- 1German Center for Neurodegenerative Diseases (DZNE), Rostock, Germany
- 2Department of Psychosomatic Medicine, Rostock University Medical Center, Rostock, Germany
Introduction: Neurodegenerative diseases such as Alzheimer's disease (AD) or frontotemporal lobar degeneration (FTLD) involve specific loss of brain volume, detectable in vivo using T1-weighted MRI scans. Supervised machine learning approaches classifying neurodegenerative diseases require diagnostic-labels for each sample. However, it can be difficult to obtain expert labels for a large amount of data. Self-supervised learning (SSL) offers an alternative for training machine learning models without data-labels.
Methods: We investigated if the SSL models can be applied to distinguish between different neurodegenerative disorders in an interpretable manner. Our method comprises a feature extractor and a downstream classification head. A deep convolutional neural network, trained with a contrastive loss, serves as the feature extractor that learns latent representations. The classification head is a single-layer perceptron that is trained to perform diagnostic group separation. We used N = 2,694 T1-weighted MRI scans from four data cohorts: two ADNI datasets, AIBL and FTLDNI, including cognitively normal controls (CN), cases with prodromal and clinical AD, as well as FTLD cases differentiated into its phenotypes.
Results: Our results showed that the feature extractor trained in a self-supervised way provides generalizable and robust representations for the downstream classification. For AD vs. CN, our model achieves 82% balanced accuracy on the test subset and 80% on an independent holdout dataset. Similarly, the Behavioral variant of frontotemporal dementia (BV) vs. CN model attains an 88% balanced accuracy on the test subset. The average feature attribution heatmaps obtained by the Integrated Gradient method highlighted hallmark regions, i.e., temporal gray matter atrophy for AD, and insular atrophy for BV.
Conclusion: Our models perform comparably to state-of-the-art supervised deep learning approaches. This suggests that the SSL methodology can successfully make use of unannotated neuroimaging datasets as training data while remaining robust and interpretable.
1 Introduction
Neurodegenerative diseases such as Alzheimer's disease (AD) and frontotemporal dementia (FTD) are characterized by specific brain volume loss, which can be assessed in-vivo using structural magnetic resonance imaging (MRI). The usual radiological evaluation of MRI scans is performed mainly by visual examination, which is often time-consuming. Assistance systems for the automated detection of disease-specific patterns could be useful for better clinical diagnosis, as they can significantly decrease the evaluation time for radiologists and neurologists, and help them focus on relevant brain regions. Convolutional neural networks (CNNs) models can automatically identify neurodegenerative diseases from MRI scans and achieve state-of-the-art results in medical imaging tasks. Recent developments in the CNN architectures have in turn shaped the neuroimaging community, which is interested in automatic discovery of image features pertinent to neurological illnesses. Various tasks, such as disease diagnosis, pathology localization, anatomical region segmentation, etc., now rely on the use of CNNs (Dyrba et al., 2021; Qiu et al., 2020; Eitel et al., 2021; Wen et al., 2020; Han et al., 2022). CNN models are primarily trained in a supervised manner by using an external ground-truth label. Generating such labels for data samples is often burdensome and costly. Furthermore, CNN models require a large amount of training data to achieve competitive results. Such large datasets are not easily available within the medical domain due to the high cost of data collection and the rarity of experts for annotations.
These constraints led us to reconsider the training of CNN models in a supervised manner, and to explore self-supervised learning (SSL) approaches. The SSL methods learn without any sample labels by utilizing the internal structure of the data to generate representative features. Architectures trained in a self-supervised manner are biologically plausible, provide extensive feature space, and can compete with supervised approaches (Orhan et al., 2020).
Moreover, post hoc explanation methods have been developed within the field of eXplainable Artificial Intelligence (XAI) to interpret how deep neural networks make decisions. The XAI methods for explaining CNN models rely on local feature attribution methods, which assign a relevance score to input regions for a given input, model, and resulting output. However, only a handful of studies have explored attribution-based XAI methods within the field of self-supervised learning (SSL) applications, e.g., in the medical imaging domain (Chen et al., 2023).
The main goal of our study was to explore, in a proof-of-concept study, SSL method's ability to learn generalizable features for dementia stage and type detection from structural MRI data. We hypothesized that SSL methods could learn meaningful structural representations, and resulting models could have comparable performances to supervised models. In this paper, we trained a CNN model with the SSL setup and then evaluated it on downstream classification tasks, binary and multi-class. We also explored a saliency mapping technique for highlighting relevant input regions. The main research questions were defined as: How does the contrastive SSL paradigm compare to the supervised learning paradigm in terms of predictive power? Are the models trained in contrastive self-supervised way on neuroimaging data interpretable?
2 Background
2.1 Self-supervised learning
Self-supervised learning (SSL) methods learn generalizable features without any data labels or ground truth information by solving an initial auxiliary task. The pretrained SSL models are then used for specific downstream tasks, e.g., identification of neurodegenerative disorders. Models trained under the SSL approach have found application in different domains, that is, image processing (Jing and Tian, 2020), video processing (Schiappa et al., 2023), and audio processing (Liu et al., 2022a). Within the imaging domain, multiple auxiliary or so-called “pretext” tasks have been suggested previously: identifying data augmentations (Reed et al., 2021; Chen et al., 2020), rotation prediction (Chen et al., 2019), patch position prediction (Doersch et al., 2015; Noroozi and Favaro, 2016; Wei et al., 2019), image colorization (Larsson et al., 2017, 2016), and contrastive learning (Jaiswal et al., 2020).
SSL methods could be thought of as an alternative to pre-training or automated feature learning step and are related to the way how young children learn (Orhan et al., 2020). Particularly, contrastive SSL methods try to learn the general structure present within the data, by using supervisory signals extracted from the data itself independently of the ground truth for any specific use-case. In our study, we used contrastive learning due to its widespread application as a pretext task (Shurrab and Duwairi, 2022; VanBerlo et al., 2024).
2.1.1 Formal definition of contrastive SSL
Contrastive learning tasks have received considerable attention within the SSL methods. Contrastive learning tasks aim to learn a latent space in which embeddings of similar data samples are pulled together, and embeddings of dissimilar data samples are pushed apart (Gutmann and Hyvärinen, 2010; Weng, 2021; Chopra et al., 2005). Various loss functions have been suggested to increase the quality of learned embeddings, and expedite the training. These include contrastive loss (Gutmann and Hyvärinen, 2010), triplet loss (Chechik et al., 2010; Schroff et al., 2015), N-pair loss (Sohn, 2016), InfoNCE loss (Oord et al., 2019), and Neighborhood-based loss (Sabokrou et al., 2019) etc. Contrastive learning is based on the use of positive and negative data pairs (Grill et al., 2020; Chen et al., 2020), where a positive pair (i, j) consists of two similar data instances or views. In many studies, a data sample is paired with its own augmented variations to create such positive pairs. A negative pair generally contains two different data samples. The contrastive loss ℓ for a positive pair is formally defined as follows.
Where τ is a scaling factor called temperature, 𝟙 is an indicator function with output values being 0 or 1, N is the number of training samples, exp(·) is the exponential function, and cos(·) is the cosine similarity function, over different z latent representation of the input.
The Nearest-Neighbor Contrastive Learning (NNCLR) method (Dwibedi et al., 2021) extends the common contrastive loss by keeping a record of recent embeddings of augmented views in a queue Q. Thus, the pairs are not directly compared, rather a projection embedding that is most similar to a view is selected from Q for the comparison with another view. The NNCLR contrastive loss ℓn is defined as:
where S(z, Q) is the nearest neighbor function:
2.1.2 Self-supervised learning in medical imaging
Recent advancements in self-supervised learning (SSL) facilitate the training of models capable of effectively acquiring feature representations relevant to downstream tasks (Thomas et al., 2024; VanBerlo et al., 2024). When applied to imaging data, SSL methodologies primarily focus on image reconstruction (Hu et al., 2021a; Zhou et al., 2023), segmentation (Taleb et al., 2020; Sun et al., 2023), denoising (Pfaff et al., 2024), and disease classification (Dufumier et al., 2021; Jiang and Miao, 2022; Gorade et al., 2023). For example, the study by Taleb et al. (2020) introduces SSL pretext tasks, including patch-based prediction of latent representations and the augmentation prediction. In contrast, Hu et al. (2021a) suggests an alternative pretext task leveraging two parallel networks to minimize reconstruction loss. Additional research has used SSL on longitudinal Alzheimer's Disease (AD) MRI datasets to explore methods to integrate information from multiple imaging modalities (Fedorov et al., 2021) or to predict the trajectory of cognitive performance and/or cognitive decline (Ouyang et al., 2022; Zhao et al., 2021).
Contrary to the aforementioned studies, which aimed at applying SSL techniques for the learning of feature representations within broader application area, our work assesses the effectiveness of these representations acquired through SSL in differentiating neurodegenerative disorders with an emphasis on the interpretability of the models.
