
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr., 04 June 2021
Sec. Nutrigenomics
Volume 8 - 2021 | https://doi.org/10.3389/fnut.2021.689419
This article is part of the Research TopicMicronutrients and Fatty Acids in Precision Nutrition StrategiesView all 12 articles
 Nikola Kotur1
Nikola Kotur1 Anita Skakic1
Anita Skakic1 Kristel Klaassen1
Kristel Klaassen1 Vladimir Gasic1
Vladimir Gasic1 Branka Zukic1
Branka Zukic1 Vesna Skodric-Trifunovic2,3
Vesna Skodric-Trifunovic2,3 Mihailo Stjepanovic2,3
Mihailo Stjepanovic2,3 Zorica Zivkovic4,5
Zorica Zivkovic4,5 Olivera Ostojic4
Olivera Ostojic4 Goran Stevanovic3,6
Goran Stevanovic3,6 Lidija Lavadinovic6
Lidija Lavadinovic6 Sonja Pavlovic1
Sonja Pavlovic1 Biljana Stankovic1*
Biljana Stankovic1*Background: COVID-19 pandemic has proved to be an unrelenting health threat for more than a year now. The emerging amount of data indicates that vitamin D, zinc and selenium could be important for clinical presentation of COVID-19. Here, we investigated association of genetic variants related to the altered level and bioavailability of vitamin D, zinc and selenium with clinical severity of COVID-19.
Methods: We analyzed variants in genes significant for the status of vitamin D (DHCR7/NADSYN1 rs12785878, GC rs2282679, CYP2R1 rs10741657, and VDR rs2228570), zinc (PPCDC rs2120019) and selenium (DMGDH rs17823744) in 120 Serbian adult and pediatric COVID-19 patients using allelic discrimination. Furthermore, we carried out comparative population genetic analysis among European and other worldwide populations to investigate variation in allelic frequencies of selected variants.
Results: Study showed that DHCR7/NADSYN rs12785878 and CYP2R1 rs10741657 variants were associated with severe COVID-19 in adults (p = 0.03, p = 0.017, respectively); carriers of DHCR7/NADSYN TG+GG and CYP2R1 GG genotypes had 0.21 and 5.9 the odds for developing severe disease, OR 0.21 (0.05–0.9) and OR 5.9 (1.4–25.2), respectively. There were no associations between selected genetic variants and disease severity in pediatric patients. Comparative population genetic analysis revealed that Serbian population had the lowest frequency of CYP2R1 rs10741657 G allele compared to other non-Finish Europeans (0.58 compared to 0.69 and 0.66 in Spanish and Italian population, respectively), suggesting that other populations should also investigate the relationship of CYP2R1 variant and the COVID-19 disease course.
Conclusion: The results of the study indicated that vitamin D related genetic variants were implicated in severe COVID-19 in adults. This could direct prevention strategies based on population specific nutrigenetic profiles.
COVID-19 pandemic has proved to be an unrelenting health threat for more than a year now. More contagious and more deadly SARS-CoV-2 variants are a major concern, especially because effective, causal therapy is still unavailable, and vaccination coverage rates are lower than anticipated. Moreover, protection from vaccination is likely to last only for a season or two. One of the strategies to save lives is to identify groups at risk of severe COVID-19 disease and to implement measures aimed at those groups. The elderly, male, obese, chronic disease patients, patients with malignancies, immunocompromised, dark-skinned, socioeconomically disadvantaged and tobacco users suffered the most death toll from COVID-19 (1, 2). Also, interindividual genetic variations might be implicated in more severe COVID-19 (3, 4).
Another strategy directed toward reducing severe COVID-19 symptoms relies on high dose vitamins and trace elements (micronutrients) supplementation. Among micronutrients important for adequate immune function, vitamin D, zinc and selenium are of particular importance for coping with viral, respiratory infections, such as COVID-19 (5). Adequate status of these micronutrients is not only important for immune function and viral clearance, but also might mitigate life-threatening complications of SARS-CoV-2 infection, such as thrombosis and uncontrolled inflammation which leads to cytokine storm. Namely, vitamin D via vitamin D receptor (VDR) mediates immune function and regulation, strengthening of epithelial barriers and antioxidant defense, and also regulates expression of SARS-CoV-2 receptor, ACE2 (6–8). Zinc exerts direct anti-viral effects and also serves as a cofactor of dozens of proteins important for immune function and regulation, and antioxidative defense (9). Selenium via selenoproteins regulates immune function and provides antioxidant defense and vaso-protection (10).
Although important for various processes in our body, a lot of people worldwide have inadequate status of vitamin D, zinc or selenium. There is a pandemic of vitamin D deficiency, especially in higher latitudes due to inadequate sun exposure necessary for the synthesis of this vitamin (11). Selenium deficiency is prevalent in Europe and other parts of the world where soil is poor of this element, while zinc status is compromised predominantly in underdeveloped regions (12). Vitamin D, zinc and selenium deficiency is associated with factors of COVID-19 severity, such as old age, obesity, diabetes, dyslipidemia and chronic and acute inflammation. COVID-19 patients have lower status of vitamin D, zinc and selenium than healthy individuals (13). Also, patients with severe COVID-19 disease have lower status of vitamin D, zinc and selenium than mild and moderate disease patients (14–16). Preliminary evidence from intervention studies suggests that supplementation with these micronutrients protects from severe COVID-19 disease (17, 18).
Apart from chronic and acute health conditions, lifestyle and nutrition, genetic factors also play a role in bioavailability and status of vitamin D, zinc and selenium. Several large genome-wide association studies (GWAS) identified variants located near genes involved in synthesis, transport and metabolism of vitamin D, namely DHCR7/NADSYN1, GC and CYP2R1, respectively (19–21). Variants in these genes are associated with serum level of 25OHD, which is a measure of vitamin D status. Also, genetic variants in VDR gene, which codes for vitamin D receptor that regulates expression of more than 200 human genes (22), have been shown to increase susceptibility to acute lower respiratory infections in children, as well as several inflammatory and autoimmune diseases (23–26). GWAS studies comprising three large independent cohorts associated selenium status measured in whole blood and erythrocytes (27), as well as nails (reflects longer duration of selenium exposure) (28) with a locus near DMGDH gene. The same locus was implicated in a response to selenium supplementation in another GWAS (29). Zinc nutrigenetics is less studied, but a few candidate variants associated with status of this trace elements are identified in a large GWAS, and the most significant association was noted for rs2120019 variant near PPCDC gene (27).
