
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Neurosci. , 25 August 2022
Sec. Molecular Signalling and Pathways
Volume 15 - 2022 | https://doi.org/10.3389/fnmol.2022.974208
Neuronal communication requires precise connectivity of neurite projections (axons and dendrites). Developing neurites express cell-surface receptors that interpret extracellular cues to enable correct guidance toward, and connection with, target cells. Spatiotemporal regulation of neurite guidance molecule expression by transcription factors (TFs) is critical for nervous system development and function. Here, we review how neurite development is regulated by TFs in the Caenorhabditis elegans nervous system. By collecting publicly available transcriptome and ChIP-sequencing data, we reveal gene expression dynamics during neurite development, providing insight into transcriptional mechanisms governing construction of the nervous system architecture.
Animals have developed nervous systems to enable the transmission of information over long distances. Information transfer between neurons relies on neurite projections called axons and dendrites. Correct neurite development is therefore critical for efficient signal transduction between neurons within a neuronal circuit. During development, neurites are guided to their targets by attractive and repulsive cues from surrounding cells/tissues and the extracellular matrix (ECM) (Adler et al., 2006; Wang et al., 2008; Dong et al., 2016; Miller and Suter, 2018). Many conserved ligands and receptors involved in neurite guidance have been identified over the last three decades (Tables 1, 2). However, diverse neuron sub-types within a nervous system have specific neurite projection patterns that are guided by distinct gene expression programs. Hence, neurite development requires precisely controlled spatiotemporal expression of intrinsic and extrinsic factors.
Transcription Factors (TFs) are key regulators of gene expression that control multiple processes of neuronal development, including polarization, migration and neurite guidance (Shirasaki and Pfaff, 2002; Santiago and Bashaw, 2014; Hayashi et al., 2015; Masserdotti et al., 2016). TFs regulate neurite development by controlling signaling, cell-adhesion and junction molecules, and cytoskeleton modifiers (Nobrega-Pereira and Marin, 2009; Santiago and Bashaw, 2014; Masserdotti et al., 2016). TFs can regulate neurite development cell-autonomously or regulate the expression of cues and receptors from surrounding tissues (Rauthan et al., 2007; Yoshimura et al., 2008). The complexity and dynamic nature of the nervous system makes in vivo analysis of regulatory mechanisms governing neurite development challenging. Animal models have been used to study neurite development, including mouse (Kuwajima et al., 2017), rat (Fontanet et al., 2013), zebrafish (Koh et al., 2020), fruit fly (Contreras et al., 2018), and Caenorhabditis elegans (Chisholm et al., 2016).
The unparalleled detail of anatomical and molecular maps available for C. elegans renders it an exceptional model for studying fundamental requirements of nervous system development (White et al., 1986; Durbin, 1987; Cao et al., 2017; Cook et al., 2019; Packer et al., 2019; Taylor et al., 2021). C. elegans is a free-living nematode, with a short life span (∼3 weeks) and a transparent body that enables in vivo visualization of cellular structures at all stages of development. The C. elegans nervous system is small, containing 302 neurons and 56 glial cells (Figure 1; White et al., 1986). Importantly, neuronal positions are invariant and the synaptic connectivity (connectome) of these neurons has been mapped (Cook et al., 2019). Furthermore, single-cell transcriptomes of C. elegans neurons at distinct developmental stages are available (Cao et al., 2017; Packer et al., 2019; Taylor et al., 2021), enabling the study of how genes, including neurite development regulators and TFs, are expressed during neuronal development. Taken together, the depth of knowledge and ease of experimentation allow complex regulatory mechanisms required for neurite development to be studied in C. elegans.
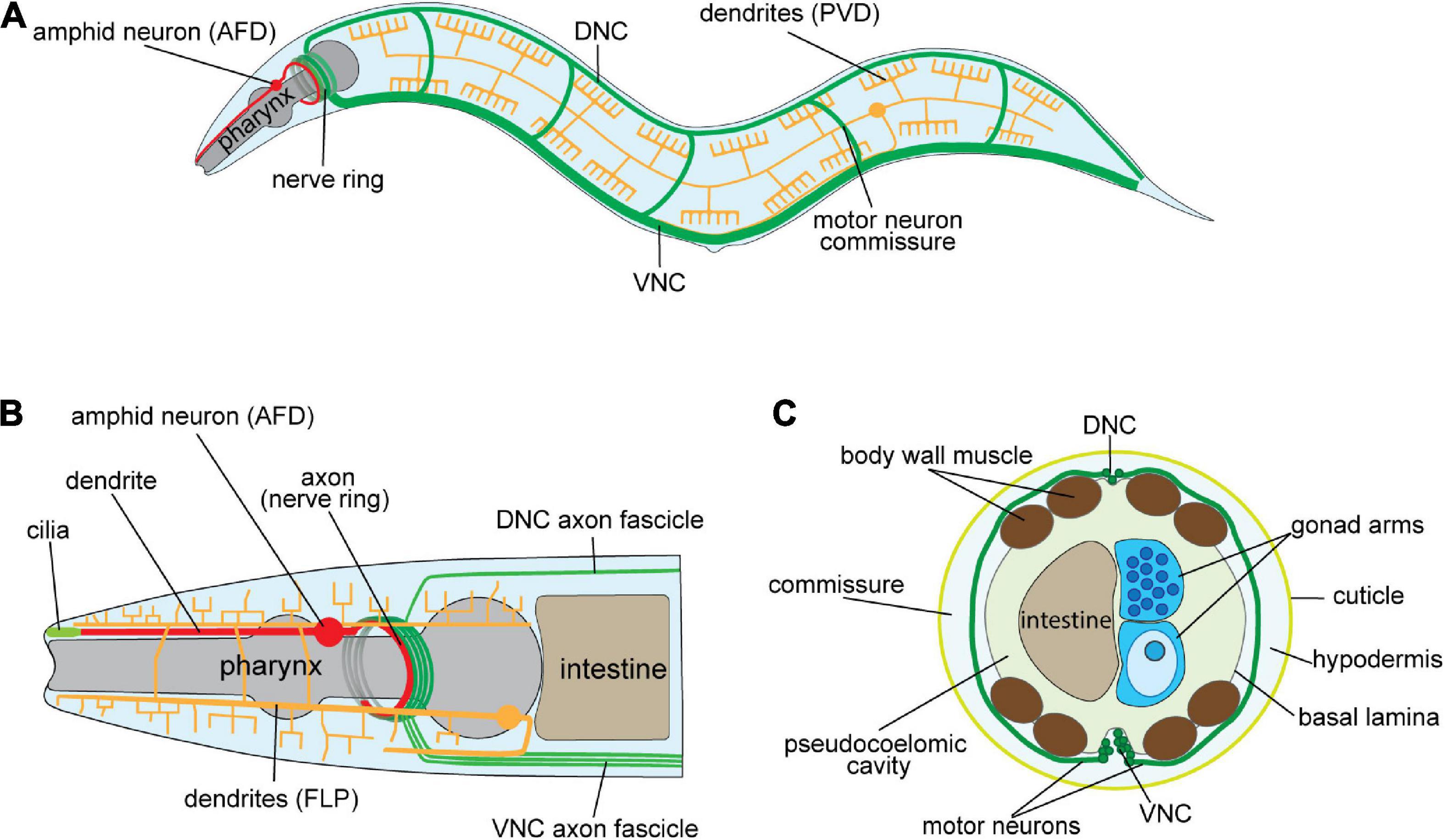
Figure 1. Architecture of the Caenorhabditis elegans nervous system. (A) Schematic of the adult C. elegans hermaphrodite nervous system (lateral view). Examples of major axo-dendritic processes: dorsal/ventral nerve cords, and circumferential commissures (DNC/VNC—green), amphid neurons, e.g., AFD (red), and the multi-dendritic PVD neurons (yellow) are shown. (B) Schematic of the C. elegans hermaphrodite anterior nervous system. Axons from the DNC, VNC and amphid neurons enter the nerve ring for integrating information and governing behavior. Amphid neurons (e.g., AFD) extend dendrites to the nose tip where cilia sense environmental cues. The FLP neurons have extensive dendritic arbors that, as with the PVD neurons in (A), enable mechano- and thermo-sensation. (C) Schematic cross-section of the C. elegans mid-body showing longitudinal axon tracts, motor neurons circumferential commissures (DNC/VNC—green), and the location of other major tissues. The circumferential commissures are placed between the hypodermis and basal lamina.
Here, we reviewed all published neurite development-related TFs in C. elegans. In addition, we collected and integrated bulk and single-cell transcriptome data to extract expression patterns of ligands, receptors and TFs involved in neurite development (Cao et al., 2017; Packer et al., 2019; Papatheodorou et al., 2020; Sun and Hobert, 2021; Taylor et al., 2021). By analyzing ChIP-seq data we also constructed a network of potential TFs that bind upstream of genes encoding ligands and receptors that control neurite development (Gerstein et al., 2010). Thus, we provide an integrated view of neurite developmental regulators providing insight into mechanisms governing nervous system development.
Neurites are neuronal projections that transmit information within the nervous system and to non-neuronal cells and tissues (Cook et al., 2019). Neurites are classified as axons and dendrites that transfer information from and to the cell body (soma), respectively (Goaillard et al., 2019). In most vertebrates, axons are insulated by a protective myelin sheath that enables rapid electrical conductance, whereas some animals such as C. elegans lack such structures (Oikonomou and Shaham, 2011). Depending on the neuron type, dendrites may be a single projection or highly arborized to receive information from multiple neurons (Agostinone and Di Polo, 2015). Neurites directly transmit information to target cells through synapses that release specific neurotransmitters, or through electrical junctions that allow ion flow (Pereda, 2014; Sudhof, 2018).
Correct neurite development requires precise regulation of intrinsic cytoskeleton-driven mechanisms and extrinsic molecular interactions between cell-surface receptors and the surrounding environment. In the absence of external factors, neurite development of mammalian hippocampal cells in culture is classified into five main stages: (I) membrane ruffling by protrusion of cell-surface lamellipodia or filopodia, (II) emergence of short immature neurites, (III) axon establishment through stochastic growth of one neurite, (IV) conversion of all other neurites into dendrites, (VI) generation of dendritic spines and axonal synapses to establish a neuronal circuit (Dotti et al., 1988; Goslin and Banker, 1989; Figure 2A). The same scenario is expected to occur in vivo; however, initiation and growth of axons and dendrites is directed by gradients of guidance cues in the environment and by physical barriers (Hutter, 2003; Adler et al., 2006; Figure 2B).
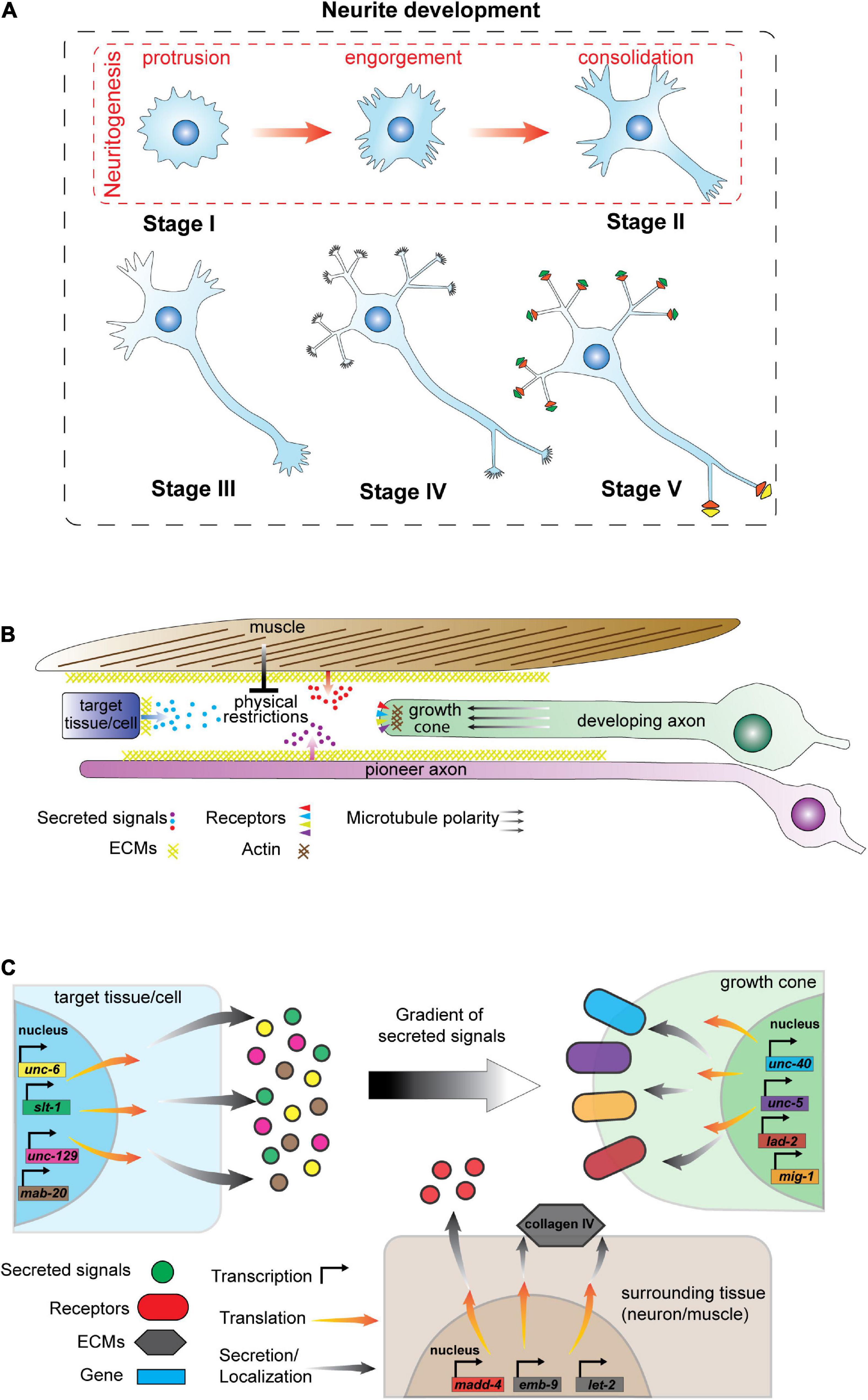
Figure 2. Neurite development and regulatory mechanisms. (A) Stages of neurite development in vitro in the absence of environmental cues. Neurite development consists of five stages: (I) lamellipodia or filopodia formation, (II) emergence of short and dynamic neurites, (III) axon establishment (IV) dendrite formation, and (V) formation of dendritic spines, axonal synapses and neuron circuits. Stage I and II are defined as neuritogenesis that has three phases: (1) protrusions that form lamellipodia or filopodia on the membrane; (2) engorgement of cytoskeletal components that enter lamellipodia and filopodia, and (3) consolidation of neurite growth cones by establishing a cylindrical cytoskeletal shaft. (B) Schematic of signaling events impacting C. elegans neurite development. Axo-dendritic guidance is controlled by signals released from other cells/tissues, the extracellular matrix (ECM) and physical barriers such as muscle. In addition, pioneer axons guide follower axons along specific tracks. (C) Schematic of signals and receptors that regulate axon development in C. elegans. Signal molecules (left) are synthesized by target and surrounding tissues/cells and are released from the cells or localized into the plasma membrane as a component of the ECM. Gradients of secreted signals guide growing axons. Elongating neurites (right) express receptors that localize at the growth cone to detect and respond to external signals. Neurons express distinct gene batteries and here we present candidate signals and receptors that may not expressed in the same cell at a given time.
