
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurol., 02 July 2024
Sec. Headache and Neurogenic Pain
Volume 15 - 2024 | https://doi.org/10.3389/fneur.2024.1356974
Objective: A growing body of evidence underscores a significant association between neurological disorders, particularly migraines, and the gut microbiota. However, a research gap persists in understanding the cause-and-effect dynamics between these elements. Therefore, we employed robust methodologies aimed at thoroughly exploring the causal relationship between the gut microbiome and migraines.
Methods: Employing bidirectional Two Sample Mendelian Randomization (TSMR) analysis, we investigated the causal association between the composition of the gut microbiota and migraines. Data summarizing the relationship between gut microbiota and migraines were extracted from one or more genome-wide association studies. The TSMR analysis employed five methods to assess the correlation between the gut microbiota and migraines, with the inverse variance-weighted method serving as the primary approach for analyzing causal links. Sensitivity analyses were applied to address horizontal pleiotropy and heterogeneity. Simultaneously, a meta-analysis was performed to strengthen the robustness of the findings. Additionally, a reverse TSMR was carried out to explore potential occurrences of reverse causal relationships.
Results: The ongoing TSMR analysis identified a collection of 14 bacterial taxa connected to migraines. Among these, 8 taxa exhibited a protective effect, while 5 taxa had a detrimental impact, and 1 taxon maintained a neutral relationship. The reverse Mendelian randomization analysis highlighted stable outcomes for only one bacterial taxonomic group.
Conclusion: The study confirms a causal relationship between the gut microbiota and migraines, offering a new perspective for migraine research. Strategically targeting specific bacterial taxa with dysregulation may be effective in both preventing and treating migraines, thus opening new avenues for therapeutic strategies.
Migraine, widely recognized as a prevalent neurological challenge with a significant impact on human well-being (1, 2), is categorized by the International Headache Society into two principal types: those accompanied by aura and those without (3). While the intricate pathological details remain elusive, there exists a consensus among researchers concerning the significant involvement of the trigeminal nervous system in the pathophysiology of migraines (4–7).
Numerous studies have explored an association between migraines and disorders of the digestive system, such as irritable bowel syndrome and gastroesophageal reflux (8–10). The increasing recognition of how dietary choices impact migraine episodes sparks consideration of a potential link to the microbiota in the gastrointestinal tract (11, 12). As our understanding deepens on the role of microbiota in neurological conditions like Alzheimer’s and Parkinson’s diseases (13, 14), attention is directed toward its potential interplay with migraines, involving the gut-brain axis (15).
The gut-brain axis, a bidirectional link between the gut and the brain, involves the gastrointestinal tract exerting influence on the central nervous system through various mechanisms (15, 16). Components contributing to this intricate relationship include the immune system, hormonal activity, inflammatory mediators, neuropeptides, and the dynamic world of gut microbiota (16–18). The microbial community not only plays a pivotal role in providing essential nutrients and safeguarding gut integrity but also affects pain perception through intricate endocrine and metabolic signaling pathways (19, 20). Recent research suggests a potential causal association between the gut microbiome and the onset of migraines, highlighting specific bacterial taxa such as the genus Coprococcus3 and genus Anaerotruncus (21). Nevertheless, it is imperative to emphasize the limited scope of these studies, necessitating future research endeavors to authenticate and illuminate the underlying mechanisms.
The ongoing research within this conceptual framework aims to unravel the intricate association between gut microbiota and migraines. Using Mendelian randomization (MR) as a methodological tool and genetic variations as instrumental variables, this study aimed to investigate the potential cause-and-effect relationship between variables such as lifestyle choices or the composition of gut microbiota and the occurrence of specific diseases (22, 23). By leveraging the inherent randomness in the inheritance of genotypes from parents to offspring, MR aims to mitigate the impact of confounding variables that might distort results in observational studies. This enhances the reliability of establishing causal relationships without being influenced by biases from reverse causation (24).
In the pursuit of a comprehensive evaluation of the evidence surrounding the causal relationship between gut microbiota and migraines, we conducted a systematic review of existing Mendelian Randomization (MR) studies Due to the limited literature in this specific domain, we performed a re-examination using consolidated summary statistics from the FinnGen study and other publicly available Genome-Wide Association Studies (GWAS). Subsequently, we conducted a meta-analysis to compile and present a concise overview of the findings derived from the MR analyses.
Investigating the potential association between gut microbiota and migraines necessitated a comprehensive examination using TSMR analysis. To enhance the credibility of the findings, we performed a subsequent meta-analysis. Information for this investigation was sourced from diverse sources including published articles and digital repositories, with strict adherence to the MRSTROBE checklist ensuring a thorough investigation (25).
The extraction of instrumental variables related to human gut microbiota involved navigating the MiBioGen Alliance website,1 established by Kurilshikov et al. in 2021. This consortium predominantly comprises extensive data, including whole-genome genotype and 16S fecal microbiota from 24 cohorts, involving a total of 18,340 individuals (26). In our investigation, we conducted a meta-analysis, specifically focusing on autosomal human genetic variation and its association with the gut microbiome. The study also investigated the changes in microbiome composition and methodological variations in microbiome data. Employing a standardized pipeline, our objective was to elucidate the specific loci responsible for microbial traits (mbTL). These include genetic loci influencing the relative abundance of microbial taxa (mbQTL) or their presence (microbial binary trait loci, or mbBTLs). This comprehensive analysis included 211 taxa, extending across 35 families, 20 orders, 16 classes, 9 phyla, and 131 genera.
We obtained summary data on migraines from three distinct studies. The primary dataset, centered on migraines, originated from the FinnGen study, which commenced in Finland in 2017, aiming to explore novel targets and methodologies for diverse diseases through genetic research. We retrieved Version 9 data, released on 11 May 2023, from their official website.2 This dataset comprised 18,477 migraine cases and 287,837 controls, with an average onset age of 40.27 years, predominantly among females (82.07% of cases). Migraine cases were identified using International Classification of Diseases (ICD) codes from the 8th, 9th, and 10th editions. Specifically, summary data for migraines with aura encompassed 7,917 cases and 287,837 controls, while migraines without aura involved 6,730 cases and 287,837 controls.
The second dataset originated from a study conducted by Choquet et al. (27). In this investigation, data were obtained from the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort (71,602 individuals, including 11,320 migraine cases) and the UK Biobank (UKB) cohort (482,967 individuals, with 17,532 migraine cases). In both cohorts, the predominant ancestry was European, constituting 94.19 and 81.53%, respectively. Notably, in the UK Biobank, there is a significant 75.30% overlap between the exposure dataset and our study sample, minimizing bias to approximately 1% (28).
The third study is grounded in the research of Dönertaş et al. (29). This study identified four distinct disease clusters among 116 diseases in the UK Biobank data. The clusters were delineated based on the age of onset, confirming genetic associations with age-related diseases through data analysis. Access to data for this study can be obtained in the GWAS Catalog (GCST90038646), including 13,971 migraine cases and 470,627 controls. A comprehensive presentation of information from the outcome databases is provided in Table 1.
The procedural sequence of the investigation is illustrated in Figure 1. To explore the potential causal relationship between gut microbiota and migraines, we conducted a Two-Sample Mendelian Randomization (TSMR) analysis During the selection of instrumental variables, we adhered to three essential three essential prerequisites to ensure the reliability of the outcomes (30) (Figure 2):
1. A fundamental requirement is a close association between gut microbiota and the instrumental variables (IVs) ultimately incorporated.
2. There must be no reciprocal dependence between the included IVs and confounding factors that may affect both the categorization of gut microbiota and migraines.
3. Migraines should be solely influenced by the categorization of gut microbiota through the IVs, without any indication of horizontal pleiotropy.
