- 1Key Laboratory of Meat Processing and Quality Control, Ministry of Education, Nanjing Agricultural University, Nanjing, China
- 2Jiangsu Collaborative Innovation Center of Meat Production and Processing, Quality and Safety Control, Nanjing Agricultural University, Nanjing, China
- 3Key Laboratory of Meat Processing, Ministry of Agriculture and Rural Affairs, Nanjing Agricultural University, Nanjing, China
- 4Experimental Teaching Center of Life Science, Nanjing Agricultural University, Nanjing, China
- 5National Center for International Research on Animal Gut Nutrition, Nanjing Agricultural University, Nanjing, China
- 6Joint International Research Laboratory of Animal Health and Food Safety, Ministry of Education, Nanjing Agricultural University, Nanjing, China
In this work, we investigated changes in protein structures in vacuum-packed pork during chill storage and its impact on the in vitro protein digestion. Longissimus dorsi muscles were vacuum packed and stored at 4°C for 3 days. Samples were subjected to Raman spectroscopy, in vitro digestion and nano LC-MS/MS. The 3 d samples had lower α-helix content, but higher β-sheet, β-turn, and random coil contents than the 0 d samples (P < 0.05). SDS-PAGE revealed significant protein degradation in the 3 d samples and the differences in digested products across the storage time. Proteome analysis indicated that the 3 d samples had the higher susceptibility to digestion. Increasing protein digestibility was mainly attributed to the degradation of myofibrillar proteins. Thus, exposure of more enzymatic sites in loose protein structure during chill storage could increase protein degradation in meat.
Introduction
Postmortem aging and its impact on eating quality of fresh meat has been widely concerned (1–3). Physicochemical and structural changes in meat proteins occurring in postmortem aging and processing may affect the protein digestion and nutritional value of meat (4–6). Postmortem aging for relatively long time may produce bioactive peptides in beef that exhibit DPPH radical scavenging, ACE- and renin-inhibitory activities (7). However, little is known about whether bioactive peptides are produced in a short-term aging of pork.
At early postmortem time, the dephosphorylation of several metabolic enzymes has been shown to affect the rate of glycolysis and pH decline, and subsequently the activation of μ-calpain, the release of lysosomal enzymes and meat tenderization (8). In addition, protein oxidation may occur in fresh meat during chill storage (9), which may cause the formation of disulfide bonds, and a decrease in protein hydrophilicity and water holding capacity. To avoid such changes, vacuum packaging and chill storage have been widely applied (10). However, protein oxidation and structural changes may still occur in vacuum-packed and chilled meat because radical oxygen species widely existing in living muscle tissues could be retained in postmortem muscles (11). Furthermore, protein oxidation also alters the protein structures and digestibility of meat (5). An increase in digestion can be attributed to protein unfolding and increased susceptibility to digestion (4). Previous studies have focused how processing and physicochemical changes affect protein structure and digestion (12, 13). However, few data are available on how structural changes of meat proteins enhanced digestibility of fresh meat from a proteomic perspective, in terms of the evolution of peptides from the stomach to the small intestine. In this study, we investigated the structural changes of meat proteins in pork during chill storage and their impacts on protein digestibility and the release of bioactive peptides.
Materials and Methods
Reagents
Ellman's reagent (4 mg/mL 5,5′-dithiobis-2-nitrobenzoic acid in Tris-Gly) and Tris-Gly (10.4 g Tris, 6.9 g glycine, and 1.2 g EDTA per liter, pH 8.0) were obtained from Sigma Aldrich (St. Louis, MO, USA), as were porcine gastric pepsin (Cat. No. P7125) and porcine pancreatic trypsin (Cat. No. T7409). BCA protein assay kit (No. 23225) and protein marker (No. 26619) were obtained from Thermo Scientific (Rockford, IL, USA). Amicon Ultracel-3 membrane (UFC500396) and Zip Tip C18 pipette tips (ZTC18S096) were obtained from Millipore (Billerica, MA, USA).
Sample Preparation
Longissimus dorsi muscles (size: 10 × 10 × 5 cm) were obtained from 8 native Suhuai pig carcasses at the same line in a commercial slaughterhouse. Each muscle was cut into four 2.5 × 10 × 5 cm pieces (weight: 50 to 65 g each), vacuum-packed and stored at 4°C for 3 days. Samples were taken on 0, 1, 2, and 3 d for further analyses.
Raman Spectroscopy
Raman spectroscopy was performed to evaluate changes in protein secondary structure (LabRAM HR Evolution, Horiba/Jobin, Yvon, Longjumeau, France) as previously described (5). Laser (excitation wavelength: 785 nm, power: 100 mW) was applied and the backscattering Raman signals ranging from 400 to 3,200 cm−1 were collected. Raman spectra were normalized against the band at 1,003 cm−1. The 1,685–1,645 and 1,309–1,229 cm−1 bands correspond to amide I and III vibrational modes of α-helix, random coil, and β-sheet. The 1,341 and 940 cm−1 bands reflect CH-bending and C-C stretching respectively. The range of 1,658–1,650 cm−1 in the amide I vibrational mode mainly reflects C=O stretching vibrations, C-N stretching, and N-H in-plane bending of peptide groups. The 1,003, 830–850, and 760 cm−1 bands correspond to phenylalanine, tyronsine, and tryptophan, respectively. The α-helix, β-sheet, β-turn, and random coil were quantified (14).
