).
Angiogenin-like protein 4 (ANGPTL4), also known as a fasting induction factor (FIAF), plays an important role in lipid deposition by inhibiting lipoprotein lipase (LPL) to regulate lipid metabolism (11, 12). ANGPTL4 can be secreted in intestines, adipose tissue, liver, skeletal muscle, heart and other tissues, and is subsequently cleaved into N-terminal and C-terminal fragments. The N-terminal of ANGPTL4 acts as a LPL inhibitor (13, 14). LPL is transported by the GPIHBP1 protein to the lumen side of capillary endothelial cells and catalyzes the hydrolysis of triglycerides (TG) into fatty acids. This process allows lipids transported from the circulation into skeletal muscle, heart and adipose tissue after absorption (15, 16). In addition, ANGPTL4 is an endogenous inhibitor for intestinal fat digestive enzymes, especially pancreatic lipase, to prevent excessive fatty acids intake and lipid overload in intestinal cells (17, 18). Furthermore, ANGPTL4 has been considered as a circulating mediator between gut microbiota and fat storage (19). The germ-free (GF) mice with a normal microbiota harvested from the cecum of conventionally raised mice could improve the TG storage in fat cells with inhibition of ANGPTL4 expression in gut, suggesting gut microbiota might promote fat deposition by modulating ANGPTL4 expression (19). The expression of lipogenic genes in the abdominal fat of mice receiving the fecal microbiota of Jinhua pigs (obese) was higher than that in the mice receiving the fecal microbiota of Landrace pigs (lean) with reduction of ANGPTL4 expression in the gastrointestinal tract (20). Therefore, the ANGPTL4 expression might be one of the key regulators for obesity induced by the dysfunction of gut microbiota.
ANGPTL4 acts as a circulating medium for the gut microbiota and fat deposition in the body, so it is of great significance to explore how the gut microbiota affects the expression of ANGPTL4. However, the research on the regulation of ANGPTL4 expression by gut microbiota in obesity is limited. Accordingly, the objective of the present study is to explore the relationship among the gut microbiota structure, the ANGPTL4 expression and fat deposition.
Ten specific pathogen-free (SPF) C57BL/6J male mice weaned at the age of 28 days were purchased from GemPharmatech Co., Ltd (Nanjing, China). The mice were raised in cages at 25 ± 2°C for 12 h light/dark cycles with free access to water and mouse chow. After acclimatization for 1 week, the mice were weighed and randomly divided into Control group and high-fat diet (HFD) group. Mice in the Control and HFD groups were fed with a commercial standard diet with 10% fat content and the standard diet supplemented with 60% fat, respectively for 12 weeks (21, 22). At the end of 12-week study, all mice were weighed individually and sacrificed by decapitation following a CO2 stun. The epididymal fat and perirenal fat of each mice were isolated and weighed. The ileal and colonic contents were collected and stored at −20°C until DNA isolation and 16S rRNA gene sequencing. Segments of liver, ileum and colon were collected and fixed in 4% paraformaldehyde for further analysis. Ileum and colon segments were isolated, rinsed with 0.9% NaCl, immediately frozen in liquid nitrogen, and stored at −80°C until RNA extraction.
Microbial genomic DNA was extracted from ileal and colonic contents using QIAamp DNA Stool Mini Kit (QIAGEN, Valencia, CA, US). The V4~V5 region of bacterial 16S rRNA gene was amplified from each genomic DNA sample by using the barcode-fusion primers 515F and 907R. Sequencing libraries were then constructed using TruSeq DNA PCR-Free Library Preparation Kit (Illumina) and sequenced on an Illumina HiSeq platform at Mingke Biotechnology (Hangzhou) Co., Ltd. The sequencing data were analyzed using QIIME software package (23). The non-repeating sequences were analyzed by operational classification unit (OTU) with 97% similarity. Species matching was performed for all representative sequences of OTU using RPD databases (24). Pie charts were generated to show taxa distribution at the phylum and genus levels. Principal coordinate analysis (PCoA) was performed to analyze the beta diversity. For further identify the specific genera related to fat deposition, this study also analyzed the raw data of gut microbiota combining with other two similar studies on mice fed with high-fat diets (DDBJ Accession Number: PRJDB7523) (25) (NCBI Accession Number: SRP113647) (26).
Total RNA was isolated using TRIzol®Plus RNA Purification Kit (Invitrogen) and RNase-Free DNase Set (Qiagen) followed by reverse transcription using the SuperScript™ III First-Strand Synthesis SuperMix (Invitrogen) strictly according to the manufacturer’s instructions. Real-time qPCR was performed in triplicate on an ABI Prism 7700 Sequence Detector system (Applied Biosystems, Foster City, CA, USA) using an annealing temperature of 63°C and gene-specific primers listed in Table 1. The data were normalized to GAPDH or 18S rRNA and calculated by 2−ΔΔCT method (27).
The liver egments were fixed in 4% paraformaldehyde for 1 h at room temperature, cryoprotected in 20% sucrose at 4°C overnight, and embedded in OCT. The prepared series of 12-μm cryosections were stained with Oil Red O (Sigma-Aldrich, St. Louis, MO, United States).
The ileal and colonic sections were deparaffinized and microwaved in sodium citrate buffer for antigen retrieval. After being rinsed with phosphate buffered saline (PBS), slides were incubated with PBS containing normal goat serum. After blocking, sections were incubated with primary antibody (proteintech ANGPTL4 18374-1-AP) at 4°C overnight. After wash with PBS and incubation with secondary antibody for 50 min at room temperature, slides were rinsed with PBS for three times and added with a few drops of chromogenic substrate DAB with incubation for 15 min. Finally, slides were rinsed in water and counterstained using hematoxylin solution for picture caption.
All statistical analyses were performed by SPSS 23.0 (IBM, New York, NY, United States) using unpaired two-tailed Student’s t-test (20). Data are presented as the mean ± SEM. Results were considered significant when P < 0.05.
To confirm whether obesity was induced by the HFD, we weighed each mouse, liver, epididymal fat, and perirenal fat individually in both of Control and HFD groups. The body weight, liver weight, epididymal fat, and perirenal fat in the HFD-treated mice were significantly higher than those of the Control group by 82.31% (P < 0.0001), 106.58% (P = 0.0234), 194.08% (P = 0.0011), and 627.91% (P = 0.0046), respectively (Figures 1A–D). To observe the fat content in the liver, we stained liver segments of mice in the Control and HFD groups. The number of lipid droplets in the liver of HFD group (Figure 1F) was obviously more and larger than that in Control group (Figure 1E), suggesting obesity had been induced by HFD.