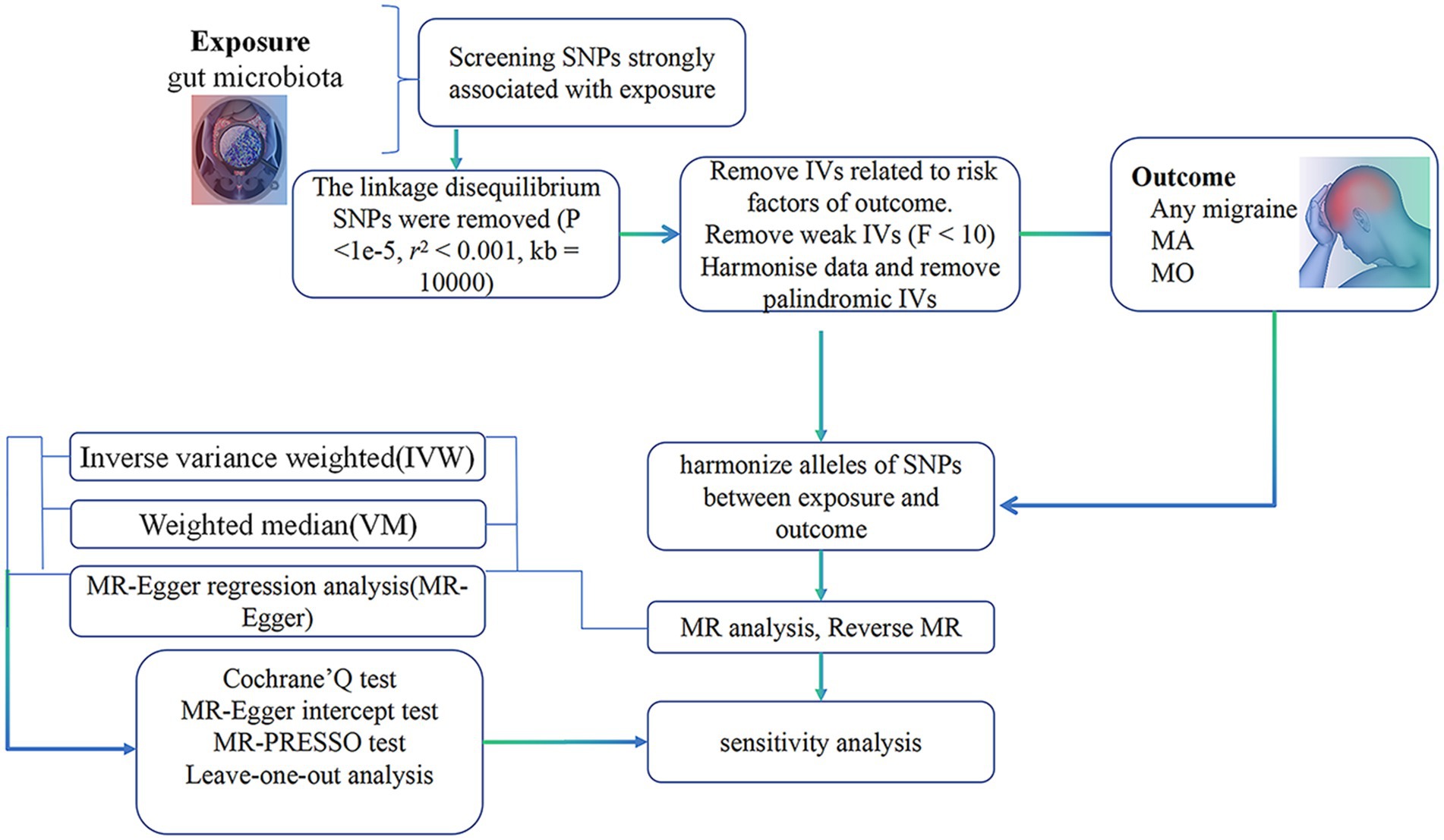
Figure 1. Summary of current Mendelian randomization processes. MA, migraine with aura; MO, migraine without aura.
To explore the causal impact of human gut microbiota on migraines and avoid overlooking potentially significant findings, we set a correlation threshold of 1 × 10−5. This choice was deliberate, especially in cases where SNP selection fell below the standard threshold of 5 × 10−8 (26). Furthermore, in the context of various MR investigations related to the gut microbiota, the threshold of 1 × 10−5 is considered optimal, aimed to increase the number of SNPs meeting the criteria for subsequent analysis (31, 32).
To ensure no linkage disequilibrium among instrumental variables (IVs) associated with gut microbiota, a clumping procedure was implemented (r2 < 0.001, clumping window size = 10,000 kb), leveraging European 1,000 Genomes Project sequencing data as the reference panel. Palindrome SNPs were removed during the allele harmonization process to eliminate any distortion in chain orientation or allele coding.
Following this, the F-statistic for each SNP was computed to assess its association strength with gut microbiota. The F-statistic formula is F = R2*(n − k − 1)/(k*(1 − R2)), where “R2” signifies explained variance, “n” denotes the exposure sample size, and “k” represents the number of SNPs (33). SNPs with an F-statistic below 10 were considered weak instrumental variables and subsequently excluded.
Afterward, MR-PRESSO tests and MR-Egger regression tests were employed to address horizontal pleiotropy and detect outliers. MR-PRESSO, recognized for its efficacy in detecting horizontal pleiotropy (34), involved calculating the p-value for overall horizontal pleiotropy for each SNP. SNPs were then arranged based on ascending p-values from the MR-PRESSO outlier test, and one SNP at a time was systematically removed. After each removal, the MR-PRESSO global test was conducted on the remaining SNPs, until the p-value of the global test was no longer significant (p > 0.05). Only SNPs that passed through this meticulous filtering process were considered for subsequent MR analysis. All these procedures were performed using the “Two Sample MR” R package (version 0.5.7).
To investigate the correlation between gut microbiota and migraines, we employed five techniques within the “Two Sample MR” toolbox. These included Inverse Variance Weighted (IVW) examination (35), MR-Egger regression (36), Weighted Median Estimate (WM) (37), Simple mode, and Weighted mode. Existing scholarly literature emphasizes the superiority of IVW as the primary analytical instrument (37). IVW produces reliable outcomes under the assumption of no horizontal pleiotropy, a condition duly validated (35). The MR-Egger method identifies potential breaches in the instrumental variable selection assumption (38), considering a MR-Egger intercept p-value > 0.05 as acceptable. SNP heterogeneity was assessed using Cochran’s Q test, with a p-value < 0.05 signifying significant diversity. The “RadialMR” R package (version 1.0) was employed to identify anomalous SNPs and rectify results in cases of significant heterogeneity (39). Horizontal pleiotropy was examined using MR-Egger intercept and MR-PRESSO, with p > 0.05 signifying its nonexistence (34, 40). The leave-one-out method identified individual SNP impact on the IVW total effect and established the directionality of the causal relationship (41).
A p-value < 0.05 was considered statistically significant. To enhance result robustness, Bonferroni correction was implemented at each taxonomic level (phylum, class, order, family, genus). The Bonferroni-corrected significance threshold was computed as 0.05 divided by the effective number of independent bacterial taxa at each corresponding taxonomic level: genera (0.05/131, 3.81 × 10−4), families (0.05/35, 1.4 × 10−3), orders (0.05/20, 2.5 × 10−3), classes (0.05/16, 3.1 × 10−3), and phyla (0.05/9, 5.5 × 10−3) (42). p-values ranging from 0.05 to the corrected threshold denoted nominal causal effects.
In conclusion, a meta-analysis of IVW results for comparable migraine outcomes was executed using the “meta” R package (version 6.5-0). In instances of heterogeneity (p < 0.05), a random-effects model was engaged; otherwise, a fixed-effects model was applied. All statistical analyses were executed in R (version 4.3.1).