Building on this premise, we hypothesize that genetic variants linked to suboptimal level or decreased bioavailability of vitamin D, zinc and selenium, may also serve as markers of disease course in COVID-19 patients, influenced by the level of these micronutrients. Therefore, we evaluated genetic determinants of vitamin D (DHCR7/NADSYN1 rs12785878, GC rs2282679, CYP2R1 rs10741657, VDR rs2228570), zinc (PPCDC rs2120019) and selenium (DMGDH rs17823744) as risk factors for severe forms of COVID-19. Furthermore, we attempted to investigate the inter-population diversity of the selected genetic variants. Inter-population nutrigenetic variability could suggest different relevance of nutrigenetic factors in populations exposed to different nutritional habits and specific environmental factors. These data should be taken into account when evaluating the possible relationship of nutrigenetic variants with severe COVID-19 forms in other populations struck by the pandemic.
Present study included 120 patients with diagnosis of COVID-19 (adult and pediatric) who were treated in tertiary healthcare institutions, the Clinic of Pulmonology, Clinical Center of Serbia and Children's Hospital for Lung Diseases and Tuberculosis, Medical Center “Dr Dragiša Mišović,” Belgrade, Serbia, between April and June of 2020. For all patients, a COVID-19 infection was defined as a positive SARS-CoV-2 laboratory test (viral RNA RT-PCR test from nasopharyngeal swabs). From each patient, peripheral blood sample was collected for DNA isolation and further genetic analyses.
Clinical data were assessed using electronic health records. From a total of 120 COVID-19 patients included in the study, clinical data were available for 115 patients, 73 adults and 42 pediatric.
Adult COVID-19 patients were categorized into mild, moderate or severe clinical course according to the National Institutes of Health (NIH) Definition of COVIID-19 Disease Severity (COVID-19 Treatment Guidelines Panel. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. 2020. Available at: http://www.covid19treatmentguidelines.nih.gov), as follows: mild – patients with symptoms such as fever, fatigue, cough, myalgia, and headache, but without dyspnea or pneumonia; moderate- those with evidence of pneumonia based on imaging showing up to 50% of lung involvement, who had blood oxygen saturation >93% on room air; severe- patients who demonstrated pneumonia with >50% of lung involvement on imaging or had blood oxygen saturation level equal to or <93% on room air and required supportive oxygen therapy.
Pediatric patients were enrolled during the first peak of the COVID-19. At that time, SARS-CoV-2 positive pediatric patients were admitted to the hospital if they had COVID-19 symptoms or for observation due to previous health issues. Most of the recruited pediatric patients (90.5%) demonstrated mild form of the disease. Therefore, for the purpose of this study, we classified pediatric patients into 2 groups – symptomatic, who were diagnosed with pneumonia or any typical COVID-19 symptoms such as cough, fever, sore throat, fatigue, and headache or asymptomatic, patients tested positive for SARS-CoV-2 with medical history of recurrent respiratory diseases.
This study was approved by the Ethics Committee of the Institute of Molecular Genetics and Genetic Engineering University of Belgrade (approval for sample collection and biobank formation O-EO-016/2020, 06.05.2020.; approval for the genetic study O-EO-016/2020/1, 03.09.2020). Informed consent was obtained from each participant or their parents/legal guardians. The study was performed in accordance with the Declaration of Helsinki.
Genomic DNA was isolated from the whole blood samples using QIAamp DNA Blood Mini Kit (Qiagen) and stored at −20°C until analysis.
Genetic variants DHCR7/NADSYN1 rs12785878, GC rs2282679, CYP2R1 rs10741657, VDR rs2228570, PPCDC rs2120019 and DMGDH rs17823744 were analyzed using TaqMan SNP Genotyping Assays (Thermo Fisher Scientific) (C__32063037_10, C__26407519_10, C__2958430_10, C__12060045_20, C__304992_10, C__32728760_10), according to the manufacturer's instructions with modifications. A total volume of 8 μl PCR reaction contained 36 ng of DNA, 1x Taqman Genotyping Master Mix and 1x TaqMan SNP Genotyping Assays (Thermo Fisher Scientific). Genotyping was performed on a real-time PCR system (Applied Biosystems 7500, Thermo Fisher Scientific). Genotype calling was implemented with built-in 7500 System Software v1.3.1 (Thermo Fisher Scientific).
Statistical analyses were performed in software SPSS v23.0 and R v.3.6.2.
Differences in demographic and clinical data between COVID-19 mild/moderate/severe subgroups were examined using adequate statistical tests for continuous and discrete data (Mann-Whitney U test, Chi-square or Fisher exact test).
For genetic data, Hardy-Weinberg equilibrium was examined to assess the performance of the genotyping assays. Differences in genotype and allele counts distribution for each variant between COVID-19 subgroups were tested using the Chi-squared test. Impact of each genetic variable on severity of the COVID-19 was estimated by odds ratio with 95% confidence interval using a logistic regression model in order to control for confounding factors, such as age and gender.
Frequencies of the analyzed variants were compared between group of Serbian COVID-19 patients and other populations using two openly available variant databases, 1,000 Genomes Project (1KGP) and Genome Aggregation Database (gnomAD) v.3.0 (30, 31). Chi square or Fisher exact tests were used to measure significant differences in allelic distributions between Serbian and European populations (Finish and non-Finish, particularly Italian, Spanish, Britain and USA with European ancestry), and other worldwide populations (African, East and South Asian, American Latino/Admixture and Ashkenazi Jewish), applying Bonferroni correction for multiple testing.