In vitro Digestion
The protein digestibility of cooked meat from 0, 1, 2 to 3 d samples was assessed as described by Zou et al. (15). Briefly, meat pieces were packed in plastic bags and cooked in a 72°C water bath until the core temperature reached 70°C. The cooking time was around 20 min. The core temperature was tracked by a portable thermal probe (Pt 100, Testo AG, Mönchaltorf, Schweiz, Germany). After cooking, meat samples were cooled for 2 h to room temperature (22°C). Then, samples (1.00 g) were homogenized (9,600 rpm, 30 s, twice; 13,400 rpm, 30 s, twice) in PBS (10 mmol/L, pH 7.0). The homogenate was digested by gastric pepsin (32 mg/mL in 0.1 mol/L HCl, pH 1.0) at 37°C for 2 h and the digestion was stopped by adjusting the pH to 7.5 with 1 mol/L NaOH. Then the resulting mixture was digested by trypsin (24 mg/mL in 0.1 mol/L PBS, pH 7.0) at 37°C for 2 h and stopped by heating the system at 95°C for 5 min.
The undigested proteins were precipitated by adding three volumes of ethanol at 4°C for 12 h. Then the samples were centrifuged at 10,000 × g at 4°C for 20 min. Protein contents in the whole meat samples and the precipitates were quantified by a BCA protein assay kit (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions. Protein digestibility was calculated as follows:
Where W1 is the content (g) of undigested proteins precipitated by ethanol. W0 is the total protein content (g) in the whole meat before digestion.
The supernatant containing digested products that are ethanol soluble was subjected to nano LC-MS/MS analysis and the precipitate including undigested proteins was separated on the SDS-PAGE gels.
SDS-PAGE
The whole proteins were extracted according to the method of Zarkadas and Maloney (16) with some modifications. Briefly, cooked meat samples (0.3 g) were homogenized in 4.5 mL 2% SDS with 3 × 30 s at 10,000 rpm and centrifuged at 4,000 × g for 20 min at 4°C. Two milliliters of the supernatant were dialyzed for 48 h in 1 L distilled water to remove SDS. The dialyzed samples were transferred to new tubes and stored at −80°C for further analyses.
SDS-PAGE was performed to separate proteins or their fragments under reducing conditions (5). Appropriate volumes of samples were mixed with 12.5 μL sample buffer (4×) and 5 μL reducing sample agent (10×) and made up to a total volume of 50 μL with ultrapure water. The final protein concentrations were 0.50 μg/μL for all samples. The samples were heated at 70°C for 10 min. Twelve microliters of samples were loaded in triplicate into the wells of 4–12% precast gels (GenScript, Piscataway, NJ, USA). The gels were run in a total 800 mL of SDS running buffer at 150 V till the blue dye front disappeared. The gels were stained with colloidal Coomassie brilliant blue R250 (CBB) for 30 min and then destained for 20 min three times. CBB staining solution contained 0.1% CBB/45% acetic acid/10% ethanol/45% ultrapure water. Destaining buffer contained 10% acetic acid/10% ethanol/80% ultrapure water. The gel images were acquired by an image scanner (GE Healthcare, Little Chalfont, Uppsala, Sweden) and the band intensities were quantified with the Quantity One image analysis software (Bio-Rad, Hercules, CA). The relative intensities of bands were calculated by the actual band intensity divided by that of the 150 kDa band in the calibration marker lane.
Protein Identification of Digested Products by Nano LC-MS/MS
The ethanol-soluble fractions of the digested products were identified by Nano LC-MS/MS system as previously described (17) with minor modifications. Briefly, ethanol in the supernatant was removed by a vacuum concentrator (ZXJY, Beijing, China), and dried in a freeze dryer (Christ, Osterode, Germany). The dried samples were dissolved in 0.2% formic acid in ultrapure water and then centrifuged (15,000 × g, 15 min, 4°C) in ultra-0.5 mL filter tubes and desalted in ZipTip C18 tips (Millipore, Billerica, MA). Peptides (1.5 μg) were separated in a C18 column (2 cm × 200 μm, 5 μm, Thermo Fisher Scientific, Palo Alto, CA) and then sequentially a C18 chromatographic column (75 μm × 100 mm, 3 μm, Thermo Fisher Scientific, Palo Alto, CA). Elution was applied by running a mixture of buffer A (0.2% formic acid in 60% acetonitrile) and buffer B (0.2% formic acid in ultrapure water) at 300 nL/min. The elution buffers were changed by 97%A from 0 to 10 min, 92%A from 10 to 70 min, 62%A from 70 to 72 min, 2%A from 72 to 82 min, and 97%A from 82 to 90 min. The eluted peptides were identified by a hybrid quadrupole orbitrap mass spectrometer equipped with a nanoelectrospray ionization source (Thermo Fisher Scientific, Palo Alto, CA). A full-scan mode was selected from 300 to 1,800 amu.
MS/MS spectra were matched using the Proteome Discoverer-1.4 (Thermo Fisher Scientific, Palo Alto, CA). Pepsin and pepsin/trypsin were selected in peptic and peptic/tryptic peptides database search, respectively. The parameters for searching were set as follows: MS/MS tolerance: 10 ppm; main search: 4.5 ppm; missed cleavage: 2; searching database: Sus scrofa under Uniprot (Uniprot-Sus scrofa.fasta, www.uniprot.org/); de-isotopic: TRUE; fixed modification: carbamidomethyl (cys); variable modification: oxidation (met), acetyl (protein N-term); label free quantification (LFQ): TRUE; decoy database pattern: reverse; LFQ min ratio count: 1; match between runs: 2 min; peptide false discovery rate (FDR): 0.01; and protein FDR: 0.01. Proteins that could not be annotated were not further used. BIOPEP database was used in the search of similar sequences previously identified showing ACE inhibitory and DPPIV inhibitory activity (http://www.uwm.edu.pl/biochemia/index.php/pl/biopep).
Statistical Analysis
The effects of storage time on measured variables (α-helix, β-sheet, β-turn, random coil, / and /) were evaluated by Student's t-test using GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla). The effects of storage time on other measured variables were evaluated by analysis of variance (ANOVA) and the least significant means were compared by Tukey's post-hoc t-test. Data were presented as means and standard deviations. The means were considered significantly different if the P-value was smaller than 0.05. Venn diagrams (http://bioinformatics.psb.ugent.be/webtools/Venn/) were applied to analyze the differences of matched proteins among the four time points.
