
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Nutr. , 06 August 2021
Sec. Nutrition and Microbes
Volume 8 - 2021 | https://doi.org/10.3389/fnut.2021.690073
This article is part of the Research Topic The Interaction between Digestive Tract Microbes and Hosts in Poultry View all 25 articles
A correction has been applied to this article in:
Corrigendum: How Early-Life Gut Microbiota Alteration Sets Trajectories for Health and Inflammatory Bowel Disease?
 Feilong Guo1,2,3†
Feilong Guo1,2,3† Demin Cai1,4†
Demin Cai1,4† Yanwei Li4
Yanwei Li4 Haotian Gu4
Haotian Gu4 Huan Qu4
Huan Qu4 Qiufang Zong4
Qiufang Zong4 Wenbin Bao4*
Wenbin Bao4* Aoxue Chen5*
Aoxue Chen5* Hao-Yu Liu1,4*
Hao-Yu Liu1,4*Inflammatory bowel disease (IBD) is a recurrent chronic inflammatory condition of the intestine without any efficient therapeutic regimens. Gut microbiota, which plays an instrumental role in the development and maturation of the immune system, has been implicated in the pathogenesis of IBD. Emerging evidence has established that early-life events particularly maternal influences and antibiotic treatment are strongly correlated with the health or susceptibility to disease of an individual in later life. Thus, it is proposed that there is a critical period in infancy, during which the environmental exposures bestow a long-term pathophysiological imprint. This notion sheds new light on the development of novel approaches for the treatment, i.e., early interventions, more precisely, the prevention of many uncurable chronic inflammatory diseases like IBD. In this review, we have integrated current evidence to describe the feasibility of the “able-to-be-regulated microbiota,” summarized the underlying mechanisms of the “microbiota-driven immune system education,” explored the optimal intervention time window, and discussed the potential of designing early-probiotic treatment as a new prevention strategy for IBD.
Inflammatory bowel disease (IBD), including ulcerative colitis and Crohn's disease, is a life-long disease once onset. It is characterized by recurrent intestinal inflammation, causes cumulative and progressive damages to the bowel wall, and consequently leads to intestinal dysfunction, obstruction, or perforation that require surgical intervention (1). IBD affects millions of people in North America and Europe, and the incidence is increasing alarmingly with the progression of global urbanization, particularly in the newly industrialized areas, such as Asia (2). Though the last decades have witnessed a great advancement of medicines for IBD, especially the successive launch of different biologics anti-TNF, anti-adhesion agents, and anti-IL-12/23 p40 antibody, etc., a proportion of patients are still not amenable to any medications. Some lose response over time, some have to frequently change drugs due to severe adverse effects, and none of the regimens is available to reverse the intestinal damages (3). Therefore, new therapeutic strategies are in great demand.
Several epidemiological studies have established a positive correlation between the early-life exposures and the future risks of IBD, including the mode of delivery (4), the feeding types (5), the childhood hygiene (6), and the antibiotic use (7). All of which are important environmental factors associated with gut microbiota alterations. In infancy, the gut microbiota is less diverse and resilient and more sensitive to the modulation. The succession of gut microbiota parallels with the development of the gut immune system and directs the health outcome via the education of immunity (8). As unstable and sensitive as the infant bacterial community structure is, it also opens up a “window of opportunity” for the associated intervention (8, 9). From this perspective, reducing the environmental incursions or facilitate the gut microbiota equilibrium within an optimal time frame can be a new therapeutic strategy for IBD. In this review, we will first describe the postnatal development of gut microbiota and the intestinal immune system, taking macrophages (MP) as an example. Then, we will summarize current evidence depicting the “window of opportunity” for the gut microbiota modulation, and finally discuss the potential of designing early probiotic treatment as a new prevention strategy for IBD.
The mammalian gastrointestinal (GI) tract accommodates the highest density of the microbial community on earth, more precisely in the distal part where up to 100 trillion microorganisms inhabit. It is far beyond the number of eukaryotic cells of the host that holds itself (10). A healthy adult intestine harbors 100s of bacterial species, including the dominant genera Bacteroides, Clostridium, Prevotella, Faecalibacterium, Eubacterium, Ruminococcus, Peptococcus, Peptostreptococcusm, and Bifidobacterium, which are affiliated to the four major phyla, i.e., Bacteroidetes, Firmicutes, Proteobacteria, and Actinobacteria (11, 12). Despite the considerable variation among individuals detected in the microbiome, Arumugam et al. (10) have identified three distinct clusters of gut microbiota driven by discriminative genera such as enterotype1-Bacteroides, enterotype2-Prevotella, and enterotype3-Ruminococcus, which applies to the global microbiome of human beings and is generally stable over time and in geography (13).
