
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CLINICAL TRIAL article
Front. Nutr., 19 February 2021
Sec. Nutrition and Microbes
Volume 8 - 2021 | https://doi.org/10.3389/fnut.2021.587974
 Xue-Jun Kong1,2*
Xue-Jun Kong1,2* Guobin Wan3
Guobin Wan3 Ruiyi Tian1
Ruiyi Tian1 Siyu Liu1
Siyu Liu1 Kevin Liu1
Kevin Liu1 Cullen Clairmont1
Cullen Clairmont1 Xiaojing Lin4
Xiaojing Lin4 Xiaoying Zhang5
Xiaoying Zhang5 Hannah Sherman1
Hannah Sherman1 Junli Zhu6
Junli Zhu6 Yelan Wang7
Yelan Wang7 Michelle Fong1
Michelle Fong1 Alice Li1
Alice Li1 Bryan K. Wang8
Bryan K. Wang8 Jinghan Wang9
Jinghan Wang9 Jun Liu1,10
Jun Liu1,10 Zhehao Yu11
Zhehao Yu11 Chen Shen11
Chen Shen11 Xianghua Cui11
Xianghua Cui11 Hanyu Cao11
Hanyu Cao11 Ting Du11
Ting Du11 Xia Cao11
Xia Cao11Background: Prader-Willi Syndrome (PWS) is a rare genetic disorder associated with developmental delay, obesity, and neuropsychiatric comorbidities. Bifidobacterium animalis subsp. lactis has demonstrated anti-obesity and anti-inflammatory effects in previous studies.
Aim: To evaluate the effects of Bifidobacterium animalis subsp. lactis probiotics supplementation on anthropometric growth, behavioral symptoms, and gut microbiome composition in patients with PWS.
Methods: Ethical Approval was issued by the Internal Review Board (IRB) of the Second Affiliated Hospital of Kunming Medical University (Review-YJ-2016-06). We conducted a 12-week, randomized, double-blind, placebo-controlled trial in 68 patients with Prader-Willi syndrome aged 11 months−16 years (mean = 4.2 years old) who were randomly assigned to receive daily B. lactis-11 probiotics (6 × 1010 CFUs) or a placebo sachet. Weight, height, ASQ-3, ABC, SRS-2, and CGI-I were compared between the two groups at baseline and at 6 and 12 weeks into treatment. Gut microbiome data were analyzed with the QIIME 2 software package, and functional gene analysis was conducted with PICRUSt-2.
Results: We found a significant increase in height (mean difference = 2.68 cm, P < 0.05) and improvement in CGI-I (P < 0.05) in the probiotics group compared to the placebo group. No significant change in weight or psychological measures were observed. Probiotic treatment altered the microbiome composition to favor weight loss and gut health and increased the abundance of antioxidant production-related genes.
Conclusions: The findings suggest a novel therapeutic potential for Bifidobacterium animalis subsp. lactis probiotics in Prader-Willi syndrome patients, although further investigation is warranted.
Prader-Willi Syndrome (PWS) is a rare genetic imprinting disorder with an estimated prevalence of 1/10,000–1/30,000 (1). Three mechanisms cause this genetic disorder: deletion (DEL) of the 15q11.2-q13 region from the paternal chromosome, maternal uniparental disomy (UPD) from the mother, and imprinting defect (2). PWS is characterized by severe hypotonia and feeding difficulties in early infancy, and subsequent hyperphagia and morbid obesity starting during early childhood (1). PWS patients also typically experience generalized neurodevelopmental delays and numerous neuropsychiatric comorbidities (3).
Gut microbiota has been implicated in the etiology of obesity and associated comorbidities in PWS subjects (4). The gut microbiome from normal-weight patients has been found to have higher phylogenetic diversity than that from overweight and obese patients (5). In previous studies, people with diet-induced obesity and obese PWS patients were found to have similar gut dysbiosis (5). Dysbiotic gut microbiota transplanted from PWS patients to rats caused impacted expression of GLP-1 and decreased insulin-receptor signaling 2 weeks prior to an increase in body fat composition, indicating that the gut microbiome dysbiosis may play a role in the development of obesity (6). Administration of probiotics has shown improvement of metabolic disturbance and normalization of gut microbiome composition in diet-induced obese mice and in a randomized controlled trial of weight management in overweight adults (7, 8). Microbiome dysbiosis is not only related to obesity but also closely associated with neuropsychiatric conditions (9). Altered microbiome could serve as a biomarker for diagnosis and subtyping of Autism Spectrum Disorder (ASD) (10). Probiotics treatments have been broadly used to help people with neuropsychiatric conditions (11, 12).
Bifidobacterium animalis subsp. lactis (B. lactis) is a rod-shaped, anaerobic bacteria that can be found in the gastrointestinal tract of most mammals, including humans (13). Many strains of B. lactis are considered to be health-promoting and are commonly formulated into fermented dairy foods. Anti-obesity effects have been linked to the administration of some strains of B. lactis, such as A6, CECT 8145, Bf141, B420, and BB-12, mostly in animals (14–19). Anti-inflammatory effects of some strains of B. lactis, such as HN019 and BB-12, have also been reported in recent years (20, 21). Furthermore, a recent publication reported that B.lactis BPL1 improve abdominal adiposity and insulin sensitivity in children and adolescents with PWS (22).
In this study, we conducted a randomized, double-blind, placebo-controlled trial to test our hypothesis that probiotics consumption has beneficial effects on obesity, mental health, and inflammation associated with gut microbiome changes in PWS. We enrolled a cohort of PWS patients to evaluate the efficacy of a B. lactis strain (BL-11) on their weights, heights, psychological measurements, and gut microbiome compositions and functions relative to placebo controls. In addition to potentially supporting a new intervention for patients with PWS, the microbiome composition data collected from this study may shed light on the underlying mechanisms of PWS pathology and the gut-brain axis.
We designed and conducted a randomized, double-blinded, placebo-controlled clinical trial (flowchart, Figure 1). In this trial, we randomly assigned the eligible PWS participants, with a 1:1 ratio, to either the probiotics or placebo group. We hypothesize that a 12-week treatment period is sufficient for probiotics supplementation to induce detectable changes. To achieve a statistical power of 80% for primary outcomes with a large effect size of 0.8 (Cohen's d) assumed, a total of 52 participants (26 in each arm) were required.
Ethical Approval was issued by the Internal Review Board (IRB) of the Second Affiliated Hospital of Kunming Medical University (Review-YJ-2016-06). Clinical Trial of Probiotics was registered at the Chinese Clinical Trial Registry (ChiCTR), with a number ChiCTR1900022646. Signed informed consent was obtained from the parents or legal guardians of the subjects according to the IRB requirements. The study was conducted in accordance with the Declaration of Helsinki.
We enrolled 68 subjects aged 50.5 ± 37.3 months (69.1% male, 30.9% female) with genetically confirmed diagnosis of Prader-Willi syndrome. Study participants were recruited through the PWS Care & Support Center, located in Zhejiang, China. Participants were included if they met the following criteria: they had been genetically confirmed to have PWS; had not been on any forms of probiotics for at least 4 weeks; had stable medications for at least 4 weeks; had no planned changes in medications or psychosocial interventions during the trial; had a willingness to provide stool samples in a timely manner; and had a willingness to collaborate with interviews and study procedures. Potential participants were excluded if they had other known genetic disorders, or if they were pregnant or breast-feeding before the study.
Randomization and allocation concealment were performed by a statistician who was not part of the research team. Randomization sampling numbers were electronically generated for each de-identified subject. Coded probiotics and placebo of identical appearance were prepared by the Beijing Huayuan Academy of Biotechnology to ensure allocation concealment. Both the participants and the research staff/investigators who collected and analyzed the outcome data were blinded to treatment status. Blinding was also maintained by making the probiotics package appear identical to the placebo sachet.
Probiotics BL-11 (Beijing Huayuan Academy of Biotechnology) was used in the study in the format of a sachet containing the probiotic BL-11 in powder form. Each sachet of probiotics supplement contained 3 × 1010 colony forming units (CFUs). The placebo was maltodextrin in the sachet with similar color, flavor, and taste as the probiotic sachets. Subjects received one sachet twice a day of either probiotics or a placebo for a duration of 12 weeks and were instructed to consume the sachet contents orally with water.
1. Weight and height measurements were obtained by parents using standard scales and collected by the research staff. Weight, height, and BMI were converted to z-score using age growth references provided by WHO (22).
2. Psychological measurements
1) Ages and Stages Questionnaires, 3rd Edition (ASQ-3) (23). ASQ-3 is one of the most widely available development screening tools for young children. The ASQ-3 has five domains: communication, gross motor, fine motor, problem-solving, and personal-social. Total scores were calculated. We interviewed all subjects younger than 5 years old.
