- 1Research Center of the University of Montreal Hospital Center (CRCHUM), University of Montreal, Montreal, QC, Canada
- 2Department of Biochemistry, University of Montreal, Montreal, QC, Canada
- 3Department of Neuroscience, University of Montreal, Montreal, QC, Canada
During development of the zebrafish embryo, glycine signaling promotes the differentiation of neural stem cells (NSCs). We found that glycine signaling suppresses the expression of Ligand of Numb X1 (lnx1, Ligand of numb protein-x1), a gene of unknown function during NSC differentiation that is selectively expressed in the embryonic central nervous system (CNS). As a consequence, Numb levels were stabilized and Notch activity (measured as her4.1 expression) was reduced, promoting NSC differentiation. These consequent actions were blocked by knockdown of lnx1. In contrast, lnx1 overexpression increased NSC proliferation and led to defects of neural tube closure at the early stages of development. Thus, our data provide evidence that glycine/lnx1 signaling modulates NSC proliferation by regulation of Notch signaling.
Introduction
During neuronal development an early spontaneous electrical activity is generated in neural stem cells (NSCs) as an essential step for their proliferation, migration and differentiation (Spitzer, 2006) and involves several neurotransmitters including glutamate, GABA and glycine (Demarque et al., 2002; Scain et al., 2010). Here we investigated the role of glycine signaling during neuronal development in the zebrafish embryo. We demonstrated previously that glycine signaling regulates NSC proliferation (Mcdearmid et al., 2006) and differentiation (Cote and Drapeau, 2012) by promoting survival of a subpopulation of NSCs (Bekri and Drapeau, 2018). An RNA sequencing analysis revealed that glycine signaling regulates several pathways in NSC development (Samarut et al., 2016) as well as some outlying genes, with Ligand of numb protein-x1 (lnx1) among the most affected.
Lnx1 protein is a RING-type E3 ubiquitin ligase (De Bie and Ciechanover, 2011; Flynn et al., 2011) that degrades Numb (Dho et al., 1998), a cell fate determinant (Uemura et al., 1989). Furthermore, Numb is associated with Shh signaling (Di Marcotullio et al., 2011) and P53 signaling (Colaluca et al., 2008), both participating in glycine-dependent neurogenesis in zebrafish models (Samarut et al., 2016; Bekri and Drapeau, 2018). Importantly, Numb is well-known to be an inhibitor of Notch signaling (Roegiers and Jan, 2004; Mcgill et al., 2009), but further elucidations are required to understand how Notch and lnx1 activity correlates with other pathways to fine-tune neuronal development.
We report here that glycine signaling suppressed lnx1 expression in NSCs and consequently modulated Notch activity by controlling Numb protein degradation.
Materials and Methods
More information about materials and methods is provided in Supplementary Materials.
Zebrafish
Zebrafish (Danio rerio) were maintained at 28°C under a 12-h light/dark cycle in the crCHUM Zebrafish Facility and they were raised and manipulated as per guidelines of the Canadian Council for Animal Care and protocol approved (N15018PDz) by the crCHUM ethics committee. To knockdown gene expression, embryos were microinjected with morpholino (MO) as described previously (Bekri and Drapeau, 2018).
FACS and RT-qPCR
Tg(GFAP:GFP) embryos were injected with glycine receptor-MO (Glr-MO) or Ctrl-MO. At 20 hpf, GFAP-NSCs were sorted by FACS. Then, total RNA was extracted and gene expression was quantified as described previously (Samarut et al., 2016). Sequence of each primer was designed by Snapgene software®.
Whole-Mount in situ Hybridization and Immunostaining
Embryos were injected with Glr-MO or Ctrl-M, then subjected to in situ hybridization or immunostaining as described previously (Bekri and Drapeau, 2018).
Western Blotting
Embryos were injected with lnx1-6myc or gal4 mRNA, then total protein was extracted at desired stages. Western blotting was performed as previously described (Swaminathan et al., 2018).
Probes and mRNA Synthesis
To make probes or mRNA, total RNA was extracted from 24 h post fertilization (hpf) of zebrafish embryos. Total RNA was reverse transcribed to cDNA. Then, used to make probes and full length lnx1 as described previously (Brustein et al., 2013).