Results and Discussion
Changes in Raw Meat Protein Structures Detected by Raman Spectroscopy
Raman spectroscopy provides powerful information on secondary structural modifications in proteins (18). The α-helix content decreased from 77.41% on 0 d to 67.89% on 3 d (P < 0.05, Figure 1), while β-sheet, β-turn, and random coil increased from 3.39, 11.72, and 8.92% on 0 d to 9.75, 13.21, and 9.49% on 3 d, respectively (P < 0.05, Figure 1). This indicates that denaturation and unfolding occurred in meat proteins from 0 to 3 d. During aging, meat proteins were degraded by endogenous enzymes into fragments. Degradation of myofibrillar proteins is modulated by protein oxidation and nitrosylation (19).
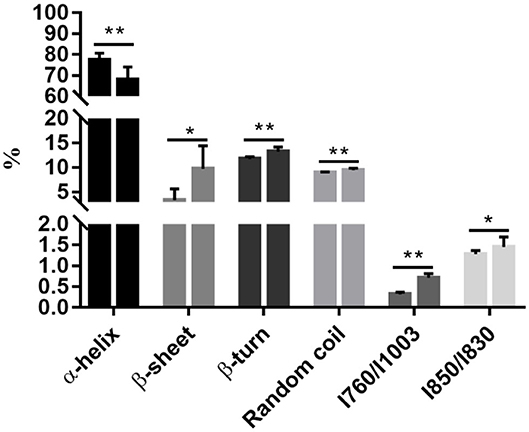
Figure 1. Postmortem aging caused a reduction of the α-helix content but an increase in the β-sheet, β-turn, and random coil contents, accompanying with the exposure of tryptophan. All data were presented as means and standard deviations. Asterisks denote that the contents of α-helix, β-sheet, β-turn, random coil, and the ratios of / and / differ significantly between the two time points. *P < 0.05; **P < 0.05.
The side chains of tryptophan and tyrosine residues may change under a polar microenvironment, which results in modifications in the tertiary structure of proteins. The ratios of / and / were related to the exposed or buried status of tryptophan and tyrosine residues, respectively (20). A substantial increase in the ratio of / representing changes of tryptophan residues was observed during chill storage (P < 0.05, Figure 1), indicating transformation of the tryptophan residues from a buried, hydrophobic state to a polar aqueous state (20). The ratio of / significantly increased from 0 to 3 d (P < 0.05, Figure 1), indicating an increasing number of exposed tyrosine residues and tryptophan residues. In addition, the ratio of / is also a good indicator for the microenvironment and hydrogen bonds of the ionization of the phenolic hydroxyl group (20). It could be the exposure of polar amino acid residues at the surface of the protein molecules. The ordered and steady structure of the protein was broken into disordered and loose fragments during storage, from a buried status of tryptophan and tyrosine residues to an exposed status, which exposed cleavage sites of digestive enzymes. Thus, we postulate that this disordered and hydrophobic status would increase the degree of protein degradation by exposing more cleavage sites to digestive enzymes.
The Degree of Protein Degradation in Cooked Meat
No significant difference was observed in the degree of protein degradation after pepsin digestion among the four time points (P > 0.05, Figure 2A). However, the degree of protein degradation increased after two-step digestion, and the 3 d samples had the highest values (P < 0.05, Figure 2B). In a previous study, postmortem storage did not improve the in vitro digestion parameters of pork (4), in which the structural organization of the muscle cell and the extracellular matrix were not taken into account. To a certain extent, the storage-induced differences in the degree of protein degradation could be attributed to changes in protein secondary or tertiary structures. Such a structural change may further affect protein digestion after meat cooking. Different cooking conditions may affect the structure of meat protein, and the digestibility of meat (17). To avoid it, we cooked meat samples with a similar size for the same time in a water bath. The proteolysis during chill storage may increase protein unfolding and expose more sites to bind to pepsin and trypsin. This is in agreement with the Raman data that reflected structural changes in side chains of proteins, including hydrophobic and electrostatic interactions.
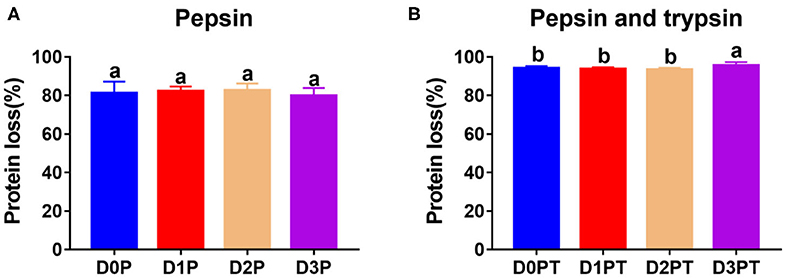
Figure 2. Protein digestibility did not differ after pepsin treatment by differ significantly after pepsin and trypsin treatments. (A) D0P, D1P, D2P, and D3P represent 0, 1, 2, and 3 d samples treated by pepsin, respectively. No significant difference was observed among groups (P > 0.05). (B) D0P/T, D1P/T, D2P/T, and D3P/T represent 0, 1, 2, and 3 d samples treated by pepsin and trypsin, respectively. a, b denotes that protein digestibility differ significantly (P < 0.05, n = 8 each).
