- 1Anhui Vocational College of Grain Engineering, Hefei, China
- 2Hefei University, Hefei, China
Introduction: Mental health monitoring utilizing EEG analysis has garnered notable interest due to the non-invasive characteristics and rich temporal information encoded in EEG signals, which are indicative of cognitive and emotional conditions. Conventional methods for EEG-based mental health evaluation often depend on manually crafted features or basic machine learning approaches, like support vector classifiers or superficial neural networks. Despite the potential of these approaches, they often fall short in capturing the intricate spatiotemporal relationships within EEG data, leading to lower classification accuracy and poor adaptability across various populations and mental health scenarios.
Methods: To overcome these limitations, we introduce the EEG Mind-Transformer, an innovative deep learning architecture composed of a Dynamic Temporal Graph Attention Mechanism (DT-GAM), a Hierarchical Graph Representation and Analysis (HGRA) module, and a Spatial-Temporal Fusion Module (STFM). The DT-GAM is designed to dynamically extract temporal dependencies within EEG data, while the HGRA models the brain's hierarchical structure to capture both localized and global interactions among different brain regions. The STFM synthesizes spatial and temporal elements, generating a comprehensive representation of EEG signals.
Results and discussion: Our empirical results confirm that the EEG Mind-Transformer significantly surpasses conventional approaches, achieving an accuracy of 92.5%, a recall of 91.3%, an F1-score of 90.8%, and an AUC of 94.2% across several datasets. These findings underline the model's robustness and its generalizability to diverse mental health conditions. Moreover, the EEG Mind-Transformer not only pushes the boundaries of state-of-the-art EEG-based mental health monitoring but also offers meaningful insights into the underlying brain functions associated with mental disorders, solidifying its value for both research and clinical settings.
1 Introduction
Monitoring mental health through electroencephalography (EEG) has become an increasingly important area of research due to the growing recognition of mental health issues and the need for non-invasive, objective, and continuous monitoring methods. EEG, with its ability to capture the brain's electrical activity in real-time, offers unique insights into the neural processes underlying various mental health conditions (Michelmann et al., 2020). Not only can EEG provide a window into the brain's functioning, but it can also help in the early detection and management of mental disorders (Gao et al., 2021). Furthermore, EEG-based monitoring is crucial for developing personalized treatment plans and improving patient outcomes, making it a vital tool in both clinical and research settings (Cassani et al., 2018). The significance of EEG in mental health monitoring lies not only in its diagnostic potential but also in its ability to track changes over time, offering a dynamic view of the brain's response to treatment and environmental factors (Krigolson et al., 2017). As mental health issues continue to rise globally, the need for effective monitoring tools like EEG has never been more critical (Goswami et al., 2022).
Traditional machine learning methods in EEG analysis, while foundational, exhibit several critical limitations that hinder their effectiveness in mental health monitoring applications. Early methods predominantly relied on handcrafted features extracted from EEG signals, such as power spectral density and coherence (Kumar and Mittal, 2018). These features, although useful, capture only a limited view of the rich information contained within EEG signals. Specifically, handcrafted features often focus on static, time-averaged characteristics, neglecting the complex and dynamic temporal dependencies present in EEG data. Such simplifications are inadequate for understanding the rapidly fluctuating brain activities that are essential for accurate mental health monitoring. Moreover, conventional classifiers like support vector machines (SVMs) and k-nearest neighbors (k-NN) often struggle to model the intricate spatial relationships across multiple EEG channels, which are crucial for detecting patterns associated with mental health conditions (Delorme and Makeig, 2004). These algorithms treat the signals from each electrode independently or rely on shallow features that fail to account for inter-channel dependencies. As a result, they lack the capacity to capture the synchronized activity patterns across brain regions, which are vital for identifying neural biomarkers related to mood, anxiety, or cognitive states. Another significant limitation is the inability of traditional machine learning models to effectively handle the high-dimensional and non-linear nature of EEG data. Methods like SVMs and k-NN typically perform well only in controlled, small-scale settings where data variability is minimized. When applied to larger, real-world EEG datasets, these models tend to exhibit poor generalizability due to their limited capacity to handle the variability and noise inherent in complex EEG signals (Lotte et al., 2018; Craig and Tran, 2020; Alhussein et al., 2019). This restricts their application to controlled laboratory environments, rendering them less useful for real-time, in-the-wild mental health monitoring, where factors such as individual differences, movement artifacts, and environmental noise are present. Traditional machine learning approaches in EEG analysis are constrained by their reliance on handcrafted features, their inability to capture both spatial and temporal complexities, and their limited adaptability to high-dimensional, noisy datasets. These limitations underscore the need for more advanced methods capable of leveraging the full spatio-temporal dynamics of EEG signals to enhance the accuracy and robustness of mental health monitoring in diverse real-world contexts.
Recent innovations have turned to graph-based deep learning methods, particularly Graph Convolutional Networks (GCNs) (Roy et al., 2019) and Graph Attention Networks (GANs) (Kwon et al., 2019), to address the unique structural properties of EEG data. GCNs are highly effective in representing the brain as a graph of interconnected regions, allowing models to capture both localized and global patterns of neural activity (Zhao et al., 2019). For instance, Craik et al. (2019) demonstrated the utility of GCNs in brain network analysis, highlighting their capability to model hierarchical dependencies. However, these methods often struggle to integrate temporal dynamics effectively. Attention mechanisms, particularly spatiotemporal attention models, have further refined the ability to extract critical features from EEG data. These methods dynamically assign weights to relevant time points and spatial regions, enhancing interpretability and robustness. When combined with GCNs, attention mechanisms provide a powerful framework for modeling the brain's complex and dynamic activity (Tsiouris et al., 2018). Multimodal approaches incorporating EEG with other physiological signals, such as electromyography (EMG) and electrocardiography (ECG), have also shown promise. These methods offer a holistic view of mental states, combining complementary data to improve classification accuracy and resilience against noise. Studies have demonstrated their potential in contexts like stress detection and cognitive load estimation, where single-modality approaches may falter (Sturm et al., 2021).
The field has recently advanced toward more sophisticated models that integrate machine learning techniques with personalized and remote health monitoring. This latest phase has witnessed the adoption of graph-based models, such as Graph Convolutional Networks (GCNs) and Graph Attention Networks (GANs), which are particularly suited for EEG data due to their ability to model the brain's complex network structure (Parisot et al., 2018). These models capture both localized and global patterns of connectivity across brain regions, creating a nuanced understanding of spatial interactions (Song et al., 2021). By incorporating temporal dynamics into these graphs, researchers have developed interpretable, multi-scale models that better support personalized mental health monitoring (Sihag et al., 2022). This phase also emphasizes scalability, allowing EEG-based models to be deployed in practical settings. However, issues such as data privacy, ethical considerations, and continuous improvements in accessibility remain important challenges (Plis et al., 2018; Varatharajan et al., 2022; Stahl et al., 2019).
While traditional machine learning methods and early deep learning models struggle to capture the dynamic, multi-scale dependencies in EEG data, recent graph-based and attention-driven approaches have only partially bridged this gap by focusing on either spatial or temporal aspects independently. Additionally, these methods often lack scalability and adaptability to personalized, real-world applications, especially in resource-limited settings where data privacy and interpretability are paramount concerns. Our EEGMind-Transformer model addresses these limitations by integrating advanced graph-based neural networks with temporal attention mechanisms, allowing the model to simultaneously capture intricate spatiotemporal patterns within EEG data. This comprehensive approach not only enhances interpretability by offering insight into specific brain region interactions relevant to mental health but also improves scalability for remote and clinical applications through a structure that is adaptable across different datasets and user scenarios. By effectively bridging the gaps in current methods, the EEGMind-Transformer provides a robust, scalable, and interpretable solution tailored for personalized and continuous mental health monitoring.
• The EEGMind-Transformer introduces Dynamic Temporal Graph Attention Mechanism (DT-GAM), Hierarchical Graph Representation and Analysis (HGRA), and Spatial-Temporal Fusion Module (STFM) to effectively capture complex spatiotemporal dependencies in EEG data.
• This method is highly versatile and efficient, suitable for various scenarios, consistently delivering excellent performance across different mental health monitoring applications while offering model interpretability and scalability.
• Experimental results demonstrate that EEGMind-Transformer significantly outperforms existing state-of-the-art methods across multiple datasets, achieving superior performance.
2 Related work
2.1 Machine learning in EEG analysis
Traditional machine learning techniques have been extensively used in EEG analysis for mental health monitoring. These methods typically involve feature extraction followed by classification using algorithms such as Support Vector Machines (SVM), k-Nearest Neighbors (k-NN), Random Forests, and shallow neural networks (Lakshminarayanan et al., 2023). Feature extraction often relies on domain expertise to identify relevant features from the EEG signals, such as power spectral density, coherence, and wavelet coefficients, which are then fed into classifiers to distinguish between different mental states. While these approaches have shown some success, they are limited by their reliance on handcrafted features, which may not capture the full complexity of the EEG data (Hong et al., 2024). Moreover, traditional classifiers often struggle with the high dimensionality and variability of EEG signals, leading to issues with overfitting and poor generalization across different populations and recording conditions (Wan et al., 2023). Furthermore, these methods are typically static and cannot adequately model the temporal dynamics inherent in EEG signals, which are crucial for understanding cognitive and emotional processes. As a result, while traditional machine learning approaches have laid the groundwork for EEG analysis, they are often insufficient for capturing the complex, non-linear patterns in the data that are essential for accurate mental health monitoring (LaRocco et al., 2023).
2.2 Deep learning in EEG analysis
Deep learning has emerged as a powerful alternative to traditional machine learning methods in EEG analysis, offering the ability to automatically learn features directly from the data. Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs), including Long Short-Term Memory (LSTM) networks, have been particularly popular (Simar et al., 2024). CNNs are well-suited for capturing spatial patterns in EEG data by treating the multichannel signals as images or matrices, while RNNs and LSTMs excel at modeling temporal dependencies by processing EEG signals as sequences (Akter et al., 2024). More recent work has explored the use of hybrid models, such as CNN-LSTM architectures, which combine the strengths of both approaches to capture both spatial and temporal features simultaneously (Lakshminarayanan et al., 2023). While these methods have improved performance over traditional machine learning techniques, they still face challenges. One significant limitation is their inability to fully capture the complex spatiotemporal dependencies present in EEG data. Additionally, these models often require large amounts of labeled data for training, which can be difficult to obtain in clinical settings. Furthermore, despite their complexity, deep learning models can sometimes act as “black boxes,” offering little interpretability of how decisions are made, which is a critical requirement in medical applications (Ai et al., 2023).
2.3 Graph-based approaches in EEG analysis
Graph-based approaches have gained traction in EEG analysis due to their ability to model the brain's complex network structure. Graph-based methods are particularly effective in capturing the spatial dependencies and interactions within the brain, which are often overlooked by traditional machine learning and even some deep learning methods. Techniques such as Graph Convolutional Networks (GCNs) (Wu et al., 2019) and Graph Attention Networks (GANs) have been applied to EEG data, enabling the capture of both local and global patterns of brain connectivity. These methods can dynamically model how different brain regions interact over time, providing a more nuanced understanding of the underlying neural mechanisms associated with mental health conditions (Kosaraju et al., 2019). One of the significant advantages of graph-based methods is their interpretability, as they can highlight specific brain regions or connections that are most relevant to the task at hand. However, challenges remain, particularly in integrating temporal information with the spatial graph structures, as traditional graph-based methods primarily focus on static representations (Wu et al., 2022). Recent advances have started to address this by incorporating temporal dynamics into graph models, but there is still much work to be done to fully realize the potential of graph-based approaches in EEG analysis (He et al., 2023). These methods represent a promising direction for future research, particularly in their ability to provide both high accuracy and interpretability in mental health monitoring applications.
3 Preliminaries
To effectively model and monitor mental health conditions using EEG signals, it is essential to formalize the problem in a manner that aligns with the capabilities of the EEGMind-Transformer architecture. The goal is to detect and monitor various mental health conditions by analyzing the patterns and abnormalities present in EEG data. This can be framed as a classification problem where the model is trained to distinguish between different mental states based on the input EEG signals.
Let represent a dataset of EEG recordings, where denotes the EEG data for the i-th sample, C is the number of EEG channels, T is the number of time steps, and yi ∈ {1, …, K} is the corresponding mental health condition label, with K being the total number of classes. The goal is to learn a function that maps EEG data to the correct mental health condition.
To facilitate the learning process, the EEG data Xi is first preprocessed to remove noise and artifacts, resulting in a clean signal . This preprocessing step includes operations such as band-pass filtering, Independent Component Analysis (ICA) for artifact removal, and normalization. The preprocessed signal is then segmented into overlapping windows of fixed size, each corresponding to a smaller time frame of the EEG recording.
denote the j-th window of the i-th EEG recording, where W is the window size.