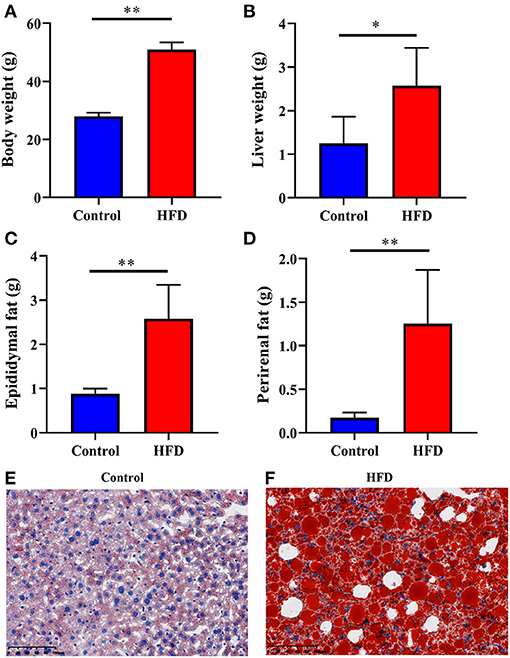
Figure 1. Characterization of the obesity induced by HFD. The Control and HFD groups mice were analyzed for body weight (A), liver weight (B), epididymal fat weight (C), perirenal fat weight (D). Oil-red O stains of paraformaldehyde-fixed liver sections prepared from the Control (E) and HFD (F) group mice. Data were expressed as mean ± SEM and statistically analyzed by using unpaired two-tailed Students’ t-test (n = 5). Asterisks (* and **) represent significant differences with P < 0.05 and P < 0.01, respectively.
To examine whether the ANGPTL4 expression is involved in fat deposition, we determined the ANGPTL4 expression in the gastrointestinal tract by RT-qPCR analysis following RNA isolation and reverse transcription. Compared with the Control group, the relative expression of ANGPTL4 in the ileum (P = 0.0452) and colon (P = 0.0409) of mice in HFD group was increased significantly (Figures 2A,B). Furthermore, immunohistochemistry revealed the ANGPTL4-positive cells seemed to be more abundant in the HFD group (Figures 2D,F) than the Control group (Figures 2C,E), which was consistent with the trend of the relative expression of ANGPTL4 in the ileum and colon of mice in two groups.
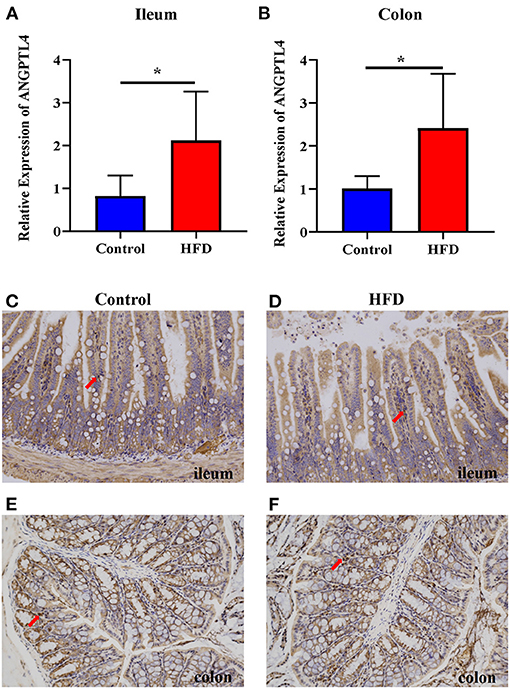
Figure 2. The expression of ANGPTL4 in the ileum and colon of the Control and HFD groups. The mRNA expression level of ANGPTL4 was measured in the ileum (A) and colon (B) of mice in the Control and HFD groups using RT-qPCR analysis followed by RNA isolation. Sections of the ileum (C,D) and colon (E,F) in Control group (C,E) and HFD group (D,F) were stained with rabbit polyclonal antibodies to mouse ANGPTL4. The arrowhead points in the direction where ANGPTL4-positive cells are present. Data were expressed as mean ± SEM and statistically analyzed by using unpaired two-tailed Students’ t-test (n = 5). Asterisks (*) represent significant differences with P < 0.05.
To investigate the structure of gut microbiota in mice of the Control and HFD groups, we collected the ileal and colonic contents from mice of the Control and HFD groups and analyzed the alpha -diversity of gut microbiota. The alpha-diversity indicated that the number of OTU, Shannon index, and Chao1 index in the Control group were significantly higher than that in the HFD group (P < 0.05) while the Simpson index was significantly lower of Control group compared to HFD group (Table 2).
Next, we analyzed the composition of gut microbiota in the ileum and colon of mice in the Control and HFD groups. Taxonomic analysis showed that the dominant bacteria phyla were Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Deferribacteres, and Cyanobacteria in both of ileum and colon, accounting for more than 99.12% of the total sequences in most samples (Figures 3A, 4A). The ratio of Firmicutes to Bacteroidetes, that was associated with the obesity phenotype, was remarkably higher in the ileum and colon of mice in the HFD group than those of the Control group (Figures 3, 4). Compared to the Control group, the relative abundance of Firmicutes in the HFD group was increased by 38.81 and 15.00% in ileum and colon (P < 0.05), respectively. However, the relative abundance of Bacteroidetes in the HFD group was decreased by 48.45 and 20.02% in ileal and colonic samples relative to the Control group (P < 0.05), respectively (Figures 3A, 4A). Cyanobacteria was less abundant in both of the ileum and colon of HFD group compared to Control group.