2) Aberrant Behavior Checklist (ABC) (24). ABC is a 58-item behavior rating scale used to measure behavior problems across five subscales: irritability, lethargy/social withdrawal, stereotypic behavior, hyperactivity/non-compliance, and inappropriate speech. Total scores were calculated. We interviewed all subjects older than 5 years old.
3) Social Responsiveness Scale (SRS)(25). SRS consists of 65 items used for quantitative assessment of the severity of social behaviors. Total scores were calculated. We interviewed all subjects older than 5 years old.
4) Restricted and repetitive behaviors (RRB) is based on a 4-point scale (0–3) adopted from the Gilliam Autism Rating Scale, Third Edition (GARS-3) (26). Total scores were calculated. We interviewed all subjects older than 3 years old.
1. Fecal microbiome
1) Sample Handling and Collection
Stool samples were collected at three study timepoints: prior to intervention (0-weeks), 6 and 12-weeks. Sample collection was performed with DNA/RNA shield fecal collection tubes (Zymo, Cat#R1101) containing 1 mL preservation solution and were transported to the laboratory by ice bags and then frozen at −80°C. TIANmap stool DNA kit was used to extract DNA (TIANGEN, Cat#DP328) according to the manufacturer's instructions, and DNA samples were carefully quantified with a Nanodrop Spectrophotometer. A260/A280 ratios were also measured to confirm high-purity DNA yield. DNA samples were frozen at −20°C until use.
2) 16S rRNA Gene Amplicon Sequencing
The 16S rRNA V3-V4 library was constructed by two rounds of PCR with the following primers: 341F:5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGAGGCAGCAGCCTACGGGNBGCASCAG3′ and 805R:5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTGACTACNVGGGTATCTAATCC3′ via reaction procedure (95°C for 2 min, followed by 25 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and a final extension at 72°C for 5 min). PCR products were purified with 1x KAPA AMPure beads (KAPA, Cat#KK8002). Then, products were put through a second PCR reaction procedure (95°C for 2 min, followed by eight cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and a final extension at 72°C for 5 min). PCR products were purified with 1x KAPA AMPure beads and analyzed using a Bioanalyzer DNA kit, followed by quantification with real-time PCR. DNA libraries were pooled and sequenced on Illumina MiSeq (Illumina; CA) using a 2 × 250 bp paired-end protocol with overlapping reads.
2. Clinical Global Impression (CGI) was developed for use in clinical trials to provide a brief, stand-alone assessment of the clinician's view of the patient's global functioning prior to and after initiating a study medication. The CGI comprises two companion one-item measures evaluating the following: (a) severity of psychopathology from 1 to 7 (CGI-S) and (b) change from the initiation of treatment on a similar seven-point scale (CGI-I)31.
3. GI symptoms were assessed based on the total number of existing GI symptoms at baseline, including constipation, diarrhea, abdominal pain, excessive flatulence, bloody stool, nausea, difficulty swallowing, poor appetite, indigestion, and acid reflux.
All raw data were recorded and processed in Microsoft Excel 2007 and R. The presentation of data follows the CONSORT recommendations for reporting results of Randomized Clinical Trials (RCTs). Statistical procedures were carried out using α = 0.05 as the significance level.
We applied the Wilcoxon rank-sum test to explore the intergroup differences in the z-scores of weight, height, total scores and sub-scores of ASQ-3, ABC, and SRS at baseline, per-subject changes from 0 to 6 weeks, and per-subject changes from 6 to 12 weeks. Linear mixed models were also used to account for repeated measures.
Due to having several primary outcomes, false discovery rate (FDR) was used to adjust for multiple comparisons. Secondary outcomes were analyzed using similar methods as that of primary outcomes. In addition, linear regression was performed to check for correlations between clinical indices and microbiome compositions.
The sequencing reads were filtered using the QIIME2 (v2019.10) based on quality scores (27). Deblur was used to denoise with default parameters and obtain an abundance table of samples by amplicon sequence variants (ASVs) (28).
Alpha diversities were calculated with QIIME2. Bray-Curtis distance was used to characterize microbiome beta diversity. Taxonomies for ASVs were assigned using the sklearn-based taxonomy classifier trained on the sequences at 99% similarity level from Greengenes v13.8. Significant differences in the relative abundance of microbial phyla, genera, and alpha diversity between placebo and probiotics groups were identified by Kruskal–Wallis tests. A false discovery rate (FDR) based on the Benjamini–Hochberg (BH) adjustment was applied for multiple comparisons (29).
PICRUSt2 was used to infer microbial functional content based on ASVs' abundant tables and then produced the Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologs (KO), Enzyme Classification numbers, and pathway abundance table (30, 31). The differential analyses were performed on the fold ratios between probiotics and placebo groups with a permutation-based non-parametric test, and the top differential features were rendered and plotted with Calour (32). All raw data from 16s rRNA Illumina amplicon sequencing have been deposited in The National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA, PRJNA643297).
A total of 68 subjects with genetically confirmed diagnosis of Prader-Willi syndrome were enrolled. Of which, 33 subjects aged 47.2 ± 32.4 months were randomized to receive active probiotic, BB-11, while 35 subjects aged 53.9 ± 42.0 months were randomized to receive placebo. Groupwise comparisons of baseline age and gender distributions did not indicate any significant differences (P > 0.05). Detailed demographic characteristics and co-morbid GI symptoms of the enrolled participants are summarized in Table 1. The overall severity presented by CGI-S scores at baseline compared between groups is presented in Supplementary Figure 1. No group differences were observed (P > 0.05). 47.5% of subjects display one or more GI symptoms within the study population.
No serious or severe adverse events were observed. All the observed adverse events and major causes of drop out are listed in Supplementary Table 1. There was no significant difference found between the two groups (P > 0.05).
Anthropometric measures were collected and analyzed throughout the treatment course. No significant groupwise differences in mean height (z-score) were observed at baseline (week 0, Figure 2A). Furthermore, we did not observe any significant groupwise difference in the mean change in height (z-score) between 0- to 6-weeks (Figure 2B). The height increase from 6 to 12 weeks was significantly greater in the probiotics group than the placebo group (mean difference = 2.58 cm, P < 0.05, Figure 2C). No significant changes in weight over time were observed in either group (Figures 2D–F).
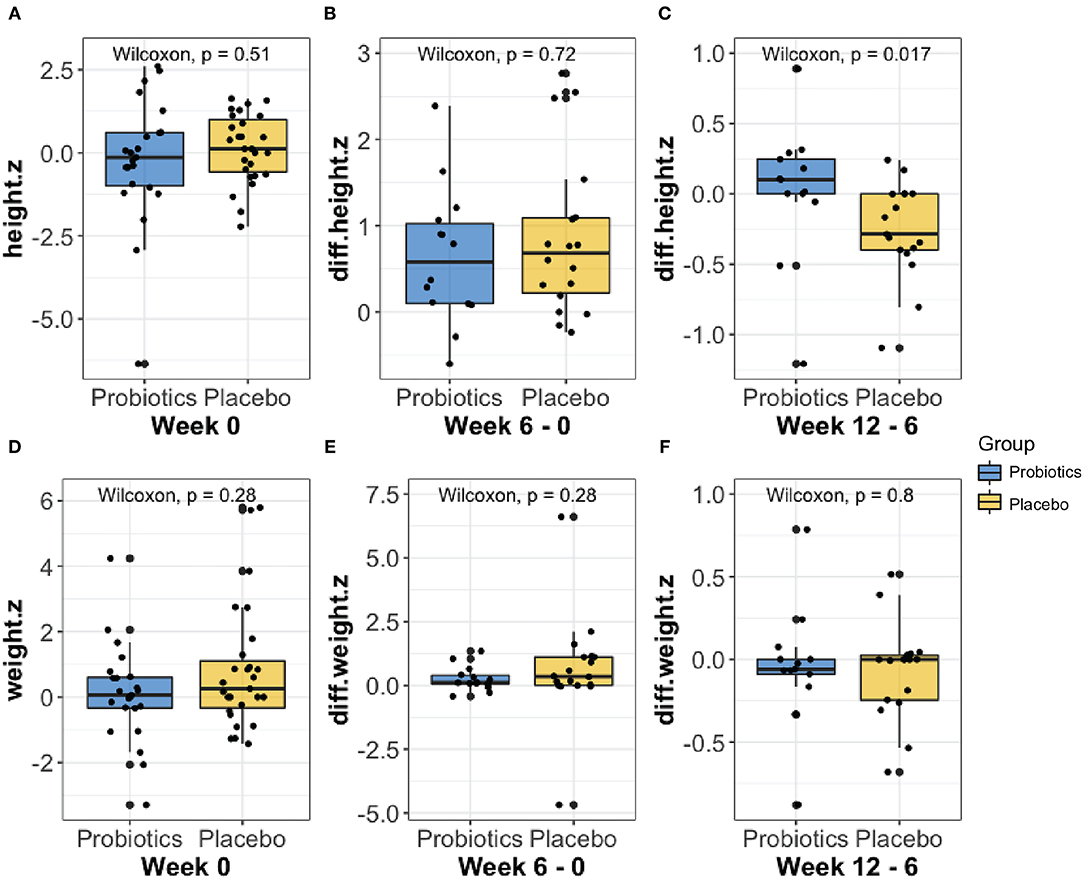
Figure 2. Comparison of the height (A–C) and weight (D–F) z-score changes at baseline, from week 0 to 6, and from week 6 to 12 between probiotic groups (blue) and placebo (yellow) using Wilcoxon rank-sum test. As shown in (C), the probiotics group had a significantly greater height (P < 0.05) increase than the placebo group from week 6 to 12.