We examined the level of population genetic variability at each genetic loci using the maximal global difference in risk allele frequency (delta AF), which was calculated by subtracting the maximum and the minimum allele frequency across analyzed populations.
All tests were bi-directional. The threshold for statistical significance was defined using appropriate multiple comparisons correction methods, such as Bonferroni or Benjamini-Hochberg false discovery rate (FDR).
In the selected study group, 73 were adults while 42 were pediatric patients. Among adults 65.8% were females and the median age was 43 (range 20–82); among pediatric patients 38.1% were females and median age was 9 (range 0.3–17). Demographic and clinical descriptions of COVID-19 patients are presented in Table 1.
In the group of adult COVID-19 patients, 10 (13.7%) required supportive oxygen therapy and 3 (4%) had COVID-19 related death outcomes. Examining available clinical data, patients were categorized into mild (n = 35), moderate (n = 21) or severe (n = 17) disease groups. Age distribution of the patients significantly varied between groups; gradual increase in participants' age has been observed across mild, moderate and severe groups (p < 0.001). In the severe group, a significantly higher number of patients suffered from hypertension, diabetes and were using ACE inhibitors, compared to mild and moderate groups (p < 0.001, p < 0.001, p < 0.001, respectively). The highest percent of lymphopenia and thrombocytopenia events were observed in the severe group, 93.8 and 56.3%, respectively. Also, patients with severe COVID-19 had higher levels of CRP, compared to patients with moderate and mild disease (p < 0.001).
Pediatric COVD-19 patients were categorized into two groups – asymptomatic (n = 20) and symptomatic (n = 22). There was no significant difference in distribution of age, gender, nor in the presence of comorbidity between the two groups, except past respiratory diseases. The level of CRP was significantly higher in COVID-19 symptomatic compared to asymptomatic pediatric patients (p = 0.003).
COVID-19 patients were genotyped for variants in DHCR7/NADSYN1, GC, CYP2R1, VDR, PPCDC and DMGDH genes (Table 2). Distributions of genotypes of all analyzed variants were in accordance with Hardy-Weinberg equilibrium.
We compared the distribution of analyzed genotypes between different COVID-19 severity groups. In the group of adult patients, two outcomes were considered in the logistic regression model – first included both mild and moderate disease, and second was severe disease (Table 3). Logistic regression used genotypes of analyzed variants as variables, controlled for age and gender in multivariate models. To increase the statistical power of the study, a dominant genetic model has been applied, in particular, a group of homozygous carriers of more frequent allele were compared to heterozygous and homozygous carriers of the less frequent, minor allele. Analysis showed that carriers of DHCR7/NADSYN1 G allele (TG+GG), associated with lower levels of 25OHD were 4.9 times less likely (or had 0.21 the odds) to develop a severe form of COVID-19, compared with carriers of TT genotype. When adjusted for age and gender, observed association was still significant (OR 0.21, 95% CI 0.05–0.9, p = 0.03). Association has also been observed between severe COVID-19 and CYP2R1 GG genotype, previously related to decreased levels of 25OHD. In the logistic regression model, CYP2R1 GG carriers had 5.9 times higher odds to develop severe disease, adjusted for age and gender (OR 5.9, 95% CI 1.4–25.2, p = 0.017). Additionally, we applied adjustment for obesity in multivariate logistic regression models, showing that DHCR7/NADSYN1 rs12785878 and CYP2R1 rs10741657 variants remained significant predictors of severe COVID-19 in adults (OR 0.22, 95% CI 0.06-0.93, p= 0.02; OR 4.9, 95% CI 1.5-16.4, p= 0.009, respectively). Our study did not show association of GC and VDR variants with higher risk of severe COVID-19 in adults.
Homozygous carriers of DMGDH A allele, which has been related to lower levels of selenium, in our study were less likely to present severe form of COVID-19 (OR 0.2, 95% CI 0.09–0.9, p = 0.03). However, this association was not significant after correction for age and gender. Regarding zinc-related PPCDC variant, we observed no significant association with severe COVID-19 outcome in adult patients.
In pediatric COVID-19 cases, we applied a logistic regression model to predict two outcomes – asymptomatic and symptomatic disease, using genotype data (Table 4). No significant associations with increased risk for symptomatic pediatric COVID-19 disease were found with any of the analyzed genetic variants.
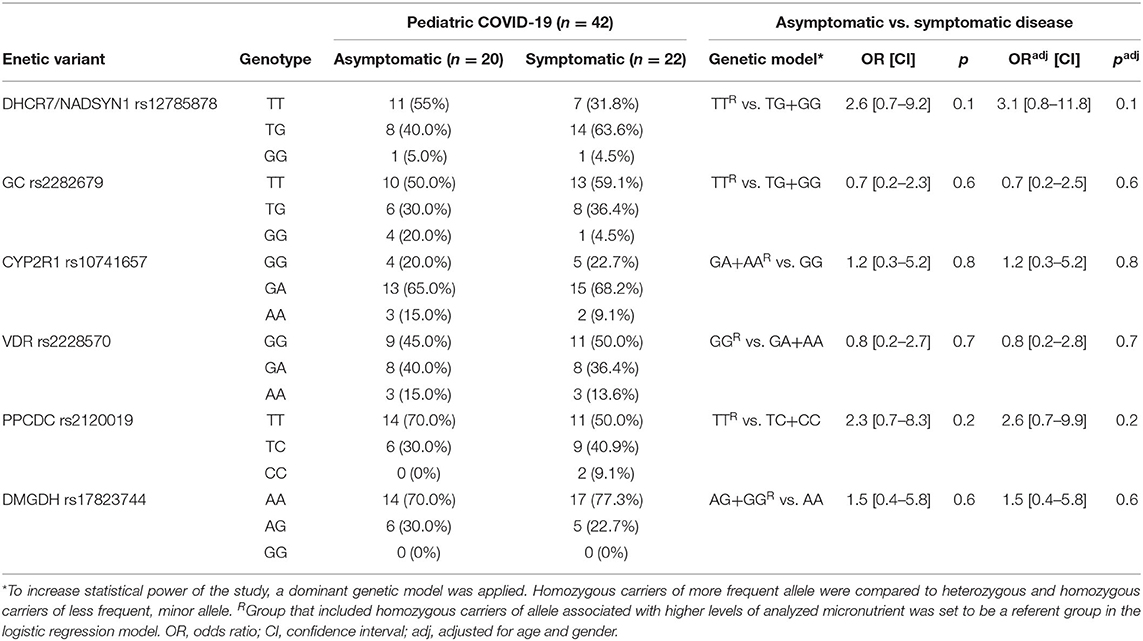
Table 4. Association of the analyzed genotypes with the risk of symptomatic COVID-19 disease in children.
