
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Neurol., 16 April 2025
Sec. Neuro-Oncology and Neurosurgical Oncology
Volume 16 - 2025 | https://doi.org/10.3389/fneur.2025.1532398
This article is part of the Research TopicArtificial Intelligence in Neurosurgical Practices: Current Trends and Future OpportunitiesView all articles
 Kivanc Yangi1
Kivanc Yangi1
 Jinpyo Hong1
Jinpyo Hong1
 Arianna S. Gholami1
Arianna S. Gholami1
 Thomas J. On1
Thomas J. On1
 Alexander G. Reed1
Alexander G. Reed1
 Pravarakhya Puppalla1
Pravarakhya Puppalla1
 Jiuxu Chen1
,2
Jiuxu Chen1
,2
 Carlos E. Calderon Valero1
Carlos E. Calderon Valero1
 Yuan Xu1
Yuan Xu1
 Baoxin Li2
Baoxin Li2
 Marco Santello3
Marco Santello3
 Michael T. Lawton1
Michael T. Lawton1
 Mark C. Preul1
*
Mark C. Preul1
*
Objective: This study systematically reviewed deep learning (DL) applications in neurosurgical practice to provide a comprehensive understanding of DL in neurosurgery. The review process included a systematic overview of recent developments in DL technologies, an examination of the existing literature on their applications in neurosurgery, and insights into the future of neurosurgery. The study also summarized the most widely used DL algorithms, their specific applications in neurosurgical practice, their limitations, and future directions.
Materials and methods: An advanced search using medical subject heading terms was conducted in Medline (via PubMed), Scopus, and Embase databases restricted to articles published in English. Two independent neurosurgically experienced reviewers screened selected articles.
Results: A total of 456 articles were initially retrieved. After screening, 162 were found eligible and included in the study. Reference lists of all 162 articles were checked, and 19 additional articles were found eligible and included in the study. The 181 included articles were divided into 6 categories according to the subspecialties: general neurosurgery (n = 64), neuro-oncology (n = 49), functional neurosurgery (n = 32), vascular neurosurgery (n = 17), neurotrauma (n = 9), and spine and peripheral nerve (n = 10). The leading procedures in which DL algorithms were most commonly used were deep brain stimulation and subthalamic and thalamic nuclei localization (n = 24) in the functional neurosurgery group; segmentation, identification, classification, and diagnosis of brain tumors (n = 29) in the neuro-oncology group; and neuronavigation and image-guided neurosurgery (n = 13) in the general neurosurgery group. Apart from various video and image datasets, computed tomography, magnetic resonance imaging, and ultrasonography were the most frequently used datasets to train DL algorithms in all groups overall (n = 79). Although there were few studies involving DL applications in neurosurgery in 2016, research interest began to increase in 2019 and has continued to grow in the 2020s.
Conclusion: DL algorithms can enhance neurosurgical practice by improving surgical workflows, real-time monitoring, diagnostic accuracy, outcome prediction, volumetric assessment, and neurosurgical education. However, their integration into neurosurgical practice involves challenges and limitations. Future studies should focus on refining DL models with a wide variety of datasets, developing effective implementation techniques, and assessing their affect on time and cost efficiency.
Deep learning (DL), a subset of machine learning (ML), is an artificial intelligence (AI) method based on artificial neural networks that includes multiple layers of data processing to produce higher-level features. Artificial neural networks can combine various inputs to create a single input (1). Such technology holds substantial potential for improved pattern recognition and problem-solving in different medical disciplines, including neurosurgery. With the recent advancements in AI technologies, DL algorithms have begun to be integrated into neurosurgical practice in various ways.
For instance, DL technologies can improve surgical workflow analysis through real-time monitoring and video segmentation (2–7). DL can also potentially provide diagnostic support to surgeons by monitoring for adverse events or complications due to pathophysiological events during procedures (8). DL technologies may enhance the safety of neurosurgical procedures and provide a sense of reassurance to clinicians and patients by potentially diminishing intraoperative adverse events (9–11). On a related note, DL algorithms could allow for surgical instrument and motion tracking, allowing for more precise feedback intraoperatively and in teaching applications (12).
DL could also strengthen the ability to visualize and recognize complex anatomical structures by improving the accuracy of detection methods, including magnetic resonance imaging (MRI) and neuronavigation, and by identifying hemorrhages, spinal pathologies, and neuro-oncological conditions (13–17). Moreover, DL methods can be used to identify and classify intracranial lesions and perform volumetric assessments (18).
In this study, we elucidate prominent applications of DL algorithms in neurosurgery and provide evidence and examples of their current use within the field by conducting a systematic review of the existing literature. We also address future directions and limitations of these technologies. Because not all studies can be specifically expounded upon in such a review, we used representative articles to illustrate specific concepts and applications.
A systematic search was conducted in PubMed, Embase, and Scopus databases on 8 November 2024 using the following keywords: (Neurosurgical Procedure) OR (neurosurgery) OR (neurologic surgery) OR (neurological surgery) OR (Procedure, Neurosurgical) OR (Procedures, Neurosurgical) OR (Surgical Procedures, Neurologic) OR (Neurologic Surgical Procedure) OR (Neurologic Surgical Procedures) OR (Procedure, Neurologic Surgical) OR (Procedures, Neurologic Surgical) OR (Surgical Procedure, Neurologic) AND ((Deep learning) OR (Learning, Deep) OR (Hierarchical Learning) OR (Learning, Hierarchical)).
These medical subject heading terms were linked with Boolean operators “AND” and “OR” to maximize the extent of coverage. An advanced search was conducted in PubMed using these medical subject heading terms. Then, the search was expanded by including Scopus and Embase database searches using the exact keywords. No time restrictions were applied. Our search words and articles were filtered by title or abstract. Duplications were excluded, and 2 independent neurosurgically experienced reviewers (A.G. and J.H.) screened the articles and examined all the full texts. A strict selection process was employed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines (19). Most publications included in the study were original research papers focused on DL applications in neurosurgical practice. Review articles, editorials, letters, and errata were excluded. Articles not published in English and articles for which the full text was unavailable were excluded. Studies that did not use DL algorithms and studies that used DL algorithms but did not apply them to neurosurgical practice were excluded. Finally, the reference lists of these articles were checked by 3 independent reviewers (J.H., A.S.G., K.Y.). Ultimately, all reviewers (J.H., A.S.G., K.Y., A.G.R., P.P.) agreed on the articles included in our study.
Initially, 456 articles were retrieved. Eighty-seven duplicated papers were excluded. Twenty-two articles for which the full text was unavailable, 2 articles whose texts were unavailable in English, and 6 retracted articles were removed. Ninety-four articles determined to be letters, reviews, editorials, and errata were removed. Eighty-three articles that did not meet the inclusion criteria were excluded. The reference lists of the selected articles were screened by 3 independent reviewers (J.H., A.S.G., K.Y.), and 19 articles were found that were within the scope of our review and were included in the study. Upon completion of the screening, 181 articles were found eligible for the study and included (2, 3, 7, 9–11, 13–18, 20–188). The selection process is depicted in the PRISMA flowchart in Figure 1.
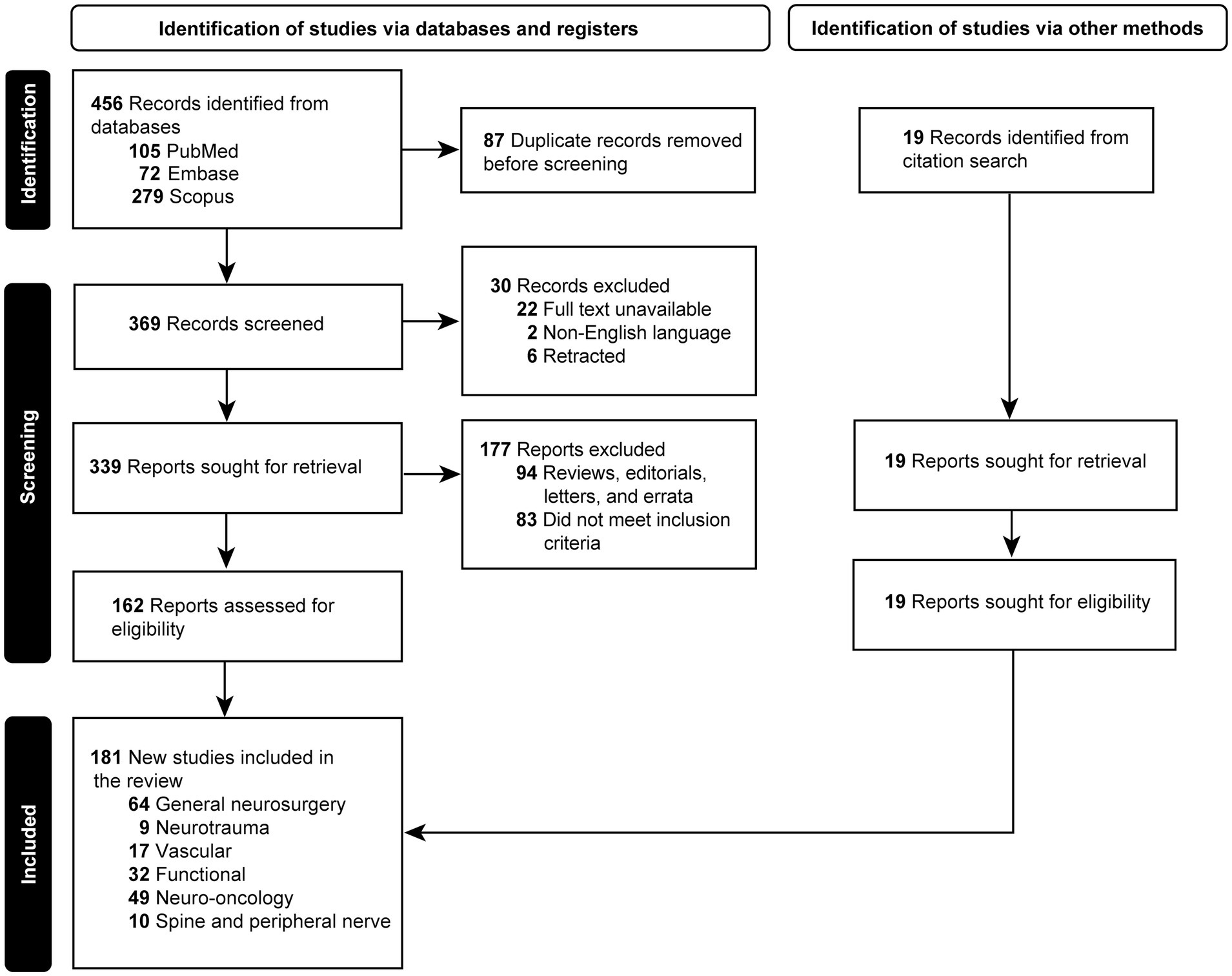
Figure 1. Flow diagram documenting the study selection process. Used with permission from Barrow Neurological Institute, Phoenix, Arizona.
Included articles were divided into 6 groups according to the subspecialty: spinal surgery and peripheral nerve (n = 10) (Table 1) (14, 20–26, 168, 182), neurotrauma (n = 9) (Table 2) (27–35), vascular neurosurgery (n = 17) (Table 3) (9, 10, 13, 36–45, 166, 169, 179, 186), functional neurosurgery (n = 32) (Table 4) (46–76, 187), neuro-oncology (n = 49) (Table 5) (2, 3, 7, 15, 16, 18, 77–114, 175–178, 188), and general neurosurgery (n = 64) (Table 6) (11, 17, 115–165, 167, 170–174, 180, 181, 183–185).
Although DL applications have been widely used in different subspecialties of neurosurgery, our findings indicate that there are leading subspecialties that are at the forefront of this technological advancement. General neurosurgery (n = 64), functional neurosurgery (n = 32), and neuro-oncology (n = 49) are the leading subspecialties, as shown in Figure 2.
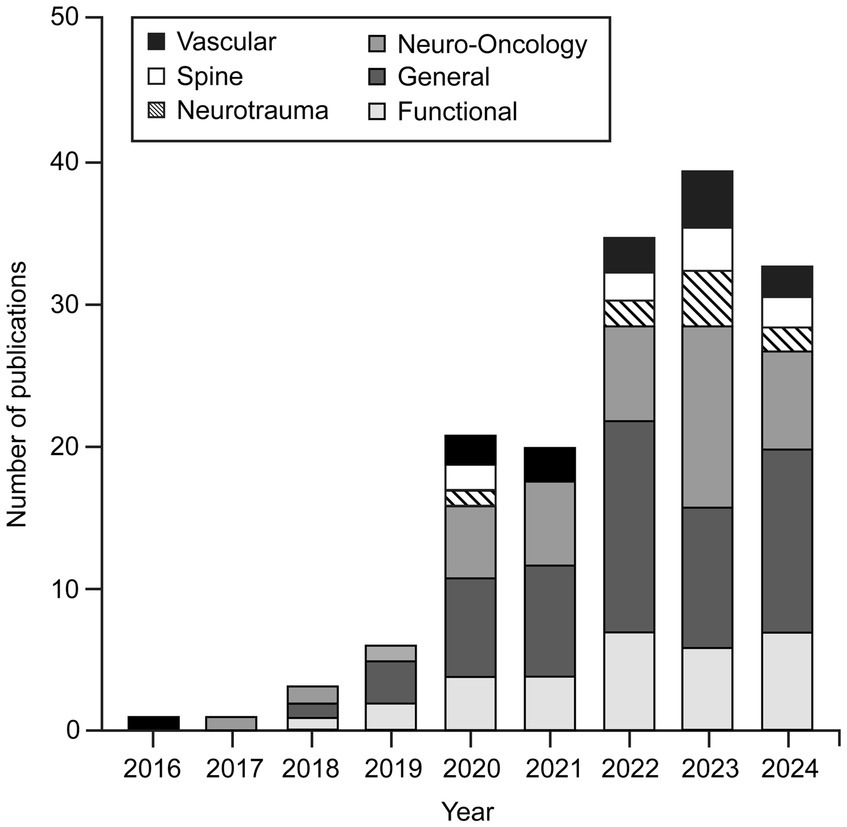
Figure 2. Number of publications focused on applying various deep learning (DL) algorithms in neurosurgical procedures by year and subspecialty. This bar chart shows that applications of DL algorithms in neurosurgery were little studied in 2016. Such studies began to gain popularity in 2019, and the trend has continued to grow. By November 2024, the publication numbers had nearly matched those for 2023. Although these algorithms have been applied across almost all neurosurgery subspecialties, general neurosurgery, neuro-oncology, and functional neurosurgery are the leading subspecialties in using DL algorithms. Used with permission from Barrow Neurological Institute, Phoenix, Arizona.
The procedures that most commonly used DL algorithms were deep brain stimulation (DBS) and subthalamic and anterior thalamic nuclei localization (n = 24) in the functional neurosurgery group (Table 4); segmentation, identification, classification, and diagnosis of various brain tumors (n = 29) in the neuro-oncology group (Table 5), and neuronavigation and image-guided neurosurgery (n = 13) in general neurosurgery group (Table 6). Computerized tomography (CT), MRI, and ultrasonography images (n = 79) were found to be the most widely used datasets in all groups to train DL architectures, followed by surgical videos and various types of image datasets, including radiography, digital subtraction angiography, surgical microscopy, and neuroendoscopy images (n = 51).
To the best of our knowledge, interest in applying DL algorithms in neurosurgery began in the 2010s, when the first studies were published. Although very few articles were published on this topic in 2016, more studies were published beginning in 2019, and this trend has continued to grow. The publication numbers during 2024 nearly matched those for 2023 (Figure 2).
ML is an AI method that enables computers to process data and learn valuable patterns, facilitating better user decision-making. DL is a subset of ML that focuses on mimicking the complex neuron structures found in the human brain by using multiple layers of latent units to compose a neural network. It is widely recognized as an effective tool for data analysis due to its ability to understand the intricate nature of data.
There are various types of DL neural network models designed to simulate different real-world scenarios: multilayer perceptron, convolutional neural network (CNN) (189), recurrent neural network (RNN) (190), graph neural network (191), generative adversarial network (192), transformer (193), and diffusion model (194, 195), among others. Many variant neural network models, such as long short-term memory (196) and denoising diffusion probabilistic model (195), have been developed to address specific limitations and meet the challenge of significantly changing real-world use cases. Some variations, such as the vision transformer, are designed as fusions of different neural network modules to leverage their superior performance (197). Motivated by this flexibility in neural network architecture design, researchers can better facilitate their specific purposes.
In recent years, with the continuing advancements in the fields of AI and medicine, various architectural designs of DL have started to be used in neurosurgery (Table 7) (37, 38, 41, 73, 74, 81–83, 88, 115, 116, 123, 125, 137). This study focuses on the applications of the most widely used deep neural architecture designs within the neurosurgical practice.
Over the past several years, the development of AI, specifically DL techniques, has shown considerable potential in neurosurgery (198). Various DL architectural designs are used in neurosurgical practice for purposes that include operative video analysis, predicting outcomes, microsurgical skill assessments, diagnostic support, volumetric assessment, and neurosurgical education. This section will examine the most commonly used applications of DL in neurosurgical training.
DL CNNs can analyze surgical videos through computer vision (CV) (199). This approach analyzes operative video to define surgical workflows by establishing start and end points for various stages of a procedure and annotating clinically relevant details, such as anatomical landmarks and instrument detection, aiding in assessment, real-time monitoring, and surgical coaching (2–6).
Many CV pipelines share common elements, beginning with creating a dataset composed of individual surgical images (frames) annotated with overlays that highlight tools, anatomical structures, or the stage of the operation. These data are often generated manually and require expert evaluation. CV models are trained on this ground-truth data and tested on new images or videos. The successful implementation of CV models relies heavily on the quality, quantity, and accuracy of these annotated video sets (167).
Neurosurgeons often use exoscopes, microscope-exoscope hybrid systems, and endoscopes, all equipped with cameras to capture surgical video. Neurosurgical procedures are frequently recorded, resulting in surgeons accumulating hundreds of hours of surgical video. Surgeons use this video for educational purposes, cropping it to highlight important parts of the surgery and discuss critical aspects. To date, CV has primarily been applied to neurosurgery with 2 main focuses: workflow analysis and video segmentation.