Neurite development is therefore composed of projection, polarization and extension of neurites that are controlled by dedicated signaling pathways and cytoskeletal rearrangement. Detailed reviews on neurite developmental processes have recently been published: neurite polarity (Yogev and Shen, 2017; Armakola and Ruvkun, 2019); the cytoskeleton in neurite formation (Sainath and Gallo, 2015; Miller and Suter, 2018) and neurite development and repair (Richardson and Shen, 2019). Mechanisms governing axon guidance in C. elegans have also been reviewed by Chisholm and colleagues (Chisholm et al., 2016). In general, polarization determines how the cytoskeleton rearranges to direct projection and neurite formation (Schelski and Bradke, 2017), whereas, cytoskeletal stability regulates neurite growth by providing mechanical support, reviewed by Miller and Suter (2018). For example, neurite initiation, also known as neuritogenesis, begins with protrusion of lamellipodia or filopodia on the cell surface and movement of microtubules and other components into these structures, reviewed by Flynn (2013). Neurites are consolidated through cytoskeletal reorganization into a cylindrical structure at their base. Subsequently, polarization is shaped by the gradient(s) of cues that determine the direction of axo-dendritic growth (Schelski and Bradke, 2017; Miller and Suter, 2018).
Neuron polarization is the result of a combination of internal and external cues that impact cytoskeletal rearrangement (Miller and Suter, 2018). Internal cues are inherited from the existing apical-basal polarity of progenitor cells (Zolessi et al., 2006; Pollarolo et al., 2011), while extracellular cues can be produced by surrounding neuronal or non-neuronal tissues. External cues found to regulate neurite development in C. elegans are listed at Table 1. Polarity-related signaling molecules, such as UNC-34/Ena/VASP in C. elegans, and Cdc42 in mammals predominantly function by localizing at axonal growth cones and rearranging the cytoskeleton (Gitai et al., 2003; Schwamborn and Puschel, 2004; Fleming et al., 2010). UNC-34 is a downstream effector of the UNC-6/Netrin cue and its receptor UNC-40/DCC that are critical conserved regulators of axon guidance (Adler et al., 2006).
Following axo-dendritic growth, neurons communicate with each other by forming synaptic connections (Sudhof, 2018). Multiple processes are involved in synapse formation, including dendrite branching and pruning (Riccomagno and Kolodkin, 2015). Dendrite branching is a crucial step for establishing connections between different cells that also requires cytoskeletal rearrangement and cell surface molecule interactions, reviewed by Jin and Kim (2020). Additionally, neurite development requires other important processes including axon arborization (Gibson and Ma, 2011) and neurite pruning (Schuldiner and Yaron, 2015). With the latter, a critical step that removes superfluous neurites formed during development (Lu and Mizumoto, 2019).
In summary, neurite development is a multi-step process that can initiate during embryogenesis and continue over the lifespan of an organism. In C. elegans most neurons develop during embryogenesis (222 of the 302 neurons in the hermaphrodites), while others (80 of the 302 neurons) develop post-embryonically (White et al., 1986; Durbin, 1987). Here, we focus on axo-dendritic development in C. elegans and describe the mechanisms and TFs that regulate these processes.
Correct neurite development is controlled by coordinated intrinsic and extrinsic molecular mechanisms. In C. elegans, the majority of molecules identified to regulate neurite development were found by studying axon guidance. Multiple factors impact axon development including physical restrictions imposed by surrounding tissues, cues from other cells, cell surface molecules and receptors (Durbin, 1987; Baum and Garriga, 1997; Kim and Wadsworth, 2000; Levy-Strumpf and Culotti, 2014; Figure 2C). Tables 1, 2 lists the cues and membrane proteins shown to control C. elegans neurite development. In addition to these cues and receptors, there are multiple intracellular proteins important for cytoskeletal rearrangement and growth cone navigation that are not reviewed here (more details from Chisholm et al., 2016). Instead, we focus on the cues and receptors involved in the neurite development, with a focus on axon guidance.
Axon guidance cues can be secreted from surrounding tissues or presented within the ECM (Table 1). The ECM is a complex structure containing laminins, collagen IV and nidogen that provides a substratum for axon guidance by localizing guidance cues (Forrester and Garriga, 1997; Kim and Wadsworth, 2000; Huang et al., 2003; Kao et al., 2006). ECM components are sensed by receptors such as integrins (Baum and Garriga, 1997), dystroglycan (Johnson et al., 2006; Lindenmaier et al., 2019) and Discoidin domain receptors (Unsoeld et al., 2013) to control axon development. The ECM consists of multiple distinct proteins (Yue, 2014) and cells/tissues may express different combinations of ECM molecules. Additionally, post-translational regulation of ECM proteins impact axon guidance. For example, the prolyl 4-hydroxylase DPY-18 regulates HSN axon guidance by impacting the folding of collagen IV proteins encoded by emb-9 and let-2 (Torpe and Pocock, 2014). Therefore, the specific effects of ECM regulation on neurite development may be highly context- and structure-dependent.
Secreted guidance cues may act as axon guidance attractants or repellents. Most cues in C. elegans were identified in reverse and forward genetic screens for ventral nerve cord (VNC) and circumferential axon guidance defects (Kim and Wadsworth, 2000; Hao et al., 2001; Boulin et al., 2006). The C. elegans VNC axons are present in fascicles that extend in the posterior-anterior axis (White et al., 1986). Some VNC axons, known as pioneer axons, extend prior to others and provide a platform for follower axons in the same tract by producing extracellular cues, such as UNC-6/Netrin and SAX-3/Robo (Klose and Bentley, 1989; Wadsworth et al., 1996; Hutter, 2003). In contrast, ventral-dorsal circumferential axons are not guided by pioneer axons and grow in response to trophic cues that act as attractants or repellents. Trophic guidance cues are classified in different groups, including Netrins, Transforming growth factor-β, Wnts, Slits and Semaphorins. We will briefly discuss each group in the following sections.
UNC-6/Netrin and SLT-1/Slit are well-studied and conserved guidance molecules. UNC-6/Netrin is expressed by ventral cells and possibly forms a gradient to direct axon guidance (Kulkarni et al., 2008; Hao et al., 2010; Smith et al., 2012; Levy-Strumpf and Culotti, 2014). Depending on Netrin receptor expression, UNC-6/Netrin can be an attractive or repulsive cue. For example, UNC-40/DCC expression directs axon attraction toward the UNC-6/Netrin, whereas, expression of UNC-5, either alone or in combination with UNC-40/DCC, repels axons from high levels of UNC-6 (Norris and Lundquist, 2011). SLT-1/Slit, a cue expressed in dorsal muscle, is interpreted by receptors such as SAX-3/Robo and EVA-1 that coordinate dorso-ventral guidance of the AVM axon in parallel with UNC-6/Netrin (Hao et al., 2001; Fujisawa et al., 2007).
Growth factors, such as TGF-β ligands, are prominent guidance cues during axon development (Table 1). UNC-129/TGF-β is secreted by dorsal body wall muscle and directs axon guidance through unknown non-canonical TGF-β receptors (MacNeil et al., 2009; Baltaci et al., 2022). Notably, UNC-129 binds to the Netrin receptor UNC-5 and facilitates UNC-6/Netrin repulsive guidance by enhancing UNC-5 and UNC-40 signaling (MacNeil et al., 2009). The precise roles of growth factors in axon guidance have recently been reviewed by Onesto et al. (2021).
The C. elegans Wnt ligands EGL-20, CWN-1, CWN-2 and LIN-44 also regulate axon development (Hilliard and Bargmann, 2006; Pan et al., 2006; Kennerdell et al., 2009; Kirszenblat et al., 2011). EGL-20 and CWN-1 regulate anterior-posterior axon guidance of the AVM and PVM neurons by binding to Frizzled receptors MIG-1 and MOM-5 (Pan et al., 2006). While LIN-44 and the Frizzled receptor LIN-17 regulate axon and dendrite development of PLM and PQR neurons, respectively (Hilliard and Bargmann, 2006; Kirszenblat et al., 2011).
Some members of the Semaphorin family and ECM components are released from cells to guide axons. For example, MAB-20/Sema2 is a secreted semaphorin that interacts with PLX-2/Plexin and LAD-2/L1CAM to guide axons of the DA, DB and SDQL neurons (Roy et al., 2000; Wang et al., 2008; Dong et al., 2016). While ECM components, such as the metalloprotease MADD-4, are secreted from dorsal and VNC and cooperate with UNC-6/Netrin and SLT-1/Slit to control both muscle arm extension and AVM axon guidance following interaction with the UNC-40/DCC receptor (Seetharaman et al., 2011).
Neurite guidance proteins, expressed in neurons or surrounding tissues, must be precisely expressed to enable correct neurite growth. Such multi-layered regulatory processes involving multiple tissues require precisely controlled gene expression to support development of both pan-neuronal and neuron-specific neurite characteristics. These include fine navigation of neurites to establish synaptic connections, or the exit of neurites of specific neurons from nerve fascicles. The limited number of regulators compared to the diversity of the nervous system suggest that these regulators work in combinatorial patterns to regulate neuronal guidance decisions in a context-dependent manner. For example, SLT-1/Slit and UNC-6/Netrin cooperate in AVM ventral guidance such that removing both ligands causes more severe axon guidance defects compared to each single mutation (Hao et al., 2001). Notably, in C. elegans most studies of axon guidance have been performed on the VNC and circumferential axons and information is limited for more structurally complex regions such as the nerve ring. In the next section, we discuss how specific TFs regulate neurite development in C. elegans.
TF function in neurite development has been examined for both dendritic and axonal development. Here, we review the TFs with experimentally validated roles in neurite development in C. elegans. All TFs shown to regulate neurite development are listed in Table 3 but due to space limitations we only discuss the role of some key TFs below.
In C. elegans, most mechanistic information for dendrite development originates from studies of the PVD sensory neurons that develop post embryonically, and exhibit extensive and clearly visible dendritic branches (Figure 1A; Smith et al., 2010, 2013; O’Brien et al., 2017). PVD dendrites are classified into two groups: (i) pioneer dendrites that attach the epidermis, and (ii) commissural dendrites that fasciculate with motor neurons (White et al., 1986; Halevi et al., 2002; Tsalik et al., 2003). Several TFs regulate development of these dendritic structures. Among them, MEC-3, a LIM homeodomain TF, is a central regulator of PVD function and dendrite development (Tsalik et al., 2003; Smith et al., 2010). MEC-3 function is dose-dependent—with low levels driving dendritic branching of the PVDs, and high levels correlated with the simple dendritic morphology of the AVM mechanosensory neurons (Smith et al., 2013). AVM dendritic morphology is regulated by the aryl hydrocarbon receptor TF AHR-1, which elevates MEC-3 expression and suppresses MEC-3 target genes such as the HPO-30/Claudin protein (Smith et al., 2013). MEC-3 is also regulated by the POU TF UNC-86 and its loss phenocopies the mec-3 mutant dendrite developmental defects (Xue et al., 1992; Smith et al., 2010). Additionally, UNC-86 is required for IL2Q dendrite arborization (Schroeder et al., 2013). In contrast, PVD commissural dendrite development is regulated by EGL-46, a zinc-finger TF, and its binding partner, EGL-44, a TEA/YAP domain TF (O’Brien et al., 2017). Interestingly, EGL-46 is itself a target of MEC-3 (O’Brien et al., 2017). Therefore, MEC-3 regulates dendrite development of the PVDs through two parallel pathways: HPO-30/Claudin for pioneer dendrites and EGL-46/EGL-44 for commissural dendrites.
Axons can also regulate dendrite development of other neurons. For example, the ALA axon regulates PVD dendrite development in a contact-dependent manner. Here, MIG-6/Papilin, an ECM protein, UNC-6/Netrin and UNC-40/DCC regulate ALA axon development in early developmental stages and this axon controls PVD dendrite development in the later stages (Ramirez-Suarez et al., 2019).
Most C. elegans studies of axon development were conducted on neurons with easily observable axons to facilitate mutant isolation (Figure 1, a general view of the C. elegans nervous system). These neurons include the ventral motor neurons (DA, DB, DD, and VD), VNC interneurons (AVG, PVQ, and PVP) and the HSN motor neurons (Jin et al., 1994; Pujol et al., 2000; Wacker et al., 2003; Doonan et al., 2008; Pocock et al., 2008b; Weinberg et al., 2013; Table 3).
TFs can regulate axon development both cell-autonomously and non-autonomously. TFs may also indirectly regulate axon guidance of some neurons by regulating pioneer axon development. For example, LIN-11 and UNC-30 affect VNC axon patterning by regulating AVG and PVP pioneer axon development, respectively (Hutter, 2003). TFs can also regulate development of a particular neuron when expressed in another neuron. For example, MNM-2, a zinc finger domain TF, controls axon guidance of the M2 neurons while its expression occurs in the M3, sister neuron of M2. Apparently, MNM-2 regulates axon guidance by functioning alongside genes involved in cytoskeleton or membrane dynamics, and Netrin/TGF-β signaling pathways (Rauthan et al., 2007). Glial cells also regulate axon development. Yoshimura and colleagues showed that CEPsh glial cells control AWC and AFD axon guidance within the nerve ring, dendrite development and nerve ring assembly (Yoshimura et al., 2008). Further, the MLS-2 and VAB-3 TFs, non-autonomously regulate axon guidance by regulating CEPsh development (Yoshimura et al., 2008).
TFs also regulate neurite developmental factors such as ligands, receptors and plasma membrane components in neuronal and non-neuronal cells. For example, UNC-130, a Forkhead TF, regulates motor neuron axon development through the dorso-ventral axis in parallel to the Netrin signaling pathway (Nash et al., 2000). UNC-130 represses expression of the UNC-129 TGF-β ligand in ventral, but not dorsal muscles, leading to a dorso-ventral biased gradient of UNC-129 and resulting in dorsal axon guidance (Nash et al., 2000). Further, UNC-42, a homeodomain TF expressed in 15 classes of neurons, regulates axon outgrowth and guidance of multiple neurons including ASH, AVH, AVA, AVD, and HSN (Wightman et al., 1997; Brockie et al., 2001; Berghoff et al., 2021). UNC-42 regulates axon guidance molecules such as UNC-6/Netrin in command interneurons, and LAD-2/L1CAM, RIG-6/IGCAM and NCAM-1 in a subset of UNC-42 expressing neurons (Berghoff et al., 2021). Further, the CTBP-1 transcriptional corepressor regulates multiple features of SMD axonal development, including outgrowth, guidance and termination (Reid et al., 2015; Sherry et al., 2020). CTBP-1 represses expression of SAX-7/L1CAM, a fibronectin type-III protein critical for neuronal development and maintenance (Wang et al., 2005; Ramirez-Suarez et al., 2019). It was revealed that ectopic overexpression of SAX-7 in neurons causes defects in the SMD axon development, revealing the importance of precisely controlling the expression of axon guidance molecules (Sherry et al., 2020).
