
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Neurosci., 19 March 2025
Sec. Neurodevelopment
Volume 19 - 2025 | https://doi.org/10.3389/fnins.2025.1552790
This article is part of the Research TopicNeurodevelopmental Disorders and Cancer - Similarities and Differentiating FactorsView all 5 articles
Genome DNA of neurons in the brain is unstable, and mutations caused by inaccurate repair can lead to neurodevelopmental and neurodegenerative disorders. Damage to the neuronal genome is induced both exogenously and endogenously. Rapid cell proliferation of neural stem cells during embryonic brain development can lead to errors in genome duplication. Electrical excitations and drastic changes in gene expression in functional neurons cause risks of damaging genomic DNA. The precise repair of DNA damages caused by events making genomic DNA unstable maintains neuronal functions. The maintenance of the DNA sequence and structure of the genome is known as genomic integrity. Molecular mechanisms that maintain genomic integrity are critical for healthy neuronal function. In this review, we describe recent progress in understanding the genome integrity in functional neurons referring to their disruptions reported in neurological diseases.
The integrity of genomic DNA including DNA sequence and chromatin structure, is essential for cell survival and normal physiological function. Additionally, biochemical modifications such as DNA methylation, histone-associated epigenomic modifications, and other three-dimensional DNA structures play significant roles in maintaining genomic integrity. The genomic integrity must be preserved especially in stem cells, which produce the differentiated cells that constitute each organ, to maintain the physiological functions of each organ and for the health of individuals.
Genomic instability can occur even under normal physiological conditions. DNA damage which can be caused by mutations affect physiological cellular function and systemic health deterioration (Aguilera and García-Muse, 2013). For example, genomic integrity is disrupted in cancer cells, which results in the inability to maintain normal cellular function, as well as uncontrolled proliferation and metastasis. Therefore, the mechanisms that maintain genomic integrity, such as chromatin regulation and DNA repair systems, are crucial for maintaining normal cellular function. A better understanding of these mechanisms, along with strategies for repairing genomic damage, will be essential for disease prevention and treatment (Scheijen and Wilson, 2022).
It has been suggested that genomic integrity is not preserved in mature brain neurons (Zolzaya et al., 2024). 13–41% of human cortical neurons exhibit copy number variants (CNVs) of genes (McConnell et al., 2013), and recent next-generation sequencing studies have revealed that many smaller DNA sequence variants occur in the neuronal genome (Lodato et al., 2018; Luquette et al., 2022). Given that neurons in the brain have an extremely long lifespan and the production of new neurons from neural stem cells is limited, the mechanism preserving the normal function of neurons from genome instability is essential for healthy brain function.
Neurons must maintain high metabolic activity to transmit information effectively within the nervous system. The brain consumes approximately 25% of the body's glucose to produce the energy needed for this activity (Steiner, 2019; Trigo et al., 2022). Mature neurons produce 4.7 billion molecules of adenosine triphosphate (ATP) per second in their mitochondria. During this process, 1–3% of the oxygen is converted to reactive oxygen species (ROS), which can destabilize the genomic DNA (Salehi et al., 2018). As a result, neurons face higher risks of genome instability than other somatic cells (Zhu et al., 2012; Magistretti and Allaman, 2015). Above all, genomic DNA damage can also occur as a part of normal physiological brain functions. For example, double-strand breaks (DSBs) increase in the entorhinal cortex, parietal cortex, and dentate gyrus during the exploration of a novel environment (Suberbielle et al., 2013). Additionally, DSBs increase in the primary visual cortex when the eyes are illuminated for 15 min (Suberbielle et al., 2013) and in the hippocampus during memory formation in mice (Castro-Pérez et al., 2016).
In addition to physiological activity, neuronal genomics is highly susceptible to damage from drug toxicities (Sanchez-Aceves et al., 2024; Torre et al., 2021; Calls et al., 2021). Alcohol administration has been shown to cause DSBs accumulation in neurons (Rulten et al., 2008). Repeated cocaine administration to mice causes histone hyperacetylation at 1,696 loci in the nucleus accumbens (Renthal et al., 2009), resulting in DNA damage in multiple brain areas (de Souza et al., 2014). Methamphetamine, an indirect adrenergic receptor stimulator, also induces genomic DNA damage in neurons (Johnson et al., 2015; Tokunaga et al., 2008).
Neuronal activity induces the expression of immediate early genes (IEGs) which play important roles in neuronal plasticity (Yap and Greenberg, 2018). Dysregulation of IEGs leads to various neurological disorders (Ebert and Greenberg, 2013). The rapid regulation of IEGs is mediated by physical contact between the enhancer and promoter by single-strand breaks (SSBs) or DSBs in genomic DNA (Madabhushi et al., 2015; Wu et al., 2021; Delint-Ramirez et al., 2022). However, repeated SSBs and DSBs may induce physiological dysfunction of neurons when the 3D structure of the genome is altered (Dileep et al., 2023).
Epigenomic modifications in neurons are required for memory and learning functions in the brain (Zovkic et al., 2013). DNA methylation is modified to cytosines on CpG islands during brain development. The DNA methyltransferase DNMT1 is responsible for the methylation of newly synthesized strands after DNA replication (Li et al., 1992), while DNMT3A regulates gene expression in response to cellular conditions (Wei et al., 2021; Li et al., 2022). DNMT3B, on the other hand, is involved in methylation of X-chromosome-specific genes (Yagi et al., 2020). Proteins that bind to methylated DNA can induce chromatin structural transformation, repressing transcription of downstream genes. Because DNA methylation patterns are disrupted in various malignancies of cancers, DNMT mutations can destabilize the genome (Valencia and Kadoch, 2019). For example, contextual fear conditioning increases DNMT expression in hippocampal neurons, and DNMT inhibition reduces conditioned memory and suppresses long-term potentiation (LTP) in Schaeffer's lateral branch (Levenson et al., 2006). Therefore, DNA methylation regulation plays an important role in neuronal plasticity. Additionally, differences in DNA methylation levels are observed among neurons with different projection sites in the central nervous system (Zhang et al., 2021; Zhou et al., 2023).
The regulatory process of epigenomic modification involves DNA repair with base substitutions by the ten-eleven translocation (TET) enzyme family, TET1, TET2, and TET3. TET enzyme is important to activate the demethylation of DNA involving the process of 5-methylcytosine oxidation to 5-hydroxymethylcytosine (Zhang et al., 2023). It has been shown that spatial learning and short-term memory are impaired in TET1 knockout mice (Zhang et al., 2013), suggesting that maintenance of genomic integrity by regulation of epigenomic modifications is essential for normal neuronal function.
In most organs, malfunctioning cells are removed through apoptosis or phagocytosis, with the replacement of lost cells by regenerative mechanisms. However, in the brain, postnatal neurogenesis is limited only to a few regions, such as the dentate gyrus of the hippocampal formation. Microglia can phagocytose degenerated neurons (Butler et al., 2021), however, the mechanism of removal of neurons as a response to mutations in their genome remains unclear. Genomic mutations in many cell types often result in aberrant cellular characteristics, typically either undergoing apoptosis in a p53-dependent manner or being eliminated by the immune system (Attardi, 2005; Szybińska and Leśniak, 2017). However, mutated cells that break through these protective systems can become proliferative, ultimately leading to cancer. In contrast, neurons rarely acquire proliferative potential. Thus, it is likely that abnormal neurons will continue to reside in the brain contributing to neural activity, throughout the individual's neuronal lifespan.