2.2 Convolutional neural network backbones
Convolutional neural networks (CNN) have been the state-of-the-art solutions for computer vision tasks for almost a decade. In the last few years, numerous approaches on the advancement of CNNs were proposed: introduction of skip connections (He et al., 2016; Huang et al., 2017), experimentation with model hyper-parameters such as kernel size (Ganjdanesh et al., 2023), normalization strategies (Ioffe and Szegedy, 2015) and activation functions (Dubey et al., 2022; Apicella et al., 2021), depthwise convolutions (Howard et al., 2017), and model's block architecture (Sandler et al., 2018).
With the introduction of attention priors, vision transformers (ViT) (Dosovitskiy et al., 2020) soon became a viable alternative to purely convolutional models, and currently represent the state-of-the-art model architecture as generic vision backbones. ViTs were inspired by the transformer models applied to language processing tasks. To the best of our knowledge, there weren't attempts of systematically comparing attention priors with convolutional priors. However, in their study Liu et al. (2022b) culminated many of the CNN advancements proposed over the years, and compared the resulting ConvNeXt model with comparable vision transformers. ConvNeXt (Liu et al., 2022b) was proposed as a purely convolutional model, which achieved favorable results on common vision benchmarks such as the ImageNet (Deng et al., 2009) and the COCO (Lin et al., 2014) datasets, sometimes even providing higher accuracy than competing ViT models. Notably, ConvNeXt achieved these results while maintaining the computational simplicity and efficiency of standard CNN models, highlighting the importance of convolutional priors for vision tasks.
2.3 Feature attribution
With the growing popularity of CNN models and these models becoming the off-the-shelf baselines, there has also been a growing need to understand them. Multiple studies have attempted to explain and interpret black-box CNN models. Within the domain of explainable AI (XAI), there are various methods to derive the importance of input features, i.e., the importance scores with respect to each prediction. These importance scores can be visualized by superimposing them on the input scans (Van der Velden et al., 2022). Certain preferred methods of importance scoring are Layer-wise Relevance Propagation (LRP) (Montavon et al., 2019; Kohlbrenner et al., 2020), Gradient-weighted Class Activation Mapping (Grad-CAM) (Selvaraju et al., 2020), and Integrated Gradients (IG) (Sundararajan et al., 2017). Multiple studies have been conducted mapping importance scores to input regions, particularly within the neuroscience application of dementia detection (Dyrba et al., 2021; Singh and Dyrba, 2023; Böhle et al., 2019; Leonardsen et al., 2024; Wang et al., 2023).
3 Methods
3.1 Neuroimaging datasets
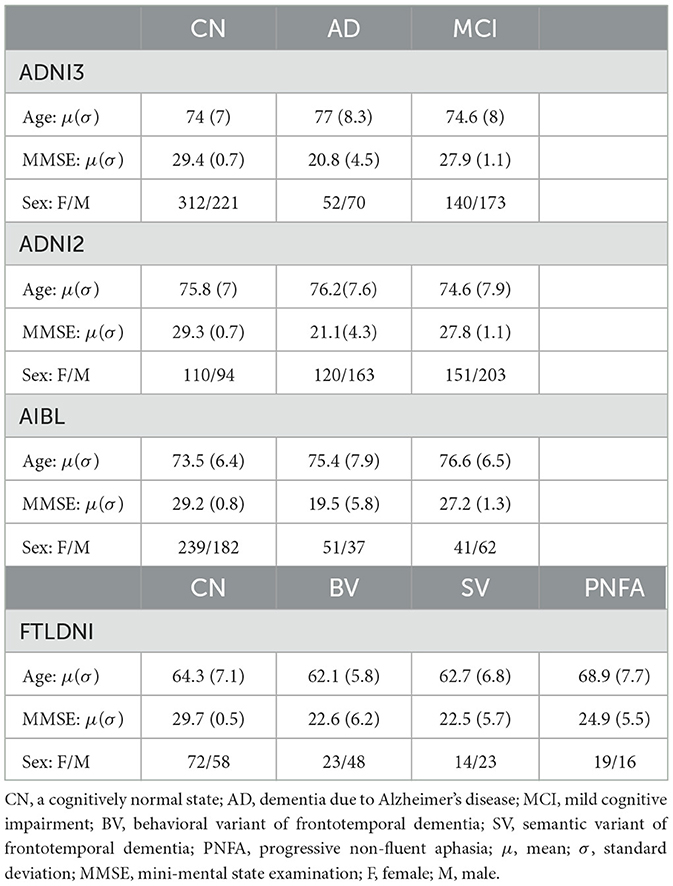
We used T1-weighted brain MRI scans from publicly available neuroimaging repositories. The data scans in our study were pooled from the following data repositories: (i) the Alzheimer's Disease Neuroimaging Initiative (ADNI),1 study phases ADNI2 and ADNI3, (ii) the Australian Imaging, Biomarker & Lifestyle Flagship Study of Aging (AIBL),2 collected by the AIBL study group, and (iii) the Frontotemporal Lobar Degeneration Neuroimaging Initiative (FTLDNI).3 In our study, the cognitively normal (CN) scan samples were consolidated from all three data cohorts. The ADNI and AIBL data cohorts provided samples with dementia due to Alzheimer's disease (AD) and mild cognitive impairment (MCI). While, FTLDNI was the only data cohort with samples categorized into different frontotemporal lobar degeneration (FTLD) phenotypes, i.e., the behavioral variant of frontotemporal dementia (BV), the semantic variant of frontotemporal dementia (SV), and the progressive non-fluent aphasia (PNFA). Notably, the data from ADNI3, ADNI2 and FTLDNI was used for training all models, and AIBL was used as independent test dataset.
We applied the “t1-linear pipeline” of the Clinica Python library (Routier et al., 2021; Wen et al., 2020) to preprocess the raw MRI scans. The pipeline uses the N4ITK method for bias field correction and the SyN algorithm from ANTs to perform an affine registration for alignment of each scan with the Montreal Neurological Institute (MNI) reference space. However, more advanced steps such as brain extraction, tissue segmentation, and non-linear warping were not performed. Some MRI scans were excluded due to severe quality issues, i.e., the presence of imaging artifacts such as blurring or ghosting, or missing diagnostic information.
Additionally, each scan was cropped to the size of 169 × 208 × 179 voxels with 1 mm isotropic resolution. After applying preprocessing methods, our study includes 841 scans from the ADNI2, 968 scans from the ADNI3, 612 scans from AIBL and 273 scans from FTLDNI. Table 1 summarizes the sample statistics of the different data sources.
3.2 Proposed self-supervised learning pipeline
Our proposed method consists of two modules: a feature extractor and a classification head. The feature extractor is a convolutional neural network trained without any sample labels in a self-supervised manner. The classification head is a simple neural network subsequently trained in a supervised way. The proposed architecture is shown in Figure 1.
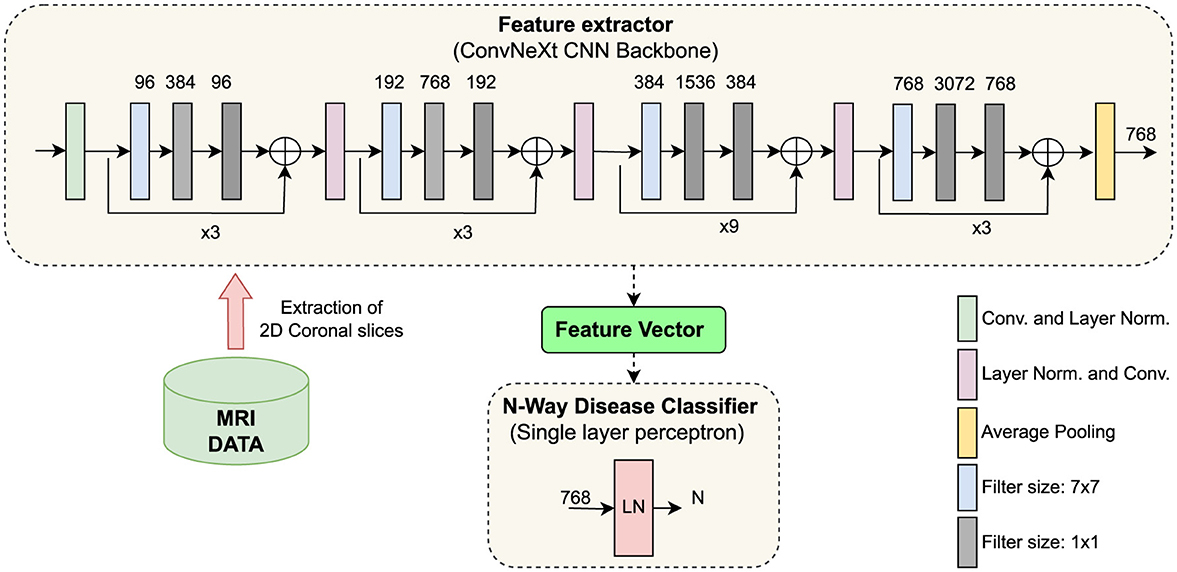
Figure 1. Illustration of the proposed architecture. (Top) ConvNeXt, a CNN model, trained under a self-supervised learning paradigm, extracts features from coronal brain slices. (Bottom center) The classification head learns to classify neurodegenerative disorders from the extracted features. CNN, convolutional neural network; LN, layer normalization; Conv, convolutional operation; LN, Layer Normalization.
