
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr. , 20 July 2021
Sec. Nutrition and Microbes
Volume 8 - 2021 | https://doi.org/10.3389/fnut.2021.646819
Inflammatory bowel disease (IBD) is a serious hazard to public health, but the precise etiology of the disease is unclear. High intake of red meat diet is closely related to the occurrence of IBD. In this study, we investigated whether the high intake of red meat can increase the sensitivity of colitis and the underlying mechanism. Mice were fed with different levels of red meat for 8 weeks and then the colonic contents were analyzed by 16S rRNA sequencing. Then 3% dextran sulfate sodium was used to induce colitis in mice. We observed the severity of colitis and inflammatory cytokines. We found that high-dose red meat caused intestinal microbiota disorder, reduced the relative abundance of Lachnospiraceae_NK4A136_group, Faecalibaculum, Blautia and Dubosiella, and increased the relative abundance of Bacteroides and Alistipes. This in turn leads to an increase in colitis and inflammatory cytokine secretion. Moreover, we found that high red meat intake impaired the colon barrier integrity and decreased the expression of ZO-1, claudin, and occludin. We also found high red meat intake induced the production of more inflammatory cytokines such as IL-1β, TNF-α, IL-17, and IL-6 and inflammatory inducible enzymes such as COX-2 and iNOS in dextran sulfate sodium-induced colitis. These results suggest that we should optimize the diet and reduce the intake of red meat to prevent the occurrence of IBD.
The gut microbiota is an indispensable part of the human body. It evolves with the host and acts as a natural barrier to help the body digest food, absorb nutrients and maintain health (1). After cessation of breastfeeding during childhood, the gut microbiota begins to stabilize and become a stable system. Studies have clarified the importance of relationships between microbiota and host in early life, and these interactions have been further confirmed in later specific disease states (2), such as diabetes, asthma (3) and colitis (4). Intestinal microbiota imbalance can affect host metabolism, immunity and health, and induce many diseases such as inflammatory bowel disease (IBD), colorectal cancer (5), obesity (6), type 1 and type 2 diabetes (6), and weakened immunity among the elderly.
Crohn's disease and ulcerative colitis are the most common subtypes of inflammatory bowel disease and can be differentiated based on their clinical presentation (including disease location and symptoms) as well as their histopathology. The incidence of IBD is increasing worldwide, but the exact cause of the disease remains unclear. Studies have confirmed that excessive intake of red meat is associated with an increased incidence of IBD (7–9). A cohort study of 67,511 people in France showed that high-protein, high-fat diets of red meat were significantly associated with the onset of IBD (8). Another nutritional study suggests that high meat intake, especially red meat, increases the risk of UC and CD (9). In a prospective study on cancer and nutrition in Europe, red meat, margarine, sunflower oil and corn oil were found to contain large amounts of omega-6 unsaturated fatty acids, while omega-6 unsaturated fatty acids have been shown to increase UC risk (10). In another prospective cohort study, red meat and processed meat were associated with a risk of UC recurrence (11). We also found the incidence of IBD was higher than other places in Xinjiang of China, which was believed to be related with their custom of daily high red meat intake for a long time.
Red meat mainly contains protein, fat and heme. In general, high protein intake causes disturbances in the gut microbiota, which in turn leads to changes in metabolites such as branched-chain fatty acids, amino acids and hydrogen sulfide. These all affect the viability and proliferation of epithelial cells, intestinal barrier function and immune response. In addition, high fat intake also changes the intestinal microbiota, thereby increasing intestinal mucosal permeability, reducing the expression of tight junction proteins in the intestinal epithelium, and exacerbating intestinal inflammation. Heme can mediate inflammatory stress responses in Gram-negative bacteria (12). Constante M et al. reported dietary heme can affect the gut microbial community structure and contribute to microbiota dysbiosis (13). Therefore, we hypothesize that high intake of red meat can cause damage to the gut microbiota, which in turn exacerbates inflammation.
In this study, we assessed the effects of red meat diet on gut microbiota composition in mice and their subsequent sensitivity to DSS (dextran sulfate sodium)-induced colitis.
Three-week-old male and female BALB/c mice were purchased from Beijing Vital River Laboratory (Beijing, China). All animals were housed in specific pathogen-free conditions and in a 12-h light/dark cycle. All animal experiments complied with the ethical guidelines of People's Republic of China. The experimental protocol was approved by the Ethics Committee of Xinjiang Medical University (approval no. IACUC20180411-04).
Red meat diet was made from beef powder (Weifang meibaolai animal protein Co., Ltd. China), containing about 70–78% protein, 10–12% fat, 10% water and others. All diets were prepared synthetically from Jiangsu Xietong Pharmaceutical Bio-engineering Co., Ltd (Jiangsu, China). The ingredients of the experimental diets were shown on Table 1.
Shortly after weaning and 1 week of adaption, the mice were randomly divided into four groups (half male and half female in each group) according to the red meat-based diet. The normal control group (n = 20) was given a standard diet (AIN-93G diet, without red meat, Table 1). A low-dose red meat group (n = 20) was given a diet with 25% red meat (Table 1), a medium-dose red meat group (n = 20) was given a diet with 50% red meat (Table 1), a high-dose red meat group (n = 20) was given a diet with 75% red meat (Table 1). All mice were fed ad libitum. Body weight was recorded weekly for each mouse. After 8 weeks, 10 mice per group were randomly anesthetized and sacrificed and colonic contents were immediately stored at −80°C for 16S rRNA gene sequencing.
The remaining mice in each group (n = 10) received drinking water containing 3% DSS for 9 days. The drinking bottles were checked every 2 days and the DSS solution was reconfigured to ensure that the concentration of DSS was the same over time. During the 9-day feeding, all the mice were continually fed the uniform standard diets (AIN-93G diet, without red meat) ad libitum. Body weight, food intakes, the presence of occult blood or gross blood in the feces, and stool consistency were recorded daily for each mouse during DSS (M.W 20000, Shanghai Aladdin Biochemical Technology Co., Ltd. China) induced colitis model. Mice of all DSS groups behaved in the same way. The disease activity index (DAI) was determined as previously described (14). Occult blood in feces was evaluated with Urine Fecal Occult Blood Test Kit (Yeasen Biotech Co., Ltd. China). On the 9th day, all of the mice were sacrificed, and whole colons were collected and measured length by a ruler. 16S rRNA gene sequencing
Total bacteria genome DNA from colonic contents was extracted with a Qiagen DNA Mini Kit (Qiagen, USA) and 16S rRNA genes were amplified used the specific primers of V3-V4: 341 Forward Primer: 5′-ACTCCTACGGGAGGCAGCAG-3′, 806 Reverse Primer: 5′-GGACTACHVGGGTWTCTAAT-3′. To remove contamination of samples with exogenous DNA or PCR products, PCR water instead of sample/template was used as negative controls. All PCR reactions were carried out in 30 μL reactions with 15 μL of Phusion High-Fidelity PCR Master Mix (New England Biolabs, USA), 0.2 μM of forward and reverse primers, and about 10 ng template DNA. Thermal cycling consisted of initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and extension at 72°C for 60 s, and a final extension at 72°C for 5 min. The PCR products were subjected to electrophoresis on 2% agarose gel. Samples with bright main strip between 400 and 450bp were chosen for further experiments. Then, the PCR products were purified with Gene JET Gel Extraction Kit (Thermo Scientific, USA). Sequencing libraries were generated using NEB Next®Ultra™DNA Library Prep Kit for Illumina (New England Biolabs, USA) following manufacturer's recommendations and index codes were added. The library quality was assessed on the Qubit@2.0 Fluorometer (Thermo Scientific, USA) and Agilent Bioanalyzer 2100 system. At last, the library was sequenced on an Illumina MiSeq platform and 250bp/300bp paired-end reads were generated. The data presented in the study are deposited in the NCBI SRA repository, accession number SRP310208.