Considering the potential impact of host diseases on gut microbiota, we conducted a reverse MR analysis. Initially, each of the five migraine databases served as individual exposures, with the gut microbiota designated as the outcome. The ensuing MR analysis yielded significant findings. Differing from the earlier phase, we adjusted the threshold for tool variable selection to p < 5 × 10−8. All other parameters were maintained in accordance with the prior iteration, and the outcomes underwent both Bonferroni correction and sensitivity analysis for validation.
Following pre-established criteria, the selection of instrumental variables (IVs) resulted in the categorization of SNPs for comprehensive analysis within the Two-Sample Mendelian Randomization (TSMR) framework, shedding light on the intricate relationship between gut microbiota and migraines. Detailed information can be found in Supplementary Tables S1–S5. All SNPs considered in the analysis exhibited F-statistics surpassing 10, confirming the exclusion of weak IVs (Supplementary Tables S1–S5). After harmonization, the count of SNPs associated with each pair of bacterial taxa and migraines exceeded 3.
To unravel the causal dynamics between each bacterial taxon and migraines, we employed five TSMR methodologies (Supplementary Tables S6–S10). Furthermore, we established potential causal relationships between gut microbiota and migraines using three TSMR approaches (Figures 3, 4). Cross-validation unfolded through the Inverse Variance Weighted (IVW) and Weighted Median (WM) techniques, with a preference for the IVW method as the principal tool for result analysis. A comprehensive examination of the outcomes involved Bonferroni correction to enhance dependability.
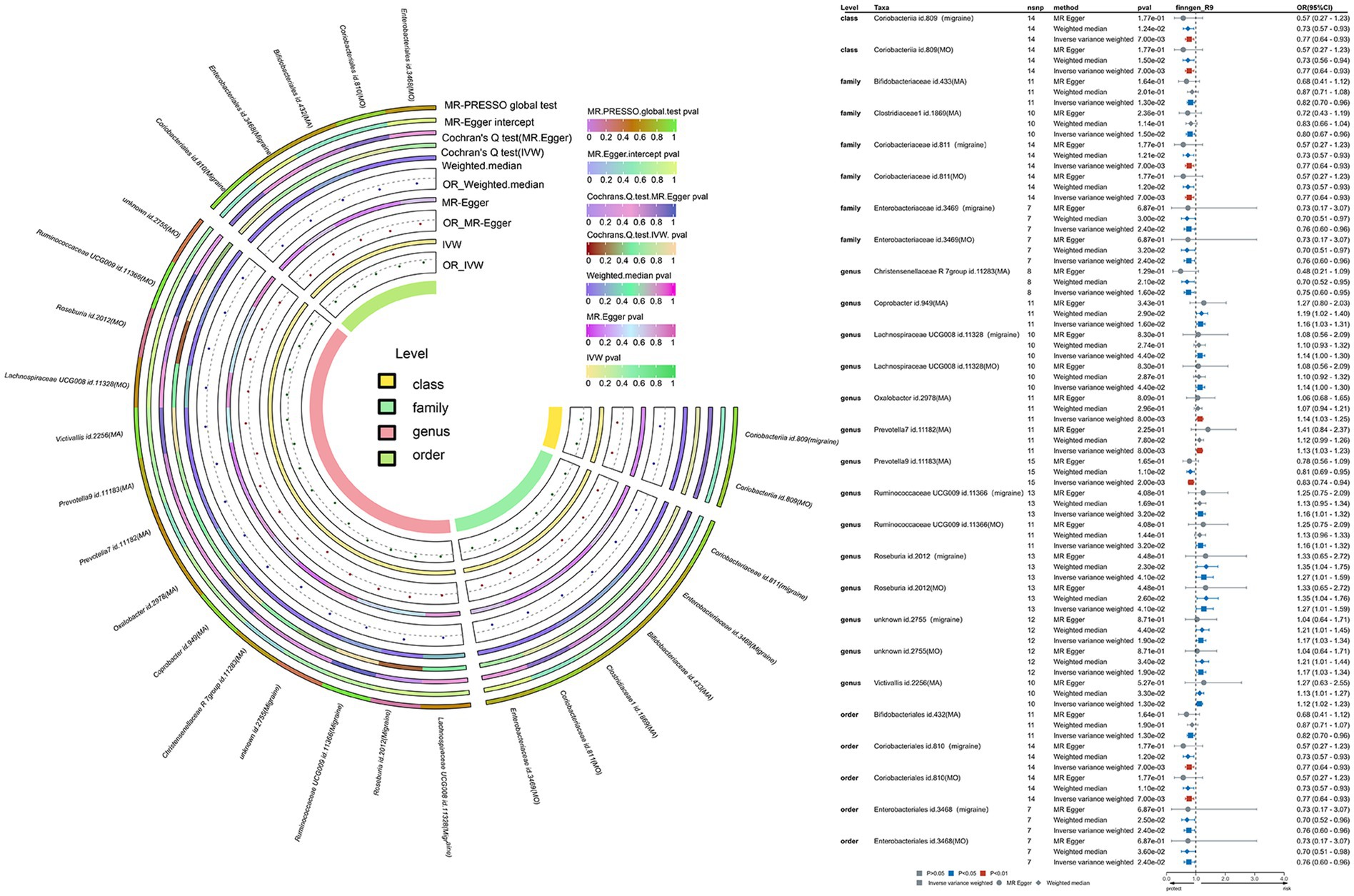
Figure 3. This is a causal analysis of the gut microbiota and migraine disease obtained through a study using the FinnGen study (significant at all loci, p < 1 × 10−5). MR, Mendelian randomization; IVW, inverse-variance-weighted; MR-PRESSO, MR pleiotropy residual sum and outlier.

Figure 4. This involves a causal analysis examining the correlation between gut microbiota and migraine disease, as indicated by investigations by Choquet et al. and Dönertaş et al. (statistically significant across all loci, p < 1 × 10–5). The abbreviations encompass MR for Mendelian randomization, IVW for inverse-variance-weighted, and MR-PRESSO for MR pleiotropy.
Cochran’s Q test was applied to assess heterogeneity among SNPs, focusing on results with a p-value exceeding 0.05. Additionally, the outcomes included were required to meet the criteria of MR-PRESSO global test p > 0.05 and MR Egger regression p > 0.05 to mitigate the risk of horizontal pleiotropy (Supplementary Tables S11, S12). Emphasis was placed on the imperative nature of consistency in the direction of Beta values across all methodologies, as detailed in Supplementary Tables S11, S12. To establish the reliability of our findings, a sensitivity analysis utilizing the leave-one-out approach was executed on identified key bacterial taxa.
The IVW methodology was used to establish causative links between migraines and nine bacterial taxonomic groups. By consolidating these discoveries with cross-validation, the constancy of outcomes for seven bacterial taxonomic groups was confirmed (Supplementary Table S13). Specifically, TSMR analysis revealed a protective impact against migraines for Class Coriobacteriia (OR: 0.77, 95% CI: 0.64–0.93, p = 0.007), Family Coriobacteriaceae (OR: 0.77, 95% CI: 0.64–0.93, p = 0.007), Family Enterobacteriaceae (OR: 0.76, 95% CI: 0.60–0.96, p = 0.024), Order Coriobacteriales (OR: 0.77, 95% CI: 0.64–0.93, p = 0.007), and Order Enterobacteriales (OR: 0.76, 95% CI: 0.60–0.96, p = 0.024). Conversely, Genus Roseburia (OR: 1.27, 95% CI: 1.01–1.59, p = 0.041) and Unknown Genus (OR: 1.17, 95% CI: 1.03–1.34, p = 0.019) were identified as significantly amplifying the risk of migraines. After Bonferroni correction, these revelations maintained their causal effects. In the sensitivity analysis, Cochran’s Q test revealed no indications of heterogeneity for the seven identified bacterial taxa. Both the MR-Egger intercept test and MR-PRESSO global test were oblivious to horizontal pleiotropy for these bacterial taxa (Supplementary Table S13). The robustness of the outcomes was further validated through a sensitivity analysis using the leave-one-out approach (Supplementary Figures S1A–F).
