- 1Department of Internal Medicine-Molecular Medicine and Department of Microbiology and Immunology, Wake Forest School of Medicine, Winston-Salem, NC, United States
- 2Department of Pathology-Comparative Medicine, Wake Forest School of Medicine, Winston-Salem, NC, United States
- 3Department of Epidemiology and Prevention, Wake Forest School of Medicine, Winston-Salem, NC, United States
The mammalian gastrointestinal tract harbors a highly diverse and dynamic community of bacteria. The array of this gut bacterial community, which functions collectively as a fully unified organ in the host metabolism, varies greatly among different host species and can be shaped by long-term nutritional interventions. Non-human primates, our close phylogenetic relatives and ancestors, provide an excellent model for studying diet-microbiome interaction; however, compared to clinical and rodent studies, research targeting primate gut microbiome has been limited. Herein, we analyze the gut microbiome composition in female cynomolgus macaques (Macaca fascicularis; n = 20) after the long-term (2.5 years) consumption of diets designed to mimic recent human Western- (WD; n = 10) or Mediterranean-type (MD; n = 10) diets. Microbiome diversity in MD consumers was significantly higher by the Shannon diversity index compared to the WD consumers, with similar but non-significant trends noted for the diversity metrics of species richness (Chao 1), observed operational taxonomic units (OTUs) and phylogenetic diversity (PD) whole Tree. Compared to the MD, the WD group demonstrated a higher Firmicutes-Bacteroides ratio and a significantly higher abundance of families Clostridiacea and Lactobacillaceae. Further analyses reveal significantly higher abundance of genera Lactobacillus, Clostridium, Faecalibacterium, and Oscillospira and lower abundance of Ruminococcus and Coprococcus in MD consumers relative to WD consumers. OTUs belonging to several species also show significant differences between the two groups, with Lactobacillus species demonstrating a prominently higher abundance in the MD consumers. The data reveal several differences in the gut microbiome of primates consuming the two different diets and should be useful for further studies aimed at understanding the diet-microbiome-health interactions in primates.
Introduction
The past decade has been remarkable in revealing the fundamental role of the gut microbiome (a highly complex and diverse community of microbes living within gastrointestinal tract) in human health and diseases [1, 2]. Among various intrinsic and extrinsic factors that affect gut microbiome composition and diversity, diet has received appreciable attention because of its potential influence on host health and metabolism [3, 4]. Diet shapes the gut microbiome spectrum by providing substrates that differentially promote the growth and activities of specific microbial communities [5–9]. Diet-microbiome interactions are consistently reproducible in clinical and animal studies [5, 8, 10, 11]. Specifically, many microbiome studies have focused on the effects of high-fat and/or high-sugar vs. low-fat and/or low-sugar diets on the gut microbial populations, particularly with respect to risk of chronic diseases and disorders such as obesity, type 2 diabetes, cardiovascular disease, and psychiatric disorders [12–15].
It is difficult to study long-term diet effects in human beings, as such studies rely on self-reported dietary intake collected using food frequency questionnaires which are not comprehensive or standardized, and nutrient intakes are estimated from these self-reports utilizing food composition tables. Therefore, actual nutrient intake is unknown. Several animal models including mice, rats, guinea pigs, and zebra fish have been used for studying diet-microbiome interactions. However, considering the potential health impact of diet effects on gut microbiome, the use of animal models demonstrably useful for studies of obesity, type 2 diabetes, cardiovascular disease, and psychiatric disorders [16] would be most helpful. Several population studies have indicated a beneficial effect of consumption of a Mediterranean diet on chronic diseases, and some evidence suggests that gut microbiota-mediated production of metabolites influencing metabolic health are involved. However, effects of Mediterranean diets have not been studied in animal models.
Cynomolgus monkeys (Macaca fascicularis) are widely used as models of diet-induced obesity, type 2 diabetes, coronary artery disease, and mood disorders [17–19]. Here we report the gut microbiome composition in healthy adult NHPs after long-term (2.5 years) consumption of Western- vs. Mediterranean-type diets.
Materials and Methods
Subjects
The current work was conducted in socially housed adult female cynomolgus macaques maintained at the Wake Forest University Primate Center (established 1956; http://www.wakehealth.edu/ccpr). All animals were in apparently good health, free from gastrointestinal infections. The monkeys were randomized to one of two diet groups: Western or Mediterranean diet. Earlier studies of microbiome [20–23] used 3–15 animals to achieve >80% power and an α of 0.05, hence we selected n = 20 animals (n = 10 in each group), and no subjects were excluded from analysis. The monkeys were fed the experimental diets for 30 months (2.5 years), and water was accessible ad libitum.
Diet
The western diet (hereafter, WD) consisted of lard, beef tallow, butter, egg, cholesterol, casein, lactalbumin, dextrin, high-fructose corn syrup, and sucrose; while the Mediterranean diet (hereafter, MD) comprised fish oil, olive oil, fish meal, butter, egg, black and garbanzo bean flour, wheat flour, V-8 juice, fruit puree, and sucrose. The detailed composition in terms of the percent of calories in these diets is provided in Table 1.