Operative workflow analysis deconstructs operations into distinct steps and phases. Each operative video is labeled with timestamps corresponding to these steps and stages. These labeled data and the video are fed into DL models, enabling automatic recognition and analysis of these components. This process allows for standardized skill assessment, automated operative note generation, and the development of enhanced educational tools. This type of work in neurosurgery has been primarily limited to endoscopic procedures (2, 169). Khan et al. (2) conducted a study using ML to develop and validate an automated workflow analysis model for endoscopic transsphenoidal pituitary surgery, achieving high accuracy in recognizing surgical phases and steps (Figure 3). Pangal et al. analyzed the phases of endonasal endoscopic surgery using a validated cadaveric simulator of internal carotid artery injury (169).
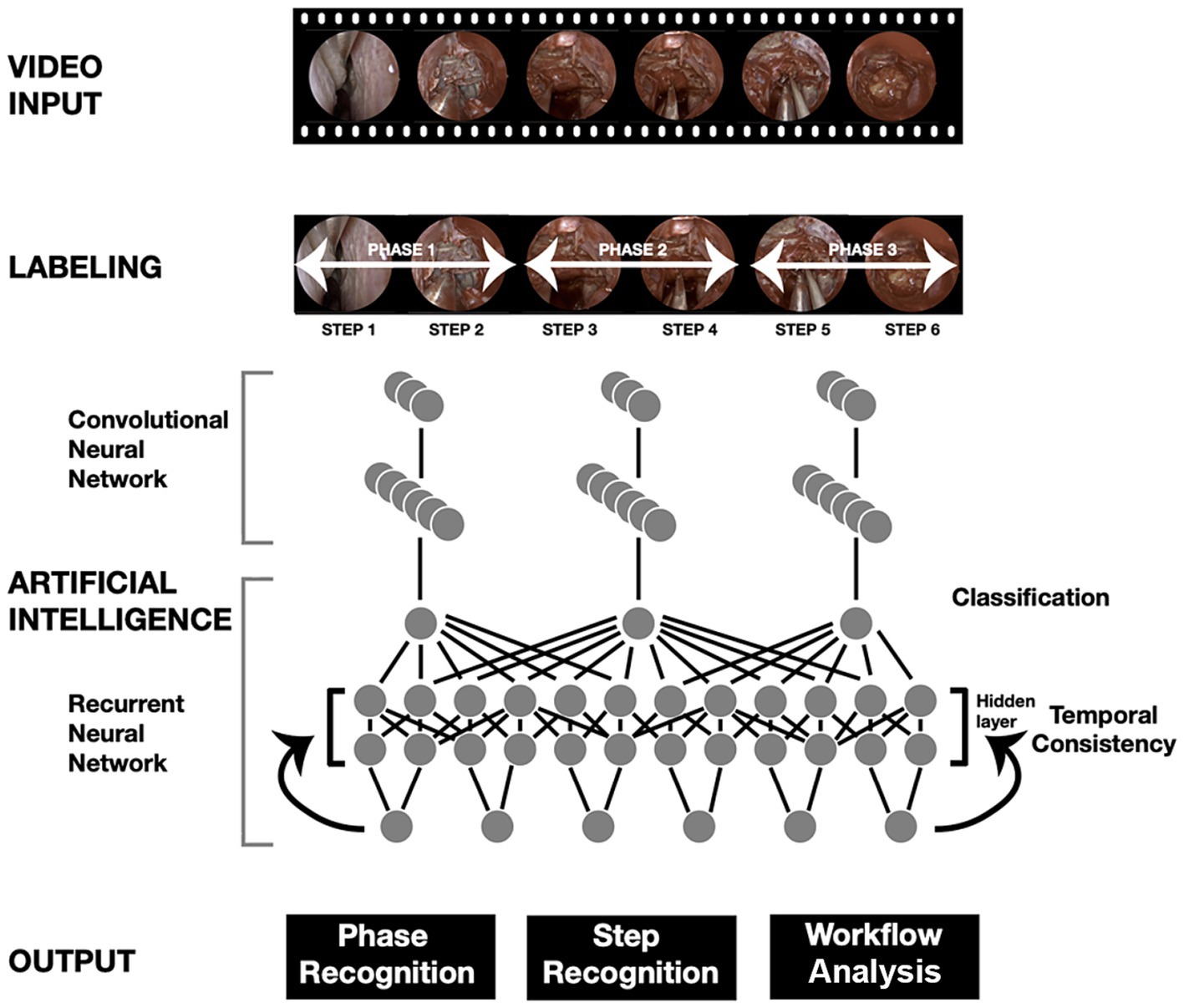
Figure 3. Use of deep learning (DL) algorithms in surgical workflow analysis. The workflow diagram adapted from Khan et al. (2) illustrates the phase and step recognition process in surgical videos using artificial intelligence. The process begins with a video input that undergoes labeling into phases and steps. A convolutional neural network (CNN) extracts features from the labeled video frames, whereas a recurrent neural network (RNN) ensures temporal consistency across the video sequence. The combined CNN and RNN architecture enables accurate classification of surgical phases, recognition of specific steps, and comprehensive workflow analysis. The final output provides detailed insight into phase recognition, step recognition, and overall surgical workflow. Used with permission from Khan DZ, Luengo I, Barbarisi S, et al. Automated operative workflow analysis of endoscopic pituitary surgery using machine learning: development and preclinical evaluation (IDEAL stage 0). J Neurosurg. 2022;137 (1):51–58. doi: 10.3171/2021.6.JNS21923.
In neurosurgery, CV has been increasingly applied to annotate or segment instruments and anatomical landmarks within operative videos (7). For instance, Jawed et al. developed a video annotation methodology for microdiscectomy, creating a standardized workflow to facilitate the annotation of surgical videos (168). Their method involved labeling surgical tools, anatomy, and phases in microdiscectomy videos. Similarly, Staartjes et al. conducted a proof-of-concept study evaluating machine vision algorithms for identifying anatomic structures in endoscopic endonasal approaches using only the endoscopic camera (170). The DL algorithm, trained on videos from 23 patients, significantly improved nasal structure detection compared to a baseline model, which is established using the average positions of the training ground truth labels within a semi-quantitative 3-tiered system.
Our ongoing work analyzing operative videos of middle cerebral artery aneurysm surgery to categorize, discriminate, and label surgical maneuvers or events with an ML methodology, however, suggests that the accuracy outcome depends on the information load presented to the analytical system (unpublished data). We have found that highly detailed, unique annotation and labeling are less accurate in identifying surgical maneuvers and events than more general labeling, suggesting that ML analytics has limits. This finding is intuitive and reflects the unique nature of each individual surgical procedure; the specific pathology, structure, site, and surgical maneuvers are different for each middle cerebral artery aneurysm.
In another study, Pangal et al. evaluated video-based metrics to predict task success and blood loss during endonasal endoscopic surgery in a cadaveric simulator (169). They manually annotated videos of 73 surgeons’ trials, focusing on instruments and anatomical landmarks. The study found that these metrics, derived from expert analysis, predicted performance more accurately than training level or experience, with a regression model effectively predicting blood loss.
Furthermore, in a recent study by Park et al. (17), computer algorithms including structural similarity, mean squared error, and DL were used to analyze the distortion, color, sharpness, and depth of field of the images collected from an advanced hybrid operating exoscope-microscope platform to improve the quality of the images for neurosurgical procedures. Operating microscopes are becoming sophisticated imaging platforms, incorporating fluorescence imaging, robotics, exoscopic vision, and now various proprietary functions for the enhancement of images to better define and visualize anatomy. These functions themselves operate on the basis of DL algorithms to enhance image sharpness and color and provide the operator with visual environments that allow improvements in capturing the depth of the operative field. Furthermore, while improving image quality, exoscopes do not significantly distort the images (17). These systems can provide high-definition images and help improve the recognition of structures during surgery.
Neurosurgery involves high-risk procedures, making the use of AI for predicting outcomes a potentially important tool for improving surgical planning, patient counseling, and postoperative care. Outcome prediction in neurosurgery has dramatically benefited from advances in DL. These models can predict functional recovery and overall quality of life by analyzing preoperative imaging and intraoperative data, aiding in surgical planning and patient expectation management (25). DL models, such as CNNs and RNNs, are particularly effective in analyzing large datasets that may not yield meaningful patterns through traditional statistical methods. These datasets can include imaging, clinical records, and operative notes, allowing for more accurate outcome predictions.
The adaptability of DL makes it a valuable tool for personalizing neurosurgical care and improving long-term outcomes. For example, Jumah et al. (200) explored surgical phase recognition using AI and DL to improve outcome prediction in neurosurgery. Surgical phase recognition analyzes visual and kinematic data from surgeries to identify different phases of a procedure in real time. This capability enhances decision-making by providing surgeons with critical insights and alerts during high-risk phases, thereby reducing complications and improving surgical precision. This innovative approach contributes significantly to outcome prediction in neurosurgery. Additionally, Danilov et al. (136) used RNNs to predict the duration of postoperative hospital stays based on unstructured operative reports. This model demonstrated the potential of using narrative medical texts for making meaningful predictions, further illustrating the usefulness of DL in neurosurgery.
Wang et al. (85) developed a DL model to predict short-term postoperative facial nerve function in patients with acoustic neuroma. The study integrated clinical and radiomic features from multisequence MRI scans to enhance prediction accuracy. The CNN model achieved an area under the curve of 0.89, demonstrating superior predictive performance compared to traditional models in which various subtypes of ML are used (e.g., Nomogram, Light Gradient Boosting Machine). This predictive capability aids in surgical decision-making and patient counseling, allowing surgeons to anticipate facial nerve functional outcomes and tailor surgical approaches to minimize nerve damage.
In the context of traumatic brain injury (TBI), DL models used to enhance TBI triage have shown promise in predicting outcomes at hospital discharge, especially in low-resource settings where decision-making support is needed (30). These models offer a significant advantage in patient care and resource allocation, highlighting the growing role of AI in neurosurgical practice.
Furthermore, several studies have also demonstrated the potential of DL in predicting adverse outcomes, such as postoperative complications or prolonged hospital stays. Biswas et al. (31) introduced the ANCHOR model, an artificial neural network designed to predict referral outcomes for patients with chronic subdural hematoma. Validated using data from 1713 patient referrals at a tertiary neurosurgical center, the ANCHOR model demonstrated high accuracy (92.3%), sensitivity (83.0%), and specificity (96.2%) in predicting which referrals would be accepted for neurosurgical intervention. By integrating clinical features such as patient age, hematoma size, and medical history, the model supports clinical decision-making and potentially reduces adverse events by ensuring appropriate surgical referrals.
Pangal et al. (11) explored the SOCALNet model to predict surgical outcomes during hemorrhage events in neurosurgery. This neural network analyzes surgical videos to estimate the likelihood of achieving hemostasis, outperforming expert surgeons in accuracy. This model is valuable for real-time decision support, enhancing patient safety and surgical outcomes.
Survival prediction in neurosurgery has also been enhanced by DL approaches that integrate multimodal data, including imaging and genomic information. By accurately predicting survival probabilities, these models support clinicians in making informed decisions regarding treatment strategies and patient counseling (83). Di Ieva et al. (93) applied a DL model to predict outcomes in brain tumor management, focusing on survival prediction. The integration of imaging and genomic data enhances the prediction of patient survival, enabling better surgical planning and prognosis.
DL models enhance decision-making, optimize resource allocation, and improve patient outcomes by leveraging complex data from imaging, clinical records, and surgical videos. As neurosurgery continues to embrace AI, integrating DL technologies will be crucial in personalizing treatment strategies, minimizing risks, and ultimately elevating the standard of care in this high-stakes discipline.
The application of DL in hand motion and instrument tracking in neurosurgery is an innovative and transformative topic. DL is capable of tracking both relevant and redundant surgical motions in real time and serves as an educational and forecasting tool for training the next generation of neurosurgeons. On the other hand, DL algorithms have also been used to track patient motor movements during functional neurosurgery procedures. In a study by Baker et al. (54), a DL-based CV algorithm was used for markerless tracking to evaluate the motor behaviors of patients undergoing DBS surgery. Intraoperative kinematic data were extracted using the Phyton-based CV suite DeepLabCut from the surgical videos of 5 patients who underwent DBS electrode implantation surgery, with comparison to manual data acquisition. The automated DL-based model showed 80% accuracy. Furthermore, a support vector machine model was also used in this study to classify patient movements. Classification by a support vector machine had 85.7% accuracy, including for 2 common upper limb movements: 92.3% accuracy for arm chain pulls, and 76.2% accuracy for hand clenches. This study emphasized the application of DL-based algorithms in DBS surgery to accurately detect and classify the movements of the patients undergoing surgery. Although the accuracy of a support vector machine for a specific type of movement was found to be low, the results are promising for future studies (54).
Koskinen et al. (12) investigated the use of a CV model to properly train an algorithm to accurately detect the movement of microsurgical instruments correlated with eye tracking. A DL approach using YOLOv5-1 composed of Cross Stage Partial Network allowed analysis of 6 specific metrics of path length, velocity, acceleration, jerk, curvature, and the intertool tip distance in 4 categories of surgical movements of dissection (to use a microscissor for vessel dissection), enhancement of the visual scene (to move objects away from the visual field to expose the dissection area), exploration with the tools (to find a new dissection plane), and intervention (to clean the bleeding from the field of focus). This novel DL application for an intracranial vessel dissection task was a successful proof-of-concept that demonstrated surgical movement tracking without sensors attached to hands or digits (12).
DL is widely used as a tool for educators and academicians, and its role in neurosurgical education is also prevalent. In a DL-based analysis of the surgical performance of surgeons of varying degrees of expertise, Davids et al. (131) analyzed videos of 19 surgeons by using a CNN and evaluated the skill levels using the area under the curve and accuracy. Novice surgeons showed a significantly higher median dissector velocity and a more considerable intertool tip distance, both of which served as discernment points between the experts and the beginners.
Microvascular anastomosis is another procedure used in neurosurgery that is highly dependent on the precision and accuracy of the surgeon’s intraoperative moves. Sugiyama et al. (171) employed a similar approach to Davids et al. (131) in using a YOLOv2 training system to train with microvascular anastomosis videos of surgeons of various experience levels ranging from novice to expert. Although experts could complete the tasks faster than the novice, nondominant hand results were observable only in certain phases of the simulated practice surgeries. This study introduced a DL-based video analysis algorithm (171).
Xu et al. (119) also studied the skill levels of surgeons performing microsurgery, developing a novel sensorized surgical glove by applying a piezoresistive sensor on the thumb of the glove. Additionally, this study used force-based data, meaning that the interaction between the surgical tools served as the basis for analyzing surgeon skillfulness. Similar to the studies mentioned above, this study also encompassed surgeons of varying degrees of expertise and compared various DL models, including long short-term memory and gate recurrent unit for movement detection. The study asserted that force data can yield discrimination between experts and novices, which means this concept could be used as an educational tool (119).
In a recent study conducted by Gonzalez-Romo et al. (174), a novel hand-digit motion detector incorporating an open-source ML model (https://ai.google.dev/edge/mediapipe/solutions/vision/hand_landmarker; MediaPipe, Google, Inc.) was developed using the Python programming language (Python Software Foundation, https://www.python.org/) to track operators’ hands during a microvascular anastomosis simulation (201). The model tracked hand and digit motion with 21 hand landmarks without physical sensors attached to the operators’ hands. Hand motion during the microanastomosis simulation was recorded with a neurosurgical operating microscope and an external camera. Six expert neurosurgeons performed the simulation, with interesting elucidation and comparison of their technical commonalities and variances using time series analysis. The hand-tracking system employed in this study is a promising example of tracking motion during surgical procedures in a sensorless manner (174). On et al. (172) used the same DL-based sensorless motion detection system (MediaPipe) to track operators’ hand motions during a cadaveric mastoidectomy. The procedures were recorded using an external camera, and the video output was processed to assess surgical performance and provide feedback.
Research on DL technology applicable to various neurosurgical educational settings is continuously growing, with the development of various scalable open-source solutions aimed at the goal of molding the next generation of competent neurosurgeons. The paradigm shift in neurosurgery, whereby real-time piecewise intraoperative motion can be evaluated, may ultimately improve postsurgical patient outcomes and optimize neurosurgeons’ efficiency in carrying out preplanned tasks and improvising to overcome unexpected surgical situations (172–174).
Due to various factors, fast and accurate diagnosis in neurosurgery can be challenging (202, 203). Thus, DL algorithms have been employed to provide diagnostic support. This support includes segmenting urgent intracranial pathologies on CT to help with fast decision-making, identifying and classifying intracranial tumors and spinal pathologies for accurate treatment planning, and enhancing neuronavigation systems to achieve better surgical outcomes.
Intracranial hemorrhages (ICH) are among the most challenging pathologies for neurosurgeons, particularly in emergency settings (204–206). Selecting the optimal treatment modality for ICH can be controversial; however, timing is crucial for choosing the appropriate treatment. DL algorithms could be used to segment ICHs on CT, aiding in selecting the most effective surgical strategy.
In a study by Tong et al. (13), the authors developed a 3-dimensional (3D) U-Net embedded DL model to segment intraparenchymal and intraventricular hemorrhages on CT (13). This study aimed to improve understanding of the boundaries, volume, and centroid deviation of each type of hematoma. By achieving this, the authors hoped to aid clinicians in selecting the most accurate catheter puncture path for treatment (13).
Previously, the diagnostic accuracy of ICHs using DL models was tested in a retrospective study by Voter et al. (38). This study used a US Food and Drug Administration–approved DL model, Aidoc, to assess the diagnostic accuracy of ICHs using 3,605 noncontrast CT of adults (Figure 4). This study showed a decreased sensitivity and positive predictive value of the model compared to their expectations and previous studies, with specific patient features such as previous neurosurgery, hemorrhage type, and number of hemorrhages further reducing diagnostic accuracy. The authors raised concerns regarding the generalizability of these DL models. They additionally stressed the need to include patients with a prior history of a neurosurgical procedure when training these models and a more stringent standardization of study parameters in future studies (38).