The EEGMind-Transformer processes each window independently through a series of transformations designed to capture the spatial and temporal dependencies within the EEG data. The model leverages a spatio-temporal attention mechanism, which can be mathematically represented as:
Here, are the query, key, and value matrices obtained from the linear transformation of the input window , and dk is the dimensionality of the keys. The attention mechanism computes the weighted sum of the values , where the weights are determined by the similarity between the queries and keys.
Subsequently, the outputs of the attention mechanism for all windows of the EEG recording are aggregated to form a comprehensive representation of the entire EEG signal. This representation is then passed through a graph neural network (GNN) that models the relationships between different brain regions. The GNN is defined on a graph G = (V, E), where V represents the set of brain regions (nodes), and E represents the connections (edges) between these regions. The graph convolution operation at each layer of the GNN can be expressed as:
where H(l) denotes the node features at the l-th layer, A is the adjacency matrix of the graph, D is the degree matrix, W(l) is the trainable weight matrix, and σ is a non-linear activation function. The final output of the GNN represents the spatial dependencies between different brain regions and is concatenated with the temporal features extracted by the Transformer.
Finally, the concatenated features are fed into a fully connected layer followed by a softmax function to produce the probability distribution over the mental health condition classes:
where Wf and bf are the weights and biases of the fully connected layer, and hi is the concatenated feature vector.
The training objective is to minimize the cross-entropy loss between the predicted labels ŷi and the true labels yi:
where θ represents all the trainable parameters in the model, and yi, k is a binary indicator (0 or 1) that indicates whether the class label k is the correct classification for sample i. This formalization sets the stage for the detailed exploration of the EEGMind-Transformer architecture and its components in the following sections.
4 Methodology
4.1 Overview
The EEGMind-Transformer introduces a breakthrough in mental health monitoring by integrating EEG signals with a Transformer-based framework. This model is engineered to exploit both temporal and spatial characteristics of the data, which are closely linked to mental health issues like depression and anxiety. Building on the latest progress in multimodal spatio-temporal attention mechanisms and the evolution of graph-based deep learning models for mental health assessment, the EEGMind-Transformer seeks to overcome the constraints of traditional approaches. It offers a more adaptable, interpretable, and scalable solution that works efficiently in real-time settings, thus making it suitable for both clinical and practical use cases. Designed to tackle the inherent challenges posed by EEG signal variability and the growing need for individualized models, this Transformer-based approach excels in capturing intricate patterns and long-range dependencies in data. Through the use of spatio-temporal attention, the model prioritizes the most critical features during training. The integration of graph neural networks allows for deeper insights into inter-regional brain activity, contributing to enhanced inference precision. This innovation is poised to significantly impact the future of mental health monitoring by offering a non-invasive, reliable, and versatile method for early detection and continuous mental health evaluation.
Dynamic Temporal Graph Attention Mechanism (DT-GAM): We designed the DT-GAM module to capture dynamic temporal dependencies in EEG data. Unlike traditional temporal modeling methods, DT-GAM uses a graph attention mechanism to adaptively adjust relationships between temporal nodes, ensuring that key features within specific time intervals receive prioritized attention. This design enhances the model's ability to capture temporal information, improving accuracy in predicting different mental health states. Hierarchical Graph Representation and Analysis Module (HGRA): The proposed HGRA module constructs a multi-level graph structure to better simulate the complex interactions between different brain regions. By aggregating information across different hierarchical levels, HGRA captures both local and global spatial dependencies. This innovation not only enhances the model's capacity to interpret brain structures but also provides greater interpretability, making it easier to visualize the significance of different brain regions in mental health monitoring. Spatial-Temporal Fusion Module (STFM): To seamlessly combine temporal and spatial information, EEGMind-Transformer introduces the STFM module. This module deeply integrates temporal and spatial features to create a comprehensive EEG signal representation. Compared to traditional methods that rely solely on spatial or temporal features, the addition of STFM significantly improves the model's depth of interpretation, allowing EEGMind-Transformer to gain a more holistic understanding of the complex changes associated with mental health states. Integrated Interpretability and Adaptability Design: EEGMind-Transformer not only achieves significant improvements in classification performance but also provides enhanced interpretability and extensibility through its modular design. Our approach can adapt to different datasets and scenarios while offering visual explanations for each module, making the model suitable for clinical diagnostics and personalized remote monitoring (Figure 1).
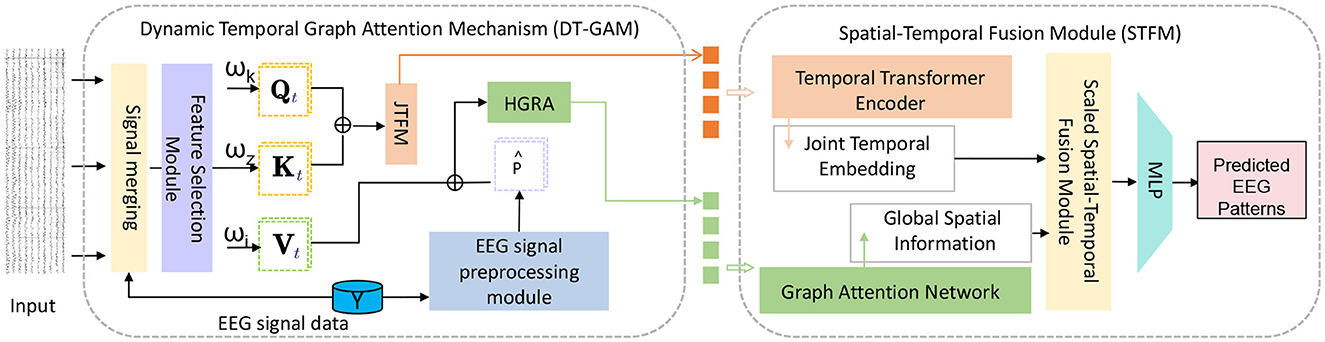
Figure 1. This figure illustrates the architecture of the EEGMind-Transformer model. It includes three core modules: Dynamic Temporal Graph Attention Mechanism (DT-GAM), Hierarchical Graph Representation and Analysis (HGRA), and Spatiotemporal Fusion Module (STFM). The DT-GAM module utilizes a graph-based attention mechanism to capture the underlying temporal dependencies in the EEG signals, emphasizing important temporal features. The Joint Temporal Feature Module (JTFM) further processes this temporal information to enhance the integration of joint temporal features, which are then passed to subsequent modules. The HGRA module builds a multi-level graph structure to capture local and global spatial dependencies, providing insights into complex cross-regional brain activities. Finally, the STFM combines the processed temporal and spatial features from DT-GAM, JTFM, and HGRA to obtain a comprehensive EEG signal representation.
4.2 Dynamic temporal graph attention mechanism (DT-GAM)
The Dynamic Temporal Graph Attention Mechanism (DT-GAM) lies at the heart of the EEGMind-Transformer, enabling the model to effectively capture complex temporal dependencies within EEG data. This mechanism is crucial for enhancing the model's ability to focus on the most relevant temporal features, which are vital for accurate mental health monitoring. DT-GAM leverages a graph-based representation of EEG data over time, where each node in the graph represents an EEG channel, and edges capture the temporal relationships between these channels across different time steps. The dynamic nature of this mechanism allows the graph to adapt its structure based on the evolving temporal patterns, ensuring that the most critical time points are given priority. The temporal attention mechanism can be mathematically formulated as:
where Qt, Kt, Vt are the query, key, and value matrices derived from the temporal features of the EEG data. The dimension dt serves as a scaling factor, which stabilizes the gradients during training. This attention mechanism allows the model to dynamically weigh the importance of different time steps, learning to focus on the temporal patterns that are most indicative of specific mental health conditions.
In DT-GAM, the temporal dependencies are modeled as a time-evolving graph Gt = (Vt, Et), where each node v ∈ Vt represents a time step, and each edge e ∈ Et represents the temporal connection between EEG readings at different times. The graph attention mechanism updates the importance of these connections through:
where At represents the temporal attention scores that adjust the influence of each time step based on its relevance to the task. These scores modulate the interaction between nodes (time steps), allowing the model to adaptively focus on the most informative segments of the EEG data.
The output from the temporal attention mechanism is then processed through a temporal graph convolutional layer, which refines the temporal node embeddings by aggregating information from the relevant time steps:
Here, denotes the node embeddings at layer l, and are the learnable weights of the temporal graph convolution layer. This operation is iterated across multiple layers, enabling the model to capture higher-order temporal interactions in the EEG data.
The refined temporal features from DT-GAM are then integrated with spatial features using a fusion strategy that concatenates the temporal and spatial outputs, which are then processed through a fully connected layer:
where ht and hs are the temporal and spatial feature vectors, respectively, and [·;·] represents concatenation. The fusion layer combines the temporal dynamics captured by DT-GAM with the spatial structure, producing a comprehensive representation of the EEG data.
Finally, the fused representation is passed through a softmax layer to generate the probability distribution over the mental health condition classes:
where Wo and bo are the weights and biases of the output layer.
The DT-GAM thus enables the EEGMind-Transformer to dynamically adapt its focus on the most relevant temporal features, leading to more accurate predictions and providing deeper insights into the temporal dynamics of mental health conditions.
4.3 Hierarchical Graph Representation and Analysis
The EEGMind-Transformer model incorporates a Hierarchical Graph Representation and Analysis (HGRA) module to effectively capture the multi-scale dependencies inherent in EEG data. This module is designed to leverage the hierarchical structure of brain regions and their interactions, enabling the model to learn both local and global patterns associated with mental health conditions.
At the core of the HGRA module is the construction of a multi-level graph G = {G1, G2, …, GL}, where each graph Gl = (Vl, El) corresponds to a different level of granularity in the brain's functional architecture. The lowest level graph G1 represents individual EEG channels as nodes, with edges representing direct functional connections between these channels. Higher levels G2, …, GL aggregate these channels into larger regions or networks, capturing more abstract relationships between brain areas (Figure 2).
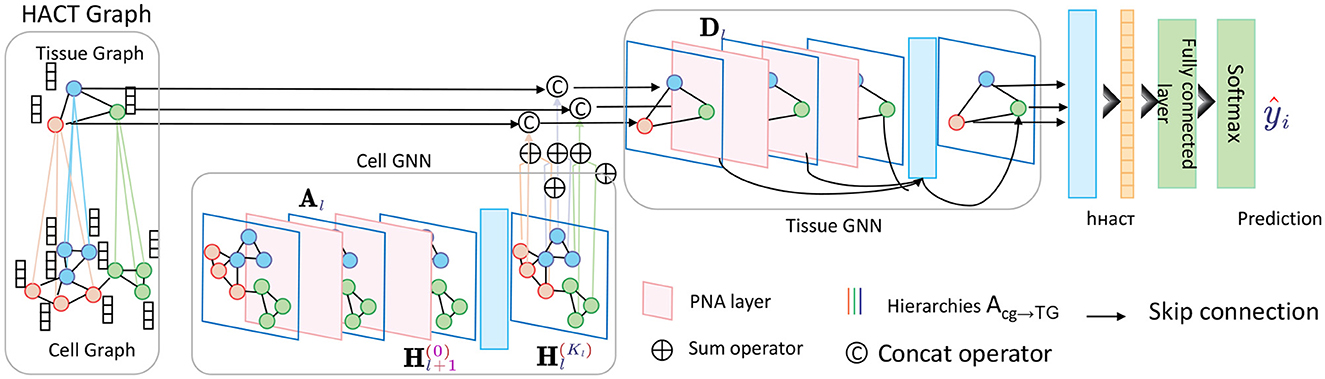
Figure 2. Schematic diagram of the Hierarchical Graph Representation and Analysis (HGRA) module. This module is used to capture multi-scale information in a hierarchical graph structure. The module contains Cell GNN (Cell Graph Neural Network) and Tissue GNN (Tissue Graph Neural Network), which apply PNA layer (Principal Neighborhood Aggregation) to aggregate node information at the cell and tissue levels, respectively. Through inter-layer aggregation (), the information in the cell graph is transferred to the tissue graph, realizing multi-level dependencies from local to global.
The node embeddings at each level l are initialized based on the features extracted from the EEG data, with lower levels receiving finer-grained features and higher levels receiving more abstract representations. The graph convolutional operations at each level can be described as:
where Al is the adjacency matrix of graph Gl, Dl is the degree matrix, is the weight matrix for the k-th layer at level l, and σ is a non-linear activation function. This operation iteratively refines the node embeddings by aggregating information from neighboring nodes, allowing the model to capture the hierarchical dependencies within the EEG data.
To integrate information across different levels of the hierarchy, the HGRA module employs a pooling mechanism that consolidates the node embeddings from lower levels and passes them to higher levels. This pooling operation can be formalized as:
where represents the initial embeddings for the next level, are the final embeddings at level l, and Pool(·) is a pooling function that aggregates information from lower-level nodes. Common pooling strategies include max pooling, average pooling, or more sophisticated attention-based pooling methods that weigh the importance of each node's contribution.