Gut microbiota shows an age-dependent succession difference. Unlike the stable enterotypes in adults and the deterioration occurring in old age (13), the gut microbial ecosystem of newborns is characterized by rapid changes in bacterial abundance, diversity, and large interindividual variability of community composition (14). Conventionally, the human GI tract is first colonized by bacteria from the immediate environment. Often, by the maternal vagina and feces-associated microbes including Lactobacillus and Prevotella in infants of vaginal birth, or by bacteria from maternal skin and the surroundings represented by Staphylococcus in babies of cesarean delivery (15). Of note, recent studies have also reported the microorganisms detections in the placenta (16) or amniotic fluid (17). Nevertheless, facultative anaerobes, predominantly Escherichia coli and other bacteria that belong to Gammaproteobacteria are the first gut colonizers. Subsequently, the oxygen in the intestinal microenvironment is exhausted by these bacteria and becomes anaerobic that favors the growth of strict anaerobes such as Bifidobacterium, Clostridium, and Bacteroides, which constitute an early community of low diversity. The aerobic to anaerobic transition seems to occur very rapidly, as the obligate anaerobes can be detected to occupy the intestine at the first week after birth (18). Thereafter, the Bifidobacterium genus presents its diversity and prevalence in the gut microbiome of young infants at the age of 7–42 days (19–21). Given that, human milk is enriched with non-digestible human milk oligosaccharides (HMOs) and the infant Bifidobacteria are proficient at utilizing HMOs as the sole carbon source, by dedicating a sizable fraction of the genome to encode the proteins associated with the HMOs metabolism (12). Finally, the introduction of solid foods during weaning (at about 6 months old) primes the infant gut microbiota to adapt to an adult diet, which results in a significant increase in the bacterial community diversity and complexity, accompanied by climbing of Bacteroidetes and a decline of Bifidobacterium (22). However, a recent study suggests that the cessation of breastfeeding, rather than the solid food introduction, determines the maturation of the microbiome into an adult-like phenotype. It is usually around 1 year of age for human beings, thereafter, the gut microbiota becomes relatively resilient and stable at the age of 3 (8, 14). Notably, two longitudinal cohort studies of Spanish and Swedish infants revealed an overall directionality of gut microbiota changes toward their mothers, both compositionally and functionally within the first 12 months (18, 23).
Constantly exposed to the extremely complicated gut microbiota, the intestine also prepares itself with the largest compartment of the immune system (24, 25), which consists of the organized lymphoid tissue [Peyer's patches (PPs), solitary isolated lymphoid tissues (SILTs), and the draining lymph nodes], along with the scattered effector cells distributed in the lamina propria (LP) and the epithelium (26). Interestingly, although partially programmed before birth, the LP and the epithelium are devoid of mononuclear cells at birth (26).
Among the effector immune cells, the intestinal MPs are the most plastic population. Located directly underneath the surface epithelium in the LP, they serve as sentinels of the first line of defense, are responsible for the bacterial antigen presentation, and discriminating the innocuous agents from the pathogenic insults in the gut lumen, and clearing apoptotic and senescent epithelial cells (27). MP are important players in maintaining gut homeostasis and contribute to IBD pathogenesis (28). Discrete populations of MPs are also found in the submucosa and muscularis mucosa throughout the GI tract, together which exhibit a variety of functions to ensure the proper immune reactions (29). Apart from being highly phagocytic and producing TNF-α, IL-1, and IL-6 in response to inflammation, the intestinal MPs also mediate mucosal tolerance by secreting the anti-inflammatory cytokine IL-10, thereby promote the regulatory T-cell population (28, 30), suggesting a case of innate immunity that controls adaptive immunity. The introduction of the “macrophage-waterfall” of Ly6Chi monocytes, which develop progressively into CX3CR1hi-MHCIIhi resident cells based on CD64 expression, makes it possible to more accurately identify the intestinal resident MPs (28). The gut MPs represent a mixed population, constantly replenished by the blood monocytes originated from the bone marrow (29). During the neonatal period, the MPs derived from embryo and yolk sac predominate the cell pool (31), but only the latter is found to be replaced by cells from a series of development including the chemokine receptor CCR2-dependent recruitment of Ly6Chi monocytes, downregulation of Ly6C, and upregulation of F4/80, CD64, and CX3CR1 expressions, as well as obtaining the MHCII phenotype. This process-driven largely by the microbiota, occurs during weaning and peaks during the third week of life in mice, whereas germ-free mice are found to have fewer MP in the gut wall, and its turnover can be suppressed by antibiotics (29).