Age-related genomic instability accumulates in neurons. For example, comparative genomic DNA sequencing of the prefrontal cortex and hippocampal neurons of individuals aged 4 months to 82 years reveals an age-dependent mutation (Lodato et al., 2018). Neurons in the brain with age-related neurodegeneration exhibit more mutations than those in healthy brains (Li et al., 2023). The accumulation of genomic DNA damage in neurons results in various neurodegenerative diseases (Rass et al., 2007). Neurons of Alzheimer's disease patients, for example, exhibit an increase in DSBs accumulation in the early stages, as observed in postmortem brain studies (Suberbielle et al., 2013; Wu et al., 2021; Madabhushi et al., 2015; Reid et al., 2021). Multiple reports have suggested that age-related loss of genomic integrity presumably contributes to Alzheimer's pathogenesis (Weissman et al., 2009; Kruman et al., 2004; Iourov et al., 2009; Herrup et al., 2013).
Mutations in the main component of DNA damage response (DDR) molecules are associated with human chromosomal instability syndromes (McKinnon, 2017; Shiloh, 2003). For example, telangiectatic ataxia is caused by mutations in ATM gene, which is a DDR molecule in DSBs (Shiloh and Rotman, 1996). Mutations in the MRE11, which act in SSBs detection have also been reported to occur in telangiectasia ataxia-like syndrome (Stewart et al., 1999). Similarly, mutations in the XRCC1 gene involved in SSBs repair cause oculomotor palsy, axonal neuropathy, and progressive cerebellar ataxia (Hoch et al., 2017; O'Connor et al., 2018).
As described above, functional neurons are frequently damaged by cellular metabolism, neurotransmission, and the regulation of gene expression, which are responsible for the physiological function of a healthy brain. To ensure the long-term maintenance of genomic integrity and neuronal functionality, neurons may rely on specific maintenance mechanisms [57]. Recent evidence from disease associations and experimental studies has shown that DNA structures such as R-loops, G-quadruplexes, and “long genes” influence the neuronal genomic integrity, and these structures are tightly regulated by enzymes such as topoisomerases and helicases (Figure 1).
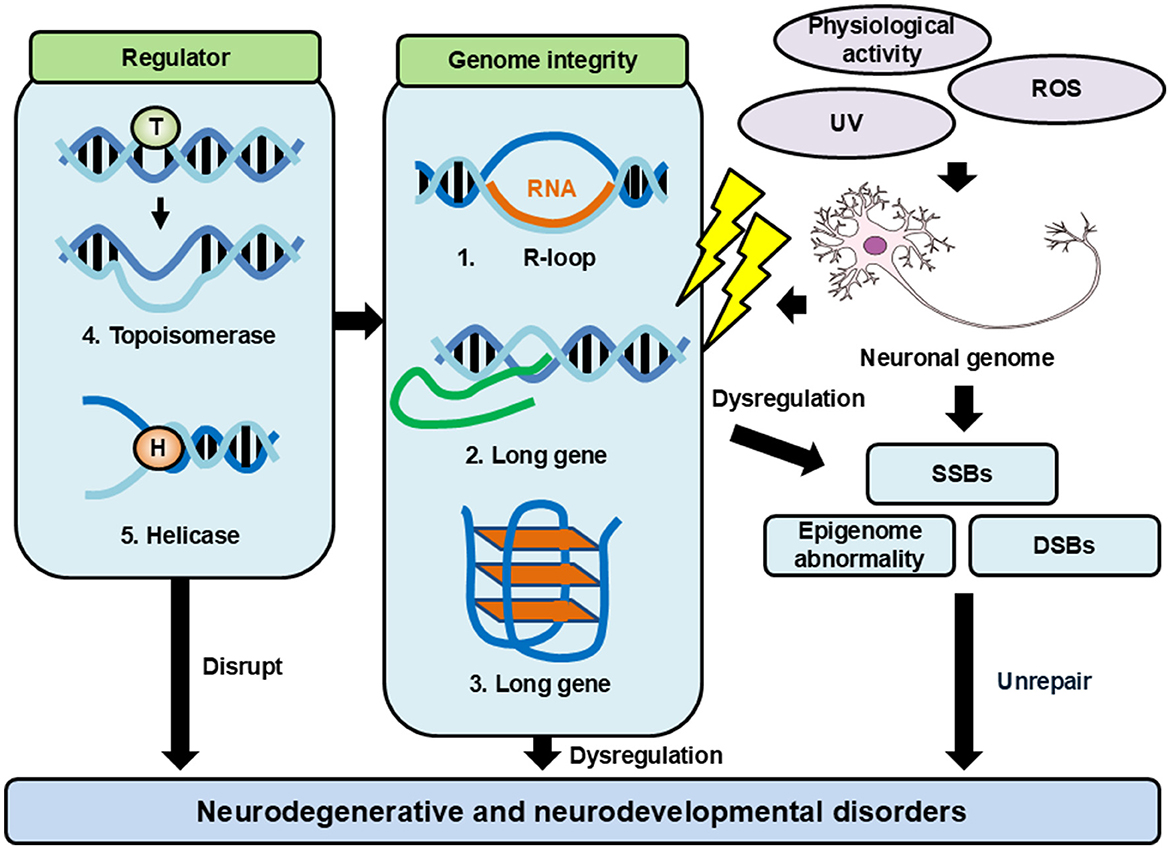
Figure 1. Factors affecting genomic integrity and regulatory mechanisms. Neurons have a lot of events that give damage to genome DNA. These damages cause genomic DNA aberrations and genomic instability. Neurons have many mechanisms to maintain their genomic integrity. When these mechanisms are failed, they lead to diseases such as neurodegenerative and neurodevelopmental disorders.
During gene expression, a DNA:RNA hybrid is formed between the template genomic DNA and the nascent RNA transcript, leaving non-template DNA single-stranded. This structure is called R-loop. If R-loop is not properly deleted by Senataxin or RNase H, it causes aberrant of replication forks and lead to DNA damage accumulation. In addition, R-loops affect various biological processes, including transcription, translation, and DNA repair mechanisms. Dysregulation of R-loop is involved in several neurological diseases (Groh and Gromak, 2014; Skourti-Stathaki et al., 2011; Sollier and Cimprich, 2015; Loomis et al., 2014). Abnormal accumulation of R-loop contributes to disorders such as telangiectatic ataxia (Groh and Gromak, 2014; García-Muse and Aguilera, 2019), amyotrophic lateral sclerosis (ALS; Salvi and Mekhail, 2015), ataxia-oculomotor apraxia (Fogel et al., 2014; Becherel et al., 2015), and spinal muscular atrophy (Kannan et al., 2018; Hensel et al., 2020). Recent studies have shown that R-loop contributes to the regulation of NPAS4 expression in response to chronic psychosocial stress or cocaine exposure (Akiki et al., 2024). Thus, R-loop also functions in the immediate response of neural activity, and other physiological functions of R-loop will be elucidated in the future.
The single-stranded DNA within R-loop is vulnerable to SSBs due to its susceptibility to nucleases (Allison and Wang, 2019). Additionally, R-loops create regions of negative and positive supercoiled DNA structures near transcription start sites, forming barriers to transcription elongation (Zardoni et al., 2021), participating in epigenomic regulation (Ginno et al., 2012), and influencing the DNA repair pathway (Keskin et al., 2014). To overcome this barrier, cells transiently cleave and rejoin DNA strands by topoisomerases to relieve torsional stress (Saunders et al., 2006; Le et al., 2019).