SDS-PAGE of Proteins in Cooked Meat and Their Digested Products
SDS-PAGE was applied to separate soluble meat proteins or their digested products before or after pepsin and trypsin treatments (Figure 3). Before digestion (untreated samples), a reduction in band intensity of several protein bands was observed in cooked pork as the storage time of raw meat increased, indicating fragmentation and breakdown of meat proteins. This result was in agreement with our previous study (15). The appearance of the 30 kDa component (Troponin-T fragment) has been considered a good indicator of meat tenderization (21). During postmortem storage, high molecular weight proteins (65–200 kDa) may be degraded into smaller fragments (<25 kDa) (22).
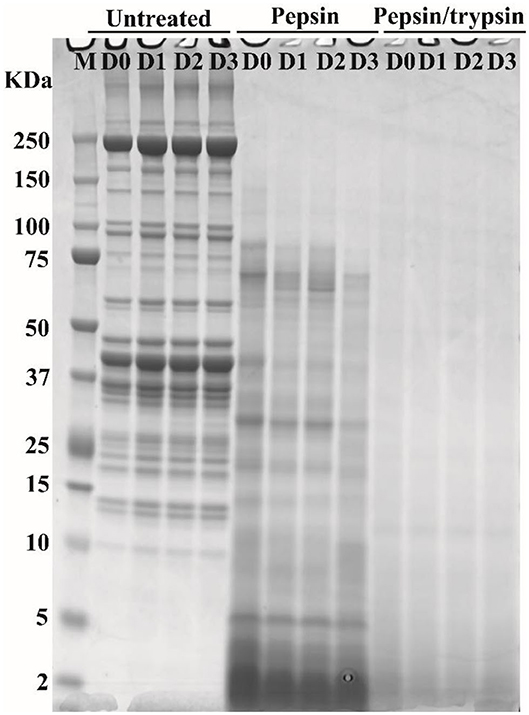
Figure 3. Typical SDS-PAGE patterns of pork proteins before and after digestion. “Untreated” represents total proteins in cooked pork before digestion. Pepsin represents ethanol-insoluble fraction after pepsin digestion. Pepsin/trypsin represents ethanol-insoluble fraction after pepsin and trypsin treatments. Lanes M, D0, D1, D2, and D3 represent calibration marker, 0, 1, 2, and 3 d samples.
In pepsin treated samples, a substantial decrease in band intensity was observed for some large-molecular-weight protein bands (Figure 3), which could be due to degradation of proteins into smaller peptides or free amino acids. In addition, some insoluble proteins could become soluble after enzymatic digestion (23). Intensity loss or even disappearance of bands was observed for pepsin and trypsin treated samples (Figure 3). Most of large molecular weight bands disappeared because proteins or peptides were further degraded into smaller ones after two-step digestion (Figure 3). Similar phenomena have been observed in different pork cuts and pork products (14, 24). However, SDS-PAGE could not separate smaller peptides well that were characterized by nano LC-MS/MS (15).