After executing the t1-linear pipeline of the Clinica library, we obtained a 3D image for the brain of each participant. However, we only used 2D convolutional operations, as they reduce the CNN parameter space and model complexity. We selected only the coronal plane for the present study. In each MRI sample, there were in total 208 coronal slices; however, we considered only 120 coronal slices in the middle. The slices from the middle contain the relevant regions, such as the hippocampus and the temporal lobe, which are reported to be affected already in the earliest stages of Alzheimer's disease (Whitwell et al., 2008).
Feature extractor: We used the ConvNeXt model (Liu et al., 2022b) as the backbone for the SSL framework. It was trained with the NNCLR loss ℓn to learn visual representations of input data (see Equation 2). We chose the NNCLR method as it provides a more generalizable learning paradigm by sampling semantic variations in the latent space and being less reliant on transformation from specific pretext tasks (Dwibedi et al., 2021). We applied a series of random augmentations to a randomly selected coronal slice for the creation of positive pairs, as exemplified in Figure 2.
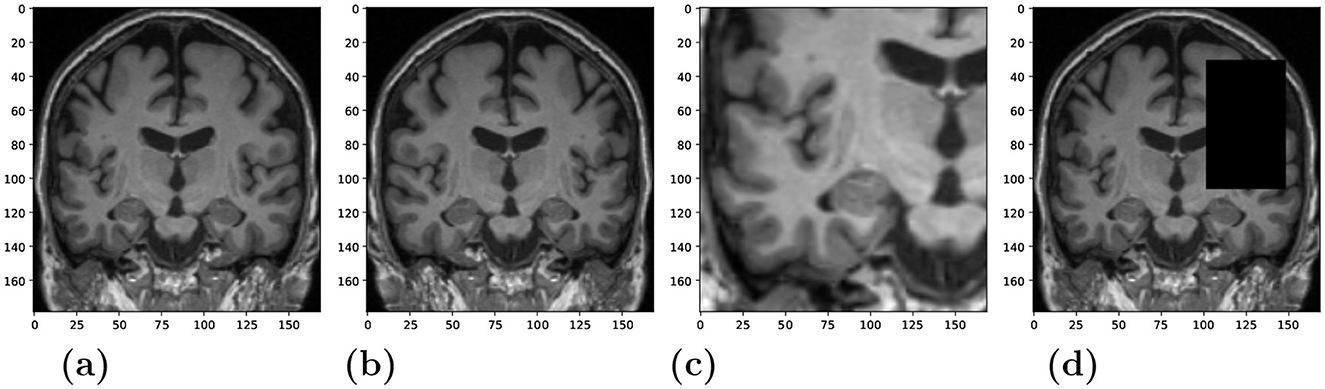
Figure 2. Randomly applied data augmentations to the input during training. (A) Original. (B) Horizontal flip (C) Crop and resize. (D) Occlusion.
The loss optimized for a data batch was:
where ℓn is the NNCLR loss from Equation 2, 2k − 1 and 2k represent the indices of the same augmented slice, and N is the total number of training samples.
Specifically, we used the “tiny” variant ConvNeXt model (Liu et al., 2022b) as our backbone model. It has a configuration with sequential blocks set to (3, 3, 9, 3) and the number of output channels equalling to (96, 192, 384, 768). ConvNeXt culminates in many architectural advancements such as larger 7x7 kernel sizes, skip connections, inverted bottleneck, Gaussian error linear units (GELU) as activation function, layer-wise normalization (LN) strategy instead of batch normalisations (BN), etc. The ConvNeXt model and pretrained model weights can be downloaded from the publicly available PyTorch library (Paszke et al., 2019).
Classification head: While using the ConvNeXt model as a feature extractor, we considered the output produced by a 2D adaptive average pooling layer after the last convolutional block as input for the subsequent “classification head” (Figure 1). That means the classification head takes as input the latent feature representations of the MRI scans that where processed by the backbone CNN model. The dimension of the extracted feature vector per MRI slice is 768. Our classification head is a simple neural network consisting of a single fully-connected layer preceded by a layer normalization operation (Figure 1 bottom). A single-layer perceptron was chosen as the classification head to leverage the features extracted from the ConvNeXt feature extractor directly, and not transforming the features by applying multiple levels of nonlinearities. This design choice aims to preserve the integrity of the extracted features. Employing a single-layer perceptron is a widely recognized methodology, commonly referred to as linear evaluation or linear probing (Dubois et al., 2023; Scheibenreif et al., 2024; Kalibhat et al., 2024).
3.3 Feature attribution
Integrated gradients (IG) can be applied to various data modalities, such as text, images, or structured data (Sundararajan et al., 2017). IG was chosen over other feature-attribution methods because of its strong theoretical justifications, such as the completeness property of the integrated gradients. IG considers a straight path from some baseline to the input, and computes the gradients along that path. These accumulated gradients are called integrated gradients. However, this accumulation is an approximation of the actual integration of the gradients, and the number of steps taken between the baseline to the input determines the quality of this approximation. In our study, we set N = 50 as the number of integration steps taken between the baseline image and the input image. To calculate IG importance scores, a mean CN image was used as a baseline for the IG attribution method. We used the IG implementation provided by the Captum library (Kokhlikyan et al., 2020) to calculate importance maps for MRI scans with respect to the classification task.
3.4 Experimental setup
Training the feature extractor: We trained a feature extraction model (ConvNeXt) using the NNCLR method on ADNI3, ADNI2 and FTLDNI data for three learning trials. For each trial, we created random training and test sets. These sets were held constant for all experiments. If more than one MRI recording was available per participant, then we assigned all participant's MRI scans only to one set, thus avoiding data leakage. This resulted in 10% of data belonging to the test set.
The model was trained for 1,000 epochs using a batch size of 180 samples. The size of the NNCLR queue Q was set to 8,192. We applied three different data augmentation techniques with a probability of 0.5 to produce views visualized in Figures 2B–D: horizontal flip, cropping and resizing, and occlusion. We experimented with different data sources to train the feature extractor, i.e., utilizing in-domain medical images vs. training with out-of-domain natural images. More details about model training and results could be found in the supplementary.
Training the classification head: To determine if a 3D MRI scan belongs to a specific diagnostic group, we first derive the latent representation vectors for 2D coronal slices using the ConvNeXt feature extractor and then make a prediction for each slice using the classification head. For evaluation with the test data, we applied a majority voting procedure in which the group label that occurs the most frequently determined the final group assignment. We trained the classification head for 100 epochs, on the same three training trials that were used to train the feature extractors. We used a batch size of 64 samples and decayed the learning rate with cosine annealing after every 20 epochs.
We experimented with various setups for training a classification head while keeping the weights of the feature extractor frozen vs. unfrozen, i.e., letting the weights change during the classification head training. For the downstream task, we compared different multi-class classification heads, i.e., predicting four (CN, MCI, AD, BV) or three classes—(CN, MCI, AD) and (CN, AD, BV), and binary classification heads—(CN, AD), (CN, BV), and (AD, BV). Furthermore, we evaluated our models on the independent AIBL dataset, which was not used during training. The independent test dataset enabled us to assess the generalizability of our approach.
We used balanced accuracy, sensitivity (true positive rate), specificity (true negative rate), and the Matthews correlation coefficient (MCC) as evaluation metrics. Due to the class imbalance in our dataset, we have chosen balanced accuracy over simple accuracy in our study. Balanced accuracy is the average of the true positive rate and the true negative rate, and thus avoids the overestimation of model quality that (simple) accuracy generally shows in class imbalance scenarios. With the true positives TP, true negatives TN, false positives FP, and false negatives FN, the balanced accuracy is defined as:
As shown in Chicco and Jurman (2020), the MCC should be preferred over the (simple) accuracy and the F1 score, as they could generate misleading results in unbalanced data sets. The MCC ranges between [−1, 1]. To achieve a high MCC score, the classifier would have to make correct predictions on both the majority and minority classes. The MCC is formally defined as:
4 Results
4.1 Diagnostic group separation
We evaluated the manner in which the classification head could be configured. We compared multi-class vs. binary classification heads. Table 2 shows the results achieved with our proposed architecture for the identification of neurodegenerative disorders, using a frozen ConvNeXt feature extractor trained under the NNCLR SSL paradigm on brain images. The reported numbers were averaged over three learning trials. For the binary (AD vs. CN) classification model, the balance accuracy reached 82% for the cross-validation test sets and 80% for the independent AIBL data cohort.