In fission yeast (Ohle et al., 2016) and human immortalized cell lines (RPE-hTERT cells; Yasuhara et al., 2018), RAD52 recognizes the R-loop as a landmark for repair, while XPG helicase removes it to activate homologous recombination repair (HRR). However, neurons lack efficient HRR mechanisms for DNA repair. For example, DSBs repair is reduced in sporadic ALS due to abnormal subcellular localization of TDP-43 in motoneurons. TDP-43 is rapidly accumulated at sites of DSBs in neurons and assembles factors that act on DNA repair, particularly non-homologous end-joining (NHEJ; Orii et al., 2006; Mitra et al., 2019). This observation suggests that the molecular machinery is one of the DNA repair mechanisms in R-loop metabolism in neurons.
G-quadruplex (G4) is the higher-order structure of nucleic acids which is formed in Hoogsteen hydrogen bonds of guanine (Monsen et al., 2022). These structures have high structural stability and play important functions in the regulation of transcription, replication, DSB site determination, genome stability, and RNA metabolism (Hänsel-Hertsch et al., 2017; Fay et al., 2017). Especially in the neurons, G4 functions to regulate the expression of downstream molecules of genes, such as the promoter of Tyrosine hydroxylase, the rate-limiting enzyme for catecholamine neurotransmitter biosynthesis (Banerjee et al., 2014). In fact, selective disruption of these G4 by mutating promoter DNA sequences affects Tyrosine hydroxylase transcription (Banerjee et al., 2014). Immunohistochemical analysis using adult mouse brains reveals that G4 is widely distributed in neurons throughout the brain regions, including the olfactory bulb, pyramidal cells in the hippocampus, granule cells in the dentate gyrus, and Purkinje cells in the cerebellum (Asamitsu et al., 2020; Comptdaer et al., 2024). Interestingly, G4 distribution in the nucleus of neurons is highly dynamic. G4 immunostaining revealed a lower positive number of G4 in glial cells than in neurons, suggesting that G4 formation is particularly active in neurons (Asamitsu et al., 2020).
Structural analysis indicated that DHX36 helicase, which belongs to the DExD/H box family, resolves the G4 structure (Chen et al., 2018). Genome-wide detection of G4 structure by G4-DNA sequencing, DHX36 unravels the G4 structure and restores the expression of genes suppressed during fear memory in mice exposed to electric shocks paired with sound stimuli, followed by the subsequent fear memory induced upon exposure to sound stimuli alone (Marshall et al., 2024). However, in neuronal progenitor cells, G4 stabilization promotes apoptosis due to DNA damage (Watson et al., 2013). Similarly, in rat neurons, G4 stabilization suppresses Brca1 gene expression, which is essential for DNA repair, and causes the accumulation of DSBs (Moruno-Manchon et al., 2017). Mutations in the ATRX gene stabilize G4 and accumulate DNA damage (Wang et al., 2019). G4 can be detected by gel-shift assay. ATRX helicase binds togenomic DNA and has been suggested to play a role in resolvinggenome-wide G4 and alleviating their adverse effects of G4 (Law et al., 2010).
Defects in MECP2, a protein that represses gene expression by binding to methylated DNA, result in impaired synaptic function and cause Rett syndrome. Studies using mouse models and brains of Rett syndrome patients have revealed that the MECP2 deficiency increased the expression of “long genes” spanning more than 100 kilobases across the genome, which encode synapse-related genes (Sugino et al., 2014; Gabel et al., 2015). Notably, neurons exhibit significantly higher expression of “long genes” than other cell types (Gabel et al., 2015). “Long genes” are strongly expressed in the frontal lobe and amygdala, which are associated with neurodevelopmental disorders such as autism (Gabel et al., 2015). Also, the expression of “longer genes” is specific to neurons among the cells that compose the brain (Zylka et al., 2015).
“Long genes” have also been implicated in other neurological diseases. TDP-43 and FUS/TLS loci are the genes responsible for amyotrophic lateral sclerosis and transcribe RNA products exceeding 100 kilobases in length (Lagier-Tourenne et al., 2012; Polymenidou et al., 2011). Similarly, CNTNAP2, another “long gene,” has been implicated in autism (Peñagarikano and Geschwind, 2012). Topoisomerase inhibitor topotecan treatment reverses overexpression of “long genes” in Rett syndrome models (Mabb et al., 2014; King et al., 2013).
In Drosophila neurons, aging causes an accumulation of R-loop in the “long gene” and this topological stress is resolved by Top3B (Jauregui-Lozano et al., 2022). “Long genes” make particularly complex DNA and RNA tangles in the regulation of the genome which is a characteristic of neurons. To maintain the genomic integrity of neurons, topoisomerase and various helicase complexes work together regulating “long gene” inducing SSBs and DSBs (Zagnoli-Vieira and Caldecott, 2020).
Topoisomerase (Top) works extensively to stabilize the genome and relieve topological stress on the DNA strand. A significant amount of research highlights the Top function in resolving DNA strand breaks during DNA replication to eliminate the super helix structure. Top inhibitors are widely studied for their anticancer effects because they produce DNA breaks that are lethal to proliferative cells (Pommier et al., 2022). Even in non-proliferative neurons, the Top is important for genomic integrity that works to resolve R-loop and G4 structures formed during gene expression. Dysregulation of various Top enzymes is involved in neurodegenerative and neurodevelopmental disorders such as autism, intellectual disorder, schizophrenia, and dementia (Katyal et al., 2014; Neale et al., 2012; Stoll et al., 2013; Tiwari and Wilson, 2019; Fragola et al., 2020; Milano et al., 2024; Crewe and Madabhushi, 2021). For example, Top3B mutations are associated with autism (Stoll et al., 2013; Iossifov et al., 2012), mental disorders (Ahmad et al., 2017a; Stoll et al., 2013), schizophrenia (Xu et al., 2012), and cognitive dysfunction (Kaufman et al., 2016).
Topoisomerase 2β (Top2B) also plays a unique role in neurogenesis. While deletion of Top2B does not affect neuronal production, it disrupts axon outgrowth of ventral horn motor neurons in the spinal cord (Yang et al., 2000). Consistently, Top2B deficient embryonic stem cells show no defects in proliferation or neuronal differentiation (Tiwari et al., 2012). Top2B inhibitor treatment increases the expression of 18% of genes expression in cerebellar granule cells (Tsutsui et al., 2001). In stimulated neurons, Top2B regulates gene expression by inducing DBS into IEGs (Delint-Ramirez et al., 2022). Therefore, regulation of genomic DNA structure by Top is essential for neuronal differentiation and functions.
Topoisomerase 1 (Top1) plays roles during transcription by resolving DNA supercoil, which promotes R-loop formation (Drolet et al., 1995; El Hage et al., 2010). However, Top1 deficiency also increases topological stress and promotes R-loop formation, suggesting that the Top enzyme acts in both R-loop formation and resolution (Promonet et al., 2020). Top3B, which is classified as a Type I Top like Top1, is unique as it can act on both DNA and RNA (Ahmad et al., 2017a,b; Saha et al., 2020). It is known that the functional inhibition of Top3B impairs the R-loop, resulting in a decrease in neuronal function, whereas overexpression of Top3B results in an increase in neuronal function (Skourti-Stathaki and Proudfoot, 2014). Mutant Top3B with reduced enzyme activity causes R-loop accumulation in the genome (Huang et al., 2018). Once engaged with R-loop, Top3B interacts with DDX5 to dissolve the structure (Saha et al., 2022a).