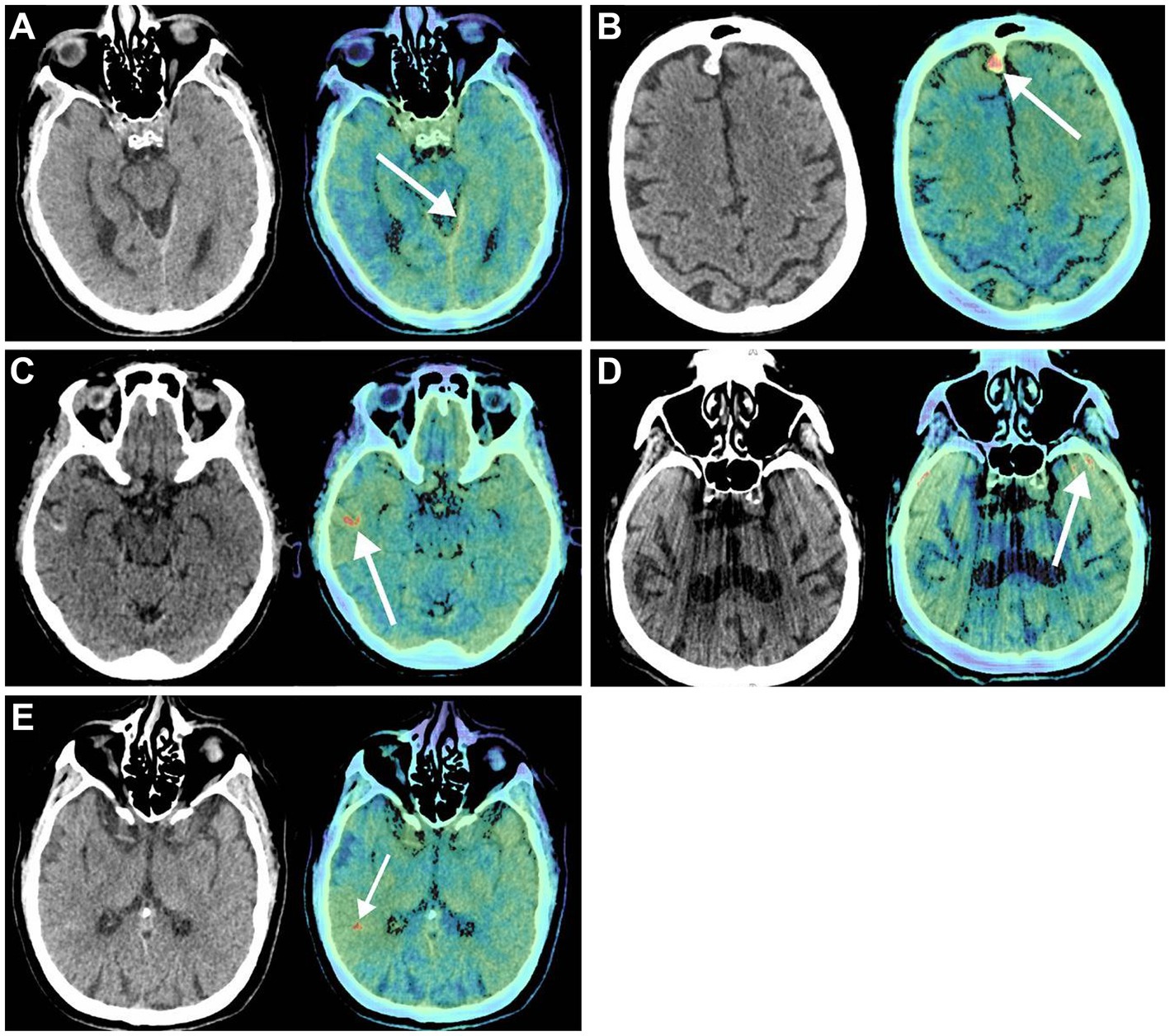
Figure 4. Deep learning (DL) in diagnostic support in neurosurgery. Voter et al. (38) focused on the application of a US Food and Drug Administration–approved DL, Aidoc, to determine its ability to recognize intracranial hemorrhages on noncontrast computed tomography (CT) accurately. Noncontrast CT (left) and the key images (right) in which the Aidoc identified the pathology (white arrows). (A) True positive finding in which the intracranial hemorrhage was not identified by the neuroradiologist. (B) Image of a meningioma that was incorrectly identified as an intracranial hemorrhage by Aidoc. (C) Cortical laminar necrosis incorrectly identified as an intracranial hemorrhage by Aidoc. (D) An artifact misidentified by the Aidoc. (E) Failure mode with the absence of a clear pathology. Used with permission from Voter AF, Meram E, Garrett JW, Yu JJ. Diagnostic Accuracy and Failure Mode Analysis of a Deep Learning Algorithm for the Detection of Intracranial Hemorrhage. J Am Coll Radiol. 2021 Aug;18 (8):1143–1,152. Doi:10.1016/j.jacr.2021.03.005. Epub 2021 Apr 3.33819478; PMCID: PMC8349782.
The recognition of subarachnoid hemorrhages (SAHs) using DL has also been investigated. Nishi et al. recognized the difficulty in diagnosing patients with SAH (39). An AI system using a deep neural network architecture segmented noncontrast CT images from 757 patients with 3D U-net. Of these 757 patients, 419 had SAH confirmed by 2 neurosurgical specialists. Of these 419 cases, 392 were used to train the DL model, and 27 were used for validation. Image interpretation was conducted on 331 cases, which included 135 SAH cases and 196 non-SAH cases. The AI system demonstrated a high accuracy in diagnosing SAH, almost comparable to that of the neurosurgical specialists. Importantly, the system was useful in aiding in the diagnosis of SAH when used by physicians who were not specialists in neurosurgery, reflecting its potential use as a screening tool in settings such as the emergency room (39).
Proper identification and classification of intracranial tumors are crucial for determining early management but might not always be successful on initial imaging (207, 208). Thus, auxiliary methods to improve this process are essential. DL algorithms have been widely employed in adult and pediatric neuro-oncology to accurately identify and classify intracranial tumors (175, 176, 209). These algorithms facilitate rapid diagnostic estimation and support the determination of the most appropriate treatment strategy (210). For this purpose and to thereby enhance their effectiveness, DL algorithms can be trained with MRI datasets to accurately predict tumor classification, or they can be applied to overcome the limitations of advanced real-time, cellular-scale imaging technologies, such as confocal laser endomicroscopy (CLE) and Raman scattering microscopy. As neurosurgery rapidly advances into an era in which such imaging technologies are increasingly employed intraoperatively, DL algorithms serve as a critical tool for improving diagnostic workflows, accelerating treatment selection, and ultimately optimizing patient outcomes.
CLE, a real-time in vivo intraoperative fluorescence-based cellular-resolution imaging technique used in brain tumor surgery, has the potential to revolutionize the surgical workflow in that it essentially provides a digital optical biopsy without requiring tissue extraction (176, 177, 209). Although promising, CLE devices are hand-held, extremely movement sensitive, and have a small field of view, making them prone to motion artifacts; they also provide images in grayscale only. To address the colorization limitation, Izadyyazdanabadi et al. (175) studied image style transfer, which is a neural network method used to integrate or rationalize the content of 2 distinct images in an attempt to transform the fluorescence-based grayscale CLE image into a familiar histology-standard hematoxylin and eosin–like image (Figure 5). Evaluation of the images by neurosurgeons and neuropathologists found that the transformed images had fewer artifacts and more prominent critical structures when compared to the original grayscale fluorescence-based images. This study emphasized an important application of DL technologies in neuro-oncology that enhances the diagnostic quality of intraoperative imaging techniques for better precision, which is particularly crucial for malignant and invasive brain tumors (175).
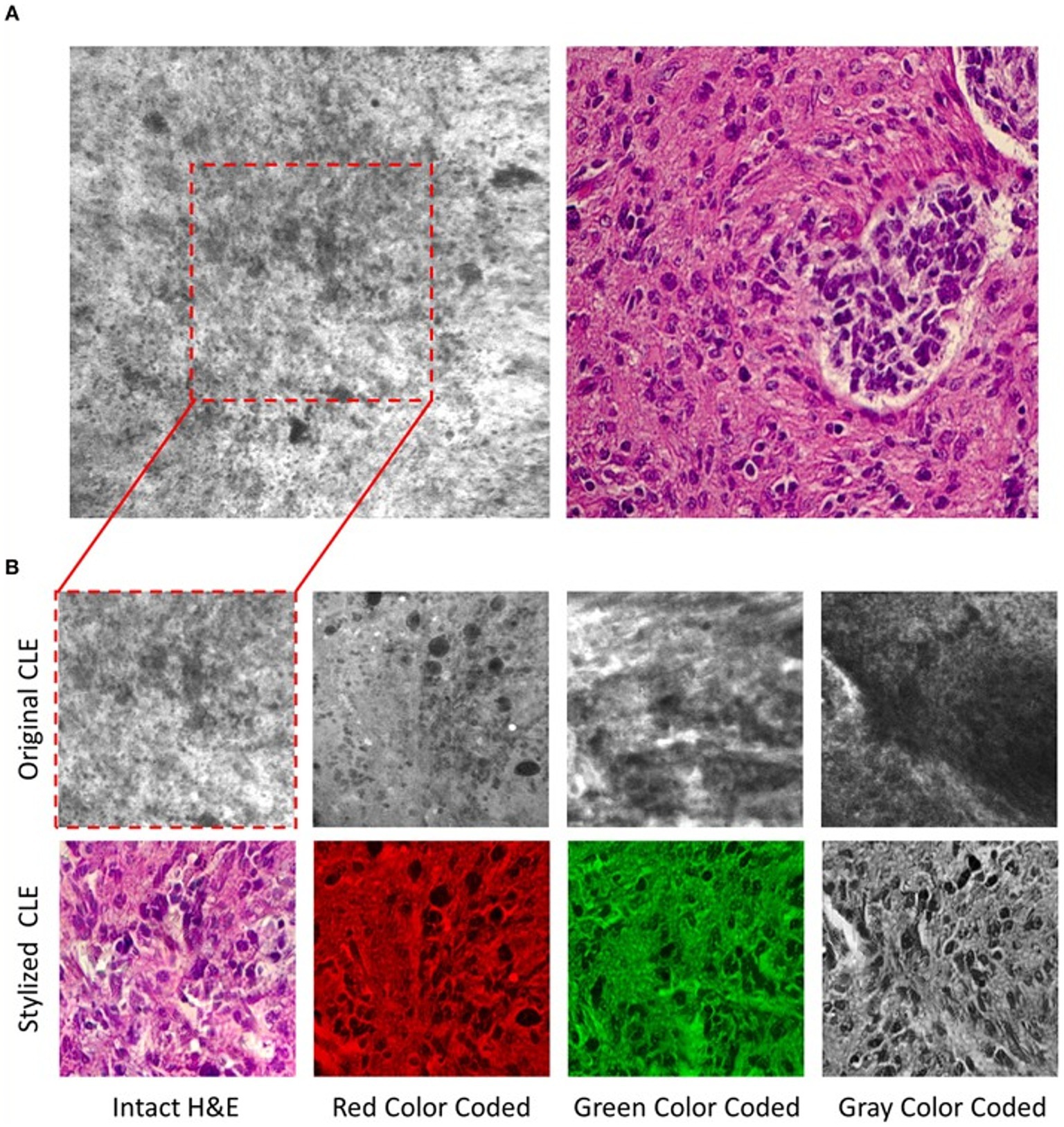
Figure 5. Deep learning (DL)–based image style transfer method to improve the diagnostic quality of confocal laser endomicroscopy (CLE) images. Image style transfer is a neural network–based model used in a study by Izadyyazdanabadi et al. (175) to integrate the content and style of 2 distinct images to transform fluorescence-based grayscale CLE images into familiar histologic hematoxylin and eosin (H&E)–like images. (A) Representative CLE (Optiscan 5.1, Optiscan Pty., Ltd.) and H&E images from human glioma tissues. The difference between original and stylized images of human gliomas can be seen on (B), in 4 distinct color scales: intact H&E, red, green, and gray. Used with permission from Izadyyazdanabadi M, Belykh E, Zhao X, Moreira LB, Gandhi S, Cavallo C, Eschbacher J, Nakaji P, Preul MC, Yang Y. Fluorescence Image Histology Pattern Transformation Using Image Style Transfer. Front Oncol. 2019 Jun 25;9:519. Doi: 10.3389/fonc.2019.00519.31293966; PMCID: PMC6603166.
Because CLE imaging is sensitive to motion artifacts, it produces many images with nondiagnostic findings or limited surgical information (176). To address this discriminatory challenge, Izadyyazdanabadi et al. developed and used a DL method to detect diagnostic images among many nondiagnostic ones (176). AlexNet, a DL architecture, was trained with CLE image datasets collected from CLE-aided brain tumor surgeries, with all images verified by a pathologist. The mean accuracy of the model in detecting the diagnostic images was 91%; sensitivity and specificity were each also 91%. The results of this study showed that image detection and discrimination techniques based on a CNN have the potential to quickly and reliably identify informative or actionable CLE images. Incorporation of such techniques into the CLE operating system has the potential to aid the surgeon or pathologist in making an informed surgical decision on the fly when imaging with CLE.
Moreover, in another study (177), different CNN algorithms were trained using the CLE images of patients with different intracranial neoplasms to detect diagnostic images automatically. Accuracies of distinct CNN models were compared, and the study found that a combination of deep fine-tuning and creating an ensemble of models reached the maximum accuracy (0.788 for an arithmetic ensemble and 0.818 for a geometric ensemble). Using DL algorithms in intraoperative imaging techniques such as CLE has yielded promising results that may be a focus of future research.
Other intraoperative imaging has used DL image identification results, such as Raman scattering microscopy, which allows the generation of digitally stained histological images. Reinecke et al., using the residual CNN ResNet50v2, correctly identified tissue as tumor, nontumor, or low-quality tissue imaged with stimulated Raman histological images of intraoperative tissue samples (15). This study reported that the Raman histology-based residual network was 90.2% accurate in correctly classifying the different tissues compared to the classification conducted by neuropathologists, thereby aiding in surgical and clinical decision-making (15).
Danilov et al. focused on predicting tumor classification by using contrast-enhanced T1 axial MRI images of World Health Organization (WHO)–grade verified glial tumors from 1,280 patients to train a DL model to accurately classify each tumor according to the WHO grading system (89). Two methods were used to achieve this goal: a 3D classification method in which the whole-brain MRI was used to predict tumor type and a 2-dimensional (2D) classification method using individual slices of each MRI scan. For model training, the processing of the 3D set was performed by the Dense-Net architecture, whereas the processing of the 2D model was performed by the Resnet200e architecture. The authors of this study concluded that the accuracy of their DL model in separating glial tumors based on the WHO grading system was similar to the results of other studies found in the literature (89).
Accurate classification of intracranial tumors is highly important because treatment strategies may vary accordingly, particularly in the pediatric age group. In this context, the use of DL for radiological diagnostic support has also been demonstrated in pediatric neuro-oncology to classify sellar and parasellar tumors accurately. In a study by Castiglioni et al., the ability of a DL model to determine the presence or absence of craniopharyngiomas on MRI of pediatric patients was assessed (80). To achieve this goal, the authors developed a CNN. They trained this model with sagittal MRI slices of the sellar-suprasellar regions of 3 groups: controls, craniopharyngiomas, and differentials (80).
The use of DL algorithms in neuro-oncology is crucial in enhancing early diagnostic capabilities for adult and pediatric patients. In this context, DL algorithms can be trained conventionally using MRI or employed to address the limitations of intraoperative methods such as CLE and Raman scattering microscopy, thereby enhancing their effectiveness.
Neuronavigation systems aim to track surgical tools to ensure their real-time locations are precisely aligned with the patient’s anatomy. Various DL algorithms can also enhance these systems to increase accuracy and improve surgical outcomes. For example, NeuroIGN is a navigation system that integrates trained DL algorithms to recognize and segment brain tumors from MRI while including explainable AI techniques (Figure 6) (16). This study’s authors tested this system’s utility and accuracy to complete specific tasks such as registration and tracking, tumor segmentation, and real-time ultrasound imaging capabilities. They also evaluated how user-friendly this system was when used by individuals who had received only a short presentation on its use and were novices in using the NeuroIGN system (16). When evaluating the accuracy of the segmentation model, the authors observed that the system demonstrated good accuracy, making it an ideal candidate for image-guided neurosurgery (16).