Results obtained from psychological measurements, including the ASQ-3, ABC, SRS, and RRB, are shown in Figure 3. No significant difference was found with the linear mixed effect model (P > 0.05) for ASQ-3, ABC, SRS, and RRB scores.
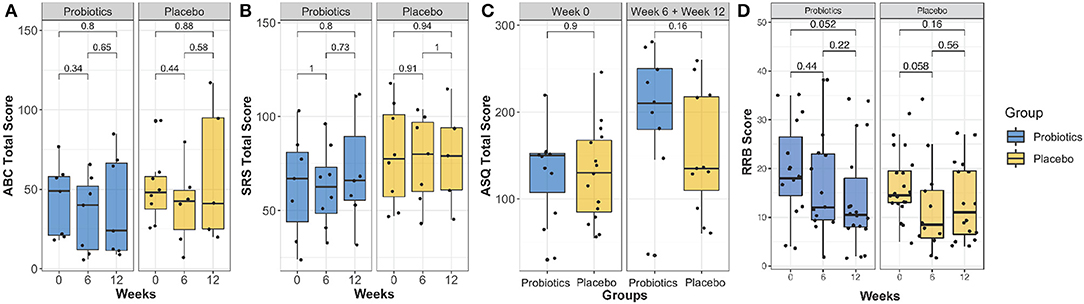
Figure 3. Comparison of the ABC total score (A), SRS-2 total score (B), ASQ-3 total score (C) and RRB score (D) over the intervention course between probiotics group (blue) and placebo group (brown). There was no group significance found (P > 0.05).
The overall improvement of symptoms during the treatment course was measured using the CGI-I scale. We observed significantly greater symptom improvement in the probiotics group compared to the placebo group (Figure 4, P < 0.05).
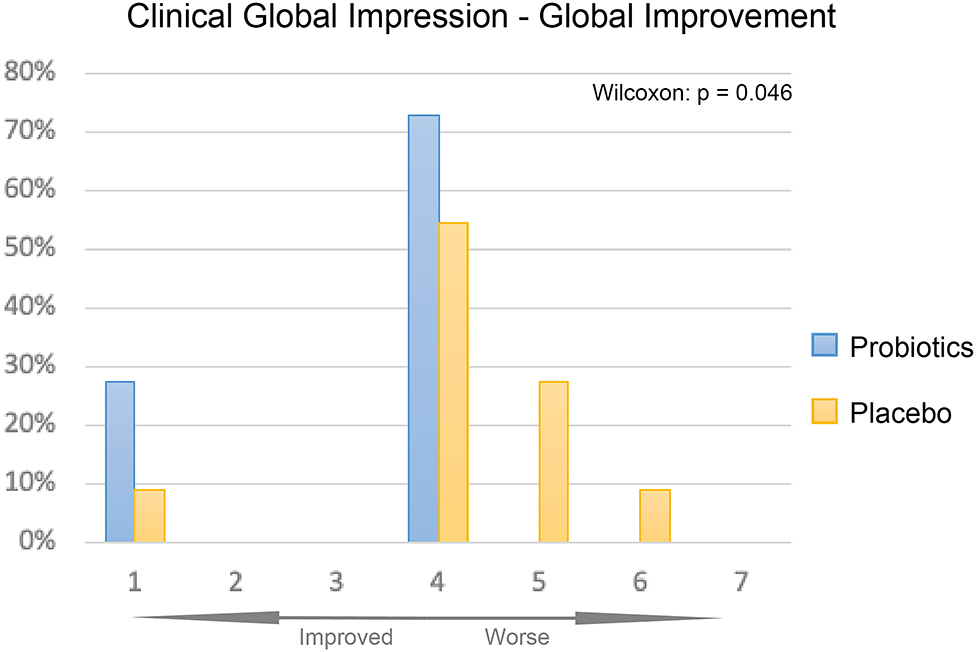
Figure 4. CGI-I of probiotics and placebo at 12 weeks. Percentage of participants given each improvement level was displayed as bar plot, probiotics group (blue) had overall significantly better improvement than the placebo group (yellow, P < 0.05).
After sequencing, we obtained a total of 3,088,722 raw reads and an average of 49,818 reads per sample (range = 29,329–119,440 reads). Overall phylum and genus level variations in gut microbiota composition over the intervention course are shown in Figure 5 for both probiotics and placebo groups.
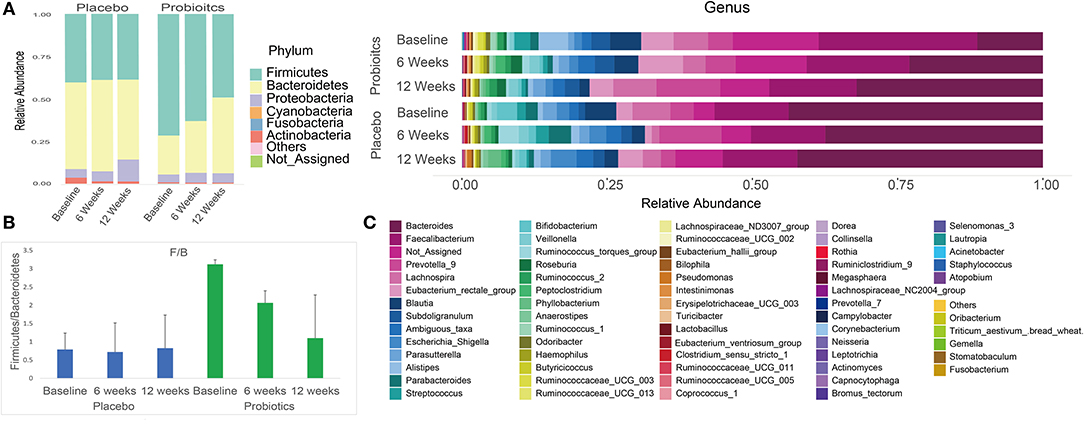
Figure 5. Summary of phylum and genus level gut microbiota relative abundances in both probiotics and placebo group subjects at baseline, 6 and 12 weeks. (A) Phylum level gut microbiota relative abundance per group at each study visit. (B) Firmicutes/Bacteroidetes ratio per group at each study visit. (C) Genus level gut microbiota relative abundance per group at each study visit.
α diversity slightly but significantly increased in the probiotics group compared with the placebo group after 6 weeks (Figures 6A–D). β-diversity, analyzed by a permutational multivariate ANOVA (PERMANOVA), showed a significant separation with probiotics treatment (F-statistic = 2.2526; R2 = 0.035613; P < 0.05, NMDS Stress = 0.19048, Figure 6E).