Using the IVW methodology, we identified causal associations between migraine with aura and nine bacterial taxonomic groups. Integrating these results with cross-validation, we confirmed the stability of outcomes for four bacterial taxonomic groups (Supplementary Table S14). Specifically, Genus Christensenellaceae R-7 group (OR: 0.75, 95% CI: 0.60–0.95, p = 0.016) and Genus Prevotella 9 (OR: 0.83, 95% CI: 0.74–0.94, p = 0.002) exhibited a protective effect against migraine with aura, while Genus Coprobacter (OR: 1.16, 95% CI: 1.03–1.31, p = 0.016) and Genus Victivallis (OR: 1.12, 95% CI: 1.02–1.23, p = 0.013) had an adverse impact. The p-values for these findings adhered to Bonferroni’s correction. In the sensitivity analysis, Cochran’s Q test revealed no signs of heterogeneity for the four identified bacterial taxa. Both the MR-Egger intercept test and MR-PRESSO global test were oblivious to horizontal pleiotropy for these bacterial taxa (Supplementary Table S14). The leave-one-out sensitivity analysis plots underscored the robustness of the conclusions (Supplementary Figures S2A–D).
The IVW methodology revealed a causal association between migraine without aura and nine bacterial taxonomic groups. Integrating these findings with cross-validation, we confirmed the stability of outcomes for seven bacterial taxonomic groups (Supplementary Table S15). TSMR analysis revealed a protective impact against migraine without aura for Class Coriobacteriia (OR: 0.77, 95% CI: 0.64–0.93, p = 0.007), Family Coriobacteriaceae (OR: 0.77, 95% CI: 0.64–0.93, p = 0.007), Family Enterobacteriaceae (OR: 0.76, 95% CI: 0.60–0.96, p = 0.024), Order Coriobacteriales (OR: 0.77, 95% CI: 0.64–0.93, p = 0.007), and Order Enterobacteriales (OR: 0.76, 95% CI: 0.60–0.96, p = 0.024). Conversely, Genus Roseburia (OR: 1.27, 95% CI: 1.01–1.59, p = 0.041) and Unknown Genus (OR: 1.17, 95% CI: 1.03–1.34, p = 0.019) emerged as significantly increasing the risk of migraine without aura. These outcomes align with those derived from the initial migraine Finnish database, further supporting the causal linkage of these seven bacterial taxa with migraine. In the sensitivity analysis, no indications of heterogeneity or horizontal pleiotropy for the seven identified bacterial taxa were detected (Supplementary Table S15). The leave-one-out sensitivity analysis underscored the robustness of the conclusions (Supplementary Figures S3A–G).
Applying the IVW methodology, we established causal associations between migraines and six bacterial taxonomic groups. Through the integration of these findings with cross-validation, we confirmed the stability of outcomes for two bacterial taxonomic groups (Supplementary Table S16). Specifically, Family Streptococcaceae (OR: 1.00, 95% CI: 0.99–1.00, p = 0.007) exhibited a neutral causal connection with migraines, while Genus Collinsella (OR: 0.99, 95% CI: 0.99–1.00, p = 0.006) demonstrated a protective impact against migraines.
The p-values for the mentioned bacterial taxa fall within the corrected p-value range. In the sensitivity analysis, Cochran’s Q test showed no indications of heterogeneity for Family Streptococcaceae (p = 0.756) and Genus Collinsella (p = 0.779). The MR-Egger intercept test failed to identify horizontal pleiotropy for Family Streptococcaceae (p = 0.637) and Genus Collinsella (p = 0.159). Similarly, the MR-PRESSO global test did not detect horizontal pleiotropy for Family Streptococcaceae (p = 0.81) and Genus Collinsella (p = 0.91; Supplementary Table S16). The durability of the outcomes was underscored through leave-one-out sensitivity analysis (Supplementary Figures S4A,B).
Utilizing the IVW methodology, we identified causal associations between migraines and four bacterial taxonomic groups. Integrating these findings with cross-validation, we confirmed that only the results for one bacterial taxonomic group exhibited uniformity (Supplementary Table S17). Our TSMR analysis reveals a clear and statistically significant causal association between Genus Parabacteroides (OR: 1.19, 95% CI: 1.02–1.39, p = 0.031) and migraines. Importantly, this conclusion remains robust even after applying Bonferroni correction. In the sensitivity analysis, the absence of indications of heterogeneity for Genus Parabacteroides (p = 0.191) in Cochran’s Q test and the failure of the MR-Egger intercept test to identify horizontal pleiotropy (p = 0.792). Similarly, the MR-PRESSO global test did not reveal horizontal pleiotropy (p = 0.26) for Genus Parabacteroides (Supplementary Table S17). The robustness of the findings is further supported by a leave-one-out sensitivity analysis (Supplementary Figure S5A).
The outcomes of the reverse Mendelian randomization are outlined in Supplementary Tables S18–S31. Upon review of cross-validation findings and sensitivity analysis, it became apparent that the stability of outcomes is retained for a single bacterial taxonomic group (refer to Supplementary Tables S27, S31). Specifics of the leave-one-out sensitivity analysis are illustrated in Supplementary Figure S5B.
Employing the IVW methodology and integrating cross-validation outcomes, while excluding redundant entries from the Finnish database, a total of 14 observations from five datasets were included in our meta-analysis. The assess heterogeneity among the four datasets, we conducted the I2 test, revealing significant heterogeneity (I2 = 84%, p < 0.001). Consequently, a random-effects model was applied for the meta-analysis. Our results illuminated an adverse impact of gut microbiota on the likelihood of gut microbiota occurrence (OR = 0.97, 95% CI = 0.87–1.08, p < 0.001), underscoring the influence of MD on GERD (Figure 5).
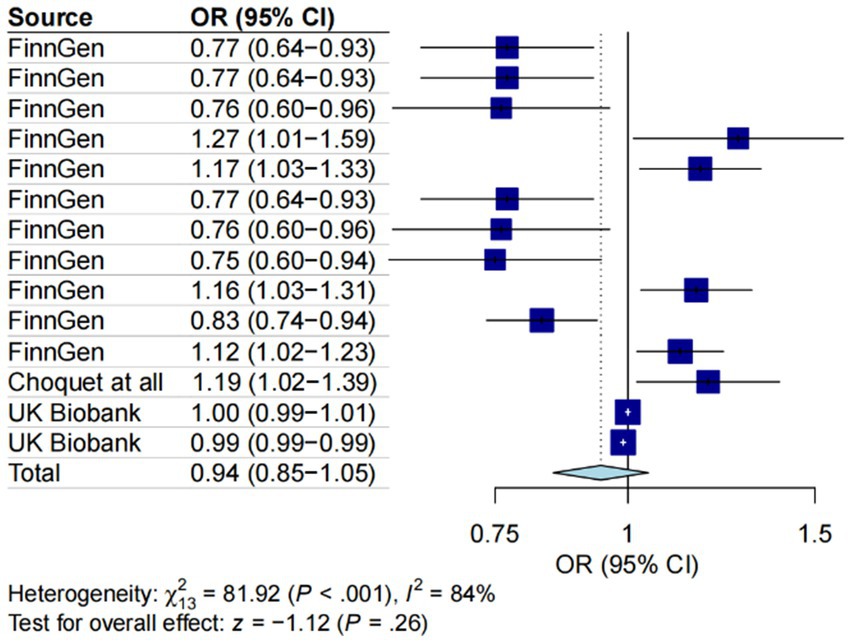
Figure 5. A forest plot of the meta-analysis including five different migraine datasets. The plot illustrates the average genetic predictive effect of Gut microbiota on migraine. The presented odds ratio (OR) and confidence interval (CI) correspond to the average impact of Gut microbiota on migraine. The I2 statistic and chi-square-based Q test were used to assess heterogeneity among studies.
