- 1Department of Clinical Laboratory, The First Affiliated Hospital of Henan University of Science and Technology, Luoyang, China
- 2Department of Clinical Laboratory, Affiliated Xiaoshan Hospital, Hangzhou Normal University, Hangzhou, China
- 3Department of Clinical Laboratory, Second Affiliated Hospital of Zhejiang University, Hangzhou, China
Linezolid-resistant enterococci have increased in recent years due to the worldwide spread of acquired resistance genes (cfr, optrA, and poxtA) in clinical, animal, and environmental settings. This study investigated the carriage of optrA-positive enterococci among patients in the anorectal surgery ward in Hangzhou, China, and characterized the genetic context of optrA. A total of 173 wound secretion samples were obtained to screen optrA-positive enterococci. Of the 173 samples, 15 (8.67%) were positive for optrA, including 12 Enterococcus faecalis, two E. faecium, and one E. hirae. Multilocus sequence type analysis revealed that 12 optrA-positive E. faecalis isolates belonged to eight different sequence types (STs), of which ST16 was the main type. Eight optrA variants were identified, whose optrA flanking regions with a fexA gene downstream were bounded by different mobile genetic elements. Furthermore, the optrA gene in 8 out of 15 optrA-positive enterococci could be successfully transferred through conjugation. The findings revealed a high carriage rate of optrA in enterococci from one anorectal surgery ward in China. The dissemination of optrA-positive enterococci isolates in clinical settings should be continually monitored.
1 Introduction
Enterococci are important commensal bacteria found in the intestines of humans and many animals. However, they can also cause hospital-acquired infections, including meningitis, bacteremia, pneumonia, surgical wound infections, and urinary tract infections (Yi et al., 2022). The emergence of multidrug-resistant strains has complicated the treatment of enterococcal infections.
Linezolid is an oxazolidinone antimicrobial agent that is exclusively used for the treatment of severe infections caused by vancomycin-resistant enterococci (VRE), methicillin-resistant Staphylococcus aureus (MRSA), and penicillin-resistant pneumococci (Echeverria-Esnal et al., 2019). However, the extensive use of linezolid results in the emergence of resistance. The main mechanism mediating resistance to linezolid has been attributed to mutations in the central loop of the domain V region of the 23 s rRNA gene. In addition, acquired resistance genes (cfr, poxtA, and optrA) were identified (Zhang et al., 2023; Partridge et al., 2018). Since its first description in 2015, optrA has been frequently reported in enterococci of human, animal, and environmental origins from many countries (Andrea et al., 2015; Mendes et al., 2016; Flamm et al., 2016; Gawryszewska et al., 2017; Na et al., 2020; Han et al., 2020; Li et al., 2020). Furthermore, optrA has been reported in Gram-positive bacteria including Enterococcus, Staphylococcus, and Streptococcus as well as Gram-negative bacteria such as Campylobacter and Salmonella (Zhang et al., 2023; Liu et al., 2020; Schwarz et al., 2021). The optrA gene is often located on chromosomes or plasmids and can be transmitted by mobile genetic elements such as transposons and insertion sequences (Chen et al., 2018).
Enterococci can readily acquire or transfer multidrug resistance genes via mobile genetic elements and are the most predominant source of the spread of optrA. Enterococci harboring optrA have been widely detected in clinical, farm, and environmental settings worldwide (Gagetti et al., 2023). The dissemination of optrA is a serious concern and poses a potential public health threat. The prevalence of optrA-positive E. faecalis was 0.2% in Austria (Kerschner et al., 2021) and 0.7% in Spain (Rodríguez-Lucas et al., 2022). A 3-year survey in Korea showed that 0.23% of clinical E. faecalis isolates harbored the optrA gene (Park et al., 2020). However, in China, the positive rate of optrA has increased from 0.4% in 2004 to 3.9% in 2014 (Cui et al., 2016). Therefore, the prevalence and spread of optrA-carrying enterococci should be monitored carefully. The optrA-positive E. faecalis emerged in a tertiary care hospital (Park et al., 2020). Several studies have reported fecal carriage rates of 3.53% for optrA-positive enterococci in healthy individuals and 15.1% in patients who underwent anorectal surgery (Cai et al., 2019; Cai et al., 2016). optrA-positive enterococci may cause transmission of resistance genes in the intestine. However, reports of optrA-positive enterococci from anorectal infections have been limited. In this study, we investigated the prevalence of the optrA-positive enterococci isolates from an anorectal surgery ward in a Chinese hospital. We utilized whole-genome sequencing (WGS) to further describe the optrA genetic context.
2 Materials and methods
2.1 Sample collection and bacterial isolation
A total of 173 non-duplicated wound secretion samples were collected from an anorectal surgery ward (118 male and 55 female patients) in a tertiary care hospital in Hangzhou, China. Each sample was collected from a different patient: 108 samples were taken from patients with perianal abscess and 65 samples were taken from patients with appendicitis. All the samples were processed to screen for isolates harboring optrA as we previously described (Shen et al., 2022). Briefly, 20 mg of each wound secretion sample was inoculated into 5 ml of Luria-Bertani (LB) broth (Beijing AOBOX Biotechnology, Beijing, China) within 4 h of collection and incubated at 37°C for 24 h. A volume of 100 ml from each enriched sample was transferred to 5 ml fresh LB broth containing 5% NaCl and 10 mg/L florfenicol (Shanghai Aladdin Biochemical Technology, Shanghai, China) and subcultured for 24 h. Then, 20 ul of each resulting culture was streaked onto a selective medium consisting of Columbia agar (Autibio, Henan, China) base supplemented with 5% (v/v) sheep blood (Biolife Italiana S.r.l., Milan, Italy) and 10 mg/L florfenicol and incubated at 37°C for 24 h.
2.2 Species identification and detection of oxazolidinone-resistance genes
Based on the colony morphology, putative target isolates were selected from the developed colonies. Species identification was performed using matrix-assisted laser desorption ionization–time of flight mass spectrometry (Bruker Daltonik GmbH, Bremen, Germany). All florfenicol-resistant enterococci isolates were again subcultured for purification and then screened for the presence of optrA, poxtA, cfr, and cfrD genes using PCR and Sanger sequencing, following previously described procedures (Li et al., 2020). The following primers were used (Shang et al., 2019; Antonelli et al., 2018; Kehrenberg and Schwarz, 2006; Coccitto et al., 2023; optrA-F: GCACCAGACCAATACGATACAA, optrA-R: TCCTTCTTAACCTTCTCCTTCTCA, poxtA-F: GGTCTGACTGGCTTGTTTTGCT, poxtA-R: ATAAGGTCGGTATTGTCGGCGT, cfr-F: TAAGAAGTAATAATGAGC, cfr-R: TATAGAAAGTCTACGAGG, cfr(D)-F: TGCGCTACTGGAAAAATTGGC, and cfr(D)-R: GCTTGAACGTTCTTGGTGCAT).