To confirm differences in the abundances of individual taxonomy between groups, Metastats software was utilized. LEfSe was used for the quantitative analysis of biomarkers within different groups. To identify differences of microbial communities between groups, ANOSIM and MRPP (multi-response permutation procedure) were performed based on the Bray-Curtis dissimilarity distance matrices.
The colon tissues were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned with the thickness of 5 μm. Then sections were stained with HE according to routine procedure. The histological score was evaluated by two independent investigators in a blinded fashion as previously described (15).
Immunohistochemistry evaluation of the colon was performed as previously described (16). Briefly, the tissue sections were incubated with a primary antibody against inducible claudin1 (1:250, Abcam, USA) and evaluated after the incubation with a biotin-conjugated secondary antibody (1:800, ZSJQB Co., Ltd. Beijing).
Total RNA was extracted from mice colon tissues using Trizol reagent (Takara, Japan). The cDNA was synthesized using a Revert Aid First Strand cDNA Synthesis kit (Thermo Scientific, USA). The mRNA levels of COX2, iNOS, NF-kB, ZO-1, claudin, and occludin were detected. The sequences of all primers are listed in Table 2. Quantitative real time PCR (RT-qPCR) was carried out on a CFX96 real-time PCR system (Bio-Rad, USA) by TB Green (Takara Bio, Japan). The two-step PCR amplification standard procedure was used: 95°C 30 s and 40 cycles of 95°C 5 s, 60°C 30 s and 72°C 30 s. To remove contamination of samples with exogenous DNA or PCR products, PCR water instead of sample/template was used as negative controls. Relative expression was calculated with 2−ΔΔCt method using β-actin for normalization.
The colonic tissue was lysed in RIPA buffer (Thermo Scientific, USA) containing a protease inhibitor (Thermo Scientific, USA). Protein concentrations were measured with BCA protein assay kit (Thermo Scientific, USA). Equal amounts of protein samples were loaded to SDS-PAGE, and proteins were transferred to polyvinylidene fluoride membranes (Immobilon, USA). Following blocking with 5% non-fat milk for 1.5 h at room temperature, membranes were incubated with primary antibodies of IL-1β, TNF-α, IL-17, IL-6, NF-kB, and β-actin (Abcam, USA) overnight at 4°C. Then the membranes were incubated with HRP-conjugated secondary antibody (1:1000, ZSJQB Co., Ltd. Beijing) for 2 h at room temperature. Finally, enhanced chemiluminescence detection kit (Thermo Scientific, USA) was used for color development. The bands were detected with a Bio-Rad Western blot detection system (Bio-Rad ChemiDoc XRS+ imager, Bio-Rad Laboratories, USA), and β-actin was used as the internal control.
All statistical analysis was performed using IBM SPSS software (IBM SPSS Statistics for Windows, version 21.0). Measurement data were presented as mean ± standard deviation (SD). Differences between groups were evaluated using univariate analysis of variance (ANOVA) and least significant difference post-hoc tests. Each experiment was repeated three times. A p-value < 0.05 was considered statistically significant.
Mice were fed with different levels of red meat diets for 8 weeks and the colonic contents were analyzed by 16S rRNA gene sequencing. The measured values of the abundance of microbiota (Chao1) and the microbiota diversity (Shannon) were shown in Figure 1A. We found that the microbiota abundance and diversity did not change significantly. Principal Component Analysis (PCA), which reflects the difference in gut microbiota between groups based on different levels of red meat diet (Figure 1B), showed that red meat diet altered the gut microbiota. Microbial community barplot showed that the microbiota composition of phylum and genus changed significantly (Figures 2A,B). At the phylum level, the major phyla of gut microbiota were Firmicutes, Bacteroidetes, Desulfobacterota, Proteobacteria, and Actinobacteria. Meanwhile, we found the relative abundance of phylum Firmicutes decreased gradually and the phylum Bacteroidetes increased gradually in the high-dose red meat groups compared to the controls or low-dose red meat group, but there was no statistically significant difference (Figures 2C,D). At the genus level, the relative abundance of genera Bacteroides and Alistipes increased significantly in the high-dose red meat groups compared to the controls and low-dose red meat groups (Figures 2E,F, P < 0.01), and the genera of Lachnospiraceae_NK4A136_group decreased significantly in the high-dose red meat groups compared to the controls (Figure 2G, P < 0.05). Meanwhile, we found that the genera of Faecalibaculum, Blautia and Dubosiella decreased significantly in the high-dose red meat groups compared to the controls (Figures 2H–J, P < 0.05). The changes of gut microbiota within different levels of red meat diets groups were further analyzed by LEfSe. In our study, a total of 29 differentially abundant taxons (from phylum to genus level) were found between the four groups (Figure 3).
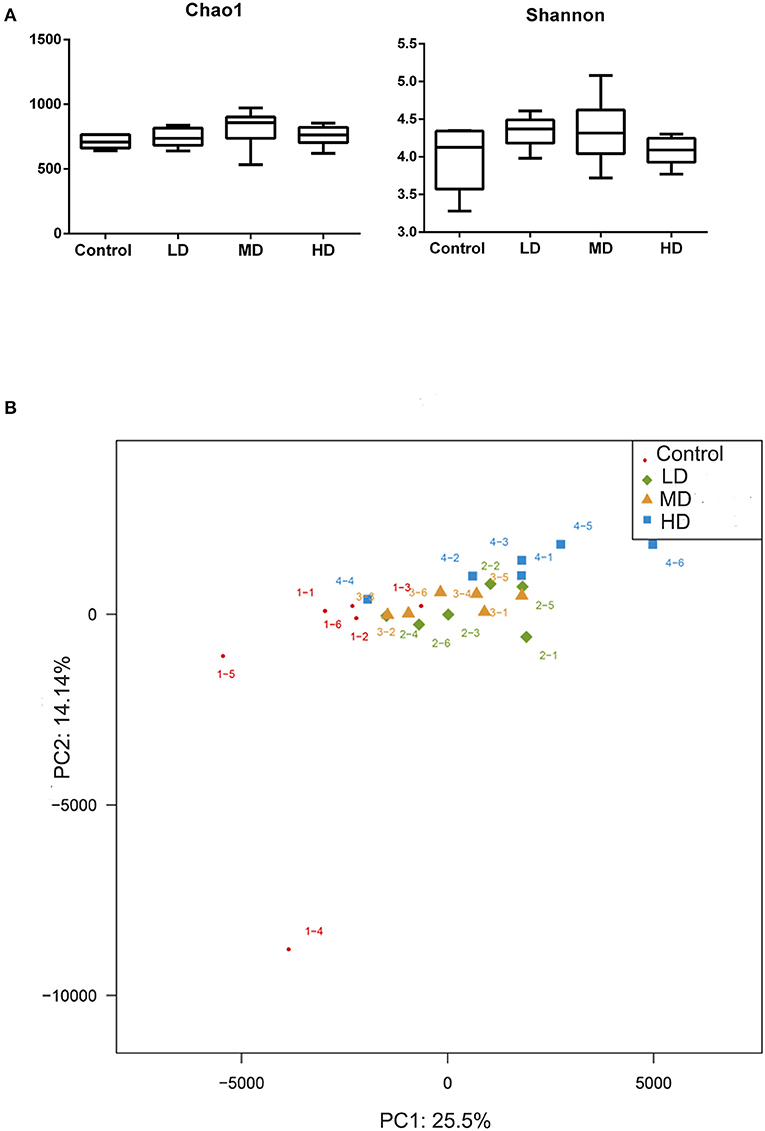
Figure 1. Correlation of high red meat intake with gut microbiota. (A) The abundance and diversity of microbiota (Chao 1 and shannon index) in mice fed with different levels of red meat diets. (B) Principal Component Analysis (PCA) of microbiota in mice fed with different levels of red meat diets. LD, low-dose red meat group; MD, medium-dose red meat group; HD, high-dose red meat group.