During TSMR analysis, 14 bacterial features associated with migraines were identified by leveraging extensive aggregated GWAS data (Figure 6).
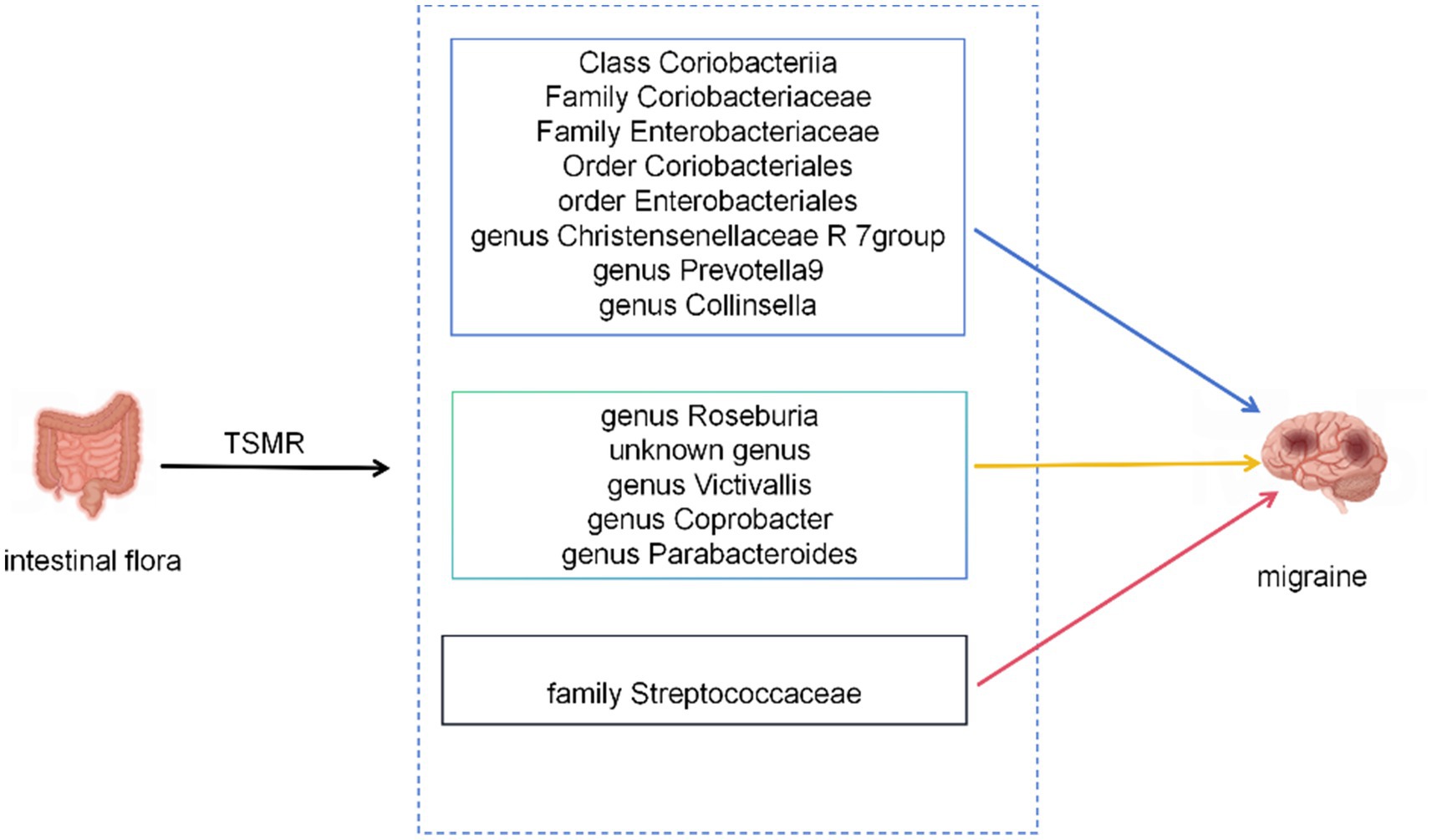
Figure 6. The current MR analysis identified gut microbiota associated with migraines. Blue arrows represent bacterial taxa that are protective factors for the outcome, yellow arrows represent bacterial taxa that are risk factors for the outcome. Red arrows indicate taxa that might be both protective and risk factors.
The current understanding of the mechanisms underlying the interaction between the gastrointestinal tract and the brain in individuals experiencing migraines is currently insufficient. Multiple investigations suggest that this association is influenced by a variety of factors, including inflammatory mediators, gut microbiota, neuropeptides, and other components (43). Of particular significance is the function fulfilled by gut microbiota, impacting the system through the generation of neurotransmitters, inflammatory molecules, and hormones. Moreover, it can directly interact with the nerve endings of the vagus nerve (43). Similarly, the central nervous system can regulate gut microbiota by releasing neuroendocrine peptides, signifying a bidirectional mechanism (44). Substances such as calcitonin gene-related peptide (CGRP), substance P (SP), vasoactive intestinal peptide (VIP), and other compounds are believed to contribute to the bidirectional association between the gastrointestinal tract and the brain. These substances are theorized to exhibit antimicrobial effects on diverse bacterial strains within the gastrointestinal tract, including Escherichia coli, Enterococcus faecalis, and Lactobacillus acidophilus (44). CGRP, playing a pivotal role in the pathophysiology of migraines, is abundantly present in neurons of the trigeminal ganglion, released from peripheral and central nerve endings, and secreted within the trigeminal ganglion. Treatments targeting the functional aspects of CGRP in the peripheral trigeminal system have demonstrated efficacy in managing migraines (45, 46). Consequently, it can be inferred that gut microbiota may confer a significant effect against migraines, aligning with the findings of our investigation.
Numerous studies indicate an elevation in pro-inflammatory cytokines, including IL-1β, IL-6, IL-8, and TN-αduring the interictal phase of migraines. Consequently, the relationship between pro-inflammatory factors and migraines is currently under scrutiny (47, 48). Hirayama et al. discovered that the Collinsella genus can stimulate the production of ursodeoxycholic acid and other secondary bile acids. Ursodeoxycholic acid, in turn, can inhibit pro-inflammatory cytokines such as TNF-α, IL-1β, IL-2, IL-4, and IL-6, at both mRNA and protein levels (49). Moreover, ursodeoxycholic acid exhibits antioxidant and anti-apoptotic properties (49). The order Coriobacteriales, encompassing the genus Collinsella, plays a crucial role in this context (50). From these findings, we deduce that both the genus Collinsella and the order Coriobacteriales may exert a protective effect against migraines. This inference aligns seamlessly with the results of our investigation. Liu et al.’s study, which revealed a comparatively diminished abundance of the Collinsella genus in the gut of migraine patients, provides additional support for this conclusion (51).
At present, there is no direct and well-established evidence connecting the class Coriobacteriia with migraines. Nonetheless, the hierarchical relationship among Class Coriobacteriia, Order Coriobacteriales, and Family Coriobacteriaceae is well recognized. The classification begins broadly with Class Coriobacteriia and becomes more specific through Order Coriobacteriales to Family Coriobacteriaceae, each being nested within the previous, revealing a tight genetic and functional connection (52). Therefore, we propose that Class Coriobacteriia and Family Coriobacteriaceae could have a relationship with migraines. Previous studies suggest that Coriobacteriaceae contributes to host bile acid and lipid metabolism (53), further reinforcing our conclusions.