TFs also regulate asymmetric and sex-specific axon development (Bertrand et al., 2011; Serrano-Saiz et al., 2017). In C. elegans, some neurons show asymmetric gene expression in either the left or right member of a bilaterally symmetric neuron pair (Johnston and Hobert, 2003; Bertrand et al., 2011; Cochella et al., 2014). Bertrand and colleagues found that the HLH-16 TF exhibits higher expression in some neurons on the left side compared to those on the right. The authors showed that HLH-16 controls axon projection of both left and right AIY interneurons, but the left AIY is more dependent on HLH-16 expression (Bertrand et al., 2011). This suggests a role for TFs in determining the asymmetric development of axon projections. Furthermore, specific TFs might also be responsible for sex-specific neurite development. Among neurons that are present in both hermaphrodites and males, some exhibit sex-specific connectivity and differential gene expression (Oren-Suissa et al., 2016; Serrano-Saiz et al., 2017). For example, sexual dimorphic development of neurites has been documented for the PHC sensory neurons where male axons are longer, and dendrites are retracted. This phenotype depends on sexual maturity, as both sexes show similar PHC anatomy in immature larval stages. DMD-3, a member of Doublesex TFs family, cell-autonomously regulates PHC neurite morphology (Serrano-Saiz et al., 2017). In turn, DMD-3 expression is itself regulated by the sex-determination pathway, through the TRA-1 TF that represses DMD-3 expression in hermaphrodites (Goodwin and Ellis, 2002; Serrano-Saiz et al., 2017).
In addition to TFs, other types of gene regulatory proteins, including chromatin remodeling and histone modifying enzymes, regulate neurite development. Polycomb Group proteins (PcG) are histone modifiers that control gene expression (Di Croce and Helin, 2013). In C. elegans, SOP-2, a polycomb protein, and SOR-3, a protein containing histone-binding domain MBT, show PcG-like function and repress the expression of homeotic genes (Zhang et al., 2003; Yang et al., 2007). Both SOP-2 and SOR-3 are required for the development of dopaminergic and serotonergic neurons and also control axon guidance of B-type ray neurons in the C. elegans male tail (Yang et al., 2007). MIG-32 and SPAT-3, members of RING-type domain-containing proteins, regulate HSN migration and axon development. In addition, MIG-32, a PRC1-like protein, controls axon guidance in the VD and PVQ neurons (Karakuzu et al., 2009). Another example is ham-3 that encodes a subunit of the SWI/SNF chromatin-remodeling complex. HAM-3 controls serotonergic identity, migration and axon guidance of the HSN neurons (Weinberg et al., 2013). Therefore, epigenetic regulators and TFs control neurite development, likely by regulating the expression of specific genes.
The bias in study of neurite development toward those with clearly observable axons raises the question of how neurite development is regulated in the neurons within densely packed fascicles, such as those located in the C. elegans nerve ring. Another important question is whether pan-neuronal or neuron-specific gene regulatory systems control neurite development in all and specific neurons, respectively. As neurite development is controlled by multiple tissues, the existence of coordinated gene expression programs is likely. Therefore, TFs must orchestrate gene batteries in different cells to achieve this goal. To address these questions, analyzing the expression of the neurite development regulators (ligands, receptors and TFs) at a cell/tissue level throughout development would be a useful discovery tool for neurite specific developmental mechanisms. Here, we analyzed the expression of neurite development regulators through C. elegans development. Using bulk and single-neuron level transcriptomic datasets we examined how the expression of these genes change during neurite development.
The availability of single-cell and whole animal transcriptome data throughout C. elegans development enables tracking of neuronal developmental regulator expression dynamics (Papatheodorou et al., 2020). Bulk expression of ligands (19), receptors (31 including other cell-surface molecules) and neurite development-related TFs (35) shows the majority of these genes are expressed in the elongating and 3-fold embryonic stages, which is consistent with the timing of neurite development (Durbin, 1987; Figure 3A). Generally, most neurite regulatory genes show higher expression during embryogenesis and lower levels at the final larval stage (L4) and adult stages (Figures 3A,B). However, only a few genes show a positive pairwise correlation with synchronous expression patterns during development (Figure 3C). For example, CEH-14, SEM-4 TFs and LIN-18 Wnt receptor show a high correlation of expression (Figure 3C). The other cluster of co-expressed genes includes the TFs HAM-3, HLH-3, HLH-16, MIG-32, MNM-2, UNC-39, and VAB-7, and the receptors LON-2 and MOM-5 (Figure 3C). To the best of our knowledge, there is no study showing a regulatory relationship between the receptors and TFs in these clusters. Therefore, it would be interesting to investigate possible biological connections between them. Bulk gene expression from whole animals does not however, represent neuron-specific expression patterns. To obtain a more precise view, we tracked the expression of neurite development regulators in single neurons.
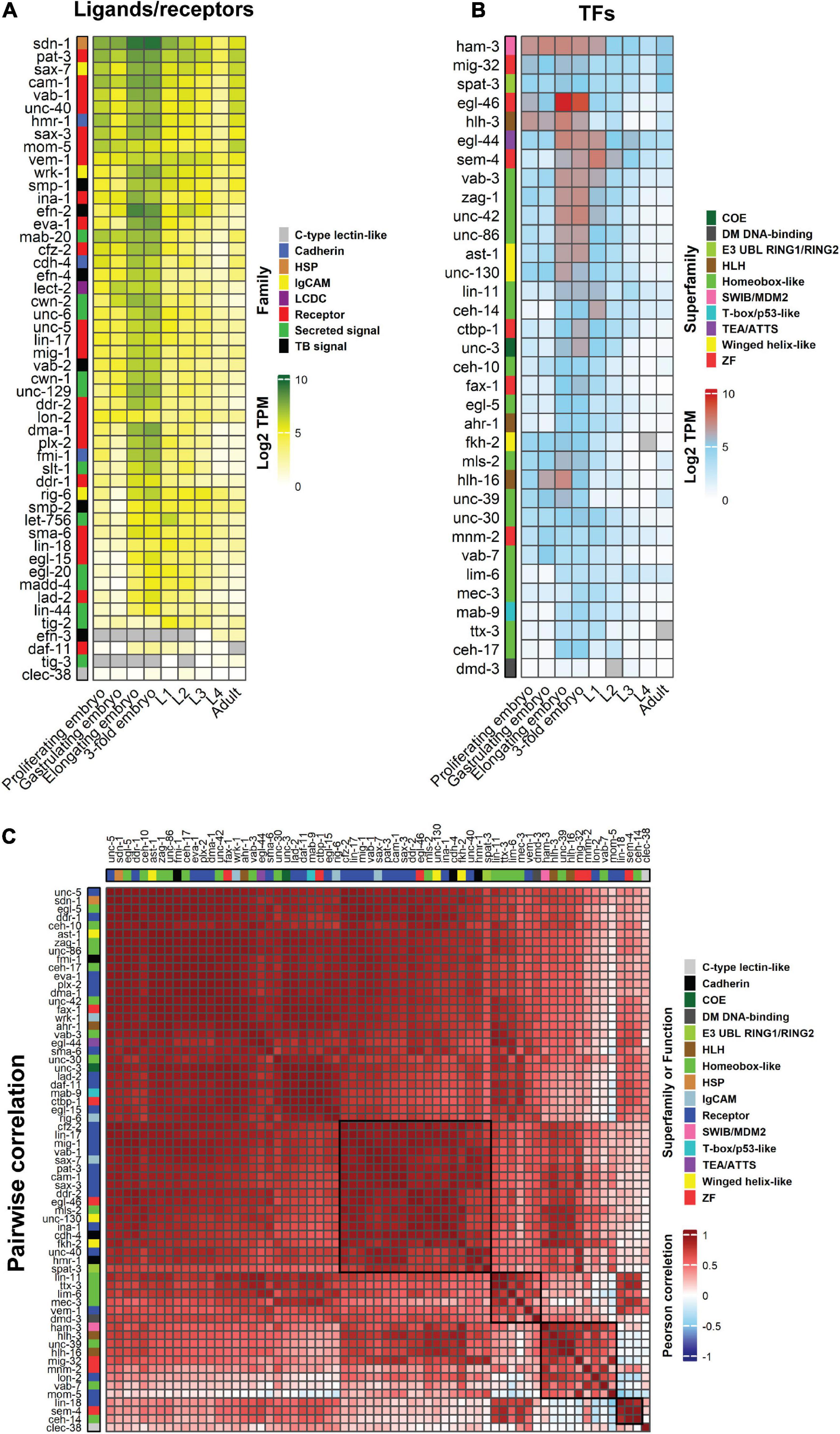
Figure 3. Heatmaps showing the temporal, quantitative and pairwise correlation of neurite development regulators. (A,B) Bulk expression of C. elegans ligands and receptors (A) and TFs (B) during embryogenesis, larval stages and adulthood. (C) Pairwise correlation between guidance receptors and TFs to identify expression correlations through animal development. Guidance ligands were excluded as they can be expressed in non-neuronal tissues. Genes are clustered according to Euclidean distance. Data are presented as Log2 TPM (transcripts per million) to normalize inter-sample differences. Legends show the ligand and receptor families or superfamily of TFs. Two histone modifier proteins, MIG-32 and SPAT-3, were also included in the TF lists. Gray cells = data not available. Black boxes delineate gene clusters. Gene expression values were obtained from Expression Atlas (ebi.ac.uk/gxa/home). HSP, heparan sulfate proteoglycans; LCDC, leukocyte cell-derived chemotaxin; TB, signal transmembrane-bound signal.
Single-cell C. elegans transcriptomes have been obtained from different stages of development, from embryonic through to larval stages (Cao et al., 2017; Packer et al., 2019; Sun and Hobert, 2021; Taylor et al., 2021). We focused on four neurons that extend neurites either in the embryonic (AFDs, DVA and CANs) or post-embryonic (PVDs) periods (Sundararajan et al., 2019; Figure 4A). More than half of the genes involved in neurite development are expressed at high levels during embryonic development of these neurons. The gene expression patterns of the AFDs, DVA and CANs are similar, though some genes exhibit a neuron-specific expression pattern. For example, the FAX-1 and CEH-10 TFs are highly expressed in DVA and CAN, respectively (Figure 4B). Interestingly, the MLS-2 and VAB-3 TFs, which control axon guidance of several neurons including the AFDs, are highly expressed in these neurons during embryonic development. However, these TFs regulate AFD axon guidance by functioning in cephalic sheath glia (Yoshimura et al., 2008). MAB-20/Sema2 semaphorin, which is a secreted cue, is expressed in all four neurons throughout development, suggesting that semaphorin may act as a common signal for neuron-neuron interactions. Likewise, the expression of many guidance receptor-encoding genes is detected in these neurons during embryogenesis, suggesting a role in neuron/neurite development. The PVD neurons, which extend dendrites post-embryonically (Sundararajan et al., 2019), express many of TFs including MEC-3, EGL-44, EGL-46, and UNC-86 that regulate PVD dendrite development (Smith et al., 2010, 2013; O’Brien et al., 2017; Figure 4C). Similarly, many guidance receptors are expressed in the PVD neurons at the L2 and L4 stages, highlighting their potential importance for dendrite development. For example, DMA-1 (a leucine-rich repeat protein), FMI-1/Flamingo, and SAX-3/Robo are required for PVD development (Liu and Shen, 2012; Hsu et al., 2020). SAX-7/L1CAM expression in hypodermal cells and the ALA neuron is also required for PVD development (Dong et al., 2013; Chen et al., 2019). However, this gene is also expressed in the PVDs, suggesting cell-autonomous and non-autonomous modes of action.
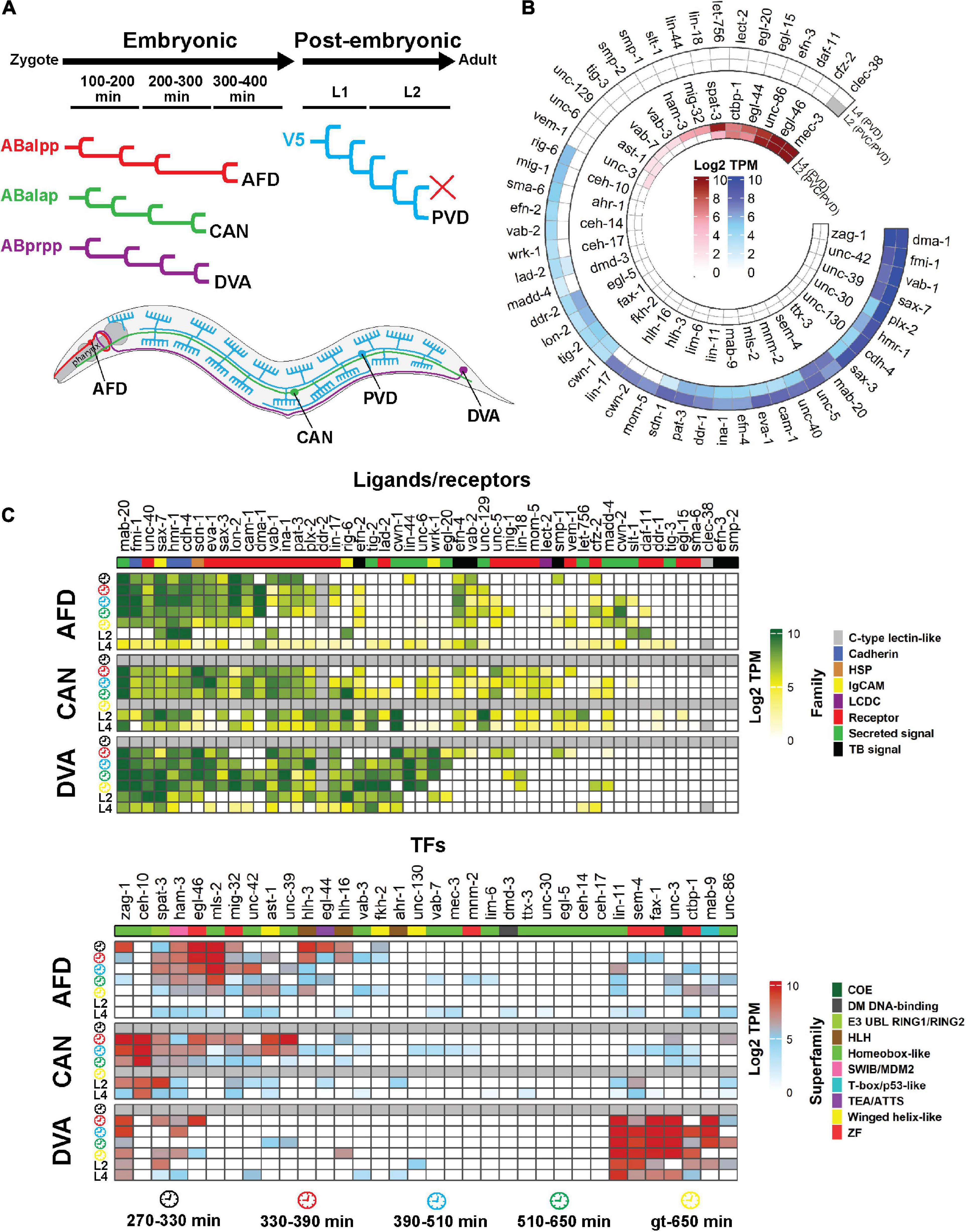
Figure 4. Single-cell expression analysis of neurite development regulators throughout development. (A) Four neurons (AFD, CAN, DVA and PVD) were selected based the availability of single-cell gene expression data and diversity of neuron type. The AFDs are sensory amphid neurons located in the head that extend anteriorly-directed dendrites and axons into the nerve ring (red). The CANs extend long neurites both anteriorly and posteriorly from the central cell body (green). DVA is a posterior sensory neuron that extends an anterior axon into the nerve ring (purple). PVDs are polymodal sensory neurons that extend elaborate dendritic arbors throughout the animal (blue). The AFDs, CANs and DVA develop embryonically while the PVD develops post-embryonically. (B) Expression of neurite developmental regulator ligands/receptors and TFs in the PVD (and PVC) neurons at the L2 and the PVDs at the L4 stage of development. The outer and inner heatmaps show the expression of ligands/receptors and TFs, respectively. (C) Single-cell expression of neurite development regulators in the AFD, CAN and DVA neurons during embryonic and larval development. Genes are clustered according to Euclidean distance. Data are present as Log2 TPM to normalize inter-sample differences. Legends show the family of the ligand and receptor proteins or superfamily of the TFs. Gray cells = data not available. Gene expression values were obtained from multiple studies: AFD, CAN and DVA at embryonic stages (Packer et al., 2019); AFD, CAN, DVA and PVD at L2 stage (Cao et al., 2017); AFD, CAN, DVA and PVD at L4 stage (Taylor et al., 2021). Clock faces denote time following bleaching of adult hermaphrodites. HSP, heparan sulfate proteoglycans; LCDC, leukocyte cell-derived chemotaxin; TB, signal Transmembrane-bound signal; gt, greater or equal to.