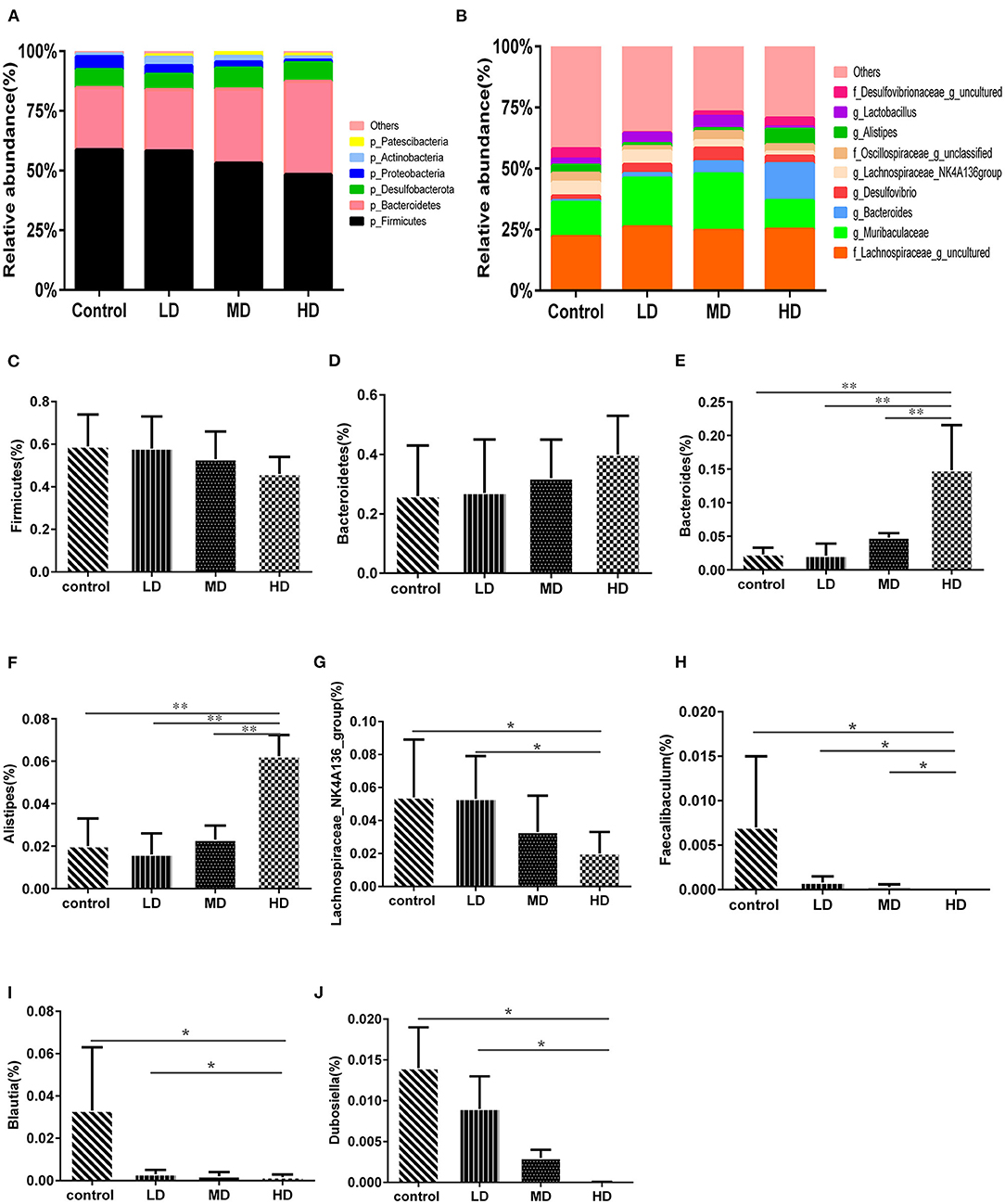
Figure 2. High red meat intake alters gut microbiota. (A) The phylum level of microbiota composition and comparison of microbiota in mice fed with different levels of red meat diets: (C) Firmicutes, (D) Bacteroidetes. (B) The genus level of composition and comparison of microbiota in mice fed with different levels of red meat diets: (E) Bacteroides, (F) Alistipes, (G) Lachnospiraceae_NK4A136_group, (H) Faecalibaculum, (I) Blautia, (J) Dubosiella. The chart data are presented as mean ± SD, and *p < 0.05, **p < 0.01. LD, low-dose red meat group; MD, medium-dose red meat group; HD, high-dose red meat group.
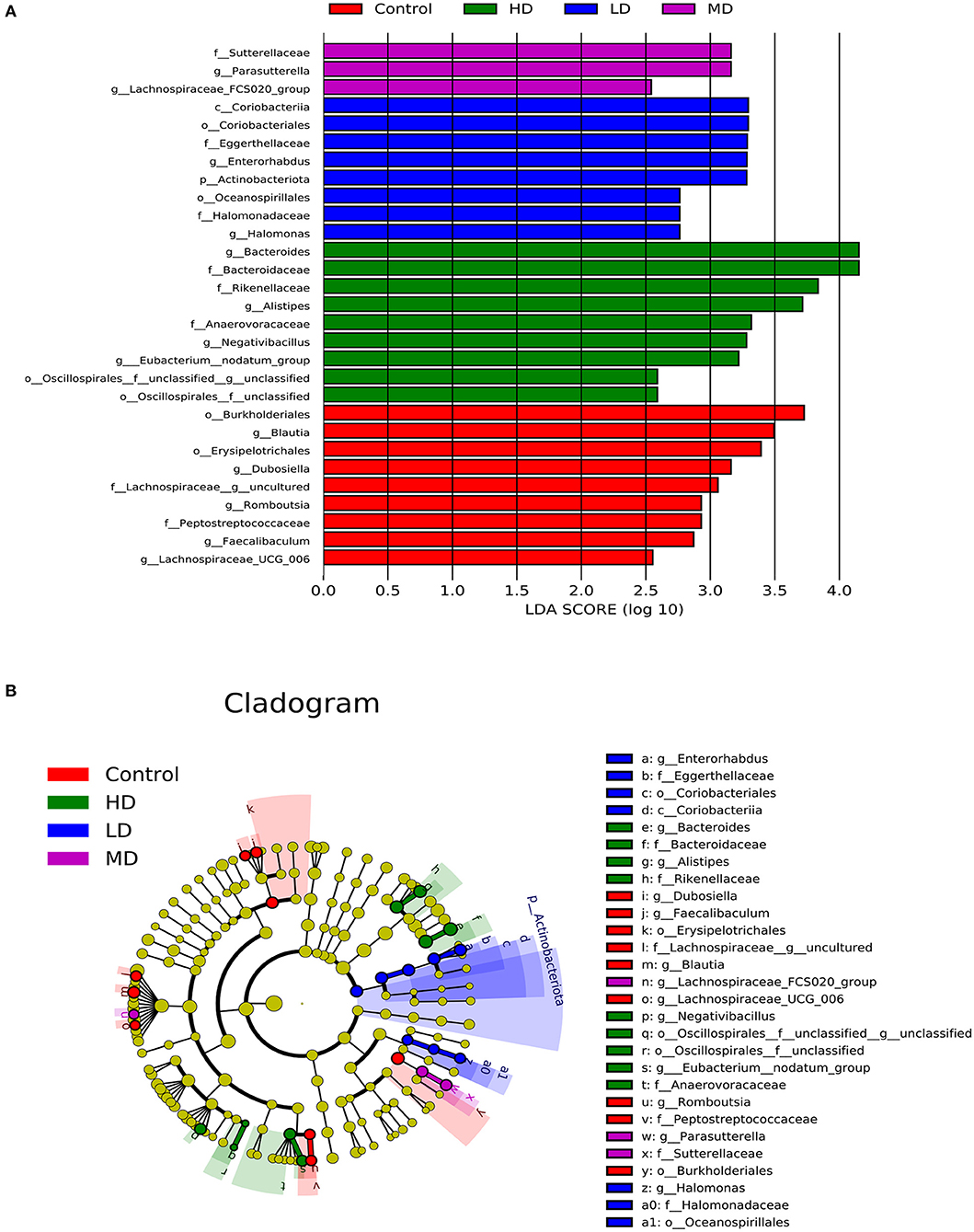
Figure 3. Characteristics of microbial community composition using LEfSe (LDA Effect Size) analysis. (A) The differential bacteria of the gut microbiota meeting a significant LDA threshold value of >2.5 between groups with different levels of red meat diet. (B) The cladogram based on LEfSe shows differential bacteria of the gut microbiota between groups with different levels of red meat diet.
