- Department of Materials, Imperial College London, London, United Kingdom
In situ imaging for direct visualization is important for physical and biological sciences. Research endeavors into elucidating dynamic biological and nanoscale phenomena frequently necessitate in situ and time-resolved imaging. In situ liquid cell electron microscopy (LC-EM) can overcome certain limitations of conventional electron microscopies and offer great promise. This review aims to examine the status-quo and practical challenges of in situ LC-EM and its applications, and to offer insights into a novel correlative technique termed microfluidic liquid cell electron microscopy. We conclude by suggesting a few research ideas adopting microfluidic LC-EM for in situ imaging of biological and nanoscale systems.
Introduction
Central to nanoscale characterization of materials, especially those in biological milieu, is direct visualization. Electron microscopy is a well-established technique that offers structural information about a sample across meso- to subnanometer scales of spatial resolution (Williams and Carter, 2009). Since its development, most samples examined have been thin, solid samples in a static environment (Williams and Carter, 2009), and challenges arise for characterizing biological samples in aqueous media using electron microscopies regardless of the modality. New methodologies (de Jonge et al., 2010; Nishiyama et al., 2010; de Jonge and Ross, 2011; Yuk et al., 2012; Yu, 2020) enjoy advantages such as minimized perturbances to biological specimens and high spatial and temporal resolutions, thus opening new doors for in situ characterization.
In this review, we discuss recent developments in LC-EMs with a focus on biological and nanometer scale samples. LC-EM equipment, spatial and temporal resolutions, and practical considerations are discussed, with some applications highlighted. We discuss the advances in microfluidic platforms in other imaging and scattering techniques and examine the possibility of adopting microfluidic channels as liquid cells, creating microfluidic LC-EMs. We conclude with an outlook in terms of the future applications of microfluidic LC-EMs specifically for imaging complex biological and nanoscale systems.
In situ Liquid Cell Electron Microscopy
The Liquid Cell
In situ LC-EM is a type of liquid-phase electron microscopy (de Jonge and Ross, 2011). Advances in the past few decades have given new opportunities to image samples in liquids (de Jonge et al., 2010; de Jonge and Ross, 2011; Yuk et al., 2012) via their encapsulation in electron-transparent, hermetically sealed enclosures, known as liquid cells. The liquid cell enjoys high adaptability, where scanning, transmission, and scanning transmission electron microscopy (SEM, TEM, and STEM, respectively) modalities can be exploited (Figures 1A–C).
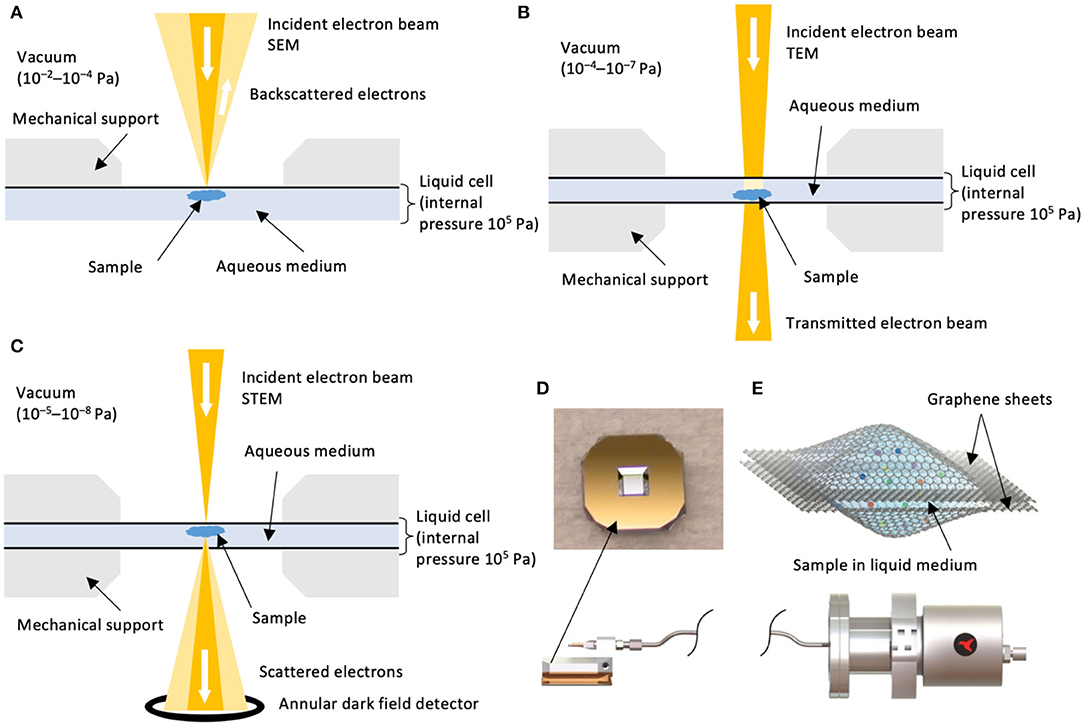
Figure 1. Configurations of LC-EMs. Schematic configurations of (A) LC-SEM, (B) LC-TEM, and (C) LC-STEM. Schematics are not to scale and are adapted from de Jonge and Ross (2011) with permission from Macmillan Publishers Ltd (Springer Nature). Typical chamber vacuum values are reported (Williams and Carter, 2009). (D) (Top) Photograph of an example of microchip with Si3N4 liquid cell window by Norcada Inc. (www.norcada.com). Printed with permission. The window shown has a frame size of 5 mm × 5 mm and a window size of 0.25 mm × 0.25 mm, although other dimensions are available. (Bottom) A microfluidic liquid flow system by Hummingbird ScientificTM (www.hummingbirdscientific.com). Printed with permission. (E) Schematic illustration of a static liquid cell where the liquid sample is encapsulated by two graphene sheets. Figure reprinted from Yuk et al. (2012) with permission from AAAS.
A typical liquid cell is composed of two membranes (or “windows”) encapsulating the sample in an aqueous medium. A commercial example is shown in Figure 1D. Situated in the electron microscope, the membrane faces stringent materials requirements: electron transparency, homogeneity in thickness (de Jonge and Ross, 2011), stability under high vacuum (Williams and Carter, 2009), as well as minimal susceptibility to charging or thermal effects (Leijten et al., 2017). Typical materials for liquid cells include thin films of silicon nitride (Si3N4) of 10–50 nm in thickness (Mirsaidov et al., 2012; de Jonge and Peckys, 2016; Fu et al., 2017; Wu et al., 2019) and multilayer graphene or graphene oxide sheets of a few nm in thickness (Park et al., 2015; Textor and de Jonge, 2018) (Figure 1E).
Spatial and Temporal Resolutions
LC-EMs suffer reduced resolutions (Figure 2) compared to conventional electron microscopies due to exacerbated scattering and absorption induced by the liquid cell membrane, the membrane/liquid interface, and the aqueous medium. For LC-SEM (Figure 1A), theoretical calculations (de Jonge and Ross, 2011) showed functional relationships between spatial resolution d and liquid cell parameters:
where t is the thickness of the membrane, Z is the atomic number, E is the electron energy (in eV), ρ is the density of the sample. For TEM and STEM, respectively,
and
where Cc is the chromatic aberration factor attributable to inelastic scattering of electrons by the liquid, T is the thickness of the liquid layer [1–10 μm (de Jonge et al., 2009; de Jonge and Ross, 2011; Keskin et al., 2019)], and lobject is the dimension of the object being imaged. Evidently, strategies to improve spatial resolutions for LC-EMs include appropriate materials selection for the liquid cell membrane, chromatic aberration correction, and energy filtering. However, such strategies must be weighed against electron dosage which should remain sufficiently low as not to damage biological samples. Successes from time-resolved imaging of inorganic materials using LC-EM show promise for its use for in situ bioimaging. Step-by-step dynamics of the nucleation and growth of metal nanoparticles (Yuk et al., 2012; Loh et al., 2017) have been captured, along with studies on interfacial dynamics of nanostructures during catalysis (Matsubu et al., 2017) and dissolution/degradation processes in battery materials (Leenheer et al., 2015). Millisecond-timescale events, which cover most biologically relevant timescales, can be resolved. The aforementioned list readily demonstrates the adequate spatial and temporal resolutions of LC-EM which can allow us to develop a mechanistic understanding of dynamic nanoscale phenomena. Lessons learned from these studies can be adapted for or shed light on characterizing soft, biological matters.
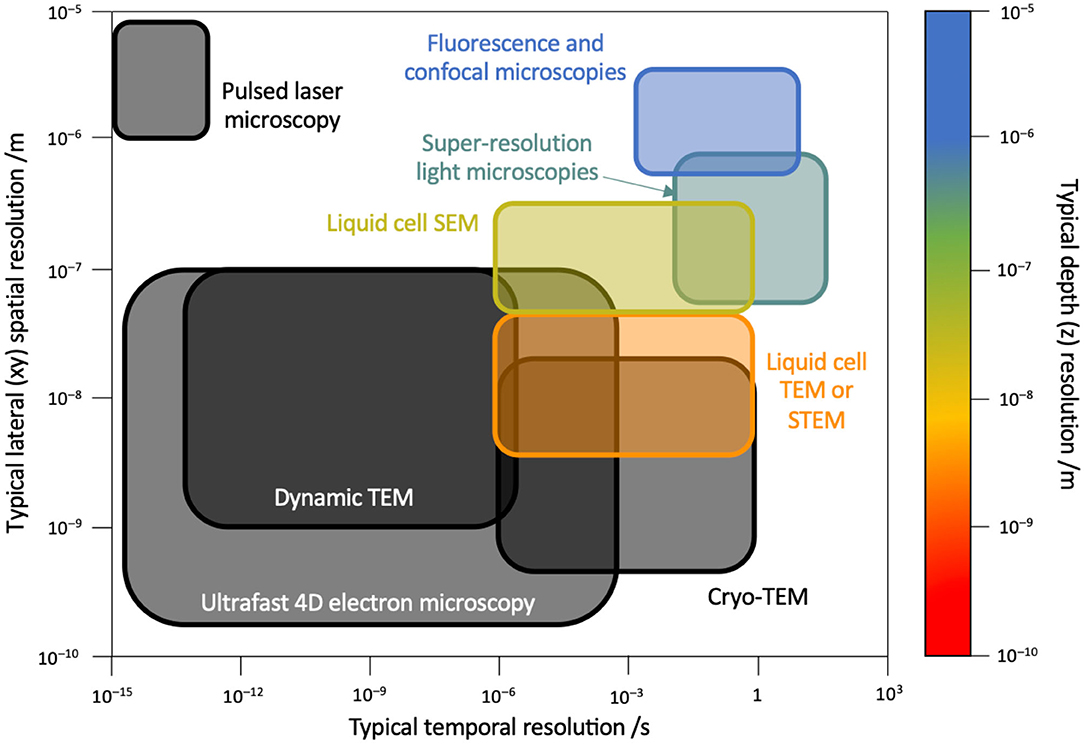
Figure 2. Spatial and temporal resolutions. Map of spatial and temporal resolutions of various types of microscopies with sub-differentiation of electron microscopy modalities. Shown right is the color scale for depth resolution. Black solid lines suggest either unavailability of depth resolution data or non-applicability to the technique. Corresponding data references are as follows: conventional fluorescence and confocal microscopies (Pawley, 2006), pulsed laser microscopy (Bové et al., 2016), super-resolution light microscopies (Göttfert et al., 2013; Agarwal and Machán, 2016; Chen et al., 2019), LC-SEM (Thiberge et al., 2004), LC-TEM/STEM (de Jonge et al., 2019), cryogenic TEM (Diebolder et al., 2012; Fu et al., 2019), dynamic TEM (Lagrange et al., 2008; Egan et al., 2018), and ultrafast 4D EM (Fu et al., 2017; Zhu et al., 2020).