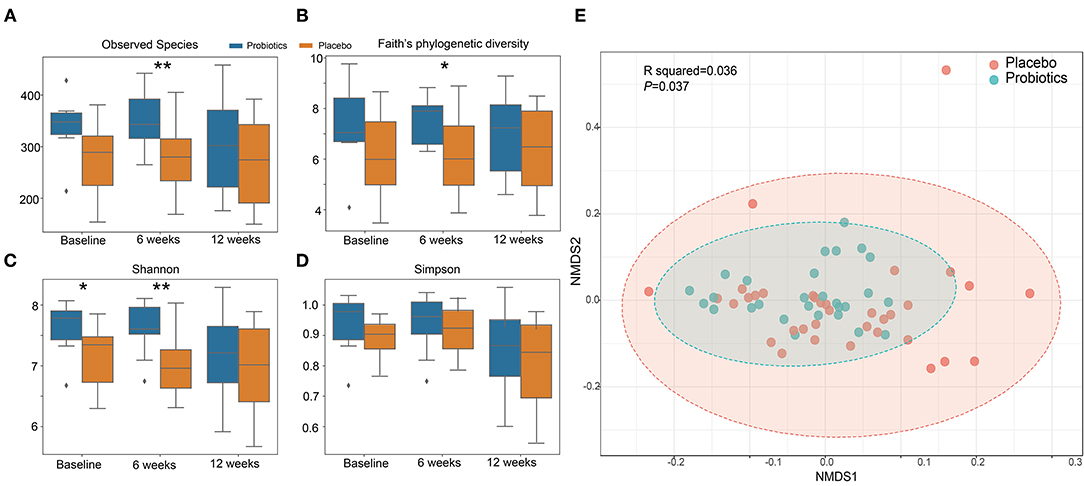
Figure 6. α and β diversity index changes from probiotics intervention. (A): observed species index; (B): faith's phylogenetic diversity; (C): Shannon index; (D): Simpson index. *P < 0.05; **P < 0.01, via t-test. (E): β diversity with Non-metric multidimensional scaling (NMDS) score plots of gut microbial data based on a Bray–Curtis dissimilarity matrix. Placebo (red dots) and probiotics (blue dots).
In order to characterize the change in abundance of potentially clinically significant bacteria over the intervention course, we presented the fold changes of several selected bacterial genera and families in Figure 7. The relative abundances of Lachnospiraceae ND3007, Ruminococcaceae UCG-003, Streptococcus mutans, Comamonadaceae, Alistipes, and Rothia showed decreasing trends from baseline levels in the probiotics group at both 6 and 12 weeks (Figures 7A–F). Among such bacterial taxa, only Comamonadaceae showed a significant decrease among probiotic group subjects at 6 weeks compared to baseline levels (Figure 7D, P < 0.05). In contrast, Bifidobacterium, Lactobacillus, and Prevotella 9 were increased from baseline at 12 weeks in the probiotics group (Figures 7G–I). At the family level, we found similar trends in both groups (Supplementary Figure 2).
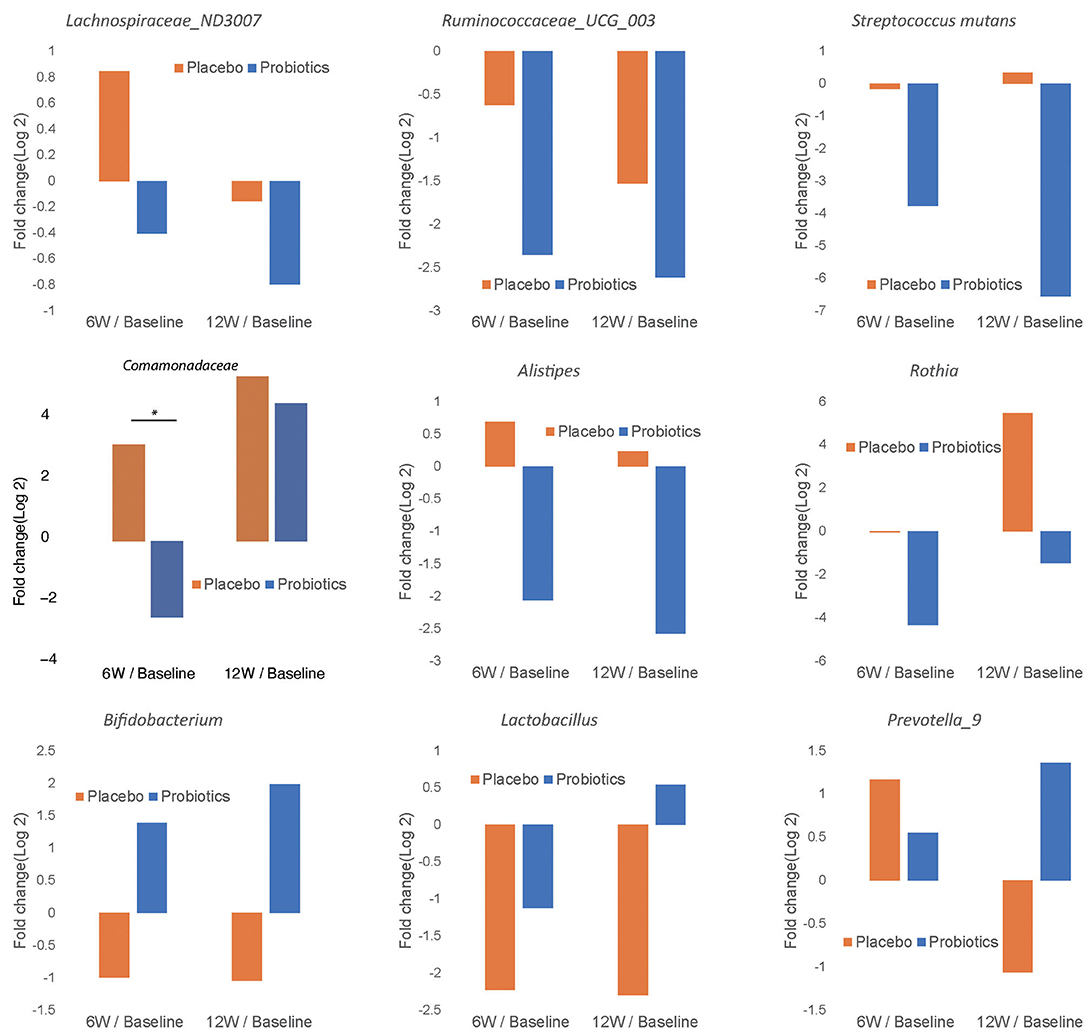
Figure 7. Fold change of relative abundance at genus/species level over the course of intervention for the probiotics group (blue) and placebo (orange). Each bar represents the log 2 transferred relative change of gut microbial abundance of 6 and 12 weeks compared with the baseline. Significant differences are marked with * to indicate P < 0.05.
Functional gene predictive analysis indicated that several genes had different abundances in the probiotics group after the 12-week treatment period. Notably, genes encoding the ubiquinone biosynthesis protein (ubiB, k03688), phytoene desaturase (EC:1.3.99.29), phytoene desaturase (lycopene-forming) (EC:1.3.99.31), and all-trans-zeta-carotene desaturase (EC:1.3.99.26) were all upregulated, while the genes encoding dimethylargininase (k01482) and acid phosphatase (phoN, k09474, EC:3.1.3.2) were downregulated (Figure 8). These findings do not meet the false discovery criteria for significance with multiple comparisons. The analysis results from the predicted KEGG pathway, shown in Supplementary Figure 3, and the predicted KO, shown in Supplementary Figure 4, further compare the gene expression of the probiotics and placebo groups.