2.3 Antimicrobial susceptibility testing
The minimum inhibitory concentrations (MICs) of seven antimicrobial agents (Shanghai Aladdin Biochemical Technology, Shanghai, China) were determined using the broth microdilution method (CLSI, 2018; Tang et al., 2022). The antimicrobial agents were linezolid, chloramphenicol, penicillin G, vancomycin, ciprofloxacin, erythromycin, and tetracycline. Broth microdilution was performed in Brucella broth supplemented with 2% fetal calf serum. Twofold dilutions of each antimicrobial agent ranging from 0.125 to 256 μg/ml were used. To each plate, 100 μl was added with an inoculum concentration of approximately 5 × 107 colony-forming units (CFUs)/ml. The plates were incubated for 24–48 h at 37°C. The MIC was defined as the lowest concentration of the drug. The results were interpreted according to the Clinical and Laboratory Standards Institute standard (CLSI M100-Ed32; CLSI, 2022).
2.4 Conjugation experiment
To investigate the transferability of optrA, poxtA, and cfrD, conjugation experiments were performed using the filter-mating method with rifampicin-resistant E. faecalis JH2-2 as a recipient (Hao et al., 2019; Xie et al., 2021). Briefly, the donor and recipient were cultured in fresh LB broth at 37°C for 4 h to reach the logarithmic phase. Then, the donor and recipient were mixed at a ratio of 1:4 and then incubated on a 0.45-μm membrane placed on an LB agar plate for 24 h at 37°C. Transconjugants were selected on LB agar (TSA) plates supplemented with 30 mg/L rifampicin and 10 mg/L florfenicol. Colonies that grew on these selective plates were chosen after incubation for 16–24 h at 37°C. The presence of optrA, poxtA, and cfrD and species identification of the transconjugants were confirmed using PCR and MALDI TOF/MS, respectively. Conjugation frequency was determined as the number of transconjugants/the number of recipients.
2.5 Whole-genome sequencing and genome analysis
Total genomic DNA was extracted from overnight cultures of 15 isolates using the PureLink Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA) according to the provided instructions. WGS was performed using the NovaSeq 6,000 platform (Illumina, San Diego, CA, USA). The sequencing data were de novo assembled into contigs by SPAdes v.3.13.1 (Hölzer and Marz, 2019). Antimicrobial resistance genes were analyzed using the ResFinder2.1 bioinformatic database.1 Plasmid replicons were identified using PlasmidFinder (2.1).2 A heatmap of antimicrobial resistance genes was performed using Morpheus.3 Multilocus sequence typing (MLST) of strains was performed using the PubMLST tool.4 Plasmid sequences were initially annotated5 using a subsystem technology (RAST version 2.0) server and curated manually using the BLASTn and BLASTp algorithms.6 Easyfig (v2.2.2) was used to visualize the linear alignment of the genetic environment of the optrA gene in different isolates.7
2.6 Phylogenetic analysis using core-genome single-nucleotide polymorphism
Trimmed and quality-filtered assembly sequences of 12 optrA-E. faecalis were aligned with the reference strain GZ86, and the phylogenetic trees of the isolates were constructed using Parsnp v2.0.3 based on core genomic single-nucleotide polymorphism (cgSNPs).8 The phylogenetic tree was visualized and retouched using iTOL.9
3 Results
3.1 Characteristics of enterococci harboring optrA
In this study, 15 florfenicol-resistant enterococci were obtained from 173 wound discharge samples. All samples were positive for optrA with a carriage rate of 8.67% (15/173). Among the optrA-carrying isolates, E. faecalis had the highest frequency (80%, 12/15), while two E. faecium and one E. hirae were also isolated.
The carriage rates of optrA for male patients and female patients were 8.47% (10/118) and 9.09% (5/55), respectively. The median age of the patients was 32 (IQR: 26–47) years (Table 1). Among the differentially diagnosed diseases, the optrA carriage rates varied. In total, 7.4% (8/108) of the patients had perianal abscess and 10.77% (7/65) of the patients had appendicitis. Cephalosporins were used during the treatment period. All patients were discharged.
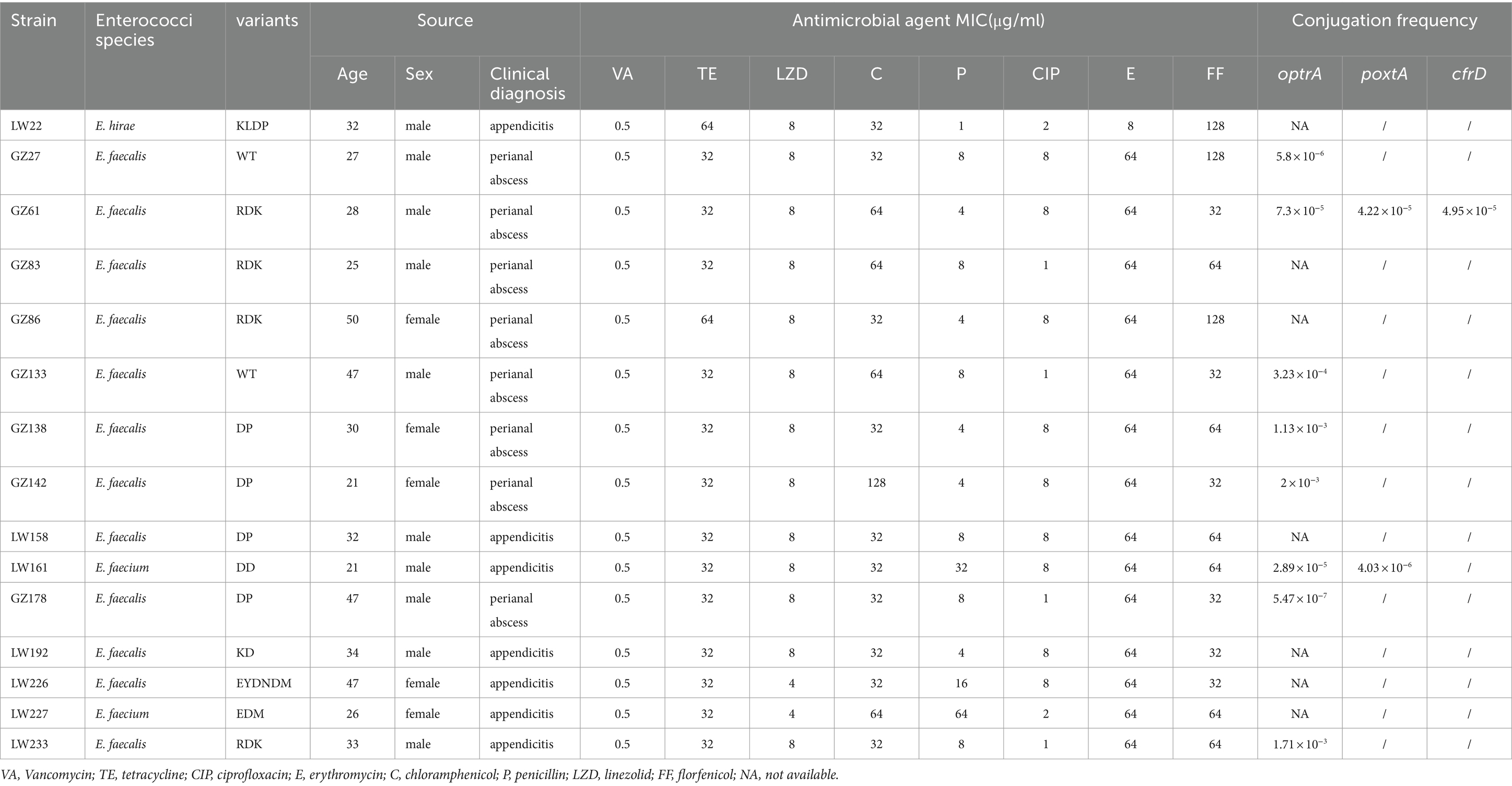
Table 1. Antimicrobial susceptibility results (μg/ml), variants, and clinical information of 15 optrA-positive enterococci isolates.