During the 8 weeks of feeding with red meat diets, the body weights of mice decreased a little compared to the controls, but there was no statistical significance (Figure 4A). To evaluate effects of dietary red meat on colonic inflammation, we exposed mice fed with different levels of red meat-based diets to DSS (17), which is a widely used to induce colitis resembling IBD. The food intake variation of mice after 3.0% DSS solution of drinking water in different groups is shown in Figure 4B. When the mice were exposed to DSS in early3 days, the food intake changed slightly. However, during the last 5 days, the food intake reduced significantly in high-dose red meat group compared with the control group (p < 0.05). The body weight variation of mice after 3.0% DSS solution of drinking water is shown in Figure 4C. During the first 5 days, the body weight changed slightly, but during the last 3 days, the body weight reduced significantly in high-dose red meat group compared with the control group (p < 0.05). Significant reductions in food intake and body weight seem to indicate malnutrition in mice. DAI score is a comprehensive score to assess the severity of colitis. We found at the last 4 days of the experiment, the DAI score of high-dose red meat group increased significantly compared with the control group after 3.0% DSS induced colitis (p < 0.05, Figure 4D). Our results also showed that high-dose red meat group significantly shortened the colonic length compared with the control group after 3.0% DSS induced colitis (p < 0.05, Figure 4E). By HE staining, we observed that the high and medium-dose red meat groups exhibited more severe epithelial damage and higher levels of inflammation infiltration compared with the control groups (Figure 5A). In conclusion, our data demonstrates that high intake of red meat diet enhances colonic inflammation of DSS-induced colitis.
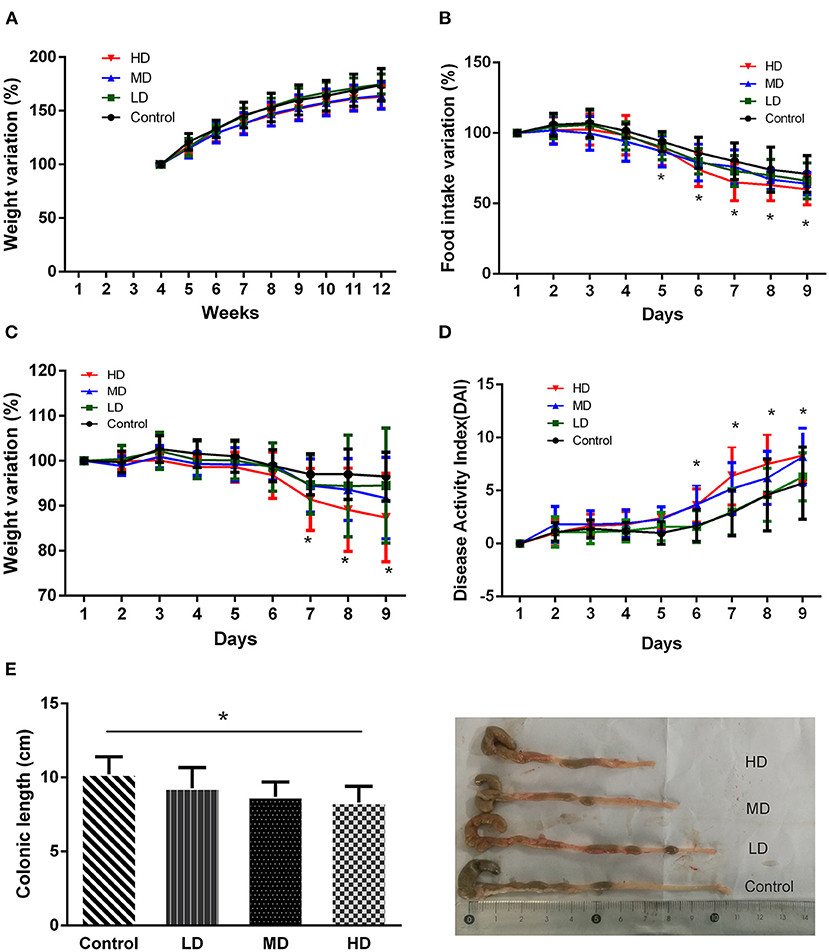
Figure 4. Effects of different levels of red meat diets. (A) Body weight variation during 8 weeks of feeding with different levels of red meat diets. (B) Food intake variation of different levels of red meat diets on DSS colitis. (C) Body weight variation of different levels of red meat diets on DSS colitis. (D) Disease activity index of different levels of red meat diets on DSS colitis. (E) Comparison of different red meat diets on colonic length and representative images of DSS colitis mice. Data are presented as mean ± SD, and *indicates p < 0.05.

Figure 5. HE staining and immunohistochemical analysis in the colon tissue of DSS colitis. (A) Representative images of HE staining of the colon tissues and histologic injury scored for HE staining in mice fed with various levels of red meat diets. Magnifications: × 100 and × 200. (B) Immunohistochemical analysis of Claudin-1 expression in the colon tissue of mice fed with various levels of red meat diets. Magnifications: × 400. Data are presented as mean ± SD, and *p < 0.05.
The epithelial cells and tight junctions contribute to intestinal barrier integrity in conjunction with the mucus secreted from the goblet cells and antimicrobial factors from the immune system cells (18). Tight junctions are critical in preventing the entry of the microbial toxins, antigens and other harmful substances (19). Immunohistochemistry evaluation showed that the expression of tight junction proteins-claudin1 was significantly reduced in high-dose red meat groups compared with the control and low-dose red meat groups (Figure 5B). Meanwhile, we found the relative mRNA expression of claudin, occludin, and ZO-1 was also decreased in high-dose red meat groups compared with the control or low-dose red meat groups (Figures 6A–C). These experiments indicate that high-dose red meat diet could impair colonic barrier integrity in DSS-induced colitis.
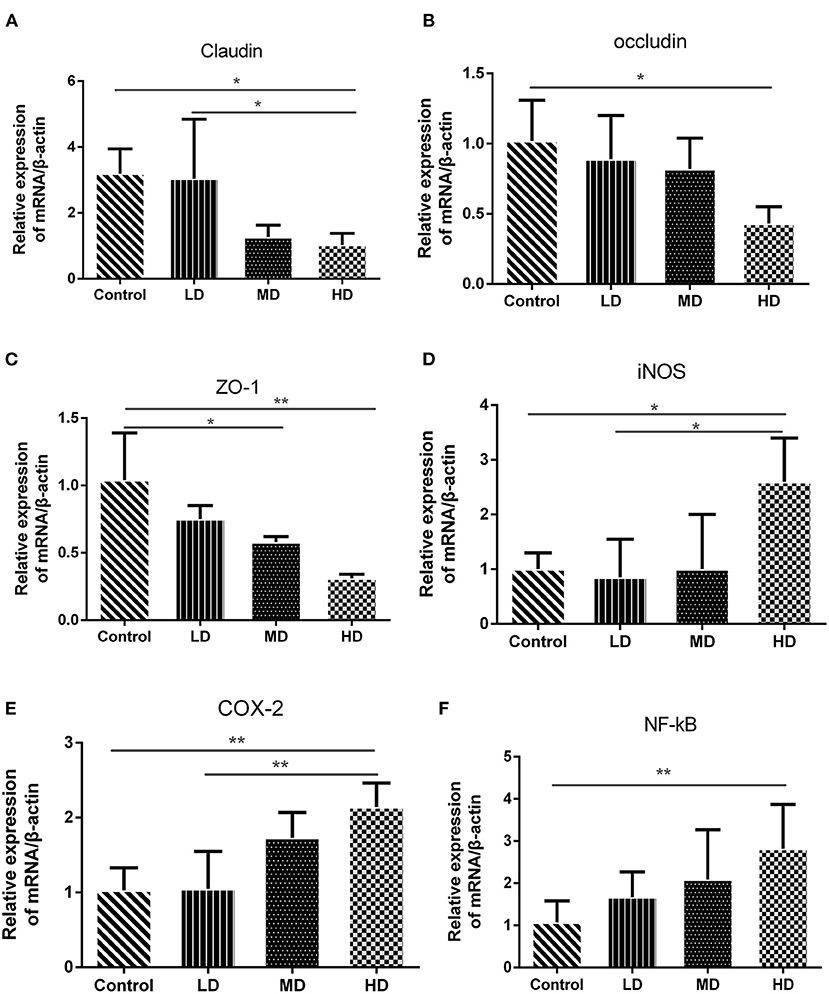
Figure 6. Gene expression levels determined by RT-PCR. The mRNA level of (A) Claudin, (B) Occludin, (C) ZO-1, (D) COX-2, (E) iNOS, and (F) NF-kB. Data are presented as mean ± SD, and *p < 0.05, **p < 0.01.