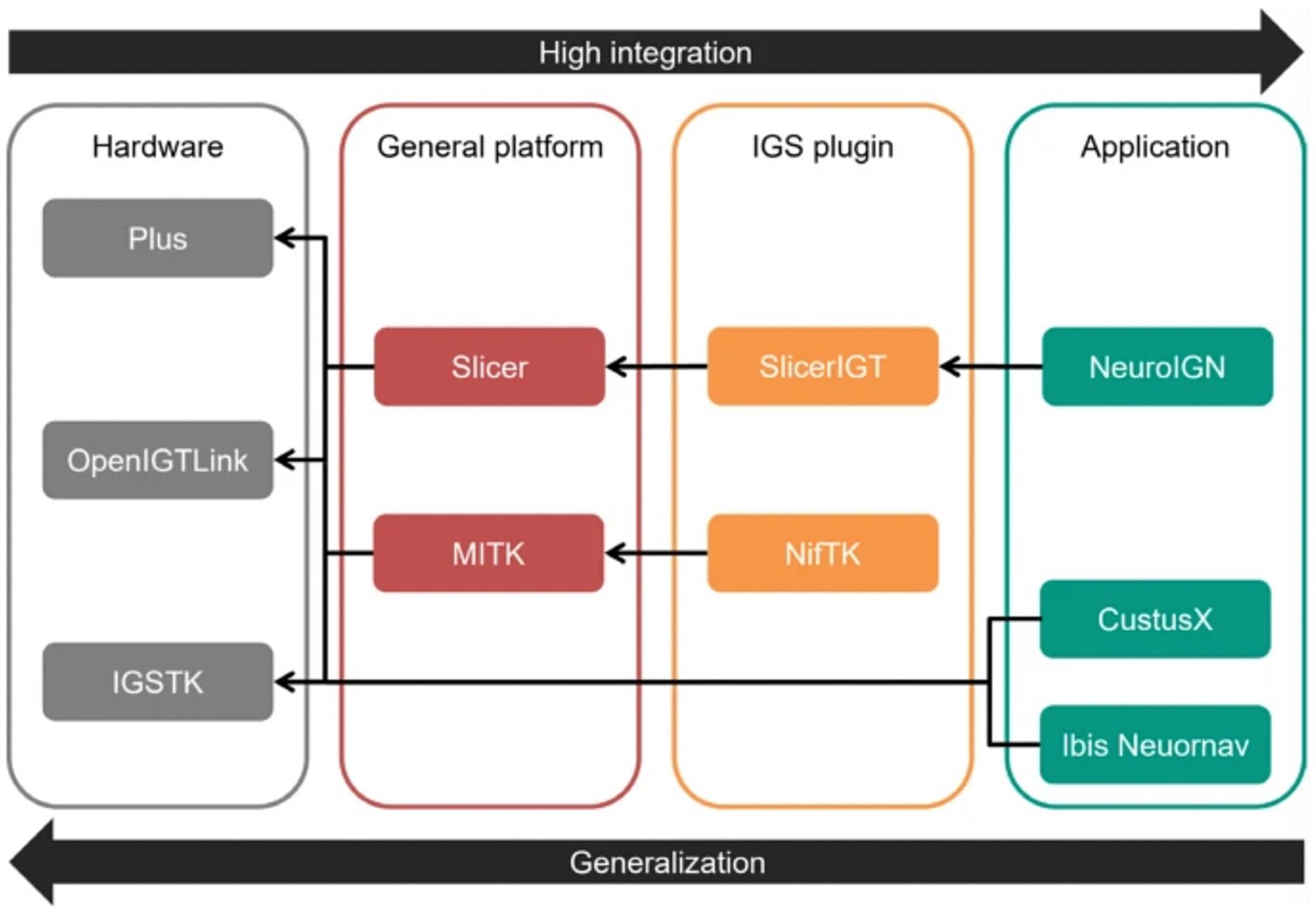
Figure 6. Role of deep learning (DL) in neuroimaging. The study by Zeineldin et al. (16) focused on developing a DL algorithm for tumor segmentation that includes explainable artificial intelligence techniques. This figure shows each level of the Neuro Image-Guided Neurosurgery (IGN) system, separated into hardware, general platform, image-guided surgery (IGS) plugin, and application. The hardware section is made up of 3 platforms: the Public software Library for UltraSound (Plus) platform, the Open Image-Guided Therapy Link (OpenIGTLink), and the Image-Guided Surgery Toolkit (IGSTK). The general platform level comprises the Slicer and Medical Imaging Interaction Toolkit (MITK). The IGS plugin level comprises the Slicer image-guided therapy (SlicerIGT) and the NiftyLink Toolkit (NifTK; NiftyLink). Finally, the application level includes the Neuro IGN system, CustusX, and Intraoperative Brain Imaging System Neuronavigation (Ibis Neuornav). The large arrow to the right shows increasing integration from left to right, and the large arrow to the left shows increasing generalization from right to left. The small arrows illustrate the dependency direction. Used with permission from Zeineldin, R.A., Karar, M.E., Burgert, O. et al. NeuroIGN: Explainable Multimodal Image-Guided System for Precise Brain Tumor Surgery. J Med Syst 48, 25 (2024). https://doi.org/10.1007/s10916-024-02037-3.
Neuronavigation during surgery is typically limited by brain shift following the incision and opening of the dura, which reduces the ability of neurosurgeons to identify their location intracranially during procedures. Shimamoto et al. used a CNN to design updated MRI to help remediate this issue (123). Preoperative and intraoperative MRI data from 248 patients was used, with the preoperative images serving as the training data for the CNN and the intraoperative images serving as the ground truth. This method allowed the model to learn how the brain shifted after the dural opening and adjust accordingly based only on preoperative images (Figure 7) (123).
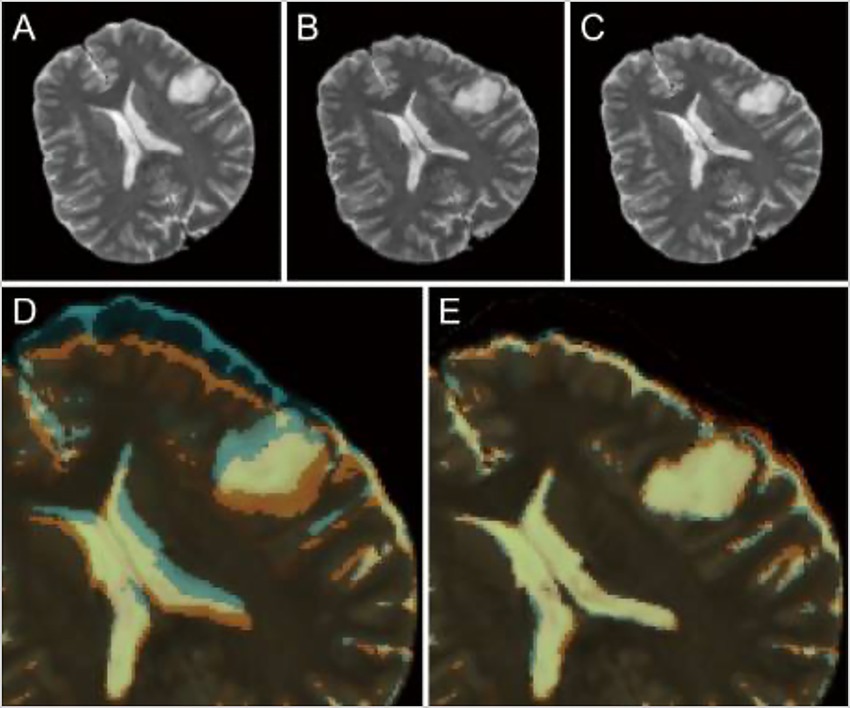
Figure 7. Role of deep learning (DL) in enhancing neuronavigation systems. A study by Shimamoto et al. (123) focused on the ability of a convolutional neural network (CNN) to adjust for and predict brain shifts based on preoperative and intraoperative magnetic resonance imaging (MRI). This figure illustrates the process of predicting brain shift intraoperatively compared to preoperatively in a selected case from the study (case 47). (A) Preoperative T2-weighted MRI. (B) Intraoperative image of the corresponding T2-weighted MRI. (C) The corresponding updated MRI. (D) The overlay of both the preoperative and intraoperative MRIs. (E) The updated MRI and the intraoperative MRI. The purpose of this image is to show the ability of the W-Net DL system to compensate for the brain shift. Reproduced from Shimamoto et al., Neurol Med Chir (Tokyo), 2023. Licensed under CC BY-NC-ND 4.0.
Drakopoulos et al. aimed to tackle the issue of image distortion due to brain shift with the added obstacle of tissue resection by studying the utility of the Adaptive Physics-Based Non-Rigid Registration method (137). This study highlighted the limitations of neuronavigation systems that use rigid transformation and the need for a more accurate method to map patient coordinates once the brain has shifted. Preoperative images of 30 glioma patients were segmented into brain, tumor, and nonbrain regions, first by removing the skull and outer tissue with the Brain Extraction Tool (Oxford Center for Functional MRI of the Brain) and then segmenting with the 3D Slicer Software. The Adaptive Physics-Based Non-Rigid Registration method had superior accuracy in detecting tissue deformation compared to other systems for modeling deformation (137).
Zufiria et al. proposed a feature-based CNN to improve the accuracy of real-time interventional MRI during procedures such as electrode placements for DBS (129). This CNN was trained by simulating an interventional needle superimposed on 2,560 coronal and axial slices of T1 and T2 weighted MRIs from 1,200 patients. Using this feature-based reconstruction process to reconstruct interventional MRIs, this study aimed to increase real-time understanding of the exact locations of brain structures after brain shift in procedures such as electrode placements during DBS or biopsy procedures (129).
Zhang et al. focused on enhancing intraoperative cone-beam CT (CBCT) quality using a 3D DL reconstruction framework (153). The purpose of this study, similar to those mentioned above, was to improve the accuracy of neuroimaging following a brain shift. Although intraoperative CBCT is a cost and time-efficient imaging method, its drawbacks include a reduced soft-tissue contrast resolution, limiting its utility in intraoperative use. After training with simulated brain lesions from CBCT images, the 3D DL reconstruction framework’s efficacy was tested in neurosurgery patients to assess its reliability using clinical data (153).
DL methods have also been applied to improve the accurate identification and recognition of different shunt valves of cerebrospinal fluid shunts using radiographs (115). In a study by Rhomberg et al. (115), a CNN was trained using a dataset of 2,070 radiographs and CT scout images of shunt valves to recognize and correctly identify the model type. This study found that their CNN had a high accuracy in correctly identifying standard shunt models. The utility of this system stems from the necessity of recognizing different shunt models in patients with an unknown medical history (115).
Recognition of objects intracranially using DL methods has also been applied to identify foreign bodies left behind following a neurosurgical procedure. Abramson et al. proposed an ultrasound-based system harnessing the powers of DL to help solve this issue (125). This study demonstrated the capabilities of their ultrasound system (using the Philips EPIQ 7 ultrasound machine) by first acquiring data by capturing images of a cotton ball implanted within porcine brains. In addition, the algorithm was tested to recognize a latex glove fragment measuring 5 mm in diameter, a stainless steel rod, and an Eppendorf tube. Following the success of these tests, cotton balls were placed within the resection cavities of 2 patients, with images captured by ultrasound used to train a DL detection algorithm (Figure 8). A custom version of the VGG16 CNN model demonstrated 99% accuracy in detecting foreign bodies within the brain (125).
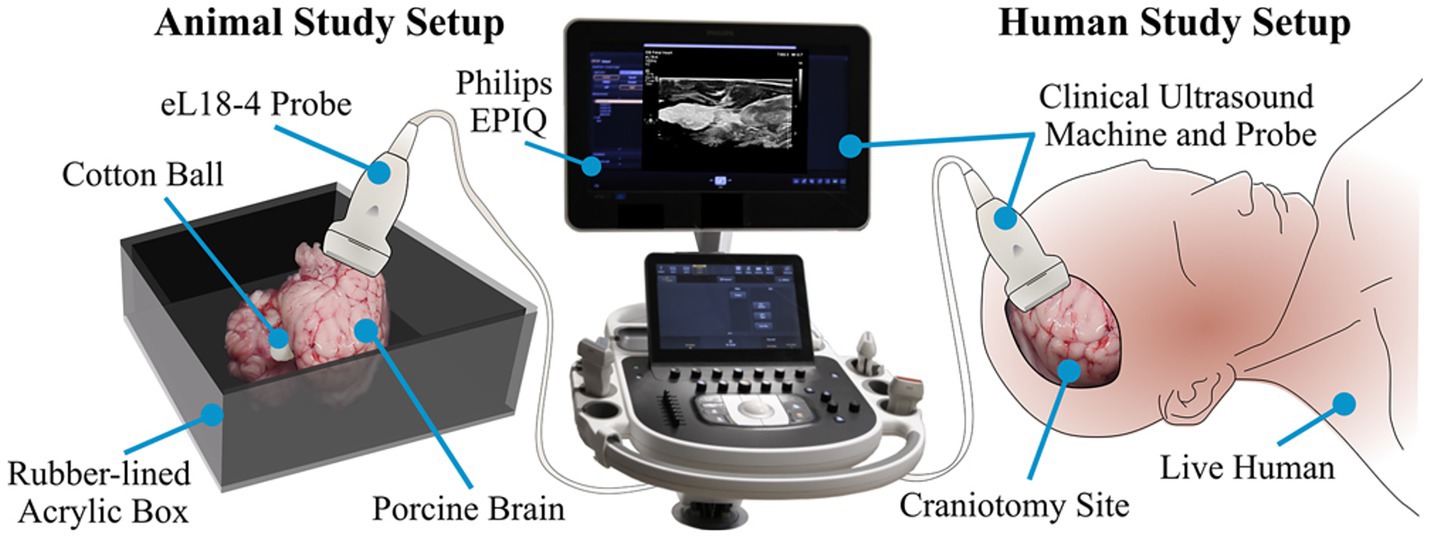
Figure 8. Deep learning (DL) in recognition of intracranial objects. The study by Abramson et al. (125) focused on developing an ultrasound-guided DL system that could ultimately recognize foreign objects left behind in patients’ surgical resection cavities. This figure illustrates the experimental design of this study. On the left, the authors started with testing the ability of the ultrasound DL system to recognize a cotton ball placed in porcine brains. When this test had been completed successfully, the researchers tested the ability of this DL system to recognize the presence of cotton balls within a human in vivo resection cavity. The ultrasound used in this case was the Philips EPIQ 7 using an eL 18–4 probe. Used with permission from Abramson et al. Automatic detection of foreign body objects in neurosurgery using a deep learning approach on intraoperative ultrasound images: from animal models to first in-human testing. Copyright of Frontiers in Surgery and made available under the CC BY 2.0 (http://creativecommons.org/licenses/by-nc-sa/2.0/).
The use of DL to identify specific abnormalities in neuroimaging modalities has also been examined in a study by Jiang et al. (28). This study analyzed noncontrast CT of patients with suspected TBI (28). This study focused on the ability of physicians with variable levels of expertise in neuroradiology or neurosurgery to detect a TBI compared with the ability of the DL model of icobrain TBI. Parameters used to detect TBIs included the presence of midline shift, hydrocephalus, hematomas, and Neuroimaging Radiological Interpretation System scores. The DL system’s ability to correctly categorize TBIs was similar to that of the attending physicians’ diagnoses. On the other hand, trainees had a lower level of agreement with the ground truth when compared to attending physicians. Moreover, they also demonstrated that, although the trainees had a substantial level of agreement on their initial review, using the DL algorithm as a supportive tool increased their agreement with the ground truth to an almost perfect level on their second review (28).
Although there are some limitations and controversies regarding their accuracy (27), DL algorithms can enhance diagnostic support. DL can facilitate surgical and clinical decision-making and aid in selecting the most appropriate treatment modality on a patient-specific basis, with particular auxiliary use in controversial cases and emergency settings.
Volumetric assessments are among the frequently used methods in neurosurgical studies for various purposes (211). However, since these assessments may require technical expertise, accurate measurements are not always achievable. Therefore, DL algorithms have increasingly been used to improve volumetric analyzes.
Measuring tumor burden is essential for evaluating treatment responses, particularly in neuro-oncology. Conventional 2D techniques are often unsuccessful in accurately measuring the volume of intracranial tumors, particularly gliomas, due to their irregular borders. Although still in development, DL models can address this challenge by providing more precise volumetric measurements for intracranial lesions.
An example of an intracranial lesion to which this method has been applied is pituitary adenoma (82). A study by Da Mutten et al. (82) created an automated volumetry pipeline to segment T1 contrast-enhanced MRIs of pituitary adenomas both preoperatively and postoperatively. This pipeline was developed by training a group of CNNs with 2D U-Net as the model architecture using manually segmented scans as training material. The model accurately segmented and completed a volumetric assessment preoperatively; however, the technique had difficulty achieving favorable results when assessing postoperative images. The authors hypothesized this may be partly due to interrater disagreement in ground truth segmentation of residual tumor tissue and image downsampling (82).
Tumor burden and volumetric assessment are often complex and limited due to the heterogenic and multifaceted nature of intracranial tumors. Chang et al. (18) studied automatic evaluation of the level of glioma burden employing the 3D U-Net architecture, specialized for detailed segmentation, by leveraging preoperative and postoperative MRI. A total of 239 patients were included in this study, which used automated processes to carry out tumor volumetric calculations. The study acknowledged that brain extraction, a step to discerning nonbrain tissue, was a rate-limiting step that created room for error during tumor segmentation (18).
A study assessing the feasibility of the application of DL in the extent of resection volumetric assessment of brain tumors was conducted by Zanier et al. (86). Single-institutional pre- and postoperative MRIs were manually labeled and combined with the Brain Tumor Segmentation Challenge 2015 and 2021 data from 1,053 patients. Use of U-Net architecture allowed the DL system to achieve faster and more accurate estimation of intracranial tumor volume (86).
Kang et al. (178) leveraged 12 DL models to automate the MRI segmentation process and obtainment of meningioma volumetric data. U-Net and nnU-Net–based DL training followed the manual segmentation. Although smaller than the Zanier et al. (86) study, the Kang et al. (178) study contained 459 subjects’ MRIs. nnU-Net, known to be better at image segmentation than U-Net, superseded the meningioma segmentation performance of U-Net, and 2D nnU-Net was the best performer. This study foresees the clinical applicability of this technology to manage specific meningioma cases (178).
The essential nature of accurately measuring tumor volumes warrants further research on the intersection between DL and volumetric assessment to optimize patient neuro-oncological surgical outcomes.
The contribution of DL algorithms to reducing human errors and accelerating diagnostic processes in clinical practice is undeniable. However, their integration into medical and surgical practices faces several limitations.
One significant technical limitation is the heterogeneity of image datasets used for training DL algorithms (210). For instance, when MRI datasets are used to train these algorithms, the similarity of MRI signal intensities is crucial. Variations in signal intensity caused by differences in scanners can hinder the creation of homogeneous datasets, which can affect the reliability of outputs. To address this challenge, efforts must focus on standardizing the datasets to ensure more reliable and reproducible outputs (212, 213).