Practical Considerations
Beam Effects
Biological systems are highly sensitive to electron beam damage, and structural and chemical alterations are commonly observed (Mirsaidov et al., 2012; Schneider et al., 2014; de Jonge and Peckys, 2016; Moser et al., 2018). This section, relevant to LC-EMs, examines the beam effects to the liquid cell membrane, on the liquid layer, as well as on the specimen.
For the liquid layer, electron-water interactions are inevitable and should be minimized or at least understood to correctly interpret collected images. Irradiating electrons can react with water molecules to produce transient species such as solvated electrons, hydrogen radicals H·, and hydroxyl radicals OH· (Schneider et al., 2014) (Figure 3A, left and middle). Further reactions also lead to pH variation (Figure 3A, right), which proved detrimental for samples in an unbuffered medium even though the pH change is localized within a sub-micrometer region (Schneider et al., 2014). Radicals are highly reactive, leading to alterations of the subcellular structures of the biological specimen (Mirsaidov et al., 2012; Schneider et al., 2014; de Jonge and Peckys, 2016). Bubbles may also form and even grow during imaging (Klein et al., 2011; Wu et al., 2019), compromising the spatial resolution and affecting the behavior of the biological or nanoscale system (Mirsaidov et al., 2012; Schneider et al., 2014; Tomo et al., 2017; Wu et al., 2019) (Figure 3B). This makes retaining the spatial resolution challenging when trying to image a crowded environment. Tomo et al. (2017) have elucidated how bubbles—of either water vapor or hydrogen gas—nucleate under irradiation by the electron beam. They have found that higher electron energy density leads to the formation of larger bubbles, as nanobubbles increase in nucleation density which consequently coalesce via Ostwald ripening. Strategies to minimize bubble formation should focus on (i) uniform heat dissipation in the liquid cell to minimize water vapor bubble formation and (ii) minimizing radical formation or removing radical species in a sufficiently fast manner to minimize hydrogen bubble formation.
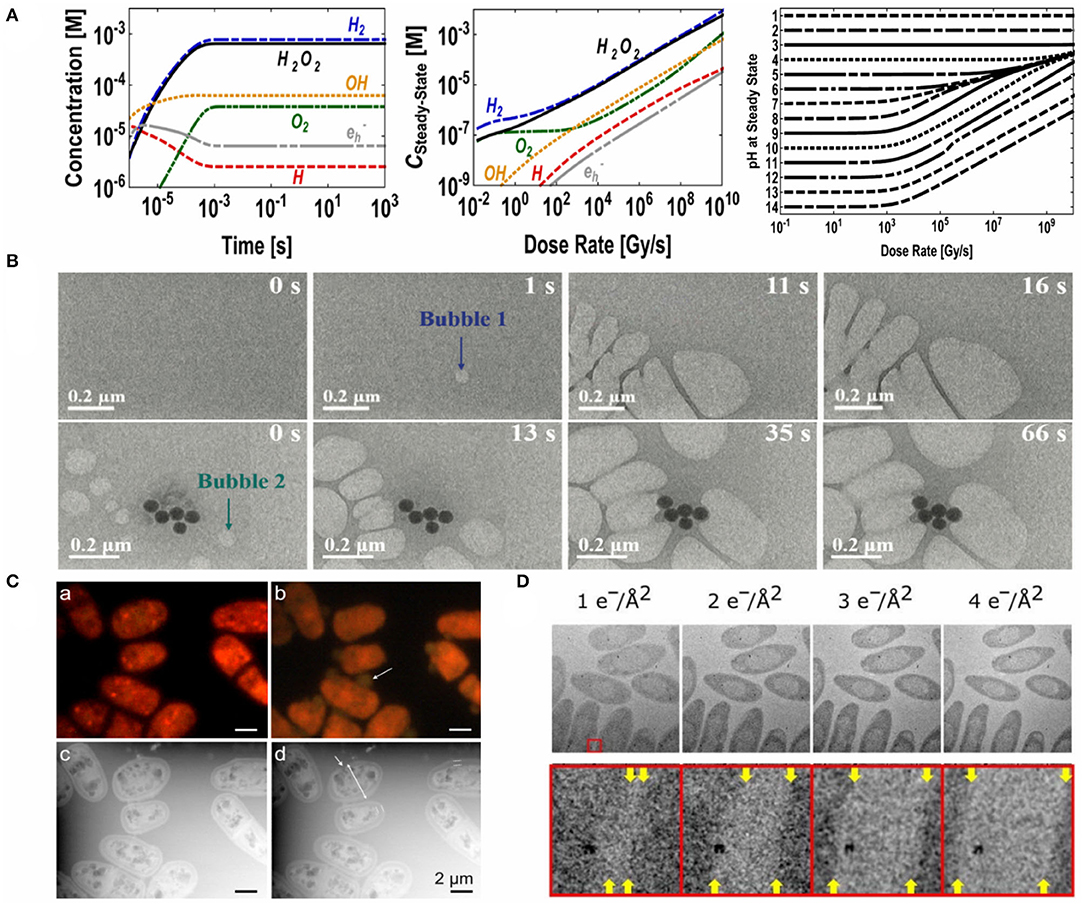
Figure 3. Classification and evaluation of electron beam damage to biological and nanoscale systems. (A,B) Damage to the liquid layer. (A) (Left) Concentrations of beam-generated species as a function of imaging time at a dose rate of 7.5 × 107 Gy s−1. (Middle) Steady-state concentrations of beam-generated species in water as a function of dose rate. (Right) Steady-state pH as a function of dose rate. Reprinted with permission from Schneider et al. (2014). Copyright 2014, American Chemical Society. (B) In situ LC-TEM micrographs illustrating bubble formation: (top) a bubble forms and grows in an organic solution; (bottom) the bubbles interact with a Au-CdS nanocluster. Reproduced from Wu et al. (2019) with permission from the Center National de la Recherche Scientifique (CNRS) and The Royal Society of Chemistry. (C,D) Damages to the sample. (C) (a–b) Fluorescence image of S. pombe cells recorded (a) prior to electron beam exposure and (b) 5 min into electron beam exposure. (c–d) Corresponding LC-STEM images. Reprinted with permission from de Jonge and Peckys (2016). Copyright 2016, American Chemical Society. (D) (Top) Electron micrographs of bacterium C. Metallidurans, imaged using increasing electron fluxes. The time between each acquisition was 20 min. (Bottom) ×10 magnifications of the region boxed in red, where the edge of the cells (indicated using yellow arrows) move relative to the metal cluster with increasing electron influx used. From Moser et al. (2018). Reprinted with permission from AAAS.
Liquid cell membranes made of Si3N4, graphene and graphene oxide sheets possess relatively high stability under high vacuum and electron irradiation (Park et al., 2015; Cho et al., 2016). The use of graphene sheets, in particular, reduces beam damage by radical scavenging (Cho et al., 2016). In fact, while it is typically advised to maintain a low imaging time for biological samples, graphene sheets ensure prolonged in situ imaging times of up to tens of minutes (Yuk et al., 2012; Park et al., 2015; Keskin and de Jonge, 2018; Narayanan et al., 2020). The use of both graphene sheets as liquid cell membranes and free radical scavengers (Schneider et al., 2014) can partially remediate one major damaging mechanism to the liquid layer.
Direct specimen damages due to high-energy electrons can be compositional or structural (Bloebaum et al., 2006; Mirsaidov et al., 2012; Moser et al., 2018). Mirsaidov et al. (2012) report in situ carbonization of acrosomal bundles in water under high electron dosage (Figure 3C). Past research in bioimaging and quantitative elemental mapping of bone tissues also revealed that there were significant changes in calcium, phosphorus and carbon concentrations in specimens under conventional imaging conditions (Bloebaum et al., 2006).
Acceptable Electron Dosage
All aforementioned beam effects dictate the appropriate electron dosage to be used to minimize specimen damage. Structural damage to biological samples can begin to occur at a sub-nanometer scale at a dosage of 1 e− Å−2 (de Jonge and Ross, 2011; Moser et al., 2018) (Figure 3D), which is significant for imaging nanoscale structures such as small globular proteins. As there exists no consensus on the acceptable electron dosage across biological structures, it is generally suggested that samples be imaged below an electron dosage of 10 e− Å−2, a typical tolerance value for proteins (Kempner and Schlegel, 1979), in order to resolve single-protein features. For tissues, room-temperature chemical fixation can improve sample stability under relative high dosages to around 100 e− Å−2 (Thiberge et al., 2004; de Jonge and Ross, 2011). Yet it is not uncommon to observe altered ultrastructure in tissues that have been chemically fixed, and a decrease in extracellular space has been reported (Korogod et al., 2015). Cryo-fixation can help preserve the ultrastructural features of tissues (Korogod et al., 2015) whilst allowing the similar extent of dosage delivery, i.e., around 100 e− Å−2 (Hurbain and Sachse, 2011). In any case, there is a compromise to be made between damage and spatial resolution, as dictated by the electron energy (Equations 1–2) (Williams and Carter, 2009).