Finally, we aimed to compare allele frequencies of selected genetic variants in Serbian COVID-19 study group with their frequencies in other populations. Therefore, we retrieved variant site data from the two genetic databases, 1KGP and gnomAD. Distributions of alleles frequencies (AF) associated with decreased levels of selected micronutrients in European and worldwide populations have been presented in Figure 1 (data used for calculations are provided in Supplementary Table 1). For all analyzed genetic variants, observed AF in Serbian study group did not diverge from overall AF in European non-Finish populations. When comparing Serbian study group with populations of non-Finish Europeans, we noticed a difference in the frequency of DHCR7/NADSYN1 allele G between Italian compared to Serbian population (0.22 and 0.3, respectively, p = 0.07). Difference was also observed in the frequency of CYP2R1 rs10741657 allele G between Serbian (0.58) and Spanish (0.69) as well as Italian population (0.66) (p = 0.02, p = 0.08, respectively). Serbian population had the lowest frequency of the CYP2R1 rs10741657 G variant among European non-Finish populations, while Spanish and Italians demonstrated the highest. However, noted differences were not statistically significant. As expected, significant differences were mostly seen between Serbian and African, South and East Asian, American Latino/Admixed, and Ashkenazi populations.
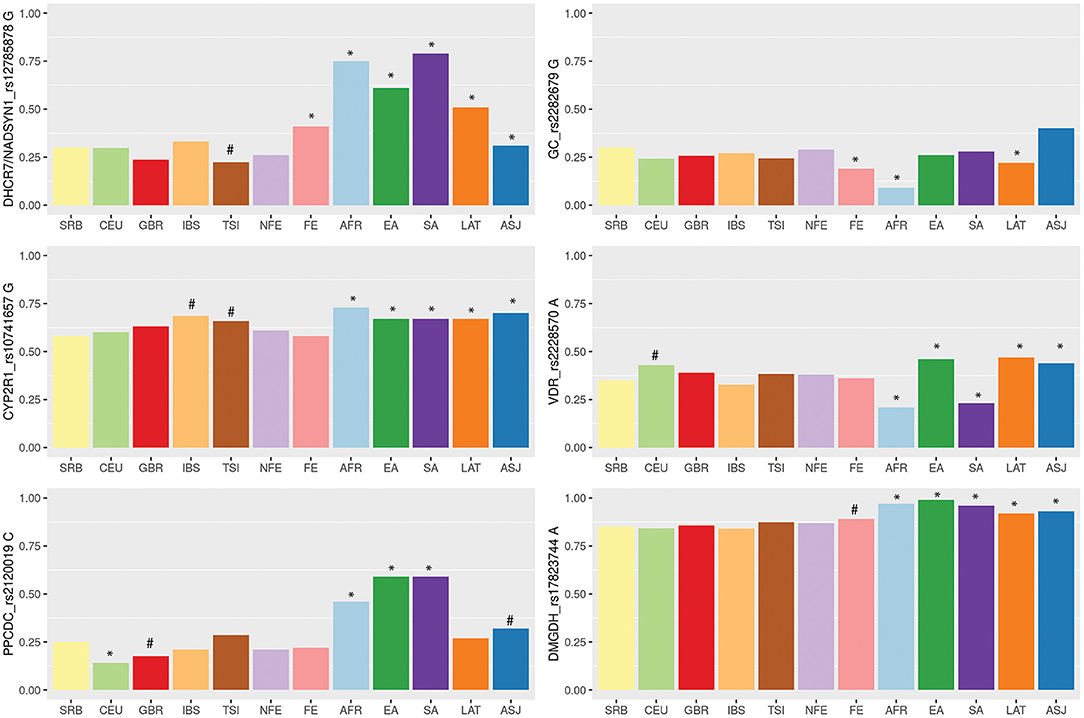
Figure 1. Variability of vitamin D, zinc and selenium related genetic loci among wordwide populations based on the 1,000 Genome project (1KGP), Genome Aggregation Database (gnomAD) and Serbian COVID-19 study group data. Populations: SRB, Serbian; CEU, Utah residents with Northern and Western European ancestry (1KGP); GBR, British in England and Scotland (1KGP); TSI, Tuscany in Italy (1KGP); IBS, Iberian populations in Spain (1KGP); FIN-Finnish European (gnomAD); NFE-non-Finnish European, including Northwestern European, Bulgarian, Estonian, Swedish, Southern European, and Other non-Finnish European (gnomAD); AFR, African (gnomAD); EA, East Asian (gnomAD); SA, South Asian (gnomAD); LAT, Latino/Admixed American (gnomAD); ASJ, Ashkenazi Jewish (gnomAD). Barplots are showing frequencies of the alleles associated with lower levels of selected micronutrients. Differences in allelic distributions were analyzed between Serbian and other populations using Chi square test followed by Bonferroni correction for multiple testing. Statistical significance was marked with *(p = < 0.008), while statistical trend with #(0.1 < p < 0.008).
The level of population genetic variability at each selected loci was assessed using delta AF (dAF), representing the difference between maximum and minimum AF value across analyzed populations. The highest dAF was observed for the DHCR7/NADSYN1 rs12785878 variant (dAF 0.56). The lowest dAF was detected for CYP2R1 rs10741657 and DMGDH rs17823744 variants (dAF 0.15 and 0.15, respectively).