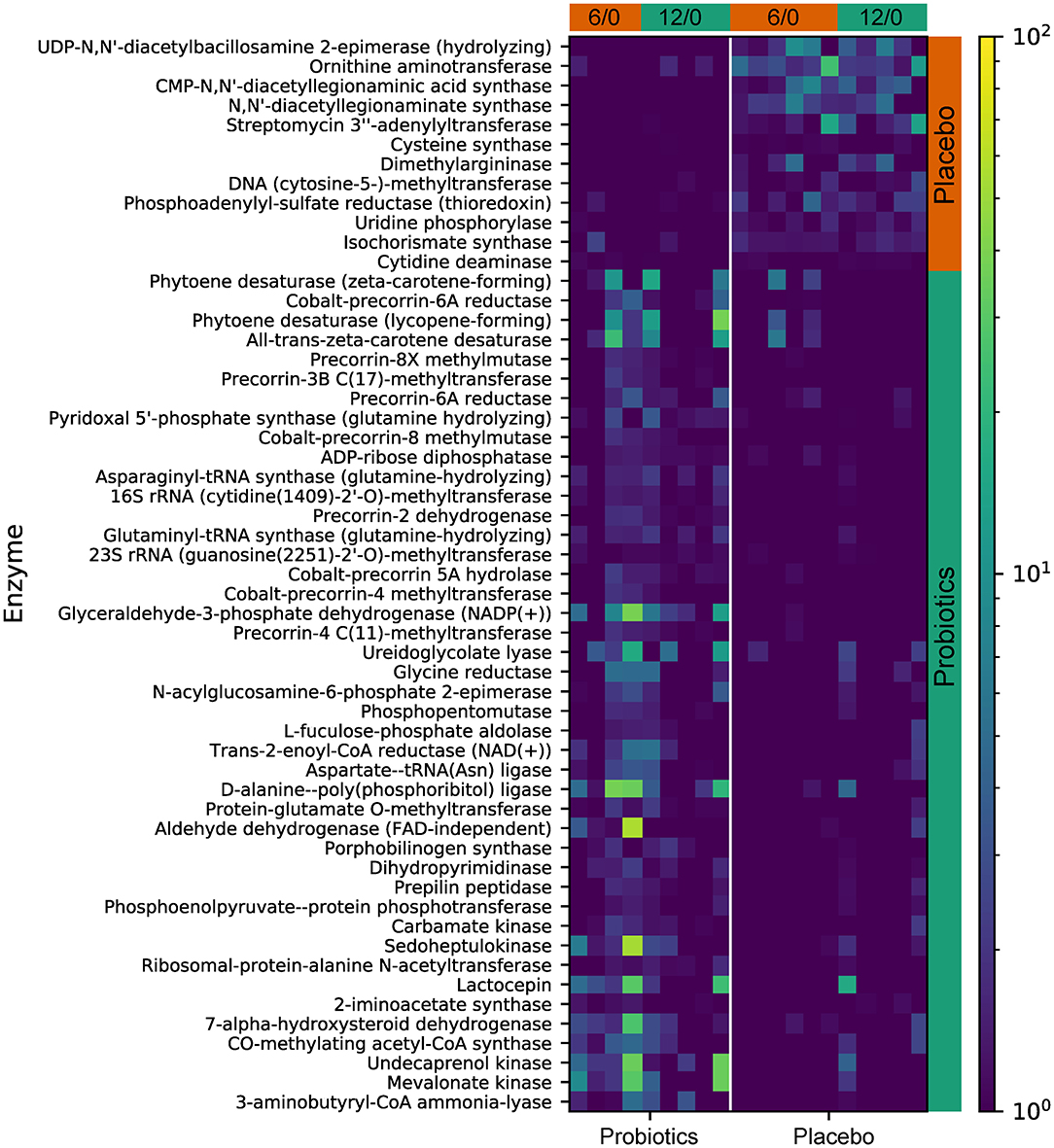
Figure 8. The predicted KEGG enzyme abundance based on PICRUSt2 functional gene analysis for the probiotics and placebo groups. The average abundance of KEGG enzyme differentially enriched in placebo and probiotics according to level 3.
Clinical indices were correlated with the abundance of bacterial genera; one correlation was found to be significant in the probiotics group while no significant correlations were observed in the placebo group. Specifically, a positive correlation was discovered between the RRB scores and Rothia in the probiotics group at week 6 (Figure 9, R = 0.97, P < 0.005).
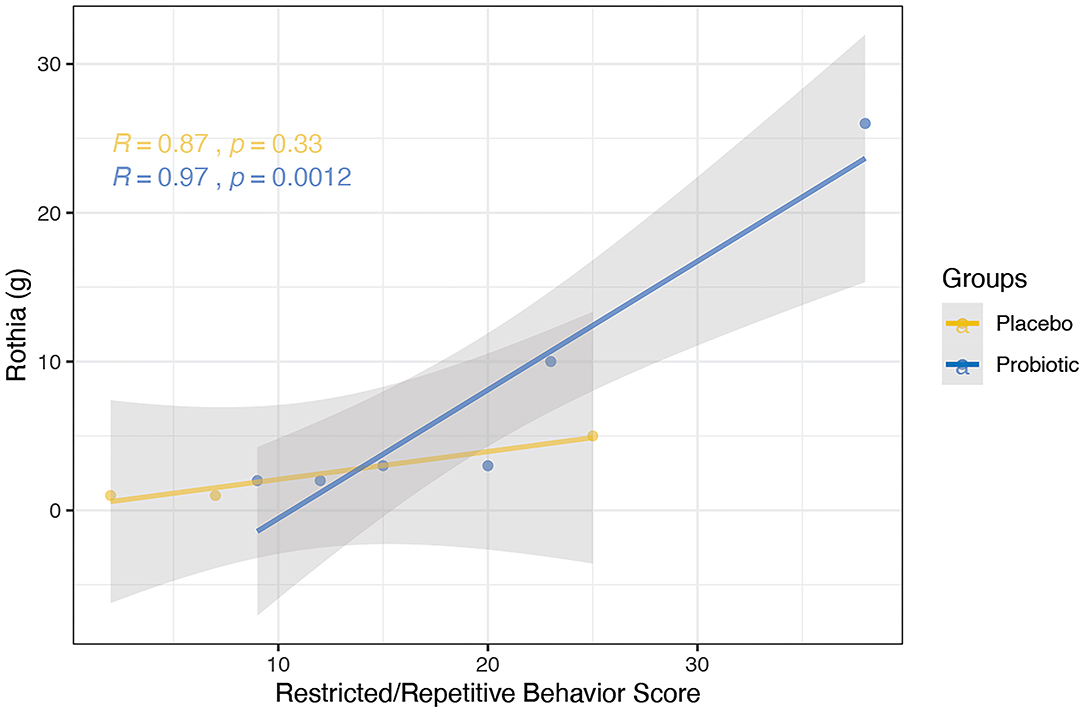
Figure 9. Correlation between the abundance of bacterial genera and clinical indices using spearman's method was performed for the probiotics (blue) and placebo (yellow) group at the 6-week time point. Probiotic group showed positive correlation between RRB scores and Rothia (R = 0.97, P < 0.005). No significant correlation was observed in the placebo group.
In our 12-week, randomized, double-blind, placebo-controlled trial of 68 PWS patients, BL-11 increased height in PWS subjects without changing weight. We observed that, during the treatment period, the probiotics group had a significantly greater height increase than the placebo group (P < 0.05). Interventions with other probiotics in the past have failed to elicit improvement in height (33). This study provides novel evidence for the use of BL-11 as an early intervention for patients with PWS. Interventions that lead to increased height in PWS may benefit patients at early developmental stages most and substantially improve long-term prognosis. PWS individuals were found to have absolute or functional Growth Hormone (GH) deficiency, and GH replacement is currently the most effective treatment for PWS (34, 35). GH was found not only to increase height, but also decrease body fat and improve cognition, motor, and mental function. With earlier initiation of GH treatment, better efficacy and prognostic benefit have been observed (35). One study found probiotics L. reuteri could increase growth hormone level in mice (36), which reveals a potential mechanism by which probiotics can enhance height and treat PWS patients: promotion of endogenous growth hormone release. Our findings warrant further investigation into the biological mechanisms of probiotics, a promising intervention for PWS with better tolerance and convenience than GH replacement (33).
We did not observe significant weight reduction within the intervention period, possibly due to the majority of our participants being < 5 years old, an age range at which obesity is not yet a major problem. Interestingly, the microbiome composition changes we observed with the intervention of B. lactis have been previously linked to weight or adiposity reduction (14–19, 22), improve fasting insulin sensitivity (22) and inflammatory attenuation (20, 21). Notably, we found a significant separation of the gut microbiome β-diversity between the probiotics and the placebo group after treatment. Baseline β-diversity has been directly correlated with long-term weight loss when adhering to a controlled diet (37). Therefore, probiotics supplementation may have preventative effects or may facilitate diet-induced weight reduction.
After administration of BL-11, we also noted reduction in the abundance of several bacterial genera and species that have been implicated in the pathology of obesity and associated inflammation. Ruminococcaceae UCG-003, associated with VLDL and metabolic syndrome, has also been implicated in inflammatory bowel diseases (38, 39). Lachnospiraceae ND3007 has been linked to elevated cholesterol, signs of insulin resistance, and infant obesity (40–42). Elevated Streptococcus has been associated with inflammatory GI disorders, maternal inflammation, bacteremia, and antibiotic use during pregnancy (43, 44). Rothia was found to have a higher abundance in a gestational diabetic cohort than a healthy pregnant cohort (45). The Comamonadaceae family is generally regarded as pathogenic in humans (46).
Conversely, Bifidobacterium, Lactobacillus, and Prevotella were each found to be considerably increased in the gut after BL-11 treatment. Bifidobacterium, the genus to which the interventional probiotic belongs, is widely regarded as beneficial to gut health and weight reduction (14–18, 20, 47–51). Lactobacillus, in addition to having protective effects against weight gain in humans, has been found to inhibit the activity of proinflammatory interleukins, which have been linked to obesity and poor obesity-related outcomes (52–54). The effect of Prevotella in the gut microbiome remains uncertain, as evidence linking this genus to health benefit and disease have both been reported. Wang et al. (55) reported that Prevotella-9 was found to be significantly decreased in both mice put on a high-fat diet and Zeng et al. (56) reported the same in women with PCOS who were insulin-resistant (55, 56). Further, Park et al. (57) reported an increased abundance of Prevotella in obesity improved mice and Kovatcheva-Datchary et al. (58) reported dietary fiber-induced improvements in post-prandial blood glucose and insulin were found to be positively associated with the abundance of Prevotella (57, 58). On the other hand, one study found that the Prevotellaceae family had greater relative abundance in 3 obese patients, compared to 3 normal weight patients (59). Another study, investigating fecal bacteria composition in HIV-positive patients, found that Prevotella was positively correlated with BMI, although most participants in this study had a BMI within the normal range (60). The conflicting findings about Prevotella in gut health and obesity may indicate the importance of balancing the abundance of this genus within the microbiome.