3.2 Antimicrobial susceptibility and identification of optrA variants
All optrA-positive enterococcal strains were resistant to erythromycin and chloramphenicol (Table 1). All the isolates were either intermediate or resistant to linezolid and exhibited MICs of 4 or 8 μg/ml. Two E. faecium isolates were resistant to penicillin, as was one E. faecalis isolate with an MIC of 16 μg/ml. The isolate E. faecalis GZ27 was susceptible to tetracycline with an MIC of 1ug/ml. No vancomycin-resistant enterococcal strains were isolated.
A total of 8 different optrA variants (including the WT) were identified among the 15 optrA-positive enterococci (Table 1). The RDK and DP variants, were the common variants (n = 4), followed by WT (n = 2). The RDK and DP (n = 4) variants were common among E. faecalis. One each of the DD, EDM, and KLDP variants was detected in E. faecium and E.hirae. Among the seven optrA variants, two variants only (EDM and EYDNDM) showed intermediate resistance to linezolid (MIC = 4 μg/ml), whereas the remaining variants (RDK, DP, DD, EDM, and KLDP) showed resistance to linezolid (MIC = 8 μg/ml).
3.3 Transferability of optrA, poxtA, and cfrD
To investigate the transferability of optrA, poxtA, and cfrD, all 15 optrA-positive isolates were subjected to conjugation experiments. The optrA gene in eight isolates could be successfully transferred to E. faecalis JH2-2 but could not be in the remaining seven isolates. Conjugation efficiency in the transconjugants of strains differed substantially, ranging from 10−3 to 10−7 (Table 1). For poxtA, the conjugation efficiency in the transconjugants of strains GZ61 and LW161 was 4.22 × 10−5 and 4.03 × 10−6, respectively. For cfrD, the conjugation efficiency in transconjugants of strain GZ61 was 4.95 × 10−5.
3.4 Genotyping and phylogenetic analysis of optrA-positive enterococcal isolates
MLST analysis revealed 8 different sequence types (STs) among the 12 optrA-positive E. faecalis isolates (Figure 1), including ST16 (n = 5), ST1022 (n = 1), ST179 (n = 1), ST824 (n = 1), ST58 (n = 1), ST403 (n = 1), ST1938 (n = 1), and ST239 (n = 1). One E. faecium isolate belonged to ST885, and another was divided into ST1818 and 244 according to two different parting systems. In this study, it was assigned to ST1818. Nevertheless, phylogenetic analysis based on the SNPs showed that the distribution of E. faecalis in an anorectal surgery ward was highly diverse.
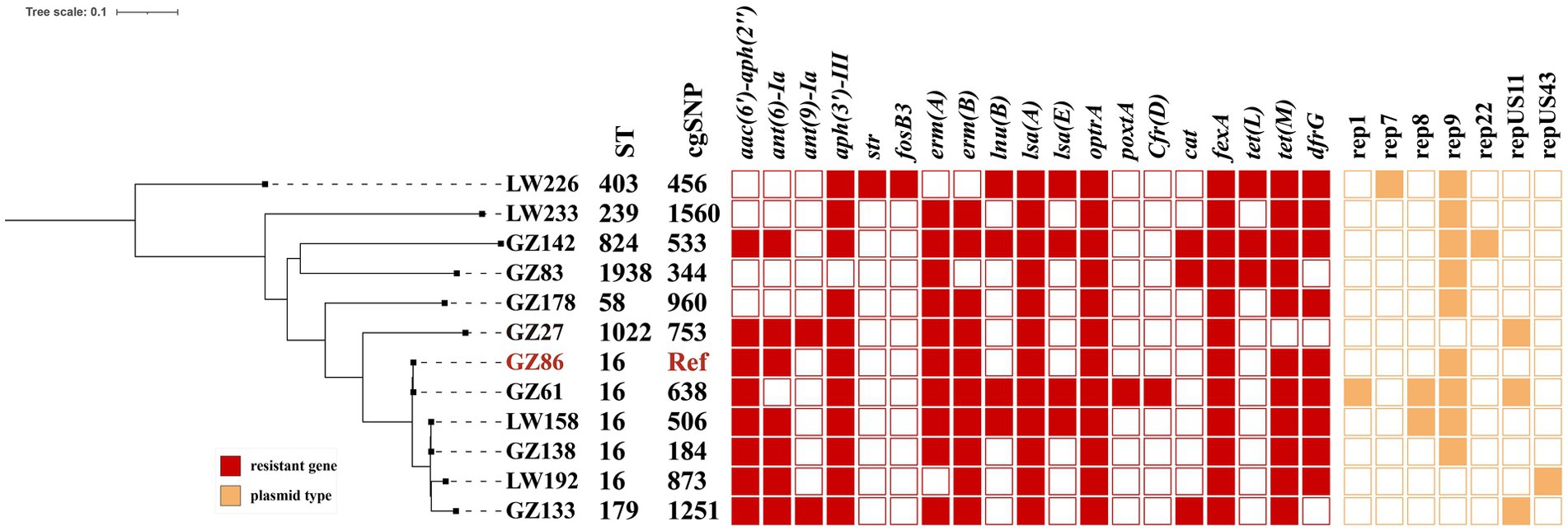
Figure 1. Phylogenetic tree analysis and heatmap of 12 optrA-positive E. faecalis strains A core-genome Journal Pre-proof phylogenetic tree. Antimicrobial resistance genes, ST, and plasmid types in optrA-positive strains are labeled. Colored cells in each column denote the presence of a particular resistance gene or plasmid replicon as labeled at the top. Antimicrobial resistance genes and plasmid replicon types are shown in red and orange, respectively.