In this study, we focused our attention to genetic variants related to altered level, bioavailability or mechanism of action of vitamin D, zinc and selenium, aiming to investigate their association with different COVID-19 presentation in children and adults. Serbia, like much of Europe and the world, faces micronutrient deficiency/insufficiency which is in part influenced by genetic variability. The prevalence of suboptimal levels of vitamin D and selenium in Serbian population is estimated to be around 50% (32, 33), while zinc insufficiency is around 7% (12). Bearing in mind that even a mild insufficiency of these micronutrients can cause suboptimal immune function, implications for COVID-19 pandemic can be formidable.
Results of our study showed association of DHCR7/NADSYN1 rs12785878 and CYP2R1 rs10741657 variants with severe form of COVID-19 in adult patients. In the pediatric COVID-19 group, we did not detect any associations between analyzed genetic factors and disease severity.
Variant rs12785878 is located 8 kilobases upstream of the DHCR7 gene on chromosome 11q12. DHCR7 codes for 7-dehydrocholesterol reductase, a key enzyme in cholesterol biosynthesis and vitamin D cutaneous production. Particularly, the activity of DHCR7 enzyme decreases the level of vitamin D precursor availability, 7-dehydorcholesterol, shunting it in the direction of cholesterol biosynthesis (34). DHCR7 rs12785878 variant was firstly noted in a large GWAS of 25OHD concentrations in 33 996 individuals of European descent by (21), and subsequently confirmed in other numerous studies, where the G allele was shown to be associated with lower 25OHD serum levels. In our comparative genetic population analysis, we showed that frequency of G allele markedly varied across different populations; it ranged from 0.75 in African and Asian populations to 0.22 in European populations. Considerable differences in allele frequencies between populations may indicate a locus that has undergone positive selection in a specific geographical area. It has been suggested that positive selection influenced the DHCR7 gene, increasing the frequency of reduced activity variant (allele T) in European populations (35). Reduced activity of DHCR7 leads to increased availability of 7-dehydrocholesterol for vitamin D synthesis allowing Europeans to avoid deficiency in northern latitudes. Our results suggested an association of the rs12785878 variant with the severe form of COVID-19. However, in our study, carriers of the TT genotype which has been linked to higher circulating level of 25OHD, were more likely to develop a severe form of COVID-19. Given the immune-enhancing aspects of vitamin D, one might expect the opposite. However, it should be noted that the roll of DHCR7 variants to vitamin D status is constrained to vitamin D synthesis in the skin exposed to UV light and probably does not influence contribution of dietary sources and supplementation to the level of this vitamin. COVID-19 patients enrolled in our study were hospitalized with this disease in the spring of 2020 during or right after lockdown measures were in place. That means that vitamin D was scarcely synthetized in skin during that time and, therefore, DHCR7 variants had limited influence on vitamin D level. Regardless of vitamin D production, DHCR7 variants could influence immune function mediated through cholesterol metabolites. Namely, reduced activity of DHCR7 leads to increased 7-dehydrocholesterol levels which influences interferon production and viral clearance (36).
After initial UV-light mediated production of vitamin D3 in the skin, the second step in the synthesis of active vitamin D is catalyzed in the liver by cytochrome P450 (CYP) enzyme which hydroxylates carbon 25, producing the intermediate 25-hydroxyvitamin D3, or 25OHD, the major circulatory form of the vitamin D (37). This enzyme is encoded by the CYP2R1 (cytochrome P450, family 2, subfamily R, polypeptide 1) gene. Variant rs10741657 located near the CYP2R1 gene was linked by several studies to 25OHD serum concentrations (21, 38), with the allele G shown to be associated with lower 25OHD serum levels, and with the GG homozygotes having the lowest levels. Our results indicated an association between severe COVID-19 and CYP2R1 GG genotype. In the logistic regression model, CYP2R1 rs10741657 GG carriers had 5.9 times higher odds to develop severe disease. We noticed low level of variability of allele G frequency for CYP2R1 rs10741657 variant across analyzed populations (dAF 0.15); G allele frequencies ranged from 0.58 in Serbian, which was the lowest among European populations, to 0.73 in African population. The highest frequency of the G allele among Europeans was observed in Italian and Spanish populations (0.66 and 0.69, respectively). Recent study that reported negative correlations between mean levels of vitamin D in various European countries with COVID-19 cases per million people, suggested that Spain, Italy and Switzerland are the most vulnerable countries since they have very low levels of vitamin D in the aging population (39). Our results could be of particular interest for these populations. Relatively high occurrence of the CYP2R1 rs10741657 G variant in different populations, reproducible association with decreased 25OHD levels and observed relationship with severe COVID-19 in our group of patients, gives a solid ground for future studies to examine relationship of this CYP2R1 variant with the clinical course of COVID-19 in other populations.
A role of vitamin D in the pathogenesis of COVID-19 has been extensively studied since the beginning of the pandemic. Calcitriol (1, 25-dihydroxyvitamin D3) has an important role in regulating renin angiotensin system by enhancing the expression of ACE2, which is the main target of SARS-CoV2 cell entry (7, 8). Also, vitamin D has an immunomodulatory effect and it can prevent macrophages to release excessive proinflammatory cytokines and chemokines (6, 40). A recent study showed that the intake levels of relevant micronutrients such as vitamin D are inversely associated with higher COVID-19 incidence and/or mortality (41).
