- 1College of Animal Science and Technology, Shandong Agricultural University, Taian, China
- 2National Engineering Research Center for Gelatin-Based Traditional Chinese Medicine, Dong-E E-Jiao Co., Ltd, Liaocheng, China
- 3Institute of Animal Husbandry and Veterinary Medicine, Beijing Academy of Agricultural and Forestry Sciences, Beijing, China
- 4Shandong Donkey Industry, Technology Collaborative Innovation Center, Liaocheng University, Liaocheng, China
- 5College of Animal Science and Technology, Qingdao Agricultural University, Qingdao, China
Previous studies have found donkey milk (DM) has the similar compositions with human milk (HM) and could be used as a potential hypoallergenic replacement diet for babies suffering from cow's milk allergy. Milk fat globule membrane (MFGM) proteins are involved in many biological functions, behaving as important indicators of the nutritional quality of milk. In this study, we used label-free proteomics to quantify the differentially expressed MFGM proteins (DEP) between DM (in 4–5 months of lactation) and HM (in 6–8 months of lactation). In total, 293 DEP were found in these two groups. Gene Ontology (GO) enrichment analysis revealed that the majority of DEP participated in regulation of immune system process, membrane invagination and lymphocyte activation. Several significant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were determined for the DEP, such as lysosome, galactose metabolism and peroxisome proliferator-activated receptor (PPAR) signaling pathway. Our study may provide valuable information in the composition of MFGM proteins in DM and HM, and expand our knowledge of different biological functions between DM and HM.
Introduction
Donkey milk is more similar to human milk because of its total protein and lactose contents, similar fatty acid and protein profiles (1). It has been indicated to be more suitable for children and elderly people due to its remarkable nutritional value and less allergenic. Besides, milk fat globules in DM are smaller and more easily digested and absorbed by infants (2). With its obvious advantages, the demand of direct consumption for DM has increased.
Milk fat globule membrane (MFGM), a three-layer membrane, covers on the surface of the milk fat globule (3). MFGM proteins make up only 1–2% of the total milk proteins, but they are thought to play important roles in biological processes, including cell growth promotion, cell activity regulation and defense mechanisms against bacteria and viruses in infants (4). Investigations on MFGM proteome have primarily focused on profiling analyses of MFGM fractions from different mammals. For example, Yang et al. identified 232 differentially expressed MFGM proteins in HM and CM across different lactation stages using the iTRAQ proteomic approach (5). To reveal the differences in the formation of MFGM in different mammals, the MFGM proteins of cow, yak, buffalo, goat, horse, camel and human were also compared by iTRAQ proteomics (6). Recently, Li et al. hoped to explore the changes in the regulation mechanism of different lactation stages by analyzing the differences of MFGM proteins between donkey colostrum and mature milk (2). However, studies of the DM MFGM proteome are relatively sparse and less comprehensive, especially fully comparative analyses of MFGM protein compositions and potential biological activities between DM and other species.
The aim of our study was to compare the expression of MFGM proteins between DM and HM by label-free quantification and to explore the biological processes they were involved in. The results are helpful for us to better understand the differences between DM and HM in the composition of MFGM and provide strong support for the future development of formula milk using donkey milk as nutritional provider.
Materials and Methods
Sample Collection and Treatment
HM samples were donated by twelve healthy mothers with the lactation of 6–8 months with written informed consent which indicated that the milk would be used in research. Twelve DM samples were obtained from a local farm breeding Dezhou donkeys (6–9 years old) with the lactation of 4–5 months in Liaocheng City of Shandong province, China. All procedures involving donkeys were performed by the Shandong Agricultural University Animal Care and Use Committee (approval number, SDAUA-2020-053). Four samples of each group were randomly mixed and stored at −80°C.
MFGM proteins separation was done as described by previous report (2). All samples were centrifuged at 4°C and 10,000 g for 15 min to obtain the upper fat portion containing MFGM. The upper fat layer was washed with 10 ml cold phosphate-buffered saline (0.24 g KH2PO4, 1.44 g Na2HPO4, 8 g NaCl and 0.2 g KCl were dissolved in 800 ml deionized water, the PH was adjusted to 7.4 and the volume was fixed to 1,000 mL) and homogenized by ultrasound (80 W, 15 s) for 3 times. The mixture was centrifuged at 4°C and 10,000 g for 1 h. Then, acetone was added to the collected supernatant to segregate the fat globules overnight. After centrifugation at 15,000 g at 4°C for 30 min, the supernatant was poured away. Finally, lysate was added to the protein particle sample and ultrasonicated for 15 s at 80 W for 10 cycles.
MFGM Protein Digestion
Thirty microliters protein solution were taken from each sample and dithiothreitol (DTT) were added to the final concentration of 100 mM. The samples were incubated in boiling water for 5 min and cooled to room temperature. Then, 200 μL UA buffer (8 M urea, 150 mM TrisHCl, pH 8.0) was added and mixed thoroughly. After that, each sample was transferred into a 10 kDa ultrafiltration centrifuge tube (Sartorius, Germany) and centrifuged at 14,000g for 15 min, then discarded the filtrate (repeat this step once). 100 μL IAA buffer (100 mM IAA in UA) was added to the protein mixture in the tube and shaken at 600 rpm for 1 min. The mixture was left at room temperature in dark for 30 min and centrifuged at 14,000 g for 15 min and the process was repeated twice. Hundred microliters 25mm NH4HCO3 solution was added to the tube and centrifuged at 14,000 g for 15min and repeated the procedure twice. Subsequently, 40 μL trypsin buffer (4 μg trypsin in 40 μL 100mM NH4HCO3) were added to the mixture and sample was shaken at 600 rpm for 1 min. The mixture was left at 37°C for 16–18 h and then transferred to a new collecting tube and centrifuged at 14,000 g for 15min. Then, 40 μL 25 mm NH4HCO3 was added in the mixture and sample was centrifuged at 14,000 g for 15 min to collect the filtrate. The peptide was desalted by C18 cartridge (Sigma, USA). After freeze-drying, the peptide was dissolved in 40 μL 0.1% formic acid solution.