Furthermore, by using predictive functional gene analysis, we found the enhancement of antioxidant production-related pathways that exert anti-inflammatory and anti-obesity effects. The gene encoding the ubiquinone biosynthesis protein (ubiB, k03688), responsible for the biosynthesis of ubiquinone (CoQ10), was found to have increased abundance following probiotics treatment. CoQ10 supplementation can be useful in the treatment of a variety of chronic cardiovascular, inflammatory, and obesity-related disease (61). We also found an elevated abundance of genes encoding phytoene desaturase (EC:1.3.99.29), phytoene desaturase (lycopene-forming), (EC:1.3.99.31), and all-trans-zeta-carotene desaturase (EC:1.3.99.26), which all contribute to the biosynthesis of carotenoids, previously found to have beneficial effects on obesity and obesity-associated pathologies (62–64). We also found downregulation of two enzymes, dimethylargininase (k01482) and acid phosphatase (phoN, k09474, EC:3.1.3.2), which have been linked to obesity development and elevated cholesterol and triglyceride levels in human patients (65–67).
Taken together, the microbiome composition data and predictive functional gene analysis indicate that the diversity separation caused by BL-11 probiotics treatment favors protection against obesity and obesity-related pathology.
Although we did not find a significant change in psychological measurements (ASQ-3, ABC, SRS, and RRB), CGI-I showed significant overall improvement in the probiotics group after the treatment period compared with the placebo group (P < 0.05).
Interestingly, we found that the RRB score was positively correlated with Rothia at the genus level (P < 0.005). RRB is one of the core symptoms of ASD, which has been reported in as many as 25–40% of PWS cases (3, 68). Rothia, in addition to being linked to diabetes (45), has been reported to be more prevalent in children with ASD than typically-developing children (12.2-fold-change; FDR, P < 0.05) (69). While the mechanism by which the BL-11 improved clinical impression of PWS patients is unknown, both the correlation found between Rothia and RRB and alterations in the gut microbiome composition may be associated with the administration of BL-11. Such findings may further implicate modulatory effects on brain functions or host metabolism via signaling through the gut-brain axis. Further investigation of Rothia and other microbiome markers may reveal potent and feasible targets for neuropsychiatric therapies.
Our randomized trial showed that treatment with probiotic B. Lactis strain (BL-11) for 12 weeks significantly increased height, a novel finding with vital implications for early treatment in PWS. Probiotic treatment may improve overall psychological clinical symptoms, as suggested by CGI-I results. In conjunction with its observed effects in inducing favorable alterations in the gut microbiome composition and functional profile, our results suggest that B. lactis is a viable probiotic candidate for facilitating the improvement in obesity-related gut microbiome dysbiosis in individuals with PWS, thereby potentially inducing a reduction of obesity in such populations. There are some limitations to the study that deserve consideration. First, despite our adoption of proper subject recruitment procedures, subject retention strategies, and data collection plans, the enrollment, retention, and data collection of PWS participants were challenging, thus leading to reduced sample sizes as anticipated and missing data, which limited further subgroup analysis. Second, although there was no statistical difference in clinical indices between the probiotics and placebo groups at baseline, the broad age range of enrolled participants in this study resulted in high subject population heterogeneity and may contribute to the variability of the treatment efficacy. Third, assessment of fecal microbiome was not controlled for dietary habits, which may influence the microbial abundances at the individual level. Thus, future studies with larger sample sizes, improved control for environmental factors, and subgroup stratification are warranted. Due to the limitations of the study listed above, further studies are warranted to investigate the mechanism and efficacy of BL-11 probiotics treatment in PWS.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA643297.
The studies involving human participants were reviewed and approved by Internal Review Board (IRB) of the Second Affiliated Hospital of Kunming Medical University. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
X-JK conceived the concept, developed experimental design and trial protocol, provided resources and major funding, led the trial conduction, finished the first draft, and major revisions of the manuscript. XL, XZ, RT, SL, JZ, JW, YW, ZY, CS, XCu, TD, MF, and HC contributed on subject interview and data collection. XCa and GW helped admin works. RT and KL contributed on data analysis and creation of figures and tables while JL provided assistance. KL, CC, BW, HS, and AL contributed writing and editing. All authors have read and agreed to the published version of the manuscript.
This study was primarily funded by Massachusetts General Hospital funding (233263) awarded to X-JK, which funded the compensation of Kong lab staff for data collection, training of non-MGH staff, coordination, and data analysis; additionally, the Shenzhen Sanming Project of Medicine (SZSM201512009) and Yunnan Province Li Bin Expert Work Station Foundation (2019IC034) helped fund the compensation of staff hours.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We highly appreciate Drs. Gang Fu, Qing Song, Peter Huge, Yuanyuan Liang, Hui Tan from Baheal Pharmaceutical Group for their generous lab support, we highly appreciate Dr. Heng Yang from Institute of Basic Medical Sciences of Chinese Academy of Medical Sciences and Ms. Yanan Wang from Suzhou Func Biotech Inc for their great helps of 16S rRNA Gene Amplicon Sequencing, we highly appreciate Dr. Yunfeng Duan from Institute of Microbiology of Chinese Academy of Science for his generous help in blinded microbiome analysis, we also appreciate Ms. Ye Zhang from Hebei Xiongnan Biotechnology Co., Ltd. for her help on probiotics and placebo manufacturing and transportation.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.587974/full#supplementary-material
1. Cassidy SB, Schwartz S, Miller JL, Driscoll DJ. Prader-Willi syndrome. Genet Med. (2012) 14:10–26. doi: 10.1038/gim.0b013e31822bead0
2. Cassidy SB, Forsythe M, Heeger S, Nicholls RD, Schork N, Benn P, et al. Comparison of phenotype between patients with Prader-Willi syndrome due to deletion 15q and uniparental disomy 15. Am J Med Genet. (1997) 68:433–40. doi: 10.1002/(SICI)1096-8628(19970211)68:4<433::AID-AJMG12>3.0.CO;2-T
3. Salehi P, Herzig L, Capone G, Lu A, Oron AP, Kim S. Comparison of aberrant behavior checklist profiles across Prader-Willi syndrome, Down syndrome, and autism spectrum disorder. Am J Med Genet A. (2018) 176:2751–9. doi: 10.1002/ajmg.a.40665
4. Olsson LM, Poitou C, Tremaroli V, Coupaye M, Aron-Wisnewsky J, Bäckhed F, et al. Gut microbiota of obese subjects with Prader-Willi syndrome is linked to metabolic health. Gut. (2020) 69:1229–38. doi: 10.1136/gutjnl-2019-319322
5. Zhang C, Yin A, Li H, Wang R, Wu G, Shen J, et al. Dietary modulation of gut microbiota contributes to alleviation of both genetic and simple obesity in children. Ebiomedicine. (2015) 2:968–84. doi: 10.1016/j.ebiom.2015.07.007