Results
Glycine Signaling Suppresses lnx1 Expression and Regulates Neural Tube Development
We identified that expression of lnx1 was strongly suppressed by glycine signaling during zebrafish development (Samarut et al., 2016). To confirm our transcriptomic study, we analyzed the expression level of lnx1 upon disruption of glycine signaling by RT-qPCR and in situ hybridization. We used the tg(GFAP:GFP) line that expresses GFP under the gfap promoter (Bernardos and Raymond, 2006), which is an early marker of NSCs. Embryos from this line were treated with a Glr-MO to disturb glycine signaling, or with control Ctrl-MO or in uninjected eggs as control conditions. Embryos at 18 hpf were dissociated and GAFP+ NSCs were sorted, total RNA was extracted and lnx1 expression was analyzed by RT-qPCR. Disruption of glycine signaling confirmed a significant increase of lnx1 expression compared with Ctrl-MO or uninjected embryos condition (Figure 1A). To further validate these results, lnx1 expression was visualized by whole-mount in situ hybridization, revealing a strong expression of lnx1 upon Glr knockdown especially in the central nervous system (CNS) at 18 and 24 hpf stages (Figure 1B; right side, asterisk), compared with control condition which showed only a slight expression of lnx1 in the brain (Figure 1B; left side). Taken together, these results confirm that glycine signaling suppresses lnx1 expression into NSC at early stage of development.
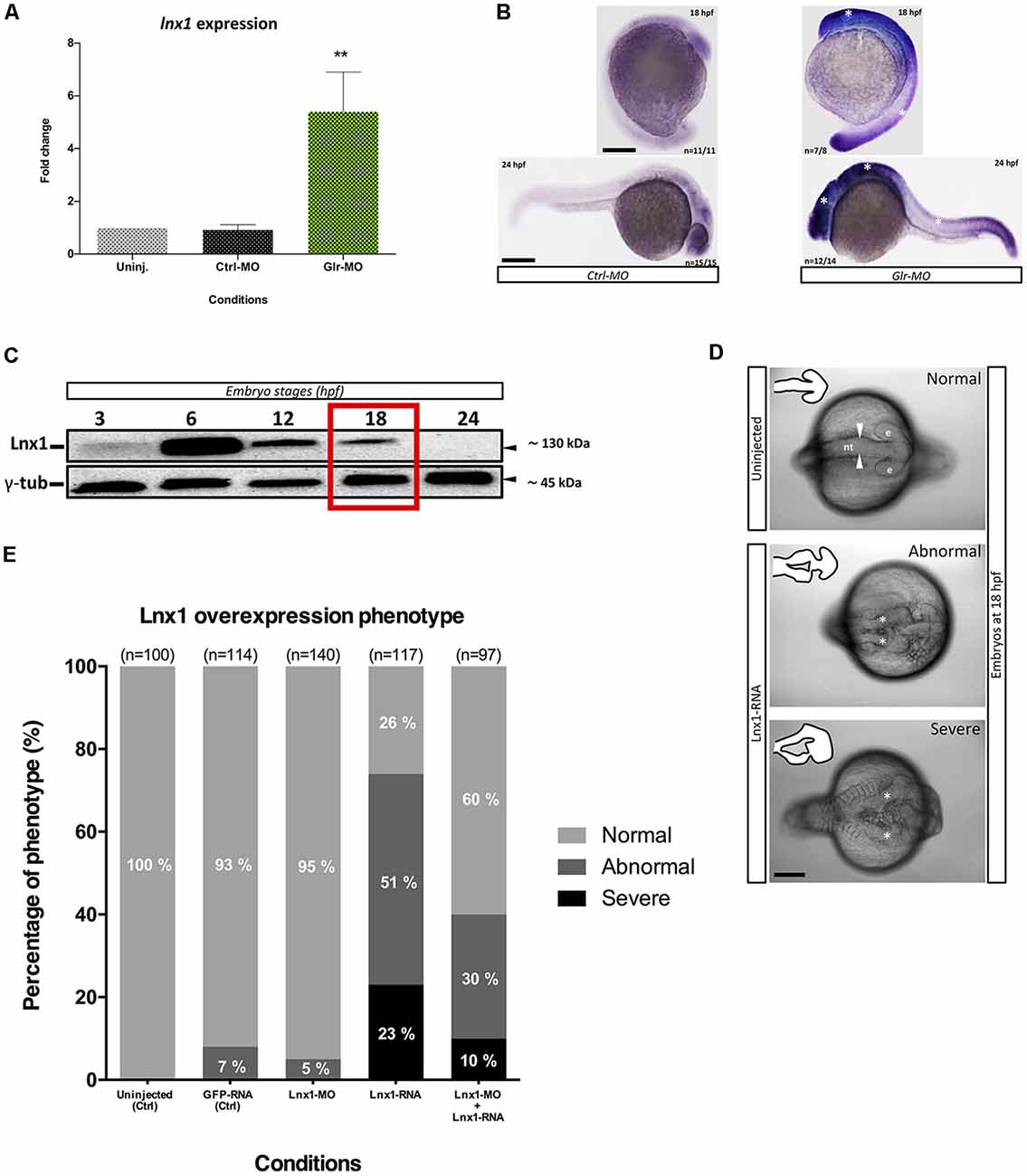
Figure 1. Glycine signaling regulates Ligand of numb protein-x1 (lnx1) expression during neural tube development. (A) Quantification of lnx1 expressions into sorted GFAP+-neural stem cell (NSC) by RT-qPCR revealed a significant up-regulation of lnx1 expression upon glycine signaling disruption compared with uninjected and Ctrl-morpholino (MO) conditions. One-way ANOVA statistical analysis was performed (n = 3, **p-value < 0.01). (B) Whole-mount in situ hybridization at 18 and 24 hours post fertilization (hpf) revealed that disruption of glycine signaling by glycine receptor morpholino (Glr-MO) induces an overexpression of lnx1 into central nervous system (CNS; right) compared with control condition (left; Scale bar, 200 μm). (C) Time course of transient lnx1 overexpression revealed by Western blot; embryos were injected with lnx1-6myc RNA then in vivo expression of lnx1 protein was detected by anti-myc antibodies and followed during five-time point including, 3, 6, 12, 18 and 24 hpf, and anti-γ-tub antibody was used as loading protein control. (D) Neural tube closes defects upon lnx1 overexpression; embryos were injected