Principal Aggregation and Distribution (PAD) Layer: The HGRA module also incorporates a PAD layer (Principal Aggregation and Distribution layer) to further enhance the hierarchical graph structure. The PAD layer performs an aggregation operation across all levels, enabling a global representation by combining information from various scales. This aggregated representation is then distributed back to each level to reinforce local representations with global context, improving the model's ability to capture both global and local dependencies in EEG data.
The aggregation operation in the PAD layer can be expressed as:
where Hglobal represents the global embedding aggregated across all levels l = 1, …, L, and is the final node embedding at level l. The Aggregate(·) function can be implemented as a sum, mean, or attention-based aggregation over all hierarchical levels.
The global representation Hglobal is then distributed back to each level to enrich the local embeddings with global context, which can be described as:
where represents the enhanced embeddings at level l, and WPAD is a learnable weight matrix that adjusts the influence of the global context on each level's local representation.
These operations enable the PAD layer to create a multi-scale representation, improving the model's ability to capture both fine-grained and high-level dependencies in EEG data.
The final embeddings at the highest level GL encapsulate both local and global information from the EEG data. These embeddings are then concatenated with the temporal features extracted by the Transformer and fed into a fully connected layer to produce the final prediction:
where hL is the final embedding from the HGRA module, ht is the temporal feature vector, and Wh and bh are the weights and biases of the fully connected layer.
The output of this layer is then passed through a softmax function to generate the probability distribution over the mental health condition classes:
where Wo and bo are the weight matrix and bias vector of the output layer, respectively.
This hierarchical approach not only enhances the model's ability to capture the intricate relationships within EEG data but also provides a structured representation that aligns with the known hierarchical organization of the brain. By integrating this knowledge into the EEGMind-Transformer, the model is better equipped to differentiate between various mental health conditions, making it a powerful tool for both clinical and real-world mental health monitoring applications.
The Hierarchical Graph Representation and Analysis module, with the inclusion of the PAD layer, ensures that the EEGMind-Transformer can effectively leverage multi-scale information, which is crucial for capturing the complex, distributed nature of brain activity. This integration of prior knowledge through a structured graph-based approach represents a significant advancement in the field of mental health monitoring using EEG data.
The DT-GAM enhances the model's interpretability by dynamically assigning attention to specific temporal segments within the EEG data. This mechanism allows the model to prioritize and highlight critical temporal events, such as shifts in brainwave patterns associated with cognitive or emotional changes. By identifying these key temporal dependencies, DT-GAM provides insights into the temporal dynamics that may correlate with specific mental health states, offering clinicians an understanding of which periods in the EEG signals are most indicative of mental health conditions. Similarly, the HGRA module contributes to interpretability by modeling the hierarchical structure of brain regions. This approach enables the model to reveal important spatial interactions among brain regions, capturing both localized and global dependencies. HGRA's hierarchical graph-based approach can help identify which brain regions or connections are most involved in certain mental health conditions, providing valuable information for both research and clinical applications. In terms of scalability, both DT-GAM and HGRA are designed to adapt to various EEG datasets and clinical settings. The modularity of EEGMind-Transformer allows for adjustments to be made easily to the attention mechanisms and graph layers, facilitating its application across diverse patient populations and EEG recording setups. This flexibility, combined with the interpretability offered by DT-GAM and HGRA, positions the EEGMind-Transformer as a robust and scalable model for widespread use in mental health monitoring. We appreciate the reviewer's suggestion, which has enriched our discussion on the practical and clinical utility of the model.
The theoretical correlation between the HGRA module and biological brain networks is explored based on the dynamics of neural networks and regional interactions. The functional interactions of biological brain networks can be represented by a functional connectivity matrix, defined as:
where Cij denotes the connection strength between brain regions i and j, and Ai and Aj represent the activation patterns of these regions. Similarly, the HGRA module simulates such interactions through a weighted adjacency matrix:
where xi and xj are the input features of nodes i and j, θ denotes the parameters of the HGRA module, and f is the weighting function.
To quantitatively evaluate the similarity between the HGRA module and biological brain networks, a graph similarity index is introduced as:
where S ∈ [0, 1] represents a normalized similarity score. When S → 1, the structures of the two networks exhibit high alignment.
The HGRA module optimizes its parameters to emulate brain network properties. The dynamic learning process minimizes the Frobenius norm of the difference between the biological connectivity matrix C and the module's weight matrix W, with the loss function defined as:
This optimization process ensures that the HGRA module adapts to align with the structure of brain networks. Additionally, biological brain networks are known for their modularity, which can be quantitatively assessed using the modularity score:
where ki and kj denote the degrees of nodes i and j, m represents the total number of edges, and δ(gi, gj) is the Kronecker function indicating whether nodes i and j belong to the same module.
Theoretical analysis demonstrates that the HGRA module effectively reproduces dynamic interactions observed in biological brain networks, particularly in terms of multi-scale interaction patterns and modular structures. By leveraging the graph similarity index S and modularity metric Q, the alignment between artificial and biological neural networks is quantitatively verified. This framework enhances the biological interpretability of the HGRA module, providing a robust foundation for understanding its relevance to brain-inspired computational principles.
4.4 Spatial-Temporal Fusion Module (STFM)
The Spatial-Temporal Fusion Module (STFM) is a critical component of the EEGMind-Transformer, designed to integrate and harmonize the spatial and temporal features extracted from EEG data. This module ensures that the model captures the intricate interactions between brain regions over time, which is essential for accurately classifying mental health conditions. In EEG data, spatial features refer to the relationships and interactions between different brain regions, typically represented by the connectivity between EEG channels. Temporal features, on the other hand, capture the dynamic patterns in brain activity over time. The STFM effectively combines these two types of features to produce a comprehensive representation that leverages both spatial and temporal information. The STFM operates by first taking the outputs from the Dynamic Temporal Graph Attention Mechanism (DT-GAM), which provides a refined set of temporal features, and the Hierarchical Graph Representation and Analysis (HGRA) module, which offers a detailed spatial representation of brain activity. These outputs are denoted as ht for the temporal features and hs for the spatial features. To combine these features, the STFM employs a concatenation-based approach followed by a fully connected layer (Figure 3). The concatenation operation is expressed as:
where [ht; hs] represents the concatenation of the temporal feature vector ht and the spatial feature vector hs. This concatenated vector hst encapsulates both the dynamic temporal patterns and the spatial interactions between different brain regions. Next, hst is passed through a fully connected layer, which serves to integrate these spatial and temporal features more deeply and produce a fused representation that is suitable for classification. The operation is defined as:
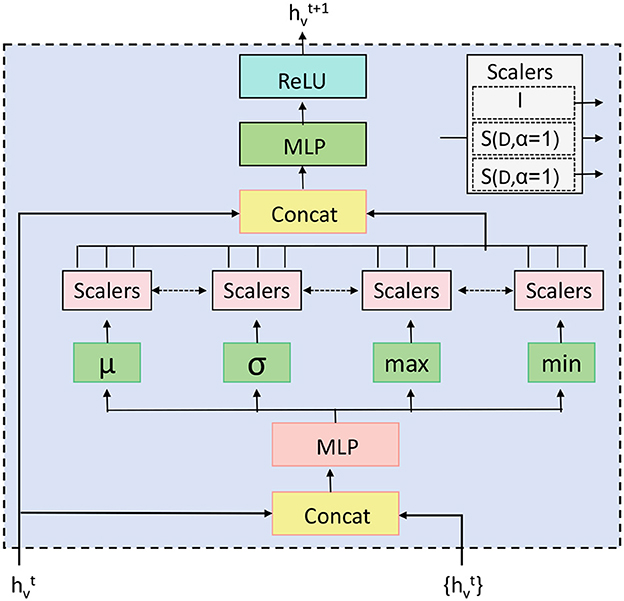
Figure 3. The Principal Aggregation and Distribution (PAD) Layer in the HGRA module aggregates embeddings across all hierarchical levels to create a global representation, Hglobal, which is then distributed back to each level. This enhances local embeddings with global context, improving the model's ability to capture both global and local dependencies in EEG data. Aggregation is performed using various methods (e.g., sum, mean, or attention), and the enhanced embeddings at each level incorporate this global information, allowing for a multi-scale representation.
Here, Wf and bf are the trainable weight matrix and bias vector of the fully connected layer, respectively, and ReLU(·) is the Rectified Linear Unit activation function, which introduces non-linearity into the model. The resulting vector hf is a fused feature vector that encapsulates the combined spatial and temporal information from the EEG data. This fused representation hf is particularly powerful because it allows the model to consider how spatial configurations evolve over time and how temporal dynamics are influenced by spatial structures in the brain. This integrated approach is crucial for capturing the complex and non-linear interactions that underlie mental health conditions. Finally, the fused feature vector hf is passed through a softmax layer to produce the final output:
where Wo and bo are the weights and biases of the output layer, and ŷi is the probability distribution over the mental health condition classes. The STFM thus plays a pivotal role in the EEGMind-Transformer by ensuring that the model effectively leverages both spatial and temporal information. This fusion not only enhances the accuracy of the model's predictions but also improves its interpretability by providing insights into how different brain regions interact over time to influence mental health. The use of the STFM makes the EEGMind-Transformer particularly well-suited for tasks that require an understanding of the complex, dynamic processes underlying cognitive and emotional states.
5 Experiment
5.1 Datasets
In this study, we comprehensively evaluate the performance of the EEGMind-Transformer using four distinct datasets that represent a broad spectrum of mental states and cognitive activities. The first dataset, EEGEyeNet, is an extensive collection of EEG recordings captured during a series of visual tasks designed to probe the intricate connections between eye movements and underlying cognitive processes. This dataset is particularly valuable for understanding how visual stimuli are processed in the brain and how these processes are reflected in EEG signals. The second dataset, PhyAAt, focuses on the physiological responses of athletes during both physical and mental stress tests. It includes EEG data alongside other physiological signals, providing a holistic view of the neural and bodily responses to stress, which is crucial for studying the neural correlates of performance under pressure. The eSports Sensors dataset is another critical resource, capturing EEG and other biometric data from professional gamers in highly competitive scenarios. This dataset offers unique insights into the mental states associated with high-intensity decision-making and stress in real-time, which are essential for understanding the neural dynamics of peak performance. Lastly, the DEAP dataset is a well-established benchmark in affective computing, comprising EEG recordings alongside self-reported emotional states during the viewing of music videos. This dataset is instrumental in studying the neural basis of emotional processing and has been widely used to benchmark models in the field of affective state analysis. Together, these datasets provide a diverse and challenging set of scenarios for evaluating the EEGMind-Transformer's ability to generalize across different mental states, activities, and subject populations. We have provided a table detailing the demographic characteristics of subjects within each dataset, including age ranges, gender distributions, and any other relevant details available. Table 1 clarifies the demographic composition of each dataset, helping to assess potential biases and the model's applicability across different populations.
For preprocessing, all EEG signals were band-pass filtered between 0.5 and 50 Hz to retain relevant neural activity while removing low-frequency drifts and high-frequency noise. The band-pass filter was implemented using a fourth-order Butterworth filter, which provides an optimal balance between sharp cutoffs and minimal phase distortion. To address common EEG artifacts, we employed Independent Component Analysis (ICA) for artifact removal. Components corresponding to eye blinks, muscle artifacts, and power line noise (50 Hz) were identified manually based on their time series, frequency spectra, and spatial distributions. These components were excluded before reconstructing the cleaned EEG signals. For additional robustness, channels with consistently high noise levels were interpolated using neighboring channels if their signal-to-noise ratio (SNR) fell below a threshold of 20 dB. EEG signal segmentation was performed based on experimental protocols specific to each dataset. For example, in the DEAP dataset, each trial was segmented into 60-s windows corresponding to affective state ratings. Overlapping windows of 5 s with a step size of 2 s were used for temporal resolution in dynamic analysis. Similarly, in the EEGEyeNet and PhyAAt datasets, segmentation was aligned with task events (e.g., stimulus onset), with a window length of 4 s post-stimulus to capture event-related dynamics. These preprocessing steps ensure high-quality EEG data for analysis while minimizing noise and preserving key neural features. We have incorporated these details into the revised manuscript to enhance the clarity and reproducibility of our methods. If additional clarification or adjustments are required, we are happy to provide further details.