LC- MS/MS Experiments
Each sample was separated by the Easy-nLCTM (Proxeon Biosystems, Thermo Fisher Scientific) orbitrap HPLC system with nano-flow. Buffer A was 0.1% formic acid, and buffer B was 84% acetonitrile in 0.1% formic acid. The chromatographic column was equilibrated with 95% buffer A. The sample was loaded by an automatic sampler to the sample column (Thermo Scientific Acclaim Pepmap 100, 100 μm × 2 cm, nanoViper C18), and separated by the analytical column (Thermo scientific EASY column, 10 cm, ID75 μm, 3 μm, C18-A2). The flow rate was 300 nL/min. Two-hour gradient: 0–110 min, buffer B linear gradient from 0 to 55%; 110–115 min, buffer B linear gradient from 55 to 100%; 115–120 min, buffer B maintained at 100%.
The samples were analyzed on a Q ExactiveTM mass spectrometer (Thermo Fisher Scientific). Parameters were set as follows: analysis time 120 min; detection mode: positive ion; scanning range of parent ion: 300–1,800 m/z; first-order mass spectrometry resolution: 70,000 at 200 m/z; AGC (automatic gain control) target: 1e6; maximum IT (inject time): 50 ms; dynamic exclusion: 60 s. The mass charge ratio of polypeptide and polypeptide fragments was collected according to the following methods: after each full scan, 20 fragments (MS2 scan) were collected. The MS2 activation type was HCD, the isolation width was 2 m/z, the secondary mass spectrum resolution was 17,500 at 200 m/z. Normalized collision energy was 30 eV, and the underfill ratio was defined as 0.1%.
Sequence Database Searching
Relative intensity-based label-free quantification (LFQ) was analyzed by MaxQuant version 1.5.3.17 (Max Planck Institute of Biochemistry in Martinsried, Germany) and searched against the UniProtKB Equus asinus database (47,825 total entries, downloaded on August 12, 2019) and Homo sapiens (included 20,422 series, downloaded on May 22, 2019). Proteins with p < 0.05 and fold change >2 or <0.5 were deemed significantly expressed between groups by t-test. The false discovery rate (FDR) for peptide and protein identification was set to 1%.
Bioinformatics Analysis
Quantified MFGM samples were used to performing hierarchical clustering analysis. For this purpose, Cluster3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) and the Java Treeview software (http://jtreeview.sourceforge.net) were used. GO enrichment on three ontologies (biological process, molecular function, and cellular component) and KEGG pathway enrichment analyses were applied based on the Fisher' exact test, considering the whole quantified protein annotations as background dataset using DAVID (https://david.ncifcrf.gov/). Benjamini-Hochberg correction for multiple testing was further applied to adjust derived P-values, and only functional categories and pathways with P-values under a threshold of 0.05 were considered as significant.
Results
Quantitative Overview of Identified MFGM Proteins in DM and HM
As shown in Figures 1A, 454 MFGM proteins were identified in DM and HM. Proteins having at least two replicates and fold changes >2.0 or <0.5 and P < 0.05 were defined as differentially expressed proteins (DEP). In addition, the proteins with at least two replicates in one group and in another group with null values were also defined as DEP. A volcano plot was used to show significant differences between the two groups based on the fold change and P-value (Figure 1B). Compared with MFGM proteins in HM, 204 MFGM proteins in DM were upregulated and 89 MFGM proteins were downregulated. DEP detailed information is shown in Supplementary Table 1.
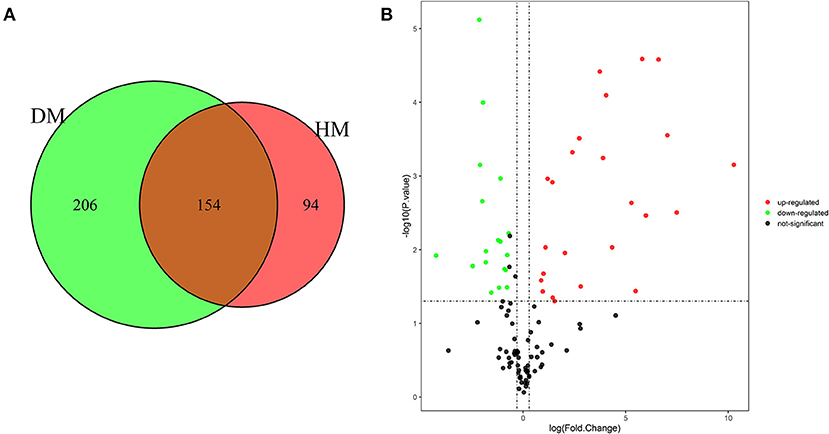
Figure 1. Venn diagram of MFGM proteins identified from DM and HM group (A); Volcano plot of proteins identified from DM and HM group (B). Red dot, up-regulated proteins in DM and HM MFGM; Green dot, up-regulated proteins in DM and HM MFGM; Black dot, not significant different proteins between DM and HM. MFGM, milk fat ball membrane; DM, donkey milk; HM, human milk.
Cluster Analysis
Meanwhile, a hierarchical cluster analysis of MEGM proteins in DM and HM group was shown in Figure 2. The upregulated MFGM proteins in DM compared with HM mainly included alpha-s2-casein, lysozyme, alpha-s1-casein and secreted phosphoprotein 1 (SPP1), and the downregulated MFGM proteins were lipoprotein lipase, fatty acid-binding protein 3 and nucleobindin 1.