with lnx1-6myc RNA, then neural tube was imaged at 18 hpf. Phenotype of neural tube defect closure caused by lnx1 overexpression was divided into three classes: normal neural tube (arrowheads), abnormal neural tube and neural tube with severe defects (asterisks) from top to down respectively. Structure of neural tube was delineated in the corner of each image. (e, eye; nt, neural tube. Scale bar, 250 μm). (E) Quantification of lnx1 overexpression phenotype in each condition including uninjected, GFP-mRNA, lnx1-MO, lnx1-RNA or lnx1-MO and lnx1-RNA embryos.
We next tested the effects of early overexpression of lnx1. First, due to the unavailability of efficient antibodies against lnx1, we created a construct which expressed lnx1 with myc-tag (lnx1-myc) to reveal lnx1 expression by myc-tag antibodies. Then, we overexpressed lnx1 by injecting lnx1-myc mRNA. Result showed a low expression level at 3 hpf and strong expression at 6 hpf, followed by degradation from 12 to 18 hpf until 24 hpf (midway through embryonic development), when lnx1 expression was no longer detected (Figure 1C). Based on these results, we defined 18 hpf, near the start of neurogenesis, as the best time point to analyze the effect of early lnx1 expression on zebrafish development. Control embryos showed normal brain and neural tube development (Figure 1D, in the top), whereas those injected with lnx1 mRNA showed a major defect of neural tube closure, especially during head development (Figure 1D, in the middle and bottom, asterisk). We then tested several doses of lnx1 mRNA and determined that 40 pg was the lowest dose that consistently produced an effect. We classified the defective neural tube phenotype into three classes: normal, abnormal and severe (Figure 1D). Control embryos uninjected or injected with GFP mRNA or lnx1-MO showed normal development of the neural tube (Figure 1E). However, upon lnx1 mRNA injection, many of the embryos showed defective neural tube closure (Figure 1E). To verify whether the defect of neural tube closure was caused by overexpression of lnx1 and was not an artifact caused by toxicity of mRNA injections, we tested for rescue of the defect of neural tube closure by co-injection of lnx1 mRNA with lnx1-MO to block translation of lnx1 mRNA. The results revealed a partial rescue, with a doubling of the normal phenotype and reduction by half in the two classes of defective phenotypes (Figure 1E). Taken together, these results provide evidence that overexpression of lnx1 induced a defect of neural tube closure, accruing in a major malformation of the head region during zebrafish embryogenesis.