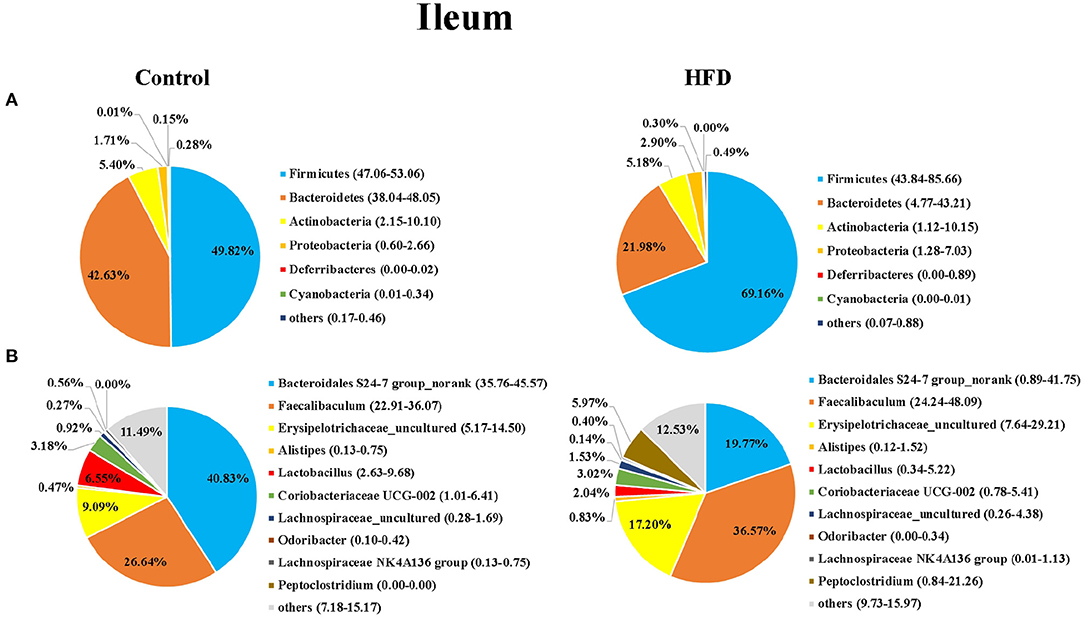
Figure 3. The bacterial community in ileum of the Control group and HFD group mice at the phylum (A) and genus (B) levels. The top 6 phyla and top 10 genera are shown.
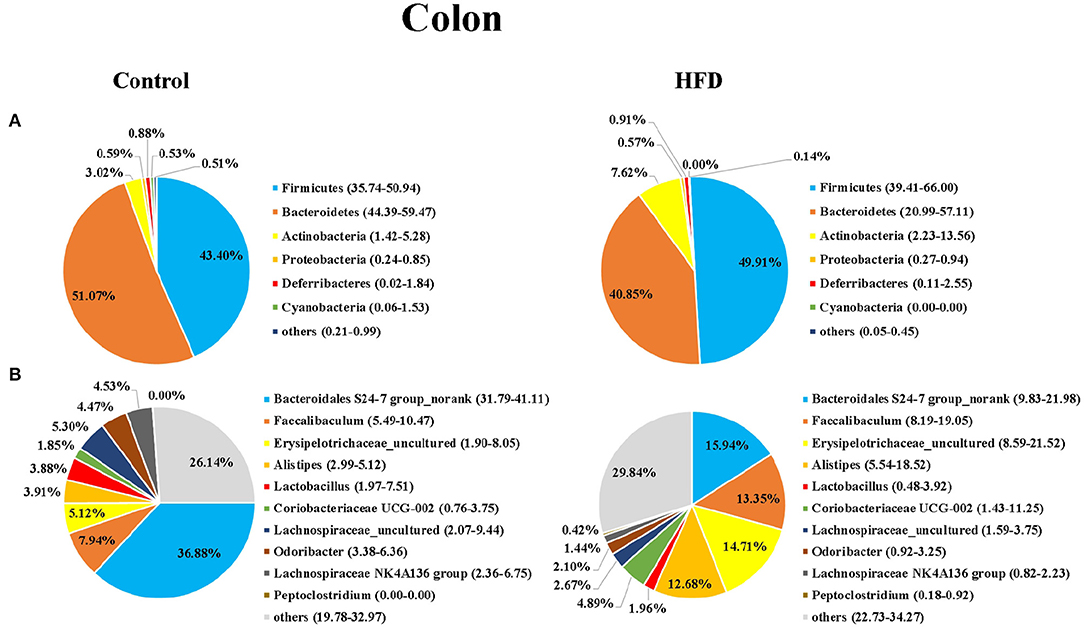
Figure 4. The bacterial community in colon of mice in the Control group and HFD group at the phylum (A) and genus (B) levels. The top 6 phyla and top 10 genera are shown.
At the genus level, Bacteroidales S24-7 group_norank, Faecalibaculum, Erysipelotrichaceae _uncultured, Alistipes, Lactobacillus, Coriobacteriaceae UCG-002, Lachnospiraceae_uncultured, Odoribacter, Lachnospiraceae NK4A136dium, and Peptoclostri were the most abundant genera in both of the ileum and colon (Figures 3B, 4B). Compared to the Control group, the relative abundance of Bacteroidales S24-7 group_norank in the HFD group was decreased by 51.58% and 56.76% in ileum and colon (P < 0.05), respectively, while the relative abundance of Faecalibaculum was increased by 37.27 and 68.18% in the ileal and colonic samples (P < 0.05), respectively (Figures 3B, 4B). The relative abundance of Lactobacillus and Odoribacter was decreased while Alistipes and Peptoclostridium were increased in the ileum and colon (Figures 3B, 4B) of the HFD group as compared to the Control group.
Hierarchical clustering together with a heat-map was performed to reveal the distinct characteristics of the significantly different bacterial genera (including the 21 from 248 total ileal bacterial genera, and 46 from 248 total colonic bacterial genera) based on the abundance of the identified bacterial genera in the Control group and HFD group (Figure 5). For example, the relative abundance of Allobaculum in ileum and colon of mice in the HFD group was significantly higher than that in the Control group (P < 0.05) (Figure 5). The relative abundance of Adlercreutzia, Bifidobacterium, Prevotellaceae UCG-001 and Ruminococcus in the ileum and colon in HFD group was significantly lower than those in Control group (P < 0.05; Figure 5).