Furthermore, other studies sought for more detailed nutrigenetic markers as factors that might contribute to the bioavailability of vitamin D and therefore influence COVID-19 susceptibility and/or clinical course. A study which aimed to assess the association between variants in the GC gene that encodes vitamin D binding/transport protein (DBP), and the prevalence and mortality rates of COVID-19, pointed out to rs7041 variant (42). Another recent study (not peer-reviewed yet) performed in Portuguese population of COVID-19 patients found an association of GC rs2282679 variant with COVID-19 disease severity (43). Our study indicated an association between DHCR7/NADSYN1 rs12785878 and CYP2R1 rs10741657 variants and the severity of COVID-19, but the variants GC rs2282679 and VDR rs2228570 were not linked to a higher risk of severe COVID-19 in adults. In a recent Mendelian randomization study (not peer-reviewed yet) on vitamin D and COVID-19 in individuals of European ancestry, genetically increased 25OHD concentrations did not protect against COVID-19 susceptibility, but increased the odds for hospitalization and severe disease (44). Another Mendelian randomization study failed to find evidence of a linkage between vitamin D deficiency and COVID-19 infection rates or severe disease (45). However, described studies did not consider true vitamin D deficiency in participants, and it can be possible that vitamin D supplementation could benefit insufficient/deficient patients (44, 45).
Nutrigenetics of zinc and selenium were discussed in the context of COVID-19 as a promising strategy to implement personalized approach to strengthening antioxidant and antiviral defense (41). However, the present study is the first study on genetic determinants of zinc and selenium that included COVID-19 patients. Our results did not show association of these variants with severe COVID-19, however, our univariate analysis pointed out to DMGDH rs17823744 variant, which warrants further investigation. DMGDH is implicated in homocysteine metabolism and it is postulated that there is a connection between selenium exposure and the homocysteine metabolic pathways (28). Apart from locus near DMGDH gene, which was noted in several GWAS cohorts focused on selenium status, other studies analyzed genes coding for selenoproteins. Candidate gene approach studies or studies that analyzed protein activity, associated glutathione peroxidase 1 (GPX1), GPX4 and selenoprotein P (SELENOP) with response to selenium supplementation (46–48). Although important for selenium action, these genes were not associated with selenium status in GWAS studies.
Apart from nutrigenetics, other studies also dealt with genetic factors associated with COVID-19 severity, but the results were not consistent. A GWAS study of almost 4,000 healthy volunteers and severely ill Italian and Spanish COVID-19 patients associated loci on chromosomes 3 and chromosome 9 (near ABO locus) (3), while another GWAS study of 2,244 critically ill UK patients associated variants located on chromosomes 12, 19 and 21 with more severe disease or poor outcome (4). None of these loci are located near nutrigenetic variants analyzed in the present study.
Results of this study should be evaluated in larger cohorts of COVID-19 patients. Small number of patients is the main limitation of this study, although, both mild and moderate, as well as severe groups are well-represented among adult patients. Among pediatric patients, the majority of patients had mild symptoms so we could not investigate risk factors of severe disease in children. Instead, genetic variants were analyzed as risk factors of symptomatic disease.
Another limitation of this study is lack of micronutrient measurement needed to assess actual status of vitamin D, zinc and selenium. We focused solely on the genetic variants associated with altered levels of vitamin D, zinc and selenium. Status of the micronutrient is highly confounded variable that depends on numerous factors. Beside genetics, nutrition habits, individual lifestyle and presence of comorbidities are also important. Therefore, comprehensive nutrigenetic studies complemented with measures of nutritional intake, micronutrient's serum level and detailed lifestyle evaluations are needed in the future.
This study highlights the importance of personalized as well as population based strategies directed toward reduction of COVID-19 burden. These strategies can be implemented through employment of established nutrigenetic markers, such as genetic variants involved in vitamin D disposition. Our results pointed out to DHCR7/NADSYN1 rs12785878 and CYP2R1 rs10741657 variants, both involved in vitamin D synthesis, as potential risk factors of severe COVID-19. Holistic approach comprising nutrigenetics, micronutrient, status, lifestyle and clinical parameters could effectively combat micronutrient deficiency, and potentially improve anti-viral defense. This approach is especially important in populations at risk of micronutrient deficiency.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
This study was approved by the Ethics Committee of the Institute of Molecular Genetics and Genetic Engineering University of Belgrade (approval for sample collection and biobank formation O-EO-016/2020, 06.05.2020.; approval for the genetic study O-EO-016/2020/1, 03.09.2020).