High meat protein diet induces colonic inflammation and upregulates several key cytokines including IL-1β, TNF-α, IL-6 (20). High fat diet impairs the intestinal immune system and increases sensitivity to DSS (21). High meat heme reduces fecal butyrate and further exacerbates DSS-induced colitis (13). To determine whether high red meat intake exacerbates colonic inflammation in DSS-induced colitis, RT-PCR and Western blot were performed. We observed that relative mRNA expression of iNOS and COX-2 was significantly higher in the high-dose red meat group than in the control group or the low-dose red meat group (Figures 6D,E). We also noticed that NF-kB p65 mRNA was expressed at higher levels in the high-dose red meat groups than control group (Figure 6F). Besides, pro-inflammatory cytokines including IL-1β, TNF-α, IL-17, and IL-6 were significantly increased in high-dose red meat groups compared with the control group (Figures 7A–E). Moreover, western blotting also showed an increase of NF-kB p65 protein in the high and medium-dose red meat groups than control group (Figures 7A,F). All of above results suggest that high-dose red meat diet significantly increases colonic inflammation in the DSS-induced colitis mouse model.
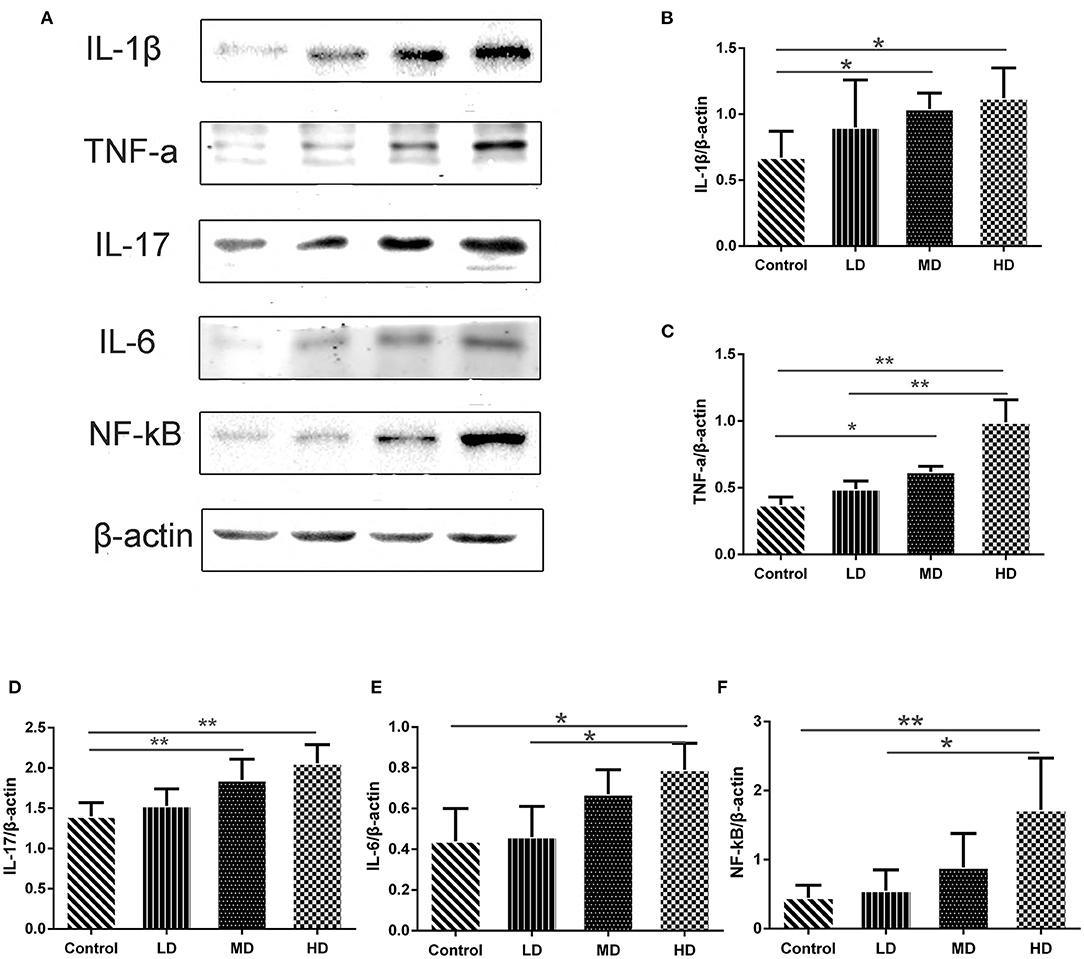
Figure 7. Gene expression levels determined by Western blot. (A) Representative pictures of Western blot bands and Western blot analysis showed the protein expression comparison of (B) IL-1β, (C) TNF-α, (D) IL-17, (E) IL-6, and (F) NF-kB, and β-actin was used as the internal control. Data are presented as mean ± SD, and *p < 0.05, **p < 0.01.
Epidemiological study has shown that the prevalence and incidence of IBD is highest in Western countries (22). In particular, the prevalence and incidence of IBD in immigrants of Western countries increases significantly, far exceeding those in their country of origin (23). More evidence suggests that Western diets, such as high red meat intake, are associated with increased incidence of IBD (7). Eating habits can affect microbiota in the human gut, and gut microbiota can respond to altered diets (24). Different dietary components, such as protein (25), fat, fiber (26), salt, amino acids, heme, etc., can alter the intestinal microbiota, thereby affecting intestinal permeability and inflammation. However, how the red meat diet affects the gut microbiota is still not clear.
In this study, we found that the mostly increased nutrient in high red meat diets was protein. The total protein content of MD (36%) and HD (54%) compared to the control group (20%) almost increased exponentially. It has been shown that long-term high-protein diets can reduce body weight, alert metabolism and intestinal function (27). Most excessive dietary protein can lead to significant weight loss in mice with colitis, especially casein, isolated soy protein and whey (26). Similarly, our study also found that long-term high red meat diets led to reduced intake and body weight significantly during DSS-induced colitis, which further led to malnutrition. Malnutrition is highly prevalent in IBD patients and the reported prevalence of malnutrition in IBD patients ranges between 20 and 85% (28). Malnutrition often indicates more severe colitis and poor clinical outcomes (29).
High-protein diets can also affect the intestinal microbiota. In this study, we analyzed the gut microbiota in mice fed with different levels of red meats by 16S rRNA gene sequencing. We found that there was no difference in bacterial alpha diversity between the four groups, but the composition of microbiota changed significantly. This is consistent with the results of Klara Kostovcikova et al., which found that a protein-rich diet led to gut microbiota disorders and promoted colitis in mice (30).
The importance of the intestinal microbiota in the early life has been established. During childhood, different dietary habits can alert intestinal microbiota, and these intestinal microbiota stabilizes and becomes a balanced system in adulthood (31, 32). The intestinal microbiota of mice fed with different levels of red meat in early developmental stage changes and becomes stable. Therefore, we provided a uniform diet during the DSS-induced colitis.