Taken together, many neurite development regulators are expressed during embryogenesis, when the majority of neurons develop. Interestingly, individual developing neurons show high expression of multiple neurite development regulators revealing the potential occurrence of combinatorial regulatory systems in each neuron. This hypothesis is supported by observations that removing individual neurite guidance regulators causes partially penetrant defects (Hao et al., 2001). Thus, our analysis of single-neuron transcriptome data through development reveals potential multifactor-controlled processes that requires regulatory tuning.
Although genetic screens have identified several TFs (Table 3) that regulate neurite development, there are likely other potential regulators controlling this process. Analysis of ChIP-seq data enables the identification of putative TF target genes (Furey, 2012). Using this approach, we identified binding sites for TFs within the regulatory regions of neurite development regulators. The availability of ChIP-seq data (modENCODE) for multiple C. elegans TFs (74) enabled us to generate a regulatory network of TFs that bind upstream of genes encoding ligands and receptors involved in neurite development (Gerstein et al., 2010; Figure 5A). Among these TFs, EGL-5, ZAG-1, and UNC-3 are known to regulate neurite development (Wightman et al., 1997; Prasad et al., 1998; Wacker et al., 2003; Hartin et al., 2017). As one would predict, we found that multiple TFs can bind within 5kb upstream of genes encoding ligands and receptors (Figure 5A). Notably, the number of targets that are regulated by these TFs vary. Some TFs such as UNC-62, HLH-1, and BLMP-1 may bind to the upstream region of several ligand and receptor encoding genes, while others bind to specific regions, such as AHA-1, NHR-67 which have only one target gene each (Figures 5A,C). In addition, the upstream regions of genes encoding ligands and receptors are occupied with different TFs (Figure 5C). The expression of about half of these TFs is enriched in neurons throughout larval and adult stages (Figure 5B). However, expression level analysis shows no direct correlation between the expression of TFs and their targets. For example, despite the fact that HLH-1 and BLMP-1 bind to the upstream region of numerous target genes (Figure 5A), they do not show pan-neuronal expression (Figure 5B), however, low-level expression in specific neurons is possible. In contrast, DAF-16, a FOXO TF that binds to the upstream region of 30 ligand/receptor encoding genes, shows high neuronal expression during development (Figure 5B). This TF regulates multiple biological processes such as lifespan, metabolism, and axon regeneration (Lee et al., 2003; Murphy et al., 2003; Basu et al., 2021). Similarly, TFs with known functions in neurite development (Table 3), such as EGL-5, ZAG-1, UNC-3, and SEM-4, also have binding sites within the upstream regions of genes encoding ligands and receptors.
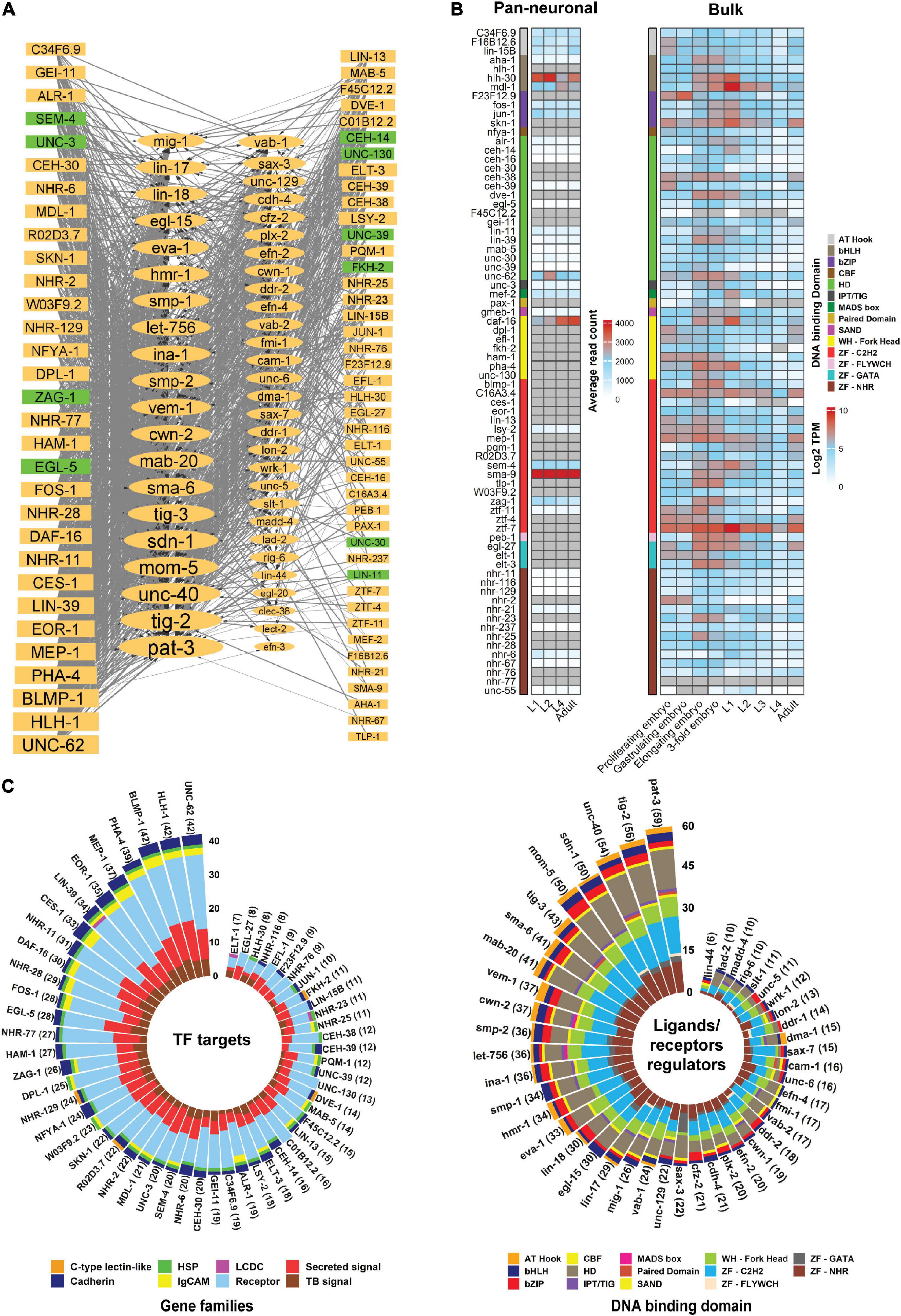
Figure 5. Transcriptional regulatory network of ligands and receptors that regulate neurite development.(A) Regulatory network of TFs with a binding peak upstream of genes that encode ligands and receptors involved in neurite development. The network was manually constructed by identifying statistically significant peaks (q-value < 0.01) within 2 and 5 kb upstream of the transcription start site (TSS) of each gene. For the genes with multiple isoforms the longest isoform was analyzed. Data was obtained from modENCODE (Gerstein et al., 2010; Yu et al., 2017; Li et al., 2020). The network was visualized via Cytoscape 3.7.1 (Shannon et al., 2003). Nodes represent genes in which the rectangles and ellipses show TFs and ligands/receptors, respectively. Node size relates to number of connections. Nodes in green are TFs with experimentally verified roles in neurite development. The edges show interactions between two nodes, where the thick and thin lines represent binding peaks at 2 and 5 kb from the TSS, respectively. Arrowheads depict TFs binding upstream of a gene. (B) Expression level of TFs (modENCODE) in the nervous system (pan-neuronal) or whole body (bulk) during animal development. Genes are not clustered. Gray cells = expression not detected. Pan-neuronal gene expression was obtained from Sun and Hobert (2021) and bulk gene expression was fetched from Expression Atlas (ebi.ac.uk/gxa/home). (C) Stacked bar plot showing target families for each TF and the superfamily of TFs binding upstream of neurite regulatory genes. TFs may have binding peaks upstream of multiple genes and a gene promoter can be occupied by multiple types of TFs. The left stacked bar plot shows the family of genes occupied by the TFs. The right bar plot represents which superfamily of TFs binding upstream of each ligand/receptor gene. In both plots, TFs and genes with more than 5 TF-gene interactions were visualized. The number next to the gene names shows the interaction counts. Interaction data were extracted from the network (A) and visualized using R language programming. HSP, heparan sulfate proteoglycans; LCDC, leukocyte cell-derived chemotaxin; TB, signal Transmembrane-bound signal.
Promoters of ligand and receptor encoding genes may be occupied by multiple TFs. For example, pat-3, tig-2 and unc-40 promoters are occupied by over 50 TFs, however, the binding of these TFs could be cell-specific, or at particular developmental stages (Van Buskirk and Sternberg, 2010; Smith et al., 2013; Serrano-Saiz et al., 2017). In addition, some neurite development regulators function in non-neuronal tissues. For example, VAB-1/Eph receptor is required for epidermal morphogenesis (George et al., 1998). Therefore, coordinated cell/tissue transcriptional regulation of ligands/receptors by multiple TFs can orchestrate the function of these genes in distinct cellular contexts.
We found that some parts of the genome are hot spots for TF binding (Figure 6) (Araya et al., 2014; Joshi, 2014). However, linking these hot spots with gene expression and functionality need to be studied in more detail to identify how multiple TFs can cooperate with, or compensate for each other (Stefanakis et al., 2015; Leyva-Diaz and Hobert, 2022). In addition, promoter regions of some genes overlap. For example, the unc-40 and npp-7 promoters overlap with over 50 TFs binding within this region (Figure 6D). In this case, as both genes could be simultaneously expressed, it is challenging to identify which gene is the real target of each TF. Gene-specific regulation by each TF may require additional independent regulatory proteins, such as co-repressors and co-activators (O’Brien et al., 2018). Finally, some genes encode multiple isoforms that are controlled by alternative Transcription Start Sites (TSSs) (Rojas-Duran and Gilbert, 2012; Craig et al., 2013; Reyes and Huber, 2018). For example, sax-7 encodes two short and long isoforms that regulate neuronal development and maintenance (Sasakura et al., 2005; Pocock et al., 2008a; Sherry et al., 2020). The upstream region of each isoform is occupied by different sets of TFs with several to dozens for longer and shorter isoforms, respectively (data not shown). This highlights the potential importance for transcriptional regulation of isoform-specific expression through binding of specific TFs.
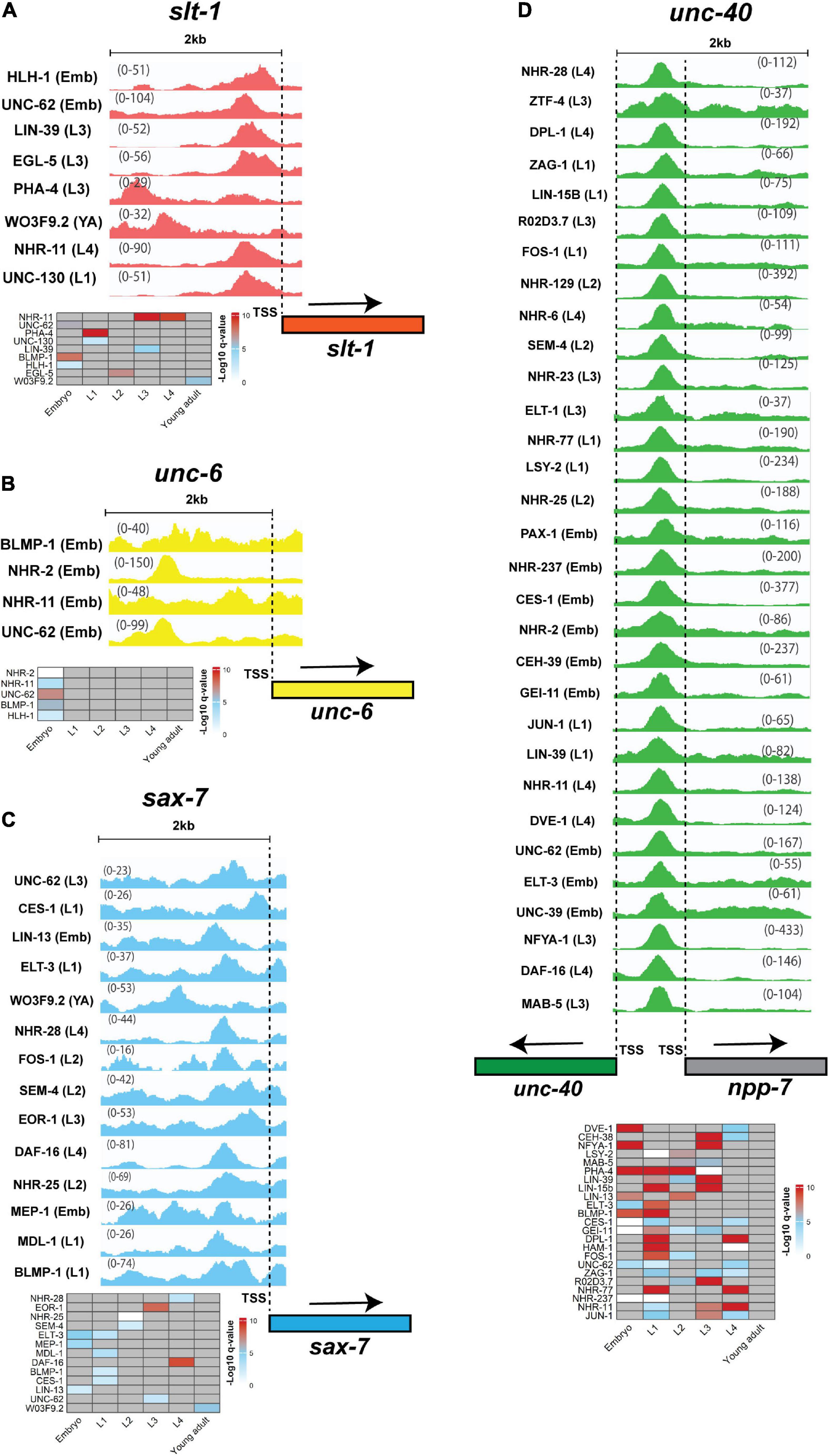
Figure 6. TFs binding peaks within 2 kb upstream of neurite development regulators. Genes encoding guidance cues (A) SLT-1, (B) UNC-6, and the guidance receptors (C) SAX-7, (D) UNC-40 were chosen to analyze ChIP-peak density. The heatmap below each group of peaks represent stages in which peaks were detected. Only a few TFs occupy 2 kb upstream region of unc-6 and slt-1, however, the number of TFs that bind at 5 kb region is larger (Figure 5). Multiple TFs occupy the sax-7 promoter. This gene has two short and long isoforms with the promoter region of short isoform occupied by 38 TFs (not shown). In contrast to the other genes, the unc-40 promoter is occupied with several dozen TFs. Interestingly, the promoter region of unc-40 overlaps npp-7 promoter and as such some TFs may regulate the expression of both or either gene. All peaks are statistically significant (q-value < 0.01). The numbers on the peaks represent the scale of data within that region that shows the lowest and highest levels of the detected peaks within the region. Input levels are not shown or subtracted from the treatment peaks. The dashed lines show the location of TSSs. In the heatmaps, gray cells = no peak identified. Due to large number of peaks for unc-40 and space limitation only some of the candidates are presented. The heatmap for unc-40 includes genes with peaks at multiple stages. The peaks were visualized using Integrative Genomics Viewer 2.12.0 (Robinson et al., 2011). The data was obtained from modENCODE (Gerstein et al., 2010).