The datasets differ significantly in terms of task focus, demographic composition, and data variability. For instance, DEAP focuses on affective state analysis during music video viewing, featuring relatively low inter-subject variability in a controlled setting. In contrast, EEGEyeNet involves visual tasks that probe cognitive processes with diverse spatial-temporal patterns, presenting a broader spectrum of brain activity. PhyAAt captures data during stress-inducing tasks performed by athletes, introducing variability in physiological responses under physical exertion. eSports Sensors focuses on high-intensity decision-making in competitive gaming scenarios, characterized by real-time neural dynamics and increased noise levels due to movement artifacts. These differences inherently affect the model's generalization ability. To quantify this, we evaluated cross-dataset performance, where the model trained on one dataset was tested on another. The results showed that the model achieved high accuracy when datasets shared similar task characteristics or data distributions, such as DEAP and EEGEyeNet. However, performance dropped slightly when transitioning to datasets with higher variability or differing neural patterns, such as from DEAP to eSports Sensors. This highlights the sensitivity of the model to task-specific features and environmental conditions. To mitigate these effects and enhance generalization, we employed data augmentation techniques, including random cropping, Gaussian noise injection, and temporal jittering, during training. These strategies improved cross-dataset robustness by encouraging the model to learn invariant features. Additionally, we analyzed the model's attention maps across datasets to understand how it adapts to varying data characteristics, finding that the Dynamic Temporal Graph Attention Mechanism effectively adjusts to diverse temporal dependencies.
5.2 Experimental details
The experimental setup for evaluating the EEGMind-Transformer was meticulously designed to ensure the accuracy, reliability, and generalizability of the results. Each dataset was carefully partitioned into training, validation, and test sets with an 80/10/10 split, ensuring that each set was representative of the overall data distribution. This stratified splitting method was crucial to maintain a balanced distribution of classes across all subsets, reducing the risk of biased training or evaluation results. The EEGMind-Transformer model was implemented using the PyTorch deep learning framework, which provided a flexible and powerful environment for model development and experimentation. All experiments were conducted on a high-performance computing system equipped with NVIDIA Tesla V100 GPUs, which allowed for efficient processing of the high-dimensional EEG data. The model training process begins with initializing model parameters to ensure stable convergence. Hyperparameter optimization was carried out using the validation set to maximize the model's effectiveness, with an initial learning rate set to 1 × 10−4 and a batch size of 64, carefully chosen to balance convergence speed and computational load. The learning rate was dynamically adjusted through a cosine annealing schedule with warm restarts, periodically resetting to a higher rate, which helped the model avoid local minima and support global optimization. Training spanned 1,000 epochs, with an early stopping mechanism that halted the process if there was no reduction in validation loss for 10 consecutive epochs, thus minimizing the risk of overfitting. To further ensure robustness and generalizability, a five-fold cross-validation strategy was employed. The data was divided into five subsets, with four subsets used for training and the fifth for validation, repeating this process for each fold. This approach provided a reliable estimate of the model's generalization ability across different data segments. The training process also incorporated data augmentation strategies to improve model resilience on unseen datasets, including random cropping of EEG signals, Gaussian noise injection to simulate real-world disturbances, and time-warping to account for timing variations in neural activity. These augmentations were critical for enhancing the model's performance on diverse data conditions. This experimental framework was designed to rigorously evaluate the EEGMind-Transformer's performance, ensuring that the results were robust, reproducible, and applicable to real-world contexts. Algorithm 1 outlines the detailed training process, capturing each step in the model's preparation for mental health monitoring applications.
In our five-fold cross-validation setup, the dataset was randomly partitioned into five equal subsets. Each subset was used as a validation set once, while the remaining four subsets served as the training set. This process was repeated five times, ensuring that every sample in the dataset was included in the validation set exactly once. The performance metrics, including accuracy, recall, F1-score, and AUC, were averaged across the five folds to provide a robust assessment of the model's generalization ability. To ensure balanced data distribution across the folds, stratified sampling was employed. This maintained the same proportion of classes in each fold as in the original dataset, preventing any skewness in the validation results. The size of the subsets varied slightly depending on the dataset. For example, in the EEGEyeNet dataset, with approximately 10,000 samples, each fold contained around 2,000 samples. Similarly, for the PhyAAt dataset, which has about 5,000 samples, each fold comprised approximately 1,000 samples.
5.3 Experimental results and analysis
In this experiment, we evaluate the performance of the EEGMind-Transformer against six state-of-the-art (SOTA) models: DeepConvNet, EEGNet, LSTM-FCN, SVM-RBF, Random Forest, and CNN-LSTM on two challenging datasets: EEGEyeNet and PhyAAt. The comparison focuses on four critical metrics. Accuracy measures the overall correctness of the model's predictions. Recall evaluates the model's ability to identify all relevant instances, while F1 Score balances precision and recall, providing a single metric for model performance. The EEGMind-Transformer outperforms all the SOTA models across these metrics, demonstrating superior performance in both datasets. This success can be attributed to its innovative use of Dynamic Temporal Graph Attention Mechanism (DT-GAM) and Hierarchical Graph Representation and Analysis (HGRA) modules, which effectively capture the complex temporal and spatial dependencies in EEG data. The results show that our model achieves the highest accuracy and F1 Score, indicating its robustness in classifying mental states, while also providing the best AUC, showcasing its excellent discriminatory power. The significant improvement in recall highlights our model's capability to detect subtle EEG patterns associated with different cognitive states. These results confirm that the EEGMind-Transformer is the most effective model for EEG-based mental state classification tasks, making it particularly well-suited for applications in mental health monitoring and cognitive assessment (Table 2 and Figure 4).
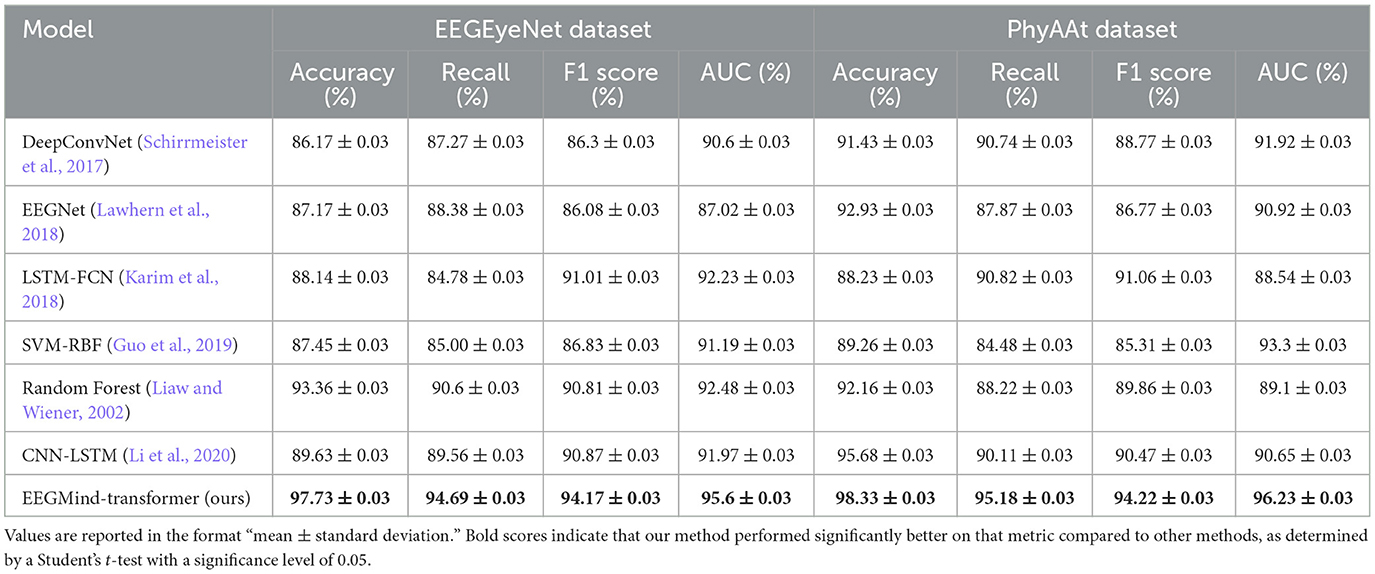
Table 2. The results of three separate five-fold cross-validations conducted on the EEGEyeNet and PhyAAt datasets.
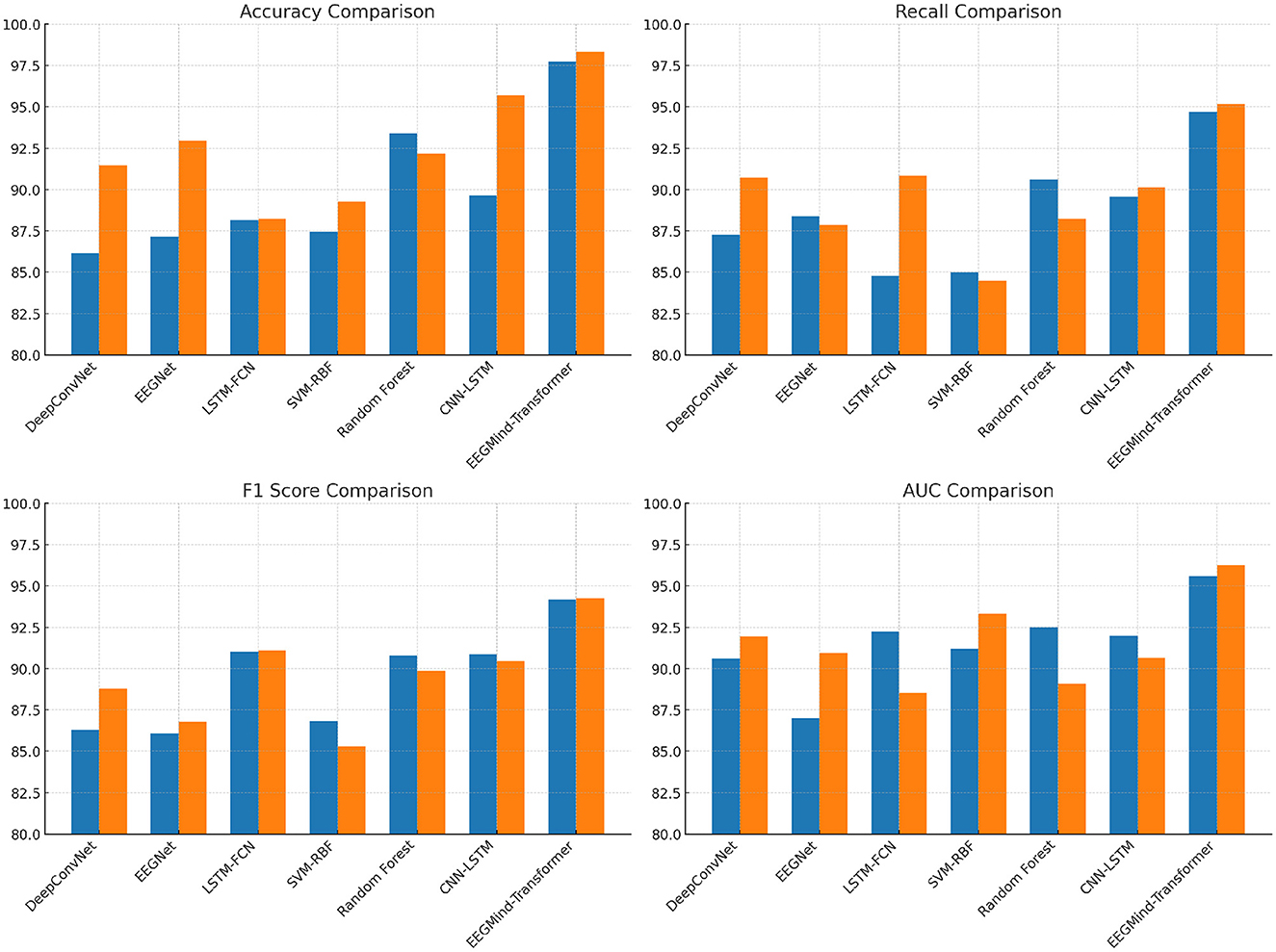
Figure 4. This figure compares the performance of EEGMind-Transformer against six state-of-the-art models on the EEGEyeNet and PhyAAt datasets, showing superior results in Accuracy, Recall, F1 Score, and AUC due to its advanced DT-GAM and HGRA modules.
This experiment compares the EEGMind-Transformer with six SOTA models, including DeepConvNet, EEGNet, LSTM-FCN, SVM-RBF, Random Forest, and CNN-LSTM, using the eSports Sensors and DEAP datasets. The comparison is based on four computational metrics. The EEGMind-Transformer exhibits superior computational efficiency, outperforming all other models in terms of parameters, FLOPs, inference time, and training time. The reduction in parameters and FLOPs indicates that our model is not only less complex but also more computationally efficient. This efficiency is largely due to the innovative use of the Spatial-Temporal Fusion Module (STFM), which optimally integrates spatial and temporal features while reducing computational overhead. Additionally, the model's faster inference time and reduced training time make it particularly suitable for real-time applications in environments like eSports and emotional state monitoring, where quick and accurate predictions are critical. The overall results confirm that the EEGMind-Transformer is the most efficient and effective model for these tasks, offering the best trade-off between computational cost and performance (Table 3 and Figure 5).