6. Deng L, Wang R, Li H, Zhang C, Zhao L, Zhang M. miRNA-gene regulatory network in gnotobiotic mice stimulated by dysbiotic gut microbiota transplanted from a genetically obese child. Front Microbiol. (2019) 10:1517. doi: 10.3389/fmicb.2019.01517
7. Ke X, Walker A, Haange S-B, Lagkouvardos I, Liu Y, Schmitt-Kopplin P, et al. Synbiotic-driven improvement of metabolic disturbances is associated with changes in the gut microbiome in diet-induced obese mice. Mol Metab. (2019) 22:96–109. doi: 10.1016/j.molmet.2019.01.012
8. Hibberd AA, Yde CC, Ziegler ML, Honoré AH, Saarinen MT, Lahtinen S, et al. Probiotic or synbiotic alters the gut microbiota and metabolism in a randomised controlled trial of weight management in overweight adults. Benef Microbes. (2019) 10:121–35. doi: 10.3920/BM2018.0028
9. Vindegaard N, Speyer H, Nordentoft M, Rasmussen S, Benros ME. Gut microbial changes of patients with psychotic and affective disorders: a systematic review. Schizophr Res. (2020) 19:30584–5. doi: 10.1016/j.schres.2019.12.014
10. Kong X, Liu J, Cetinbas M, Sadreyev R, Koh M, Huang H, et al. New and preliminary evidence on altered oral and gut microbiota in individuals with Autism Spectrum Disorder (ASD): implications for ASD diagnosis and subtyping based on microbial biomarkers. Nutrients. (2019) 11:2128. doi: 10.3390/nu11092128
11. Liu J, Wan G, Huang M, Agyapong G, Zou T, Zhang X, et al. Probiotic therapy for treating behavioral and gastrointestinal symptoms in autism spectrum disorder: a systematic review of clinical trials. Curr Medical Sci. (2019) 39:173–84. doi: 10.1007/s11596-019-2016-4
12. Dickerson F, Adamos M, Katsafanas E, Khushalani S, Origoni A, Savage C, et al. Adjunctive probiotic microorganisms to prevent rehospitalization in patients with acute mania: a randomized controlled trial. Bipolar Disord. (2018) 20:614–21. doi: 10.1111/bdi.12652
13. Ventura M, Zink R. Rapid identification, differentiation, and proposed new taxonomic classification of bifidobacterium lactis. Appl Environ Microb. (2002) 68:6429–34. doi: 10.1128/AEM.68.12.6429-6434.2002
14. Huo Y, Lu X, Wang X, Wang X, Chen L, Guo H, et al. Bifidobacterium animalis subsp. lactis A6 alleviates obesity associated with promoting mitochondrial biogenesis and function of adipose tissue in mice. Molecules. (2020) 25:1490. doi: 10.3390/molecules25071490
15. Carreras NL, Martorell P, Chenoll E, Genovés S, Ramón D, Aleixandre A. Anti-obesity properties of the strain Bifidobacterium animalis subsp. lactis CECT 8145 in Zücker fatty rats. Benef Microbes. (2018) 9:629–41. doi: 10.3920/BM2017.0141
16. Pedret A, Valls RM, Calderón-Pérez L, Llauradó E, Companys J, Pla-Pagà L, et al. Effects of daily consumption of the probiotic Bifidobacterium animalis subsp. lactis CECT 8145 on anthropometric adiposity biomarkers in abdominally obese subjects: a randomized controlled trial. Int J Obesity. (2019) 43:1863–8. doi: 10.1038/s41366-018-0220-0
17. Barz ML, Daniel N, Varin TV, Naimi S, Demers-Mathieu V, Pilon G, et al. In vivo screening of multiple bacterial strains identifies Lactobacillus rhamnosus Lb102 and Bifidobacterium animalis ssp. lactis Bf141 as probiotics that improve metabolic disorders in a mouse model of obesity. Faseb J. (2019) 33:4921–35. doi: 10.1096/fj.201801672R
18. Uusitupa H-M, Rasinkangas P, Lehtinen MJ, Mäkelä SM, Airaksinen K, Anglenius H, et al. Bifidobacterium animalis subsp. lactis 420 for metabolic health: review of the research. Nutrients. (2020) 12:892. doi: 10.3390/nu12040892
19. Mekkes MC, Weenen TC, Brummer RJ, Claassen E. The development of probiotic treatment in obesity: a review. Benef Microbes. (2014) 5:19–28. doi: 10.3920/BM2012.0069
20. Ibarra A, Latreille-Barbier M, Donazzolo Y, Pelletier X, Ouwehand AC. Effects of 28-day Bifidobacterium animalis subsp. lactis HN019 supplementation on colonic transit time and gastrointestinal symptoms in adults with functional constipation: A double-blind, randomized, placebo-controlled, and dose-ranging trial. Gut Microbes. (2017) 9:236–51. doi: 10.1080/19490976.2017.1412908
21. Meng H, Ba Z, Lee Y, Peng J, Lin J, Fleming JA, et al. Consumption of Bifidobacterium animalis subsp. lactis BB-12 in yogurt reduced expression of TLR-2 on peripheral blood-derived monocytes and pro-inflammatory cytokine secretion in young adults. Eur J Nutr. (2017) 56:649–61. doi: 10.1007/s00394-015-1109-5
22. GROUP WMGRS, Onis M. WHO child growth standards based on length/height, weight and age. Acta Paediatr. (2006) 95:76–85. doi: 10.1111/j.1651-2227.2006.tb02378.x
23. Bricker DD, Squires J, Potter L. Ages & Stages Questionnaires. Baltimore, MD: Paul H. Brookes Publishing Co. (2009).
24. Aman MG, Singh NN, Stewart AW, Field CJ. The aberrant behavior checklist: a behavior rating scale for the assessment of treatment effects. Am J Ment Retard. (1985) 89:485–91. doi: 10.1037/t10453-000
25. Constantino JN, Gruber CP. Social Responsive Scale Manual (2nd ed., SRS-2). Los Angeles, CA: Western Psychological Services (2012).
27. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. (2019) 37:852–7. doi: 10.1038/s41587-019-0209-9
28. Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Xu ZZ, et al. Deblur rapidly resolves single-nucleotide community sequence patterns. Msystems. (2017) 2:e00191–16. doi: 10.1128/mSystems.00191-16
29. Jiang L, Amir A, Morton JT, Heller R, Arias-Castro E, Knight R. Discrete false-discovery rate improves identification of differentially abundant microbes. Msystems. (2017) 2:e00092–17. doi: 10.1128/mSystems.00092-17
30. Douglas GM, Maffei VJ, Zaneveld J, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2: an improved and customizable approach for metagenome inference. Biorxiv. [Preprint]. (2020) 672295. doi: 10.1101/672295
31. Czech L, Barbera P, Stamatakis A. Genesis and Gappa: processing, analyzing and visualizing phylogenetic (placement) data. Bioinformatics. (2020) 36:3263–65. doi: 10.1093/bioinformatics/btaa070
32. Xu ZZ, Amir A, Sanders J, Zhu Q, Morton JT, Bletz MC, et al. Calour: an interactive, microbe-centric analysis tool. Msystems. (2019) 4:e00269–18. doi: 10.1128/mSystems.00269-18
33. Onubi OJ, Poobalan AS, Dineen B, Marais D, McNeill G. Effects of probiotics on child growth: a systematic review. J Heal Popul Nutrition. (2015) 34:8. doi: 10.1186/s41043-015-0010-4
34. Zhu J, Kong X. Current treatments of prader-willi syndrome: a systematic review. N A J Med Sci. (2018) 10:156–63. doi: 10.7156/najms.2017.1004156
35. Bakker NE, Lindberg A, Heissler J, Wollmann HA, Camacho-Hübner C, Hokken-Koelega AC, et al. Growth hormone treatment in children with prader-willi syndrome: three years of longitudinal data in prepubertal children and adult height data from the KIGS database. J Clin Endocrinol Metabolism. (2017) 102:1702–11. doi: 10.1210/jc.2016-2962
36. Varian BJ, Gourishetti S, Poutahidis T, Lakritz JR, Levkovich T, Kwok C, et al. Correction: Beneficial bacteria inhibit cachexia. Oncotarget. (2018) 9:29536. doi: 10.18632/oncotarget.25750
37. Grembi JA, Nguyen LH, Haggerty TD, Gardner CD, Holmes SP, Parsonnet J. Gut microbiota plasticity is correlated with sustained weight loss on a low-carb or low-fat dietary intervention. Sci Rep-uk. (2020) 10:1405. doi: 10.1038/s41598-020-68280-z
38. Vojinovic D, Radjabzadeh D, Kurilshikov A, Amin N, Wijmenga C, Franke L, et al. Relationship between gut microbiota and circulating metabolites in population-based cohorts. Nat Commun. (2019) 10:5813. doi: 10.1038/s41467-019-13721-1
39. Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. (2017) 9:103. doi: 10.1186/s13073-017-0490-5
40. Wang J, Xiong K, Zhao S, Zhang C, Zhang J, Xu L, et al. Long-term effects of multi-drug-resistant tuberculosis treatment on gut microbiota and its health consequences. Front Microbiol. (2020) 11:53. doi: 10.3389/fmicb.2020.00053