Recent developments demonstrated a significantly increased acceptable electron dosage when imaging biological samples. Using multilayered graphene as static liquid cell membranes, Keskin and de Jonge (2018) have shown that the structural features of microtubule proteins can be resolved at the nanometer resolution for an electron density of up to 720 e− Å−2. We envision that, when employing a flow cell where the sample under imaging is constantly replenished, damage will not be cumulative such as for a static cell. This has been validated by a few studies (Yang et al., 2011a,b). The potential offered by microfluidic platforms will be discussed in the later sections. Pulsed electron beams for modulated electron dosage (VandenBussche and Flannigan, 2019) can also be implemented into LC-EM to further reduce beam effects on biological specimens.
Applications of in situ Liquid Cell Electron Microscopy
In situ LC-EM has been used to study the biological structures of proteins (Evans et al., 2012; Mirsaidov et al., 2012) (Figures 4A,C) and even whole cells (de Jonge et al., 2009; Park et al., 2015) (Figure 4B). One advantage of in situ LC-EM is the ability to locate and image unstained and unlabeled samples, as demonstrated by a few studies (Mirsaidov et al., 2012; Hoppe et al., 2013). This results in minimized chemical perturbance, approaching imaging samples in their native state. Additionally, assemblies of macromolecular complexes can be resolved. While the spatial resolution of cryo-EM exceeds most achievable values of LC-EM modalities (Figure 2), freezing may affect gross morphologies or sub-cellular structures. One particular instance is the supposedly spurious stacking of nanolipoprotein particles (Figure 4C) (Schneider et al., 2014), a result of self-assembly due to cryo-EM sample preparation and subsequently resolved by LC-EM. With recent advances in research investigating the bio/nano-interfaces, dynamic studies using LC-TEM have also been performed: Park et al. (2015) imaged the translocation of gold nanoparticles within a canine kidney cell (Figure 4D). Albeit that such dynamics are inferred from static images, in situ LC-EM imaging can already shed light on the internalization process of engineered nanomaterials, such as nanoparticles, by cells. It should be noted that these studies, while promising, remain less-than-ideal: using pure water as the liquid layer is unphysiological as compared to cellular environments. Taking proteins for example, most proteins do not adopt the same conformation in water as intracellularly (Bellissent-Funel et al., 2016). Moreover, to image protein structures and to probe protein-protein interactions in their native environment, additives such as hydrotropes or cosolvents (Patel et al., 2017) are required to achieve high, intracellular protein concentrations. These strategies will lead to a step closer to imaging samples in the native state; nevertheless, we must also consider the complications that arise such as a decrease in spatial resolution due to extensive scattering.
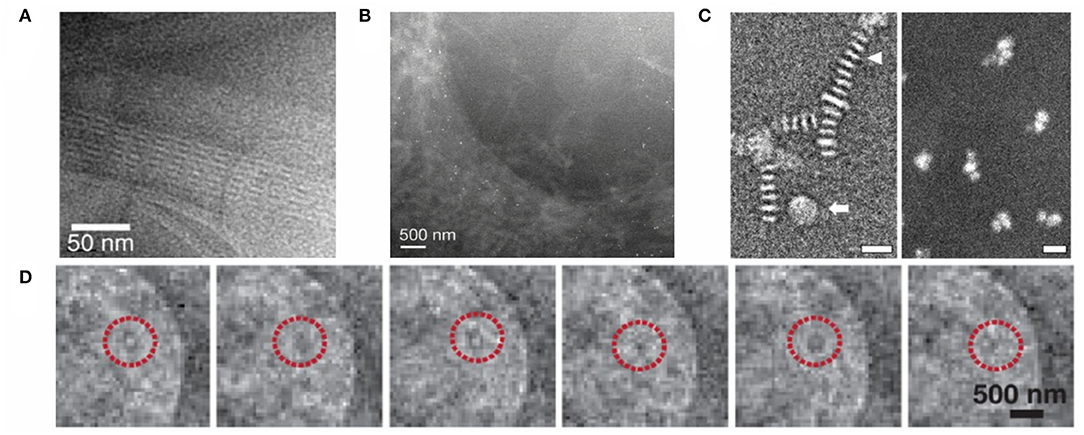
Figure 4. LC-EMs in imaging biological and nanoscale systems. (A) LC-TEM dark-field image of acrosomal bundles in water under an electron dosage of 35 e− Å−2. Image reprinted from Mirsaidov et al. (2012) with permission from Cell Press. (B) LC-STEM dark-field image of the edge of a fibroblast cell, labeled with gold-tagged epidermal growth factors. The labels are visible as bright punctate spots. Image reprinted from de Jonge et al. (2009) with permission from National Academy of Sciences. (C) (Left) Cryogenic negative stain TEM image of nanolipoprotein particles; the triangular arrow indicates particle stacking. (Right) LC-STEM dark-field image of nanolipoprotein particles. Both scale bars represent 20 nm. Images reprinted from Evans et al. (2012) with permission from Elsevier. (D) LC-TEM snapshots of a gold nanoparticle in a hydrated kidney cell. Time interval between frames is 0.1 s. Reprinted with permission from Park et al. (2015). Copyright 2015, American Chemical Society.
Caveats on Drawing Conclusions
In situ LC-EMs promise great opportunities for direct visualization of biological structures as well as imaging dynamic processes given the appropriate temporal resolution. However, due to low-contrast and various beam damages, one may easily fall into an “Einstein from noise” trap (Henderson, 2013): spuriously attributing noise or beam-induced bubbles (Klein et al., 2011; Wu et al., 2019) to a structural feature. In general, one should always generate a statistically significant number of images and evaluate possible beam damage mechanisms before arriving at a specific conclusion. It is helpful to assess the time-dependency of observed features and possibly correlate to another technique. Consider imaging the cellular uptake of gold nanoparticles depicted in Figure 4D. One must examine the electron irradiation history (Moser et al., 2018) in order to ascertain that the dynamics imaged is due to intracellular movements rather than a radiation-induced artifact. This is applicable for studying a range of dynamic biological processes from biomineralization to virus-cell interactions. In addition, thermal effects which influence the kinetics of dynamic processes must be considered. Prolonged imaging can increase the temperature of the liquid cell by a few degrees (Park et al., 2015), which is sufficient to modify the Brownian motion of small biological and nanoscale species at play. One may hence falsely attribute “dynamics” due to enhanced Brownian motion to an effect of added analyte(s). In this case, a temperature-controlled chamber can be used to minimize relatively large temperature variations.
We are still at an inchoate stage in understanding complex biological systems in their native state. Since the minimum electron dose to obtain contrast exceeds the lethal dose for cell-lysis, which lies in the order of 10−3-10−4 e− Å−2, it is unfeasible to image live cells using a LC-EM (de Jonge and Peckys, 2016), whereas the viability of multiple cell lines has been confirmed in microfluidic chambers upon an hour-long exposure (Peckys and de Jonge, 2014). While in situ bio-imaging has immense potential, we must keep in mind that in situ does not equate in vivo or in operando. At this stage, it is best that we relate additional in vivo characterization techniques with in situ imaging.
Microfluidics: Opportunities and Challenges for LC-EM
Design of Microfluidic Systems
LC-EMs can use either static (de Jonge et al., 2010; Nishiyama et al., 2010; Yuk et al., 2012) or flow cells (Ring and de Jonge, 2010; Klein et al., 2011; Yu, 2020), hinting at the potential of implementing microfluidic setups as part of the LC-EM for dynamic process imaging. In fact, the prospects of microfluidics in biological sciences has been suggested for over a decade now (Whitesides, 2006). While much focus has been on microfluidic-assisted fabrication and processing (Martin et al., 2016), the potential of microfluidics in LC-EM remains to be further explored.
Microfluidic devices have inlets and channels with at least one dimension being <1 mm (Amato et al., 2017; Wu et al., 2020). These devices can be broadly classified into chip-based microfluidics and capillary microfluidics (Solsona et al., 2019) (Figure 5A) or can be categorized based on the channel geometries to induce specific flow patterns such as T-junction, co-flow, or flow-focusing (Li W. et al., 2018). Numerous interesting geometries and designs have been developed from microfluidic-based fabrication and processing of multicompartmental nanoparticles (Wu et al., 2020) or complex microcapsules (Amato et al., 2017). For LC-EM, chip-based microfluidics would be of practical interest to most bioimaging applications. The microfluidic design can be customized to allow controlled mixing of multiple input materials and to accommodate biological structures of different length-scales. For miscible liquids, the microchannel allows a continuous flow of analytes via diffusional mixing (Arter et al., 2020), i.e., achieved via passive mixing based on the physical design of the microchannel alone (Figure 5B). Active mixing is possible by applying suitable external stimuli such as electric or magnetic fields, pressure, and acoustics (Solsona et al., 2019). Evidently, the compatibility and applicability of these modes of mixing with biological specimens will need to be assessed for individual samples. Opportunities also arise where, by controlling the fluidic dynamics, we can intentionally design the flow profiles of liquids within the microchannels. Both laminar and turbulent flows can be achieved (Figure 5C). Such has been exploited to mimic the flow in biological environments, such as the blood flow within brain capillaries (Shao et al., 2016) and flow in proximal tubules (Duan et al., 2010). Besides analyte mixing, using immiscible liquids may also hold applications. For instance, when two immiscible liquids are introduced into the main microfluidic channel, along with a third inlet with samples to be imaged (Figure 5D), their behavior at the liquid-liquid interface can be addressed under the correct conditions. Overall, control over the microfluidic setup means spatial and temporal control of the microenvironment containing the sample, proving advantageous for imaging applications.