Another critical challenge is methodology bias. If the training datasets for these algorithms come from only one or a few centers, the DL model might unintentionally favor the imaging protocols unique to those centers rather than focus on the tumor’s more critical pathological features (210, 214). This bias can result in the DL model failing to generalize when presented with data from other centers with different imaging protocols. Considering the heterogeneity of brain tumors, this bias is a significant limitation. To mitigate this issue, datasets used for training DL algorithms should be extensive and, if possible, sourced from multiple centers with different patient populations. Additionally, incorporating imaging sequences with varying protocols during model training can enhance the assessment and segmentation capabilities of these algorithms (210, 214).
Performance evaluation presents another challenge. In some studies, the performance of DL algorithms in image segmentation is evaluated against the ground truth. However, the ground truth is typically determined through manual segmentation by radiologists or neurosurgeons, introducing subjectivity that can cause fluctuations in model performance. To address this, multiple manual segmentations should be performed, and their averages should be used as a reference to reduce human error (188, 210).
On the other hand, using imaging datasets composed exclusively of high-quality images to train DL algorithms raises the issue of selection bias. Although training with high-quality images may improve performance during the training process, real-world clinical scenarios often involve suboptimal-quality images in clinical settings. For this reason, the datasets used to train these algorithms can indirectly affect the clinical applicability of the outputs. Therefore, careful attention must be paid to the selection of datasets, and the final purpose of the algorithm should be thoroughly evaluated (188, 210).
In summary, large datasets with diverse images should be used for model training to maximize the effectiveness of DL algorithms in clinical practice. Selection and methodology biases must be carefully considered and minimized to ensure reliable and generalizable outcomes, and DL algorithms should be trained on datasets that accurately represent the target patient population. It should be noted that there is no guarantee that using DL algorithms in clinical settings will yield more accurate or efficient data, reduce process costs, accelerate results, or decrease latency for result appearance. Implementing DL processes requires substantial computing power and infrastructure. Furthermore, the results and outputs of DL algorithms are entirely dependent on the quality of the informational inputs.
The increasing use of DL algorithms in medicine and surgery presents significant ethical challenges. Standardized guidelines are required to evaluate the scientific integrity and clinical applicability of studies using DL algorithms. Ensuring reliability and reproducibility is crucial for guiding future research. Collaborative teamwork among surgeons, data scientists, and ethicists can play a pivotal role in creating robust standards and addressing these challenges (210, 215).
One of the critical issues is the “black-box problem,” which refers to the difficulty in understanding the connection between the input data used by DL algorithms and the output they generate. This lack of interpretability stems from the complexity of the processes within DL systems, often comprising numerous hidden layers. Despite this complexity, understanding these processes is essential to building trust in and refining these models and advancing their clinical applications (210, 216).
Patient privacy and data protection are also critical concerns. Although these algorithms are trained using extensive datasets, it is crucial to ensure the security and privacy of the data as the dataset size increases. DL algorithms must be trained with deidentified datasets to prevent the possibility of linking imaging data to the individuals they belong to. Institutions must prioritize privacy by applying strict deidentification protocols and fostering trust with patients through transparent communication (210, 217).
Addressing these ethical considerations through standardized guidelines, interpretability-focused advancements, and privacy-focused practices is essential for the responsible integration of DL in clinical settings (210, 218, 219).
In light of the challenges associated with the application of DL algorithms in clinical and surgical practice, future research should focus on several key points. Consolidated models can be developed to integrate various AI algorithms across the pre-, intra-, and postoperative spectrum to improve reliability. Achieving this will require standardized guidelines and effective collaborative teamwork (218).
In addition, integrating DL as a broad and widely available resource for clinicians will likely require significant monetary and infrastructural investment. Training programs will need to be implemented to inform clinicians about the use of these technologies and their diagnostic and computational limitations.
One avenue of future research could be to investigate the quantitative advantages of using DL in surgical practice from a time-saving or cost-saving perspective (218). Furthermore, the computational errors that these technologies may have could lead to technical errors that would directly affect a patient’s well-being. Future research should be directed toward not only how these technical errors can be minimized but also developing safeguards to avoid complete reliance on these technologies if errors do arise.
Despite the challenges, integrating DL technologies into surgical practice has the potential to significantly improve the surgical workflow from both operative and diagnostic perspectives. The ability to intraoperatively monitor a patient’s condition for adverse events or precisely determine tumor borders via neuronavigation could be invaluable for clinicians. The technologies discussed in this paper provide a general outline of how DL can be used. As research into these applications progresses, it is crucial for clinicians and patients to understand how and when to use these technologies. Simultaneously, the development of standardized guidelines and privacy considerations should be prioritized as the technical capabilities of DL continue to evolve.
Employing DL and other AI algorithms in neurosurgical practice requires a collaborative team composed of neurosurgeons, data and computer scientists, and bioengineers. Expertise in these fields is necessary for using DL algorithms in neurosurgical studies. It is advantageous for future research on this topic that the fields of computer and data sciences are among the most rapidly evolving fields in the scientific community.
DL technologies can potentially enhance neurosurgical practice in various beneficial ways. These include improving the surgical workflow through real-time monitoring and detection of adverse events and pathophysiological conditions in a diagnostic fashion. Moreover, DL can also potentially aid in training novice neurosurgeons by learning from the techniques of experienced neurosurgeons.
Future studies should focus on developing mechanisms to improve the ease of use and access to these technologies within the neurosurgical workflow and training physicians to understand their benefits and current limitations. Furthermore, future research should be guided toward training DL models using more diverse and robust data so that the diagnostic applications of these technologies can be expanded further.
The data supporting this article will be made available by the authors on reasonable request.
KY: Data curation, Writing – original draft, Conceptualization, Methodology, Writing – review & editing. JH: Data curation, Writing – original draft, Formal analysis. ASG: Data curation, Formal analysis, Methodology, Writing – original draft. TJO: Data curation, Writing – original draft. AR: Data curation, Writing – original draft. PP: Formal analysis, Methodology, Writing – original draft. JC: Methodology, Validation, Writing – original draft. CECV: Formal analysis, Writing – original draft. YX: Data curation, Writing – original draft. BL: Conceptualization, Validation, Writing – review & editing. MS: Conceptualization, Validation, Writing – review & editing. MTL: Writing – review & editing. MCP: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by funds from the Newsome Chair of Neurosurgery Research held by Dr. Preul and from the Barrow Neurological Foundation.
We thank the staff of Neuroscience Publications at Barrow Neurological Institute for assistance with manuscript preparation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
AI, artificial intelligence; CBCT, cone-beam computed tomography; CLE, confocal laser endomicroscopy; CNN, convolutional neural network; CT, computed tomography; CV, computer vision; DBS, deep brain stimulation; DL, deep learning; IGS, image-guided surgery; ML, machine learning; MRI, magnetic resonance imaging; RNN, recurrent neural network; SAH, subarachnoid hemorrhage; TBI, traumatic brain injury; WHO, World Health Organization; 2D, 2-dimensional; 3D, 3-dimensional.
1. Kriegeskorte, N, and Golan, T. Neural network models and deep learning. Curr Biol. (2019) 29:R231–6. doi: 10.1016/j.cub.2019.02.034
2. Khan, DZ, Luengo, I, Barbarisi, S, Addis, C, Culshaw, L, Dorward, NL, et al. Automated operative workflow analysis of endoscopic pituitary surgery using machine learning: development and preclinical evaluation (IDEAL stage 0). J Neurosurg. (2022) 137:51–8. doi: 10.3171/2021.6.JNS21923
3. Khan, DZ, Newall, N, Koh, CH, Das, A, Aapan, S, Layard Horsfall, H, et al. Video-based performance analysis in pituitary surgery – part 2: artificial intelligence assisted surgical coaching. World Neurosurg. (2024) 190:e797–808. doi: 10.1016/j.wneu.2024.07.219
4. Hashimoto, DA, Rosman, G, Witkowski, ER, Stafford, C, Navarette-Welton, AJ, Rattner, DW, et al. Computer vision analysis of intraoperative video: automated recognition of operative steps in laparoscopic sleeve gastrectomy. Ann Surg. (2019) 270:414–21. doi: 10.1097/SLA.0000000000003460
5. Hung, AJ, Chen, J, Ghodoussipour, S, Oh, PJ, Liu, Z, Nguyen, J, et al. A deep-learning model using automated performance metrics and clinical features to predict urinary continence recovery after robot-assisted radical prostatectomy. BJU Int. (2019) 124:487–95. doi: 10.1111/bju.14735
6. Volkov, M, Hashimoto, DA, Rosman, G, Meireles, OR, and Rus, D. Machine learning and coresets for automated real-time video segmentation of laparoscopic and robot-assisted surgery. 2017 IEEE International Conference on Robotics and Automation (ICRA); Singapore, Singapore: IEEE Press (2017). p. 754–759.
7. Fischer, E, Jawed, KJ, Cleary, K, Balu, A, Donoho, A, Thompson Gestrich, W, et al. A methodology for the annotation of surgical videos for supervised machine learning applications. Int J Comput Assist Radiol Surg. (2023) 18:1673–8. doi: 10.1007/s11548-023-02923-0
8. Eppler, MB, Sayegh, AS, Maas, M, Venkat, A, Hemal, S, Desai, MM, et al. Automated capture of intraoperative adverse events using artificial intelligence: a systematic review and meta-analysis. J Clin Med. (2023) 12:1687. doi: 10.3390/jcm12041687
9. Su, R, van der Sluijs, M, Cornelissen, SAP, Lycklama, G, Hofmeijer, J, Majoie, C, et al. Spatio-temporal deep learning for automatic detection of intracranial vessel perforation in digital subtraction angiography during endovascular thrombectomy. Med Image Anal. (2022) 77:102377. doi: 10.1016/j.media.2022.102377
10. Kugener, G, Zhu, Y, Pangal, DJ, Sinha, A, Markarian, N, Roshannai, A, et al. Deep neural networks can accurately detect blood loss and hemorrhage control task success from video. Neurosurgery. (2022) 90:823–9. doi: 10.1227/neu.0000000000001906
11. Pangal, DJ, Kugener, G, Zhu, Y, Sinha, A, Unadkat, V, Cote, DJ, et al. Expert surgeons and deep learning models can predict the outcome of surgical hemorrhage from 1 min of video. Sci Rep. (2022) 12:8137. doi: 10.1038/s41598-022-11549-2
12. Koskinen, J, Torkamani-Azar, M, Hussein, A, Huotarinen, A, and Bednarik, R. Automated tool detection with deep learning for monitoring kinematics and eye-hand coordination in microsurgery. Comput Biol Med. (2022) 141:105121. doi: 10.1016/j.compbiomed.2021.105121
13. Tong, G, Wang, X, Jiang, H, Wu, A, Cheng, W, Cui, X, et al. A deep learning model for automatic segmentation of intraparenchymal and intraventricular hemorrhage for catheter puncture path planning. IEEE J Biomed Health Inform. (2023) 27:4454–65. doi: 10.1109/JBHI.2023.3285809
14. Ghauri, MS, Reddy, AJ, Tak, N, Tabaie, EA, Ramnot, A, Riazi Esfahani, P, et al. Utilizing deep learning for X-ray imaging: detecting and classifying degenerative spinal conditions. Cureus. (2023) 15:e41582. doi: 10.7759/cureus.41582
15. Reinecke, D, von Spreckelsen, N, Mawrin, C, Ion-Margineanu, A, Fürtjes, G, Jünger, ST, et al. Novel rapid intraoperative qualitative tumor detection by a residual convolutional neural network using label-free stimulated Raman scattering microscopy. Acta Neuropathol Commun. (2022) 10:109. doi: 10.1186/s40478-022-01411-x
16. Zeineldin, RA, Karar, ME, Burgert, O, and Mathis-Ullrich, F. NeuroIGN: explainable multimodal image-guided system for precise brain tumor surgery. J Med Syst. (2024) 48:25. doi: 10.1007/s10916-024-02037-3
17. Park, W, Abramov, I, On, TJ, Xu, Y, Castillo, AL, Gonzalez-Romo, NI, et al. Computational image analysis of distortion, sharpness, and depth of field in a next-generation hybrid exoscopic and microsurgical operative platform. Front Surg. (2024) 11:1418679. doi: 10.3389/fsurg.2024.1418679
18. Chang, K, Beers, AL, Bai, HX, Brown, JM, Ly, KI, Li, X, et al. Automatic assessment of glioma burden: a deep learning algorithm for fully automated volumetric and bidimensional measurement. Neuro-Oncology. (2019) 21:1412–22. doi: 10.1093/neuonc/noz106
19. Moher, D, Liberati, A, Tetzlaff, J, and Altman, DGPRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. (2009) 6:e1000097. doi: 10.1371/journal.pmed.1000097
20. Yu, Q, Shi, L, Xu, Z, Ren, Y, Yang, J, Zhou, Y, et al. Therapeutic effectiveness of percutaneous endoscopic spinal surgery for intraspinal cement leakage following percutaneous vertebroplasty: an early clinical study of 12 cases. Pain Physician. (2020) 23:E377–88. doi: 10.36076/ppj.2020/23/E377
21. Massalimova, A, Timmermans, M, Cavalcanti, N, Suter, D, Seibold, M, Carrillo, F, et al. Automatic breach detection during spine pedicle drilling based on vibroacoustic sensing. Artif Intell Med. (2023) 144:102641. doi: 10.1016/j.artmed.2023.102641
22. Agaronnik, ND, Kwok, A, Schoenfeld, AJ, and Lindvall, C. Natural language processing for automated surveillance of intraoperative neuromonitoring in spine surgery. J Clin Neurosci. (2022) 97:121–6. doi: 10.1016/j.jocn.2022.01.015
23. Bakaev, MA, Kobzev, DA, Piletskii, DK, Yakimenko, AA, and Gladkov, AV. Feasibility of spine segmentation in ML-based recognition of vertebrae in X-ray images. Proceedings of the 2023 IEEE 16th International Scientific and Technical Conference Actual Problems of Electronic Instrument Engineering, APEIE 2023; (2023). IEEE.
24. Kim, I-H, Kang, J, Jeong, J, Kim, J-S, Nam, Y, Ha, Y, et al. A fully automated landmark detection for spine surgery planning with a cascaded convolutional neural net. Inform Med Unlocked. (2022) 32:101045. doi: 10.1016/j.imu.2022.101045
25. Martino Cinnera, A, Morone, G, Iosa, M, Bonomi, S, Calabro, RS, Tonin, P, et al. Artificial neural network analysis of factors affecting functional independence recovery in patients with lumbar stenosis after neurosurgery treatment: an observational cohort study. J Orthop. (2024) 55:38–43. doi: 10.1016/j.jor.2024.04.003
26. Zhong, Y, Zhu, Y, Li, F, Zhu, S, and Qi, J. Generic approach to obtain contours of fascicular groups from microCT images of the peripheral nerve. J Image Graph. (2020) 25:354–65. doi: 10.11834/jig.190243
27. Agrawal, D, Joshi, S, Bahel, V, Poonamallee, L, and Agrawal, A. Three dimensional convolutional neural network-based automated detection of midline shift in traumatic brain injury cases from head computed tomography scans. J Neurosci Rural Pract. (2024) 15:293–9. doi: 10.25259/JNRP_490_2023
28. Jiang, B, Ozkara, BB, Creeden, S, Zhu, G, Ding, VY, Chen, H, et al. Validation of a deep learning model for traumatic brain injury detection and NIRIS grading on non-contrast CT: a multi-reader study with promising results and opportunities for improvement. Neuroradiology. (2023) 65:1605–17. doi: 10.1007/s00234-023-03170-5
29. Agrawal, D, Poonamallee, L, Joshi, S, and Bahel, V. Automated intracranial hemorrhage detection in traumatic brain injury using 3D CNN. J Neurosci Rural Pract. (2023) 14:615–21. doi: 10.25259/JNRP_172_2023
30. Adil, SM, Elahi, C, Patel, DN, Seas, A, Warman, PI, Fuller, AT, et al. Deep learning to predict traumatic brain injury outcomes in the low-resource setting. World Neurosurg. (2022) 164:e8–e16. doi: 10.1016/j.wneu.2022.02.097
31. Biswas, S, Mac Arthur, J, Pandit, A, Sarkar, V, and Joshi, GK. 294 evaluation of the clinical utility of ANCHOR (an artificial neural network for chronic subdural hematoma referral outcome prediction): an external validation study of 1713 patient referrals. Br J Surg. (2023) 110:znad258-074. doi: 10.1093/bjs/znad258.074
32. Vargas, J, Pease, M, Snyder, MH, Blalock, J, Wu, S, Nwachuku, E, et al. Automated preoperative and postoperative volume estimates risk of retreatment in chronic subdural hematoma: a retrospective, multicenter study. Neurosurgery. (2024) 94:317–24. doi: 10.1227/neu.0000000000002667
33. Gençtürk, TH, Kaya, İ, and Gülağız, FK. A comparative study on subdural brain hemorrhage segmentation. In: Lecture notes in networks and systems (2023), Eds. Fausto Pedro Garcia Marquez, Akhtar Jamil, Suleyman Eken, Alaa Ali Hameed. Springer.
34. Koschmieder, K, Paul, MM, van den Heuvel, TLA, van der Eerden, AW, van Ginneken, B, and Manniesing, R. Automated detection of cerebral microbleeds via segmentation in susceptibility-weighted images of patients with traumatic brain injury. Neuroimage Clin. (2022) 35:103027. doi: 10.1016/j.nicl.2022.103027
35. Matzkin, F, Newcombe, V, Stevenson, S, Khetani, A, Newman, T, Digby, R, et al. Self-supervised skull reconstruction in brain CT images with decompressive craniectomy. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics). arXiv, (2020).