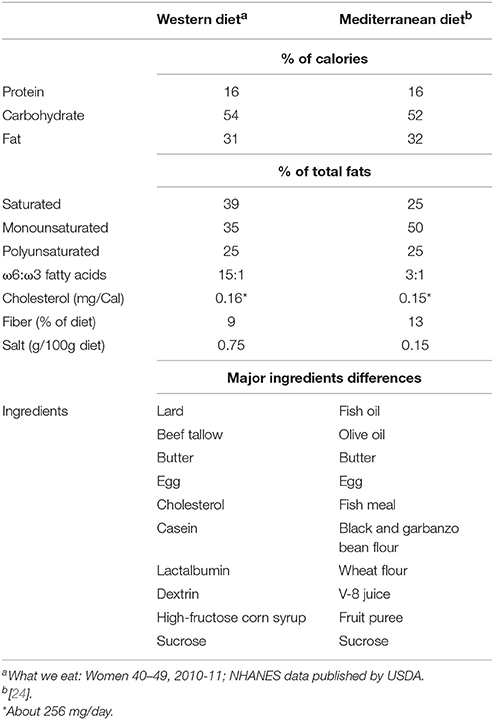
Table 1. Dietary composition of Western- and Mediterranean-style diets fed to the primates enrolled in this study.
Feeding
Individual feeding cages were fabricated and placed inside social group pens. The monkeys were taught to run into their individual feeding cages on voice command, were given 2 h to consume their diet, and released back into the social group pen. The monkeys consumed most of the diet in the first 30 min. Each monkey was offered 100 Cal/kg of diet.
Sample Collection
The NHPs were euthanized after the 30-month intervention period. Samples of rectal/anal contents of 10 randomly chosen monkeys from each of the diet groups (n = 20 total) were collected at necropsy and immediately placed in sterile tubes under aseptic conditions and stored at −80°C until further processing. All protocols related to the sampling, care and management of animals were reviewed and approved by the Institutional Animal Care and Use Committee at the Wake Forest School of Medicine.
Microbiome Analysis
Nearly 200 mg (wet weight) of each sample was used to extract genomic DNA by using the Qiagen DNA Stool Mini Kit (Qiagen, CA, USA) per manufacturer instructions. The bacterial 16S rRNA gene was amplified using the primers 515F (barcoded) and 806R, which flanked the V4 hypervariable region of bacterial 16S rRNA, in accordance with the Earth Microbiome Project protocol [25, 26] with the following minor modification. The PCR reaction consisted 25 μl of SYBR® Premix ExTaqTMII (Takara Bio, Shiga, Japan), 1 μl of each of the primers, 5 μl of DNA template, and 18 μl of RNase-free water (Invitrogen, Eugene, OR, USA). PCR conditions comprised an initial step at 50°C for 2 min and at 95°C for 10 min, with subsequent amplification steps at 95°C for 30 s, at 55°C for 30 s, and at 72°C for 90 s for repeated cycles on an Applied Biosystems® 7500 Real Time PCR System (Applied Biosystems). The amplification step was stopped before the fluorescent intensity reached a plateau. The resulting amplicons were purified using Agencourt® AMPure® XP (Beckman Coulter), quantified using the Qubit-3 fluorimeter (InVitrogen), normalized to an equal concentration (4 nm) and pooled together for 16S Miseq analysis. The sample pool was denatured, diluted to 8 pM and sequenced on an Illumina MiSeq platform, using a MiSeq Reagent Kit v3 (Illumina, San Diego, CA, USA) as described previously [27]. Sequencing procedure was monitored on the Illumina BaseSpace® website wherein analysis of the data generated on the Miseq platform was executed using the BaseSpace® 16S Metagenomics App (Illumina). Operational taxonomic unit (OTU) assignment to the Greengenes database was performed using the Quantitative Insights Into Microbial Ecology (QIIME) pipeline software package that enables microbial community analysis [26]. Demultiplexed R1 and R2 sequencing read files were obtained from the Illumina BaseSpace® website and the Illumina reads were quality-filtered, clustered and analyzed using default parameters in QIIME. A total of 1.33 million reads (mean = 51665.50; standard deviation = 21791.5) were generated after filtering. Paired-end reads were joined together with fastq_join_paired_ends.py and split_libraries_fastq.py scripts. The assembled sequences were grouped into OTUs at a sequence similarity of 97% identity and sequences were classified into the taxonomical levels based on the Greengenes 16S rRNA gene database (http://greengenes.secondgenome.com). Representative OTU sequences were aligned to a Greengenes reference alignment [25]. De novo OTUs were classified using RDP classifier and the Greengenes reference set with a minimum 80% confidence threshold [28]. Samples were rarified at an even sequencing depth of 10,000 reads per sample for subsequent downstream analyses.
Data Analysis
Taxonomy assignment and diversity analyses were computed through QIIME with default settings to compare bacterial species richness between the two diet groups. Bacterial composition of each sample was measured at various taxonomic levels using QIIME. Alpha diversity (rarefaction curve for observed OTUs, Chao1, PD_Whole_Tree and Shannon) indices were computed with core_diversity_analysis.py script. Beta diversity was generated within QIIME by using weighted and unweighted Unifrac distance matrices. UniFrac distances are appraised as the distance between bacterial communities explaining phylogenetic relationship between bacteria. Principal components analysis (PCA), an unsupervised analysis that allows estimation and visualization of sample distribution based on UniFrac distance patterns, was performed to determine the influence of dietary treatments on the overall microbiome composition of the samples. PCA plots were visualized using EMPeror version 0.9.3-dev. Unweighted PCA was used to determine if the NHP microbiome phenotypes clustered by the type of diet. The data of bacterial diversity and abundance between the two diet groups were compared by using non-parametric analyses in R statistical software package (version 3.4.3; https://www.r-project.org/). Statistically significant differences between two groups were calculated by Mann–Whitney U-test with Bonferroni correction. Statistical significance in alpha-diversity was calculated by non-parametric two-sample t-tests with 9999 Monte-Carlo permutations. Difference in beta-diversity was examined by permutational multivariate analysis of variance (PERMANOVA), a permutation-based extension of multivariate analysis of variance to a matrix of pairwise distance that partitions the inter-group and intra-group distances. Hierarchical clustering maps based on average linkage on Euclidean distance were constructed in R using “ggplots” library and ward.D2 method. Results are expressed as mean ± SEM. Unless otherwise stated, a value of P < 0.05 was considered statistically significant.