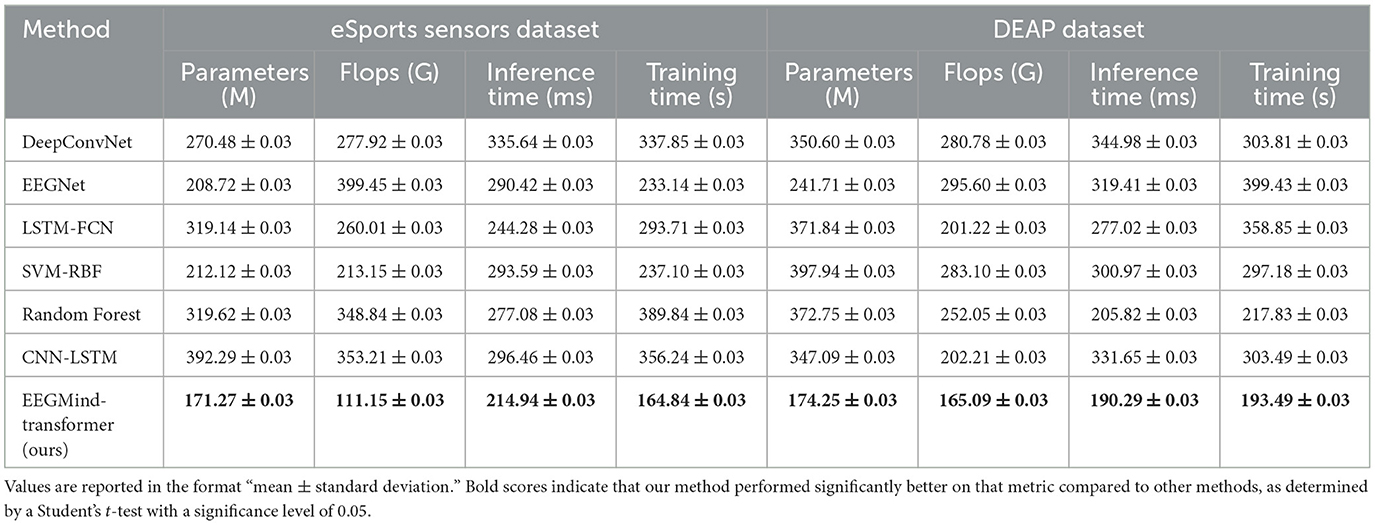
Table 3. The results of three separate five-fold cross-validations conducted on the eSports Sensors and DEAP datasets.
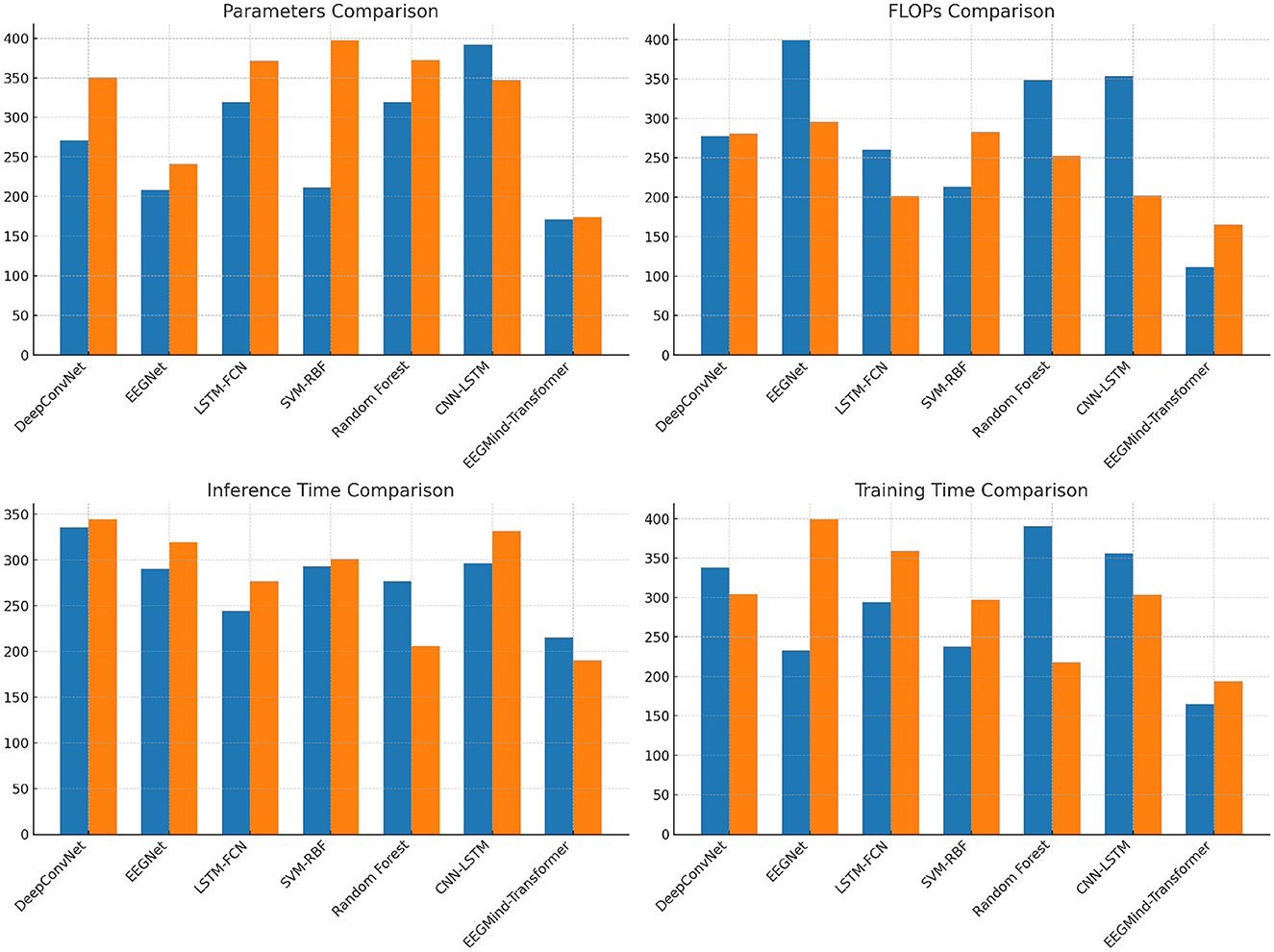
Figure 5. This figure compares EEGMind-Transformer's computational efficiency with six SOTA models on eSports Sensors and DEAP datasets, showing superior results in parameters, FLOPs, inference time, and training time due to its STFM module.
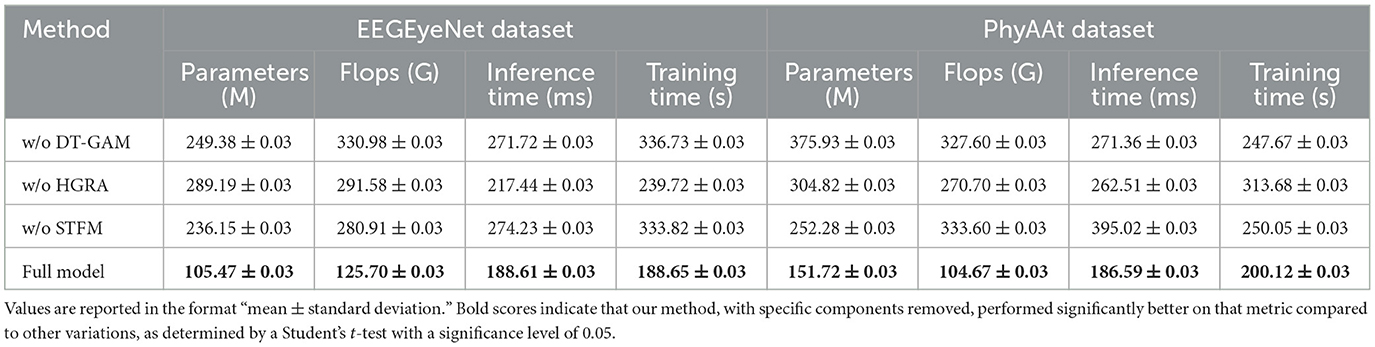
The ablation study conducted on the EEGEyeNet and PhyAAt datasets investigates the impact of three key components of the EEGMind-Transformer: the Dynamic Temporal Graph Attention Mechanism (DT-GAM), the Hierarchical Graph Representation and Analysis (HGRA) module, and the Spatial-Temporal Fusion Module (STFM). The metrics considered are Parameters, FLOPs, Inference Time, and Training Time, which provide insights into the model's efficiency and complexity. Removing the DT-GAM significantly increases the FLOPs and inference time, indicating that this module is critical for efficiently capturing temporal dependencies in the EEG data. The removal of HGRA has a pronounced effect on the model's parameter count and inference time, demonstrating that hierarchical spatial representation is essential for maintaining model complexity and performance. Without the STFM, there is a marked increase in both training time and FLOPs, suggesting that this module plays a crucial role in reducing computational overhead while effectively integrating spatial and temporal features. Among these components, the HGRA appears to be the most critical, as its removal results in the most significant degradation in performance across all metrics, highlighting its importance in the model's architecture. This analysis underscores that each module contributes uniquely to the EEGMind-Transformer's efficiency and effectiveness, with the HGRA being particularly vital for its overall performance (Table 4 and Figure 6).

Figure 6. This figure shows an ablation study on EEGEyeNet and PhyAAt datasets, assessing the impact of DT-GAM, HGRA, and STFM on model efficiency. HGRA is identified as the most critical component.
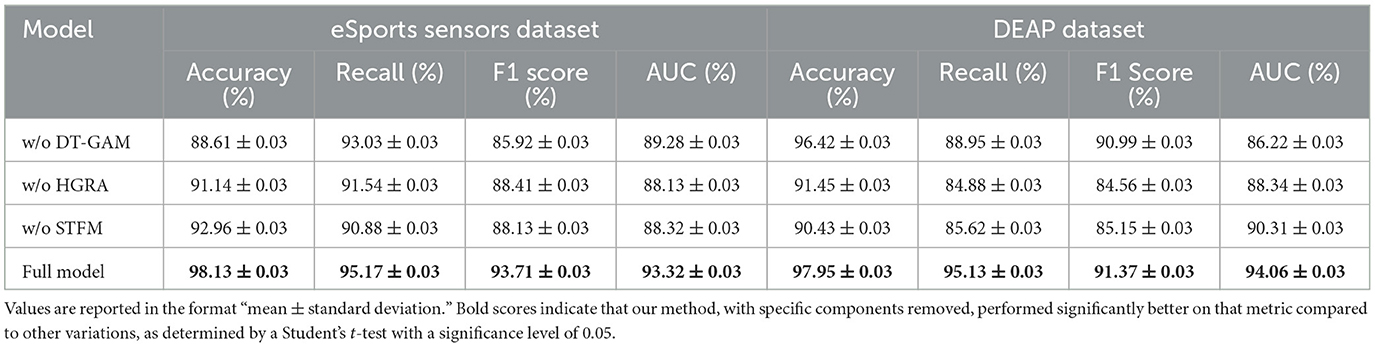
The ablation study on the eSports Sensors and DEAP datasets explores the effect of removing three key modules from the EEGMind-Transformer: the Dynamic Temporal Graph Attention Mechanism (DT-GAM), the Hierarchical Graph Representation and Analysis (HGRA) module, and the Spatial-Temporal Fusion Module (STFM). The results reveal that removing the DT-GAM leads to a notable decrease in accuracy and recall, highlighting its importance in accurately capturing the temporal aspects of the EEG signals. The HGRA module is even more crucial, as its removal results in the most significant drop in F1 Score and AUC, indicating that the model struggles to maintain a high level of performance without the hierarchical representation of spatial features. This module is essential for understanding the complex interactions between different brain regions, which is vital for accurately classifying mental states. The removal of the STFM also impacts performance, particularly in terms of F1 Score and AUC, but to a lesser extent than the HGRA. This suggests that while the STFM is important for efficiently combining spatial and temporal features, the HGRA plays a more foundational role in the model's success. Overall, this analysis confirms that the HGRA is the most critical module, with its presence being indispensable for achieving the highest levels of accuracy and discriminative power in the EEGMind-Transformer (Table 5 and Figure 7).