Recent studies have revealed a significant connection between migraines and irritable bowel syndrome (IBS) (54). Among individuals with IBS, approximately 25%–50% report encountering migraines or headaches, a notable contrast to the 4%–19% prevalence in the control group (55). The odds ratio for the simultaneous occurrence of IBS and migraines or headaches among IBS individuals stands at 2.66 (10), indicating an elevated likelihood of migraines in this subgroup. Several investigations have highlighted that, within the healthy control cohort, the proportional prevalence of Christensenellaceae surpasses that observed in individuals with IBS. This distinction may be linked to inflammatory processes and an expedited transit time in IBS patients (56). Additionally, a heightened proportional prevalence of Christensenellaceae has been documented in patients diagnosed with Parkinson’s disease and multiple sclerosis relative to their healthy counterparts (57–60). This nuanced observation supports the proposition that Christensenellaceae, by shaping the gut milieu in individuals with IBS, potentially contributes to the manifestation of migraines.
In alignment with the aforementioned investigations, the research of Ornello et al. (61) implies an increased susceptibility to migraines and chronic migraines in individuals grappling with obesity. Goodrich et al. (62) observed a significant enrichment of Christensenellaceae in individuals maintaining a normal BMI (18.5–24.9) compared to those classified as obese (BMI ≥ 30). Furthermore, the prevalence of Christensenellaceae exhibited a surge following weight loss induced by dietary modifications (63). This suggests a plausible association between Christensenellaceae and migraines, presenting a novel perspective on migraine management for individuals contending with obesity. Our study similarly posits a causal connection between Christensenellaceae and migraines, underscoring the imperative for supplementary experimental affirmation.
Research into obesity-related conditions indicates an increase in the abundance of Enterobacteriaceae within the gut microbiota of obese subjects. When weight loss occurs, the Enterobacteriaceae population decreases. In contrast, transferring Enterobacteriaceae from obese humans into mice led to increased fat formation in the animals (64). This finding implies a link between Enterobacteriaceae and obesity. Given the earlier discussion on the correlation between obesity and migraines, it can be hypothesized that Enterobacteriaceae could also be associated with migraines.
He et al.’s investigation uncovers a causal link between the genera Prevotella9 (OR = 0.82, 95%CI = 0.72–0.94, p = 0.004), Roseburia (OR = 1.15, 95%CI = 1.02–1.30, p = 0.0234), and Coprobacter (OR = 1.09, 95%CI = 1.01–1.17, p = 0.0347) and migraines. Prevotella9 is suggested to confer a protective effect against migraines, while Roseburia and Coprobacter are implicated in an increased risk of migraines (21). These outcomes resonate with our study findings. Nonetheless, the precise mechanisms remain elusive, necessitating additional investigation for validation.
Parabacteroides, a collection of anaerobic Gram-negative bacteria commonly found in the human gastrointestinal tract, possess the ability to induce pro-inflammatory responses through lipopolysaccharide (LPS) and the metabolic byproduct succinic acid (65). Research has illuminated an increased prevalence of Parabacteroides in individuals experiencing migraines in conjunction with irritable bowel syndrome (IBS) (51). Therefore, we propose that Parabacteroides may exert an unfavorable influence on migraines, aligning with our investigation. While current research has not specifically examined the associations between the family Streptococcaceae, unidentified genera, and the genus Victivallis with migraines, this gap presents a valuable opportunity for future studies.
Our experiments have yielded several notable advantages. Firstly, this investigation reaffirms the causal link between gut microbiota and migraines through bidirectional TSMR analysis. This analysis remains robust against confounding factors or reverse causation. Secondly, we set stringent criteria for selecting instrumental variables, differentiating our approach from similar studies. We only consider causation plausible when two or more TSMR methods support the relationship. Thirdly, we utilized data from five GWAS datasets, distinct from those databases previously used in studies on gut microbiota and migraines. This not only strengthens the credibility and stability of our MR research but also marks a significant enhancement in evaluating the causal connection between gut microbiota and migraines. Moreover, we conducted the first-ever meta-analysis of results for this type of study, further reinforcing the robustness of our research outcomes. Fourthly, this work establishes the foundation for scrutinizing the gut-brain axis from a genetic standpoint. Through TSMR analysis, we identified 14 bacterial taxa linked to migraines. These crucial bacterial taxa could pave the way for future research on migraine treatment by promoting beneficial bacteria and inhibiting the growth of harmful bacteria to prevent and treat migraines.
This study has specific limitations. Firstly, the gut microbiota GWAS statistics include a limited number of instrumental variables, and there is an insufficient count of associated loci for bacterial taxa. To address this, we selected a more lenient association threshold to ensure an adequate number of SNPs. Secondly, confirming overlapping participants in the GWAS data related to exposures and outcomes central to this study is impossible. Thirdly, the original studies mainly focused on European blood populations and lacked demographic data, hindering subgroup analyses based on factors such as age. Moreover, our findings require additional validation through clinical and foundational research. In future studies, we aim to increase the sample size, conduct collaborative experiments, and more precisely explore the relationship between gut microbiota and migraines at the species level.
Recent studies involving European populations have shown that modifications to dietary habits can influence the gut microbiome, impacting the production of short-chain fatty acids (SCFAs) (66). The research indicates that, compared to the Canadian Diet (CanDiet), the Mediterranean Diet (MedDiet) might produce higher levels of SCFAs, which are known for their neuroprotective effects (43). This leads to the hypothesis that dietary changes could help reduce migraine episodes. Additional research points to the potential benefits of a high omega-3/low omega-6 diet, a low glycemic index diet, and supplementation of probiotics and vitamin D, as these interventions may improve migraines by promoting SCFAs production and managing inflammation (43).
In summary, our use of MR analysis provides new evidence supporting the causal link between gut microbiota and migraines. Our study suggests that specific taxonomic groups, such as Class Coriobacteriia, Family Coriobacteriaceae, Family Enterobacteriaceae, Order Coriobacteriales, and others, might exert a notable influence on migraines. The implications of our findings extend to the potential development of innovative interventions and treatments for migraines. Nevertheless, additional validation is required through clinical trials or foundational experiments.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
XZ: Writing – review & editing, Writing – original draft, Visualization, Formal analysis, Data curation. YD: Writing – review & editing, Data curation. MJ: Writing – review & editing, Data curation. SZ: Writing – review & editing, Data curation. LW: Writing – review & editing, Formal analysis, Data curation. XH: Writing – review & editing, Visualization, Formal analysis, Data curation.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Project 20240402020GH, Jilin Provincial Department of Science and Technology International Cooperation Project and Project 2023SCZ13, Jilin Provincial Health Research Talent Special.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2024.1356974/full#supplementary-material
1. Steiner, TJ, Stovner, LJ, and Vos, T. GBD 2015: migraine is the third cause of disability in under 50s. J Headache Pain. (2016) 17:104. doi: 10.1186/s10194-016-0699-5
2. Steiner, TJ, Stovner, LJ, Vos, T, Jensen, R, and Katsarava, Z. Migraine is first cause of disability in under 50s: will health politicians now take notice? J Headache Pain. (2018) 19:17. doi: 10.1186/s10194-018-0846-2
3. Headache Classification Committee of the International Headache Society (IHS). The international classification of headache disorders, 3rd edition (beta version). Cephalalgia. (2013) 33:629–808. doi: 10.1177/0333102413485658
4. Ashina, M, Hansen, JM, Do, TP, Melo-Carrillo, A, Burstein, R, and Moskowitz, MA. Migraine and the trigeminovascular system-40 years and counting. Lancet Neurol. (2019) 18:795–804. doi: 10.1016/S1474-4422(19)30185-1
5. Cernuda-Morollón, E, Larrosa, D, Ramón, C, Vega, J, Martínez-Camblor, P, and Pascual, J. Interictal increase of CGRP levels in peripheral blood as a biomarker for chronic migraine. Neurology. (2013) 81:1191–6. doi: 10.1212/WNL.0b013e3182a6cb72
6. Moskowitz, MA, Reinhard, JF, Romero, J, Melamed, E, and Pettibone, DJ. Neurotransmitters and the fifth cranial nerve: is there a relation to the headache phase of migraine? Lancet. (1979) 2:883–5.