36. Wang, B, Liao, X, Ni, Y, Zhang, L, Liang, J, Wang, J, et al. High-resolution medical image reconstruction based on residual neural network for diagnosis of cerebral aneurysm. Front Cardiovasc Med. (2022) 9:1013031. doi: 10.3389/fcvm.2022.1013031
37. Angkurawaranon, S, Sanorsieng, N, Unsrisong, K, Inkeaw, P, Sripan, P, Khumrin, P, et al. A comparison of performance between a deep learning model with residents for localization and classification of intracranial hemorrhage. Sci Rep. (2023) 13:9975. doi: 10.1038/s41598-023-37114-z
38. Voter, AF, Meram, E, Garrett, JW, and Yu, JJ. Diagnostic accuracy and failure mode analysis of a deep learning algorithm for the detection of intracranial hemorrhage. J Am Coll Radiol. (2021) 18:1143–52. doi: 10.1016/j.jacr.2021.03.005
39. Nishi, T, Yamashiro, S, Okumura, S, Takei, M, Tachibana, A, Akahori, S, et al. Artificial intelligence trained by deep learning can improve computed tomography diagnosis of nontraumatic subarachnoid hemorrhage by nonspecialists. Neurol Med Chir (Tokyo). (2021) 61:652–60. doi: 10.2176/nmc.oa.2021-0124
40. Danilov, G, Kotik, K, Negreeva, A, Tsukanova, T, Shifrin, M, Zakharova, N, et al. Classification of intracranial hemorrhage subtypes using deep learning on CT scans. In: Studies in health technology and informatics (2020), Eds. John Mantas, Arie Hasman, Mowafa S. Househ, Parisis Gallos, Emmanouil Zoulias. IOS Press Ebooks.
41. Venugopal, A, Moccia, S, Foti, S, Routray, A, Mac Lachlan, RA, Perin, A, et al. Real-time vessel segmentation and reconstruction for virtual fixtures for an active handheld microneurosurgical instrument. Int J Comput Assist Radiol Surg. (2022) 17:1069–77. doi: 10.1007/s11548-022-02584-5
42. Xu, J, Wu, J, Lei, Y, and Gu, Y. Application of Pseudo-three-dimensional residual network to classify the stages of Moyamoya disease. Brain Sci. (2023) 13:742. doi: 10.3390/brainsci13050742
43. Wang, S, Li, Y, Xu, Y, Song, S, Lin, R, Xu, S, et al. Resection-inspired histopathological diagnosis of cerebral cavernous malformations using quantitative multiphoton microscopy. Theranostics. (2022) 12:6595–610. doi: 10.7150/thno.77532
44. Hoffmann, N, Koch, E, Steiner, G, Petersohn, U, and Kirsch, M. Learning thermal process representations for intraoperative analysis of cortical perfusion during ischemic strokes. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) (2016) Springer.
45. Balu, A, Pangal, DJ, Kugener, G, and Donoho, DA. Pilot analysis of surgeon instrument utilization signatures based on Shannon entropy and deep learning for surgeon performance assessment in a cadaveric carotid artery injury control simulation. Oper Neurosurg (Hagerstown). (2023) 25:e330–7. doi: 10.1227/ons.0000000000000888
46. Al-Jaberi, F, Moeskes, M, Skalej, M, Fachet, M, and Hoeschen, C. 3D-visualization of segmented contacts of directional deep brain stimulation electrodes via registration and fusion of CT and FDCT. EJNMMI Rep. (2024) 8:17. doi: 10.1186/s41824-024-00208-6
47. Eid, MM, Chinnaperumal, S, Raju, SK, Kannan, S, Alharbi, AH, Natarajan, S, et al. Machine learning-powered lead-free piezoelectric nanoparticle-based deep brain stimulation: a paradigm shift in Parkinson’s disease diagnosis and evaluation. AIP Adv. (2024) 14. doi: 10.1063/5.0194094
48. Chen, J, Xu, H, Xu, B, Wang, Y, Shi, Y, and Xiao, L. Automatic localization of key structures for subthalamic nucleus-deep brain stimulation surgery via prior-enhanced multi-object magnetic resonance imaging segmentation. World Neurosurg. (2023) 178:e472–9. doi: 10.1016/j.wneu.2023.07.103
49. Zheng, YQ, Akram, H, Smith, S, and Jbabdi, S. A transfer learning approach to localising a deep brain stimulation target. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) (2023) Eds. Hayit Greenspan, Anant Madabhushi, Parvin Mousavi, Septimiu Salcudean, James Duncan, Tanveer Syeda-Mahmood, Russell Taylor. Springer.
50. Joseph, AS, Lazar, AJP, Sharma, DK, Maria, AB, Ganesan, N, and Sengan, S. ConvNet-based deep brain stimulation for attack patterns. In: Artificial intelligence for smart healthcare. Cham: Springer Innovations in Communication and Computing Part F632, Eds. Parul Agarwal, Kavita Khanna, Ahmed A. Elngar, Ahmed J. Obaid, Zdzislaw Polkowski. (2023). 275–92.
51. Chen, K, Li, C, Aronson, J, Fan, X, and Paulsen, K. Estimating shift at deep brain targets in deep brain stimulation: a comparison between a machine learning approach and a biomechanical model. Progress in Biomedical Optics and Imaging – Proceedings of SPIE ; (2023). SPIE Digital Library.
52. Rui-Qiang, L, Xiao-Dong, C, Ren-Zhe, T, Cai-Zi, L, Wei, Y, Dou-Dou, Z, et al. Automatic localization of target point for subthalamic nucleus-deep brain stimulation via hierarchical attention-UNet based MRI segmentation. Med Phys. (2023) 50:50–60. doi: 10.1002/mp.15956
53. Zhang, J, Zhou, C, Xiao, X, Chen, W, Jiang, Y, Zhu, R, et al. Magnetic resonance imaging image analysis of the therapeutic effect and neuroprotective effect of deep brain stimulation in Parkinson's disease based on a deep learning algorithm. Int J Numer Method Biomed Eng. (2022) 38:e3642. doi: 10.1002/cnm.3642
54. Baker, S, Tekriwal, A, Felsen, G, Christensen, E, Hirt, L, Ojemann, SG, et al. Automatic extraction of upper-limb kinematic activity using deep learning-based markerless tracking during deep brain stimulation implantation for Parkinson's disease: a proof of concept study. PLoS One. (2022) 17:e0275490. doi: 10.1371/journal.pone.0275490
55. Hosny, M, Zhu, M, Gao, W, and Fu, Y. A novel deep learning model for STN localization from LFPs in Parkinson’s disease. Biomed Signal Process Control. (2022) 77:103830. doi: 10.1016/j.bspc.2022.103830
56. Baxter, JSH, and Jannin, P. Combining simple interactivity and machine learning: a separable deep learning approach to subthalamic nucleus localization and segmentation in MRI for deep brain stimulation surgical planning. J Med Imaging (Bellingham). (2022) 9:045001. doi: 10.1117/1.JMI.9.4.045001
57. Gao, Q, Schmidt, SL, Kamaravelu, K, Turner, DA, Grill, WM, and Pajic, M. Offline policy evaluation for learning-based deep brain stimulation controllers. Proceedings – 13th ACM/IEEE international conference on cyber-physical systems, ICCPS 2022 ; (2022).
58. Liu, H, Holloway, KL, Englot, DJ, and Dawant, BM. A multi-rater comparative study of automatic target localization methods for epilepsy deep brain stimulation procedures. Progress in Biomedical Optics and Imaging – Proceedings of SPIE ; (2022). SPIE.
59. Chen, K, Li, C, Fan, X, Khan, T, Aronson, J, and Paulsen, K. Estimating shift at brain surface in deep brain stimulation using machine learning based methods. Progress in Biomedical Optics and Imaging – Proceedings of SPIE ; (2022). SPIE Digital Library.
60. Jiang, Z, Harati, S, Crowell, A, Mayberg, HS, Nemati, S, and Clifford, GD. Classifying major depressive disorder and response to deep brain stimulation over time by analyzing facial expressions. IEEE Trans Biomed Eng. (2021) 68:664–72. doi: 10.1109/TBME.2020.3010472
61. Cui, C, Liu, H, Englot, DJ, and Dawant, BM. Brain vessel segmentation in contrast-enhanced T1-weighted MR images for deep brain stimulation of the anterior thalamus using a deep convolutional neural network. Progress in Biomedical Optics and Imaging – Proceedings of SPIE ; (2021). SPIE Digital Library.
62. Liu, H, Cui, C, Englot, DJ, and Dawant, BM. Uncertainty estimation in medical image localization: towards robust anterior thalamus targeting for deep brain stimulation. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics). arXiv, (2020).
63. Baxter, JSH, Maguet, E, and Jannin, P. Localisation of the subthalamic nucleus in MRI via convolutional neural networks for deep brain stimulation planning. Proceedings of SPIE – the International Society for Optical Engineering ; (2020). SPIE Digital Library.
64. Bermudez, C, Rodriguez, W, Huo, Y, Hainline, AE, Li, R, Shults, R, et al. Towards machine learning prediction of deep brain stimulation (DBS) intra-operative efficacy maps. Progress in Biomedical Optics and Imaging – Proceedings of SPIE ; (2019). SPIE DIgital Library.
65. Peralta, M, Bui, QA, Ackaouy, A, Martin, T, Gilmore, G, Haegelen, C, et al. SepaConvNet for localizing the subthalamic nucleus using one second micro-electrode recordings. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS ; (2020). IEEE.
66. Souriau, R, Vigneron, V, Lerbet, J, and Chen, H. Probit latent variables estimation for a Gaussian process classifier: application to the detection of high-voltage spindles. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) (2018) Eds. Yannick Deville, Sharon Gannot, Russell Mason, Mark D. Plumbley, Dominic Ward. Springer.
67. Maged, A, Zhu, M, Gao, W, and Hosny, M. Lightweight deep learning model for automated STN localization using MER in Parkinson’s disease. Biomed Signal Process Control. (2024) 96:106640. doi: 10.1016/j.bspc.2024.106640
68. Makaram, N, Gupta, S, Pesce, M, Bolton, J, Stone, S, Haehn, D, et al. Deep learning-based visual complexity analysis of electroencephalography time-frequency images: can it localize the epileptogenic zone in the brain? Algorithms. (2023) 16:567. doi: 10.3390/a16120567
69. Courtney, MR, Sinclair, B, Neal, A, Nicolo, JP, Kwan, P, Law, M, et al. Automated segmentation of epilepsy surgical resection cavities: comparison of four methods to manual segmentation. NeuroImage. (2024) 296:120682. doi: 10.1016/j.neuroimage.2024.120682
70. Onofrey, JA, Staib, LH, and Papademetris, X. Segmenting the brain surface from CT images with artifacts using locally oriented appearance and dictionary learning. IEEE Trans Med Imaging. (2019) 38:596–607. doi: 10.1109/TMI.2018.2868045
71. Caredda, C, Ezhov, I, Sdika, M, Lange, F, Giannoni, L, Tachtsidis, I, et al. Pixel-wise and real-time estimation of optical mean path length using deep learning: application for intraoperative functional brain mapping. Proceedings of SPIE – the International Society for Optical Engineering ; (2024). SPIE.
72. Li, C, Fan, X, Duke, R, Chen, K, Evans, LT, and Paulsen, K. Intraoperative stereovision cortical surface segmentation using fast segment anything model. Progress in Biomedical Optics and Imaging – Proceedings of SPIE ; (2024). SPIE Digital LIbrary.
73. Uneri, A, Wu, P, Jones, CK, Vagdargi, P, Han, R, Helm, PA, et al. Deformable 3D-2D registration for high-precision guidance and verification of neuroelectrode placement. Phys Med Biol. (2021) 66:215014. doi: 10.1088/1361-6560/ac2f89
74. Edwards, CA, Goyal, A, Rusheen, AE, Kouzani, AZ, and Lee, KH. DeepNavNet: automated landmark localization for neuronavigation. Front Neurosci. (2021) 15:670287. doi: 10.3389/fnins.2021.670287
75. Yokota, T, Maki, T, Nagata, T, Murakami, T, Ugawa, Y, Laakso, I, et al. Real-time estimation of electric fields induced by transcranial magnetic stimulation with deep neural networks. Brain Stimul. (2019) 12:1500–7. doi: 10.1016/j.brs.2019.06.015
76. Martineau, T, He, S, Vaidyanathan, R, Brown, P, and Tan, H. Optimizing time-frequency feature extraction and channel selection through gradient backpropagation to improve action decoding based on subthalamic local field potentials. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS; (2020). IEEE.
77. Madhogarhia, R, Fathi Kazerooni, A, Arif, S, Ware, JB, Familiar, AM, Vidal, L, et al. Automated segmentation of pediatric brain tumors based on multi-parametric MRI and deep learning. Progress in Biomedical Optics and Imaging – Proceedings of SPIE; (2022). SPIE Digital Library.
78. Carton, FX, Chabanas, M, BKR, Munkvold, Reinertsen, I, and Noble, JH. Automatic segmentation of brain tumor in intraoperative ultrasound images using 3D U-Net. Proceedings of SPIE – the International Society for Optical Engineering ; (2020). SPIE Digital Library.
79. Fabelo, H, Halicek, M, Ortega, S, Shahedi, M, Szolna, A, Pineiro, JF, et al. Deep learning-based framework for in vivo identification of glioblastoma tumor using hyperspectral images of human brain. Sensors (Basel). (2019) 19:920. doi: 10.3390/s19040920
80. Castiglioni, G, Vallejos, J, Intriago, J, Hernandez, MI, Valenzuela, S, Fernandez, J, et al. Diagnostic support in pediatric craniopharyngioma using deep learning. Childs Nerv Syst. (2024) 40:2295–300. doi: 10.1007/s00381-024-06400-0
81. Zhu, E, Wang, J, Jing, Q, Shi, W, Xu, Z, Ai, P, et al. Individualized survival prediction and surgery recommendation for patients with glioblastoma. Front Med (Lausanne). (2024) 11:1330907. doi: 10.3389/fmed.2024.1330907
82. Da Mutten, R, Zanier, O, Ciobanu-Caraus, O, Voglis, S, Hugelshofer, M, Pangalu, A, et al. Automated volumetric assessment of pituitary adenoma. Endocrine. (2024) 83:171–7. doi: 10.1007/s12020-023-03529-x
83. Luckett, PH, Olufawo, M, Lamichhane, B, Park, KY, Dierker, D, Verastegui, GT, et al. Predicting survival in glioblastoma with multimodal neuroimaging and machine learning. J Neuro-Oncol. (2023) 164:309–20. doi: 10.1007/s11060-023-04439-8
84. Puustinen, S, Vrzakova, H, Hyttinen, J, Rauramaa, T, Falt, P, Hauta-Kasari, M, et al. Hyperspectral imaging in brain tumor surgery-evidence of machine learning-based performance. World Neurosurg. (2023) 175:e614–35. doi: 10.1016/j.wneu.2023.03.149
85. Wang, MY, Jia, CG, Xu, HQ, Xu, CS, Li, X, Wei, W, et al. Development and validation of a deep learning predictive model combining clinical and Radiomic features for short-term postoperative facial nerve function in acoustic neuroma patients. Curr Med Sci. (2023) 43:336–43. doi: 10.1007/s11596-023-2713-x
86. Zanier, O, Da Mutten, R, Vieli, M, Regli, L, Serra, C, and Staartjes, VE. DeepEOR: automated perioperative volumetric assessment of variable grade gliomas using deep learning. Acta Neurochir. (2023) 165:555–66. doi: 10.1007/s00701-022-05446-w
87. Wu, J, Wang, T, Uckermann, O, Galli, R, Schackert, G, Cao, L, et al. Learned end-to-end high-resolution lensless fiber imaging towards real-time cancer diagnosis. Sci Rep. (2022) 12:18846. doi: 10.1038/s41598-022-23490-5
88. Fang, A, Hu, J, Zhao, W, Feng, M, Fu, J, Feng, S, et al. Extracting clinical named entity for pituitary adenomas from Chinese electronic medical records. BMC Med Inform Decis Mak. (2022) 22:72. doi: 10.1186/s12911-022-01810-z
89. Danilov, G, Korolev, V, Shifrin, M, Ilyushin, E, Maloyan, N, Saada, D, et al. Noninvasive glioma grading with deep learning: a pilot study. Stud Health Technol Inform. (2022) 290:675–8. doi: 10.3233/SHTI220163
90. Danilov, GV, Pronin, IN, Korolev, VV, Maloyan, NG, Ilyushin, EA, Shifrin, MA, et al. MR-guided non-invasive typing of brain gliomas using machine learning. Zh Vopr Neirokhir Im N N Burdenko. (2022) 86:36–42. doi: 10.17116/neiro20228606136
91. Lee, CC, Lee, WK, Wu, CC, Lu, CF, Yang, HC, Chen, YW, et al. Applying artificial intelligence to longitudinal imaging analysis of vestibular schwannoma following radiosurgery. Sci Rep. (2021) 11:3106. doi: 10.1038/s41598-021-82665-8
92. Rahmat, R, Saednia, K, Haji Hosseini Khani, MR, Rahmati, M, Jena, R, and Price, SJ. Multi-scale segmentation in GBM treatment using diffusion tensor imaging. Comput Biol Med. (2020) 123:103815. doi: 10.1016/j.compbiomed.2020.103815
93. Di Ieva, A, Russo, C, Al Suman, A, and Liu, S. IOTG-01. computational neurosurgery in brain tumors: a paradigm shift on the use of artificial intelligence and connectomics in pre-and intra-operative imaging. Neuro-Oncology. (2021) 23:vi227. doi: 10.1093/neuonc/noab196.910
94. Wang, J, Wang, S, Liu, J, Zhang, J, Liu, B, Li, Z, et al. The application value of pre-trained deep learning neural network model in differentiating central nervous system tumors. Chin J Neurosurg. (2024) 40:378–84. doi: 10.3760/cma.j.cn112050-20230930-00095
95. Hsu, SPC, Lin, MH, Lin, CF, Hsiao, TY, Wang, YM, and Sun, CW. Brain tumor grading diagnosis using transfer learning based on optical coherence tomography. Biomed Opt Express. (2024) 15:2343–57. doi: 10.1364/BOE.513877
96. Cekic, E, Pinar, E, Pinar, M, and Dagcinar, A. Deep learning-assisted segmentation and classification of brain tumor types on magnetic resonance and surgical microscope images. World Neurosurg. (2024) 182:e196–204. doi: 10.1016/j.wneu.2023.11.073
97. Prathaban, K, Wu, B, Tan, CL, and Huang, Z. Detecting tumor infiltration in diffuse gliomas with deep learning. Adv Intell Syst. (2023) 5:2300397. doi: 10.1002/aisy.202300397
98. Zhu, E, Shi, W, Chen, Z, Wang, J, Ai, P, Wang, X, et al. Reasoning and causal inference regarding surgical options for patients with low-grade gliomas using machine learning: a SEER-based study. Cancer Med. (2023) 12:20878–91. doi: 10.1002/cam4.6666
99. Li, Z, Shu, H, Liang, R, Goodridge, A, Sahu, M, Creighton, FX, et al. Tatoo: vision-based joint tracking of anatomy and tool for skull-base surgery. Int J Comput Assist Radiol Surg. (2023) 18:1303–10. doi: 10.1007/s11548-023-02959-2
100. Pirhadi, A, Salari, S, Ahmad, MO, Rivaz, H, and Xiao, Y. Robust landmark-based brain shift correction with a Siamese neural network in ultrasound-guided brain tumor resection. Int J Comput Assist Radiol Surg. (2023) 18:501–8. doi: 10.1007/s11548-022-02770-5
101. Salari, S, Rasoulian, A, Rivaz, H, and Xiao, Y. FocalErrorNet: uncertainty-aware focal modulation network for inter-modal registration error estimation in ultrasound-guided neurosurgery. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) (2023) arXiv.