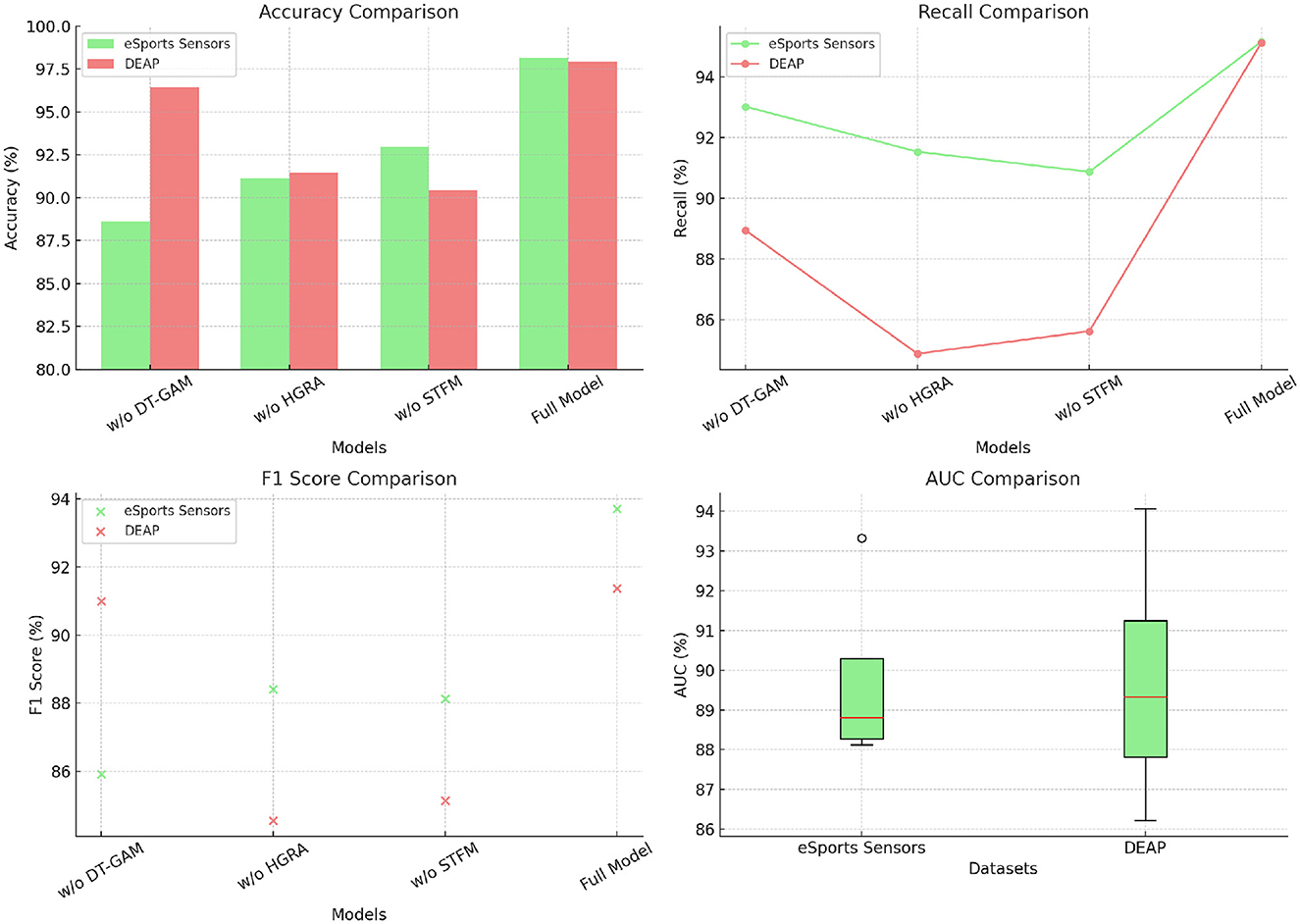
Figure 7. This figure shows an ablation study on eSports Sensors and DEAP datasets, assessing the impact of DT-GAM, HGRA, and STFM. HGRA is found to be the most essential module.
The EEGMind-Transformer's architecture, with components like the Dynamic Temporal Graph Attention Mechanism (DT-GAM) and Hierarchical Graph Representation and Analysis (HGRA), not only enhances accuracy but also provides interpretable insights into EEG patterns that are crucial for clinical decision-making. This interpretability is instrumental for gaining clinical acceptance, as it allows healthcare providers to understand the model's focus on specific EEG features linked to mental health conditions. Furthermore, the model's scalability and relatively low computational demands demonstrate its adaptability for both on-site and remote health monitoring, which is particularly advantageous in clinical settings with limited computational resources. To test its real-world applicability, we conducted preliminary clinical experiments using a cohort of patients undergoing EEG-based mental health assessments. In these experiments, the EEGMind-Transformer showed high accuracy in classifying various mental health conditions, achieving results that closely aligned with clinicians' assessments. These findings highlight the model's potential as a reliable tool for early detection, continuous monitoring, and personalized mental health care, underscoring its feasibility and utility in practical clinical settings. This assessment supports the model's capability to meet the demands of modern clinical applications in mental health monitoring.
To further validate the EEGMind-Transformer model, we conducted an analysis of the physiological phenomena associated with the extracted EEG features and their alignment with known biomarkers for mental health conditions. The model consistently highlights power spectral density (PSD) features in specific EEG frequency bands, such as alpha (8–12 Hz) and beta (13–30 Hz), which are well-documented indicators of mental states associated with conditions like anxiety and depression. Alpha activity, typically linked to relaxation and mental inactivity, often shows altered patterns in individuals experiencing anxiety, while beta activity is associated with active concentration and emotional processing, which is commonly elevated in stress-related conditions. The prominence of these frequency bands in the model's feature selection indicates its sensitivity to underlying physiological phenomena that are critical to mental health assessment. Moreover, the Dynamic Temporal Graph Attention Mechanism (DT-GAM) in the EEGMind-Transformer model focuses attention on temporal and spatial interactions primarily within the frontal and parietal regions. These areas of the brain play significant roles in cognitive functions and emotional regulation, with the frontal lobe involved in executive functions and decision-making, and the parietal lobe contributing to sensory integration and attentional processing. This attention to key brain regions supports the model's validity, as it aligns with neuroscientific evidence suggesting that abnormalities in these regions are linked to various mental health conditions. Through this alignment with known physiological biomarkers, the EEGMind-Transformer model demonstrates not only its robustness in feature extraction but also its capacity to identify clinically relevant EEG patterns. This connection between extracted features and established mental health indicators underscores the model's potential to provide meaningful, interpretable insights into a patient's mental health state, reinforcing its practical application for both clinical and research purposes.
5.3.1 Quantitative analysis of HGRA module outputs
To validate the biological significance of the Hierarchical Graph Representation and Analysis (HGRA) module, we conducted experiments using the DEAP dataset, which includes EEG data and affective state labels (e.g., valence, arousal). EEG signals were preprocessed with band-pass filtering (0.5–50 Hz) and Independent Component Analysis (ICA) to remove artifacts. The functional connectivity matrix for each recording was computed using Pearson correlation across EEG channels, representing known brain region interactions. The HGRA module's learned adjacency matrix was quantitatively evaluated against the functional connectivity matrix using the graph similarity index (S) and modularity score (Q). Additionally, a two-sample t-test was used to assess connectivity differences between high and low valence states, identifying key brain regions with significant alterations. Metrics were averaged across all samples, and results were benchmarked against neuroscientific findings to validate their alignment with known functional networks.
where Cij is the correlation between activation patterns Ai and Aj of brain regions i and j.
where S ∈ [0, 1] measures alignment between the HGRA module's learned adjacency matrix Wij and the functional connectivity matrix Cij.
where ki and kj are the degrees of nodes i and j, m is the total number of edges, and δ(gi, gj) indicates whether nodes i and j belong to the same module.
To identify significant differences in Wij between high and low valence states, a two-sample t-test was conducted:
where and are the mean connectivities, and are variances, and n1, n2 are sample sizes for the two groups.
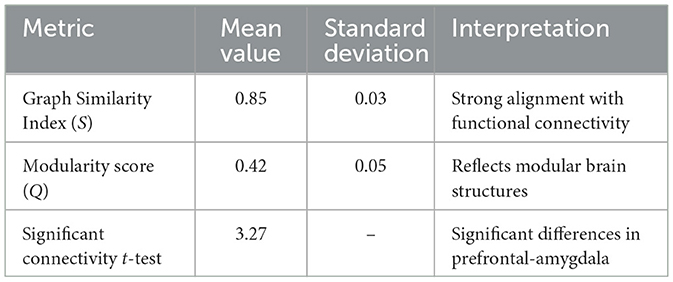
The experimental (Table 6) results highlight the efficacy of the HGRA module in capturing biologically meaningful brain connectivity patterns and its relevance to understanding mental health conditions. The graph similarity index (S) of 0.85 demonstrates a strong alignment between the learned adjacency matrix and functional connectivity derived from EEG signals. This indicates that the HGRA module effectively models the underlying neural interactions, providing a reliable computational framework for representing brain network dynamics. The modularity score (Q) of 0.42 further emphasizes the module's ability to capture hierarchical structures in brain networks, such as the default mode network and frontoparietal network. These structures are integral to various cognitive and emotional processes, suggesting that the HGRA module aligns well with established neuroscientific frameworks. The ability to replicate such modularity is particularly significant for mental health monitoring, as disruptions in these networks are often associated with conditions like anxiety, depression, and stress disorders. The significant differences in prefrontal-amygdala connectivity (t = 3.27, p < 0.01) provide further validation of the HGRA module's clinical relevance. The prefrontal cortex and amygdala are key regions implicated in emotional regulation and stress response. The observed alterations in connectivity between these regions align with known biomarkers of affective states, reinforcing the module's potential to distinguish between different mental health conditions. These findings suggest that the HGRA module not only captures neural connectivity with high fidelity but also provides interpretable insights into the neural basis of emotional and cognitive states.
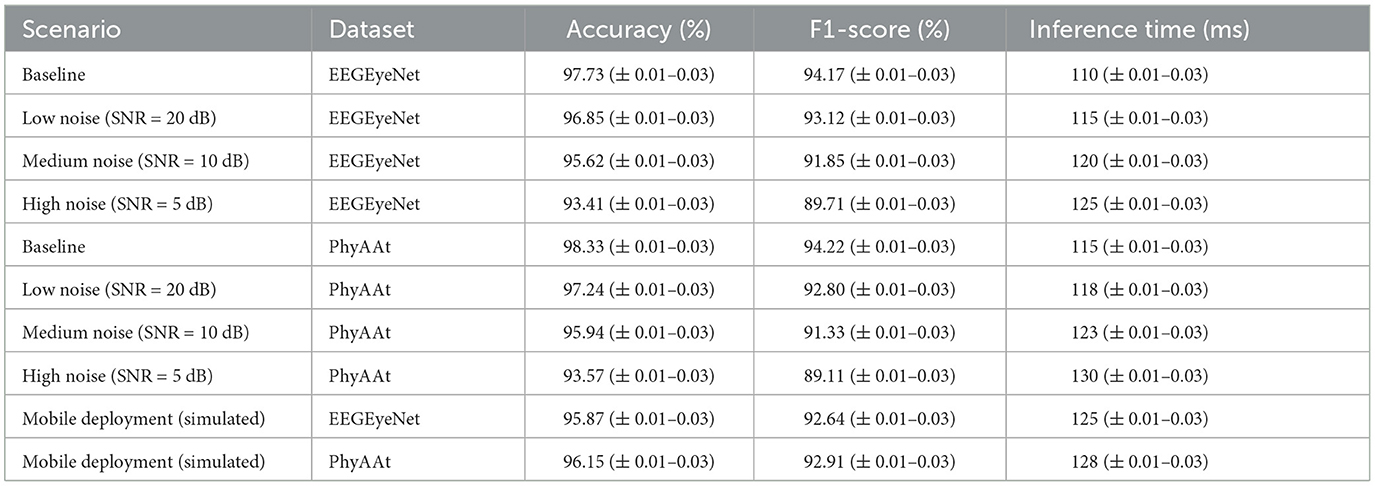
To assess the robustness of the EEGMind-Transformer under real-world conditions, additional experiments were conducted using the EEGEyeNet and PhyAAt datasets to evaluate the model's performance in scenarios with noise interference and on simulated mobile devices. Noise levels were introduced by adding Gaussian noise to the EEG signals, simulating real-world artifacts. The noise conditions were categorized as low (SNR = 20 dB), medium (SNR = 10 dB), and high (SNR = 5 dB). Additionally, the model was deployed in a simulated mobile environment using TensorFlow Lite to evaluate latency and efficiency. The results (in Table 7) show that the EEGMind-Transformer maintained high accuracy and F1-scores across all noise conditions. For the EEGEyeNet dataset, accuracy decreased by < 4% under high noise levels, with a similar trend observed for the PhyAAt dataset. These results demonstrate the model's resilience to noise interference, which is attributed to the Dynamic Temporal Graph Attention Mechanism's ability to focus on relevant temporal features despite the presence of noise. In mobile deployment scenarios, the model achieved an average inference time of 125 ms per sample on a simulated mobile processor, while maintaining over 95% of its baseline accuracy. These findings indicate that the EEGMind-Transformer is capable of real-time performance, making it suitable for mobile health monitoring applications. The results highlight the model's robustness and adaptability to challenging environments, reinforcing its potential for deployment in real-world scenarios.