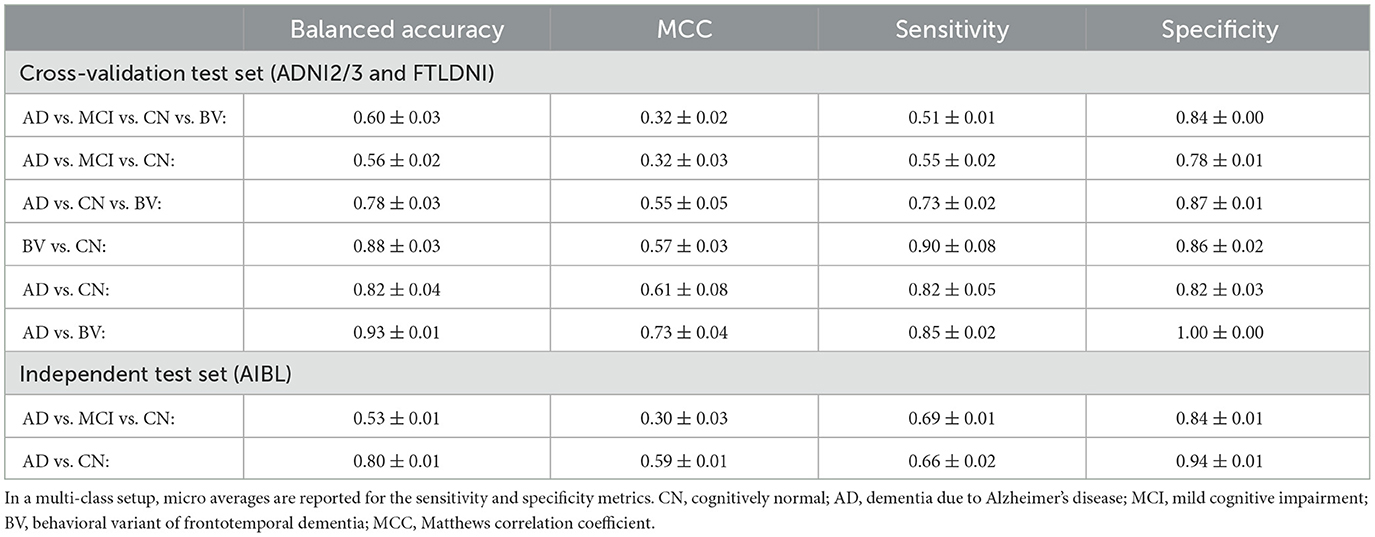
Table 2. Classification results of our proposed architecture, consisting of a frozen feature extractor trained under a SSL paradigm, and a single-layer neural network as the downstream classification head.
Upon comparing results from various settings of classification heads trained over a frozen feature extractor, we can observe a general trend, i.e., the binary classification for separating cognitively normal (CN) and Alzheimer's disease (AD) samples is a much simpler task than the 4-way multi-class classification of CN, mild cognitive impairment (MCI), AD and behavioral variant of frontotemporal dementia (BV) samples. This finding has often been reported in other studies in the field.
In the multi-class classification setting, the AD vs. MCI vs. CN model, often confuses MCI samples with CN or AD samples. This reflects the progressive nature of the Alzheimer's dementia, with MCI being intermediate stage between CN and AD. Interestingly, we found that the AD vs. MCI vs. CN vs. BV model is substantially better at separating BV samples from the other CN, MCI and AD samples, with the recall (=sensitivity) of the BV class being 0.89, compared to the average micro recall of the same model being 0.51. This finding points toward the model being sensitive to different underlying pathologies of different dementia diseases—frontotemporal dementia and AD. The same fact could also be corroborated from the high performance metrics of the binary AD vs. BV model. In Section 5.1 below, we discuss the achieved results and compare them with the state of the art.
4.2 Model interpretability
To highlight the input regions that were found to be useful by the SSL model, we used the Integrated Gradients (IG) attribution method. IG calculates the importance scores for the input regions for a specified prediction label. The IG importance scores were calculated for every sample of the test data set (from ADNI2/3 and FTLDNI), on which our multi-class model (AD vs. CN vs. BV) makes a correct classification. Figure 3 presents mean IG importance scores for the disease types AD and BV, visualized over the brain scan of a healthy sample chosen from the ADNI cohort. While making a prediction toward the diseased classes, the red regions in the image highlight input regions representing the evidence for the diseased class, while the green regions in the image highlight input regions representing the evidence against the diseased class. The mean importance scores were thresholded to visualize the most relevant findings.
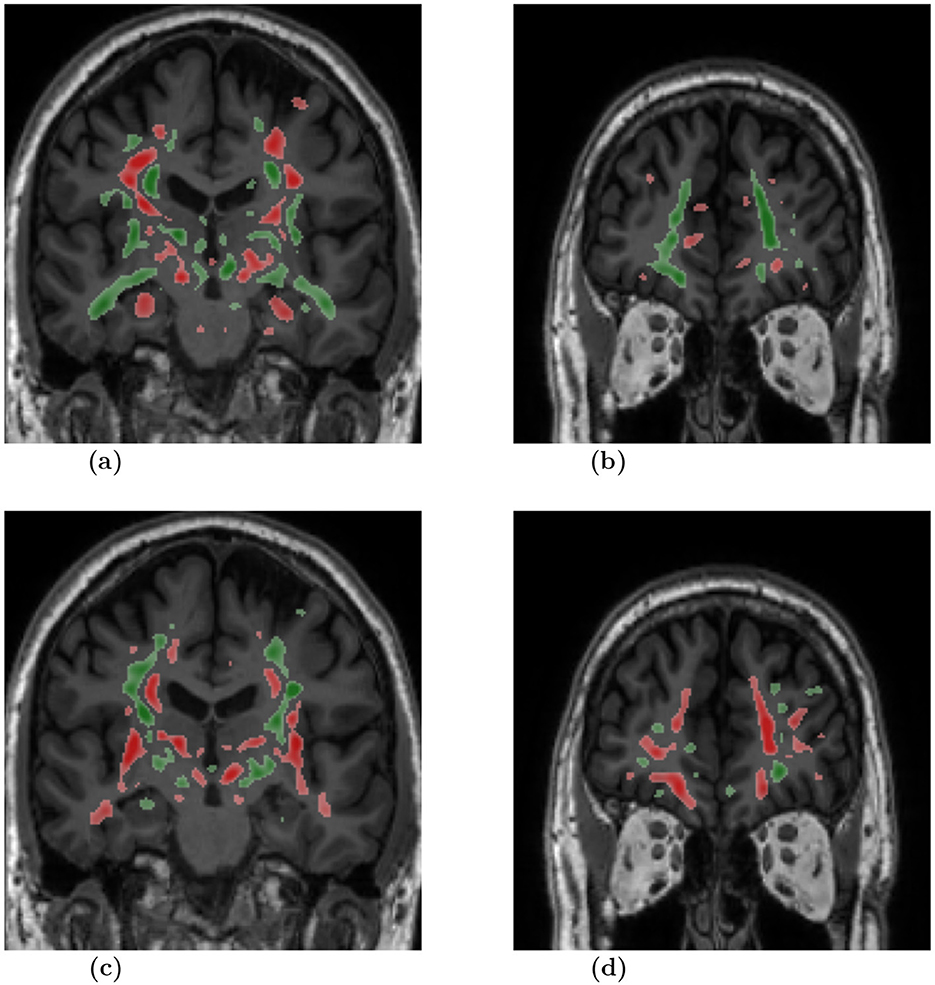
Figure 3. Mean attribution maps derived from the Integrated Gradients method for correctly identified AD and BV samples. Green and red color highlight pixel contributions to the model's prediction. Here, red highlights evidence for the respective disease classification and green indicates evidence against it. The attribution map overlay image was smoothed and thresholded to highlight relevant findings and improve visualization. AD, dementia due to Alzheimer's disease; BV, behavioral variant of frontotemporal dementia. (A) Slice: 0, Diagnosis: AD. (B) Slice: 60, Diagnosis: AD. (C) Slice: 0, Diagnosis: BV. (D) Slice: 60, Diagnosis: BV.
5 Discussion
5.1 Feature learning
In our proposed SSL framework, we rely on signals that are derived from the data itself rather than on external classification target labels to train a feature extractor. We trained our SSL model while restricting input to a subset of 2D coronal slices. It should be noted that other SSL studies also avoided training 3D CNN with high input resolution and followed similar 2D approaches as our study (Couronné et al., 2021) or alternatively needed to drastically downscale the 3D images to a very low 64 × 64 × 64 resolution to reduce computing time (Ouyang et al., 2022; Fedorov et al., 2021).