Results
Diversity metrics viz. Shannon, Chao1, observed OTU, and phylogenetic diversity indices demonstrated varying but similar trend for higher microbiome diversity in MD vs. WD consumers (Figure 1; Supplementary Figure S1). Chao1 index, observed species and phylogenetic diversity were insignificantly higher in MD group compared with WD group; while the Shannon Diversity index, which constitutes both microbial richness and abundance, was found to be significantly (P < 0.05) higher in MD vs. WD consumers (Figure 1).
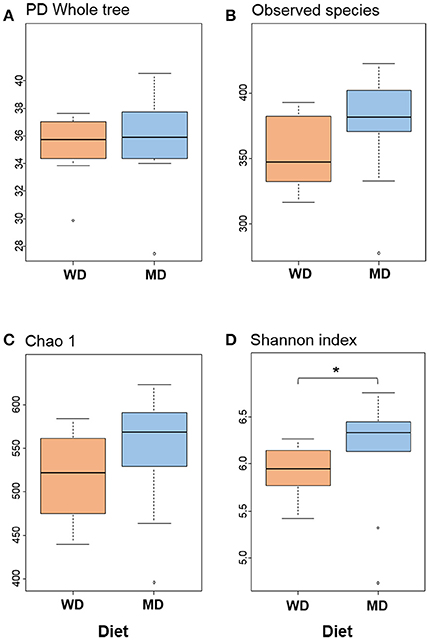
Figure 1. Bacterial diversity indices (A–D) in the distal gut samples of non-human primates consuming either a Western-style diet (WD; n = 10) or a Mediterranean-style diet (MD; n = 10) for a period of 30 months. *P < 0.05; Mann–Whitney U-test (Monte Carlo permutation).
Overall, at phyla level, the microbiome of these NHP was predominated by Bacteroidetes (42%), Firmicutes (38%), and Proteobacteria (5%), followed by Verrucomicrobia, Fibrobacteres, Actinobacteria, Cyanobacteria, Spirochaetes, Tenericutes, and Elusibacteria (Figure 2A). Compared with WD group, MD group demonstrated slightly higher abundance of Bacteroides (44 vs. 41%), Proteobacetria (6.2 vs. 4.9%), Fibrobacteres (4.5 vs. 2.4%), and Spirochaetes (4.6 vs. 2.3%) and lower abundance of Firmicutes (37 vs. 40%) and Verrucomicrobia (0.7 vs. 4.6%) (Figures 2A,B). Accordingly, the Firmicutes to Bacteroides ratio was found to be relatively lower in MD group (0.86) compared to that in WD group (0.97) (Figure 2C). Within the three major phyla (i.e., Bacteroidetes, Firmicutes, and Proteobacteria), Paraprevotellaceae, Prevotellaceae, Lactobacillaceae, Clostridiaceae, Lachnospiraceae, Ruminococcaceae, Veillonellaceae, Erysipelotrichaceae, Alcaligenaceae, Desulfovibrionaceae, Helicobacteraceae, and Succinivibrionaceae represented the top 12 most abundant families (Figure 2B). Among these, Clostridiaceae and Lactobacillaceae were found to be significantly more abundant in MD vs. WD group (Figures 2D,E). The details of other major taxa detected at the level of phyla, class and order are provided in Supplementary Figure S2.
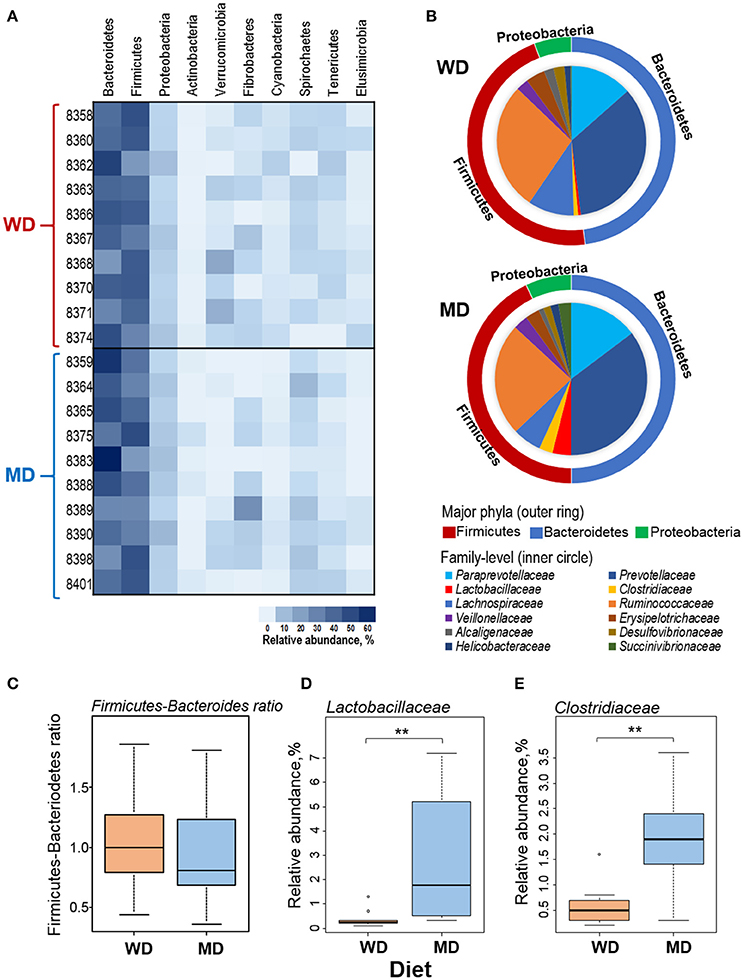
Figure 2. (A) Heat-map depicting the relative abundance of major phyla observed in 20 primates consuming either a Western-style diet (WD; n = 10) or a Mediterranean-style diet (MD; n = 10) for a period of 30 months. (B) Pie-charts showing the comparison in the relative abundance of top three phyla and major families detected within these phyla. (C–E) Box-plots showing the comparison in the Firmicutes to Bacteroidetes ratio (C) and the relative abundance of families Lactobacillaceae (D) and Clostridiaceae (E) between WD and MD groups. **P < 0.001; Mann–Whitney U-test (Monte Carlo permutation).