Although the detection of TF binding upstream of a gene does not necessarily mean that the TF is a regulator of that gene, there is a possibility of regulation. To determine direct regulation, the expression of target genes in TF mutant backgrounds, TF-promoter binding assays and mutation of endogenous TF binding sites followed by ChIP-PCR would be necessary (Van Nostrand and Kim, 2013; Angelini and Costa, 2014; Pai and Gilad, 2014). Additionally, the difference in the number of regulators for different genes and its impact on the development and physiology must be investigated. Taken together, ligands and receptors involved in neurite development are potentially regulated by multiple factors.
Many TFs have DNA binding domains, such as zinc-finger and homeodomains, that are highly conserved across species (Narasimhan et al., 2015; Nitta et al., 2015). The majority of TFs involved in C. elegans neurite development have orthologs in other animal models such as fruit flies and mice. However, genes have different levels of conservation among species or may have multiple orthologs in other animals (Table 4). Interestingly, many C. elegans TF orthologs in mice are also required for nervous system development. For example, axon guidance is regulated by mouse Pax6 and Lhx2 TFs that are orthologs of worm neurite development regulators VAB-3 and TTX-3, respectively (Altun-Gultekin et al., 2001; Hevner et al., 2002; Lakhina et al., 2007; Yoshimura et al., 2008). This high level of evolutionary and functional conservation shows the importance of neurite development-related TFs. However, increasing complexity of the mammalian tissues may have caused alterations in specific TF functions from those observed in worms.
Similarly, many of the neurite development TFs in C. elegans have human orthologs, with ∼50% of them associated with mental or neurodegenerative disorders (Table 4). Developmental defects in human neurons, particularly those related to eye disorders, such as Fuchs endothelial dystrophy, aniridia, and microphthalmia are linked to multiple TFs with a worm ortholog (Verma and Fitzpatrick, 2007; Nanda and Alone, 2019; Landsend et al., 2021; Table 4). Further, central hypoventilation syndrome and pituitary hormone deficiency are due to defects in the nervous system development caused by mutations in human orthologs of HLH-3 and UNC-42 (Correa et al., 2019; Trang et al., 2020; Table 4). In addition to developmental defects, neurodegenerative and psychological disorders also show association to orthologs of worm neurite development regulators. For example, amyotrophic lateral sclerosis and schizophrenia are associated to FLI1 (AST-1) and PCGF6 (MIG-32), respectively (Weinberger, 2019). However, the roles of these genes in human neuronal development are unknown. Psychological disorders, such as major depressive disorder and attention-deficit/hyperactivity disorder are caused by a combination of genetic and environmental factors (Otte et al., 2016; Posner et al., 2020; Table 4). These diseases may be caused by neurodegeneration or defects during development, including neurite guidance.
Conservation of TFs involved in neurite development across evolution suggest common regulatory mechanisms in the compact C. elegans and complex mammalian nervous systems. This similarity provides an opportunity to decipher fundamental mechanisms of neurite and nervous system development in model organisms, and to extend the knowledge to higher animals for potential therapeutic applications.
The nervous system contains complex neuronal circuits comprised of highly regulated neurite architecture. Correct nervous system development depends on precisely controlled gene expression patterns, and interactions of gene products in the surrounding environment. TFs are key regulators of gene expression that perform critical roles in neurite development. Most neurite regulators we examined show similarities in their expression patterns during embryonic and early larval stages in C. elegans (Figures 3A,B). Individual signaling pathways are shown to control neurite development (Hao et al., 2001, 2010; Fujisawa et al., 2007; MacNeil et al., 2009; Smith et al., 2012), however, multiple pathways also combine to provide robustness. For example, double mutants for slt-1 (Slit) and unc-6 (Netrin) exhibit increased defects in axon guidance compared to single mutation of these genes (Hao et al., 2001). We found that individual neurons express many neurite regulators at each developmental stage (Figure 4), suggesting the existence of multiple redundant mechanisms controlling neurite development. This hypothesis is highlighted in complicated axon growth behaviors. For example, axon guidance toward intermediate targets is controlled by precise expression of the Netrin signaling components UNC-5, UNC-6, and UNC-40 (Hedgecock et al., 1990; Wadsworth et al., 1996; Dickson and Zou, 2010). Another less explored example is the guidance of individual axons in a compact axon fascicle, such as nerve ring (Figure 1), where neuron-specific development of axons must be precisely controlled to enable synapse establishment. Hence, combinatorial signaling mechanisms seem to be inevitable, due to the existence of a limited number of the signal molecules and receptors that control neurite development.
Combinatorial interactions require highly regulated pan-neuronal and also neuron specific signaling mechanisms that are controlled by TFs. Neuron-specific regulatory mechanisms have been widely studied for neuron fate determination in C. elegans (Hobert, 2008; Stefanakis et al., 2015). Some TFs have been identified as “terminal selectors” that are essential for neuron fate determination throughout animal life (Hobert, 2008). Because of the neuron-specific features of neurites, such as growth to specific targets, there is a possibility that terminal regulator concepts for neurite development can be applied. Here, the limitation is that neurite development is a multi-tissue regulated process and, as such, requires co-regulation of genes in multiple tissues (Hao et al., 2001; Fujisawa et al., 2007; Norris and Lundquist, 2011). Further, the timing of TF and target expression is essential for proper neurite development. For example, the correct timing of UNC-86 expression is critical for axon initiation and activation of ventral guidance responses (Olsson-Carter and Slack, 2011). Alongside neurite growth, maintenance of neurite position is controlled by proteins such as SAX-7/L1CAM (Pocock et al., 2008a). The identification of regulatory mechanisms to ensure expression of “maintenance factors” may be critical for inducing axon growth following injury.
Combinatorial gene regulation requires the activity of multiple TFs (Reiter et al., 2017). We identified several TF binding sites present in the upstream region of genes encoding neurite guidance ligands and receptors, the relevance of which could be explored further using functional and mechanistic studies (Gerstein et al., 2010). Furthermore, the involvement of the reported TFs in neurite development has been studied in a limited number of neurons, with their regulatory roles in the majority of neurons unclear. In addition, TFs execute their functions via either protein-protein interaction with other factors or indirectly through other regulatory processes, suggesting a need to identify potential cooperative components for these TFs (Altun-Gultekin et al., 2001; Smith et al., 2013; O’Brien et al., 2017).
In addition to TF-dependent gene regulation, other gene expression regulatory mechanisms, such as epigenetic modification (Abay-Norgaard et al., 2020) and post-translational regulation via microRNAs (Zou et al., 2012; Hong et al., 2013; Pedersen et al., 2013) are involved in neuron development. Indeed, a combination of epigenetic modifications, transcriptional regulation by TFs and post-transcriptional modifications likely converge to precisely control nervous system development.
Finally, the dynamic and combinatorial behavior of neurite development regulators is reflected in the transcriptome, particularly from single-cell level analysis. This information may shed light on how to manipulate these regulators to overcome neuronal deficits and neurodegenerative decline.
RG wrote the manuscript. HF and RP edited the manuscript. All authors contributed to the article and approved the submitted version.
RP was supported by the National Health and Medical Research Council (GNT1137645, GNT2000766, and GNT1183908), the Australian Research Council (DP200103293) and veski (VIF23). This research was supported by Monash International Tuition Scholarship (MITS) and Monash Graduate Scholarships (MGS) to RG.
We thank members of our laboratories for discussion of this review. We apologize to colleagues whose important primary studies could not be cited due to space constraints.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abay-Norgaard, S., Attianese, B., Boreggio, L., and Salcini, A. E. (2020). Regulators of H3K4 methylation mutated in neurodevelopmental disorders control axon guidance in Caenorhabditis elegans. Development 147:dev190637. doi: 10.1242/dev.190637
Adler, C. E., Fetter, R. D., and Bargmann, C. I. (2006). UNC-6/Netrin induces neuronal asymmetry and defines the site of axon formation. Nat. Neurosci. 9, 511–518. doi: 10.1038/nn1666
Agostinone, J., and Di Polo, A. (2015). Retinal ganglion cell dendrite pathology and synapse loss: Implications for glaucoma. Prog. Brain Res. 220, 199–216. doi: 10.1016/bs.pbr.2015.04.012
Altun-Gultekin, Z., Andachi, Y., Tsalik, E. L., Pilgrim, D., Kohara, Y., and Hobert, O. (2001). A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 128, 1951–1969. doi: 10.1242/dev.128.11.1951
Angelini, C., and Costa, V. (2014). Understanding gene regulatory mechanisms by integrating ChIP-seq and RNA-seq data: Statistical solutions to biological problems. Front. Cell Dev. Biol. 2:51. doi: 10.3389/fcell.2014.00051
Araya, C. L., Kawli, T., Kundaje, A., Jiang, L., Wu, B., Vafeados, D., et al. (2014). Regulatory analysis of the C. elegans genome with spatiotemporal resolution. Nature 512, 400–405. doi: 10.1038/nature13497
Armakola, M., and Ruvkun, G. (2019). Regulation of Caenorhabditis elegans neuronal polarity by heterochronic genes. Proc. Natl. Acad. Sci. U.S.A. 116, 12327–12336. doi: 10.1073/pnas.1820928116
Baltaci, O., Pedersen, M. E., Sherry, T., Handley, A., Snieckute, G., Cao, W., et al. (2022). Atypical TGF-beta signaling controls neuronal guidance in Caenorhabditis elegans. iScience 25:103791. doi: 10.1016/j.isci.2022.103791
Basson, M., and Horvitz, H. R. (1996). The Caenorhabditis elegans gene sem-4 controls neuronal and mesodermal cell development and encodes a zinc finger protein. Genes. Dev. 10, 1953–1965. doi: 10.1101/gad.10.15.1953
Basu, A., Behera, S., Bhardwaj, S., Dey, S., and Ghosh-Roy, A. (2021). Regulation of UNC-40/DCC and UNC-6/Netrin by DAF-16 promotes functional rewiring of the injured axon. Development 148:dev198044. doi: 10.1242/dev.198044
Baum, P. D., and Garriga, G. (1997). Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants. Neuron 19, 51–62. doi: 10.1016/s0896-6273(00)80347-5
Berghoff, E. G., Glenwinkel, L., Bhattacharya, A., Sun, H., Varol, E., Mohammadi, N., et al. (2021). The Prop1-like homeobox gene unc-42 specifies the identity of synaptically connected neurons. Elife 10:e64903. doi: 10.7554/eLife.64903
Bertrand, V., Bisso, P., Poole, R. J., and Hobert, O. (2011). Notch-dependent induction of left/right asymmetry in C. elegans interneurons and motoneurons. Curr. Biol. 21, 1225–1231. doi: 10.1016/j.cub.2011.06.016
Blanchette, C. R., Perrat, P. N., Thackeray, A., and Benard, C. Y. (2015). Glypican is a modulator of netrin-mediated axon guidance. PLoS Biol. 13:e1002183. doi: 10.1371/journal.pbio.1002183
Boulin, T., Pocock, R., and Hobert, O. (2006). A novel Eph receptor-interacting IgSF protein provides C. elegans motoneurons with midline guidepost function. Curr. Biol. 16, 1871–1883. doi: 10.1016/j.cub.2006.08.056
Broadbent, I. D., and Pettitt, J. (2002). The C-elegans hmr-1 gene can encode a neuronal classic cadherin involved in the regulation of axon fasciculation. Curr. Biol. 12, 59–63. doi: 10.1016/S0960-9822(01)00624-8
Brockie, P. J., Madsen, D. M., Zheng, Y., Mellem, J., and Maricq, A. V. (2001). Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J. Neurosci. 21, 1510–1522. doi: 10.1523/Jneurosci.21-05-01510.2001
Bulow, H. E., Boulin, T., and Hobert, O. (2004). Differential functions of the C. elegans FGF receptor in axon outgrowth and maintenance of axon position. Neuron 42, 367–374. doi: 10.1016/s0896-6273(04)00246-6
Cao, J., Packer, J. S., Ramani, V., Cusanovich, D. A., Huynh, C., Daza, R., et al. (2017). Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667. doi: 10.1126/science.aam8940
Chan, K. K. M., Seetharaman, A., Bagg, R., Selman, G., Zhang, Y. Q., Kim, J., et al. (2014). EVA-1 functions as an UNC-40 co-receptor to enhance attraction to the MADD-4 guidance cue in Caenorhabditis elegans. PLoS Genet. 10:e1004521. doi: 10.1371/journal.pgen.1004521
Chen, C.-H., Hsu, H.-W., Chang, Y.-H., and Pan, C.-L. (2019). Adhesive L1CAM-robo signaling aligns growth cone F-actin dynamics to promote axon-dendrite fasciculation in C. elegans. Develop. Cell 48, 215–228. doi: 10.1016/j.devcel.2018.10.028
Chien, J., Devkota, R., Yosef, N., and Morck, C. (2017). Regulation of axon guidance by the wnt receptor Ror/CAM-1 in the PVT Guidepost Cell in Caenorhabditis elegans. Genetics 207, 1533–1545. doi: 10.1534/genetics.117.300375
Chisholm, A. D., Hutter, H., Jin, Y., and Wadsworth, W. G. (2016). The genetics of axon guidance and axon regeneration in Caenorhabditis elegans. Genetics 204, 849–882. doi: 10.1534/genetics.115.186262
Coburn, C. M., Mori, I., Ohshima, Y., and Bargmann, C. I. (1998). A cyclic nucleotide-gated channel inhibits sensory axon outgrowth in larval and adult Caenorhabditis elegans: A distinct pathway for maintenance of sensory axon structure. Development 125, 249–258. doi: 10.1242/dev.125.2.249
Cochella, L., Tursun, B., Hsieh, Y. W., Galindo, S., Johnston, R. J., Chuang, C. F., et al. (2014). Two distinct types of neuronal asymmetries are controlled by the Caenorhabditis elegans zinc finger transcription factor die-1. Genes. Dev. 28, 34–43. doi: 10.1101/gad.233643.113
Colavita, A., and Culotti, J. G. (1998). Suppressors of ectopic UNC-5 growth cone steering identify eight genes involved in axon guidance in Caenorhabditis elegans. Dev. Biol. 194, 72–85. doi: 10.1006/dbio.1997.8790
Contreras, E. G., Palominos, T., Glavic, A., Brand, A. H., Sierralta, J., and Oliva, C. (2018). The transcription factor SoxD controls neuronal guidance in the Drosophila visual system. Sci. Rep. 8:13332. doi: 10.1038/s41598-018-31654-5
Cook, S. J., Jarrell, T. A., Brittin, C. A., Wang, Y., Bloniarz, A. E., Yakovlev, M. A., et al. (2019). Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 571, 63–71. doi: 10.1038/s41586-019-1352-7
Correa, F. A., Nakaguma, M., Madeira, J. L. O., Nishi, M. Y., Abrao, M. G., Jorge, A. A. L., et al. (2019). Combined pituitary hormone deficiency caused by PROP1 mutations: Update 20 years post-discovery. Arch. Endocrinol. Metab. 63, 167–174. doi: 10.20945/2359-3997000000139
Craig, H. L., Wirtz, J., Bamps, S., Dolphin, C. T., and Hope, I. A. (2013). The significance of alternative transcripts for Caenorhabditis eleganstranscription factor genes, based on expression pattern analysis. BMC Genom. 14:249. doi: 10.1186/1471-2164-14-249
Dalpe, G., Zhang, L. W., Zheng, H., and Culotti, J. G. (2004). Conversion of cell movement responses to Semaphorin-1 and Plexin-1 from attraction to repulsion by lowered levels of specific RAC GTPases in C. elegans. Development 131, 2073–2088. doi: 10.1242/dev.01063
Desai, C., Garriga, G., McIntire, S. L., and Horvitz, H. R. (1988). A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons. Nature 336, 638–646. doi: 10.1038/336638a0
Di Croce, L., and Helin, K. (2013). Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20, 1147–1155. doi: 10.1038/nsmb.2669
Díaz-Balzac, C. A., Rahman, M., Lázaro-Peña, M. I., Hernandez, L. A. M., Salzberg, Y., Aguirre-Chen, C., et al. (2016). Muscle-and skin-derived cues jointly orchestrate patterning of somatosensory dendrites. Curr. Biol. 26, 2379–2387.