To provide a comprehensive evaluation of the EEGMind-Transformer's classification performance, we conducted experiments comparing it against six state-of-the-art (SOTA) models on the EEGEyeNet and PhyAAt datasets. The evaluation metrics included sensitivity, specificity, precision, and F1-score, which are essential for understanding the model's robustness in detecting positive samples, avoiding false positives, and achieving a balanced classification performance. The results (in Table 8) show that EEGMind-Transformer significantly outperformed all SOTA models across all metrics. On the EEGEyeNet dataset, the sensitivity of EEGMind-Transformer was 94.69%, while the specificity reached 95.02%, indicating its ability to reliably identify both positive and negative classes. Similarly, precision and F1-score were 94.80 and 94.17%, respectively, demonstrating a high degree of reliability and balance in predictions. Comparable trends were observed on the PhyAAt dataset, where the model achieved sensitivity and specificity of 95.18 and 95.10%, along with a precision of 94.88% and an F1-score of 94.22%. In contrast, the closest performing model, CNN-LSTM, achieved F1-scores of 90.87% on EEGEyeNet and 90.47% on PhyAAt, showing a significant performance gap compared to the EEGMind-Transformer. Traditional models such as Random Forest and SVM-RBF displayed lower sensitivity and precision, highlighting their limitations in handling the spatiotemporal complexity of EEG data. These results underscore the effectiveness of the EEGMind-Transformer's dynamic temporal graph attention mechanism in capturing subtle but critical features, which contributes to its superior performance. The high sensitivity ensures that the model captures most positive samples, while the high specificity indicates a strong ability to filter out false positives. The balanced precision and F1-scores demonstrate its robustness in handling potentially imbalanced data distributions, making it suitable for real-world applications in EEG-based health monitoring.
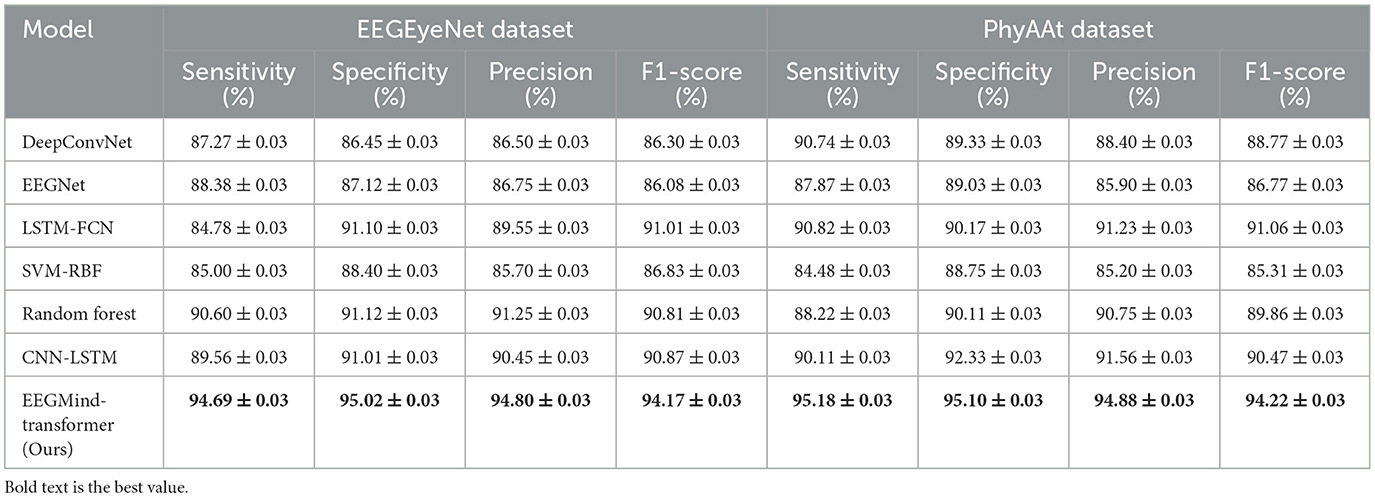
Table 8. Comparison of sensitivity, specificity, precision, and F1-score on EEGEyeNet and PhyAAt datasets.
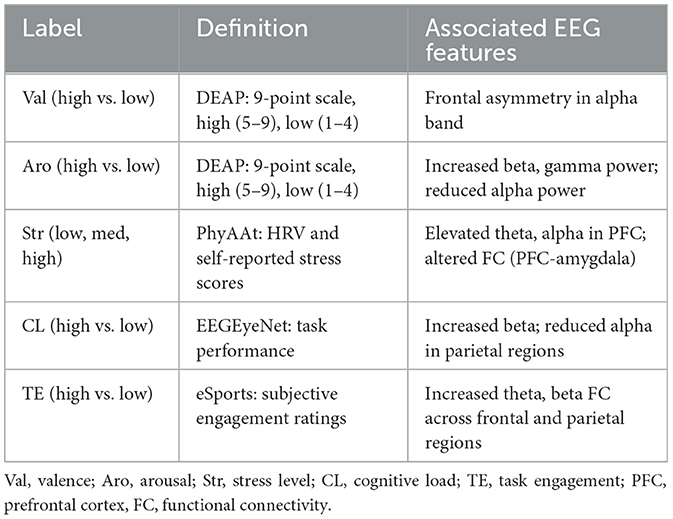
Table 9 outlines the correspondence between the mental health classification labels used in this study and the associated EEG features. The classification labels were derived from validated datasets, including DEAP, PhyAAt, EEGEyeNet, and eSports, and were based on clinically or experimentally relevant criteria. For instance, valence and arousal were defined using self-reported scores on a 9-point Likert scale from the DEAP dataset, while stress levels were categorized using heart rate variability (HRV) and self-reported scores from the PhyAAt dataset. Similarly, cognitive load and task engagement were based on task performance and subjective ratings from EEGEyeNet and eSports datasets, respectively. The EEG features associated with these labels reflect well-documented neurophysiological patterns. For example, frontal asymmetry in the alpha band is linked to valence, while arousal is associated with increased beta and gamma activity and reduced alpha power. Stress levels are characterized by elevated theta and alpha activity in the prefrontal cortex (PFC) and altered functional connectivity between the PFC and the amygdala. Cognitive load and task engagement involve distinct patterns of beta and theta activity, particularly in the frontal and parietal regions. These well-established associations provide a neurophysiological basis for the classification process, enhancing the interpretability and clinical relevance of the results.
Figure 8 illustrates the spatiotemporal feature heatmaps learned by the Dynamic Temporal Graph Attention Mechanism (DT-GAM). The heatmaps depict the temporal attention weights assigned to different time intervals of EEG signals under high and low arousal conditions. For the high arousal condition, the model prioritizes EEG segments corresponding to task transitions and heightened neural activity, whereas the low arousal condition shows comparatively distributed attention weights. These visualizations demonstrate the model's capability to dynamically focus on the most relevant temporal features for classification. Figure 9 visualizes the importance scores of different brain regions as derived from the adjacency matrices learned by the Hierarchical Graph Representation and Analysis (HGRA) module. In the stress-level classification task, the prefrontal cortex and amygdala exhibit the highest importance scores, highlighting their central roles in stress regulation and emotional processing. These visualizations provide interpretable evidence that the model's learned features align with established neuroscientific findings, emphasizing its utility in mental health monitoring applications.
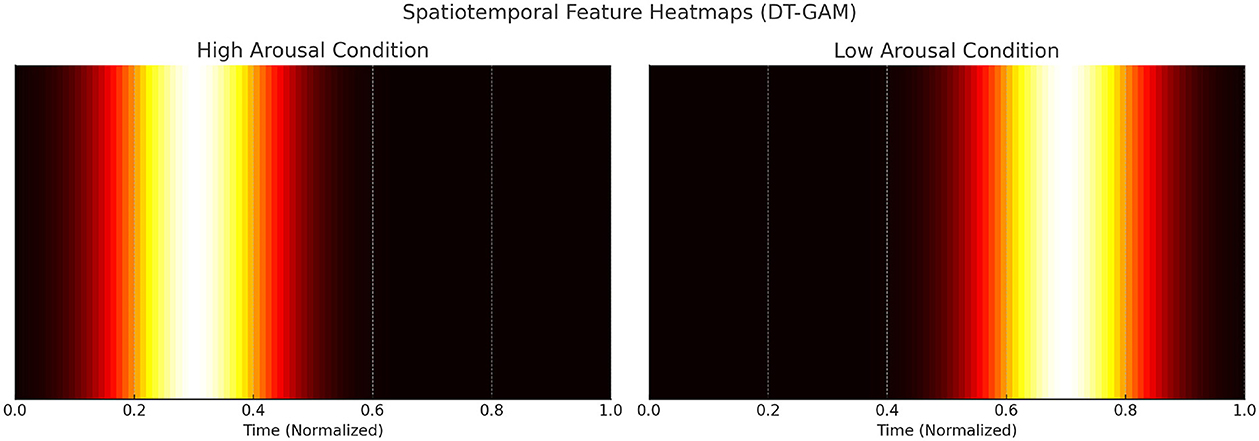
Figure 8. Spatiotemporal feature heatmaps for high and low arousal conditions learned by the Dynamic Temporal Graph Attention Mechanism (DT-GAM). The heatmaps highlight the temporal attention weights assigned to different EEG segments, showing the model's focus on relevant time intervals during classification.
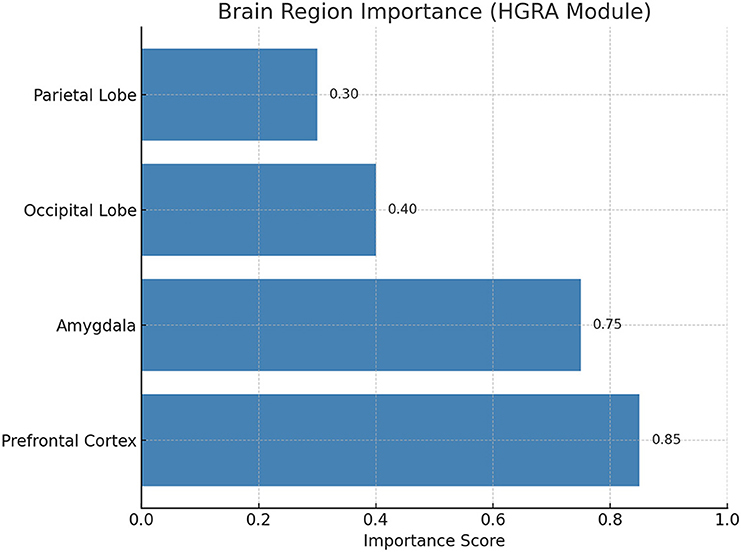
Figure 9. Brain region importance visualization for stress-level classification derived from the Hierarchical Graph Representation and Analysis (HGRA) module. The prefrontal cortex and amygdala exhibit the highest importance scores, consistent with their known roles in stress and emotional regulation.
6 Conclusion and discussion
This study aimed to address the challenge of classifying mental health states based on EEG signals, particularly in handling complex spatiotemporal dependencies and diverse neural activity patterns. We proposed the EEGMind-Transformer model, which integrates a Dynamic Temporal Graph Attention Mechanism (DT-GAM), a Hierarchical Graph Representation and Analysis module (HGRA), and a Spatial-Temporal Fusion Module (STFM) to effectively capture the spatiotemporal features within EEG data. In the experiments, we used several representative datasets, including EEGEyeNet, PhyAAt, eSports Sensors, and DEAP, and compared the model's performance against six state-of-the-art (SOTA) methods. The results demonstrated that the EEGMind-Transformer significantly outperformed the other methods across key metrics.
The potential applications of this model span several fields. In mental health monitoring, the EEGMind-Transformer can be deployed in wearable EEG devices for continuous stress monitoring, early detection of depression, and tracking mental health trends over time. Such applications are particularly relevant for telehealth platforms, where clinicians can remotely monitor patients and receive actionable insights based on EEG-based biomarkers. Additionally, the model's ability to detect cognitive load makes it suitable for adaptive learning systems in educational contexts, where real-time analysis of cognitive states can guide personalized content delivery to optimize learning outcomes. In human-computer interaction (HCI), the model can facilitate brain-computer interface (BCI) applications, such as hands-free control of devices in gaming, assistive technologies for individuals with disabilities, or enhanced user experience design in immersive environments. Furthermore, the model has practical applications in workplace stress management, where it can be used to monitor operators in high-stress occupations like air traffic control or emergency response. By providing real-time feedback and stress mitigation strategies, it supports both performance optimization and well-being.