Top enzymes also interact with DNA via cleavage of complexes. For example, Top1 covalently binds to the 3′ phosphate terminus of DNA when DNA is untwisted, and this reaction intermediate (complex with DNA) is called Top1 cleavage complex (Topoisomerase1 cleavage complex; Top1cc), and the complex of Top2 and DNA is called Top2cc (Wojtaszek and Williams, 2024). If unresolved during DNA repair, these complexes can lead to additional DNA damage. Top1cc is removed from DNA by the enzymatic activity of tyrosyl DNA phosphodiesterase 1 (TDP1), and mutations in TDP1 cause spinocerebellar degeneration (El-Khamisy et al., 2005; Takashima et al., 2002). Similarly, mutations in TDP2 which remove Top2cc, are also linked to spinocerebellar degeneration (Gómez-Herreros et al., 2014).
Helicase is an enzyme that cuts the hydrogen bonds between the bases of DNA and RNA chains in an ATP-dependent manner and dissociates the nucleic acid strands. In neurons, specific helicases such as DHX36 and ATRX are responsible for the G4 structure in the genome, while DDX5 works to resolve the R-loop (Saha et al., 2022a). Notably, reduced expression of DDX21 in primarily cultured cortical neurons has been shown to accumulate G4 and genomic DNA damage (Lyu et al., 2022). DDX21 also plays a role in eliminating R-loops (Song et al., 2017). Since helicases can unwind both DNA-DNA and DNA-RNA hybrids, they are important factors for maintaining genomic integrity. However, the functional specificity of approximately 100 helicases in the human genome is still largely unknown. Many helicases work in cooperation with Top, suggesting that these complexes form to preserve genomic integrity (Tsukada et al., 2024; Tan et al., 2023; Saha et al., 2022b; Gupta et al., 2022; Yang et al., 2020). Unique and/or specific functions of each helicase should be unraveled in the future.
The genomic integrity that sustains neuronal function must be maintained for the healthy brain. However, genomic destabilization is an ongoing challenge, arising from daily stress, the physiological activity of neurons, and the accumulation of DNA damage (Zolzaya et al., 2024). Mutations can also occur in the genomes of neurons in normal growth and aging of individuals. In other words, the repair of the neuronal genomic DNA is often incomplete, causing unavoidable genomic damage and possible effects on neuronal function. This ongoing struggle with long-lived neurons during the growth and aging process contributes to the risk of neurodegenerative and psychiatric diseases, where repeated DNA damage and structural genomic changes impair neural function.
Recent studies have suggested that the protective mechanism for neuron-specific genomic integrity may be the action of a large molecular network of topoisomerases, helicases, and factors involved in DNA repair in response to changes in the genomic DNA structure of neurons, including R-loop, G4, and “long gene” regulation. Technological innovations have made it possible to analyze the function of these molecules in a whole genome, and advances in mass spectrometry-based methods have made it possible to identify larger molecular networks comprehensively. By analyzing molecules essential for genomic integrity, we can examine how the molecular networks are responsible for neuronal function throughout the genome, thereby revealing the dynamism of the neuronal genome and linking malfunctions of this network to a variety of neurological diseases.
DI: Writing – original draft, Writing – review & editing. NR: Writing – review & editing. YK: Writing – original draft, Writing – review & editing.
The author(s) declare that no financial support was received for the research and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aguilera, A., and García-Muse, T. (2013). Causes of genome instability. Annu. Rev. Genet. 47, 1–32. doi: 10.1146/annurev-genet-111212-133232
Ahmad, M., Shen, W., Li, W., Xue, Y., Zou, S., Xu, D., et al. (2017a). Topoisomerase 3β is the major topoisomerase for mRNAs and linked to neurodevelopment and mental dysfunction. Nucleic. Acids Res. 45, 2704–2713. doi: 10.1093/nar/gkw1293
Ahmad, M., Xu, D., and Wang, W. (2017b). Type IA topoisomerases can be “magicians” for both DNA and RNA in all domains of life. RNA Biol. 14, 854–864. doi: 10.1080/15476286.2017.1330741
Akiki, R. M., Cornbrooks, R. G., Magami, K., Greige, A., Snyder, K. K., Wood, D. J., et al. (2024). A long noncoding eRNA forms R-loops to shape emotional experience-induced behavioral adaptation. Science. 386, 1282–1289. doi: 10.1126/science.adp1562
Allison, D. F., and Wang, G. G. (2019). R-loops: formation, function, and relevance to cell stress. Cell Stress. 3, 38–46. doi: 10.15698/cst2019.02.175
Asamitsu, S., Imai, Y., Yabuki, Y., Ikenoshita, S., Takeuchi, M., Kashiwagi, H., et al. (2020). Identification and immunohistochemical characterization of G-quadruplexes in mouse brain. Biochem. Biophys. Res. Commun. 531, 67–74. doi: 10.1016/j.bbrc.2020.01.145
Attardi, L. D. (2005). The role of p53-mediated apoptosis as a crucial anti-tumor response to genomic instability: lessons from mouse models. Mutat. Res. 569, 145–157. doi: 10.1016/j.mrfmmm.2004.04.019
Banerjee, K., Wang, M., Cai, E., Fujiwara, N., Baker, H., Cave, J. W., et al. (2014). Regulation of tyrosine hydroxylase transcription by hnRNP K and DNA secondary structure. Nat. Commun. 5:5769. doi: 10.1038/ncomms6769
Becherel, O. J., Sun, J., Yeo, A. J., Nayler, S., Fogel, B. L., Gao, F., et al. (2015). A new model to study neurodegeneration in ataxia oculomotor apraxia type 2. Hum. Mol. Genet. 24, 5759–5774. doi: 10.1093/hmg/ddv296
Butler, C. A., Popescu, A. S., Kitchener, E. J. A., Allendorf, D. H., Puigdellívol, M., Brown, G. C., et al. (2021). Microglial phagocytosis of neurons in neurodegeneration, and its regulation. J. Neurochem. 158, 621–639. doi: 10.1111/jnc.15327
Calls, A., Torres-Espin, A., Navarro, X., Yuste, V. J., Udina, E., Bruna, J., et al. (2021). Cisplatin-induced peripheral neuropathy is associated with neuronal senescence-like response. Neuro Oncol. 23, 88–99. doi: 10.1093/neuonc/noaa151
Castro-Pérez, E., Soto-Soto, E., Pérez-Carambot, M., Dionisio-Santos, D., Saied-Santiago, K., Ortiz-Zuazaga, H. G., et al. (2016). Identification and characterization of the V(D)J recombination activating gene 1 in long-term memory of context fear conditioning. Neural. Plast. 2016:1752176. doi: 10.1155/2016/1752176