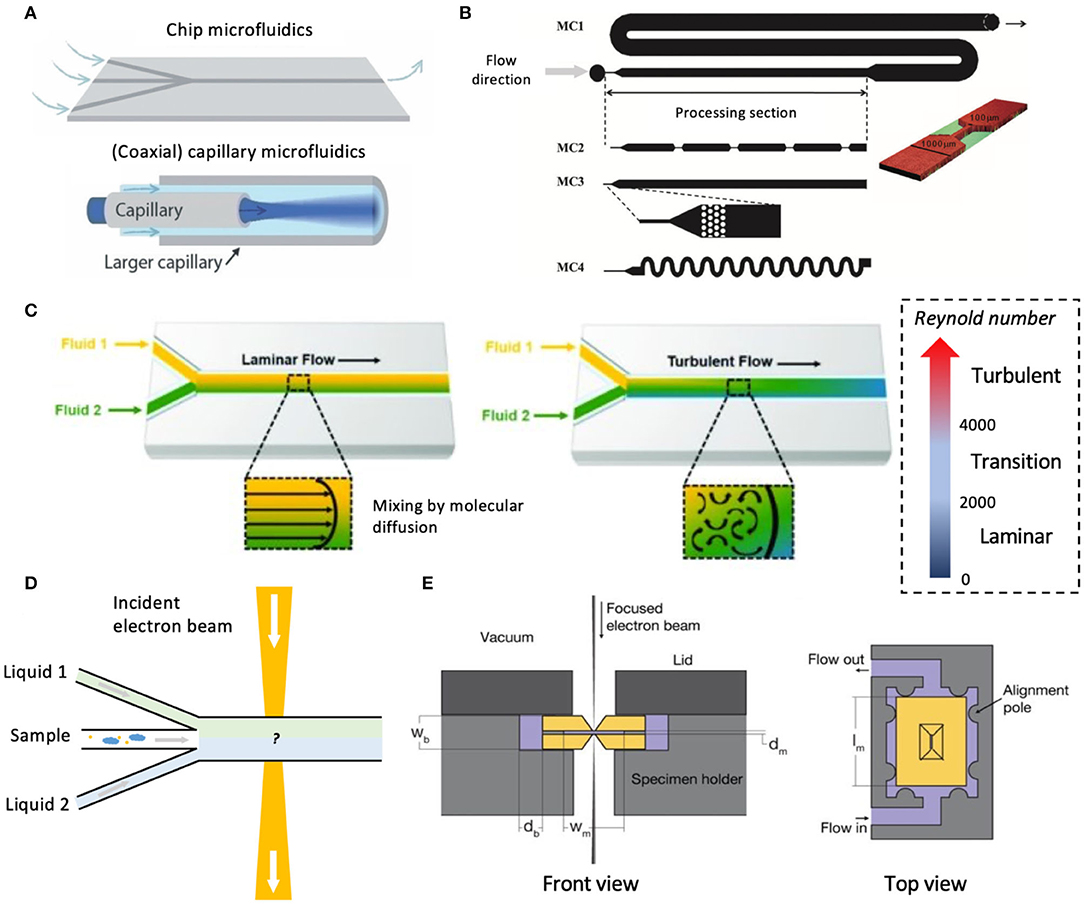
Figure 5. Microfluidic systems and microfluidic LC-EM. (A) Chip-based and capillary microfluidic designs. Figure reprinted from Solsona et al. (2019), published by The Royal Society of Chemistry. (B) Schematic showing different possible geometries of the microfluidic channel. Reproduced from Martin et al. (2016) with permission from The Royal Society of Chemistry. (C) Flow behaviors in microfluidic channels. Shown on the right is a rough scale for categorizing the Reynold number which characterizes the fluid mechanics of the liquid in the microchannel. Figure reprinted from Saliba et al. (2018), MDPI. (D) Schematic of a hypothetical design of microfluidic channel containing two immiscible liquids for studying liquid-liquid interfacial phenomena. (E) Schematic setup of a microfluidic chamber for LC-TEM/STEM (not to scale). The two Si3N4 chips are shown in a golden yellow color. Figure reprinted from Ring and de Jonge (2010) with permission from Microscopy Society of America and Cambridge University Press.
Implications and Consequences for Biological Specimens
Downscaling to micro or sub-micrometer scales in microfluidics allows for miniaturization, providing apparent advantages such as reduced volumetric materials input and enhanced mass and heat transfer due to increased surface-area-to-volume ratio (Whitesides, 2006). Moreover, such downscaling inevitably leads to pronounced effects from properties such as viscosity, shear stress, and surface tension. Their interplay directly determines the transport phenomenon and sample behavior inside the microchannel. Viscosity should be of little concern as the concentration for biological samples can be carefully chosen for imaging. At low concentrations, clustering of biological and nanoscale entities is unlikely provided an appropriate liquid medium has been selected. Thus, shear thickening is unlikely even under confinement (Bian et al., 2014). Shear stress, especially near the channel walls, may impact the behavior of biological samples under flow. Without physical alterations, this can offer opportunities for mimicking flow in blood capillaries. For instance, a threshold of shear stress exists to trigger the adhesion of von Willebrand factor fibers (Schneider et al., 2007). Microfluidics can be designed to mimic and image similar phenomena in situ in a time-resolved fashion. For other applications, varying physical design of the channel may allow different flow profiles to emerge, whereas chemically modifying the inner walls of microchannels can impart properties such as anti-fouling properties to resist protein adsorption under shear, a common problem that has been observed in microfluidic devices (Koc et al., 2008).
In situ Imaging Using Microfluidic Platforms
Shown in Figure 5E is the Ring and de Jonge's design of microfluidic system for LC-TEM/STEM (Ring and de Jonge, 2010). Two oppositely-facing Si3N4 chips were used to assemble the microfluidic chamber, “sandwiching” the liquid specimen and allowing liquid flow. More intricate designs to manipulate the liquid flow, as discussed above, can be applied to study complex biological and nanoscale systems. One particular advantage of the deployment of microfluidic-based imaging is the capability to image dynamic processes, which renders it far superior to conventional microscopies. The liquid chemistry via the microfluidic tubings can be precisely controlled (Knoška et al., 2020) and altered (Mattern et al., 2020) in real-time to examine the kinetics of complex biological processes. In introducing species in aliquots, such as aggregation-inducers (Zheng et al., 2003) or fluorescent dyes (Chen and Lee, 2010), kinetic processes can be studied in detail. Moreover, since the sample is constantly replenished, the beam damage will not be cumulative; this reduces the overall sample damage and increases the imaging times as needed. For optical microscopies and X-ray scattering, tubing materials are typically polydimethylsiloxane with 1-μm thickness (Firpo et al., 2015), inappropriate for LC-EMs. Multilayered graphene oxide membranes used in LC-EMs are both electron- and optically-transparent (Yulaev et al., 2016), serving as a great candidate for microfluidic EM, rendering the technique feasible.
Additional functionalization of the microchannels can provide prospective venues for experimental design. For example, functionalizing the inner walls of the microchannels with biotin, leveraging its well-known strong interaction with streptavidin, can help understand similar multivalent interactions in biomaterials and at bio/nano-interfaces. Strategies for creating such functional microchannels, by chemically or physically modifying the inner walls of the microchannels, have been summarized in a recent review (Wang et al., 2019). As the field progresses, additional functionalities of the microfluidic platform such as electrophoretic manipulation (Huh et al., 2008; Yasukawa et al., 2008; Unocic et al., 2014; Arter et al., 2020) can be incorporated for more advanced characterization.
Outlook
Toward Correlative and Multimodal Imaging
Much progress has been made in the field of structural biology via X-ray or neutron scattering characterizations (Neylon, 2008), cryo-EM (Mäeots et al., 2020), model fitting, and molecular dynamics simulations (Karplus and McCammon, 2002). The gap between biophysicochemical analysis and direct visualization can be further bridged using microfluidic LC-EM, leading to correlative characterization. The microfluidic channel houses a statistical distribution of all orientations of biological and nanoscale structures; such allows a 3D model to be reconstructed using a computer algorithm, such as demonstrated by Chen et al. (2013) on the imaging of DNA-gold nanoconjugates on a second timescale. This is analogous to the well-developed single-particle image processing algorithms for cryo-EM (Punjani et al., 2017). To study the dynamics of proteins (Zheng et al., 2003; Chen and Lee, 2010; Lutz-Bueno et al., 2016; Knoška et al., 2020) and nanoparticles (Ghazal et al., 2016), microfluidic platforms have been coupled to optical (Zheng et al., 2003; Sakai et al., 2019), fluorescence microscopies (Chen and Lee, 2010; Ding et al., 2016; Hua et al., 2016; Tian et al., 2019), cryo-EM (Fuest et al., 2019; Mäeots et al., 2020), and small-angle X-ray scattering (Lutz-Bueno et al., 2016; Anaraki et al., 2020). Some examples include capturing the transient conformations of DNA during self-assembly (Wang et al., 2020), imaging protein crystallization (Zheng et al., 2003) (Figure 6A) or adsorption (Yu et al., 2020), probing intracellular communication via gap junctions (Chen and Lee, 2010) (Figure 6B), mapping the dynamics of amyloid formation (Lutz-Bueno et al., 2016; Saar et al., 2016) (Figure 6C), chemical and molecular mapping of live biofilms (Ding et al., 2016) (Figure 6D), as well as imaging biomineralization and bio-sedimentation (He et al., 2019; Narayanan et al., 2020). Hydrated single cells can also be manipulated in a microfluidic chamber and imaged in a correlative fashion using super-resolution microscopy and secondary ion mass spectrometry (Hua et al., 2016). Microfluidic LC-EM can be coupled with other microscopies (such as confocal microscopy), spectroscopies (such as X-ray spectroscopy), or scattering techniques (such as lab-sized SAXS) to offer potentially continuous and time-resolved correlative characterization of dynamic biological processes and interactions between biological entities and nanoparticles. Kinetic information may also be extracted (Mäeots et al., 2020).