Our AD vs. CN vs. BV multi-class model achieves a balanced accuracy of 78%. Certain fully supervised methods solve the same task, achieving performance metrics as—Ma et al. (2020) reports (simple) accuracy of 86.0% from a model comparable to ours and 88.3% from a model with multimodal information sources and generative data augmentation, and Hu et al. (2021b) reports (simple) accuracy of 66.8% on a larger diverse dataset, and 91.8% on a smaller cleaner dataset. While our BV vs. CN binary model achieves a balanced accuracy of 88.2%. For the same task Moguilner et al. (2023) reports (simple) accuracy of 80% and 95% on MRI scans with 1.5T and 3T strength, respectively.
There are other SSL studies that report AD vs. CN group separation results on the ADNI dataset. Dufumier et al. (2021) reported an AUC score around 0.96. Ouyang et al. (2022) achieved a balanced accuracy between 81.9% and 83.6%, pre and post model finetuning. Seyfioğlu et al. (2022) using a vision transformer reported a mean simple accuracy of 83.4%. While there also other SSL applications that reported sub-optimal results, Chen et al. (2023) reported a balanced accuracy between 68.23% and 77.5% depending on model architecture used, while Jiang and Miao (2022) reported a balanced accuracy between 73.1% and 74% depending on the pretext task used. For the same task reported in these studies, our model with a frozen feature extractor, achieves a balanced accuracy of 82% on ADNI dataset, which is competitive with metrics reported in other studies. And on a holdout independent test set (AIBL), our model achieves a balanced accuracy of 80%, which is only a two-percent drop from the cross-validation testing of the model, highlighting the robustness of the model. It should noted that many studies don't evaluate their models on a holdout independent test set, which makes it is difficult to access their generalizability.
In Table 3, we compare our model evaluation results with the state-of-the-art studies that also used AIBL as an independent test dataset. Here, we compare our SSL model with other models trained in a supervised manner. Qiu et al. (2020), reports manual expert rating scores, with a simple accuracy metric of 82.3%. This performance level is comparable to that of our SSL models, which achieved the simple accuracy measure of 89.9% on the AIBL independent test set. It should be noted that some papers did not report the balanced accuracy measure, thus, their “simple” accuracy results might be biased toward the majority class of cognitively normal people who comprise 80% in the AIBL dataset for the group comparison AD vs. sCN.
With regard to our achieved level of performance, we can conclude that the ConvNeXt model trained under a SSL paradigm learns generalizable features for the subsequent downstream classification tasks without requiring data sampling techniques or sophisticated data augmentations, and consequently achieving competitive results in comparison to other supervised approaches. The reported results show that our model learned meaningful feature representations in a self-supervised manner, which can be used successfully to separate different stages and types of dementia.
5.2 Neural network interpretability
We chose the SSL paradigm to extract more generalizable image features independently of a downstream task. However, the SSL paradigm also allows the backbone model to learn features of the brain that may correlate with a specific neurodegenerative disorder. We applied the Integrated Gradients (IG) method to interpret the models and provide insights into the significance of input regions for the predictions. The IG importance scores were calculated for samples from the test dataset for which our AD vs. CN vs. BV multi-class model makes correct classifications. Figure 3 illustrates the mean IG importance scores for classifying samples into the AD or BV group. In Figures 3A, B, we see the hippocampus region highlighted in red for AD classification. Temporal lobe atrophy, specifically hippocampus atrophy, is a hallmark sign of Alzheimer's disease. In Figures 3C, D, we see the insula and frontal lobe regions being highlighted in red. Insular atrophy is associated with the behavioral variant of frontotemporal dementia (Moguilner et al., 2023; Seeley, 2010; Luo et al., 2020; Mandelli et al., 2016). It is of great interest to see the IG maps separately highlighting regions, which in the literature are often associated with AD and BV pathology.
Furthermore, to our knowledge, only one previous study, Dadsetan et al. (2022), has systematically compared different pretext methods for training SSL models for AD progression prediction, while also employing an XAI method, i.e., GradCAM, to generate relevance maps to evaluate the learned features. However, the reported relevance maps were particularly diffuse and widespread, offering limited interpretability. In addition, as an ablation study, we investigated different XAI methods beyond IG, but the results of these experiments also produced diffuse, spiky and unspecific relevance maps. This highlights that the application of XAI methods to SSL methods remains an open area of research.
Notably, our model successfully learned to not consider tissue outside of the brain or regions outside of the skull. However, the derived attributions provide a rather general indication of important input regions throughout the brain, including primarily gray matter and white matter tissue. Few studies have pointed out the complex nature of IG importance scores that highlight multiple image features, both for and against a class instance, making their comprehension non-trivial (Adebayo et al., 2018; Kakogeorgiou and Karantzalos, 2021; Hiller et al., 2025).
5.3 Limitations and future work
Our study uses only a subset of coronal slices to make sample-level classifications. We acknowledge that the selection of the full slice set along the coronal axis or selection of the full 3D MRI data could have a positive effect on classification performance; however, the main goal of the study was to investigate the application of SSL and to compare it with traditional supervised approaches; thus only a subset of slices along the coronal axis was chosen as input. Learning a 3D CNN is a computationally expensive problem for self-supervised learning, as it relies on (a) very large data corpus, (b) data augmentation algorithms which are markedly more computationally expensive in 3D due to the cubic time-complexity of the algorithms, and (c) many learning iterations as training typically converges much slower than in supervised learning. More specifically, training our models for 1,000 epochs on a single NVIDIA Quadro RTX 6000 GPU took on average 27 h. In the future, to train better feature extractors, we will incorporate more spatial neuroanatomical information, by combining three CNNs, i.e., one trained along each orthogonal planes—axial, coronal, and sagittal, and hence learning feature representations for the full 3D MRI data, as was proposed for supervised models (Qiao et al., 2021). Alternatively, a vision transformer model could also be explored to efficiently process smaller 3D patches of the brain (Qiu et al., 2020; Wen et al., 2020; Han et al., 2022; Wolf et al., 2023).
With regard to neural network interpretability and feature attribution, a comprehensive analysis of the salient features and feature attribution methods lies outside the scope of our current work. Although it remains to be seen whether the somewhat dispersed attribution maps we see in the current study are due to a difference in the training paradigm, i.e., SSL vs. supervised learning. To the best of our knowledge, no systematic efforts have been undertaken to compare the effects of training paradigm and attribution methods in highlighting disease-specific brain structures known in the clinical literature for different types of dementia. Additional experiments are required to holistically understand our SSL model and the informative importance of the generated maps. In our future work, we will explore other methods for feature attribution and methods to summarize attributions per brain region to assess if specific disease patterns emerge.
We also intend to include additional datasets in our future studies to learn more robust models. Specifically, we intend to add FTLD data cohorts.
5.4 Conclusion
We presented an architecture for the identification of neurodegenerative diseases from MRI data, consisting of a feature extractor and a classification head. The feature extractor used the ConvNeXt architecture as a backbone, which was trained under a self-supervised learning paradigm with nearest-neighbor contrastive learning (NNCLR) loss on brain MRI scans. The feature extractor model was used for subsequent downstream tasks by training only an additional single-layer neural network component which performs the classification. From our experiments, we show that CNN models trained under SSL paradigm have comparable performance to state-of-the-art CNN models trained in a supervised manner. With this presented approach, we provide a practical application of self-supervised learning on MRI data, as well as also demonstrate the application of attribution mapping methods for such systems to improve interpretability of the model's decision.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: Alzheimer's Disease Neuroimaging Initiative (ADNI) (http://adni.loni.usc.edu/data-samples/access-data), Australian Imaging Biomarkers and Lifestyle flagship study of aging (AIBL) (https://aibl.csiro.au), and Frontotemporal Lobar Degeneration Neuroimaging Initiative (FTLDNI) (https://memory.ucsf.edu/research-trials/research/allftd). Our source code for data processing, model training and evaluation, and creating attribution maps will be made publicly available at: (https://github.com/VadymV/clinic-net).
Ethics statement
The studies involving humans were approved by the respective neuroimaging initiatives internal review boards of each of the participating study sites. See https://adni.loni.usc.edu and https://aibl.csiro.au for details. All initiatives met common ethical standards in the collection of the data such as the Declaration of Helsinki. Analysis of the data was approved by the internal review board of the Rostock University Medical Center, reference number A 2020-0182. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants' legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
VG: Conceptualization, Data curation, Methodology, Software, Visualization, Writing – original draft. DS: Conceptualization, Software, Visualization, Writing – original draft. ST: Conceptualization, Methodology, Supervision, Validation, Writing – review & editing. MD: Conceptualization, Methodology, Supervision, Visualization, Writing – original draft, Writing – review & editing.