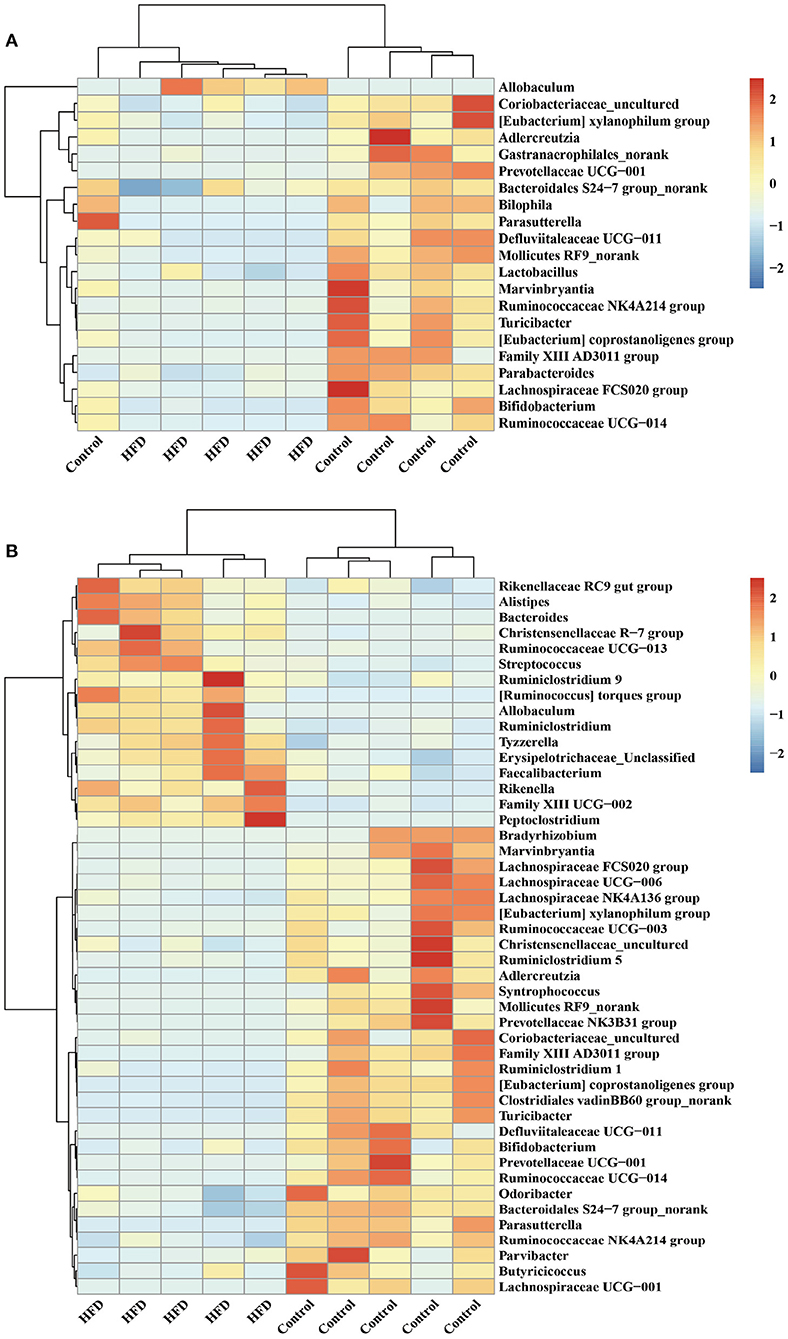
Figure 5. Hierarchically clustered heat map for the significantly different bacterial genera in ileum (A) and colon (B) of mice.
To evaluate the degree of discrepancy between the bacterial community structures of the Control group and HFD group mice, a principal coordinate analysis (PCoA) was performed. The ileal and colonic microbiota in the HFD group and the Control group were clearly separated from each other (Figure 6A). There was also a differently clustering of the bacterial community structure between the Control and HFD groups in ileum and colon (Figures 6B,C), indicating that the structure of the ileum and colon microbiota of the mice had a great difference between the Control and HFD groups.
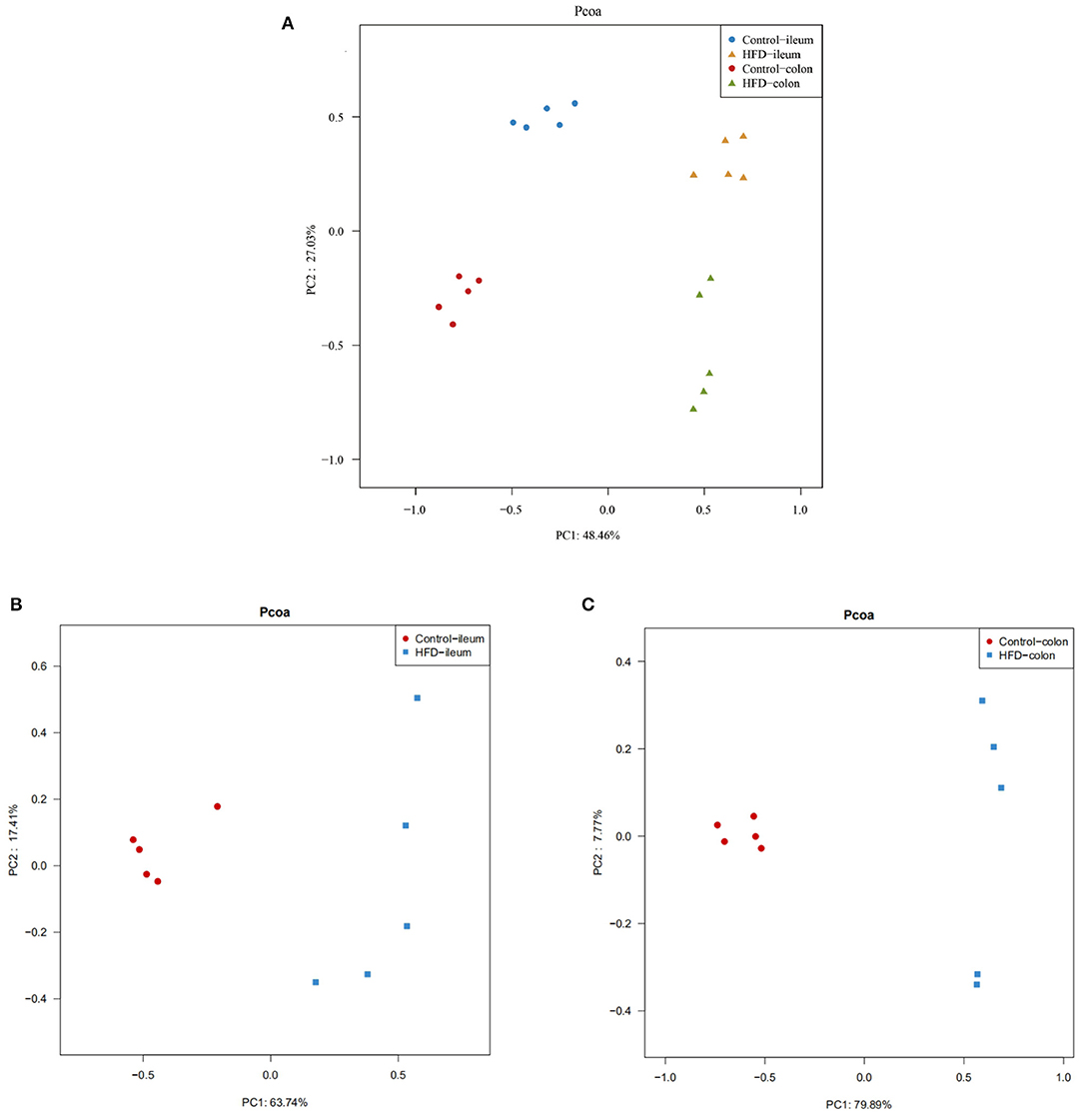
Figure 6. PCoA of the ileum and colon microbial community composition of the Control and HFD mice based on weighted unifrac distance. (A) The overall characteristics of all 20 samples were analyzed by a principal coordinate analysis. The differences in ileum (B) and colon (C) microbial community structure between the Control group and HFD group.