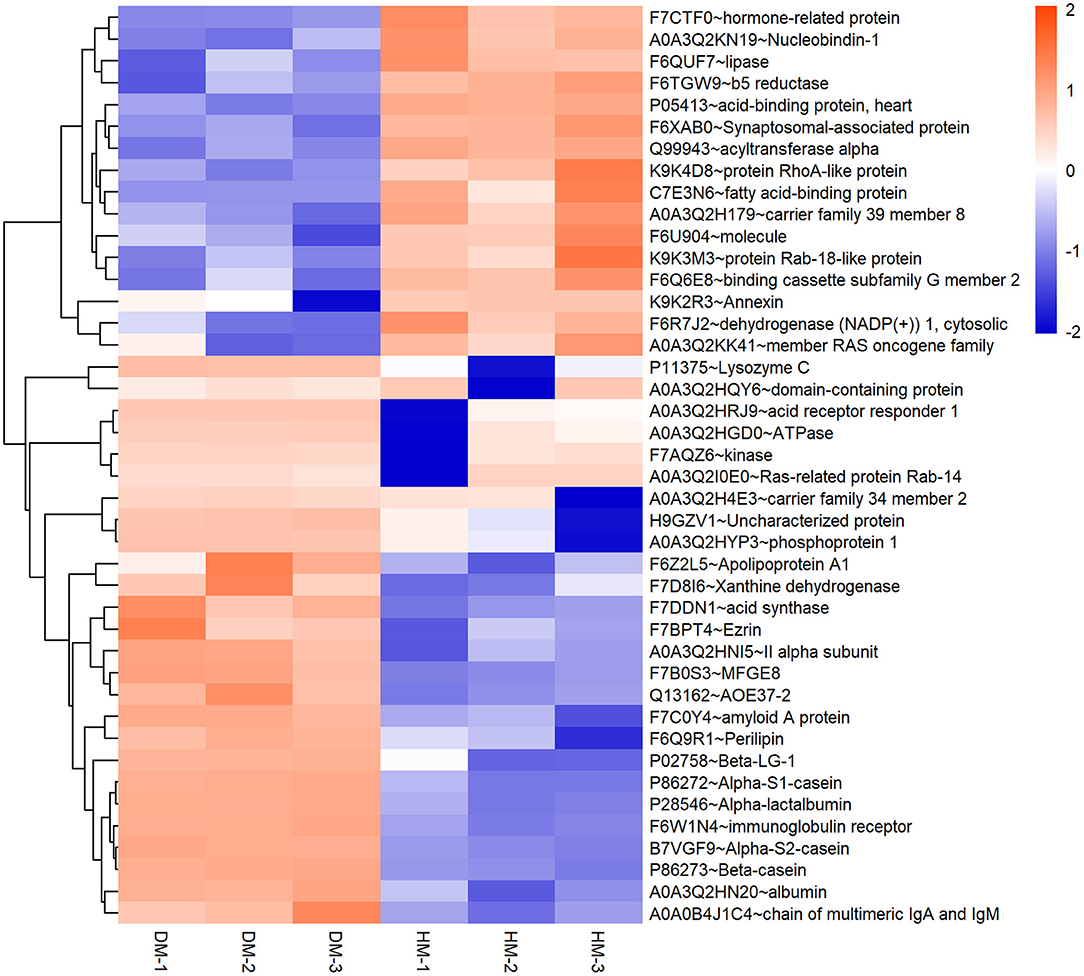
Figure 2. Hierarchical clustering of DEP in DM and HM group. Bar color represents a logarithmic scale from −2 to 2. DEP, differentially expressed proteins; DM, donkey milk. HM,: human milk.
GO Analysis of DEP in DM Compared With HM and CM
DEP were then classified into the GO enrichment analysis of three distinctive functional sets, cellular component, molecular function, and biological process. In terms of molecular function, DEP in DM and HM group were primarily related to immunoglobulin receptor binding, protein homodimerization activity and antigen binding (Figure 3). In the category of biological process, DEP in these two groups were mainly involved in regulation of immune system process, membrane invagination and lymphocyte activation. GO enrichment of DEP information is listed in Supplementary Table 2.
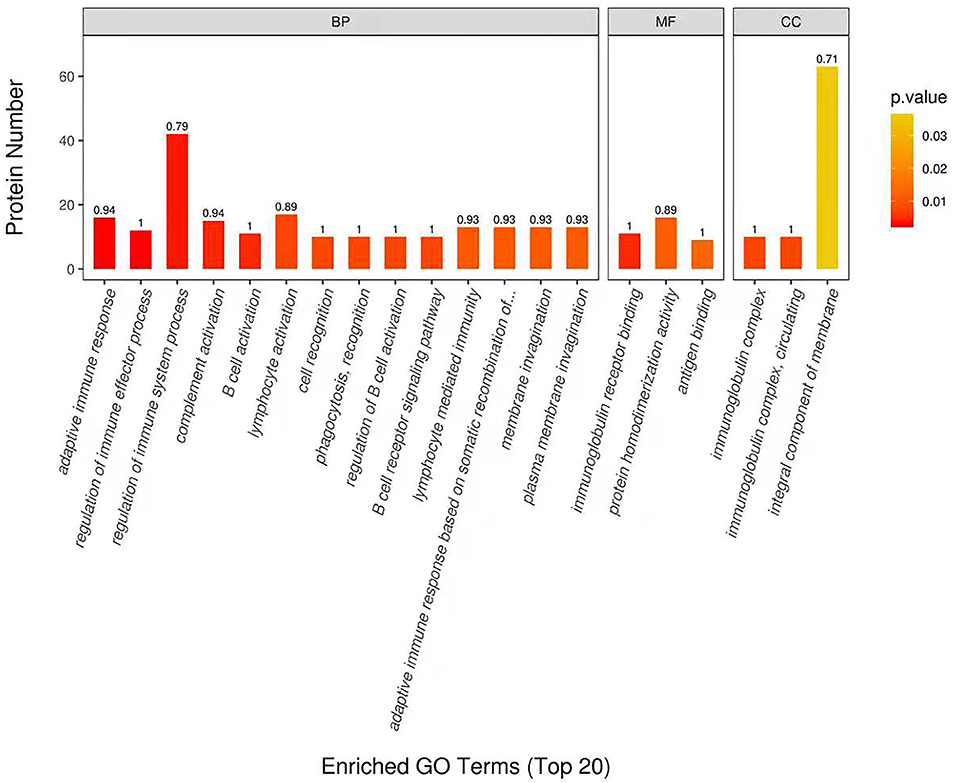
Figure 3. Enriched Gene Ontology (GO) Terms of the DEP in DM and HM group. GO enrichment of DEP on three categories. BP, biological processes; MF, molecular functions; CC, cellular components; DEP, differentially expressed proteins; DM, donkey milk; HM, human milk.