For the ADNI, AIBL, FTLDNI study groups
A complete listing of ADNI investigators can be found at https://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf. AIBL researchers are listed at https://www.aibl.csiro.au.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG) under grant DY151/2-1, project ID 454834942. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (https://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. Data collection and sharing for this project was funded by the Frontotemporal Lobar Degeneration Neuroimaging Initiative (National Institutes of Health Grant R01 AG032306). The study is coordinated through the University of California, San Francisco, Memory and Aging Center. FTLDNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. Data used in the preparation of this article was also obtained from the Australian Imaging Biomarkers and Lifestyle flagship study of aging (AIBL) funded by the Commonwealth Scientific and Industrial Research Organization (CSIRO). The AIBL researchers contributed data but did not participate in the analysis or writing of this report. AIBL study methodology has been reported previously (Ellis et al., 2009).
Conflict of interest
ST served as member of advisory boards of Lilly, Eisai, and Biogen, and is member of the independent data safety and monitoring board of the study ENVISION (Biogen).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fninf.2025.1527582/full#supplementary-material
Abbreviations
AD, Alzheimer's disease; ADNI, Alzheimer's Disease Neuroimaging Initiative; AIBL, Australian Imaging, Biomarker & Lifestyle Flagship Study of Aging; BN, Batch normalization; BV, behavioral variant of frontotemporal dementia; CN, Cognitively normal participants; CNN, Convolutional neural network; ConvNeXT, A highly optimized CNN model architecture recently introduced by Liu et al. (2022b); DZNE, Deutsches Zentrum for Neurodegenerative Erkrankungen (German Center for Neurodegenerative Diseases); FTLD, Frontotemporal lobar degeneration; FTLDNI, Frontotemporal Lobar Degeneration Neuroimaging Initiative; GELU, Gaussian error linear units; Grad-CAM, Gradient-weighted class activation mapping; IG, Integrated gradients; InfoNCE, A form a contrastive loss metric, where NCE stands for Noise-Contrastive Estimation; LN, Layer-wise normalization; LRP, Layer-wise relevance propagation; MCC, Matthews correlation coefficient; MCI, Mild cognitive impairment; MNI, Montreal Neurological Institute; MRI, Magnetic resonance imaging; NNCLR, Nearest-Neighbor Contrastive Learning; PNFA, Progressive non-fluent aphasia; SL, Supervised learning; SSL, Self-supervised learning; SV, semantic variant of frontotemporal dementia; ViT, Vision Transformers; XAI, Explainable artificial intelligence.
Footnotes
1. ^ADNI: https://adni.loni.usc.edu/.
2. ^AIBL: https://aibl.csiro.au/.
3. ^FTLDNI: https://memory.ucsf.edu/research-trials/research/allftd.
References
Adebayo, J., Gilmer, J., Muelly, M., Goodfellow, I., Hardt, M., and Kim, B. (2018). “Sanity checks for saliency maps,” in Proceedings of the 32nd International Conference on Neural Information Processing Systems, 9525–9536.
Apicella, A., Donnarumma, F., Isgró, F., and Prevete, R. (2021). A survey on modern trainable activation functions. Neural Netw. 138, 14–32. doi: 10.1016/j.neunet.2021.01.026
Böhle, M., Eitel, F., Weygandt, M., and Ritter, K. (2019). Layer-wise relevance propagation for explaining deep neural network decisions in MRI-based Alzheimer's disease classification. Front. Aging Neurosci. 11:194. doi: 10.3389/fnagi.2019.00194
Chechik, G., Sharma, V., Shalit, U., and Bengio, S. (2010). Large scale online learning of image similarity through ranking. J. Mach. Learn. Res. 11, 1109–1135. doi: 10.1007/978-3-642-02172-5_2
Chen, T., Kornblith, S., Norouzi, M., and Hinton, G. (2020). “A simple framework for contrastive learning of visual representations,” in International Conference on Machine Learning (PMLR), 1597–1607.
Chen, T., Zhai, X., Ritter, M., Lucic, M., and Houlsby, N. (2019). “Self-supervised GANs via auxiliary rotation loss,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 12154–12163. doi: 10.1109/CVPR.2019.01243
Chen, Y., Lou, S., Shuai, M., He, K., and An, Z. (2023). “CLCA: contrastive learning using combined additional information for Alzheimer's diagnosis,” in 2023 3rd International Conference on Neural Networks, Information and Communication Engineering (NNICE), 316–323. doi: 10.1109/NNICE58320.2023.10105726
Chicco, D., and Jurman, G. (2020). The advantages of the matthews correlation coefficient (MCC) over f1 score and accuracy in binary classification evaluation. BMC Genomics 21, 1–13. doi: 10.1186/s12864-019-6413-7
Chopra, S., Hadsell, R., and LeCun, Y. (2005). “Learning a similarity metric discriminatively, with application to face verification,” in 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR'05) (IEEE), 539–546. doi: 10.1109/CVPR.2005.202
Couronné, R., Vernhet, P., and Durrleman, S. (2021). “Longitudinal self-supervision to disentangle inter-patient variability from disease progression,” in Medical Image Computing and Computer Assisted Intervention-MICCAI 2021: 24th International Conference, Strasbourg, France, September 27-October 1, 2021, Proceedings, Part II 24 (Springer), 231–241. doi: 10.1007/978-3-030-87196-3_22
Dadsetan, S., Hejrati, M., Wu, S., and Hashemifar, S. (2022). Cross-domain self-supervised deep learning for robust Alzheimer's disease progression modeling. arXiv preprint arXiv:2211.08559.
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-Fei, L. (2009). “ImageNet: a large-scale hierarchical image database,” in 2009 IEEE Conference on Computer Vision and Pattern Recognition (IEEE), 248–255. doi: 10.1109/CVPR.2009.5206848
Doersch, C., Gupta, A., and Efros, A. A. (2015). “Unsupervised visual representation learning by context prediction,” in Proceedings of the IEEE International Conference on Computer Vision, 1422–1430. doi: 10.1109/ICCV.2015.167
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., et al. (2020). An image is worth 16x16 words: transformers for image recognition at scale. arXiv preprint arXiv:2010.11929.
Dubey, S. R., Singh, S. K., and Chaudhuri, B. B. (2022). Activation functions in deep learning: a comprehensive survey and benchmark. Neurocomputing. 503, 92–108. doi: 10.1016/j.neucom.2022.06.111
Dubois, Y., Hashimoto, T., and Liang, P. (2023). “Evaluating self-supervised learning via risk decomposition,” in Proceedings of the 40th International Conference on Machine Learning, ICML'23.
Dufumier, B., Gori, P., Victor, J., Grigis, A., Wessa, M., Brambilla, P., et al. (2021). “Contrastive learning with continuous proxy meta-data for 3D MRI classification,” in Medical Image Computing and Computer Assisted Intervention-MICCAI 2021: 24th International Conference, Strasbourg, France, September 27-October 1, 2021, Proceedings, Part II 24 (Springer), 58–68. doi: 10.1007/978-3-030-87196-3_6
Dwibedi, D., Aytar, Y., Tompson, J., Sermanet, P., and Zisserman, A. (2021). “With a little help from my friends: nearest-neighbor contrastive learning of visual representations,” in 2021 IEEE/CVF International Conference on Computer Vision (ICCV), 9588–9597. doi: 10.1109/ICCV48922.2021.00945
Dyrba, M., Hanzig, M., Altenstein, S., Bader, S., Ballarini, T., Brosseron, F., et al. (2021). Improving 3D convolutional neural network comprehensibility via interactive visualization of relevance maps: evaluation in Alzheimer's disease. Alzheimer's Res. Ther. 13, 1–18. doi: 10.1186/s13195-021-00924-2
Eitel, F., Schulz, M.-A., Seiler, M., Walter, H., and Ritter, K. (2021). Promises and pitfalls of deep neural networks in neuroimaging-based psychiatric research. Exp. Neurol. 339:113608. doi: 10.1016/j.expneurol.2021.113608
Ellis, K. A., Bush, A. I., Darby, D., De Fazio, D., Foster, J., Hudson, P., et al. (2009). The Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging: methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer's disease. Int. Psychoger. 21, 672–687. doi: 10.1017/S1041610209009405
Fedorov, A., Wu, L., Sylvain, T., Luck, M., DeRamus, T. P., Bleklov, D., et al. (2021). “On self-supervised multimodal representation learning: an application to Alzheimer's disease,” in 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) (IEEE), 1548–1552. doi: 10.1109/ISBI48211.2021.9434103
Ganjdanesh, A., Gao, S., and Huang, H. (2023). “EffConv: efficient learning of kernel sizes for convolution layers of CNNs,” in Thirty Seventh AAAI Conference on Artificial Intelligence (AAAI 2023), 7604–7612. doi: 10.1609/aaai.v37i6.25923
Gorade, V., Mittal, S., and Singhal, R. (2023). Pacl: patient-aware contrastive learning through metadata refinement for generalized early disease diagnosis. Comput. Biol. Med. 167:107569. doi: 10.1016/j.compbiomed.2023.107569
Grill, J.-B., Strub, F., Altché, F., Tallec, C., Richemond, P., Buchatskaya, E., et al. (2020). “Bootstrap your own latent-a new approach to self-supervised learning,” in Proceedings of the 34th International Conference on Neural Information Processing Systems (Curran Associates Inc.), 21271–21284.