The phenotypic and functional identity of the MPs is imprinted by the local environment. Microbiota as a whole is an important stimulus for monocyte-MP differentiation (32). While the number and the subsets diversity change during intestinal inflammation and infection (27, 29). It is suggested that the gut MPs are susceptible to environmental (re)programming, even with the matured phenotype. There is a causal link between the intestinal MPs and IBD, where the altered monocyte-macrophage differentiation impairs the resolution of intestinal inflammation in patients with IBD, leading to the chronic relapse in individuals with the genetic predisposition (33). On the other hand, we postulate in this study that the targeted MPs polarization of the intestinal microenvironment, by therapeutic strategies like the probiotic usage at an early stage, may induce sustained intestinal immune protection. This is supported by a recent study by Danne et al. (34) that Helicobacter hepaticus-polysaccharides induced an anti-inflammatory MP response.
As discussed above, the microbial colonization of the gut of an infant represents the de novo assembly of a bacterial community with functional attributes similar to adults. It usually takes 2–3 years for humans and 4–6 weeks for mice (22). Important changes are witnessed during weaning when the solid food is gradually introduced to a suckling mammal while withdrawing the milk of its mother (9, 35). This leads to a “weaning reaction” of the immune system against the microbiota alterations. A proper “weaning reaction” is associated with changes in the global gene expression in the intestine, such as genes encoding defensins, chemokine receptors, and mucins. Furthermore, commensal microbiota induces gene upregulations of the pro-inflammatory cytokines TNF-α and IFN-γ during weaning at 3 weeks of age in mice (9). An elevation of IL-1β expression was also observed in 21-days old rats at weaning (36). In contrast, the inhibition of “weaning reaction” in mice using antibiotics led to a “pathological imprinting” of the immune system, which cannot be reversed after weaning (9).
It is, therefore, instrumental that the microbiota establishes its mutually beneficial cohabitation with the host in the time window of early life, whereas the perturbations may result in potentially persistent immune abnormalities (8, 37). Several genome-wide association analyses and large-scale cohort studies reveal that exposure to antibiotics in childhood especially during the first year of life is associated with increased susceptibility to IBD and allergy (7, 37, 38). Furthermore, a meta-analysis demonstrates that breastfeeding is associated with lower risks of IBD, with even lower disease incidence in infants under 12 months breastfeeding regimen than 6 months (39). A study employing surrogate markers of childhood hygiene (e.g., fewer siblings and living in urban areas) revealed that individuals raised in a sanitary environment are at a higher risk of IBD later in life (6). In contrast, children raised close to farm animals develop less inflammation and allergies, which supports the “hygiene hypothesis” (40). In germ-free mice, the immunological abnormality can be reversed by introducing the commensal bacteria from a healthy counterpart but only as an early-life intervention (41). Furthermore, Cox et al. (35) found that temporal antibiotics administration causes a transient microbiota perturbation in mice infancy but induces a long-term metabolic phenotype. These results highlight the importance of the early-life microbial perturbations vs. the early-life intervention, with the emphasis on gut microbiota modulation (8, 37). There is a saying that “who started the trouble should end it.” It appears that several aspects of immune development are indeed more permissive to the microbial-mediated changes during early life, and that specific bacterial taxa are crucial in these interactions.
Because gut microbiota perturbation is one of the major causes of IBD and the modulation is a goal, it is relevant to consider using probiotics as a therapeutic strategy. Indeed, live microorganisms given personally produce health benefits, are defined as “probiotics” and have intrigued humans for centuries (42). In particular, the probiotic effects in IBD treatment have been studied to reverse the dysbiosis-associated inflammation (43). Although having been added widely in snacks, drinks, infant formulas, and consumed as health supplements, the clinical use of probiotics as therapeutics and their effects are still less demonstrated (42). The protective effects in animal models of IBD and in vitro studies were robust as seen by several conventional probiotic strains, e.g., Bifidobacterium infantis, Lactobacillus plantarum, and Lactobacillus reuteri, as well as verified by novel strains Lactobacillus helveticus PI5 and Lactobacillus salivarius LA307 (44). All of them point to the efficacy and proficiency in strengthening the intestinal epithelial barrier and improving the immune functions and the microbiome (45). However, their clinical effects with patients with IBD were poorly verified. In a meta-analysis study, no effect was observed on probiotic-induced remission or prevention of relapse in Crohn's disease, whereas some beneficial effects were shown in patients with colitis (46).