Chen, M. C., Tippana, R., Demeshkina, N. A., Murat, P., Balasubramanian, S., Myong, S., et al. (2018). Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature. 558, 465–469. doi: 10.1038/s41586-018-0209-9
Comptdaer, T., Tardivel, M., Schirmer, C., Buée, L., and Galas, M. C. (2024). Cell redistribution of G quadruplex-structured DNA is associated with morphological changes of nuclei and nucleoli in neurons during tau pathology progression. Brain Pathol. 35:e13262. doi: 10.1111/bpa.13262
Crewe, M., and Madabhushi, R. (2021). Topoisomerase-mediated DNA damage in neurological disorders. Front. Aging Neurosci. 13:751742. doi: 10.3389/fnagi.2021.751742
de Souza, M. F., Gonçales, T. A., Steinmetz, A., Moura, D. J., Saffi, J., Gomez, R., et al. (2014). Cocaine induces DNA damage in distinct brain areas of female rats under different hormonal conditions. Clin. Exp. Pharmacol. Physiol. 41, 265–269. doi: 10.1111/1440-1681.12218
Delint-Ramirez, I., Konada, L., Heady, L., Rueda, R., Jacome, A. S. V., Marlin, E., et al. (2022). Calcineurin dephosphorylates topoisomerase IIβ and regulates the formation of neuronal-activity-induced DNA breaks. Mol. Cell 82, 3794–809. e8. doi: 10.1016/j.molcel.2022.09.012
Dileep, V., Boix, C. A., Mathys, H., Marco, A., Welch, G. M., Meharena, H. S., et al. (2023). Neuronal DNA double-strand breaks lead to genome structural variations and 3D genome disruption in neurodegeneration. Cell 186, 4404–21. e20. doi: 10.1016/j.cell.2023.08.038
Drolet, M., Phoenix, P., Menzel, R., Mass,é, E., Liu, L. F., Crouch, R. J., et al. (1995). Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc. Natl. Acad. Sci. U.S.A. 92, 3526–3530. doi: 10.1073/pnas.92.8.3526
Ebert, D. H., and Greenberg, M. E. (2013). Activity-dependent neuronal signalling and autism spectrum disorder. Nature 493, 327–337. doi: 10.1038/nature11860
El Hage, A., French, S. L., Beyer, A. L., and Tollervey, D. (2010). Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 24, 1546–1558. doi: 10.1101/gad.573310
El-Khamisy, S. F., Saifi, G. M., Weinfeld, M., Johansson, F., Helleday, T., Lupski, J. R., et al. (2005). Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434, 108–113. doi: 10.1038/nature03314
Fay, M. M., Lyons, S. M., and Ivanov, P. R. N. A. (2017). G-quadruplexes in biology: principles and molecular mechanisms. J. Mol. Biol. 429, 2127–2147. doi: 10.1016/j.jmb.2017.05.017
Fogel, B. L., Cho, E., Wahnich, A., Gao, F., Becherel, O. J., Wang, X., et al. (2014). Mutation of senataxin alters disease-specific transcriptional networks in patients with ataxia with oculomotor apraxia type 2. Hum. Mol. Genet. 23, 4758–4769. doi: 10.1093/hmg/ddu190
Fragola, G., Mabb, A. M., Taylor-Blake, B., Niehaus, J. K., Chronister, W. D., Mao, H., et al. (2020). Deletion of topoisomerase 1 in excitatory neurons causes genomic instability and early onset neurodegeneration. Nat. Commun. 11:1962. doi: 10.1038/s41467-020-15794-9
Gabel, H. W., Kinde, B., Stroud, H., Gilbert, C. S., Harmin, D. A., Kastan, N. R., et al. (2015). Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 522, 89–93. doi: 10.1038/nature14319
García-Muse, T., and Aguilera, A. R. (2019). Loops: from physiological to pathological roles. Cell 179, 604–618. doi: 10.1016/j.cell.2019.08.055
Ginno, P. A., Lott, P. L., Christensen, H. C., Korf, I., and Chédin, F. (2012). R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 45, 814–825. doi: 10.1016/j.molcel.2012.01.017
Gómez-Herreros, F., Schuurs-Hoeijmakers, J. H., McCormack, M., Greally, M. T., Rulten, S., Romero-Granados, R., et al. (2014). TDP2 protects transcription from abortive topoisomerase activity and is required for normal neural function. Nat. Genet. 46, 516–521. doi: 10.1038/ng.2929
Groh, M., and Gromak, N. (2014). Out of balance: R-loops in human disease. PLoS Genet. 10:e1004630. doi: 10.1371/journal.pgen.1004630
Gupta, P., Majumdar, A. G., and Patro, B. S. (2022). Non-enzymatic function of WRN RECQL helicase regulates removal of topoisomerase-I-DNA covalent complexes and triggers NF-κB signaling in cancer. Aging Cell 21:e13625. doi: 10.1111/acel.13625
Hänsel-Hertsch, R., Di Antonio, M., and Balasubramanian, S. (2017). DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 18, 279–284. doi: 10.1038/nrm.2017.3
Hensel, N., Detering, N. T., Walter, L. M., and Claus, P. (2020). Resolution of pathogenic R-loops rescues motor neuron degeneration in spinal muscular atrophy. Brain 143, 2–5. doi: 10.1093/brain/awz394
Herrup, K., Li, J., and Chen, J. (2013). The role of ATM and DNA damage in neurons: upstream and downstream connections. DNA Repair. 12, 600–604. doi: 10.1016/j.dnarep.2013.04.012
Hoch, N. C., Hanzlikova, H., Rulten, S. L., Tétreault, M., Komulainen, E., Ju, L., et al. (2017). XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature 541, 87–91. doi: 10.1038/nature20790
Huang, L., Wang, Z., Narayanan, N., and Yang, Y. (2018). Arginine methylation of the C-terminus RGG motif promotes TOP3B topoisomerase activity and stress granule localization. Nucleic Acids Res. 46, 3061–3074. doi: 10.1093/nar/gky103
Iossifov, I., Ronemus, M., Levy, D., Wang, Z., Hakker, I., Rosenbaum, J., et al. (2012). De novo gene disruptions in children on the autistic spectrum. Neuron 74, 285–299. doi: 10.1016/j.neuron.2012.04.009
Iourov, I. Y., Vorsanova, S. G., Liehr, T., and Yurov, Y. B. (2009). Aneuploidy in the normal, Alzheimer's disease and ataxia-telangiectasia brain: differential expression and pathological meaning. Neurobiol. Dis. 34, 212–220. doi: 10.1016/j.nbd.2009.01.003
Jauregui-Lozano, J., Escobedo, S., Easton, A., Lanman, N. A., Weake, V. M., Hall, H., et al. (2022). Proper control of R-loop homeostasis is required for maintenance of gene expression and neuronal function during aging. Aging Cell 21:e13554. doi: 10.1111/acel.13554
Johnson, Z., Venters, J., Guarraci, F. A., and Zewail-Foote, M. (2015). Methamphetamine induces DNA damage in specific regions of the female rat brain. Clin. Exp. Pharmacol. Physiol. 42, 570–575. doi: 10.1111/1440-1681.12404
Kannan, A., Bhatia, K., Branzei, D., and Gangwani, L. (2018). Combined deficiency of Senataxin and DNA-PKcs causes DNA damage accumulation and neurodegeneration in spinal muscular atrophy. Nucleic Acids Res. 46, 8326–8346. doi: 10.1093/nar/gky641