Glycine/lnx1 Signaling Regulates Notch Activity and NSCs Proliferation
Lnx1/2 are E3 ubiquitin ligases which promote the degradation of Numb and modulate Numb/Notch signaling during neurogenesis (Nie et al., 2002; Kageyama et al., 2007), though the role of lnx1 in NSCs is unknown. To test whether disruption of glycine signaling in zebrafish NSCs, with elevated lnx1 expression (Figure 1), modulates Notch signaling, we injected Tg(gfap:GFP) embryos at the one-cell stage with Glr-MO or Ctrl-MO, which were then sorted at 18 hpf GFAP+-NSCs, followed by RNA extraction. Using RT-qPCR we quantified Her4.1 expression, a reporter of Notch activity in zebrafish (Takke et al., 1999). The results showed a significant increase of her4.1 expression in GFAP+-NSCs upon glycine disruption, compared with uninjected and Ctrl-MO controls conditions (Figure 2A). This suggests that disruption of glycine signaling promotes Notch activity in NSCs.
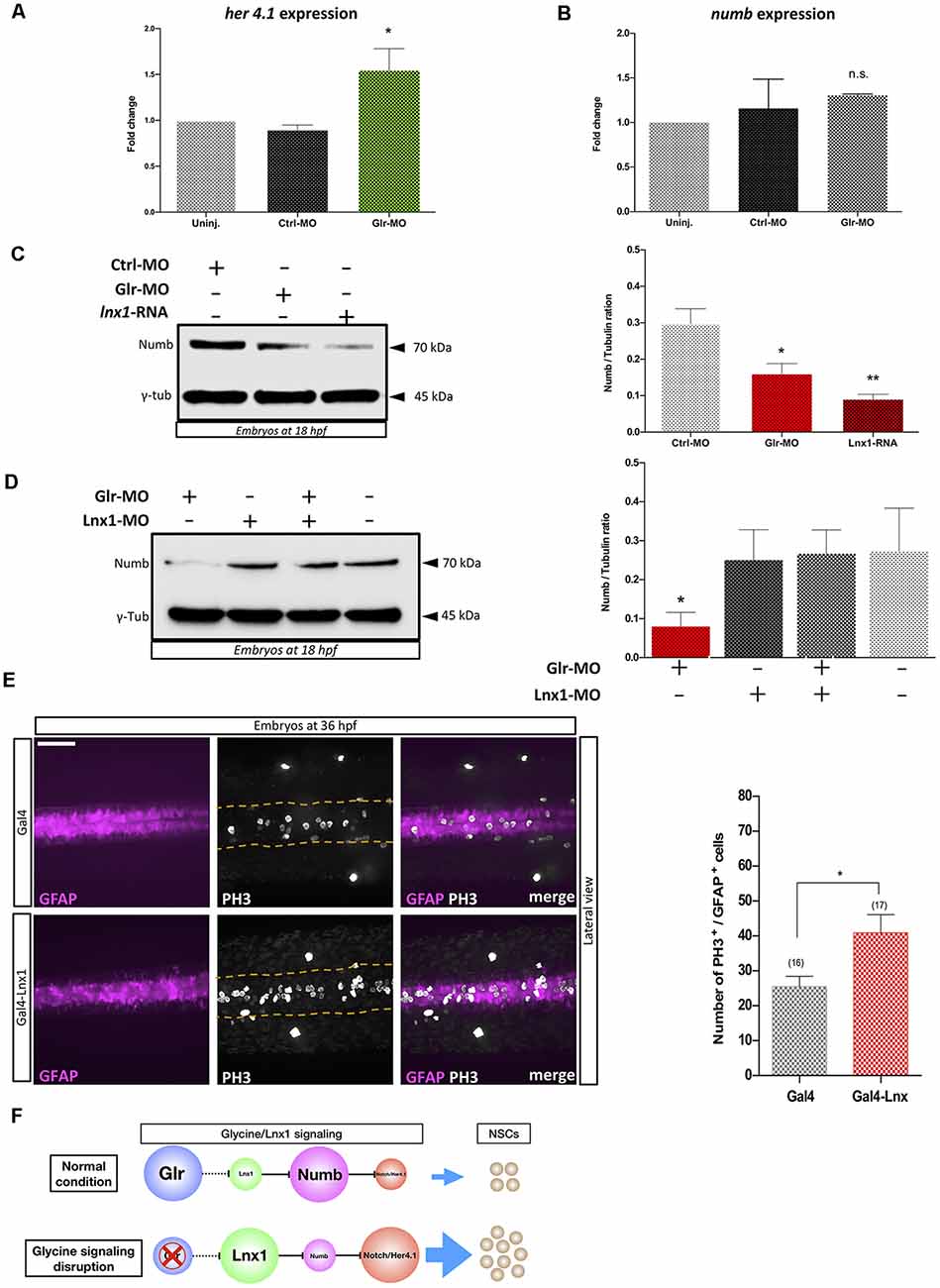
Figure 2. Glycine signaling modulates Notch activity via lnx1 expression and promotes NSCs proliferation. Quantification of her4.1 mRNA (A) and numb mRNA (B) level into sorted GFAP+-NSC by RT-qPCR revealed a significant increase of her4.1 expression upon disruption of glycine signaling by Glr-MO compared with uninjected and Ctrl-MO conditions. However, no significant changing of numb expression. One-way ANOVA statistical analysis was performed [n = 3, *p-value < 0.05, not significant (n.s.)]. (C) Expression of Numb protein was revealed by western bolt at 18 hpf showing a significant degradation of numb protein upon disruption of glycine signaling by Glr-MO and overexpression of lnx1 by lnx1 mRNA injection compared with Ctrl-MO injections which were used as control condition. However, Co-injection of Glr-MO and lnx1-MO (D) rescued Numb protein degradation. One-way ANOVA statistical analysis was performed (n = 3, *p-value < 0.05, **p-value < 0.01). (E) Proliferation of GFAP+-NSCs (pink) in spinal cord by PH3 immunostaining (white) into tg(GFAP:Gal4, UAS:RED; top panel), and tg(GFAP:Gal4, UAS, lnx1, UAS:RED; bottom panel) revealed a significant increase of GFAP+-NSCs proliferation in lnx1 overexpression embryos (right panel). One-way ANOVA statistical analysis was performed (n = 17, **p-value < 0.0001, scale bar, 250 μm). (F) The schematic model of regulation of Notch activity by glycine/lnx1 signaling into zebrafish NSCs during early development.