One enigma is whether or not the probiotic bacteria could colonize the intestine by peroral administration, if so for how long they could stay in place after the administration is removed. This would challenge the actions of probiotics when regarded as contact-dependent (47). As aforementioned, the established microbiota in adulthood remains quite stable albeit the fluctuation under drastic changes (10). A recent study demonstrates that human targeted probiotics exhibited low-level mucosal colonization in SPF mice, due to resistance from the indigenous microbiome (48). Several studies found a decline of probiotic bacteria detection in the fecal microbiota shortly after the administration cessation (49, 50). In human beings, the probiotics differentially affect the “permissive” and “resistant” individuals, which features a personal effect of combined factors including microbiota variation and host gene signatures associated with immune responses and metabolism (48). This could be a possible explanation for the ambiguous effects achieved in clinical trials of IBD-probiotic treatments. In both humans and mice, the probiotics are seen to induce changes in microbiome and host GI transcriptome, despite the limited colonization (48). The mechanisms behind this and the causality of the microbiome in IBD onset warrant further studies. In this regard, approaches combining high-resolution single-cell RNA sequencing/scRNA-seq of inflammatory lesions with the clinical characterization of patients with IBD may provide deeper insights (51). Interestingly, using scRNA-seq, Aschenbrenner et al. (52) revealed that bacteria exposure induces functional IL-10 resistance in monocytes and a hyperinflammation-associated IL-23 production in patients with severe ulcerative colitis.
Surprisingly, several recent studies have described enhanced probiotic colonization within or post-antibiotic treatment, suggesting that probiotics were more likely to act in the “microorganism less abundant” niches (53, 54), as a property that can be found in the gut microbiota during early life. Furthermore, Schultz et al. (55) have shown that administration of Lactobacillus rhamnosus GG to pregnant women causes infantile colonization for up to 24 months and increased the bifidobacteria diversity in neonates. During the postnatal period, L. reuteri DSM 17938 treatment is shown to promote lactobacilli growth while inhibiting that of E. coli in infants with colic (56). This is confirmed to be effective as an infantile colic intervention. The probiotic treatment is also considered well-tolerated and safe in preterm infants, to prevent necrotizing enterocolitis and all-cause mortality (57). However, there are very few longitudinal studies concerning the probiotic intervention in infants and track their IBD susceptibility growing up. It is recently shown that infants born to IBD mothers exhibited altered gut microbiome from the first week after birth to at least 3 months old, causing aberrant adaptive immune responses (58). This again underlines the urgency of early microbiota modulation for IBD prevention.
We suggest that a better understanding of the gut microbiome signature in infant exposure to maternal influences changes of diets and medications using the readily available next-generation sequencing tools could help to develop predictive markers and guide the selection of the probiotic strain fitted for the potential early-life intervention. For instance, giving probiotic Bifidobacterium strains to the IBD mother-delivered babies, who displayed a depletion of bifidobacteria in the first week of life (58) to promote their health, or use a combination of probiotic strains to enrich the gut microbiota with low diversity, like in the cesarean section-delivered newborns (15) and formula-fed infants (39). For early antibiotic perturbations associated with the increase of opportunistic pathogens, a probiotic designed to target the corresponding bacterial taxa (e.g., Gammaproteobacteria) should be considered. In addition, probiotics can also be tailored for metabolic reprogramming, for instance, to restore a lipid dysregulation caused by dysbiosis during a disrupted “weaning reaction.” For those breast-fed, virginally delivered, full-term healthy babies, who are not exposed to antibiotics but could still suffer from IBD, the probiotic strains should be selected according to the normal gut microbiota succession trend discussed above. For example, with the probiotic strains from Actinobacteria given before weaning and those from Firmicutes afterward. Meanwhile, the characteristics of the gut microbiome in adult patients with IBD should be used as a reference for early intervention.
Because gut microbiota is implicated in human and animal health and disease, an essential goal of this review is to better understand the assembly and community composition of the microbiota with a special emphasis on the early-life period. Unlike the adult microbiota, which is relatively resistant to perturbations and stable over time, the gut microbial ecosystem of the newborn is characterized by rapid changes in bacterial community composition, with lower diversity and lower complexity. Therefore, a convergence in gut microbiota succession, environmental stimuli for the time being, and maturation of the immune system decide the disease susceptibility in later life (Figure 1). Especially for IBD that is life-long and is featured by recurrent chronic intestinal inflammation and dysregulation of the immunity toward commensal bacteria. Since the immune system is more permissive to microbial-mediated changes during the early-life period, specific probiotic bacteria could be the key to the potential modulation. This early-life period (i.e., from born till the microbiota reach an adult phenotype) opens up an exciting “window of opportunity.” However, it should be carefully defined, categorized, and evaluated. Stable and resilient gut microbiota in an adult but with pathological imprinting can be detrimental in many aspects. This may explain why there are “permissive” and “resistant” patients with IBD to medications.