Katyal, S., Lee, Y., Nitiss, K. C., Downing, S. M., Li, Y., Shimada, M., et al. (2014). Aberrant topoisomerase-1 DNA lesions are pathogenic in neurodegenerative genome instability syndromes. Nat. Neurosci. 17, 813–821. doi: 10.1038/nn.3715
Kaufman, C. S., Genovese, A., and Butler, M. G. (2016). Deletion of TOP3B is associated with cognitive impairment and facial dysmorphism. Cytogenet. Genome Res. 150, 106–111. doi: 10.1159/000452815
Keskin, H., Shen, Y., Huang, F., Patel, M., Yang, T., Ashley, K., et al. (2014). Transcript-RNA-templated DNA recombination and repair. Nature 515, 436–439. doi: 10.1038/nature13682
King, I. F., Yandava, C. N., Mabb, A. M., Hsiao, J. S., Huang, H. S., Pearson, B. L., et al. (2013). Topoisomerases facilitate transcription of long genes linked to autism. Nature 501, 58–62. doi: 10.1038/nature12504
Kruman, I. I., Wersto, R. P., Cardozo-Pelaez, F., Smilenov, L., Chan, S. L., Chrest, F. J., et al. (2004). Cell cycle activation linked to neuronal cell death initiated by DNA damage. Neuron 41, 549–561. doi: 10.1016/S0896-6273(04)00017-0
Lagier-Tourenne, C., Polymenidou, M., Hutt, K. R., Vu, A. Q., Baughn, M., Huelga, S. C., et al. (2012). Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 15, 1488–1497. doi: 10.1038/nn.3230
Law, M. J., Lower, K. M., Voon, H. P., Hughes, J. R., Garrick, D., Viprakasit, V., et al. (2010). ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 143, 367–378. doi: 10.1016/j.cell.2010.09.023
Le, T. T., Gao, X., Park, S. H., Lee, J., Inman, J. T., Lee, J. H., et al. (2019). Synergistic coordination of chromatin torsional mechanics and topoisomerase activity. Cell 179, 619–31. e15. doi: 10.1016/j.cell.2019.09.034
Levenson, J. M., Roth, T. L., Lubin, F. D., Miller, C. A., Huang, I. C., Desai, P., et al. (2006). Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J. Biol. Chem. 281, 15763–15773. doi: 10.1074/jbc.M511767200
Li, E., Bestor, T. H., and Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926. doi: 10.1016/0092-8674(92)90611-F
Li, J., Pinto-Duarte, A., Zander, M., Cuoco, M. S., Lai, C. Y., Osteen, J., et al. (2022). Dnmt3a knockout in excitatory neurons impairs postnatal synapse maturation and increases the repressive histone modification H3K27me3. Elife 11:e66909. doi: 10.7554/eLife.66909
Li, Z., Min, S., Alliey-Rodriguez, N., Giase, G., Cheng, L., Craig, D. W., et al. (2023). Single-neuron whole genome sequencing identifies increased somatic mutation burden in Alzheimer's disease related genes. Neurobiol. Aging. 123, 222–232. doi: 10.1016/j.neurobiolaging.2022.12.002
Lodato, M. A., Rodin, R. E., Bohrson, C. L., Coulter, M. E., Barton, A. R., Kwon, M., et al. (2018). Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 359, 555–559. doi: 10.1126/science.aao4426
Loomis, E. W., Sanz, L. A., Chédin, F., and Hagerman, P. J. (2014). Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLoS Genet. 10:e1004294. doi: 10.1371/journal.pgen.1004294
Luquette, L. J., Miller, M. B., Zhou, Z., Bohrson, C. L., Zhao, Y., Jin, H., et al. (2022). Single-cell genome sequencing of human neurons identifies somatic point mutation and indel enrichment in regulatory elements. Nat. Genet. 54, 1564–1571. doi: 10.1038/s41588-022-01180-2
Lyu, J., Shao, R., Kwong Yung, P. Y., and Elsässer, S. J. (2022). Genome-wide mapping of G-quadruplex structures with CUTandTag. Nucleic. Acids Res. 50:e13. doi: 10.1093/nar/gkab1073
Mabb, A. M., Kullmann, P. H., Twomey, M. A., Miriyala, J., Philpot, B. D., Zylka, M. J., et al. (2014). Topoisomerase 1 inhibition reversibly impairs synaptic function. Proc. Natl. Acad. Sci. U S A. 111, 17290–17295. doi: 10.1073/pnas.1413204111
Madabhushi, R., Gao, F., Pfenning, A. R., Pan, L., Yamakawa, S., Seo, J., et al. (2015). Activity-Iinduced DNA breaks govern the expression of neuronal early-response genes. Cell 161, 1592–1605. doi: 10.1016/j.cell.2015.05.032
Magistretti, P. J., and Allaman, I. (2015). A cellular perspective on brain energy metabolism and functional imaging. Neuron 86, 883–901. doi: 10.1016/j.neuron.2015.03.035
Marshall, P. R., Davies, J., Zhao, Q., Liau, W. S., Lee, Y., Basic, D., et al. (2024). DNA G-quadruplex is a transcriptional control device that regulates memory. J. Neurosci. 44:e0093232024. doi: 10.1523/JNEUROSCI.0093-23.2024
McConnell, M. J., Lindberg, M. R., Brennand, K. J., Piper, J. C., Voet, T., Cowing-Zitron, C., et al. (2013). Mosaic copy number variation in human neurons. Science 342, 632–637. doi: 10.1126/science.1243472
McKinnon, P. J. (2017). Genome integrity and disease prevention in the nervous system. Genes Dev. 31, 1180–1194. doi: 10.1101/gad.301325.117
Milano, L., Gautam, A., and Caldecott, K. W. (2024). DNA damage and transcription stress. Mol. Cell 84, 70–79. doi: 10.1016/j.molcel.2023.11.014
Mitra, J., Guerrero, E. N., Hegde, P. M., Liachko, N. F., Wang, H., Vasquez, V., et al. (2019). Motor neuron disease-associated loss of nuclear TDP-43 is linked to DNA double-strand break repair defects. Proc. Natl. Acad. Sci. U S A. 116, 4696–4705. doi: 10.1073/pnas.1818415116
Monsen, R. C., Trent, J. O., and Chaires, J. B. (2022). G-quadruplex DNA: a longer story. Acc. Chem. Res. 55, 3242–3252. doi: 10.1021/acs.accounts.2c00519
Moruno-Manchon, J. F., Koellhoffer, E. C., Gopakumar, J., Hambarde, S., Kim, N., McCullough, L. D., et al. (2017). The G-quadruplex DNA stabilizing drug pyridostatin promotes DNA damage and downregulates transcription of Brca1 in neurons. Aging 9, 1957–1970. doi: 10.18632/aging.101282
Neale, B. M., Kou, Y., Liu, L., Ma'ayan, A., Samocha, K. E., Sabo, A., et al. (2012). Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245. doi: 10.1038/nature11011
O'Connor, E., Vandrovcova, J., Bugiardini, E., Chelban, V., Manole, A., Davagnanam, I., et al. (2018). Mutations in XRCC1 cause cerebellar ataxia and peripheral neuropathy. J. Neurol. Neurosurg. Psychiatry 89, 1230–1232. doi: 10.1136/jnnp-2017-317581
Ohle, C., Tesorero, R., Schermann, G., Dobrev, N., Sinning, I., Fischer, T., et al. (2016). Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell 167, 1001–13. e7. doi: 10.1016/j.cell.2016.10.001
Orii, K. E., Lee, Y., Kondo, N., and McKinnon, P. J. (2006). Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc. Natl. Acad. Sci. U S A 103, 10017–10022. doi: 10.1073/pnas.0602436103
Peñagarikano, O., and Geschwind, D. H. (2012). What does CNTNAP2 reveal about autism spectrum disorder? Trends Mol. Med. 18, 156–163. doi: 10.1016/j.molmed.2012.01.003