Unique Peptides Could Be a Good Indicator for Chill Storage Time
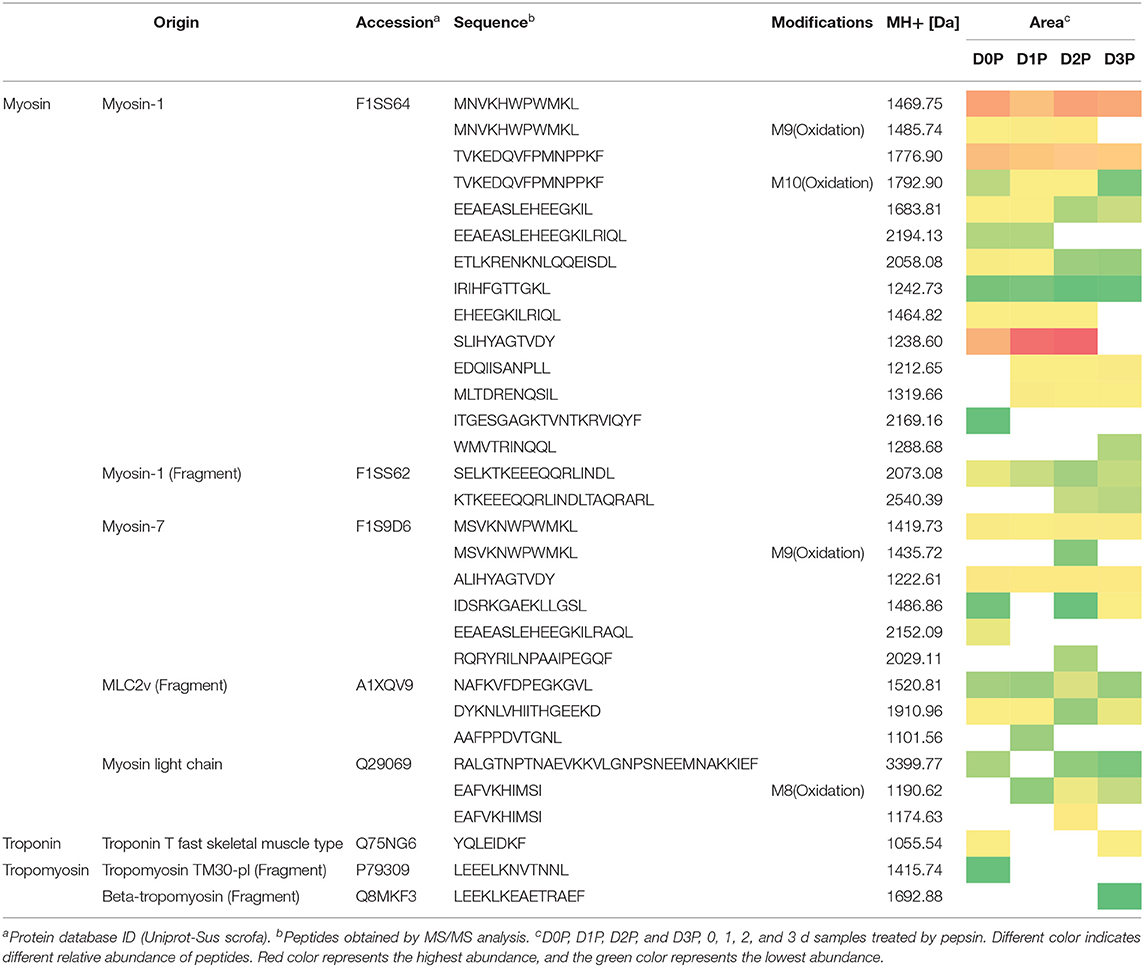
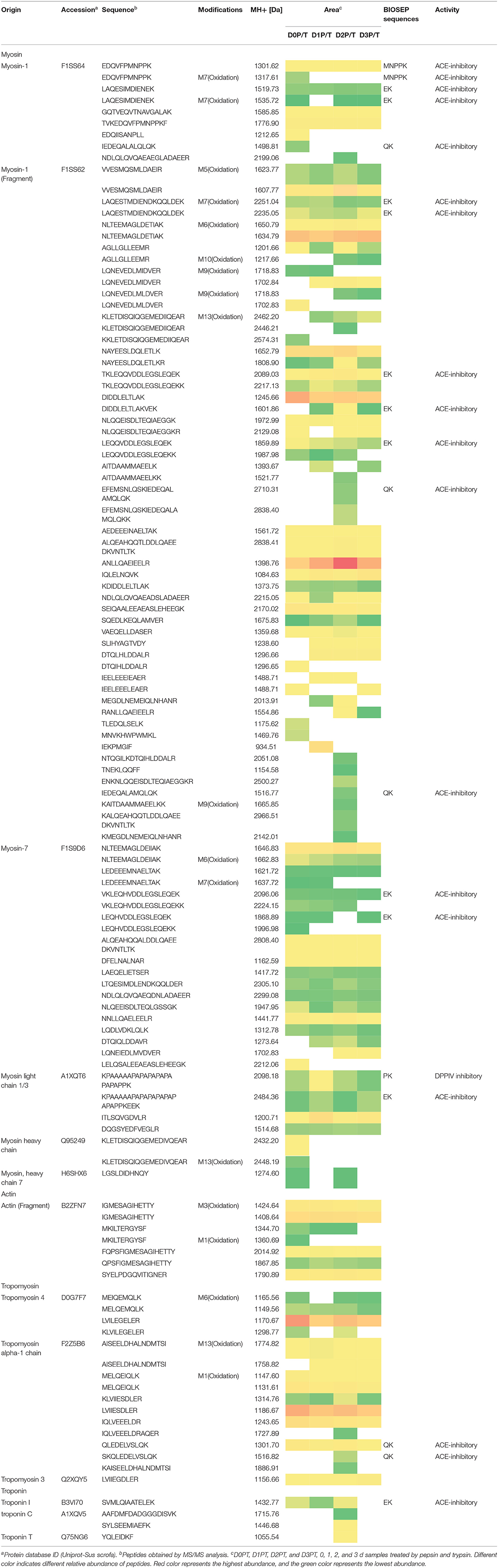
Pepsin or two-step digestion products were identified by nano LC-MS/MS and matched to proteins. All matched proteins are listed in Supplementary Files 1, 2. Venn diagrams revealed that 97 peptides were matched in all pepsin digested samples and pepsin and trypsin digested samples, respectively (Figures 4A,B). Most of these peptides came from myofibrillar proteins (myosin-1, myosin-7, actin) and sarcoplasmic proteins (phosphoglycerate kinase, creatine kinase M-type). In general, peptide abundances decreased as the storage time of raw meat increased, indicating that these peptides were degraded into smaller ones during digestion. Furthermore, 9 peptides matched to myofibrillar proteins, and 52 peptides matched to sarcoplasmic proteins in pepsin-treated samples (Supplementary File 1). More peptides matched to myofibrillar proteins (48/97) than to sarcoplasmic proteins (22/97) (Supplementary File 2). Taken together, the increased protein degradation could be mainly attributed to the degradation of myofibrillar proteins.
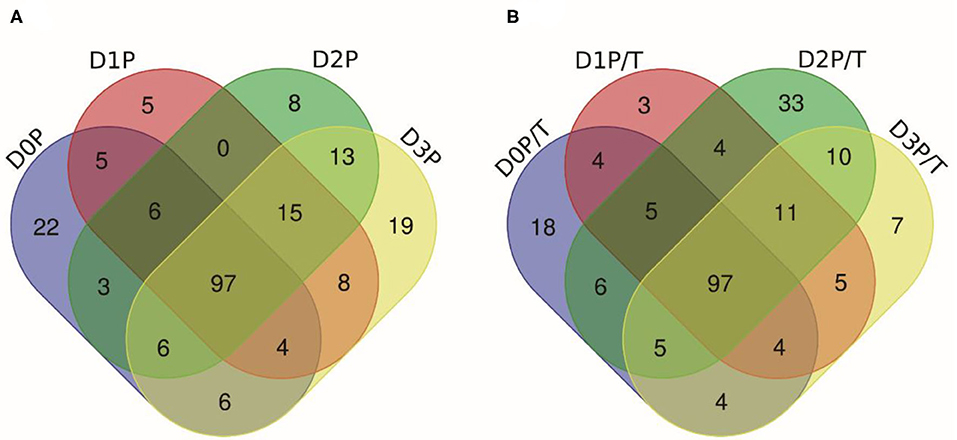
Figure 4. The numbers of unique peptides after pepsin and trypsin treatments. (A) D0P, D1P, D2P, and D3P represent 0, 1, 2, and 3 d pepsin treated samples, respectively; (B) D0P/T, D1P/T, D2P/T, and D3P/T represent 0, 1, 2, and 3 d pepsin and trypsin treated samples, respectively.