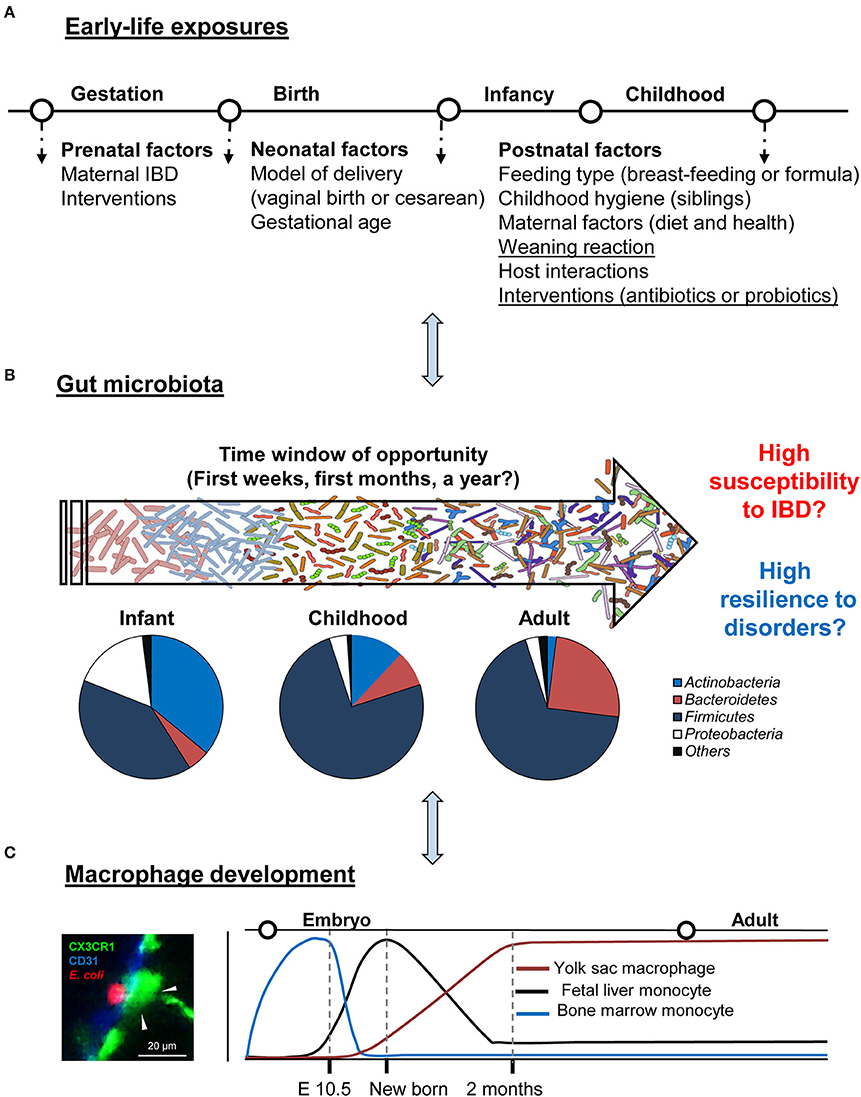
Figure 1. Critical time window: gut development and regulation. (A,B) Schematic summary of early-life environmental factors that impact the gut microbiota establishment of an individual and drive its phenotype toward health or IBD. Typical phylum-level of microbiota composition in healthy individuals in different life stages and the window of opportunity for possible modulation. (C) Gut resident macrophage ontogeny in the steady-state. A representative image of colon macrophage (green), interacting with Escherichia coli (red, left panel).
H-YL, WB, and FG conceived this review. FG, DC, AC, YL, HG, HQ, and QZ wrote the manuscript. H-YL, WB, and AC supervised and approved the final version. All the authors have read, revised, and approved the final manuscript.
This study was supported by the National Natural Science Foundation of China (32002243), Natural Science Foundation of Jiangsu Province (BK20200932), Natural Science Foundation of the Higher Education Institutions of Jiangsu Province (20KJB230001), Breeding and Reproduction Innovation Team of Jiangsu Province Modern Agriculture (Swine) Industry Technical System (JATS [2020] 4400), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The work of FG was partly supported by the China Scholarship Council (201706190200).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The author specially thank to Dr. Xiaobo Chen (Center for Environment and Sustainability, University of Surrey) for the help of proofreading.