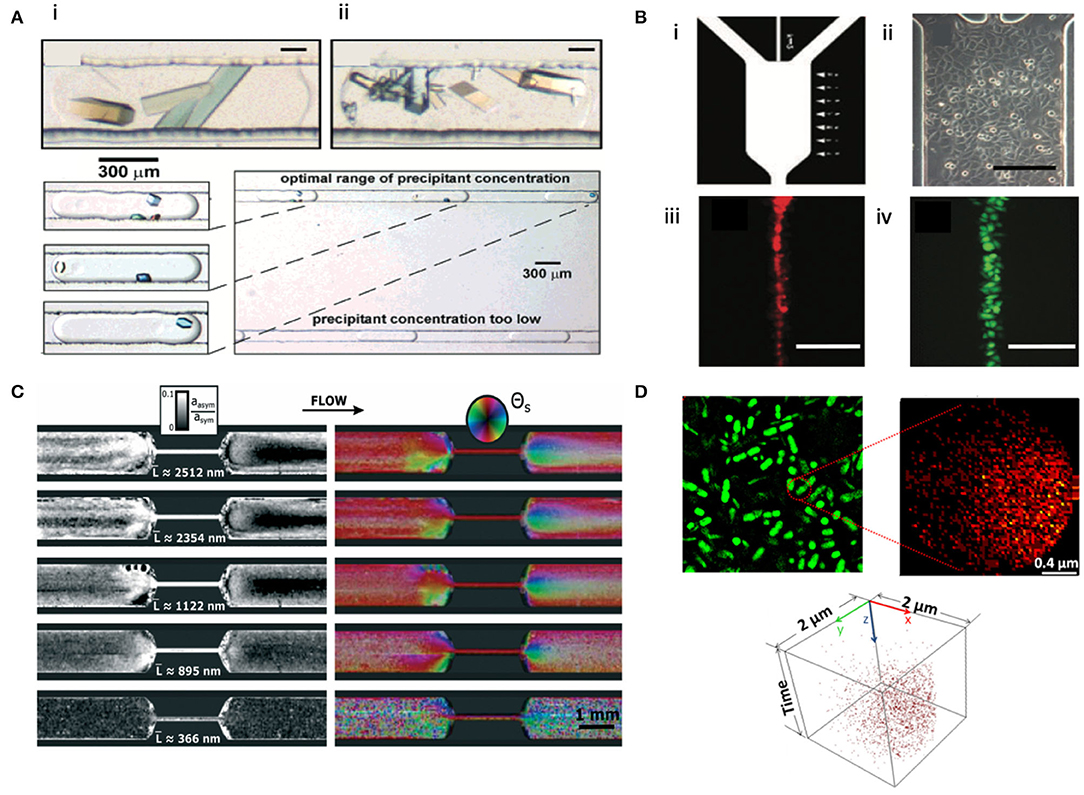
Figure 6. Microfluidic-based microscopy, scattering, and spectrometry techniques. (A) (Top, i–ii) Polarized light micrographs of bovine liver catalase (i) and glucose isomerase (ii) in a microfluidic setup. (Bottom) Polarized light micrograph illustrating the crystallization of lysozyme inside microfluidic droplets. Reprinted with permission from Zheng et al. (2003). Copyright 2003, American Chemical Society. (B) (i) Photolithographic mask of a microfluidic channel. (ii) Optical photograph of HeLa cells in the microfluidic channel. (iii–iv) Fluorescence images of dye focusing over HeLa cells at the center of the channel using calcein/AM (iii) and DiI (iv). Scale bar represents 200 μm. Reproduced from Chen and Lee (2010) with permission from The Royal Society of Chemistry. (C) Nanostructure mapping of amyloid fibrils with different average contour lengths L using microfluidic-based SAXS. Color saturation denotes asymmetric amplitude, white denotes symmetric amplitude, and the color wheel categorizes molecular alignment. Reproduced from Lutz-Bueno et al. (2016) with permission from The Royal Society of Chemistry. (D) S. oneidensis MR-1 biofilms characterized by tight-of-flight secondary ion mass spectrometry (TOF-SIMS) in a microfluidic chamber. (Top, left) SIM image; (Top, right) correlated 2D image of palmitic acid from the biofilm. (Bottom) 3D cluster analysis of palmitic acid from the biofilm. Reprinted with permission from Ding et al. (2016). Copyright 2016, American Chemical Society.
Appropriate sample preparation is essential to enable multimodal imaging of the same sample across techniques. When using the same sample for correlative imaging, which is often sought after if sample preparation proves time-consuming and laborious, attention should be paid to determine the sequence in which serial imaging or multimode characterizations are carried out on a case-by-case basis. At the very least, destructive imaging or analytical techniques should come last, and detailed planning is contingent upon the structural level on which the damage can be caused.
Although this review specifically focuses on liquid cell EM, where the sample is encapsulated or “sandwiched” in a liquid medium, open specimen holders (Nishiyama et al., 2010) used in atmospheric or environmental EM can also benefit from the use of microfluidic platforms for in situ imaging. Several studies have employed atmospheric or environmental EM for correlative imaging of biological systems, including neuronal networks (Sato et al., 2019a), secretory glands (Yamazawa et al., 2016), and bacterial chains in a biofilm matrix (Raab and Bachelet, 2017). The open-cell EM configurations use either low-vapor-pressure liquid or differential pumping (Takeda et al., 2015), and the microfluidics can adopt an open-channel-like design to implement into the EM stages. One example of such a hybrid microfluidic platform has been designed to couple with secondary ion mass spectrometry (Yang et al., 2011a; Yu et al., 2020). Due to the open-cell design, atmospheric or environmental EM would face the general problems of having limited options of the liquid medium and increased susceptibility to radiation damage compared to LC-EM. Yet an improved resolution may be possible due to the disuse of multiple layers of liquid cell membrane materials. Recent work by Sato et al. (2019b) utilized a microfluidic chamber to directly image calcium phosphate mineralization in fixed, unstained bone tissues. Thus, there do exist possibilities for in situ bioimaging when integrating microfluidic systems with open cells, not just liquid cells.
Suggested below are a few areas of interest at the interface between nanotechnology and biological sciences which can benefit from the development of and advancement in microfluidic LC-EM.
Future Work Applying Microfluidic LC-EM
Hydrotropic Mechanism in Protein Solubilization
Recently, Patel et al. (2017) reported that adenosine triphosphate (ATP), a ubiquitous energy supplier within cells, behaves as a hydrotrope which facilitates protein dissolution. In fact, amphiphilic proteins maintain soluble and stable at high concentrations inside cells (Brown, 1991), and its origin is poorly understood. Microfluidic LC-EMs can be used to provide direct visualization for the hydrotropic stabilization phenomenon. This will facilitate the understanding of the operative mechanism of hydrotropes as more biological hydrotropes become discovered. It can have further implications in understanding protein-protein interactions, or interactomes (Tian et al., 2011; Arter et al., 2020), in vivo.
Cellular Uptake of Nanoparticles
There is a surge of interests in understanding the pharmacokinetics of engineered nanomaterials, particularly nanoparticles which are used in everyday cosmeceuticals or deliberately introduced into the body as nanotherapeutics (Cagno et al., 2017; Rees et al., 2019; Jones et al., 2020). Microfluidic LC-EMs can be designed to accommodate a constant flow of cells and image extracellular processes (e.g., nanoparticle aggregation, physical degradation and settling rates in culture) and cellular uptake of relatively large nanoparticles (e.g., >50 nm in size) with ms temporal resolution and without much damage. Yet limitations in imaging resolution remain as nanoparticle size cannot be too small, and sub-cellular features may not be fully resolved. If necessary, confocal microscopy can be coupled to microfluidic LC-EM to image stained cells or antibody-decorated nanomaterials while obtaining a secondary, fluorescence signal from proteins of interest. This design can offer appreciable information about the dynamics of cellular interactions of nanomaterials, allowing correlation with existing studies on their pharmacokinetics (Rees et al., 2019).
Mechanism of Virucidal Nanoparticles
Monolayer-protected metal nanoparticles can be designed to be protein-mimetic and serve as a prospective candidate for non-toxic nanomedicines (Cagno et al., 2017; Jones et al., 2020). More importantly, these nanoparticles are reported to be broad-spectrum (Cagno et al., 2017; Jones et al., 2020), hinting potential applications in treating patients afflicted by emerging, novel viruses such as SARS-CoV-2 (Weiss et al., 2020). Understanding fundamental supramolecular interactions between viruses and biomimetic antiviral nanoparticles can greatly assist materials design and their employment for biomedical applications, tackling viral pandemics/epidemics such as the COVID-19 pandemic. Microfluidic LC-EMs can be used to introduce aliquots of such nanoparticle suspension to a main channel containing viruses (Dukes et al., 2013), allowing direct and real-time visualization of purported virucidal mechanisms.
Intrinsically Disordered Proteins and Their Self-Assembly
Intrinsically disordered proteins (IDPs) and regions (IDRs) represent an emerging and paradigm-shifting field of research in life science and soft matter physics. In terms of individual proteins, IDPs differ from folded, globular proteins in that they enjoy high conformational flexibility and complexity (Oldfield and Dunker, 2014; Li B. et al., 2018). This translates to enhanced interaction densities and strengths (Uversky, 2013) through multivalency effects, as well as a much larger conformational energy landscape for a particularly-sequenced protein to explore. In terms of their self-assembled structures, the self-assembly of IDPs/IDRs is believed to drive intracellular liquid-liquid phase separation (Uversky et al., 2008; Shin et al., 2018), i.e., formation of membraneless organelles, a process that is responsible for cellular compartmentalization. Moreover, this self-assembling process bears pathological implications [such as Alzheimer's disease, Parkinson disease, and amyotrophic lateral sclerosis (ALS)] (Uversky et al., 2008) as it links closely to the formation of protein aggregates. Both areas remain largely unexplored. While significant strides have been made in visualizing liquid-liquid phase separation on a cellular level using optogenetics, both static and dynamic characterization of IDPs and their self-assembly at the single-protein level are still needed. Microfluidic LC-EM can be used to control specific liquid environments to trigger IDP self-assembly and to complement studies involving single-molecule Förster resonance energy transfer measurements (Ferreon et al., 2010) or cryo-EM imaging (Li B. et al., 2018). This way, it may be possible to identify transient states of IDPs, to capture conformational switching (Choi et al., 2019) in IDPs and extract its kinetic information, and to characterize the hierarchical organization and dynamics during the self-assembly of IDPs. Consequently, an improved understanding of IDPs and IDRs can better inform the design of supramolecular nanomaterials to interact with complex biological entities.
Perspectives
The examples described above demonstrate the potential wide applicability of microfluidic LC-EM in biology and nanomaterials science. Evident advantages of the technique include ease of sample preparation (such as compared to cryo-EM), ability to image dynamic processes and extraction of kinetic information via precise control of the liquid environment, as well as ease of implementation to other imaging or scattering techniques for multimodal analysis. Yet certain challenges remain and will need to be addressed. First, data acquisition and processing should be optimized for the respective imaging modality, especially for video recording of dynamic processes, as this dictates the eventual spatial and temporal resolution of images/videos during analysis. Using an automated process, 2D projection of image snapshots can be grouped to create class averages—a common procedure in structural biology—and 3D reconstruction of single particles or macromolecular complexes may be possible. Secondly, to better interpret imaging results, respective electron beam damage to the sample and the liquid layer must be understood and, more importantly, quantified on a case-by-case basis. Control groups with only the liquid medium should be imaged, and electron-dosage-dependency of the dynamics imaged by microfluidic LC-EM should be examined. Thirdly, alternative liquid cell membranes could help. Besides graphene and graphene oxide sheets, it has been suggested that various 2D layered nanomaterials can be investigated for their suitability and applicability as liquid cell membranes (Yu, 2020) with a focus on their electron-transparency, vacuum stability, inertness, and radical-scavenging capabilities. Such materials selection can improve imaging resolution and possibly lessen electron beam damage. From a research standardization and reproducibility point of view, researchers must be comprehensive in reporting imaging conditions in works using microfluidic LC-EM. At a minimum, we recommend reporting information about the sample, about the liquid layer (e.g., thickness and chemical composition), about the microfluidic setup (e.g., membrane material, chamber temperature if monitored and controlled, and flow rate), as well as the electron microscopy imaging conditions (i.e., electron energy, electron energy density, irradiation time, and accumulated dosage).