We hypothesized that disruption of glycine signaling could modulate Numb protein expression, the main mediator between lnx and Notch signaling (Nie et al., 2002). We therefore used total RNA extracted from GFAP+-NSCs upon glycine signaling disruption to quantify numb expression by RT-qPCR. The results showed no significant change in numb mRNA level between disruption of glycine signaling (Glr-MO) and control conditions (Glr-MO or uninjected embryos) in NSCs (Figure 2B). However, analysis of Numb protein expression by western blot using anti-Numb antibody revealed a decrease in Numb protein level upon disruption of glycine signaling (Glr-MO) compared with control condition (Ctrl-MO; Figure 2C). This result suggests that while the numb mRNA level was unaffected by disruption of glycine signaling, Numb protein was degraded, likely via up-regulation of lnx1 expression. To confirm that overexpression of lnx1 in zebrafish embryos could mimic the disruption of glycine signaling and degrade Numb protein expression, we overexpressed lnx1 and analyzed Numb protein expression at 18 hpf. The results showed an important decrease of Numb protein (Figure 2C). Finally, to verify whether degradation of Numb protein by glycine signaling was due specifically to lnx1 overexpression, we tested whether down regulation of lnx1 upon disruption of glycine signaling rescued Numb expression. To do so, we injected embryos with Glr-MO, lnx1-MO or both Glr-MO and lnx1-MO and evaluated Numb protein expression in each condition compared with uninjected embryos. The results showed a significant reduction of Numb protein level upon disruption of glycine signaling by Glr-MO compared with control whereas co-injection of Glr-MO and lnx1-MO together rescued the Numb protein level (Figure 2D). These results provide evidence that glycine/lnx1 signaling modulates Notch activity by controlling Numb protein degradation in NSCs.