1. Weimers P, Munkholm P. The natural history of IBD: lessons learned. Curr Treat Options Gastroenterol. (2018) 16:101–11. doi: 10.1007/s11938-018-0173-3
2. Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. (2017) 390:2769–78. doi: 10.1016/S0140-6736(17)32448-0
3. Gomollón F, Dignass A, Annese V, Tilg H, Van Assche G, Lindsay JO, et al. 3rd European evidence-based consensus on the diagnosis and management of Crohn's disease 2016: Part 1: diagnosis and medical management. J Crohns Colitis. (2017) 11:3–25. doi: 10.1093/ecco-jcc/jjw168
4. Bager P, Simonsen J, Nielsen NM, Frisch M. Cesarean section and offspring's risk of inflammatory bowel disease: a national cohort study. Inflamm Bowel Dis. (2012) 18:857–62. doi: 10.1002/ibd.21805
5. Xu L, Lochhead P, Ko Y, Claggett B, Leong RW, Ananthakrishnan AN. Systematic review with meta-analysis: breastfeeding and the risk of Crohn's disease and ulcerative colitis. Aliment Pharmacol Ther. (2017) 46:780–9. doi: 10.1111/apt.14291
6. Klement E, Lysy J, Hoshen M, Avitan M, Goldin E, Israeli E. Childhood hygiene is associated with the risk for inflammatory bowel disease: a population-based study. Am J Gastroenterol. (2008) 103:1775–82. doi: 10.1111/j.1572-0241.2008.01905.x
7. Kronman MP, Zaoutis TE, Haynes K, Feng R, Coffin SE. Antibiotic exposure and IBD development among children: a population-based cohort study. Pediatrics. (2012) 130:e794–e803. doi: 10.1542/peds.2011-3886
8. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. (2016) 352:539–44. doi: 10.1126/science.aad9378
9. Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Dejardin F, et al. A Weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity. (2019) 50:1276–88.e5. doi: 10.1016/j.immuni.2019.02.014
10. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. (2011) 473:174–80. doi: 10.1038/nature09944
11. Hu Y, Wang L, Shao D, Wang Q, Wu Y, Han Y, et al. Selectived and reshaped early dominant microbial community in the cecum with similar proportions and better homogenization and species diversity due to organic acids as AGP alternatives mediate their effects on broilers growth. Front Microbiol. (2019) 10:2948. doi: 10.3389/fmicb.2019.02948
12. Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. (2017) 81:e00036–17. doi: 10.1128/MMBR.00036-17
13. Costea PI, Hildebrand F, Arumugam M, Bäckhed F, Blaser MJ, Bushman FD, et al. Enterotypes in the landscape of gut microbial community composition. Nat Microbiol. (2018) 3:8–16. doi: 10.1038/s41564-017-0072-8
14. Matamoros S, Gras-Leguen C, Le Vacon F, Potel G, de La Cochetiere M-F. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. (2013) 21:167–73. doi: 10.1016/j.tim.2012.12.001
15. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. (2010) 107:11971–5. doi: 10.1073/pnas.1002601107
16. Satokari R, Grönroos T, Laitinen K, Salminen S, Isolauri E. Bifidobacterium and Lactobacillus DNA in the human placenta. Lett Appl Microbiol. (2009) 48:8–12. doi: 10.1111/j.1472-765X.2008.02475.x
17. DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One. (2008) 3:e3056. doi: 10.1371/journal.pone.0003056
18. Valles Y, Artacho A, Pascual-Garcia A, Ferrus ML, Gosalbes MJ, Abellan JJ, et al. Microbial succession in the gut: directional trends of taxonomic and functional change in a birth cohort of Spanish infants. PLoS Genet. (2014) 10:e1004406. doi: 10.1371/journal.pgen.1004406
19. Aires J, Thouverez M, Allano S, Butel MJ. Longitudinal analysis and genotyping of infant dominant bifidobacterial populations. Syst Appl Microbiol. (2011) 34:536–41. doi: 10.1016/j.syapm.2011.02.007
20. Sjögren YM, Tomicic S, Lundberg A, Böttcher MF, Björkstén B, Sverremark-Ekström E, et al. Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin Exp Allergy. (2009) 39:1842–51. doi: 10.1111/j.1365-2222.2009.03326.x