We found that at the phylum level, high red meat intake reduced the relative abundance of Firmicutes, and increased the abundance of Bacteroidetes. Our findings about phylum level changes were consistent with Matijašić's study (33), which revealed that the abundance of Firmicutes significantly decreased and Bacteroidetes significantly increased in patients with UC and CD. In another experimental study on diet and gut microbiota (30), it was also found that high intake of animal protein diet reduced the relative abundance of Firmicutes, which was in agreement with our findings.
In this study, we found at the genus level, high red meat intake significantly increased the relative abundance of Bacteroides and Alistipes genera in colon, but reduced the abundance of Lachnospiraceae_NK4A136 group, Faecalibaculum, Blautia, and Dubosiella genera. Previous studies have shown that the genera of Bacteroides and Alistipes can affect amino acid transport, metabolism, energy production and transport, and inflammation (34–36). Bacteroides can secret metalloprotease toxin both in vivo and in vitro (37), which stimulate the cleavage of intercellular adhesion protein E-cadherin in colonic epithelial cells, resulting in increased colonic mucosal permeability and activating NF-kB pathway. The genus Alistipes is considered as a kind of pathogenic bacteria. It has been shown that the genus Alistipes was correlated with obesity (38), diabetes (39), constipation, colitis and colorectal cancer (40). Schirmer et al. (41) found that the relative abundance of Bacteroides and Alistipes in IBD patients was increased.
Lachnospiraceae_NK4A136 group is butyrate-producing bacteria (42), and butyrate is a beneficial substance in the gut, which protects the intestinal mucosa, regulates immunity, and reduces inflammation. So, the reduction of Lachnospiraceae_NK4A136– group will be detrimental to the integrity of the intestinal mucosa and can easily lead to intestinal inflammation. Some studies have found that the genus Lachnospiraceae_NK4A136 group have effects on the prevention and treatment of depressive symptoms (43), acute colitis (44), type 2 diabetes and obesity. In our experiment, we found that high intake of red meat diet reduced the relative abundance of Lachnospiraceae_NK4A136_group compared with the control group.
Both Faecalibaculum and Blautia are considered to be potential probiotics, and both of them can produce short-chain fatty acids (SCFAs). Faecalibaculum can produce butyrate, which promotes the differentiation of Treg cells in the colon to reduce inflammation (45). It has been found that the relative abundance of Faecalibaculum in patients with IBD was increased (46). Some studies have also found the reduction of Faecalibaculum is related with the occurrence of colorectal cancer (47), allergies, asthma (48), and rheumatoid arthritis (49). In our study, we also found high intake of red meat diet reduced the relative abundance of Faecalibaculum.
The genus Blautia has been confirmed to kill harmful microorganisms in the intestines, thereby greatly preventing infections (50). Studies have found that the reduction of genus Blautia is associated with IBD (51), colorectal cancer (52), diabetes and obesity (53), and major depressive disorder (54). Our results showed that high intake of red meat diet reduced the relative abundance of Blautia. Meanwhile, high intake of red meat diet reduced the relative abundance of Dubosiella. Qiu et al. (55) showed that Dubosiella played a novel role in regulating SCFAs production and obesity.
Microbiota composition in the gut not only affects the production of the metabolites produced by the microbiota but also affects the immune system, and then aggravates the colonic inflammation (56). Study on colitis of animal models has suggested that colonic inflammation relies heavily on a triggering event mediated by the gut microbiota (57). Recent study has identified over 200 IBD risk loci in the IBD patients (58). It has been postulated that gut microbiota interacts with these risk loci, resulting in gut microbiota disorders and subsequently leading to the occurrence of IBD. The association of gut microbial disorders and the subsequent dysregulated immune response leads to ThI/Th2 immune response and the progression of IBD (58). In our study, we found that high intake of red meat diet led to gut microbiota disorders. Furthermore, we found that the sensitivity of mice with higher red meat intake to DSS-induced colitis was enhanced, resulting in faster body weight loss, shortened colonic length, severe histological colon damage and colon barrier integrity loss. Moreover, it also increased pro-inflammatory cytokines including IL-1β, TNF-α, IL-17, IL-6, and inflammatory inducible enzymes such as COX-2 and iNOS.
Apart from the gut microbiota, other crucial factors that are also involved in the maintenance of intestinal homeostasis include intestinal barrier integrity and permeability, because dysfunction of intestinal barrier integrity and permeability are common symptoms in patients with IBD (59). Tight junction proteins are critical for intestinal barrier integrity and can regulate paracellular permeability (60). The tight junction protein is mainly composed of claudin, occludin and ZO-1 (61). Studies have found that patients with UC have reduced expression of tight junction proteins (claudin, occludin and ZO-1) in the colonic epithelium (62), suggesting that tight junction proteins may be potential markers of colonic epithelial cell integrity. In our study, we also found that high intake of red meat diet decreased the expression of claudin, occludin and ZO-1 and upregulated of nuclear transcription factor NF-kB. The upregulation of NF-kB correlated significantly with the severity of intestinal inflammation.
In colonic inflammation, a number of pro-inflammatory cytokines are produced, such as IL-1β, TNF-α, IL-17, and IL-6 (63). The nuclear transcription factor NF-kB affects the pathogenesis of colitis and regulates the expression of some pro-inflammatory cytokines (64) and inducible enzymes such as COX-2 and iNOS (65). These results indicate that high intake of red meat diet exacerbates experimental colitis in mice. Our study is consistent with the recently published articles (25, 30), which demonstrates that increased animal protein diet promotes the susceptibility to colitis by gut microbiota disorders.
In summary, our results suggest that high red meat intake can induce the destruction of gut microbiota, which in turn impairs the colon barrier integrity and aggravates DSS-induced colitis. Our results may provide more insight into the relationship between dietary interventions with IBD and suggest that an optimized diet, reasonable nutrients, and appropriate red meat intake may prevent the occurrence of IBD.
The data presented in the study are deposited in the NCBI SRA repository, accession number SRP310208.
The animal study was reviewed and approved by the Ethics Committee of Xinjiang Medical University (approval no. IACUC20180411-04).
D-pL: study Design, data collection, and manuscript preparation. MC: study design and data collection. FT: statistical analysis and data interpretation. X-yL: literature search. PY: study design and funds collection. All authors contributed to the article and approved the submitted version.