Polymenidou, M., Lagier-Tourenne, C., Hutt, K. R., Huelga, S. C., Moran, J., Liang, T. Y., et al. (2011). Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 14, 459–468. doi: 10.1038/nn.2779
Pommier, Y., Nussenzweig, A., Takeda, S., and Austin, C. (2022). Human topoisomerases and their roles in genome stability and organization. Nat. Rev. Mol. Cell Biol. 23, 407–427. doi: 10.1038/s41580-022-00452-3
Promonet, A., Padioleau, I., Liu, Y., Sanz, L., Biernacka, A., Schmitz, A. L., et al. (2020). Topoisomerase 1 prevents replication stress at R-loop-enriched transcription termination sites. Nat. Commun. 11:3940. doi: 10.1038/s41467-020-17858-2
Rass, U., Ahel, I., and West, S. C. (2007). Defective DNA repair and neurodegenerative disease. Cell 130, 991–1004. doi: 10.1016/j.cell.2007.08.043
Reid, D. A., Reed, P. J., Schlachetzki, J. C., Nitulescu, I. I., Chou, G., Tsui, E. C., et al. (2021). Incorporation of a nucleoside analog maps genome repair sites in postmitotic human neurons. Science 372, 91–94. doi: 10.1126/science.abb9032
Renthal, W., Kumar, A., Xiao, G., Wilkinson, M., Covington, H. E., 3rd, Maze, I., et al. (2009). Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron 62, 335–348. doi: 10.1016/j.neuron.2009.03.026
Rulten, S. L., Hodder, E., Ripley, T. L., Stephens, D. N., and Mayne, L. V. (2008). Alcohol induces DNA damage and the Fanconi anemia D2 protein implicating FANCD2 in the DNA damage response pathways in brain. Alcohol Clin. Exp. Res. 32, 1186–1196. doi: 10.1111/j.1530-0277.2008.00673.x
Saha, S., Sun, Y., Huang, S.-yN, Baechler SA, Pongor LS, Agama K, et al. (2020). DNA and RNA cleavage complexes and repair pathway for TOP3B RNA-and DNA-protein crosslinks. Cell Rep. 33:108569. doi: 10.1016/j.celrep.2020.108569
Saha, S., Yang, X., Huang, S. N., Agama, K., Baechler, S. A., Sun, Y., et al. (2022a). Resolution of R-loops by topoisomerase III-β (TOP3B) in coordination with the DEAD-box helicase DDX5. Cell Rep. 40:111067. doi: 10.1016/j.celrep.2022.111067
Saha, S., Yang, X., Huang, S.-y. N., Agama, K., Baechler, S. A., Sun, Y., Zhang, H., et al. (2022b). Resolution of R-loops by topoisomerase III-β (TOP3B) in coordination with the DEAD-box helicase DDX5. Cell Rep. 40:111067. doi: 10.2139/ssrn.4046140
Salehi, F., Behboudi, H., Kavoosi, G., and Ardestani, S. K. (2018). Oxidative DNA damage induced by ROS-modulating agents with the ability to target DNA: A comparison of the biological characteristics of citrus pectin and apple pectin. Sci. Rep. 8:13902. doi: 10.1038/s41598-018-32308-2
Salvi, J. S., and Mekhail, K. (2015). R-loops highlight the nucleus in ALS. Nucleus 6, 23–29. doi: 10.1080/19491034.2015.1004952
Sanchez-Aceves, L. M., Pérez-Alvarez, I., Onofre-Camarena, D. B., Gutiérrez-Noya, V. M., Rosales-Pérez, K. E., Orozco-Hernández, J. M., et al. (2024). Prolonged exposure to the synthetic glucocorticoid dexamethasone induces brain damage via oxidative stress and apoptotic response in adult Daniorerio. Chemosphere 364:143012. doi: 10.1016/j.chemosphere.2024.143012
Saunders, A., Core, L. J., and Lis, J. T. (2006). Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7, 557–567. doi: 10.1038/nrm1981
Scheijen, E. E. M., and Wilson, D. M. (2022). 3rd. Genome integrity and neurological disease. Int. J. Mol. Sci. 23:4142. doi: 10.3390/ijms23084142
Shiloh, Y. (2003). ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3, 155–168. doi: 10.1038/nrc1011
Shiloh, Y., and Rotman, G. (1996). Ataxia-telangiectasia and the ATM gene: linking neurodegeneration, immunodeficiency, and cancer to cell cycle checkpoints. J. Clin. Immunol. 16, 254–260. doi: 10.1007/BF01541389
Skourti-Stathaki, K., and Proudfoot, N. J. (2014). A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 28, 1384–1396. doi: 10.1101/gad.242990.114
Skourti-Stathaki, K., Proudfoot, N. J., and Gromak, N. (2011). Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42, 794–805. doi: 10.1016/j.molcel.2011.04.026
Sollier, J., and Cimprich, K. A. (2015). Breaking bad: R-loops and genome integrity. Trends Cell. Biol. 25, 514–522. doi: 10.1016/j.tcb.2015.05.003
Song, C., Hotz-Wagenblatt, A., Voit, R., and Grummt, I. (2017). SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 31, 1370–1381. doi: 10.1101/gad.300624.117
Steiner, P. (2019). Brain fuel utilization in the developing brain. Ann. Nutr. Metab. 75, 8–18. doi: 10.1159/000508054
Stewart, G. S., Maser, R. S., Stankovic, T., Bressan, D. A., Kaplan, M. I., Jaspers, N. G., et al. (1999). The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99, 577–587. doi: 10.1016/S0092-8674(00)81547-0
Stoll, G., Pietiläinen, O. P. H., Linder, B., Suvisaari, J., Brosi, C., Hennah, W., et al. (2013). Deletion of TOP3β, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 16, 1228–1237. doi: 10.1038/nn.3484
Suberbielle, E., Sanchez, P. E., Kravitz, A. V., Wang, X., Ho, K., Eilertson, K., et al. (2013). Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat. Neurosci. 16, 613–621. doi: 10.1038/nn.3356
Sugino, K., Hempel, C. M., Okaty, B. W., Arnson, H. A., Kato, S., Dani, V. S., et al. (2014). Cell-type-specific repression by methyl-CpG-binding protein 2 is biased toward long genes. J. Neurosci. 34, 12877–12883. doi: 10.1523/JNEUROSCI.2674-14.2014
Szybińska, A., and Leśniak, W. (2017). P53 dysfunction in neurodegenerative diseases—the cause or effect of pathological changes? Aging Dis. 8, 506–518. doi: 10.14336/AD.2016.1120
Takashima, H., Boerkoel, C. F., John, J., Saifi, G. M., Salih, M. A., Armstrong, D., et al. (2002). Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 32, 267–272. doi: 10.1038/ng987
Tan, Y., Yao, L., Gamliel, A., Nair, S. J., Taylor, H., Ohgi, K., et al. (2023). Signal-induced enhancer activation requires Ku70 to read topoisomerase1-DNA covalent complexes. Nat. Struct. Mol. Biol. 30, 148–158. doi: 10.1038/s41594-022-00883-8
Tiwari, V., and Wilson, D. M. (2019). 3rd. DNA damage and associated DNA repair defects in disease and premature aging. Am. J. Hum. Genet. 105, 237–257. doi: 10.1016/j.ajhg.2019.06.005
Tiwari, V. K., Burger, L., Nikoletopoulou, V., Deogracias, R., Thakurela, S., Wirbelauer, C., et al. (2012). Target genes of Topoisomerase IIβ regulate neuronal survival and are defined by their chromatin state. Proc. Natl. Acad. Sci. U S A 109, E934–E943. doi: 10.1073/pnas.1119798109