WGS revealed multiple antimicrobial resistance genes and plasmid replicons in 12 optrA-positive E. faecalis strains (Figure 1) and 3 non-E. faecalis isolates (Table 2). Each strain was found to carry resistance genes (7–17) and a diverse range of plasmid replicons (1–4). These included aminoglycoside-inactivating enzyme genes [aac(6′)-Iid, aac(6′)-aph(2″), aadD, ant(6)-Ia, ant(9)-Ia, aph(3′)-III, and str], macrolide resistance genes (ermA, ermB, and msrC), phenicol resistance genes (cfrD, cat, fexA, and fexB), tetracycline resistance genes [tet(L) and tet(M)], trimethoprim resistance gene (dfrG), oxazolidinone resistance genes (optrA and poxtA), and fosfomycin resistance gene (fosB3). The fosB3 gene was detected in only one E. faecalis LW226. Concerning the phenicol resistance genes, fexA was present in all optrA-positive strains. Genes less frequently present included fexB and poxtA (n = 2), cat (n = 3), and cfrD (n = 1).In addition, other resistance genes including aac(6′)-aph(2″) and dfrG (n = 10), aph(3′)-III and ermA (n = 13), ermB and lsaA (n = 12), and tet(M) (n = 14) were detected.

Table 2. Resistance genes and plasmid replicon types of optrA-positive non-E. faecalis enterococcal isolates.
3.5 Genomic context of optrA
For the 12 E. faecalis isolates, identical genetic structures were found in two and three isolates, respectively (Figure 2). The two isolates (GZ61 and GZ142) carried the shortest contig in which only optrA was identified.
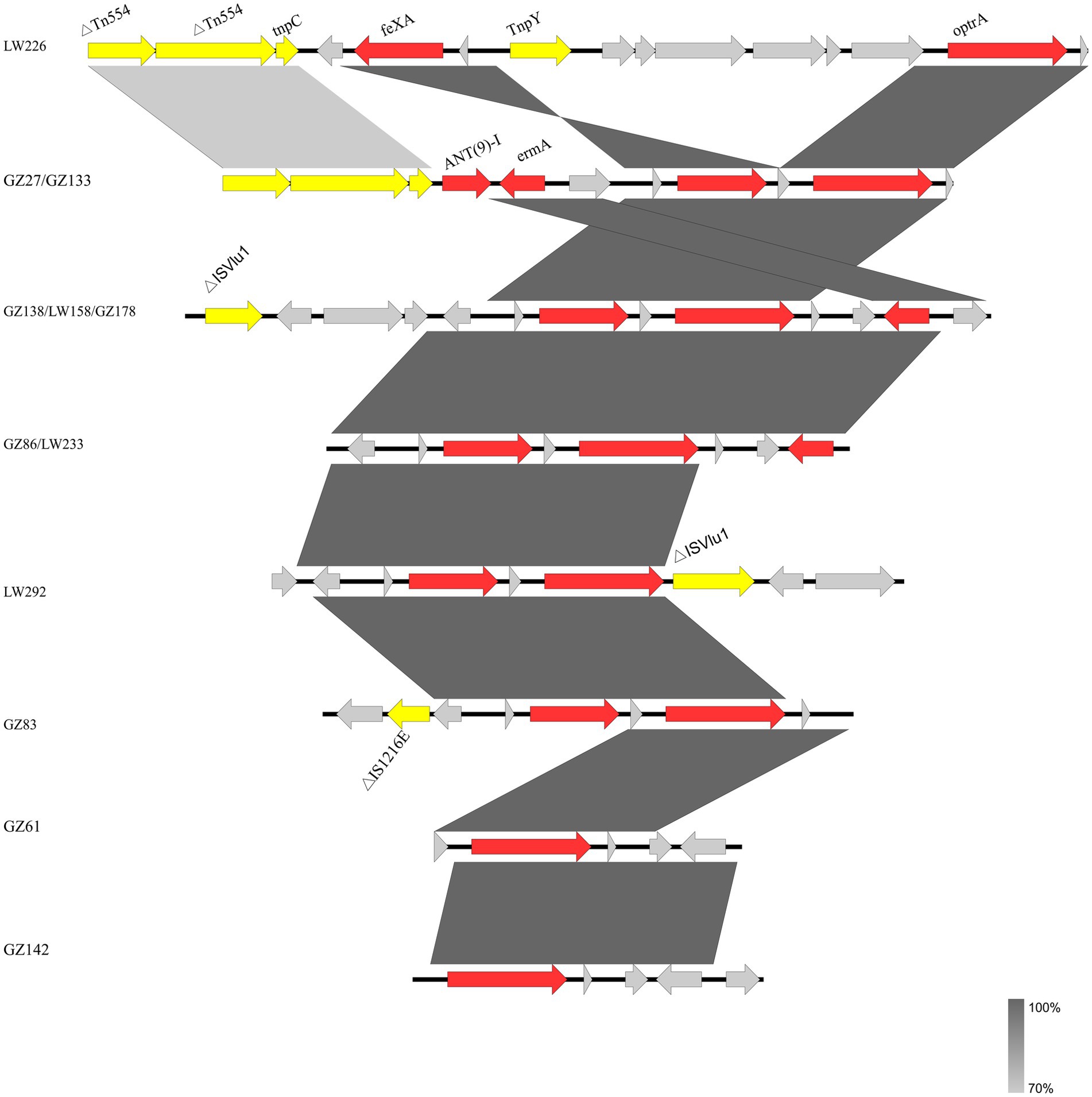
Figure 2. Genetic environment of 12 optrA-positive E. faecalis isolates in this study. The genes of different functions are labeled with different colors. Δ indicates a truncated gene.