7. Sarchielli, P, Alberti, A, Codini, M, Floridi, A, and Gallai, V. Nitric oxide metabolites, prostaglandins and trigeminal vasoactive peptides in internal jugular vein blood during spontaneous migraine attacks. Cephalalgia. (2000) 20:907–18. doi: 10.1046/j.1468-2982.2000.00146.x
8. Aamodt, AH, Stovner, LJ, Hagen, K, and Zwart, JA. Comorbidity of headache and gastrointestinal complaints. The head-HUNT study. Cephalalgia. (2008) 28:144–51. doi: 10.1111/j.1468-2982.2007.01486.x
9. Cámara-Lemarroy, CR, Rodriguez-Gutierrez, R, Monreal-Robles, R, and Marfil-Rivera, A. Gastrointestinal disorders associated with migraine: a comprehensive review. World J Gastroenterol. (2016) 22:8149–60. doi: 10.3748/wjg.v22.i36.8149
10. Chang, FY, and Lu, CL. Irritable bowel syndrome and migraine: bystanders or partners? J Neurogastroenterol Motil. (2013) 19:301–11. doi: 10.5056/jnm.2013.19.3.301
11. Moschen, AR, Wieser, V, and Tilg, H. Dietary factors: major regulators of the Gut's microbiota. Gut Liver. (2012) 6:411–6. doi: 10.5009/gnl.2012.6.4.411
12. Peatfield, RC, Glover, V, Littlewood, JT, Sandler, M, and Clifford Rose, F. The prevalence of diet-induced migraine. Cephalalgia. (1984) 4:179–83. doi: 10.1046/j.1468-2982.1984.0403179.x
13. Keshavarzian, A, Engen, P, Bonvegna, S, and Cilia, R. The gut microbiome in Parkinson's disease: a culprit or a bystander? Prog Brain Res. (2020) 252:357–450. doi: 10.1016/bs.pbr.2020.01.004
14. Zhuang, ZQ, Shen, LL, Li, WW, Fu, X, Zeng, F, Gui, L, et al. Gut microbiota is altered in patients with Alzheimer's disease. J Alzheimers Dis. (2018) 63:1337–46. doi: 10.3233/JAD-180176
15. Rutsch, A, Kantsjö, JB, and Ronchi, F. The gut-brain Axis: how microbiota and host Inflammasome influence brain physiology and pathology. Front Immunol. (2020) 11:604179. doi: 10.3389/fimmu.2020.604179
16. Russo, R, Cristiano, C, Avagliano, C, De Caro, C, La Rana, G, Raso, GM, et al. Gut-brain Axis: role of lipids in the regulation of inflammation, pain and CNS diseases. Curr Med Chem. (2018) 25:3930–52. doi: 10.2174/0929867324666170216113756
17. Guo, R, Chen, LH, Xing, C, and Liu, T. Pain regulation by gut microbiota: molecular mechanisms and therapeutic potential. Br J Anaesth. (2019) 123:637–54. doi: 10.1016/j.bja.2019.07.026
18. Lima-Ojeda, JM, Rupprecht, R, and Baghai, TC. "I am I and my bacterial circumstances": linking gut microbiome, neurodevelopment, and depression. Front Psych. (2017) 8:153. doi: 10.3389/fpsyt.2017.00153
19. Lin, B, Wang, Y, Zhang, P, Yuan, Y, Zhang, Y, and Chen, G. Gut microbiota regulates neuropathic pain: potential mechanisms and therapeutic strategy. J Headache Pain. (2020) 21:103. doi: 10.1186/s10194-020-01170-x
20. Ustianowska, K, Ustianowski, Ł, Machaj, F, Gorący, A, Rosik, J, Szostak, B, et al. The role of the human microbiome in the pathogenesis of pain. Int J Mol Sci. (2022) 23:13267. doi: 10.3390/ijms232113267
21. He, Q, Wang, W, Xiong, Y, Tao, C, Ma, L, Ma, J, et al. International headache genetics consortium. A causal effects of gut microbiota in the development of migraine. J Headache Pain. (2023) 24:90. doi: 10.1186/s10194-023-01609-x
22. Greenland, S. An introduction to instrumental variables for epidemiologists. Int J Epidemiol. (2000) 29:722–9. doi: 10.1093/ije/29.4.722
23. Lawlor, DA, Harbord, RM, Sterne, JA, Timpson, N, and Davey Smith, G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. (2008) 27:1133–63. doi: 10.1002/sim.3034
24. Larsson, SC, and Burgess, S. Appraising the causal role of smoking in multiple diseases: a systematic review and meta-analysis of Mendelian randomization studies. EBioMedicine. (2022) 82:104154. doi: 10.1016/j.ebiom.2022.104154
25. Skrivankova, VW, Richmond, RC, Woolf, BAR, Yarmolinsky, J, Davies, NM, Swanson, SA, et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the STROBE-MR statement. JAMA. (2021) 326:1614–21. doi: 10.1001/jama.2021.18236
26. Kurilshikov, A, Medina-Gomez, C, Bacigalupe, R, Radjabzadeh, D, Wang, J, Demirkan, A, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. (2021) 53:156–65. doi: 10.1038/s41588-020-00763-1
27. Choquet, H, Yin, J, Jacobson, AS, Horton, BH, Hoffmann, TJ, Jorgenson, E, et al. New and sex-specific migraine susceptibility loci identified from a multiethnic genome-wide meta-analysis. Commun Biol. (2021) 4:864. doi: 10.1038/s42003-021-02356-y
28. Niu, PP, Wang, X, and Xu, YM. Higher circulating vitamin D levels are associated with decreased migraine risk: a Mendelian randomization study. Front Nutr. (2022) 9:907789. doi: 10.3389/fnut.2022.907789
29. Dönertaş, HM, Fabian, DK, Valenzuela, MF, Partridge, L, and Thornton, JM. Common genetic associations between age-related diseases. Nat Aging. (2021) 1:400–12. doi: 10.1038/s43587-021-00051-5
30. Davey Smith, G, and Hemani, G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. (2014) 23:R89–98. doi: 10.1093/hmg/ddu328
31. Brown, K, Godovannyi, A, Ma, C, Zhang, Y, Ahmadi-Vand, Z, Dai, C, et al. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. ISME J. (2016) 10:321–32. doi: 10.1038/ismej.2015.114
32. Sanna, S, van Zuydam, NR, Mahajan, A, Kurilshikov, A, Vich Vila, A, Võsa, U, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. (2019) 51:600–5. doi: 10.1038/s41588-019-0350-x
33. Burgess, S, and Thompson, SG. Bias in causal estimates from Mendelian randomization studies with weak instruments. Stat Med. (2011) 30:1312–23. doi: 10.1002/sim.4197
34. Verbanck, M, Chen, CY, Neale, B, and Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. (2018) 50:693–8. doi: 10.1038/s41588-018-0099-7
35. Burgess, S, Butterworth, A, and Thompson, SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. (2013) 37:658–65. doi: 10.1002/gepi.21758
36. Bowden, J, Davey Smith, G, and Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int J Epidemiol. (2015) 44:512–25. doi: 10.1093/ije/dyv080
37. Bowden, J, Davey Smith, G, Haycock, PC, and Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. (2016) 40:304–14. doi: 10.1002/gepi.21965
38. Xue, F, He, Z, Zhuang, DZ, and Lin, F. The influence of gut microbiota on circulating inflammatory cytokines and host: a Mendelian randomization study with meta-analysis. Life Sci. (2023) 332:122105. doi: 10.1016/j.lfs.2023.122105
39. Bowden, J, Spiller, W, Del Greco, MF, Sheehan, N, Thompson, J, Minelli, C, et al. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the radial plot and radial regression. Int J Epidemiol. (2018) 47:1264–78. doi: 10.1093/ije/dyy101