Gutmann, M., and Hyvärinen, A. (2010). “Noise-contrastive estimation: a new estimation principle for unnormalized statistical models,” in Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics (JMLR Workshop and Conference Proceedings), 297–304.
Han, K., He, M., Yang, F., and Zhang, Y. (2022). Multi-task multi-level feature adversarial network for joint Alzheimer's disease diagnosis and atrophy localization using sMRI. Phys. Med. Biol. 67:085002. doi: 10.1088/1361-6560/ac5ed5
He, K., Zhang, X., Ren, S., and Sun, J. (2016). “Deep residual learning for image recognition,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 770–778. doi: 10.1109/CVPR.2016.90
Hiller, B. C., Bader, S., Singh, D., Kirste, T., Becker, M., and Dyrba, M. (2025). “Evaluating the fidelity of explanations for convolutional neural networks in alzheimer's disease detection,” in Bildverarbeitung für die Medizin 2025, Informatik aktuell, eds. A. Maier, T. M. Deserno, H. Handels, K. Maier-Hein, C. Palm, and T. Tolxdorff (Wiesbaden: Springer Fachmedien Wiesbaden).
Howard, A. G., Zhu, M., Chen, B., Kalenichenko, D., Wang, W., Weyand, T., et al. (2017). Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:1704.04861.
Hu, C., Li, C., Wang, H., Liu, Q., Zheng, H., and Wang, S. (2021a). “Self-supervised learning for MRI reconstruction with a parallel network training framework,” in Medical Image Computing and Computer Assisted Intervention-MICCAI 2021: 24th International Conference, Strasbourg, France, September 27-October 1, 2021, Proceedings, Part VI 24 (Springer), 382–391. doi: 10.1007/978-3-030-87231-1_37
Hu, J., Qing, Z., Liu, R., Zhang, X., Lv, P., Wang, M., et al. (2021b). Deep learning-based classification and voxel-based visualization of frontotemporal dementia and alzheimer's disease. Front. Neurosci. 14:626154. doi: 10.3389/fnins.2020.626154
Huang, G., Liu, Z., Van Der Maaten, L., and Weinberger, K. Q. (2017). “Densely connected convolutional networks,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (IEEE Computer Society), 2261–2269. doi: 10.1109/CVPR.2017.243
Ioffe, S., and Szegedy, C. (2015). “Batch normalization: accelerating deep network training by reducing internal covariate shift,” in International Conference on Machine Learning (PMLR), 448–456.
Jaiswal, A., Babu, A. R., Zadeh, M. Z., Banerjee, D., and Makedon, F. (2020). A survey on contrastive self-supervised learning. Technologies 9:2. doi: 10.3390/technologies9010002
Jiang, H., and Miao, C. (2022). “Pre-training 3D convolutional neural networks for prodromal Alzheimer's disease classification,” in 2022 International Joint Conference on Neural Networks (IJCNN) (IEEE), 1–8. doi: 10.1109/IJCNN55064.2022.9891966
Jing, L., and Tian, Y. (2020). Self-supervised visual feature learning with deep neural networks: a survey. IEEE Trans. Pattern Anal. Mach. Intell. 43, 4037–4058. doi: 10.1109/TPAMI.2020.2992393
Kakogeorgiou, I., and Karantzalos, K. (2021). Evaluating explainable artificial intelligence methods for multi-label deep learning classification tasks in remote sensing. Int. J. Appl. Earth Observ. Geoinform. 103:102520. doi: 10.1016/j.jag.2021.102520
Kalibhat, N., Narang, K., Firooz, H., Sanjabi, M., and Feizi, S. (2024). “Measuring self-supervised representation quality for downstream classification using discriminative features,” in Proceedings of the AAAI Conference on Artificial Intelligence, 13031–13039. doi: 10.1609/aaai.v38i12.29201
Kohlbrenner, M., Bauer, A., Nakajima, S., Binder, A., Samek, W., and Lapuschkin, S. (2020). “Toward best practice in explaining neural network decisions with LRP,” in 2020 International Joint Conference on Neural Networks (IJCNN) (IEEE), 1–7. doi: 10.1109/IJCNN48605.2020.9206975
Kokhlikyan, N., Miglani, V., Martin, M., Wang, E., Alsallakh, B., Reynolds, J., et al. (2020). Captum: A unified and generic model interpretability library for pytorch. arXiv:2009.07896.
Larsson, G., Maire, M., and Shakhnarovich, G. (2016). “Learning representations for automatic colorization,” in Computer Vision-ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, October 11–14, 2016, Proceedings, Part IV 14 (Springer), 577–593. doi: 10.1007/978-3-319-46493-0_35
Larsson, G., Maire, M., and Shakhnarovich, G. (2017). “Colorization as a proxy task for visual understanding,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 840–849. doi: 10.1109/CVPR.2017.96
Leonardsen, E. H., Persson, K., Grødem, E., Dinsdale, N., Schellhorn, T., Roe, J. M., et al. (2024). Constructing personalized characterizations of structural brain aberrations in patients with dementia using explainable artificial intelligence. NPJ Dig. Med. 7:110. doi: 10.1038/s41746-024-01123-7
Lin, T.-Y., Maire, M., Belongie, S., Hays, J., Perona, P., Ramanan, D., et al. (2014). “Microsoft coco: common objects in context,” in Computer Vision-ECCV 2014: 13th European Conference, Zurich, Switzerland, September 6–12, 2014, Proceedings, Part V 13 (Springer), 740–755. doi: 10.1007/978-3-319-10602-1_48
Liu, S., Mallol-Ragolta, A., Parada-Cabaleiro, E., Qian, K., Jing, X., Kathan, A., et al. (2022a). Audio self-supervised learning: a survey. Patterns 3:100616. doi: 10.1016/j.patter.2022.100616
Liu, Z., Mao, H., Wu, C.-Y., Feichtenhofer, C., Darrell, T., and Xie, S. (2022b). “A convnet for the 2020s,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 11976–11986. doi: 10.1109/CVPR52688.2022.01167
Luo, C., Hu, N., Xiao, Y., Zhang, W., Gong, Q., and Lui, S. (2020). Comparison of gray matter atrophy in behavioral variant frontal temporal dementia and amyotrophic lateral sclerosis: a coordinate-based meta-analysis. Front. Aging Neurosci. 12:14. doi: 10.3389/fnagi.2020.00014
Ma, D., Lu, D., Popuri, K., Wang, L., Beg, M. F., Initiative, A. D. N., et al. (2020). Differential diagnosis of frontotemporal dementia, alzheimer's disease, and normal aging using a multi-scale multi-type feature generative adversarial deep neural network on structural magnetic resonance images. Front. Neurosci. 14:853. doi: 10.3389/fnins.2020.00853
Mandelli, M. L., Vitali, P., Santos, M., Henry, M., Gola, K., Rosenberg, L., et al. (2016). Two insular regions are differentially involved in behavioral variant FTD and nonfluent/agrammatic variant PPA. Cortex 74, 149–157. doi: 10.1016/j.cortex.2015.10.012
Moguilner, S., Whelan, R., Adams, H., Valcour, V., Tagliazucchi, E., and Ibáñez, A. (2023). Visual deep learning of unprocessed neuroimaging characterises dementia subtypes and generalises across non-stereotypic samples. EBioMedicine 90:104540. doi: 10.1016/j.ebiom.2023.104540
Montavon, G., Binder, A., Lapuschkin, S., Samek, W., and Müller, K.-R. (2019). Layer-Wise Relevance Propagation: An Overview. Cham: Springer International Publishing, 193–209. doi: 10.1007/978-3-030-28954-6_10
Noroozi, M., and Favaro, P. (2016). “Unsupervised learning of visual representations by solving Jigsaw puzzles,” in European Conference on Computer Vision (Springer), 69–84. doi: 10.1007/978-3-319-46466-4_5
Oord, A. V. D., Li, Y., and Vinyals, O. (2019). Representation learning with contrastive predictive coding. arXiv preprint arXiv:1807.03748.
Orhan, E., Gupta, V., and Lake, B. M. (2020). “Self-supervised learning through the eyes of a child,” in NIPS'20: Proceedings of the 34th International Conference on Neural Information Processing Systems, 9960–9971.
Ouyang, J., Zhao, Q., Adeli, E., Zaharchuk, G., and Pohl, K. M. (2022). Self-supervised learning of neighborhood embedding for longitudinal MRI. Med. Image Anal. 82:102571. doi: 10.1016/j.media.2022.102571
Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J., Chanan, G., et al. (2019). “Pytorch: an imperative style, high-performance deep learning library,” in Advances in Neural Information Processing Systems (Curran Associates, Inc.), 8024–8035.