Various strategies exist to lessen the beam damage for biological samples, allowing their structural features to be resolved at the nanometer scale in situ. Microfluidic designs, allowing liquid flow, may further reduce the beam damage and can offer additional opportunities to create functional microenvironments for manipulation of biological structures. We anticipate that, when microfluidic platforms become incorporated into LC-EMs, important dynamic processes in biological and nanoscale systems can be addressed in a time-resolved fashion. Microfluidic LC-EM could have the potential to help elucidate fundamental phenomena in nature, such as how naturally-occurring biological hydrotropes work and how IDRs self-assemble in vivo and its implication for the development of neurodegenerative diseases. At the bio/nano-interface, the interactions between biological entities and engineered nanomaterials can also be better understood via correlative imaging with microfluidic LC-EM. Fluorescent tags can be added to help image the binding process between antiviral nanoparticles and different viruses. At a cellular level, cellular uptake of nanomaterials and relevant extracellular processes can be imaged, and their concerted effect can be better appreciated in vivo using microfluidic LC-EM. With further perfection of imaging systems and software, a future with imaging biological and nanoscale systems in operando using microfluidic LC-EM may be possible for a myriad of applications.
Author Contributions
ZH wrote this paper. AP supervised this work and revised the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
We thank support from the Natural Environment Research Council project: NE/N006402/1 and an EPSRC pump priming grant from the Rosalind Franklin Institute: EP/S001999/1.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Agarwal, K., and Machán, R. (2016). Multiple signal classification algorithm for super-resolution fluorescence microscopy. Nat. Commun. 7:13752. doi: 10.1038/ncomms13752
Amato, D. V., Lee, H., Werner, J. G., Weitz, D. A., and Patton, D. L. (2017). Functional microcapsules via thiol–ene photopolymerization in droplet-based microfluidics. ACS Appl. Mater. Interfaces 9, 3288–3293. doi: 10.1021/acsami.6b16382
Anaraki, N. I., Sadeghpour, A., Iranshahi, K., Toncelli, C., Cendrowska, U., Stellacci, F., et al. (2020). New approach for time-resolved and dynamic investigations on nanoparticles agglomeration. Nano Res. 13, 2847–2856. doi: 10.1007/s12274-020-2940-4
Arter, W. E., Levin, A., Krainer, G., and Knowles, T. P. (2020). Microfluidic approaches for the analysis of protein–protein interactions in solution. Biophys. Rev. 12, 575–585. doi: 10.1007/s12551-020-00679-4
Bellissent-Funel, M., Hassanali, A., Havenith, M., Henchman, R., Pohl, P., Sterpone, F., et al. (2016). Water determines the structure and dynamics of proteins. Chem. Rev. 116, 7673–7697. doi: 10.1021/acs.chemrev.5b00664
Bian, X., Litvinov, S., Ellero, M., and Wagner, N. J. (2014). Hydrodynamic shear thickening of particulate suspension under confinement. J. Nonnewton Fluid Mech. 213, 39–49. doi: 10.1016/j.jnnfm.2014.09.003
Bloebaum, R. D., Holmes, J. L., and Skedros, J. G. (2006). Mineral content changes in bone associated with damage induced by the electron beam. Scanning 27, 240–248. doi: 10.1002/sca.4950270504
Bové, H., Steuwe, C., Fron, E., Slenders, E., D'Haen, J., Fujita, Y., et al. (2016). Biocompatible label-free detection of carbon black particles by femtosecond pulsed laser microscopy. Nano Lett. 16, 3173–3178. doi: 10.1021/acs.nanolett.6b00502
Brown, G. C. (1991). Total cell protein concentration as an evolutionary constraint on the metabolic control distribution in cells. J. Theor. Biol. 153, 195–203. doi: 10.1016/S0022-5193(05)80422-9
Cagno, V., Andreozzi, P., D'Alicarnasso, M., Silva, P. J., Mueller, M., Galloux, M., et al. (2017). Broad-spectrum non-toxic antiviral nanoparticles with a virucidal inhibition mechanism. Nat. Mater. 17, 195–203. doi: 10.1038/nmat5053
Chen, Q., Smith, J. M., Park, J., Kim, K., Ho, D., Rasool, H. I., et al. (2013). 3D motion of DNA-Au nanoconjugates in graphene liquid cell electron microscopy. Nano Lett. 13, 4556–4561. doi: 10.1021/nl402694n
Chen, S., and Lee, L. P. (2010). Non-invasive microfluidic gap junction assay. Integr. Biol. 2, 130–8. doi: 10.1039/b919392h
Chen, S., Wang, J., Xin, B., Yang, Y., Ma, Y., Zhou, Y., et al. (2019). Direct observation of nanoparticles within cells at subcellular levels by super-resolution fluorescence imaging. Anal. Chem. 91, 5747–5752. doi: 10.1021/acs.analchem.8b05919
Cho, H., Jones, M. R., Nguyen, S. C., Hauwiller, M. R., Zettl, A., and Alivisatos, A. P. (2016). The use of graphene and its derivatives for liquid-phase transmission electron microscopy of radiation-sensitive specimens. Nano Lett. 17, 414–420. doi: 10.1021/acs.nanolett.6b04383
Choi, U., Sanabria, H., Smirnova, T., Bowen, M., and Weninger, K. (2019). Spontaneous switching among conformational ensembles in intrinsically disordered proteins. Biomolecules 9:114. doi: 10.3390/biom9030114
de Jonge, N., Houben, L., Dunin-Borkowski, R. E., and Ross, F. M. (2019). Resolution and aberration correction in liquid cell transmission electron microscopy. Nat. Rev. Mater. 4, 61–78. doi: 10.1038/s41578-018-0071-2
de Jonge, N., and Peckys, D. B. (2016). Live cell electron microscopy is probably impossible. ACS Nano 10, 9061–9063. doi: 10.1021/acsnano.6b02809
de Jonge, N., Peckys, D. B., Kremers, G. J., and Piston, D. W. (2009). Electron microscopy of whole cells in liquid with nanometer resolution. Proc. Natl. Acad. Sci. U.S.A. 106, 2159–2164. doi: 10.1073/pnas.0809567106
de Jonge, N., Poirier-Demers, N., Demers, H., Peckys, D. B., and Drouin, D. (2010). Nanometer-resolution electron microscopy through micrometers-thick water layers. Ultramicroscopy 110, 1114–1119. doi: 10.1016/j.ultramic.2010.04.001
de Jonge, N., and Ross, F. M. (2011). Electron microscopy of specimens in liquid. Nat. Nanotechnol. 6, 695–704. doi: 10.1038/nnano.2011.161
Diebolder, C. A., Koster, A. J., and Koning, R. I. (2012). Pushing the resolution limits in cryo electron tomography of biological structures. J. Microsc. 248, 1–5. doi: 10.1111/j.1365-2818.2012.03627.x
Ding, Y., Zhou, Y., Yao, J., Szymanski, C., Fredrickson, J., Shi, L., et al. (2016). In situ molecular imaging of the biofilm and its matrix. Anal. Chem. 88, 11244–11252. doi: 10.1021/acs.analchem.6b03909
Duan, Y., Weinstein, A. M., Weinbaum, S., and Wang, T. (2010). Shear stress-induced changes of membrane transporter localization and expression in mouse proximal tubule cells. Proc. Natl. Acad. Sci. U.S.A. 107, 21860–21865. doi: 10.1073/pnas.1015751107
Dukes, M., Mcdonald, S., and Kelly, D. (2013). In-situ transmission electron microscopy of biological specimens in liquid. Microsc. Microanal. 19, 258–259. doi: 10.1017/S1431927613003280
Egan, G. C., Li, T. T., Roehling, J. D., McKeown, J. T., and Campbell, G. H. (2018). In situ dynamic TEM characterization of unsteady crystallization during laser processing of amorphous germanium. Acta Mater. 143, 13–19. doi: 10.1016/j.actamat.2017.10.003
Evans, J. E., Jungjohann, K. L., Wong, P. C., Chiu, P., Dutrow, G. H., Arslan, I., et al. (2012). Visualizing macromolecular complexes with in situ liquid scanning transmission electron microscopy. Micron 43, 1085–1090. doi: 10.1016/j.micron.2012.01.018
Ferreon, A. C., Moran, C. R., Gambin, Y., and Deniz, A. A. (2010). Single-molecule fluorescence studies of intrinsically disordered proteins. Methods Enzymol Single Mol Tools. 472, 179–204. doi: 10.1016/S0076-6879(10)72010-3
Firpo, G., Angeli, E., Repetto, L., and Valbusa, U. (2015). Permeability thickness dependence of polydimethylsiloxane (PDMS) membranes. J. Membrane Sci. 481, 1–8. doi: 10.1016/j.memsci.2014.12.043
Fu, X., Chen, B., Tang, J., Hassan, M. T., and Zewail, A. H. (2017). Imaging rotational dynamics of nanoparticles in liquid by 4D electron microscopy. Science 355, 494–498. doi: 10.1126/science.aah3582
Fu, Z., Indrisiunaite, G., Kaledhonkar, S., Shah, B., Sun, M., Chen, B., et al. (2019). The structural basis for release factor activation during translation termination revealed by time-resolved cryogenic electron microscopy. Biophys. J. 116, 574a−575a. doi: 10.1016/j.bpj.2018.11.3090
Fuest, M., Schaffer, M., Nocera, G. M., Galilea-Kleinsteuber, R. I., Messling, J.-E., Heymann, M., et al. (2019). In situ microfluidic cryofixation for cryo focused ion beam milling and cryo electron tomography. Sci. Rep. 9:19133. doi: 10.1038/s41598-019-55413-2
Ghazal, A., Gontsarik, M., Kutter, J. P., Lafleur, J. P., Ahmadvand, D., Labrador, A., et al. (2016). Microfluidic platform for the continuous production and characterization of multilamellar vesicles: a synchrotron small-angle X-ray scattering (SAXS) study. J. Phys. Chem. Lett. 8, 73–79. doi: 10.1021/acs.jpclett.6b02468