40. Burgess, S, and Thompson, SG. Interpreting findings from Mendelian randomization using the MR-egger method. Eur J Epidemiol. (2017) 32:377–89. doi: 10.1007/s10654-017-0255-x
41. Hemani, G, Tilling, K, and Davey Smith, G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. (2017) 13:e1007081. doi: 10.1371/journal.pgen.1007081
42. Li, N, Wang, Y, Wei, P, Min, Y, Yu, M, Zhou, G, et al. Causal effects of specific gut microbiota on chronic kidney diseases and renal function-a two-sample Mendelian randomization study. Nutrients. (2023) 15:360. doi: 10.3390/nu15020360
43. Arzani, M, Jahromi, SR, Ghorbani, Z, Vahabizad, F, Martelletti, P, Ghaemi, A, et al. School of Advanced Studies of the European headache federation (EHF-SAS). Gut-brain Axis and migraine headache: a comprehensive review. J Headache Pain. (2020) 21:15. doi: 10.1186/s10194-020-1078-9
44. Holzer, P, and Farzi, A. Neuropeptides and the microbiota-gut-brain axis. Adv Exp Med Biol. (2014) 817:195–219. doi: 10.1007/978-1-4939-0897-4_9
45. Iyengar, S, Johnson, KW, Ossipov, MH, and Aurora, SK. CGRP and the trigeminal system in migraine. Headache. (2019) 59:659–81. doi: 10.1111/head.13529
46. Russo, AF, and Hay, DL. CGRP physiology, pharmacology, and therapeutic targets: migraine and beyond. Physiol Rev. (2023) 103:1565–644. doi: 10.1152/physrev.00059.2021
47. Ramachandran, R. Neurogenic inflammation and its role in migraine. Semin Immunopathol. (2018) 40:301–14. doi: 10.1007/s00281-018-0676-y
48. Theoharides, TC, Donelan, J, Kandere-Grzybowska, K, and Konstantinidou, A. The role of mast cells in migraine pathophysiology. Brain Res Brain Res Rev. (2005) 49:65–76. doi: 10.1016/j.brainresrev.2004.11.006
49. Hirayama, M, Nishiwaki, H, Hamaguchi, T, Ito, M, Ueyama, J, Maeda, T, et al. Intestinal Collinsella may mitigate infection and exacerbation of COVID-19 by producing ursodeoxycholate. PLoS One. (2021) 16:e0260451. doi: 10.1371/journal.pone.0260451
50. Han, KI, Kim, JS, Eom, MK, Lee, KC, Suh, MK, Kim, HS, et al. Collinsella acetigenes sp. nov., an anaerobic Actinobacterium isolated from human feces, and emended description of the genus Collinsella and Collinsella aerofaciens. Curr Microbiol. (2021) 78:3667–73. doi: 10.1007/s00284-021-02625-3
51. Liu, J, Tang, W, Hou, L, Wang, J, Wang, R, Zhang, Y, et al. Alteration of gut microbiota in migraine patients with irritable bowel syndrome in a Chinese Han population. Front Neurol. (2022) 13:899056. doi: 10.3389/fneur.2022.899056
52. Parte, A, Sardà Carbasse, J, Meier-Kolthoff, JP, Reimer, LC, and Göker, M. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. (2020) 70:5607–12. doi: 10.1099/ijsem.0.004332
53. Liu, H, Zhang, H, Wang, X, Yu, X, Hu, C, and Zhang, X. The family Coriobacteriaceae is a potential contributor to the beneficial effects of roux-en-Y gastric bypass on type 2 diabetes. Surg Obes Relat Dis. (2018) 14:584–93. doi: 10.1016/j.soard.2018.01.012
54. Chen, J, Chen, X, Xie, Y, Sun, Y, Wang, X, and Hesketh, T. Irritable bowel syndrome and migraine: evidence from Mendelian randomization analysis in the UK biobank. Expert Rev Gastroenterol Hepatol. (2021) 15:1233–9. doi: 10.1080/17474124.2021.1949290
55. Cole, JA, Rothman, KJ, Cabral, HJ, Zhang, Y, and Farraye, FA. Migraine, fibromyalgia, and depression among people with IBS: a prevalence study. BMC Gastroenterol. (2006) 6:26. doi: 10.1186/1471-230X-6-26
56. Waters, JL, and Ley, RE. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. (2019) 17:83. doi: 10.1186/s12915-019-0699-4
57. Barichella, M, Severgnini, M, Cilia, R, Cassani, E, Bolliri, C, Caronni, S, et al. Unraveling gut microbiota in Parkinson's disease and atypical parkinsonism. Mov Disord. (2019) 34:396–405. doi: 10.1002/mds.27581
58. Hill-Burns, EM, Debelius, JW, Morton, JT, Wissemann, WT, Lewis, MR, Wallen, ZD, et al. Parkinson's disease and Parkinson's disease medications have distinct signatures of the gut microbiome. Mov Disord. (2017) 32:739–49. doi: 10.1002/mds.26942
59. Petrov, VA, Saltykova, IV, Zhukova, IA, Alifirova, VM, Zhukova, NG, Dorofeeva, YB, et al. Analysis of gut microbiota in patients with Parkinson's disease. Bull Exp Biol Med. (2017) 162:734–7. doi: 10.1007/s10517-017-3700-7
60. Tremlett, H, Fadrosh, DW, Faruqi, AA, Zhu, F, Hart, J, Roalstad, S, et al. Gut microbiota in early pediatric multiple sclerosis: a case-control study. Eur J Neurol. (2016) 23:1308–21. doi: 10.1111/ene.13026
61. Ornello, R, Ripa, P, Pistoia, F, Degan, D, Tiseo, C, Carolei, A, et al. Migraine and body mass index categories: a systematic review and meta-analysis of observational studies. J Headache Pain. (2015) 16:27. doi: 10.1186/s10194-015-0510-z
62. Goodrich, JK, Waters, JL, Poole, AC, Sutter, JL, Koren, O, Blekhman, R, et al. Human genetics shape the gut microbiome. Cell. (2014) 159:789–99. doi: 10.1016/j.cell.2014.09.053
63. Alemán, JO, Bokulich, NA, Swann, JR, Walker, JM, De Rosa, JC, Battaglia, T, et al. Fecal microbiota and bile acid interactions with systemic and adipose tissue metabolism in diet-induced weight loss of obese postmenopausal women. J Transl Med. (2018) 16:244. doi: 10.1186/s12967-018-1619-z
64. Moreira de Gouveia, MI, Bernalier-Donadille, A, and Jubelin, G. Enterobacteriaceae in the human gut: dynamics and ecological roles in health and disease. Biology. (2024) 13:142. doi: 10.3390/biology13030142
65. Ezeji, JC, Sarikonda, DK, Hopperton, A, Erkkila, HL, Cohen, DE, Martinez, SP, et al. Parabacteroides distasonis: intriguing aerotolerant gut anaerobe with emerging antimicrobial resistance and pathogenic and probiotic roles in human health. Gut Microbes. (2021) 13:1922241. doi: 10.1080/19490976.2021.1922241
Keywords: Mendelian randomization, meta-analysis, migraine, gut microbiome, migraine with aura, migraine without aura
Citation: Zang X, Du Y, Jiang M, Zhou S, Wang L and Han X (2024) A thorough investigation into the correlation between migraines and the gut microbiome: an in-depth analysis using Mendelian randomization studies. Front. Neurol. 15:1356974. doi: 10.3389/fneur.2024.1356974
Received: 12 March 2024; Accepted: 29 May 2024;
Published: 02 July 2024.
Edited by:
Sabina Cevoli, IRCCS Institute of Neurological Sciences of Bologna (ISNB), ItalyReviewed by:
Claudia Altamura, Fondazione Policlinico Campus Bio-Medico, ItalyCopyright © 2024 Zang, Du, Jiang, Zhou, Wang and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Libo Wang, d2FuZ19saWJvQGpsdS5lZHUuY24=; Xuemei Han, aHhtQGpsdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.