Several common peptides appeared in 1, 2, and/or 3 days samples, which were derived from large molecular weight peptides or proteins. Specifically, 22, 5, 8, and 19 peptides were uniquely matched in pepsin-treated samples on 0, 1, 2, and 3 days respectively, and 18, 3, 33, and 6 peptides were unique for pepsin/trypsin-treated samples at the four-time points (Figures 4A,B). Unique peptides could be a good indicator for chill storage time.
Digested Products From Myofibrillar Proteins in Pork Evolved With Storage Time
All peptides matched to myofibrillar proteins were listed in Table 1. Previous studies have shown that actin, myosin-1, myosin-4, myosin-7, and many glycolytic enzymes are highly abundant proteins in pork muscle (15, 25). These proteins are involved in muscle contraction and energy production. In the present study, most of differently abundant peptides are derived from these proteins (Tables 1, 2).
Oxidative modification happened in four peptides (MNVKHWPWMKL, EAFVKHIMSI TVKEDQVFPMNPPKF, MSVKNWPWMKL) originating from myosin. Long-chain peptides (e.g., EEAEASLEHEEGKILRIQL) were less abundant than short-chain peptides (e.g., EEAEASLEHEEGKIL) and some long-chain peptides disappeared in 2nd days samples (Table 1). After gastric digestion, unique peptides were further hydrolyzed by trypsin. Abundances of common peptides (ALIHYAGTVDY, TVKEDQVFPMNPPKF, LEQQVDDLEGSLEQEK, VKLEQHVDDLEGSLEQEK, KPAAAAAPAPAPAPAPAPAPAPPK, EDQVFPMNPPK) decreased as the storage time of raw meat increased, indicating that these peptides were digested by pepsin or/ and trypsin (Tables 1, 2). A total of 16 peptides, including KPAAAAAPAPAPAPAPAPAPAPPK, KPAAAAAPAPAPAPAPAPAPAPPKEEK, TKLEQQVDDLEGSLEQEK, and TKLEQQVDDLEGSLEQEKK were cleaved into 1 to 4 amino acids peptides by trypsin (Table 2).
From the digested products, 117 peptides were selected to match the BIOPEP database showing ACE-inhibitory (angiotensin-converting-enzyme inhibitory) and dipeptidyl peptidase IV (DPPIV) inhibitory activities (Table 2). Eighteen peptides were estimated to have ACE-inhibitory and DPPIV inhibitory activities, which have been identified in the database (26). ACE inhibitors are known to beneficial to treat arterial hypertension (27), diabetes mellitus, ischemic heart disease, chronic heart failure, angioedema, chronic kidney disease, and Parkinson's disease (28, 29). DPPIV inhibitor may be beneficial for NASH subjects (30). Four peptides (TKLEQQVDDLEGSLEQEKK, LEQQVDDLEGSLEQEKK, VKLEQHVDDLEGSLEQEKK, LEQHVDDLEGSLEQEKK) were identified, which contain sequences VDDLEGSLEQEKK and DDLEGSLEQEKK that were previously reported to have antioxidant peptides (31). It is of note that some identified peptides maintain a long sequence after the two-step digestion. However, almost all the peptides would be hydrolyzed by peptidases into smaller fragments (dipeptides and tripeptides) locating the outer layer of intestinal epithelium before reaching the blood stream (32). Thus, moderate chill storage may enhance the production of antioxidant, ACE inhibitor and DPP-IV inhibitory peptides.
Table 2 illustrates dynamic evolution of 117 peptides derived from cooked meat samples. The potential bioactive peptides were predominantly originated from myosin, actin, tropomyosin and troponin. Peptides EDQVFPMNPPK (Oxidation) and IEDEQALALQLQK appeared in 0 day samples, whereas peptides EFEMSNLQSKIEDEQALAMQLQK, IEDEQALAMQLQK, and SKQLEDELVSLQK appeared only in 2 day samples. Abundances of LAQESIMDIENEK, LAQESTMDIENDKQQLDEK, and their oxidative peptides and KPAAAAAPAPAPAPAPAPAPAPPKEEK were the lowest in 2 day samples. Abundances of SVMLQIAATELEK, DIDDLELTLAKVEK, and TKLEQQVDDLEGSLEQEK were the highest in 2 day samples. This could be mainly attributed to the degradation of proteins and accumulation of peptides in the first stage, which would be further degraded into smaller peptides that have ACE and DPPIV inhibitory bioactivities. Thus, 2 days could be a key time point for chill storage of pork in terms of meat nutrition.
Two peptides were observed to have the same sequence but their molecular weights were different, which is attributed to protein oxidation based on the database. Eighteen peptides from two-step digestion were observed to have oxidation modification. It is known that protein oxidation occurs during storage and the extent of protein oxidation may be enhanced as meat storage time increases. In the present study, pork samples were vacuum packed and stored at low temperature (4°C) for 3 days. In such a condition, protein oxidation could be alleviated. And thus more cleavage sites could be exposed in loose protein structure and protein degradation increased. The number of bioactive peptides may increase after the two-step digestion.
The above results could be associated with rigor mortis and subsequent aging of muscle (33). It is well-known that glycolysis occurs in skeletal muscles after slaughter (34, 35). This process is accompanied by the dephosphorylation of energy metabolic enzymes, the formation of actomyosin, and the shortening of sarcomeres (6, 36). Such activities may make some energy metabolic enzymes and myofibrillar proteins less susceptible to pepsin and trypsin digestion under in vitro condition on day 1. However, prolonged storage can tenderize meat, and make the myofibrillar proteins and bound energy metabolic enzymes more digestible. This is because the endogenous enzymes, e.g., μ-calpain and cathepsins, catalyze the degradation of myofibrillar proteins into smaller fragments (33), which will expose the cleavage sites for digestive enzymes. It is notable that, too long-time storage of fresh meat may have the problems of food spoilage, discoloration, and the decreased digestibility of meat proteins due to oxidation (37).