To investigate the effect of gut microbiota on the expression of ANGPTL4, the correlation analysis was conducted. The microorganism positively correlated with ANGPTL4 expression was Allobaculum, and others were negatively correlated with ANGPTL4 expression including Adlercreutzia, Bacteroidales S24–7 group_norank, Bifidobacterium, Coriobacteriaceae_uncultured, Defluviitaleaceae UCG−011, Family XIII AD3011 group, Lachnospiraceae FCS020 group, Marvinbryantia, Mollicutes RF9_norank, Parasutterella, Prevotellaceae UCG−001, Ruminococcaceae NK4A214 group, Ruminococcaceae UCG−014, Turicibacter, (Eubacterium) coprostanoligenes group, (Eubacterium) xylanophilum group (Figure 7). Combining with the raw data from other similar studies, it was found that the relative abundance of Allobaculum in feces, ileum, cecum, and colon was increased significantly in HFD group compared to Control group (Figure 8A). Correlation analysis confirmed the positive association between the abundance of Allobaculum and the expression of ANGPTL4 in the ileum and colon, wherein, the Allobaculum was much more relevant to the ANGPL4 expression in colon (R = 0.8928, P = 0.0005) (Figure 8B).
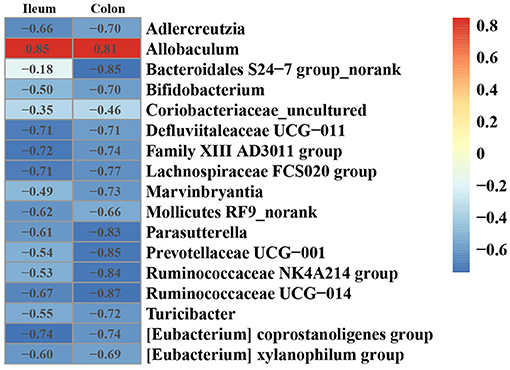
Figure 7. Heatmap of Spearman correlation between the relative expression of ANGPTL4 and microbial genus in ileum and colon of mice.
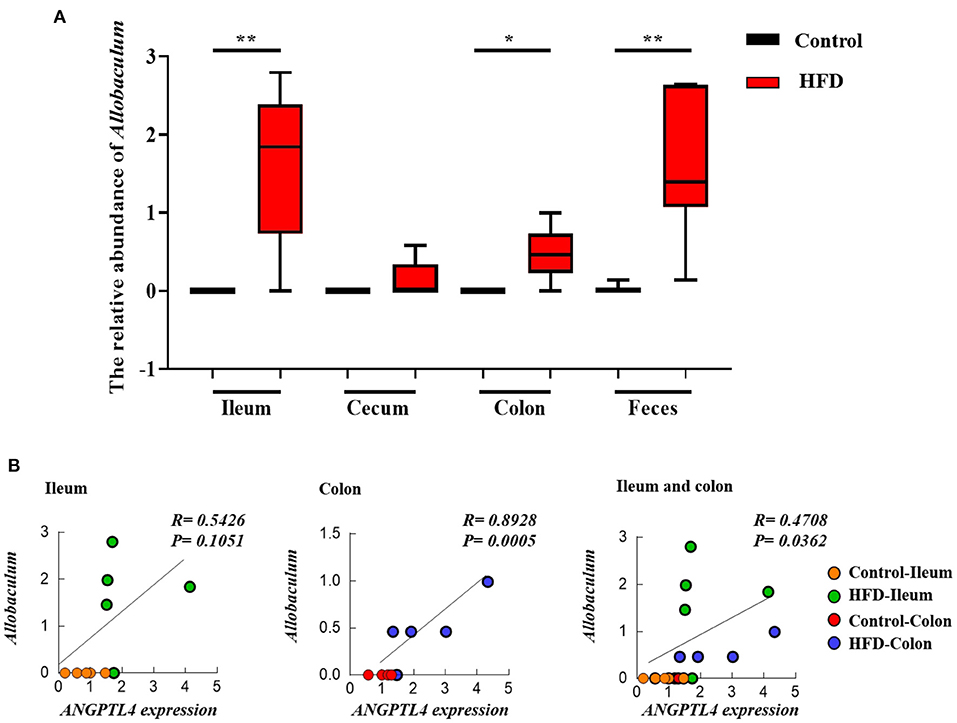
Figure 8. The relative abundance of Allobaculum in ileum, cecum, colon, and feces of Control group and HFD group mice (A), and the correlation between the Allobaculum and ANGPTL4 expression (B) in ileum and colon. The data of cecum and feces were obtained from the NCBI database (DDBJ Accession Number: PRJDB7523 and NCBI Accession Number: SRP113647) (A). Asterisks (* and **) represent significant differences with P < 0.05 and P < 0.01, respectively.