Dickson, B. J., and Zou, Y. (2010). Navigating intermediate targets: The nervous system midline. Cold Spring Harb. Perspect Biol. 2:a002055. doi: 10.1101/cshperspect.a002055
Dong, B., Moseley-Alldredge, M., Schwieterman, A. A., Donelson, C. J., McMurry, J. L., Hudson, M. L., et al. (2016). EFN-4 functions in LAD-2-mediated axon guidance in Caenorhabditis elegans. Development 143, 1182–1191. doi: 10.1242/dev.128934
Dong, X., Liu, O. W., Howell, A. S., and Shen, K. (2013). An extracellular adhesion molecule complex patterns dendritic branching and morphogenesis. Cell 155, 296–307. doi: 10.1016/j.cell.2013.08.059
Doonan, R., Hatzold, J., Raut, S., Conradt, B., and Alfonso, A. (2008). HLH-3 is a C. elegans Achaete/Scute protein required for differentiation of the hermaphrodite-specific motor neurons. Mech. Dev. 125, 883–893. doi: 10.1016/j.mod.2008.06.002
Dotti, C. G., Sullivan, C. A., and Banker, G. A. (1988). The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 8, 1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988
Durbin, R. M. (1987). Studies on the development and organisation of the nervous system of Caenorhabditis elegans. Cambridge: University of Cambridge.
Esmaeili, B., Ross, J. M., Neades, C., Miller, D. M. III, and Ahringer, J. (2002). The C. elegans even-skipped homologue, vab-7, specifies DB motoneurone identity and axon trajectory. Development 129, 853–862. doi: 10.1242/dev.129.4.853
Fleming, T., Chien, S. C., Vanderzalm, P. J., Dell, M., Gavin, M. K., Forrester, W. C., et al. (2010). The role of C. elegans Ena/VASP homolog UNC-34 in neuronal polarity and motility. Dev. Biol. 344, 94–106. doi: 10.1016/j.ydbio.2010.04.025
Flynn, K. C. (2013). The cytoskeleton and neurite initiation. Bioarchitecture 3, 86–109. doi: 10.4161/bioa.26259
Fontanet, P., Irala, D., Alsina, F. C., Paratcha, G., and Ledda, F. (2013). Pea3 transcription factor family members Etv4 and Etv5 mediate retrograde signaling and axonal growth of DRG sensory neurons in response to NGF. J. Neurosci. 33, 15940–15951. doi: 10.1523/JNEUROSCI.0928-13.2013
Forrester, W. C., and Garriga, G. (1997). Genes necessary for C. elegans cell and growth cone migrations. Development 124, 1831–1843. doi: 10.1242/dev.124.9.1831
Forrester, W. C., Dell, M., Perens, E., and Garriga, G. (1999). A C. elegans Ror receptor tyrosine kinase regulates cell motility and asymmetric cell division. Nature 400, 881–885. doi: 10.1038/23722
Fujisawa, K., Wrana, J. L., and Culotti, J. G. (2007). The slit receptor EVA-1 coactivates a SAX-3/Robo mediated guidance signal in C. elegans. Science 317, 1934–1938. doi: 10.1126/science.1144874
Furey, T. S. (2012). ChIP-seq and beyond: New and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet. 13, 840–852. doi: 10.1038/nrg3306
George, S. E., Simokat, K., Hardin, J., and Chisholm, A. D. (1998). The VAB-1 Eph receptor tyrosine kinase functions in neural and epithelial morphogenesis in C. elegans. Cell 92, 633–643. doi: 10.1016/s0092-8674(00)81131-9
Gerstein, M. B., Lu, Z. J., Van Nostrand, E. L., Cheng, C., Arshinoff, B. I., Liu, T., et al. (2010). Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330, 1775–1787. doi: 10.1126/science.1196914
Gibson, D. A., and Ma, L. (2011). Developmental regulation of axon branching in the vertebrate nervous system. Development 138, 183–195. doi: 10.1242/dev.046441
Ginzburg, V. E., Roy, P. J., and Culotti, J. G. (2002). Semaphorin 1a and semaphorin 1b are required for correct epidermal cell positioning and adhesion during morphogenesis in C. elegans. Development 129, 2065–2078. doi: 10.1242/dev.129.9.2065
Gitai, Z., Yu, T. W., Lundquist, E. A., Tessier-Lavigne, M., and Bargmann, C. I. (2003). The netrin receptor UNC-40/DCC stimulates axon attraction and outgrowth through enabled and, in parallel, Rac and UNC-115/AbLIM. Neuron 37, 53–65. doi: 10.1016/S0896-6273(02)01149-2
Goaillard, J. M., Moubarak, E., Tapia, M., and Tell, F. (2019). Diversity of axonal and dendritic contributions to neuronal output. Front. Cell Neurosci. 13:570. doi: 10.3389/fncel.2019.00570
Goodwin, E. B., and Ellis, R. E. (2002). Turning clustering loops: Sex determination in Caenorhabditis elegans. Curr. Biol. 12, R111–R120. doi: 10.1016/s0960-9822(02)00675-9
Goslin, K., and Banker, G. (1989). Experimental observations on the development of polarity by hippocampal neurons in culture. J. Cell Biol. 108, 1507–1516. doi: 10.1083/jcb.108.4.1507
Halevi, S., McKay, J., Palfreyman, M., Yassin, L., Eshel, M., Jorgensen, E., et al. (2002). The C-elegans ric-3 gene is required for maturation of nicotinic acetylcholine receptors. EMBO J. 21, 1012–1020. doi: 10.1093/emboj/21.5.1012
Hamelin, M., Zhou, Y. W., Su, M. W., Scott, I. M., and Culotti, J. G. (1993). Expression of the Unc-5 guidance receptor in the touch neurons of C-elegans steers their axons dorsally. Nature 364, 327–330. doi: 10.1038/364327a0
Hao, J. C., Adler, C. E., Mebane, L., Gertler, F. B., Bargmann, C. I., and Tessier-Lavigne, M. (2010). The tripartite motif protein MADD-2 functions with the receptor UNC-40 (DCC) in Netrin-mediated axon attraction and branching. Dev. Cell 18, 950–960. doi: 10.1016/j.devcel.2010.02.019
Hao, J. C., Yu, T. W., Fujisawa, K., Culotti, J. G., Gengyo-Ando, K., Mitani, S., et al. (2001). C. elegans slit acts in midline, dorsal-ventral, and anterior-posterior guidance via the SAX-3/Robo receptor. Neuron 32, 25–38. doi: 10.1016/s0896-6273(01)00448-2
Hartin, S. N., Kurland, M., and Ackley, B. D. (2017). Syndecan functions to regulate Wnt-dependent axon guidance in C. elegans. BioRxiv [Preprint]. doi: 10.1101/046094
Hayashi, K., Kubo, K., Kitazawa, A., and Nakajima, K. (2015). Cellular dynamics of neuronal migration in the hippocampus. Front. Neurosci. 9:135. doi: 10.3389/fnins.2015.00135
Hedgecock, E. M., Culotti, J. G., and Hall, D. H. (1990). The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron 4, 61–85. doi: 10.1016/0896-6273(90)90444-k
Hevner, R. F., Miyashita-Lin, E., and Rubenstein, J. L. R. (2002). Cortical and thalamic axon pathfinding defects in Tbr1, Gbx2, and Pax6 mutant mice: Evidence that cortical and thalamic axons interact and guide each other. J. Comp. Neurol. 447, 8–17. doi: 10.1002/cne.10219
Hilliard, M. A., and Bargmann, C. I. (2006). Wnt signals and frizzled activity orient anterior-posterior axon outgrowth in C. elegans. Dev. Cell. 10, 379–390. doi: 10.1016/j.devcel.2006.01.013
Hobert, O. (2008). Regulatory logic of neuronal diversity: Terminal selector genes and selector motifs. Proc. Natl. Acad. Sci. U.S.A. 105, 20067–20071. doi: 10.1073/pnas.0806070105
Hobert, O., Tessmar, K., and Ruvkun, G. (1999). The Caenorhabditis elegans lim-6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons. Development 126, 1547–1562. doi: 10.1242/dev.126.7.1547
Hong, J., Zhang, H. J., Kawase-Koga, Y., and Sun, T. (2013). MicroRNA function is required for neurite outgrowth of mature neurons in the mouse postnatal cerebral cortex. Front. Cell. Neurosci. 7:151. doi: 10.3389/fncel.2013.00151
Hsu, H. W., Liao, C. P., Chiang, Y. C., Syu, R. T., and Pan, C. L. (2020). Caenorhabditis elegans Flamingo FMI-1 controls dendrite self-avoidance through F-actin assembly. Development 147:dev179168. doi: 10.1242/dev.179168
Hu, Y., Comjean, A., Roesel, C., Vinayagam, A., Flockhart, I., Zirin, J., et al. (2017). FlyRNAi.org-the database of the Drosophila RNAi screening center and transgenic RNAi project: 2017 update. Nucleic Acids Res. 45, D672–D678. doi: 10.1093/nar/gkw977
Hu, Y., Flockhart, I., Vinayagam, A., Bergwitz, C., Berger, B., Perrimon, N., et al. (2011). An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12:357. doi: 10.1186/1471-2105-12-357
Huang, C.-c, Hall, D. H., Hedgecock, E. M., Kao, G., Karantza, V., Vogel, B. E., et al. (2003). Laminin α subunits and their role in C. elegans development. Development 130, 3343–3358. doi: 10.1242/dev.00481
Hutter, H. (2003). Extracellular cues and pioneers act together to guide axons in the ventral cord of C. elegans. Development 130, 5307–5318. doi: 10.1242/dev.00727
Jin, H., and Kim, B. (2020). Neurite branching regulated by neuronal cell surface molecules in Caenorhabditis elegans. Front. Neuroanat. 14:59. doi: 10.3389/fnana.2020.00059
Jin, Y., Hoskins, R., and Horvitz, H. R. (1994). Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature 372, 780–783. doi: 10.1038/372780a0
Johnson, R. P., Kang, S. H., and Kramer, J. M. (2006). C. elegans dystroglycan DGN-1 functions in epithelia and neurons, but not muscle, and independently of dystrophin. Development 133, 1911–1921. doi: 10.1242/dev.02363
Johnston, R. J., and Hobert, O. (2003). A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426, 845–849. doi: 10.1038/nature02255
Joshi, A. (2014). Mammalian transcriptional hotspots are enriched for tissue specific enhancers near cell type specific highly expressed genes and are predicted to act as transcriptional activator hubs. BMC Bioinformatics 15:412. doi: 10.1186/s12859-014-0412-0
Kagoshima, H., Cassata, G., Tong, Y. G., Pujol, N., Niklaus, G., and Burglin, T. R. (2013). The LIM homeobox gene ceh-14 is required for phasmid function and neurite outgrowth. Dev. Biol. 380, 314–323. doi: 10.1016/j.ydbio.2013.04.009
Kao, G., Huang, C. C., Hedgecock, E. M., Hall, D. H., and Wadsworth, W. G. (2006). The role of the laminin beta subunit in laminin heterotrimer assembly and basement membrane function and development in C. elegans. Dev. Biol. 290, 211–219. doi: 10.1016/j.ydbio.2005.11.026
Karakuzu, O., Wang, D. P., and Cameron, S. (2009). MIG-32 and SPAT-3A are PRC1 homologs that control neuronal migration in Caenorhabditis elegans. Development 136, 943–953. doi: 10.1242/dev.029363
Katidou, M., Tavernarakis, N., and Karagogeos, D. (2013). The contactin RIG-6 mediates neuronal and non-neuronal cell migration in Caenorhabditis elegans. Dev. Biol. 373, 184–195. doi: 10.1016/j.ydbio.2012.10.027
Kennerdell, J. R., Fetter, R. D., and Bargmann, C. I. (2009). Wnt-Ror signaling to SIA and SIB neurons directs anterior axon guidance and nerve ring placement in C. elegans. Development 136, 3801–3810. doi: 10.1242/dev.038109
Kim, S., and Wadsworth, W. G. (2000). Positioning of longitudinal nerves in C. elegans by nidogen. Science 288, 150–154. doi: 10.1126/science.288.5463.150
Kirszenblat, L., Pattabiraman, D., and Hilliard, M. A. (2011). LIN-44/Wnt Directs Dendrite Outgrowth through LIN-17/Frizzled in C. elegans Neurons. PLoS Biol. 9:e1001157. doi: 10.1371/journal.pbio.1001157
Klose, M., and Bentley, D. (1989). Transient pioneer neurons are essential for formation of an embryonic peripheral nerve. Science 245, 982–984. doi: 10.1126/science.2772651
Koh, A., Tao, S. J., Goh, Y. J., Chaganty, V., See, K., Purushothaman, K., et al. (2020). A Neurexin2aa deficiency results in axon pathfinding defects and increased anxiety in zebrafish. Hum. Mol. Genet. 29, 3765–3780. doi: 10.1093/hmg/ddaa260
Kulkarni, G., Li, H., and Wadsworth, W. G. (2008). CLEC-38, a transmembrane protein with C-type lectin-like domains, negatively regulates UNC-40-mediated axon outgrowth and promotes presynaptic development in Caenorhabditis elegans. J. Neurosci. 28, 4541–4550. doi: 10.1523/JNEUROSCI.5542-07.2008
Kuwajima, T., Soares, C. A., Sitko, A. A., Lefebvre, V., and Mason, C. (2017). SoxC transcription factors promote contralateral retinal ganglion cell differentiation and axon guidance in the mouse visual system. Neuron 93, 1110–1125. doi: 10.1016/j.neuron.2017.01.029
Lakhina, V., Falnikar, A., Bhatnagar, L., and Tole, S. (2007). Early thalamocortical tract guidance and topographic sorting of thalamic projections requires LIM-homeodomain gene Lhx2. Dev. Biol. 306, 703–713. doi: 10.1016/j.ydbio.2007.04.007