Several mobile genetic elements including IS1216E, ISVlu1, and transposase genes tnpC and tnpY, were inserted into the flank structure of fexA-optrA. Insertion sequences ISVlu1 and IS1216E, belonging to family member ISL3, were detected upstream and/or downstream of the fexA-optrA fragment. The IS1216E element was located upstream of the fexA-optrA segment in E. faecalis GZ83. The genetic segment fexA-optrA-ISVlu1 was identified in isolate LW192. Three isolates (GZ138, GZ178, and LW158) shared the same genetic segments ISVlu1-fexA-optrA-erm(A). Truncated transposon Tn554 and transposase genes tnpC and tnpY were located upstream of the fexA-optrA segments in E. faecalis isolates LW226, GZ27, and GZ133. In addition, for the two same genetic environment isolates (GZ27 and GZ133), the resistance genes ant9-1 and erm(A) were present in the region between transposon Tn554 and fexA-optrA fragments.
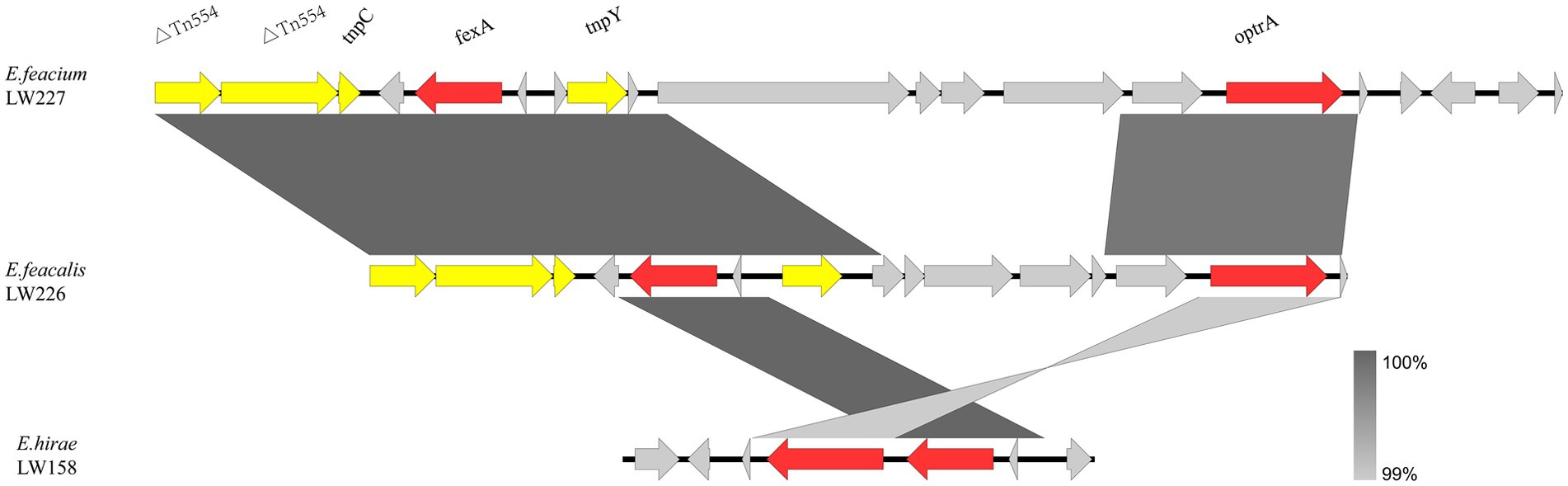
The optrA flanking regions in three non-E. faecalis enterococci are shown in Figure 3. Two E. faecium LW227 and LW161 isolates shared the genetic environment Tn544-tnpC-fexA-tnpY-optrA. No mobile genetic elements were identified in the E. hirae strain.
4 Discussion
The prevalence of linezolid resistance has rapidly increased. The spread of optrA-mediated linezolid resistance in Enterococcus could be an imminent threat.
In this study, the prevalence of optrA-positive enterococci in wound secretion samples obtained from patients in an anorectal surgery ward of a Chinese hospital was 8.65%. It was 1.1% at a tertiary care hospital in Nanjing and Beijing, China (Zhou et al., 2019; Wang et al., 2024). Several studies showed that fecal carriage rates of optrA from healthy humans varied from 3.53% in 2019 to 5.89% in 2022 (Cai et al., 2019; Shen et al., 2022). A previous study reported a human intestinal carriage rate of optrA at 19.3%, utilizing an optimized high-sensitivity screening approach (Shen et al., 2022). We used this method to screen for samples. As a result, we discovered an unexpectedly high prevalence. Nevertheless, a study suggested that the prevalence of optrA is higher in enterococci from animals than those from humans (Wang et al., 2015; Shen et al., 2024). The isolates from shared bicycles were identified for the presence of optrA gene (Han et al., 2020). The optrA gene has been widespread in animal, environmental, and clinical isolates, indicating that the horizontal transfer of optrA plays a crucial role in the human–animal–environment interfaces (Shen et al., 2024; Gouliouris et al., 2018). In the present study, the patients had no history of linezolid use during hospitalization, indicating that infections were not associated with linezolid use.
In this study, STs and SNPs of optrA-positive E. faecalis isolated were genetically highly diverse. STs belonged to eight different types, mostly for ST16. ST16, with the GCTGAACC SNP profile, has often been identified in humans, animals, and surface water in various countries (Zhou et al., 2019; Freitas et al., 2020; Bender et al., 2018; Rathnayake et al., 2011). Previous studies reported ST480 as one of the predominant types in France and Germany (Egan et al., 2020). These findings demonstrated the non-clonal dissemination and the widespread presence of optrA-positive E. faecalis in hospitals.
All strains harbored multiple resistance genes and showed a multidrug-resistant phenotype, which indicated a broad antibiotic resistance spectrum of enterococcal isolates. To our surprise, the fosfomycin resistance gene foB3 was detected in a single optrA-carrying E. faecalis. In 2021, the coexistence of foB3 and optrA was the first reported in E. faecalis from pigs (Wang et al., 2021). Given that fosfomycin and linezolid are the last-resort antibiotics for treating infections caused by VRE, the co-occurrence of fosB and optrA in clinical strains may seriously compromise the effectiveness of clinical therapy and is another potential threat to public health.