KEGG Pathway Analysis of DEP in DM Compared With HM and CM
In Figure 4, the DEP were mainly involved in lysosome, galactose metabolism and PPAR signaling pathway. KEGG pathway enrichment of DEP information was listed in Table 1.
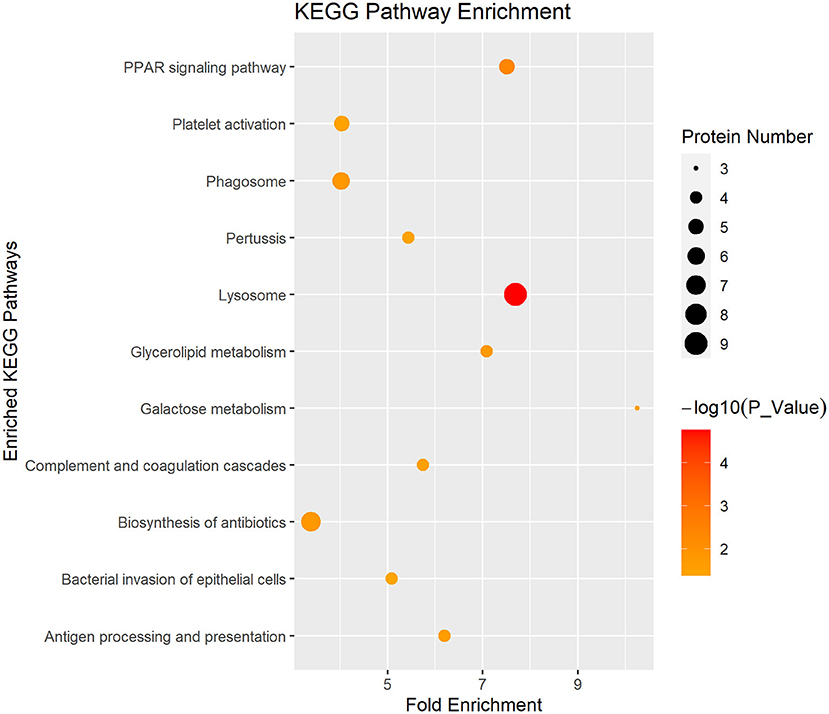
Figure 4. Enriched KEGG pathway analysis of the DEP in DM and HM group. KEGG, Kyoto Encyclopedia of Genes and Genomes pathways; DEP, differentially expressed proteins; DM, donkey milk; HM, human milk.
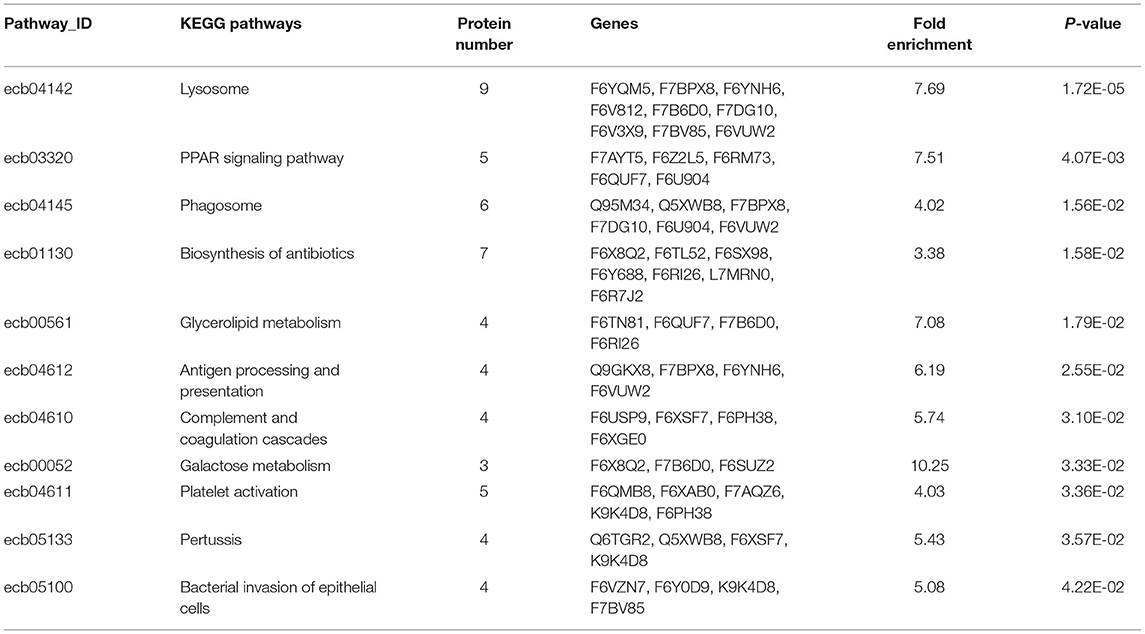
Table 1. Detailed information of KEGG pathway-based enrichment of the differentially expressed proteins between DM and HM.