Göttfert, F., Wurm, C., Mueller, V., Berning, S., Cordes, V., Honigmann, A., et al. (2013). Coaligned dual-channel STED nanoscopy and molecular diffusion analysis at 20 nm resolution. Biophys. J. 105, L01–L03. doi: 10.1016/j.bpj.2013.05.029
He, K., Liu, C., Yuan, Y., Song, B., Lu, Y.-, Shokuhfar, T., and Shahbazian-Yassar, R. (2019). In situ liquid cell transmission electron microscopy study of hydroxyapatite mineralization process. Microsc. Microanal. 25:1502. doi: 10.1017/S1431927619008249
Henderson, R. (2013). Avoiding the pitfalls of single particle cryo-electron microscopy: einstein from noise. Proc. Natl. Acad. Sci. U.S.A. 110, 18037–18041. doi: 10.1073/pnas.1314449110
Hoppe, S. M., Sasaki, D. Y., Kinghorn, A. N., and Hattar, K. (2013). In-situ transmission electron microscopy of liposomes in an aqueous environment. Langmuir 29, 9958–9961. doi: 10.1021/la401288g
Hua, X., Szymanski, C., Wang, Z., Zhou, Y., Ma, X., Yu, J., et al. (2016). Chemical imaging of molecular changes in a hydrated single cell by dynamic secondary ion mass spectrometry and super-resolution microscopy. Integr. Biol. 8, 635–644. doi: 10.1039/c5ib00308c
Huh, Y. S., Park, T. J., Yang, K., Lee, E. Z., Hong, Y. K., Lee, S. Y., et al. (2008). Advanced cleanup process of the free-flow microfluidic device for protein analysis. Ultramicroscopy 108, 1365–1370. doi: 10.1016/j.ultramic.2008.04.085
Hurbain, I., and Sachse, M. (2011). The future is cold: cryo-preparation methods for transmission electron microscopy of cells. Biol. Cell 103, 405–420. doi: 10.1042/BC20110015
Jones, S. T., Cagno, V., Janeček, M., Ortiz, D., Gasilova, N., Piret, J., et al. (2020). Modified cyclodextrins as broad-spectrum antivirals. Sci. Adv. 6:9318. doi: 10.1126/sciadv.aax9318
Karplus, M., and McCammon, J. A. (2002). Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 9, 646–652. doi: 10.1038/nsb0902-646
Kempner, E., and Schlegel, W. (1979). Size determination of enzymes by radiation inactivation. Anal. Biochem. 92, 2–10. doi: 10.1016/0003-2697(79)90617-1
Keskin, S., and de Jonge, N. (2018). Reduced radiation damage in transmission electron microscopy of proteins in graphene liquid cells. Nano Lett. 18, 7435–7440. doi: 10.1021/acs.nanolett.8b02490
Keskin, S., Kunnas, P., and de Jonge, N. (2019). Liquid-phase electron microscopy with controllable liquid thickness. Nano Lett. 19, 4608–4613. doi: 10.1021/acs.nanolett.9b01576
Klein, K., Anderson, I., and de Jonge, N. (2011). Transmission electron microscopy with a liquid flow cell. J. Microsc. 242, 117–123. doi: 10.1111/j.1365-2818.2010.03484.x
Knoška, J., Adriano, L., Awel, S., Beyerlein, K. R., Yefanov, O., Oberthuer, D., et al. (2020). Ultracompact 3D microfluidics for time-resolved structural biology. Nat. Commun. 11:657. doi: 10.1038/s41467-020-14434-6
Koc, Y., Mello, A. J., Mchale, G., Newton, M. I., Roach, P., and Shirtcliffe, N. J. (2008). Nano-scale superhydrophobicity: suppression of protein adsorption and promotion of flow-induced detachment. Lab Chip. 8, 582–6. doi: 10.1039/b716509a
Korogod, N., Petersen, C. C. H., and Knott, G. W. (2015). Ultrastructural analysis of adult mouse neocortex comparing aldehyde perfusion with cryo fixation. Elife 4:05793. doi: 10.7554/eLife.05793
Lagrange, T., Campbell, G. H., Reed, B., Taheri, M., Pesavento, J. B., Kim, J. S., et al. (2008). Nanosecond time-resolved investigations using the in situ of dynamic transmission electron microscope (DTEM). Ultramicroscopy 108, 1441–1449. doi: 10.1016/j.ultramic.2008.03.013
Leenheer, A. J., Jungjohann, K. L., Zavadil, K. R., Sullivan, J. P., and Harris, C. T. (2015). Lithium electrodeposition dynamics in aprotic electrolyte observed in situ via transmission electron microscopy. ACS Nano 9, 4379–4389. doi: 10.1021/acsnano.5b00876
Leijten, Z. J., Keizer, A. D., With, G. D., and Friedrich, H. (2017). Quantitative analysis of electron beam damage in organic thin films. J. Phys. Chem. C 121, 10552–10561. doi: 10.1021/acs.jpcc.7b01749
Li, B., Ge, P., Murray, K. A., Sheth, P., Zhang, M., Nair, G., et al. (2018). Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun. 9:3609. doi: 10.1038/s41467-018-05971-2
Li, W., Zhang, L., Ge, X., Xu, B., Zhang, W., Qu, L., et al. (2018). Microfluidic fabrication of microparticles for biomedical applications. Chem. Soc. Rev. 47, 5646–5683. doi: 10.1039/C7CS00263G
Loh, N. D., Sen, S., Bosman, M., Tan, S. F., Zhong, J., Nijhuis, C. A., et al. (2017). Multistep nucleation of nanocrystals in aqueous solution. Nat. Chem. 9, 77–82. doi: 10.1038/nchem.2618
Lutz-Bueno, V., Zhao, J., Mezzenga, R., Pfohl, T., Fischer, P., and Liebi, M. (2016). Scanning-SAXS of microfluidic flows: nanostructural mapping of soft matter. Lab Chip. 16, 4028–4035. doi: 10.1039/C6LC00690F
Mäeots, M., Lee, B., Nans, A., Jeong, S., Esfahani, M. M., Ding, S., et al. (2020). Modular microfluidics enables kinetic insight from time-resolved cryo-EM. Nat. Commun. 11:3465. doi: 10.1038/s41467-020-17230-4
Martin, H. P., Brooks, N. J., Seddon, J. M., Luckham, P. F., Terrill, N. J., Kowalski, A. J., et al. (2016). Microfluidic processing of concentrated surfactant mixtures: online SAXS, microscopy and rheology. Soft Matter. 12, 1750–1758. doi: 10.1039/C5SM02689J
Matsubu, J. C., Zhang, S., DeRita, L., Marinkovic, N. S., Chen, J. G., Graham, G. W., et al. (2017). Adsorbate-mediated strong metal–support interactions in oxide-supported Rh catalysts. Nat. Chem. 9, 120–127. doi: 10.1038/nchem.2607
Mattern, K., Trotha, J. W., Erfle, P., Köster, R. W., and Dietzel, A. (2020). NeuroExaminer: an all-glass microfluidic device for whole-brain in vivo imaging in zebrafish. Commun. Biol. 3:311. doi: 10.1038/s42003-020-1029-7
Mirsaidov, U., Zheng, H., Casana, Y., and Matsudaira, P. (2012). Imaging protein structure in water at 2.7 nm resolution by transmission electron microscopy. Biophys. J. 102, L15–L17. doi: 10.1016/j.bpj.2012.01.009
Moser, T. H., Mehta, H., Park, C., Kelly, R. T., Shokuhfar, T., and Evans, J. E. (2018). The role of electron irradiation history in liquid cell transmission electron microscopy. Sci. Adv. 4:1202. doi: 10.1126/sciadv.aaq1202
Narayanan, S., Shahbazian-Yassar, R., and Shokuhfar, T. (2020). In situ visualization of ferritin biomineralization via graphene liquid cell-transmission electron microscopy. ACS Biomater. Sci. Eng. 6, 3208–3216. doi: 10.1021/acsbiomaterials.9b01889
Neylon, C. (2008). Small angle neutron and X-ray scattering in structural biology: recent examples from the literature. Eur. Biophys. J. 37, 531–541. doi: 10.1007/s00249-008-0259-2
Nishiyama, H., Suga, M., Ogura, T., Maruyama, Y., Koizumi, M., Mio, K., et al. (2010). Atmospheric scanning electron microscope observes cells and tissues in open medium through silicon nitride film. J. Struct. Biol. 169, 438–449. doi: 10.1016/j.jsb.2010.01.005
Oldfield, C. J., and Dunker, A. K. (2014). Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 83, 553–584. doi: 10.1146/annurev-biochem-072711-164947
Park, J., Park, H., Ercius, P., Pegoraro, A. F., Xu, C., Kim, J. W., et al. (2015). Direct observation of wet biological samples by graphene liquid cell transmission electron microscopy. Nano Lett. 15, 4737–4744. doi: 10.1021/acs.nanolett.5b01636
Patel, A., Malinovska, L., Saha, S., Wang, J., Alberti, S., Krishnan, Y., et al. (2017). ATP as a biological hydrotrope. Science 356, 753–756. doi: 10.1126/science.aaf6846
Pawley, J. (2006). Handbook of Biological Confocal Microscopy. New York, NY: Springer. doi: 10.1007/978-0-387-45524-2
Peckys, D. B., and de Jonge, N. (2014). Liquid scanning transmission electron microscopy: imaging protein complexes in their native environment in whole eukaryotic cells. Microsc. Microanal. 20, 346–365. doi: 10.1017/S1431927614000099
Punjani, A., Rubinstein, J. L., Fleet, D. J., and Brubaker, M. A. (2017). CryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. doi: 10.1038/nmeth.4169
Raab, N., and Bachelet, I. (2017). Resolving biofilm topography by native scanning electron microscopy. J. Biol. Methods 4:e70. doi: 10.14440/jbm.2017.173
Rees, P., Wills, J. W., Brown, M. R., Barnes, C. M., and Summers, H. D. (2019). The origin of heterogeneous nanoparticle uptake by cells. Nat. Commun. 10:2341. doi: 10.1038/s41467-019-10112-4
Ring, E. A., and de Jonge, N. (2010). Microfluidic system for transmission electron microscopy. Microsc. Microanal. 16, 622–629. doi: 10.1017/S1431927610093669
Saar, K., Yates, E. V., Müller, T., Saunier, S., Dobson, C. M., and Knowles, T. P. (2016). Automated ex situ assays of amyloid formation on a microfluidic platform. Biophys. J. 110, 555–560. doi: 10.1016/j.bpj.2015.11.3523