Figure 3A shows the relative abundance of top 20 most abundant genera observed in this cohort of NHPs. The overall spectrum of these genera appeared to be slightly different between the two groups wherein several genera showed differences between the two diet groups. Among these, Lactobacillus, Faecalibacterium, Clostridium, Oscillospira, and Prevotella exhibited the most prominent difference with significantly or insignificantly higher abundance in MD vs. WD group (Figure 3B) whereas Ruminococcus and Coprococcus exhibited an opposite trend, i.e., a significantly lower relative abundance in MD vs. WD group (Figure 3B). However, PCA analysis did not show any considerable clustering of the gut bacterial communities between the two diet groups (Supplementary Figure S4).
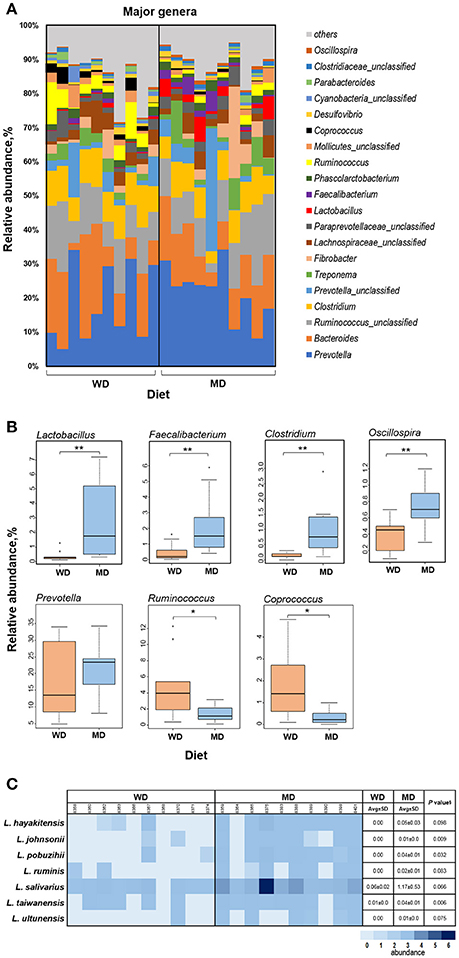
Figure 3. (A) Bar graph showing the relative abundance of major genera observed in the gut microbiome of 20 primates consuming either a Western-style diet (WD; n = 10) or a Mediterranean-style diet (MD; n = 10) for a period of 30 months. (B) Box-plots showing the comparison of relative abundance of major genera that showed a notable difference between WD and MD groups. *P < 0.05; **P < 0.005; Mann–Whitney U-test (Monte Carlo permutation). (C) Relative abundance of major species belonging to the genus Lactobacillus in WD and MD groups. §Unpaired t-test.
The two diet groups also exhibited differences in the abundance of several species belonging to these genera (Supplementary Table S1), among which Lactobacillus species demonstrated the most prominent difference (Figure 3C). Although L. salivarius represented the most abundant Lactobacillus species, the abundance of all of the Lactobacillus species detected was higher in MD compared to WD group (Figure 3C).
Discussion
The gut of NHPs is inhabited by a complex and dynamic bacterial community and the compositional as well as functional array of this community can be influenced by various intrinsic and extrinsic elements such as host diet, physiology, geography, and clinical health [29]. The gut microbiome of NHPs has been found to be more similar to those of human primates than to other animals [30]. For instance, the human gut is inhabited by microorganisms belonging to nine different divisions of Bacteria: Firmicutes and Bacteroides (the predominant and most abundant), Actinobacteria, Fusobacteria, Proteobacteria, Verrucomicrobia, Cyanobacteria, Spirochaetes and VadinBE97 [31, 32]. In the present study, we identified Bacteroidetes, Firmicutes, (the most abundant), Proteobacteria, Actinobacteria, Verrucomicrobia, Fibrobacteres, Cyanobacteria, Spirochaetes, and Tenericutes as the top nine bacterial phyla in these NHPs. Hence, investigation of microbes dwelling in the gastrointestinal tract of these NHPs might provide important clues about the characteristics of these bacterial groups in the human gut. It is also likely that the microbiome composition of these captive monkeys is more humanized than that of wild free-living NHPs consuming more native diets due to the human-like diets they were fed [33].