By analogy to lnx2 (Won et al., 2015; Yin et al., 2015), we hypothesized that glycine/lnx1 signaling controls NSC proliferation and that its disruption would cause a developmental phenotype with stabilized NSCs. To test this hypothesis, we expressed lnx1 specifically in NSCs by generating a stable zebrafish line expressing lnx1 (UAS:lnx1) in the Tg(UAS:RFP) reporter background, thus generating the double-Tg(UAS:lnx1;UAS;RFP) effector-line (Supplementary Figure S1A). First, to validate the transcriptional activation of lnx1 in the Tg(UAS:lnx1;UAS;RFP) line, we induced ubiquitous expression of lnx1 by injections of Gal4-activator mRNA (20 pg) into Tg(UAS:lnx1;UAS:RFP) or Tg(UAS:RFP) embryos, with the latter as controls. At 18 hpf, neural tube development was evaluated and lnx1 mRNA level was analyzed by semi-quantitative RT-qPCR. The results showed a drastic defect of neural tube closure in Tg(UAS:lnx1;UAS;RFP) embryos compared with Tg(UAS:RFP) embryos, a phenotype similar to that of lnx1 mRNA injection (data not shown). Moreover, quantification of lnx1 mRNA levels demonstrated a strong transcriptional activity of lnx1 in Tg(UAS:lnx1;UAS;RFP) compared with Tg(UAS:RFP) control. However, no significant change in transcriptional activity was observed in rpl13a and ef1a, used as reference genes (Supplementary Figure S1B,C). These results replicated the defect of neural tube closure observed by lnx1 mRNA injections and confirmed the phenotype upon ubiquitous early expression of lnx1 (Figure 1).
Next, in order to test the effect of lnx1 overexpression on NSC proliferation, we specifically overexpressed lnx1 in NSCs by crossing Tg(UAS:lnx1;UAS:RFP) with Tg(GFAP:Gal4) adult zebrafish. Embryos were fixed at 36 hpf and proliferation was assayed by PH3 immunostaining. The results revealed similar GFAP+-NSC populations (pink color) in both conditions including Tg(GFAP:Gal4;UAS:lnx1, UAS:RFP) and Tg(GFAP:Gal4;UAS:RFP; Figure 2E, in the left). However, in vivo overexpression of lnx1 in NSCs in the Tg(GFAP:Gal4;UAS:lnx1, UAS:RFP) line revealed a large increase of PH3+-NSCs compared to the Tg(UAS:RFP) control line (Figure 2E, in the middle). This result indicates that early expression of lnx1 in NSCs promotes their proliferation. Taken together, these results provide evidence that glycine/lnx1 signaling modulates NSC proliferation through regulation of Notch activity (Figure 2F).
Discussion
During neuronal development, several molecular changes take place in NSCs when glycine signaling is disrupted (Samarut et al., 2016). We demonstrated with different approaches that disruption of glycine signaling induced an overexpression of lnx1 in NSCs (Figure 1). While regulation of lnx2 transcription has been related to Gli3 and RunX2 (Pregizer et al., 2007; Wang et al., 2014), no transcription factors or pathways have been related to lnx1 expression, leaving it as somewhat of an orphaned gene. However, increased lnx1 expression reduces expression of the glycine transporter 2 (GlyT2) and impairs glycine transport in cortical neurons (Núñez et al., 2017). We showed that disruption of glycine signaling by knockdown of glycine receptors (Glrs) induced an overexpression of lnx1 in NSCs. Furthermore, we demonstrated that GFAP+-NSCs up-regulated lnx1 upon disruption of glycine signaling (Figure 1). Thus, glycine signaling suppresses lnx1, which appears to increase GlyT2, possibly as a homeostatic mechanism to regulate glycine levels. On other hand, a few studies have highlighted the potential role of the lnx protein family during developmental stages. Investigation of the Shh signaling component “Gli3” revealed that in knockout mice (Gli3−/−) there is an increased expression of lnx2 and a dramatic decrease of Numb protein level in NSCs. These Gli3−/− mice exhibit hydrocephaly and reduced cortical thickness as well (Wang et al., 2011, 2014). While lnx2 signaling during neurogenesis is well explored, the role of lnx1 in NSC development remained unknown. We demonstrated that transient expression of lnx1 at an early stage of development caused a severe defect of neural tube closure in the head region, probably caused by the early loss of Numb proteins during embryogenesis. In support of our results, Numb−/− null mice exhibit a severe defect in cranial neural tube closure and die around embryonic day 11.5 (E11.5; Zhong et al., 2000). These neural tube defects could be caused by disruption of neuronal development by affecting NSC proliferation.
We demonstrated that disruption of glycine signaling modulated Notch activity by increasing her4.1 expression. While it did not affect numb transcription yet, it reduced the Numb protein level as a consequence of lnx1 overexpression (Figure 2). We reported in zebrafish that disruption of glycine signaling increased NSC proliferation (Mcdearmid et al., 2006; Cote and Drapeau, 2012). Herein, by using a novel transgenic Tg(UAS:lnx1;UAS;RFP), we showed that overexpression of lnx1 in NSCs promotes their proliferation (Figure 2). These results provide compelling evidence that glycine signaling controls degradation of Numb via regulation of lnx1 expression and modulate Notch activity and proliferation of NSCs. Over all, in this study we suggest that glycine/lnx1 signaling controls NSC proliferation and differentiation by modulating the Notch pathway.