Sakai, K., Charlot, F., Le Saux, T., Bonhomme, S., Nogué, F., Palauqui, J.-C., et al. (2019). Design of a comprehensive microfluidic and microscopic toolbox for the ultra-wide spatio-temporal study of plant protoplasts development and physiology. Plant Methods 15:79. doi: 10.1186/s13007-019-0459-z
Saliba, J., Daou, A., Damiati, S., Saliba, J., El-Sabban, M., and Mhanna, R. (2018). Development of microplatforms to mimic the in vivo architecture of CNS and PNS physiology and their diseases. Genes 9:285. doi: 10.3390/genes9060285
Sato, C., Yamazaki, D., Sato, M., Takeshima, H., Memtily, N., Hatano, Y., et al. (2019b). Calcium phosphate mineralization in bone tissues directly observed in aqueous liquid by atmospheric SEM (ASEM) without staining: microfluidics crystallization chamber and immuno-EM. Sci. Rep. 9:7352. doi: 10.1038/s41598-019-43608-6
Sato, C., Yamazawa, T., Ohtani, A., Maruyama, Y., Memtily, N., Sato, M., et al. (2019a). Primary cultured neuronal networks and type 2 diabetes model mouse fatty liver tissues in aqueous liquid observed by atmospheric SEM (ASEM): staining preferences of metal solutions. Micron 118, 9–21. doi: 10.1016/j.micron.2018.11.005
Schneider, N. M., Norton, M. M., Mendel, B. J., Grogan, J. M., Ross, F. M., and Bau, H. H. (2014). Electron–water interactions and implications for liquid cell electron microscopy. J. Phys. Chem. C 118, 22373–22382. doi: 10.1021/jp507400n
Schneider, S. W., Nuschele, S., Wixforth, A., Gorzelanny, C., Alexander-Katz, A., Netz, R. R., et al. (2007). Shear-induced unfolding triggers adhesion of von willebrand factor fibers. Proc. Natl. Acad. Sci. U.S.A. 104, 7899–7903. doi: 10.1073/pnas.0608422104
Shao, X., Gao, D., Chen, Y., Jin, F., Hu, G., Jiang, Y., et al. (2016). Development of a blood-brain barrier model in a membrane-based microchip for characterization of drug permeability and cytotoxicity for drug screening. Anal. Chim. Acta 934, 186–193. doi: 10.1016/j.aca.2016.06.028
Shin, Y., Chang, Y., Lee, D. S., Berry, J., Sanders, D. W., Ronceray, P., et al. (2018). Liquid nuclear condensates mechanically sense and restructure the genome. Cell 175, 1481–1491. doi: 10.1016/j.cell.2018.10.057
Solsona, M., Vollenbroek, J. C., Tregouet, C. B. M., Nieuwelink, A.-E., Olthuis, W., van den Berg, A., et al. (2019). Microfluidics and catalyst particles. Lab Chip. 19, 3575–3601. doi: 10.1039/C9LC00318E
Takeda, S., Kuwauchi, Y., and Yoshida, H. (2015). Environmental transmission electron microscopy for catalyst materials using a spherical aberration corrector. Ultramicroscopy 151, 178–190. doi: 10.1016/j.ultramic.2014.11.017
Textor, M., and de Jonge, N. (2018). Strategies for preparing graphene liquid cells for transmission electron microscopy. Nano Lett. 18, 3313–3321. doi: 10.1021/acs.nanolett.8b01366
Thiberge, S., Nechushtan, A., Sprinzak, D., Gileadi, O., Behar, V., Zik, O., et al. (2004). Scanning electron microscopy of cells and tissues under fully hydrated conditions. Proc. Natl. Acad. Sci. U.S.A. 101, 3346–3351. doi: 10.1073/pnas.0400088101
Tian, R., Hoa, X. D., Lambert, J., Pezacki, J. P., Veres, T., and Figeys, D. (2011). Development of a multiplexed microfluidic proteomic reactor and its application for studying protein–protein interactions. Anal. Chem. 83, 4095–4102. doi: 10.1021/ac200194d
Tian, R., Li, K., Shi, W., Ding, C., and Lu, C. (2019). In situ visualization of hydrophilic spatial heterogeneity inside microfluidic chips by fluorescence microscopy. Lab Chip. 19, 934–940. doi: 10.1039/C8LC01336E
Tomo, Y., Takahashi, K., Nishiyama, T., Ikuta, T., and Takata, Y. (2017). Nanobubble nucleation studied using fresnel fringes in liquid cell electron microscopy. Int. J. Heat Mass Transf. 108, 1460–1465. doi: 10.1016/j.ijheatmasstransfer.2017.01.013
Unocic, R. R., Sacci, R. L., Brown, G. M., Veith, G. M., Dudney, N. J., More, K. L., et al. (2014). Quantitative electrochemical measurements using in situ ec-S/TEM devices. Microsc. Microanal. 20, 452–461. doi: 10.1017/S1431927614000166
Uversky, V. N. (2013). A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Sci. 22, 693–724. doi: 10.1002/pro.2261
Uversky, V. N., Oldfield, C. J., and Dunker, A. K. (2008). Intrinsically disordered proteins in human diseases: introducing the D2 concept. Ann. Rev. Biophys. 37, 215–246. doi: 10.1146/annurev.biophys.37.032807.125924
VandenBussche, E. J., and Flannigan, D. J. (2019). Reducing radiation damage in soft matter with femtosecond-timed single-electron packets. Nano Lett. 19, 6687–6694. doi: 10.1021/acs.nanolett.9b03074
Wang, H., Li, B., Kim, Y.-J., Kwon, O.-H., and Granick, S. (2020). Intermediate states of molecular self-assembly from liquid-cell electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 117, 1283–1292. doi: 10.1073/pnas.1916065117
Wang, S., Yang, X., Wu, F., Min, L., Chen, X., and Hou, X. (2019). Inner surface design of functional microchannels for microscale flow control. Small 16:1905318. doi: 10.1002/smll.201905318
Weiss, C., Carriere, M., Fusco, L., Capua, I., Regla-Nava, J. A., Pasquali, M., et al. (2020). Toward nanotechnology-enabled approaches against the COVID-19 pandemic. ACS Nano 14, 6383–6406. doi: 10.1021/acsnano.0c03697
Whitesides, G. M. (2006). The origins and the future of microfluidics. Nature 442, 368–373. doi: 10.1038/nature05058
Williams, D. B., and Carter, C. B. (2009). The Transmission Electron Microscopy. Boston, MA: Springer. doi: 10.1007/978-0-387-76501-3_1
Wu, Y., Chen, X., Li, C., Fang, J., and Liu, H. (2019). In situ liquid cell TEM observation of solution-mediated interaction behaviour of Au/CdS nanoclusters. New J. Chem. 43, 12548–12554. doi: 10.1039/C9NJ03520F
Wu, Z., Zheng, Y., Lin, L., Mao, S., Li, Z., and Lin, J. (2020). Controllable synthesis of multicompartmental particles using 3D microfluidics. Angew. Chem. 132, 2245–2249. doi: 10.1002/ange.201911252
Yamazawa, T., Nakamura, N., Sato, M., and Sato, C. (2016). Secretory glands and microvascular systems imaged in aqueous solution by atmospheric scanning electron microscopy (ASEM). Microsc. Res. Tech. 79, 1179–1187. doi: 10.1002/jemt.22773
Yang, L., Yu, X.-Y., Zhu, Z., Iedema, M. J., and Cowin, J. P. (2011a). Probing liquid surfaces under vacuum using SEM and ToF-SIMS. Lab Chip. 11, 2481–4. doi: 10.1039/c0lc00676a
Yang, L., Yu, X.-Y., Zhu, Z., Thevuthasan, T., and Cowin, J. P. (2011b). Making a hybrid microfluidic platform compatible for in situ imaging by vacuum-based techniques. J. Vac. Sci. Technol. A 29:061101. doi: 10.1116/1.3654147
Yasukawa, T., Nagamine, K., Horiguchi, Y., Shiku, H., Koide, M., Itayama, T., et al. (2008). Electrophoretic cell manipulation and electrochemical gene-function analysis based on a yeast two-hybrid system in a microfluidic device. Anal. Chem. 80, 3722–3727. doi: 10.1021/ac800143t
Yu, J., Zhou, Y., Engelhard, M., Zhang, Y., Son, J., Liu, S., et al. (2020). In situ molecular imaging of adsorbed protein films in water indicating hydrophobicity and hydrophilicity. Sci. Rep. 10:3695. doi: 10.1038/s41598-020-60428-1
Yu, X. (2020). In situ, in vivo, and in operando imaging and spectroscopy of liquids using microfluidics in vacuum. J. Vac. Sci. Technol. A 38:040804. doi: 10.1116/1.5144499
Yuk, J. M., Park, J., Ercius, P., Kim, K., Hellebusch, D. J., Crommie, M. F., et al. (2012). High-resolution EM of colloidal nanocrystal growth using graphene liquid cells. Science 336, 61–64. doi: 10.1126/science.1217654
Yulaev, A., Lipatov, A., Lu, A. X., Sinitskii, A., Leite, M. S., and Kolmakov, A. (2016). Imaging and analysis of encapsulated objects through self-assembled electron and optically transparent graphene oxide membranes. Adv. Mater. Interfaces 4:1600734. doi: 10.1002/admi.201600734
Zheng, B., Roach, L. S., and Ismagilov, R. F. (2003). Screening of protein crystallization conditions on a microfluidic chip using nanoliter-size droplets. J. Am. Chem. Soc. 125, 11170–11171. doi: 10.1021/ja037166v
Keywords: in situ, liquid cell electron microscopy, microfluidic, bioimaging, nanoscale, dynamics
Citation: Han Z and Porter AE (2020) In situ Electron Microscopy of Complex Biological and Nanoscale Systems: Challenges and Opportunities. Front. Nanotechnol. 2:606253. doi: 10.3389/fnano.2020.606253
Received: 14 September 2020; Accepted: 13 November 2020;
Published: 03 December 2020.
Edited by:
Tamilselvan Mohan, Graz University of Technology, AustriaReviewed by:
Andrea Lassenberger, Institut Laue-Langevin, FranceChikara Sato, National Institute of Advanced Industrial Science and Technology (AIST), Japan
Copyright © 2020 Han and Porter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexandra E. Porter, YS5wb3J0ZXJAaW1wZXJpYWwuYWMudWs=
 Zexiang Han
Zexiang Han Alexandra E. Porter*
Alexandra E. Porter*