41. Liang Y, Ming Q, Liang J, Zhang Y, Zhang H, Shen T. Gut microbiota dysbiosis in Polycystic ovary syndrome (PCOS): association with obesity - a preliminary report. Can J Physiol Pharm. (2019) 98:803–9. doi: 10.1139/cjpp-2019-0413
42. Tun HM, Bridgman SL, Chari R, Field CJ, Guttman DS, Becker AB, et al. Roles of birth mode and infant gut microbiota in intergenerational transmission of overweight and obesity from mother to offspring. Jama Pediatr. (2018) 172:368. doi: 10.1001/jamapediatrics.2017.5535
43. Iakovlev AA, Volkov A, Bashtovaya O, Shcherbakova I, Ghazudin T, Leonova G, et al. P081 The structure of the proteomic profile and gut microbiota in patients with primary immunodeficiency, common variable immunodeficiency and inflammatory bowel disease. J Crohn's Colitis. (2020) 14:S175–6. doi: 10.1093/ecco-jcc/jjz203.210
44. Li N, Yang J, Zhang J, Liang C, Wang Y, Chen B, et al. Correlation of gut microbiome between ASD children and mothers and potential biomarkers for risk assessment. Genom Proteom Bioinform. (2019) 17:26–38. doi: 10.1016/j.gpb.2019.01.002
45. Crusell MKW, Hansen TH, Nielsen T, Allin KH, Rühlemann MC, Damm P, et al. Gestational diabetes is associated with change in the gut microbiota composition in third trimester of pregnancy and postpartum. Microbiome. (2018) 6:89. doi: 10.1186/s40168-018-0472-x
46. Willems A. The Prokaryotes, Alphaproteobacteria and Betaproteobacteria. Berlin; Heidelberg: Springer (2014). p. 777–851. doi: 10.1007/978-3-642-30197-1_238
47. Dimidi E, Zdanaviciene A, Christodoulides S, Taheri S, Louis P, Duncan PI, et al. Randomised clinical trial: Bifidobacterium lactis NCC2818 probiotic vs placebo, and impact on gut transit time, symptoms, and gut microbiology in chronic constipation. Aliment Pharm Therap. (2019) 49:251–64. doi: 10.1111/apt.15073
48. Alyousif Z, Miller JL, Sandoval MY, MacPherson CW, Nagulesapillai V, Dahl WJ. The effects of Bifidobacterium animalis ssp. lactis B94 on gastrointestinal wellness in adults with Prader-Willi syndrome: study protocol for a randomized controlled trial. Trials. (2018) 19:256. doi: 10.1186/s13063-018-2648-x
49. Oliveira LFF, Salvador SL, Silva PHF, Furlaneto FAC, Figueiredo L, Casarin R, et al. Benefits of Bifidobacterium animalis subsp. lactis Probiotic in Experimental Periodontitis. J Periodontol. (2017) 88:197–208. doi: 10.1902/jop.2016.160217
50. Taipale TJ, Pienihäkkinen K, Isolauri E, Jokela JT, Söderling EM. Bifidobacterium animalis subsp. lactis BB-12 in reducing the risk of infections in early childhood. Pediatr Res. (2016) 79:65–9. doi: 10.1038/pr.2015.174
51. Li S-C, Hsu W-F, Chang J-S, Shih C-K. Combination of Lactobacillus acidophilus and Bifidobacterium animalis subsp. lactis shows a stronger anti-inflammatory effect than individual strains in HT-29 cells. Nutrients. (2019) 11:969. doi: 10.3390/nu11050969
52. Rosing JA, Walker KC, Jensen BAH, Heitmann BL. Oral Lactobacillus counts predict weight gain susceptibility: a 6-year follow-up study. Obesity Facts. (2017) 10:473–82. doi: 10.1159/000478095
53. Ayyanna R, Ankaiah D, Arul V. Anti-inflammatory and antioxidant properties of probiotic bacterium Lactobacillus mucosae AN1 and Lactobacillus fermentum SNR1 in Wistar Albino rats. Front Microbiol. (2018) 9:3063. doi: 10.3389/fmicb.2018.03063
54. Cox AJ, West NP, Cripps AW. Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endocrinol. (2015) 3:207–15. doi: 10.1016/S2213-8587(14)70134-2
55. Wang X, Chen D, Li Y, Zhao S, Chen C, Ning D. Alleviating effects of walnut green husk extract on disorders of lipid levels and gut bacteria flora in high fat diet-induced obesity rats. J Funct Food. (2019) 52:576–86. doi: 10.1016/j.jff.2018.11.022
56. Zeng B, Lai Z, Sun L, Zhang Z, Yang J, Li Z, et al. Structural and functional profiles of the gut microbial community in polycystic ovary syndrome with insulin resistance (IR-PCOS): a pilot study. Res Microbiol. (2018) 170:43–52. doi: 10.1016/j.resmic.2018.09.002
57. Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. (2013) 17:141–52. doi: 10.1016/j.cmet.2012.12.007
58. Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of prevotella. Cell Metab. (2015) 22:971–82. doi: 10.1016/j.cmet.2015.10.001
59. Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, et al. Human gut microbiota in obesity and after gastric bypass. Proc National Acad Sci USA. (2009) 106:2365–70. doi: 10.1073/pnas.0812600106
60. Pinto-Cardoso S, Lozupone C, Briceño O, Alva-Hernández S, Téllez N, Adriana A, et al. Fecal Bacterial Communities in treated HIV infected individuals on two antiretroviral regimens. Sci Rep-uk. (2017) 7:43741. doi: 10.1038/srep43741
61. Zozina VI, Covantev S, Goroshko OA, Krasnykh LM, Kukes VG. Coenzyme Q10 in cardiovascular and metabolic diseases: current state of the problem. Curr Cardiol Rev. (2018) 14:164–74. doi: 10.2174/1573403X14666180416115428
62. Paes-Silva RP, Gadelha PCFP, Lemos M da CC de, Castro CMMB de, Arruda IKG de, Diniz A da S. Adiposity, inflammation and fat-soluble vitamins in adolescents. J Pediat. (2018) 95:575–83. doi: 10.1016/j.jpedp.2018.05.011
63. Mounien L, Tourniaire F, Landrier J-F. Anti-Obesity effect of carotenoids: direct impact on adipose tissue and adipose tissue-driven indirect effects. Nutrients. (2019) 11:1562. doi: 10.3390/nu11071562
64. Wiese M, Bashmakov Y, Chalyk N, Nielsen DS, Krych Ł, Kot W, et al. Prebiotic effect of lycopene and dark chocolate on gut microbiome with systemic changes in liver metabolism, skeletal muscles and skin in moderately obese persons. Biomed Res Int. (2019) 2019:1–15. doi: 10.1155/2019/4625279
65. Arlouskaya Y, Sawicka A, Głowala M, Giebułtowicz J, Korytowska N, Tałałaj M, et al. Asymmetric Dimethylarginine (ADMA) and Symmetric Dimethylarginine (SDMA) concentrations in patients with obesity and the risk of obstructive Sleep Apnea (OSA). J Clin Medicine. (2019) 8:897. doi: 10.3390/jcm8060897
66. Lång P, Zakaroff-Girard A, Wåhlén K, Andersson J, Olsson T, Bambace C, et al. Expression and secretion of the novel adipokine tartrate-resistant acid phosphatase from adipose tissues of obese and lean women. Int J Obesity. (2011) 35:1502–10. doi: 10.1038/ijo.2011.17
67. Bottini N, MacMurray J, Peters W, Rostamkhani M, Comings DE. Association of the acid phosphatase (ACP1) gene with triglyceride levels in obese women. Mol Genet Metab. (2002) 77:226–9. doi: 10.1016/S1096-7192(02)00120-8
68. Bennett JA, Germani T, Haqq AM, Zwaigenbaum L. Autism spectrum disorder in Prader-Willi syndrome: a systematic review. Am J Med Genet A. (2015) 167:2936–44. doi: 10.1002/ajmg.a.37286
Keywords: Prader-Willi syndrome, microbiome, probiotics, Bifidobacterium animalis subsp lactis, obesity, inflammation, height, microbiota (microorganism)
Citation: Kong X-J, Wan G, Tian R, Liu S, Liu K, Clairmont C, Lin X, Zhang X, Sherman H, Zhu J, Wang Y, Fong M, Li A, Wang BK, Wang J, Liu J, Yu Z, Shen C, Cui X, Cao H, Du T and Cao X (2021) The Effects of Probiotic Supplementation on Anthropometric Growth and Gut Microbiota Composition in Patients With Prader-Willi Syndrome: A Randomized Double-Blinded Placebo-Controlled Trial. Front. Nutr. 8:587974. doi: 10.3389/fnut.2021.587974
Received: 29 July 2020; Accepted: 02 February 2021;
Published: 19 February 2021.
Edited by:
Silvia Turroni, University of Bologna, ItalyReviewed by:
Funmilola Abidemi Ayeni, University of Ibadan, NigeriaCopyright © 2021 Kong, Wan, Tian, Liu, Liu, Clairmont, Lin, Zhang, Sherman, Zhu, Wang, Fong, Li, Wang, Wang, Liu, Yu, Shen, Cui, Cao, Du and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xue-Jun Kong, eGtvbmcxQG1naC5oYXJ2YXJkLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.