Obesity is one of the most important health topics worldwide. The incidence rate of obesity keeps increasing for many years due to the improvement of living standards, excessive calorie intake and lack of physical exercise (28). High-fat diet is the direct cause of obesity (6). Extensive researches showed ANGPTL4 was an important regulator of TG metabolism by inhibiting LPL and pancreatic lipase (29, 30). This study found that the body weight of the mice was increased by 82.31% (P < 0.05) with the HFD treatment. Compared to the Control group, the expression of ANGPTL4 in the ileum and colon of the HFD group was increased significantly. Similar studies showed that the expression of ANGPTL4 mRNA in small intestine (17) and liver (31) of mice was significantly increased with the HFD intervention. With the treatment of HFD, the absence of any regulation on TG digestion may lead to excess fatty acid uptake into enterocytes. The excess fatty acids would beyond the ability of re-esterification and chyle granule secretion, resulting in the lipid overload of intestinal cells. Therefore, the HFD regulated the expression of ANGPTL4 through a negative feedback mechanism to inhibit the excessive intake of TG. This feedback mechanism aimed to match the lipid uptake amounts of intestinal cells with the capacity for TG secretion. Similarly, the protection of ANGPTL4 against lipid overload in cardiomyocytes (32) and macrophages (18) had also been found. Additionally, Wostmann first discovered that the growth rate of GF mice was slower than normal mice. Gut microbiota also contributes to fat deposition (33). Bäckhed et al. (19) proved that gut microbiota could inhibit the expression of LPL cycle inhibitor, ANGPTL4, in the intestine. The ANGPTL4 expression level of GF mice were higher resulting in slower body weight gain of mice. The ANGPTL4 knock-out GF mice were more obese than the wild-type mice (34). These findings indicate that ANGPTL4 was a circulating medium between gut microbiota and fat deposition. However, the specific mechanism in the microbial regulation by ANGPTL4 expression is still unclear. Meantime, research on the modulation mechanism of ANGPTL4 expression have never stopped. Alex et al. (35) found that short chain fatty acids (SCFAs), as the main metabolites of gut microbiota, could effectively induce the expression of ANGPTL4 in human colon cancer cells T84 and HT29. Aronsson’s research showed that by being fed with the high-fat diet supplemented with Lactobacillus paraliquefaciens (F19), the body fat of mice was decreased significantly with the significant increase in the expression of ANGPTL4 (36). All of these prove that ANGPTL4 is involved in the regulation of gut microbiota on obesity.
Extensive studies have shown that the gut microbiota affects the body’s immune response (37), neural signal (38), and bone density (39), regulates intestinal endocrine function (40), provides energy (41), synthesizes vitamins (42) and many other compounds (43). Therefore, the changes of gut microbiota structure will induce gastrointestinal diseases even a series of diseases in other tissues or systems such as liver, heart, nervous system, respiratory system, and so on (44). In recent years, studies have found that gut microbiota are widely involved in host lipid metabolism and obesity (7, 45). By comparing the response of GF and normal mice to the HFD treatment, researchers found that GF mice had a resistance to HFD, but microbial remodeling led to obesity for GF mice (46, 47). Therefore, gut microbiota is an important factor regulating fat deposition. In this study, the relative abundance ratio of Firmicutes to Bacteroidetes was increased significantly in the ileum and colon of mice in the HFD group. Similar studies found that the relative abundance ratio of Firmicutes to Bacteroidetes in the gut microbiota of mice showed a downward trend through exercise (48). In the obese humans and mice, the relative abundance of Firmicutes and Bacteroidetes was improved and reduced, respectively (49, 50). These findings were consistent with the results of this experiment.
In order to further explore the effect of gut microbiota on the expression of ANGPTL4, we preformed a correlation analysis between the microbial community composition and ANGTPL4 mRNA level. The results showed that the expression of ANGPTL4 was positively correlated with the relative abundance of Allobaculum, but negatively correlated with Adlercreutzia, Bifidobacterium, Prevotellaceae UCG-001, and Ruminococcus. Allobaculum cells are rod-shaped, stain Gram-positive and are arranged in pairs or chains (51). Related research showed that Allobaculum could produce butyric acid, and positively correlated with ANGPTL4 expression (52, 53). Butyric acid could transactivate PPARγ and regulated ANGPTL4, the target gene of PPARγ, in colon cells (54). Early studies have shown that SCFAs play an important role in the human gastrointestinal system, especially in regulating the host fat storage (55, 56). Janssen found that ANGPTL4−/− mice had significantly lower butyric acid levels than the wild-type mice. The butyrate-producing Allobaculum was less abundant in than the wild-type mice but Adlercreutzia abundance was significantly increased in ANGPTL4−/− mice (52). Similar alteration in Allobaculum abundance was found in other studies (25, 26). The relative abundances of Adlercreutzia (26), Bifidobacterium (57), Prevotellaceae UCG-001 and Ruminococcus (58) were significantly decreased in obese mice induced by HFD. These findings were consistent with the results of this study. On the other hand, studies revealed that the abundance of Allobaculum might be negatively correlated with inflammation, insulin resistance and obesity with the intervention of ginger (59), berberine and metformin (60), or prebiotics (61) in mice. Consistent with these studies, our results found the significant increase in the relative abundance of Allobaculum and the expression level of ANGPTL4 by the HFD treatment in mice. The increase in ANGPTL4 expression also had an inhibitory effect on the lipid absorption. These results indicate that Allobaculum could be a promising target for the strategy to control obesity.
This study demonstrated that the HFD treatment significantly increased the expression of ANGPTL4 in the ileum and colon of mice with the enhancement of body weight, liver weight, epididymal fat weight, perirenal fat weight, and the lipid contents in the liver. This might be associated with the change in the composition of gut microbiota in mice including the significantly increased abundance of Allobaculum and the significantly decreased abundance of Adlercreutzia, Bifidobacterium, Prevotellaceae UCG-001, and Ruminococcus in the HFD mice. This work identified the positive correlation between the ANGPTL4 expression and the Allobaculum abundance and highlighted their important role in the regulation of lipid metabolism. This is meaningful to explore the regulation of ANGPTL4 by gut microbiota in the treatment of obesity.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA715938.
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Zhejiang Academy of Agricultural Sciences.
ZZ and YX designed the experiment. ZZ, WL, SZ, XL, and YX conducted the animal experiments. ZZ, WL, HY, and YX wrote and revised the manuscript. ZZ, WL, YR, HY, and YX did experimental analysis, collected, and analyzed the data. All authors reviewed the manuscript.