Landsend, E. C. S., Lagali, N., and Utheim, T. P. (2021). Congenital aniridia–A comprehensive review of clinical features and therapeutic approaches. Surv. Ophthalmol. 66, 1031–1050. doi: 10.1016/j.survophthal.2021.02.011
Lee, S. S., Kennedy, S., Tolonen, A. C., and Ruvkun, G. (2003). DAF-16 target genes that control C-elegans life-span and metabolism. Science 300, 644–647. doi: 10.1126/science.1083614
Levy-Strumpf, N., and Culotti, J. G. (2014). Netrins and Wnts function redundantly to regulate antero-posterior and dorso-ventral guidance in C. elegans. PLoS Genet. 10:e1004381. doi: 10.1371/journal.pgen.1004381
Levy-Strumpf, N., Krizus, M., Zheng, H., Brown, L., and Culotti, J. G. (2015). The Wnt frizzled receptor MOM-5 Regulates the UNC-5 netrin receptor through small GTPase-dependent signaling to determine the polarity of migrating cells. PLoS Genet. 11:e1005446. doi: 10.1371/journal.pgen.1005446
Leyva-Diaz, E., and Hobert, O. (2022). Robust regulatory architecture of pan-neuronal gene expression. Curr. Biol. 171:e1718. doi: 10.1016/j.cub.2022.02.040
Li, Y., Osuma, A., Correa, E., Okebalama, M. A., Dao, P., Gaylord, O., et al. (2020). Establishment and maintenance of motor neuron identity via temporal modularity in terminal selector function. Elife 9:e59464. doi: 10.7554/eLife.59464
Lindenmaier, L. B., Parmentier, N., Guo, C. Y., Tissir, F., and Wright, K. M. (2019). Dystroglycan is a scaffold for extracellular axon guidance decisions. Elife 8:e42143. doi: 10.7554/eLife.42143
Liu, O. W., and Shen, K. (2012). The transmembrane LRR protein DMA-1 promotes dendrite branching and growth in C. elegans. Nat. Neurosci. 15, 57–63. doi: 10.1038/nn.2978
Lu, M., and Mizumoto, K. (2019). Gradient-independent Wnt signaling instructs asymmetric neurite pruning in C. elegans. Elife 8:e50583. doi: 10.7554/eLife.50583
MacNeil, L. T., Hardy, W. R., Pawson, T., Wrana, J. L., and Culotti, J. G. (2009). UNC-129 regulates the balance between UNC-40 dependent and independent UNC-5 signaling pathways. Nat. Neurosci. 12, 150–155. doi: 10.1038/nn.2256
Masserdotti, G., Gascon, S., and Gotz, M. (2016). Direct neuronal reprogramming: Learning from and for development. Development 143, 2494–2510. doi: 10.1242/dev.092163
Miller, K. E., and Suter, D. M. (2018). An integrated cytoskeletal model of neurite outgrowth. Front. Cell Neurosci. 12:447. doi: 10.3389/fncel.2018.00447
Much, J. W., Slade, D. J., Klampert, K., Garriga, G., and Wightman, B. (2000). The fax-1 nuclear hormone receptor regulates axon pathfinding and neurotransmitter expression. Development 127, 703–712. doi: 10.1242/dev.127.4.703
Mukhopadhyay, S., Lu, Y., Qin, H., Lanjuin, A., Shaham, S., and Sengupta, P. (2007). Distinct IFT mechanisms contribute to the generation of ciliary structural diversity in C. elegans. EMBO J. 26, 2966–2980. doi: 10.1038/sj.emboj.7601717
Murphy, C. T., McCarroll, S. A., Bargmann, C. I., Fraser, A., Kamath, R. S., Ahringer, J., et al. (2003). Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277–284. doi: 10.1038/nature01789
Najarro, E. H., Wong, L., Zhen, M., Carpio, E. P., Goncharov, A., Garriga, G., et al. (2012). Caenorhabditis elegans flamingo cadherin fmi-1 regulates GABAergic neuronal development. J. Neurosci. 32, 4196–4211. doi: 10.1523/JNEUROSCI.3094-11.2012
Nakao, F., Hudson, M. L., Suzuki, M., Peckler, Z., Kurokawa, R., Liu, Z., et al. (2007). The plexin PLX-2 and the ephrin EFN-4 have distinct roles in MAB-20/semaphorin 2A signaling in Caenorhabditis elegans morphogenesis. Genetics 176, 1591–1607. doi: 10.1534/genetics.106.067116
Nanda, G. G., and Alone, D. P. (2019). REVIEW: Current understanding of the pathogenesis of Fuchs’ endothelial corneal dystrophy. Mol. Vis. 25, 295–310.
Narasimhan, K., Lambert, S. A., Yang, A. W., Riddell, J., Mnaimneh, S., Zheng, H., et al. (2015). Mapping and analysis of Caenorhabditis elegans transcription factor sequence specificities. Elife 4, e06967. doi: 10.7554/eLife.06967
Nash, B., Colavita, A., Zheng, H., Roy, P. J., and Culotti, J. G. (2000). The forkhead transcription factor UNC-130 is required for the graded spatial expression of the UNC-129 TGF-β guidance factor in C. elegans. Genes Develop. 14, 2486–2500. doi: 10.1101/gad.831500
Nitta, K. R., Jolma, A., Yin, Y., Morgunova, E., Kivioja, T., Akhtar, J., et al. (2015). Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. Elife 4:e04837. doi: 10.7554/eLife.04837
Nobrega-Pereira, S., and Marin, O. (2009). Transcriptional control of neuronal migration in the developing mouse brain. Cereb Cortex 19, i107–i113. doi: 10.1093/cercor/bhp044
Norris, A. D., and Lundquist, E. A. (2011). UNC-6/netrin and its receptors UNC-5 and UNC-40/DCC modulate growth cone protrusion in vivo in C. elegans. Development 138, 4433–4442. doi: 10.1242/dev.068841
Norris, A. D., Sundararajan, L., Morgan, D. E., Roberts, Z. J., and Lundquist, E. A. (2014). The UNC-6/Netrin receptors UNC-40/DCC and UNC-5 inhibit growth cone filopodial protrusion via UNC-73/Trio. Rac-like GTPases and UNC-33/CRMP. Development 141, 4395–4405. doi: 10.1242/dev.110437
O’Brien, B. M. J., Palumbos, S. D., Novakovic, M., Shang, X., Sundararajan, L., and Miller, D. M. III (2017). Separate transcriptionally regulated pathways specify distinct classes of sister dendrites in a nociceptive neuron. Dev. Biol. 432, 248–257. doi: 10.1016/j.ydbio.2017.10.009
O’Brien, J., Hayder, H., Zayed, Y., and Peng, C. (2018). Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol (Lausanne) 9:402. doi: 10.3389/fendo.2018.00402
Oikonomou, G., and Shaham, S. (2011). The glia of Caenorhabditis elegans. Glia 59, 1253–1263. doi: 10.1002/glia.21084
Olsson-Carter, K., and Slack, F. J. (2011). The POU transcription factor UNC-86 controls the timing and ventral guidance of Caenorhabditis elegans axon growth. Dev. Dyn. 240, 1815–1825. doi: 10.1002/dvdy.22667
Onesto, M. M., Short, C. A., Rempel, S. K., Catlett, T. S., and Gomez, T. M. (2021). Growth factors as axon guidance molecules: Lessons from in vitro studies. Front. Neurosci. 15:678454. doi: 10.3389/fnins.2021.678454
Oren-Suissa, M., Bayer, E. A., and Hobert, O. (2016). Sex-specific pruning of neuronal synapses in Caenorhabditis elegans. Nature 533, 206–211. doi: 10.1038/nature17977
Otte, C., Gold, S. M., Penninx, B. W., Pariante, C. M., Etkin, A., Fava, M., et al. (2016). Major depressive disorder. Nat. Rev. Dis. Primers 2, 1–20. doi: 10.1038/nrdp.2016.65
Packer, J. S., Zhu, Q., Huynh, C., Sivaramakrishnan, P., Preston, E., Dueck, H., et al. (2019). A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science 365, 1–8. doi: 10.1126/science.aax1971
Pai, A. A., and Gilad, Y. (2014). Comparative studies of gene regulatory mechanisms. Curr. Opin. Genet. Dev. 29, 68–74. doi: 10.1016/j.gde.2014.08.010
Pan, C. L., Howell, J. E., Clark, S. G., Hilliard, M., Cordes, S., Bargmann, C. I., et al. (2006). Multiple Wnts and frizzled receptors regulate anteriorly directed cell and growth cone migrations in Caenorhabditis elegans. Dev. Cell 10, 367–377. doi: 10.1016/j.devcel.2006.02.010
Papatheodorou, I., Moreno, P., Manning, J., Fuentes, A. M., George, N., Fexova, S., et al. (2020). Expression atlas update: From tissues to single cells. Nucleic Acids Res. 48, 77–83. doi: 10.1093/nar/gkz947
Pedersen, M. E., Snieckute, G., Kagias, K., Nehammer, C., Multhaupt, H. A. B., Couchman, J. R., et al. (2013). An epidermal MicroRNA regulates neuronal migration through control of the cellular glycosylation state. Science 341, 1404–1408. doi: 10.1126/science.1242528
Pereda, A. E. (2014). Electrical synapses and their functional interactions with chemical synapses. Nat. Rev. Neurosci. 15, 250–263. doi: 10.1038/nrn3708
Pocock, R., Benard, C. Y., Shapiro, L., and Hobert, O. (2008a). Functional dissection of the C. elegans cell adhesion molecule SAX-7, a homologue of human L1. Mol. Cell. Neurosci. 37, 56–68. doi: 10.1016/j.mcn.2007.08.014
Pocock, R., Mione, M., Hussain, S., Maxwell, S., Pontecorvi, M., Aslam, S., et al. (2008b). Neuronal function of Tbx20 conserved from nematodes to vertebrates. Dev. Biol. 317, 671–685. doi: 10.1016/j.ydbio.2008.02.015
Poinat, P., De Arcangelis, A., Sookhareea, S., Zhu, X., Hedgecock, E. M., Labouesse, M., et al. (2002). A conserved interaction between β1 integrin/PAT-3 and Nck-interacting kinase/MIG-15 that mediates commissural axon navigation in C. elegans. Curr. Biol. 12, 622–631. doi: 10.1016/S0960-9822(02)00764-9
Pollarolo, G., Schulz, J. G., Munck, S., and Dotti, C. G. (2011). Cytokinesis remnants define first neuronal asymmetry in vivo. Nat. Neurosci. 14, 1525–1533. doi: 10.1038/nn.2976
Posner, J., Polanczyk, G. V., and Sonuga-Barke, E. (2020). Attention-deficit hyperactivity disorder. Lancet 395, 450–462. doi: 10.1016/S0140-6736(19)33004-1
Prasad, B. C., Ye, B., Zackhary, R., Schrader, K., Seydoux, G., and Reed, R. R. (1998). unc-3, a gene required for axonal guidance in Caenorhabditis elegans, encodes a member of the O/E family of transcription factors. Development 125, 1561–1568. doi: 10.1242/dev.125.8.1561
Pujol, N., Torregrossa, P., Ewbank, J. J., and Brunet, J. F. (2000). The homeodomain protein CePHOX2/CEH-17 controls antero-posterior axonal growth in C. elegans. Development 127, 3361–3371. doi: 10.1242/dev.127.15.3361
Qin, H., and Powell-Coffman, J. A. (2004). The Caenorhabditis elegans aryl hydrocarbon receptor, AHR-1, regulates neuronal development. Dev. Biol. 270, 64–75. doi: 10.1016/j.ydbio.2004.02.004
Ramirez-Suarez, N. J., Belalcazar, H. M., Salazar, C. J., Beyaz, B., Raja, B., Nguyen, K. C. Q., et al. (2019). Axon-Dependent Patterning and Maintenance of Somatosensory Dendritic Arbors. Dev. Cell 48, 229–244e224. doi: 10.1016/j.devcel.2018.12.015
Rauthan, M., Morck, C., and Pilon, M. (2007). The C. elegans M3 neuron guides the growth cone of its sister cell M2 via the Kruppel-like zinc finger protein MNM-2. Dev. Biol. 311, 185–199. doi: 10.1016/j.ydbio.2007.08.037
Reid, A., Sherry, T. J., Yucel, D., Llamosas, E., and Nicholas, H. R. (2015). The C-terminal binding protein (CTBP-1) regulates dorsal SMD axonal morphology in Caenorhabditis elegans. Neuroscience 311, 216–230. doi: 10.1016/j.neuroscience.2015.10.026
Reiter, F., Wienerroither, S., and Stark, A. (2017). Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev. 43, 73–81. doi: 10.1016/j.gde.2016.12.007
Reyes, A., and Huber, W. (2018). Alternative start and termination sites of transcription drive most transcript isoform differences across human tissues. Nucleic Acids Res. 46, 582–592. doi: 10.1093/nar/gkx1165
Rhiner, C., Gysi, S., Frohli, E., Hengartner, M. O., and Hajnal, A. (2005). Syndecan regulates cell migration and axon guidance in C. elegans. Development 132, 4621–4633. doi: 10.1242/dev.02042
Riccomagno, M. M., and Kolodkin, A. L. (2015). Sculpting neural circuits by axon and dendrite pruning. Annu. Rev. Cell Dev. Biol. 31, 779–805. doi: 10.1146/annurev-cellbio-100913-013038
Richardson, C. E., and Shen, K. (2019). Neurite Development and Repair in Worms and Flies. Annu. Rev. Neurosci. 42, 209–226. doi: 10.1146/annurev-neuro-070918-050208
Robinson, J. T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. doi: 10.1038/nbt.1754
Rojas-Duran, M. F., and Gilbert, W. V. (2012). Alternative transcription start site selection leads to large differences in translation activity in yeast. RNA 18, 2299–2305. doi: 10.1261/rna.035865.112
Roy, P. J., Zheng, H., Warren, C. E., and Culotti, J. G. (2000). mab-20 encodes Semaphorin-2a and is required to prevent ectopic cell contacts during epidermal morphogenesis in Caenorhabditis elegans. Development 127, 755–767. doi: 10.1242/dev.127.4.755
Runko, E., and Kaprielian, Z. (2004). Caenorhabditis elegans VEM-1, a novel membrane protein, regulates the guidance of ventral nerve cord-associated axons. J. Neurosci. 24, 9015–9026. doi: 10.1523/JNEUROSCI.2385-04.2004