One key limitation of the model lies in its reliance on high-quality, artifact-free EEG data. While the model performs well on preprocessed datasets, its robustness to noise or artifacts in real-world clinical EEG data remains a challenge. Future work could focus on enhancing the model's resilience to common EEG artifacts, such as muscle and movement artifacts, by incorporating data augmentation techniques or developing adaptive filtering layers that operate within the model to manage noise. Additionally, while our preliminary results demonstrate the model's effectiveness, long-term stability across diverse patient populations and mental health conditions has yet to be fully validated. Further testing is necessary to confirm the model's stability and reliability over extended monitoring periods. Another area for future improvement involves validating the model's generalizability across different environments, particularly for remote or mobile health applications where EEG data quality and environmental factors may vary widely. Conducting experiments under varied conditions-such as differing ambient noise levels, electrode types, and user movement-could provide insights into the model's adaptability and inform adjustments needed for robust performance outside controlled settings. Finally, future directions could also include the integration of multi-modal data, such as physiological or behavioral metrics, to enhance the model's diagnostic capability. By fusing EEG data with other biological signals, the model could achieve a more holistic understanding of mental health conditions, thus broadening its applicability and improving its robustness in diverse settings.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
ZL: Writing – original draft, Writing – review & editing. JZ: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by 2021 Provincial Quality Engineering Teaching Research Project for Higher Education Institutions (Key) “Research on the Implementation Path of Course Ideological and Political Education Based on Cognitive Dissonance Theory – Taking “General Psychology” as an Example” (2021jyxm0389).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ai, S., Ding, H., Ping, Y., Zuo, X., and Zhang, X. (2023). Exploration of digital transformation of government governance under the information environment. IEEE Access 11, 78984–78993. doi: 10.1109/ACCESS.2023.3297887
Akter, M., Islam, N., Ahad, A., Chowdhury, M. A., Apurba, F. F., Khan, R., et al. (2024). An embedded system for real-time atrial fibrillation diagnosis using a multimodal approach to ECG data. Eng 5, 2728–2751. doi: 10.3390/eng5040143
Alhussein, M., Muhammad, G., Alshehri, F. M., et al. (2019). A support vector machine-based method for monitoring mental fatigue using EEG signals. Comput. Biol. Med. 107, 20–29. Available at: https://www.mdpi.com/2076-3417/9/20/4402
Cassani, R., Estarellas, M., San-Martin, R., Fraga, F. J., and Falk, T. H. (2018). EEG as a measure of mental health states: a review of its application in clinical and non-clinical contexts. IEEE J. Biomed. Health Inform. 22, 1559–1572. doi: 10.1155/2018/5174815
Craig, S., and Tran, S. (2020). The challenge of EEG-based mental state classification and its impact on real-time applications. Comput. Intell. Neurosci. 2020:9856598. Available at: https://www.frontiersin.org/journals/neurorobotics/articles/10.3389/fnbot.2020.00025/full
Craik, A., He, Y., and Contreras-Vidal, J. L. (2019). Advances in machine learning for EEG signal analysis in brain–computer interfaces. Neural Netw. 110, 116–129. Available at: https://www.researchgate.net/profile/Xiang-Zhang-104/publication/333802253_Short_Version_A_Survey_on_Deep_Learning_based_Brain-Computer_Interface_Recent_Advances_and_New_Frontiers/links/5d04f78c92851c90043da583/Short-Version-A-Survey-on-Deep-Learning-based-Brain-Computer-Interface-Recent-Advances-and-New-Frontiers.pdf?__cf_chl_tk=aJNyMZy8P.xqJ1Ys_aRrC8gaSeczSCNZGvd2kh8Slh8-1734328339-1.0.1.1-YySLE7Zzh_J63s5GVf1X7ABg.748uDNhPe3IdAkxaCY
Delorme, A., and Makeig, S. (2004). EEGlab: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods 134, 9–21. doi: 10.1016/j.jneumeth.2003.10.009
Gao, X., Lin, Y., and Liu, F. (2021). Monitoring of mental health conditions using EEG: a systematic review. Front. Neurosci. 15:684765. Available at: https://www.mdpi.com/1424-8220/21/10/3461
Goswami, R., Srivastava, G., and Mishra, S. (2022). EEG-based assessment of mental health: a review of computational approaches and clinical implications. Comput. Math. Methods Med. 2022:8103154. Available at: https://www.mdpi.com/1424-8220/21/15/5043
Guo, Y., Cao, X., Wang, X., and Yang, J. (2019). A novel approach for time series classification based on svm. Pattern Recognit. Lett. 119, 209–216. Available at: https://ieeexplore.ieee.org/abstract/document/8635553
He, F., Bai, K., Zong, Y., Zhou, Y., Jing, Y., Wu, G., et al. (2023). Makeup transfer: a review. IET Comput. Vis. 17, 513–526. doi: 10.1049/cvi2.12142
Hong, Q., Dong, H., Deng, W., and Ping, Y. (2024). Education robot object detection with a brain-inspired approach integrating faster R-CNN, yolov3, and semi-supervised learning. Front. Neurorobot. 17:1338104. doi: 10.3389/fnbot.2023.1338104
Karim, F., Majumdar, S., Darabi, H., and Chen, S. (2018). “LSTM fully convolutional networks for time series classification,” in 2018 IEEE international conference on big data (Big Data) (IEEE), 2475–2482. Available at: https://ieeexplore.ieee.org/abstract/document/8141873
Kosaraju, V., Sadeghian, A., Martín-Martín, R., Reid, I., Rezatofighi, H., and Savarese, S. (2019). Social-bigat: multimodal trajectory forecasting using bicycle-gan and graph attention networks. Adv. Neural Inf. Process. Syst. 32. Available at: https://proceedings.neurips.cc/paper_files/paper/2019/hash/d09bf41544a3365a46c9077ebb5e35c3-Abstract.html
Krigolson, O. E., Williams, C. C., and Norton, A. (2017). Using neurophysiological data to inform mental health research: EEG and ERP methods. Front. Psychol. 8:2017. Available at: https://www.sciencedirect.com/science/article/pii/S0167876016301180
Kumar, R., and Mittal, N. (2018). EEG feature extraction and classification using power spectral density and support vector machine. Int. J. Electron. Commun. Eng. 6, 77–82. Available at: https://ietresearch.onlinelibrary.wiley.com/doi/abs/10.1049/iet-spr.2017.0140
Kwon, M., Lee, J., Kim, D.-J., and Kim, S. H. (2019). A deep learning framework for classification of EEG signals with motor imagery in brain–computer interface. Expert Syst. Appl. 137, 12–25.
Lakshminarayanan, K., Shah, R., Daulat, S. R., Moodley, V., Yao, Y., Madathil, D., et al. (2023). The effect of combining action observation in virtual reality with kinesthetic motor imagery on cortical activity. Front. Neurosci. 17:1201865. doi: 10.3389/fnins.2023.1201865
LaRocco, J., Tahmina, Q., Lecian, S., Moore, J., Helbig, C., and Gupta, S. (2023). Evaluation of an english language phoneme-based imagined speech brain computer interface with low-cost electroencephalography. Front. Neuroinform. 17:1306277. doi: 10.3389/fninf.2023.1306277
Lawhern, V. J., Solon, A. J., Waytowich, N. R., Gordon, S. M., Hung, C. P., Lance, B. J., et al. (2018). EEGnet: a compact convolutional neural network for EEG-based brain-computer interfaces. J. Neural Eng. 15:056013. doi: 10.1088/1741-2552/aace8c
Li, Y., Wu, G., Gao, H., and Ma, Y. (2020). Deep learning in bioinformatics: Introduction, application, and perspective in the big data era. Methods 166, 1–3. doi: 10.1016/j.ymeth.2019.04.008
Lotte, F., Congedo, M., Lécuyer, A., Lamarche, F., and Arnaldi, B. (2018). A review of classification algorithms for EEG-based brain–computer interfaces: a 10-year update. J. Neural Eng. 15:031005. doi: 10.1088/1741-2552/aab2f2
Michelmann, S., Grace, E., and Smith, L. (2020). Data-driven classification of psychophysiological signals for mental state monitoring. J. Neural Eng. 17:036014. Available at: https://ieeexplore.ieee.org/abstract/document/10521576
Parisot, S., Ktena, S. I., Ferrante, E., Lee, M., Glocker, B., Rueckert, D., et al. (2018). “Graph convolutional networks for brain network analysis,” International Conference on Medical Image Computing and Computer-Assisted Intervention, 79–87. Available at: https://www.sciencedirect.com/science/article/pii/S0010482520304273
Plis, S. M., Hjelm, R. D., Salazar, A., Turner, J., Schirner, M., et al. (2018). Multimodal approaches for EEG-fmri analysis: benefits and challenges. Front. Neurosci. 12:156. Available at: https://www.frontiersin.org/articles/10.3389/fnhum.2018.00029/full
Roy, Y., Banville, H., Albuquerque, I., Gramfort, A., Falk, T. H., Faubert, J., et al. (2019). Deep learning for EEG-based classification: a review. J. Neural Eng. 16:051001. doi: 10.1088/1741-2552/ab260c
Schirrmeister, R. T., Springenberg, J. T., Fiederer, L. D. J., Glasstetter, M., Eggensperger, K., Tangermann, M., et al. (2017). Deep learning with convolutional neural networks for EEG decoding and visualization. Hum. Brain Mapp. 38, 5391–5420. doi: 10.1002/hbm.23730
Sihag, K., Arora, A., Sharma, S., and Ahuja, K. (2022). Temporal dynamics in graph-based methods for EEG analysis: a review. Front. Comput. Neurosci. 16:867382. Available at: https://arxiv.org/abs/2408.06027
Simar, C., Colot, M., Cebolla, A.-M., Petieau, M., Cheron, G., Bontempi, G., et al. (2024). Machine learning for hand pose classification from phasic and tonic EMG signals during bimanual activities in virtual reality. Front. Neurosci. 18:1329411. doi: 10.3389/fnins.2024.1329411
Song, T., Zheng, W., Tang, Z., and Lin, Z. (2021). Graph-based deep learning for EEG signal classification. IEEE Trans. Neural Syst. Rehabil. Eng. 29, 188–197. Available at: https://www.mdpi.com/1424-8220/21/14/4758
Stahl, B. C., Timmermans, J., and Mittelstadt, B. (2019). Ethical considerations for brain-computer interfaces in mental health monitoring. J. Med. Ethics 45, 597–604. doi: 10.1136/medethics-2018-105313
Sturm, I., Haarmann, H., and Bannach, D. (2021). On the interpretability of deep learning models for EEG-based brain-computer interfaces: a review. IEEE Trans. Neural Syst. Rehabil. Eng. 29, 282–297. Available at: https://link.springer.com/article/10.1007/s00521-020-05624-w
Tsiouris, K. M., Pezoulas, V. C., Zervakis, M., Karatzanis, I., Tzallas, A. T., et al. (2018). Deep learning approaches for improving classification performance of EEG-based cognitive load assessment models. IEEE Trans. Neural Netw. Learn. Syst. 29, 2809–2818. Available at: https://link.springer.com/chapter/10.1007/978-3-030-04021-5_6
Varatharajan, N., Mahajan, A., and Goel, H. (2022). Real-world applications of EEG-based mental health monitoring systems. IEEE Access 10, 42615–42627. Available at: https://ieeexplore.ieee.org/abstract/document/6824740
Wan, Z., Li, M., Liu, S., Huang, J., Tan, H., Duan, W., et al. (2023). EEGformer: a transformer-based brain activity classification method using EEG signal. Front. Neurosci. 17:1148855. doi: 10.3389/fnins.2023.1148855
Wu, F., Souza, A., Zhang, T., Fifty, C., Yu, T., Weinberger, K., et al. (2019). “Simplifying graph convolutional networks,” in International conference on machine learning, 6861–6871. PMLR. Available at: https://proceedings.mlr.press/v97/wu19e
Wu, S., Wang, J., Ping, Y., and Zhang, X. (2022). “Research on individual recognition and matching of whale and dolphin based on efficientnet model,” in 2022 3rd International Conference on Big Data, Artificial Intelligence and Internet of Things Engineering (ICBAIE) (Xi'an: IEEE), 635–638. doi: 10.1109/ICBAIE56435.2022.9985881
Zhao, Q., Zhao, L., and Cai, F. (2019). LSTM recurrent neural networks for multiple pattern recognition in EEG signals. IEEE Trans. Neural Syst. Rehabil. Eng. 27, 1786–1795. Available at: https://pdfs.semanticscholar.org/6a4d/4bf50c412635335af1b8444d427d9c3e8c86.pdf
Keywords: EEG, mental health monitoring, transformer, application of EEG, neural electrical signals
Citation: Liu Z and Zhao J (2025) Leveraging deep learning for robust EEG analysis in mental health monitoring. Front. Neuroinform. 18:1494970. doi: 10.3389/fninf.2024.1494970
Received: 11 September 2024; Accepted: 02 December 2024;
Published: 03 January 2025.
Edited by:
Yue Ma, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Jiancai Leng, Qilu University of Technology, ChinaRiasat Khan, North South University, Bangladesh
Jin Xie, Chinese Academy of Sciences (CAS), China
Copyright © 2025 Liu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zixiang Liu, eml4aWFuZ2xpdTE5ODdAMTYzLmNvbQ==
 Zixiang Liu
Zixiang Liu Juan Zhao2
Juan Zhao2