This study was funded by the National Natural Science Foundation of China (31972999), the Open Project of Hubei Key Laboratory of Animal Nutrition and Feed Science (201806), and State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products (2010DS700124-ZZ1905).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Aron-Wisnewsky J, Warmbrunn MV, Nieuwdorp M, Clément K. Metabolism and metabolic disorders and the microbiome: the intestinal microbiota associated with obesity, lipid metabolism, and metabolic health-pathophysiology and therapeutic strategies. Gastroenterology. (2021) 160:573–99. doi: 10.1053/j.gastro.2020.10.057
2. Caballero B. Humans against obesity: who will win? Adv Nutr. (2019) 10:S4–9. doi: 10.1093/advances/nmy055
3. Chang CJ, Lin CS, Lu CC, Martel J, Ko YF, Ojcius DM, et al. Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat Commun. (2015) 6:7489. doi: 10.1038/ncomms8489
4. Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. (2010) 90:859–904. doi: 10.1152/physrev.00045.2009
5. He X, Zheng N, He J, Liu C, Feng J, Jia W, et al. Gut microbiota modulation attenuated the hypolipidemic effect of simvastatin in high-fat/cholesterol-diet fed mice. J Proteome Res. (2017) 16:1900–10. doi: 10.1021/acs.jproteome.6b00984
6. Kaya SD, Sinen O, Bülbül M. Gastric motor dysfunction coincides with the onset of obesity in rats fed with high-fat diet. Clin Exp Pharmacol Physiol. (2021) 48:553–62. doi: 10.1111/1440-1681.13448
7. Abenavoli L, Scarpellini E, Colica C, Boccuto L, Salehi B, Sharifi-Rad J, et al. Gut microbiota and obesity: a role for probiotics. Nutrients. (2019) 11:2690. doi: 10.3390/nu11112690
8. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. (2013) 341:1241214. doi: 10.1126/science.1241214
9. Yin J, Li Y, Han H, Ma J, Liu G, Wu X, et al. Administration of exogenous melatonin improves the diurnal rhythms of the gut microbiota in mice fed a high-fat diet. mSystems. (2020) 5:e00002–20. doi: 10.1128/mSystems.00002-20
10. Gérard P. Gut microbiota and obesity. Cell Mol Life Sci. (2016) 73:147–62. doi: 10.1007/s00018-015-2061-5
11. Li J, Li L, Guo D, Li S, Zeng Y, Liu C, et al. Triglyceride metabolism and angiopoietin-like proteins in lipoprotein lipase regulation. Clin Chim Acta. (2020) 503:19–34. doi: 10.1016/j.cca.2019.12.029
12. Fernández-Hernando C, Suárez Y. ANGPTL4: a multifunctional protein involved in metabolism and vascular homeostasis. Curr Opin Hematol. (2020) 27:206–13. doi: 10.1097/MOH.0000000000000580
13. Lichtenstein L, Kersten S. Modulation of plasma TG lipolysis by angiopoietin-like proteins and GPIHBP1. Biochim Biophys Acta. (2010) 1801:415–20. doi: 10.1016/j.bbalip.2009.12.015
14. Chen YQ, Pottanat TG, Siegel RW, Ehsani M, Qian YW, Roell WC, et al. Angiopoietin-like protein 4(E40K) and ANGPTL4/8 complex have reduced, temperature-dependent LPL-inhibitory activity compared to ANGPTL4. Biochem Biophys Res Commun. (2021) 534:498–503. doi: 10.1016/j.bbrc.2020.11.053
15. Dijk W, Kersten S. Regulation of lipid metabolism by angiopoietin-like proteins. Curr Opin Lipidol. (2016) 27:249–56. doi: 10.1097/MOL.0000000000000290
16. Kersten Sander. Physiological regulation of lipoprotein lipase. Biochim Biophys Acta. (2014) 1841:919–33. doi: 10.1016/j.bbalip.2014.03.013
17. Mattijssen F, Alex S, Swarts HJ, Groen AK, Van Schothorst EM, Kersten S. Angptl4 serves as an endogenous inhibitor of intestinal lipid digestion. Mol Metab. (2013) 3:135–44. doi: 10.1016/j.molmet.2013.11.004
18. Lichtenstein L, Mattijssen F, Wit NJD, Georgiadi A, Hooiveld GJ, Meer RVD, et al. Angptl4 protects against severe proinflammatory effects of saturated fat by inhibiting fatty acid uptake into mesenteric lymph node macrophages. Cell Metab. (2010) 12:580–92. doi: 10.1016/j.cmet.2010.11.002
19. Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. (2004) 101:15718–23. doi: 10.1073/pnas.0407076101
20. Yang H, Xiang Y, Robinson K, Wang J, Zhang G, Zhao J, et al. Gut microbiota is a major contributor to adiposity in pigs. Front Microbiol. (2018) 9:3045. doi: 10.3389/fmicb.2018.03045
21. Shi X, Zhou X, Chu X, Wang J, Xie B, Ge J, et al. Allicin improves metabolism in high-fat diet-induced obese mice by modulating the gut microbiota. Nutrients. (2019) 11:2909. doi: 10.3390/nu11122909
22. An Y, Li Y, Wang X, Chen Z, Xu H, Wu L, et al. Cordycepin reduces weight through regulating gut microbiota in high-fat diet-induced obese rats. Lipids Health Dis. (2018) 17:276. doi: 10.1186/s12944-018-0910-6
23. Lawley B, Tannock GW. Analysis of 16S rRNA gene amplicon sequences using the QIIME software package. Methods Mol Biol. (2017) 1537:153–63. doi: 10.1007/978-1-4939-6685-1_9
24. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. (2007) 73:5261–7. doi: 10.1128/AEM.00062-07
25. Ushiroda C, Naito Y, Takagi T, Uchiyama K, Mizushima K, Higashimura Y, et al. Green tea polyphenol (epigallocatechin-3-gallate) improves gut dysbiosis and serum bile acids dysregulation in high-fat diet-fed mice. J Clin Biochem Nutr. (2019) 65:34–46. doi: 10.3164/jcbn.18-116
26. Jiao X, Wang Y, Lin Y, Lang Y, Li E, Zhang X, et al. Blueberry polyphenols extract as a potential prebiotic with anti-obesity effects on C57BL/6 J mice by modulating the gut microbiota. J Nutr Biochem. (2019) 64:88–100. doi: 10.1016/j.jnutbio.2018.07.008
27. Xiao Y, Wu C, Li K, Gui G, Zhang G, Yang H. Association of growth rate with hormone levels and myogenic gene expression profile in broilers. J Anim Sci Biotechnol. (2017) 8:43. doi: 10.1186/s40104-017-0170-8
28. NCD-RisC. Trends in adult body-mass index in 200 countries from 1975 to 2014: a pooled analysis of 1698 population-based measurement studies with 19·2 million participants. Lancet. (2016) 387:1377–96. doi: 10.1016/S0140-6736(16)30054-X
29. Aryal B, Price NL, Suarez Y, Fernández-Hernando C. ANGPTL4 in metabolic and cardiovascular disease. Trends Mol Med. (2019) 25:723–34. doi: 10.1016/j.molmed.2019.05.010