21. Zhang S, Zhong G, Shao D, Wang Q, Hu Y, Wu T, et al. Dietary supplementation with Bacillus subtilis promotes growth performance of broilers by altering the dominant microbial community. Poultry Sci. (2021) 100:100935. doi: 10.1016/j.psj.2020.12.032
22. Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. (2011) 108(Suppl. 1):4578–85. doi: 10.1073/pnas.1000081107
23. Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. (2015) 17:852. doi: 10.1016/j.chom.2015.05.012
24. Wu S, Shen Y, Zhang S, Xiao Y, Shi S. Salmonella interacts with autophagy to offense or defense. Front Microbiol. (2020) 11:721. doi: 10.3389/fmicb.2020.00721
25. Shi S, Wu S, Shen Y, Zhang S, Xiao Y, He X, et al. Iron oxide nanozyme suppresses intracellular Salmonella Enteritidis growth and alleviates infection in vivo. Theranostics. (2018) 8:6149–62. doi: 10.7150/thno.29303
27. Kühl AA, Erben U, Kredel LI, Siegmund B. Diversity of intestinal macrophages in inflammatory bowel diseases. Front Immunol. (2015) 6:613. doi: 10.3389/fimmu.2015.00613
28. Bain CC, Scott CL, Uronen-Hansson H, Gudjonsson S, Jansson O, Grip O, et al. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. (2013) 6:498–510. doi: 10.1038/mi.2012.89
29. Bain CC, Bravo-Blas A, Scott CL, Gomez Perdiguero E, Geissmann F, Henri S, et al. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol. (2014) 15:929–37. doi: 10.1038/ni.2967
30. Klug F, Prakash H, Huber PE, Seibel T, Bender N, Halama N, et al. Low-dose irradiation programs macrophage differentiation to an iNOS+/M1 phenotype that orchestrates effective T cell immunotherapy. Cancer Cell. (2013) 24:589–602. doi: 10.1016/j.ccr.2013.09.014
31. Hashimoto D, Chow A, Noizat C, Teo P, Beasley Mary B, Leboeuf M, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. (2013) 38:792–804. doi: 10.1016/j.immuni.2013.04.004
32. Schridde A, Bain CC, Mayer JU, Montgomery J, Pollet E, Denecke B, et al. Tissue-specific differentiation of colonic macrophages requires TGFβ receptor-mediated signaling. Mucosal Immunol. (2017) 10:1387–99. doi: 10.1038/mi.2016.142
33. Gren ST, Grip O. Role of monocytes and intestinal macrophages in Crohn's disease and ulcerative colitis. Inflamm Bowel Dis. (2016) 22:1992–8. doi: 10.1097/MIB.0000000000000824
34. Danne C, Ryzhakov G, Martinez-Lopez M, Ilott NE, Franchini F, Cuskin F, et al. A large polysaccharide produced by Helicobacter hepaticus induces an anti-inflammatory gene signature in macrophages. Cell Host Microbe. (2017) 22:733–45.e5. doi: 10.1016/j.chom.2017.11.002
35. Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. (2014) 158:705–21. doi: 10.1016/j.cell.2014.05.052
36. Mengheri E, Ciapponi L, Vignolini F, Nobili F. Cytokine gene expression in intestine of rat during the postnatal developmental period: increased IL-1 expression at weaning. Life Sci. (1996) 59:1227–36. doi: 10.1016/0024-3205(96)00446-8
37. Gollwitzer ES, Marsland BJ. Impact of early-life exposures on immune maturation and susceptibility to disease. Trends Immunol. (2015) 36:684–96. doi: 10.1016/j.it.2015.09.009
38. Theochari NA, Stefanopoulos A, Mylonas KS, Economopoulos KP. Antibiotics exposure and risk of inflammatory bowel disease: a systematic review. Scand J Gastroenterol. (2018) 53:1–7. doi: 10.1080/00365521.2017.1386711
39. Klement E, Cohen RV, Boxman J, Joseph A, Reif S. Breastfeeding and risk of inflammatory bowel disease: a systematic review with meta-analysis. Am J Clin Nutr. (2004) 80:1342–52. doi: 10.1093/ajcn/80.5.1342
40. Riedler J, Braun-Fahrländer C, Eder W, Schreuer M, Waser M, Maisch S, et al. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet. (2001) 358:1129–33. doi: 10.1016/S0140-6736(01)06252-3
41. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. (2012) 336:489–93. doi: 10.1126/science.1219328
42. Suez J, Zmora N, Segal E, Elinav E. The pros, cons, and many unknowns of probiotics. Nat Med. (2019) 25:716–29. doi: 10.1038/s41591-019-0439-x
43. Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. (2017) 14:573–84. doi: 10.1038/nrgastro.2017.88
44. Alard J, Peucelle V, Boutillier D, Breton J, Kuylle S, Pot B, et al. New probiotic strains for inflammatory bowel disease management identified by combining in vitro and in vivo approaches. Benef Microbes. (2018) 9:317–31. doi: 10.3920/BM2017.0097
45. van Baarlen P, Wells JM, Kleerebezem M. Regulation of intestinal homeostasis and immunity with probiotic lactobacilli. Trends Immunol. (2013) 34:208–15. doi: 10.1016/j.it.2013.01.005
46. Derwa Y, Gracie DJ, Hamlin PJ, Ford AC. Systematic review with meta-analysis: the efficacy of probiotics in inflammatory bowel disease. Aliment Pharmacol Ther. (2017) 46:389–400. doi: 10.1111/apt.14203
47. Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. (2015) 163:367–80. doi: 10.1016/j.cell.2015.08.058
48. Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. (2018) 174:1388–405.e21. doi: 10.1016/j.cell.2018.08.041
49. Charbonneau D, Gibb RD, Quigley EM. Fecal excretion of Bifidobacterium infantis 35624 and changes in fecal microbiota after eight weeks of oral supplementation with encapsulated probiotic. Gut Microbes. (2013) 4:201–11. doi: 10.4161/gmic.24196
50. Smith TJ, Anderson D, Margolis LM, Sikes A, Young AJ. Persistence of Lactobacillus reuteri DSM17938 in the human intestinal tract: response to consecutive and alternate-day supplementation. J Am Coll Nutr. (2011) 30:259–64. doi: 10.1096/fasebj.24.1_supplement.340.1
51. Corridoni D, Chapman T, Antanaviciute A, Satsangi J, Simmons A. Inflammatory bowel disease through the lens of single-cell RNA-seq technologies. Inflamm Bowel Dis. (2020) 26:1658–68. doi: 10.1093/ibd/izaa089
52. Aschenbrenner D, Quaranta M, Banerjee S, Ilott N, Jansen J, Steere B, et al. Deconvolution of monocyte responses in inflammatory bowel disease reveals an IL-1 cytokine network that regulates IL-23 in genetic and acquired IL-10 resistance. Gut. (2021) 70:1023–36. doi: 10.1136/gutjnl-2020-321731
53. Grazul H, Kanda LL, Gondek D. Impact of probiotic supplements on microbiome diversity following antibiotic treatment of mice. Gut Microbes. (2016) 7:101–14. doi: 10.1080/19490976.2016.1138197
54. Suez J, Zmora N, Zilberman-Schapira G, Mor U, Dori-Bachash M, Bashiardes S, et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. (2018) 174:1406–23.e16. doi: 10.1016/j.cell.2018.08.047
55. Schultz M, Göttl C, Young RJ, Iwen P, Vanderhoof JA. Administration of oral probiotic bacteria to pregnant women causes temporary infantile colonization. J Pediatr Gastroenterol Nutr. (2004) 38:293–7. doi: 10.1097/00005176-200403000-00012
56. Savino F, Cordisco L, Tarasco V, Palumeri E, Calabrese R, Oggero R, et al. Lactobacillus reuteri DSM 17938 in infantile colic: a randomized, double-blind, placebo-controlled trial. Pediatrics. (2010) 126:e526–e33. doi: 10.1542/peds.2010-0433
57. AlFaleh K, Anabrees J. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Evid Based Child Health. (2014) 9:584–671. doi: 10.1002/ebch.1976
Keywords: early-life intervention, gut microbiota, probiotics, weaning, inflammatory bowel disease
Citation: Guo F, Cai D, Li Y, Gu H, Qu H, Zong Q, Bao W, Chen A and Liu H-Y (2021) How Early-Life Gut Microbiota Alteration Sets Trajectories for Health and Inflammatory Bowel Disease? Front. Nutr. 8:690073. doi: 10.3389/fnut.2021.690073
Received: 02 April 2021; Accepted: 05 July 2021;
Published: 06 August 2021.
Edited by:
Shourong Shi, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Haibo Sun, University of San Francisco, United StatesCopyright © 2021 Guo, Cai, Li, Gu, Qu, Zong, Bao, Chen and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao-Yu Liu, aGFveXUubGl1MDUyM0BvdXRsb29rLmNvbQ==; Aoxue Chen, YW94dWUuY2hlbkBjYW1wdXMubG11LmRl; Wenbin Bao, d2JiYW9AeXp1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.