Western- and Mediterranean-type diets are generally studied and exemplified as unhealthy and healthy dietary habits, respectively. Mounting evidence shows that diets rich in fiber and unsaturated fatty acids are important for maintenance of a diverse and healthy gut microbiome. Although we did not observe a remarkable difference in the overall microbiome signature between the two groups; we noted several interesting differences at family, genus, and species levels. Consistent with previous findings in different animal species including humans [3, 4, 6, 34], this demonstrates that diet can have a strong influence on the gut microbiome communities in NHPs and supports the notion that diet can affect the gut microbiota composition without causing dramatic changes in the overall microbial diversity [30, 34]. The higher Shannon diversity index in MD vs. WD group is interesting and could be ascribed to higher proportion of fiber in MD [3, 33, 35, 36]. The responses of NHP gut microbiome toward these diets might differ from those of human microbiome [37]; although we found several patterns of diet-induced differences similar to those reported previously in humans or small animals. This may be exemplified by higher abundance of Oscillospira (a genus belonging to the Ruminococcaceae family) in MD vs. WD group. Oscillospira species are generally prevalent in the gut of ruminants consuming diets rich in complex plant polysaccharides and are considered to be bacteria adapted to subsisting on vegetable-rich diet such as the MD [38]. Interestingly, a higher intestinal carriage of Oscillospira has also been reported in humans consuming a MD [39].
The relatively lower Firmicutes-Bacteroides ratio and higher abundance of genera Clostridium and Prevotella might be expected, in agreement with previous studies reporting that consumption of MD is associated with positive alterations in the gut microbiota composition with increased abundance of Bacteroides, Clostridium, and Prevotella [6, 40, 41]. Particularly, a lower Firmicutes-Bacteroides ratio can also be ascribed to high fiber proportion in MD. Hierarchical clustering also generated two clusters of Firmicutes and Bacteroides abundance wherein the Firmicutes-rich cluster had higher proportion of WD subjects (64%; 7 out of 11) while Bacteroides-type cluster was dominated by MD subjects (67%; 6 out of 9) (Chi-square, P = 0.17) (Supplementary Figure S3). Interestingly, high prevalence of Bacteroidetes and reduced abundance of Firmicutes has also been demonstrated in humans and other mammals and are linked to higher fiber intake and less intake of high-glycemic index sugars [6, 42, 43]. Species of Bacteroides and Prevotella, the major constituents of gut microbiota, have been linked with high-polysaccharide diets [33, 43], whereas WD is linked with increased Firmicutes and reduced Bacteroides population [6]. The higher abundance of Faecalibacterium genus (Figure 3B) as well as Faecalibacterium prausnitzii (Supplementary Table S1) also coincides with previous human studies reporting increased abundance of several butyrate-producing bacteria including Faecalibacterium in human subjects consuming a Mediterranean-style diet [39, 44]. Interestingly, the abundance of F. prausnitzii is found to be decreased and negatively correlated with inflammatory markers in type-2 diabetes patients [39, 45, 46]; whereas MD has been found to increased in abundance in patients with metabolic syndrome[39]. This indicates that the beneficial effects of specific Mediterranean-style diets on host health might be conveyed at least partly via amelioration of gut dysbiosis. This, in turn, might also suggest a preventative effect of MD against type-2 diabetes and should be an interesting topic for prospective studies to further investigate diet-induced alterations in gut microbiome and metabolome in particular context to the prevention/ amelioration of metabolic disorders.
Generally, Mediterranean-style diets favor the intestinal population of lactic acid bacteria through higher proportion of fermented foods (e.g., yogurt) in these diets. Although the diets used in the present study did not include any fermented ingredient per se, we still observed significantly higher abundance of genus Lactobacillus in MD group. Diets containing complex carbohydrates, e.g., prebiotics, have been shown to favor the proliferation of beneficial bacteria including Lactobacillus sp. [47, 48] and Faecalibacterium sp. [49] in the gut, as reported in the present study. Furthermore, omega-3 fatty acids, which are present in higher proportion in MD, are also known to promote the population of several beneficial bacterial groups including lactobacilli that populate the distal gut, a site for the metabolism of mono- and poly-unsaturated fatty acids [50–52]. On the other hand, western-style high-fat diets have been shown to reduce the gut Lactobacillus population [12, 53, 54].
Despite being present in subdominant abundance; lactobacilli represent an important bacterial group within the human gut bacterial community. Ecological studies have validated their occurrence in the gastrointestinal tract of a wide variety of animals including mammals, rodents, birds, ungulates, lagomorphs, and insects. However, data on the composition of Lactobacillus community in NHPs is lacking. Lactobacilli are highly prevalent in food and dairy products, although primitive sources might have been fecal contamination and/or deliberate probiotic addition [55]. For that reason, Lactobacillus species are generally considered as host species-specific and could also be categorized as human- or non-human type [56]. In these contexts, the investigation of Lactobacillus community in the gut of NHPs could be an interesting area for further studies, particularly owing to their long and close evolutionary connection with the human gastrointestinal tract [57]. In addition, such exploration might also pave way for discovery of novel strains that could also be exploited as probiotics for NHPs. Nevertheless, more inclusive and broader studies are required to corroborate this relationship as well as to identify the core Lactobacillus taxa for specific NHP species and to validate whether and how these taxa are related to the host health, nutrition and disease. Based on our data, L. salivarius appears to be the major Lactobacillus group in these monkeys; however further investigation by both culture-dependent and -independent methods is needed to establish this finding as well as to examine whether and how Lactobacillus flora varies during different stages of NHP's lifespan as it does in human hosts [9, 58].