To date, at least 69 optrA variants have been detected (Schwarz et al., 2021), and 7 OptrA variants were detected in this study. We found the RDK and DP variants were the common variants in E. faecalis. Previous studies suggested that the different OptrA variants might have an impact on the relative linezolid susceptibility/resistance of the respective isolates (Schwarz et al., 2021). A previous study demonstrated that enterococci strains (isolated from asymptomatic healthy humans) carrying the wild-type optrA gene or the RDK variant exhibited relatively high levels of resistance to linezolid compared to other variants (Cai et al., 2019). Moreover, another study demonstrated that strains with the RKD variant had linezolid MICs of 8–32 μg/ml, while those with the wild-type optrA gene showed MICs of 8–48 μg/ml (Wang et al., 2024). The RDK variant increased the MIC of linezolid (Li et al., 2020). We observed that isolates harboring the EDM and EYDNDM variants for linezolid MICs were 4 μg/ml, as observed in previous studies (Cai et al., 2015). The wild-type and other variants (RDK, DP, DD, EDM, and KLDP) were linezolid MICs of 8 μg/ml. Thus, distinct variants of the optrA gene may confer differential resistance to linezolid in enterococci.
In this study, the core structure fexA-optrA was found, which was also identified in various bacteria from humans, wastewater, and animals (Tang et al., 2021; Yang et al., 2024; Freitas et al., 2017; Abdullahi et al., 2023). Variations were distinguished by various flanking IS elements and other genes located between these elements. Mobile genetic elements, including IS1216E and ISVlu1, contribute significantly to the transmission of optrA (Partridge et al., 2018). Tn554 mediation of optrA transfer has been identified (Kang et al., 2019). The optrA flanking structures were observed from different species, suggesting that the optrA cluster may jump via mobile elements, including transposon (Tn554), insertion sequences (IS1216E and ISVlu1), and transposase genes (tnpY and tnpC). The findings suggested that transposable elements, including ISVlu1, IS1216E, and Tn554, may be important in the transmission of optrA in the anorectal surgery ward.
Due to the limitations of second-generation short reads, we were unable to obtain and analyze the complete genetic environment including complete plasmids.
The intestine is a reservoir of drug-resistant genes. The detection of optrA-positive E. faecium from bile suggests that drug-resistant bacteria can also exist in the gallbladder upstream of the intestine (Deng et al., 2023). In the present study, the high carriage of optrA-positive Enterococcus in anorectal disease not only makes the treatment difficult but also may pose a potential human health risk. Hence, optrA-carrying enterococcal from the intestine needs further attention.
In conclusion, we report the high carriage rate of the optrA gene isolates from anorectal disease patients and present the genetic diversity. Different mobile genetic elements including Tn554, IS1216E, and ISVlu1 mediated the dissemination of optrA. The prevalence and spread of optrA-carrying enterococci among patients in the anorectal surgery ward should be actively monitored.
Data availability statement
Genome sequences of 15 Enterococci strains tested in this study have been deposited in the NCBI database under the BioProject accession number PRJNA1115951.
Ethics statement
The studies involving humans were approved by Approval Letter of Medical Ethics Committee of Zhejiang Xiaoshan Hospital (Approval ID: K2022027). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
YL: Conceptualization, Methodology, Software, Investigation, Formal Analysis, Writing – original draft, Writing – review & editing. TJ: Data Curation, Supervision, Writing – original draft, Writing – review & editing. JM: Resources, Writing – original draft, Writing – review & editing. FX: Resources, Writing – original draft, Writing – review & editing. RZ: Supervision, Writing – original draft, Writing – review & editing. JY: Funding acquisition, Writing – original draft, Writing – review & editing. JC: Supervision, Writing – original draft, Writing – review & editing. YX: Conceptualization, Data curation, Resources, Supervision, Funding acquisition, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Health Commission of Zhejiang Province (2023RC254) and the Science and Technology Commission of Hangzhou City (B20220073).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1. ^https://cge.cbs.dtu.dk/services/all.php
2. ^https://cge.cbs.dtu.dk/services/PlasmidFinder/
3. ^https://software.broadinstitute.org/morpheus/
6. ^http://blast.ncbi.nlm.nih.gov/blast
References
Abdullahi, I. N., Lozano, C., Juárez-Fernández, G., Höfle, U., Simón, C., Rueda, S., et al. (2023). Nasotracheal enterococcal carriage and resistomes: detection of optrA-, poxtA- and cfrD-carrying strains in migratory birds, livestock, pets, and in-contact humans in Spain. Europ. J. Clin. Microbiol. Infect. Dis. 42, 569–581. doi: 10.1007/s10096-023-04579-9
Andrea, B., Gianluca, M., Chiara, V., Manso, E., Mingoia, M., Giovanetti, E., et al. (2015). Detection in Italy of two clinical Enterococcus faecium isolates carrying both the oxazolidinone and phenicol resistance gene optrA and a silent multiresistance gene cfr. J. Antimicrob. Chemother. 71, 1118–1119. doi: 10.1093/jac/dkv438
Antonelli, A., D’Andrea, M. M., Brenciani, A., Galeotti, C. L., Morroni, G., Pollini, S., et al. (2018). Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J. Antimicrob. Chemother. 73, 1763–1769. doi: 10.1093/jac/dky088
Bender, J. K., Fleige, C., Lange, D., Klare, I., and Werner, G. (2018). Rapid emergence of highly variable and transferable oxazolidinone and phenicol resistance gene optrA in German Enterococcus spp. clinical isolates. Int. J. Antimicrob. Agents 52, 819–827. doi: 10.1016/j.ijantimicag.2018.09.009
Cai, J., Schwarz, S., Chi, D., Wang, Z., Zhang, R., and Wang, Y. (2019). Faecal carriage of optrA-positive enterococci in asymptomatic healthy humans in Hangzhou, China. Clin. Microbiol. Infect. 25, 630.e1–630.e6. doi: 10.1016/j.cmi.2018.07.025
Cai, J., Wang, Y., Schwarz, S., Lv, H., Li, Y., Liao, K., et al. (2015). Enterococcal isolates carrying the novel oxazolidinone resistance gene optrA from hospitals in Zhejiang, Guangdong, and Henan, China, 2010-2014. Clin. Microbiol. Infect. 21, 1095.e1–1095.e4. doi: 10.1016/j.cmi.2015.08.007
Cai, J., Wang, Y., Schwarz, S., Zhang, G., Chen, S., Gu, D., et al. (2016). High detection rate of the oxazolidinone resistance gene optrA in Enterococcus faecalis isolated from a Chinese anorectal surgery ward. Int. J. Antimicrob. Agents 48, 757–759. doi: 10.1016/j.ijantimicag.2016.08.008
Chen, W., Mandali, S., Hancock, S. P., Kumar, P., Collazo, M., Cascio, D., et al. (2018). Multiple serine transposase dimers assemble the transposon-end synaptic complex during IS607-family transposition. eLife 7:7. doi: 10.7554/eLife.39611
CLSI (Ed.) (2018). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically,11th ed. Clinical and Laboratory Standards Institute, 950.