Discussion
In this study, 293 MFGM proteins were found to be significantly different between DM and HM. These DEP were involved in regulation of immune system process, complement activation and integral component of membrane. These results may provide valuable information in the MFGM composition of DM and HM, especially for low abundant components, and expand our knowledge of different biological functions between DM and HM.
Caseins in milk exert multifunctional effects including amino acid and calcium supply (5), cellular immune functions stimulation (7, 8), and chemotactic properties (9). Studies of human, bovine, goat, and camel MFGM proteomes identified milk caseins in the MFGM fractions (2, 6, 10). In our data, four caseins (α-s1-casein, α-s2-casein, β-casein, κ-casein) were detected in DM MFGM fractions and significantly higher than that in HM. The main role of human α-s1-casein is to serve as an amino acid source to the newborns. Beyond nutritional aspects, α-s1-casein could contribute to the development of immune system. Moreover, α-s1-casein in nursed individuals gives rise to sustained specific IgG production (8). Cocco et al. found that single amino acid substitution in the specific linear epitopes of alpha s1-casein can significantly reduce the binding ability of serum IgE in patients with CM allergy (11). Donkey and cow milk alpha s1-casein share a low sequence homology, and particularly their IgE binding linear epitopes have remarkable differences in amino acid sequences (12). Moreover, Bertino et al. found that donkey alpha-s1-casein appears as both phosphorylated and glycosylated forms, but neither human nor bovine alpha-s1-caseins have been reported to be glycosylated (13). Studies have shown that in many cases milk from donkey represents a safe and alternative food in both IgE-mediated and non-IgE-mediated cow's milk protein allergy (14, 15). These differences in alpha s1-casein amino acid sequence or post-translational modifications might be related to the low allergenic properties of donkey milk. As for β-casein and kappa-casein, both of them from HM and DM are more closely related to each other than the cow counterparts (16). β-casein is the major casein constituent in human MFGM and generates smaller casein phosphopeptides upon digestion to aid the absorption of calcium (17). κ-casein is the only glycosylated in the four casein families. DM κ-casein carries a higher number of potential O-glycosylation than that cow milk κ-casein. Therefore, DM similar to HM may contribute to inhibit the adhesion of Helicobacter pylori to gastric mucosa in infant (18).
In general, β-lactoglobulin can be found in the majority of milks, but not in human's milk. However, in this study we detected β-lactoglobulin in HM. It has been found that beta-lactoglobulin can be detected in the human milk within 7 days after ingestion of milk (19). We hypothesized that the mothers who donated breast milk in this study were likely to have taken other milks before providing milk samples.
According to the GO enrichment analysis, DEP in DM vs. HM group were mainly involved immune response, such as complement activation, defense response or positive regulation of B cell activation (Supplementary Table 2). It is well-known that milk could provide large amounts of bioactive components to the infants in the critical phase of immunological immaturity. Mature breast milk could enhance B cell proliferation and antibody secretion (20). Among the DEP involved in the positive regulation of B cell activation, 9 proteins were all upregulated in DM, including immunoglobulin heavy constant gamma 3 (IGHG3) and immunoglobulin kappa constant (IGKC), which all function in B cell selection or antigen recognition (21). In addition, 49 DEP were involved in defense response, among which 32 proteins were upregulated in the DM MFGM, namely semaphorin 7A, complement 3 (C3), joining (J) chain. Semaphorin 7A (also known as CD108) plays a key role in innate immune regulation. Semaphorin 7A could induce proinflammatory cytokines production (22). Semaphorin 7A-deficient mice are defective in T cell-mediated inflammatory responses, indicating the role of semaphorin 7A in evoking inflammatory immune reactions (23). In addition to its role in the immune response, semaphorin 7A also functions as a chemoattractant and stimulates neuronal migration, which is an essential process in central nervous system development (24). The defect of neuron migration may lead to nervous system disorder (25). J chain is a small polypeptide contained in dimeric IgA and pentameric IgM, which plays an important role in the generation of secretory antibodies (26). This peptide can be produced by immunocytes of all Ig isotypes, but it becomes incorporated only into IgA and pentameric IgM (27). Moreover, the expression of J chain may be a marker of B cell clone in mucosa associated lymphoid tissue, as there is a positive correlation between the production of polymeric IgA, IgG or IgD-producing cells and J chain (28). C3 is an important part of the innate immune system. It combines with other complement proteins to form the main host mechanism for detecting and eliminating potential pathogens (29). Complement proteins contribute to the establishment of natural immune system in newborns (2). The presence of abundant immunological factors in DM MFGM proteins are helpful for the newborns to establish an immune system against microbial infection to adapt to the new environment to prevent diseases.
In this study, a solute carrier (SLC) superfamily, namely, SLC34A2, SLC36A1, SLC4A9, and SLC9A3R1, was more abundant in DM MFGM proteins. This superfamily is a major membrane transporter group that controls the uptake and excretion of nutrients, neurotransmitters, metabolites, drugs and toxins (30). SLC34A2 is a member of SLC34 family, a group of phosphate transporters, which are responsible for transporting inorganic phosphate. Phosphate is an essential nutrient for life and a key component of bone formation (31). Recently, it was concluded that SLC34A2 was responsible for the sodium-dependent component of intestinal phosphate absorption (32). SLC36A1, an amino acids transporter in small intestinal enterocytes, could regulate cell growth and sense the availability of amino acids in other cell types. SLC36A1 also could be a target for rapamycin complex 1 (TORC1) activation (33). TORC1 regulates some metabolic pathways and adapts cells to applied bioenergy and anabolic conditions (34). SLC9A3R1 is a multifunctional scaffold protein, which is involved in cell activities and affects many protein functions, including ion channels, receptors, signaling and nuclear proteins. In addition, SLC9A3R1 has potential antitumor effects in breast cancer (35). SLCs also play an important role in the function of the central nervous system. A total of 287 SLC genes were identified in the brain, especially in the barrier cells. SLCs expressed in neurons and glial cells play irreplaceable roles in maintaining brain homeostasis (36). Moreover, we also found some other abundant proteins in DM MFGM that are expressed in the brain, such as neuro plastin (NP). NP is a cell adhesion molecule rich in synaptic membranes and belongs to immunoglobulin superfamily. Np plays a role in synaptic plasticity and neurite growth (37, 38). In recent years, NP has attracted much attention because of its correlation with adolescent cortical thickness and intelligence (39). Therefore, the abovementioned functional components in DM MFGM may play a more important role in promoting the growth and learning of newborn infants.