30. Alex S, Lichtenstein L, Dijk W, Mensink RP, Tan NS, Kersten S. ANGPTL4 is produced by entero-endocrine cells in the human intestinal tract. Histochem Cell Biol. (2014) 141:383–91. doi: 10.1007/s00418-013-1157-y
31. Gao J, Ding G, Li Q, Gong L, Huang J, Sang Y. Tibet kefir milk decreases fat deposition by regulating the gut microbiota and gene expression of Lpl and Angptl4 in high fat diet-fed rats. Food Res Int. (2019) 121:278–87. doi: 10.1016/j.foodres.2019.03.029
32. Georgiadi A, Lichtenstein L, Degenhardt T, Boekschoten MV, van Bilsen M, Desvergne B, et al. Induction of cardiac Angptl4 by dietary fatty acids is mediated by peroxisome proliferator-activated receptor beta/delta and protects against fatty acid-induced oxidative stress. Circ Res. (2010) 106:1712–21. doi: 10.1161/CIRCRESAHA.110.217380
33. Wostmann BS, Larkin C, Moriarty A, Bruckner-Kardoss E. Dietary intake, energy metabolism, and excretory losses of adult male germfree Wistar rats. Lab Anim Sci. (1983) 33:46–50. doi: 10.2307/3808211
34. Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. (2007) 104:979–84. doi: 10.1073/pnas.0605374104
35. Alex S, Lange K, Amolo T, Grinstead JS, Haakonsson AK, Szalowska E, et al. Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor γ. Mol Cell Biol. (2013) 33:1303–16. doi: 10.1128/MCB.00858-12
36. Aronsson L, Huang Y, Parini P, Korach-André M, Håkansson J, Gustafsson J, et al. Decreased fat storage by Lactobacillus paracasei is associated with increased levels of angiopoietin-like 4 protein (ANGPTL4). PLoS ONE. (2010) 5:e13087. doi: 10.1371/journal.pone
37. Fulde M, Hornef MW. Maturation of the enteric mucosal innate immune system during the postnatal period. Immunol Rev. (2014) 260:21–34. doi: 10.1111/imr.12190
38. Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. (2015) 161:264–76. doi: 10.1016/j.cell.2015.02.047
39. Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. (2012) 488:621–6. doi: 10.1038/nature11400
40. Neuman H, Debelius JW, Knight R, Koren O. Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiol Rev. (2015) 39:509–21. doi: 10.1093/femsre/fuu010
41. Canfora EE, Jocken JW, Blaak EE: Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol. (2015) 11:577–91. doi: 10.1038/nrendo.2015.128
42. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. (2012) 486:222–7. doi: 10.1038/nature11053
43. Devlin AS, Fischbach MA. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol. (2015) 11:685–90. doi: 10.1038/nchembio.1864
44. Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. (2016) 375:2369–79. doi: 10.1056/NEJMra1600266
45. Schoeler M, Caesar R. Dietary lipids, gut microbiota and lipid metabolism. Rev Endocr Metab Disord. (2019) 20:461–72. doi: 10.1007/s11154-019-09512-0
46. Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. (2008) 3:213–23. doi: 10.1016/j.chom.2008.02.015
47. Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu P, Yetukuri L, Islam S, et al. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res. (2010) 51:1101–12. doi: 10.1194/jlr.M002774
48. Evans CC, LePard KJ, Kwak JW, Stancukas MC, Laskowski S, Dougherty J, et al. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS ONE. (2014) 9:e92193. doi: 10.1371/journal.pone.0092193
49. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. (2006) 444:1027–31. doi: 10.1038/nature05414
50. Parnell JA, Reimer RA. Prebiotic fiber modulation of the gut microbiota improves risk factors for obesity and the metabolic syndrome. Gut Microbes. (2012) 3:29–34. doi: 10.4161/gmic.19246
51. Greetham HL, Gibson GR, Giffard C, Hippe H, Merkhoffer B, Steiner U, et al. Allobaculum stercoricanis gen. nov., sp. nov., isolated from canine feces. Anaerobe. (2004) 10:301–7. doi: 10.1016/j.anaerobe
52. Janssen AWF, Katiraei S, Bartosinska B, Eberhard D, Willems van Dijk K, Kersten S. Loss of angiopoietin-like 4 (ANGPTL4) in mice with diet-induced obesity uncouples visceral obesity from glucose intolerance partly via the gut microbiota. Diabetologia. (2018) 61:1447–58. doi: 10.1007/s00125-018-4583-5
53. Li S, Qi Y, Ren D, Qu D, Sun Y. The structure features and improving effects of polysaccharide from astragalus membranaceus on antibiotic-associated diarrhea. Antibiotics (Basel). (2019) 9:8. doi: 10.3390/antibiotics9010008
54. Kumar J, Rani K, Datt C. Molecular link between dietary fibre, gut microbiota and health. Mol Biol Rep. (2020) 47:6229–37. doi: 10.1007/s11033-020-05611-3
55. Galvez J, Rodríguez-Cabezas ME, Zarzuelo A. Effects of dietary fiber on inflammatory bowel disease. Mol Nutr Food Res. (2005) 49:601–8. doi: 10.1002/mnfr.200500013
56. Murphy EF, Cotter PD, Healy S, Marques TM, O’Sullivan O, Fouhy F, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. (2010) 59:1635–42. doi: 10.1136/gut.2010.215665
57. Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. (2007) 50:2374–83. doi: 10.1007/s00125-007-0791-0
58. Kong C, Gao R, Yan X, Huang L, Qin H. Probiotics improve gut microbiota dysbiosis in obese mice fed a high-fat or high-sucrose diet. Nutrition. (2019) 60:175–84. doi: 10.1016/j.nut.2018.10.002
59. Wang J, Wang P, Li D, Hu X, Chen F. Beneficial effects of ginger on prevention of obesity through modulation of gut microbiota in mice. Eur J Nutr. (2020) 59:699–718. doi: 10.1007/s00394-019-01938-1
60. Zhang X, Zhao Y, Xu J, Xue Z, Zhang M, Pang X, et al. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci Rep. (2015) 5:14405. doi: 10.1038/srep14405
Keywords: gut microbiota, ANGPTL4, fat deposition, high-fat diet, mice
Citation: Zheng Z, Lyu W, Ren Y, Li X, Zhao S, Yang H and Xiao Y (2021) Allobaculum Involves in the Modulation of Intestinal ANGPTLT4 Expression in Mice Treated by High-Fat Diet. Front. Nutr. 8:690138. doi: 10.3389/fnut.2021.690138
Received: 02 April 2021; Accepted: 26 April 2021;
Published: 19 May 2021.
Edited by:
Jie Yin, Hunan Agricultural University, ChinaReviewed by:
Shiyu Tao, Huazhong Agricultural University, ChinaCopyright © 2021 Zheng, Lyu, Ren, Li, Zhao, Yang and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingping Xiao, eXB4aWFvempAaG90bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.