Tokunaga, I., Ishigami, A., Kubo, S., Gotohda, T., and Kitamura, O. (2008). The peroxidative DNA damage and apoptosis in methamphetamine-treated rat brain. J. Med. Invest. 55, 241–245. doi: 10.2152/jmi.55.241
Torre, M., Dey, A., Woods, J. K., and Feany, M. B. (2021). Elevated oxidative stress and DNA damage in cortical neurons of chemotherapy patients. J. Neuropathol. Exp. Neurol. 80, 705–712. doi: 10.1093/jnen/nlab074
Trigo, D., Avelar, C., Fernandes, M., Sá, J., and Cruz da, E. S. O. (2022). Mitochondria, energy, and metabolism in neuronal health and disease. FEBS Lett. 596, 1095–1110. doi: 10.1002/1873-3468.14298
Tsukada, K., Jones, S. E., Bannister, J., Durin, M. A., Vendrell, I., Fawkes, M., et al. (2024). BLM and BRCA1-BARD1 coordinate complementary mechanisms of joint DNA molecule resolution. Mol. Cell 84, 640–58. e10. doi: 10.1016/j.molcel.2023.12.040
Tsutsui, K., Tsutsui, K., Sano, K., Kikuchi, A., and Tokunaga, A. (2001). Involvement of DNA topoisomerase IIbeta in neuronal differentiation. J. Biol. Chem. 276, 5769–5778. doi: 10.1074/jbc.M008517200
Valencia, A. M., and Kadoch, C. (2019). Chromatin regulatory mechanisms and therapeutic opportunities in cancer. Nat. Cell. Biol. 21, 152–161. doi: 10.1038/s41556-018-0258-1
Wang, Y., Yang, J., Wild, A. T., Wu, W. H., Shah, R., Danussi, C., et al. (2019). G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat. Commun. 10:943. doi: 10.1038/s41467-019-08905-8
Watson, L. A., Solomon, L. A., Li, J. R., Jiang, Y., Edwards, M., Shin-Ya, K., et al. (2013). Atrx deficiency induces telomere dysfunction, endocrine defects, and reduced life span. J. Clin. Inves. 123, 2049–2063. doi: 10.1172/JCI65634
Wei, J., Cheng, J., Waddell, N. J., Wang, Z. J., Pang, X., Cao, Q., et al. (2021). DNA Methyltransferase 3A is involved in the sustained effects of chronic stress on synaptic functions and behaviors. Cereb. Cortex 31, 1998–2012. doi: 10.1093/cercor/bhaa337
Weissman, L., de Souza-Pinto, N. C., Mattson, M. P., and Bohr, V. A. (2009). DNA base excision repair activities in mouse models of Alzheimer's disease. Neurobiol. Aging 30, 2080–2081. doi: 10.1016/j.neurobiolaging.2008.02.014
Wojtaszek, J. L., and Williams, R. S. (2024). From the TOP: formation, recognition and resolution of topoisomerase DNA protein crosslinks. DNA Repair 142:103751. doi: 10.1016/j.dnarep.2024.103751
Wu, W., Hill, S. E., Nathan, W. J., Paiano, J., Callen, E., Wang, D., et al. (2021). Neuronal enhancers are hotspots for DNA single-strand break repair. Nature 593, 440–444. doi: 10.1038/s41586-021-03468-5
Xu, B., Ionita-Laza, I., Roos, J. L., Boone, B., Woodrick, S., Sun, Y., et al. (2012). De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet. 44, 1365–1369. doi: 10.1038/ng.2446
Yagi, M., Kabata, M., Tanaka, A., Ukai, T., Ohta, S., Nakabayashi, K., et al. (2020). Identification of distinct loci for de novo DNA methylation by DNMT3A and DNMT3B during mammalian development. Nat. Commun. 11, 3199. doi: 10.1038/s41467-020-16989-w
Yang, X., Garnier, F., Débat, H., Strick, T. R., and Nadal, M. (2020). Direct observation of helicase-topoisomerase coupling within reverse gyrase. Proc. Natl. Acad. Sci. U S A. 117, 10856–10864. doi: 10.1073/pnas.1921848117
Yang, X., Li, W., Prescott, E. D., Burden, S. J., and Wang, J. C. D. N. A. (2000). topoisomerase IIbeta and neural development. Science 287, 131–134. doi: 10.1126/science.287.5450.131
Yap, E. L., and Greenberg, M. E. (2018). Activity-regulated transcription: bridging the gap between neural activity and behavior. Neuron 100, 330–348. doi: 10.1016/j.neuron.2018.10.013
Yasuhara, T., Kato, R., Hagiwara, Y., Shiotani, B., Yamauchi, M., Nakada, S., et al. (2018). Human Rad52 promotes XPG-mediated R-loop processing to initiate transcription-associated homologous recombination repair. Cell 175, 558–70. e11. doi: 10.1016/j.cell.2018.08.056
Zagnoli-Vieira, G., and Caldecott, K. W. (2020). Untangling trapped topoisomerases with tyrosyl-DNA phosphodiesterases. DNA Repair 94:102900. doi: 10.1016/j.dnarep.2020.102900
Zardoni, L., Nardini, E., Brambati, A., Lucca, C., Choudhary, R., Loperfido, F., et al. (2021). Elongating RNA polymerase II and RNA:DNA hybrids hinder fork progression and gene expression at sites of head-on replication-transcription collisions. Nucleic Acids Res. 49, 12769–12784. doi: 10.1093/nar/gkab1146
Zhang, R. R., Cui, Q. Y., Murai, K., Lim, Y. C., Smith, Z. D., Jin, S., et al. (2013). Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 13, 237–245. doi: 10.1016/j.stem.2013.05.006
Zhang, X., Zhang, Y., Wang, C., and Wang, X. (2023). TET (Ten-eleven translocation) family proteins: structure, biological functions and applications. Signal Transduct. Target Ther. 8:297. doi: 10.1038/s41392-023-01537-x
Zhang, Z., Zhou, J., Tan, P., Pang, Y., Rivkin, A. C., Kirchgessner, M. A., et al. (2021). Epigenomic diversity of cortical projection neurons in the mouse brain. Nature 598, 167–173. doi: 10.1038/s41586-021-03223-w
Zhou, J., Zhang, Z., Wu, M., Liu, H., Pang, Y., Bartlett, A., et al. (2023). Brain-wide correspondence of neuronal epigenomics and distant projections. Nature 624, 355–365. doi: 10.1038/s41586-023-06823-w
Zhu, X. H., Qiao, H., Du, F., Xiong, Q., Liu, X., Zhang, X., et al. (2012). Quantitative imaging of energy expenditure in human brain. Neuroimage 60, 2107–2117. doi: 10.1016/j.neuroimage.2012.02.013
Zolzaya, S., Narumoto, A., and Katsuyama, Y. (2024). Genomic variation in neurons. Dev. Growth Differ. 66, 35–42. doi: 10.1111/dgd.12898
Zovkic, I. B., Guzman-Karlsson, M. C., and Sweatt, J. D. (2013). Epigenetic regulation of memory formation and maintenance. Learn Mem. 20, 61–74. doi: 10.1101/lm.026575.112
Keywords: genome integrity, R-loop, G-quadruplex, DNA repair, topoisomerase
Citation: Ihara D, Rasli NR and Katsuyama Y (2025) How do neurons live long and healthy? The mechanism of neuronal genome integrity. Front. Neurosci. 19:1552790. doi: 10.3389/fnins.2025.1552790
Received: 29 December 2024; Accepted: 17 February 2025;
Published: 19 March 2025.
Edited by:
Sigrid A. Langhans, Nemours Children's Health, United StatesReviewed by:
Partha Sarathi Sarkar, University of Texas Medical Branch at Galveston, United StatesCopyright © 2025 Ihara, Rasli and Katsuyama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Katsuyama, a2F0c0BiZWxsZS5zaGlnYS1tZWQuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.