Data Availability
All datasets generated for this study are included in the manuscript and the supplementary files.
Author Contributions
AB conceived and performed most of experiments, generated the transgenic line with assistance from ML, wrote the manuscript. ML provided expertise and feedback. AB and PD reviewed and edited the manuscript. PD was responsible for supervision and funding acquisition.
Funding
This project was supported by funding from the Canadian Institutes of Health Research (Grant no: MOP-130448; to PD).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Karl Fernandes and Lamia Sid-otmane for their critical reading and valuable comments on the manuscript. we also thank Marina Drits for her assistance with fish maintenance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2019.00044/full#supplementary-material
References
Bekri, A., and Drapeau, P. (2018). Glycine promotes the survival of a subpopulation of neural stem cells. Front Cell Dev. Biol. 6:68. doi: 10.3389/fcell.2018.00068
Bernardos, R. L., and Raymond, P. A. (2006). GFAP transgenic zebrafish. Gene Expr. Patterns 6, 1007–1013. doi: 10.1016/j.modgep.2006.04.006
Brustein, E., Cote, S., Ghislain, J., and Drapeau, P. (2013). Spontaneous glycine-induced calcium transients in spinal cord progenitors promote neurogenesis. Dev. Neurobiol. 73, 168–175. doi: 10.1002/dneu.22050
Colaluca, I. N., Tosoni, D., Nuciforo, P., Senic-Matuglia, F., Galimberti, V., Viale, G., et al. (2008). NUMB controls p53 tumour suppressor activity. Nature 451, 76–80. doi: 10.3410/f.1097644.553745
Cote, S., and Drapeau, P. (2012). Regulation of spinal interneuron differentiation by the paracrine action of glycine. Dev. Neurobiol. 72, 208–214. doi: 10.1002/dneu.20972
De Bie, P., and Ciechanover, A. (2011). Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 18, 1393–1402. doi: 10.1038/cdd.2011.16
Demarque, M., Represa, A., Becq, H., Khalilov, I., Ben-Ari, Y., and Aniksztejn, L. (2002). Paracrine intercellular communication by a Ca2+- and SNARE-independent release of GABA and glutamate prior to synapse formation. Neuron 36, 1051–1061. doi: 10.1016/s0896-6273(02)01053-x
Dho, S. E., Jacob, S., Wolting, C. D., French, M. B., Rohrschneider, L. R., and Mcglade, C. J. (1998). The mammalian numb phosphotyrosine-binding domain. Characterization of binding specificity and identification of a novel PDZ domain-containing numb binding protein, LNX. J. Biol. Chem. 273, 9179–9187. doi: 10.1074/jbc.273.15.9179
Di Marcotullio, L., Greco, A., Mazza, D., Canettieri, G., Pietrosanti, L., Infante, P., et al. (2011). Numb activates the E3 ligase Itch to control Gli1 function through a novel degradation signal. Oncogene 30, 65–76. doi: 10.1038/onc.2010.394
Flynn, M., Saha, O., and Young, P. (2011). Molecular evolution of the LNX gene family. BMC Evol. Biol. 11:235. doi: 10.1186/1471-2148-11-235
Kageyama, R., Ohtsuka, T., and Kobayashi, T. (2007). The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243–1251. doi: 10.1242/dev.000786
Mcdearmid, J. R., Liao, M., and Drapeau, P. (2006). Glycine receptors regulate interneuron differentiation during spinal network development. Proc. Natl. Acad. Sci. U S A 103, 9679–9684. doi: 10.1073/pnas.0504871103
Mcgill, M. A., Dho, S. E., Weinmaster, G., and Mcglade, C. J. (2009). Numb regulates post-endocytic trafficking and degradation of Notch1. J. Biol. Chem. 284, 26427–26438. doi: 10.1074/jbc.m109.014845
Nie, J., Mcgill, M. A., Dermer, M., Dho, S. E., Wolting, C. D., and Mcglade, C. J. (2002). LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J. 21, 93–102. doi: 10.1093/emboj/21.1.93