The work was supported by National Natural Science Foundation of China (No: 81760100).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Adak A, Khan MR. An insight into gut microbiota and its functionalities. Cell Mol Life Sci. (2019) 76:473–93. doi: 10.1007/s00018-018-2943-4
2. Borbet TC, Blaser MJ. Host genotype and early life microbiota alterations have additive effects on disease susceptibility. Mucosal Immunol. (2019) 12:586–8. doi: 10.1038/s41385-019-0157-1
3. Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Trans Med. (2015) 7:307ra152. doi: 10.1126/scitranslmed.aab2271
4. Goethel A, Turpin W, Rouquier S, Zanello G, Robertson SJ, Streutker CJ, et al. Nod2 influences microbial resilience and susceptibility to colitis following antibiotic exposure. Mucosal Immuno. (2019) 12:720–32. doi: 10.1038/s41385-018-0128-y
5. Fong W, Li Q, Yu J. Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Oncogene. (2020) 39:4925–43. doi: 10.1038/s41388-020-1341-1
6. Delzenne NM, Rodriguez J, Olivares M, Neyrinck AM. Microbiome response to diet: focus on obesity and related diseases. Rev Endocr Metab Disord. (2020) 21:369–80. doi: 10.1007/s11154-020-09572-7
7. Khalili H, Chan SSM, Lochhead P, Ananthakrishnan AN, Hart AR, Chan AT. The role of diet in the aetiopathogenesis of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. (2018) 15:525–35. doi: 10.1038/s41575-018-0022-9
8. Jantchou P, Morois S, Clavel-Chapelon F, Boutron-Ruault MC, Carbonnel F. Animal protein intake and risk of inflammatory bowel disease: the E3N prospective study. Am J Gastroenterol. (2010) 105:2195–201. doi: 10.1038/ajg.2010.192
9. Hou JK, Abraham B, El-Serag H. Dietary intake and risk of developing inflammatory bowel disease: a systematic review of the literature. Am J Gastroenterol. (2011) 106:563–73. doi: 10.1038/ajg.2011.44
10. Tjonneland A, Overvad K, Bergmann MM, Nagel G, Linseisen J, Hallmans G, et al. Linoleic acid, a dietary n-6 polyunsaturated fatty acid, and the aetiology of ulcerative colitis: a nested case-control study within a European prospective cohort study. Gut. (2009) 58:1606–11. doi: 10.1136/gut.2008.169078
11. Jowett SL, Seal CJ, Pearce MS, Phillips E, Gregory W, Barton JR, et al. Influence of dietary factors on the clinical course of ulcerative colitis: a prospective cohort study. Gut. (2004) 53:1479–84. doi: 10.1136/gut.2003.024828
12. van der Logt EM, Blokzijl T, van der Meer R, Faber KN, Dijkstra G. Westernized high-fat diet accelerates weight loss in dextran sulfate sodium-induced colitis in mice, which is further aggravated by supplementation of heme. J Nutr Biochem. (2013) 24:1159–65. doi: 10.1016/j.jnutbio.2012.09.001
13. Constante M, Fragoso G, Calvé A, Samba-Mondonga M, Santos MM. Dietary heme induces gut dysbiosis, aggravates colitis, and potentiates the development of adenomas in mice. Front Microbiol. (2017) 8:1809. doi: 10.3389/fmicb.2017.01809
14. Han Y, Song M, Gu M, Ren D, Zhu X, Cao X, et al. Dietary intake of whole strawberry inhibited colonic inflammation in dextran-sulfate-sodium-treated mice via restoring immune homeostasis and alleviating gut microbiota dysbiosis. J Agric Food Chem. (2019) 67:9168–77. doi: 10.1021/acs.jafc.8b05581
15. Nunes NS, Chandran P, Sundby M, Visioli F, da Costa Gonçalves F, Burks SR, et al. Therapeutic ultrasound attenuates DSS-induced colitis through the cholinergic anti-inflammatory pathway. EBioMedicine. (2019) 45:495–510. doi: 10.1016/j.ebiom.2019.06.033
16. Wang Y, He G, Tang H, Shi Y, Kang X, Lyu J, et al. Aspirin inhibits inflammation and scar formation in the injury tendon healing through regulating JNK/STAT-3 signalling pathway. Cell Prolif. (2019) 52:e12650. doi: 10.1111/cpr.12650
17. Chassaing B, Aitken JD, Malleshappa M, Vijay-Kumar M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr Protoc Immunol. (2014) 104:15.25.1–.14. doi: 10.1002/0471142735.im1525s104
18. France MM, Turner JR. The mucosal barrier at a glance. J Cell Sci. (2017) 130:307–14. doi: 10.1242/jcs.193482
19. Vaziri ND, Yuan J, Nazertehrani S, Ni Z, Liu S. Chronic kidney disease causes disruption of gastric and small intestinal epithelial tight junction. American J Nephrol. (2013) 38:99–103. doi: 10.1159/000353764
20. Hussain M, Umair Ijaz M, Ahmad MI, Khan IA, Brohi SA, Shah AU, et al. Meat proteins in a high-fat diet have a substantial impact on intestinal barriers through mucus layer and tight junction protein suppression in C57BL/6J mice. Food Funct. (2019) 10:6903–14. doi: 10.1039/C9FO01760G
21. Tanaka S, Nemoto Y, Takei Y, Morikawa R, Oshima S, Nagaishi T, et al. High-fat diet-derived free fatty acids impair the intestinal immune system and increase sensitivity to intestinal epithelial damage. Biochem Biophys Res Commun. (2020) 522:971–7. doi: 10.1016/j.bbrc.2019.11.158
22. Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. (2015) 12:720–7. doi: 10.1038/nrgastro.2015.150
23. Benchimol EI, Manuel DG, Mojaverian N, Mack DR, Nguyen GC, To T, et al. Health services utilization, specialist care, and time to diagnosis with inflammatory bowel disease in immigrants to ontario, canada: a population-based cohort study. Inflamm Bowel Dis. (2016) 22:2482–90. doi: 10.1097/MIB.0000000000000905
24. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. (2014) 505:559–63. doi: 10.1038/nature12820
25. Vidal-Lletjós S, Andriamihaja M, Blais A, Grauso M, Lepage P. Dietary protein intake level modulates mucosal healing and mucosa-adherent microbiota in mouse model of colitis. Nutrients. (2019) 11:514. doi: 10.3390/nu11030514
26. Llewellyn SR, Britton GJ, Contijoch EJ, Vennaro OH, Mortha A, Colombel JF, et al. Interactions between diet and the intestinal microbiota alter intestinal permeability and colitis severity in mice. Gastroenterology. (2018) 154:1037–46.e2. doi: 10.1053/j.gastro.2017.11.030
27. Beaumont M, Portune K, Steuer N, Lan A, Cerrudo V, Audebert M, et al. Quantity and source of dietary protein influence metabolite production by gut microbiota and rectal mucosa gene expression: a randomized, parallel, double-blind trial in overweight humans. Am J Clin Nutr. (2017) 106:1005–19. doi: 10.3945/ajcn.117.158816
28. Chiu E, Oleynick C. Optimizing inpatient nutrition care of adult patients with inflammatory bowel disease in the 21st century. Nutrients. (2021) 13:1581. doi: 10.3390/nu13051581
29. Balestrieri P, Ribolsi M, Guarino MPL, Emerenziani S, Altomare A. Nutritional aspects in inflammatory bowel diseases. Nutrients. (2020) 12:372. doi: 10.3390/nu12020372
30. Kostovcikova K, Coufal S, Galanova N, Fajstova A, Hudcovic T, Kostovcik M, et al. Diet rich in animal protein promotes pro-inflammatory macrophage response and exacerbates colitis in mice. Front Immunol. (2019) 10:919. doi: 10.3389/fimmu.2019.00919
31. Kashtanova DA, Popenko AS, Tkacheva ON, Tyakht AB, Alexeev DG, Boytsov SA. Association between the gut microbiota and diet: fetal life, early childhood, and further life. Nutrition. (2016) 32:620–7. doi: 10.1016/j.nut.2015.12.037
32. Tanaka M, Nakayama J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol Int. (2017) 66:515–22. doi: 10.1016/j.alit.2017.07.010
33. Matijašić M, Meštrović T, Perić M, Cipčić Paljetak H, Panek M, Vranešić Bender D, et al. Modulating composition and metabolic activity of the gut microbiota in IBD patients. Int J Mol Sci. (2016) 17:578. doi: 10.3390/ijms17040578