NK: conceptualization, investigation, statistical analysis, writing - draft preparation, and editing. AS: methodology, investigation, and writing – draft preparation. KK: investigation, methodology, and writing – draft preparation. VG: data analysis and interpretation, investigation, and writing – draft preparation. BZ: methodology, investigation, writing — review, and editing, VS-T, MS, ZZ, OO, GS, and LL: methodology, sample collection, and clinical data analysis. SP: concept and design of the study, writing –review, and editing, BS: concept and design of the study, statistical analysis, results interpretation, drafting, and review of final manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Ministry of Education, Science and Technological Development, Republic of Serbia (Grant No. 451-03-9/2021-14/200042).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.689419/full#supplementary-material
1. Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. (2020) 584:430–6. doi: 10.1038/s41586-020-2521-4
2. Reddy RK, Charles WN, Sklavounos A, Dutt A, Seed PT, Khajuria A. The effect of smoking on COVID-19 severity: a systematic review and meta-analysis. J Med Virol. (2021) 93:1045–56. doi: 10.1002/jmv.26389
3. Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, et al. Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med. (2020) 383:1522–34. doi: 10.1056/NEJMoa2020283
4. Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, et al. Genetic mechanisms of critical illness in Covid-19. Nature. (2020) 591:92–8. doi: 10.1101/2020.09.24.20200048
5. Alexander J, Tinkov A, Strand TA, Alehagen U, Skalny A, Aaseth J. Early nutritional interventions with zinc, selenium and vitamin D for raising anti-viral resistance against progressive COVID-19. Nutrients. (2020) 12:1–12. doi: 10.3390/nu12082358
6. Teymoori-Rad M, Marashi SM. Vitamin D and Covid-19: from potential therapeutic effects to unanswered questions. Rev Med Virol. (2020) 31:e2159. doi: 10.1002/rmv.2159
7. Cui C, Xu P, Li G, Qiao Y, Han W, Geng C, et al. Vitamin D receptor activation regulates microglia polarization and oxidative stress in spontaneously hypertensive rats and angiotensin II-exposed microglial cells: role of renin-angiotensin system. Redox Biol. (2019) 26:101295. doi: 10.1016/j.redox.2019.101295
8. Xu J, Yang J, Chen J, Luo Q, Zhang Q, Zhang H. Vitamin D alleviates lipopolysaccharide-induced acute lung injury via regulation of the renin-angiotensin system. Mol Med Rep. (2017) 16:7432–8. doi: 10.3892/mmr.2017.7546
9. Iddir M, Brito A, Dingeo G, Del Campo SSF, Samouda H, La Frano MR, et al. Strengthening the immune system and reducing inflammation and oxidative stress through diet and nutrition: considerations during the covid-19 crisis. Nutrients. (2020) 12:1562. doi: 10.3390/nu12061562
10. Gröber U, Holick MF. The coronavirus disease (COVID-19) - a supportive approach with selected micronutrients. Int J Vitam Nutr Res. (2021). doi: 10.1024/0300-9831/a000693
11. Cashman KD, Dowling KG, Škrabáková Z, Gonzalez-Gross M, Valtueña J, De Henauw S, Moreno L, et al. Vitamin D deficiency in Europe: pandemic? Am J Clin Nutr. (2016) 103:1033–44. doi: 10.3945/ajcn.115.120873
12. Wessells KR, Brown KH. Estimating the global prevalence of zinc deficiency: results based on zinc availability in national food supplies and the prevalence of stunting. PLoS ONE. (2012) 7. doi: 10.1371/journal.pone.0050568
13. Im JH, Je YS, Baek J, Chung MH, Kwon HY, Lee JS. Nutritional status of patients with COVID-19. Int J Infect Dis. (2020) 100:390–3. doi: 10.1016/j.ijid.2020.08.018
14. Liu N, Sun J, Wang X, Zhang T, Zhao M, Li H. Low vitamin D status is associated with coronavirus disease 2019 outcomes: a systematic review and meta-analysis. Int J Infect Dis. (2021) 104:58–64. doi: 10.1016/j.ijid.2020.12.077
15. Jothimani D, Kailasam E, Danielraj S, Nallathambi B, Ramachandran H, Sekar P, et al. COVID-19: poor outcomes in patients with zinc deficiency. Int J Infect Dis. (2020) 100:343–9. doi: 10.1016/j.ijid.2020.09.014
16. Moghaddam A, Heller RA, Sun Q, Seelig J, Cherkezov A, Seibert L, et al. Selenium deficiency is associated with mortality risk from COVID-19. Nutrients. (2020) 12:1–13. doi: 10.3390/nu12072098
17. Shah K, Saxena D, Mavalankar D. Vitamin D supplementation, COVID-19 and disease severity: a meta-analysis. QJM An Int J Med. (2021). doi: 10.1093/qjmed/hcab009
18. Carlucci PM, Ahuja T, Petrilli C, Rajagopalan H, Jones S, Rahimian J. Zinc sulfate in combination with a zinc ionophore may improve outcomes in hospitalized COVID-19 patients. J Med Microbiol. (2020) 69:1228–34. doi: 10.1099/jmm.0.001250
19. Jiang X, O'Reilly PF, Aschard H, Hsu YH, Richards JB, Dupuis J, et al. Genome-wide association study in 79,366 European-ancestry individuals informs the genetic architecture of 25-hydroxyvitamin D levels. Nat Commun. (2018) 9:260–71. doi: 10.1038/s41467-017-02662-2
20. Moy KA, Mondul AM, Zhang H, Weinstein SJ, Wheeler W, Chung CC, et al. Genome-wide association study of circulating vitamin D-binding protein. Am J Clin Nutr. (2014) 99:1424–31. doi: 10.3945/ajcn.113.080309
21. Wang TJ, Zhang F, Richards JB, Kestenbaum B, Van Meurs JB, Berry D, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. (2010) 376:180–8. doi: 10.1016/S0140-6736(10)60588-0
22. Ramagopalan S V., Heger A, Berlanga AJ, Maugeri NJ, Lincoln MR, Burrell A, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. (2010) 20:1352–60. doi: 10.1101/gr.107920.110
23. Roth DE, Jones AB, Prosser C, Robinson JL, Vohra S. Vitamin D receptor polymorphisms and the risk of acute lower respiratory tract infection in early childhood. J Infect Dis. (2008) 197:676–80. doi: 10.1086/527488
24. Gallone G, Haerty W, Disanto G, Ramagopalan SV, Ponting CP, Berlanga-Taylor AJ. Identification of genetic variants affecting vitamin D receptor binding and associations with autoimmune disease. Hum Mol Genet. (2017) 26:2164–76. doi: 10.1093/hmg/ddx092
25. Tizaoui K, Kaabachi W, Hamzaoui A, Hamzaoui K. Association between Vitamin D receptor polymorphisms and multiple sclerosis: systematic review and meta-analysis of case-control studies. Cell Mol Immunol. (2015) 12:243–52. doi: 10.1038/cmi.2014.47