Coccitto, S. N., Cinthi, M., Simoni, S., Vignaroli, C., Massacci, F. R., Albini, E., et al. (2023). Identification of plasmids co-carrying cfr(D)/optrA and cfr(D2)/poxtA linezolid resistance genes in two Enterococcus avium isolates from swine brain. Vet. Microbiol. 282:109749. doi: 10.1016/j.vetmic.2023.109749
Cui, L., Wang, Y., Lv, Y., Wang, S., Song, Y., Li, Y., et al. (2016). Nationwide surveillance of novel Oxazolidinone resistance gene optrA in Enterococcus isolates in China from 2004 to 2014. Antimicrob. Agents Chemother. 60, 7490–7493. doi: 10.1128/AAC.01256-16
Deng, L., Zhen, W., Wang, J., and Lin, D. (2023). Bile carriage of optrA-positive Enterococcus faecium in a patient with Choledocholith. Microbiol. Spect. 11:e0285222. doi: 10.1128/spectrum.02852-22
Echeverria-Esnal, D., Sorli, L., Prim, N., Conde-Estévez, D., Mateu-de Antonio, J., Martin-Ontiyuelo, C., et al. (2019). Linezolid vs glycopeptides in the treatment of glycopeptide-susceptible Enterococcus faecium bacteraemia: a propensity score matched comparative study. Int. J. Antimicrob. Agents 54, 572–578. doi: 10.1016/j.ijantimicag.2019.08.018
Egan, S. A., Shore, A. C., O’Connell, B., Brennan, G. I., and Coleman, D. C. (2020). Linezolid resistance in Enterococcus faecium and Enterococcus faecalis from hospitalized patients in Ireland: high prevalence of the MDR genes optrA and poxtA in isolates with diverse genetic backgrounds. J. Antimicrob. Chemother. 75, 1704–1711. doi: 10.1093/jac/dkaa075
Flamm, R. K., Mendes, R. E., Hogan, P. A., Streit, J. M., Ross, J. E., and Jones, R. N. (2016). Linezolid surveillance results for the United States (LEADER surveillance program 2014). Antimicrob. Agents Chemother. 60, 2273–2280. doi: 10.1128/AAC.02803-15
Freitas, A. R., Elghaieb, H., León-Sampedro, R., Abbassi, M. S., Novais, C., Coque, T. M., et al. (2017). Detection of optrA in the African continent (Tunisia) within a mosaic Enterococcus faecalis plasmid from urban wastewaters. J. Antimicrob. Chemother. 72, 3245–3251. doi: 10.1093/jac/dkx321
Freitas, A. R., Tedim, A. P., Novais, C., Lanza, V. F., and Peixe, L. (2020). Comparative genomics of global optrA-carrying Enterococcus faecalis uncovers a common chromosomal hotspot for optrA acquisition within a diversity of core and accessory genomes. Microb. Genom. 6:350. doi: 10.1099/mgen.0.000350
Gagetti, P., Faccone, D., Ceriana, P., Lucero, C., Menocal, A., Argentina, G. L. R. E., et al. (2023). Emergence of optrA-mediated linezolid resistance in clinical isolates of Enterococcus faecalis from Argentina. J. Glob. Antimicrob. Resist. 35, 335–341. doi: 10.1016/j.jgar.2023.10.014
Gawryszewska, I., Żabicka, D., Hryniewicz, W., and Sadowy, E. (2017). Linezolid-resistant enterococci in polish hospitals: species, clonality and determinants of linezolid resistance. Eur. J. Clin. Microbiol. Infect. Dis. 36, 1279–1286. doi: 10.1007/s10096-017-2934-7
Gouliouris, T., Raven, K. E., Ludden, C., Blane, B., Corander, J., Horner, C. S., et al. (2018). Genomic surveillance of Enterococcus faecium reveals limited sharing of strains and resistance genes between livestock and humans in the United Kingdom. MBio 9, e01780–e01718. doi: 10.1128/mBio.01780-18
Han, R., Shi, Q., Wu, S., Yin, D., Peng, M., Dong, D., et al. (2020). Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among Carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front. Cell. Infect. Microbiol. 10:314. doi: 10.3389/fcimb.2020.00314
Hao, W., Shan, X., Li, D., Schwarz, S., Zhang, S. M., Li, X. S., et al. (2019). Analysis of a poxtA- and optrA-co-carrying conjugative multiresistance plasmid from Enterococcus faecalis. J. Antimicrob. Chemother. 74, 1771–1775. doi: 10.1093/jac/dkz109
Hölzer, M., and Marz, M. (2019). De novo transcriptome assembly: a comprehensive cross-species comparison of short-read RNA-Seq assemblers. GigaScience 8:giz039. doi: 10.1093/gigascience/giz039
Kang, Z. Z., Lei, C. W., Kong, L. H., Wang, Y. L., Ye, X. L., Ma, B. H., et al. (2019). Detection of transferable oxazolidinone resistance determinants in Enterococcus faecalis and Enterococcus faecium of swine origin in Sichuan Province, China. J. Glob. Antimicrob. Resist. 19, 333–337. doi: 10.1016/j.jgar.2019.05.021
Kehrenberg, C., and Schwarz, S. (2006). Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50, 1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006
Kerschner, H., Rosel, A. C., Hartl, R., Hyden, P., Stoeger, A., Ruppitsch, W., et al. (2021). Oxazolidinone resistance mediated by optrA in clinical Enterococcus faecalis isolates in Upper Austria: first report and characterization by whole genome sequencing. Microbial. Drug Resist. 27, 685–690. doi: 10.1089/mdr.2020.0098
Li, P., Yang, Y., Ding, L., Xu, X., and Lin, D. (2020). Molecular investigations of linezolid resistance in enterococci OptrA variants from a Hospital in Shanghai. Infect. Drug Resist. 13, 2711–2716. doi: 10.2147/IDR.S251490
Liu, B. G., Yuan, X. L., He, D. D., Hu, G. Z., Miao, M. S., and Xu, E. P. (2020). Research progress on the oxazolidinone drug linezolid resistance. Eur. Rev. Med. Pharmacol. Sci. 24, 9274–9281. doi: 10.26355/eurrev_202009_23009
Mendes, R. E., Hogan, P. A., Jones, R. N., Sader, H. S., and Flamm, R. K. (2016). Surveillance for linezolid resistance via the Zyvox® annual appraisal of potency and Spectrum (ZAAPS) programme (2014): evolving resistance mechanisms with stable susceptibility rates. J. Antimicrob. Chemother. 71, 1860–1865. doi: 10.1093/jac/dkw052