Mediterranean- and Western-type diets are well known to promote different metabolic phenotypes in several animal and human studies, wherein perturbations in the gut microbiome induced by western-style diets have been shown to play causal role in several gut-related diseases including adiposity, type 2 diabetes, and other metabolic syndromes [3, 12]. MD and WD subjects in the present study differed in food consumption, insulin sensitivity, BMI and several physiological markers between the two groups (data not shown here). Future studies (in-progress) will more closely evaluate relationships between the microbiome characteristics and an array of metabolic phenotypes along with metabolomic signature being evaluated in the parent study.
The strengths of the present study are; (a) the intervention period (2.5 years) is controlled and relatively much longer compared to most other dietary interventions in either animal model or randomized human trials, (b) relative to rodent diets, the formulation of diets given to NHPs is quite similar in composition to that of typical human diet, and (c) the method of fecal sample collection, which involved near sterile collection of rectal/colonic contents at the time of necropsy, provided a fresh specimen representing the distal gut microbiome while evading the possibility of contamination from skin or environmental flora as might occur with opportunistic fecal collections following defecation. However, the study also had some limitations, including the lack of baseline data which would allow assessments of microbiome changes occurring due to the transition from a chow diet to more human like diets. Given that the animals were randomized before enrollment to the two different diets, chances of any potential bias in results are minimal. The number of subjects (n = 20) is modest compared to large epidemiological studies, however well controlled and reasonable with respect to many rodent studies where the number typically ranges from 5 to <10.
In summary, the differences observed here in the gut microbiome of NHPs consuming either a Western or Mediterranean diet suggest that this model will be useful in further studies aimed at understanding the diet-microbiome-health interactions in primates.
Author Contributions
CS, TR, HY: conceived and designed study; RN: performed microbiome experiments; RN, HY: analyzed data; RN, HY: interpreted data; CS, SA, TR, KM: coordinated animal studies and sample collections; RN, HY, CS, TR: wrote and revised manuscript; RN, CS, SA, TR, KM, MV, HY: approved final version of manuscript.
Conflict of Interest Statement
The authors declare that there are no competing interests, either financial or otherwise, with regard to this manuscript.
Acknowledgments
We thank all the participating researchers, technicians, and the staff members of the primate facility for their consistent help during the study. This work was supported by National Institutes of Health grant R01 HL087103 (CS), R01 HL122393 (TR), T32-OD010957 and the Pepper Older Americans for Independence Center (P30 AG21332), as well funds and services provided from Center for Diabetes, Obesity and Metabolism, Wake Forest Baptist Medical Center and National Center for Advancing Translational Sciences (NCATS), National Institutes of Health funded Wake Forest Clinical and Translational Science Institute (WF CTSI) through Grant Award Number UL1TR001420.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2018.00028/full#supplementary-material
References
1. Nagpal R, Yadav H, Marotta F. Gut microbiota: the next-gen frontier in preventive and therapeutic medicine? Front Med. (2014) 1:15. doi: 10.3389/fmed.2014.00015
2. Nagpal R, Kumar M, Yadav AK, Hemalatha R, Yadav H, Marotta F, et al. Gut microbiota in health and disease: an overview focused on metabolic inflammation. Benef Microbes (2016) 7:181–94. doi: 10.3920/bm2015.0062
3. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature (2014) 505:559–63. doi: 10.1038/nature12820
4. Carmody RN, Gerber GK, Luevano JM Jr, Gatti DM, Somes L, Svenson KL, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe (2015) 17:72–84. doi: 10.1016/j.chom.2014.11.010
5. Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. (2009) 1:6ra14. doi: 10.1126/scitranslmed.3000322
6. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. (2010) 107:14691–6. doi: 10.1073/pnas.1005963107
7. Moschen AR, Wieser V, Tilg H. Dietary factors: major regulators of the gut's microbiota. Gut Liver (2012) 6:411–6. doi: 10.5009/gnl.2012.6.4.411
8. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature (2012) 486:222–7. doi: 10.1038/nature11053
9. Nagpal R, Kurakawa T, Tsuji H, Takahashi T, Kawashima K, Nagata S, et al. Evolution of gut Bifidobacterium population in healthy Japanese infants over the first three years of life: a quantitative assessment. Sci Rep. (2017a) 7:10097. doi: 10.1038/s41598-017-10711-5
10. Ou J, Carbonero F, Zoetendal EG, DeLany JP, Wang M, Newton K, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr. (2013) 98:111–20. doi: 10.3945/ajcn.112.056689
11. Suzuki Y, Ikeda K, Sakuma K, Kawai S, Sawaki K, Asahara T, et al. Association between yogurt consumption and intestinal microbiota in healthy young adults differs by host gender. Front Microbiol. (2017) 8:847. doi: 10.3389/fmicb.2017.00847
12. Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes (2008) 57:1470–81. doi: 10.2337/db07-1403
13. Kisidayova S, Varadyova Z, Pristas P, Piknova M, Nigutova K, Petrzelkova KJ, et al. Effects of high- and low-fiber diets on fecal fermentation and fecal microbial populations of captive chimpanzees. Am J Primatol. (2009) 71:548–57. doi: 10.1002/ajp.20687
14. Flowers SA, Ellingrod VL. The microbiome in mental health: potential contribution of gut microbiota in disease and pharmacotherapy management. Pharmacotherapy (2015) 35:910–6. doi: 10.1002/phar.1640
15. Miele L, Giorgio V, Alberelli MA, De Candia E, Gasbarrini A, Grieco A. Impact of gut microbiota on obesity, diabetes, and cardiovascular disease risk. Curr Cardiol Rep. (2015) 17:120. doi: 10.1007/s11886-015-0671-z
16. Tosti V, Bertozzi B, Fontana L. Health benefits of the mediterranean diet: metabolic and molecular mechanisms. J Gerontol A Biol Sci Med Sci. (2018) 73:318–26. doi: 10.1093/gerona/glx227
17. Shively CA, Register TC, Clarkson TB. Social stress, visceral obesity, and coronary artery atherosclerosis in female primates. Obesity (2009) 17:1513–20. doi: 10.1038/oby.2009.74
18. Bauer SA, Arndt TP, Leslie KE, Pearl DL, Turner PV. Obesity in rhesus and cynomolgus macaques: a comparative review of the condition and its implications for research. Comp Med. (2011) 61:514–26.