Núñez, E. A.-G. E., López-corcuera, B., Aragón, C., and De Juan-Sanz, J. (2017). Ubiquitin ligase LNX1 is a major regulator of glycine recapture by the presynaptic transporter GlyT2. doi: 10.1101/233213
Pregizer, S., Barski, A., Gersbach, C. A., Garcia, A. J., and Frenkel, B. (2007). Identification of novel Runx2 targets in osteoblasts: cell type-specific BMP-dependent regulation of Tram2. J. Cell Biochem. 102, 1458–1471. doi: 10.1002/jcb.21366
Roegiers, F., and Jan, Y. N. (2004). Asymmetric cell division. Curr. Opin. Cell Biol. 16, 195–205. doi: 10.1016/j.ceb.2004.02.010
Samarut, E., Bekri, A., and Drapeau, P. (2016). Transcriptomic Analysis of Purified Embryonic Neural Stem Cells from Zebrafish Embryos Reveals Signaling Pathways Involved in Glycine-Dependent Neurogenesis. Front. Mol. Neurosci. 9:22. doi: 10.3389/fnmol.2016.00022
Scain, A. L., Le Corronc, H., Allain, A. E., Muller, E., Rigo, J. M., Meyrand, P., et al. (2010). Glycine release from radial cells modulates the spontaneous activity and its propagation during early spinal cord development. J. Neurosci. 30, 390–403. doi: 10.1523/JNEUROSCI.2115-09.2010
Spitzer, N. C. (2006). Electrical activity in early neuronal development. Nature 444, 707–712. doi: 10.1038/nature05300
Swaminathan, A., Hassan-Abdi, R., Renault, S., Siekierska, A., Riche, R., Liao, M., et al. (2018). Non-canonical mTOR-independent role of depdc5 in regulating gabaergic network development. Curr. Biol. 28, 1924.e5–1937.e5. doi: 10.1016/j.cub.2018.04.061
Takke, C., Dornseifer, P., V Weizsacker, E., and Campos-Ortega, J. A. (1999). her4, a zebrafish homologue of the Drosophila neurogenic gene E(spl), is a target of NOTCH signalling. Development 126, 1811–1821.
Uemura, T., Shepherd, S., Ackerman, L., Jan, L. Y., and Jan, Y. N. (1989). numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 58, 349–360. doi: 10.1016/0092-8674(89)90849-0
Wang, H., Ge, G., Uchida, Y., Luu, B., and Ahn, S. (2011). Gli3 is required for maintenance and fate specification of cortical progenitors. J. Neurosci. 31, 6440–6448. doi: 10.1523/JNEUROSCI.4892-10.2011
Wang, H., Kane, A. W., Lee, C., and Ahn, S. (2014). Gli3 repressor controls cell fates and cell adhesion for proper establishment of neurogenic niche. Cell Rep. 8, 1093–1104. doi: 10.1016/j.celrep.2014.07.006
Won, M., Ro, H., and Dawid, I. B. (2015). Lnx2 ubiquitin ligase is essential for exocrine cell differentiation in the early zebrafish pancreas. Proc. Natl. Acad. Sci. U S A 112, 12426–12431. doi: 10.1073/pnas.1517033112
Yin, F. T., Futagawa, T., Li, D., Ma, Y. X., Lu, M. H., Lu, L., et al. (2015). Caspr4 interaction with LNX2 modulates the proliferation and neuronal differentiation of mouse neural progenitor cells. Stem Cells Dev. 24, 640–652. doi: 10.1089/scd.2014.0261
Keywords: LNX1, NSCs, glycine signaling, neurogenesis, Notch activity
Citation: Bekri A, Liao M and Drapeau P (2019) Glycine Regulates Neural Stem Cell Proliferation During Development via Lnx1-Dependent Notch Signaling. Front. Mol. Neurosci. 12:44. doi: 10.3389/fnmol.2019.00044
Received: 23 November 2018; Accepted: 01 February 2019;
Published: 18 February 2019.
Edited by:
Abdul R. Asif, University of Göttingen, GermanyReviewed by:
Raman M. Das, University of Manchester, United KingdomDaniele Bottai, University of Milan, Italy
Copyright © 2019 Bekri, Liao and Drapeau. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pierre Drapeau, cC5kcmFwZWF1QHVtb250cmVhbC5jYQ==
† Lead contact
 Abdelhamid Bekri
Abdelhamid Bekri Meijiang Liao1
Meijiang Liao1 Pierre Drapeau
Pierre Drapeau