Na, S. H., Moon, D. C., Kim, M. H., Kang, H. Y., Kim, S. J., Choi, J. H., et al. (2020). Detection of the phenicol-oxazolidinone resistance gene poxtA in Enterococcus faecium and Enterococcus faecalis from food-producing animals during 2008-2018 in Korea. Microorganisms 8:1839. doi: 10.3390/microorganisms8111839
Park, K., Jeong, Y. S., Chang, J., Sung, H., and Kim, M. N. (2020). Emergence of optrA-mediated linezolid-nonsusceptible Enterococcus faecalis in a tertiary care hospital. Ann. Lab. Med. 40, 321–325. doi: 10.3343/alm.2020.40.4.321
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e00088–e00017. doi: 10.1128/CMR.00088-17
Rathnayake, I. U., Hargreaves, M., and Huygens, F. (2011). Genotyping of Enterococcus faecalis and Enterococcus faecium isolates by use of a set of eight single nucleotide polymorphisms. J. Clin. Microbiol. 49, 367–372. doi: 10.1128/JCM.01120-10
Rodríguez-Lucas, C., Fernández, J., Vázquez, X., de Toro, M., Ladero, V., Fuster, C., et al. (2022). Detection of the optrA gene among polyclonal linezolid-susceptible isolates of Enterococcus faecalis recovered from community patients. Microbial. Drug Resist. 28, 773–779. doi: 10.1089/mdr.2021.0402
Schwarz, S., Zhang, W., du, X. D., Krüger, H., Feßler, A. T., Ma, S., et al. (2021). Mobile oxazolidinone resistance genes in gram-positive and gram-negative bacteria. Clin. Microbiol. Rev. 34:e0018820. doi: 10.1128/CMR.00188-20
Shang, Y., Li, D., Hao, W., Schwarz, S., Shan, X., Liu, B., et al. (2019). A prophage and two ICESa2603-family integrative and conjugative elements (ICEs) carrying optrA in Streptococcus suis. J. Antimicrob. Chemother. 74, 2876–2879. doi: 10.1093/jac/dkz309
Shen, W., Cai, C., Dong, N., Chen, J., Zhang, R., and Cai, J. (2024). Mapping the widespread distribution and transmission dynamics of linezolid resistance in humans, animals, and the environment. Microbiome 12:52. doi: 10.1186/s40168-023-01744-2
Shen, W., Huang, Y., and Cai, J. (2022). An optimized screening approach for the Oxazolidinone resistance gene optrA yielded a higher fecal carriage rate among healthy individuals in Hangzhou, China. Microbiol. Spect. 10:e0297422. doi: 10.1128/spectrum.02974-22
Tang, X., Shen, Y., Song, X., Benghezal, M., Marshall, B. J., Tang, H., et al. (2022). Reassessment of the broth microdilution method for susceptibility testing of Helicobacter pylori. J. Infect. Dis. 226, S486–s492. doi: 10.1093/infdis/jiac389
Tang, B., Wang, Y., Luo, Y., Zheng, X., Qin, X., Yang, H., et al. (2021). Coexistence of optrA and fexA in Campylobacter. mSphere 6, e00125–e00121. doi: 10.1128/mSphere.00125-21
Wang, X., Gao, Y., Liu, X., Sun, N., Huang, J., and Wang, L. (2021). First report of the plasmid-mediated fosB gene in Enterococcus faecalis from pigs. Genes 12:1684. doi: 10.3390/genes12111684
Wang, Z., Liu, D., Zhang, J., Liu, L., Zhang, Z., Liu, C., et al. (2024). Genomic epidemiology reveals multiple mechanisms of linezolid resistance in clinical enterococci in China. Ann. Clin. Microbiol. Antimicrob. 23:41. doi: 10.1186/s12941-024-00689-0
Wang, Y., Lv, Y., Cai, J., Schwarz, S., Cui, L., Hu, Z., et al. (2015). A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 70, 2182–2190. doi: 10.1093/jac/dkv116
Xie, M., Yang, X., Xu, Q., Ye, L., Chen, K., Zheng, Z., et al. (2021). Clinical evolution of ST11 carbapenem resistant and hypervirulent Klebsiella pneumoniae. Commun. Biol. 4:650. doi: 10.1038/s42003-021-02148-4
Yang, Q., Wang, X., Schwarz, S., Zhu, Y., and Zhang, W. (2024). Identification of ISVlu1-derived translocatable units containing optrA and/or fexA genes generated by homologous or illegitimate recombination in Lactococcus garvieae of porcine origin. Vet. Microbiol. 292:110048. doi: 10.1016/j.vetmic.2024.110048
Yi, M., Zou, J., Zhao, J., Tang, Y., Yuan, Y., Yang, B., et al. (2022). Emergence of optrA-mediated linezolid resistance in Enterococcus faecium: a molecular investigation in a tertiary Hospital of Southwest China from 2014-2018. Infect. Drug Resist. 15, 13–20. doi: 10.2147/IDR.S339761
Zhang, J., Yang, Y., Sun, H., Luo, X., Cui, X., Miao, Q., et al. (2023). Prevalence of the optrA gene among Streptococcus suis isolates from diseased pigs and identification of a novel integrative conjugative element ICESsu988S. Res. Microbiol. 174:104078. doi: 10.1016/j.resmic.2023.104078
Keywords: optrA, linezolid resistance, enterococci, genetic context, hospital
Citation: Li Y, Jiang T, Mao J, Xu F, Zhang R, Yan J, Cai J and Xie Y (2024) Prevalence and genetic diversity of optrA-positive enterococci isolated from patients in an anorectal surgery ward of a Chinese hospital. Front. Microbiol. 15:1481162. doi: 10.3389/fmicb.2024.1481162
Edited by:
Jens Andre Hammerl, Bundesinstitut für Risikobewertung, GermanyReviewed by:
Sunghyun Yoon, National Center for Toxicological Research (FDA), United StatesAmira Awad Moawad, Friedrich Loeffler Institut, Germany
Peter Kinnevey, Dublin Dental University Hospital, Ireland
Copyright © 2024 Li, Jiang, Mao, Xu, Zhang, Yan, Cai and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanjun Xie, d2N5ODkyMTVAMTI2LmNvbQ==
 Yuanyuan Li
Yuanyuan Li Tao Jiang1
Tao Jiang1 Rong Zhang
Rong Zhang Jiachang Cai
Jiachang Cai