Among these DEP, a family of apolipoproteins (Apos), namely, apolipoprotein A1 (ApoA1), ApoA2, ApoC3, ApoD, and ApoE, were significantly upregulated in DM MFGM. ApoA1 and ApoA2 are the major proteins in high-density lipoproteins (HDL) (40). ApoA1 has multiple beneficial functions, including potent antioxidant, anti-inflammatory, antiviral and antibacterial activities in blood (41, 42). Recently, ApoA1 was found in HM and DM. Kim et al. identified that ApoA1 interacts with cholesterol in HM, provides antioxidant activity and improves embryo survivability (43). In DM MFGM, ApoA1 was upregulated in colostrum compared with mature milk (2). ApoD is a glycosylated protein involved in lipid transport, food intake, inflammation, antioxidant response and development. In humans, ApoD levels rise considerably in association with aging possibly in response to accumulated damage (44). Overexpression of human ApoD could protect drosophila against various extrinsic stresses and extend its normal lifespan (45). ApoE is an important element in the lipoprotein metabolism and cholesterol transport (46). Cholesterol plays a key role in vitamin D and steroid hormones synthesis, which is critical to the development of the newborns (47). In our study, DM MFGM provides a higher level of cholesterol transporters, which may help the newborns acquire a large amount of cholesterol.
In conclusion, 204 up-regulated proteins were identified in the lipid globules of donkey milk. Through GO functional annotation and KEGG pathway enrichment analysis, we found that these upregulated proteins not only have nutritional effects, but also promote the improvement of the immune system, cognitive learning and anti-oxidation of newborns. Therefore, donkey milk might be used as a nutritional provider in infant formula. In addition, the comparison of the fat globular membrane protein between donkey milk and cow milk was missing in this study, which was a limitation of our research and will be carried out in the future.
Conclusion
A quantitative proteomic method was used to investigate the MFGM proteins proteome in DM and HM. DEP were analyzed by multivariate statistical methods and found mainly involved in regulation of immune system process, membrane invagination and lymphocyte activation. Our findings also provided more in-depth reference for the dairy food industry and infant health.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Animal Care and Use Committee of Shangdong Agricultural University. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Shangdong Agricultural University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
FZ designed the experiment and revised the manuscript. XZ completed the experiment and drafted the manuscript. BJ and CJ participated in improving the experiment. HL, LY, GJ, YW, GuaL, and GuiL participated in collecting and preparing the samples. LM helped to revise the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by The Open Project of Shandong Collaborative Innovation Center for Donkey Industry Technology (No. 3193308) and Agriculture Improved Varieties Project of Shandong Province, China (2017LZGC020). Beijing Agricultural Forestry Academy Youth Fund (No. QNJJ201930).
Conflict of Interest
XZ, CJ, HL, LY, GJ, YW, GuaL, and FZ were employed by the company Dong-E E-Jiao Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.670099/full#supplementary-material
References
1. Medhammar E, Wijesinha-Bettoni R, Stadlmayr B, Nilsson E, Charrondiere UR, Burlingame B. Composition of milk from minor dairy animals and buffalo breeds: a biodiversity perspective. J Sci Food Agric. (2012) 92:445–74. doi: 10.1002/jsfa.4690
2. Li W, Li M, Cao X, Yang M, Han H, Kong F, et al. Quantitative proteomic analysis of milk fat globule membrane (MFGM) proteins from donkey colostrum and mature milk. Food Funct. (2019) 10:4256–68. doi: 10.1039/C9FO00386J
3. Argov N, Lemay DG, German JB. Milk fat globule structure & function; nanosciece comes to milk production. Trends Food Sci Technol. (2008) 19:617–23. doi: 10.1016/j.tifs.2008.07.006
4. Reinhardt TA, Lippolis JD. Bovine milk fat globule membrane proteome. J Dairy Res. (2006) 73:406–16. doi: 10.1017/S0022029906001889
5. Yang M, Cong M, Peng X, Wu J, Wu R, Liu B, et al. Quantitative proteomic analysis of milk fat globule membrane (MFGM) proteins in human and bovine colostrum and mature milk samples through iTRAQ labeling. Food Funct. (2016) 7:2438–50. doi: 10.1039/C6FO00083E
6. Yang Y, Zheng N, Zhao X, Zhang Y, Han R, Ma L, et al. Proteomic characterization and comparison of mammalian milk fat globule proteomes by iTRAQ analysis. J Proteomics. (2015) 116:34–43. doi: 10.1016/j.jprot.2014.12.017