19. Shively CA, Register TC, Appt SE, Clarkson TB. Effects of long-term sertraline treatment and depression on coronary artery atherosclerosis in premenopausal female primates. Psychosom Med. (2015) 77:267–78. doi: 10.1097/PSY.0000000000000163
20. Yildirim S, Yeoman CJ, Sipos M, Torralba M, Wilson BA, Goldberg TL, et al. Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS ONE (2010) 5:e13963. doi: 10.1371/journal.pone.0013963
21. Ardeshir A, Narayan NR, Mendez-Lagares G, Lu D, Rauch M, Huang Y, et al. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci Transl Med. (2014) 6:252ra120. doi: 10.1126/scitranslmed.3008791
22. Narayan NR, Mendez-Lagares G, Ardeshir A, Lu D, Van Rompay KK, Hartigan-O'Connor DJ. Persistent effects of early infant diet and associated microbiota on the juvenile immune system. Gut Microbes (2015) 6:284–9. doi: 10.1080/19490976.2015.1067743
23. Springer A, Fichtel C, Al-Ghalith GA, Koch F, Amato KR, Clayton JB, et al. Patterns of seasonality and group membership characterize the gut microbiota in a longitudinal study of wild Verreaux's sifakas (Propithecus verreauxi). Ecol Evol. (2017) 7:5732–45. doi: 10.1002/ece3.3148
24. Bedard A, Riverin M, Dodin S, Corneau L, Lemieux S. Sex differences in the impact of the Mediterranean diet on cardiovascular risk profile. Br J Nutr. (2012) 108:1428–34. doi: 10.1017/S0007114511006969
25. Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics (2010a) 26:266–7. doi: 10.1093/bioinformatics/btp636
26. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods (2010b) 7:335–6. doi: 10.1038/nmeth.f.303
27. Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Isme J. (2012) 6:1621–4. doi: 10.1038/ismej.2012.8
28. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. (2007) 73:5261–7. doi: 10.1128/AEM.00062-07
29. Ochman H, Worobey M, Kuo CH, Ndjango JB, Peeters M, Hahn BH, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. (2010) 8:e1000546. doi: 10.1371/journal.pbio.1000546
30. Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science (2008) 320:1647–51. doi: 10.1126/science.1155725
31. Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science (2005) 307:1915–20. doi: 10.1126/science.1104816
32. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell (2006) 124:837–48. doi: 10.1016/j.cell.2006.02.017
33. Clayton JB, Vangay P, Huang H, Ward T, Hillmann BM, Al-Ghalith GA, et al. Captivity humanizes the primate microbiome. Proc Natl Acad Sci USA. (2016) 113:10376–81. doi: 10.1073/pnas.1521835113
34. Hale VL, Tan CL, Niu K, Yang Y, Knight R, Zhang Q, et al. Diet versus phylogeny: a comparison of gut microbiota in captive colobine monkey species. Microb Ecol. (2017) 75:515–27. doi: 10.1007/s00248-017-1041-8
35. Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A, et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. (2013) 7:1344–53. doi: 10.1038/ismej.2013.16
36. Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature (2016) 529:212–5. doi: 10.1038/nature16504
37. Amato KR, Leigh SR, Kent A, Mackie RI, Yeoman CJ, Stumpf RM, et al. The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouatta pigra). Microb Ecol. (2015) 69:434–43. doi: 10.1007/s00248-014-0554-7
38. Mackie RI, Aminov RI, Hu W, Klieve AV, Ouwerkerk D, Sundset MA, et al. Ecology of uncultivated Oscillospira species in the rumen of cattle, sheep, and reindeer as assessed by microscopy and molecular approaches. Appl Environ Microbiol. (2003) 69:6808–15. doi: 10.1128/AEM.69.11.6808-6815.2003
39. Haro C, Montes-Borrego M, Rangel-Zuniga OA, Alcala-Diaz JF, Gomez-Delgado F, Perez-Martinez P, et al. Two healthy diets modulate gut microbial community improving insulin sensitivity in a human obese population. J Clin Endocrinol Metab. (2016) 101:233–42. doi: 10.1210/jc.2015-3351
40. Sanchez-Villegas A, Delgado-Rodriguez M, Alonso A, Schlatter J, Lahortiga F, Serra Majem L, et al. Association of the Mediterranean dietary pattern with the incidence of depression: the Seguimiento Universidad de Navarra/University of Navarra follow-up (SUN) cohort. Arch Gen Psychiatry (2009) 66:1090–8. doi: 10.1001/archgenpsychiatry.2009.129
41. Marlow G, Ellett S, Ferguson IR, Zhu S, Karunasinghe N, Jesuthasan AC, et al. Transcriptomics to study the effect of a Mediterranean-inspired diet on inflammation in Crohn's disease patients. Hum Genomics (2013) 7:24. doi: 10.1186/1479-7364-7-24
42. Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. (2010) 26:5–11. doi: 10.1097/MOG.0b013e328333d751
43. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science (2011) 334:105–8. doi: 10.1126/science.1208344
44. Miquel S, Martin R, Rossi O, Bermudez-Humaran LG, Chatel JM, Sokol H, et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. (2013) 16:255–61. doi: 10.1016/j.mib.2013.06.003
45. Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes (2010) 59:3049–57. doi: 10.2337/db10-0253
46. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature (2012) 490:55–60. doi: 10.1038/nature11450
47. Videla S, Vilaseca J, Antolin M, Garcia-Lafuente A, Guarner F, Crespo E, et al. Dietary inulin improves distal colitis induced by dextran sodium sulfate in the rat. Am J Gastroenterol. (2001) 96:1486–93. doi: 10.1111/j.1572-0241.2001.03802.x
48. Costabile A, Klinder A, Fava F, Napolitano A, Fogliano V, Leonard C, et al. Whole-grain wheat breakfast cereal has a prebiotic effect on the human gut microbiota: a double-blind, placebo-controlled, crossover study. Br J Nutr. (2008) 99:110–20. doi: 10.1017/S0007114507793923
49. Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr. (2009) 101:541–50. doi: 10.1017/S0007114508019880
50. Pusceddu MM, El Aidy S, Crispie F, O'Sullivan O, Cotter P, Stanton C, et al. N-3 Polyunsaturated Fatty Acids (PUFAs) reverse the impact of early-life stress on the gut microbiota. PLoS ONE (2015) 10:e0139721. doi: 10.1371/journal.pone.0139721
51. Kaliannan K, Wang B, Li XY, Bhan AK, Kang JX. Omega-3 fatty acids prevent early-life antibiotic exposure-induced gut microbiota dysbiosis and later-life obesity. Int J Obes. (2016) 40:1039–42. doi: 10.1038/ijo.2016.27
52. Robertson RC, Seira Oriach C, Murphy K, Moloney GM, Cryan JF, Dinan TG, et al. Omega-3 polyunsaturated fatty acids critically regulate behaviour and gut microbiota development in adolescence and adulthood. Brain Behav Immun. (2017) 59:21–37. doi: 10.1016/j.bbi.2016.07.145
53. Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Backhed F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. (2015) 22:658–68. doi: 10.1016/j.cmet.2015.07.026
54. Lecomte V, Kaakoush NO, Maloney CA, Raipuria M, Huinao KD, Mitchell HM, et al. Changes in gut microbiota in rats fed a high fat diet correlate with obesity-associated metabolic parameters. PLoS ONE (2015) 10:e0126931. doi: 10.1371/journal.pone.0126931
55. Nagpal R, Kumar A, Kumar M, Behare PV, Jain S, Yadav H. Probiotics, their health benefits and applications for developing healthier foods: a review. FEMS Microbiol Lett. (2012) 334:1–15. doi: 10.1111/j.1574-6968.2012.02593.x
56. Claesson MJ, van Sinderen D, O'Toole PW. The genus Lactobacillus–a genomic basis for understanding its diversity. FEMS Microbiol Lett. (2007) 269:22–8. doi: 10.1111/j.1574-6968.2006.00596.x
57. Sanders ME. Probiotics: considerations for human health. Nutr Rev. (2003) 61:91–9. doi: 10.1301/nr.2003.marr.91-99
58. Nagpal R, Tsuji H, Takahashi T, Nomoto K, Kawashima K, Nagata S, et al. Ontogenesis of the gut microbiota composition in healthy, full-term, vaginally born and breast-fed infants over the first 3 years of life: a quantitative bird's-eye view. Front Microbiol. (2017b) 8:1388. doi: 10.3389/fmicb.2017.01388
Keywords: diet, microbiome, mediterranean, primates, cynomolgus macaque, western, metabolism, microbiota
Citation: Nagpal R, Shively CA, Appt SA, Register TC, Michalson KT, Vitolins MZ and Yadav H (2018) Gut Microbiome Composition in Non-human Primates Consuming a Western or Mediterranean Diet. Front. Nutr. 5:28. doi: 10.3389/fnut.2018.00028
Received: 22 December 2017; Accepted: 06 April 2018;
Published: 25 April 2018.
Edited by:
Marco Falasca, Curtin University, AustraliaReviewed by:
Angel Lanas, University of Zaragoza, SpainRex Gaskins, University of Illinois at Urbana-Champaign, United States
Copyright © 2018 Nagpal, Shively, Appt, Register, Michalson, Vitolins and Yadav. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hariom Yadav, aHlhZGF2QHdha2VoZWFsdGguZWR1
 Ravinder Nagpal
Ravinder Nagpal Carol A. Shively2
Carol A. Shively2 Thomas C. Register
Thomas C. Register Hariom Yadav
Hariom Yadav