Pfaff, L., Darwish, O., Wagner, F., Thies, M., Vysotskaya, N., Hossbach, J., et al. (2024). Enhancing diffusion-weighted prostate MRI through self-supervised denoising and evaluation. Sci. Rep. 14:24292. doi: 10.1038/s41598-024-75007-x
Qiao, H., Chen, L., and Zhu, F. (2021). “A fusion of multi-view 2D and 3D convolution neural network based MRI for Alzheimer's disease diagnosis,” in 2021 43rd Annual International Conference of the IEEE Engineering in Medicine &Biology Society (EMBC) (IEEE), 3317–3321. doi: 10.1109/EMBC46164.2021.9629923
Qiu, S., Joshi, P. S., Miller, M. I., Xue, C., Zhou, X., Karjadi, C., et al. (2020). Development and validation of an interpretable deep learning framework for Alzheimer's disease classification. Brain 143, 1920–1933. doi: 10.1093/brain/awaa137
Reed, C. J., Metzger, S., Srinivas, A., Darrell, T., and Keutzer, K. (2021). “Selfaugment: automatic augmentation policies for self-supervised learning,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2674–2683. doi: 10.1109/CVPR46437.2021.00270
Routier, A., Burgos, N., Díaz, M., Bacci, M., Bottani, S., El-Rifai, O., et al. (2021). Clinica: an open-source software platform for reproducible clinical neuroscience studies. Front. Neuroinform. 15:689675. doi: 10.3389/fninf.2021.689675
Sabokrou, M., Khalooei, M., and Adeli, E. (2019). “Self-supervised representation learning via neighborhood-relational encoding,” in Proceedings of the IEEE/CVF International Conference on Computer Vision, 8009–8018. doi: 10.1109/ICCV.2019.00810
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., and Chen, L.-C. (2018). “Mobilenetv2: inverted residuals and linear bottlenecks,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 4510–4520. doi: 10.1109/CVPR.2018.00474
Scheibenreif, L., Mommert, M., and Borth, D. (2024). “Parameter efficient self-supervised geospatial domain adaptation,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 27841–27851. doi: 10.1109/CVPR52733.2024.02630
Schiappa, M. C., Rawat, Y. S., and Shah, M. (2023). Self-supervised learning for videos: a survey. ACM Comput. Surv. 55, 1–37. doi: 10.1145/3577925
Schroff, F., Kalenichenko, D., and Philbin, J. (2015). “Facenet: a unified embedding for face recognition and clustering,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 815–823. doi: 10.1109/CVPR.2015.7298682
Seeley, W. W. (2010). Anterior insula degeneration in frontotemporal dementia. Brain Struct. Funct. 214, 465–475. doi: 10.1007/s00429-010-0263-z
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., and Batra, D. (2020). Grad-cam: visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 128, 336–359. doi: 10.1007/s11263-019-01228-7
Seyfioğlu, M. S., Liu, Z., Kamath, P., Gangolli, S., Wang, S., Grabowski, T., et al. (2022). “Brain-aware replacements for supervised contrastive learning in detection of Alzheimer's disease,” in International Conference on Medical Image Computing and Computer-Assisted Intervention (Springer), 461–470. doi: 10.1007/978-3-031-16431-6_44
Shurrab, S., and Duwairi, R. (2022). Self-supervised learning methods and applications in medical imaging analysis: a survey. PeerJ Comput. Sci. 8:e1045. doi: 10.7717/peerj-cs.1045
Singh, D., and Dyrba, M. (2023). “Comparison of CNN architectures for detecting Alzheimer's disease using relevance maps,” in Bildverarbeitung für die Medizin 2023 (Springer), 238–243. doi: 10.1007/978-3-658-41657-7_51
Sohn, K. (2016). “Improved deep metric learning with multi-class n-pair loss objective,” in Proceedings of the 30th International Conference on Neural Information Processing Systems (Curran Associates Inc.), 1857–1865.
Sun, Y., Wang, L., Gao, K., Ying, S., Lin, W., Humphreys, K. L., et al. (2023). Self-supervised learning with application for infant cerebellum segmentation and analysis. Nat. Commun. 14:4717. doi: 10.1038/s41467-023-40446-z
Sundararajan, M., Taly, A., and Yan, Q. (2017). “Axiomatic attribution for deep networks,” in Proceedings of the 34th International Conference on Machine Learning (PMLR), 3319–3328.
Taleb, A., Loetzsch, W., Danz, N., Severin, J., Gaertner, T., Bergner, B., et al. (2020). “3D self-supervised methods for medical imaging,” in Proceedings of the 34th International Conference on Neural Information Processing Systems (Curran Associates Inc.), 18158–18172.
Thomas, A. W., Ré, C., and Poldrack, R. A. (2024). “Self-supervised learning of brain dynamics from broad neuroimaging data,” in Proceedings of the 36th International Conference on Neural Information Processing Systems, NIPS '22, Red Hook, NY, USA (Curran Associates Inc.).
Van der Velden, B. H., Kuijf, H. J., Gilhuijs, K. G., and Viergever, M. A. (2022). Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med. Image Anal. 79:102470. doi: 10.1016/j.media.2022.102470
VanBerlo, B., Hoey, J., and Wong, A. (2024). A survey of the impact of self-supervised pretraining for diagnostic tasks in medical x-ray, CT, MRI, and ultrasound. BMC Med. Imaging 24:79. doi: 10.1186/s12880-024-01253-0
Wang, D., Honnorat, N., Fox, P. T., Ritter, K., Eickhoff, S. B., Seshadri, S., et al. (2023). Deep neural network heatmaps capture alzheimer's disease patterns reported in a large meta-analysis of neuroimaging studies. Neuroimage 269:119929. doi: 10.1016/j.neuroimage.2023.119929
Wei, C., Xie, L., Ren, X., Xia, Y., Su, C., Liu, J., et al. (2019). “Iterative reorganization with weak spatial constraints: solving arbitrary Jigsaw puzzles for unsupervised representation learning,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 1910–1919. doi: 10.1109/CVPR.2019.00201
Wen, J., Thibeau-Sutre, E., Diaz-Melo, M., Samper-González, J., Routier, A., Bottani, S., et al. (2020). Convolutional neural networks for classification of Alzheimer's disease: overview and reproducible evaluation. Med. Image Anal. 63:101694. doi: 10.1016/j.media.2020.101694
Weng, L. (2021). Contrastive representation learning. Available at: https://lilianweng.github.io/posts/2021-05-31-contrastive/ (accessed January 9, 2024).
Whitwell, J. L., Shiung, M. M., Przybelski, S. A., Weigand, S. D., Knopman, D. S., Boeve, B. F., et al. (2008). MRI patterns of atrophy associated with progression to AD in amnestic mild cognitive impairment. Neurology 70, 512–520. doi: 10.1212/01.wnl.0000280575.77437.a2
Wolf, D., Payer, T., Lisson, C. S., Lisson, C. G., Beer, M., Götz, M., et al. (2023). Self-supervised pre-training with contrastive and masked autoencoder methods for dealing with small datasets in deep learning for medical imaging. Sci. Rep. 13:20260. doi: 10.1038/s41598-023-46433-0
Zhao, Q., Liu, Z., Adeli, E., and Pohl, K. M. (2021). Longitudinal self-supervised learning. Med. Image Anal. 71:102051. doi: 10.1016/j.media.2021.102051
Zhou, B., Dey, N., Schlemper, J., Mohseni Salehi, S. S., Liu, C., Duncan, J. S., et al. (2023). “DSFormer: a dual-domain self-supervised transformer for accelerated multi-contrast MRI reconstruction,” in 2023 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), 4955–4964, Los Alamitos, CA, USA (IEEE Computer Society). doi: 10.1109/WACV56688.2023.00494
Keywords: contrastive learning, self-supervised learning, neurodegenerative disorders, deep learning, structural magnetic resonance imaging, Alzheimer's disease, frontotemporal lobar degeneration
Citation: Gryshchuk V, Singh D, Teipel S and Dyrba M (2025) Contrastive self-supervised learning for neurodegenerative disorder classification. Front. Neuroinform. 19:1527582. doi: 10.3389/fninf.2025.1527582
Received: 13 November 2024; Accepted: 17 January 2025;
Published: 17 February 2025.
Edited by:
Hamed Honari, Stanford University, United StatesReviewed by:
Tingwei Quan, Huazhong University of Science and Technology, ChinaMailyn Moreno, Centro de Investigación en Computación, Instituto Politécnico Nacional (CIC-IPN), Mexico
Copyright © 2025 Gryshchuk, Singh, Teipel and Dyrba. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Dyrba, bWFydGluLmR5cmJhQGR6bmUuZGU=
†These authors have contributed equally to this work and share first authorship
 Vadym Gryshchuk
Vadym Gryshchuk Devesh Singh
Devesh Singh Stefan Teipel
Stefan Teipel Martin Dyrba
Martin Dyrba