Conclusions
In this study, postmortem aging was shown to have significant impacts on the structural characteristics and in vitro digestion of pork proteins. It is observed that the 3 d samples had lower α-helix content and the peak intensity at 760 cm−1 (tryptophan residues), but higher β-sheet, β-turn, and random coil contents than the 0 d samples. This is accompanied by significant protein degradation and increased susceptibility to digestion. The structural changes of proteins altered the accessibility of proteolytic enzymes to the cleavage sites, and consequently the protein digestion. Further work is necessary to evaluate the effect of postmortem aging on in vivo digestion of pork protein.
Data Availability Statement
The datasets generated for this study can be found in Proteome Xchange, Accession No. PXD020595.
Ethics Statement
The animal study was reviewed and approved by the Ethical Committee of Experimental Animal Center of Nanjing Agricultural University.
Author Contributions
CL designed the experiments. XZ, JH, DZ, MZ, YX, and CW conducted the experiments. XZ, JH, and CL wrote the paper. All authors have read and approved the manuscript.
Funding
This work was funded by 31530054 (NSFC), CARS-35 (MARA), SXGC [(2017])212, and SZ-XZ2017029.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2020.00151/full#supplementary-material
References
1. Li CB, Wu JQ, Zhang N, Zhang S, Liu J, Li JP, et al. Effects of boning method and postmortem aging on meat quality characteristics of pork loin. Anim Sci J. (2009) 80:591–6. doi: 10.1111/j.1740-0929.2009.00677.x
2. Channon HA, Kerr MG, Walker PJ. Effect of Duroc content, sex and aging period on meat and eating quality attributes of pork loin. Meat Sci. (2004) 66:881–8. doi: 10.1016/j.meatsci.2003.08.010
3. Lepper-Blilie AN, Berg EP, Buchanan DS, Berg PT. Effects of post-mortem aging time and type of aging on palatability of low marbled beef loins. Meat Sci. (2016) 112:63–8. doi: 10.1016/j.meatsci.2015.10.017
4. Bax ML, Sayd T, Aubry L, Sante-Lhoutellier V. Muscle composition slightly affects in vitro digestion of aged and cooked meat: identification of associated proteomic markers. Food Chem. (2013) 136:1249–62. doi: 10.1016/j.foodchem.2012.09.049
5. He J, Zhou GH, Bai Y, Wang C, Zhu SR, Xu XL, et al. The effect of meat processing methods on changes in disulfide bonding and alteration of protein structures: impact on protein digestion products. RSC Adv. (2018) 8:17595. doi: 10.1039/C8RA02310G
6. Santé-Lhoutellier V, Aubry L, Gatellier P. Effect of oxidation on in vitro digestibility of skeletal muscle myofibrillar proteins. J Agric Food Chem. (2007) 55:5343–8. doi: 10.1021/jf070252k
7. Fu Y, Young JF, Therkildsen M. Bioactive peptides in beef: endogenous generation through postmortem aging. Meat Sci. (2017) 123:?134–42 doi: 10.1016/j.meatsci.2016.09.015
8. Li CB, Li J, Zhou GH, Lametsch R, Ertbjerg P, Brüggemann DA, et al. Electrical stimulation affects metabolic enzyme phosphorylation, protease activation, and meat tenderization in beef. J Anim Sci. (2012) 90:1638–49. doi: 10.2527/jas.2011-4514
9. Rowe LJ, Maddock KR, Lonergan SM, Huff-Lonergan E. Influence of early postmortem protein oxidation on beef quality. J Anim Sci. (2014) 82:785–93. doi: 10.1093/ansci/82.3.785
10. Zhou GH, Xu XL, Liu Y. Preservation technologies for fresh meat - A review. Meat Sci. (2010) 86:119–28. doi: 10.1016/j.meatsci.2010.04.033
11. Warner RD, Dunshea FR, Ponnampalam EN, Cottrell JJ. Effects of nitric oxide and oxidation in vivo and postmortem on meat tenderness. Meat Sci. (2005) 71:205–17. doi: 10.1016/j.meatsci.2005.04.008
12. Rysman T, Hecke TV, Poucke CV, Smet SD, Royen GV. Protein oxidation and proteolysis during storage and in vitro digestion of pork and beef patties. Food Chem. (2016) 209:177–84. doi: 10.1016/j.foodchem.2016.04.027
13. Du XJ, Sun YY, Pan DD, Wang Y, Ou CR, Cao JX. The effect of structural change on the digestibility of sarcoplasmic proteins in Nanjing dry-cured duck during processing. Poultry Sci. (2018) 1–8. doi: 10.1002/jsfa.8815
14. Alix AJP, Pedanou G, Berjot M. Fast determination of the quantitative secondary structure of proteins by using some parameters of the Raman Amide I band. J Mol Struct. (1988) 174:159–64. doi: 10.1016/0022-2860(88)80151-0
15. Zou XY, Zhou GH, Yu XB, Bai Y, Wang C, Xu XL, et al. In vitro protein digestion of pork cuts differ with muscle type. Food Res Int. (2018) 106:344–53. doi: 10.1016/j.foodres.2017.12.070
16. Zarkadas CG, Maloney SA. Assessment of the protein quality of the smooth muscle myofibrillar and connective tissue proteins of chicken gizzard. Poultry Sci. (1998) 77:770–9. doi: 10.1093/ps/77.5.770