7. Wada Y, Lonnerdal B. Bioactive peptides derived from human milk proteins–mechanisms of action. J Nutr Biochem. (2014) 25:503–14. doi: 10.1016/j.jnutbio.2013.10.012
8. Vordenbaumen S, Saenger T, Braukmann A, Tahan T, Bleck E, Jose J, et al. Human casein alpha s1 induces proinflammatory cytokine expression in monocytic cells by TLR4 signaling. Mol Nutr Food Res. (2016) 60:1079–89. doi: 10.1002/mnfr.201500792
9. Vordenbaumen S, Braukmann A, Altendorfer I, Bleck E, Jose J, Schneider M. Human casein alpha s1 (CSN1S1) skews in vitro differentiation of monocytes towards macrophages. BMC Immunol. (2013) 14:46. doi: 10.1186/1471-2172-14-46
10. Saadaoui B, Henry C, Khorchani T, Mars M, Martin P, Cebo C. Proteomics of the milk fat globule membrane from Camelus dromedarius. Proteomics. (2013) 13:1180–4. doi: 10.1002/pmic.201200113
11. Cocco RR, Jarvinen KM, Sampson HA, Beyer K. Mutational analysis of major, sequential IgE-binding epitopes in alpha s1-casein, a major cow's milk allergen. J Allergy Clin Immunol. (2003) 112:433–7. doi: 10.1067/mai.2003.1617
12. Cunsolo V, Cairone E, Fontanini D, Criscione A, Muccilli V, Saletti R, et al. Sequence determination of alphas1-casein isoforms from donkey by mass spectrometric methods. J Mass Spectrom. (2009) 44:1742–53. doi: 10.1002/jms.1683
13. Bertino E, Gastaldi D, Monti G, Baro C, Fortunato D, Perono Garoffo L, et al. Detailed proteomic analysis on DM: insight into its hypoallergenicity. Front Biosci. (2010) 2:526–36. doi: 10.2741/e111
14. Carroccio A, Cavataio F, Montalto G, D'Amico D, Alabrese L, Iacono G. Intolerance to hydrolysed cow's milk proteins in infants: clinical characteristics and dietary treatment. Clin Exp Allergy. (2000) 30:1597–603. doi: 10.1046/j.1365-2222.2000.00925.x
15. Monti G, Bertino E, Muratore Cristina M, Coscia A, Cresi F, Silvestro L, et al. Efficacy of donkey's milk in treating highly problematic cow's milk allergic children: an in vivo and in vitro study. Pediatr Allergy Immunol. (2008) 19:90–1. doi: 10.1111/j.1399-3038.2007.00655.x
16. Cunsolo V, Saletti R, Muccilli V, Gallina S, Di Francesco A, Foti S. Proteins and bioactive peptides from donkey milk: the molecular basis for its reduced allergenic properties. Food Res Int. (2017) 99(Pt. 1):41–57. doi: 10.1016/j.foodres.2017.07.002
17. Lonnerdal B. Bioactive proteins in breast milk. J Paediatr Child Health. (2013) 49(Suppl. 1):1–7. doi: 10.1111/jpc.12104
18. Spada V, Ferranti P, Chianese L, Salimei E, Addeo F, Picariello G. Antibacterial potential of donkey's milk disclosed by untargeted proteomics. J Proteomics. (2021) 231:104007. doi: 10.1016/j.jprot.2020.104007
19. Matangkasombut P, Padungpak S, Thaloengsok S, Kamchaisatian W, Sasisakulporn C, Jotikasthira W, et al. Detection of beta-lactoglobulin in human breast-milk 7 days after cow milk ingestion. Paediatr Int Child Health. (2017) 37:199–203. doi: 10.1080/20469047.2017.1289310
20. Juto P. Human milk stimulates B cell function. Arch Dis Child. (1985) 60:610–3. doi: 10.1136/adc.60.7.610
21. Steck AJ, Kinter J, Renaud S. Differential gene expression in nerve biopsies of inflammatory neuropathies. J Peripher Nerv Syst. (2011) 16 Suppl 1:30–3. doi: 10.1111/j.1529-8027.2011.00302.x
22. Kang HR, Lee CG, Homer RJ, Elias JA. Semaphorin 7A plays a critical role in TGF-beta1-induced pulmonary fibrosis. J Exp Med. (2007) 204:1083–93. doi: 10.1084/jem.20061273
23. Suzuki K, Okuno T, Yamamoto M, Pasterkamp RJ, Takegahara N, Takamatsu H, et al. Semaphorin 7A initiates T-cell-mediated inflammatory responses through alpha1beta1 integrin. Nature. (2007) 446:680–4. doi: 10.1038/nature05652
24. Iragavarapu-Charyulu V, Wojcikiewicz E, Urdaneta A. Semaphorins in angiogenesis and autoimmune diseases: therapeutic targets? Front Immunol. (2020) 11:346. doi: 10.3389/fimmu.2020.00346
25. Rahimi-Balaei M, Bergen H, Kong J, Marzban H. Neuronal migration during development of the cerebellum. Front Cell Neurosci. (2018) 12:484. doi: 10.3389/fncel.2018.00484
26. Johansen FE, Braathen R, Brandtzaeg P. Role of J chain in secretory immunoglobulin formation. Scand J Immunol. (2000) 52:240–8. doi: 10.1046/j.1365-3083.2000.00790.x
27. Brandtzaeg P, Baekkevold ES, Farstad IN, Jahnsen FL, Johansen FE, Nilsen EM, et al. Regional specialization in the mucosal immune system: what happens in the microcompartments? Immunol Today. (1999) 20:141–51. doi: 10.1016/S0167-5699(98)01413-3
28. Erlandsson L, Akerblad P, Vingsbo-Lundberg C, Kallberg E, Lycke N, Leanderson T. Joining chain-expressing and -nonexpressing B cell populations in the mouse. J Exp Med. (2001) 194:557–70. doi: 10.1084/jem.194.5.557
29. Delanghe JR, Speeckaert R, Speeckaert MM. Complement C3 and its polymorphism: biological and clinical consequences. Pathology. (2014) 46:1–10. doi: 10.1097/PAT.0000000000000042
30. Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, Bruford EA. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteinsIntroduction. Pflugers Arch. (2004) 447:465–8. doi: 10.1007/s00424-003-1192-y
31. Hernando N, Gagnon K, Lederer E. Phosphate transport in epithelial and nonepithelial tissue. Physiol Rev. (2021) 101:1–35. doi: 10.1152/physrev.00008.2019
32. Marks J. The role of SLC34A2 in intestinal phosphate absorption and phosphate homeostasis. Pflugers Arch. (2019) 471:165–73. doi: 10.1007/s00424-018-2221-1
33. Jensen A, Figueiredo-Larsen M, Holm R, Broberg ML, Brodin B, Nielsen CU. PAT1 (SLC36A1) shows nuclear localization and affects growth of smooth muscle cells from rats. Am J Physiol Endocrinol Metab. (2014) 306:E65–74. doi: 10.1152/ajpendo.00322.2013
34. Proud CG. mTOR signalling in health and disease. Biochem Soc Trans. (2011) 39:431–6. doi: 10.1042/BST0390431
35. Liu H, Ma Y, He HW, Wang JP, Jiang JD, Shao RG. SLC9A3R1 stimulates autophagy via BECN1 stabilization in breast cancer cells. Autophagy. (2015) 11:2323–34. doi: 10.1080/15548627.2015.1074372
36. Hu C, Tao L, Cao X, Chen L. The solute carrier transporters and the brain: physiological and pharmacological implications. Asian J Pharm Sci. (2020) 15:131–44. doi: 10.1016/j.ajps.2019.09.002
37. Owczarek S, Soroka V, Kiryushko D, Larsen MH, Yuan Q, Sandi C, et al. Neuroplastin-65 and a mimetic peptide derived from its homophilic binding site modulate neuritogenesis and neuronal plasticity. J Neurochem. (2011) 117:984–94. doi: 10.1111/j.1471-4159.2011.07269.x
38. Owczarek S, Kiryushko D, Larsen MH, Kastrup JS, Gajhede M, Sandi C, et al. Neuroplastin-55 binds to and signals through the fibroblast growth factor receptor. FASEB J. (2010) 24:1139–50. doi: 10.1096/fj.09-140509
39. Desrivieres S, Lourdusamy A, Tao C, Toro R, Jia T, Loth E, et al. Single nucleotide polymorphism in the neuroplastin locus associates with cortical thickness and intellectual ability in adolescents. Mol Psychiatry. (2015) 20:263–74. doi: 10.1038/mp.2013.197
40. Mineo C, Shaul PW. HDL stimulation of endothelial nitric oxide synthase: a novel mechanism of HDL action. Trends Cardiovasc Med. (2003) 13:226–31. doi: 10.1016/S1050-1738(03)00098-7
41. Biedzka-Sarek M, Metso J, Kateifides A, Meri T, Jokiranta TS, Muszynski A, et al. Apolipoprotein A-I exerts bactericidal activity against Yersinia enterocolitica serotype O:3. J Biol Chem. (2011) 286:38211–9. doi: 10.1074/jbc.M111.249482
42. Suzuki M, Pritchard DK, Becker L, Hoofnagle AN, Tanimura N, Bammler TK, et al. High-density lipoprotein suppresses the type I interferon response, a family of potent antiviral immunoregulators, in macrophages challenged with lipopolysaccharide. Circulation. (2010) 122:1919–27. doi: 10.1161/CIRCULATIONAHA.110.961193
43. Kim SM, Kim SJ, Kim JY, Kim JR, Cho KH. Breast milk from smokers contains less cholesterol and protein and smaller size of apolipoprotein A-I resulting in lower zebrafish embryo survivability. Breastfeed Med. (2017) 12:365–72. doi: 10.1089/bfm.2016.0097
44. Kalman J, McConathy W, Araoz C, Kasa P, Lacko AG. Apolipoprotein D in the aging brain and in Alzheimer's dementia. Neurol Res. (2000) 22:330–6. doi: 10.1080/01616412.2000.11740678
45. Muffat J, Walker DW, Benzer S. Human ApoD, an apolipoprotein up-regulated in neurodegenerative diseases, extends lifespan and increases stress resistance in Drosophila. Proc Natl Acad Sci USA. (2008) 105:7088–93. doi: 10.1073/pnas.0800896105
46. Phie J, Krishna SM, Moxon JV, Omer SM, Kinobe R, Golledge J. Flavonols reduce aortic atherosclerosis lesion area in apolipoprotein E deficient mice: a systematic review and meta-analysis. PLoS ONE. (2017) 12:e0181832. doi: 10.1371/journal.pone.0181832
Keywords: human, donkey, quantitative proteomic, comparison, milk fat globule membrane
Citation: Zhang X, Jiang B, Ji C, Li H, Yang L, Jiang G, Wang Y, Liu G, Liu G, Min L and Zhao F (2021) Quantitative Label-Free Proteomic Analysis of Milk Fat Globule Membrane in Donkey and Human Milk. Front. Nutr. 8:670099. doi: 10.3389/fnut.2021.670099
Received: 20 February 2021; Accepted: 21 May 2021;
Published: 22 June 2021.
Edited by:
Aida Serra, IMDEA Food Institute, SpainReviewed by:
Tânia Rodrigues Dias, University of Aveiro, PortugalJoana Costa, University of Porto, Portugal
Céline Henry, INRAE Centre Jouy-en-Josas, France
Copyright © 2021 Zhang, Jiang, Ji, Li, Yang, Jiang, Wang, Liu, Liu, Min and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuwei Zhao, ODE5ODk5MTY0QHFxLmNvbQ==; Lingjiang Min, bWxqMjk2M0AxMjYuY29t
†These authors have contributed equally to this work
 Xinhao Zhang1,2†
Xinhao Zhang1,2† Bo Jiang
Bo Jiang Fuwei Zhao
Fuwei Zhao