34. Sun X, Shen J, Liu C, Li S, Peng Y, Chen C, et al. L-Arginine and N-carbamoylglutamic acid supplementation enhance young rabbit growth and immunity by regulating intestinal microbial community. Asian Aust J Anim Sci. (2019) 33:166–76. doi: 10.5713/ajas.18.0984
35. Su X, Yin X, Liu Y, Yan X, Zhang S, Wang X, et al. Gut dysbiosis contributes to the imbalance of Treg and Th17 cells in Graves' disease patients by propionic acid. J Clin Endocri Metab. (2020) 105:dgaa511. doi: 10.1210/clinem/dgaa511
36. Yin J, Li Y, Han H, Chen S, Gao J, Liu G, et al. Melatonin reprogramming of gut microbiota improves lipid dysmetabolism in high-fat diet-fed mice. J Pineal Res. (2018) 65:e12524. doi: 10.1111/jpi.12524
37. Wick EC, Sears CL. Bacteroides spp. and diarrhea. Curr Opin Infect Dis. (2010) 23:470–4. doi: 10.1097/QCO.0b013e32833da1eb
38. Zhu J, Kong Y, Yu J, Shao S, Mao M, Zhao M, et al. Consumption of drinking water N-Nitrosamines mixture alters gut microbiome and increases the obesity risk in young male rats. Environ Poll. (2019) 248:388–96. doi: 10.1016/j.envpol.2019.02.012
39. Remely M, Hippe B, Zanner J, Aumueller E, Brath H, Haslberger AG. Gut microbiota of obese, type 2 diabetic individuals is enriched in faecalibacterium prausnitzii, akkermansia muciniphila and peptostreptococcus anaerobius after weight loss. Endocr Metab Immune Disord Drug Targets. (2016) 16:99–106. doi: 10.2174/1871530316666160831093813
40. Borges-Canha M, Portela-Cidade JP, Dinis-Ribeiro M, Leite-Moreira AF, Pimentel-Nunes P. Role of colonic microbiota in colorectal carcinogenesis: a systematic review. Rev Esp Enferm Dig. (2015) 107:659–71. doi: 10.17235/reed.2015.3830/2015
41. Schirmer M, Franzosa EA. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat Microbiol. (2018) 3:337–46. doi: 10.1038/s41564-017-0089-z
42. Antonissen G, Eeckhaut V, Van Driessche K, Onrust L, Haesebrouck F, Ducatelle R, et al. Microbial shifts associated with necrotic enteritis. Avian Pathol. (2016) 45:308–12. doi: 10.1080/03079457.2016.1152625
43. Cheng D, Chang H, Ma S, Guo J, She G, Zhang F, et al. Tiansi liquid modulates gut microbiota composition and Tryptophan? Kynurenine metabolism in rats with hydrocortisone-induced depression. Molecules. (2018) 23:2832. doi: 10.3390/molecules23112832
44. Wang YN, Meng XC, Dong YF, Zhao XH, Qian JM, Wang HY, et al. Effects of probiotics and prebiotics on intestinal microbiota in mice with acute colitis based on 16S rRNA gene sequencing. Chin Med J. (2019) 132:1833–42. doi: 10.1097/CM9.0000000000000308
45. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. (2013) 341:569–73. doi: 10.1126/science.1241165
46. Takahashi K, Nishida A, Fujimoto T, Fujii M, Shioya M, Imaeda H, et al. Reduced abundance of butyrate-producing bacteria species in the fecal microbial community in Crohn's disease. Digestion. (2016) 93:59–65. doi: 10.1159/000441768
47. Zagato E, Pozzi C, Bertocchi A, Schioppa T, Saccheri F, Guglietta S, et al. Endogenous murine microbiota member Faecalibaculum rodentium and its human homologue protect from intestinal tumour growth. Nat Microbiol. (2020) 5:511–24. doi: 10.1038/s41564-019-0649-5
48. Galazzo G, van Best N, Bervoets L, Dapaah IO, Savelkoul PH, Hornef MW, et al. Development of the microbiota and associations with birth mode, diet, and atopic disorders in a longitudinal analysis of stool samples, collected from infancy through early childhood. Gastroenterology. (2020) 158:1584–96. doi: 10.1053/j.gastro.2020.01.024
49. Chu XJ, Cao NW, Zhou HY, Meng X, Guo B, Zhang HY, et al. The oral and gut microbiome in rheumatoid arthritis patients: a systematic review. Rheumatology. (2021) 60:1054–66. doi: 10.1093/rheumatology/keaa835
50. Caballero S, Kim S, Carter RA, Leiner IM, Susac B, Miller L, et al. Cooperating commensals restore colonization resistance to vancomycin-resistant enterococcus faecium. Cell Host Microbe. (2017) 21:592–602.e4. doi: 10.1016/j.chom.2017.04.002
51. Zhu X, Xiang S, Feng X, Wang H, Tian S, Xu Y, et al. Impact of cyanocobalamin and methylcobalamin on inflammatory bowel disease and the intestinal microbiota composition. J Agric Food Chem. (2019) 67:916–26. doi: 10.1021/acs.jafc.8b05730
52. Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PloS ONE. (2012) 7:e39743. doi: 10.1371/journal.pone.0039743
53. Wei Y, Liang J, Su Y, Wang J, Amakye WK, Pan J, et al. The associations of the gut microbiome composition and short-chain fatty acid concentrations with body fat distribution in children. Clin Nutr. (2020) 40:3379–90. doi: 10.1016/j.clnu.2020.11.014
54. Yang J, Zheng P. Landscapes of bacterial and metabolic signatures and their interaction in major depressive disorders. Sci Adv. (2020) 6:eaba8555. doi: 10.1126/sciadv.aba8555
55. Qiu X, Macchietto MG, Liu X, Lu Y, Ma Y, Guo H, et al. Identification of gut microbiota and microbial metabolites regulated by an antimicrobial peptide lipocalin 2 in high fat diet-induced obesity. Int J Obes. (2021) 45:143–54. doi: 10.1038/s41366-020-00712-2
56. Shen B, Hu J, Song H, Wang Z, Fan J, Sun Y, et al. Antibiotics exacerbated colitis by affecting the microbiota, Treg cells and SCFAs in IL10-deficient mice. Biomed Pharmacother. (2019) 114:108849. doi: 10.1016/j.biopha.2019.108849
57. Nascimento RPD, Machado A, Galvez J, Cazarin CBB, Maróstica Junior MR. Ulcerative colitis: gut microbiota, immunopathogenesis and application of natural products in animal models. Life Sci. (2020) 258:118129. doi: 10.1016/j.lfs.2020.118129
58. Eom T, Kim YS, Choi CH, Sadowsky MJ, Unno T. Current understanding of microbiota- and dietary-therapies for treating inflammatory bowel disease. J Microbiol. (2018) 56:189–98. doi: 10.1007/s12275-018-8049-8
59. Johansson ME, Gustafsson JK, Holmén-Larsson J, Jabbar KS, Xia L, Xu H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. (2014) 63:281–91. doi: 10.1136/gutjnl-2012-303207
60. Landy J, Ronde E, English N, Clark SK, Hart AL, Knight SC, et al. Tight junctions in inflammatory bowel diseases and inflammatory bowel disease associated colorectal cancer. World J Gastroenterol. (2016) 22:3117–26. doi: 10.3748/wjg.v22.i11.3117
61. Sharma D, Malik A, Guy CS, Karki R, Vogel P, Kanneganti TD. Pyrin Inflammasome Regulates Tight Junction Integrity to Restrict Colitis and Tumorigenesis. Gastroenterology. (2018) 154:948–64.e8. doi: 10.1053/j.gastro.2017.11.276
62. Tan Y, Guan Y, Sun Y, Zheng C. Correlation of intestinal mucosal healing and tight junction protein expression in ulcerative colitis patients. Am J Med Sci. (2019) 357:195–204. doi: 10.1016/j.amjms.2018.11.011
63. Friedrich M, Pohin M, Powrie F. Cytokine networks in the pathophysiology of inflammatory bowel disease. Immunity. (2019) 50:992–1006. doi: 10.1016/j.immuni.2019.03.017
64. Atreya I, Atreya R, Neurath MF. NF-kappaB in inflammatory bowel disease. J Int Med. (2008) 263:591–6. doi: 10.1111/j.1365-2796.2008.01953.x
Keywords: gut microbiota, red meat diet, inflammatory bowel disease, dextran sulfate, ulcerative colitis
Citation: Li D-p, Cui M, Tan F, Liu X-y and Yao P (2021) High Red Meat Intake Exacerbates Dextran Sulfate-Induced Colitis by Altering Gut Microbiota in Mice. Front. Nutr. 8:646819. doi: 10.3389/fnut.2021.646819
Received: 18 January 2021; Accepted: 22 June 2021;
Published: 20 July 2021.
Edited by:
Silvia Turroni, University of Bologna, ItalyReviewed by:
George Grant, University of Aberdeen, United KingdomCopyright © 2021 Li, Cui, Tan, Liu and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Yao, eWFvcGluZzEyMzIwMjBAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.