102. Salari, S, Rasoulian, A, Rivaz, H, and Xiao, Y. Towards multi-modal anatomical landmark detection for ultrasound-guided brain tumor resection with contrastive learning. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) (2023) arXiv.
103. Srikanthan, D, Kaufmann, M, Jamzad, A, Syeda, A, Santilli, A, Sedghi, A, et al. Attention based multi-instance learning for improved glioblastoma detection using mass spectrometry. Progress in Biomedical Optics and Imaging – Proceedings of SPIE ; (2023). SPIE Digital Library.
104. Zeineldin, RA, Pollok, A, Mangliers, T, Karar, ME, Mathis-Ullrich, F, and Burgert, O. Deep automatic segmentation of brain tumours in interventional ultrasound data. Curr Dir Biomed Eng. (2022) 8:133–7. doi: 10.1515/cdbme-2022-0034
105. Wu, S, Wu, Y, Chang, H, Su, FT, Liao, H, Tseng, W, et al. Deep learning-based segmentation of various brain lesions for radiosurgery. Appl Sci. (2021) 11:9180. doi: 10.3390/app11199180
106. Shen, B, Zhang, Z, Shi, X, Cao, C, Zhang, Z, Hu, Z, et al. Real-time intraoperative glioma diagnosis using fluorescence imaging and deep convolutional neural networks. Eur J Nucl Med Mol Imaging. (2021) 48:3482–92. doi: 10.1007/s00259-021-05326-y
107. Zeineldin, RA, Weimann, P, Karar, ME, Mathis-Ullrich, F, and Burgert, O. Slicer-DeepSeg: open-source deep learning toolkit for brain tumour segmentation. Curr Dir Biomed Eng. (2021) 7:30–4. doi: 10.1515/cdbme-2021-1007
108. Chen, C, Cheng, Y, Xu, J, Zhang, T, Shu, X, Huang, W, et al. Automatic meningioma segmentation and grading prediction: a hybrid deep-learning method. J Pers Med. (2021) 11:786. doi: 10.3390/jpm11080786
109. Li, Y, Charalampaki, P, Liu, Y, Yang, GZ, and Giannarou, S. Context aware decision support in neurosurgical oncology based on an efficient classification of endomicroscopic data. Int J Comput Assist Radiol Surg. (2018) 13:1187–99. doi: 10.1007/s11548-018-1806-7
110. Ermis, E, Jungo, A, Poel, R, Blatti-Moreno, M, Meier, R, Knecht, U, et al. Fully automated brain resection cavity delineation for radiation target volume definition in glioblastoma patients using deep learning. Radiat Oncol. (2020) 15:100. doi: 10.1186/s13014-020-01553-z
111. Colecchia, F, Ruffle, JK, Pombo, GC, Gray, R, Hyare, H, and Nachev, P. Knowledge-driven deep neural network models for brain tumour segmentation. J Phys Conf Ser. (2020) 1662:012010. doi: 10.1088/1742-6596/1662/1/012010
112. Franco, P, Wurtemberger, U, Dacca, K, Hubschle, I, Beck, J, Schnell, O, et al. SPectroscOpic prediction of bRain Tumours (SPORT): study protocol of a prospective imaging trial. BMC Med Imaging. (2020) 20:123. doi: 10.1186/s12880-020-00522-y
113. Touati, R, and Kadoury, S. A least square generative network based on invariant contrastive feature pair learning for multimodal MR image synthesis. Int J Comput Assist Radiol Surg. (2023) 18:971–9. doi: 10.1007/s11548-023-02916-z
114. Wang, Y, and Ye, X. U-Net multi-modality glioma MRIs segmentation combined with attention. 2023 International Conference on Intelligent Supercomputing and BioPharma, ISBP 2023 ; (2023). IEEE.
115. Rhomberg, T, Trivik-Barrientos, F, Hakim, A, Raabe, A, and Murek, M. Applied deep learning in neurosurgery: identifying cerebrospinal fluid (CSF) shunt systems in hydrocephalus patients. Acta Neurochir. (2024) 166:69. doi: 10.1007/s00701-024-05940-3
116. Rahmani, M, Moghaddasi, H, Pour-Rashidi, A, Ahmadian, A, Najafzadeh, E, and Farnia, P. D2BGAN: Dual Discriminator Bayesian Generative Adversarial Network for deformable MR-ultrasound registration applied to brain shift compensation. Diagnostics (Basel). (2024) 14:1319. doi: 10.3390/diagnostics14131319
117. Sastry, RA, Setty, A, Liu, DD, Zheng, B, Ali, R, Weil, RJ, et al. Natural language processing augments comorbidity documentation in neurosurgical inpatient admissions. PLoS One. (2024) 19:e0303519. doi: 10.1371/journal.pone.0303519
118. Baghdadi, A, Lama, S, Singh, R, and Sutherland, GR. Tool-tissue force segmentation and pattern recognition for evaluating neurosurgical performance. Sci Rep. (2023) 13:9591. doi: 10.1038/s41598-023-36702-3
119. Xu, J, Anastasiou, D, Booker, J, Burton, OE, Layard Horsfall, H, Salvadores Fernandez, C, et al. A deep learning approach to classify surgical skill in microsurgery using force data from a novel sensorised surgical glove. Sensors (Basel). (2023) 23:8947. doi: 10.3390/s23218947
120. Danilov, G, Kotik, K, Shifrin, M, Strunina, Y, Pronkina, T, Tsukanova, T, et al. Data quality estimation via model performance: machine learning as a validation tool. In: Studies in health technology and informatics (2023) Eds. John Mantas, Parisis Gallos, Emmanouil Zoulias, Arie Hasman, Mowafa S. Househ, Martha Charalampidou, Andriana Magdalinou. IOS Press Ebooks.
121. Chiou, SY, Liu, LS, Lee, CW, Kim, DH, Al-Masni, MA, Liu, HL, et al. Augmented reality surgical navigation system integrated with deep learning. Bioengineering (Basel). (2023) 10:617. doi: 10.3390/bioengineering10050617
122. Zhang, X, Sisniega, A, Zbijewski, WB, Lee, J, Jones, CK, Wu, P, et al. Combining physics-based models with deep learning image synthesis and uncertainty in intraoperative cone-beam CT of the brain. Med Phys. (2023) 50:2607–24. doi: 10.1002/mp.16351
123. Shimamoto, T, Sano, Y, Yoshimitsu, K, Masamune, K, and Muragaki, Y. Precise brain-shift prediction by new combination of W-Net deep learning for neurosurgical navigation. Neurol Med Chir (Tokyo). (2023) 63:295–303. doi: 10.2176/jns-nmc.2022-0350
124. Yilmaz, R, Winkler-Schwartz, A, Mirchi, N, Reich, A, Christie, S, Tran, DH, et al. Continuous monitoring of surgical bimanual expertise using deep neural networks in virtual reality simulation. NPJ Digit Med. (2022) 5:54. doi: 10.1038/s41746-022-00596-8
125. Abramson, HG, Curry, EJ, Mess, G, Thombre, R, Kempski-Leadingham, KM, Mistry, S, et al. Automatic detection of foreign body objects in neurosurgery using a deep learning approach on intraoperative ultrasound images: from animal models to first in-human testing. Front Surg. (2022) 9:1040066. doi: 10.3389/fsurg.2022.1040066
126. Han, R, Jones, CK, Lee, J, Wu, P, Vagdargi, P, Uneri, A, et al. Deformable MR-CT image registration using an unsupervised, dual-channel network for neurosurgical guidance. Med Image Anal. (2022) 75:102292. doi: 10.1016/j.media.2021.102292
127. Han, R, Jones, CK, Lee, J, Zhang, X, Wu, P, Vagdargi, P, et al. Joint synthesis and registration network for deformable MR-CBCT image registration for neurosurgical guidance. Phys Med Biol. (2022) 67:125008. doi: 10.1088/1361-6560/ac72ef
128. Su, Y, Sun, Y, Hosny, M, Gao, W, and Fu, Y. Facial landmark-guided surface matching for image-to-patient registration with an RGB-D camera. Int J Med Robot. (2022) 18:e2373. doi: 10.1002/rcs.2373
129. Zufiria, B, Qiu, S, Yan, K, Zhao, R, Wang, R, She, H, et al. A feature-based convolutional neural network for reconstruction of interventional MRI. NMR Biomed. (2022) 35:e4231. doi: 10.1002/nbm.4231
130. Lam, L, Lam, A, Bacchi, S, and Abou-Hamden, A. Neurosurgery inpatient outcome prediction for discharge planning with deep learning and transfer learning. Br J Neurosurg. (2022) 39:110–4. doi: 10.1080/02688697.2022.2151565
131. Davids, J, Makariou, SG, Ashrafian, H, Darzi, A, Marcus, HJ, and Giannarou, S. Automated vision-based microsurgical skill analysis in neurosurgery using deep learning: development and preclinical validation. World Neurosurg. (2021) 149:e669–86. doi: 10.1016/j.wneu.2021.01.117
132. Ramesh, A, Beniwal, M, Uppar, AM, Vikas, V, and Rao, M. Microsurgical tool detection and characterization in intra-operative neurosurgical videos. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS ; (2021). IEEE.
133. Han, R, Jones, CK, Ketcha, MD, Wu, P, Vagdargi, P, Uneri, A, et al. Deformable MR-CT image registration using an unsupervised synthesis and registration network for neuro-endoscopic surgery. Progress in Biomedical Optics and Imaging – Proceedings of SPIE ; (2021). SPIE Digital Library.
134. Canalini, L, Klein, J, Miller, D, and Kikinis, R. Enhanced registration of ultrasound volumes by segmentation of resection cavity in neurosurgical procedures. Int J Comput Assist Radiol Surg. (2020) 15:1963–74. doi: 10.1007/s11548-020-02273-1
135. Farnia, P, Mohammadi, M, Najafzadeh, E, Alimohamadi, M, Makkiabadi, B, and Ahmadian, A. High-quality photoacoustic image reconstruction based on deep convolutional neural network: towards intra-operative photoacoustic imaging. Biomed Phys Eng Express. (2020) 6:045019. doi: 10.1088/2057-1976/ab9a10
136. Danilov, G, Kotik, K, Shifrin, M, Strunina, U, Pronkina, T, and Potapov, A. Predicting postoperative hospital stay in neurosurgery with recurrent neural networks based on operative reports. In: Studies in health technology and informatics (2020) Eds. Louise B. Pape-Haugaard, Christian Lovis, Inge Cort Madsen, Patrick Weber, Per Hostrup Nielsen, Philip Scott. IOS Press Ebooks.
137. Drakopoulos, F, Tsolakis, C, Angelopoulos, A, Liu, Y, Yao, C, Kavazidi, KR, et al. Adaptive physics-based non-rigid registration for immersive image-guided neuronavigation systems. Front Digit Health. (2020) 2:613608. doi: 10.3389/fdgth.2020.613608
138. Danilov, G, Kotik, K, Shifrin, M, Strunina, U, Pronkina, T, and Potapov, A. Prediction of postoperative hospital stay with deep learning based on 101 654 operative reports in neurosurgery. In: Studies in health technology and informatics (2019) Eds. Amnon Shabo (Shvo), Inge Madsen, Hans-Ulrich Prokosch, Kristiina Häyrinen, Klaus-Hendrik Wolf, Fernando Martin-Sanchez, Matthias Löbe, Thomas M. Deserno. IOS Press Ebooks.
139. Moccia, S, Foti, S, Routray, A, Prudente, F, Perin, A, Sekula, RF, et al. Toward improving safety in neurosurgery with an active handheld instrument. Ann Biomed Eng. (2018) 46:1450–64. doi: 10.1007/s10439-018-2091-x
140. Tan, Y, Patel, RV, Wang, Z, Luo, Y, Chen, J, Luo, J, et al. Generation and applications of synthetic computed tomography images for neurosurgical planning. J Neurosurg. (2024) 141:742–51. doi: 10.3171/2024.1.JNS232196
141. Shi, H, Liu, J, and Liao, H. A classification and segmentation combined two-stage CNN model for automatic segmentation of brainstem. IFMBE proceedings ; (2019). Springer Nature Singapore.
142. Nitsch, J, Klein, J, Moltz, JH, Miller, D, Sure, U, Kikinis, R, et al. Neural-network-based automatic segmentation of cerebral ultrasound images for improving image-guided neurosurgery. Progress in Biomedical Optics and Imaging – Proceedings of SPIE ; (2019). SPIE Digital Library.
143. Li, J, and Egger, J. Dataset descriptor for the autoimplant cranial implant design challenge. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) (2020) Eds. Jianning Li, Jan Egger. Springer.
144. Matzkin, F, Newcombe, V, Glocker, B, and Ferrante, E. Cranial implant design via virtual craniectomy with shape priors. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) (2020), arXiv.
145. Lucena, O, Vos, SB, Vakharia, V, Duncan, J, Ourselin, S, and Sparks, R. Convolutional neural networks for fiber orientation distribution enhancement to improve single-shell diffusion MRI tractography. In: Computational diffusion MRI mathematics and visualization Eds. Elisenda Bonet-Carne, Jana Hutter, Marco Palombo, Marco Pizzolato, Farshid Sepehrband, Fan Zhang. Springer. (2020). 101–12.
146. Mahapatra, S, Balamurugan, M, Chung, K, Kuppoor, V, Curry, E, Aghabaglau, F, et al. Automatic detection of cotton balls during brain surgery: where deep learning meets ultrasound imaging to tackle foreign objects. Progress in Biomedical Optics and Imaging – Proceedings of SPIE ; (2021).
147. Zeineldin, RA, Karar, ME, Elshaer, Z, Schmidhammer, M, Coburger, J, Wirtz, CR, et al. iRegNet: non-rigid registration of MRI to interventional us for brain-shift compensation using convolutional neural networks. IEEE Access. (2021) 9:147579–90. doi: 10.1109/access.2021.3120306
148. Zeineldin, RA, Karar, ME, Mathis-Ullrich, F, and Burgert, O. A hybrid deep registration of MR scans to interventional ultrasound for neurosurgical guidance. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) (2021) Eds. Chunfeng Lian, Xiaohuan Cao, Islem Rekik, Xuanang Xu, Pingkun Yan. Springer.
149. Quon, JL, Han, M, Kim, LH, Koran, ME, Chen, LC, Lee, EH, et al. Artificial intelligence for automatic cerebral ventricle segmentation and volume calculation: a clinical tool for the evaluation of pediatric hydrocephalus. J Neurosurg Pediatr. (2021) 27:131–8. doi: 10.3171/2020.6.PEDS20251
150. Li, J, von Campe, G, Pepe, A, Gsaxner, C, Wang, E, Chen, X, et al. Automatic skull defect restoration and cranial implant generation for cranioplasty. Med Image Anal. (2021) 73:102171. doi: 10.1016/j.media.2021.102171
151. McKinley, R, Felger, LA, Hewer, E, Maragkou, T, Murek, M, Novikova, T, et al. Machine learning for white matter fibre tract visualization in the human brain via Mueller matrix polarimetric data. Proceedings of SPIE – the International Society for Optical Engineering ; (2022). SPIE Digital Library.
152. Gaur, PK, Bhardwaj, A, Venkata, PPK, Sarode, D, and Mahajan, A. Assistance technique for fiducial localization in MRI images with subslice classification neural network. Proceedings – 2022 6th International Conference on Intelligent Computing and Control Systems, ICICCS 2022 ; (2022). IEEE.
153. Zhang, X, Wu, P, Zbijewski, WB, Sisniega, A, Han, R, Jones, CK, et al. DL-Recon combining 3D deep learning image synthesis and model uncertainty with physics-based image reconstruction. Proceedings of SPIE – the International Society for Optical Engineering ; (2022). SPIE.
154. Korycinski, M, Ciecierski, KA, and Niewiadomska-Szynkiewicz, E. Neural fiber prediction with deep learning. International Conference on Wireless and Mobile Computing, Networking and Communications ; (2022). IEEE.
155. Li, L, Feng, P, Ding, H, and Wang, G. A preliminary exploration to make stereotactic surgery robots aware of the semantic 2D/3D working scene. IEEE Trans Med Robot Bionics. (2022) 4:17–27. doi: 10.1109/tmrb.2021.3124160
156. Philipp, M, Alperovich, A, Lisogorov, A, Gutt-Will, M, Mathis, A, Saur, S, et al. Annotation-efficient learning of surgical instrument activity in neurosurgery. Curr Dir Biomed Eng. (2022) 8:30–3. doi: 10.1515/cdbme-2022-0008
157. Feng, P, Li, L, Ding, H, and Wang, C. Head pose estimation of patients with monocular vision for surgery robot based on deep learning. Chin J Biomed Eng. (2022) 41:537–46. doi: 10.3969/j.issn.0258-8021.2022.05.003
158. Sarwin, G, Carretta, A, Staartjes, V, Zoli, M, Mazzatenta, D, Regli, L, et al. Live image-based neurosurgical guidance and roadmap generation using unsupervised embedding. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) (2023). arXiv.