26. Bojović K, Stanković B, Kotur N, Krstić-Milošević D, Gašić V, Pavlović S, et al. Genetic predictors of celiac disease, lactose intolerance, and vitamin D function and presence of peptide morphins in urine of children with neurodevelopmental disorders. Nutr Neurosci. (2019) 22:40–50. doi: 10.1080/1028415X.2017.1352121
27. Evans DM, Zhu G, Dy V, Heath AC, Madden PAF, Kemp JP, et al. Genome-wide association study identifies loci affecting blood copper, selenium and zinc. Hum Mol Genet. (2013) 22:3998–4006. doi: 10.1093/hmg/ddt239
28. Cornelis MC, Fornage M, Foy M, Xun P, Gladyshev VN, Morris S, et al. Genome-wide association study of selenium concentrations. Hum Mol Genet. (2015) 24:1469–77. doi: 10.1093/hmg/ddu546
29. Batai K, Trejo MJ, Chen Y, Kohler LN, Lance P, Ellis NA, et al. Genome-wide association study of response to selenium supplementation and circulating selenium concentrations in adults of European descent. J Nutr. (2021) 151:293–302. doi: 10.1093/jn/nxaa355
30. Auton A, Abecasis GR, Altshuler DM, Durbin RM, Bentley DR, Chakravarti A, et al. A global reference for human genetic variation. Nature. (2015) 526:68–74. doi: 10.1038/nature15393
31. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. (2020) 581:434–43. doi: 10.1038/s41586-020-2308-7
32. Lips P, Cashman KD, Lamberg-Allardt C, Bischoff-Ferrari HA, Obermayer-Pietsch B, Bianchi ML, et al. Current Vitamin D status in European and middle east countries and strategies to prevent Vitamin D deficiency: a position statement of the European calcified tissue society. Eur J Endocrinol. (2019) 180:P23–54. doi: 10.1530/EJE-18-0736
33. Pavlovic Z, Miletic I, Zekovic M, Nikolic M, Glibetic M. Impact of selenium addition to animal feeds on human selenium status in Serbia. Nutrients. (2018) 10:225. doi: 10.3390/nu10020225
34. Prabhu AV, Luu W, Li D, Sharpe LJ, Brown AJ. DHCR7: a vital enzyme switch between cholesterol and vitamin D production. Prog Lipid Res. (2016) 64:138–51. doi: 10.1016/j.plipres.2016.09.003
35. Kuan V, Martineau AR, Griffiths CJ, Hyppönen E, Walton R. DHCR7 mutations linked to higher vitamin D status allowed early human migration to Northern latitudes. BMC Evol Biol. (2013) 13:144. doi: 10.1186/1471-2148-13-144
36. Xiao J, Li W, Zheng X, Qi L, Wang H, Zhang C, et al. Targeting 7-Dehydrocholesterol reductase integrates cholesterol metabolism and irf3 activation to eliminate infection. Immunity. (2020) 52:109–22.e6. doi: 10.1016/j.immuni.2019.11.015
37. Christakos S, Dhawan P, Verstuyf A, Verlinden L, Carmeliet G. Vitamin D: metabolism, molecular mechanism of action, and pleiotropic effects. Physiol Rev. (2015) 96:365–408. doi: 10.1152/physrev.00014.2015
38. Ahn J, Yu K, Stolzenberg-Solomon R, Claire Simon K, McCullough ML, Gallicchio L, et al. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. (2010) 19:2739–45. doi: 10.1093/hmg/ddq155
39. Ilie PC, Stefanescu S, Smith L. The role of vitamin D in the prevention of coronavirus disease 2019 infection and mortality. Aging Clin Exp Res. (2020) 32:1195–8. doi: 10.1007/s40520-020-01570-8
40. Helming L, Böse J, Ehrchen J, Schiebe S, Frahm T, Geffers R, et al. 1α,25-dihydroxyvitamin D3 is a potent suppressor of interferon γ-mediated macrophage activation. Blood. (2005) 106:4351–8. doi: 10.1182/blood-2005-03-1029
41. Galmés S, Serra F, Palou A. Current state of evidence: influence of nutritional and nutrigenetic factors on immunity in the COVID-19 pandemic framework. Nutrients. (2020) 12:1–33. doi: 10.3390/nu12092738
42. Karcioglu Batur L, Hekim N. The role of DBP gene polymorphisms in the prevalence of new coronavirus disease 2019 infection and mortality rate. J Med Virol. (2021) 93:1409–13. doi: 10.1002/jmv.26409
43. Freitas AT, Calhau C, Antunes G, Araújo B, Bandeira M, Barreira S, et al. Vitamin D-related polymorphisms and vitamin D levels as risk biomarkers of COVID-19 infection severity. Medrxiv. (2021). doi: 10.1101/2021.03.22.21254032
44. Butler-Laporte G, Nakanishi T, Mooser V, Morrison DR, Abdullah T, Adeleye O, et al. Vitamin D and Covid-19 susceptibility and severity: a mendelian randomization study. Medrxiv [preprint]. (2020). doi: 10.1101/2020.09.08.20190975
45. Amin HA, Drenos F. No evidence that vitamin D is able to prevent or affect the severity of COVID-19 in individuals with European ancestry: a mendelian randomisation study of open data. BMJ Nutr Prev Heal. (2021). doi: 10.1136/bmjnph-2020-000151
46. Donadio JLS, Rogero MM, Guerra-Shinohara EM, Barbosa F, Desmarchelier C, Borel P, et al. Genetic variants in selenoprotein genes modulate biomarkers of selenium status in response to Brazil nut supplementation (the SU.BRA.NUT study). Clin Nutr. (2019) 38:539–48. doi: 10.1016/j.clnu.2018.03.011
47. Combs GF, Jackson MI, Watts JC, Johnson LAK, Zeng H, Idso J, et al. Differential responses to selenomethionine supplementation by sex and genotype in healthy adults. Br J Nutr. (2012) 107:1514–25. doi: 10.1017/S0007114511004715
Keywords: DHCR7, CYP2R1, nutrigenetics, COVID-19, population genetics
Citation: Kotur N, Skakic A, Klaassen K, Gasic V, Zukic B, Skodric-Trifunovic V, Stjepanovic M, Zivkovic Z, Ostojic O, Stevanovic G, Lavadinovic L, Pavlovic S and Stankovic B (2021) Association of Vitamin D, Zinc and Selenium Related Genetic Variants With COVID-19 Disease Severity. Front. Nutr. 8:689419. doi: 10.3389/fnut.2021.689419
Received: 31 March 2021; Accepted: 11 May 2021;
Published: 04 June 2021.
Edited by:
Manja Zec, University of Arizona, United StatesReviewed by:
Matina Kouvari, Harokopio University, GreeceCopyright © 2021 Kotur, Skakic, Klaassen, Gasic, Zukic, Skodric-Trifunovic, Stjepanovic, Zivkovic, Ostojic, Stevanovic, Lavadinovic, Pavlovic and Stankovic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Biljana Stankovic, YmlsamFuYS5zdGFua292aWNAaW1nZ2UuYmcuYWMucnM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.