17. Wen SY, Zhou GH, Li L, Xu XL, Yu XB, Bai Y, et al. Effect of Cooking on in vitro digestion of pork proteins: a peptidomic perspective. J Agric Food Chem. (2015) 63:250–61. doi: 10.1021/jf505323g
18. Herrero AM. Raman spectroscopy for monitoring protein structure in muscle food systems. Crit Rev Food Sci. (2008) 48:512–23. doi: 10.1080/10408390701537385
19. Liu R, Li Y, Wang M, Zhou GH, Zhang WG. Effect of protein S-nitrosylation on autolysis and catalytic ability of μ-calpain. Food Chem. (2016) 213:470–7. doi: 10.1016/j.foodchem.2016.06.104
20. Li-Chan ECY. The applications of Raman spectroscopy in food science. Trends Food Sci Tech. (1996) 7:361–70. doi: 10.1016/S0924-2244(96)10037-6
21. Macbride MA, Parrish FC. The 30,000 dalton component of tender bovine longissimus muscle. J Food Sci. (1977) 42:1627–9. doi: 10.1111/j.1365-2621.1977.tb08442.x
22. Naveena BM, Kiran M, Sudhakar RK, Ramakrishna C, Vaithiyanathan S, Devatkal SK. Effect of ammonium hydroxide on ultrastructure and tenderness of buffalo meat. Meat Sci. (2011) 88:727–32. doi: 10.1016/j.meatsci.2011.03.005
23. Diaz M, Vattem D, Mahoney RR. Production of dialysable and reduced iron by in vitro digestion of chicken muscle protein fractions. J Sci Food Agric. (2002) 82:1551–5. doi: 10.1002/jsfa.1219
24. Li L, Liu Y, Zou XY, He J, Xu XL, Zhou GH, et al. In vitro protein digestibility of pork products is affected by the method of processing. Food Res Int. (2017) 92:88–94. doi: 10.1016/j.foodres.2016.12.024
25. Kim GD, Yang HS, Jeong JY. Intramuscular variations of proteome and muscle fiber type distribution in semimembranosus and semitendinosus muscles associated with pork quality. Food Chem. (2018) 244:143–52. doi: 10.1016/j.foodchem.2017.10.046
26. Minkiewicz P, Dziuba J, Iwaniak A, Dziuba M, Darewicz M. BIOPEP database and other programs for processing bioactive peptide sequences. J AOAC Int. (2008) 91:965–80. doi: 10.1093/jaoac/91.4.965
27. Zhou M, Daubresse M, Stafford RS, Alexander GC. National trends in the ambulatory treatment of hypertension in the United States, 1997-2012. PLoS ONE. (2015) 10:e0119292. doi: 10.1371/journal.pone.0119292
28. Weir MRMD. Effects of renin-angiotensin system inhibition on end-organ protection: can we do better? Clin Ther. (2007) 29:1803–24. doi: 10.1016/j.clinthera.2007.09.019
29. Montinaro V, Cicardi M. ACE inhibitor-mediated angioedema. Int Immunopharmacol. (2020) 78:106081. doi: 10.1016/j.intimp.2019.106081
30. Balaban YH, Korkusuz P, Simsek H, Gokcan H, Tatar G. Dipeptidyl peptidase IV (DDP IV) in NASH patients. Ann Hepatol. (2007) 6:242–50. doi: 10.1016/S1665-2681(19)31905-2
31. Je JY, Qian ZJ, Kim SK. Antioxidant peptide isolated from muscle protein of bullfrog, Rana catesbeiana Shaw. J Med Food. (2007) 10:401–7. doi: 10.1089/jmf.2006.169
32. Babusiak M, Man P, Petrak J, Vyoral D. Native proteomic analysis of protein complexes in murine intestinal brush border membranes. Proteomics. (2007) 7:121–9. doi: 10.1002/pmic.200600382
33. Geesink GH, Kuchay S, Chishti AH, Koohmaraie M. μ-Calpain is essential for postmortem proteolysis of muscle proteins. J Anim Sci. (2006) 84:2834–40. doi: 10.2527/jas.2006-122
34. Hollung K, Veiseth E, Froystein T, Aass L, Langsrud O, Hildrum KI. Variation in the response to manipulation of post-mortem glycolysis in beef muscles by low-voltage electrical stimulation and conditioning temperature. Meat Sci. (2007) 77:372–83. doi: 10.1016/j.meatsci.2007.03.029
35. Wang SD, Li CB, Xu XL, Zhou GH. Effect of fasting on energy metabolism and tenderizing enzymes in chicken breast muscle early postmortem. Meat Sci. (2013) 93:865–72. doi: 10.1016/j.meatsci.2012.11.053
36. Højlund K, Brown BP, Hwang H, Flynn CR, Madreddy L, Geetha T, et al. In vivo phosphoproteome of human skeletal muscle revealed by phosphopeptide enrichment and HPLC-ESI-MS/MS. J Proteome Res. (2009) 8:4954–65. doi: 10.1021/pr9007267
Keywords: pork, chill storage, in vitro digestion, Raman spectroscopy, LC-MS/MS
Citation: Zou X, He J, Zhao D, Zhang M, Xie Y, Dai C, Wang C and Li C (2020) Structural Changes and Evolution of Peptides During Chill Storage of Pork. Front. Nutr. 7:151. doi: 10.3389/fnut.2020.00151
Received: 01 April 2020; Accepted: 28 July 2020;
Published: 22 September 2020.
Edited by:
Aida Serra, IMDEA Alimentación, SpainReviewed by:
Manuel Suarez Recio, Department of Biochemistry and Biotechnology, Rovira i Virgili University, SpainXue Guo, Nanyang Technological University, Singapore
Yangying Sun, Ningbo University, China
Copyright © 2020 Zou, He, Zhao, Zhang, Xie, Dai, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunbao Li, Y2h1bmJhby5saSYjeDAwMDQwO25qYXUuZWR1LmNu
 Xiaoyu Zou
Xiaoyu Zou Jing He1,2,3
Jing He1,2,3 Di Zhao
Di Zhao Chong Wang
Chong Wang Chunbao Li
Chunbao Li