159. Eskandari, M, Gueziri, HE, and Collins, DL. Hessian-based similarity metric for multimodal medical image registration. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) (2023) arXiv.
160. Yoon, BC, Pomerantz, SR, Mercaldo, ND, Goyal, S, L'Italien, EM, Lev, MH, et al. Incorporating algorithmic uncertainty into a clinical machine deep learning algorithm for urgent head CTs. PLoS One. (2023) 18:e0281900. doi: 10.1371/journal.pone.0281900
161. Haber, MA, Biondetti, GP, Gauriau, R, Comeau, DS, Chin, JK, Bizzo, BC, et al. Detection of idiopathic normal pressure hydrocephalus on head CT using a deep convolutional neural network. Neural Comput & Applic. (2023) 35:9907–15. doi: 10.1007/s00521-023-08225-5
162. de Boer, M, Kos, TM, Fick, T, van Doormaal, JAM, Colombo, E, Kuijf, HJ, et al. nnU-Net versus mesh growing algorithm as a tool for the robust and timely segmentation of neurosurgical 3D images in contrast-enhanced T1 MRI scans. Acta Neurochir. (2024) 166:92. doi: 10.1007/s00701-024-05973-8
163. Moriconi, S, Rodriguez-Nunez, O, Gros, R, Felger, LA, Maragkou, T, Hewer, E, et al. Near-real-time Mueller polarimetric image processing for neurosurgical intervention. Int J Comput Assist Radiol Surg. (2024) 19:1033–43. doi: 10.1007/s11548-024-03090-6
164. Matasyoh, NM, Schmidt, R, Zeineldin, RA, Spetzger, U, and Mathis-Ullrich, F. Interactive surgical training in neuroendoscopy: real-time anatomical feature localization using natural language expressions. IEEE Trans Biomed Eng. (2024) 71:2991–9. doi: 10.1109/TBME.2024.3405814
165. Bi, L, Pieper, S, Chlorogiannis, DD, Golby, AJ, and Frisken, S. Open-source, deep-learning skin surface segmentation model for cost-effective neuronavigation accessible to low-resource settings. Progress in Biomedical Optics and Imaging – Proceedings of SPIE ; (2024). SPIE Digital Library.
166. Garcia-Garcia, S, Cepeda, S, Arrese, I, and Sarabia, R. A fully automated pipeline using Swin transformers for deep learning-based blood segmentation on head computed tomography scans after aneurysmal subarachnoid hemorrhage. World Neurosurg. (2024) 190:e762–73. doi: 10.1016/j.wneu.2024.07.216
167. Pangal, DJ, Kugener, G, Shahrestani, S, Attenello, F, Zada, G, and Donoho, DA. A guide to annotation of neurosurgical intraoperative video for machine learning analysis and computer vision. World Neurosurg. (2021) 150:26–30. doi: 10.1016/j.wneu.2021.03.022
168. Jawed, KJ, Buchanan, I, Cleary, K, Fischer, E, Mun, A, Gowda, N, et al. A microdiscectomy surgical video annotation framework for supervised machine learning applications. Int J Comput Assist Radiol Surg. (2024) 19:1947–52. doi: 10.1007/s11548-024-03203-1
169. Pangal, DJ, Kugener, G, Cardinal, T, Lechtholz-Zey, E, Collet, C, Lasky, S, et al. Use of surgical video-based automated performance metrics to predict blood loss and success of simulated vascular injury control in neurosurgery: a pilot study. J Neurosurg. (2022) 137:840–9. doi: 10.3171/2021.10.JNS211064
170. Staartjes, VE, Volokitin, A, Regli, L, Konukoglu, E, and Serra, C. Machine vision for real-time intraoperative anatomic guidance: a proof-of-concept study in endoscopic pituitary surgery. Oper Neurosurg (Hagerstown). (2021) 21:242–7. doi: 10.1093/ons/opab187
171. Sugiyama, T, Sugimori, H, Tang, M, Ito, Y, Gekka, M, Uchino, H, et al. Deep learning-based video-analysis of instrument motion in microvascular anastomosis training. Acta Neurochir. (2024) 166:6. doi: 10.1007/s00701-024-05896-4
172. On, TJ, Xu, Y, Gonzalez-Romo, NI, Gomez-Castro, G, Alcantar-Garibay, O, Santello, M, et al. Detection of hand motion during cadaveric mastoidectomy dissections: a technical note. Front Surg. (2024) 11:1441346. doi: 10.3389/fsurg.2024.1441346
173. On, TJ, Xu, Y, Chen, J, Gonzalez-Romo, NI, Alcantar-Garibay, O, Bhanushali, J, et al. Deep learning detection of hand motion during microvascular anastomosis simulations performed by expert cerebrovascular neurosurgeons. World Neurosurg. (2024) 192:e217–32. doi: 10.1016/j.wneu.2024.09.069
174. Gonzalez-Romo, NI, Hanalioglu, S, Mignucci-Jimenez, G, Koskay, G, Abramov, I, Xu, Y, et al. Quantification of motion during microvascular anastomosis simulation using machine learning hand detection. Neurosurg Focus. (2023) 54:E2. doi: 10.3171/2023.3.FOCUS2380
175. Izadyyazdanabadi, M, Belykh, E, Zhao, X, Moreira, LB, Gandhi, S, Cavallo, C, et al. Fluorescence image histology pattern transformation using image style transfer. Front Oncol. (2019) 9:519. doi: 10.3389/fonc.2019.00519
176. Izadyyazdanabadi, M, Belykh, E, Martirosyan, N, Eschbacher, J, Nakaji, P, Yang, Y, et al. Improving utility of brain tumor confocal laser endomicroscopy: Objective value assessment and diagnostic frame detection with convolutional neural networks: SPIE (2017). SPIE Digitla Library.
177. Izadyyazdanabadi, M, Belykh, E, Mooney, M, Martirosyan, N, Eschbacher, J, Nakaji, P, et al. Convolutional neural networks: ensemble modeling, fine-tuning and unsupervised semantic localization for neurosurgical CLE images. J Vis Commun Image Represent. (2018) 54:10–20. doi: 10.1016/j.jvcir.2018.04.004
178. Kang, H, Witanto, JN, Pratama, K, Lee, D, Choi, KS, Choi, SH, et al. Fully automated MRI segmentation and volumetric measurement of intracranial meningioma using deep learning. J Magn Reson Imaging. (2023) 57:871–81. doi: 10.1002/jmri.28332
179. Won, SY, Kim, JH, Woo, C, Kim, DH, Park, KY, Kim, EY, et al. Real-world application of a 3D deep learning model for detecting and localizing cerebral microbleeds. Acta Neurochir. (2024) 166:381. doi: 10.1007/s00701-024-06267-9
180. Payman, AA, El-Sayed, I, and Rubio, RR. Exploring the combination of computer vision and surgical neuroanatomy: a workflow involving artificial intelligence for the identification of skull base foramina. World Neurosurg. (2024) 191:e403–10. doi: 10.1016/j.wneu.2024.08.137
181. Wodzinski, M, Kwarciak, K, Daniol, M, and Hemmerling, D. Improving deep learning-based automatic cranial defect reconstruction by heavy data augmentation: from image registration to latent diffusion models. Comput Biol Med. (2024) 182:109129. doi: 10.1016/j.compbiomed.2024.109129
182. Mehandzhiyski, A, Yurukov, N, Ilkov, P, Mikova, D, and Gabrovsky, N. Innovations in neurosurgery – a novel machine learning predictive model for lumbar disc reherniation following microsurgical discectomy. Brain Spine. (2024) 4:103614. doi: 10.1016/j.bas.2024.103614
183. Zanier, O, Da Mutten, R, Carretta, A, Zoli, M, Mazzatenta, D, Serra, C, et al. Real-time intraoperative depth estimation in transsphenoidal surgery using deep learning. Brain Spine. (2024) 4:103783. doi: 10.1016/j.bas.2024.103783
184. Zanier, O, Da Mutten, R, Ryu, S-J, Carretta, A, Palandri, G, Mazzatenta, D, et al. Synthesis of cranial CT imaging from biplanar radiographs using deep learning. Brain Spine. (2024) 4:103459. doi: 10.1016/j.bas.2024.103459
185. Bobeff, E, Puzio, T, Wiśniewski, K, Matera, K, and Jaskólski, D. Harnessing AI in neuroradiology: advantages of volumetric assessment and reproducibility. Brain Spine. (2024) 4:103820. doi: 10.1016/j.bas.2024.103820
186. Kiewitz, J, Aydin, OU, Hilbert, A, Gultom, M, Nouri, A, Khalil, AA, et al. Deep learning-based multiclass segmentation in aneurysmal subarachnoid hemorrhage. Front Neurol. (2024) 15:1490216. doi: 10.3389/fneur.2024.1490216
187. Ho, T-L, Ferreira, F, Brudfors, M, Bot, M, Ashburner, J, and Akram, H. Segmentation of the subthalamic nucleus from clinical MRI: combining registration and deep learning. Stereotact Funct Neurosurg. (2024) 102:90–1. doi: 10.1159/000540478
188. Bianconi, A, Rossi, LF, Bonada, M, Zeppa, P, Nico, E, De Marco, R, et al. Deep learning-based algorithm for postoperative glioblastoma MRI segmentation: a promising new tool for tumor burden assessment. Brain Inform. (2023) 10:26. doi: 10.1186/s40708-023-00207-6
189. LeCun, Y, Bengio, Y, and Hinton, G. Deep learning. Nature. (2015) 521:436–44. doi: 10.1038/nature14539
190. Bengio, Y, Simard, P, and Frasconi, P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans Neural Netw. (1994) 5:157–66. doi: 10.1109/72.279181
191. Scarselli, F, Gori, M, Tsoi, AC, Hagenbuchner, M, and Monfardini, G. The graph neural network model. IEEE Trans Neural Netw. (2009) 20:61–80. doi: 10.1109/TNN.2008.2005605
192. Goodfellow, I, Pouget-Abadie, J, Mirza, M, Xu, B, Warde-Farley, D, Ozair, S, et al. Generative adversarial networks. Commun ACM. (2020) 63:139–44. doi: 10.1145/3422622
193. Vaswani, A, Shazeer, NM, Parmar, N, Uszkoreit, J, Jones, L, Gomez, AN, et al. Attention is all you need. In: Neural information processing systems (2017), arXiv.
194. Sohl-Dickstein, JN, Weiss, EA, Maheswaranathan, N, and Ganguli, S. Deep unsupervised learning using nonequilibrium thermodynamics. ArXiv. (2015):abs/1503.03585. doi: 10.48550/arXiv.1503.03585
195. Ho, J, Jain, A, and Abbeel, P. Denoising diffusion probabilistic models. ArXiv. (2020):abs/2006.11239. doi: 10.48550/arXiv.2006.11239
196. Hochreiter, S, and Schmidhuber, J. Long short-term memory. Neural Comput. (1997) 9:1735–80. doi: 10.1162/neco.1997.9.8.1735
197. Dosovitskiy, A, Beyer, L, Kolesnikov, A, Weissenborn, D, Zhai, X, Unterthiner, T, et al. An image is worth 16x16 words: transformers for image recognition at scale. ArXiv. (2020):abs/2010.11929. doi: 10.48550/arXiv.2010.11929
198. Yangi, K, On, TJ, Xu, Y, Gholami, AS, Hong, J, Reed, AG, et al. Artificial intelligence integration in surgery through hand and instrument tracking: a systematic literature review. Front Surg. (2025) 12:12. doi: 10.3389/fsurg.2025.1528362
199. Harari, RE, Dias, RD, Kennedy-Metz, LR, Varni, G, Gombolay, M, Yule, S, et al. Deep learning analysis of surgical video recordings to assess nontechnical skills. JAMA Netw Open. (2024) 7:e2422520. doi: 10.1001/jamanetworkopen.2024.22520
200. Jumah, F, Raju, B, Nagaraj, A, Shinde, R, Lescott, C, Sun, H, et al. Uncharted waters of machine and deep learning for surgical phase recognition in neurosurgery. World Neurosurg. (2022) 160:4–12. doi: 10.1016/j.wneu.2022.01.020
201. Lugaresi, C, Tang, J, Nash, H, McClanahan, C, Uboweja, E, Hays, M, et al. MediaPipe: a framework for building perception pipelines. ArXiv. (2019):abs/1906.08172. doi: 10.48550/arXiv.1906.08172
202. Yangi, K, Uzunkol, A, and Celik, SE. Benign intracranial calcified lesion or a so-called brain stone: a challenging diagnosis. Cureus. (2023) 15:e39596. doi: 10.7759/cureus.39596
203. Zhu, M, Chang, W, Jing, L, Fan, Y, Liang, P, Zhang, X, et al. Dual-modality optical diagnosis for precise in vivo identification of tumors in neurosurgery. Theranostics. (2019) 9:2827–42. doi: 10.7150/thno.33823
204. Gok, H, Celik, SE, Yangi, K, Yavuz, AY, Percinoglu, G, Unlu, NU, et al. Management of epidural hematomas in pediatric and adult population: a hospital-based retrospective study. World Neurosurg. (2023) 177:e686–92. doi: 10.1016/j.wneu.2023.06.123
205. Yangi, K, Demir, DD, and Uzunkol, A. Intracranial hemorrhage after Pfizer-BioNTech (BNT162b2) mRNA COVID-19 vaccination: a case report. Cureus. (2023) 15:e37747. doi: 10.7759/cureus.37747
206. Caceres, JA, and Goldstein, JN. Intracranial hemorrhage. Emerg Med Clin North Am. (2012) 30:771–94. doi: 10.1016/j.emc.2012.06.003
207. Yangi, K, Yavuz, AY, Percinoglu, G, Aki, B, and Celik, SE. A huge calcified supratentorial ependymoma: a case report. Cureus. (2023) 15:e37493. doi: 10.7759/cureus.37493
208. Aydin, MV, Yangi, K, Toptas, E, and Aydin, S. Skull base collision tumors: giant non-functioning pituitary adenoma and olfactory groove meningioma. Cureus. (2023) 15:e44710. doi: 10.7759/cureus.44710
209. Izadyyazdanabadi, M, Belykh, E, Mooney, MA, Eschbacher, JM, Nakaji, P, Yang, Y, et al. Prospects for theranostics in neurosurgical imaging: empowering confocal laser endomicroscopy diagnostics via deep learning. Front Oncol. (2018) 8:240. doi: 10.3389/fonc.2018.00240
210. Bonada, M, Rossi, LF, Carone, G, Panico, F, Cofano, F, Fiaschi, P, et al. Deep learning for MRI segmentation and molecular subtyping in glioblastoma: critical aspects from an emerging field. Biomedicines. (2024) 12:1878. doi: 10.3390/biomedicines12081878
211. Çal, M, and Yangi, K. A single-center study comparing volumetric and morphometric measurements of Chiari malformation in the Turkish population. European. Arch Med Res. (2024) 40:50–6. doi: 10.4274/eamr.galenos.2024.24892
212. Pandey, U, Saini, J, Kumar, M, Gupta, R, and Ingalhalikar, M. Normative baseline for radiomics in brain MRI: evaluating the robustness, regional variations, and reproducibility on FLAIR images. J Magn Reson Imaging. (2021) 53:394–407. doi: 10.1002/jmri.27349
213. Orlhac, F, Lecler, A, Savatovski, J, Goya-Outi, J, Nioche, C, Charbonneau, F, et al. How can we combat multicenter variability in MR radiomics? Validation of a correction procedure. Eur Radiol. (2021) 31:2272–80. doi: 10.1007/s00330-020-07284-9
214. Renard, F, Guedria, S, Palma, N, and Vuillerme, N. Variability and reproducibility in deep learning for medical image segmentation. Sci Rep. (2020) 10:13724. doi: 10.1038/s41598-020-69920-0
215. Lambin, P, Leijenaar, RTH, Deist, TM, Peerlings, J, de Jong, EEC, van Timmeren, J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. (2017) 14:749–62. doi: 10.1038/nrclinonc.2017.141
216. Reyes, M, Meier, R, Pereira, S, Silva, CA, Dahlweid, FM, von Tengg-Kobligk, H, et al. On the interpretability of artificial intelligence in radiology: challenges and opportunities. Radiol Artif Intell. (2020) 2:e190043. doi: 10.1148/ryai.2020190043
217. Ibrahim, M, Muhammad, Q, Zamarud, A, Eiman, H, and Fazal, F. Navigating glioblastoma diagnosis and care: transformative pathway of artificial intelligence in integrative oncology. Cureus. (2023) 15:e44214. doi: 10.7759/cureus.44214
218. Awuah, WA, Adebusoye, FT, Wellington, J, David, L, Salam, A, Weng Yee, AL, et al. Recent outcomes and challenges of artificial intelligence, machine learning, and deep learning in neurosurgery. World Neurosurg X. (2024) 23:100301. doi: 10.1016/j.wnsx.2024.100301
Keywords: artificial intelligence, convolutional neural network, deep learning, machine learning, neurological surgery, neurosurgery
Citation: Yangi K, Hong J, Gholami AS, On TJ, Reed AG, Puppalla P, Chen J, Calderon Valero CE, Xu Y, Li B, Santello M, Lawton MT and Preul MC (2025) Deep learning in neurosurgery: a systematic literature review with a structured analysis of applications across subspecialties. Front. Neurol. 16:1532398. doi: 10.3389/fneur.2025.1532398
Received: 21 November 2024; Accepted: 04 March 2025;
Published: 16 April 2025.
Edited by:
Santiago Cepeda, Hospital Universitario Río Hortega, SpainReviewed by:
Pietro Fiaschi, University of Genoa, ItalyCopyright © 2025 Yangi, Hong, Gholami, On, Reed, Puppalla, Chen, Calderon Valero, Xu, Li, Santello, Lawton and Preul. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark C. Preul, TmV1cm9wdWJAYmFycm93bmV1cm8ub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.