Sainath, R., and Gallo, G. (2015). Cytoskeletal and signaling mechanisms of neurite formation. Cell Tissue Res. 359, 267–278. doi: 10.1007/s00441-014-1955-0
Santiago, C., and Bashaw, G. J. (2014). Transcription factors and effectors that regulate neuronal morphology. Development 141, 4667–4680. doi: 10.1242/dev.110817
Sasakura, H., Inada, H., Kuhara, A., Fusaoka, E., Takemoto, D., Takeuchi, K., et al. (2005). Maintenance of neuronal positions in organized ganglia by SAX-7, a Caenorhabditis elegans homologue of L1. EMBO J. 24, 1477–1488. doi: 10.1038/sj.emboj.7600621
Schelski, M., and Bradke, F. (2017). Neuronal polarization: From spatiotemporal signaling to cytoskeletal dynamics. Mol. Cell Neurosci. 84, 11–28. doi: 10.1016/j.mcn.2017.03.008
Schmid, C., Schwarz, V., and Hutter, H. (2006). AST-1, a novel ETS-box transcription factor, controls axon guidance and pharynx development in C. elegans. Dev. Biol. 293, 403–413. doi: 10.1016/j.ydbio.2006.02.042
Schmitz, C., Wacker, I., and Hutter, H. (2008). The Fat-like cadherin CDH-4 controls axon fasciculation, cell migration and hypodermis and pharynx development in Caenorhabditis elegans. Dev. Biol. 316, 249–259. doi: 10.1016/j.ydbio.2008.01.024
Schroeder, N. E., Androwski, R. J., Rashid, A., Lee, H., Lee, J., and Barr, M. M. (2013). Dauer-Specific Dendrite Arborization in C. elegans Is Regulated by KPC-1/Furin. Curr. Biol. 23, 1527–1535. doi: 10.1016/j.cub.2013.06.058
Schuldiner, O., and Yaron, A. (2015). Mechanisms of developmental neurite pruning. Cell Mol. Life Sci. 72, 101–119. doi: 10.1007/s00018-014-1729-6
Schwamborn, J. C., and Puschel, A. W. (2004). The sequential activity of the GTPases Rap1B and Cdc42 determines neuronal polarity. Nat. Neurosci. 7, 923–929. doi: 10.1038/nn1295
Seetharaman, A., Selman, G., Puckrin, R., Barbier, L., Wong, E., D’Souza, S. A., et al. (2011). MADD-4 Is a Secreted Cue Required for Midline-Oriented Guidance in Caenorhabditis elegans. Dev. Cell 21, 669–680. doi: 10.1016/j.devcel.2011.07.020
Serrano-Saiz, E., Oren-Suissa, M., Bayer, E. A., and Hobert, O. (2017). Sexually dimorphic differentiation of a C. elegans hub neuron is cell autonomously controlled by a conserved transcription factor. Curr. Biol. 27, 199–209. doi: 10.1016/j.cub.2016.11.045
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sherry, T., Handley, A., Nicholas, H. R., and Pocock, R. (2020). Harmonization of L1CAM expression facilitates axon outgrowth and guidance of a motor neuron. Development 147:dev193805. doi: 10.1242/dev.193805
Shirasaki, R., and Pfaff, S. L. (2002). Transcriptional codes and the control of neuronal identity. Annu. Rev. Neurosci 25, 251–281. doi: 10.1146/annurev.neuro.25.112701.142916
Smith, C. J., O’Brien, T., Chatzigeorgiou, M., Spencer, W. C., Feingold-Link, E., Husson, S. J., et al. (2013). Sensory neuron fates are distinguished by a transcriptional switch that regulates dendrite branch stabilization. Neuron 79, 266–280. doi: 10.1016/j.neuron.2013.05.009
Smith, C. J., Watson, J. D., Spencer, W. C., O’Brien, T., Cha, B., Albeg, A., et al. (2010). Time-lapse imaging and cell-specific expression profiling reveal dynamic branching and molecular determinants of a multi-dendritic nociceptor in C. elegans. Dev. Biol. 345, 18–33. doi: 10.1016/j.ydbio.2010.05.502
Smith, C. J., Watson, J. D., VanHoven, M. K., Colon-Ramos, D. A., and Miller, D. M. III (2012). Netrin (UNC-6) mediates dendritic self-avoidance. Nat. Neurosci 15, 731–737. doi: 10.1038/nn.3065
Song, S., Zhang, B., Sun, H., Li, X., Xiang, Y., Liu, Z., et al. (2010). A Wnt-Frz/Ror-Dsh pathway regulates neurite outgrowth in Caenorhabditis elegans. PLoS Genet. 6:e1001056. doi: 10.1371/journal.pgen.1001056
Stefanakis, N., Carrera, I., and Hobert, O. (2015). Regulatory Logic of Pan-Neuronal Gene Expression in C. elegans. Neuron 87, 733–750. doi: 10.1016/j.neuron.2015.07.031
Steimel, A., Wong, L., Najarro, E. H., Ackley, B. D., Garriga, G., and Hutter, H. (2010). The Flamingo ortholog FMI-1 controls pioneer-dependent navigation of follower axons in C. elegans. Development 137, 3663–3673. doi: 10.1242/dev.054320
Sudhof, T. C. (2018). Towards an understanding of synapse formation. Neuron 100, 276–293. doi: 10.1016/j.neuron.2018.09.040
Sun, H., and Hobert, O. (2021). Temporal transitions in the post-mitotic nervous system of Caenorhabditis elegans. Nature 600, 93–99. doi: 10.1038/s41586-021-04071-4
Sundararajan, L., Stern, J., and Miller, D. M. (2019). Mechanisms that regulate morphogenesis of a highly branched neuron in C. elegans. Dev. Biol. 451, 53–67. doi: 10.1016/j.ydbio.2019.04.002
Taylor, S. R., Santpere, G., Weinreb, A., Barrett, A., Reilly, M. B., Xu, C., et al. (2021). Molecular topography of an entire nervous system. Cell 432:e4323. doi: 10.1016/j.cell.2021.06.023
Torpe, N., and Pocock, R. (2014). Regulation of axonal midline guidance by prolyl 4-hydroxylation in Caenorhabditis elegans. J. Neurosci. 34, 16348–16357. doi: 10.1523/JNEUROSCI.1322-14.2014
Trang, H., Samuels, M., Ceccherini, I., Frerick, M., Garcia-Teresa, M. A., Peters, J., et al. (2020). Guidelines for diagnosis and management of congenital central hypoventilation syndrome. Orphanet. J. Rare Dis. 15, 252. doi: 10.1186/s13023-020-01460-2
Tsalik, E. L., Niacaris, T., Wenick, A. S., Pau, K., Avery, L., and Hobert, O. (2003). LIM homeobox gene-dependent expression of biogenic amine receptors in restricted regions of the C. elegans nervous system. Dev. Biol. 263, 81–102. doi: 10.1016/s0012-1606(03)00447-0
Unsoeld, T., Park, J. O., and Hutter, H. (2013). Discoidin domain receptors guide axons along longitudinal tracts in C. elegans. Dev. Biol. 374, 142–152. doi: 10.1016/j.ydbio.2012.11.001
Van Buskirk, C., and Sternberg, P. W. (2010). Paired and LIM class homeodomain proteins coordinate differentiation of the C. elegans ALA neuron. Development 137, 2065–2074. doi: 10.1242/dev.040881
Van Nostrand, E. L., and Kim, S. K. (2013). Integrative analysis of C. elegans modENCODE ChIP-seq data sets to infer gene regulatory interactions. Genome Res. 23, 941–953. doi: 10.1101/gr.152876.112
Verma, A. S., and Fitzpatrick, D. R. (2007). Anophthalmia and microphthalmia. Orphanet. J. Rare Dis. 2:47. doi: 10.1186/1750-1172-2-47
Wacker, I., Schwarz, V., Hedgecock, E. M., and Hutter, H. (2003). zag-1, a Zn-finger homeodomain transcription factor controlling neuronal differentiation and axon outgrowth in C. elegans. Development 130, 3795–3805. doi: 10.1242/dev.00570
Wadsworth, W. G., Bhatt, H., and Hedgecock, E. M. (1996). Neuroglia and pioneer neurons express UNC-6 to provide global and local netrin cues for guiding migrations in C. elegans. Neuron 16, 35–46. doi: 10.1016/s0896-6273(00)80021-5
Wang, J., and Ding, M. (2018). Robo and Ror function in a common receptor complex to regulate Wnt-mediated neurite outgrowth in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 115, E2254–E2263. doi: 10.1073/pnas.1717468115
Wang, X. L., Zhang, W., Cheever, T., Schwarz, V., Opperman, K., Hutter, H., et al. (2008). The C-elegans L1CAM homologue LAD-2 functions as a coreceptor in MAB-20/Sema2-mediated axon guidance. J. Cell Biol. 180, 233–246. doi: 10.1083/jcb.200704178
Wang, X., Kweon, J., Larson, S., and Chen, L. (2005). A role for the C. elegans L1CAM homologue lad-1/sax-7 in maintaining tissue attachment. Dev. Biol. 284, 273–291. doi: 10.1016/j.ydbio.2005.05.020
Weinberg, P., Flames, N., Sawa, H., Garriga, G., and Hobert, O. (2013). The SWI/SNF chromatin remodeling complex selectively affects multiple aspects of serotonergic neuron differentiation. Genetics 194, 189–198. doi: 10.1534/genetics.112.148742
Weinberger, D. R. (2019). Thinking About Schizophrenia in an Era of Genomic Medicine. Am. J. Psychiatry 176, 12–20. doi: 10.1176/appi.ajp.2018.18111275
White, J. G., Southgate, E., Thomson, J. N., and Brenner, S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 314, 1–340. doi: 10.1098/rstb.1986.0056
Wightman, B., Baran, R., and Garriga, G. (1997). Genes that guide growth cones along the C-elegans ventral nerve cord. Development 124, 2571–2580. doi: 10.1242/dev.124.13.2571
Xu, Y., Taru, H., Jin, Y., and Quinn, C. C. (2015). SYD-1C, UNC-40 (DCC) and SAX-3 (Robo) function interdependently to promote axon guidance by regulating the MIG-2 GTPase. PLoS Genet. 11:e1005185. doi: 10.1371/journal.pgen.1005185
Xue, D., Finney, M., Ruvkun, G., and Chalfie, M. (1992). Regulation of the mec-3 gene by the C.elegans homeoproteins UNC-86 and MEC-3. EMBO J 11, 4969–4979. doi: 10.1002/j.1460-2075.1992.tb05604.x
Yang, Y., Sun, Y., Luo, X., Zhang, Y., Chen, Y., Tian, E., et al. (2007). Polycomb-like genes are necessary for specification of dopaminergic and serotonergic neurons in Caenorhabditis elegans. Proc. Natl. Acad. Sci U.S.A. 104, 852–857. doi: 10.1073/pnas.0610261104
Yanowitz, J. L., Shakir, M. A., Hedgecock, E., Hutter, H., Fire, A. Z., and Lundquist, E. A. (2004). UNC-39, the C. elegans homolog of the human myotonic dystrophy-associated homeodomain protein Six5, regulates cell motility and differentiation. Dev. Biol. 272, 389–402. doi: 10.1016/j.ydbio.2004.05.010
Yogev, S., and Shen, K. (2017). Establishing neuronal polarity with environmental and intrinsic mechanisms. Neuron 96, 638–650. doi: 10.1016/j.neuron.2017.10.021
Yoshimura, S., Murray, J. I., Lu, Y., Waterston, R. H., and Shaham, S. (2008). mls-2 and vab-3 Control glia development, hlh-17/Olig expression and glia-dependent neurite extension in C. elegans. Development 135, 2263–2275. doi: 10.1242/dev.019547
Yu, B., Wang, X., Wei, S., Fu, T., Dzakah, E. E., Waqas, A., et al. (2017). Convergent transcriptional programs regulate cAMP Levels in C. elegans GABAergic motor neurons. Dev Cell 21:e217. doi: 10.1016/j.devcel.2017.09.013
Yue, B. (2014). Biology of the extracellular matrix: An overview. J. Glaucoma 23(8 Suppl. 1) S20–S23. doi: 10.1097/IJG.0000000000000108
Zallen, J. A., Kirch, S. A., and Bargmann, C. I. (1999). Genes required for axon pathfinding and extension in the C. elegans nerve ring. Development 126, 3679–3692. doi: 10.1242/dev.126.16.3679
Zhang, H., Azevedo, R. B., Lints, R., Doyle, C., Teng, Y., Haber, D., et al. (2003). Global regulation of Hox gene expression in C. elegans by a SAM domain protein. Dev. Cell. 4, 903–915. doi: 10.1016/s1534-5807(03)00136-9
Zhou, X., Gueydan, M., Jospin, M., Ji, T., Valfort, A., Pinan-Lucarre, B., et al. (2020). The netrin receptor UNC-40/DCC assembles a postsynaptic scaffold and sets the synaptic content of GABAA receptors. Nat. Commun. 11:2674. doi: 10.1038/s41467-020-16473-5
Zinovyeva, A. Y., and Forrester, W. C. (2005). The C. elegans Frizzled CFZ-2 is required for cell migration and interacts with multiple Wnt signaling pathways. Dev. Biol. 285, 447–461. doi: 10.1016/j.ydbio.2005.07.014
Zolessi, F. R., Poggi, L., Wilkinson, C. J., Chien, C. B., and Harris, W. A. (2006). Polarization and orientation of retinal ganglion cells in vivo. Neural. Dev. 1:2. doi: 10.1186/1749-8104-1-2
Keywords: neurite development, transcription factors, neuronal guidance, gene regulatory landscape, Caenorhabditis elegans
Citation: Godini R, Fallahi H and Pocock R (2022) The regulatory landscape of neurite development in Caenorhabditis elegans. Front. Mol. Neurosci. 15:974208. doi: 10.3389/fnmol.2022.974208
Received: 20 June 2022; Accepted: 26 July 2022;
Published: 25 August 2022.
Edited by:
George Smith, Temple University, United StatesReviewed by:
Nathaniel J. Szewczyk, Ohio University, United StatesCopyright © 2022 Godini, Fallahi and Pocock. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rasoul Godini, cmFzb3VsLmdvZGluaUBtb25hc2guZWR1; Roger Pocock, cm9nZXIucG9jb2NrQG1vbmFzaC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.